Introduction
Lung cancer has become the most common cause of
cancer-related mortality and a serious public health concern
worldwide. In China, the lung cancer-related mortality rate has
been multiplied by 4.6 in the past 3 decades. It will reach the
alarming 1 million per year by 2025, as estimated by WHO (1). Among all lung cancers, 85% are NSCLC
(non-small cell lung cancer). At the moment of diagnosis, most of
NSCLC patients present with locally advanced or metastatic disease,
when the surgical intervention is no longer feasible despite its
high success rate if applied earlier. For these patients, systemic
chemotherapy with concurrent radiotherapy remains a necessary
option for cure. For this reason, the identification of the
proteins playing key roles in the physiopathology of NSCLC that
could be potential targets for the chemotherapy reveals to be
important and emergent.
The flightless I protein (FLII) belongs to the
gelsolin family of actin severing proteins. At the time of its
discovery, Drosophila melanogaster FLII was shown to play an
important role in the embryonic development. The following studies
on mammalian FLII demonstrated that this protein is also involved
in the regulation of wound repair, skin barrier development, the
recovery after blistering and regulation of immune response
(2–9). Recently, studies have also
demonstrated that FLII is involved in colorectal cancer,
hepatocellular and prostate cancer (10–13).
Notably, a role of tumor suppressor in prostate cancer has been
attributed to this protein (14).
Despite the previous attempts of identifying the
FLII interactome in the cells using yeast-two hybrid system
(15,16), the systemic picture of protein
interaction network of FLII still remains elusive. In this study,
by combining co-immunoprecipitation and mass spectrometry analysis,
we have identified 132 putative FLII interactors in lung
adenocarcinoma cancer H1299 cells, and found that more than a half
of them are proteins involved in RNA post-transcriptional
regulation and protein biosynthesis. By combining cell
fractionation, mRNA-seq and translating mRNA sequencing (RNC-seq),
we evaluated the function of FLII on the transcription, nuclear
export and translation of mRNAs. We demonstrated that FLII affects
the overall nuclear export and translation of mRNAs.
Materials and methods
Antibodies
Rabbit polyclonal anti-FLII (sc-30046) antibody,
mouse polyclonal anti-HNRNPQ (sc-56703) antibody, goat polyclonal
anti-TIAL1 (sc-1749) antibody mouse monoclonal anti-actin
(sc-47778), mouse monoclonal anti-NuP88 (sc-136009) antibody,
peroxidase-conjugated AffiniPure goat anti-Mouse IgG (H+L),
peroxidase-conjugated AffiniPure goat anti-rabbit IgG (H+L), rabbit
anti-goat IgG (H+L)-HRP (sc-2768) were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA).
Cell culture
Human H1299 and HBE cells (Cell Resource Center,
Institute of Life Science Chinese Academy of Sciences, Shanghai,
China) were cultured in Dulbecco's modified Eagle's medium (DMEM,
Gibco BRL, Grand island, NY, USA) supplemented with 10% fetal
bovine serum (PAA Laboratories, Weike Biochemical Reagent,
Shanghai, China), 1% penicillin/streptomycin (Genom, Hangzhou,
China) at 37°C in a humidified atmosphere containing 5%
CO2.
Plasmid constructions
The pot-b7-FLII plasmid containing the cDNA of the
human FLII was obtained from Yingrun Biotechnologies Inc.
(Changsha, China). The cDNA fragment encoding FLII was
PCR-amplified and inserted into pCMV-N-FLAg-vector using the EcoRV
and SpeI restriction sites. A primer pair (upstream:
5′-ATTGATATCATGGAGGCCACCGGGGTGCTG-3′ and downstream:
5′-ATAACTAGTTTAGGCCAGGGCCTTGCAGAA-3′) was designed based on NCBI
GenBank NP_002009.1. All the plasmids were accuratly confirmed by
DNA sequencing.
Transfections and immunoprecipitation of
FLII
Cells were transfected with either empty pCMV-N-Flag
or pCMV-N-Flag-FLII vectors using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA). At 48 h after transfection, cells were lysed
using EBC lysis buffer. Total soluble extract (1 mg) was incubated
with anti-Flag antibody overnight at 4°C. Protein A/G-Sepharose was
added and incubated for 4 h at 4°C. After washing, the precipitated
proteins were eluted and separated using SDS-PAGE. The gel was
stained with Silver nitrate after migration, and gel lanes were
subsequently cut, destained, reduced, alkylated and digested with
gold-trypsin at 37°C overnight. The tryptic peptides were
extracted, and the peptide mixtures were concentrated by SpeedVac
centrifuge to dryness and re-dissolved with 2% ACN in 0.1% formic
acid before LC-MS/MS analysis. For the confirmation of the presence
of Flag-FLII in the immune-complexes, the immunoprecipitates were
washed with immunoprecipitation assay buffer three times, and
subjected to western blot analysis using anti-Flag antibody.
MS analysis
The peptide mixtures were analyzed by reverse-phase
liquid chromatography coupled with LTQ-Orbitrap mass spectrometer
(Thermo Electron, Bremen, Germany) as previously described with
minor modification (17). Briefly,
the peptide mixtures were firstly loaded on a C18 reverse-phase
column (100-mm i.d., 10-cm long, 5-mm resin from Michrom
Bioresources, Auburn, CA, USA) using an auto sampler. The peptide
mixtures were eluted with 0–40% gradient buffer solution (Buffer A,
0.1% formic acid, and 5% ACN; Buffer B, 0.1% formic acid and 95%
ACN) over 180 min. The eluate was then analyzed online in the
LTQ-Orbitrap mass spectrometer operated in a data-dependent mode
with capillary temperature of 2001 C and spray voltage of 1.80 kV.
A full MS scan with m/z 350–1800 was carried out in the Orbitrap at
resolution r5100,000 at m/z 400, and followed by five MS2 scans in
the LTQ with Dynamic Exclusion setting: 2 repeat counts with repeat
duration of 30 sec and exclusion duration of 90 sec. MS3 was
further performed if an ion had a neutral loss of −98.00, −58.00,
−49.00, −38.67, −32.67 or −24.50 Da in the MS2 and the ion was one
of the top five most intense ions in the MS2. Conditions with 35%
normalized collision energy, activation q of 0.25 and activation
time of 30 msec were applied for MS2 and MS3 acquisitions.
Stable FLII knockdown in HBE cells
Lentiviral shRNA vectors (GenePharma, Shanghai,
China) targeting human FLII were utilized for stable knockdown in
HBE cells. Procedures were conducted according to the
manufacturer's protocol. The FLII siRNA sense sequence
(5′-GGGCTAGACATCTACGTAT-3′) and Scramble siRNA sense sequence
(5′-TTCTCCGAACGTGTCACGTTTC-3′) were used to design the shRNA that
were inserted into PGLV3/H1/GFP+ pur Vector under H1 promoter.
Cells resistant to puromycin (1.5 µg/ml) were selected and
passaged for further study.
Cell fractionation
Six million human H1299 or HBE cells were
resuspended in 500 µl ice cold Extraction Buffer (10 mM
Tris-HCl, 10 mMKCl, 5 mM MgCl2, protease and phosphatase
inhibitors, RNase inhibitor, VRC) and incubated on ice for 10 min,
500 µl 0.6% Triton/Extraction Buffer was then added,
followed by 10 sec of vortex, and incubation on ice for further 10
min, then vortex for 10 sec again. Lysate was then subjected to
vertical centrifugation for 5 min at 600 × g speed at RT.
Supernatant corresponding to the cytoplasmic fraction was
transferred into a new tube. The pellet corresponding to the cell
nucleus was subsequently resuspended in 1 ml 0.6% Triton Extraction
Buffer by pipetting 10 times, incubated on ice for 5 min, followed
by vertical centrifugation for 5 min at 600 × g speed at RT, The
supernatant corresponded to nuclear extract was then collected.
This experiment was repeated thrice, and the cytoplasmic or nuclear
extract resulted from the three experiments were pooled together,
and served for the RNA extraction using Trizol reagent
(Invitrogen).
Ribosome-nascent chain complex (RNC)
extraction
The RNC extraction was performed as described by
Esposito et al (18) with
modifications. In brief, cells were pre-treated with 100 mg/ml
cycloheximide for 15 min, followed by pre-chilled phosphate
buffered saline washes and addition of 2 ml cell lysis buffer [1%
Triton X-100 in ribosome buffer (RB buffer) 20 mM HEPES-KOH (pH
7.4), 15 mM MgCl2, 200 mMKCl, 100 mg/ml cycloheximide
and 2 mM dithiothreitol]. After 30-min ice-bath, cell lysates were
scraped and transferred to pre-chilled 1.5 ml tubes. Cell debris
was removed by centrifuging at 13, 200 × g for 10 min at 4°C.
Supernatants were transferred on the surface of 20 ml of sucrose
buffer (30% sucrose in RB buffer). RNCs were pelleted after
ultra-centrifugation at 185,000 × g for 5 h at 4°C. RNC-RNA was
purified using TRIzol method.
Next-generation sequencing and sequence
analysis
Total RNA and RNC-RNA of human H1299 and HBE cells
were extracted as previously described (18). Equal amounts of total mRNA or
RNC-mRNA isolated from three independent cultures were pooled and
reverse-transcribed into cDNA. The yielded cDNA library was then
constructed and subjected to RNA-seq analysis using Illumina HiSeq™
2000. The sequencing data sets are deposited in Gene Expression
Omnibus database under the accession number of GSE92979. Reads were
mapped to human mRNA reference sequence (RefSeq) for GRCh37/hg19 in
UCSC genome browser (downloaded from http://hgdownload.cse.ucsc.edu/downloads, accessed
January 21, 2013) using FANSe2 mapping algorithm with the options
-L78 -S8 -I0 -E9 -B1. The reads mapped to splice variants of one
gene were summed. The mRNA abundance was normalized using both rpkM
(reads per kilobase per million reads). Differential expression was
evaluated using edgeR since it outperforms the other mainstream
differential expression models (19). Genes with >10 mapped reads were
considered as quantified genes. The cytoplasm/nucleus distribution
of each given mRNA has been quantified in all experimental groups
of cells, and the variation of such distribution between the FLII
knocked down HBE or the FLII overexpressing H1299 cells and their
respective control cells were calculated. Where Flag-FLII and
Flag-Vector refer to H1299 cells transfected with Flag-FLII and
Flag vector; shRNA-FLII and shRNA-Ctrl refer to HBE cells
expressing the shRNA against FLII and the control shRNA. 'C' for
cytoplasmic, and 'N' for nuclear.
Reverse transcription and PCR
Total RNA or RNC-RNA, isolated from both H1299 and
HBE cells, were reverse transcribed to cDNA with poly-dT primer
using Reverse Transcriptase XL (AMV) (Takara, Foster City, CA,
USA), by following the manufacturer's instructions. The
quantitative real-time PCR (qPCR) was then performed with
gene-specific primers and the SsoFast™EvaGreen Supermix (Bio-Rad,
Hercules, CA, USA) on a Bio-Rad MiniOpticon real-time PCR system
(Bio-Rad) by following the manufacturer's instructions. The
specificity of the primers was verified by both in Silico
Computation (NCBI Primer-BLAST) and melting curve measurement after
the qPCR amplification.
Computational analyses
Protein interaction network was built by using
STRING-DB (string-db.org) with default settings
(all interaction sources selected, 'medium confidence (0.400)' for
minimum required interaction score, and 'query protein only' for
max number of interactors to show). Ingenuity pathway analysis
(IPA) was performed as described previously with minor
modifications. Briefly, target proteins (DEPs) were uploaded into
www.ingenuity.com (Ingenuity Systems, Inc.,
Redwood City, CA, USA). Core analyses were performed to identify
top canonical pathways, bioprocesses and effects on functions.
Cell migration and invasion
In vitro migration and invasion assays were
performed using Boyden chambers as previously described with minor
modifications (20). In brief, for
migration assays, cells transfected with pCMV-N-Flag-FLII or
pCMV-N-Flag plasmid were resuspended in serum-free medium and
seeded in the upper transwell chamber (8.0 mM pore size, Corning),
and medium supplemented with 10% FCS was added to the bottom
chamber, and cultured under regular conditions for 6 h, cells on
the upper surface of filters were then removed and those on the
under-surface were stained with 5% crystal violet. Images were
captured from each membrane and the number of migrated cells was
counted under a microscope. For invasion assays, similar transwell
chambers coated with Matrigel (8.0 mM pore size, Chemicon) were
used to analyze the invasive potential regulated by FLII.
Statistics
The Spearman correlation coefficients were
calculated to determine bivariate relationships. The regression,
data distribution and standard deviations were calculated by using
MATLAB R2012a software package (MathWorks, Natick, MA, USA). Data
are shown as mean ± standard deviation. Statistical difference was
accepted at P<0.001.
Results
FLII is downregulated in lung carcinoma
cells
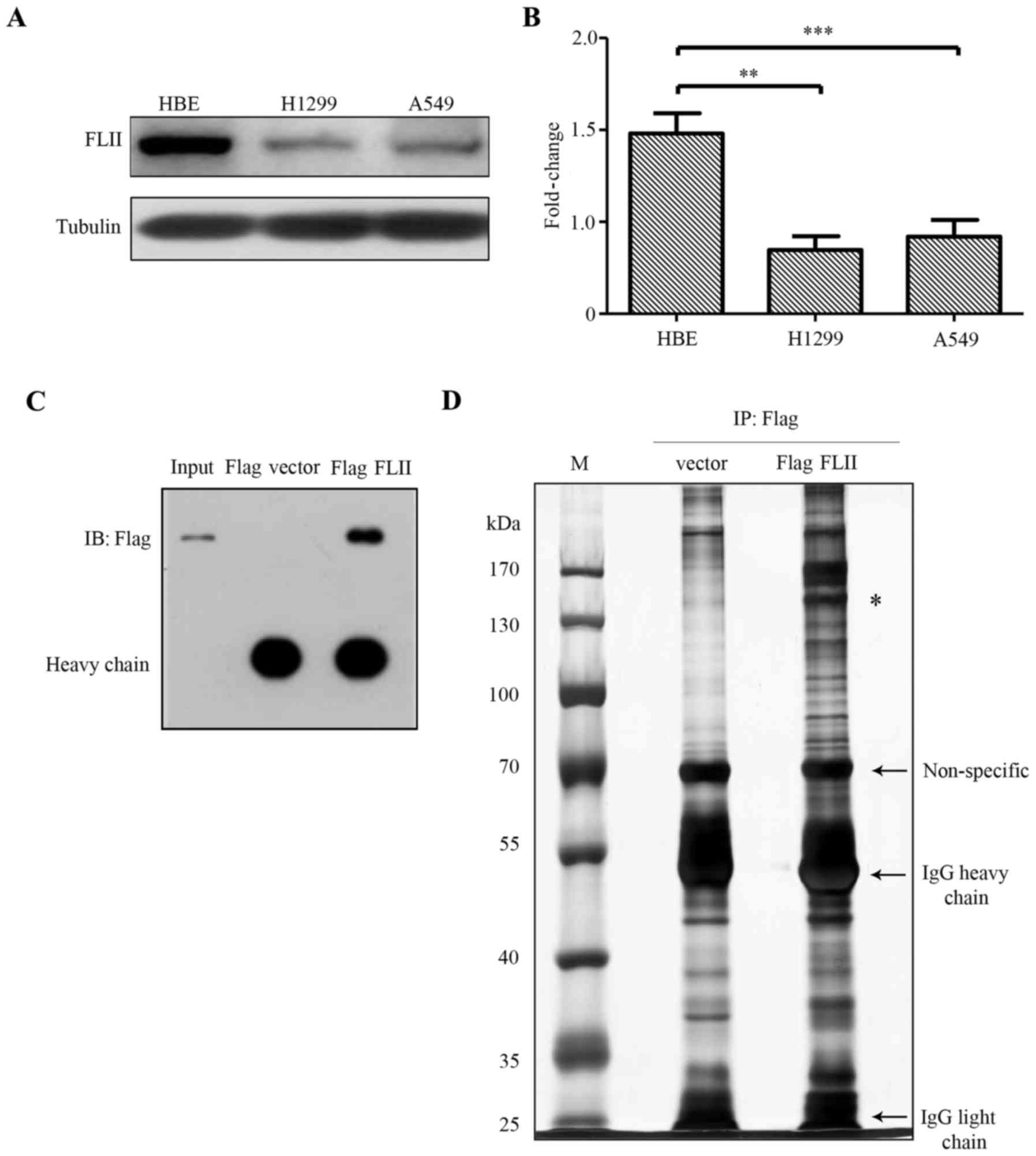
In order to assess the possible role of FLII in the
physiopathology of NSCLC, we firstly examined its expression
pattern in two lung adenocarcinoma epithelial cell lines, namely
H1299 and A549 cell lines. Of note, we observed that FLII was
significantly downregulated in A549 and H1299 lung carcinoma cells,
as compared with the human bronchial epithelial (HBE) cells
(Fig. 1A and B). This result is
consistent with the recent study reporting that FLII is a tumor
suppressor (14).
Identification of FLII interactome in
H1299 lung carcinoma cells
We then tried to identify the possible interactors
of FLII in H1299 cells. Immunoprecipitation experiment was carried
out using anti-Flag antibody in Flag-FLII transfected H1299 cells
as described in Materials and methods. The immunoprecipitation of
Flag-FLII was then confirmed by western blotting using an anti-Flag
antibody with one tenth of the immunoprecipitates from the
Flag-FLII transfected and the empty Flag vector transfected cells
(Fig. 1C). The remaining
immunoprecipitates were run on an SDS-PAGE gel that was
subsequently silver-stained (Fig.
1D). The protein bands of both Flag-FLII transfected and the
negative control lanes were extracted for tryptic in-gel digestion.
Digested peptides were then subjected to LC-MS/MS analysis for
protein identification. Proteins (263) were identified in Flag-FLII
transfected lane. One hundred and thirty of them were also found in
the negative control lane, thus considered to be unspecific and
excluded from the candidates for further analyses. Among the 133
remaining proteins, we found FLII protein with 15 unique peptides
specifically counted, further confirming the effectiveness of the
immunoprecipitation. The other 132 proteins were considered to be
the putative FLII-interacting partners.
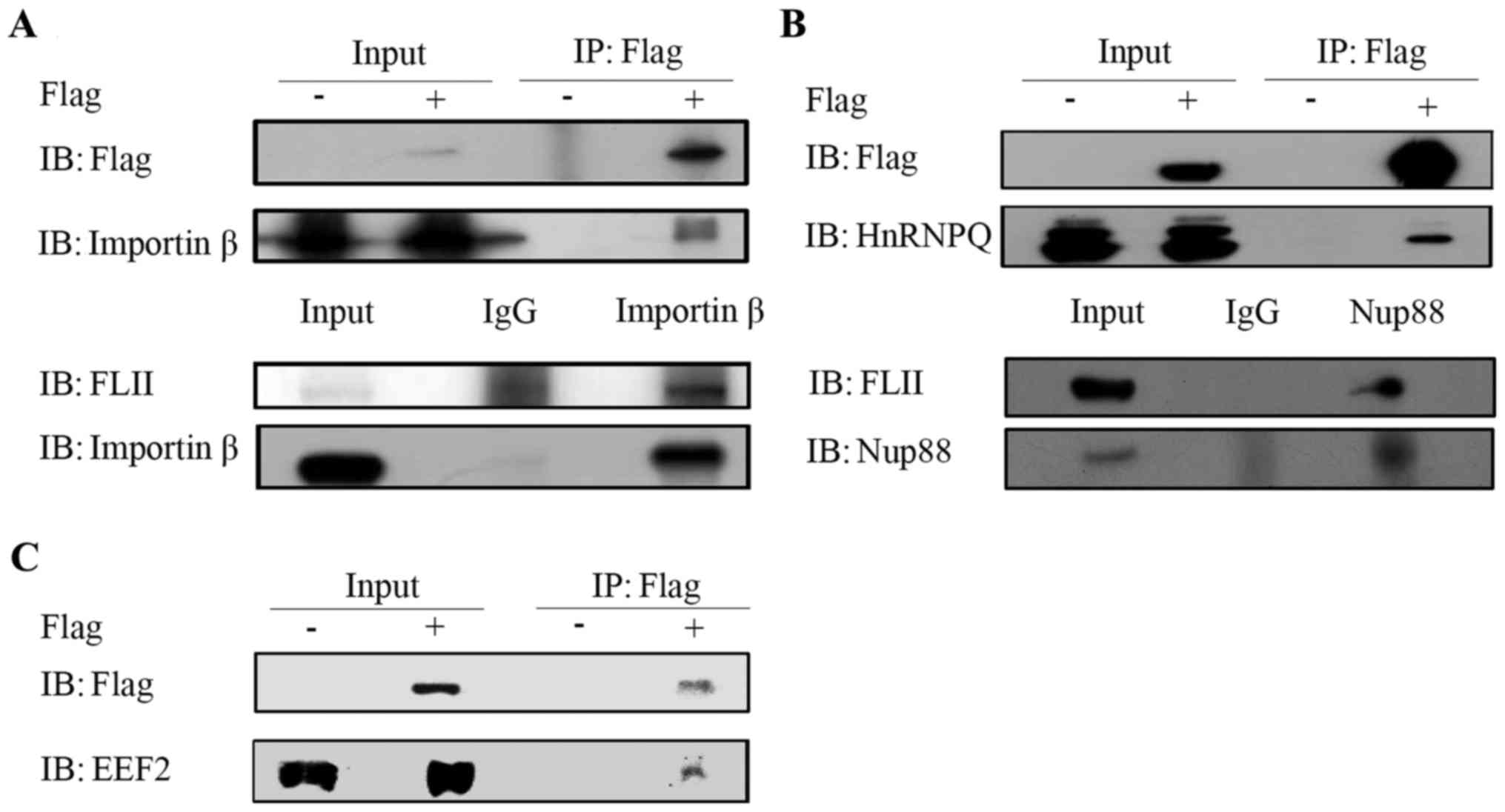
Validation of interactions between FLII
and the representative interacting partner
In order to confirm the interactions of FLII with
its putative partners identified by mass spectrometry, we carried
out experiments to confirm the co-immunoprecipitation of FLII with
its several representative partners using corresponding specific
antibodies, as shown in Fig. 2 for
Importin β (karyopherin β), Nup88, Syncrip and EEF2. Due to the
variable quality or suitability of the antibodies for
co-immunoprecipitation assays, the experiments were performed
either with endogenous or overexpressed Flag-tagged FLII as
indicated when appropriate. Fig.
2A shows the interaction of the overexpressed FLII with
Importin β using the anti-Flag antibody in Flag-FLII transfected
H1299 cells (upper panel) and the interaction between endogenous
FLII and Importin β in H1299 cells (lower panel). The interaction
of FLII with Syncrip and Nup88 has been confirmed, respectively, by
using the anti-Flag antibody in Flag-FLII transfected H1299 cells
(Fig. 2B, upper panel) and by
using the Nup88 antibody in H1299 cells (Fig. 2B, lower panel). As shown in
Fig. 2C, we confirmed the
interaction between FLII and EEF2 in Flag-FLII transfected H1299
cells. Among these confirmed representative partners, Importin β
and Nup88 are, respectively, important nuclear transport receptor
and component of the nuclear pore complex (NPC) involved in the
nucleocytoplasmic transport. Particularly, both of them have been
reported to be involved in the nuclear export of mRNA, protein and
60S ribosomal complex (21–23).
Syncrip is an RNA-binding protein involved in RNA metabolism, such
as RNA stability, splicing, and translational control (24–27).
EEF2 (eukaryotic elongation factor 2), as its name indicates, is an
essential factor for protein synthesis, promoting the GTP-dependent
translocation of the nascent peptide from the A-site to the P-site
of ribosome during the translational process (28).
Construction and functional enrichment
analysis of the protein interaction network involving FLII and its
interactors
The biological functions of all putative FLII
binders were then categorized by consulting the genecards
databases, we found that more than a half (74 out of the 132 total)
of the putative FLII interactors are associated with RNA
post-transcriptional modifications, RNA and protein
nucleocytoplasmic transport and protein biosynthesis (Table I). To further assess the relevance
of this result and to identify the biological processes and
signaling pathways involving FLII and its interacting proteins, we
used STRING program to build a protein interaction network
comprising these putative interactors (not shown due to the
impossibility of exporting a figure which could meet the
publication standard). Among the total 133 proteins analyzed
(nodes), 115 are connected with at least one other partner by at
least one kind of interactions (edges). An important number of
nodes are connected with its partner(s) by multiple edges. For the
133 nodes analyzed, the number of edges was 305, significantly
superior to the expected value that is 164, resulting in an average
node degree of 4.59, a clustering coefficient of 0.539, and a PPI
enrichment P-value of 0. These network statistics clearly
demonstrate that this protein interaction network has significantly
more interactions than expected, and is therefore statistically
reliable and functionally relevant. The functional enrichments in
this network were determined according to biological process,
molecular function, cellular component, KEGG pathways and PFAM
protein domains (Table II). As
expected, the results clearly indicate that the interactome of FLII
is associated with RNA post-transcriptional regulations and
functions, with the top ranked biological process being the gene
expression, and the top molecular functions being the RNA/nucleic
acid binding, supported by the top ranked KEGG pathways as RNA
transport, spliceosome, protein processing and mRNA surveillance
(Table II). These results, which
depict a comprehensive interactome of FLII in H1299 cells, suggest
strongly that FLII might play a role in the post-transcriptional
regulation and function of RNA through interactions with the
RNA-binding proteins and nucleoporins.
 | Table IFlightless I-interacting proteins
associated with RNA post-transcriptional regulation. |
Table I
Flightless I-interacting proteins
associated with RNA post-transcriptional regulation.
| Protein IDs | Protein names | Gene names | Function |
|---|
| IPI00893918 | Valyl-tRNA
synthetase | VARS | Protein
translation |
| IPI00001639 | Karyopherin subunit
β-1 | KPNB1 | Protein import into
nucleus |
| IPI00001738 | Nuclear pore
complex protein Nup88 | NUP88 | RNA
translocation |
| IPI00877174 |
2′-5′-oligoadenylate synthase 3 | OAS3 | RNA binding |
| IPI00003704 | RNA-binding protein
4 | RBM4 | RNA binding |
| IPI00644708 | Nucleolysin
TIAR | TIAL1 | RNA binding |
| IPI00005675 | NF-κ-B-repressing
factor | NKRF | RNA binding |
| IPI00218187 |
Serine/threonine-protein phosphatase PP1-γ
catalytic subunit | PPP1CC | RNA binding |
| IPI00006025 | Squamous cell
carcinoma antigen recognized by T-cells 3 | SART3 | mRNA splicing |
| IPI00337397 | Nuclear pore
complex protein Nup98 | NUP98 | mRNA export from
nucleus |
| IPI00007818 | Cleavage and
polyadenylation specificity factor subunit 3 | CPSF3 | mRNA export from
nucleus |
| IPI00179713 | Insulin-like growth
factor 2 mRNA-binding protein 2 | IMP2 | mRNA transport |
| IPI00010200 | Probable
ATP-dependent RNA helicase YTHDC2 | YTHDC2 | RNA processing |
| IPI00010700 | Large proline-rich
protein BAT2 | BAT2 | RNA binding |
| IPI00873899 | ATP-binding
cassette sub-family F member 1 | ABCF1 | Protein
translation |
| IPI00013877 | Heterogeneous
nuclear ribonucleoprotein H3 | HNRNPH3 | RNA splicing, RNA
processing |
| IPI00014474 | A-kinase anchor
protein 8 | AKAP8 | RNA binding |
| IPI00015952 | Eukaryotic
translation initiation factor 4γ | EIF4G2 | Translational
initiation |
| IPI00016910 | Eukaryotic
translation initiation factor 3 subunit C | EIF3C | Translational
initiation |
| IPI00017669 | SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin subfamily
E member 1 | BAF57 | RNA binding |
| IPI00018120 | 28S ribosomal
protein S29 | DAP3 | RNA binding |
| IPI00018140 | Heterogeneous
nuclear ribonucleoprotein Q | HNRPQ
(Syncrip) | RNA splicing, RNA
processing, RNA binding |
| IPI00018522 | Protein arginine
N-methyltransferase 1 | PRMT1 | RNA binding |
| IPI00019380 | Nuclear cap-binding
protein subunit 1 | NCBP1 | mRNA export from
nucleus, RNA splicing, RNA binding |
| IPI00021435 | 26S protease
regulatory subunit 7 | PSMC2 | Regulation of mRNA
stability |
| IPI00926977 | 26S protease
regulatory subunit S10B | PSMC6 | Regulation of mRNA
stability |
| IPI00025491 | Eukaryotic
initiation factor 4A-I | EIF4A1 | RNA binding,
translational initiation |
| IPI00026665 | Glutaminyl-tRNA
synthetase variant | QARS | RNA binding,
protein translation |
| IPI00026969 | SEC23-interacting
protein | SEC23IP | RNA binding |
| IPI00027107 | Elongation factor
Tu | TUFM | Translational
elongation, RNA binding |
| IPI00027415 | Probable
ATP-dependent RNA helicase DHX36 | DHX36 | RNA binding |
| IPI00847793 | Dermcidin isoform
2 | DCD | RNA binding |
| IPI00043407 | Trinucleotide
repeat-containing gene 6C protein | TNRC6C | Translation
regulation, RNA binding |
| IPI00384265 | Constitutive
coactivator of PPAR-γ-like protein 1 | FAM120A | RNA binding |
| IPI00073779 | 28S ribosomal
protein S35 | MRPS35 | RNA binding |
| IPI00100151 | 5′-3′
exoribonuclease 2 | XRN2 | RNA processing, RNA
binding |
| IPI00106567 | WD
repeat-containing protein 33 | WDR33 | mRNA processing,
RNA binding |
| IPI00141318 |
Cytoskeleton-associated protein 4 | CKAP4 | RNA binding |
| IPI00163084 | Pre-mRNA-splicing
factor SYF1 | XAB2 | mRNA processing,
mRNA splicing |
| IPI00165434 | YLP
motif-containing protein 1 | YLPM1 | RNA binding |
| IPI00168885 | Putative
ATP-dependent RNA helicase DHX57 | DHX57 | RNA binding, RNA
processing |
| IPI00171127 |
Ubiquitin-associated protein 2 | UBAP2 | RNA binding |
| IPI00646361 | Nuclear pore
complex protein Nup214 | NUP214 | mRNA export from
nucleus, regulation of mRNA stability |
| IPI00220609 | Nucleoporin
SEH1-like | SEH1L | mRNA export from
nucleus |
| IPI00186290 | Elongation factor
2 | EEF2 | RNA binding |
| IPI00217413 | ATP-dependent RNA
helicase DHX29 | DHX29 | RNA binding, RNA
processing |
| IPI00217466 | Histone H1c | HIST1H1D | RNA binding |
| IPI00375144 | Serrate RNA
effector molecule homolog | SRRT | mRNA splicing, RNA
binding |
| IPI00396171 |
Microtubule-associated protein 4 | MAP4 | RNA binding |
| IPI00941899 | Pyruvate kinase
isozymes M1/M2 | PKM2 | RNA binding |
| IPI00549248 | Nucleophosmin | NPM1 | RNA binding,
regulation of translation |
| IPI00220834 | ATP-dependent DNA
helicase 2 subunit 2 | XRCC5 | RNA binding |
| IPI00221106 | Splicing factor 3B
subunit 2 | SF3B2 | mRNA processing,
mRNA splicing, RNA binding |
| IPI00291939 | Structural
maintenance of chromosomes protein 1A | SMC1A | RNA binding |
| IPI00910194 | Nuclear pore
complex protein Nup153 | NUP153 | mRNA export from
nucleus |
| IPI00294242 | 28S ribosomal
protein S31 | MRPS31 | RNA binding |
| IPI00294536 | Serine-threonine
kinase receptor-associated protein | STRAP | mRNA processing,
mRNA splicing, RNA binding |
| IPI00296337 | DNA-dependent
protein kinase catalytic subunit | PRKDC | RNA binding |
| IPI00646058 | Scaffold attachment
factor B | SAFB | regulation of mRNA
processing, RNA binding |
| IPI00304692 | Heterogeneous
nuclear ribonucleoprotein G | RBMX | mRNA processing,
mRNA splicing |
| IPI00304925 | Heat shock 70 kDa
protein 1 | HSPA1A | regulation of mRNA
stability |
| IPI00853598 | Protein SEC13
homolog | SEC13 | mRNA transport |
| IPI00382470 | Heat shock protein
HSP 90-α | HSP90AA1 | RNA binding |
| IPI00940033 | Trinucleotide
repeat-containing gene 6B protein | TNRC6B | Translation
regulation |
| IPI00396435 | Putative
pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 | DHX15 | mRNA processing,
mRNA splicing |
| IPI00396485 | Elongation factor
1-α1 | EEF1A1 | translational
elongation |
| IPI00418497 | Mitochondrial
import inner membrane translocase subunit TIM50 | TIMM50 | RNA binding |
| IPI00453473 | Histone H4 | HIST1H4A | RNA binding |
| IPI00455134 | Heterogeneous
nuclear ribonucleoprotein A3 | HNRNPA3 | mRNA processing,
mRNA splicing, RNA binding, mRNA transport |
| IPI00644127 | Isoleucyl-tRNA
synthetase | IARS | protein
translation |
| IPI00647635 | PERQ amino
acid-rich with GYF domain-containing protein 2 | GIGYF2 | RNA binding,
regulation of translation |
| IPI00784224 | zinc finger
RNA-binding protein | ZFR | RNA binding |
| IPI00783872 | Caprin-1 | CAPRIN1 | RNA binding |
| IPI00871240 | Protein SCAF8 | RBM16 | mRNA processing,
mRNA splicing |
 | Table IIFunctional enrichments in Flightless
I-interacting network. |
Table II
Functional enrichments in Flightless
I-interacting network.
| Pathway ID | Pathway
description | Count in gene
set | False discovery
rate |
|---|
| Biological process
(GO) | | | |
| GO:0010467 | Gene
expression | 61 | 7.44e-10 |
| GO:0006807 | Nitrogen compound
metabolic process | 68 | 1.16e-07 |
| GO:0034641 | Cellular nitrogen
compound metabolic process | 65 | 1.57e-07 |
| GO:0010033 | Response to organic
substance | 41 | 9.68e-07 |
| GO:0043170 | Macromolecule
metabolic process | 77 | 9.68e-07 |
| Molecular function
(GO) | | | |
| GO:0044822 | poly(A) RNA
binding | 54 | 9.17e-30 |
| GO:0003723 | RNA binding | 57 | 4.45e-27 |
| GO:0003676 | Nucleic acid
binding | 56 | 1.66e-08 |
| GO:0000166 | Nucleotide
binding | 42 | 6.87e-08 |
| GO:0036094 | Small molecule
binding | 43 | 4.56e-07 |
| Cellular component
(GO) | | | |
| GO:0031981 | Nuclear lumen | 68 | 1.16e-18 |
| GO:0005654 | Nucleoplasm | 62 | 1.89e-18 |
| GO:0044428 | Nuclear part | 70 | 1.89e-18 |
| GO:0070013 | Intracellular
organelle lumen | 72 | 9.78e-12 |
| GO:0005634 | Nucleus | 84 | 2.02e-12 |
| KEGG pathways | | | |
| 03013 | RNA transport | 12 | 8.61e-08 |
| 03040 | Spliceosome | 8 | 0.000217 |
| 04141 | Protein processing
in endoplasmic reticulum | 7 | 0.00885 |
| 03015 | mRNA surveillance
pathway | 5 | 0.0177 |
| PFAM protein
domains | | | |
| PF00076 | RNA recognition
motif. (a.k.a. RRM, RBD, or RNP domain) | 7 | 0.0306 |
| PF03144 | Elongation factor
Tu domain 2 | 3 | 0.0466 |
Assessment of FLII functions in RNA
trafficking and translation by high throughput sequencing
The nature of the FLII binders identified by us
strongly pointed to an involvement of FLII in RNA
post-transcriptional modification and trafficking in lung
adenocarcinoma H1299 cells, we strived to verify this hypothesis.
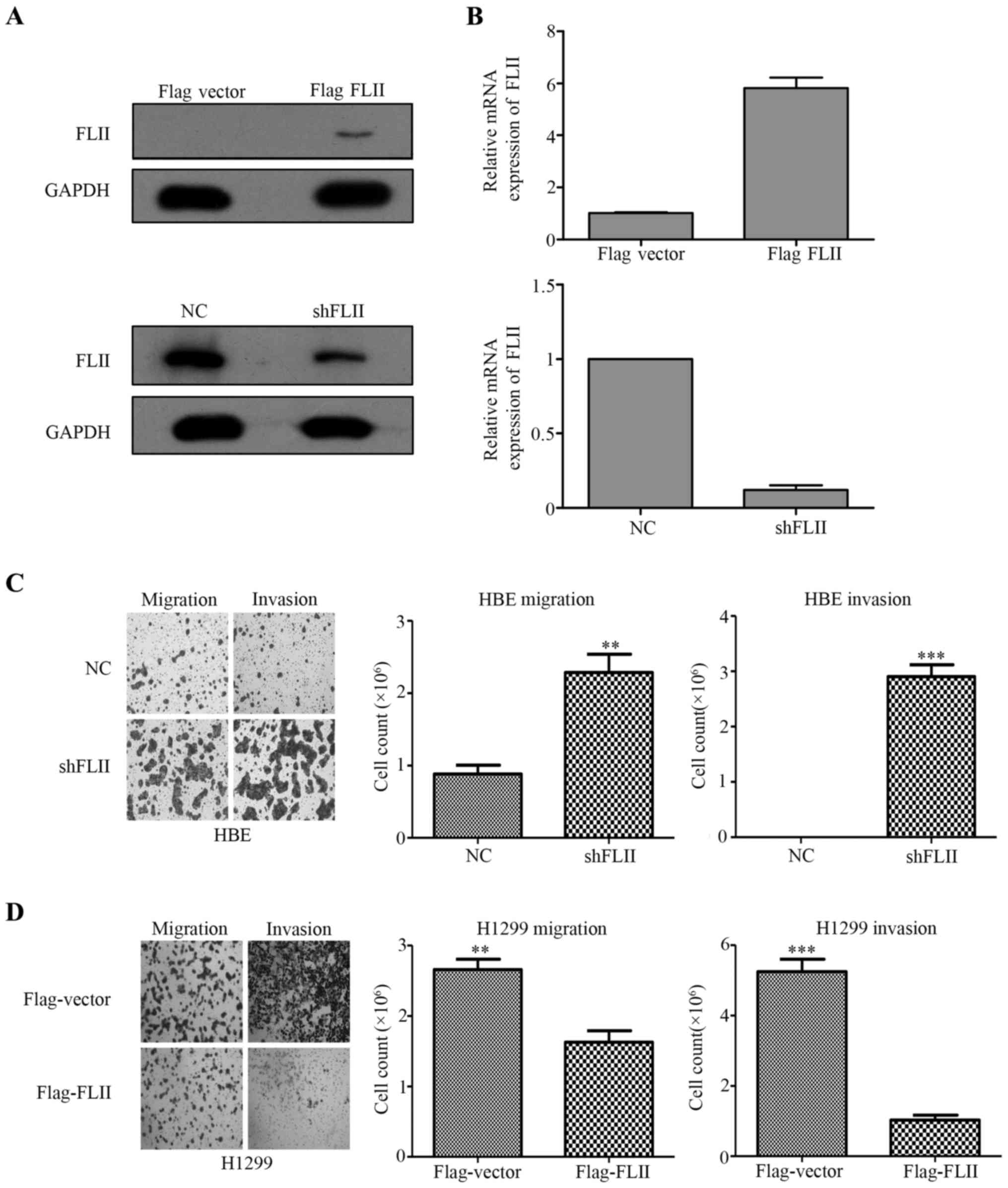
We have stably knocked down FLII in HBE cells by using a lentiviral
shRNA vector, and in parallel, overexpressed Flag-FLII in H1299
cells by transient transfection. The effectiveness of the knockdown
and overexpression of FLII respectively in HBE and H1299 cells was
confirmed by both western blotting using anti-FLII and anti-Flag
antibodies (Fig. 4A), and by qPCR
(Fig. 3B). Furthermore, Boyden
chamber assays were performed to confirm the functional impact of
the alteration of FLII expression in these cells. As shown in
Fig. 3C and D, FLII knockdown in
HBE cells considerably stimulated their migration and invasion,
whereas FLII overexpression showed inhibition ability.
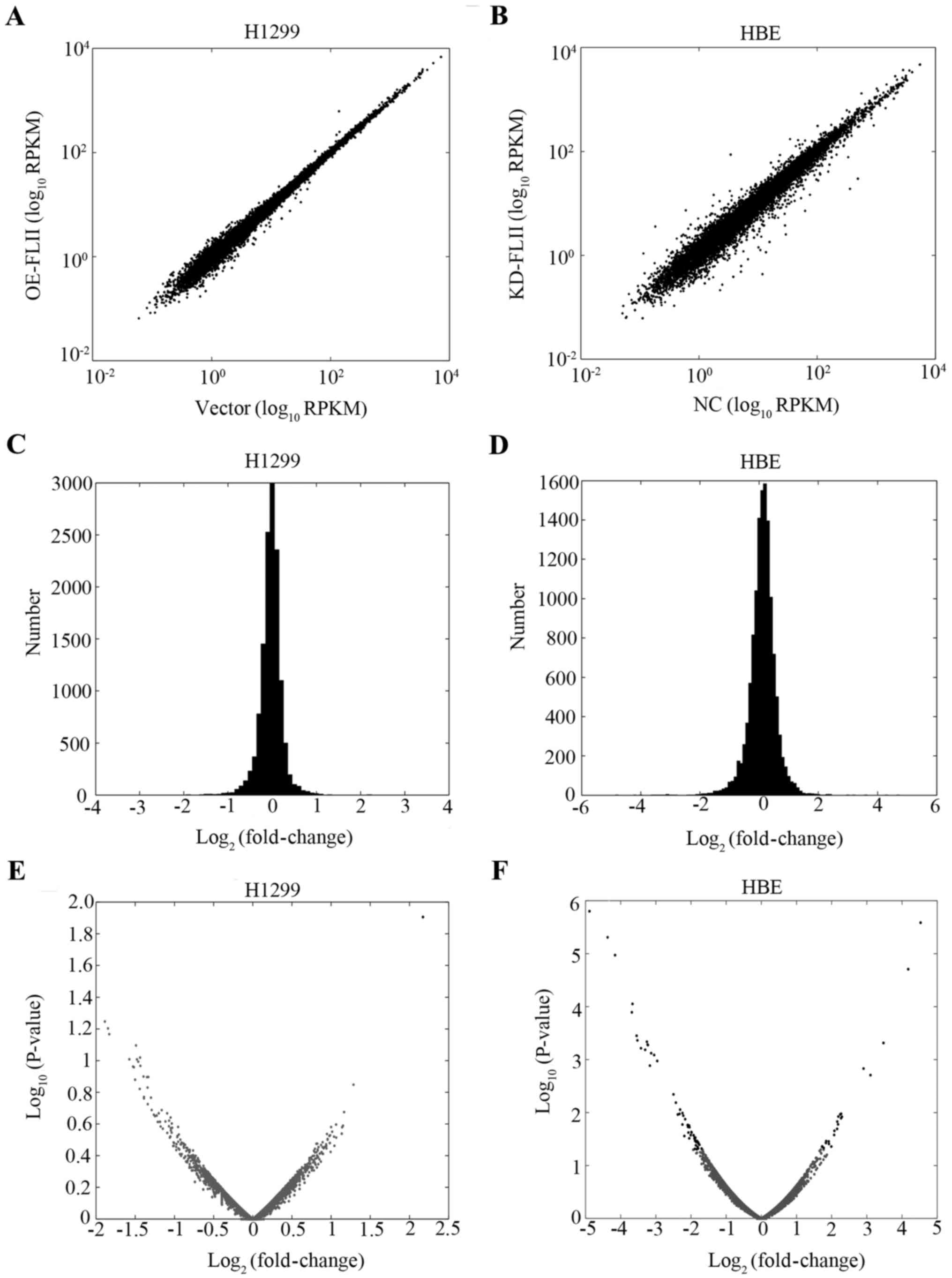
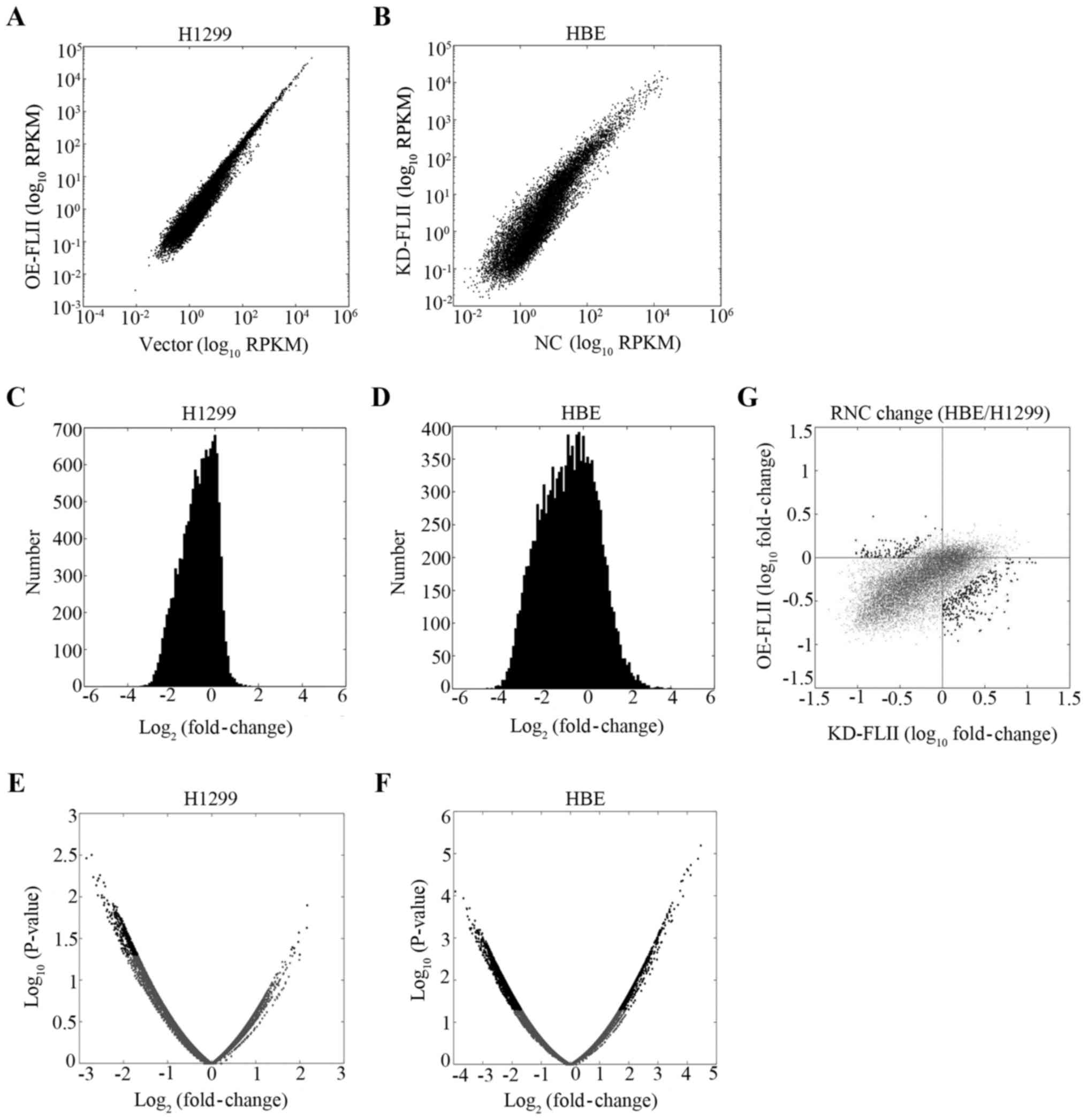
High throughput sequencing was then used to assess
the role of FLII in the overall biosynthesis and metabolism of the
mRNA. We checked the effect of FLII knockdown and overexpression on
the overall transcription of the genes. Total mRNA for both FLII
knocked down HBE cells and H1299 cells overexpressing FLII,
together with their corresponding control cells (respectively the
empty vector-transfected and control virus-infected cells), were
extracted and reverse transcribed into cDNA. High throughput
sequencing was then performed to determine the mRNA species and
levels in the various cells. Approximately 14,000 genes were
quantified. As shown in Fig. 4, no
significant effect of FLII knockdown or overexpression could be
observed on the level of total mRNA levels in HBE or H1299 cells,
suggesting that FLII does not affect the overall transcription of
genes.
We then evaluated the impact of FLII knockdown or
overexpression on the nucleocytoplasmic distribution of mRNA to
investigate the possible role of FLII in the nucleocytoplasmic
trafficking of mRNA. Cell fractionation experiments were performed
with FLII knocked down HBE cells, FLII overexpressing H1299 cells,
and the corresponding control cells described above, and the
cytoplasmic and nuclear mRNA of each type of cells were separately
extracted and subjected to high-throughput sequencing. The
variations of the cytoplasmic and nuclear mRNA levels in FLII
knockdown or over expression cells versus their control cells for
each gene were analyzed as described in Materials and methods. Our
results showed that both FLII overexpression in H1299 cells and its
knockdown in HBE cells significantly affected the general
nuclear-cytoplasmic distribution of mRNA (Fig. 6A–D). These two sets of data were
then challenged against each other to compare the quantitative
variation of mRNA for each single gene in the two experimental
conditions (Fig. 5E). Among all
genes whose nucleocytoplasmic distribution was affected by FLII
overexpression or FLII knockdown, we considered as relevant those
on which FLII knockdown and overexpression significantly exerted
opposite effects, that is, either their level was significantly
increased by FLII knockdown and decreased by FLII overexpression,
or vice versa (Fig. 5E, black
spots in the second and fourth quadrants). By this method, we
eliminated the bias introduced by the transfection and infection
systems we used in this study.
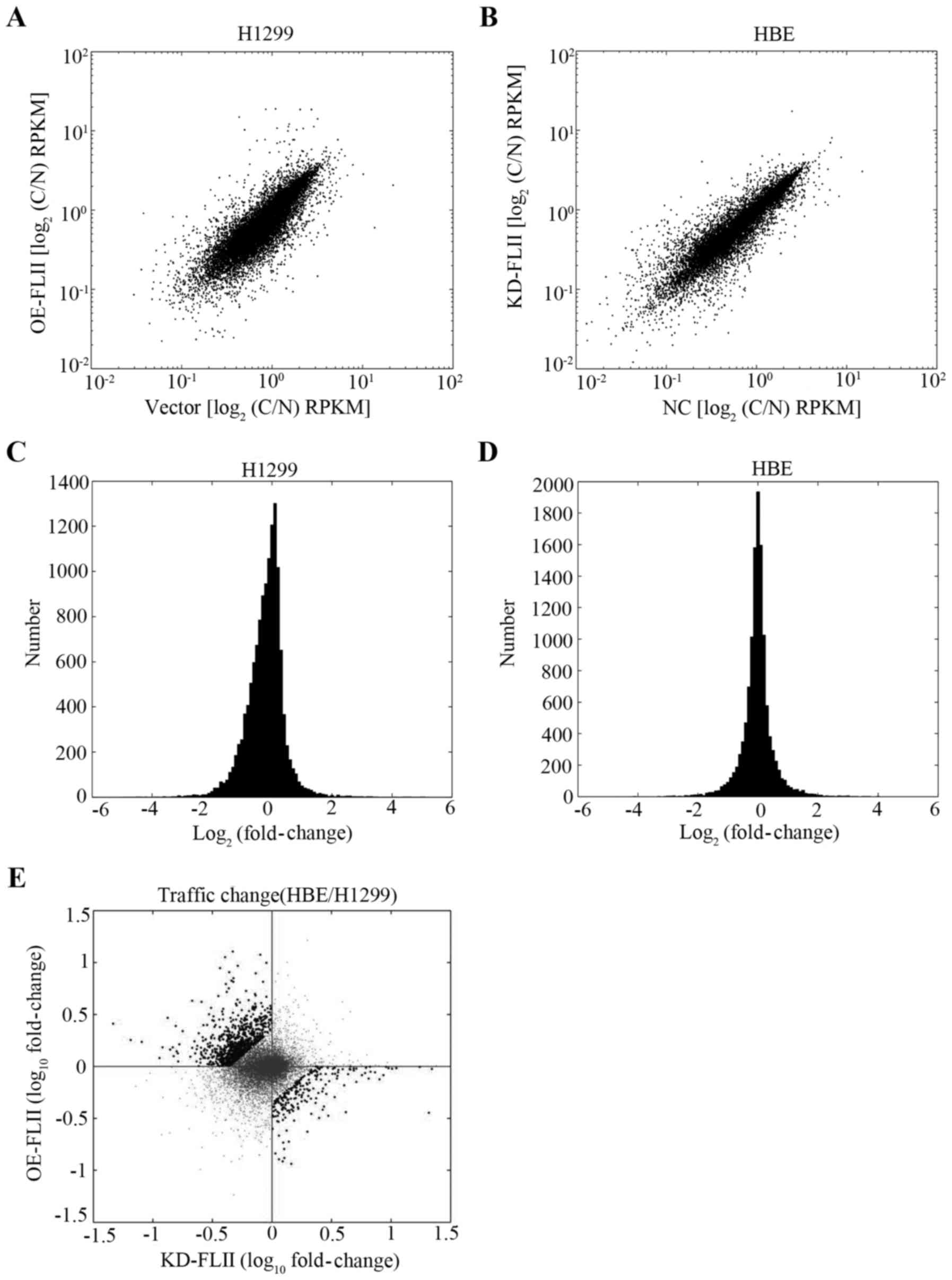
In order to examine the role of FLII on the
translational process of mRNA, ribosome-nascent chain
complex-associated mRNAs (RNC-mRNAs) from the same cells used for
the studies described above were extracted and similarly
quantified. Translatome sequencing has quantified over 12,000 genes
in translating mRNA. Contrary to the results of the total mRNA
levels, we found that FLII knockdown and overexpression
significantly affected the level of RNC-mRNAs. Similarly as
described above, we have compared the effect of FLII knockdown and
overexpression on each gene analyzed and selected as potential FLII
target genes those on which FLII knockdown and overexpression
significantly exerted opposite effects (Fig. 6E, black spots in the second and
fourth quadrants). As indicated, for 4% of the genes analyzed, the
transcription was upregulated by FLII overexpression and
downregulated by its knockdown, whereas for another 9.5%, the
transcription was inversely regulated by FLII. These genes were
then submitted to IPA program to assess their possible involvement
in various biological processes and signaling pathways. Table III shows the top ranked diseases
and biological functions, top networks and canonical pathways,
upstream regulators, and the top regulator effect networks. As
shown, the top ranked molecular and cellular functions are cellular
movement and cellular growth and proliferation, and the top
diseases and disorders associated with FLII regulated genes are
dermatological diseases and conditions, organismal injury and
abnormalities, and cancer. The top canonical pathways revealed to
be the IL-12 signaling and production in macrophages and the
VDR/RXR activation, whereas the top upstream regulator is estrogen
receptor 1, and the top regulator effector network is NF-κB. Taken
together, these results suggest that FLII might be involved in the
determination of the cell fate, the regulation of development, the
injury repair, the immune response and cancer by controlling the
mRNA metabolism and trafficking of several groups of genes.
 | Table IIIFunctional categorization of
Flightless I-regulated genes. |
Table III
Functional categorization of
Flightless I-regulated genes.
Top diseases and
bio-functions
|
|---|
| Name | P-value | # Molecules |
|---|
| Diseases and
disorders | | |
| Dermatological
diseases and conditions |
2.28E-02-7.41E-08 | 55 |
| Organismal injury
and abnormalities |
2.68E-02-7.41E-08 | 337 |
| Cancer |
2.68E-02-3.48E-06 | 329 |
| Reproductive
system disease |
2.28E-02-3.48E-06 | 142 |
| Endocrine system
disorders |
2.28E-02-5.20E-06 | 112 |
| Molecular and
cellular functions | | |
| Cellular
movement |
2.45E-02-1.42E-08 | 80 |
| Cellular growth
and proliferation |
2.73E-02-3.30E-07 | 112 |
| Cell-to-cell
signaling and interaction |
2.85E-02-8.30E-06 | 66 |
| Cell
morphology |
1.94E-02-2.46E-05 | 39 |
| Cellular
development |
2.73E-02-2.46E-05 | 99 |
| Physiological
system development and function | | |
| Cardiovascular
system development and function |
2.71E-02-2.06E-06 | 33 |
| Organismal
development |
2.71E-02-2.06E-06 | 49 |
| Tissue
development |
2.73E-02-2.46E-05 | 56 |
| Connective tissue
development and function |
2.71E-02-1.15E-04 | 22 |
| Nervous system
development and function |
2.73E-02-1.09E-03 | 12 |
Top networks
|
|---|
| ID | Associated network
functions | Score |
|---|
| 1 | Cellular movement,
cellular growth and proliferation, cell morphology | 40 |
| 2 | Cellular
development, cellular growth and proliferation, cellular function
and maintenance | 38 |
| 3 | Cellular movement,
hematological system development and function, immune cell
trafficking | 34 |
| 4 | Cellular movement,
organismal injury and abnormalities, reproductive system
disease | 24 |
| 5 | Cancer, organismal
injury and abnormalities, reproductive system disease | 19 |
Top canonical
pathways
|
|---|
| Name | P-value | Overlap |
|---|
| IL-12 signaling and
production in macrophages | 1.08E-04 | 7.6%, 11/144 |
| VDR/RXR
activation | 1.14E-04 | 10.4%, 8/77 |
| Axonal guidance
signaling | 3.68E-04 | 4.5%, 20/441 |
| Production of
nitric oxide and reactive oxygen species in macrophages | 1.26E-03 | 5.7%, 11/192 |
| LPS/IL-1 mediated
inhibition of RXR function | 2.37E-03 | 5.3%, 11/208 |
Top upstream
regulators
|
|---|
| Upstream
regulator | P-value of
overlap | False discovery
rate |
|---|
| ESR1 | 4.94E-06 | |
| Estrogen
receptor | 6.22E-05 | |
| PDGF BB | 7.15E-05 | Activated |
| TNF | 9.44E-05 | |
| SMARCA4 | 1.49E-04 | Activated |
Top regulator
effect networks
|
|---|
| ID | Regulators | Diseases and
functions | Consistency
score |
|---|
| 1 | NF-κB
(complex) | Differentiation of
cells (+1 more) | 3.13 |
| 2 | TGFB1 | Cell movement | −6.935 |
| 3 | NF-κB
(complex) | Proliferation of
cells | −7.906 |
| 4 | ERBB2 | Invasion of tumor
cell lines | −12.522 |
| 5 | PDGF BB | Cell proliferation
of tumor cell lines | −15.497 |
Discussion
Growing evidence indicates that FLII plays important
roles in multiple cellular processes (8,9,29).
The characteristic feature of FLII being composed by two
protein-protein interaction domains suggests that protein
interactions might be key elements of the mechanisms governing its
versatile functions. However, little has been done with success in
the past to specifically and systematically identify the
interacting partners of this protein. Two yeast two-hybrid
screenings, respectively reported by Liu and yin in 1998 (15), and by Fong and de Couet in 1999
(16), have led to the
identification of three homologous proteins as the interactors of
FLII LRR domain, referred to as LRRFIP1 and LRRFIP2 (LRR FLII
interacting protein 1 and 2) and FLAP (FLII associating protein).
Unfortunately, neither study was able to depict, at the systemic
level, the whole interactome of FLII, because all [4 clones,
(15)] or most [12 from the 18,
(16)] of the positive clones from
these two screenings were derived from the genes encoding these
three proteins. Besides, a few other proteins have also been
independently reported to bind FLII in various contexts and with
diverse functions, such as actin, BAF53, CARM1, ChREBP and Rac1
(10,15,30–34).
In this study, by combining immunoprecipitation and mass
spectrometry analysis, we successfully identified 132 FLII
interacting proteins and constructed the first putative interaction
network of FLII in the cell. Interestingly, our data displayed
striking directivity, in that more than a half of these candidate
interactors are associated with RNA post-transcriptional regulation
such as splicing, maturation, trafficking and protein synthesis
(Table II). These results
strongly suggested an undiscovered function of FLII in the
regulation of RNA metabolism.
The target genes of FLII that we identified by using
the RNC-bound RNA sequencing and IPA analysis encode mainly the
proteins involved in the cellular processes such as cellular
movement, cellular growth and proliferation, and the development at
cellular, tissue, and organismal levels. They are also associated
with the diseases and disorders such as dermatological diseases and
conditions, organismal injury and abnormalities, and cancer.
Moreover, the top ranked canonical pathways identified are IL-12
signaling and production in macrophages and VDR/RXR activation.
These physiological and pathological processes and functions are
very reminiscent of the reported functions of FLII, that are
embryonic development, the regulation of wound repair, skin barrier
development, the recovery after blistering and regulation of immune
response, and more recently cancer (2–13).
This confirmed, on one hand, the reliability of our approaches and
the authenticity of our results, and on the other hand, suggested a
novel mechanism of action of FLII, that is the overall
post-transcriptional regulation of mRNA.
We find that FLII knockdown or overexpression
affects at the same time the nucleus/cytoplasm ratio of mRNA and
the RNC-bound mRNA amounts and species. The result of FLII
interactome suggest that FLII might bind to both the proteins
involved in the mRNA post-transcriptional modifications and
translation, and those involved in mRNA trafficking, such as the
nucleoporins. However, it remains to be determined whether FLII is
involved in both the trafficking and the translational regulation
of mRNA, or just play a role in mRNA trafficking which might impact
on the subsequent translational process. No known RNA-binding
domain has been found on FLII protein, which suggests that its
function in the regulation of RNA trafficking or/and
post-transcriptional regulation might be mediated by its
interactions with numerous proteins involved in these biological
processes. FLII might take part in the protein complexes associated
with mRNA, and exerts its functions through direct and indirect
interactions with various partners regulating one or several steps
of mRNA post-transcriptional modification, trafficking and
metabolism. For example, its interaction with Nup88 and Nup214
might be involved in the nuclear export of mRNP complexes since
Nup214-Nup88 nucleoporin subcomplex has been shown to play an
important role in the nuclear export of the mRNA and protein
complexes (35–38). Particularly, Nup214-Nup88 complex
is required for the CRM1-mediated nuclear export of the 60S
preribosomal subunit (36),
hinting to the possible impact on the overall translation of mRNA.
On the other hand, the interactions of FLII with the proteins
playing roles in the translational regulation such as TIAL1 might
also be functionally relevant. TIAL1, also named TIAR, was found to
bind several mRNAs encoding translation factors such as eukaryotic
initiation factor 4A (eIF4A) and eIF4E (translation initiation
factors), eEF2 (a translation elongation factor), and c-Myc (which
transcriptionally controls the expression of numerous translation
regulatory proteins), suppressing their translation and
participating to the global inhibition of translation machinery in
response to low levels of short-wavelength uV irradiation (39). FLII possibly exerts its regulatory
functions by interacting with these RNA-binding proteins through
its N-terminal LRR domain, and with actin cytoskeleton via its
C-terminal gelsolin related repeats, thereby mediating the traffic
of the mRNP complexes along the actin filaments, and modulating the
translational processes. In support of this hypothesis, we have
been able to detect the interaction of the FLII LRR domain with
Nup88 by in vitro GST pull-down assay (data not shown).
Guided by the results of FLII interactome obtained
in this study, further studies allowing the confirmation and
functional demonstration of these relevant interactions will
provide new elements for the comprehensive understanding of the
precise mechanisms of action of FLII.
Abbreviations:
|
FLII
|
flightless I
|
|
LRR
|
leucine-rich repeats
|
|
HBE
|
human bronchial epithelial
|
|
IPA
|
ingenuity pathway analysis
|
|
NSCLC
|
non-small cell lung cancer
|
|
RNC
|
ribosome-nascent chain complex
|
|
NPC
|
nuclear pore complex
|
|
rpkM
|
reads per kilobase per million
reads
|
|
EEF2
|
eukaryotic elongation factor 2
|
Acknowledgments
This work was partially supported by National
Program on Key Basic Research Project (973 Program) (grant no.
2011CB910700); Natural Science Foundation of China (grant no.
81322028); Natural Science Foundation of Guangdong Province (grant
no. 2016A030313083; 2016A030313420); Guangzhou Science and
Technology Project (20160701175); Fundamental Research Funds for
the Central Universities (grant no. 21615407; 21609317); Science
and Technology Program of Huadu District of Guangzhou, Guangdong
Province (15-HDWS-016).
References
|
1
|
She J, Yang P, Hong Q and Bai C: Lung
cancer in China: Challenges and interventions. Chest.
143:1117–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Campbell HD, Schimansky T, Claudianos C,
Ozsarac N, Kasprzak AB, Cotsell JN, Young IG, de Couet HG and
Miklos GL: The Drosophila melanogaster flightless-I gene involved
in gastrulation and muscle degeneration encodes gelsolin-like and
leucine-rich repeat domains and is conserved in Caenorhabditis
elegans and humans. Proc Natl Acad Sci USA. 90:11386–11390. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Campbell HD, Fountain S, Young IG,
Claudianos C, Hoheisel JD, Chen K-S and Lupski JR: Genomic
structure, evolution, and expression of human FLII, a gelsolin and
leucine-rich-repeat family member: Overlap with LLGL. Genomics.
42:46–54. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Davy DA, Ball EE, Matthaei KI, Campbell HD
and Crouch MF: The flightless I protein localizes to actin-based
structures during embryonic development. Immunol Cell Biol.
78:423–429. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Campbell HD, Fountain S, McLennan IS,
Berven LA, Crouch MF, Davy DA, Hooper JA, Waterford K, Chen K-S,
Lupski JR, et al: Fliih, a gelsolin-related cytoskeletal regulator
essential for early mammalian embryonic development. Mol Cell Biol.
22:3518–3526. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang T, Chuang T-H, Ronni T, Gu S, Du Y-C,
Cai H, Sun H-Q, Yin HL and Chen X: Flightless I homolog negatively
modulates the TLR pathway. J Immunol. 176:1355–1362. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cowin AJ, Adams DH, Strudwick XL, Chan H,
Hooper JA, Sander GR, Rayner TE, Matthaei KI, Powell BC and
Campbell HD: Flightless I deficiency enhances wound repair by
increasing cell migration and proliferation. J Pathol. 211:572–581.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Yin HL and Yuan J: Flightless-I
regulates proinflammatory caspases by selectively modulating
intracellular localization and caspase activity. J Cell Biol.
181:321–333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kopecki Z, Yang GN, Arkell RM, Jackson JE,
Melville E, Iwata H, Ludwig RJ, Zillikens D, Murrell DF and Cowin
AJ: Flightless I over-expression impairs skin barrier development,
function and recovery following skin blistering. J Pathol.
232:541–552. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu L, Chen H, Zhu Y, Meng J, Li Y, Li M,
Yang D, Zhang P, Feng M and Tong X: Flightless I homolog negatively
regulates ChREBP activity in cancer cells. Int J Biochem Cell Biol.
45:2688–2697. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeong KW: Flightless I (Drosophila)
homolog facilitates chromatin accessibility of the estrogen
receptor α target genes in MCF-7 breast cancer cells. Biochem
Biophys Res Commun. 446:608–613. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kopecki Z, Yang GN, Jackson JE, Melville
EL, Calley MP, Murrell DF, Darby IA, O'Toole EA, Samuel MS and
Cowin AJ: Cytoskeletal protein Flightless I inhibits apoptosis,
enhances tumor cell invasion and promotes cutaneous squamous cell
carcinoma progression. Oncotarget. 6:36426–36440. 2015.PubMed/NCBI
|
|
13
|
Wang T, Song W, Chen Y, Chen R, Liu Z, Wu
L, Li M, Yang J, Wang L, Liu J, et al: Flightless I homolog
represses prostate cancer progression through targeting androgen
receptor signaling. Clin Cancer Res. 22:1531–1544. 2016. View Article : Google Scholar
|
|
14
|
Stone L: Bladder cancer: In the driving
seat - AGL loss drives tumour growth. Nat Rev Urol. 13:3. 2016.
View Article : Google Scholar
|
|
15
|
Liu Y-T and Yin HL: Identification of the
binding partners for flightless I, A novel protein bridging the
leucine-rich repeat and the gelsolin superfamilies. J Biol Chem.
273:7920–7927. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fong KS and de Couet HG: Novel proteins
interacting with the leucine-rich repeat domain of human
flightless-I identified by the yeast two-hybrid system. Genomics.
58:146–157. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo W, Slebos RJ, Hill S, Li M, Brábek J,
Amanchy R, Chaerkady R, Pandey A, Ham A-JL and Hanks SK: Global
impact of oncogenic Src on a phosphotyrosine proteome. J Proteome
Res. 7:3447–3460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Esposito AM, Mateyak M, He D, Lewis M,
Sasikumar AN, Hutton J, Copeland PR and Kinzy TG: Eukaryotic
polyribosome profile analysis. J Vis Exp. pii; pp. 19482010,
View Article : Google Scholar
|
|
19
|
Rajkumar AP, Qvist P, Lazarus R, Lescai F,
Ju J, Nyegaard M, Mors O, Børglum AD, Li Q and Christensen JH:
Experimental validation of methods for differential gene expression
analysis and sample pooling in RNA-seq. BMC Genomics. 16:5482015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ström A-C and Weis K: Importin-beta-like
nuclear transport receptors. Genome Biol. 2:S30082001. View Article : Google Scholar
|
|
22
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hutten S and Kehlenbach RH: Nup214 is
required for CRM1-dependent nuclear protein export in vivo. Mol
Cell Biol. 26:6772–6785. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vincendeau M, Nagel D, Brenke JK,
Brack-Werner R and Hadian K: Heterogenous nuclear ribonucleoprotein
Q increases protein expression from HIV-1 Rev-dependent
transcripts. Virol J. 10:1512013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Svitkin YV, Yanagiya A, Karetnikov AE,
Alain T, Fabian MR, Khoutorsky A, Perreault S, Topisirovic I and
Sonenberg N: Control of translation and miRNA-dependent repression
by a novel poly(A) binding protein, hnRNP-Q. PLoS Biol.
11:e10015642013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shimizu Y, Nishitsuji H, Marusawa H, Ujino
S, Takaku H and Shimotohno K: The RNA-editing enzyme APOBEC1
requires heterogeneous nuclear ribonucleoprotein Q isoform 6 for
efficient interaction with interleukin-8 mRNA. J Biol Chem.
289:26226–26238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Beuck C, Williamson JR, Wüthrich K and
Serrano P: The acidic domain is a unique structural feature of the
splicing factor SyNCRIP. Protein Sci. 25:1545–1550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kaul G, Pattan G and Rafeequi T:
Eukaryotic elongation factor-2 (eEF2): Its regulation and peptide
chain elongation. Cell Biochem Funct. 29:227–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kopecki Z and Cowin AJ: Flightless I: An
actin-remodelling protein and an important negative regulator of
wound repair. Int J Biochem Cell Biol. 40:1415–1419. 2008.
View Article : Google Scholar
|
|
30
|
Lee Y-H, Campbell HD and Stallcup MR:
Developmentally essential protein flightless I is a nuclear
receptor coactivator with actin binding activity. Mol Cell Biol.
24:2103–2117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Archer S, Behm C, Hooper J, Judge R,
Matthaei K, Powell B, Cowin A and Campbell H: A novel,
evolutionarily conserved role in intracellular signalling for the
gelsolin-related actin-binding protein Flightless I. The FEBS
Journal. 272:2822005.
|
|
32
|
Orloff G, Allen P, Miklos G, Young I,
Campbell H and Kwiatkowski D: Human flightless-I has actin-binding
ability. Molecular Biology of the Cell. 6:8031995.
|
|
33
|
Jeong KW, Lee Y-H and Stallcup MR:
Recruitment of the SWI/SNF chromatin remodeling complex to steroid
hormone-regulated promoters by nuclear receptor coactivator
flightless-I. J Biol Chem. 284:29298–29309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marei H, Carpy A, Woroniuk A, Vennin C,
White G, Timpson P, Macek B and Malliri A: Differential Rac1
signalling by guanine nucleotide exchange factors implicates FLII
in regulating Rac1-driven cell migration. Nat Commun. 7:106642016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van Deursen J, Boer J, Kasper L and
Grosveld G: G2 arrest and impaired nucleocytoplasmic transport in
mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J.
15:5574–5583. 1996.PubMed/NCBI
|
|
36
|
Bernad R, Engelsma D, Sanderson H,
Pickersgill H and Fornerod M: Nup214-Nup88 nucleoporin subcomplex
is required for CRM1-mediated 60 S preribosomal nuclear export. J
Biol Chem. 281:19378–19386. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carmody SR and Wente SR: mRNA nuclear
export at a glance. J Cell Sci. 122:1933–1937. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Simon DN and Rout MP: Cancer and the
nuclear pore complex. Cancer Biology and the Nuclear Envelope.
Schirmer EC and de las Heras JI: Springer; New York: pp. 285–307.
2014, View Article : Google Scholar
|
|
39
|
Mazan-Mamczarz K, Lal A, Martindale JL,
Kawai T and Gorospe M: Translational repression by RNA-binding
protein TIAR. Mol Cell Biol. 26:2716–2727. 2006. View Article : Google Scholar : PubMed/NCBI
|