Introduction
Breast cancer is the dominant cause of death in
women worldwide and it is the most common cancer in big city areas
(1). Breast cancer in its
advanced-stage has been related to the degree of metastasis
(2). The process of metastasis
seems to be regulated by a variety of gene products and EMT has
been recognized as a fundamental process of embryogenesis, it is an
important event in the metastatic cascade where the cells acquire
migratory, and invasive capabilities (3).
Curcumin, an effective component of the spice
turmeric (Curcuma longa) and a dietary chemopreventive agent
(4,5), has been shown to resist initiation of
carcinogenesis, modulation of cell survival, induction of
apoptosis, inhibition of angiogenesis and induce anti-invasive and
anti-metastatic effects (6).
Cadherins are cell adhesion molecules fundamental in
the development of multicellular organisms (7). Among them, E-cadherin is
essential for epithelial tissue integrity (8) and N-cadherin is expressed at
gastrulation stage by downregulation of E-cadherin and
undergoing EMT considered in cells of mesenchymal origin (9). Both establish cell-cell adhesion with
their extracellular domains and are connected with catenins at
their intracellular domains (10).
Both also interact with receptors for growth factors involved in
the modulation of signaling pathways, E-cadherin in relation with
receptors of epidermal growth factors (11), and N-cadherin with fibroblast
growth factor receptors (FGFR) (12). The protein B-catenin plays a role
in signaling and cell adhesion (13,14).
SLUG, a member of the SNAI family (15–17)
is involved in development of EMT (16), it is an inhibitor of apoptosis
(18), and is part of breast and
kidney development (15,16). AXL is activated through several
mechanisms, as binding of its ligand and dimerization with HER2/neu
(19–21). Since AXL is overexpressed in human
cancers, it has significant correlation with tumor stage in breast
cancer, especially in metastases (22,24).
Twist1 is another factor that induces EMT and degradation of
extracellular matrix (25–27) by promoting loosening of cell-cell
junctions of epithelial cells and becoming invasive (28).
Vimentin is the most important part of the
cytoskeletal of the cell along with microtubules and microfilaments
(29). Fibronectin exerts multiple
effects in vitro and in vivo as a component of the
extracellular matrix stimulating proliferation, migration and
differentiation (30–33). It activates various cell surface
receptors most notably integrins (34) as well as development of fibrillar
structures (35) and activation of
various growth factors (36).
An inducer of EMT in cancer metastasis is the ZEB1,
a transcription factor that also induces EMT-suppressing
microRNA-200s (miR-200s) (37).
ZEB2 belongs to the ZEB protein family (38) and it is involved in differentiation
(39,40). Enhancer of zeste homolog 2 (EZH2)
silences gene transcription by trimethylation of histone H3
(41). It is upregulated in
multiple malignancies (42,43),
and mediated by silencing tumor suppressor genes (44). It is implicated in transcriptional
activation whose mechanism is not known (45–48).
STAT3, member of the family known as signal
transducers, is involved in oncogenesis (49). Cyclins have been identified as
regulatory subunits and catalytic subunits of cell cycle-regulated
kinases. The cyclin/cdk complexes are implicated in the control of
mitosis. G1 to S transition is regulated by Cyclin D since
abnormalities involving cyclin D1 deregulate control of the G1-S
transition contributing to tumor development (50,51).
Notch proteins are a family of transmembranes with
five ligands. Notch signaling is activated in human breast cancer
with the accumulation of Notch1 intracellular domain in tissue
(52). It has been shown that
Notch1 activates Akt and survivin (53,54),
and has also been involved in chemoresistance. Increased Notch
ligands have been shown to be correlated with poor overall survival
in breast cancer patients (55).
Thus, the purpose of this study was to evaluate the
effect of curcumin on EMT when a comparison was done between a
triple-positive and a triple-negative breast cancer cell lines for
ER, PgR and Erb-B2 in relation to this process. Curcumin effect was
evaluated with triple-negative cell line the immortalized breast
epithelial cell line MCF-10F, Tumor2, a triple-positive cell line
derived from Alpha5 injected into the nude mice, and MDA-MB231, a
triple-negative for the same markers.
Materials and methods
Cell cultures and treatment
MCF-10F, Tumor2 and MDA-MB-231 human breast cell
lines were maintained in Dulbecco’s (DMEM; Gibco, USA) supplemented
with penicillin (100 U/ml), streptomycin (100 μg/ml), 0.1 mM
non-essential amino acids, 0.2 mM glutamine, 1 mM pyruvate, and 10%
heat-inactivated fetal bovine serum and incubated in a 5%
CO2 humidified atmosphere at 37°C.
RNA extraction and cDNA synthesis
Total RNA was isolated by using TRIzol reagent
(Invitrogen Corp., Carlsband, CA, USA) according to the
manufacturer’s instructions. Total RNA (2 μg) was reverse
transcribed to cDNA using High Capacity cDNA Reverse Transcription
kit (Applied Biosystems, Carlsband, CA, USA) and RNase inhibitor
(Applied Biosystems) were used in these studies.
RT-qPCR
The cDNA (2 μl) was used in 20 μl qPCR reaction
containing SYBR Green PCR Master Mix (Agilent, La Jolla, CA, USA)
and 5 μM of each primer for the target genes such as E-cadherin,
N-cadherin, β-catenin, Slug, AXL, Twist1, Vimentin, Fibronectin,
ZEB1, ZEB2, EZH2, STAT3, Cyclin D1, and Notch1. Table I shows the primers for the gene
selected to develop cDNA probes. The reaction was performed in a
CFX 96 Real-Time PCR (Bio-Rad Laboratories, Hercules, CA, USA) at
95°C for 10 min and 40 cycles of a 2-step program of 95°C for 10
sec and 61°C for 45 sec when fluorescence-reading occurs. After
amplification, PCR product was monitored through dissociation curve
analysis (measurement of fluorescence during an increasing heating
of 2°C/min from 61 to 95°C). At this step, undesirable DNA
contamination (if present) could be detected since primers were
designed to encompass an intron. Reactions were performed in
triplicate and the threshold of the cycle was obtained using
Bio-Rad CFX Manager 2.1 software and the average gene expression
was normalized with a reference housekeeping gene β-actin.
Relative expression was a normalized to the average in cells.
 | Table IPrimers for genes selected to develop
cDNA probes. |
Table I
Primers for genes selected to develop
cDNA probes.
| Gene name | Product length
(bp)a | Primer
sequenceb |
|---|
|
E-cadherin | 93 | F:
AGTGGGCACAGATGGTGTGA
R: TAGGTGGAGTCCCAGGCGTA |
|
N-cadherin | 67 | F: TCG ATT GGT TTG
ACC ACG G
R: GAC GGT TCG CCA TCC AGA C |
|
β-catenin | 94 | F:
GCAGAGTGCTGAAGGTGCTA
R: TCTGTCAGGTGAAGTCCTAAAGC |
| Slug | 72 | F:
GACCCTGGTTGCTTCAAGGA
R: TGTTGCAGTGAGGGCAAGAA |
| AXL | 121 | F:
GTTTGGAGCTGTGATGGAAGGC
R: CGCTTCACTCAGGAAATCCTCC |
| Twist1 | 118 | F:
TCCGCGTCCCACTAGCA
R: AGTTATCCAGCTCCAGAGTCTCTAGAC |
|
Vimentin | 117 | F:
TGTCCAAATCGATGTGGATGTTTC
R: TTGTACCATTCTTCTGCCTCCTG |
|
Fibronectin | 105 | F:
GGAGGAAGCCGAGGTTTTAAC
R: ACGCTCATAAGTGTCACCCA |
| ZEB1 | 141 | F:
GCACAACCAAGTGCAGAAGA
R: GCCTGGTTCAGGAGAAGATG |
| ZEB2 | 128 | F:
CAAGAGGCGCAAACAAGC
R: GGTTGGCAATACCGTCATCC |
| EZH2 | 84 | F:
CCAAGAGAGCCATCCAGACT
R: CGATGCCGACATACTTCAGG |
| STAT3 | 163 | F:
GGTTGGACATGATGCACACTAT
R: AGGGCAGACTCAAGTTTATCAG |
| Cyclin
D1 | 60 | F:
GTGGCCTCTAAGATGAAGGA
R: GGTGTAGATGCACAGCTTCT |
| Notch1 | 140 | F:
GAGGCGTGGCAGACTATGC
R: CTTGTACTCCGTCAGCGTGA |
Cell migration and invasion assays
The modified Boyden’s chambers to analyze migration
and invasiveness were used as described (56) (Corning, NY, USA). Cells
(3×105) in 100 μl of medium were used for migration and
invasion assays; 8-μm membrane pores were pre-coated with 60 μl
Matrigel matrix gel (BD Biosciences, Grand Island, NY, USA) at
least one hour before seeding the cells for upper chambers; 600 μl
of medium with 10% FBS was placed in the lower chambers as
chemoattractant. Cells treated with 30 μM curcumin were cultured
for 48 h in a humidified incubator. Then, the upper chambers were
wiped using cotton swabs. The membranes were fixed with 100%
methanol at room temperature for 15 min, visualized and quantified
using DAPI. Ten fields of each chamber were photographed (x40
magnification). This experiment was independently repeated two
times.
Statistical analysis
Results of gene expression of control and treated
group were compared with ANOVA followed by Dunnet’s test. The
average ± standard error of the mean was used to express numerical
data. A p-value <0.05 was considered statistically
significant.
Results
Effect of curcumin on growth of breast
cancer cells in vitro
We analyzed the effect of curcumin on cell
proliferation of MCF-10F, Tumor2 and MDA-MB-231 cell lines after 48
h. Graded concentrations of curcumin (0–70 μM/l) were used to
determine cell viability by MTT assay. The growth curves showed
that cell proliferation was inhibited in a dose-dependent manner by
curcumin with inhibition at doses ≥30 μM/l.
Curcumin inhibits the expression of
markers of EMT in breast cancer cells
To confirm the effects of curcumin on EMT, we
sequentially analyzed gene expression of EMT markers by RT-qPCR
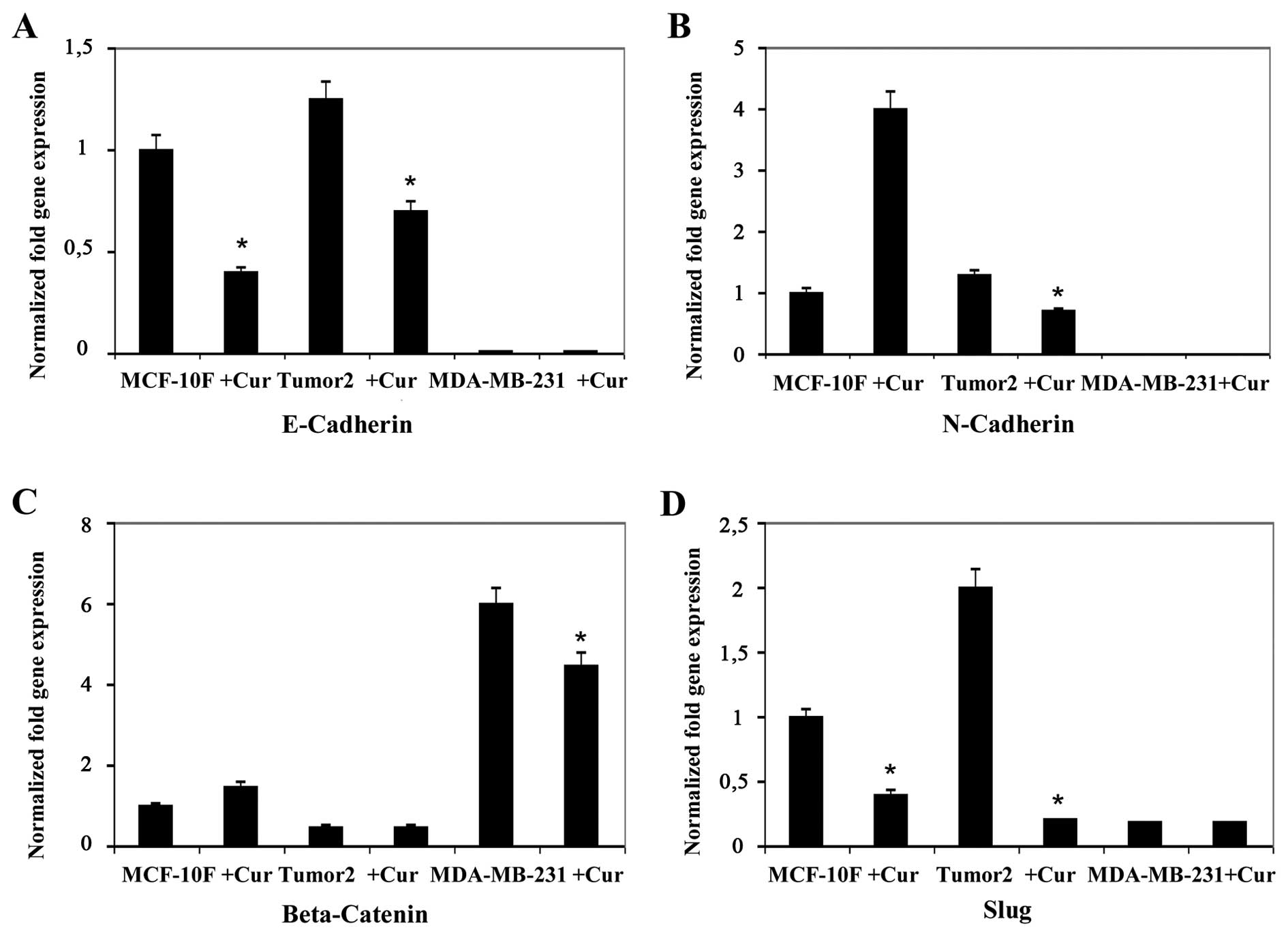
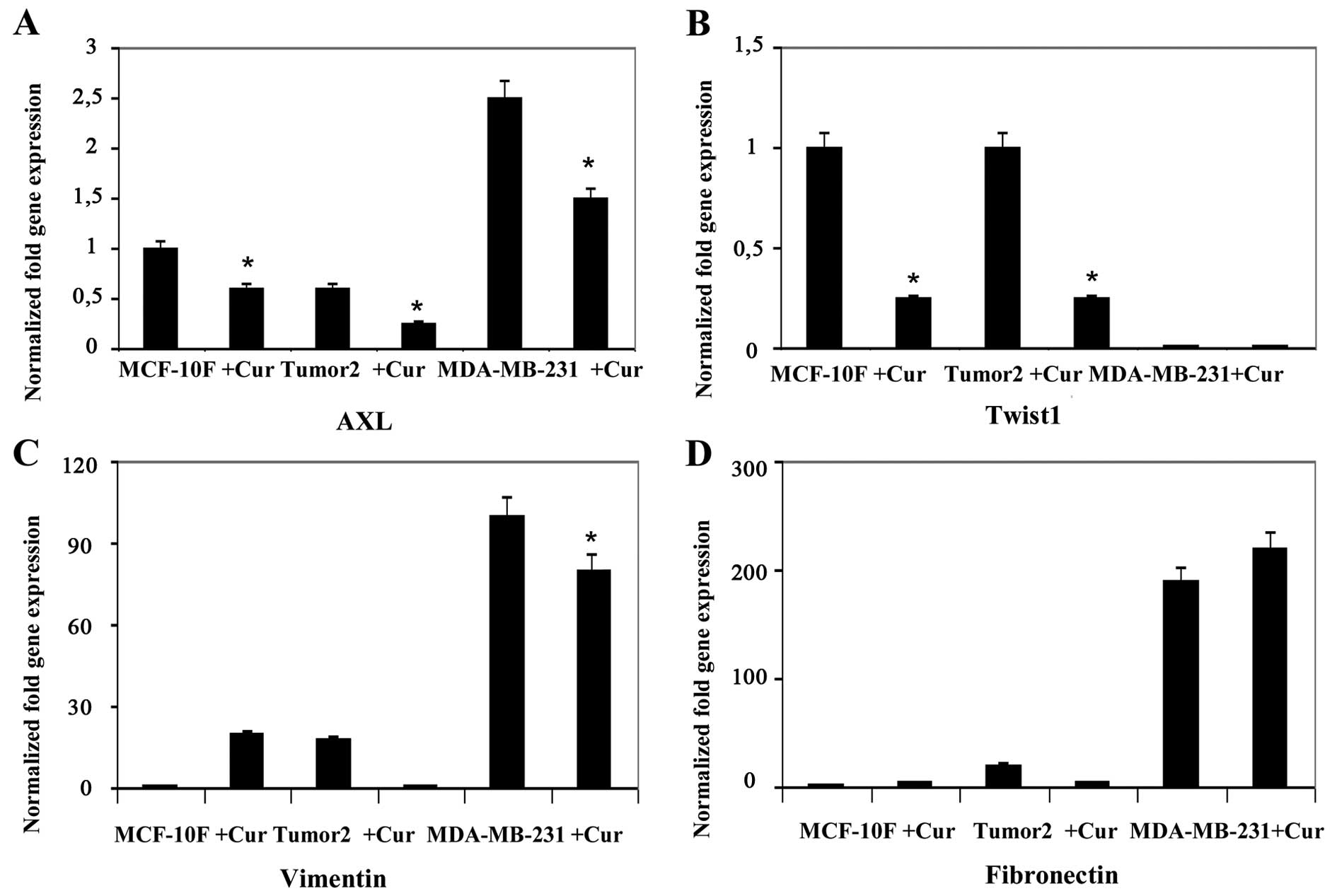
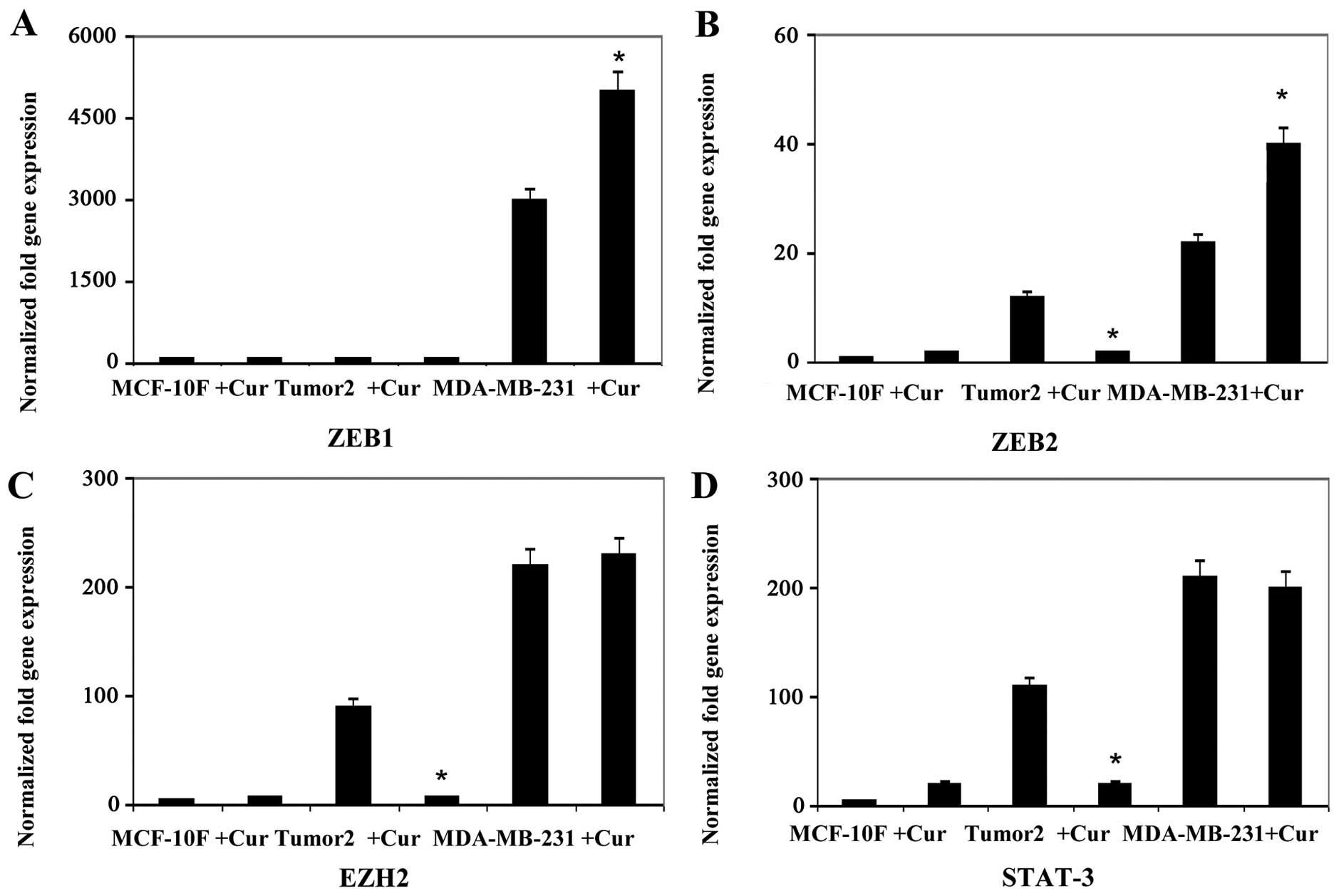
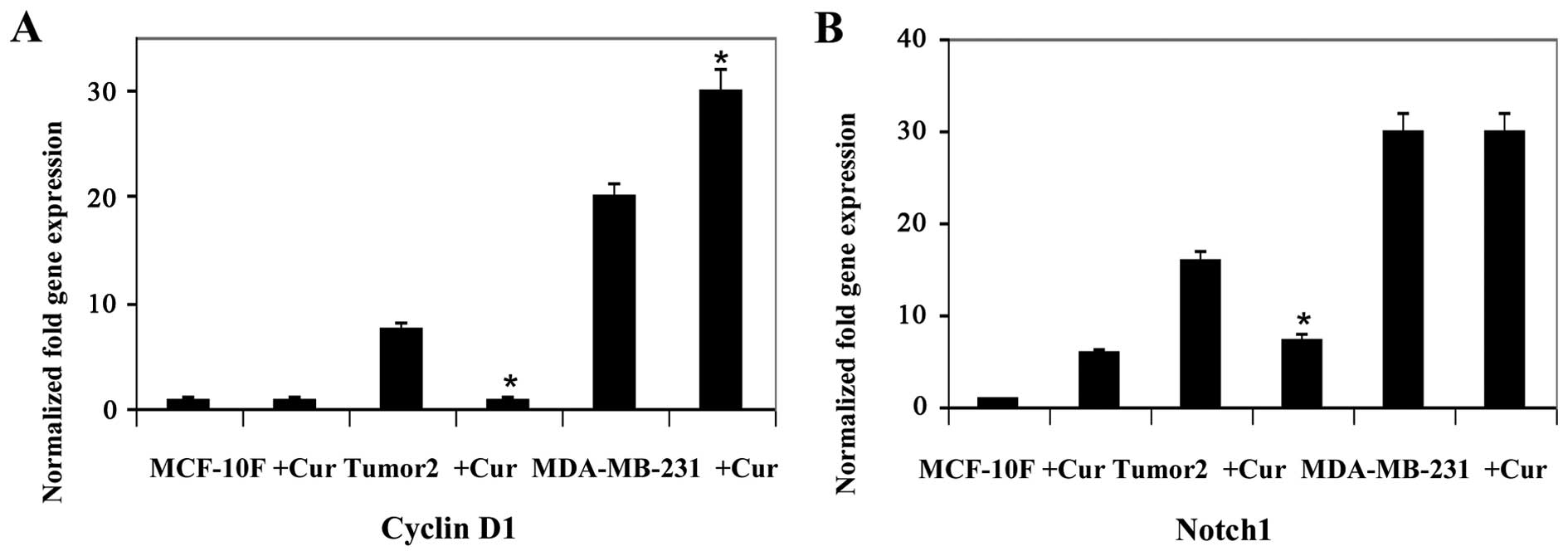
analysis. Results indicated that curcumin decreased gene expression
of twelve genes, E-Cad, N-Cad, Slug, AXL, Twist1, Vimentin,
Fibronectin, ZEB2, EZH2, STAT3, Cyclin D1 and Notch1 in Tumor2
triple-positive for ER, PgR and ErbB2 protein expression (Figs. 1Figure 2Figure 3–4). However, curcumin only decreased gene
expression of four genes, β-catenin and Slug (Fig. 1), AXL and Vimentin (Fig. 2) in MDA-MB-231 triple-negative
cells for the same markers used in clinic. Curcumin increased gene
expression of E-Cad and N-Cad (Fig.
1), ZEB1 (Fig. 3), Cyclin D1
(Fig. 4) in MDA-MB-231
triple-negative. MCF-10F decreased gene expression of four genes,
E-Cad, Slug, AXL and Twist1 (Fig.
2), and it increased gene expression of eight genes, N-Cad,
β-catenin, Vimentin, Fibronectin (Fig.
2), ZEB2, EZH2 and STAT3 (Fig.
3) and Notch1 (Fig. 4).
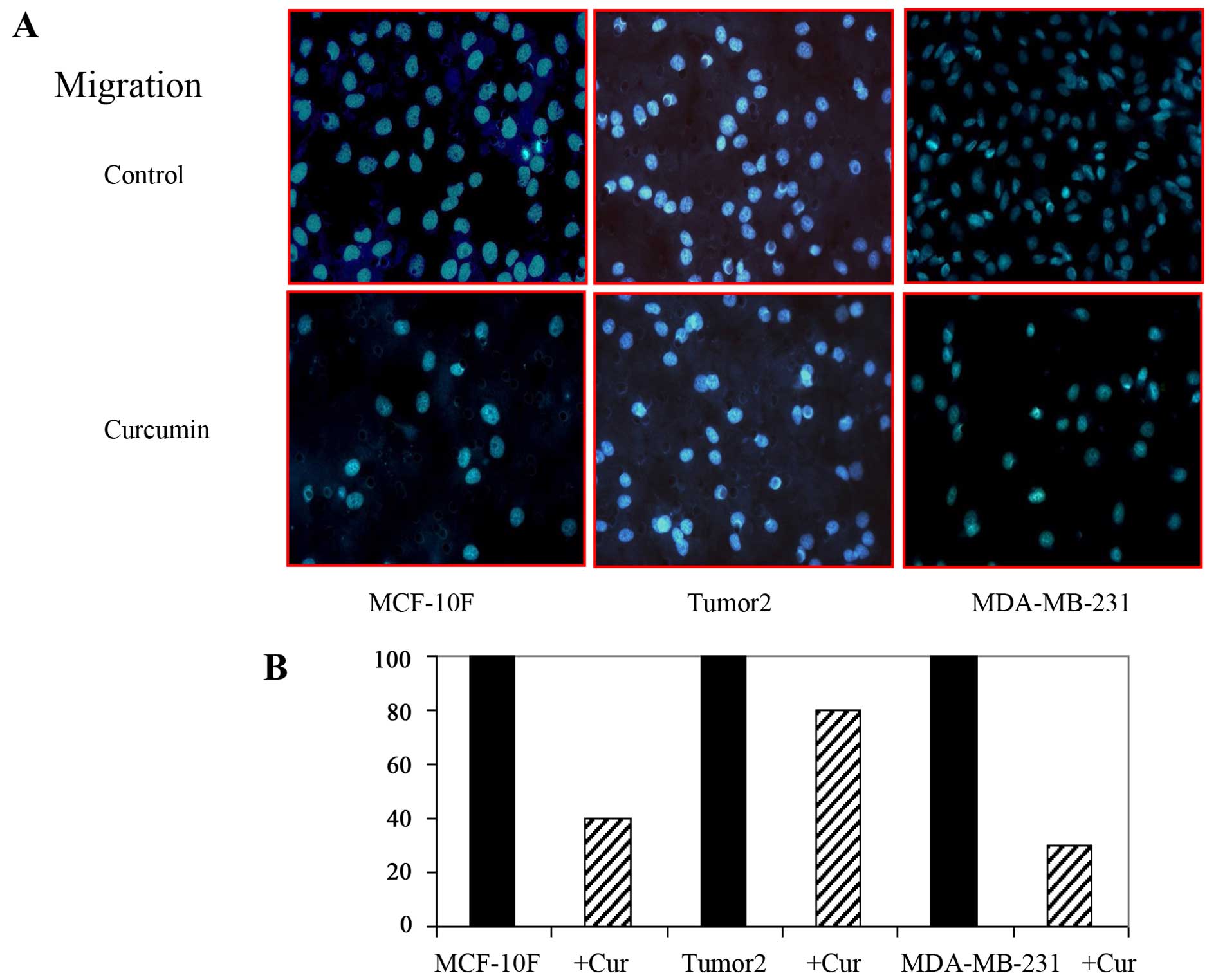
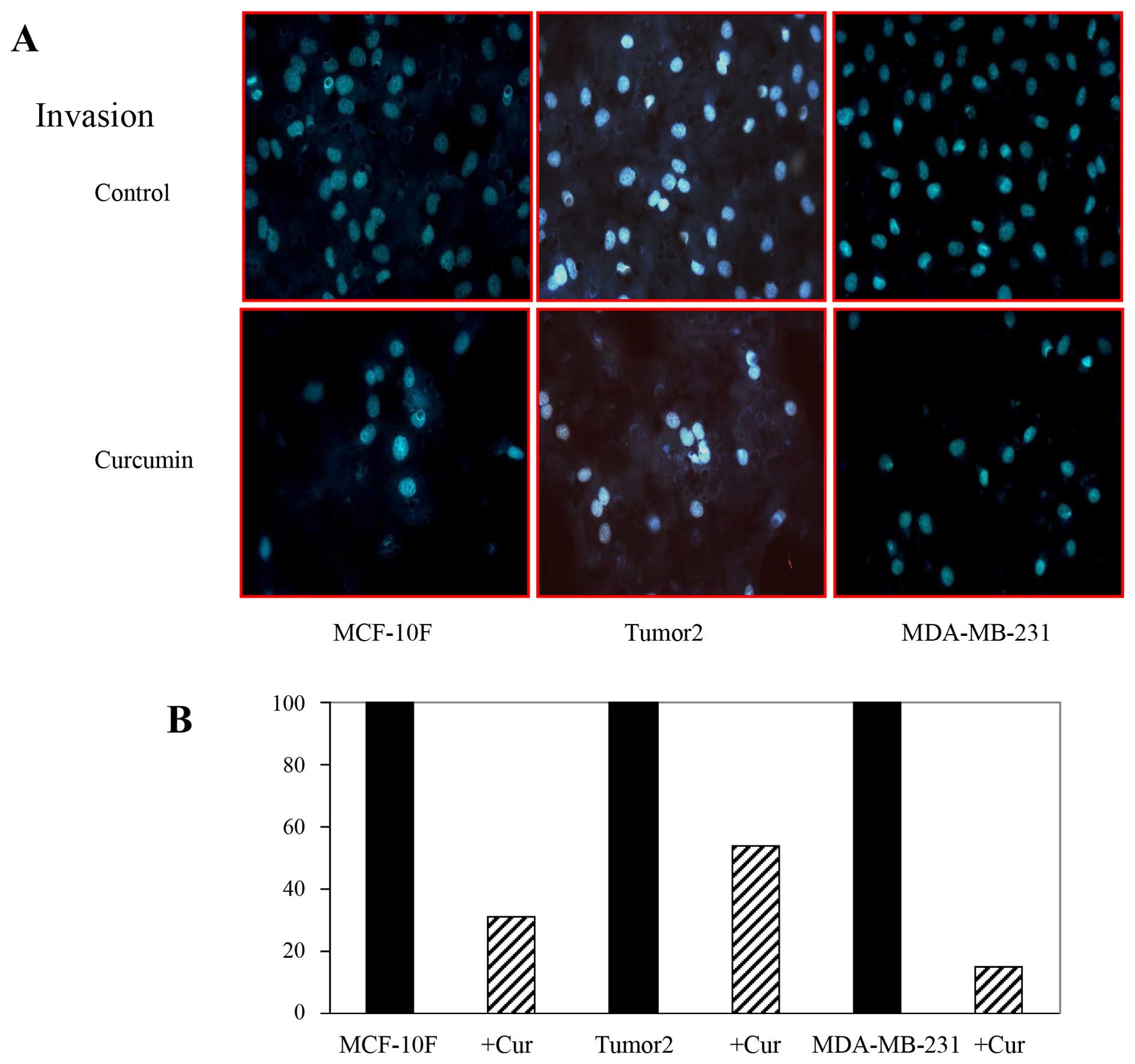
Effect of curcumin on migration and
invasion of breast cancer cells
EMT is associated with metastasis. The motile
phenotypes of cells treated with curcumin were evaluated. The
number of migratory (Fig. 5) and
invasive (Fig. 6) capabilities of
cells was significantly reduced in cells after treatment with
curcumin. Thus, our study suggested that curcumin could delay
cancer progression through its ability to disrupt EMT. It can be
concluded that curcumin influenced biochemical changes associated
with EMT.
Discussion
Curcumin has been shown to inhibit carcinogen
activation and angiogenesis, modulate cell survival and apoptosis,
with anti-invasive and anti-metastatic effects on breast, lung,
colon and prostate cancer (57).
Curcumin reduced cell proliferation of MCF-10F, Tumor2 and
MDA-MB-231 cell lines after 48 h when cell viability was measured
by MTT assay. Cell proliferation was inhibited in a dose-dependent
manner with evident inhibition at dose ≥30 μmol/l). Curcumin has
demonstrated antioxidant and antiproliferative properties in breast
cancer and seems to induce a G2/M phase arrest (58–60).
EMT has a role in embryonic development and cancer
progression, where epithelial cells acquire mesenchymal phenotypes.
It reduces cell-to-cell adhesion, loses cell polarity, enhances
migratory and invasive capabilities (61); then tumor cells migrate from their
site of origin to other tissues activating specific genetic changes
(62). During EMT, epithelial
cancer cell layers lose polarity, cell-to-cell contact and then
undergo a dramatic remodeling of the cytoskeleton. Expression of
E-cadherin and γ-catenin are lost and cells acquire mesenchymal
markers such as N-cadherin, vimentin and fibronectin enhancing the
ability for cell migration and invasion (63). Once tumor cells migrate they
re-express E-cadherin and other epithelial markers through a
process that is often referred to as mesenchymal-epithelial
transition (MET) (64). Therefore,
agents that block or reverse these processes offer a therapeutic
strategy to avoid cancer progression. Curcumin can re-establish an
epithelial phenotype from mesenchymal cells by blocking EMT-related
gene expression.
EMT induction is driven by interplay between tumor
environment and cancer cells which mechanisms may activate
different transcriptional factors such as Twist, Slug and Snail,
through multiple cellular signaling pathways (65–69).
Analysis of the expression of EMT-related genes indicated that
curcumin decreased gene expression of E-cadherin, Slug, AXL
and Twist1 in MCF-10F cell line (four genes). While the
substance decreased N-cadherin, β-catenin, Slug, AXL, Twist1,
Vimentin, Fibronectin, ZEB2, EZH2 and STAT3 in Tumor2
(ten genes) and in MDA-MB-231, triple-negative cell lines such as
E-cadherin, N-cadherin, Twist1, AXL and Fibronectin
(five genes) gene expression in comparison to its counterpart. It
is important to conclude that EMT was triggered by curcumin in
MDA-MB-231 cells since it not only decreased the expression of EMT
genes but induced morphological changes and inhibited cell motility
and invasiveness.
Curcumin did not induce significant difference in
Fibronectin gene expression in MCF-10F or Tumor2. However,
curcumin decreased gene expression of AXL, and
Fibronectin in MDA-MB-231 cell line. During cancer
progression carcinoma cells seem to enter into an EMT program,
acquiring features of mesenchymal-like cells that influenced
invasiveness (62). Evidence has
shown that EMT is involved in malignant progression by inducing
genes such as Slug, AXL and Twist1 since such genes
are expressed in numerous tumor types favoring the metastatic
process.
E-cadherin plays an important role in
epithelial cell adhesion and acts as a metastatic suppressor in
epithelial carcinomas since loss of E-cadherin is associated
with advanced diseases (70).
Vimentin is found in mesenchymal cells and its expression has been
observed in the progression of EMT, tumor cells that are highly
proliferative and invasive (71).
E-cadherin and Vimentin are markers of EMT and directly
regulated by Slug (72,73) since many preventive agents
effectively inhibit EMT by inhibiting Slug transcription factors.
In this study curcumin inhibited Slug expression, affecting
E-cadherin and vimentin to retard cancer cell invasion and
providing new mechanistic bases for therapeutic use in breast
cancer patients.
Cyclin D1 gene plays a critical role in breast
carcinogenesis. It seems that the antiproliferative effects of
curcumin are due to inhibition of Cyclin D1 expression (74). Thus, decreased expression of Cyclin
D1 was observed in Tumor2 and MDA-MB-231. Others reported a
decrease in the Cyclin D1 protein expression with curcumin
treatment (75). Curcumin
inhibited the expression of several EMT markers such as β-catenin
and Slug in both Tumor2 and MDA-MB-231. Our data demonstrated the
efficacy of curcumin since it reduced Notch1 expression suggesting
its antimetastasis function through downregulation of EMT genes and
by promoting other genes. It was also found that curcumin inhibited
migration and invasion of breast cancer cells. It was reported that
curcumin inhibits the migration and invasion of lung (76) and breast (77) cancer cells. These data provide a
new perspective on the role of curcumin in the anti-invasive
properties of breast cancer cells by its ability to interfere with
the EMT process. Most importantly, this study demonstrated and
clarified the potential effect of curcumin to inhibit EMT-related
gene expression in a triple-positive and a triple-negative cell
line.
Acknowledgements
The technical support of Guiliana Rojas, Georgina
Vargas Marchant and Leodán A. Crispin is greatly appreciated. This
study was supported by grant support FONDECYT no. 1120006 (G.M.C.)
and MINEDUC-UTA (G.M.C.).
References
|
1
|
Beiki O, Hall P, Ekbom A and Moradi T:
Breast cancer incidence and case fatality among 4.7 million women
in relation to social and ethnic background: A population-based
cohort study. Breast Cancer Res. 14:R52012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malfettone A, Saponaro C, Paradiso A,
Simone G and Mangia A: Peritumoral vascular invasion and NHERF1
expression define an immunophenotype of grade 2 invasive breast
cancer associated with poor prognosis. BMC Cancer. 12:1062012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cowin P and Welch DR: Breast cancer
progression: Controversies and consensus in the molecular
mechanisms of metastasis and EMT. J Mammary Gland Biol Neoplasia.
12:99–102. 2007. View Article : Google Scholar
|
|
4
|
Naithani R, Huma LC, Moriarty RM,
McCormick DL and Mehta RG: Comprehensive review of cancer
chemopreventive agents evaluated in experimental carcinogenesis
models and clinical trials. Curr Med Chem. 15:1044–1071. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Verschoyle RD, Steward WP and Gescher AJ:
Putative cancer chemopreventive agents of dietary origin-how safe
are they? Nutr Cancer. 59:152–162. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Basnet P and Skalko-Basnet N: Curcumin: An
anti-inflammatory molecule from a curry spice on the path to cancer
treatment. Molecules. 16:4567–4598. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takeichi M: Morphogenetic roles of classic
cadherins. Curr Opin Cell Biol. 7:619–627. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotb AM, Hierholzer A and Kemler R:
Replacement of E-cadherin by N-cadherin in the mammary gland leads
to fibrocystic changes and tumor formation. Breast Cancer Res.
13:R1042011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Radice GL, Rayburn H, Matsunami H, Knudsen
KA, Takeichi M and Hynes RO: Developmental defects in mouse embryos
lacking N-cadherin. Dev Biol. 181:64–78. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ozawa M, Baribault H and Kemler R: The
cytoplasmic domain of the cell adhesion molecule uvomorulin
associates with three independent proteins structurally related in
different species. EMBO J. 8:1711–1717. 1989.PubMed/NCBI
|
|
11
|
Hoschuetzky H, Aberle H and Kemler R:
Beta-catenin mediates the interaction of the cadherin-catenin
complex with epidermal growth factor receptor. J Cell Biol.
127:1375–1380. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Williams EJ, Furness J, Walsh FS and
Doherty P: Activation of the FGF receptor underlies neurite
outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron.
13:583–594. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ilyas M and Tomlinson IP: The interactions
of APC, E-cadherin and beta-catenin in tumour development and
progression. J Pathol. 182:128–137. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nieto MA: The snail superfamily of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barrallo-Gimeno A and Nieto MA: The Snail
genes as inducers of cell movement and survival: Implications in
development and cancer. Development. 132:3151–3161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hemavathy K, Guru SC, Harris J, Chen JD
and Ip YT: Human Slug is a repressor that localizes to sites of
active transcription. Mol Cell Biol. 20:5087–5095. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu WS, Heinrichs S, Xu D, Garrison SP,
Zambetti GP, Adams JM and Look AT: Slug antagonizes p53-mediated
apoptosis of hematopoietic progenitors by repressing puma. Cell.
123:641–653. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
O’Bryan JP, Frye RA, Cogswell PC, Neubauer
A, Kitch B, Prokop C, Espinosa R III, Le Beau MM, Earp HS and Liu
ET: axl, a transforming gene isolated from primary human myeloid
leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell
Biol. 11:5016–5031. 1991. View Article : Google Scholar
|
|
20
|
Bose R, Molina H, Patterson AS, Bitok JK,
Periaswamy B, Bader JS, Pandey A and Cole PA: Phosphoproteomic
analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci
USA. 103:9773–9778. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hafizi S and Dahlbäck B: Gas6 and protein
S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase
subfamily. FEBS J. 273:5231–5244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hutterer M, Knyazev P, Abate A, Reschke M,
Maier H, Stefanova N, Knyazeva T, Barbieri V, Reindl M, Muigg A, et
al: Axl and growth arrest-specific gene 6 are frequently
overexpressed in human gliomas and predict poor prognosis in
patients with glioblastoma multiforme. Clin Cancer Res. 14:130–138.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Ye X, Tan C, Hongo JA, Zha J, Liu J,
Kallop D, Ludlam MJ and Pei L: Axl as a potential therapeutic
target in cancer: Role of Axl in tumor growth, metastasis and
angiogenesis. Oncogene. 28:3442–3455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang YX, Knyazev PG, Cheburkin YV, Sharma
K, Knyazev YP, Orfi L, Szabadkai I, Daub H, Kéri G and Ullrich A:
AXL is a potential target for therapeutic intervention in breast
cancer progression. Cancer Res. 68:1905–1915. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Casas E, Kim J, Bendesky A, Ohno-Machado
L, Wolfe CJ and Yang J: Snail2 is an essential mediator of
Twist1-induced epithelial mesenchymal transition and metastasis.
Cancer Res. 71:245–254. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eckert MA, Lwin TM, Chang AT, Kim J, Danis
E, Ohno-Machado L and Yang J: Twist1-induced invadopodia formation
promotes tumor metastasis. Cancer Cell. 19:372–386. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Low-Marchelli JM, Ardi VC, Vizcarra EA,
van Rooijen N, Quigley JP and Yang J: Twist1 induces CCL2 and
recruits macrophages to promote angiogenesis. Cancer Res.
73:662–671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sommers CL, Skerker JM, Chrysogelos SA,
Bosseler M and Gelmann EP: Regulation of vimentin gene
transcription in human breast cancer cell lines. Cell Growth
Differ. 5:839–846. 1994.PubMed/NCBI
|
|
30
|
Manabe R, Oh-e N and Sekiguchi K:
Alternatively spliced EDA segment regulates fibronectin-dependent
cell cycle progression and mitogenic signal transduction. J Biol
Chem. 274:5919–5924. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ohnishi T, Hiraga S, Izumoto S, Matsumura
H, Kanemura Y, Arita N and Hayakawa T: Role of
fibronectin-stimulated tumor cell migration in glioma invasion in
vivo: Clinical significance of fibronectin and fibronectin receptor
expressed in human glioma tissues. Clin Exp Metastasis. 16:729–741.
1998. View Article : Google Scholar
|
|
32
|
Sakai T, Johnson KJ, Murozono M, Sakai K,
Magnuson MA, Wieloch T, Cronberg T, Isshiki A, Erickson HP and
Fässler R: Plasma fibronectin supports neuronal survival and
reduces brain injury following transient focal cerebral ischemia
but is not essential for skin-wound healing and hemostasis. Nat
Med. 7:324–330. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moursi AM, Damsky CH, Lull J, Zimmerman D,
Doty SB, Aota S and Globus RK: Fibronectin regulates calvarial
osteoblast differentiation. J Cell Sci. 109:1369–1380.
1996.PubMed/NCBI
|
|
34
|
Johansson S, Svineng G, Wennerberg K,
Armulik A and Lohikangas L: Fibronectin-integrin interactions.
Front Biosci. 2:d126–d146. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sottile J and Hocking DC: Fibronectin
polymerization regulates the composition and stability of
extracellular matrix fibrils and cell-matrix adhesions. Mol Biol
Cell. 13:3546–3559. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goerges AL and Nugent MA: pH regulates
vascular endothelial growth factor binding to fibronectin: A
mechanism for control of extracellular matrix storage and release.
J Biol Chem. 279:2307–2315. 2004. View Article : Google Scholar
|
|
37
|
Vannier C, Mock K, Brabletz T and Driever
W: Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion
molecule) expression to control cell behavior in early zebrafish
development. J Biol Chem. 288:18643–18659. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Verschueren K, Remacle JE, Collart C,
Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT,
Bodmer R, et al: SIP1, a novel zinc finger/homeodomain repressor,
interacts with Smad proteins and binds to 5′-CACCT sequences in
candidate target genes. J Biol Chem. 274:20489–20498. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Remacle JE, Kraft H, Lerchner W, Wuytens
G, Collart C, Verschueren K, Smith JC and Huylebroeck D: New mode
of DNA binding of multi-zinc finger transcription factors: deltaEF1
family members bind with two hands to two target sites. EMBO J.
18:5073–5084. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Francis NJ and Kingston RE: Mechanisms of
transcriptional memory. Nat Rev Mol Cell Biol. 2:409–421. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chang CJ and Hung MC: The role of EZH2 in
tumour progression. Br J Cancer. 106:243–247. 2012. View Article : Google Scholar :
|
|
43
|
Min J, Zaslavsky A, Fedele G, McLaughlin
SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE,
Beroukhim R, et al: An oncogene-tumor suppressor cascade drives
metastatic prostate cancer by coordinately activating Ras and
nuclear factor-kappaB. Nat Med. 16:286–294. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bracken AP, Pasini D, Capra M, Prosperini
E, Colli E and Helin K: EZH2 is downstream of the pRB-E2F pathway,
essential for proliferation and amplified in cancer. EMBO J.
22:5323–5335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu
H, Sun L, Zhang Y, Chen Y, Li R, et al: Integration of estrogen and
Wnt signaling circuits by the polycomb group protein EZH2 in breast
cancer cells. Mol Cell Biol. 27:5105–5119. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis
RT, Wu X, Stack EC, Loda M, Liu T, et al: EZH2 oncogenic activity
in castration-resistant prostate cancer cells is
Polycomb-independent. Science. 338:1465–1469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee ST, Li Z, Wu Z, Aau M, Guan P,
Karuturi RK, Liou YC and Yu Q: Context-specific regulation of NF-κB
target gene expression by EZH2 in breast cancers. Mol Cell.
43:798–810. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Asangani IA, Ateeq B, Cao Q, Dodson L,
Pandhi M, Kunju LP, Mehra R, Lonigro RJ, Siddiqui J, Palanisamy N,
et al: Characterization of the EZH2-MMSET histone methyltransferase
regulatory axis in cancer. Mol Cell. 49:80–93. 2013. View Article : Google Scholar :
|
|
49
|
Takeda K and Akira S: STAT family of
transcription factors in cytokine-mediated biological responses.
Cytokine Growth Factor Rev. 11:199–207. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou P, Jiang W, Weghorst CM and Weinstein
IB: Overexpression of cyclin D1 enhances gene amplification. Cancer
Res. 56:36–39. 1996.PubMed/NCBI
|
|
51
|
Arnold A and Papanikolaou A: Cyclin D1 in
breast cancer pathogenesis. J Clin Oncol. 23:4215–4224. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Stylianou S, Clarke RB and Brennan K:
Aberrant activation of notch signaling in human breast cancer.
Cancer Res. 66:1517–1525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mungamuri SK, Yang X, Thor AD and
Somasundaram K: Survival signaling by Notch1: Mammalian target of
rapamycin (mTOR)-dependent inhibition of p53. Cancer Res.
66:4715–4724. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee CW, Simin K, Liu Q, Plescia J, Guha M,
Khan A, Hsieh CC and Altieri DC: A functional Notch-survivin gene
signature in basal breast cancer. Breast Cancer Res. 10:R972008.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dickson BC, Mulligan AM, Zhang H, et al:
High-level JAG1 mRNA and protein predict poor outcome in breast
cancer. Mod Pathol. 20:685–693. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jagtap S, Meganathan K, Wagh V, Winkler J,
Hescheler J and Sachinidis A: Chemoprotective mechanism of the
natural compounds, epigallocatechin-3-O-gallate, quercetin and
curcumin against cancer and cardiovascular diseases. Curr Med Chem.
16:1451–1462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Calaf GM, Echiburú-Chau C, Wen G, Balajee
AS and Roy D: Effect of curcumin on irradiated and
estrogen-transformed human breast cell lines. Int J Oncol.
40:436–442. 2012.
|
|
59
|
Weissenberger J, Priester M, Bernreuther
C, Rakel S, Glatzel M, Seifert V and Kögel D: Dietary curcumin
attenuates glioma growth in a syngeneic mouse model by inhibition
of the JAK1,2/STAT3 signaling pathway. Clin Cancer Res.
16:5781–5795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Calaf GM, Echiburú-Chau C, Roy D, Chai Y,
Wen G and Balajee AS: Protective role of curcumin in oxidative
stress of breast cells. Oncol Rep. 26:1029–1035. 2011.PubMed/NCBI
|
|
61
|
Tiwari N, Gheldof A, Tatari M and
Christofori G: EMT as the ultimate survival mechanism of cancer
cells. Semin Cancer Biol. 22:194–207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mulholland DJ, Kobayashi N, Ruscetti M,
Zhi A, Tran LM, Huang J, Gleave M and Wu H: Pten loss and RAS/MAPK
activation cooperate to promote EMT and metastasis initiated from
prostate cancer stem/progenitor cells. Cancer Res. 72:1878–1889.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Moreno-Bueno G, Portillo F and Cano A:
Transcriptional regulation of cell polarity in EMT and cancer.
Oncogene. 27:6958–6969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Creighton CJ, Chang JC and Rosen JM:
Epithelial-mesenchymal transition (EMT) in tumor-initiating cells
and its clinical implications in breast cancer. J Mammary Gland
Biol Neoplasia. 15:253–260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Vuoriluoto K, Haugen H, Kiviluoto S,
Mpindi JP, Nevo J, Gjerdrum C, Tiron C, Lorens JB and Ivaska J:
Vimentin regulates EMT induction by Slug and oncogenic H-Ras and
migration by governing Axl expression in breast cancer. Oncogene.
30:1436–1448. 2011. View Article : Google Scholar
|
|
66
|
Hardy KM, Booth BW, Hendrix MJ, Salomon DS
and Strizzi L: ErbB/EGF signaling and EMT in mammary development
and breast cancer. J Mammary Gland Biol Neoplasia. 15:191–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Vincan E and Barker N: The upstream
components of the Wnt signalling pathway in the dynamic EMT and MET
associated with colorectal cancer progression. Clin Exp Metastasis.
25:657–663. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Javle MM, Gibbs JF, Iwata KK, Pak Y,
Rutledge P, Yu J, Black JD, Tan D and Khoury T:
Epithelial-mesenchymal transition (EMT) and activated extracellular
signal-regulated kinase (p-Erk) in surgically resected pancreatic
cancer. Ann Surg Oncol. 14:3527–3533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yin T, Wang C, Liu T, Zhao G and Zhou F:
Implication of EMT induced by TGF-beta1 in pancreatic cancer. J
Huazhong Univ Sci Technolog Med Sci. 26:700–702. 2006. View Article : Google Scholar
|
|
70
|
Pinho SS, Oliveira P, Cabral J, Carvalho
S, Huntsman D, Gärtner F, Seruca R, Reis CA and Oliveira C: Loss
and recovery of Mgat3 and GnT-III Mediated E-cadherin
N-glycosylation is a mechanism involved in
epithelial-mesenchymal-epithelial transitions. PLoS One.
7:e331912012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ivaska J: Vimentin: Central hub in EMT
induction? Small GTPases. 2:51–53. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wu Y and Zhou BP: Snail: More than EMT.
Cell Adhes Migr. 4:199–203. 2010. View Article : Google Scholar
|
|
73
|
Fendrich V, Waldmann J, Feldmann G,
Schlosser K, König A, Ramaswamy A, Bartsch DK and Karakas E: Unique
expression pattern of the EMT markers Snail, Twist and E-cadherin
in benign and malignant parathyroid neoplasia. Eur J Endocrinol.
160:695–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mukhopadhyay A, Banerjee S, Stafford LJ,
Xia C, Liu M and Aggarwal BB: Curcumin-induced suppression of cell
proliferation correlates with down-regulation of cyclin D1
expression and CDK4-mediated retinoblastoma protein
phosphorylation. Oncogene. 21:8852–8861. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kumaravel M, Sankar P and Rukkumani R:
Antiproliferative effect of an analog of curcumin
bis-1,7-(2-hydroxyphenyl)-hepta-1,6-diene-3,5-dione in human breast
cancer cells. Eur Rev Med Pharmacol Sci. 16:1900–1907.
2012.PubMed/NCBI
|
|
76
|
Lin SS, Lai KC, Hsu SC, Yang JS, Kuo CL,
Lin JP, Ma YS, Wu CC and Chung JG: Curcumin inhibits the migration
and invasion of human A549 lung cancer cells through the inhibition
of matrix metalloproteinase-2 and -9 and vascular endothelial
growth factor (VEGF). Cancer Lett. 285:127–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Huang T, Chen Z and Fang L: Curcumin
inhibits LPS-induced EMT through downregulation of NF-κB-Snail
signaling in breast cancer cells. Oncol Rep. 29:117–124. 2013.
|