Epithelial ovarian cancer (EOC) is a metastatic
disease with the highest mortality rate of all gynecologic cancers
(1). Serous, endometrioid,
clear-cell, and mucinous ovarian cancers are the most frequent
histotypes. These different ovarian cancer (OC) histotypes are
characterized by altered genomic and epigenetic patterns which
greatly impact oncogenic signaling-pathways, behavior and clinical
outcome. Hence, alternative therapeutic approaches are needed to
improve the patient survival and outcome. Although no molecular
predictors of clinical outcome are currently in use, several
cellular factors have been studied as potential prognostic and
predictive biomarkers (2).
MicroRNAs (miRNAs) are small non-coding RNAs that function as
negative regulators of gene expression (3). In cancer cells, miRNAs may function
either as oncogenes or tumor-suppressors and may contribute to the
heterogeneous biological behavior of ovarian tumors. One of the
most deadly hallmarks of cancer cells is the ability to metastasize
to other tissues and organs (4).
Notably, metastasis can be promoted by specific sets of miRNAs
which have the ability to target multiple genes related to cell
migration and invasion (5,6). In OC, metastasis is greatly promoted
by the epithelial-to-mesenchymal transition (EMT). A number of
studies have focused on the discovery of key players in the EMT to
identify potential targets for therapeutic intervention in
precision medicine. Here, we reviewed the current status on the
role of miRNAs in invasion and metastasis with a special emphasis
in the potential applications in clinical and translational
research in OC.
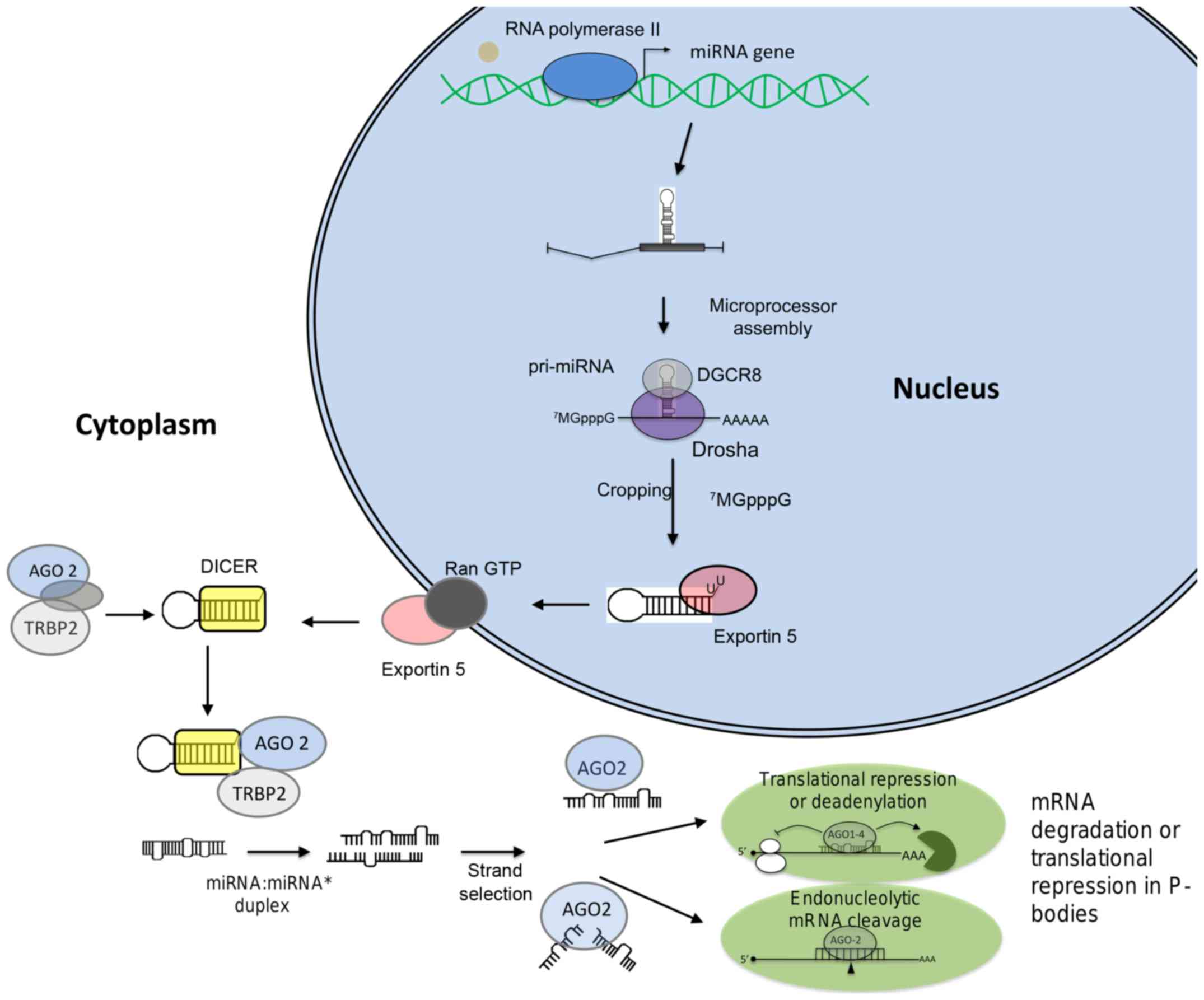
miRNAs are small non-coding RNAs of ~22 nucleotides
in length which function as negative regulators of gene expression.
These small RNAs function as guide molecules to base pairing with
target genes resulting in translation repression or messenger RNA
(mRNA) cleavage. miRNA biogenesis (Fig. 1) begins with their transcription by
the RNA polymerase II which synthesize a primary miRNA (pri-miRNA)
which contains both 5′-cap structure (7MGpppG) as well
as 3′-end poly(A) tails (7).
Pri-miRNAs are processed by the RNAse III-type protein Drosha which
is associated to DGCR8 (DiGeorge syndrome critical region gene 8),
KSRP and p68 proteins in a complex denoted as microprocessor. The
cleavage of pri-miRNA by Drosha generates a 70-nucleotide precursor
miRNA (pre-miRNA) that is exported into the cytoplasm by the
Ran-GTP-dependent exportin 5 (8).
The second step of pre-miRNA processing (dicing) is then performed
by Dicer, an RNAse III enzyme which is associated to TRBP (TAR
RNA-binding protein) or PACT, and Argonaute (AGO) proteins. This
protein complex cleaves the pre-miRNA hairpin generating a
double-stranded RNA of ~22 nucleotides in length known as the
miRNA:miRNA* duplex. Then the miRNA:miRNA*
interacts with the associated RNA-induced silencing complex (RISC),
which preferentially includes the miRNA guide strand and AGO. The
mature miRNA bound to AGO hybridizes to complementary sequence in
the 3′ untranslated region (3′-UTR) of mRNA target. If miRNA binds
to mRNA with non-perfect complementarity, it leads to translation
repression. In contrast, if miRNA binds to target with a high
complementarity, cleavage and degradation of mRNA is induced
(9). The decay of mRNA is
initiated by shortening of poly(A+) transcripts by the
canonical deadenylation machinery in cytoplasmic foci denoted as
P-bodies (10,11).
Aberrant expression of miRNAs is a common event
during carcinogenesis as they regulate key transcripts involved in
the initiation and progression of tumors, thus they have been
denominated as oncomiRs (12).
miRNAs with increased expression in tumors are frequently
considered as oncogenes as they promote tumor development by
inhibiting tumor suppressor genes. In contrast, miRNAs may function
as tumor suppressors as they prevent tumor development by
inhibiting oncogene expression or genes that controls cell
differentiation and apoptosis (13). Aberrant expression of miRNAs is
produced from genetic or epigenetic alterations represented by: i)
deletions, ii) amplifications, iii) point mutations, iv) DNA
methylation, and v) chromatin modifications. Notably, several
studies have identified miRNAs as useful biomarkers of diagnosis
and prognosis in OC patients (14,15).
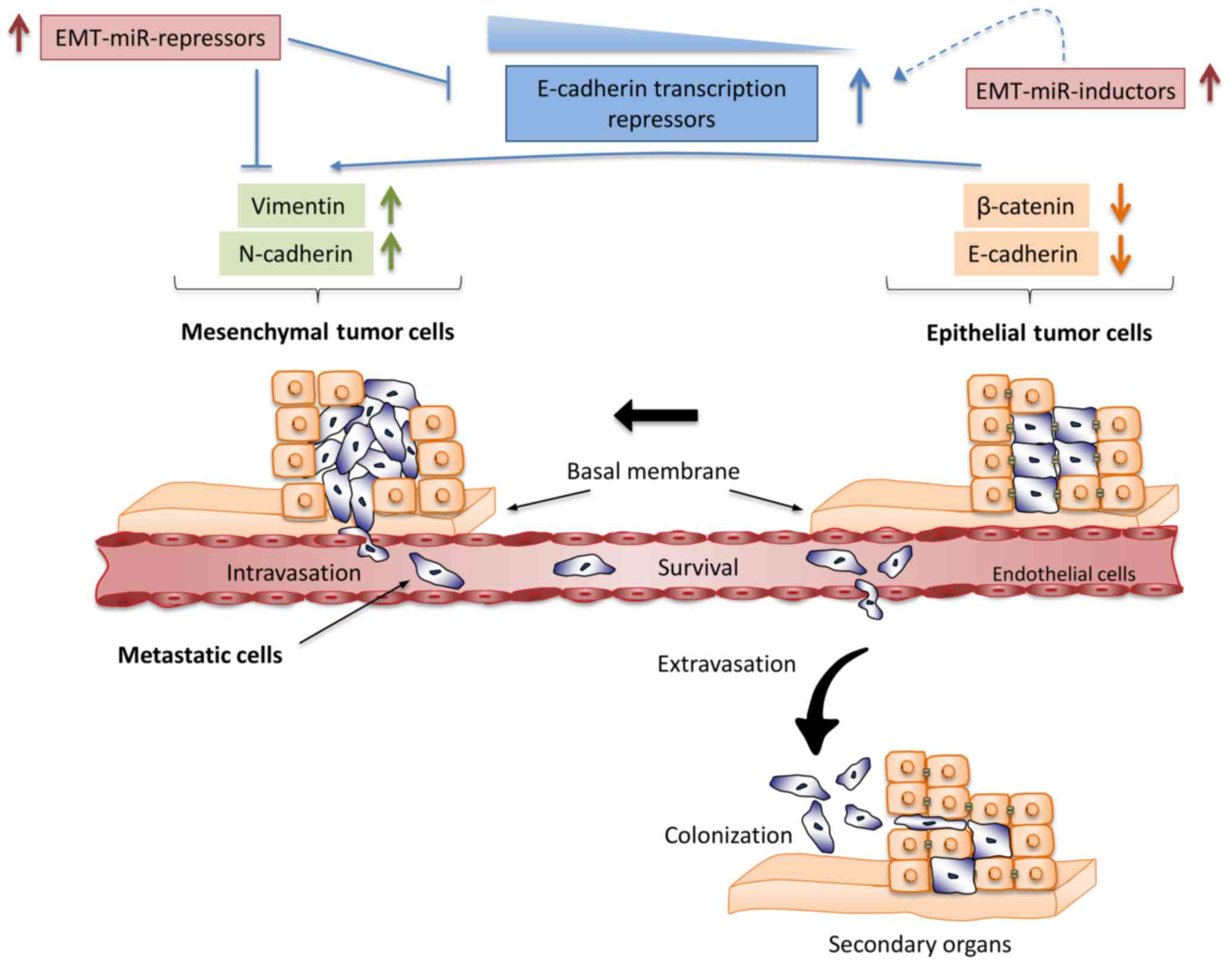
Epithelial to mesenchymal transition (EMT) is a
morphogenesis program activated during embryonic development,
tissues repair, fibrosis and cancer (16,17).
During tumor progression the EMT activation induces changes in
epithelial cells that promote loss of basal apical polarity and
cell-cell junctions. This allows that epithelial cells acquire
plasticity, invasive capacity, stem cell-like characteristics and
gain the ability to invade adjacent tissues and colonize distant
organs (18) (Fig. 2). Cells that have undergone EMT
also have the capacity to transform into epithelial cells in a
reverse process known as mesenchymal to epithelial transition
(MET). Both processes are important during embryo- and
organogenesis, but in malignant transformation they represent a
critical step to induce metastasis progression (19). Induction of EMT program is
facilitated by growth factors produced from stromal cells that
activate diverse signaling pathways including TGF-β/BMP,
WNT/β-catenin, Hedgehog, Notch, Sonic and PI3K/AKT signaling
pathways represent crucial mechanisms promoting EMT (20). Crosstalk between diverse signaling
pathways promotes the expression of repressors of E-cadherin
transcription such as: i) the Snail family of zinc-finger
transcription factors Snail, Slug and Smuc; ii) the basic
helix-loop-helix factors, Twist and E12/E47; iii) the two-handed
zinc-finger factors of δ-crystallin/E2 box factor 1 (dEF1) family
proteins, dEF1/zinc-finger E-box-binding homeobox 1 (ZEB1), and iv)
the Smad-interacting protein/ZEB2 (21). In particular, E-cadherin plays an
important role in the induction of EMT phenotype in cancer cells.
E-cadherin is a membrane glycoprotein that mediates
calcium-dependent cell-cell adhesion which is located at the
adherens junctions predominantly associated with β-catenin. Under
normal circumstances β-catenin binds E-cadherin providing a direct
link between the E-cadherin cell-adhesion complex and the
intracellular cytoskeleton (22).
During EMT activation, β-catenin influences the transcriptional
repressors Snail, Slug, and Twist to inhibit E-cadherin
transcription (23).
Studies related to the characterization of
EMT-induced signaling pathways have demonstrated that WNT signaling
pathway is predominantly activated in EOC. Additional evidence
reported that WNT signaling is negatively regulated by E-cadherin
which in turn suppresses β-catenin expression and suppresses
transcriptional activation of the EMT-related genes in OC (24). A number of studies have shown that
Sonic Hedgehog signaling pathway also participate in the process of
EMT. Upregulation of hedgehog glioma-associated oncogene 1
(shh-Gli1) promotes the activation of EMT event by crosstalk with
PI3K-AKT pathways in OC (25). In
this context, another mechanism has been described that sustains
EMT by cooperative signaling. Rac-1, a specific guanine nucleotide
exchange factor induce EMT in OC through simultaneous activation of
mitogen-activated extracellular signal-regulated kinase 1/2
(MEK1/2) and Src signaling pathways (26). Another sophisticated regulatory
network that regulates EMT is exerted by TGF-β family. Recently, it
was found that Smad4 mutations increased homodimerization of Smad4
with its receptor to promote nuclear localization leading to
reduction of E-cadherin expression and increases of N-cadherin
(27). In addition, crosstalk
between TGF-β and stem-cell promoting pathways facilitates the
formation of Smad complex during EMT. Interestingly, the molecular
basis of this cooperative mechanism indicates that many EMT
signaling molecules interact with Smads to form complexes engaged
in both repressing epithelial genes and activating mesenchymal
genes (28). A number of studies
have showed that microRNAs are important in the regulation of
signaling proteins involved in the control of EMT. miRNAs are
frequently engaged in feedback loops with signaling molecules, thus
alterations in miRNAs expression may result in an imbalance of
signals that promotes the activation of EMT program in cancer
(29).
Expression of genes responsible for EMT is mainly
under control of ZEB1 (TCF8/δ-EF1) and ZEB2 (SIP1/ZFXH1B)
transcription factors which repress E-cadherin whereas enhance
vimentin and N-cadherin. These changes in ZEB1 and ZEB2 expression
constitute a signal for starting transition from epithelial to
mesenchymal phenotype. Remarkably, it has been well demonstrated
that miR-200 family members orchestrate the initiation of EMT by
controlling the expression of ZEB1 and ZEB2 genes. The miR-200
family is expressed as two separate polycistronic pri-miRNAs: i)
miR-200a, miR-200b, and miR-429 located on chromosome 1, and ii)
miR-200c and miR-141 at chromosome 12 (30–34).
Interestingly, miR-200 family members are differentially expressed
in the bulk tumor. For instance, miR-200 members are expressed at
low or negligible levels in normal ovarian surface cells and
increased their expression in OC, whereas expression of ZEB1 and
ZEB2 shows the opposite pattern. Ovarian surface cells acquire a
more epithelial phenotype as they undergo transformation switching
from a miR-200 family low and ZEB1/2 high state to a miR-200 family
high and ZEB1/2 low phenotype (34). An early study which identified the
relationship between miR-200 family and EMT process in EOC was
carried out in the panel of 60 human cancer cell lines (NC160). One
the most relevant findings of this study was the identification of
miR-200 as a marker for E-cadherin-positive and vimentin-negative
cancer cells in both the NC160 cells as well as in tumors from OC
patients. This outstanding report showed that miR-200 targets the
E-cadherin transcriptional repressors ZEB1 and ZEB2. Furthermore
ectopic expression of miR-200 induced upregulation of E-cadherin in
diverse cancer cell lines and reduction in their motility, while
inhibition of miR-200 reduced E-cadherin expression and induced EMT
associated to increased ZEB1 levels (33). On the other hand Chen et al
performed a genome expression profile of two mesenchymal-like OC
cell lines with different metastatic potential and demonstrated
that expression of miR-429, a member of the miR-200 family,
resulted in the reversal of the mesenchymal phenotype through
activation of the MET which was associated to changes in
E-cadherin, ZEB1 and ZEB2 expression and migration and invasiveness
(35). In addition, restoration of
individual miR-200 members (miR-200a, miR-141, miR-200b and
miR-200c) in HEY ovarian cancer cells also resulted in the
acquisition of morphological epithelial characteristics, elevated
expression of KRT8, KRT18 and KRT7 epithelial markers and reduced
levels of ZEB1 and ZEB2. Remarkably, ectopic expression of miR-200
family members influenced the sensitivity of EOC to platinum-based
drugs, which was associated to the inhibition of EMT that is known
to reduce the sensitivity of chemotherapy (36). Besides these findings Chen et
al reported that overexpression of miR-429 in a primary cell
line (OCI-984) isolated from the ascites fluid an advanced stage OC
patients also induced morphological, functional and molecular
changes consistent with MET (35).
Overexpression of miR-429 in OCI-984 cells also increased the
sensitivity to cisplatin. Another study indicates that upregulation
of miR-200c in ovarian cancer tissues was inversely associated with
advanced clinical stage. miR-200c inhibited E-2 cells migration and
invasion capacity through repression of ZEB2 (37). Moreover, miR-200c showed a
potential function in stemness as it was downregulated in
CD44+/CD117+ enriched stem-like cells derived
from SKOV3 cells. Ectopic expression of miR-200c resulted in the
inhibition of migration and invasion by downregulation of ZEB1 and
vimentin, which in consequence upregulated E-cadherin in
CD44+/CD117+ subpopulations. In addition
miR-200c overexpression inhibited the EMT process and subsequently
metastasis in CD44+/CD117+ xenograft in nude
mice (38).
On the other hand, ZEB1 was upregulated in
high-grade serous ovarian carcinoma (SOC) specimens compared to
normal fallopian tube tissue. In addition, miR-1236-3p suppressed
migration and invasion abilities through targeting of 3′-UTR of
ZEB1 in A2780 and SKOV3 cell lines (39). On the other hand Wu et al
identified that ZEB2 is overexpressed in CD133/1+
ovarian cancer stem cells by downregulation of its repressor
miR-200a. Conversely, ectopic expression of miR-200a in
CD133/1+ resulted in suppression of ZEB2 causing the
increase of E-cadherin expression. Noteworthy these data show that
loss of miR-200a expression may play a critical role in the
repression of E-cadherin by ZEB2, thereby enhancing migration and
invasion in CD133/1+ cells possibly through the
acquisition of EMT phenotype (40).
Another microRNA that targets ZEB1 and control the
EMT process is miR-150 which was downregulated in primary EOC
tissues (41). Lower expression of
miR-150 was associated with advanced clinical stage (III-IV) as
well as with poor prognosis. Ectopic expression of miR-150 inhibits
cell proliferation, invasion and metastasis (42). Targeted silencing of ZEB1 using
shRNAs in SKOV3 cells resulted in enhanced expression of miR-200c
and reduced EMT and metastasis. Downregulation of ZEB1 also
decreased the growth of tumors in xenograft mouse model (43). Likewise, overexpression of miR-101
suppressed cell migration, invasion and EMT phenotypes by targeting
ZEB1 and ZEB2 in SKOV3 cells. These phenotypes were associated to a
significant increase in E-cadherin levels and decreases in
mesenchymal markers (fibronectin, N-cadherin and vimentin). In
other related studies, Zhou et al reported that miR-153 also
inhibits the EMT by targeting ZEB2 and Set7 (Set domain containing
7) genes (45). Set7 is a
methyltransferase enzyme that trimethylates lysine 4 on histone 3
and epigenetic mark that has been found enriched in promoters of
activated genes (44). In
agreement with these findings and using an RNA mimic approach, it
was demonstrated that miR-153 also promotes degradation of
SET7/ZEB2 transcripts which in turn leads to the inhibition of cell
proliferation and EMT reducing the invasive potential of OC cells.
In addition, miR-153 expression was downregulated in ovarian tumors
and correlated with poor survival rates in EOC patients (45).
SNAI family of transcription factors consists of
SNAI1 (often designated as SNAIL), SNAI2 (also known as SLUG) and
SNAI3 (also known as SMUC), and they also suppress the expression
of epithelial genes such as E-cadherin and activate the expression
of mesenchymal genes including N-cadherin and fibronectin (46). Yang et al performed an
integrated genomic analysis and found that miR-506 inhibited cell
migration and invasion and prevented TGF-β-induced EMT by targeting
SNAI2 in SOC. In addition, miR-506 expression was associated with
decreased SNAI2 and VIM, as well with increased E-cadherin levels.
Interestingly, using a nanoparticle delivery system for miR-506 in
orthotopic mouse models, a significant suppression of SNAI2/VIM and
upregulation of E-cadherin expression was found (47). Functionally, miR-506 directly binds
to the 3′-UTR of vimentin and N-cadherin genes in EOC (48). In addition using tissue microarrays
including 204 EOC subtypes it was found that miR-506 expression was
positively associated to E-cadherin and negatively correlated with
vimentin and N-cadherin. Noteworthy, all these findings confirms
that loss of miR-506 expression resulted in mesenchymal phenotype
in EOC. Thus, miR-506 is a potent regulator of both E-cadherin and
vimentin/N-cadherin to suppress and control the EMT process and
represents an innovative therapy against most aggressive ovarian
cancers (49).
Another miRNA that regulates SNAI genes is the tumor
suppressor miR-203. In human SOC tissues, miR-203 was found
repressed whereas its target SNAI2 was upregulated. In addition,
overexpression of miR-203 inhibited cell proliferation, migration
and invasion in SKOV3 and OVCAR3 cells by suppression of EMT which
was associated to E-cadherin upregulation and vimentin and SNAI2
downregulation. Outstandingly, through subcutaneously injection of
miR-203-expressing SKOV3 and OVCAR3 cells transduced with a
lentiviral luciferase vector into immunodeficient NSG female mice,
Zhao et al revealed that miR-203 inhibited EMT process and
tumor growth in a xenograft mouse model (50).
On the contrary, miR-520h regulates Twist an
oncogenic transcription factor that plays a key role in
EMT-mediated intravasation of tumor cells. Forced expression of
miR-520h in SKOV3ip1 cells enhanced invasion and metastasis. In
addition, E-cadherin was downregulated by miR-520h and associated
to increased expression of Twist (53).
Several reports have identified diverse EMT-related
genes as targets of microRNAs. For instance, miR-187 is an
independent prognostic factor in OC and was associated with
recurrence-free survival. Overexpression of miR-187 induced cell
proliferation and inhibited cell migration. On the other hand,
inhibition of miR-187 resulted in the upregulation of Dab2,
repression of E-cadherin and increased vimentin levels which in
turn promote the EMT phenotype. This means that miR-187 has a dual
function; during the initial steps of tumorigenesis miR-187 may
induce proliferation, but in the late stages it inhibits the EMT
and tumor invasiveness by the suppression of Dab2. Another miRNA
involved in the posttranscriptional regulation of EMT-related genes
is miR-29b. This small RNA modulates the EMT by targeting the high
mobility group A2 gene (HMGA2), a non-histone DNA binding protein
that plays a very important role in fetal development and
carcinogenesis. As an oncofetal gene it is overexpressed in tumors
of both epithelial and mesenchymal tissues (54). Overexpression of HMGA2 in OSE cells
induced spindle-like cell morphology, whereas its repression
restored their epithelioid phenotype. In addition, HMGA2
overexpressing xenograft tumors resulted in the loss of E-cadherin
and the increase of vimentin. Global expression profiling indicated
that HMGA2-mediated tumorigenesis was associated with changes in
the expression of miRNAs and target genes involved in EMT process.
Among these, lumican (LUM) a tumor suppressor that inhibits EMT was
found transcriptionally repressed by HMGA2 in OSE cell lines
(55).
Furthermore, miR-373 was downregulated in
cholangiocarcinoma and colon cancer and inversely correlated with
clinical stage and histological grade in human EOC (43,56).
Ectopic expression of miR-373 in EOC cells suppressed cell
invasion, metastasis and EMT process. Functional assays identified
small GTPase Rab22a as a target of miR-373. Interestingly knockdown
of Rab22a inhibits invasion, migration, and suppress N-cadherin in
EOC cells. Metastasis was suppressed after restoration of miR-373
in xenograft ovarian carcinoma metastatic nude mice (57).
In another study miR-125, which is a tumor
suppressor in different types of cancer also inhibited EMT process
by targeting the AT-rich interactive domain 3B (ARID3B) gene
(63). AIRD3B belongs to a family
of AT-rich interactive domain (ARID) proteins that are involved in
chromatin remodeling and gene expression regulation. ARID genes
participate in development, tissue-specific gene expression and its
aberrant expression is involved in tumorigenesis (64). Interestingly, miR-125 targets
ARID3B and induced the conversion of highly invasive ovarian cancer
cells from a mesenchymal to an epithelial morphology. Using
multiple biochemical approaches, authors revealed that EGFR
signaling leads to transcriptional repression of miR-125a through
the ETS family transcription factor PEA3, which in consequence
releases ARID3B to promote EMT (63).
Growing evidence indicates that activation of AKT,
ERK1/2 and TGF-β signaling pathways are essential for the induction
of EMT genetic program. Since EMT-signaling pathways largely
control metastasis, the investigations have been focused in the
study of the interplay between microRNAs and novel genes modulating
the EMT phenotype. Zhou et al reported that miR-7 plays a
major role in the reversion of EMT by the inactivation of AKT and
ERK1/2 pathways in EOC. Ectopic expression of miR-7 induced
morphological changes from an elongated, spindle-shaped,
mesenchymal phenotype to a rounded epithelial-like phenotype in
highly metastatic HO-8910 and ES-2 cell lines. In addition, miR-7
expression was significantly downregulated in EOC cell lines and
tissues, whereas EGFR expression correlated positively with
metastasis in both EOC patients and cell lines. Moreover, miR-7
overexpression leads to the upregulation of CK-18 and B-catenin,
and downregulation of vimentin by inactivation of EGFR/AKT and
AKT/ER1/2 pathways. These changes were enough to inhibit cell
migration, invasion and EMT events indicating that miR-7 is a tumor
suppressor in EOC (65).
Another miRNA that controls EMT related pathways to
suppress tumor cells invasion is miR-17-5p which was previously
found increased in chemotherapy resistant colorectal tumors
(69). In OC cells, miR-17-5p
enhances cellular invasion, migration and EMT by targeting both
PTEN and AKT (70). Introduction
of miR-17-5p in OC cell lines repressed E-cadherin while increased
N-cadherin, Snail and vimentin expression. In addition, forced
expression of miR-17-5p decreased PTEN while increased p-AKT in
OVCAR3 and SKOV3 cell lines (70).
Other studies have focused on the role of miRNAs and
metastasis in cancer stem cells. For instance, downregulation of
miR-155 in OC stem-like cells (CD44+/CD117+)
resulted in downregulation of claudin-1 (CLD-1). In vitro
and in vivo assays showed that restoration of miR-155
expression inhibits the migration and invasion in
CD44+/CD117+ subpopulations by suppression of
CLD-1. In addition, transfection of miR-155 did not induce changes
in MMP-2 and MMP-9 expression, but a significantly increase of
E-cadherin and β-catenin was found in agreement with acquisition of
epithelial phenotype (71). All
these reports evidence the alterations in the regulation network of
miRNAs and EMT-related genes which are summarized in Fig. 3.
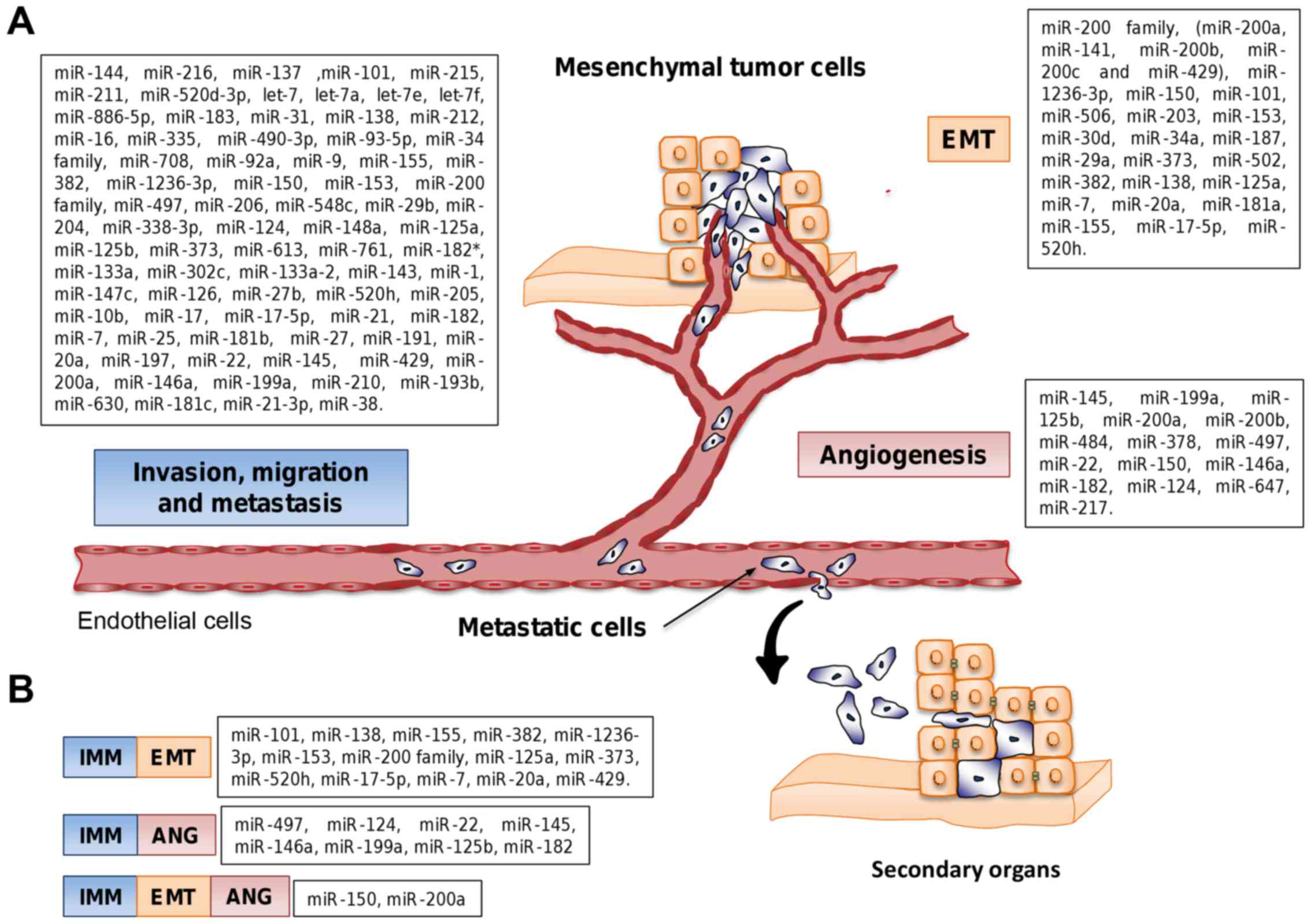
During the last decade a number of aberrantly
expressed miRNAs have been identified as key promoters of cell
migration and invasion in OC (Fig.
3). In early studies, Iorio et al reported a miRNA
expression profile in different histotypes and clinical stages of
EOC. They reported the downregulation of miR-144 and miR-216 in
lymphovascular invasion; and miR137, miR-101, miR-215 and miR-211
in OC with tubal involvement. In contrast, upregulation of miR-101,
miR-182*, miR-22 and miR-133a was found in OC with
ovarian surface involvement, miR-302c in pelvic peritoneum
involvement, and miR-133a-2, miR-143, miR-145, miR-1, miR-147 and
miR-126 in OC with uterus involvement. Subsequent functional
studies gave insights into the role of some deregulated miRNAs in
invasion and metastasis in OC (72).
Degradation of the extracellular matrix is a complex
process that stimulates cell migration, invasion and metastasis in
many types of cancer. In endometrial carcinoma histotype, 28% of
cases exhibit metastasis at diagnosis. Some miRNAs functioning as a
tumor suppressor have been identified as deregulated in this
particular OC. For instance, Habata et al found that
BCL2-associated athanogene (BAG3) inhibited the expression of
miR-29b in endometroid adenocarcinoma cells. Congruently, knockdown
of BAG3 led to increased levels of miR-29b in Ishikawa and HEC108
cells. Restoration of miR-29b using RNA-mimics decreased migration
and invasion presumably by inhibition of metallopeptidase 2 (MMP2)
gene (73). On the other hand, Sun
et al demonstrated that Twist regulates miR-548c expression.
Ectopic expression of miR-548c inhibited cell proliferation,
migration and invasion, while Twist restoration abrogates the
suppressive effect of miR-548c in cell migration and invasion in
Rl95-2 and HEC-1 ovarian cells (74).
A number of miRNAs activates the cancer hallmarks
thus functionioning as oncogenes in OC. For instance, miR-205 is
frequently over-expressed in endometrial carcinoma (EC) and it is
related to metastasis. Experimental inhibition of miR-205 decreased
cell proliferation, migration and invasion in Ishikawa cells by
targeting the estrogen-related receptor-γ (ESRRG) tumor suppressor
gene (75). Another study showed
that upregulation of miR-27 was associated with myometrial invasion
in ECC. In addition, FOXO1 transcription factor was found
down-regulated in invasive ECC indicating that miR-27-FOXO1 axis
inhibits invasiveness and apoptosis in tumors (76). Moreover, Dong et al showed
that miR-191 had a negative correlation with tissue inhibitor of
metalloprotease 3 (TIMP3) expression in tissues and serum of
patients with endometriosis-associated ovarian cancer (EAOC).
Notably, miR-191 was overexpressed in EAOC and its knockdown
significantly inhibited cell invasion by upregulation of TIMP3 in
CRL-11731 cells. These data indicate that miR-191-TIMP3 axis is
involved in the malignant transformation of endometriosis to EAOC
(77).
Hormonal receptors interplaying with miRNAs regulate
invasion and metastasis of tumor cells. Expression of estrogen
receptor-α (ER-α) and downregulation of miR-22 were associated with
cell invasion in ER-α positive EC subtype. In addition, it was
demonstrated that ER-α is a direct target of miR-22. Likewise,
ectopic expression of miR-22 in Rl95-2 and Ishikawa cells abrogates
the invasion induced by 17β-E2 (E2) through downregulation of
cyclin D1 and MMP2 and MMP9. Thus, E2-ER-α axis represents a
potential candidate for endocrine therapy in OC (78). On the contrary, it was reported
that miR-206 was significantly downregulated in ER-α positive EC
and that its suppression was modulated by E2. Ectopic expression of
miR-206 decreased ER-α positive-dependent proliferation and
invasion in EC cells. Thus, miR-206 also could be a potential tool
for endocrine therapy in ER-α positive EC patient (79,80).
Recent findings from diverse research groups pointed
out that altered miRNAs expression was associated with aggressive
phenotypes of OC. Downregulation of tumor suppressors let-7a,
let-7e, let-7f, miR-886-5p and miR-22 have been associated with
aggressive behavior of tumors and represents potential markers of
invasion and metastasis in EOC (81). Particularly, miR-22 has a critical
role on malignancy and metastasis in all subtypes of OC. Li et
al showed that overexpression of Tiam1, a guanine nucleotide
exchange factor, correlated with metastatic phenotypes of OC.
Authors showed that miR-22 is downregulated in ovarian cancer and
negatively associated with metastasis in SOC cells. Intriguingly,
the manipulation of miR-22 expression had inhibitory effects on
cell migration and invasion, but not in cell viability and
apoptosis (82). In agreement with
these findings, miR-22, miR-183 and miR-31 suppressed cell
migration and invasion of SOC cells, at least in part, by
down-regulation of Tiam1 (83).
Furthermore, miR-124 was downregulated in highly
metastatic OC cells. Ectopic expression of miR-124 suppressed the
expression of sphingosine kinase 1 (SphK1), a protein associated
with tumor cell invasion and metastasis. Knockdown of SphK1 using
miR-124 inhibited the cell motility in SKOV3 and HO8910 cells
(86). Thereafter, Wen et
al found that sphin-gosine-1-phosphate receptor 1 (S1PR1) was
also suppressed by miR-148a. Overexpression of miR-148a resulted in
the inhibition of cell migration and invasion in SKOV3 cells
(87).
Moreover, two reports showed that low levels of
miR-138 were associated with invasion and metastasis without
affecting growth of tumors (62,88).
In the first study, ectopic expression of miR-138 reduced the cell
migration and metastasis ability by targeting SOX4 and
hypoxia-inducible factor-1α (HIF-1α). Knockdown of SOX4 and HIF-1α
in SKOV-I6 cells abolished the invasion through the signaling
downstream of EGFR and Slug. Thus low levels of miR-138 and high
expression of SOX4 could represent a prognostic marker and a target
for intervention of OC metastasis (62). In the second report, suppression of
the expression of miR-138 was found in highly malignant phenotypes
of OC tissues. Notably, miR-138 inhibits cell invasion and
metastasis by targeting LIM kinase 1 (Limk1) via
Limk1/cofilin/p-cofilin axis and modulation of PCNA and Bcl-2
(88).
Another report indicates that EPH receptor A2/B2
(EphA2/B2) is implicated in malignancy of OC. Knockdown of EphA2
expression or ectopic expression of miR-520d-3p in vitro
abolished the migration and invasion of cancer cells and decreased
the tumor growth in vivo (14). It has been indicated that motility
of the ovarian cancer cells can be promoted by ubiquitin ligases.
Wang et al reported that higher levels of pro-metastatic
factor SMAD specific E3 ubiquitin protein ligase 1 (SMURF1) were
associated with shorter overall survival, whereas downregulation of
miR-497 correlated with aggressive phenotypes of OC. Exogenous
expression of miR-497 abolished the cell migration and invasion
through direct inhibition of SMURF1 activity. In agreement with
these data, restoration of miR-497 decreased invasiveness and was
associated to better survival of patients (89). On the other hand, lee et al
elucidated a link between translation machinery and miRNAs. Ectopic
expression of miR-125a and miR-125b decreasd cell invasion and
migration by targeting the eukaryotic translation initiation factor
4E binding protein 1 (EIF4EBP1) gene (90).
Some miRNAs could have a dual role as tumor
suppressor or oncogene in cancer cells. For instance, miR-7
expression was identified downregulated in EOC tumors (65). However, Meng et al
identified that miR-7 and miR-429 were overexpressed in serum of
EOC patients. In vitro miR-429 inhibited the cell migration
and invasion. Authors proposed that miR-429 could have a dual role
acting as tumor suppressor and oncogene because miR-429 levels were
high in patients with primary EOC while they diminished in patients
with distant migration and metastasis (91).
Actin cytoskeleton regulates the formation of
F-actin-rich membrane protrusions leading to motility and cellular
adhesion. Cao et al found that overexpression of miR-335
inhibited cell migration and invasion through the depolymerization
of F-actin in OC cells. Moreover, miR-335 suppressed B-cell
CLL/lymphoma w (BCL-w) and its effector MMP2. Lack of miR-335
expression resulted in the accumulation of BCL-w and acquisition of
a more invasive phenotype (92).
Loss or dysfunction of tumor suppressor p53 greatly
contributes to tumorigenesis in more than half of all types of
cancers. Chen et al demonstrated that restoration of
miR-490-3p in EOC metastatic cells resulted in increased levels of
p53. Overexpression of miR-490-3p in vitro suppressed cell
migration and invasion as well as cell proliferation, and in
vivo decreased the tumor development. Notably, miR-490-3p
targets cyclin-dependent kinase 1 (CDK1) which participates
together with Bcl-xL, CCND1, and MMP-2/9 enhancing tumorigenesis in
OC. By decreasing CDK1 expression through miR-490-3p and
stimulating the expression of p53, miR-490-3p potentially may
suppress the tumorigenesis and metastasis progression in EOC
(99). In addition, Chen et
al suggested that the expression of tumor suppressor miR-93-5p
activates p53 while represses poly (ADP-ribose) polymerase (PARP)
expression through inhibition of RAS homolog gene family member C
(RhoC), P70S6 kinase, Bcl-xL and MMP-9. Functional studies
confirmed that RhoC is a target of miR-93-5p. In addition,
overexpression of miR-93-5p in vitro inhibited invasion and
proliferation of cancer cells, and the in vivo tumor
development (100). Moreover, Liu
et al showed that the expression of p53 and p21 was
decreased because downregulation of serine/threonine kinase 11
(LKB1), which was inhibited by miR-17 in
CD44−/CD117− cancer cells. Expression of
miR-17 modulated the migration and invasion in
CD44−/CD117− cells through suppression of the
LKB1-p53-p21/WAF1 pathway. The suppression of LKB1-p53-p21/WAF1 by
miR-17 resulted in increased invasiveness of
CD44+/CD117+ cells (101).
The miR-34 family plays a key role in cell
proliferation and invasion of EOC cells. Corney et al showed
that miR-34 family is downregulated in OC with or without
p53-mutated. They found that miR-34b*/c levels are lower
in advanced OC and the expression of miR-34a was negatively
associated with the expression of the proto-oncogene receptor
tyrosine kinase (MET). Ectopic expression of miR-34 family members
into SKOV-3 cells decreased MET significantly and resulted in the
inhibition of cell motility and invasion (102). A further study highlighted that
miR-34 family is activated in the presence of functional p53. MET
can be suppressed through miR-34 in a p53-dependent way (103,104). In addition, Li et al
reported that miR-34a inhibits cell invasion and proliferation by
downregulating the AXL receptor tyrosine kinase (105).
Downregulation of miR-145 has been reported in tumor
tissues, serum and OC cancer cell lines (106). Additional reports showed that p53
regulated the expression of miR-145 and p53 is dysfunctional in
high-grade SOC (107,108). Functional studies using RNA
mimics revealed a suppressive role of miR-145 in cell viability,
invasion, cell growth, cell proliferation and colony formation in
OC cells. Moreover, cell colony formation and invasion were
mediated by P70S6K1 and MUC1, respectively. Both genes were
confirmed as true targets of miR-145 (106). Another study found that miR-145
also targets the motif containing 2 (TRIM2) gene. Inhibition of
TRIM2-BIM pathway through antagomiR-145 in EOC resulted in
decreased tumorigenicity (109).
Moreover, Dong et al found that metadherin (MTDH) oncogene,
which was overexpressed in SOC, is a target of miR-145. Inhibition
of MTDH by miR-145 abolished the cell migration and invasion, and
reduced tumor growth and metastasis (108). In other studies, Kim et al
found an inverse expression between miR-145 and HMGA2 during
ovarian cancer metastasis. In vitro, ectopic expression of
miR-145 inhibited cell growth and migration. Finally, low levels of
miR-145 and high expression of HMGA2 represent biomarkers of poor
prognosis in ovarian cancer (110).
Long non-coding RNA can act as molecular sponge to
suppress miRNAs in cancer cells. Yan et al identified that
H19 long non-coding RNA suppressed the activity of let-7 in A2780
cells. Silencing of H19 decreased the cell migration and invasion
processes by restoration of let-7 activity and inhibition of its
targets genes including HMGA2, c-Myc and Igf2bp3. A positive
correlation between the expression of H19 and Hmga2, c-Myc and
Igf2bp3 was also identified. Suppression of let-7 activity by H19
may explain why the overexpression of let-7 is associated with poor
prognosis in ovarian cancer (111). On the other hand, Gao et
al reported that HOST2 long non-coding RNA is overexpressed in
EOC resulting in the inhibition of let-7b. Targeting of HOST2 leads
to decreased cell migration and invasion in OVCAR3 cells (112).
Biogenesis of miRNAs is epigenetically and
transcriptionally controlled resulting in tissue specific miRNA
expression patterns. However, alterations in the basic components
of the miRNAs biogenesis machinery can promote carcinogenesis. Guo
et al identified that knockdown of DiGeorge critical region
8 (DGCR8), a component of the microprocessor complex, reduced cell
migration and invasion through attenuation of ERK1/2, PI3-K and AKT
pathways, as well as sensitizes cells to apoptosis induced by the
chemotherapeutic drug cisplatin. In addition, DGCR8 knockdown
resulted in dysregulated miRNA gene expression. miR-27b was
identified as the most highly downregulated miRNA in DGCR8
knockdown cells and promoted cell proliferation in OC cancer cells
(113). On the contrary,
Rupaimoole et al showed that low levels of Dicer during
hypoxia conditions were due to upregulation of miR-630 in ovarian
cancer. In addition, using luciferase reporter assays Dicer was
confirmed as target of miR-630. The delivery of miR-630 in
nano-liposomes in an orthotopic mouse model of OC resulted in
increased tumor growth and metastasis (114).
Growth factors are key promoters of tumor cell
motility, invasiveness and metastasis. For instance, Tang et
al demonstrated in vitro that miR-16 suppressed invasion
through downregulation of vascular endothelial growth factor
(VEGF), MMP-2 and BCl-2 genes (115). Another study showed that the
heparin binding epidermal growth factor (HBEGF) was repressed by
miR-212. This miRNA was found downregulated in tumor tissues and
patient's serum with EOC. Overexpression of miR-212 in SKOV3 cells
suppressed cell proliferation, migration and invasion (116). Recently, Li et al
identified a correlation between downregulation of miR-217 with
metastasis in EOC. Ectopic expression of miR-217 in EOC cells
decreased cell proliferation, migration and invasion in
vitro, as well tumor growth in vivo. miR-217 may exert
its antitumor effects by targeting the insulin-like growth factor 1
receptor (IGF1R) (117).
VEGF is able to stimulate the expression of miRNAs
leading to enhanced invasion and metastasis. For instance, Li et
al showed that VEGF induced the expression of oncogene miR-205
in OC cells, and this in turn downregulated Ezrin and Lamin A/C.
miR-205 promoted invasiveness, proliferation and inhibited
apoptosis of OC cells (118). In
addition, the expression levels of VEGF and miR-205 were found
increased in the serum of EOC patients (119).
Some studies showed that overexpression of miR-25
and miR-181b promotes cell migration and invasion of OC cells by
targeting the large tumor suppressor 2 (LATS2). The restoration of
LATS2 expression attenuated the oncogenic effects of miR-25 and
miR-181b (124,125). Other studies showed that miR-21
repressed PTEN in EOC. Moreover, knockdown of miR-21 significantly
inhibited cell proliferation and migration presumably through
inhibition of programmed cell death 4 (PDCD4), because PDCD4 was
correlated negatively with miR-21 expression (126,127). Wang et al identified that
PDCD4 is the target of miR-182. In addition, miR-182 expression
induced migration and invasion whereas its silencing inhibited cell
viability and colony formation in OC cells due to the recovery of
PDCD4 (128).
There is increased evidence that miRNAs play
crucial roles as innovative therapeutic targets in OC. Due to their
extreme stability and its potent activities that allow modulating
relevant gene networks impacting tumor biology, miRNAs has also
been studied as a prospective antitumor approach. The miRNAs can
limit tumor growth and dissemination of tumor cells. These effects
can be induced by inactivating oncogenic miRNAs or restoring the
expression of miRNAs with tumor suppressor functions (134). The potential usefulness of a
miRNA-based therapy has been exploited by different approaches such
as: i) antisense oligonucleotides and synthetic analogues of miRNAs
(antagomirs) to silence oncogenic miRNAs (135,136); and ii) synthetic miRNAs (mimics),
chemically modified oligonucle-otides or adeno-associated
virus-based vector system to restore the expression of
downregulated tumor suppressive miRNAs (137,138). Garzon et al showed that
the effects of upregulated oncomiRs could be suppressed using
antagomirs (139). For instance,
Lu et al using a modified anti-miRNA antisense
olideoxyribonucleotide (AMOs) revealed that the use of this
innovative strategy silenced multiple-target miRNAs. This
technology was used to targets oncogenic miR-21, miR-155, and
miR-17-5 (140). In other report,
Dai et al showed that using a chimera composed by a mucin 1
(MUC1) aptamer targeting the tumor cell surface MUC1 protein and
miR-29b mimics inhibiting DNA methyltransferases, resulted in the
expression of PTEN tumor suppressor. Interestingly, these findings
provide evidence that delivery of miRNAs in a tissue-specific
manner is possible and that it may exert potent antitumor effects
(141). Another example of
application of miRNAs as therapeutic tools is the miR-200 family.
Restoration of miR-200c in combination with paclitaxel treatment
decreased tumor development. miR-200c therapy before conventional
chemotherapy reduces the effective dose of drugs resulting in
increased response (142). Other
miRNAs involved in cellular mechanisms that promote the development
and progression of metastasis in OC have tumor suppressive effects
when delivered in vivo into experimental models. For
instance, miR-200c, miR-506, miR-203, miR-373, miR-138, miR-181a
and miR-155 gained attention by suppressing EMT process and
metastasis. In Tables I and
II the miRNAs that have showed
efficacy in decreasing the metastasis of tumor cells and that
target EMT with potential clinical applications are listed. Such
miRNAs are potential candidates for translational medicine in OC
therapies (143).
The recent discovery of the role of miRNAs as
tumor-suppressor genes and oncogenes has added an additional level
of complexity to the mechanisms leading to tumorigenesis. The
identification of miRNAs involved in invasion and metastasis will
provide insights into molecular mechanisms underlying
tumorigenesis. These miRNAs could be useful as clinical strategies
to customize therapeutic targets that act in spatial and temporal
manner during malignancy progression and metastasis in ovarian
cancer. The updated review presented here, showed that metastamiRs
have emerged as new molecular players to regulate invasion and
metastasis events in ovarian cancer. Some of these miRNAs are
common regulators of cell motility and invasion in distinct types
of cancers while others appear to be cancer specific. Understanding
how metastamiRs are involved in regulating tumor invasion and
metastasis processes will provide data to establish potential
strategies for the development of new metastamiR-based treatments.
However, further investigations and clinical trials challenging the
potential applications of metastamiRs are required in order to use
them as targets for personalized therapies, prognosis and diagnosis
in the near future.
This study was supported by CONACyT (grant nos.
233370 and 222335). The funders had no role in study design, data
collection and analysis, decision to publish, or preparation of the
manuscript.
|
1
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ledermann JA, Marth C, Carey MS, Birrer M,
Bowtell DD, Kaye S, McNeish I, Oza A, Scambia G, Rustin G, et al
Gynecologic Cancer InterGroup: Role of molecular agents and
targeted therapy in clinical trials for women with ovarian cancer.
Int J Gynecol Cancer. 21:763–770. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nguyen DX, Bos PD and Massagué J:
Metastasis: From dissemination to organ-specific colonization. Nat
Rev Cancer. 9:274–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lopez-Camarillo C, Marchat LA,
Arechaga-Ocampo E, Perez-Plasencia C, Del Moral-Hernandez O,
Castaneda-Ortiz EJ and Rodriguez-Cuevas S: MetastamiRs: Non-coding
MicroRNAs driving cancer invasion and metastasis. Int J Mol Sci.
13:1347–1379. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
White NM, Fatoohi E, Metias M, Jung K,
Stephan C and Yousef GM: Metastamirs: A stepping stone towards
improved cancer management. Nat Rev Clin Oncol. 8:75–84. 2011.
View Article : Google Scholar
|
|
7
|
Cai X, Hagedorn CH and Cullen BR: Human
microRNAs are processed from capped, polyadenylated transcripts
that can also function as mRNAs. RNA. 10:1957–1966. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eulalio A, Behm-Ansmant I and Izaurralde
E: P bodies: At the crossroads of post-transcriptional pathways.
Nat Rev Mol Cell Biol. 8:9–22. 2007. View Article : Google Scholar
|
|
11
|
Sen GL and Blau HM: Argonaute 2/RISC
resides in sites of mammalian mRNA decay known as cytoplasmic
bodies. Nat Cell Biol. 7:633–636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
14
|
Nishimura M, Jung E-J, Shah MY, Lu C,
Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, et al:
Therapeutic synergy between microRNA and siRNA in ovarian cancer
treatment. Cancer Discov. 3:1302–1315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Mezencev R, Švajdler M, Benigno BB
and McDonald JF: Ectopic over-expression of miR-429 induces
mesenchymal-to-epithelial transition (MET) and increased drug
sensitivity in meta- stasizing ovarian cancer cells. Gynecol Oncol.
134:96–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Micalizzi DS, Farabaugh SM and Ford HL:
Epithelial-mesenchymal transition in cancer: Parallels between
normal development and tumor progression. J Mammary Gland Biol
Neoplasia. 15:117–134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Corallino S, Malabarba MG, Zobel M, Di
Fiore PP and Scita G: Epithelial-to-mesenchymal plasticity
harnesses endocytic circuitries. Front Oncol. 5:452015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oskarsson T, Batlle E and Massagué J:
Metastatic stem cells: Sources, niches, and vital pathways. Cell
Stem Cell. 14:306–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garg M: Targeting microRNAs in
epithelial-to-mesenchymal transition-induced cancer stem cells:
Therapeutic approaches in cancer. Expert Opin Ther Targets.
19:285–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abba ML, Patil N, Leupold JH and Allgayer
H: MicroRNA regulation of epithelial to mesenchymal transition. J
Clin Med. 5:82016. View Article : Google Scholar :
|
|
22
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heuberger J and Birchmeier W: Interplay of
cadherin-mediated cell adhesion and canonical Wnt signaling. Cold
Spring Harb Perspect Biol. 2:a0029152010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ke Z, Caiping S, Qing Z and Xiaojing W:
Sonic hedgehog-Gli1 signals promote epithelial-mesenchymal
transition in ovarian cancer by mediating PI3K/AKT pathway. Med
Oncol. 32:3682015. View Article : Google Scholar
|
|
26
|
Fang D, Chen H, Zhu JY, Wang W, Teng Y,
Ding HF, Jing Q, Su SB and Huang S: Epithelial-mesenchymal
transition of ovarian cancer cells is sustained by Rac1 through
simultaneous activation of MEK1/2 and Src signaling pathways.
Oncogene Sep. 12:2016Epub ahead of print. View Article : Google Scholar
|
|
27
|
Talbot LJ, Bhattacharya SD and Kuo PC:
Epithelial-mesenchymal transition, the tumor microenvironment, and
metastatic behavior of epithelial malignancies. Int J Biochem Mol
Biol. 3:117–136. 2012.PubMed/NCBI
|
|
28
|
Fuxe J, Vincent T and Garcia de Herreros
A: Transcriptional crosstalk between TGF-β and stem cell pathways
in tumor cell invasion: Role of EMT promoting Smad complexes. Cell
Cycle. 9:2363–2374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moustakas A and Heldin CH: Signaling
networks guiding epithelial-mesenchymal transitions during
embryogenesis and cancer progression. Cancer Sci. 98:1512–1520.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH,
Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y,
Pertsemlidis A, et al: Contextual extracellular cues promote tumor
cell EMT and metastasis by regulating miR-200 family expression.
Genes Dev. 23:2140–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bendoraite A, Knouf EC, Garg KS, Parkin
RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY,
Drescher CW, et al: Regulation of miR-200 family microRNAs and ZEB
transcription factors in ovarian cancer: Evidence supporting a
mesothelial-to-epithelial transition. Gynecol Oncol. 116:117–125.
2010. View Article : Google Scholar :
|
|
35
|
Chen J, Wang L, Matyunina LV, Hill CG and
McDonald JF: Overexpression of miR-429 induces
mesenchymal-to-epithelial transition (MET) in metastatic ovarian
cancer cells. Gynecol Oncol. 121:200–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jabbari N, Reavis AN and McDonald JF:
Sequence variation among members of the miR-200 microRNA family is
correlated with variation in the ability to induce hallmarks of
mesenchymal-epithelial transition in ovarian cancer cells. J
Ovarian Res. 7:122014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu YM, Shang C, Ou YL, Yin D, Li YN, Li X,
Wang N and Zhang SL: miR-200c modulates ovarian cancer cell
metastasis potential by targeting zinc finger E-box-binding
homeobox 2 (ZEB2) expression. Med Oncol. 31:1342014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen D, Zhang Y, Wang J, Chen J, Yang C,
Cai K, Wang X, Shi F and Dou J: MicroRNA-200c overexpression
inhibits tumorige-nicity and metastasis of
CD117+CD44+ ovarian cancer stem cells by
regulating epithelial-mesenchymal transition. J Ovarian Res.
6:502013. View Article : Google Scholar
|
|
39
|
Wang Y, Yan S, Liu X, Zhang W, Li Y, Dong
R, Zhang Q, Yang Q, Yuan C, Shen K, et al: miR-1236-3p represses
the cell migration and invasion abilities by targeting ZEB1 in
high-grade serous ovarian carcinoma. Oncol Rep. 31:1905–1910.
2014.PubMed/NCBI
|
|
40
|
Wu Q, Guo R, Lin M, Zhou B and Wang Y:
MicroRNA-200a inhibits CD133/1+ ovarian cancer stem
cells migration and invasion by targeting E-cadherin repressor
ZEB2. Gynecol Oncol. 122:149–154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vang S, Wu HT, Fischer A, Miller DH,
Maclaughlan S, Douglass E, Comisar L, Steinhoff M, Collins C, Smith
PJ, et al: Identification of ovarian cancer metastatic miRNAs. PloS
One. 8:e582262013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin M, Yang Z, Ye W, Xu H and Hua X:
MicroRNA-150 predicts a favorable prognosis in patients with
epithelial ovarian cancer, and inhibits cell invasion and
metastasis by suppressing transcriptional repressor ZEB1. PloS One.
9:e1039652014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Y, Luo J, Tian R, Sun H and Zou S:
miR-373 negatively regulates methyl-CpG-binding domain protein 2
(MBD2) in hilar cholangiocarcinoma. Dig Dis Sci. 56:1693–1701.
2011. View Article : Google Scholar
|
|
44
|
Oppenheimer H, Kumar A, Meir H, Schwartz
I, Zini A, Haze A, Kandel L, Mattan Y, Liebergall M and
Dvir-Ginzberg M: Set7/9 impacts COL2A1 expression through binding
and repression of SirT1 histone deacetylation. J Bone Miner Res.
29:348–360. 2014. View Article : Google Scholar
|
|
45
|
Zhou J, Xie M, Shi Y, Luo B, Gong G, Li J,
Wang J, Zhao W, Zi Y, Wu X, et al: MicroRNA-153 functions as a
tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer
cells. Oncol Rep. 34:111–120. 2015.PubMed/NCBI
|
|
46
|
Kim YS, Yi BR, Kim NH and Choi KC: Role of
the epithelial-mesenchymal transition and its effects on embryonic
stem cells. Exp Mol Med. 46:e1082014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot
CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al: Integrated
analyses identify a master microRNA regulatory network for the
mesenchymal subtype in serous ovarian cancer. Cancer Cell.
23:186–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun Y, Mezzanzanica D and Zhang W:
MiR-506: A Multitasker in suppression of the
epithelial-to-mesenchymal transition. RNA Dis.
1:e4472014.PubMed/NCBI
|
|
49
|
Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y,
Rupaimoole R, Rodriguez-Aguayo C, Lopez-Berestein G, Ji P, Chen K,
et al: MiR-506 inhibits multiple targets in the
epithelial-to-mesenchymal transition network and is associated with
good prognosis in epithelial ovarian cancer. J Pathol. 235:25–36.
2015. View Article : Google Scholar
|
|
50
|
Zhao G, Guo Y, Chen Z, Wang Y, Yang C,
Dudas A, Du Z, Liu W, Zou Y, Szabo E, et al: miR-203 functions as a
tumor suppressor by inhibiting epithelial to mesenchymal transition
in ovarian cancer. J Cancer Sci Ther. 7:34–43. 2015.
|
|
51
|
Ye Z, Zhao L, Li J, Chen W and Li X:
miR-30d blocked transforming growth factor β1-induced
epithelial-mesenchymal transition by targeting Snail in ovarian
cancer cells. Int J Gynecol Cancer. 25:1574–1581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Siemens H, Jackstadt R, Hünten S, Kaller
M, Menssen A, Götz U and Hermeking H: miR-34 and SNAIL form a
double-negative feedback loop to regulate epithelial-mesenchymal
transitions. Cell Cycle. 10:4256–4271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Su JL, Chen PB, Chen YH, Chen SC, Chang
YW, Jan YH, Cheng X, Hsiao M and Hung MC: Downregulation of
microRNA miR-520h by E1A contributes to anticancer activity. Cancer
Res. 70:5096–5108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu J and Wei JJ: HMGA2 and high-grade
serous ovarian carcinoma. J Mol Med (Berl). 91:1155–1165. 2013.
View Article : Google Scholar
|
|
55
|
Wu J, Liu Z, Shao C, Gong Y, Hernando E,
Lee P, Narita M, Muller W, Liu J and Wei JJ: HMGA2
overexpression-induced ovarian surface epithelial transformation is
mediated through regulation of EMT genes. Cancer Res. 71:349–359.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tanaka T, Arai M, Wu S, Kanda T, Miyauchi
H, Imazeki F, Matsubara H and Yokosuka O: Epigenetic silencing of
microRNA-373 plays an important role in regulating cell
proliferation in colon cancer. Oncol Rep. 26:1329–1335.
2011.PubMed/NCBI
|
|
57
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hsiao CP, Araneta M, Wang XM and Saligan
LN: The association of IFI27 expression and fatigue intensification
during localized radiation therapy: Implication of a
para-inflammatory bystander response. Int J Mol Sci.
14:16943–16957. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng
G, Yin X, Xia H and Tao X: Interferon alpha-inducible protein 27
promotes epithelial-mesenchymal transition and induces ovarian
tumorigenicity and stemness. J Surg Res. 193:255–264. 2015.
View Article : Google Scholar
|
|
60
|
Baskar S, Wiestner A, Wilson WH, Pastan I
and Rader C: Targeting malignant B cells with an immunotoxin
against ROR1. MAbs. 4:349–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tan H, He Q, Gong G, Wang Y, Li J, Wang J,
Zhu D and Wu X: miR-382 inhibits migration and invasion by
targeting ROR1 through regulating EMT in ovarian cancer. Int J
Oncol. 48:181–190. 2016.
|
|
62
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cowden Dahl KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bobbs A, Gellerman K, Hallas WM, Joseph S,
Yang C, Kurkewich J and Cowden Dahl KD: ARID3B directly regulates
ovarian cancer promoting genes. PloS One. 10:e01319612015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang
W, Di W and Qiu L: MicroRNA-7 inhibits tumor metastasis and
reverses epithelial-mesenchymal transition through AKT/ERK1/2
inactivation by targeting EGFR in epithelial ovarian cancer. PloS
One. 9:e967182014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yuan ZQ, Sun M, Feldman RI, Wang G, Ma X,
Jiang C, Coppola D, Nicosia SV and Cheng JQ: Frequent activation of
AKT2 and induction of apoptosis by inhibition of
phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer.
Oncogene. 19:2324–2330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Luo X, Dong Z, Chen Y, Yang L and Lai D:
Enrichment of ovarian cancer stem-like cells is associated with
epithelial to mesenchymal transition through an miRNA-activated AKT
pathway. Cell Prolif. 46:436–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Parikh A, Lee C, Joseph P, Marchini S,
Baccarini A, Kolev V, Romualdi C, Fruscio R, Shah H, Wang F, et al:
microRNA-181a has a critical role in ovarian cancer progression
through the regulation of the epithelial-mesenchymal transition.
Nat Commun. 5:29772014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang Y, Xu C and Fu Y: MicroRNA-17-5p
induces drug resistance and invasion of ovarian carcinoma cells by
targeting PTEN signaling. J Biol Res (Thessalon). 22:122015.
View Article : Google Scholar
|
|
71
|
Qin W, Ren Q, Liu T, Huang Y and Wang J:
MicroRNA-155 is a novel suppressor of ovarian cancer-initiating
cells that targets CLDN1. FEBS Lett. 587:1434–1439. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Habata S, Iwasaki M, Sugio A, Suzuki M,
Tamate M, Satohisa S, Tanaka R and Saito T: BAG3 increases the
invasiveness of uterine corpus carcinoma cells by suppressing
miR-29b and enhancing MMP2 expression. Oncol Rep. 33:2613–2621.
2015.PubMed/NCBI
|
|
74
|
Sun X, Cui M, Zhang A, Tong L, Wang K, Li
K, Wang X, Sun Z and Zhang H: MiR-548c impairs migration and
invasion of endo-metrial and ovarian cancer cells via
downregulation of Twist. J Exp Clin Cancer Res. 35:102016.
View Article : Google Scholar
|
|
75
|
Su N, Qiu H, Chen Y, Yang T, Yan Q and Wan
X: miR-205 promotes tumor proliferation and invasion through
targeting ESRRG in endometrial carcinoma. Oncol Rep. 29:2297–2302.
2013.PubMed/NCBI
|
|
76
|
Mozos A, Catasús L, D'Angelo E, Serrano E,
Espinosa I, Ferrer I, Pons C and Prat J: The FOXO1-miR27 tandem
regulates myometrial invasion in endometrioid endometrial
adenocarcinoma. Hum Pathol. 45:942–951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Dong M, Yang P and Hua F: MiR-191
modulates malignant transformation of endometriosis through
regulating TIMP3. Med Sci Monit. 21:915–920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li S, Hu R, Wang C, Guo F, Li X and Wang
S: miR-22 inhibits proliferation and invasion in estrogen receptor
α-positive endometrial endometrioid carcinomas cells. Mol Med Rep.
9:2393–2399. 2014.PubMed/NCBI
|
|
79
|
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang
Y, Liu X and Wan X: Expression of the tumor suppressor miR-206 is
associated with cellular proliferative inhibition and impairs
invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett.
314:41–53. 2012. View Article : Google Scholar
|
|
80
|
Li S, Li Y, Wen Z, Kong F, Guan X and Liu
W: microRNA-206 overexpression inhibits cellular proliferation and
invasion of estrogen receptor α-positive ovarian cancer cells. Mol
Med Rep. 9:1703–1708. 2014.PubMed/NCBI
|
|
81
|
Liang SH, Li J, Al-beit M, Zhang J, Ma D
and Lu X: Screening and identification of potential miRNA involved
in ovarian cancer invasion and metastasis. Zhonghua Zhong Liu Za
Zhi. 32:650–654. 2010.In Chinese. PubMed/NCBI
|
|
82
|
Li J, Liang S, Yu H, Zhang J, Ma D and Lu
X: An inhibitory effect of miR-22 on cell migration and invasion in
ovarian cancer. Gynecol Oncol. 119:543–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li J, Liang S, Jin H, Xu C, Ma D and Lu X:
Tiam1, negatively regulated by miR-22, miR-183 and miR-31, is
involved in migration, invasion and viability of ovarian cancer
cells. Oncol Rep. 27:1835–1842. 2012.PubMed/NCBI
|
|
84
|
Wen C, Liu X, Ma H, Zhang W and Li H:
miR-338-3p suppresses tumor growth of ovarian epithelial carcinoma
by targeting Runx2. Int J Oncol. 46:2277–2285. 2015.PubMed/NCBI
|
|
85
|
Lin K-T, Yeh Y-M, Chuang C-M, Yang SY,
Chang JW, Sun SP, Wang YS, Chao KC and Wang LH: Glucocorticoids
mediate induction of microRNA-708 to suppress ovarian cancer
metastasis through targeting Rap1B. Nat Commun. 6:59172015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang H, Wang Q, Zhao Q and Di W: MiR-124
inhibits the migration and invasion of ovarian cancer cells by
targeting SphK1. J Ovarian Res. 6:842013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wen Z, Zhao S, Liu S, Liu Y, Li X and Li
S: MicroRNA-148a inhibits migration and invasion of ovarian cancer
cells via targeting sphingosine-1-phosphate receptor 1. Mol Med
Rep. 12:3775–3780. 2015.PubMed/NCBI
|
|
88
|
Chen P, Zeng M, Zhao Y and Fang X:
Upregulation of Limk1 caused by microRNA-138 loss aggravates the
metastasis of ovarian cancer by activation of Limk1/cofilin
signaling. Oncol Rep. 32:2070–2076. 2014.PubMed/NCBI
|
|
89
|
Wang W, Ren F, Wu Q, Jiang D, Li H, Peng
Z, Wang J and Shi H: MicroRNA-497 inhibition of ovarian cancer cell
migration and invasion through targeting of SMAD specific E3
ubiquitin protein ligase 1. Biochem Biophys Res Commun.
449:432–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lee M, Kim EJ and Jeon MJ: MicroRNAs 125a
and 125b inhibit ovarian cancer cells through post-transcriptional
inactivation of EIF4EBP1. Oncotarget. 7:8726–8742. 2016.
|
|
91
|
Meng X, Joosse SA, Müller V, Trillsch F,
Milde-Langosch K, Mahner S, Geffken M, Pantel K and Schwarzenbach
H: Diagnostic and prognostic potential of serum miR-7, miR-16,
miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer
patients. Br J Cancer. 113:1358–1366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Cao J, Cai J, Huang D, Han Q, Yang Q, Li
T, Ding H and Wang Z: miR-335 represents an invasion suppressor
gene in ovarian cancer by targeting Bcl-w. Oncol Rep. 30:701–706.
2013.PubMed/NCBI
|
|
93
|
Imam JS, Plyler JR, Bansal H, Prajapati S,
Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, et
al: Genomic loss of tumor suppressor miRNA-204 promotes cancer cell
migration and invasion by activating AKT/mTOR/Rac1 signaling and
actin reorganization. PloS One. 7:e523972012. View Article : Google Scholar
|
|
94
|
Chung TK, Lau TS, Cheung TH, Yim SF, Lo
KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al: Dysregulation
of microRNA-204 mediates migration and invasion of endometrial
cancer by regulating FOXC1. Int J Cancer. 130:1036–1045. 2012.
View Article : Google Scholar
|
|
95
|
Vimalraj S, Miranda PJ, Ramyakrishna B and
Selvamurugan N: Regulation of breast cancer and bone metastasis by
microRNAs. Dis Markers. 35:369–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tang H, Yao L, Tao X, Yu Y, Chen M, Zhang
R and Xu C: miR-9 functions as a tumor suppressor in ovarian serous
carcinoma by targeting TLN1. Int J Mol Med. 32:381–388.
2013.PubMed/NCBI
|
|
97
|
Ohyagi-Hara C, Sawada K, Kamiura S, Tomita
Y, Isobe A, Hashimoto K, Kinose Y, Mabuchi S, Hisamatsu T,
Takahashi T, et al: miR-92a inhibits peritoneal dissemination of
ovarian cancer cells by inhibiting integrin α5 expression. Am J
Pathol. 182:1876–1889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Doberstein K, Bretz NP, Schirmer U, Fiegl
H, Blaheta R, Breunig C, Müller-Holzner E, Reimer D, Zeimet AG and
Altevogt P: miR-21-3p is a positive regulator of L1CAM in several
human carcinomas. Cancer Lett. 354:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumorigenesis and progression. Cancer Lett. 362:122–130.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chen X, Chen S, Xiu Y-L, Sun K-X, Zong Z-H
and Zhao Y: RhoC is a major target of microRNA-93-5P in epithelial
ovarian carcinoma tumorigenesis and progression. Mol Cancer.
14:312015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu T, Qin W, Hou L and Huang Y:
MicroRNA-17 promotes normal ovarian cancer cells to cancer stem
cells development via suppression of the LKB1-p53-p21/WAF1 pathway.
Tumour Biol. 36:1881–1893. 2015. View Article : Google Scholar
|
|
102
|
Corney DC, Hwang CI, Matoso A, Vogt M,
Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH,
Hermeking H, et al: Frequent downregulation of miR-34 family in
human ovarian cancers. Clin Cancer Res. 16:1119–1128. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hwang C-I, Choi J, Zhou Z, Flesken-Nikitin
A, Tarakhovsky A and Nikitin AY: MET-dependent cancer invasion may
be preprogrammed by early alterations of p53-regulated feedforward
loop and triggered by stromal cell-derived HGF. Cell Cycle.
10:3834–3840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hwang C-I, Matoso A, Corney DC,
Flesken-Nikitin A, Körner S, Wang W, Boccaccio C, Thorgeirsson SS,
Comoglio PM, Hermeking H, et al: Wild-type p53 controls cell
motility and invasion by dual regulation of MET expression. Proc
Natl Acad Sci USA. 108:14240–14245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li R, Shi X, Ling F, Wang C, Liu J, Wang W
and Li M: MiR-34a suppresses ovarian cancer proliferation and
motility by targeting AXL. Tumour Biol. 36:7277–7283. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wu H, Xiao Z, Wang K, Liu W and Hao Q:
MiR-145 is downreg-ulated in human ovarian cancer and modulates
cell growth and invasion by targeting p70S6K1 and MUC1. Biochem
Biophys Res Commun. 441:693–700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ahmed AA, Etemadmoghadam D, Temple J,
Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, Defazio
A, et al: Driver mutations in TP53 are ubiquitous in high grade
serous carcinoma of the ovary. J Pathol. 221:49–56. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen X, Dong C, Law PT, Chan MT, Su Z,
Wang S, Wu WK and Xu H: MicroRNA-145 targets TRIM2 and exerts
tumor-suppressing functions in epithelial ovarian cancer. Gynecol
Oncol. 139:513–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kim TH, Song JY, Park H, Jeong JY, Kwon
AY, Heo JH, Kang H, Kim G and An HJ: miR-145, targeting
high-mobility group A2, is a powerful predictor of patient outcome
in ovarian carcinoma. Cancer Lett. 356B:937–945. 2015. View Article : Google Scholar
|
|
111
|
Yan L, Zhou J, Gao Y, Ghazal S, Lu L,
Bellone S, Yang Y, Liu N, Zhao X, Santin AD, et al: Regulation of
tumor cell migration and invasion by the H19/let-7 axis is
antagonized by metformin-induced DNA methylation. Oncogene.
34:3076–3084. 2015. View Article : Google Scholar
|
|
112
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar
|
|
113
|
Guo Y, Tian P, Yang C, Liang Z, Li M, Sims
M, Lu L, Zhang Z, Li H, Pfeffer LM, et al: Silencing the
double-stranded RNA binding protein DGCR8 inhibits ovarian cancer
cell proliferation, migration, and invasion. Pharm Res. 32:769–778.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Rupaimoole R, Ivan C, Yang D, Gharpure KM,
Wu SY, Pecot CV, Previs RA, Nagaraja AS, Armaiz-Pena GN, McGuire M,
et al: Hypoxia-upregulated microRNA-630 targets Dicer, leading to
increased tumor progression. Oncogene. 35:4312–4320. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tang R, Cui ZM and Lou YH: MicroRNA-16
regulates the proliferation, invasion and apoptosis of ovarian
epithelial carcinoma cells in vitro. Zhonghua Fu Chan Ke Za Zhi.
47:846–850. 2012.In Chinese.
|
|
116
|
Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y
and She MC: MiR-212 exerts suppressive effect on SKOV3 ovarian
cancer cells through targeting HBEGF. Tumour Biol. 35:12427–12434.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Li J, Li D and Zhang W: Tumor suppressor
role of miR-217 in human epithelial ovarian cancer by targeting
IGF1R. Oncol Rep. 35:1671–1679. 2016.
|
|
118
|
Li J, Li L, Li Z, Gong G, Chen P, Liu H,
Wang J, Liu Y and Wu X: The role of miR-205 in the VEGF-mediated
promotion of human ovarian cancer cell invasion. Gynecol Oncol.
137:125–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhou J, Liu H, Chen Y, Wen J, Li L and Wu
X: Expression and significance of VEGF, miR-205 and target protein
Ezrin and Lamin A/C in ovarian cancer. Zhong Nan Da Xue Xue Bao Yi
Xue Ban. 39:142–150. 2014.0 (In Chinese). PubMed/NCBI
|
|
120
|
Fu X, Cui Y, Yang S, Xu Y and Zhang Z:
MicroRNA-613 inhibited ovarian cancer cell proliferation and
invasion by regulating KRAS. Tumour Biol. 37:6477–6483. 2016.
View Article : Google Scholar
|
|
121
|
Zhang L, Li Z, Gai F and Wang Y:
MicroRNA-137 suppresses tumor growth in epithelial ovarian cancer
in vitro and in vivo. Mol Med Rep. 12:3107–3114. 2015.PubMed/NCBI
|
|
122
|
Shi C and Zhang Z: miR-761 inhibits tumor
progression by targeting MSI1 in ovarian carcinoma. Tumour Biol.
37:5437–5443. 2016. View Article : Google Scholar
|
|
123
|
Yao L, Wang L, Li F, Gao X, Wei X and Liu
Z: MiR181c inhibits ovarian cancer metastasis and progression by
targeting PRKCD expression. Int J Clin Exp Med. 8:15198–15205.
2015.PubMed/NCBI
|
|
124
|
Feng S, Pan W, Jin Y and Zheng J: MiR-25
promotes ovarian cancer proliferation and motility by targeting
LATS2. Tumour Biol. 35:12339–12344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Lou Y, Yang X, Wang F, Cui Z and Huang Y:
MicroRNA-21 promotes the cell proliferation, invasion and migration
abilities in ovarian epithelial carcinomas through inhibiting the
expression of PTEN protein. Int J Mol Med. 26:819–827. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lou Y, Cui Z, Wang F, Yang X and Qian J:
miR-21 down-regulation promotes apoptosis and inhibits invasion and
migration abilities of OVCAR3 cells. Clin Invest Med.
34:E2812011.PubMed/NCBI
|
|
128
|
Wang YQ, Guo RD, Guo RM, Sheng W and Yin
LR: MicroRNA-182 promotes cell growth, invasion, and
chemoresis-tance by targeting programmed cell death 4 (PDCD4) in
human ovarian carcinomas. J Cell Biochem. 114:1464–1473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Xu X, Ayub B, Liu Z, Serna VA, Qiang W,
Liu Y, Hernando E, Zabludoff S, Kurita T, Kong B, et al:
Anti-miR182 reduces ovarian cancer burden, invasion, and
metastasis: An in vivo study in orthotopic xenografts of nude mice.
Mol Cancer Ther. 13:1729–1739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Liu Z, Liu J, Segura MF, Shao C, Lee P,
Gong Y, Hernando E and Wei JJ: MiR-182 overexpression in
tumourigenesis of high-grade serous ovarian carcinoma. J Pathol.
228:204–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Nakayama I, Shibazaki M, Yashima-Abo A,
Miura F, Sugiyama T, Masuda T and Maesawa C: Loss of HOXD10
expression induced by upregulation of miR-10b accelerates the
migration and invasion activities of ovarian cancer cells. Int J
Oncol. 43:63–71. 2013.PubMed/NCBI
|
|
132
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin (Shanghai). 42:318–324. 2010. View Article : Google Scholar
|
|
133
|
Zou D, Wang D, Li R, Tang Y, Yuan L, Long
X and Zhou Q: MiR-197 induces Taxol resistance in human ovarian
cancer cells by regulating NLK. Tumour Biol. 36:6725–6732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Taylor MA and Schiemann WP: Therapeutic
opportunities for targeting microRNAs in cancer. Mol Cell Ther.
2:1–13. 2014. View Article : Google Scholar
|
|
135
|
Krützfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with 'antagomirs'. Nature. 438:685–689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Elmén J, Lindow M, Silahtaroglu A, Bak M,
Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF,
Straarup EM, et al: Antagonism of microRNA-122 in mice by
systemically administered LNA-antimiR leads to up-regulation of a
large set of predicted target mRNAs in the liver. Nucleic Acids
Res. 36:1153–1162. 2008. View Article : Google Scholar :
|
|
137
|
Hutvágner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Kota J, Chivukula RR, O'Donnell KA,
Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P,
Torbenson M, Clark KR, et al: Therapeutic microRNA delivery
suppresses tumorigenesis in a murine liver cancer model. Cell.
137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z
and Yang B: A single anti-microRNA antisense
oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers
an improved approach for microRNA interference. Nucleic Acids Res.
37:e242009. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Dai F, Zhang Y, Zhu X, Shan N and Chen Y:
Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial
ovarian carcinoma cells through regulation of PTEN methylation.
Target Oncol. 7:217–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Cittelly DM, Dimitrova I, Howe EN,
Cochrane DR, Jean A, Spoelstra NS, Post MD, Lu X, Broaddus RR,
Spillman MA, et al: Restoration of miR-200c to ovarian cancer
reduces tumor burden and increases sensitivity to paclitaxel. Mol
Cancer Ther. 11:2556–2565. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Gambari R, Brognara E, Spandidos DA and
Fabbri E: Targeting oncomiRNAs and mimicking tumor suppressor
miRNAs: New trends in the development of miRNA therapeutic
strategies in oncology (Review). Int J Oncol. 49:5–32.
2016.PubMed/NCBI
|