Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common cancer in the world, and it consists of a
heterogeneous group of malignancies arising from the oral cavity,
paranasal sinus, pharynx, larynx and salivary glands (1). Most of oral squamous cell carcinoma
(OSCC) occurs from oral cavity (accounts for >95%) and is the
most common type of HNSCC (2).
Despite recent advances in various treatment modalities, including
surgery, radiotherapy, chemotherapy and molecularly targeted
therapy, the survival rate of patients with OSCC has not markedly
improved (5-year survival is <50%) due to the high rate of
locoregional recurrence and distinct metastasis (3). We suggest that it would be possible
to significantly improve diagnosis, therapy, and prevention of OSCC
through a better understanding of the molecular oncogenic processes
and metastatic pathways underlying the disease. We further suggest
that this could be achieved through the use of current genome-based
approaches.
The discovery of microRNA (miRNA) in the human
genome provided new directions in cancer study. miRNAs are
endogenous small non-coding RNAs (19–22 bases long) that regulate
protein-coding/non protein-coding gene expression by repressing
translation or degradation of RNA transcripts in a
sequence-specific manner (4). A
growing body of studies have shown that miRNAs are aberrantly
expressed in many human cancers. Thus, they act pivotal roles in
the initiation, progression and metastasis of such cancers
(5). Moreover, normal RNA networks
can be disrupted by the aberrant expression of tumor-suppressive or
oncogenic miRNAs in cancer cells. Therefore, identifying aberrantly
expressed miRNAs is an important first step toward understanding
miRNA-mediated RNA networks.
Based on this proposal, we have constructed miRNA
expression signatures through genetic analysis of
hypopharyngeal-SCC, maxillary sinus-SCC and OSCC clinical specimens
(6–9). Using these miRNA expression
signatures, we have identified molecular pathways in HNSCC that are
mediated by aberrantly expressed miRNAs. For example,
downregulation of tumor-suppressive miR-375 inhibited cancer
cell apoptosis through dysregulation of AEG-1/MTDH in HNSCC
cells (10). Moreover,
downregulation of miR-874 is a frequent event in HNSCC and
miR-874 acted as a tumor suppressor that directly targets
HDAC1 (11). More recently,
we found that miR-26a and miR-26b function as tumor
suppressors through regulating of TMEM184B based on the OSCC
signature (9).
Our miRNA expression signatures of human cancers,
including OSCC, revealed that clustered miRNAs, miR-23b and
miR-27b were frequently downregulated in several types of
cancer tissues (9,12–14).
Several studies showed that these miRNAs act as tumor suppressive
miRNAs through their targeting of oncogenic genes (15–17).
Up to now, few reports have provided functional analyses of these
clustered miRNAs in OSCC. The aims of the study were to investigate
the functional roles of miR-23b and miR-27b in OSCC
and to identify novel miR-23b/27b-mediated cancer pathways
and target genes involved in OSCC oncogenesis and metastasis. We
expect that this analysis will provide novel insights into the
pivotal molecular mechanisms of OSCC oncogenesis and metastasis.
This new knowledge will facilitate the development of therapeutic
strategies for the treatment of the disease.
Materials and methods
Clinical specimens in patients with OSCC
and cell lines
A total of 37 pairs of cancer tissues and
corresponding normal epithelial tissues were obtained from patients
with OSCC at Chiba University Hospital from 2008 to 2013. The
patients were classified according to the 2002 Union for
International Cancer Control (UICC) staging criteria before
treatment. Prior written informed consent and approval were
obtained from all patients. The patients’ backgrounds and
clinicopathological characteristics are shown in Table I. The following human OSCC cell
lines were used: SAS (derived from a primary tongue SCC) and HSC3
(derived from a lymph node metastasis of tongue SCC).
 | Table IClinical features of 37 OSCC
patients. |
Table I
Clinical features of 37 OSCC
patients.
| No. | Age | Sex | Location | T | N | M | Stage |
Differentiation |
|---|
| 1 | 66 | M | Tongue | 2 | 0 | 0 | II | Moderate |
| 2 | 65 | M | Oral floor | 4a | 1 | 0 | IVA | Moderate |
| 3 | 67 | M | Tongue | 4a | 2c | 0 | IVA | Moderate |
| 4 | 36 | F | Tongue | 3 | 1 | 0 | III | Moderate |
| 5 | 73 | M | Tongue | 3 | 2b | 0 | IVA | Poor |
| 6 | 63 | F | Oral floor | 2 | 2b | 0 | IVA | Basaloid SCC |
| 7 | 77 | M | Gum | 2 | 0 | 0 | II | Moderate |
| 8 | 68 | M | Tongue | 2 | 0 | 0 | II | Well |
| 9 | 76 | F | Tongue | 1 | 0 | 0 | I | Well |
| 10 | 69 | M | Tongue | 1 | 0 | 0 | I | Well |
| 11 | 73 | F | Tongue | 1 | 0 | 0 | I | Well |
| 12 | 64 | M | Tongue | 1 | 0 | 0 | I | Well |
| 13 | 64 | M | Tongue | 1 | 0 | 0 | I | Well |
| 14 | 82 | M | Oral floor | 1 | 0 | 0 | I | Well |
| 15 | 67 | M | Oral floor | 4a | 2b | 0 | IVA | Well |
| 16 | 67 | M | Tongue | 3 | 0 | 0 | III | Moderate |
| 17 | 64 | M | Tongue | 3 | 2b | 0 | IVA | Moderate |
| 18 | 59 | M | Tongue | 1 | 2a | 0 | IVA | Moderate |
| 19 | 47 | M | Oral floor | 1 | 0 | 0 | I | Moderate |
| 20 | 67 | M | Tongue | 2 | 0 | 0 | II | Poor-moderate |
| 21 | 70 | M | Tongue | 1 | 0 | 0 | I | Well |
| 22 | 38 | M | Tongue | 1 | 0 | 0 | I | Well |
| 23 | 70 | M | Tongue, oral
floor | 2 | 0 | 0 | II | Well |
| 24 | 51 | M | Tongue | 1 | 0 | 0 | I | Well |
| 25 | 81 | M | Tongue | is | 0 | 0 | 0 | Extremely well |
| 26 | 34 | F | Tongue | 1 | 0 | 0 | I | Poor |
| 27 | 42 | M | Gum | 4a | 0 | 0 | IVA | Moderate |
| 28 | 70 | M | Tongue | 1 | 0 | 0 | I | Moderate |
| 29 | 71 | M | Tongue | 1 | 0 | 0 | I | Well |
| 30 | 60 | F | Tongue | 2 | I | 0 | III | Well |
| 31 | 77 | M | Tongue | 2 | 2b | 0 | IVA | Poorly |
| 32 | 64 | F | Oral floor | 4a | 2c | 0 | IVA | Moderate |
| 33 | 68 | M | Tongue | 1 | 0 | 0 | I | Well |
| 34 | 39 | M | Tongue | 4a | 0 | 0 | IVA | Well |
| 35 | 29 | F | Tongue | 1 | 0 | 0 | I | Poorly |
| 36 | 71 | M | Buccal mucosa | 2 | 1 | 0 | III | Poorly |
| 37 | 39 | M | Tongue | 4a | 0 | 0 | IVA | Moderate |
RNA isolation
Tissues were immersed in RNAlater (Ambion, Austin,
TX, USA), and stored at 4°C until RNA was extracted. Total RNA was
isolated using TRIzol reagent according to the manufacturer’s
instructions.
Quantitative of miRNAs and messenger RNA
by real-time RT-PCR
The procedure for PCR quantification was described
previously (6–11). The expression levels of
miR-23b (assay ID: 000400) and miR-27b (assay ID:
000409) were analyzed by TaqMan quantitative real-time PCR and
normalized to RNU48 (assay ID: 001006). TaqMan probes and
primers for MET (P/N: Hs01565584_m1), GUSB (P/N: Hs
00939627_ml) and GAPDH (P/N: Hs02758991_g1) as an internal
control were obtained from Applied Biosystems.
Function assays by miRNA and
small-interfering RNA transfection
The following miRNAs mimics were used in this study:
mirVana miRNA mimic for hsa-miR-23b (product ID: PM10711)
and hsa-miR-27b (product ID: PM10750). The transfection
procedures and transfection efficiencies of miRNA for SAS and HSC3
cells were reported in previous studies (6–9,11,15,18).
To investigate the functional significance of miR-23b,
miR-27b and si-MET, we performed cell proliferation,
migration and invasion assays using OSCC cell lines. The
experimental procedures were described in previous studies
(8,9,15,18).
Identification of target genes regulated
by miR-23b, miR-27b by using genome-wide gene expression and in
silico analysis
The miRNA public database (TargetScan) was used for
in silico identification of candidate target genes that
contained miR-23b and miR-27b binding sites in their
3′-untranslated region. These genes were then categorized into KEGG
pathways using the GeneCodis program (http://genecodis.dacya.ucm.es). To identify
upregulated genes in OSCC, we analyzed a publicly available gene
expression data set in GEO (accession no. GSE6631).
Western blotting
Cells were harvested 72 h after transfection and
lysates were prepared. From each lysate, an aliquot containing 20
μg of protein was separated on Mini-PROTEAN TGX Gels (Bio-Rad,
Hercules, CA, USA) and transferred to PVDF membranes.
Immunoblotting was performed with rabbit anti-MET antibodies
(1:1,000); mouse anti-GAPDH antibodies (1:4,000) were used as an
internal loading control. The experimental procedures were
performed as described in our previous studies (6–9,11,15,18).
Immunohistochemistry
Two OSCC clinical specimens were immunostained
following the manufacturer’s protocol with the Ultra-Vision
detection system (Thermo Scientific, Fremont, CA, USA). Primary
rabbit polyclonal antibodies against MET were diluted 1:300. The
slides were treated with biotinylated goat antibodies.
Plasmid construction and dual-luciferase
reporter assays
The partial wild-type sequence of the MET
3′-untranslated region or those with mutated miR-23b/miR-27b
target sites were inserted between the XhoI-PmeI
restriction sites in the 3′-UTR of the hRluc gene in the
psiDHECK-2 vector (C8021; Promega, Madison, WI, USA). The procedure
for the dual-luciferase reporter assay was described previously
(6–9,11,15,18).
Statistical analysis
The relationships between two groups and numerical
values obtained by real-time RT-qPCR were analyzed using
Mann-Whitney U tests. Spearman’s rank test was used to evaluate the
correlation between the expression levels of miR-23b,
miR-27b and MET mRNA. The relationships among more than
three variables and numerical values were analyzed using the
Mann-Whitney U test after Bonferroni adjustment. All analyses were
performed using Expert Stat View (version 5, SAS Institute Inc.,
Cary, NC, USA).
Results
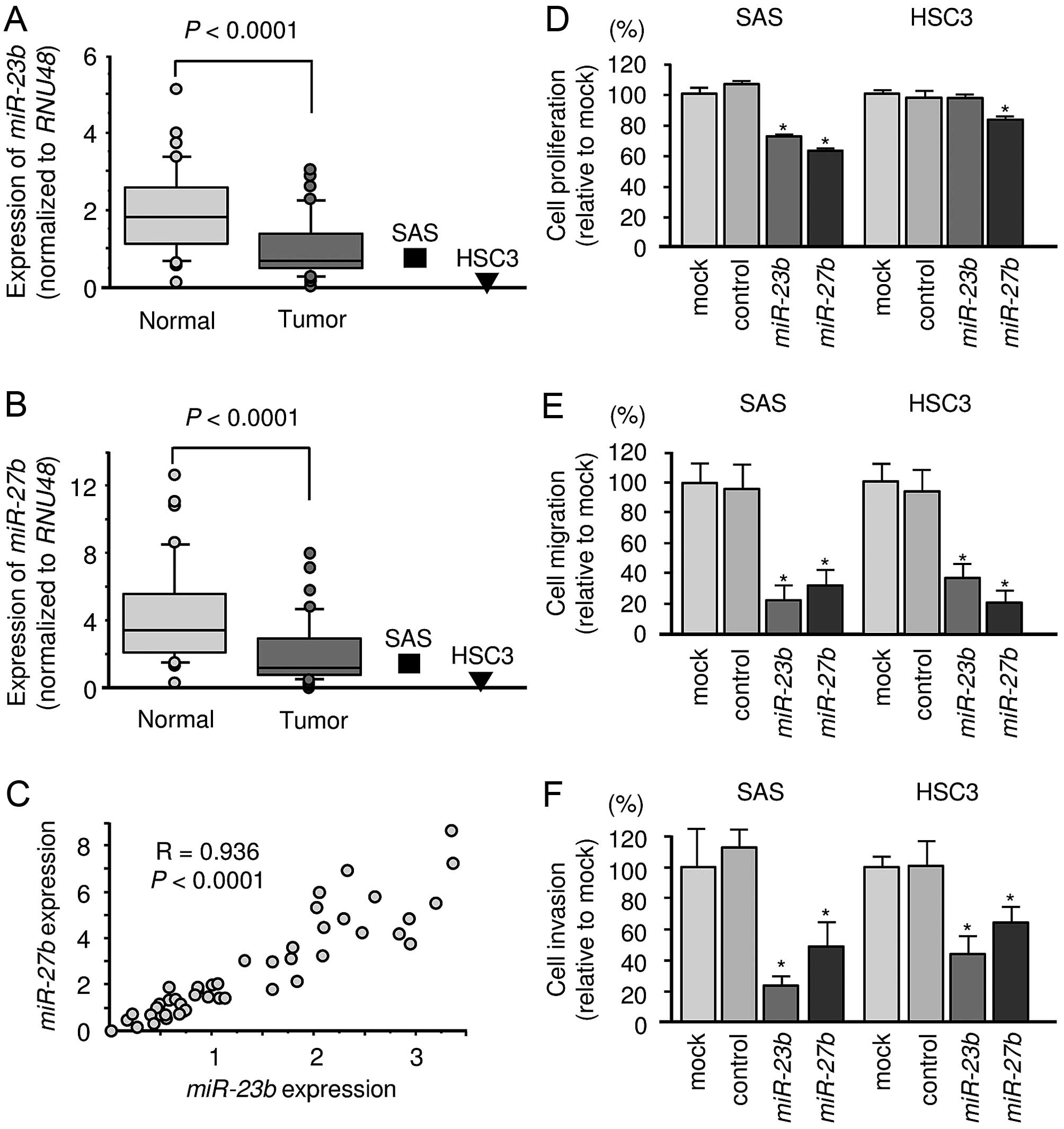
Expression levels of miR-23b and miR-27b
in OSCC tissues and cell lines
We evaluated the expression levels of the clustered
miRNAs in 37 OSCC clinical specimens and two cell lines. The
expression levels of miR-23b and miR-27b were
significantly lower in tumor tissues and cell lines than in
corresponding normal tissues (Fig. 1A
and B). Spearman’s rank test showed a positive correlation
between the expression levels of miR-23b and miR-27b
(Fig. 1C).
Gain-of-function assay of miR-23b and
miR-27b in OSCC cell lines: effects on cell proliferation,
migration and invasion
The functional significance of miR-23b and
miR-27b were investigated using miRNA transfection of OSCC
cell lines. XTT assays demonstrated that SAS cell proliferation was
significantly inhibited in miR-23b- and
miR-27b-transfectants compared with the mock or miR-control
transfected SAS cells. On the other hand, proliferation was
inhibited only in miR-27b transfectant in HSC3 (Fig. 1D). Migration and invasion assays
demonstrated that cell migration and invasion activity were
significantly inhibited in miR-23b and miR-27b
transfectants compared with the mock or miR-control transfectants
in OSCC cell lines (Fig. 1E and
F).
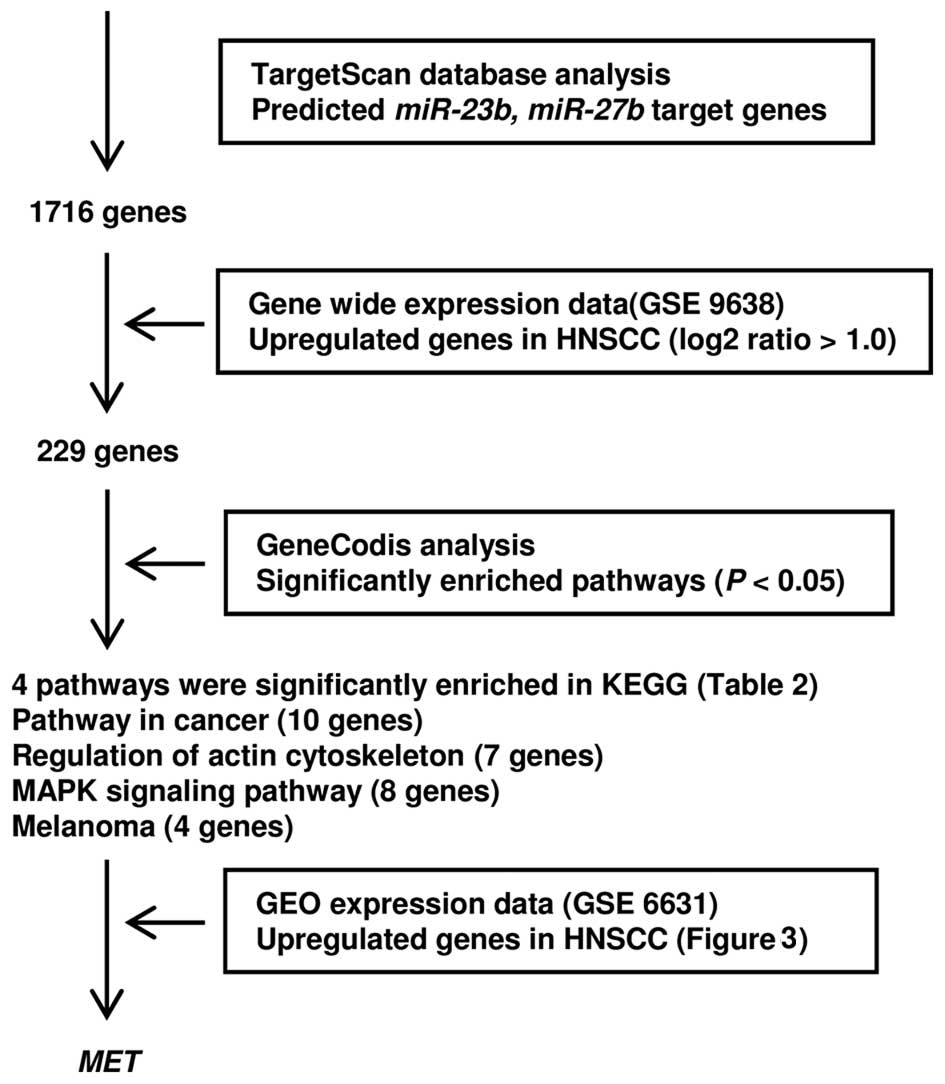
Selection of genes targeted by miR-23b
and miR-27b in OSCC
To identify genes targeted by miR-23b and
miR-27b, we use in silico analyses and genome-wide
expression analyses. Our strategy for identification of target
genes is shown in Fig. 2. First,
we screened putative candidate target genes using the TargetScan
database and identified 229 potential targets. These genes were
classified into KEGG pathways using GeneCodis analysis and four
pathways and 18 genes were identified as significantly enriched
pathways (Table IIA) and genes
(Table IIB–E). The gene set was
then analyzed with a publicly available gene expression data set in
GEO (accession no. GSE6631). In this group of genes, we focused on
the hepatocyte growth factor receptor (MET) because it was
the most significantly upregulated in HNSCC (Fig. 3).
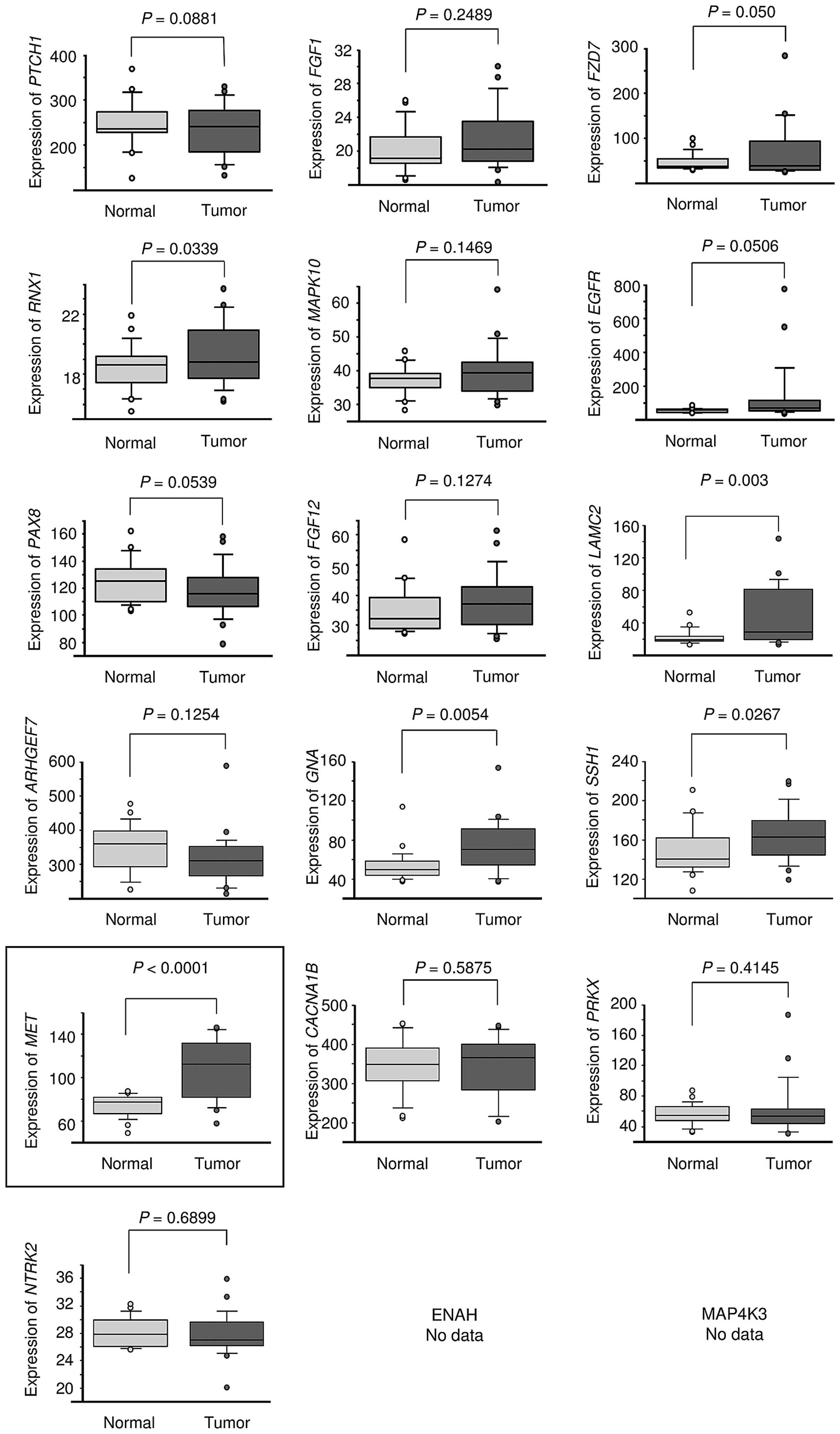
 | Table IIThe KEGG pathways. |
Table II
The KEGG pathways.
| A, Significantly
enriched KEGG pathway regulated by miR-23b/27b cluster |
|---|
|
|---|
| No. of genes | Annotations | P-value |
|---|
| 10 | (KEGG) 05200:
Pathways in cancer | 0.0082 |
| 7 | (KEGG) 04810:
Regulation of actin cytoskeleton | 0.0210 |
| 8 | (KEGG) 04010: MAPK
signaling pathway | 0.0235 |
| 4 | (KEGG) 05218:
Melanoma | 0.0372 |
|
| B, Pathway in
cancer |
|
| Gene symbol | Gene name | HNSCC log2
ratio |
|
| LAMC2 | Laminin, γ2 | 2.33 |
| FGF1 | Fibroblast growth
factor 1 (acidic) | 2.32 |
| PTCH1 | Patched 1 | 2.24 |
| FZD7 | Frizzled family
receptor 7 | 2.18 |
| PAX8 | Paired box 8 | 1.43 |
| FGF12 | Fibroblast growth
factor 12 | 1.39 |
| RUNX1 | Runt-related
transcription factor 1 | 1.27 |
| MET | Met proto-oncogene
(hepatocyte growth factor receptor) | 1.26 |
| MAPK10 | Mitogen-activated
protein kinase 10 | 1.22 |
| EGFR | Epidermal growth
factor receptor | 1.15 |
|
| C, Regulation of
actin cytoskeleton |
|
| Gene symbol | Gene name | HNSCC log2
ratio |
|
| FGF1 | Fibroblast growth
factor 1 (acidic) | 2.32 |
| FGF12 | Fibroblast growth
factor 12 | 1.39 |
| ARHGEF7 | Rho guanine
nucleotide exchange factor (GEF) 7 | 1.34 |
| SSH1 | Slingshot homolog 1
(Drosophila) | 1.20 |
| GNA13 | Guanine nucleotide
binding protein (G protein), α13 | 1.18 |
| EGFR | Epidermal growth
factor receptor | 1.15 |
| ENAH | Enabled homolog
(Drosophila) | 1.09 |
|
| D, MAPK signaling
pathway |
|
| Gene symbol | Gene name | HNSCC log2
ratio |
|
| NTRK2 | Neurotrophic
tyrosine kinase, receptor, type 2 | 3.33 |
| CACNA1B | Calcium channel,
voltage-dependent, N type, α1B subunit | 2.86 |
| FGF1 | Fibroblast growth
factor 1 (acidic) | 2.32 |
| FGF12 | Fibroblast growth
factor 12 | 1.39 |
| MAP4K3 | Mitogen-activated
protein kinase kinase kinase kinase 3 | 1.24 |
| MAPK10 | Mitogen-activated
protein kinase 10 | 1.22 |
| PRKX | Protein kinase,
X-linked | 1.16 |
| EGFR | Epidermal growth
factor receptor | 1.15 |
|
| E, Melanoma |
|
| Gene symbol | Gene name | H average |
|
| FGF1 | Fibroblast growth
factor 1 (acidic) | 2.32 |
| FGF12 | Fibroblast growth
factor 12 | 1.39 |
| MET | Met proto-oncogene
(hepatocyte growth factor receptor) | 1.26 |
| EGFR | Epidermal growth
factor receptor | 1.15 |
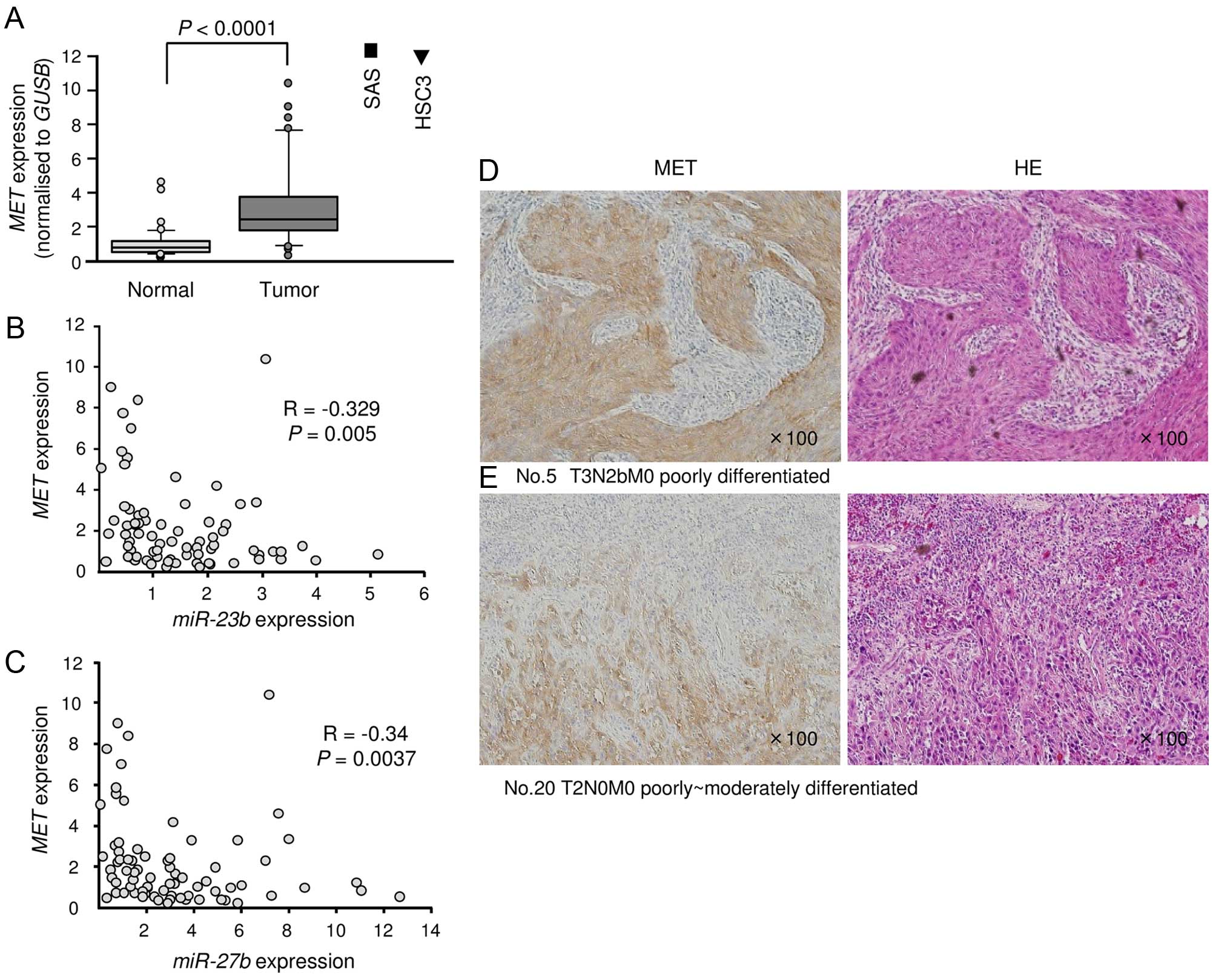
Expression of MET in OSCC clinical
specimens and cell lines
We investigated the expression levels of MET
in 37 OSCC clinical specimens and cell lines. First, qRT-PCR
revealed that MET was significantly upregulated in cancer
tissues and cell lines compared with normal tissues (Fig. 4A). Spearman’s rank test showed
negative correlations between the expression levels of
miR-23b/miR-27b and MET (Fig. 4B and
C). Next, immunohistochemistry revealed that MET was strongly
expressed in cancer tissues, while low expression was observed in
normal tissues (Fig. 4D and
E).
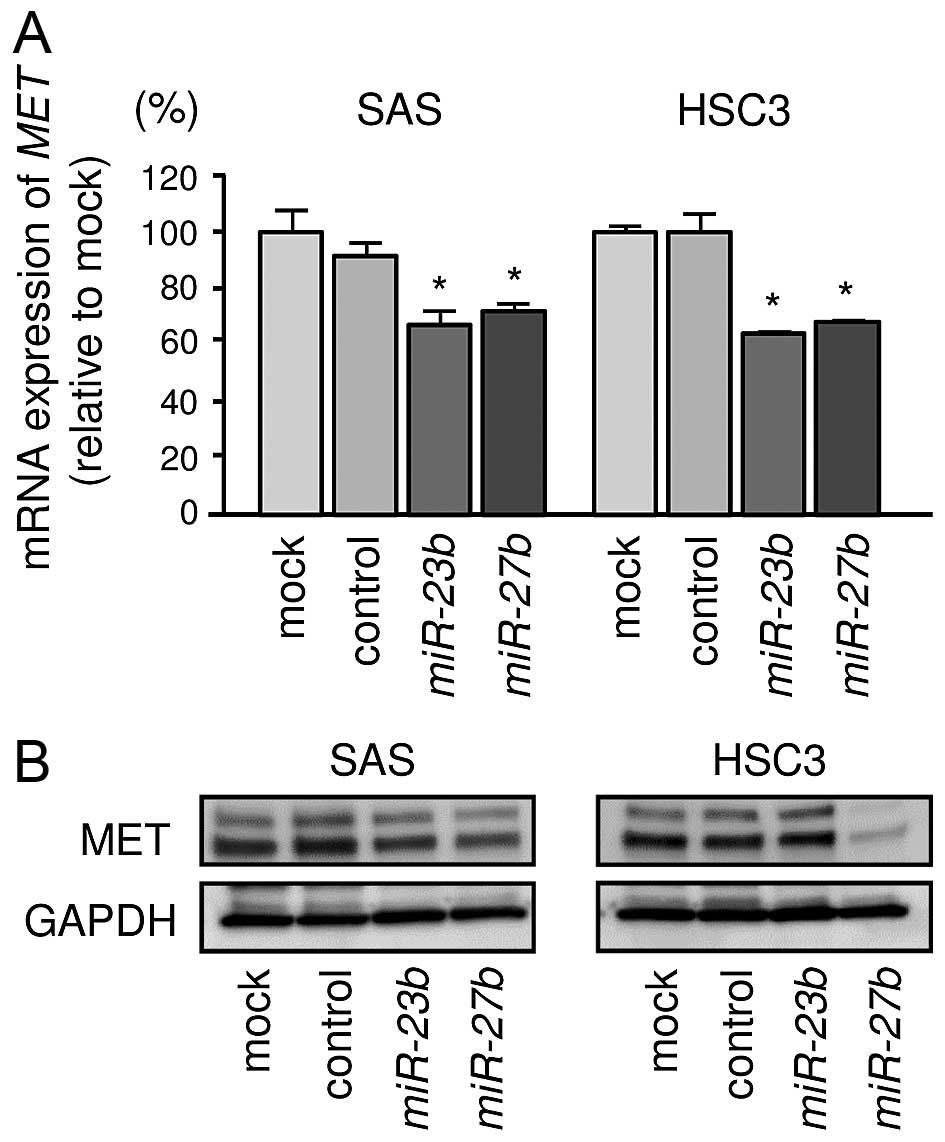
Direct regulation of MET gene by miR-23b
and miR-27b in OSCC cells
We investigated the expression levels of MET
in OSCC cell lines. We performed quantitative real-time RT-PCR and
western blotting in OSCC cell lines to investigate whether
restoration of miR-23b or miR-27b altered MET
gene and protein expression. mRNA expression levels of MET
were significantly repressed in miR-23b and miR-27b
transfectants compared with mock or miR-control transfectant in
OSCC cell lines (Fig. 5A). Protein
expression levels of MET were repressed in miR-23b and
miR-27b transfectants compared with mock or miR-control in
SAS. Although restoration of miR-27b significantly
suppressed MET protein expression, no significant downregulation of
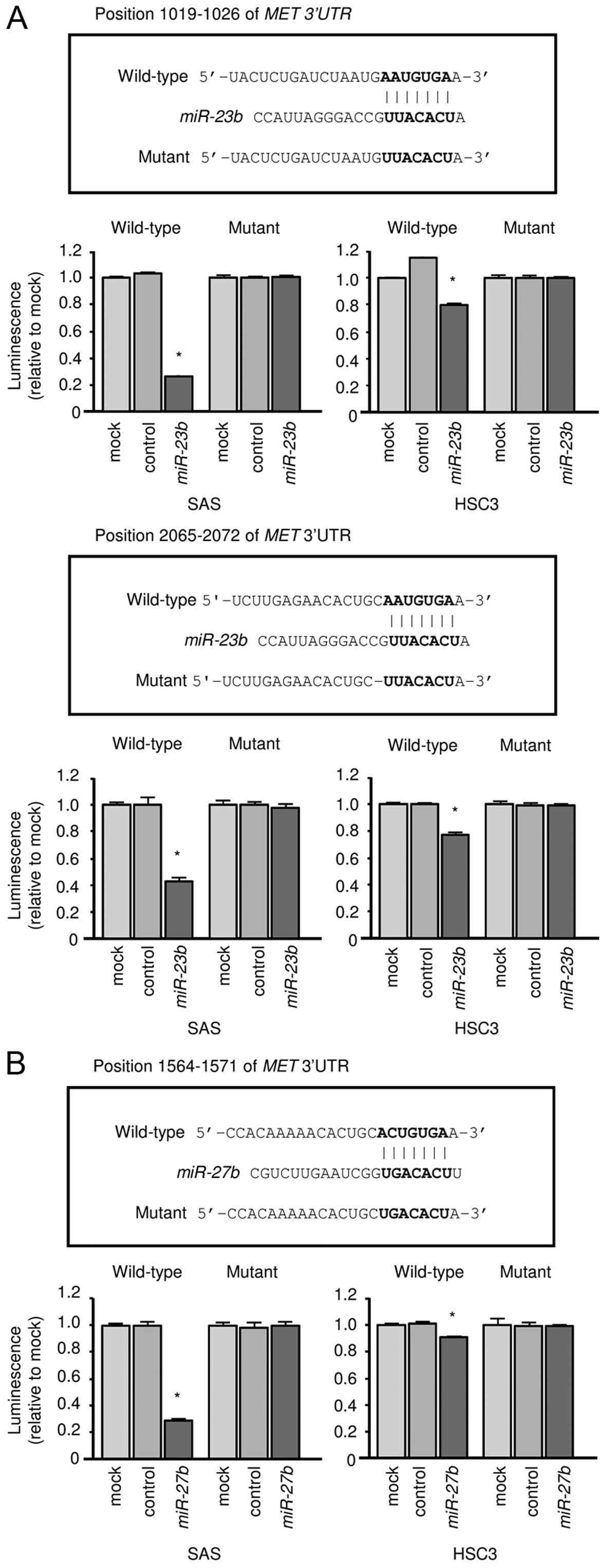
MET was observed in miR-23b transfectant in HSC3 (Fig. 5B). Next, we performed luciferase
reporter assays in OSCC cell lines to determine whether MET
mRNA contained target sites for miR-23b and miR-27b.
We used vectors encoding either a partial wild-type sequence or a
sequence in which the miRNA binding site had been mutated from the
3′-UTR of MET mRNA. Our data showed that the luminescence
intensity was significantly reduced by co-transfection with
miR-23b/miR-27b and the vector carrying the wild-type 3′-UTR
of MET mRNA (Fig. 6A and
B).
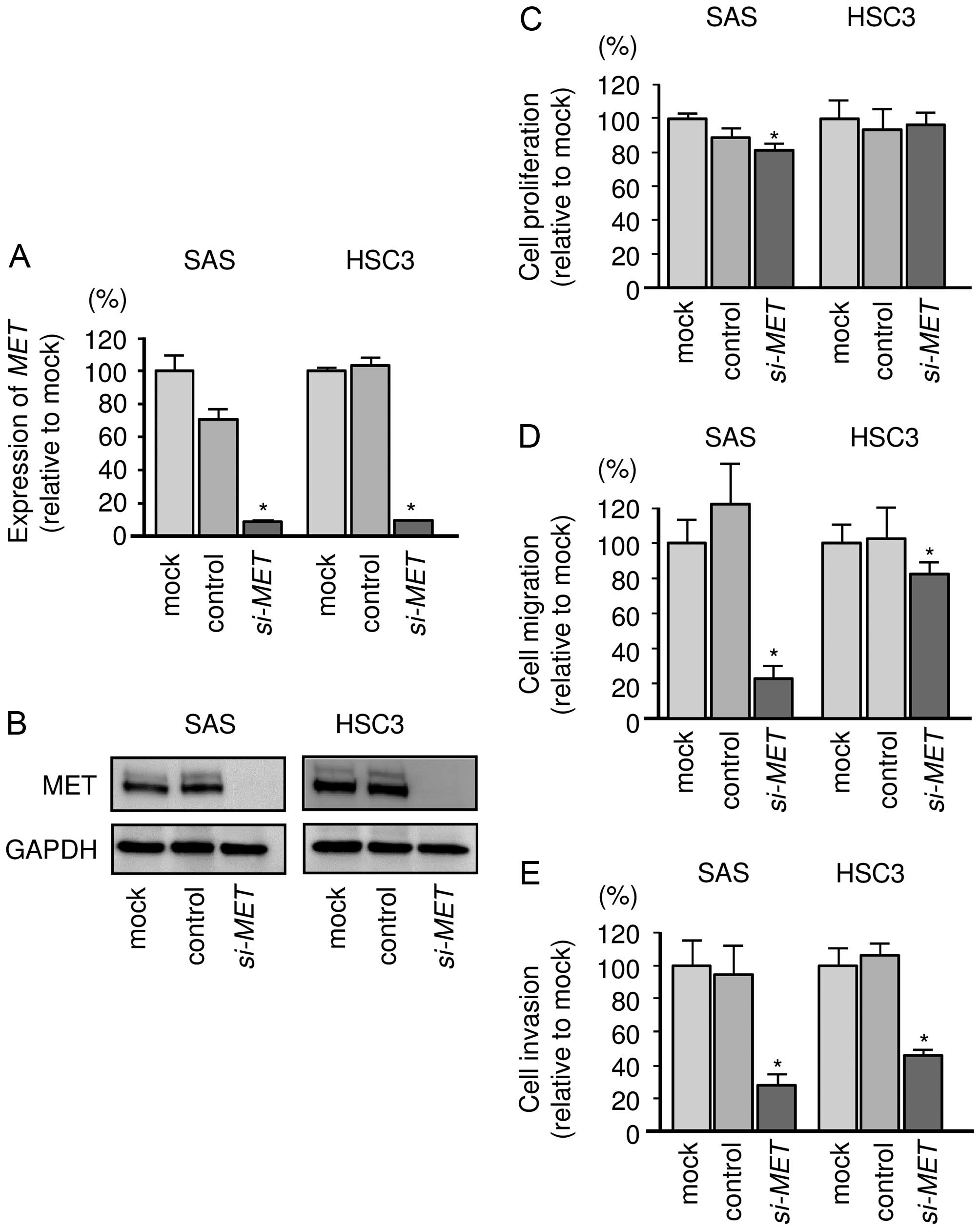
Effect of silencing MET gene on cell
proliferation, migration, and invasion in OSCC cells
To investigate the functional role of MET in OSCC,
we performed loss-of-function studies using si-MET
transfectants. First, we checked the knockdown efficiency of
si-MET transfection. Western blotting and qRT-PCR revealed
that the si-RNA effectively reduced the expression levels of MET in
OSCC cell lines (Fig. 7A and B).
Cell proliferation assays showed that SAS cell viability was
significantly inhibited in si-RNA transfectants compared with mock
or si-control. On the other hand, proliferation was not inhibited
in HSC3 cells (Fig. 7C). Migration
and invasion assays showed that cell migration activity was
significantly inhibited in OSCC cells (Fig. 7D and E).
Discussion
A significant amount of evidence suggests that
aberrantly expressed miRNAs are closely involved in human
oncogenesis, metastasis and drug resistance (19). The cause of the poor prognosis of
OSCC is distant metastasis of the cancer cells. Thus,
identification of tumor-suppressive miRNAs that regulate novel
metastatic pathways and metastasis-promoting genes may improve our
understanding of OSCC progression and metastasis. We have
sequentially identified tumor-suppressive miRNA-mediated novel
cancer pathways in HNSCC and OSCC (18,20–24).
We hypothesize that identification of novel metastatic pathways and
targets regulated by tumor-suppressive miRNAs could lead to a
better understanding of OSCC and the development of new therapeutic
strategies to treat this disease.
Here, we focused on two clustered miRNAs,
miR-23b and miR-27b, based on miRNA expression
signatures. Thus, we investigated the functional significance of
these miRNAs in OSCC cells. We found that miR-23b and
miR-27b were downregulated in cancer specimens and that
restoration of miR-23b and miR-27b significantly
inhibited cancer cell migration and invasion. These results
strongly suggested that these miRNAs functioned as tumor
suppressors in OSCC cells. Our previous studies of prostate cancer,
renal cell carcinoma and bladder cancer showed that miR-23b
and miR-27b act as tumor suppressors regulating several
oncogenic genes (15–17). In renal cell carcinoma,
significantly poor prognosis was observed in patients with lower
expression levels of the miR-23b/miR-27b cluster, suggesting
that low expression of these miRNAs increased the risk of disease
progression and predicted poor survival (18).
Other research groups have shown tumor-suppressive
roles of miR-23b and miR-27b in several cancers
(25–28). For example, miR-23b directly
controls the proto-oncogenes SRC and AKT, and
overexpression of miR-23b suppresses cell viabilities, cell
cycle arrest, and apoptosis (29).
Another report has shown that miR-23b and miR-27b are
downregulated in metastatic and castration-resistant prostate
cancer (CRPC) tumors and that ectopic expression of these miRNAs
suppresses cell invasion and migration in CRPC cell lines (30). Contrary to our data showing tumor
suppressive roles of miR-23b and miR-27b in human
cancers, expression levels of these miRNAs were significantly
upregulated in human breast cancer, and miR-23b and
miR-27b knockdown substantially represses breast cancer cell
growth. These results indicate that these miRNAs function as
oncogenes in this cellular context (31). Elucidation of the molecular
mechanisms controlling expression of the miR-23b/27b cluster
is an important theme in this field.
Identification of miRNA-regulated pathways and
targets is important to elucidate the molecular functions of
tumor-suppressive miR-23b and miR-27b in OSCC cells.
Towards that end, we combined expression data from OSCC clinical
specimens and in silico miRNA database analysis. In this
screening, several putative pathways and targets were annotated to
be subject to miR-23b and miR-27b in OSCC cells.
Among them, we focused on the MET oncogene because
overexpression of MET was indicated by gene expression data
and it is well known that MET activates signaling that contributes
to cancer cell proliferation, metastasis and drug resistance
(32). One study reported that
overexpression of MET was observed in 90% of HNSCC cell
lines and 84% of HNSCC patient samples (33). Moreover, HGF overexpression has
also been described in ~60% of HNSCC, and co-expression of MET/HGF
has been correlated with more aggressive disease behavior (33). Thus, the control of HGF/MET
oncogenic signaling is the pivotal treatment target of the
disease.
Cetuximab, a monoclonal antibody directed against
the EGFR, is now available for targeted molecular therapy in HNSCC,
including OSCC (34). Cetuximab is
currently approved in combination with radiation therapy as a
first-line treatment in combination with platinum and
5-fluorouracil in recurrent or metastatic disease (35,36).
However, the curative effects of these treatments are limited, and
it is difficult to recover completely from this disease. Many
studies have suggested different mechanisms that may be
contributing to targeted EGFR resistance (37). A recent study showed that
cetuximab-induced MET activation enhanced to cetuximab-resistance
in colon cancer cells (38).
Aberrant MET expression and hepatocyte growth factor (HGF)
signaling might be contributing as salvage pathways for EGFR
blockade-resistant cancer cells. Therefore, dual blocking
therapeutic strategies of EGFR and MET oncogenic signaling are
indispensable for HNSCC and OSCC treatment.
In conclusion, miR-23b and miR-27b
were frequently reduced in OSCC clinical specimens and appeared to
act as tumor suppressors through targeting of the MET
oncogene in this disease. Elucidation of novel target genes and
pathways regulating by tumor-suppressive miR-23b/27b cluster
may provide new information of OSCC and the development of new
treatment strategies of this disease.
Acknowledgements
This study was supported by the KAKENHI(C),
15K10800, 15K10801, 25462676 and 26462596.
References
|
1
|
Min A, Zhu C, Peng S, Rajthala S, Costea
DE and Sapkota D: MicroRNAs as important players and biomarkers in
oral carcinogenesis. BioMed Res Int. 2015:1869042015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhattacharya A, Roy R, Snijders AM,
Hamilton G, Paquette J, Tokuyasu T, Bengtsson H, Jordan RC, Olshen
AB, Pinkel D, et al: Two distinct routes to oral cancer differing
in genome instability and risk for cervical node metastasis. Clin
Cancer Res. 17:7024–7034. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang L, Zhang T, Kong Q, Liang J and Liao
G: A meta-analysis on selective versus comprehensive neck
dissection in oral squamous cell carcinoma patients with clinically
node-positive neck. Oral Oncol. 51:1076–1081. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
6
|
Kikkawa N, Hanazawa T, Fujimura L, Nohata
N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y and Seki
N: miR-489 is a tumour-suppressive miRNA target PTPN11 in
hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer.
103:877–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nohata N, Hanazawa T, Kikkawa N, Sakurai
D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H,
Nakagawa M, et al: Tumour suppressive microRNA-874 regulates novel
cancer networks in maxillary sinus squamous cell carcinoma. Br J
Cancer. 105:833–841. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukumoto I, Kinoshita T, Hanazawa T,
Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R,
Nakagawa M, et al: Identification of tumour suppressive
microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. Br J Cancer. 111:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukumoto I, Hanazawa T, Kinoshita T,
Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida
H, Nakagawa M, et al: MicroRNA expression signature of oral
squamous cell carcinoma: Functional role of microRNA-26a/b in the
modulation of novel cancer pathways. Br J Cancer. 112:891–900.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nohata N, Hanazawa T, Kikkawa N, Mutallip
M, Sakurai D, Fujimura L, Kawakami K, Chiyomaru T, Yoshino H,
Enokida H, et al: Tumor suppressive microRNA-375 regulates oncogene
AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J Hum
Genet. 56:595–601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nohata N, Hanazawa T, Kinoshita T, Inamine
A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto
Y, et al: Tumour-suppressive microRNA-874 contributes to cell
proliferation through targeting of histone deacetylase 1 in head
and neck squamous cell carcinoma. Br J Cancer. 108:1648–1658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fuse M, Kojima S, Enokida H, Chiyomaru T,
Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M,
et al: Tumor suppressive microRNAs (miR-222 and miR-31) regulate
molecular pathways based on microRNA expression signature in
prostate cancer. J Hum Genet. 57:691–699. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goto Y, Kojima S, Nishikawa R, Kurozumi A,
Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M,
et al: MicroRNA expression signature of castration-resistant
prostate cancer: The microRNA-221/222 cluster functions as a tumour
suppressor and disease progression marker. Br J Cancer.
113:1055–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Itesako T, Seki N, Yoshino H, Chiyomaru T,
Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa
M, et al: The microRNA expression signature of bladder cancer by
deep sequencing: The functional significance of the miR-195/497
cluster. PLoS One. 9:e843112014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goto Y, Kojima S, Nishikawa R, Enokida H,
Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T and Seki
N: The microRNA-23b/27b/24-1 cluster is a disease progression
marker and tumor suppressor in prostate cancer. Oncotarget.
5:7748–7759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chiyomaru T, Seki N, Inoguchi S, Ishihara
T, Mataki H, Matsushita R, Goto Y, Nishikawa R, Tatarano S, Itesako
T, et al: Dual regulation of receptor tyrosine kinase genes EGFR
and c-Met by the tumor-suppressive microRNA-23b/27b cluster in
bladder cancer. Int J Oncol. 46:487–496. 2015.
|
|
17
|
Ishihara T, Seki N, Inoguchi S, Yoshino H,
Tatarano S, Yamada Y, Itesako T, Goto Y, Nishikawa R, Nakagawa M,
et al: Expression of the tumor suppressive miRNA-23b/27b cluster is
a good prognostic marker in clear cell renal cell carcinoma. J
Urol. 192:1822–1830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y, et al: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kinoshita T, Hanazawa T, Nohata N, Kikkawa
N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto
Y, et al: Tumor suppressive microRNA-218 inhibits cancer cell
migration and invasion through targeting laminin-332 in head and
neck squamous cell carcinoma. Oncotarget. 3:1386–1400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fukumoto I, Kikkawa N, Matsushita R, Kato
M, Kurozumi A, Nishikawa R, Goto Y, Koshizuka K, Hanazawa T,
Enokida H, et al: Tumor-suppressive microRNAs (miR-26a/b,
miR-29a/b/c and miR-218) concertedly suppressed
metastasis-promoting LOXL2 in head and neck squamous cell
carcinoma. J Hum Genet. 61:109–118. 2016. View Article : Google Scholar
|
|
22
|
Cano A, Santamaría PG and Moreno-Bueno G:
LOXL2 in epithelial cell plasticity and tumor progression. Future
Oncol. 8:1095–1108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peinado H, Moreno-Bueno G, Hardisson D,
Pérez-Gómez E, Santos V, Mendiola M, de Diego JI, Nistal M,
Quintanilla M, Portillo F, et al: Lysyl oxidase-like 2 as a new
poor prognosis marker of squamous cell carcinomas. Cancer Res.
68:4541–4550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harisi R and Jeney A: Extracellular matrix
as target for antitumor therapy. Onco Targets Ther. 8:1387–1398.
2015.PubMed/NCBI
|
|
25
|
Huang TT, Ping YH, Wang AM, Ke CC, Fang
WL, Huang KH, Lee HC, Chi CW and Yeh TS: The reciprocal regulation
loop of Notch2 pathway and miR-23b in controlling gastric
carcinogenesis. Oncotarget. 6:18012–18026. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li W, Liu Z, Chen L, Zhou L and Yao Y:
MicroRNA-23b is an independent prognostic marker and suppresses
ovarian cancer progression by targeting runt-related transcription
factor-2. FEBS Lett. 588:1608–1615. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee JJ, Drakaki A, Iliopoulos D and Struhl
K: MiR-27b targets PPARγ to inhibit growth, tumor progression and
the inflammatory response in neuroblastoma cells. Oncogene.
31:3818–3825. 2012. View Article : Google Scholar
|
|
28
|
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z,
Chen Z, Qiu F, Xu J and Huang J: miRNA-27b targets vascular
endothelial growth factor C to inhibit tumor progression and
angiogenesis in colorectal cancer. PLoS One. 8:e606872013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Majid S, Dar AA, Saini S, Arora S,
Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, et
al: miR-23b represses proto-oncogene Src kinase and functions as
methylation-silenced tumor suppressor with diagnostic and
prognostic significance in prostate cancer. Cancer Res.
72:6435–6446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ishteiwy RA, Ward TM, Dykxhoorn DM and
Burnstein KL: The microRNA -23b/-27b cluster suppresses the
metastatic phenotype of castration-resistant prostate cancer cells.
PLoS One. 7:e521062012. View Article : Google Scholar
|
|
31
|
Ell B, Qiu Q, Wei Y, Mercatali L, Ibrahim
T, Amadori D and Kang Y: The microRNA-23b/27b/24 cluster promotes
breast cancer lung metastasis by targeting metastasis-suppressive
gene prosaposin. J Biol Chem. 289:21888–21895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Blumenschein GR Jr, Mills GB and
Gonzalez-Angulo AM: Targeting the hepatocyte growth factor-cMET
axis in cancer therapy. J Clin Oncol. 30:3287–3296. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seiwert TY, Jagadeeswaran R, Faoro L,
Janamanchi V, Nallasura V, El Dinali M, Yala S, Kanteti R, Cohen
EE, Lingen MW, et al: The MET receptor tyrosine kinase is a
potential novel therapeutic target for head and neck squamous cell
carcinoma. Cancer Res. 69:3021–3031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sacco AG and Cohen EE: Current treatment
options for recurrent or metastatic head and neck squamous cell
carcinoma. J Clin Oncol. 33:3305–3313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bonner JA, Harari PM, Giralt J, Azarnia N,
Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vermorken JB, Mesia R, Rivera F, Remenar
E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol
D, et al: Platinum-based chemotherapy plus cetuximab in head and
neck cancer. N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bertotti A and Sassi F: Molecular
pathways: Sensitivity and resistance to anti-EGFR antibodies. Clin
Cancer Res. 21:3377–3383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song N, Liu S, Zhang J, Liu J, Xu L, Liu Y
and Qu X: Cetuximab-induced MET activation acts as a novel
resistance mechanism in colon cancer cells. Int J Mol Sci.
15:5838–5851. 2014. View Article : Google Scholar : PubMed/NCBI
|