Introduction
Tamoxifen, a selective estrogen receptor (ER)
modulator, is the most common endocrine therapy worldwide to women
with ER-positive metastatic breast cancer or as adjuvant therapy
for early stages of the disease (1). Acquired tamoxifen resistance remains
the major obstacle to breast cancer endocrine therapy, which may
associate with anti-apoptosis during the process of TAM-resistance
(2). The multiplicity and complex
regulation of mammalian BH3-only proteins allows exquisite control
over apoptosis. Bcl-2 interacting mediator of cell death (Bim), a
BH3-only protein, have the most prominent roles (3). Recent studies have demonstrated that
suppression of Bim, provided at least partial protection from
apoptosis (4–6). The strong association between Bim and
the presence of tumor and drug resistance has been proven (7–10).
By knocking down the pro-apoptotic BCL-2 family member BIM,
Follin-Arbelet et al (11)
proved this protein to be involved in the synergistic induction of
apoptosis by dexamethasone and forskolin. Reginato et al
(12) find that Bim is a critical
regulator of luminal apoptosis during mammary acinar morphogenesis
in vitro and may be an important target of oncogenes that
disrupt glandular epithelial architecture. Thus, Bim is not only
associated with drug response but also tumorigenesis. Research
recognized pro-apoptotic BH3-only proteins (BCL2L11/Bim), another
class of BCL2 family proteins, critically determine therapeutic
responses by dual-agent regulating autophagy and apoptosis, and
contribute to acquired drug resistance in standard chemotherapy or
novel targeted therapy (13). The
expression of Bim is repressed by transcriptional regulation,
post-translational modifications and ubiquitination deregulation
(13).
The tripartite motif (TRIM) proteins, one of the
subfamilies of the RING type E3 ubiquitin ligases), are involved in
a broad range of biological processes and their alterations are
associated with disease incidence and progression relevant to the
development of common human cancers (14–16).
Most of the TRIM proteins function as E3 ubiquitin ligases, and
several TRIM family members are involved in various tumor
development by governing cell proliferation, apoptosis and
transcriptional regulation (17–19).
Abundant evidence shows that TRIM2 is aberrantly expressed in
several diseases, such as breast cancer, follicular carcinomas and
the nervous system diseases (20–22).
Research demonstrates that TRIM2 is an ubiquitin ligase and point
to a mechanism triggering neuro-degeneration through a
ubiquitination pathway (23).
Another study supports a role of TRIM2 in mediating the p42/p44
MAPK-dependent ubiquitination of Bim in rapid ischemic tolerance.
It was found that TRIM2 binds to Bim when it is phosphorylated by
p42/p44 MAPK following preconditioning ischemia (22). Thus, the level of TRIM2 mediating
the ubiquitination of Bim may act as a critical role in rapid
neuronal ischemic tolerance. Interestingly, our previous cDNA array
analysis showed that a couple of apoptosis-associated genes
including Trim2, and Bim are aberrantly expressed in TAM-resistant
MCF-7R cells. This raises a question whether Trim2 and Bim are
involved in the process of TAM resistance in ER-positive breast
cancer.
G protein-coupled estrogen receptor (GPER), a novel
estrogen receptor, has been reported to be activated by TAM and the
pure anti-estrogen fulvestrant and play an important role in the
acquired TAM-resistance (24,25).
Our previous studies have shown that GPER contributes to tamoxifen
resistance via the epidermal growth factor receptor/extracellular
regulated protein kinase (EGFR/ERK) signaling pathway (2). Other studies have shown that GPER is
involved in the acquired TAM-resistance through mitogen-activated
protein kinase/extracellular regulated protein kinase (MAPK/ERK)
signaling pathway or phosphatidylinositol 3-kinase/protein kinase B
(PI3K/Akt) (26–29). Notably, Bim is upregulated by
inhibition of signaling pathways (MAPK/ERK and/or PI3K/AKT) while
repressed its expression through transcriptional regulation and/or
post-translational modifications (14,30,31).
However, whether GPER contributes to TAM-resistance via regulating
the expression of Bim in breast cancer remains unknown.
In the present study, we elucidated that the
degradation of Bim in MCF-7R plays a key role in GPER-mediated
tamoxifen resistance in ER. Knockdown Bim in TAM-sensitive MCF-7
cells or overexpression of Bim in TAM-resistant MCF-7 cells
significantly changed its sensibility to TAM. The activation of
GPER acts as a direct mediator of the transcription of TRIM2 and
the binding between TRIM2 and Bim, which regulates the expression
of Bim in TAM-resistant breast cancer cells. These findings may
provide a novel insight to understand the mechanism of GPER in
acquired TAM-resistance in ER+ breast cancer.
Materials and methods
Reagents and cell culture
The primary antibodies against Bim, ERK and
phosphorylated ERK1/2 (pT202/Y204) were purchased from Bioworld
Technology, Inc. (St. Louis Park, MN, USA). The primary antibody of
goat anti-Trim2 was obtained from Sigma-Aldrich (Steinheim,
Germany). Antibodies against AKT and phosphorylated AKT (pS473)
were from Cell Signaling Technology (New England Biolabs,
Hertfordshire, UK). The antibody against GPER was purchased from
Abcam (Cambridge, MA, USA). β-actin antibody, goat antimouse
IgG-HRP, and goat antirabbit IgG-HRP were obtained from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). The reagents of GPER-specific
agonists G1 and specific inhibitor G15 were purchased from Tocris
Bioscience (St. Louis, MO, USA) and 4-hydroxytamoxifen (TAM),
(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
from Sigma-Aldrich. Lipofectamine™ 2000 was purchased from Life
Technologies (Carlsbad, CA, USA).
Human breast cancer cells (MCF-7) were routinely
grown in RPMI-1640 (Gibco, Mulgrave VIC, Australia) containing 5%
fetal bovine serum (FBS; Gibco), 10 μg/ml insulin, 100 IU/ml
penicillin and 100 μg/ml streptomycin. The TAM-resistant
cells (MCF-7R) (26) were derived
from MCF-7 by continuous exposure to TAM (1 μM) diluted in
0.1% ethanol. The MCF-7R cells were then grown in RPMI-1640 medium
with 5% FBS plus 100 nM TAM. Before all experiments, cells were
switched to phenol red-free DMEM containing 0.5%
charcoal-dextran-stripped FBS for 24 h, except where otherwise
noted. Culture of MDA-MB-468, MDA-MB-231, BT549 and Hs578T cells
were previously described (32).
Plasmid construct, siRNA and
transfection
The expression vector encoding Bim was constructed
by inserting human Bim cDNA into pcDNA3.0 vector (Promega, Madison,
WI, USA). The pcDNA-Bim and its control vector were transfected
into TAM-resistant MCF-7R cell lines using Lipofectamine 2000 (Life
Technologies). The siRNAs used in the present study were obtained
from Shanghai GenePharma, Co., Ltd. (Shanghai, China). Bim
(Bcl2-L11)-specific siRNA or control siRNA (100 nM) were
transfected into MCF-7; Trim2-specific siRNA and control siRNA
transfected into MCF-7R using Lipofectamine 2000 according to the
manufacturer's instructions. The target sequences for Bim
(Bcl2-L11) siRNA are 5′-GACAGAGCCACAAGGUAAUTT-3′ and 5′-AUUA
CCUUGUGGCUCUGUCTT-3′. The control siRNA sequences are
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′
(antisense). The efficiency of gene knockdown was determined by
qRT-PCR and western blot analysis.
Measurement of cell growth
Cell growth was determined by
3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl tetrazoliumbromide (MTT)
assay. The cells were plated at 5×103 cells/well in
96-well microtiter plates. After incubation and treated with
designed concentration of TAM for the specified time, MTT (5 mg/ml)
was added to each well and incubated for 4 h. The absorbance was
recorded on a digital spectrophotometer at a wavelength of 570 nm.
The experiment was repeated three times.
Tissue samples, immunohistochemistry
(IHC) staining
A total of 77 breast cancer specimens and their
matched primary tumor tissues (PTs) were obtained from patients
with breast tumors resected at the First Affiliated Hospital of
Chongqing Medical University under permission by the ethics
committee of Chongqing Medical University (Chongqing, China). All
of the patient details and exclusion criteria have been previously
described (26).
Immunohistochemistry staining was performed using an SP900 kit
(Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd., Beijing,
China) according to the manufacturer's protocol. Briefly,
deparaffinized tissue sections of 4 μm thickness were heated
for antigen retrieval at 95°C for 15 min in 10 mM citric acid
buffer (pH 6.0). After treatment with 3% H2O2
for 10 min to quench endogenous peroxidase activity, the sections
were blocked using goat serum and then incubated with primary
antibodies targeting GPER and Bim at a 1:200 dilution at 4°C for 16
h. Following treatment of horseradish peroxidase-conjugated goat
anti-rabbit IgG for 30 min at 37°C, sections were developed using
diaminobenzidine (DAB) (Beijing Zhongshan Golden Bridge
Biotechnology) and nuclei were counterstained with the Mayer's
modified hematoxylin.
GPER scores were assigned as follows: the percentage
of positive cells was categorized as 0 (negative staining in all
cells), 1 (<1% cells stained), 2 (1–10% of cells stained), 3
(11–40% cells stained), 4 (41–70% cells stained) or 5 (71–100%
cells stained), and staining intensity was categorized as 0
(negative), 1 (weak), 2 (moderate) or 3 (strong). Percentage and
intensity scores were added to give total immunohistochemical
scores, ranging from 0 to 8. Specimens that scored ≥2 were defined
as GPER+.
The Bim expression was scored based on intensity
(0–3) and extent (0, <10%, 1, 10–25%, 2, 26–50% and 3, >50%).
The individual categories were multiplied to give a total
immunohistochemical score ranging between 0 and 9. Samples that
scored ≥3 were defined as positive immunohistochemical results.
Reverse transcription and real-time
PCR
Total RNA was extracted using TRizol®
reagent (Takara Biotechnology, Dalian, China) from MCF-7 and MCF-7R
cells and reverse transcription was performed using the PrimeScript
RT reagent kit (Takara Biotechnology) following the manufacturer's
instruction. Quantitative real-time PCR was performed with SYBR
Premix Ex Taq™ II (Takara Biotechnology). The specific primers for
Bim are: 5′-CCTTTCTTGGCCCTT GTTCC-3′ (sense) and
5′-TTGTGGCTCTGTCTGTAGGG-3′ (antisense); the specific primers for
Trim2: 5′-CACCAAGGA CAAAGACGGTG-3′ (sense) and 5′-ATCAGCGGATCGGAT
CACTT-3′ (antisense). β-actin was used as internal control for
normalizing different samples. The primer sequences for α-actin
are: 5′-TGACGTGGACATCCGCAAAG-3′ (sense) and
5′-CTGGAAGGTGGACAGCGAGG-3′ (antisense). All experiments were
performed at least three times.
Western blotting
The total protein was acquired using RIPA protein
extraction buffer with protease inhibitor (Beyotime Institute of
Biotechnology, Haimen, China). Cell lysates were electrophoresed
with 10 or 12% SDS-PAGE, and the specific primary antibody
targeting to each indicated protein was incubated with the
membranes at 4°C overnight. The membranes were then incubated with
the appropriate secondary antibody conjugated to horseradish
peroxidase for 1 h and visualized by using the enhanced
chemiluminescence imaging system (Amersham Pharmacia Biotech,
Tokyo, Japan). The gray level of each band was quantified using the
Quantity One 4.62 software (Bio-Rad Laboratories, Hercules, CA,
USA). The results were expressed as fold change relative to the
loading control (β-actin).
Apoptosis assay
Cells were seeded into 6-wells plates and grown to
60% confluence. After cultured in FBS- and phenol-free medium for
24 h, the cells were then treated with or without TAM for another
24 h. At the end of the treatment, cells were washed with
phosphate-buffered saline (PBS) twice and stained with 5 ml Annexin
V-FITC and 5 ml propidium iodide following the manufacturer's
instructions (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China).
Apoptotic cells were determined using a BD FACScan flow cytometer
(BD Biosciences, Mansfield, MA, USA).
Immunoprecipitation western blotting
Coimmunoprecipitation was performed as previously
described (33,34). MCF-7 and MCF-7R cells were lysed in
cell lysis buffer containing 50 mM HEPES (pH 7.2), 150 mM NaCl, 50
mM Tris-HCl, 1.0% Nonidet P-40, 1 mM EDTA containing protease
inhibitors (1.0 g/ml aprotinin, 0.5 g/ml leupeptin, 1.0 g/ml
pepstatin and 10 g/ml PMSF). The extracts were pre-cleared by
rocking them at 4°C with washed protein G-agarose beads (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The pre-cleared extracts
were immunoprecipitated with 5 mg of with Bim antibody or Trim2
antibody and 30 ml of protein G-agarose for 12 h at 4°C. Equivalent
amounts of immunoglobulin G (IgG) were used as the control. The
beads were washed five times with lysis buffer and boiled in SDS
sample buffer, and the released proteins were resolved by SDS-PAGE.
The gel were transferred to nitrocellulose and western blotting was
performed.
Statistical analysis
Statistical analysis was performed using the SPSS
standard version 19.0 software (SPSS, Inc., Chicago, IL, USA). Data
are represented as mean ± standard deviation from at least three
independent determinations. The independent Student's t-test was
calculated to compare the results between the two groups. P<0.05
was considered to be statistically significant.
Results
GPER protein levels are conversely
correlated with Bim proteins in the recurred breast tumors treated
with TAM
A total of 114 breast cancer tissues were eligible
for analysis according to our previous inclusion criteria; of
these, 106 recurrent breast tumor samples (66 local and 40 distant
metastases) were GPER positive identified by IHC staining. Among
the 106 GPER+ specimens, GPER expression was increased
in 73.58% (78/106), decreased in 5.66% (6/106) and unchanged in
20.76% (22/106) compared with the matched primary tumor tissues
(PTs). All these GPER+ recurred tumor tissues (RTs) and
the paired PTs were used to determine the expression of Bim
(Bcl2-L11), an aberrant downregulated gene identified by cDNA-array
in TAM-resistant breast cancer cells. Bim and GPER were shown to be
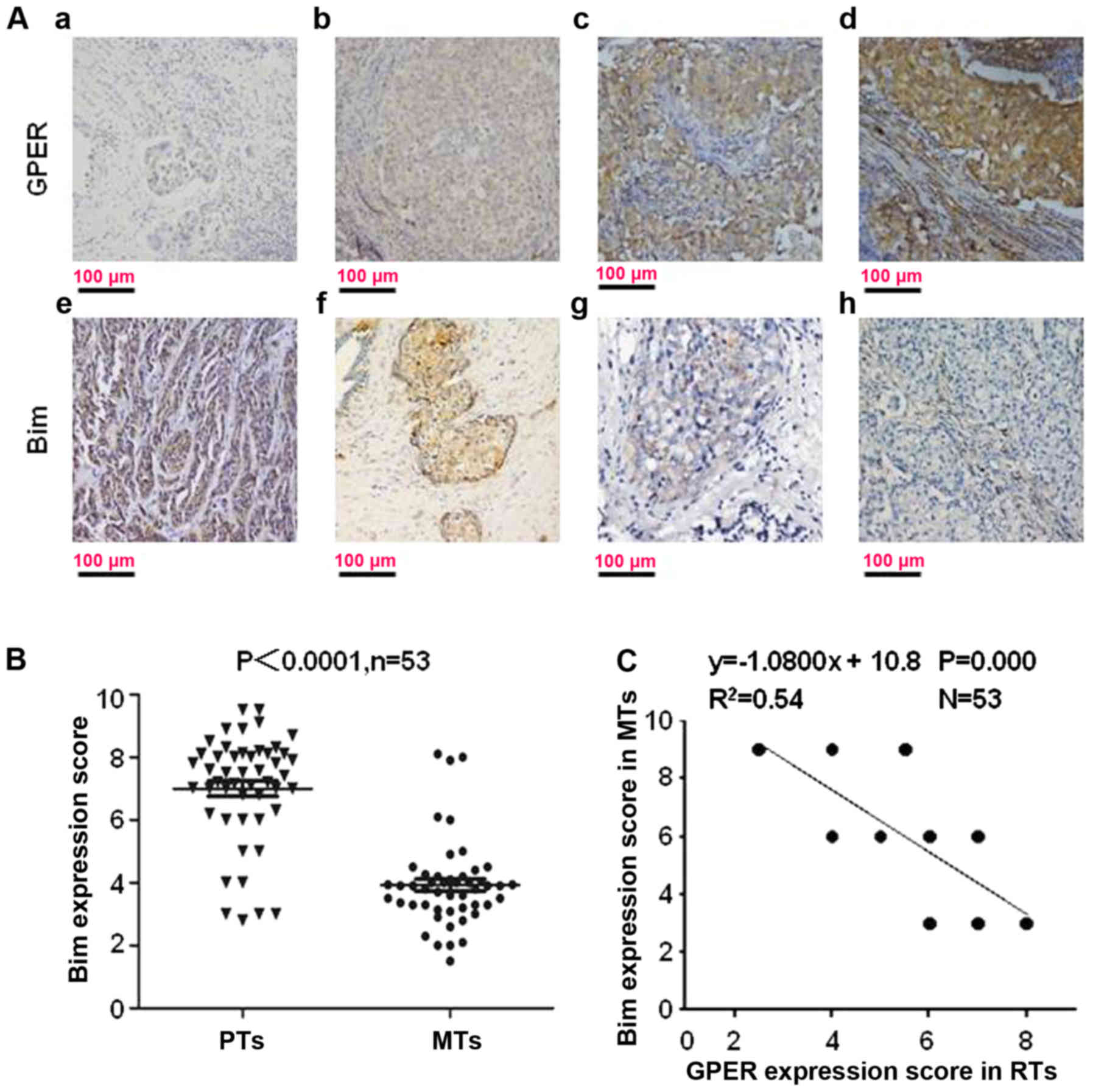
located in the cell cytoplasm (Fig.
1A).
As shown in our reports, the mean IHC score for GPER
is significantly increased in RTs (6.23±0.91) compared with that in
PTs (3.46±1.07) (P=0.001). To understand whether Bim is involved in
GPER-mediated TAM-resistance, Bim expression was scored in PTs and
the paired RTs. Bim expression was decreased in 83.02%, 44/53),
increased in 3.78% (4/106) and unchanged in 13.21% (14/106) of
these 53 GPER+ tumors which relapsed during TAM
treatment (Fig. 1B). The mean IHC
score for Bim was 7.27±0.56 in PTs and 4.02±0.49 in RTs (Fig. 1B; P<0.0001). Among the 53
GPER+ RTs, Bim expression had a converse correlation
with GPER expression (Fig. 1C;
R2=0.54).
Bim is a pro-apoptosis effector of
TAM-induced apoptosis in breast cancer cells
It has been reported that Bim plays an important
role in cell apoptosis (35–37).
To investigate the potential role of Bim protein in the process of
TAM-induced apoptosis, we further determined the Bim expression in
TAM-sensitive MCF-7 cells (MCF-7) and TAM-resistant MCF-7 cells
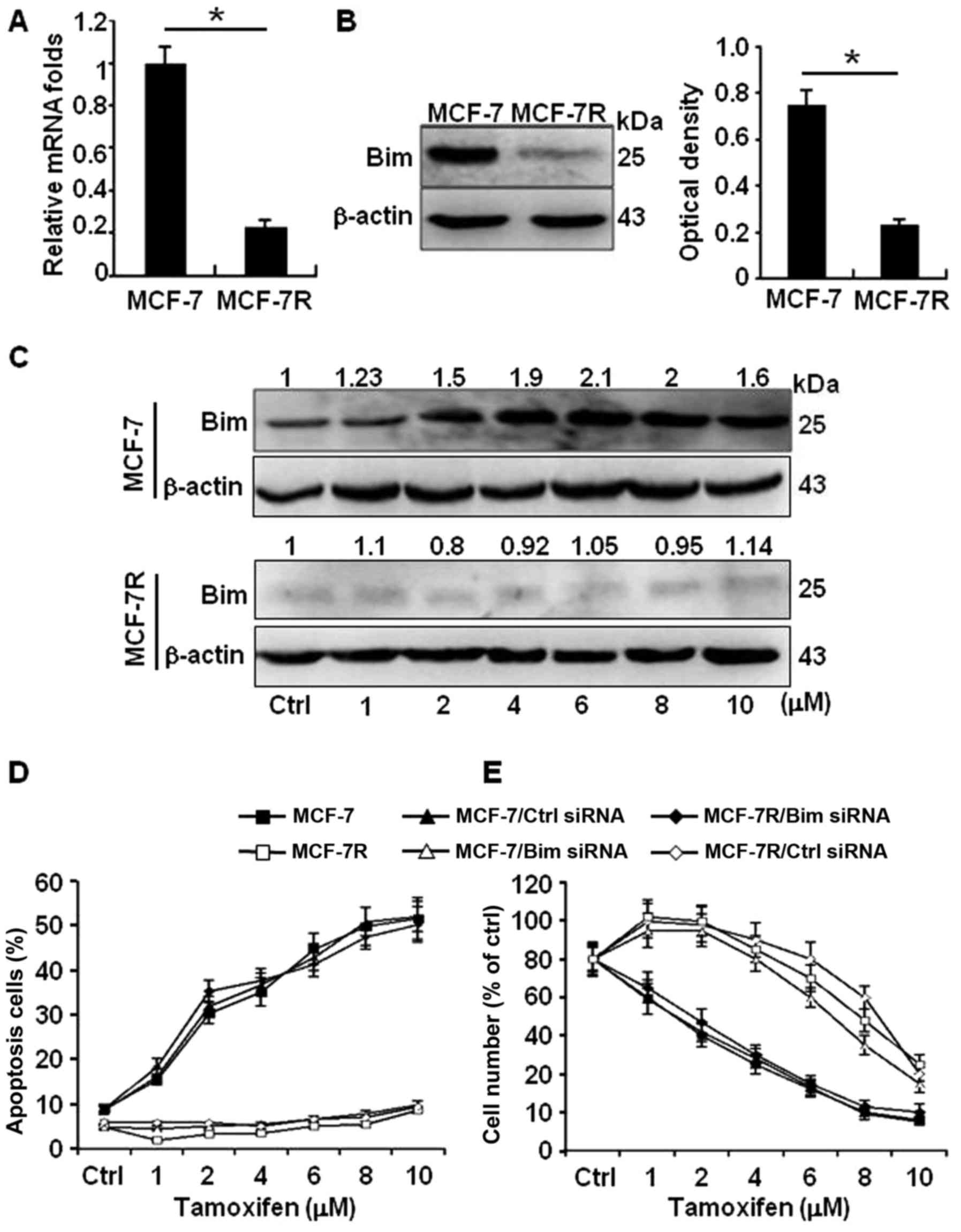
(MCF-7R) by Q-PCR and western blot analyses. The transcription
activity of Bim was not changed by Q-PCR. In contrast to their
protein levels, high level of Bim in MCF-7 and obviously reduced
Bim in MCF-7R were confirmed in western blot analyses (Fig. 2A and B). Moreover, the protein
levels of Bim in MCF-7 were gradually increased in a TAM-dose
dependent pattern (Fig. 2C, upper
panel); however, the weak Bim protein had no response to TAM
stimulation in MCF-7R (Fig. 2C,
down panel). Consistent with these findings, more apoptotic cells
by TAM were detected in MCF-7 cells than in MCF-7R cells; knockdown
of Bim in MCF-7 cells significantly decreased the apoptosis cell
ratio; however, ectopic expression of Bim in MCF-7R notably
increased their sensitivity to TAM (Fig. 2D). In addition, similar cell
survival results were acquired by MTT assays (Fig. 2E). These data suggest high level of
Bim protein in MCF-7 is an effector to TAM treatment.
Bim induces apoptosis through activating
PARP and caspase-3 to regulate the response to TAM
To further confirm the role of Bim on apoptosis in
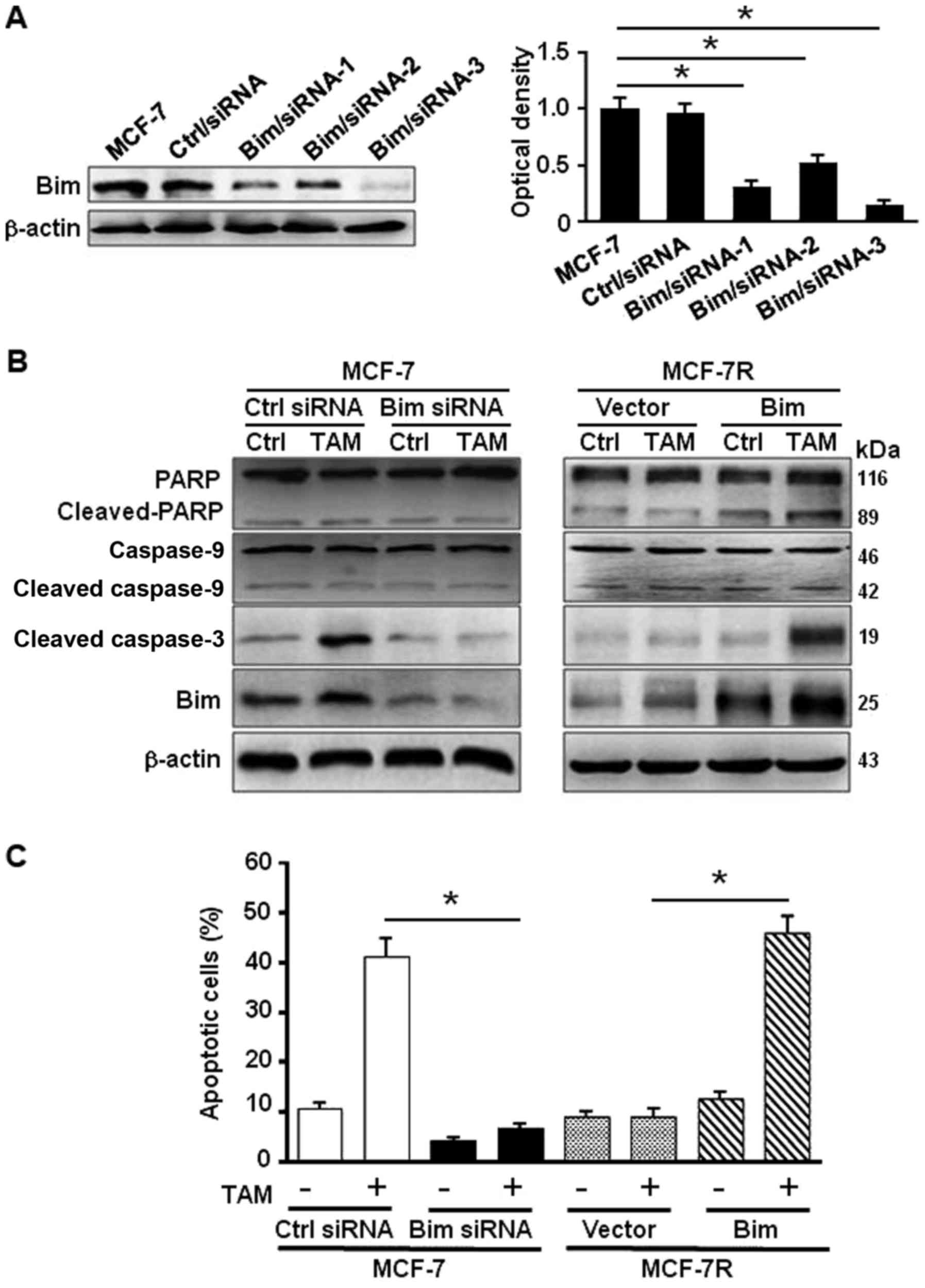
response to TAM in MCF-7, specific siRNAs against Bim (Fig. 3A) were employed. Knockdown of Bim
in MCF-7 attenuated the activated PARP and cleaved caspase-3 in
exposure to TAM treatment (Fig.
3B, left panel). Overexpression of Bim in MCF-7R increased the
levels of activated PARP and cleaved caspase-3 under treatment of
TAM (Fig. 3B, right panel).
Consistently, the percentage of apoptosis cells was 40.5% in MCF-7
and 6.75% in Bim-knocked down MCF-7 cells, whereas the apoptotic
cells were 6.8% in MCF-7R and 44.6% in MCF-7R with ectopic Bim
under TAM treatment (Fig. 3C).
These data indicate that Bim plays a critical role in TAM-induced
apoptosis in MCF-7 cells through activation of PARP and
caspase-3.
TRIM2 promotes Bim degradation in
TAM-resistant breast cancer cells
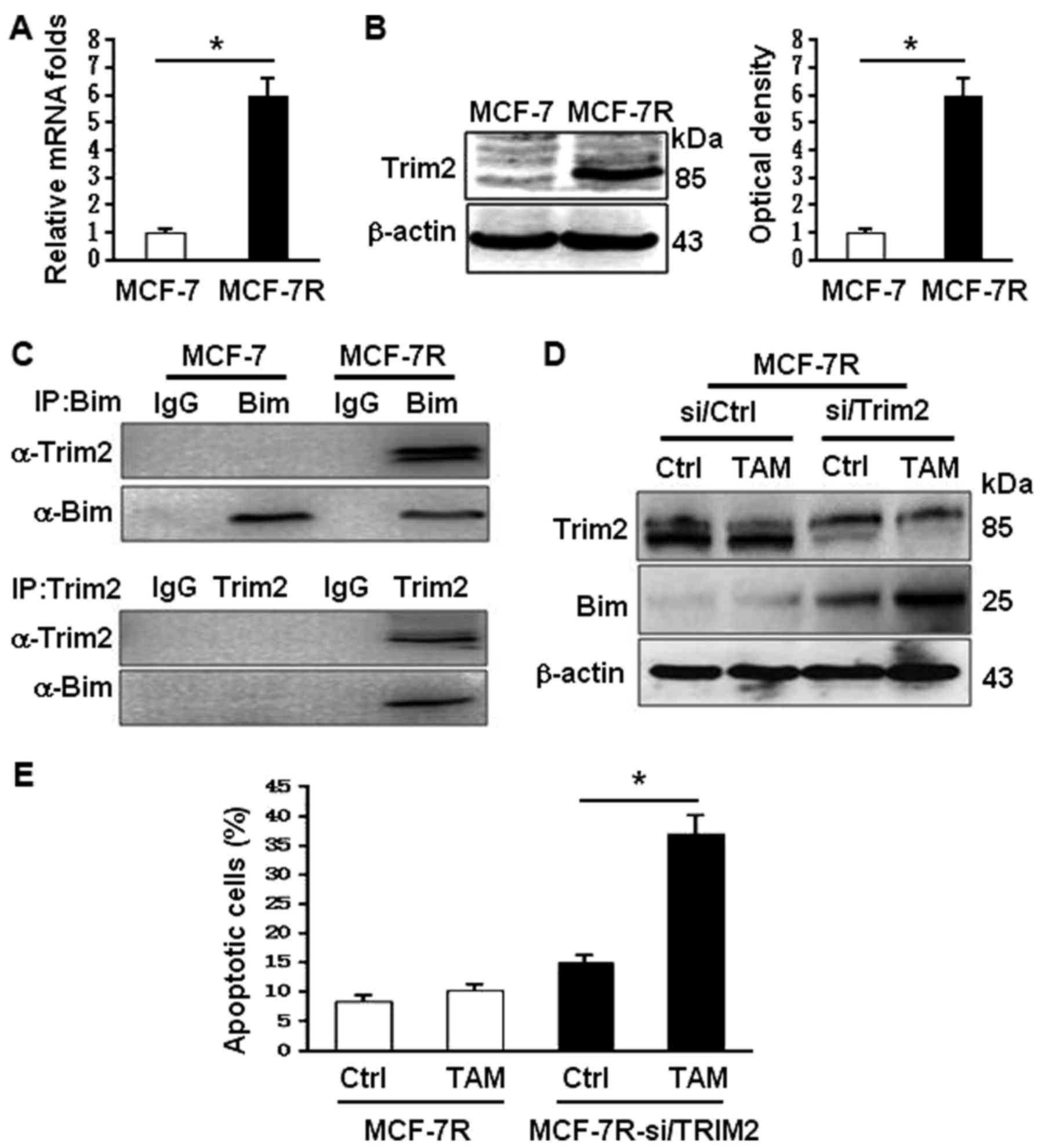
To understand why Bim was decreased in MCF-7R,
bioinformatics was applied. It was found that a tripartite motif
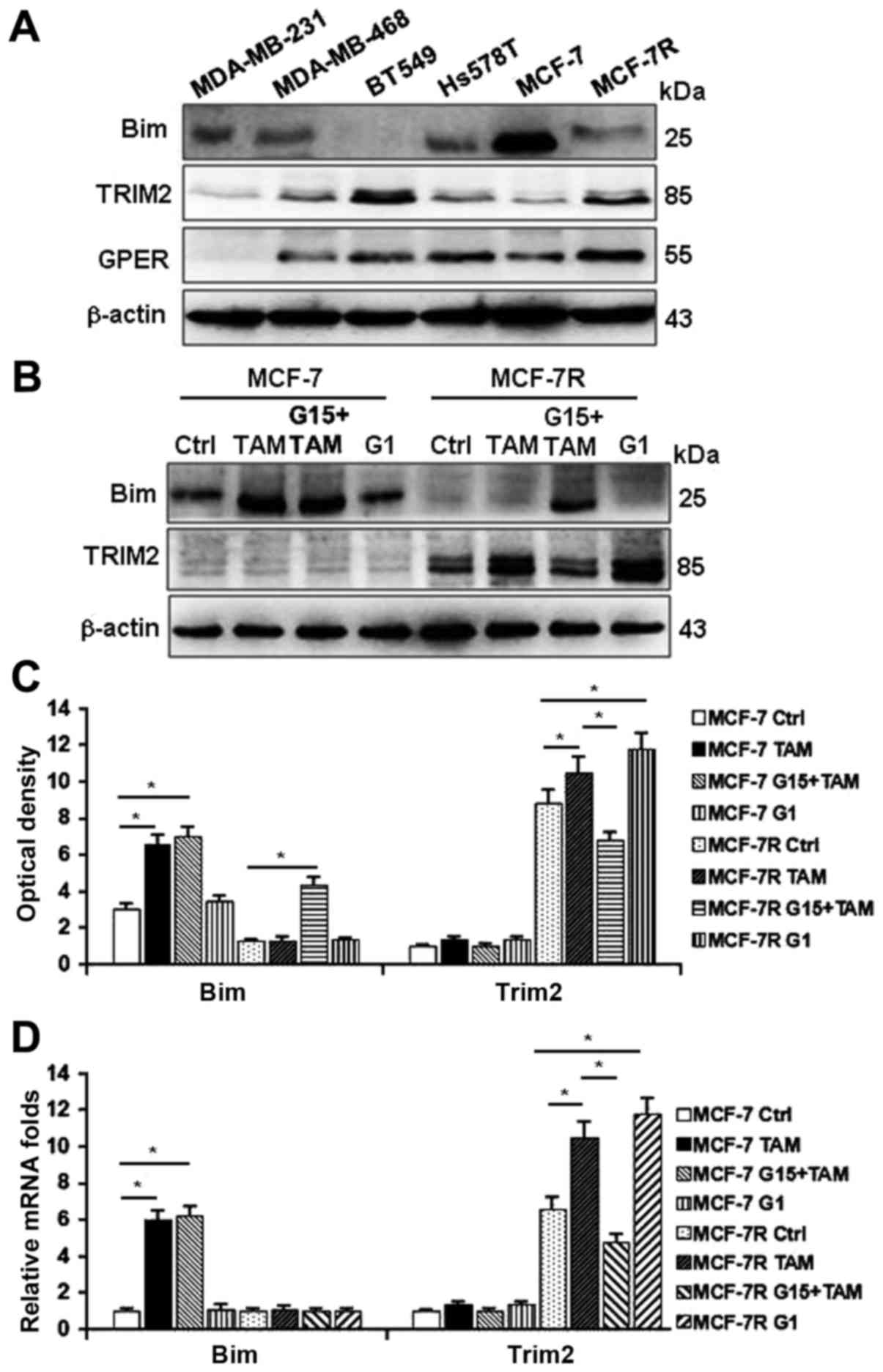
protein 2 (TRIM2) upregulated in MCF-7R cells (Fig. 4A and B) may interact with Bim.
TRIM2 has been reported to be function as the E3 ligase to enhance
Bim ubiquitinylation (23,24). Indeed, the binding of TRIM2 and Bim
in MCF-7R was confirmed using co-immunoprecipitation analysis
(Fig. 4C). Knockdown of Trim2
using specific siRNA in MCF-7R, the levels of Bim were notably
increased and its expression could be regulated by TAM stimulation
(Fig. 4D). Upregulation of Bim
expression by knocking down Trim2 in MCF-7R rescued its potential
to TAM sensitivity, suggesting the TRIM2-mediated ubiquitinylation
degradation of Bim may contribute to TAM-resistance of MCF-7.
Activation of GPER leads to enhanced
Trim2 expression in TAM-resistant breast cancer cells
Previous reports have revealed an activated GPER in
TAM-resistant breast cancer cells (2,26),
and the above data also display an adverse relationship between Bim
and GPER in TAM-resistant breast tumors. The expression of Bim,
TRIM and GPER were further determined in ER-negative breast cancer
cells and TAM-resistant MCF-7R cells. As shown in Fig. 5A, lower levels of Bim, and higher
levels of TRIM2 and GPER were detected in most of ER-breast cancer
cells and MCF-7R cells compared with TAM-sensitive MCF-7 cells.
Treating cells with TAM, G1 (the GPER-specific agonist) or TAM
combined with G15 (the GPER-specific antagonist), Bim could be
stimulated by TAM, but not by G1 in MCF-7 cells (Fig. 5B and C, left panel). However, the
levels of Bim were further decreased and TRIM2 expression was
enhanced by TAM and G1 in MCF-7R cells (Fig. 5B and C, right panel), suggesting
that activation of GPER may govern the levels of TRIM2 and Bim. In
contrast to their protein levels, the transcription activity of Bim
was changed under the treatments of TAM in MCF-7R cells (Fig. 5D, left panel); however, mRNA
expression of Trim2 were obviously increased under the treatment of
TAM and G1, but attenuated by G15 (Fig. 5D, right panel). These data suggest
that activation of GPER is necessary for TRIM2 expression.
The MAPK/ERK signaling pathway is
directly responsible for decreased Bim protein levels in MCF-7R
cells
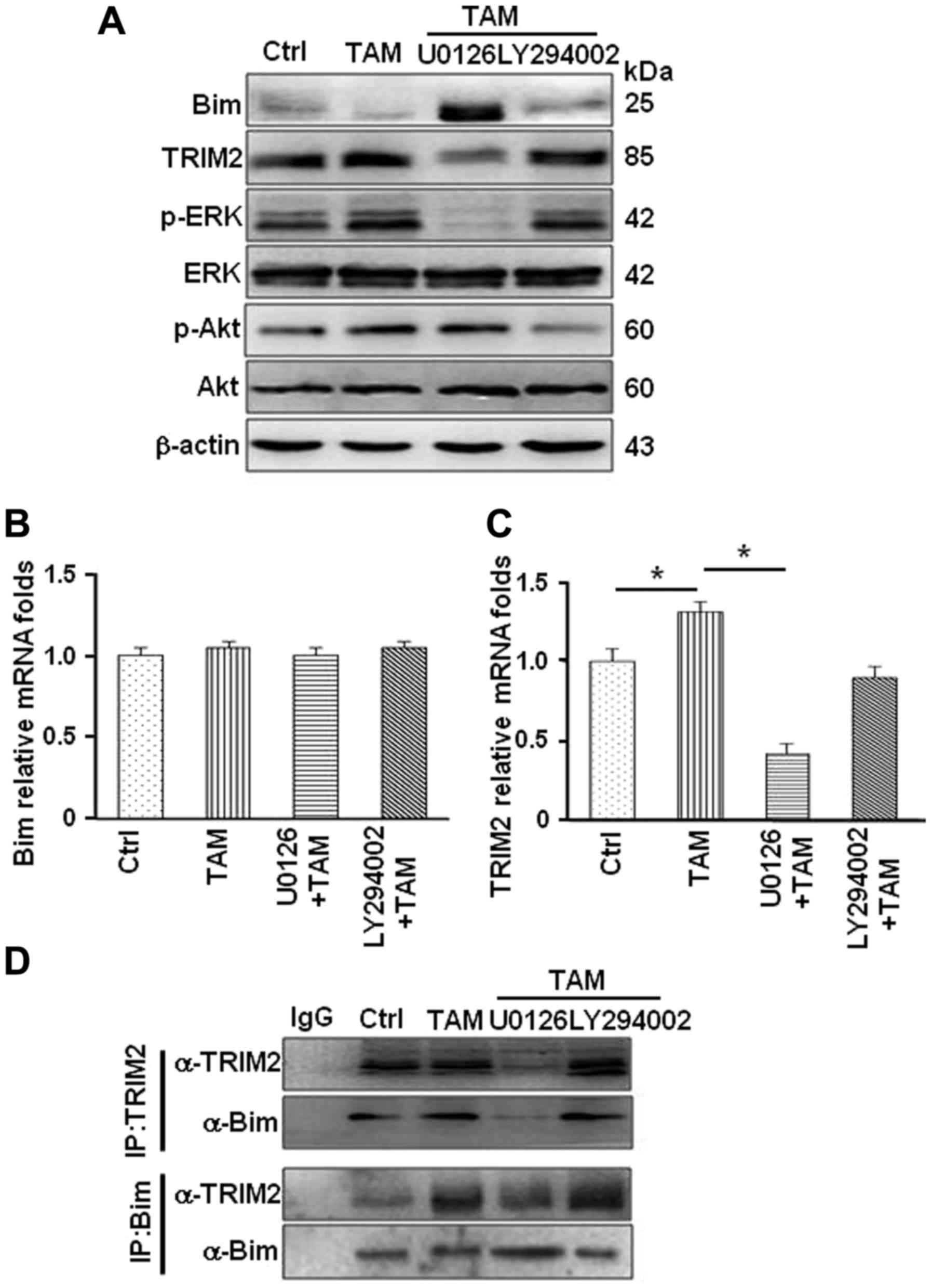
It has been previously demonstrated that TAM
activates the MAPK/ERK and PI3K/AKT pathways through GPER/EGFR
signaling axis in MCF-7R cells (2,26–29).
To investigate whether these path ways are involved in regulating
Bim protein levels, MCF-7R cells were treated with TAM alone and
TAM combined with U0126 (the MAPK/ERK inhibitor) or LY294002 (the
PI3K inhibitor). The protein levels of Bim were notably increased
in MCF-7R treated with TAM combined with U0126, not LY294002
(Fig. 6A), indicating that
MAPK/ERK signaling pathway may involve in regulation of Bim protein
levels. Treating cells with TAM alone or TAM combined with specific
inhibitor, mRNA expression of Bim was not altered (Fig. 6B). However, mRNA expression of
Trim2 was significantly reduced after inhibiting MAPK/ERK
activation in MCF-7R cells (Fig.
6C). This led to decrease in the interaction between TRIM2 and
Bim under the treatment of U0126 in MCF-7R (Fig. 6D). Thus, GPER and its downstream
MAPK/ERK signaling pathway are involved in regulation of Trim2
expression and affect the binding between TRIM2 and Bim resulting
in reduced Bim in TAM-resistant breast cancer cells.
Discussion
Estrogen plays a crucial role in ER+
breast cancer development and ER is the main mediator of the
estrogenic effect. Tamoxifen, an estrogen receptor antagonist, has
been widely used in clinical endocrine therapy (1,2).
However, breast cancer endocrine resistance is still the main
obstacle in ER+ breast cancer recurrence and metastasis
(38,39). To overcome endocrine resistance is
one of the important aims for hormone-dependent breast cancer
patients (2,40). GPER, a novel membrane-bound
estrogen receptor, has been demonstrated to contribute to the
development of TAM resistance (26–28).
However, the detail mechanism has not been fully elucidated. In our
previous study, we demonstrated that translocation of GPER from the
cytoplasm to cell membrane play a key role in mediating crosstalk
between GPER and EGFR in tamoxifen resistant breast cancer cells
(26,28), which further stimulates the
activation of MAPK and AKT signaling pathways and regulates
associated gene transcription in the tamoxifen tolerance (28). In the present study, we found that
GPER-MAPK/ERK signaling upregulates Trim2 gene expression, and
enhances TRIM2 protein binding to pro-apoptotic protein Bim, which
leads to ubiquitinylation degradation of Bim and thus plays a
critical role against apoptosis in TAM-resistant breast cancer
cells.
Bim is the BH3-only protein that may function as
death sensor that mediates the activation of the mitochondrial or
caspase-independent apoptosis pathway. It has been demonstrated
that Bim is a drug-induced apoptosis regulatory protein in breast
cancer (9,11). Here, we found that low level of Bim
was detected in TAM-resistant breast tumor tissues and breast
cancer cell lines. However, how Bim was downregulated and
contributed to TAM-resistance in ER+ breast carcinoma
are still not unveiled. Using MCF-7 and TAM-resistant MCF-7R as
cell models, we found Bim was upregulated in TAM-sensitive MCF-7
cells and triggered apoptosis in response to TAM stimulation via
activation of caspase-3 and PARP in MCF-7 cells. Knockdown of Bim
in MCF-7 cells decreased the sensitiveness of MCF-7 to TAM
treatment; overexpression of Bim in TAM-resistant MCF-7R cells
increased its sensitivity to TAM. Our data further verified the
important role of Bim in drug-induced apoptosis (9,11).
TRIM2 functions as a direct regulator to Bim
degradation in TAM-resistant breast cancer cells. TRIM2 was
increased to GPER activation in TAM-resistant MCF-7R cells. Under
treatments of TAM and the GPER-specific agonist G1, TRIM2
expression was dramatically enhanced along with reduced Bim protein
in MCF-7R cells, indicating a GPER-dependent expression of TRIM2 in
TAM-resistant breast cancer cells. Notably, the transcription
activity of Bim was not changed under these stimulators, suggesting
a post-transcription regulation of Bim in MCF-7R cells. Our data
show that the binding between TRIM2 and Bim under GPER activation
may lead to Bim degradation in TAM-resistant cells. These findings
were supported by other studies (24).
The present study provided evidence that
GPER-EGFR-MAPK/ERK signaling, not GPER-EGFR-PI3K/Akt signaling, is
the key regulator for Bim degradation in TAM-resistant breast
cancer cells. Here, we verified that GPER-MAPK/ERK is necessary for
Bim degradation. The binding between TRIM2 and Bim was dependent on
the levels of TRIM2 protein and regulated by the GPER-MAPK/ERK
signaling. Blockage of GPER-MAPK/ERK signaling using G15 or U0126
decreased TRIM2 protein levels and reduced the binding between
TRIM2 and Bim in MCF-7R cells. TRIM2 has been defined as E3
ubiquitin ligase and plays a key role in ubiquitinylation
degradation of proteins (24).
Indeed, a previous study by Thompson et al disclosed that
phosphorylation of p42/p44 MAPK is necessary for the binding of
TRIM2 to Bim in ischemic tolerance-induced neuroprotection
(22). These data highlight a
novel insight of GPER-MAPK/ERK signaling in mediating TAM-induced
drug resistance in ER+ breast cancer patients.
In conclusion, our findings provide new evidence to
GPER in TAM-induced resistance of ER+ breast cancer.
Targeting TAM/GPER/ERK signaling and TRIM2 gene may be a more
effective way in overcoming TAM-resistance for ER+
breast cancer patients. However, more prospective clinical
investigations need to be undertaken in the future.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (NSFC 81072149). Manran Liu is
supported by the Program of National Natural Science Foundation of
China (NSFC 81472476 and 31171336).
References
|
1
|
Briest S and Stearns V: Tamoxifen
metabolism and its effect on endocrine treatment of breast cancer.
Clin Adv Hematol Oncol. 7:185–192. 2009.PubMed/NCBI
|
|
2
|
Yuan J, Liu M, Yang L, Tu G, Zhu Q, Chen
M, Cheng H, Luo H, Fu W, Li Z, et al: Acquisition of
epithelial-mesenchymal transition phenotype in the
tamoxifen-resistant breast cancer cell: A new role for G
protein-coupled estrogen receptor in mediating tamoxifen resistance
through cancer-associated fibroblast-derived fibronectin and
β1-integrin signaling pathway in tumor cells. Breast Cancer Res.
17:692015. View Article : Google Scholar
|
|
3
|
Willis SN and Adams JM: Life in the
balance: How BH3-only proteins induce apoptosis. Curr Opin Cell
Biol. 17:617–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strasser A: The role of BH3-only proteins
in the immune system. Nat Rev Immunol. 5:189–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Villunger A, Scott C, Bouillet P and
Strasser A: Essential role for the BH3-only protein Bim but
redundant roles for Bax, Bcl-2, and Bcl-w in the control of
granulocyte survival. Blood. 101:2393–2400. 2003. View Article : Google Scholar
|
|
6
|
Kirschnek S, Vier J, Gautam S, Frankenberg
T, Rangelova S, Eitz-Ferrer P, Grespi F, Ottina E, Villunger A,
Häcker H, et al: Molecular analysis of neutrophil spontaneous
apoptosis reveals a strong role for the pro-apoptotic BH3-only
protein Noxa. Cell Death Differ. 18:1805–1814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang WF, Liu AH, Zhao HJ, Dong HM, Liu LY
and Cai SX: BIM gene polymorphism lowers the efficacy of EGFR-TKIs
in advanced nonsmall cell lung cancer with sensitive EGFR
mutations: A systematic review and meta-analysis. Medicine
(Baltimore). 94:e12632015.J. View Article : Google Scholar
|
|
8
|
Gu Z, Wang X, Qi R, Wei L, Huo Y, Ma Y,
Shi L, Chang Y, Li G and Zhou L: Oridonin induces apoptosis in
uveal melanoma cells by upregulation of Bim and downregulation of
Fatty Acid Synthase. Biochem Biophys Res Commun. 457:187–193. 2015.
View Article : Google Scholar
|
|
9
|
Merino D, Best SA, Asselin-Labat ML,
Vaillant F, Pal B, Dickins RA, Anderson RL, Strasser A, Bouillet P,
Lindeman GJ, et al: Pro-apoptotic Bim suppresses breast tumor cell
metastasis and is a target gene of SNAI2. Oncogene. 34:3926–3934.
2015. View Article : Google Scholar
|
|
10
|
Park SH, Ito K, Olcott W, Katsyv I,
Halstead-Nussloch G and Irie HY: PTK6 inhibition promotes apoptosis
of Lapatinib-resistant Her2+ breast cancer cells by
inducing Bim. Breast Cancer Res. 17:862015. View Article : Google Scholar
|
|
11
|
Follin-Arbelet V, Misund K, Naderi EH,
Ugland H, Sundan A and Blomhoff HK: The natural compound forskolin
synergizes with dexamethasone to induce cell death in myeloma cells
via BIM. Sci Rep. 5:130012015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reginato MJ, Mills KR, Becker EB, Lynch
DK, Bonni A, Muthuswamy SK and Brugge JS: Bim regulation of lumen
formation in cultured mammary epithelial acini is targeted by
oncogenes. Mol Cell Biol. 25:4591–4601. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai Y and Grant S: BCL2L11/Bim as a
dual-agent regulating autophagy and apoptosis in drug resistance.
Autophagy. 11:416–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zoumpoulidou G, Broceño C, Li H, Bird D,
Thomas G and Mittnacht S: Role of the tripartite motif protein 27
in cancer development. J Natl Cancer Inst. 104:941–952. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu C, Huang X, Hou S, Hu B and Li H:
Silencing of tripartite motif (TRIM) 29 inhibits proliferation and
invasion and increases chemosensitivity to cisplatin in human lung
squamous cancer NCI-H520 cells. Thorac Cancer. 6:31–37. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozato K, Shin DM, Chang TH and Morse HC
III: TRIM family proteins and their emerging roles in innate
immunity. Nat Rev Immunol. 8:849–860. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu F, Xiong JP, Deng J and Xiang XJ:
TRIM29 functions as an oncogene in gastric cancer and is regulated
by miR-185. Int J Clin Exp Pathol. 8:5053–5061. 2015.PubMed/NCBI
|
|
19
|
Wang Y and He D, Yang L, Wen B, Dai J,
Zhang Q, Kang J, He W, Ding Q and He D: TRIM26 functions as a novel
tumor suppressor of hepatocellular carcinoma and its downregulation
contributes to worse prognosis. Biochem Biophys Res Commun.
463:458–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams MD, Zhang L, Elliott DD, Perrier
ND, Lozano G, Clayman GL and El-Naggar AK: Differential gene
expression profiling of aggressive and nonaggressive follicular
carcinomas. Hum Pathol. 42:1213–1220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ivanov SV, Panaccione A, Nonaka D, Prasad
ML, Boyd KL, Brown B, Guo Y, Sewell A and Yarbrough WG: Diagnostic
SOX10 gene signatures in salivary adenoid cystic and breast
basal-like carcinomas. Br J Cancer. 109:444–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thompson S, Pearson AN, Ashley MD, Jessick
V, Murphy BM, Gafken P, Henshall DC, Morris KT, Simon RP and Meller
R: Identification of a novel Bcl-2-interacting mediator of cell
death (Bim) E3 ligase, tripartite motif-containing protein 2
(TRIM2), and its role in rapid ischemic tolerance-induced
neuroprotection. J Biol Chem. 286:19331–19339. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balastik M, Ferraguti F, Pires-da Silva A,
Lee TH, Alvarez-Bolado G, Lu KP and Gruss P: Deficiency in
ubiquitin ligase TRIM2 causes accumulation of neurofilament light
chain and neurodegeneration. Proc Natl Acad Sci USA.
105:12016–12021. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mo Z, Liu M, Yang F, Luo H, Li Z, Tu G and
Yang G: GPR30 as an initiator of tamoxifen resistance in
hormone-dependent breast cancer. Breast Cancer Res. 15:R1142013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ignatov A, Ignatov T, Weissenborn C,
Eggemann H, Bischoff J, Semczuk A, Roessner A, Costa SD and
Kalinski T: G-protein-coupled estrogen receptor GPR30 and tamoxifen
resistance in breast cancer. Breast Cancer Res Treat. 128:457–466.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ignatov A, Ignatov T, Roessner A, Costa SD
and Kalinski T: Role of GPR30 in the mechanisms of tamoxifen
resistance in breast cancer MCF-7 cells. Breast Cancer Res Treat.
123:87–96. 2010. View Article : Google Scholar
|
|
27
|
Prossnitz ER, Arterburn JB, Smith HO,
Oprea TI, Sklar LA and Hathaway HJ: Estrogen signaling through the
transmembrane G protein-coupled receptor GPR30. Annu Rev Physiol.
70:165–190. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fujiwara S, Terai Y, Kawaguchi H, Takai M,
Yoo S, Tanaka Y, Tanaka T, Tsunetoh S, Sasaki H, Kanemura M, et al:
GPR30 regulates the EGFR-Akt cascade and predicts lower survival in
patients with ovarian cancer. J Ovarian Res. 5:352012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Filardo EJ, Quinn JA, Bland KI and
Frackelton AR Jr: Estrogen-induced activation of Erk-1 and Erk-2
requires the G protein-coupled receptor homolog, GPR30, and occurs
via trans-activation of the epidermal growth factor receptor
through release of HB-EGF. Mol Endocrinol. 14:1649–1660. 2015.
View Article : Google Scholar
|
|
30
|
Lv Y, Song S, Zhang K, Gao H and Ma R:
CHIP regulates AKT/FoxO/Bim signaling in MCF7 and MCF10A cells.
PLoS One. 8:e833122013. View Article : Google Scholar :
|
|
31
|
Periyasamy-Thandavan S, Takhar S, Singer
A, Dohn MR, Jackson WH, Welborn AE, LeRoith D, Marrero M,
Thangaraju M, Huang S, et al: Insulin-like growth factor 1
attenuates antiestrogen- and antiprogestin-induced apoptosis in
ER+ breast cancer cells by MEK1 regulation of the
BH3-only pro-apoptotic protein Bim. Breast Cancer Res. 14:R522012.
View Article : Google Scholar
|
|
32
|
Yu T, Liu M, Luo H, Wu C, Tang X, Tang S,
Hu P, Yan Y, Wang Z and Tu G: GPER mediates enhanced cell viability
and motility via non-genomic signaling induced by 17β-estradiol in
triple-negative breast cancer cells. J Steroid Biochem Mol Biol.
143:392–403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu K, Li A, Rao M, Liu M, Dailey V, Yang
Y, Di Vizio D, Wang C, Lisanti MP, Sauter G, et al: DACH1 is a cell
fate determination factor that inhibits cyclin D1 and breast tumor
growth. Mol Cell Biol. 26:7116–7129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu
X, Powell MJ, Yang T, Gu W, Avantaggiati ML, et al: Hormonal
control of androgen receptor function through SIRT1. Mol Cell Biol.
26:8122–8135. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han BJ, Li W, Jiang GB, Lai SH, Zhang C,
Zeng CC and Liu YJ: Effects of daidzein in regards to cytotoxicity
in vitro, apoptosis, reactive oxygen species level, cell cycle
arrest and the expression of caspase and Bcl-2 family proteins.
Oncol Rep. 34:1115–1120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tomicic MT, Meise R, Aasland D, Berte N,
Kitzinger R, Krämer OH, Kaina B and Christmann M: Apoptosis induced
by temozolomide and nimustine in glioblastoma cells is supported by
JNK/c-Jun-mediated induction of the BH3-only protein BIM.
Oncotarget. 6:33755–33768. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang LM, Zhao XC, Sun WB, Li R and Jiang
XJ: Sevoflurane post-conditioning protects primary rat cortical
neurons against oxygen-glucose deprivation/resuscitation via
down-regulation in mitochondrial apoptosis axis of Bid, Bim,
Puma-Bax and Bak mediated by Erk1/2. J Neurol Sci. 357:80–87. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hiscox S, Jiang WG, Obermeier K, Taylor K,
Morgan L, Burmi R, Barrow D and Nicholson RI: Tamoxifen resistance
in MCF7 cells promotes EMT-like behaviour and involves modulation
of β-catenin phosphorylation. Int J Cancer. 118:290–301. 2006.
View Article : Google Scholar
|
|
39
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar
|
|
40
|
Thewes V, Simon R, Schroeter P, Schlotter
M, Anzeneder T, Büttner R, Benes V, Sauter G, Burwinkel B,
Nicholson RI, et al: Reprogramming of the ERRα and ERα target gene
landscape triggers tamoxifen resistance in breast cancer. Cancer
Res. 75:720–731. 2015. View Article : Google Scholar : PubMed/NCBI
|