Introduction
Hepatocellular carcinoma (HCC), the most frequent
primary liver cancer, and the third leading cause of cancer-related
death in the world (1). Although
the treatments have improved in recent years, metastasis and
recurrence of HCC compromise the efficiency of new therapies, and
the survival of HCC patients remain dismal (2). Better understanding of the metastasis
mechanism is required.
MicroRNAs (miRNAs), small non-coding RNAs,
post-transcriptionally regulate protein expression levels via
directly binding to mRNAs of potentially hundreds of genes at the
3′-untranslated regions (3′-UTR) (3). Dysregulated miRNAs contribute to
cancer initiation and progression by acting as proto-oncogenes or
tumor suppressor genes (4).
miR-345 were shown highly expressed in malignant mesothelioma
compared with normal samples (5).
Furthermore, miR-345 expression was increased and might play a
critical role in malignant transformation of oral carcinoma
(6). miR-345 was implicated in
cisplatin resistance of MCF-7 by targeting multidrug
resistance-associated protein 1 (MRP1) (7). In colorectal cancer, underexpression
of miR-345 conferred to lymph node metastasis and worse
histological type, and its restoration inhibited cancer cell
proliferation and invasion (8,9).
Moreover, miR-345 expression was downregulated in non-small cell
lung cancers (NSCLC) and its low expression was correlated with
malignant clinical parameters and poor prognosis (10). Loss of miR-345 was reported to
confer apoptosis resistance to pancreatic cancer (PC) cells
(11), and miR-345 inhibited
prostate cancer cell proliferation and mobility by suppressing
Smad1 (12). Rare studies reported
the correlation between miR-345 and HCC. Shiu et al reported
that hepatitis C virus (HCV) core protein upregulated the
expression of miR-345, which inhibited curcumin-induced apoptosis
by targeting p21 in Huh7 cells (13), and Jiang et al showed that
HCC patients with good survival rates had high miR-345 expression
level compared with that in cases with poor survival rates
(14). Thus, it is worth
investigating the biological role of miR-345 and its underlying
mechanisms in HCC.
This study showed that miR-345 underexpression was
observed in HCC tissues and cells. We also illustrated that loss of
miR-345 promoted HCC cell migration and invasion, and resulted in
epithelial-mesenchymal-transition (EMT) progression probably by
targeting interferon regulatory factor 1 (IRF1)-mediated
mTOR/STAT3/AKT signaling in vitro. Thus, this work supported
the first evidence that miR-345 was recognized as a potential
therapeutic target for HCC.
Materials and methods
Cell culture and transfection
HCC-derived cell lines (HepG2, SMMC-7721, MHCC97L,
MHCC97H and HCCLM3) and a normal hepatocyte cell line (LO2) were
purchased from the Cell Bank of Shanghai Institute of Cell Biology,
Chinese Academy of Medical Science (Shanghai, China). Cell lines
were cultured in DMEM with 10% fetal bovine serum (Gibco, Grand
Island, NY, USA) with antibiotics (Sigma-Aldrich, St. Louis, MO,
USA) at 37°C in 5% CO2.
miR-345 mimic (HmiR0210-MR04), miR-345 inhibitor
(HmiR-AN0437-AM04), IRF1 overexpression plasmid (pcDNA3.1-IRF1) and
corresponding negative control vectors (CmiR0001-MR04;
CmiR-AN0001-AM04) were designed and synthesized by GeneCopoeia
(Guangzhou, China). Cell transfection was performed by using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
supplier's protocol.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total RNA was extracted from the cells and tissues
with TRIzol reagent (Invitrogen). PrimeScript RT Master Mix Perfect
Real-time (Takara, Shiga, Japan) was used to assess IRF1, while
Poly-A polymerase based First-Strand Synthesis kit (Takara) was
used for miR-345 by polyadenylating the total RNA. After reverse
transcription, qRT-PCR was performed by using SYBR Premix Ex Taq II
(Takara). IRF1 was normalized to GAPDH, while U6 was used as
miR-345 endogenous control. The primers used for miR-345, U6, IRF1
and GAPDH were designed and synthesized by Sangon Biotech
(Shanghai, China).
Western blotting
Antibodies for the western blot were as follows:
IRF1 (Cell Signaling, Danvers, MA, USA), mTOR (Cell Signaling),
p-mTOR (Ser2448; Cell Signaling), STAT3 (Cell Signaling), p-STAT3
(Tyr705; Cell Signaling), AKT (Cell Signaling), p-AKT (Ser473; Cell
Signaling), E-cadherin (Abcam, Cambridge, MA, USA), N-cadherin
(Abcam), vimentin (Abcam) and GAPDH (G8140, US Biological,
Swampscott, MA, USA). The rabbit/mouse secondary antibodies were
obtained from Cell Signaling. Two days post transfection, total
protein was extracted, following by quantification with BCA protein
assay kit (Pierce, Bonn, Germany). Proteins were separated by 10%
SDS-polyacrylamide gels and subsequently transferred to PVDF
membranes. After blocking, blots were probed with specific
antibodies.
Wound healing assay
HCC cells transduced with corresponding vectors were
seeded in 6-well plates to form single confluent cell layer. The
wound were made with 100 µl tips in the confluent cell
layer. At 0 and 24 h after would scratching, the width of wound was
photographed with a phase-contrast microscope.
Transwell invasion assay
We determined the cell invasion capacities by using
Transwell chambers of pore size 8 µm (Corning Costar,
Cambridge, MA, USA). At 24 h after transfection, 5×104
cells were seeded in the 1:9 diluted Matrigel-coated (BD
Biosciences, Franklin Lakes, NJ, USA) upper chamber with 250
µl serum-free DMEM medium, while 700 µl DMEM medium
with 10% serum were added in the lower chamber. After 24 h, we
fixed cells with paraformaldehyde and the cells in upper chamber
were removed. Cells in lower chamber were then stained using 0.1%
crystal violet solution and photographed.
Dual-luciferase reporter assay
Wild-type (wt) or mutant (mt) 3′-UTR of IRF1 was
amplified and cloned into pmiR-RB-REPORT™ Luciferase. Luciferase
reporter containing the potential binding sequence of 3′-UTR of
IRF1 was co-transfected with miR-345 mimic and corresponding
negative control vector in HCCLM3 cells in 96-well plate. Two days
later, dual-luciferase reporter assay system (Promega, Madison, WI,
USA) was used to measure the alteration of luciferase. Firefly
luciferase activity was normalized to renilla luciferase
activity.
Immunofluorescence (IF)
HCC cells were seeded on chamber slides and were
fixed with 4% paraformaldehyde at room temperature (RT) for 10 min.
Then, primary antibodies against E-cadherin (Abcam) or vimentin
(Abcam) were subjected to cell incubation at 4°C overnight. Then,
the slides were incubated with matched secondary antibodies
(Invitrogen) for 1 h at RT. The nuclear of HCC cells were stained
with DAPI (Sigma) for 10 min at RT. LSM 5 Pascal Laser Scanning
Microscope was used for capturing fluorescence confocal images
(Zeiss Germany, Oberkochen, Germany).
Immunohistochemistry (IHC)
The tumor tissues that were previously
formalin-fixed and paraffin-embedded were sliced into 4 µm
sections, and underwent deparaffination and then rehydration.
Antigen retrieval, suppression of endogenous peroxidase activity
and 10% skim milk blocking were performed before primary antibody
incubation. IRF1 (Cell Signaling) antibody was used as primary
antibody overnight at 4°C. The following day, the slides were
incubated with peroxidase conjugated secondary antibody (ZSGB-BIO,
Beijing, China) for 90 min, and a peroxidase-labeled polymer, DAB
solution was used for signal development for 5 min. The sections
were counterstained with hematoxylin followed by dehydrating and
mounting.
In vivo liver metastasis assay
HCCLM3 cells (6×106) that were
transfected with miR-345 mimic or control vector were intravenously
injected into nude mice. Five weeks after injection, all mice were
euthanized, and the livers were sectioned and stained by H&E to
check if hepatic metastatic foci formed. The protocol for these
animal experiments were approved by the Ethics Review Committee of
Zhengzhou University.
Patients and tissue samples
Sixty-five pairs of human HCC and matched
tumor-adjacent tissues were obtained from HCC patients undergoing
hepatectomy at Department of Hepatobiliary Surgery, People's
Hospital of Zhengzhou University. All tissue samples were stored in
liquid nitrogen or fixed with 10% formalin after surgical excision
immediately. The patients did not receive any anti-tumor therapy
before surgical excision. Each patient was well informed and signed
the informed consent. The study was approved by the ethics
committee of Zhengzhou University in accordance with the
Declaration of Helsinki.
Statistical analysis
Data are presented as mean ± SEM and analyzed by
GraphPad Prism5 software (GraphPad Software, Inc., San Diego, CA,
USA). Chi-squared test was employed to explore the association
between two variables. The Student's t-test and ANOVA were carried
out to analyze continuous variable. Correlation curve was
constructed and differences among groups were calculated using the
Spearman's rank correlation analysis. P-value <0.05 was
considered to indicate a statistically significant difference.
Results
Loss of miR-345 promotes invasion and
migration of HCC cell lines in vitro
The mRNA expression levels of miR-345 in 65 cases of
HCC and matched tumor-adjacent specimens were evaluated by qRT-PCR.
Our data illustrated that the expression of miR-345 was
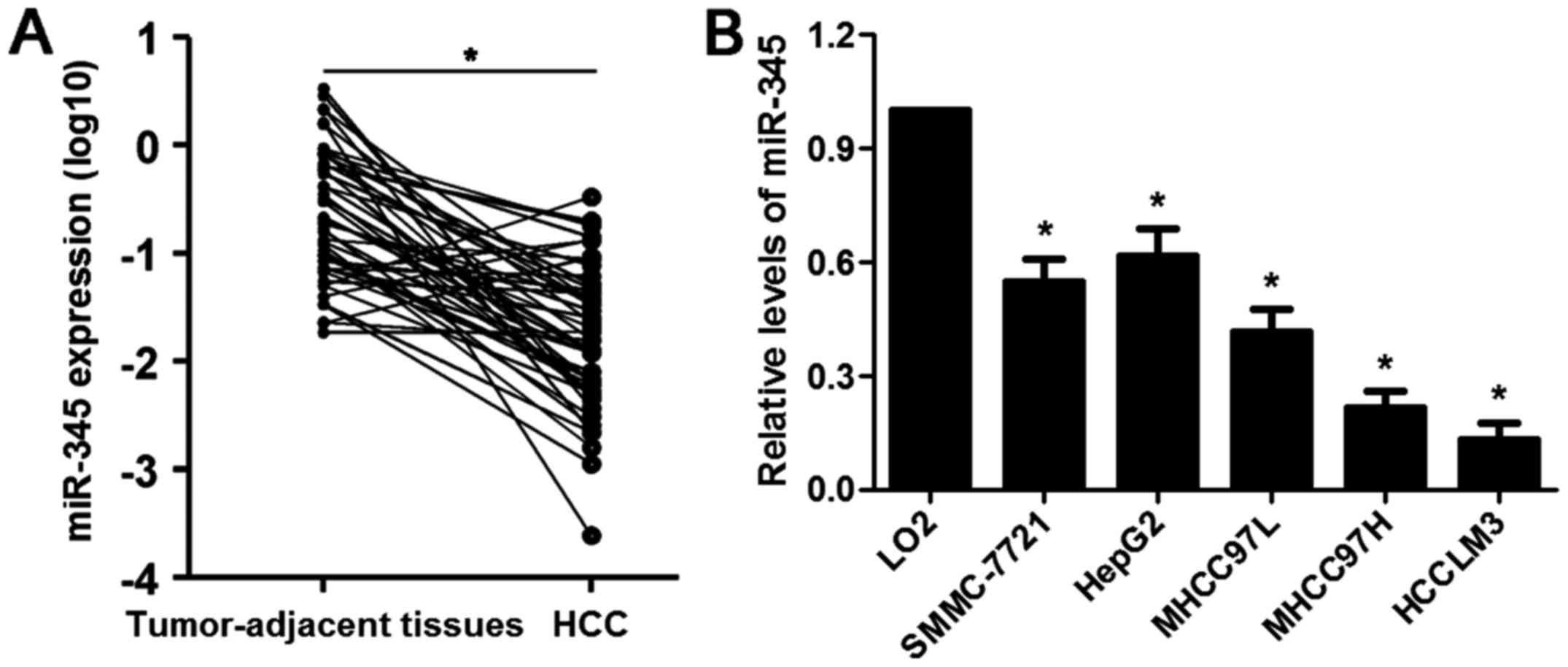
significantly downregulated in HCC cases (P<0.05, Fig. 1A). Underexpression of miR-345 was
found in HCC cell lines (SMMC-7721, HepG2, MHCC97L, MHCC97H and
HCCLM3) compared to LO2 cells (P<0.05, respectively, Fig. 1B). The median of all HCC samples
was considered as a cut-off value and the level of miR-345 was
divided into low (≤ median, n=33) or high (> median, n=32)
group. Clinical correlation analysis disclosed that the low
expression of miR-345 was associated with multiple tumor nodes,
venous infiltration and advanced tumor-node-metastasis (TNM) stage
(P<0.05, respectively, Table
I). Next, we tested the effects of miR-345 on invasion and
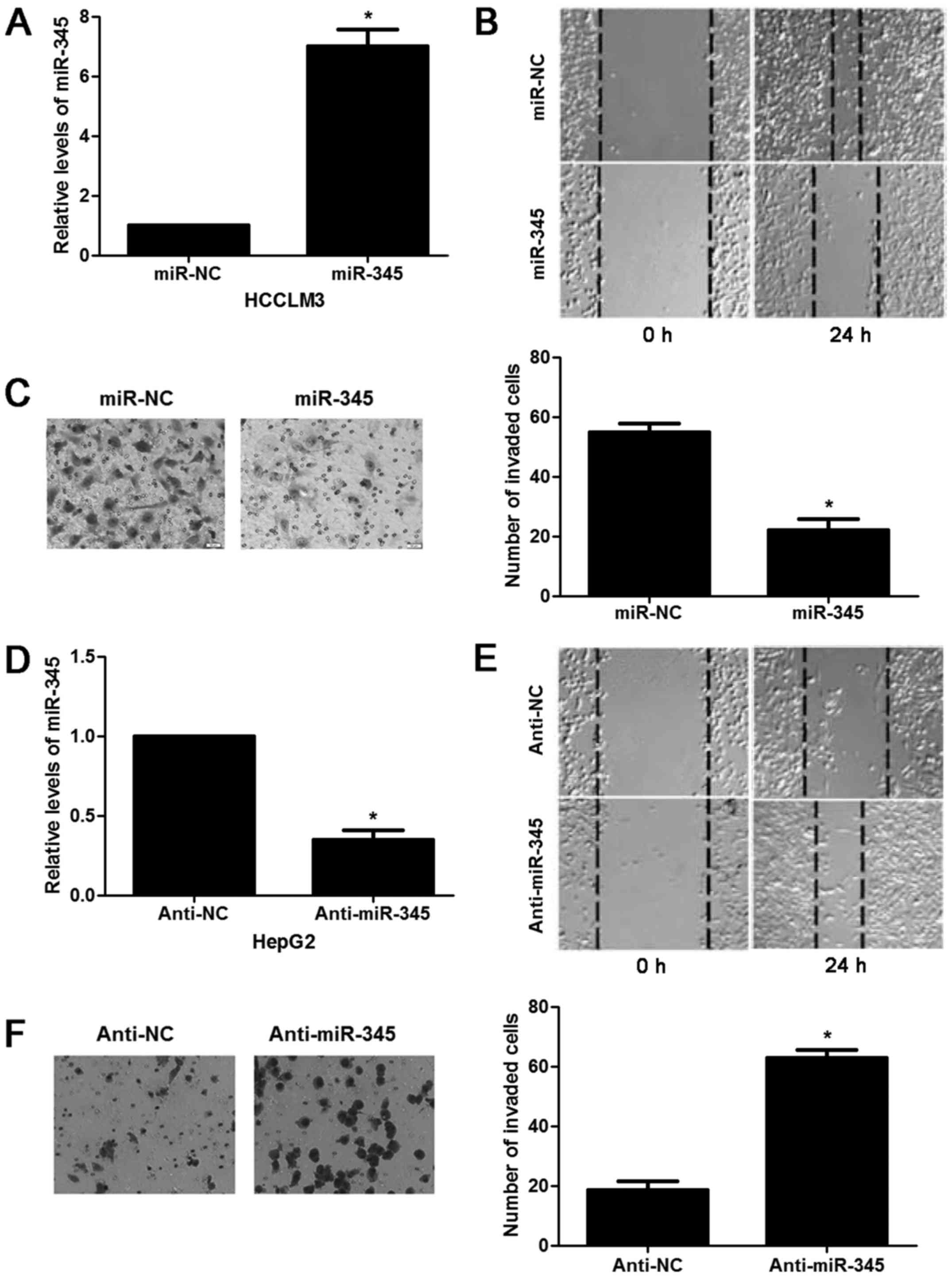
migration capacity of HCC in vitro. miR-345 mimic resulted
in significant increase of miR-345 expression in HCCLM3 cells
(P<0.05, Fig. 2A). miR-345
overexpression inhibited migration and invasion ability of HCCLM3
cells (P<0.05, respectively, Fig.
2B and C). While the opposite results were observed after
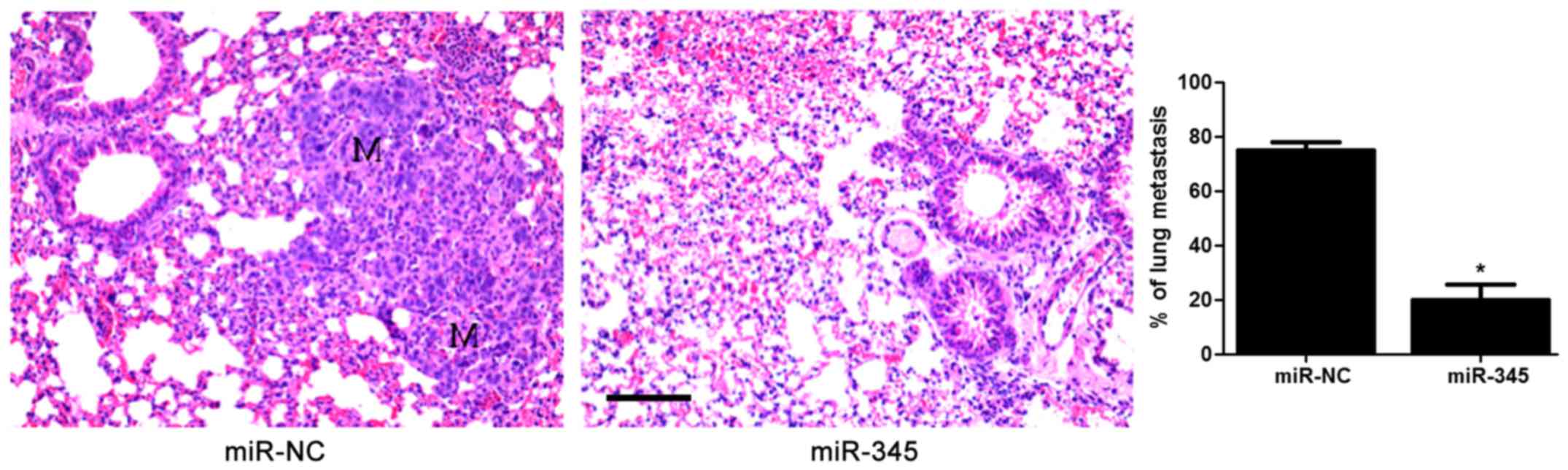
anti-miR-345 transfection in HepG2 cells (P<0.05, Fig. 2D–F). In the lung metastasis nude
mouse model, miR-345 overexpression notably reduced the lung
metastases of HCC cells in vivo (P<0.05, Fig. 3). These data uncovered an
anti-invasive role of miR-345 in HCC cell lines.
 | Table IClinicopathological correlation of
miR-345 expression in HCC. |
Table I
Clinicopathological correlation of
miR-345 expression in HCC.
| Clinicopathological
features | Cases | miR-345 expression
| P-value |
|---|
| Low (n=33) | High (n=32) |
|---|
| Age (y) | | | | 0.515 |
| ≤50 | 27 | 15 | 12 | |
| >50 | 38 | 18 | 20 | |
| Gender | | | | 0.518 |
| Male | 49 | 26 | 23 | |
| Female | 16 | 7 | 9 | |
| HBsAg | | | | 0.158 |
| Absent | 21 | 8 | 13 | |
| Present | 44 | 25 | 19 | |
| Serum AFP level
(ng/ml) | | | | 0.675 |
| ≤20 | 24 | 13 | 11 | |
| >20 | 41 | 20 | 21 | |
| Tumor size (cm) | | | | 0.170 |
| ≤5 | 25 | 10 | 15 | |
| >5 | 40 | 23 | 17 | |
| No. of tumor
nodules | | | | 0.019a |
| 1 | 51 | 22 | 29 | |
| ≥2 | 14 | 11 | 3 | |
| Cirrhosis | | | | 0.173 |
| Absent | 27 | 11 | 16 | |
| Present | 38 | 22 | 16 | |
| Venous
infiltration | | | | 0.014a |
| Absent | 48 | 20 | 28 | |
| Present | 17 | 13 | 4 | |
| Edmondson-Steiner
grading | | | | 0.098 |
| I+II | 49 | 22 | 27 | |
| III+IV | 16 | 11 | 5 | |
| TNM tumor
stage | | | | 0.010a |
| I+II | 50 | 21 | 29 | |
| III+IV | 15 | 12 | 3 | |
Underexpression of miR-345 promotes the
EMT process of HCC cells
As miR-345 harboured anti-invasive ability in HCC,
we then explored the underlying relationship between miR-345 and
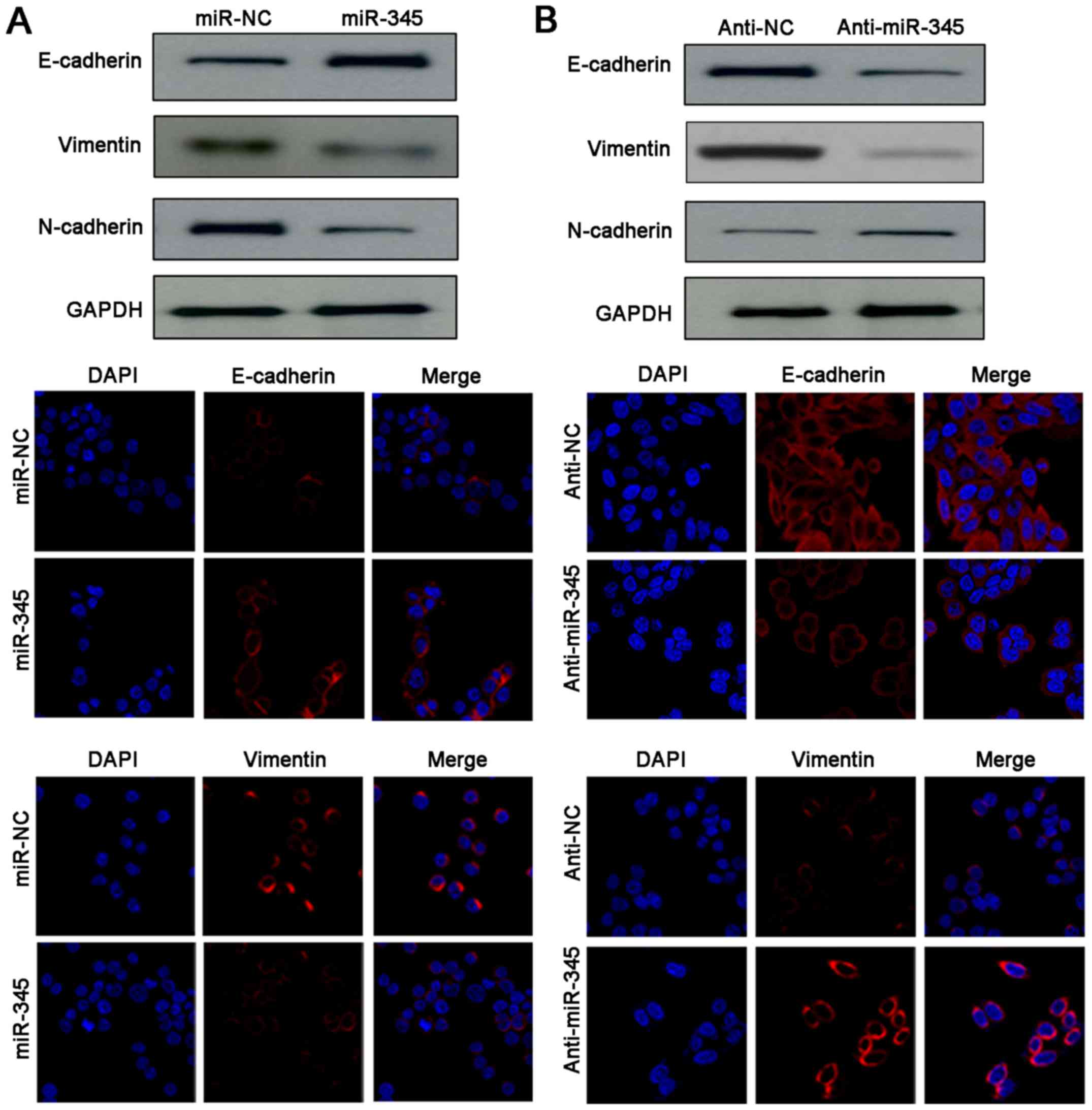
EMT. Noteworthy, western blotting and IF results indicated
remarkable gain of epithelial marker (E-cadherin) and obvious loss
of mesenchymal markers (N-cadherin, vimentin) were detected after
miR-345 restoration in HCCLM3 cells (Fig. 4A). Conversely, anti-miR-345 reduced
the expression of E-cadherin as well as upregulated the levels of
N-cadherin and vimentin in HepG2 cells (Fig. 4B). Therefore, our data demonstrated
that loss of miR-345 enhanced invasion and migration abilities of
HCC cells by promoting EMT process.
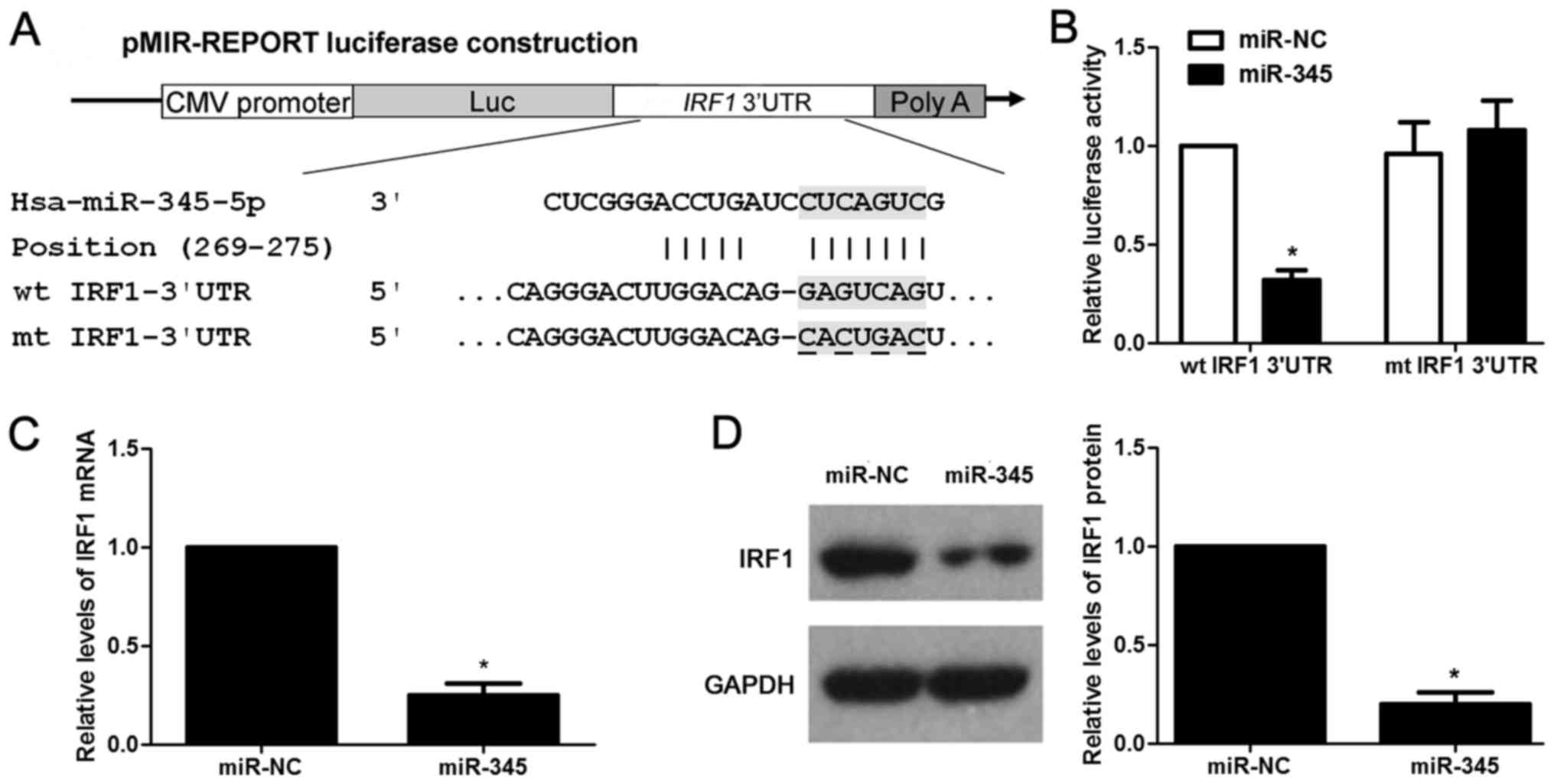
IRF1 is the direct target for miR-345 in
HCC
According to the prediction of bioinformatic
software (Targetscan, miRanda and PicTar), IRF1 is considered as
one of the candidates with which miR-345 could bind directly
(Fig. 5A). We then evaluated the
ability of miR-345 to inhibit the IRF1 expression via
dual-luciferase reporter assay. miR-345 overexpression obviously
inhibits luciferase activity of the reporter with wt 3′-UTR
sequence of IRF1 (P<0.05, Fig.
5B), miR-345 restoration showed no significant effect on
luciferase activity of mt 3′UTR of IRF1 (Fig. 5B). We next tested whether miR-345
could participate in the modulation of IRF1. The data of
gain-of-function experiments in HCCLM3 cells revealed that the
expression of IRF1 could be significantly downregulated by miR-345
overexpression at both the mRNA and protein level (P<0.05,
respectively, Fig. 5C and D).
Hence, miR-345 downregulated the expression of IRF1 in HCC cells by
binding to its 3′-UTR sequence directly according to the data in
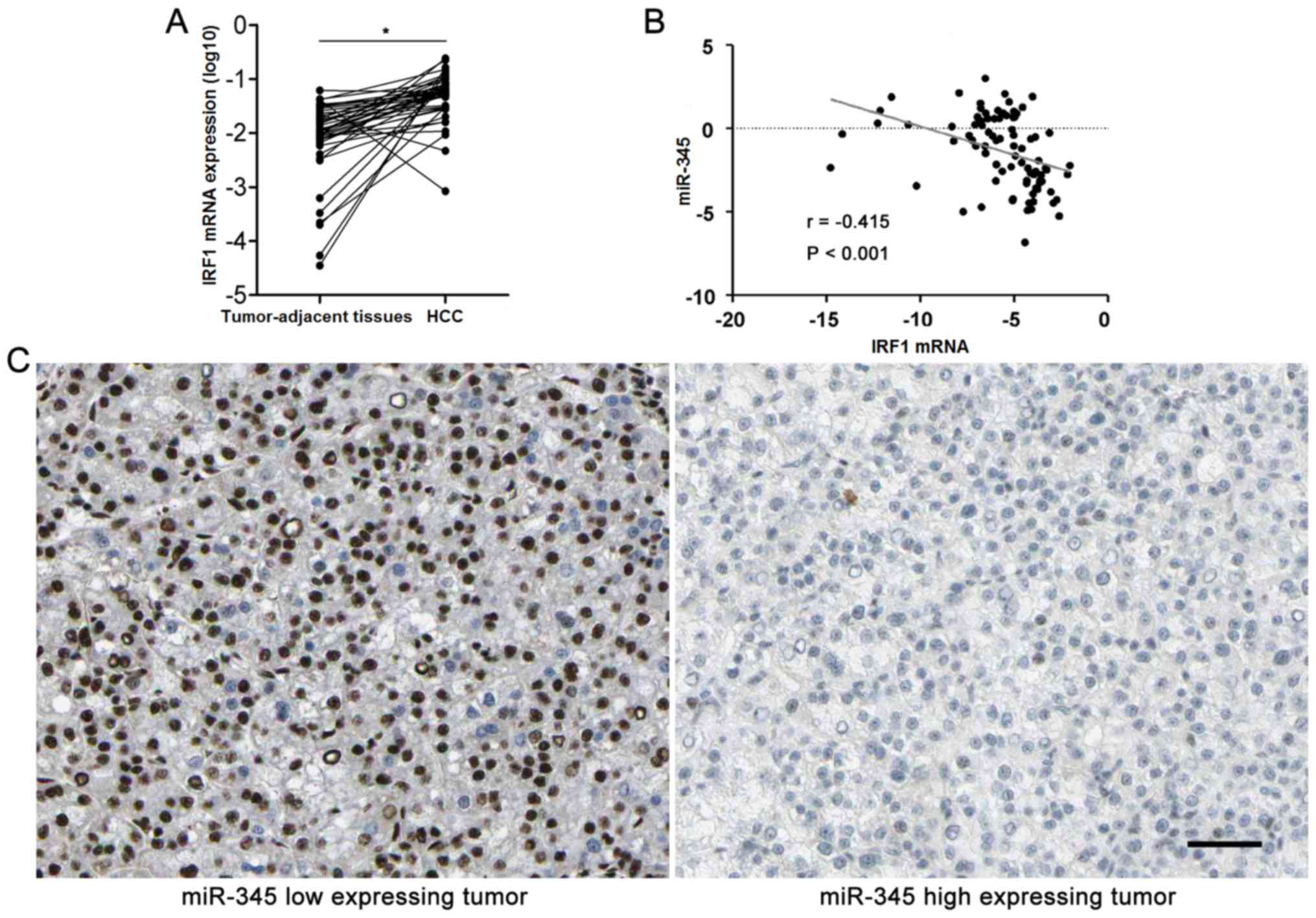
context. Furthermore, HCC cases were subjected to qRT-PCR for IRF2
mRNA expression. Quantitative data disclosed that IRF2 mRNA was
overexpressed in HCC specimens compared to matched tumor-adjacent
tissues (P<0.05, Fig. 6A). The
expression levels of miR-345 expression was negatively correlated
with the levels of IRF1 mRNA in HCC specimens analyzed by
Spearman's rank correlation analysis (r= −0.415, P<0.001,
Fig. 6B). Representative IHC
sections showed that miR-345 low expressing HCC showed strong
staining of IRF1, while weak signal was detected in miR-345 high
expressing cases (Fig. 6C).
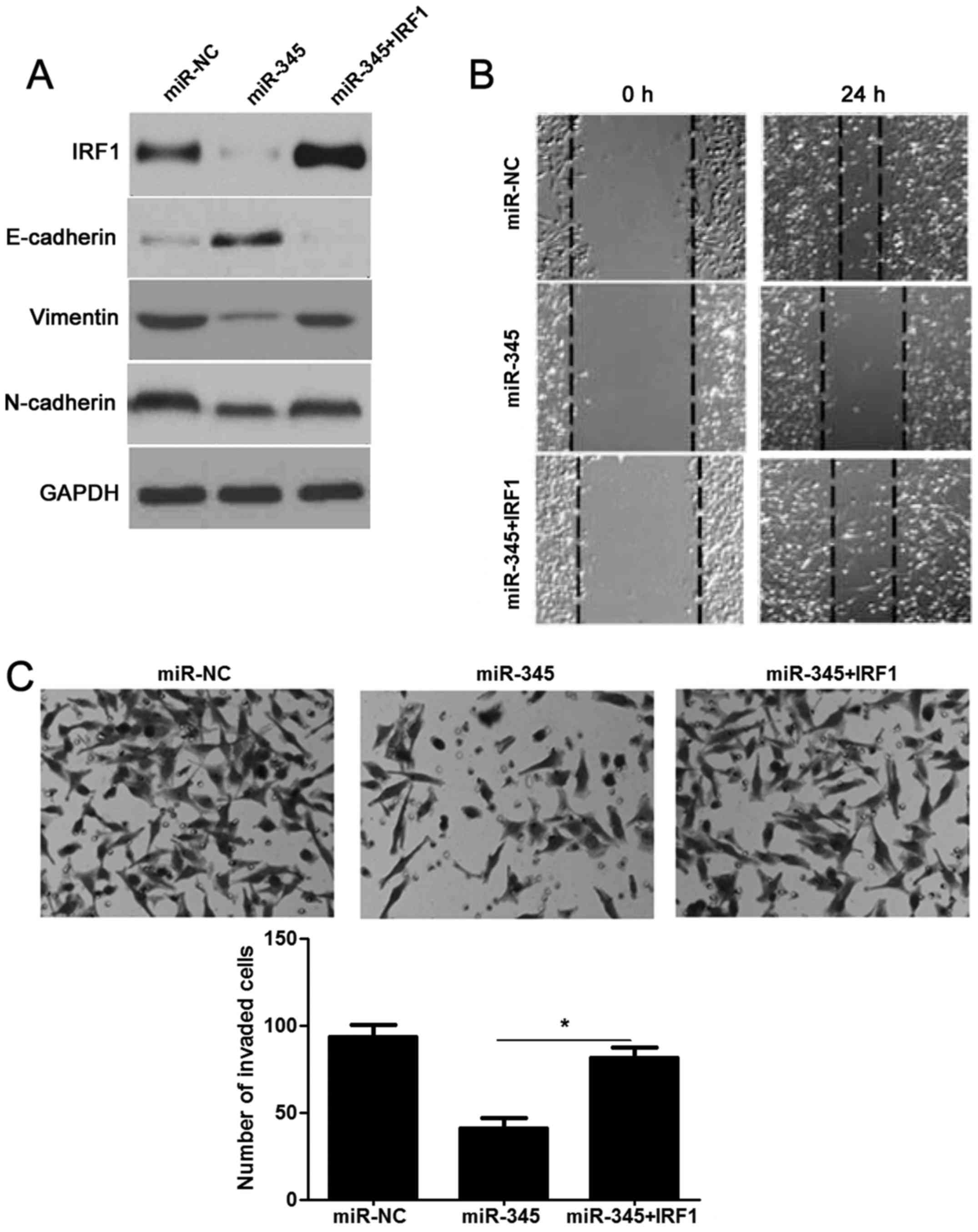
Re-expression of IRF1 reverses
miR-345-inhibited EMT process in HCC
We next detected whether re-expression of IRF1 would
rescue the effects of miR-345 in HCC. As expected, restoration of
IRF1 rescued the prohibited effect of miR-345 on IRF1, as well as
EMT-related biomarkers (E-cadherin, N-cadherin and vimentin)
(Fig. 7A). The migration and
invasion ability had also been measured and as shown in Fig. 7B and C: decreased migratory and
invasive ability caused by miR-345 was sequentially increased by
IRF1 overexpression in HCCLM3 cells (P<0.05, respectively).
Hence, these results further confirmed that loss of miR-345
promoted EMT and cell mobility of HCC by targeting IRF1.
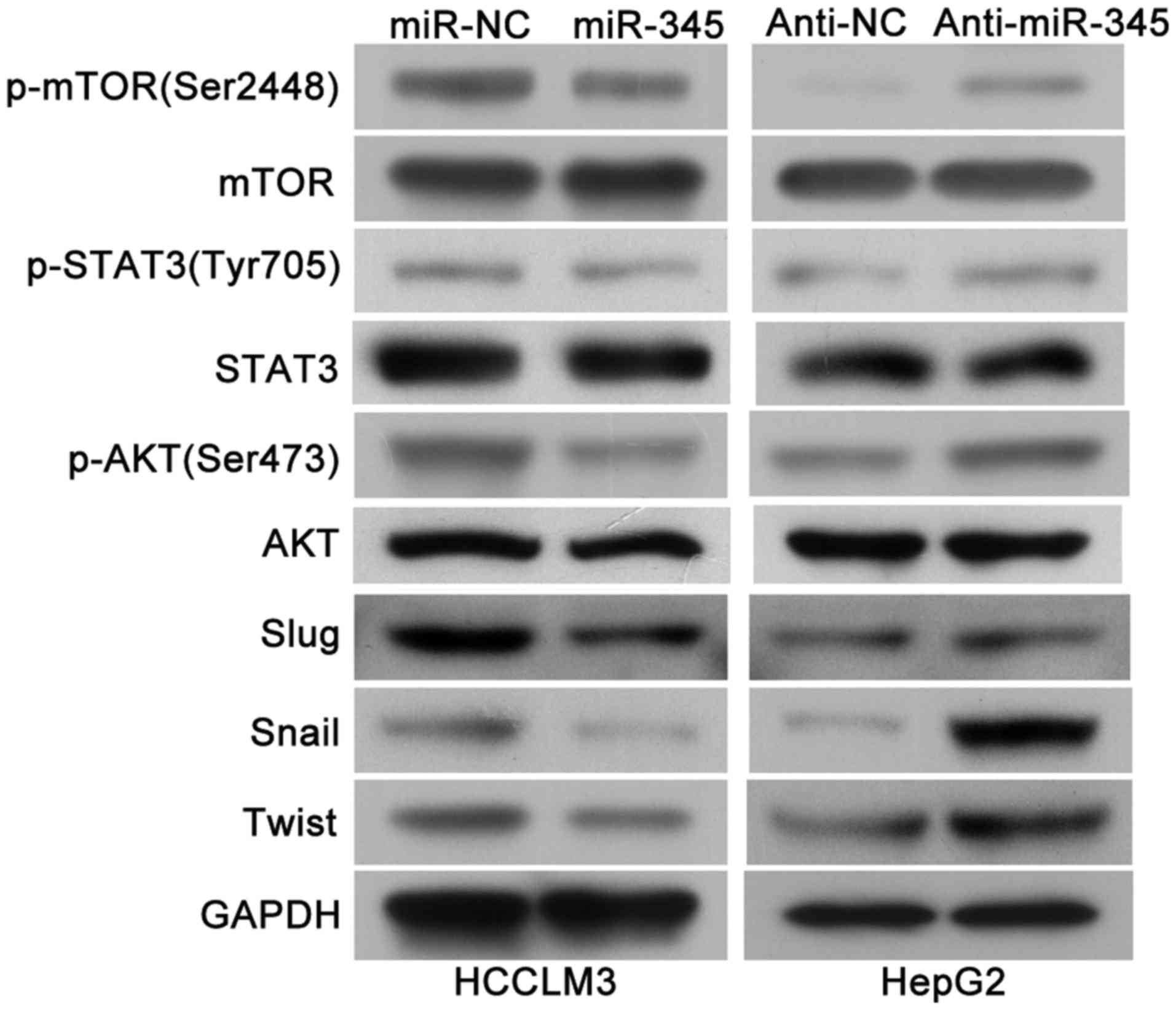
mTOR/STAT3/AKT signaling axis may
involved in the miR-345/IRF1 mediated EMT
As previous research has identified that IRF1
activated mTOR/STAT3/AKT signaling and subsequently upregulated
Slug, Snail and Twist, which could promote HCC cell migration,
invasion as well as EMT process (15) and we continued to explore whether
mTOR/STAT3/AKT signaling axis is involved in the miR-345/IRF1
mediated EMT process. Of note, we found that the levels of
phosphoryated mTOR, STAT3 and AKT as well as Slug, Snail and Twist
were remarkably decreased in the miR-345 overexpression group while
miR-345 knockdown lead to increased activity of mTOR/STAT3/AKT
signaling pathway and its downstream targets (Fig. 8). Therefore, these results
indicated that mTOR/STAT3/AKT signaling axis might be involved in
the miR-345/IRF1 mediated EMT process in HCC.
Discussion
Recent research has demonstrated that miRNAs are
involved in tumor initiation and progression as either oncogenes or
tumor suppressors by negatively regulating downstream targets
(16–18). Therefore, identifying the potential
valuable miRNAs provides novel insight for the diagnosis and
treatment of patients with HCC (3,16).
Herein, our data indicated that underexpressed miR-345 was common
in HCC tissues. In addition, the levels of miR-345 were intensively
reduced in HCC cell lines compared to a normal hepatocyte cell line
(LO2). HCC patients with multiple tumor nodes, venous infiltration
and advanced TNM stage showed significantly higher levels of
miR-345, which was consistent with poor prognosis prediction as
reported by Jiang et al (14). Although great efforts have been put
into the study of miR-345 on tumor cell growth, apoptosis and
metastasis (7,8,10–13),
the roles of miR-345 in the modulation of EMT process remain
largely unelucidated in HCC. In our recent study, gain- and
loss-of-function methods were used to determine the functional
roles of miR-345 in EMT. After restoration of miR-345, up regulated
E-cadherin (epithelial marker), suppressed N-cadherin and vimentin
(mesenchymal marker) were detected, accompanying with inhibited
cell invasion and migration ability in HCCLM3 cells. Contrary data
were obtained with anti-miR-345 treatment in HepG2 cells. These
results demonstrated that miR-345 served as an tumor suppressor by
suppressing EMT in HCC.
The role of IRF1 in HCC is still controversial
according to previous studies. Kröger et al reported that
the tumor growth of HCC was inhibited by the means of enhanced
antitumor growth effect and immune cell recognization (19). Furthermore, IRF1 signaling pathway
was implicated in interferon-gamma (IFN-γ) induced autophagy in
Huh7 cells (20). While, recent
study indicated that suppression of IRF1 was involved in miR-130b
induced inhibition of HCC cell migration and invasion (15). In this study, we found IRF1 is a
direct target of miR-345, re-expression of IRF1 rescued the miR-345
inhibited EMT process, these results demonstrated miR-345 could
inhibit the EMT process by targeting IRF1. Moriyama et al
reported that underexpression levels of IRF1 mRNA were observed in
15 of 32 HCC cases compared to matched tumor-adjacent tissues
(21). While our data revealed
that IRF1 mRNA was overexpressed in HCC specimens. We suggest that
the case number and etiology may be the causes of difference. HCV
core protein upregulated the expression of miR-345 in HCC cells
(13), while HBV infection is the
main problem in China (22).
A previous study showed that IRF1 functions as an
oncogenic factor that upregulated p-mTOR, p-STAT3 and p-AKT as well
as regulators of EMT including Slug, Snail and Twist, which
facilitate HCC cell migration and invasion (15). Hence, we then investigated the
effect of IRF1 restoration on the mTOR/STAT3/AKT signaling pathway
in order to elucidate the potential mechanism of IRF1 mediated EMT.
As expected, phosphorated AKT, STAT3 and AKT as well as EMT
regulators including Slug, Snail and Twist were downregulated in
miR-345 overexpressing cells and they were upregulated in the
miR-345 knockdown group. These results indicated that
mTOR/STAT3/AKT signaling pathway may be involved in the
miR-345/IRF1 mediated EMT process. However, further investigations
are needed to make a firm conclusion.
To conclude, the underexpression of miR-345 creates
a milieu of EMT facilitation that plays a role in HCC progression.
A mechanism by which underexpressed miR-345 promotes the EMT by
targeting IRF1 plays an important role in this process. This
finding will improve understanding of EMT progression mechanism and
provide novel targets for the molecular treatment of HCC.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81302121).
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guled M, Lahti L, Lindholm PM, Salmenkivi
K, Bagwan I, Nicholson AG and Knuutila S: CDKN2A, NF2, and JUN are
dysregulated among other genes by miRNAs in malignant mesothelioma
- A miRNA microarray analysis. Genes Chromosomes Cancer.
48:615–623. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cervigne NK, Reis PP, Machado J, Sadikovic
B, Bradley G, Galloni NN, Pintilie M, Jurisica I, Perez-Ordonez B,
Gilbert R, et al: Identification of a microRNA signature associated
with progression of leukoplakia to oral carcinoma. Hum Mol Genet.
18:4818–4829. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang JT, Wang JL, Du W, Hong J, Zhao SL,
Wang YC, Xiong H, Chen HM and Fang JY: MicroRNA 345, a
methylation-sensitive microRNA is involved in cell proliferation
and invasion in human colorectal cancer. Carcinogenesis.
32:1207–1215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schou JV, Rossi S, Jensen BV, Nielsen DL,
Pfeiffer P, Høgdall E, Yilmaz M, Tejpar S, Delorenzi M, Kruhøffer
M, et al: miR-345 in metastatic colorectal cancer: A non-invasive
biomarker for clinical outcome in non-KRAS mutant patients treated
with 3rd line cetuximab and irinotecan. PLoS One. 9:e998862014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen L, Li X and Chen X: Prognostic
significance of tissue miR-345 downregulation in non-small cell
lung cancer. Int J Clin Exp Med. 8:20971–20976. 2015.
|
|
11
|
Srivastava SK, Bhardwaj A, Arora S, Tyagi
N, Singh S, Andrews J, McClellan S, Wang B and Singh AP:
MicroRNA-345 induces apoptosis in pancreatic cancer cells through
potentiation of caspase-dependent and -independent pathways. Br J
Cancer. 113:660–668. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen QG, Zhou W, Han T, Du SQ, Li ZH,
Zhang Z, Shan GY and Kong CZ: MiR-345 suppresses proliferation,
migration and invasion by targeting Smad1 in human prostate cancer.
J Cancer Res Clin Oncol. 142:213–224. 2016. View Article : Google Scholar
|
|
13
|
Shiu TY, Huang SM, Shih YL, Chu HC, Chang
WK and Hsieh TY: Hepatitis C virus core protein down-regulates
p21(Waf1/Cip1) and inhibits curcumin-induced apoptosis through
microRNA-345 targeting in human hepatoma cells. PLoS One.
8:e610892013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of MicroRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin YH, Wu MH, Liao CJ, Huang YH, Chi HC,
Wu SM, Chen CY, Tseng YH, Tsai CY, Chung IH, et al: Repression of
microRNA-130b by thyroid hormone enhances cell motility. J Hepatol.
62:1328–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu Z, Zhang X, Wang G and Zheng H: Role
of MicroRNAs in hepatocellular carcinoma. Hepat Mon. 14:e186722014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
18
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kröger A, Ortmann D, Krohne TU, Mohr L,
Blum HE, Hauser H and Geissler M: Growth suppression of the
hepatocellular carcinoma cell line Hepa1-6 by an activatable
interferon regulatory factor-1 in mice. Cancer Res. 61:2609–2617.
2001.PubMed/NCBI
|
|
20
|
Li P, Du Q, Cao Z, Guo Z, Evankovich J,
Yan W, Chang Y, Shao L, Stolz DB, Tsung A, et al: Interferon-γ
induces autophagy with growth inhibition and cell death in human
hepatocellular carcinoma (HCC) cells through interferon-regulatory
factor-1 (IRF-1). Cancer Lett. 314:213–222. 2012. View Article : Google Scholar
|
|
21
|
Moriyama Y, Nishiguchi S, Tamori A, Koh N,
Yano Y, Kubo S, Hirohashi K and Otani S: Tumor-suppressor effect of
interferon regulatory factor-1 in human hepatocellular carcinoma.
Clin Cancer Res. 7:1293–1298. 2001.PubMed/NCBI
|
|
22
|
Tanaka M, Katayama F, Kato H, Tanaka H,
Wang J, Qiao YL and Inoue M: Hepatitis B and C virus infection and
hepatocellular carcinoma in China: A review of epidemiology and
control measures. J Epidemiol. 21:401–416. 2011. View Article : Google Scholar : PubMed/NCBI
|