Introduction
Glioblastoma multiforme (GBM) is the most common
primary brain tumor in adults with a median survival rate of only
15 months (1,2). GBM originates from the glial cells
which subsequently evolved into tumors known as glioma (3). It is the deadliest primary brain
tumor and is classified as grade IV astrocytoma by WHO criteria
(1,4,5). The
standard treatment for GBM consists of surgical removal, radiation
followed by chemotherapy. Unfortunately, these treatments only gave
minor improvements to the patients’ survival particularly for the
recurrent GBM (4).
GBM is resistant towards treatment due to the
heterogeneous nature of the disease. These are contributed by the
dysregulation of the core signaling pathways such as the
ErbB, MAPK, mTOR and p53 signaling
pathways (6–9). It was shown that the pathogenesis of
GBM requires alteration of multiple genetic pathways and each of
the primary and secondary GBMs has a unique combination of these
genetic changes (5). Primary or
de novo GBM frequently showed loss of heterozygosity (LOH)
at 10q (70% of cases), EGFR amplification (36%),
p16INK4a deletion (31%) and PTEN mutations (25%)
(10). Mutations in the
IDH1 and IDH2 genes are common in astrocytomas,
oligodendrogliomas, oligoastrocytomas and secondary glioblastoma
with prevalence of 50–80% of cases (11). In addition, TP53 and
PTEN mutations are common in primary and secondary gliomas
with a frequency of 28 and 65% for p53, 25 and 4% for
PTEN respectively (10).
Secondary GBM is generally initiated from diffuse astrocytomas.
Some common molecular lesions associated with secondary GBM are
TP53 mutations (60–65%) and gain of 7q arm (21–50%) with
MET gene gain of function (47% in primary and 44% in
secondary glioblastoma) being affected significantly and associated
with poor prognosis (12). There
are also reports on the involvement of aberrant intrinsic and
extrinsic apoptotic pathways and the overexpression of
anti-apoptotic proteins such as FLIPs, BCL2 and
survivin which contribute to apoptotic resistance (13–17).
The therapeutic strategy in GBM could be improved by
targeting the multiple pathways involved. RNA interference (RNAi)
is one of the attractive approaches and may result in the
post-transcriptional knockdown of the genes of interest.
Significant impact in in vitro experiments using RNAi has
allowed the implementation of therapeutic approach using RNAi gene
therapy in vivo (18). One
of the major problems in GBM therapy is the difficulty for the
drugs to cross the blood-brain barrier (BBB) hindering maximal drug
distribution to the tumor site. To date, there are a few strategies
being used to efficiently deliver siRNA through the BBB. The
RNAi-based nanomedicine platform has been introduced at the
pre-clinical stage (18). Based on
spherical nucleic acid gold nanoparticle conjugates, which are
densely packed, highly oriented siRNA duplexes targeting the
oncoprotein Bcl2Like12 (Bcl2L12) were used to
neutralize the oncogene expression in GBM (19). There are many delivery systems that
form complexes with siRNA including PEGylated immunoliposomes that
carry siEGFR, recombinant adeno-associated virus carrying siHec-1
and lentiviral vectors carrying siTRAIL (20–22).
An example of a molecular target that has made to clinical trial
using siRNA and showed promising results is Tenascin-C (TN-C),
which is overexpressed in the extracellular matrix (ECM) of GBM. It
has been shown that dsRNA targeting TN-C mRNA could reduce the
tumor size significantly and increase the survival rate by 11%
(23,24).
Materials and methods
Meta-analysis of microarray datasets
We performed a meta-analysis on five microarray
datasets from a cancer microarray database using an integrated
data-mining platform, the Oncomine Research Edition (25). Data were filtered based on data
source, cancer, the type of datasets and analysis. Candidate genes
were selected based on the median rank and p<0.05. Candidate
genes obtained from meta-analysis were then screened using
synthetic lethal RNAi screening and the hits were selected based on
their significant values in viability reduction. The human
glioblastoma LN18 (TP53-mutant) cells were transfected with pooled
siRNA (SMARTpool™; GE Dharmacon, Lafayette, CO, USA) targeting
against 460 genes and cultured for 48 h according to the
manufacturer’s protocol. The media were changed after 48 h
post-transfection and incubated for another 48 h. The cells were
then prepared for viability measurement using the
CellTiter-Glo® Luminescent Cell Viability Assay (Promega
Corp., Madison, WI, USA). The experiment was performed in
triplicate.
Cell culture
LN18 cells were maintained in T-75 flasks and
allowed to grow in 15 ml of Dulbecco’s modified Eagle’s medium
(DMEM) (ATCC, Manassas, VA, USA) and supplemented with 10% fetal
bovine serum (J R Scientific, Inc., Woodland, CA, USA) until 80%
confluency. The cells were incubated under 5% CO2
condition. Generally, the doubling time for LN18 cells was <24
h. Cells were harvested by removing media and cells were then
washed with 5 ml of 1X Dulbecco’s Phosphate-Buffered Saline (Gibco,
Grand Island, NY, USA) and trypsinised using 1X Trypsin EDTA 0.25%
(J R Scientific, Inc.).
Preparation of siRNA
ON-TARGETplus SMARTpool™ of PROS1 siRNA (GE
Dharmacon) consisting of four different siRNA sequences were used
in this experiment. The siRNA sequences used were: i)
GCAUGGAAGUGAAUAUUAA; ii) GCAACAGGCUUCACAAGUC; iii)
UAUUAGAGCUCACUCAUGU; and iv) GAAGAGUUGUGAGGUUGUU. Lyophilized
PROS1 siRNA was resuspended with 1X siRNA buffer (Thermo
Fisher Scientific, Inc., Rockford, IL, USA). A total of 25 nM final
concentration of PROS1 siRNA and non-targeting siRNA were used with
DharmaFECT2. All functional assays were performed 48 h
post-transfection.
RNA extraction and qPCR
RNeasy kit (Qiagen, Hilden, Germany) was used to
isolate total RNA from cells. The quality and quantity of the
isolated RNA were determined using NanoDrop (Thermo Fisher
Scientific, Inc.). Briefly, 100 ng of RNA were used to generate
cDNA using iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). qPCR was conducted using SsoFast™
EvaGreen® Supermix (Bio-Rad Laboratories, Inc.) on a
Rotor-Gene 3000 (Corbett Life Science/Qiagen, Inc., Valencia, CA,
USA) platform. The PROS1 primers used were: forward,
5′-TGCTGGCGTGTCTCCTCCTA-3′ and reverse,
5′-CAGTTCTTCGATGCATTCTCTTTCA-3′. The expression of
PROS1-related genes such as GAS6, RhoA,
FasL, Tyro-3, Axl, and Mertk was also
quantified using qPCR and results were calculated based on the ΔΔCt
method (26). ACTB gene was
used as the reference gene. Primer sequences are shown in Table I.
 | Table IPrimer sequences used for qPCR
analysis. |
Table I
Primer sequences used for qPCR
analysis.
| Gene | Primer sequence
(5′→3′) |
|---|
| PROS1 | F
TGCTGGCGTGTCTCCTCCTA
R CAGTTCTTCGATGCATTCTCTTTCA |
| Tyro-3 | F
CACGGTAGAAGGTGTGCCAT
R TGGTAACAGGTTCAGGGGGA |
| Axl | F
TTAGTGCTACGCGGAATGGG
R TGTCCATTAGCACCTCTGGG |
| Mertk | F
GTCCATCTGTCCATCCGTCC
R CCTCAGTGATAGCTCTACGCC |
| Gas6 | F
ACGACCCCGAGACGGATTA
R GCGAAGCCTGAGTTTTTGGT |
| FasL | F
CCTTGGTAGGATTGGGCCTG
R CTGTGTGCATCTGGCTGGTA |
Viability assay
The CellTiter-Glo® Luminescent Cell
Viability Assay (Promega Corp.) which is based on the
quantification of ATP present in the viable cells was used for
viability assay. Cells were cultured for 24 and 48 h
post-transfection. Subsequently, CellTiter-Glo® buffer
was added onto the CellTiter-Glo® substrate, which was
then loaded into the samples. The luminescent signal was captured
at 570 nm using SpectraMax® L Luminescence Microplate
Reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Cellular
viability was calculated based on the normalization between treated
(siPROS1) vs. non-targeting siRNA cells from three independent
experiments.
Proliferation assay
Proliferation assay was performed using the
bromodeoxyuridine (BrdU) incorporation method (Millipore Corp.,
Billerica, MA, USA). Transfected cells were cultured for 24 h in
the present of BrdU which was incorporated into newly synthesized
DNA strand of the proliferating cells. The cells were then fixed,
and incubated with anti-BrdU monoclonal antibody (Millipore Corp.)
for 1 h. Goat anti-mouse IgG peroxidase was added onto the well.
Incorporation of BrdU in the proliferating cells leads to
colorimetric changes from clear to blue which was measured using
Varioskan Flash Multimode Reader (Thermo Fisher Scientific Oy,
Vantaa, Finland) at 450 nm wavelength.
Migration assay
The effect of PROS1 gene silencing on tumor
cell invasion was investigated using QCM™ 3 μm 24-well Chemotaxis
Cell Migration Assay kit (Millipore Corp.). Cells were seeded in
the 24-well inserts at a density of 1×104 cells/well in
serum-free media for 24 h and allowed to migrate through the
membrane towards the media. The migrated cells were then lysed and
the fluorescent signal was quantified using Varioskan Flash
Multimode Reader (Thermo Fisher Scientific Oy). We also performed
wound healing scratch assay in order to observe the cellular
motility in siPROS1-treated LN18 cells. Wound closure was
observed at 0, 24, 48 and 72 h post-scratching. These assays were
performed in three independent replicates.
Invasion assay
The role of PROS1 in cell invasion was
investigated using QCM™ 24-well Cell Invasion Assay kit (Millipore
Corp.). The cells were cultured overnight in serum-free media and
allowed to invade through the ECM. The cells were harvested and
lysed prior to fluorometric quantification using Varioskan Flash
Multimode Reader (Thermo Fisher Scientific, Inc.). The invasion
assay was carried out in three independent replicates.
Apoptosis assay
Apoptosis was determined using the ssDNA Apoptosis
ELISA kit (Millipore Corp.). In total, 5×103 LN18 cells
were grown overnight in a 96-well plate. Subsequently, the cells
were transfected either with siPROS1 or non-targeting siRNA
for 48 h. Cells were then prepared for apoptosis measurement
according to the manufacturer’s protocol and the signal was
measured using ELx800 TC models 95 Microplate Reader (Biotek
Instruments, Inc., Winooski, VT, USA).
Cell cycle analysis
Cell cycle assay was performed using
1×106 LN18 cells that were transfected with siPROS1 or
non-targeting siRNA. Cells were harvested using a standard protocol
as indicated in the Cycletest™ Plus DNA Reagent Kit protocol (BD
Biosciences, Mississauga, ON, Canada). Subsequently, cells were
washed three times with wash buffer. Cells were then suspended in
solution A containing trypsin. Solution B with trypsin inhibitor
and RNase were then added into the cell suspension. Finally,
solution C which contained propidium iodide (PI) was added. Flow
cytometric analysis was performed using BD FACSAria™ (BD
Biosciences, Franklin Lakes, NJ, USA). Data were analysed using
ModFit LT software (Verity Software House, Inc., Topsham, ME, USA).
The percentage of arrested cells was measured by the percentage of
hypodiploid cells accumulated at the G0/G1, S, G2/M checkpoints of
the cell cycle.
Western blotting
Protein expression of PROS1 was assessed using
western blotting. Cells were treated with siPROS1 and proteins were
harvested and extracted using radioimmunoprecipitation assay (RIPA)
buffer. A total of 50 μg protein was loaded onto the
Mini-PROTEAN® Precast Gels (Bio-Rad Laboratories, Inc.),
and then transferred onto the Immobilon transfer membranes
(Millipore Corp.). Membranes were then incubated with
SuperBlock® (Thermo Fisher Scientific, Inc.) for 1 h at
room temperature. After that, membranes were incubated overnight
with PROS1 mouse monoclonal antibody (1:500; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 4°C. The membranes were
then washed three times with TBST. Membranes were then incubated
with goat anti-mouse secondary antibody conjugated to alkaline
phosphate (1:2,000; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. Prior to protein detection, the membranes were washed
three times. Finally, proteins were detected using Pierce ECL and
SuperSignal substrate (Thermo Fisher Scientific, Inc.). β-actin was
used as an internal control.
Protein array
Protein array was conducted using the Human
Apoptosis Array kit (RayBiotech, Norcross, GA, USA). Protein
samples were extracted from 48 h post-transfection according to the
manufacturer’s protocol. The quantity of the protein isolated was
determined using BCA Protein Assay (Thermo Fisher Scientific,
Inc.). Briefly, protein was loaded into the chamber slides coated
with 43 different types of apoptosis antibodies. Subsequently, the
slides were washed and the membranes were incubated with a cocktail
of biotin-conjugated anti-apoptotic protein antibodies. The
membranes were incubated with HRP-streptavidin prior to signal
detection.
Statistical analysis
All data were expressed as the mean ± SD of three
independent experiments. Significant differences were defined as
p<0.05. All statistical analyses were performed using Microsoft
Excel and the Statistical Package for the Social Sciences (SPSS)
software.
Results
PROS1 as a novel candidate for GBM
therapy
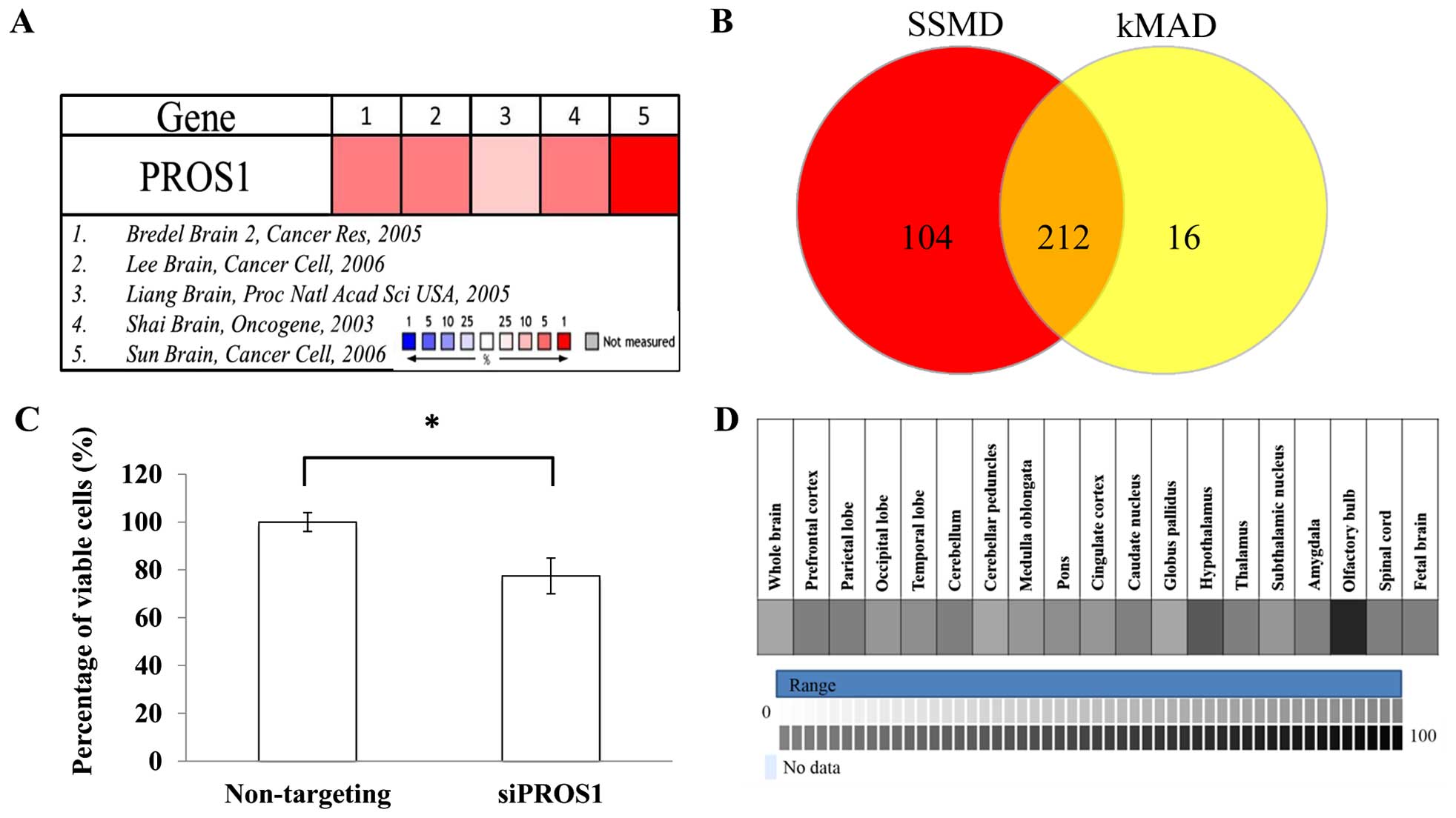
Meta-analysis on five microarray datasets (Bredel
Brain 2, Lee Brain, Liang Brain, Shai Brain, and Sun Brain)
identified 460 upregulated genes based on the median rank and
p<0.05. All datasets were normalized between cancers vs. normal
tissues. These 460 genes were used as candidates for RNAi
screening. Based on the SSMD and kMAD analyses, 212 hits were
identified. After selection, PROS1 was identified as a
target gene for validation since the role of PROS1 in GBM
has not been documented (Fig.
1).
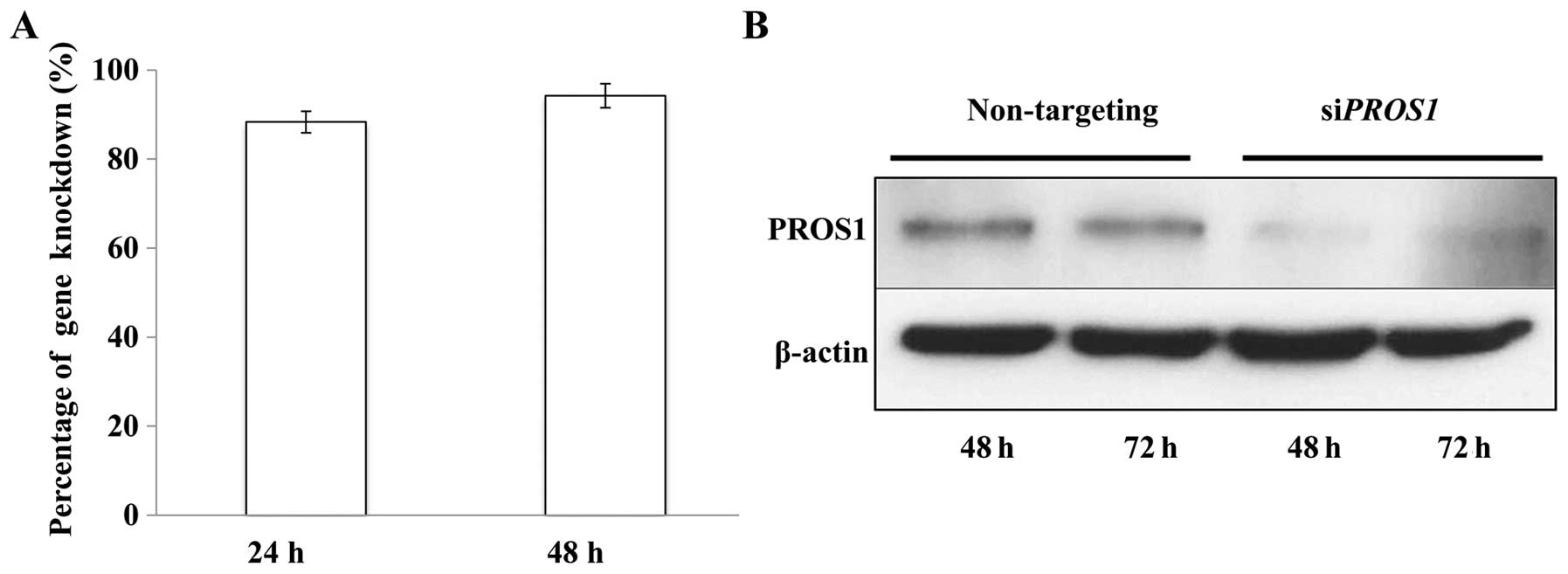
PROS1 silencing decreases PROS1 mRNA and
protein expressions
The efficiency of PROS1 silencing was
assessed using qPCR. The results showed that ~80% of the
PROS1 gene was knocked down after 24 h and it increased up
to 100% after 48 h of transfection. This was further confirmed at
protein level via western blotting at 48 and 72 h post-silencing as
the expression of the PROS1 protein was reduced significantly
compared to control (Fig. 2).
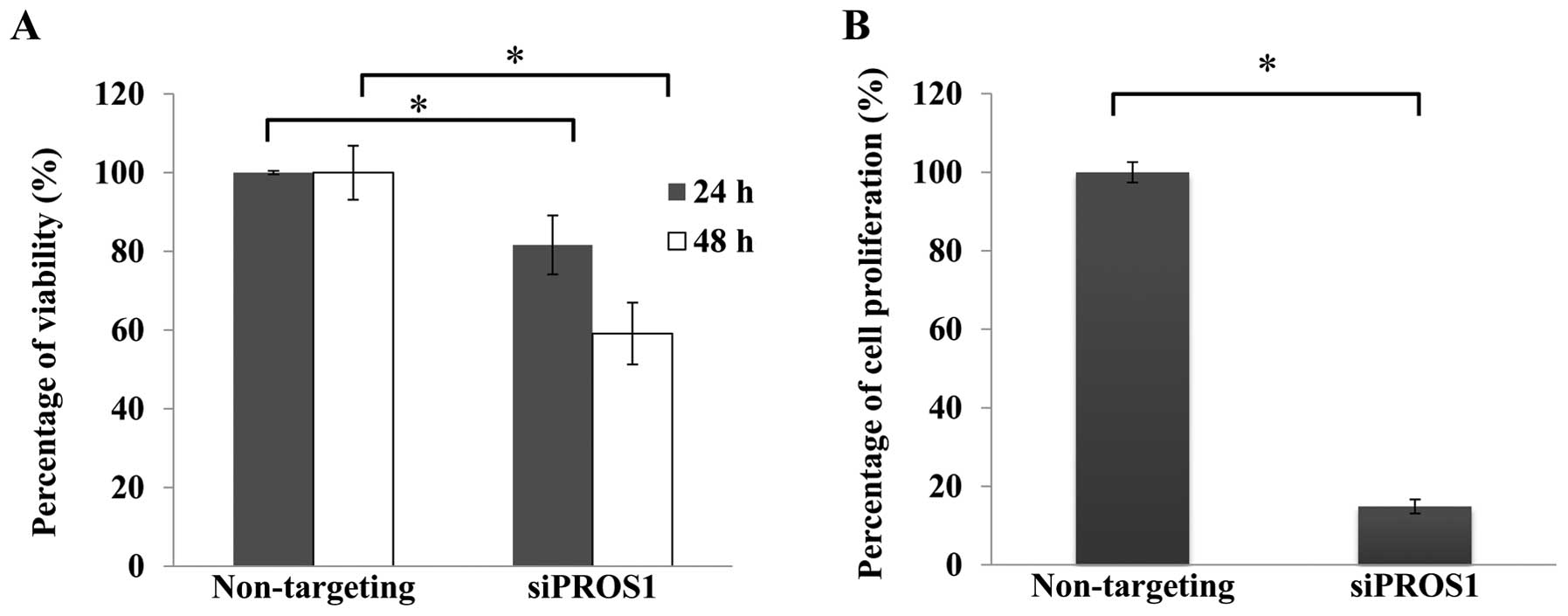
PROS1 silencing reduces cell viability
and cell proliferation
Cell viability assay was conducted using
CellTiter-Glo® Luminescent Cell Viability Assay to
determine the effect of PROS1 gene silencing. The number of
viable cells was reduced in a time-dependent manner. The
quantification of proliferating cells by BrdU showed that the
proliferation signal was decreased in siPROS1 treatment
(18%) compared to the control (Fig.
3).
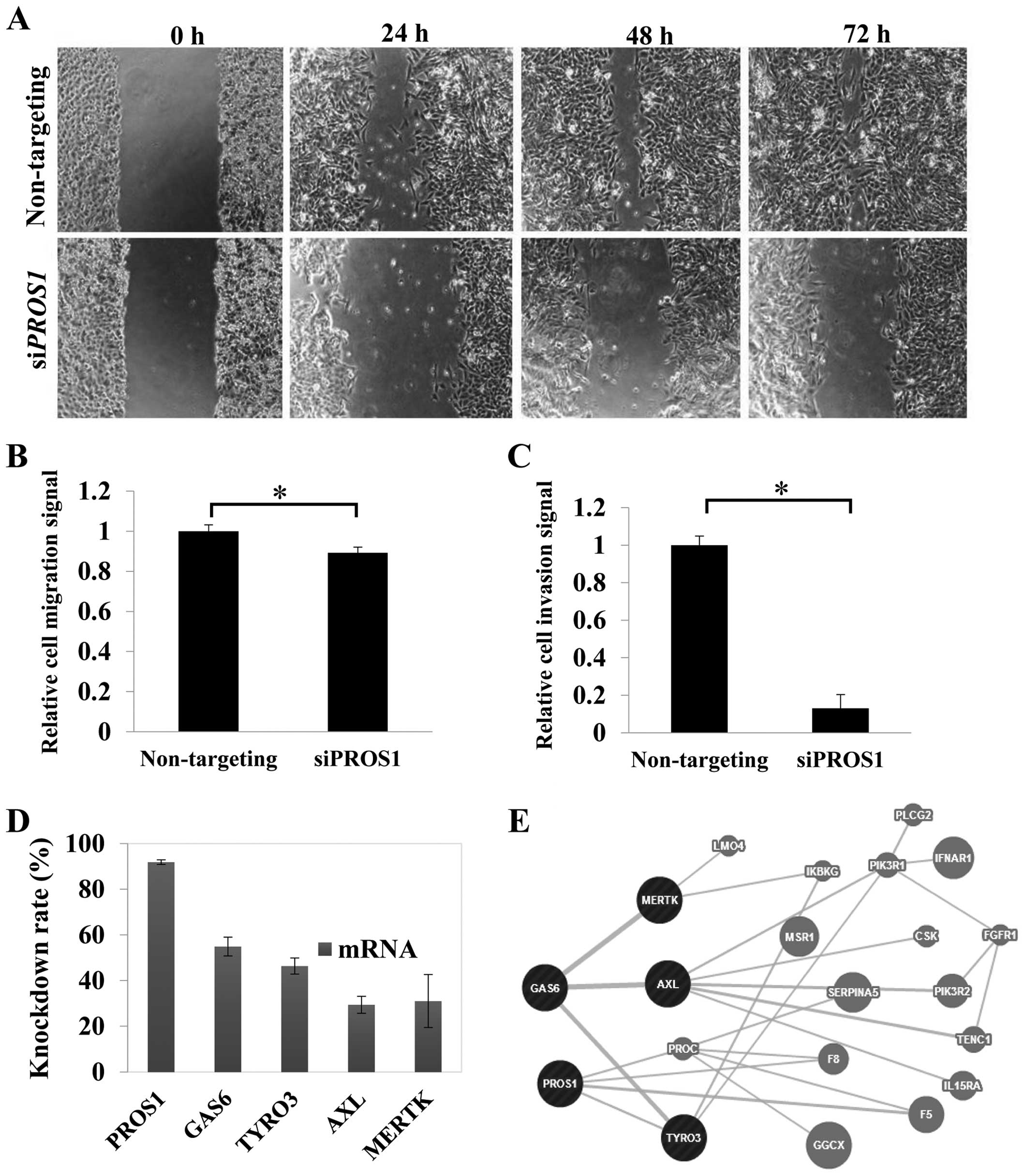
PROS1 silencing inhibits LN18 cell
migration
Migration was reduced significantly (p<0.05) in
siPROS1-treated LN18 cells. The scratch assay demonstrated
the inhibition of the migratory potential of the 24-h post-scratch
siPROS1-treated LN18 cells. Surprisingly, the size of wound
scratch remained up to 72 h. These data suggest the possible role
of PROS1 in GBM cell migration (Fig. 4A and B).
PROS1 silencing reduces GBM cell
invasion
Cell invasion assay was performed to study whether
PROS1 suppression could influence the invasion of LN18
cells. The results showed that the invasion of LN18 cells through
the ECM was inhibited with siPROS1-treated cells up to 82%
(p<0.01) compared to the control group (Fig. 4C).
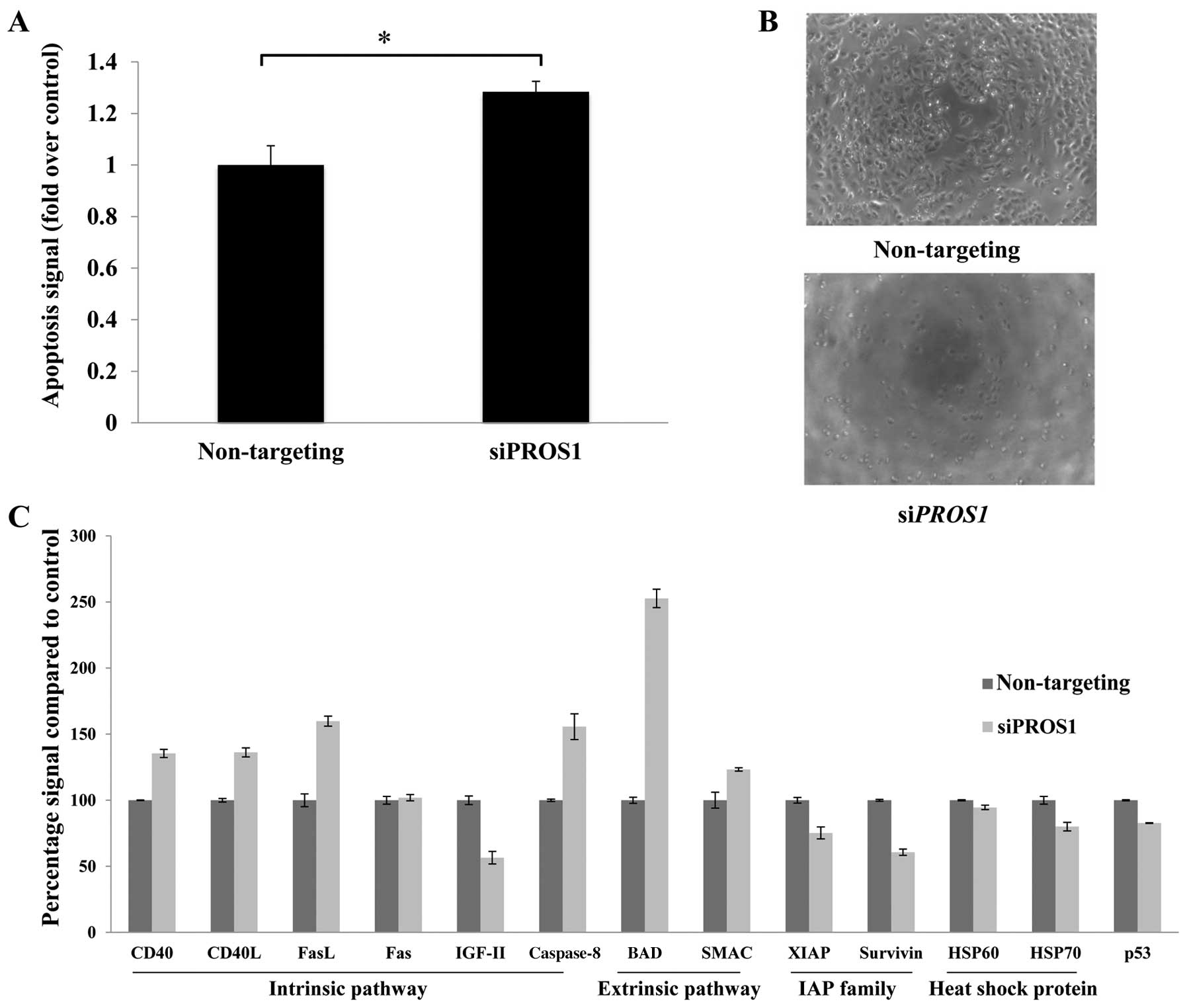
PROS1 silencing significantly induces
cell death through apoptosis
ELISA-based assay was conducted to determine the
mode of cell death in siPROS1-treated LN18 cells. Apoptosis
was increased compared to the control at 48 h post-transfection
(p<0.05) (Fig. 5A). There was
no evidence of cell cycle arrests identified from the cell cycle
assay (data not shown). Further validation was conducted using
protein array to elucidate the relevant pathways involved in this
process.
PROS1 gene silencing leads to decreased
expression of Tyro-3, Axl and Mertk
qPCR was performed to study the effect of
PROS1 silencing on its related interacting genes from the
TAM family of receptor tyrosine kinases which include GAS6,
Tyro-3, Axl, and Mertk. This was performed at
48 h post-transfection. The level of GAS6 was reduced to
68.7% compared to the control (p<0.05). PROS1 gene
silencing also reduced the expression of the tyrosine kinases,
especially the Tyro-3 where the expression was 50% reduced
compared to the control. The expression of Axl and
Mertk genes was reduced to 70.6 and 69%, respectively
(Fig. 4D and 4E).
Discussion
The main aim of this study was to understand the
functional role of PROS1 in GBM by performing silencing
experiments coupled with various functional assays. PROS1
was identified as a potential gene target for GBM from our
meta-analysis using five microarray datasets and the
loss-of-function RNAi screening of 460 upregulated genes. PROS1 is
a vitamin K-dependent plasma protein and is known to be involved in
the anticoagulant cascade. It acts as a cofactor for anticoagulant
protease in the blood coagulation system known as the activated
protein C (APC) (27). PROS1
shares ~43% of amino acid identity with GAS6, a γ-carboxyglutamic
acid (Gla)-containing protein, which stimulates cell proliferation
through activation of the Axl receptor tyrosine kinase (28–30).
PROS1 and GAS6 are ligands for Axl together with Tyro-3 and Mertk
and were reported to be overexpressed in haematological
malignancies and solid tumors, suggesting that these molecules
activate important autocrine-based oncogenic signaling events in
cancer cells (31–34). Overexpression of TAM receptors
mediates multiple oncogenic phenotypes in GBM such as in
vitro proliferation, anchorage-independent growth, xenograft
growth, resistance to apoptosis, autophagy, invasion and migration
as well as activation of the downstream PI3K and MAPK survival
pathways (33). Inhibition of
Mertk and Axl by gene knockdown in astrocytoma cells enhanced
apoptosis and improved chemosensitivity towards conventional
chemotherapeutic agents such as temozolomide, carboplatin and
vincristine (35).
Initially PROS1 was thought to be the ligand for the
TAM receptors. However, Stitt et al have shown that PROS1
has a higher affinity for the Tyro-3 receptor and can transform NIH
3T3 cells in an autocrine manner (36). One of the important findings that
changed the perspective for PROS1 was that the anticoagulant factor
played an important role in activating Tyro-3 activity as its
expression was upregulated in cultured Schwann cells and astrocytes
following nerve injury (36). This
activation of intracellular signaling cascades by specific
cell-surface receptors would promote cell proliferation for tissue
repair and growth. PROS1 was found to be highly expressed in
high-grade prostate cancers suggesting that it has an important
role in the regulation of cancer cell survival (29). Knockdown of PROS1 by shRNA
was reported to significantly reduce the number of cancerous cells
in a time-dependent manner (37).
Indeed, this is in agreement with our findings where silencing of
PROS1 using siRNA significantly reduced cell viability of
GBM cells by >40%. This was also supported by the reduction of
Brdu proliferative signals.
PROS1 is also involved in the phagocytosis of
apoptotic cells in the immune, nervous, and reproductive systems
through interaction with Tyro-3 (38). During hypoxia or ischemia,
PROS1 protects neuron cells and inhibits apoptosis by
inhibiting Fas ligand (FasL) production and inhibiting
FasL-dependent caspase-8 activation within the extrinsic apoptotic
pathway (39). Wang et al
showed that Tyro-3 silencing affected several important
signaling pathways including P13K/AKT, Wnt/β-catenin,
ERK/MAPK, PAK/JNK, JAK/Stat and TGF-β
as well as the retinoic acid receptor (RAR)
activation (40). We showed that
silencing PROS1 led to a significant increase in apoptotic
signals and this result was validated using protein array.
Silencing of PROS1 caused significant activation of the
apoptotic pathways by upregulation of CD40, CD40L, Fas, FasL and
caspase-8 of the intrinsic pathway as well as BAD and SMAC of the
extrinsic pathway. Interestingly, it significantly reduced the
expression of the inhibitor of apoptosis protein (IAP), XIAP and
survivin. PROS1 silencing also led to the downregulation of
Tyro-3, Axl, Mertk and Gas6 gene
expressions, suggesting that the GBM cells might undergo apoptosis
through the TAM receptor interaction. However, there are some
limitations in terms of the number of protein markers available in
our protein array which hindered the identification of other
downstream apoptosis proteins involved.
Another study on castration-resistant prostate
cancer cells showed that the addition of human purified
PROS1 increased the migration of these cells (29). Furthermore, the high-throughput
wound healing screening on the epithelial cells revealed the
involvement of TAM receptors in cell migration (41). Our results are consistent as we
showed that PROS1 gene silencing delayed the wound enclosure
in GBM cells and significantly reduced the capability of cells to
migrate. This might be due to the involvement of the extracellular
domains of TAM receptors that contain adhesion molecule-like motifs
which controls cell-cell contacts and actin cytoskeleton regulation
(42). Our results showed that
silencing of PROS1 expression also led to a significant
reduction in cell invasion through the ECM in GBM cells. These
findings suggest that PROS1 may provide a survival advantage
for advanced stage cancer like prostate and GBM by controlling
cancer cell migration and invasion.
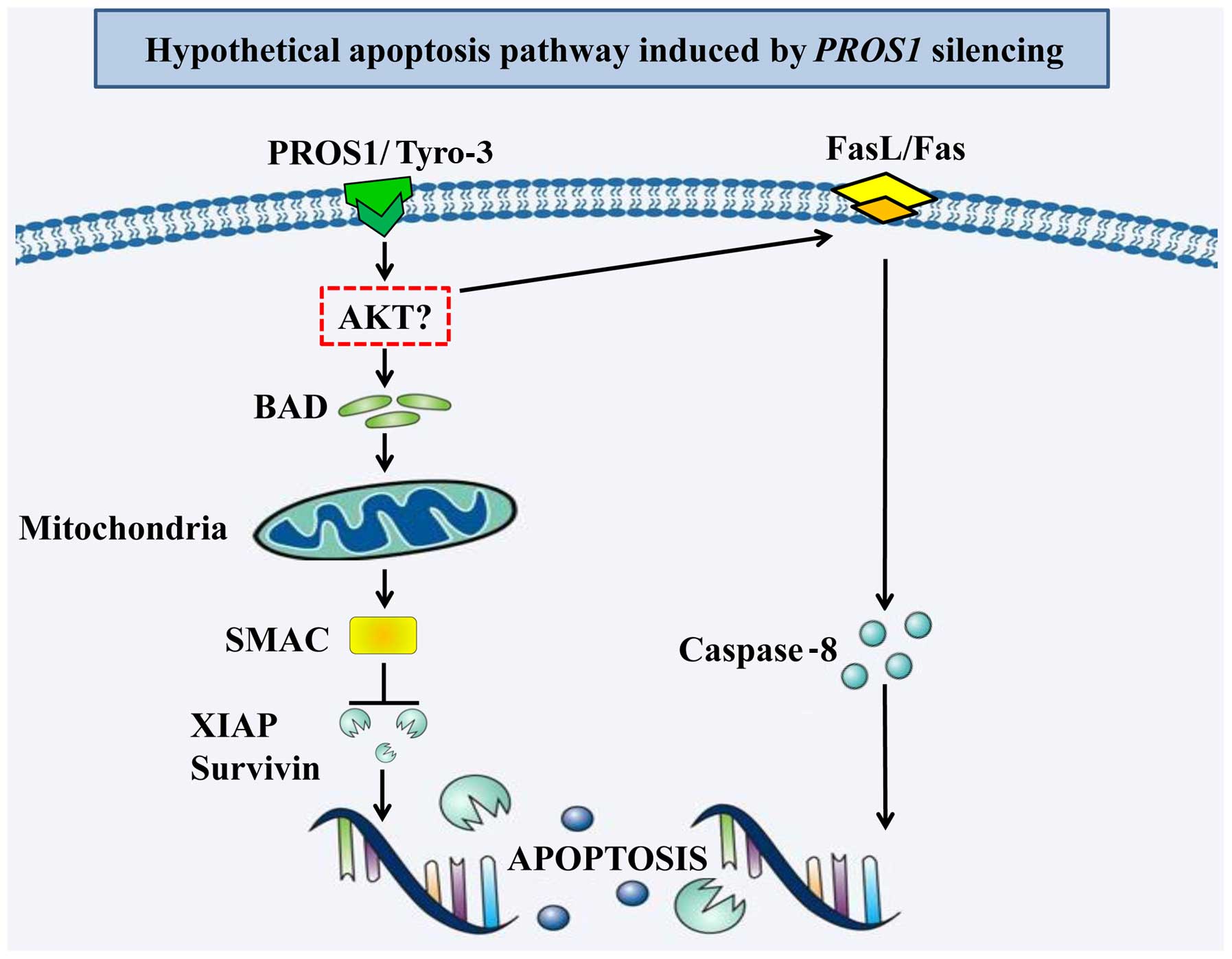
In summary, we showed that silencing PROS1
reduces survival, migration and invasion of GBM cells (as detailed
in Fig. 6). It also activates
apoptosis in GBM cells by activating the intrinsic and extrinsic
apoptotic pathways. Further validation using in vivo studies
are needed to enhance our understanding on the mechanistic role of
PROS1 in GBM cells. This will hopefully allow the
development of PROS1 gene therapy as a possible approach to
increase patient survival and improve the treatment of GBM
patients.
Acknowledgements
We would like to thank the Ministry of Education,
Malaysia for the funding. This study was funded by the Higher
Institution Centre of Excellence (HICoE) (grant no. JJ-008-2011),
Ministry of Education, Malaysia.
References
|
1
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davies AM, Weinberg U and Palti Y: Tumor
treating fields: A new frontier in cancer therapy. Ann N Y Acad
Sci. 1291:86–95. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parpura V, Heneka MT, Montana V, Oliet
SHR, Schousboe A, Haydon PG, Stout RF Jr, Spray DC, Reichenbach A,
Pannicke T, et al: Glial cells in (patho)physiology. J Neurochem.
121:4–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huse JT, Holland E and DeAngelis LM:
Glioblastoma: Molecular analysis and clinical implications. Annu
Rev Med. 64:59–70. 2013. View Article : Google Scholar
|
|
5
|
Kanu OO, Hughes B, Di C, Lin N, Fu J,
Bigner DD, Yan H and Adamson C: Glioblastoma multiforme
oncogenomics and signaling pathways. Clin Med Oncol. 3:39–52.
2009.PubMed/NCBI
|
|
6
|
Reardon DA, Conrad CA, Cloughesy T, Prados
MD, Friedman HS, Aldape KD, Mischel P, Xia J, DiLea C, Huang J, et
al: Phase I study of AEE788, a novel multitarget inhibitor of ErbB-
and VEGF-receptor-family tyrosine kinases, in recurrent
glioblastoma patients. Cancer Chemother Pharmacol. 69:1507–1518.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vitucci M, Karpinich NO, Bash RE, Werneke
AM, Schmid RS, White KK, McNeill RS, Huff B, Wang S, Van Dyke T, et
al: Cooperativity between MAPK and PI3K signaling activation is
required for glioblastoma pathogenesis. Neuro Oncol. 15:1317–1329.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akhavan D, Cloughesy TF and Mischel PS:
mTOR signaling in glioblastoma: Lessons learned from bench to
bedside. Neurooncol. 12:882–889. 2010.
|
|
9
|
Nakada M, Kita D, Watanabe T, Hayashi Y,
Teng L, Pyko IV and Hamada J: Aberrant signaling pathways in
glioma. Cancers (Basel). 3:3242–3278. 2011. View Article : Google Scholar
|
|
10
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang C, Moore LM, Li X, Yung WKA and
Zhang W: IDH1/2 mutations target a key hallmark of cancer by
deregulating cellular metabolism in glioma. Neuro Oncol.
15:1114–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pierscianek D, Kim YH, Motomura K,
Mittelbronn M, Paulus W, Brokinkel B, Keyvani K, Wrede K, Nakazato
Y, Tanaka Y, et al: MET gain in diffuse astrocytomas is associated
with poorer outcome. Brain Pathol. 23:13–18. 2013. View Article : Google Scholar
|
|
13
|
Eisele G and Weller M: Targeting apoptosis
pathways in glioblastoma. Cancer Lett. 332:335–345. 2013.
View Article : Google Scholar
|
|
14
|
Krakstad C and Chekenya M: Survival
signalling and apoptosis resistance in glioblastomas: Opportunities
for targeted therapeutics. Mol Cancer. 9:1352010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Panner A, Crane CA, Weng C, Feletti A,
Parsa AT and Pieper RO: A novel PTEN-dependent link to
ubiquitination controls FLIPS stability and TRAIL sensitivity in
glioblastoma multiforme. Cancer Res. 69:7911–7916. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ruano Y, Mollejo M, Camacho FI, Rodríguez
de Lope A, Fiaño C, Ribalta T, Martínez P, Hernández-Moneo JL and
Meléndez B: Identification of survival-related genes of the
phosphatidylinositol 3′-kinase signaling pathway in glioblastoma
multiforme. Cancer. 112:1575–1584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guvenc H, Pavlyukov MS, Joshi K, Kurt H,
Banasavadi-Siddegowda YK, Mao P, Hong C, Yamada R, Kwon CH, Bhasin
D, et al: Impairment of glioma stem cell survival and growth by a
novel inhibitor for Survivin-Ran protein complex. Clin Cancer Res.
19:631–642. 2013. View Article : Google Scholar
|
|
18
|
Catuogno S, Esposito CL, Quintavalle C,
Condorelli G, de Franciscis V and Cerchia L: Nucleic acids in human
glioma treatment: Innovative approaches and recent results. J
Signal Transduct. 2012:7351352012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jensen SA, Day ES, Ko CH, Hurley LA,
Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et
al: Spherical nucleic acid nanoparticle conjugates as an RNAi-based
therapy for glioblastoma. Sci Transl Med. 5:209ra1522013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zhang YF, Bryant J, Charles A,
Boado RJ and Pardridge WM: Intravenous RNA interference gene
therapy targeting the human epidermal growth factor receptor
prolongs survival in intracranial brain cancer. Clin Cancer Res.
10:3667–3677. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang XL, Xu R, Wu X, Gillespie D, Jensen R
and Lu ZR: Targeted systemic delivery of a therapeutic siRNA with a
multifunctional carrier controls tumor proliferation in mice. Mol
Pharm. 6:738–746. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bumcrot D, Manoharan M, Koteliansky V and
Sah DWY: RNAi therapeutics: A potential new class of pharmaceutical
drugs. Nat Chem Biol. 2:711–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rolle K, Nowak S, Wyszko E, Nowak M,
Zukiel R, Piestrzeniewicz R, Gawronska I, Barciszewska MZ and
Barciszewski J: Promising human brain tumors therapy with
interference RNA intervention (iRNAi). Cancer Biol Ther. 9:396–406.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zukiel R, Nowak S, Wyszko E, Rolle K,
Gawronska I, Barciszewska MZ and Barciszewski J: Suppression of
human brain tumor with interference RNA specific for tenascin-C.
Cancer Biol Ther. 5:1002–1007. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Dahlbäck B: The tale of protein S and
C4b-binding protein, a story of affection. Thromb Haemost.
98:90–96. 2007.PubMed/NCBI
|
|
28
|
Hafizi S and Dahlbäck B: Gas6 and protein
S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase
subfamily. FEBS J. 273:5231–5244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saraon P, Musrap N, Cretu D, Karagiannis
GS, Batruch I, Smith C, Drabovich AP, Trudel D, van der Kwast T,
Morrissey C, et al: Proteomic profiling of androgen-independent
prostate cancer cell lines reveals a role for protein S during the
development of high grade and castration-resistant prostate cancer.
J Biol Chem. 287:34019–34031. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suleiman L, Négrier C and Boukerche H:
Protein S: A multi-functional anticoagulant vitamin K-dependent
protein at the crossroads of coagulation, inflammation,
angiogenesis, and cancer. Crit Rev Oncol Hematol. 88:637–654. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lemke G: Biology of the TAM receptors.
Cold Spring Harb Perspect Biol. 5:a0090762013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lemke G and Rothlin CV: Immunobiology of
the TAM receptors. Nat Rev Immunol. 8:327–336. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Linger RMA, Keating AK, Earp HS and Graham
DK: Taking aim at Mer and Axl receptor tyrosine kinases as novel
therapeutic targets in solid tumors. Expert Opin Ther Targets.
14:1073–1090. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wimmel A, Rohner I, Ramaswamy A, Heidtmann
HH, Seitz R, Kraus M and Schuermann M: Synthesis and secretion of
the anticoagulant protein S and coexpression of the Tyro3 receptor
in human lung carcinoma cells. Cancer. 86:43–49. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keating AK, Kim GK, Jones AE, Donson AM,
Ware K, Mulcahy JM, Salzberg DB, Foreman NK, Liang X, Thorburn A,
et al: Inhibition of Mer and Axl receptor tyrosine kinases in
astrocytoma cells leads to increased apoptosis and improved
chemosensitivity. Mol Cancer Ther. 9:1298–1307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stitt TN, Conn G, Gore M, Lai C, Bruno J,
Radziejewski C, Mattsson K, Fisher J, Gies DR, Jones PF, et al: The
anticoagulation factor protein S and its relative, Gas6, are
ligands for the Tyro 3/Axl family of receptor tyrosine kinases.
Cell. 80:661–670. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saraon P, Jarvi K and Diamandis EP:
High-throughput proteomic analysis identifies protein s as a
modulator of high grade and castrate-resistant prostate cancer.
Cancer Res (AACR Annual Meeting abstracts). 72(8 Suppl):
LB-2932012.
|
|
38
|
Lemke G and Burstyn-Cohen T: TAM receptors
and the clearance of apoptotic cells. Ann N Y Acad Sci. 1209:23–29.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo H, Barrett TM, Zhong Z, Fernández JA,
Griffin JH, Freeman RS and Zlokovic BV: Protein S blocks the
extrinsic apoptotic cascade in tissue plasminogen
activator/N-methyl D-aspartate-treated neurons via Tyro3-Akt-FKHRL1
signaling pathway. Mol Neurodegener. 6:132011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Moncayo G, Morin P Jr, Xue G,
Grzmil M, Lino MM, Clément-Schatlo V, Frank S, Merlo A and Hemmings
BA: Mer receptor tyrosine kinase promotes invasion and survival in
glioblastoma multiforme. Oncogene. 32:872–882. 2013. View Article : Google Scholar
|
|
41
|
Simpson KJ, Selfors LM, Bui J, Reynolds A,
Leake D, Khvorova A and Brugge JS: Identification of genes that
regulate epithelial cell migration using an siRNA screening
approach. Nat Cell Biol. 10:1027–1038. 2008. View Article : Google Scholar
|
|
42
|
Vajkoczy P, Knyazev P, Kunkel A, Capelle
HH, Behrndt S, von Tengg-Kobligk H, Kiessling F, Eichelsbacher U,
Essig M, Read TA, et al: Dominant-negative inhibition of the Axl
receptor tyrosine kinase suppresses brain tumor cell growth and
invasion and prolongs survival. Proc Natl Acad Sci USA.
103:5799–5804. 2006. View Article : Google Scholar : PubMed/NCBI
|