Introduction
CML, a type of hematological malignancy, is
characterized by a reciprocal chromosomal translocation t(9;22)
(q34;q11) named the Philadelphia chromosome which fuses the Abelson
kinase (ABL) gene from chromosome 9 with the breakpoint cluster
region (BCR) gene on chromosome 22 (1,2).
BCR-ABL with constitutive tyrosine kinase activity can
phosphorylate and activate many signaling molecules leading to cell
proliferation and survival (3).
Clinically, tyrosine kinase inhibitors (TKIs) targeting BCR-ABL
such as imatinib, nilotinib, dasatinib and ponatinib, are frontline
drugs used in CML therapy (4,5).
Although generation of TKIs have dramatically improved the
prognosis of CML, a significant portion of patients failed
chemotherapy due to drug resistance (6,7).
TKIs resistance is a process including BCR-ABL-dependent and
-independent mechanisms (8). The
latter contains increase of intracellular TKIs efflux, intolerance
to TKIs and the development of the phenomenon known as MDR
(9). MDR is often associated with
the overexpression of ABC transporters represented by MDR1/P-gp,
MRP1 and BCRP, which powerfully extrude a wide variety of
structurally diverse chemotherapeutics across the membranes,
resulting in a decreased intracellular drug concentration and
chemotherapy bioavailability (10,11).
The HOX genes include a large family of
homeodomain-containing transcription factors, characterized by
highly conserved DNA- and co-factor binding domains (12). In mammals, there are four such
separate clusters (A–D), involving a total of 39 HOX genes, which
are key regulators of embryonic development, hematopoietic
differentiation and leukemogenesis (13). We previously found that HOXA5
knockdown in U937 human leukemia cells exerted anti-proliferative
effects, induced apoptosis and cell cycle arrest and enhanced the
cytotoxicity of cytarabine (14).
Besides, silencing of HOXA10 significantly increased the
cytotoxicity of ADR in K562/ADM cells; the effect of silencing
HOXA10 is associated with the increased intracellular accumulation
of ADR, as well as the inhibition of the expression of P-gp and
MRP1 (15). HOXB4, a member of the
HOX gene family, is essential in maintaining quiescent
hematopoietic stem cells in steady state, adult myelopoiesis and
self-renewal of the hematopoietic stem cell population (16,17).
It is worth noting that HOXB4 has been implicated as a
cancer-related gene in many malignancies, including leukemia
(18), breast (19), lung cancer (20), extra-hepatic cholangiocarcinoma
(21) and thyroid tumours
(22). The aberrant expression of
HOXB4 is often related to poor prognoses of leukemia (23), mesothelioma (24) and ovarian cancer (25). Overexpression of HOXB4 is
considered as a catalyst for the development of leukemia and
influences the leukemia phenotype (26). Furthermore, Lehne et al
(27) discovered that HOXB4 played
an important role in chemoresistance of CML.
The aberrant activation of the PI3K/Akt cell
transduction pathway has been found in different types of
neoplasms, which contributes collectively to inhibit cell
apoptosis, promote tumor proliferation, angiogenesis, invasion,
metastasis and autophagy (28,29).
An increasing amount of preclinical data suggested that the
PI3K/Akt pathway was also relevant to the MDR of different types of
human malignancies, including leukemia, breast carcinoma and
gastric cancer (30–32). Our previous study confirmed that
the MDR K562/ADM cells demonstrated higher PI3K/Akt activity than
K562 cells which was in accordance with the MDR phenotype (33). Furthermore, accumulating evidence
revealed that there was a positive relationship between the
expression of ABC proteins and the activity of PI3K/Akt signaling
pathway (33–35). Nevertheless, little is known
regarding the effect of the PI3K/Akt signaling on HOXB4,
HOXB4-mediated leukemia MDR. Besides, the efficacy and the
associated molecular mechanisms of HOXB4 suppression on the
reversal of MDR, to date, have not been examined in MDR leukemia
cells.
In the present study, the HOXB4 expression in K562
cell line and its MDR subline K562/ADM was analyzed. In addition,
the chemosensitivity and drug efflux activity of K562/ADM cells
transfected with HOXB4 short hairpin RNA (shRNA) were detected to
appraise the connection between HOXB4, MDR, and the PI3K/Akt
pathway, and evaluate HOXB4 as a potential molecular-target for
reversing MDR of human CML K562/ADM cells.
Materials and methods
Cell lines and cell culture
Human CML K562 and its MDR counterpart K562/ADM
cells were cultured as previously described (33). Human cervical carcinoma cell line
HeLa was obtained from Key Laboratory of Tumour Molecular Biology
of Binzhou Medical University (Binzhou, China). The cells were
maintained in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS; both from HyClone Laboratories, Logan, UT, USA), at
37°C containing 5% CO2.
Cell transfection
Four pairs of shRNA sequences targeting HOXB4,
termed HOXB4 shRNA-1, -2, -3 and -4 and negative control (NC) with
or without FAM-labeled were purchased from Shanghai GenePharma,
Co., Ltd. (Shanghai, China). Their target sequences are as follows:
HOXB4 shRNA-1, GC GAAGATGGATCCACGTTTC; HOXB4 shRNA-2, GGTAA
CACACACACACTCTCC; HOXB4 shRNA-3, GCCTTGACA ACTCAGGAGTGA; HOXB4
shRNA-4, AGCAGAAGCC TCTCTCCTAGA; NC shRNA, GTTCTCCGAACGTGTCA CGT.
The four shRNAs specific for HOXB4 were tested for inhibitory
activity by transient transfection into HeLa cells. Briefly, cells
were seeded at a density of 1×105/well into a 6-well
plate the day before the transfection. After incubating for 24 h,
the cells were transfected with shRNAs using Lipofectamine™ 2000
transfection reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instruction. After incubating for 6 h, the
medium was removed and replaced with the fresh one. Transfection
efficiency was observed with a fluorescence microscope. RT-PCR,
RT-qPCR and western blot analyses were performed to determine the
inhibitory efficacy. The most effective sequence was chosen to
transfect K562/ADM cells according to the manufacturer’s
instructions. After 48 h of transfection, G418 (500 ng/ml; Life
Technologies, Carlsbad, CA, USA) was added to the medium. The
stable positive clones were obtained after four weeks of
selection.
ADM accumulation assay
Cells transfected with shRNA-3 and NC shRNA were
seeded in the 6-well plate at the density of 5×105/well,
and the group without transfection was defined as a blank control.
A total of 5 μM ADM (Melone Pharmaceutical, Co., Ltd., Dalian,
China) was applied to the wells. After incubation for 1 h, the
cells were harvested by centrifugation and washed twice with
ice-cold phosphate-buffered saline (PBS). The cell-associated mean
fluorescence intensity (MFI) of ADM was detected by flow cytometer
using a FACSCalibur (Beckman Coulter, Brea, CA, USA) with
excitation/emission wavelengths of 485/580 nm.
Rhodamine-123 (Rho-123) and 5(6)-carboxyfluorescein diacetate (CFDA)
accumulation assay
Rho-123 and CFDA (both from Sigma-Aldrich, St.
Louis, MO, USA) were, respectively, used to evaluate the transport
function of P-gp and MRP1 in K562/ADM cells by flow cytometric
analysis. NC shRNA-transfected, shRNA-3-transfected and control
K562/ADM cells (5×105/well) were seeded into 6-well
plates. Subsequently, Rh-123 (5 μM) or CFDA (2 μM) was added, the
cells were incubated for 1 h, harvested, washed twice with cold PBS
and then were measured using a flow cytometer at 488 nm excitation
and 530 nm emission.
CCK-8 assay for proliferation
activity
K562/ADM cells (1×104) with or without
transfection of shRNA/well were seeded into 96-well plates and
incubated with the medium containing anticancer drugs (ADM,
etoposide, vindesine or cytarabine) in different concentrations for
24 h. Each group has five parallel wells. After that, 10 μl of
CCK-8 (Dojindo Molecular Technologies, Inc., Shanghai, China)
solution was added to every well and incubated for a further 4 h.
Then, the absorbance at 570 nm was measured using fluorescence. The
reversal fold (RF) values were calculated using the following
formula: RF=IC50 of shRNA-3 group/IC50 of
control group.
Reverse transcription polymerase chain
reaction (RT-PCR) and reverse transcription-quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen/Life Technologies) in accordance with the
manufacturer’s protocol and the purity of the total RNA was
assessed from the ratio of A260/A280 by spectrophotometer (NanoDrop
2000; NanoDrop Technologies, Inc., Wilmington, DE, USA). A total of
1–2 μg of RNA was applied for the synthesis of the first strand
cDNA. The primers (Table I) used
in this experiments were designed using Primer 5 version 5.6.0
software and synthesized by Sangon Biotech, Co., Ltd. (Shanghai,
China). The reverse transcription reaction was implemented with
PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Shiga,
Japan). The polymerase chain reaction amplification was performed
on Eppendorf Mastercycler personal (Eppendorf, Co., Ltd., Shanghai,
China) by using Premix Tap™ (Takara Bio). The reaction system
contained diethyl pyrocarbonate, forward primer, reverse primer,
Premix Tap and template cDNA. After 5 min of initial incubation at
95°C, cDNA was amplified in 35 cycles consisting of 40 sec
denaturation at 95°C, 30 sec annealing at 58°C and 40 sec
elongation at 72°C, the PCR products were separated by 1.5% agarose
gels (Takara Bio), stained with ethidium bromide for 15 min. The
images were captured by Tanon Gis systerm and β-actin served as an
internal standard for quality control and quantification of target
genes. RT-qPCR was performed on an ABI PRISM 7500 real-time PCR
system (Applied Biosystems, Foster City, CA, USA) by using
SYBR-Green reaction kit (Takara Bio). The reaction system of PCR
was: SYBR-Green reagent, forward primer, reverse primer, template
cDNA and nuclease-free distilled water. The PCR conditions were
95°C, 30 sec, followed by 50 cycles of 95°C, 5 sec, 60°C, 30 sec
and β-actin was used as an internal control.
 | Table IPrimers used in reverse
transcription-quantitative polymerase chain reaction. |
Table I
Primers used in reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer
sequence | Product length
(bp) |
|---|
| HOXB4 | Forward:
5′-GCAAAGAGCCCGTCGTCT-3′ | |
| Reverse:
5′-GAAATTCCTTCTCCAGCT-3′ | 138 |
| MDR1 | Forward:
5′-GGAGCCTACTTGGTGGCACATAA-3′ | |
| Reverse:
5′-TGGCATAGTCAGGAGCAAATGAAC-3′ | 121 |
| MRP1 | Forward:
5′-CAGCCCTTCCTGACAAGCTA-3′ | |
| Reverse:
5′-GTGGCCTCATCCAACACAAG-3′ | 133 |
| BCRP | Forward:
5′-GAAACCTGGTCTCAACGC-3′ | |
| Reverse:
5′-AGAGTGCCCATCACAACA-3′ | 189 |
| β-actin | Forward:
5′-TCCTTCCTGGGCATGGAGTC-3′ | |
| Reverse:
5′-GTAACGCAACTAAGTCATAGTC-3′ | 361 |
Western blotting assay
Cells cultured in the 6-well plates were washed with
PBS twice and proper amount of lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) was added later on. Protein
concentrations were determined by Bicinchoninic acid protein assay
kit (Beyotime Institute of Biotechnology). Protein samples were
separated with 10 or 6% SDS-PAGE gel (Beyotime Institute of
Biotechnology) and transferred onto polyvinylidine difluoride
(PVDF) membranes (EMD Millipore, Bedford, MA, USA). The membranes
were blocked by 5% skimmed dry milk in TBS containing 0.2% Tween-20
at room temperature for 2 h and incubated overnight at 4°C with
primary antibodies. Monoclonal rabbit anti-human P-gp and
polyclonal rabbit anti-human antibodies against Akt, p-Akt (Ser473)
and p-Akt (Thr308) (both 1:1,000) were purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA). Rabbit polyclonal
antibodies including HOXB4, MRP1, BCRP, NF-κB (both 1:500) and
β-actin (1:3,000) were products of Beijing Biosynthesis
Biotechnology, Co., Ltd. (Beijing, China). After three washes in
TBS/Tween buffer, the membranes were incubated in horesradish
peroxidas-elabeled goat anti-rabbit immunoglobulin G (1:5,000;
Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd., Beijing,
China) for 2 h at 37°C. Detection was performed using the FluorChem
FC2 gel imaging system (Alpha Innotech Corp., San Leandro, CA,
USA). Each band density was quantified using ImageJ image
processing program and normalized by β-actin for their respective
lanes.
Data analysis
Statistical analyses were done using SPSS 16.0
software (IBM SPSS, Armonk, NY, USA). Independent two-sample t-test
was used to compare the differences between the two groups. One-way
analysis of variance (ANOVA) with the multiple comparison test was
used to analyze the differences between three or more groups. Data
were expressed as the means ± SD. Statistical significance was
accepted at P<0.05.
Results
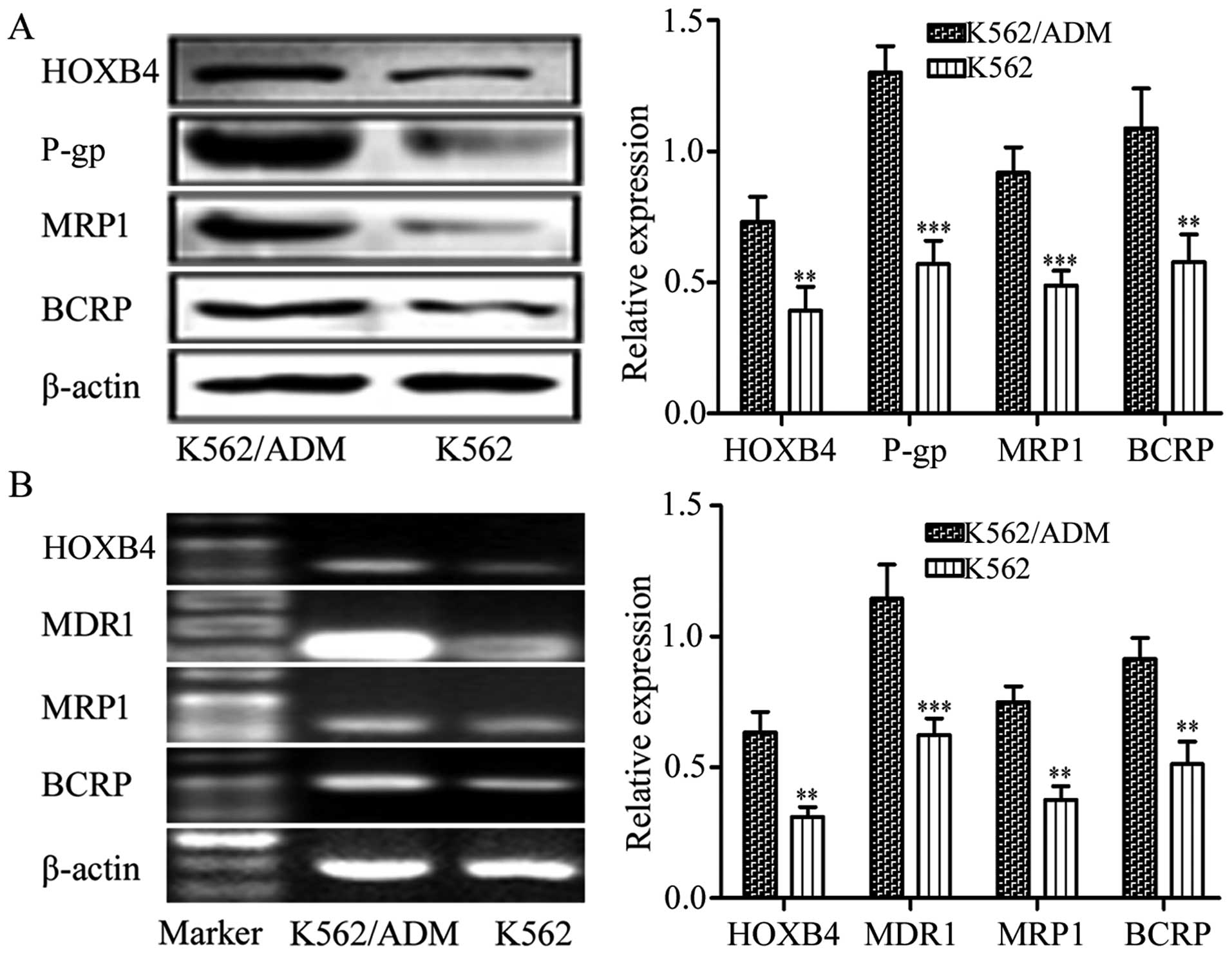
HOXB4 expression in K562 and K562/ADM
cells
We determined the expression of HOXB4 in the
sensitive K562 cells and the resistant K562/ADM cells. Results
demonstrated that the K562 and K562/ADM cells exhibited high
expression of HOXB4. The K562/ADM cells with higher expression of
P-gp, MRP1 and BCRP also showed higher HOXB4 expression than the
K562 cells (Fig. 1). Hence, the
K562/ADM cell line was chosen for the next HOXB4 interference.
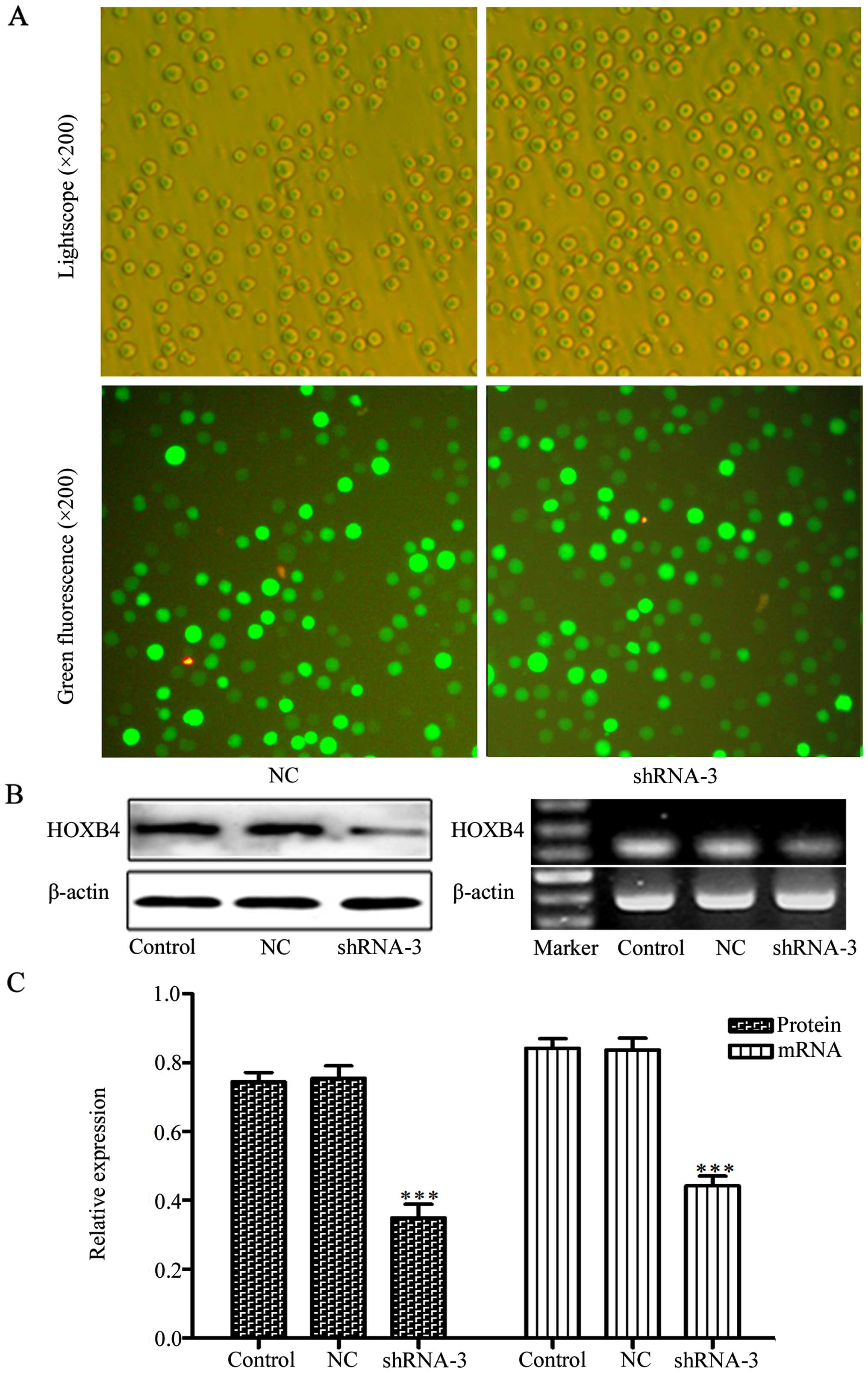
Transfection with shRNAs to silence
HOXB4
To evaluate the effects of shRNAs on knockdown of
HOXB4, we transfected four HOXB4 shRNAs and one NC shRNA into HeLa
cells, with cells untreated serving as a control. We tested
transfection efficiency through transfection of the cells with
FAM-labeled shRNAs. Green fluorescence can only be detected in the
cells that were successfully transfected with FAM-labeled shRNAs
(Fig. 2). Cells (84.96±1.32%) had
green fluorescence, indicating high efficiency of the transfection.
As shown in Fig. 2C–E, shRNA-3 was
the most efficient plasmid to downregulate HOXB4 expression and was
used to silence HOXB4 gene in K562/ADM cells. We next transfected
shRNA-3 and NC shRNA into K562/ADM cells. After four weeks of G418
selection, we successfully obtained the stable positive clones
(Fig. 3). Western blotting and
RT-PCR analyses revealed that HOXB4 expression decreased obviously
in shRNA-3 group both in protein level and mRNA level compared with
control group. While HOXB4 expression in the NC shRNA group was not
significantly different from the control group (Fig. 3B and C; P<0.001).
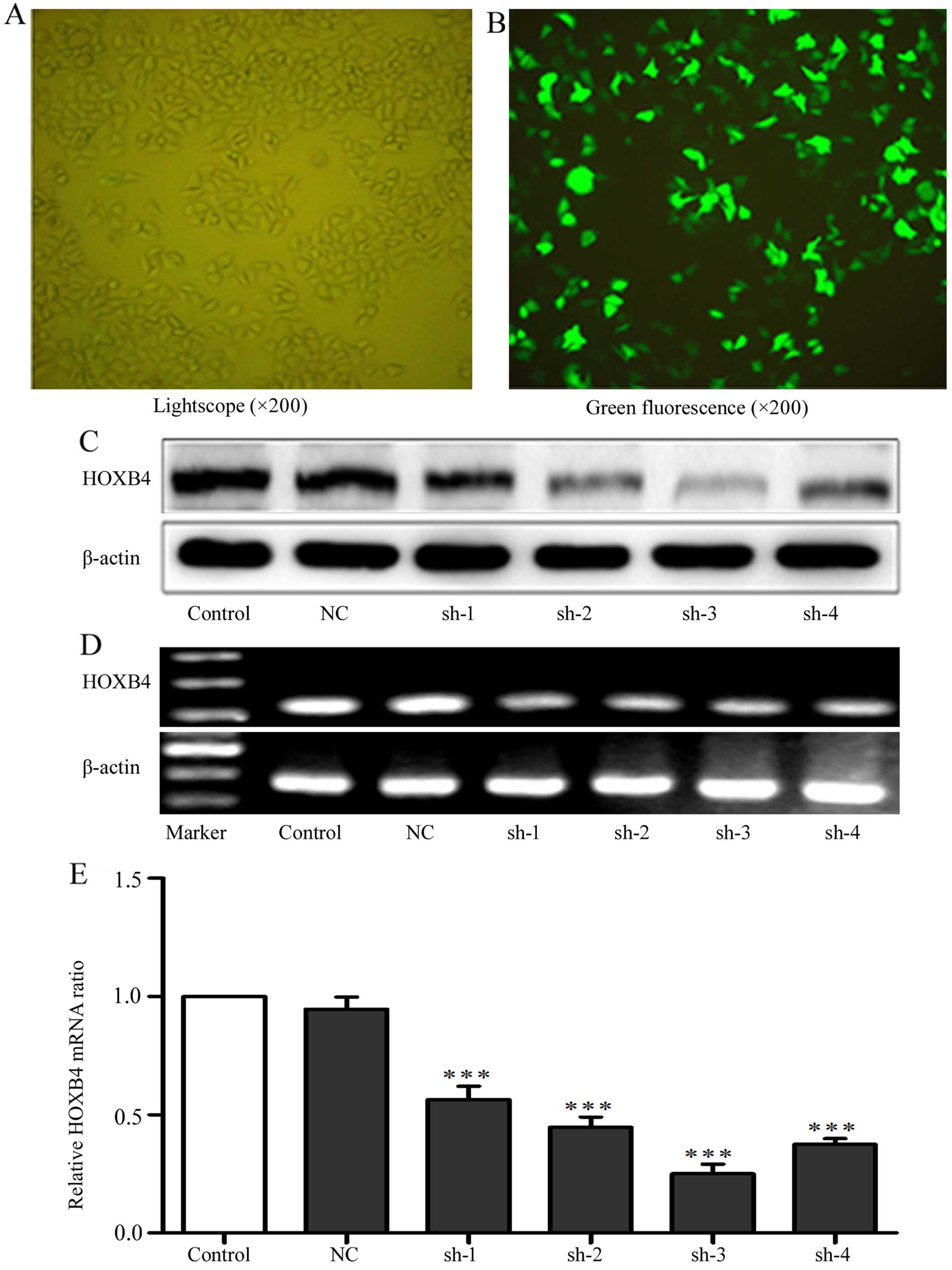 | Figure 2Transfection efficiency of HOXB4
shRNAs. (A and B) The phase contrast or the fluorescence
photomicrograph in the same vision field of the HeLa cells
transfected with shRNAs against HOXB4 at 24 h. (C–E) The HOXB4
protein expression of HeLa cells without transfection or
transfected with NC-shRNA, shRNA-1, -2, -3 and -4 were detected by
western blotting, RT-PCR and RT-qPCR (100 bp marker for HOXB4 and
β-actin). Control refers to the cells without transfection. NC,
sh-1, sh-2, sh-3 and sh-4 refer to cells tranfected with NC shRNA,
HOXB4 shRNA-1, -2, -3 and -4 respectively. Data are expressed as
means ± SD values of triplicate experiments.
***P<0.001 vs. the control group. |
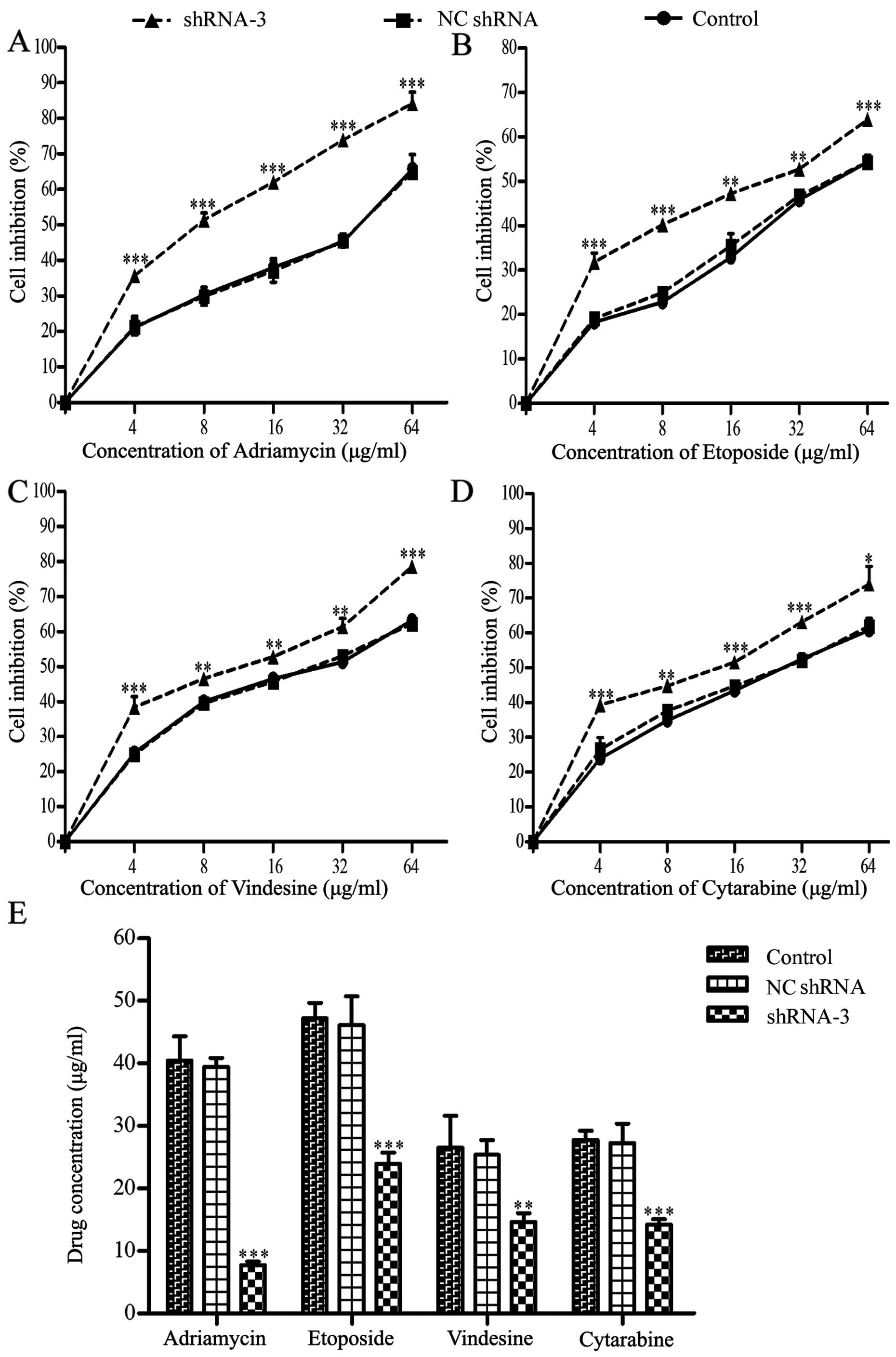
Restoration of drug sensitivity in
K562/ADM cells owing to HOXB4 knockout
As shown in Fig. 4,
the dose-response curves explained that the cell sensitivity in
shRNA-3 group was obviously enhanced, while the differences between
the NC shRNA group and the control group were not clear enough to
distinguish (Fig. 4A–D). As seen
in Fig. 4E and Table II, the IC50 values of
the four drugs decreased to different extent in shRNA-3 group,
especially for ADM.
 | Table IIEffect of silencing HOXB4 on the
sensitivity of K562/ADM cells toward four chemotherapeutic drugs by
CCK-8 assay (means ± SD of triplicate experiments). |
Table II
Effect of silencing HOXB4 on the
sensitivity of K562/ADM cells toward four chemotherapeutic drugs by
CCK-8 assay (means ± SD of triplicate experiments).
| Treatment | Control
(IC50 μg/ml) | NC shRNA
(IC50 μg/ml) | shRNA-3
(IC50 μg/ml) | RF |
|---|
| Adriamycin | 40.47±3.85 | 39.46±0.83 | 7.72±0.56a | 5.24 |
| Etoposide | 47.24±1.41 | 46.13±2.64 | 22.93±1.04a | 2.06 |
| Vindesine | 26.55±2.95 | 25.36±2.35 | 14.65±0.79b | 1.81 |
| Cytarabine | 27.79±0.82 | 27.27±1.78 | 14.21±0.50a | 1.96 |
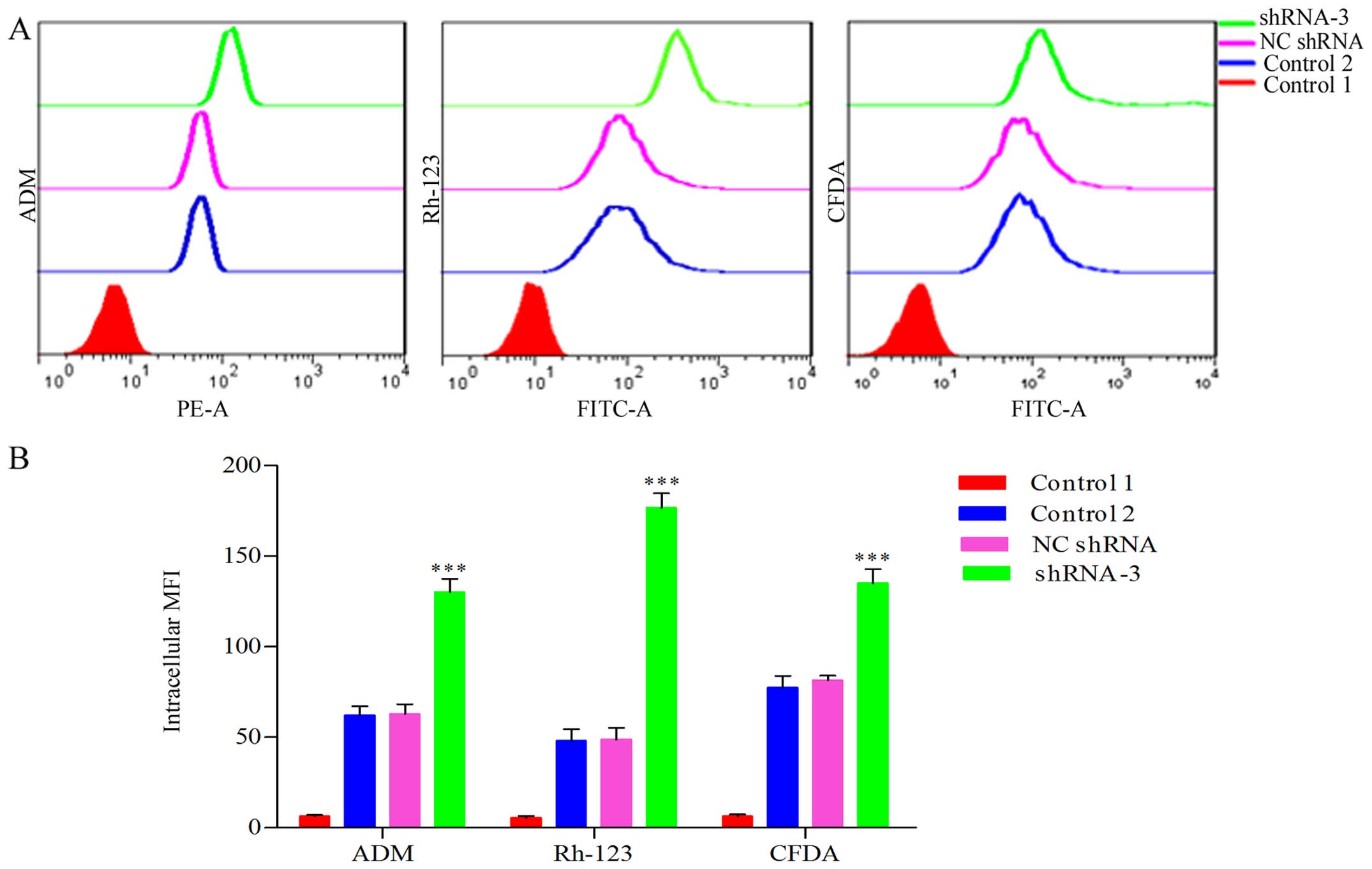
Enhancement of intracellular accumulation
of ADM for HOXB4 deletion
We previously confirmed that the intracellular
accumulation of ADM decreased significantly in K562/ADM cells
compared to the parental K562 cells (33). We determined that HOXB4 deletion
increased the intracellular accumulation of ADM in K562/ADM cells.
As shown in Fig. 5, the
accumulation value of intracellular ADM in shRNA-3 group was
elevated 1.93-fold that of NC-shRNA group and 2.07-fold that of
control group, respectively (P<0.001).
Inhibition of P-gp and MRP1-mediated
transport due to HOXB4 suppression
The impact of HOXB4 repression on the function of
P-gp and MRP1 as efflux pump in K562/ADM cells was examined by flow
cytometry. The fluorescent retention in HOXB4-silenced K562/ADM
cells obviously exceeded that of NC shRNA and control groups
(Fig. 5). Fig. 5 showed that the accumulation value
of intracellular Rh-123 in shRNA-3 group was elevated 3.59-fold
compared to NC shRNA group and 3.67-fold the control group
(P<0.001); the CFDA fluorescence in shRNA-3 group was enhanced
1.66-fold compared to NC shRNA group and 1.74-fold that of the
control (P<0.001). It indicated that the efflux functions
mediated by P-gp and MRP1 were significantly inhibited after HOXB4
suppression.
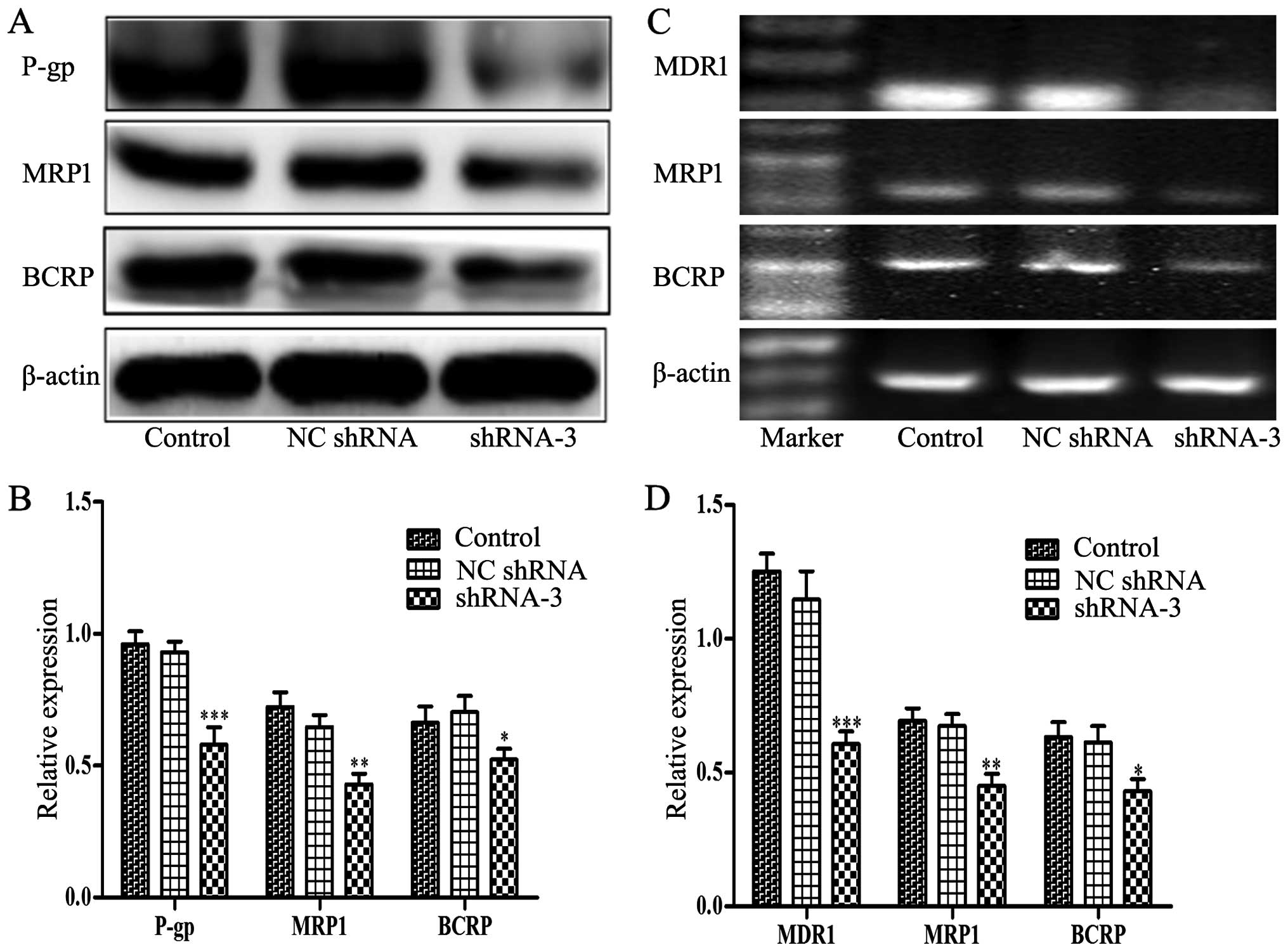
Decreased expression of P-gp, MRP1, BCRP
and blockage of PI3K/Akt signaling by HOXB4 knockdown
P-gp, MRP1 and BCRP are ABC transporters
overexpressed in many drug-resistant tumor cells which contributed
to the development of MDR. Therefore, we assessed whether HOXB4
could influence the expression of P-gp, MRP1 and BCRP. Notably,
western blotting and RT-PCR analyses (Fig. 6) illustrated that lower expression
levels of P-gp, MRP1 and BCRP were detected in shRNA-3-transfected
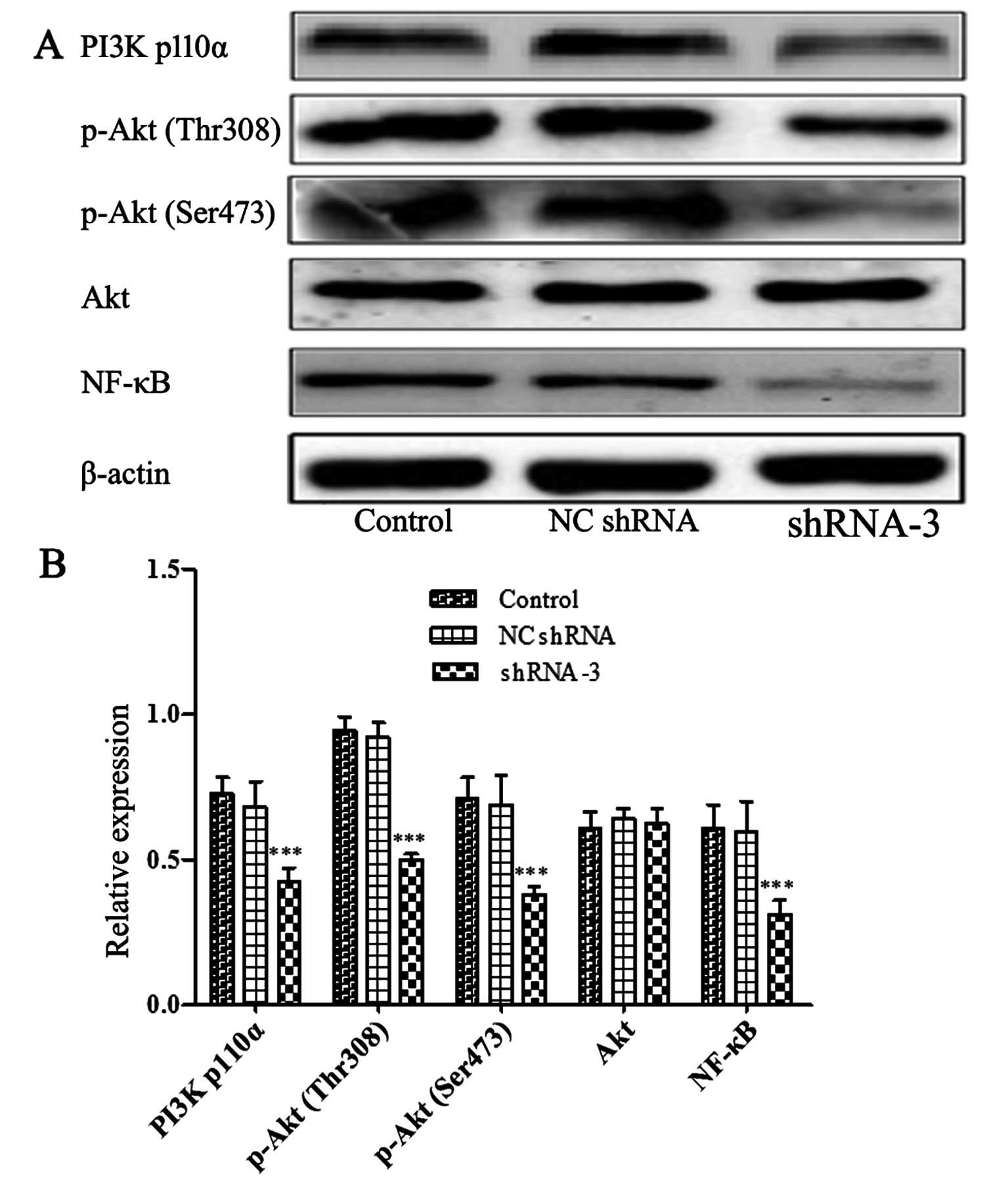
cells compared to control cells. Furthermore, Fig. 7 showed that the levels of p110α
(the catalytic subunit of PI3K), phosphorylation Akt at Ser473,
Thr308 and NF-κB were obviously reduced in shRNA-3 group. However,
there was no change in the total amount of Akt protein. The data
indicated that HOXB4 may be implicated in key steps of MDR
development in CML.
Discussion
Clinically the development of MDR remains one of the
major obstacles to effective tumor chemotherapy, and finding
strategies to overcome it is the hotspot of current cancer
research. Up to now, progress has been gained in revealing the
mechanisms of MDR including alteration of the cell environment,
inhibition of apoptosis and drug efflux mediated by ABC
transporters (36). Our present
results implied the role HOXB4 played in mediating MDR development
in human CML and its possible mechanisms. Besides, we evaluated the
efficacy of HOXB4 as a target therapy for overcoming drug
resistance of K562/ADM cells for the first time. Aberrant
expression of HOXB4 has been detected in a broad range of human
carcinomas including leukemia. A previous study proposed that
resistant K562/VCR expressing high level MDR genes ABCB1 and ABCB4
displayed significantly increased HOXB4 expression than parental
P-gp negative K562 cells (27). In
this study, upregulation of HOXB4 was also observed in MDR K562/ADM
cells with higher P-gp, MRP1 and BCRP expression, which indicated
that HOXB4 was also involved in the development of MDR in K562/ADM
cells. To explore the connection between HOXB4 and MDR,
drug-resistant K562/ADM cells were transfected with shRNA against
HOXB4 and measured in chemosensitivity. Results demonstrated that
HOXB4 knockout cells obviously restored the chemosensitivity to
anticancer drugs such as ADM, etoposide, vindesine or cytarabine.
Moreover, ADM, Rh-123 and CFDA efflux assays were exploited to
assess the inhibition of ABC transports by HOXB4 repression and to
estimate the interaction between HOXB4 and these proteins.
Consistent with the speculation, efflux pump activities mediated by
these ABC transporters were inhibited evidently after deletion of
HOXB4. Then molecular mechanism study illustrated that there was an
interrelationship between HOXB4 and these ABC proteins: HOXB4
expression in resistant K562/ADM cell line was higher than its
parent K562 cell line; notably, P-gp, MRP1 and BCRP expressions at
both the transcription and translation levels were inhibited
significantly in response to suppression of HOXB4. It might be a
reasonable explanation of chemosensitivity restoration after HOXB4
suppression when associated with the downregulation of P-gp, MRP1
and BCRP. In summary, HOXB4 repression partially reversed MDR of
K562/ADM cells by inhibiting the cellular efflux function and
downregulating the expression level of P-gp, MRP1 and BCRP, thus,
elevating intracellular chemotherapeutic accumulation.
The PI3K/Akt pathway is excessively activated in a
wide variety of hematologic malignancies including CML, diffuse
large B-cell lymphoma, acute myeloid leukemia, T-cell acute
lymphoblastic leukemia and chronic lymphoblastic leukemia; it is
considered to be responsible for tumorigenesis by simultaneously
promoting proliferation and inhibiting apoptosis (37). The PI3K/Akt pathway controls the
expression and function of many proteins that are necessary for
leukemia cell MDR and has become a promising target for systemic
therapy (36–38). Morishita et al (39) proposed that Akt phosphorylation can
induce chemotherapeutic resistance in B-pre-acute lymphoblastic
leukemia. Moreover, several lines of evidence implicated that the
maintaining of MDR in cancer cells by PI3K/Akt signaling pathway
was mostly correlated with P-gp, MRP1 and BCRP (35,36,40).
Our previous study demonstrated that the resistant cell line
K562/ADM presented higher PI3K/Akt activity than the sensitive one,
which was in accordance with the MDR phenotype; we also found a
parallel relationship between the activity of Akt and expression of
P-gp and MRP1 (33,34). In the present study, to understand
why HOXB4 inhibition decreased the expression level of P-gp, MRP1
and BCRP, we evaluated more precisely the PI3K/Akt signaling
expression alteration in the case of HOXB4 deletion. Decreased
expression of HOXB4 showed obviously lower protein expression of
the main signal molecules of PI3K/Akt signaling in K562/ADM cells.
These results indicated that HOXB4-modulated MDR in CML K562/ADM
cell line was, at least in part, PI3K/Akt-dependent.
In conclusion, HOXB4 was expressed at a higher level
in the K562/ADM cells, and knockdown of HOXB4 enhanced the
sensitivity of the K562/ADM cells to cytotoxic killing by the
therapeutic drugs, such as ADM, etoposide, vindesine or cytarabine,
as a result of the increased intracellular accumulation of
chemotherapeutics. The inhibition of drug efflux induced by HOXB4
deletion was associated with downregulation of P-gp, MRP-1 and BCRP
proteins. Moreover, the relationships between HOXB4 and PI3K/Akt
signaling pathway were further elucidated, with a conclusion that
interference of HOXB4 was able to reverse MDR in K562/ADM cells via
repression of PI3K/Akt activity. While the initial findings are
promising, more comprehensive and detailed studies still need to be
conducted, such as animal models in vivo or more precise
PI3K/Akt pathway assays. Above all, we suggest that knockdown of
HOXB4 is a novel and potent therapeutic target that could be used
for reversing MDR in human CML K562/ADM cells.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Shandong Province (nos. ZR2014HL032 and
ZR2014HQ079) and the Projects of Medical and Health Technology
Development Program in Shandong Province (no. 2014WS0183).
References
|
1
|
Cao R, Wang Y and Huang N: Discovery of
2-acylaminothiophene-3-carboxamides as multitarget inhibitors for
BCR-ABL kinase and microtubules. J Chem Inf Model. 55:2435–2442.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mathisen MS, Kantarjian HM, Cortes J and
Jabbour E: Mutant BCR-ABL clones in chronic myeloid leukemia.
Haematologica. 96:347–349. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yandim MK, Ceylan C, Elmas E and Baran Y:
A molecular and biophysical comparison of macromolecular changes in
imatinib-sensitive and imatinib-resistant K562 cells exposed to
ponatinib. Tumour Biol. 37:2365–2378. 2016. View Article : Google Scholar
|
|
4
|
O’Hare T, Shakespeare WC, Zhu X, Eide CA,
Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, et al:
AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia,
potently inhibits the T315I mutant and overcomes mutation-based
resistance. Cancer Cell. 16:401–412. 2009. View Article : Google Scholar
|
|
5
|
Sweet K and Pinilla-Ibarz J: Early switch
in tyrosine kinase inhibitor therapy for patients with chronic
myeloid leukemia: An emerging clinical question. Crit Rev Oncol
Hematol. 103:99–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hochhaus A, Ernst T, Eigendorff E and La
Rosée P: Causes of resistance and treatment choices of second- and
third-line treatment in chronic myelogenous leukemia patients. Ann
Hematol. 94(Suppl 2): S133–S140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
La Rosée P, Corbin AS, Stoffregen EP,
Deininger MW and Druker BJ: Activity of the Bcr-Abl kinase
inhibitor PD180970 against clinically relevant Bcr-Abl isoforms
that cause resistance to imatinib mesylate (Gleevec, STI571).
Cancer Res. 62:7149–7153. 2002.PubMed/NCBI
|
|
8
|
Corrêa S, Binato R, Du Rocher B,
Castelo-Branco MT, Pizzatti L and Abdelhay E: Wnt/β-catenin pathway
regulates ABCB1 transcription in chronic myeloid leukemia. BMC
Cancer. 12:3032012. View Article : Google Scholar
|
|
9
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE
and Gottesman MM: P-glycoprotein: From genomics to mechanism.
Oncogene. 22:7468–7485. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalle AM, Sachchidanand S and Pallu R:
Bcr-Abl-independent mechanism of resistance to imatinib in K562
cells: Induction of cyclooxygenase-2 (COX-2) by histone
deacetylases (HDACs). Leuk Res. 34:1132–1138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoegg S and Meyer A: Hox clusters as
models for vertebrate genome evolution. Trends Genet. 21:421–424.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cillo C, Cantile M, Faiella A and
Boncinelli E: Homeobox genes in normal and malignant cells. J Cell
Physiol. 188:161–169. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li N, Jia X, Wang J, Li Y and Xie S:
Knockdown of homeobox A5 by small hairpin RNA inhibits
proliferation and enhances cytarabine chemosensitivity of acute
myeloid leukemia cells. Mol Med Rep. 12:6861–6866. 2015.PubMed/NCBI
|
|
15
|
Yi YJ, Jia XH, Wang JY, Li YJ, Wang H and
Xie SY: Knockdown of HOXA10 reverses the multidrug resistance of
human chronic mylogenous leukemia K562/ADM cells by downregulating
P-gp and MRP-1. Int J Mol Med. 37:1405–1411. 2016.PubMed/NCBI
|
|
16
|
Kavalerchik E, Goff D and Jamieson CH:
Chronic myeloid leukemia stem cells. J Clin Oncol. 26:2911–2915.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thorén LA, Liuba K, Bryder D, Nygren JM,
Jensen CT, Qian H, Antonchuk J and Jacobsen SE: Kit regulates
maintenance of quiescent hematopoietic stem cells. J Immunol.
180:2045–2053. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang XB, Beard BC, Trobridge GD, Wood BL,
Sale GE, Sud R, Humphries RK and Kiem HP: High incidence of
leukemia in large animals after stem cell gene therapy with a
HOXB4-expressing retroviral vector. J Clin Invest. 118:1502–1510.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bodey B, Bodey B Jr, Siegel SE and Kaiser
HE: Immunocytochemical detection of the homeobox B3, B4, and C6
gene products in breast carcinomas. Anticancer Res. 20(5A):
3281–3286. 2000.PubMed/NCBI
|
|
20
|
Bodey B, Bodey B Jr, Gröger AM, Siegel SE
and Kaiser HE: Immunocytochemical detection of homeobox B3, B4, and
C6 gene product expression in lung carcinomas. Anticancer Res.
20:2711–2716. 2000.PubMed/NCBI
|
|
21
|
Hwang SH, Kim KU, Kim JE, Kim HH, Lee MK,
Lee CH, Lee SY, Oh T and An S: Detection of HOXA9 gene methylation
in tumor tissues and induced sputum samples from primary lung
cancer patients. Clin Chem Lab Med. 49:699–704. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rodríguez-Rodero S, Fernández AF,
Fernández-Morera JL, Castro-Santos P, Bayon GF, Ferrero C,
Urdinguio RG, Gonzalez-Marquez R, Suarez C, Fernández-Vega I, et
al: DNA methylation signatures identify biologically distinct
thyroid cancer subtypes. J Clin Endocrinol Metab. 98:2811–2821.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jamieson CH, Ailles LE, Dylla SJ,
Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating
A, et al: Granulocyte-macrophage progenitors as candidate leukemic
stem cells in blast-crisis CML. N Engl J Med. 351:657–667. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morgan R, Simpson G, Gray S, Gillett C,
Tabi Z, Spicer J, Harrington KJ and Pandha HS: HOX transcription
factors are potential targets and markers in malignant
mesothelioma. BMC Cancer. 16:852016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kelly Z, Moller-Levet C, McGrath S,
Butler-Manuel S, Kavitha Madhuri T, Kierzek AM, Pandha H, Morgan R
and Michael A: The prognostic significance of specific HOX gene
expression patterns in ovarian cancer. Int J Cancer. 139:1608–1617.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gordon-Keylock SA, Jackson M, Huang C,
Samuel K, Axton RA, Oostendorp RA, Taylor H, Wilson J and Forrester
LM: Induction of hematopoietic differentiation of mouse embryonic
stem cells by an AGM-derived stromal cell line is not further
enhanced by overexpression of HOXB4. Stem Cells Dev. 19:1687–1698.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lehne G, Grasmo-Wendler UH, Berner JM,
Meza-Zepeda LA, Adamsen BL, Flack A, Reiner A, Clausen OP, Hovig E
and Myklebost O: Upregulation of stem cell genes in multidrug
resistant K562 leukemia cells. Leuk Res. 33:1379–1385. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sheng Z, Ma L, Sun JE, Zhu LJ and Green
MR: BCR-ABL suppresses autophagy through ATF5-mediated regulation
of mTOR transcription. Blood. 118:2840–2848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie X, Tang B, Zhou J, Gao Q and Zhang P:
Inhibition of the PI3K/Akt pathway increases the chemosensitivity
of gastric cancer to vincristine. Oncol Rep. 30:773–782.
2013.PubMed/NCBI
|
|
30
|
Cheng L, Luo S, Jin C, Ma H, Zhou H and
Jia L: FUT family mediates the multidrug resistance of human
hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell
Death Dis. 4:e9232013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao Z, Zhou J, Luan J, Sheng W, Shen X and
Dong X: Tamoxifen reduces P-gp-mediated multidrug resistance via
inhibiting the PI3K/Akt signaling pathway in ER-negative human
gastric cancer cells. Biomed Pharmacother. 68:179–183. 2014.
View Article : Google Scholar
|
|
32
|
Tazzari PL, Cappellini A, Ricci F,
Evangelisti C, Papa V, Grafone T, Martinelli G, Conte R, Cocco L,
McCubrey JA, et al: Multidrug resistance-associated protein 1
expression is under the control of the phosphoinositide 3
kinase/Akt signal transduction network in human acute myelogenous
leukemia blasts. Leukemia. 21:427–438. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Jia XH, Chen JR, Wang JY and Li
YJ: Osthole shows the potential to overcome P-glycoprotein-mediated
multidrug resistance in human myelogenous leukemia K562/ADM cells
by inhibiting the PI3K/Akt signaling pathway. Oncol Rep.
35:3659–3668. 2016.PubMed/NCBI
|
|
34
|
Chen JR, Jia XH, Wang H, Yi YJ, Wang JY
and Li YJ: Timosaponin A-III reverses multi-drug resistance in
human chronic myelogenous leukemia K562/ADM cells via
downregulation of MDR1 and MRP1 expression by inhibiting PI3K/Akt
signaling pathway. Int J Oncol. 48:2063–2070. 2016.PubMed/NCBI
|
|
35
|
Huang FF, Wu DS, Zhang L, Yu YH, Yuan XY,
Li WJ, Chen XP, Zhao XL, Chen FP and Zeng H: Inactivation of PTEN
increases ABCG2 expression and the side population through the
PI3K/Akt pathway in adult acute leukemia. Cancer Lett. 336:96–105.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma H, Cheng L, Hao K, Li Y, Song X, Zhou H
and Jia L: Reversal effect of ST6GAL 1 on multidrug resistance in
human leukemia by regulating the PI3K/Akt pathway and the
expression of P-gp and MRP1. PLoS One. 9:e851132014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang X, Dong W, Zhou H, Li H, Wang N,
Miao X and Jia L: α-2,8-sialyltransferase is involved in the
development of multidrug resistance via PI3K/Akt pathway in human
chronic myeloid leukemia. IUBMB Life. 67:77–87. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng Z, Yang N, Liang W, Yan X, Li L and
Pan L: Effect of phosphatase and tensin homology deleted on
chromosome 10 (PTEN) gene transfection on reversal of multidrug
resistance in K562/ADM cells. Leuk Lymphoma. 53:1383–1389. 2012.
View Article : Google Scholar
|
|
39
|
Morishita N, Tsukahara H, Chayama K,
Ishida T, Washio K, Miyamura T, Yamashita N, Oda M and Morishima T:
Activation of Akt is associated with poor prognosis and
chemotherapeutic resistance in pediatric B-precursor acute
lymphoblastic leukemia. Pediatr Blood Cancer. 59:83–89. 2012.
View Article : Google Scholar
|
|
40
|
Jung KA, Choi BH and Kwak MK: The
c-MET/PI3K signaling is associated with cancer resistance to
doxorubicin and photodynamic therapy by elevating BCRP/ABCG2
expression. Mol Pharmacol. 87:465–476. 2015. View Article : Google Scholar
|