Introduction
Lung cancer is a major public health problem and the
most frequent cause of cancer-related mortality worldwide, with
estimated nearly 244,000 new cases and 158,000 deaths annually in
the United States (1). In China,
there were 733,000 new cases and 610,000 deaths in 2016 (2). In fact, non-small cell lung cancer
(NSCLC) is one of the most prevalent histologic types of lung
cancer all over the world. Despite considerable advances in early
detection and conventional therapies, the 5-year survival rate of
patients with NSCLC is <40% (3), The primary cause of mortality in
NSCLC is widespread metastasis (4). Thus, studying the molecular
mechanisms that facilitate tumor invasion and metastasis is of
great importance and could reveal new therapeutic measures for
treating lung cancer, leading to improved patient survival
rates.
Epithelial-to-mesenchymal transition (EMT) is a
cellular process that allows cells to lose their epithelial
characteristics and adopt a mesenchymal phenotype and mesenchymal
characteristics to increase motility and invasion (5). Recent studies have shown that EMT
occurs at the invasive front of tumors and is a critical event for
cancer metastasis (6). Through
EMT, tumor cells attain invasive properties and have the ability to
enter the surrounding stroma, leading to the creation of a
favorable microenvironment for metastasis (7), which is characterized by the
suppression of the adhesion protein E-cadherin (8). Thus, the cadherin family contributes
to cancer progression and metastasis. Snail, the SNAI1 gene
product, is a member of the Snail family of zinc-finger
transcription factors and triggers EMT by repressing E-cadherin
expression, ultimately enhancing cancer invasion in several
malignancies such as breast, ovarian, skin, hepatocellular and head
and neck carcinomas (9,10). In many cancers, Snail expression is
not only associated with cancer invasion, but also highly
correlated with lymph node and distant metastasis through
E-cadherin repression (11,12).
Furthermore, Snail can also function as a negative regulator of
cell growth by repressing cyclin-dependent kinases (CDKs) to block
the cell cycle, which is also involved in tumor development and
malignancy (13,14).
Our previous study indicated that the lung cancer
cell lines A549 and PG have higher Snail mRNA levels than those
with a normal epithelial phenotype [e.g., human fetal lung
fibroblasts (HLF) and a spontaneously immortalized human fetal lung
epithelial cell line (HLEC)]. As a transcriptional activator, Snail
binds E-boxes in the myosin Va promoter, promoting tumor migration
and metastasis (15). In this
study, we hypothesized that Snail could affect cell proliferation,
altering the progression and invasion of lung cancer in
vitro and in vivo. Additionally, to explore how Snail is
correlated with EMT and cell cycle progression in lung cancer
cells, we tested phenotype, cadherin expression and cell cycle
check point gene expression in our models. Interestingly, the cell
cycle regulator P21 was upregulated following Snail knockdown by
small interfering RNA (siRNA), and was correlated to decreased PCNA
expression. Recently reported as genes induced by tricostatin A,
P21 and P27 as well as Snail and Slug were upregulated in normal
epithelial cells (16). To clearly
demonstrate which is driving these phenotypes, we confirmed that
P21 was directly upregulated by Snail knockdown and suppressed the
proliferation of lung cancer cells by arresting at the G2/M phase
of the cell cycle through inhibition of Cdc2 (CDK1). These results
suggest that the loss of Snail decreased lung cancer proliferation
and invasion via contributions of EMT and cell cycle.
Materials and methods
Cell cultures
The human lung carcinoma cell lines A549 (American
Type Culture Collection) and PG (established from a lung giant cell
carcinoma by Dr Bing-Quan Wu, Department of Pathology, Peking
Medical University) were cultivated and maintained in RPMI-1640
medium (Invitrogen, Grand Island, NY, USA) under 95% humidified 5%
CO2 at 37°C incubator. Medium contains 10% fetal bovine
serum (FBS), 100 U/ml penicillin and 100 g/ml streptomycin.
Snail interfering RNA transfection
The oligonucleotides for human Snail small
interfering RNAs (siRNAs) and control were synthesized by
Invitrogen. The 19-bp sequences of Snail siRNA were designed by
OligoEngine Workstation software: 5′-CATCCGAAGCCACACGCTG-3′
(siRNA1); 5′-GCT GGCAGCCATCCCACCT-3′ (siRNA2); 5′-GCACAACAA
GCCGAATACA-3′ (Scramble). A549 cells were transfected with 200 pmol
each siRNAs by Lipofectamine 2000 (Invitrogen) according to the
manufacturer's instructions. Blank vector was transfected into
controls. At 72 h after transfection, the total RNA was extracted
and RT-PCR was performed to assess mRNA levels of Snail, in order
to confirm which siRNA was more effective.
Subsequently, 2 μg of pSuper-Snail siRNA2 was
constructed into A549 cells in 35-mm diameter culture dish. Blank
vector were transfected into control dishes. After 24 h
post-transfection, 400 μg/ml G418 was added to the culture
medium for 12 days and 15 G418-resistant cell clones were picked.
Four clones named C1-C4 were selected from the 15 to analyze the
mRNA and protein levels of Snail by RT-PCR and western blot
analysis.
Lentivirus packaging and infection
Plenti4-Snail siRNA2 plasmid was constructed by
inserting the Snail siRNA2 fragment into a plenti4 expression
vector and was verified by DNA sequencing. The mixed plasmids (3
μg of plenti4-control or plenti4-Snail siRNA2 and 9
μg of lentivirus packaging vectors) were transfected into
the 293FT packaging cells using Lipofectamine 2000.
Virus-containing supernatants were harvested 48 h later. The
supernatant was then filtered and added to A549 and PG cells.
Virus-infected cells were selected by Zeocin (200 μg/ml) 24
h after infection. The Zeocin-resistant clones were pooled and
maintained in regular RPMI-1640 media with 10% FBS containing
Zeocin (100 μg/ml). Established stable cell lines were used
in subsequent experiments.
Reverse transcription-polymerase chain
reaction
Total cellular RNA was extracted using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's protocol. For semi-quantitative reverse
transcription-polymerase chain reaction (RT-PCR), the cDNAs were
synthesized from 2 μg of total RNA in a total volume of 20
μl using the Superscript III kit (Invitrogen). The cDNA as
the template were subjected to PCR reaction. The housekeeping gene
GAPDH was used as an internal control. The final PCR products were
visualized in agarose gel electrophoresis. The PCR primers of each
gene are described in Table I.
 | Table IPCR primer sequences. |
Table I
PCR primer sequences.
| Gene name | qRT-PCR primers
|
|---|
| Sense | Antisense | Size (bp) |
|---|
| SNAI1 |
5′-CTGCAGGACTCTAATCCAG-3′ |
5′-CAAGGAAGAGACTGAAGTAG-3′; | 300 |
| SNAI2 |
5′-TCGGACCCACACATTACCTT-3′ |
5′-TGACCTGTCTGCAAATGCTC-3′ | 145 |
| TWIST1 |
5′-CCTTCTCGGTCTGGAGGAT-3 ′ |
5′-TCCATTTTCTCCTTCTCTGGAA-3′ | 127 |
| CDH1 |
5′-TCCATTTCTTGGTCTACGCCT-3 ′ |
5′-TCACCTTCAGCCATCCTGTTT-3′ | 363 |
| CDH2 |
5′-GTGCCATTAGCCAAGGGAATT-3 ′ |
5′-GGAGGAATTCCATTGTCAGAAG-3′ | 350 |
| CDKN1A |
5′-AGTCAGTTCCTTGTGGAGCC-3′ |
5′-CATGGGTTCTGACGGACAT-3′ | 108 |
| GAPDH |
5′-GACCCCTTCATTGACCTCAAC-3 ′ |
5′-CTTCTCCATGGTGGTGAAGA-3′ | 219 |
Western blot analysis
The cells were washed twice with cold PBS and
harvested with a scraper. Cells were centrifuged at 3000 g for 30
sec. The pellets were lysed in RIPA buffer (50 mM Tris, pH 7.4, 150
mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mM
EDTA, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride
and 5 μg/ml leupeptin) containing complete protease
inhibitor cocktail (Roche, Mannheim, Germany). Equal amounts of
protein lysates were resolved by SDS-PAGE, and then were
transferred onto polyvinylidene difluoride (PVDF) membrane
(Millipore, Bedford, MA, USA). Non-specific binding sites on the
membranes were blocked in Tris-buffered saline (TBS, 50 mM Tris-HCl
pH 7.5, 150 mM NaCl) containing 5% fat-free dried milk and
incubated for 1 h at room temperature. After immunoblotting with
primary antibodies at 4°C overnight, the membrane was washed 3
times with 0.05% Tween-20 in TBS. Subsequently, the membranes were
incubated with secondary antibody for 1 h at 37°C. Detection of the
reactive protein performed was detected using Immobilon™ Western
Chemiluminescent HRP substrate (Millipore). The primary antibodies
used are shown in Table II. The
HRP-conjugated antibodies were used as secondary antibody (1:50,000
dilute, Jackson ImmunoResearch, West Grove, PA, USA).
 | Table IIAntibodies used in western blots and
immunofluorescence analysis. |
Table II
Antibodies used in western blots and
immunofluorescence analysis.
| Name | Vender | Cat no. | Species | Dilution |
|---|
| Snail | R&D | 965619 | Goat polyclonal
IgG | 1:2,000 |
| E-cadherin | Santa Cruz | sc52328 | Mouse monoclonal
IgG | 1:2,000 |
| N-cadherin | Sigma | C2342 | Mouse monoclonal
IgG | 1:2,000 |
| PCNA | Santa Cruz | sc56 | Mouse monoclonal
IgG | 1:2,000 |
| P21 | Santa Cruz | sc397 | Mouse monoclonal
IgG | 1:2,000 |
| CDK1 | Abgent | Ap1497a | Rabbit polyclonal
IgG | 1:1,000 |
| CDK2 | Santa Cruz | sc163 | Rabbit polyclonal
IgG | 1:2,000 |
| CDK4 | Abgent | Ap20515b | Rabbit polyclonal
IgG | 1:2,000 |
| Actin | Roche | 1378996 | Mouse monoclonal
IgG | 1:10,000 |
cDNA microarray
Equal amounts of cDNA from A549 cultured in stable
Snail silence cell and control cells were labeled with Cy3 and Cy5,
respectively, and then hybridization was carried out according to
the manufacture's protocol using human genome cDNA microarray
consisting of 14,592 genes (Shanghai BioChip Co., Ltd., Shanghai,
China). Genes were considered to be up or downregulated when the
fluorescent intensity ratio between Cy3 and Cy5 cells was >2 or
<0.5. The experiment was repeated once. Hierarchical clustering
of regulated genes in this study was measured by Genespring
software.
Cell proliferation assay
To compare proliferation rates in vitro,
cells were seeded at a density of 1×104 cells/well in
24-well culture plates with 400 μl/well of growth medium.
Each day (5 days), cells from duplicates were collected after
trypsinization. Cells were counted with a hemocytometer under a
microscope. Cell Generation Time (GT = Tn/5; Tn = t/ln (Nf/Ni)/ln2;
Nf, final cell number at time t; Ni, initial plated cell number;
ln, natural logarithm) was calculated by plotting the graph of cell
counting versus time.
Wound healing assay
The cells were seeded in 6-well dishes with proper
concentration (~3×105 per well). Confluent cells were
scratched 24 h later by a disinfected Eppendorf Tip. The detached
cells were washed away with PBS and fresh medium was added.
Cultures were incubated at 37°C with 5% CO2 for a
further 24 h. The images of the wound width were captured by an
inverted microscope (Leica, Wetzlar, Germany) at 0 and 24 h
post-scratch, respectively. Distance of cell movement was measured
by LAF software (Leica).
Boyden chamber assay
The cell migration and invasion capacity was
analyzed by using Boyden chamber assay. Briefly, a total of
5×104 cells were re-suspended in 200 μl
serum-free RPMI-1640 media and plated in the upper chambers of
Transwell (24-well insert; pore size, 8 μm; Corning). The
lower chambers were filled with 800 μl RPMI-1640 containing
10% FBS. After cultures were incubated for 24 h at 37°C,
non-invasive cells were wiped off from the upper surface and the
invasive cells that were attached on the lower surface of the
chamber were stained with 0.5% crystal violet, followed by fixed
with methanol. The average number of cells was determined by
counting five random fields in triplicates by light microscopy.
Cell cycle analysis
Cells (5×105 cells/well) were plated in
6-well plates and incubated for 24 h. Then, the cells were
collected by trypsinization, washed with PBS twice, and fixed with
70% pre-chilled ethanol at 4°C overnight. The following day, the
cells were pelleted, treated with 50 μg/ml of RNase A
(Roche) for 30 min at room temperature followed by propidium iodide
(Invitrogen) staining at room temperature for 30 min. Finally, the
stained cells were examined by flow cytometry (BD, San Jose, CA,
USA). The percentage of cell population in different phases of cell
cycle was analyzed using CFlow software V1.0.
Immunofluorescence staining
Cells growing on coverslips were fixed with 4%
formaldehyde for 3 min at room temperature, followed by washing
with 0.5% Triton X-100 in PBS three times. Primary antibodies,
e.g., N-cadherin and E-cadherin (described above), were next
incubated for 1 h at room temperature. After being washed with 0.5%
Triton X-100 in PBS, the cells were labeled for 1 h with
TRITC-conjugated anti-mouse IgG (1:100, Jackson) or FITC-conjugated
antirabbit IgG (1:100, Jackson) secondary antibody solution at room
temperature and washed again. Afterwards, the cell nuclei were
stained by DAPI (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 3
min. Staining was detected on a Leica SP5 laser confocal
microscope.
Animal experiments
For evaluating the tumorigenicity in animals, cells
were trypsinized, centrifuged, and re-suspended in PBS. Suspensions
containing 2×105 cells in 0.2 ml PBS were implanted
subcutaneously into the under arm area of athymic nude mice
(BALB/c, Nu/Nu; 4–6-week-old, Vitalriver Laboratory Animals,
Beijing, China) (5 mice/group). Tumor formation was examined two
times each week by calipers and tumor volumes were estimated as
follows: V = the length x (the width 2)/2. After 26 days, all the
mice were euthanized and the tumors were excised and weighed.
The detection of PG cells metastasis in mouse lung
was performed as descripted (17).
Briefly, genomic DNA was extracted from fresh mouse lung tissues.
Quantitative real-time PCR was used to detect human Alu sequences
by using primers (sense, 5′-ACG CCT GTA ATC CCA GCA CTT-3′; and
antisense, 5′-TCG CCC AGG CTG GAG TGC A-3′) (18). The mouse housekeeping gene GAPDH
was used as an internal control. The level of human Alu-sequence
was normalized to amount of mouse GAPDH genomic DNA sequence
amplified. All animal studies were performed in accordance with the
policies of the Institutional Animal Care and Use Committee and
were approved by the Institutional Review Board of Peking
University School of Oncology.
Statistical analysis
Each experiment was conducted in triplicate. All
results are presented as means ± SD. The comparison between two
groups was calculated by Student's t-test with two-tails. p<0.05
was regarded as statistically significant.
Results
Snail siRNA knockdown in lung cancer
cells
We previously demonstrated that Snail mRNA
expression was increased in the highly malignant lung cancer cell
lines A549, PG and Calu6 compared with the less malignant cell line
Calu3 (15). This suggested that
Snail might be involved in cancer cell migration. To further
explore how Snail expression influences malignant behavior through
loss-of-function analysis, we inhibited Snail expression in A549
cells using two sets of siRNAs targeting Snail: siRNA1 and siRNA2.
After 48 h of growth, scramble or Snail siRNAs were transfected
into A549 cells. PCR analysis was performed to analyze the Snail
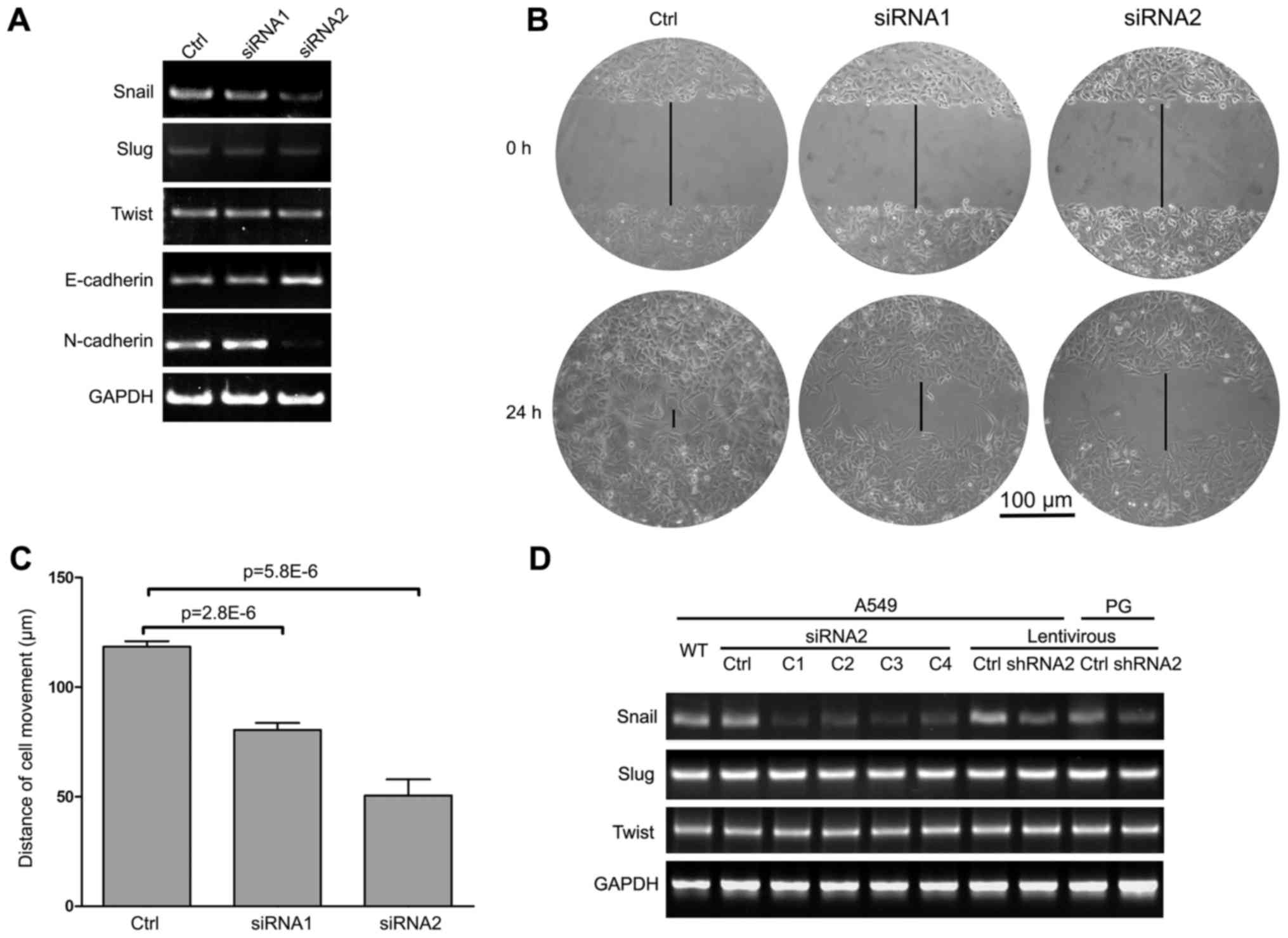
and GAPDH mRNA levels. The results showed that both Snail-targeting
siRNAs effectively repressed Snail mRNA levels. Moreover, Slug and
Twist, two close homologs of Snail, showed no significant changes
between the siRNa- or scramble-transfected cells (Fig. 1A). These results showed that siRNA2
was more effective, although both siRNAs were specific for Snail.
at the functional level, wound healing assays were performed to
assess how these siRNAs suppressed the migratory behavior of A549
cells. siRNA2 consistently showed a more significant effect on
migration (50.5±18.2 μm/day), compared with siRNAl (80.5+7.8
μm/day) or the scramble control (118.4+6.1 μm/day)
(Fig. 1B and C). Hence, siRNA2 was
selected for further experiments.
To generate stable Snail-silenced cells,
lentiviruses containing Snail-targeting constructs were used to
block Snail expression in A549 and PG cells. Four clones (C1-C4) of
Snail-silenced A549 and PG cells were isolated and screened for
Snail expression by RT-PCR. Compared with controls, Snail
expression was decreased in all four clones, but the interfering
efficiency of clone C3 was most dramatic. Similar result was shown
in Fig. 1D, the efficiency of
Snail shRNA knockdown was obtained by using the lentiviral
system.
Behavior of Snail-silenced lung cancer
cells
To clearly understand the functional changes
associated with in vitro Snail knockdown by RNA
interference, we first measured cell growth in the stable
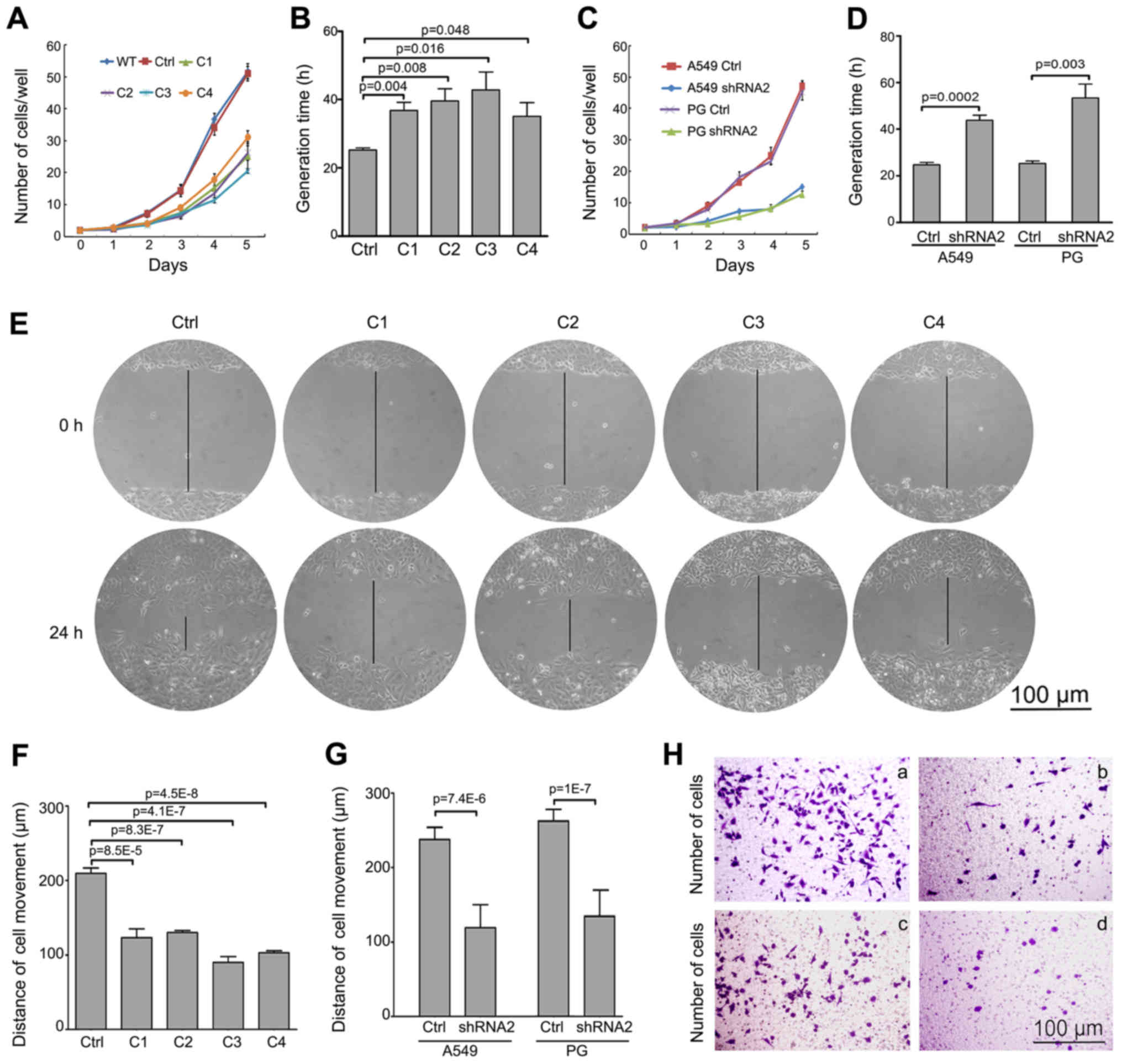
Snail-silenced cell lines by cell counting. Snail downregulation
significantly reduced growth in A549 clones (Fig. 2A), and the GT averages are shown in
Fig. 2B. Similarly, A549 and PG
cells infected with Snail-knockdown lentivirus were analyzed for
cell growth and GT (Fig 2C and D).
The data clearly shows that silencing Snail decreased proliferation
of both A549 and PG cells in vitro. Second, to analyze the
impact of Snail suppression on cell motility, we performed wounding
healing assays to test the migratory ability of each stable cell
line. The data showed that clone C3 had a highly decreased
migration capacity among the four clones and demonstrated the
weakest movement ability (Fig. 2E and
F). Furthermore, the same suppression of cell motility was
demonstrated in A549 and PG cells following Snail knockdown by
lentiviral system (Fig. 2G).
Third, we analyzed the invasive properties following Snail
knockdown using the Boyden chamber assay. As shown in Fig. 2H, the Snail knockdown A549 and PG
cells were significantly less invasive than control, A549 invasion
was decreased from 168+21 to 77±18 per well, and PG was decreased
from 158±25 to 61+14 per well. These results indicate that Snail
silencing inhibited not only proliferation, but also suppressed the
migratory behavior of lung cancer cells in vitro.
Snail downregulation induces a reversion
of EMT in lung cancer cells
In a previous study, we performed a series of
experiments exploring the phenotypes of Snail knockdown cells,
assessing how Snail knockdown suppresses cell proliferation and
migration (15). To gain further
insight into possible mechanisms underlying Snail knockdown, we
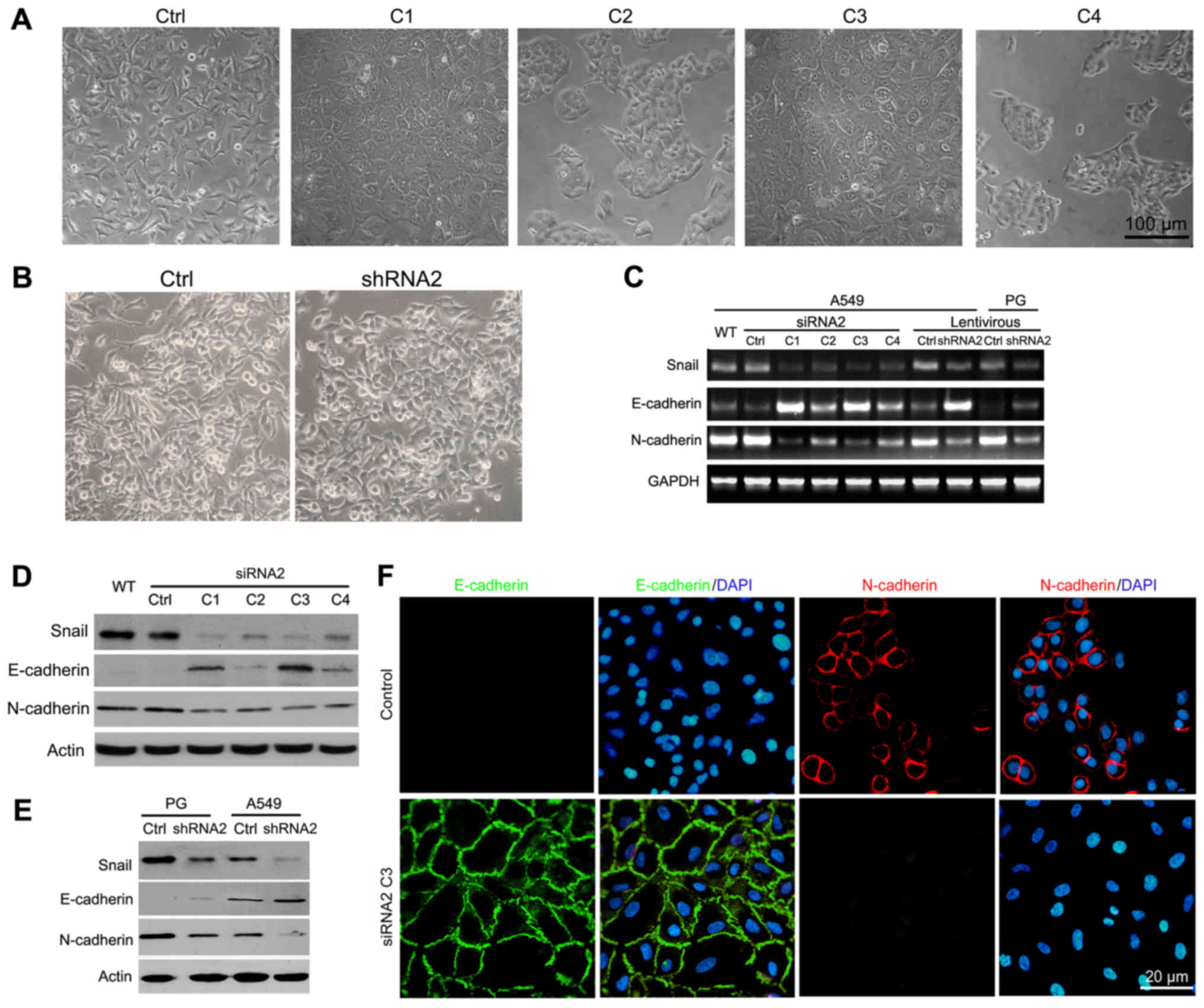
examined the morphology of A549 and PG cells following Snail
silencing. Of the four A549 clones, C1 and C3 (Fig. 3A) and PG Snail siRNA2 stable cells
(Fig. 3B) showed a sheet-like
epithelial phenotype with well-organized cell-cell adhesion,
whereas control cells had a spindle-shaped mesenchymal morphology.
Therefore, we evaluated E-cadherin and N-cadherin expression, two
important cadherin family members in cancer biology. A549 and PG
cells showed increased E-cadherin and decreased N-cadherin
expression at both the mRNA and the protein levels following Snail
knockdown (Fig. 3C–E). among the
four clones, C3 had the most significant effect on the expression
of EMT-associated markers. To confirm the hypothesis of EMT
reversion following Snail knockdown, we assessed the expression and
localization of epithelial and mesenchymal markers in clone C3
using indirect immunofluorescence staining (Fig. 3F). As expected, an increase in
E-cadherin expression was observed in clone C3. Conversely,
N-cadherin expression was absent with Snail knockdown. In addition,
compared with the control, clone C3 had a clear localization of
E-cadherin at the cell membrane. These results suggest that Snail
silencing affects lung cancer cell migration and metastasis through
the reversion of EMT.
A G2/M cell cycle arrest following Snail
knockdown
To further investigate mechanisms underlying the
suppressed tumorigenicity of Snail-silenced cells, we analyzed data
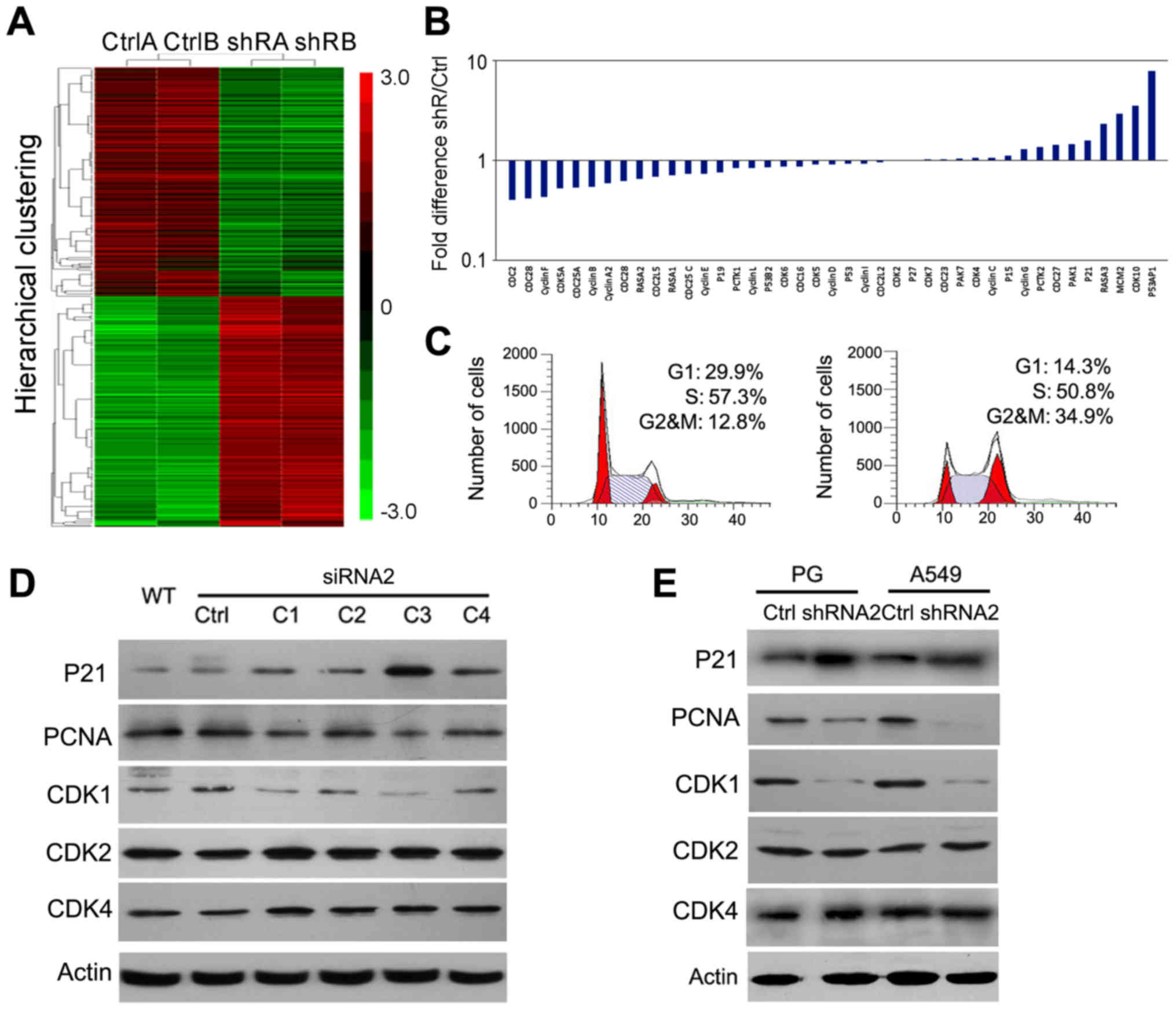
from cDNA microarray analyses to find global gene expression
changes associated with Snail knockdown (Fig. 4A). This unbiased approach allowed
us not only to find EMT-associated genes, but also genes highly
associated with the cell cycle. Associated cell cycle genes were
listed from low to high expression responded on mRNA levels
including Cdc2 (0.4-fold), CDK2 (1.01-fold),
CDK4 (1.05-fold), CDK6 (0.86-fold), P21
(1.6-fold) and their related genes (Fig. 4B). Therefore, we next performed
cell cycle analysis in A549 cells by flow cytometry. The results
showed that the G1 and S periods were down and the G2/M population
was especially overrepresented, ~3-fold higher following Snail
knockdown (Fig. 4C). To
investigate why the cell cycle was changed, we analyzed the
expression of cell cycle-related proteins, such as P21, PCNA, Cdc2,
CDK2, and CDK4 by western blot analysis. Our data showed that Snail
knockdown led to the upregulation of P21 and downregulation of
Cdc2, but did not affect CDK2 or CDK4 expression (Fig. 4D and E). These data indicated that
Snail knockdown may inhibit cell growth via causing a G2/M cell
cycle arrest.
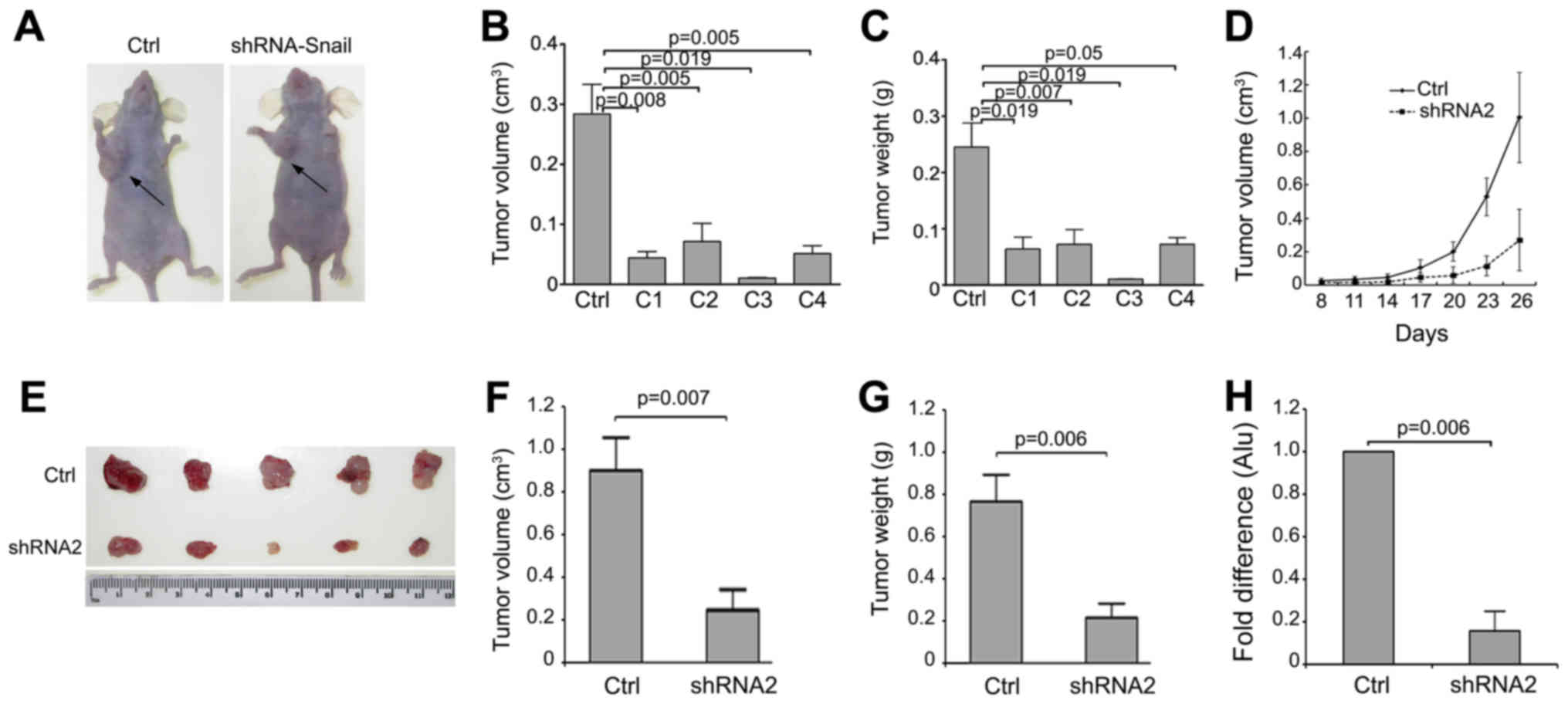
Snail knockdown inhibits tumorigenicity
and metastasis of lung cancer cells in vivo
We next investigated how Snail knockdown modulates
tumorigenicity and metastasis of lung cancer cells in vivo,
utilizing the nude mouse xenograft model (Fig. 5A). A decrease in tumor size and
weight were observed in tumors formed from the four Snail knockdown
A549 clones compared to those from control cells (Fig. 5B and C). C3 tumors tended to be the
smallest among all clones. Moreover, the plotted growth curves of
tumor formation were markedly different between PG Snail shRNA2
cells and control cells (Fig. 5D).
Compared to the controls, PG Snail shRNA2 cells had decreased tumor
size (Fig. 5E) after
26-day-injection. The size and weight measurements of tumors from
PG Snail shRNA2 cells were also markedly decreased compared with
control (Fig. 5F and G).
additionally, the quantification of human-specific alu-sequences in
the lungs was also significantly reduced following Snail knockdown
(Fig. 5H). These results suggested
that lower Snail expression reduced tumor growth and lung
metastasis in lung cancer cells.
Discussion
Cell migration and invasion is the major cause of
recurrence after surgery in lung cancer patients. A variety of
studies have focused on exploring the mechanisms underlying tumor
migration and invasion. Interestingly, during EMT, cancer cells
acquire a fibroblastic phenotype induced by decreasing their
cell-cell adhesions and become more motile (19). EMT is involved in the
proliferation, invasion and stem cell features of cancer cells
(20).
In the recent two decades, it has been appreciated
that EMT is controlled by several transcriptional factors; however,
Snail, a member of the Snail family of zinc-finger transcription
factors, is the key EMT regulator, directly repressing E-cadherin
expression by binding to E-box elements in the E-cadherin promoter
(21,22). Snail had been correlated with a
worse survival outcome in many malignancies (23–25).
Additionally, Snail has been reported to regulate stem-like
properties in thyroid and pancreatic cancer (26,27).
These findings suggest that Snail may serve as a putative molecular
target for new therapeutics. Hence, clearly understanding the
effects of Snail silencing on the biological behavior of lung
cancer cells is important. In this context, we repressed Snail
expression to investigate the role of Snail downregulation on the
progression of lung cancer. We studied the correlation between
Snail silencing and EMT processes in A549 cells, finding that Snail
knockdown induced morphological changes and a less invasive
phenotype compared with control cells. Inhibiting Snail enhanced
expression of the epithelial marker E-cadherin and repressed the
mesenchymal marker N-cadherin, which has also been reported in
melanoma (28), colon cancer
(29), and hepatic carcinoma cells
(30). These data explained the
reduced migration following Snail interference.
To assess the effects of Snail knockdown on the
biological behavior of lung cancer cells, we silenced Snail
expression in lung cancer cell lines by isolating the Snail siRNA2
transfectant clones, and characterized the impact on growth and
invasion in these lung cancer cells. Lung cancer cell lines with
Snail knockdown were more invasive than the parental cell line in a
previous study (31); however, in
our study we found that Snail knockdown markedly decreased the
proliferation and migration of A549 and PG cells. To uncover the
mechanisms of these phenotypes, we analyzed genome-wide gene
expression profiling studies. From these data, some cell
cycle-associated genes were remarkably upregulated, such as
P53AP1, CDK10, P21 and PAK1. However CDK2, CDK4,
CDK6, Cyclin D and Cyclin E, most of the keys in G1/S
phase checkpoint, were unaffected by Snail knockdown, as well as
upon P27 (upstream of CDK2), P15 and P19 (upstream of
CDK4/CDK6). On the contrary, the major G2/M phase
checkpoints, such as Cdc2, Cyclin B and Cdc25A, were
markedly downregulated by Snail knockdown. From cell cycle
analysis, we found that Snail knockdown activated the G2/M
checkpoint protein Cdc2, but did not influence the G1 phase of the
cell cycle. Moreover, the cell population was especially
overrepresented from 12.8 to 34.9% in G2/M phase following Snail
knockdown. Additionally, P21 and PCNA expression indicated that
Snail knockdown could inhibit cell proliferation by enhancing P21
expression, which in turn, caused a decrease in PCNA expression.
P21 can promote tumor cell proliferation and is dysregulated in
many human cancers (32,33), and also regulates PCNA expression
by binding to the PCNA promoter, thereby inhibiting DNA replication
and modulating DNA repair processes (34,35).
Furthermore, bioinformatics software analysis revealed that there
are four potential consensus Snail-binding sites (CAGCTG at
position −1,061 to −1,055 and −133 to −128; CACCTG at position
−1,068 to −1,063 and −466 to −461) in the P21 promoter. These
provide evidence of the relationship among Snail, P21, Cdc2 and
PCNA. Thus, we hypothesize that a Snail/P21/Cdc2/PCNA signaling
pathways exists and regulates lung cancer cell proliferation.
Because Snail overexpression promoted lung cancer
progression had been reported in vivo (36), we further examined the effects of
silencing Snail on the tumorigenic behavior of A549 and PG cells
in vivo to provide more persuasive evidence of tumor
malignancy in knockdown cells. After injecting tumor cells into
nude mice, we observed a decrease in tumor appearance and growth
rate in the Snail knockdown group. Interestingly, the frequency of
lung metastases was also reduced by Snail knockdown. This suggests
that Snail knockdown not only interferes with proliferation, but
also affects invasion and migration in vivo.
Altogether, this study showed that the inhibition of
Snail expression can repress cell growth, invasion and metastasis
of human lung cancer in vitro and in vivo.
Furthermore, this study verified that Snail silencing impacted the
malignant behavior of lung cancer cells by suppressing EMT
processes and the cell cycle. Therefore, Snail could be a potential
target for lung cancer treatment. Blocking Snail expression by
siRNA could represent a new therapeutic strategy for lung
cancer.
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EMT
|
epithelial- to-mesenchymal
transition
|
|
CDKs
|
cycl in-depended kinases
|
|
siRNA
|
small interfering RNA
|
Acknowledgments
We would like to thank the FACS Core Facility of
Peking University Cancer Hospital for performing FACS assays. This
study was supported by the National Key Research and Development
Program of China (2016 TFA 0500303), the National Natural Science
Foundation of China (grant nos. 8133005, 81372594 and 81201964),
the '863' Project (grant nos. 2014AA021606 and 2015AA020403),
Beijing Natural Science Foundation (grant nos. 5122012 and
7132051).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sher DJ, Fidler MJ, Liptay MJ and Koshy M:
Comparative effectiveness of neoadjuvant chemoradiotherapy versus
chemotherapy alone followed by surgery for patients with stage IIIA
non-small cell lung cancer. Lung Cancer. 88:267–274. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moran C: Importance of molecular features
of non-small cell lung cancer for choice of treatment. Am J Pathol.
178:1940–1948. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelialmesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010. View Article : Google Scholar
|
|
8
|
Chen A, Beetham H, Black MA, Priya R,
Telford BJ, Guest J, Wiggins GA, Godwin TD, Yap AS and Guilford PJ:
E-cadherin loss alters cytoskeletal organization and adhesion in
non-malignant breast cells but is insufficient to induce an
epithelial-mesenchymal transition. BMC Cancer. 14:5522014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Herreros AG, Peiro S, Nassour M and
Savagner P: Snail family regulation and epithelial mesenchymal
transitions in breast cancer progression. J Mammary Gland Biol
Neoplasia. 15:135–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Come C, Magnino F, Bibeau F, De Santa
Barbara P, Becker KF, Theillet C and Savagner P: Snail and slug
play distinct roles during breast carcinoma progression. Clin
Cancer Res. 12:5395–5402. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kroepil F, Fluegen G, Vallböhmer D, Baldus
SE, Dizdar L, Raffel AM, Hafner D, Stoecklein NH and Knoefel WT:
Snaill expression in colorectal cancer and its correlation with
clinical and pathological parameters. BMC Cancer. 13:1452013.
View Article : Google Scholar
|
|
13
|
Cheon MG, Kim W, Choi M and Kim JE: AK-1,
a specific SIRT2 inhibitor, induces cell cycle arrest by
downregulating Snail in HCT116 human colon carcinoma cells. Cancer
Lett. 356B:637–645. 2015. View Article : Google Scholar
|
|
14
|
Vega S, Morales AV, Ocana OH, Valdés F,
Fabregat I and Nieto MA: Snail blocks the cell cycle and confers
resistance to cell death. Genes Dev. 18:1131–1143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lan L, Han H, Zuo H, Chen Z, Du Y, Zhao W,
Gu J and Zhang Z: Upregulation of myosin Va by Snail is involved in
cancer cell migration and metastasis. Int J Cancer. 126:53–64.
2010. View Article : Google Scholar
|
|
16
|
Xiao W, Chen X, Liu X, Luo L, Ye S and Liu
Y: Trichostatin A, a histone deacetylase inhibitor, suppresses
proliferation and epithelial-mesenchymal transition in retinal
pigment epithelium cells. J Cell Mol Med. 18:646–655. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su B, Zhao W, Shi B, Zhang Z, Yu X, Xie F,
Guo Z, Zhang X, Liu J, Shen Q, et al: Let-7d suppresses growth,
metastasis, and tumor macrophage infiltration in renal cell
carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 13:2062014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zijlstra A, Mellor R, Panzarella G, Aimes
RT, Hooper JD, Marchenko ND and Quigley JP: A quantitative analysis
of rate- limiting steps in the metastatic cascade using
human-specific real-time polymerase chain reaction. Cancer Res.
62:7083–7092. 2002.PubMed/NCBI
|
|
19
|
Yuen HF, Chan YK, Grills C, McCrudden CM,
Gunasekharan V, Shi Z, Wong AS, Lappin TR, Chan KW, Fennell DA, et
al: Polyomavirus enhancer activator 3 protein promotes breast
cancer metastatic progression through Snail-induced
epithelialmesenchymal transition. J Pathol. 224:78–89. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Talbot LJ, Bhattacharya SD and Kuo PC:
Epithelial-mesenchymal transition, the tumor microenvironment, and
metastatic behavior of epithelial malignancies. Int J Biochem Mol
Biol. 3:117–136. 2012.PubMed/NCBI
|
|
21
|
Shin NR, Jeong EH, Choi CI, Moon HJ, Kwon
CH, Chu IS, Kim GH, Jeon TY, Kim DH, Lee JH, et al: Overexpression
of Snail is associated with lymph node metastasis and poor
prognosis in patients with gastric cancer. BMC Cancer. 12:5212012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuo KT, Chou TY, Hsu HS, Chen WL and Wang
LS: Prognostic significance of NBS1 and Snail expression in
esophageal squamous cell carcinoma. Ann Surg Oncol. 19(Suppl 3):
S549–S557. 2012. View Article : Google Scholar
|
|
23
|
Hotz B, Arndt M, Dullat S, Bhargava S,
Buhr HJ and Hotz HG: Epithelial to mesenchymal transition:
Expression of the regulators snail, slug, and twist in pancreatic
cancer. Clin Cancer Res. 13:4769–4776. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van Nes JG, de Kruijf EM, Putter H,
Faratian D, Munro A, Campbell F, Smit VT, Liefers GJ, Kuppen PJ,
van de Velde CJ, et al: Co-expression of SNAIL and TWIST determines
prognosis in estrogen receptor-positive early breast cancer
patients. Breast Cancer Res Treat. 133:49–59. 2012. View Article : Google Scholar
|
|
25
|
Kobayashi M, Huang CL, Sonobe M, Kikuchi
R, Ishikawa M, Imamura N, Kitamura J, Iwakiri S, Itoi K, Yasumizu
R, et al: Snail expression is associated with a poor prognosis in
malignant pleural mesotheliomas. Ann Thorac Surg. 95:1181–1188.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yasui K, Shimamura M, Mitsutake N and
Nagayama Y: SNAIL induces epithelial-to-mesenchymal transition and
cancer stem cell-like properties in aldehyde
dehydroghenase-negative thyroid cancer cells. Thyroid. 23:989–996.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou W, Lv R, Qi W, Wu D, Xu Y, Liu W, Mou
Y and Wang L: Snail contributes to the maintenance of stem
cell-like phenotype cells in human pancreatic cancer. PLoS One.
9:e874092014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Kumar SM, Martin JS, Yang R and Xu
X: Snail1 mediates hypoxia-induced melanoma progression. Am J
Pathol. 179:3020–3031. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang W, Wang J, Zou B, Sardet C, Li J,
Lam CS, Ng L, Pang R, Hung IF, Tan VP, et al: Four and a half LIM
protein 2 (FHL2) negatively regulates the transcription of
E-cadherin through interaction with Snail1. Eur J Cancer.
47:121–130. 2011. View Article : Google Scholar
|
|
30
|
Zhang JP, Zeng C, Xu L, Gong J, Fang JH
and Zhuang SM: MicroRNA-148a suppresses the epithelial-mesenchymal
transition and metastasis of hepatoma cells by targeting Met/Snail
signaling. Oncogene. 33:4069–4076. 2014. View Article : Google Scholar
|
|
31
|
Merikallio H, Turpeenniemi-Hujanen T,
Pääkkö P, Mäkitaro R, Riitta K, Salo S, Salo T, Harju T and Soini
Y: Snail promotes an invasive phenotype in lung carcinoma. Respir
Res. 13:1042012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu H, Zhou L, Shi S, Wang Y, Ni X, Xiao
F, Wang S, Li P and Ding K: Oligosaccharide G19 inhibits U-87 MG
human glioma cells growth in vitro and in vivo by targeting
epidermal growth factor (EGF) and activating p53/p21 signaling.
Glycobiology. 24:748–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Z, Zhang D, Hao J, Tian K, Wang W, Lou
H and Yuan H: Induction of DNA damage and p21-dependent senescence
by Riccardin D is a novel mechanism contributing to its growth
suppression in prostate cancer cells in vitro and in vivo. Cancer
Chemother Pharmacol. 73:397–407. 2014. View Article : Google Scholar
|
|
34
|
Rousseau D, Cannella D, Boulaire J,
Fitzgerald P, Fotedar A and Fotedar R: Growth inhibition by
CDK-cyclin and PCNA binding domains of p21 occurs by distinct
mechanisms and is regulated by ubiquitin-proteasome pathway.
Oncogene. 18:3290–3302. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao H, Ho PC, Lo YH, Espejo A, Bedford
MT, Hung MC and Wang SC: Interaction of proliferation cell nuclear
antigen (PCNA) with c-Abl in cell proliferation and response to DNA
damages in breast cancer. PLoS One. 7:e294162012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yanagawa J, Walser TC, Zhu LX, Hong L,
Fishbein MC, Mah V, Chia D, Goodglick L, Elashoff DA, Luo J, et al:
Snail promotes CXCR2 ligand-dependent tumor progression in
non-small cell lung carcinoma. Clin Cancer Res. 15:6820–6829. 2009.
View Article : Google Scholar : PubMed/NCBI
|