Introduction
Melanoma, a malignant tumor of melanocytes, is one
of the most frequent malignancies and considered to be the most
aggressive type of skin cancer (1,2).
Melanoma is characterized by rapid progression and poor prognosis,
mainly due to the limited efficiency of treatments and development
of resistance to chemotherapy (3,4).
Although melanoma accounts for <1% of skin cancer cases, it
contributes to the majority of deaths that result from skin cancer.
It is estimated that one person will die from melanoma every hour
(5). Despite of the recent
advances in anti-melanoma chemotherapies and immunotherapies,
currently available treatment strategies are only able to prolong
life for just a few months before the disease relapses, leading to
death (6,7). Therefore, there is an urgent need for
novel therapeutic strategies to improve clinical outcomes for these
patients.
Long non-coding RNAs (lncRNAs), ranging from 200
nucleotides to over 10 kb, have been found to dysregulate in many
human diseases and disorders, including various types of cancer
(8,9). lncRNAs have demonstrated potential
roles in both oncogenic and tumor suppressive pathways (10). For example, both MEG3 and GAS5
exert oncogenic activities in gastric cancer, and the
overexpression of them is often associated with poor prognosis
(11,12). MALAT1 promotes osteosarcoma growth,
also exerting an oncogenic activity (13). BRAF-activated non-coding RNA
(BANCR), a 693-bp lncRNA on chromosome 9, has been reported to
exert oncogenic acvities in many cancers under some specific
conditions, such as non-small cell lung cancer, papillary thyroid
carcinoma and colorectal cancer (14–16).
Recent studies reported that BANCR is abnormally overexpressed in
human malignant melanoma cell lines and tissues, and promotes
proliferation and migration in malignant melanoma (17,18).
These results suggest the oncogenic role and potential of BANCR as
a therapeutic target for melanoma. While these findings demonstrate
a clear correlation between BANCR and melanoma, the specific effect
of BANCR on melanoma tumorigenesis and the mechanisms involved
remain to be investigated.
MicroRNAs (miRNAs, miRs), about 22 nucleotides long,
regulate the expression of most genes in human, but we are only now
beginning to understand how they are generated, assembled into
functional complexes and degraded (19). Recent studies reported that the
interplay between miRNAs and lncRNAs plays an important role during
the tumorigenic process, providing new insight into the possible
regulatory mechanisms in cancers. For example, BANCR promotes
endometrial cancer cell proliferation and invasion by regulating
MMP2 and MMP1 via ERK/MAPK signaling pathway (20). Particularly, many lncRNA/miRNA axes
were reported to function in melanoma. lncRNA CCAT1 upregulates
proliferation and invasion in melanoma cells via suppressing
miR-33a (21). Besides,
deregulation of miR-183 promotes melanoma development via lncRNA
MALAT1 regulation (22). These
studies illustrate that the lncRNA-miRNA interaction might be
important in the process of melanoma, laying the foundation for
current study. The Notch signaling through Notch receptors
regulates cell proliferation and cell survival in several types of
cancer including melanoma. A previous study showed that the
activation of Notch may represent an early event in melanocytic
tumor growth and upregulation of this pathway may sustain tumor
progression (23). Also, other
studies showed that the activation of Notch pathway can be
controlled by microRNAs in melanoma. For example, miR-146a promotes
the initiation and progression of melanoma by activating Notch
signaling (24). In this study, we
aimed to investigate the effects of BANCR/miR-204/Notch2 axis on
the regulation of melanoma and the possible mechanisms underlying
these actions, and to provide a new aspect for development of novel
treatment strategies.
Materials and methods
Cell culture and tissues
Three human melanoma cells lines, including A375,
A875 and M14, and human epidermal melanocytes were obtained from
Regenerative Medicine Research Center/Key Laboratory of Transplant
Engineering and Immunology, Ministry of Health, West China
Hospital, Sichuan University. Cells were maintained in DMEM (Gibco,
Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS,
Gibco), 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco).
Human epidermal melanocyte was cultured in medium 245 contained
melanocyte growth supplement (Cascade Biologics, USA). All cell
lines were incubated at 37°C in 5% CO2 incubator. Fresh
melanoma tissue samples and age/gender-matched controls with
melanocytic nevus were obtained from 69 patients with advanced
melanoma who underwent surgery in the First Affiliated Hospital of
Zhengzhou University. All collected tissue samples were immediately
frozen in liquid nitrogen and stored at −80°C until further use.
The study was approved by the institutional review board of the
hospital, and written informed consent was obtained from each
patient. All diagnoses were confirmed by histopathological
examination.
Lentivirus transduction
According to the pre-experiment, the expression
level of BANCR was overexpressed in A375, A875 and M14 cells
compared with melanocytes. We chose A375 and M14 for further
investigations, for the expression of BANCR was higher in A375 and
M14 compared with cell lines A875. Recombinant lentiviruses
containing BANCR shRNAand shRNA scramble were purchased from
Shanghai GenePharma Co, Ltd (Shanghai, China). The sequences of
these shRNAs are: sh-BANCR-1: forward, 5′-GGACUCCAUGGCAAACGUUTT-3′
and reverse, 5′-AACGUUGCCAUGGAGUCCTT-3′; sh-BANCR-2, forward,
5′-GGAGUGGCGACUAUAGCAATT-3′ and reverse, 5′-UUGCUAUAGUCGCCAC
UCCTT-3′; shRNA scramble: forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
reverse, 5′-ACGUGACACGUUCGGAGAATTT-3′. According to the
pre-experiment, decreased level of BANCR was detected under
sh-BANCR-1 treatment, but a slight change was measured after
sh-BANCR-2 transfection. Thus, sh-BANCR-1 (BANCR shRNA) was chosen
for further study. The BANCR shRNAand shRNA scramble were
transfected into A375 and M14 cells cultured in six-well plates by
using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according
to the manufacturer's instructions. The MOI of these recombinant
lentiviruses in cells were at 20. Mimics/inhibitors specific for
miR-204 were designed and purchased from Shanghai GenePharma Co,
Ltd (Shanghai, China). Lentivirus transduction was performed on the
tenth day of A375 and M14 cells culture. One day before
transduction, both cells were seeded at a density of
1×105 cells/well into a 24-well culture plate. Cells
were harvested for subsequent experiments after transfection for 48
h. All above vectors were transfected into cells with Lipofectamine
2000 (Invitrogen) according to the manufacturer's instructions.
RNA extraction and real-time PCR
RNA extraction of the cells or tissue samples was
performed using TRIzol reagent (Invitrogen) according to the
manufacturer's instructions. A High Capacity cDNA Reverse
Transcription kit (Applied Biosystems, USA) was used to reverse
transcribe RNA samples. The primers used were as follows: GAPDH:
forward, 5′-AATCCCATCACCATCTTCCAG-3′ and reverse,
5′-GAGCCCCAGCCTTCTCCAT-3′; BANCR, forward,
5′-ACAGGACTCCATGGCAAACG-3′ and reverse,
5′-ATGAAGAAAGCCTGGTGCAGT-3′; miR-204, forward,
5′-CTGTCACTCGAGCTGCTGGAATG-3′ and reverse,
5′-ACCGTGTCGTGGAGTCGGCAATT-3′. To analyze the gene expression,
quantitative RT-PCR was performed using the Fast Start Universal
SYBR Green Mastermix (Roche, USA). GAPDH was used as the internal
control. Relative gene expression was analyzed by 2−ΔΔCt
method.
Cell counting kit-8 assay
Cell Proliferation assay using the cell counting
kit-8 (CCK-8; Dojindo, Japan) was performed according to the
manufacturer's instructions. Briefly, cells were seeded into
96-well plates at a density of 5×103 cells per well with
complete growth medium. After 24-h incubation, the cells were
transfected with BANCR shRNAor shRNA scramble. After 48-h of
culture, the growth medium was removed from each well, and then all
the wells were filled with 100 ml of fresh medium containing 10 ml
CCK-8 solutions. After incubated for 2 h at 37°C, cell
proliferation was assessed by absorbance detection at 450 nm with a
microplate reader (Biotek, Winooski, VT, USA).
Western blotting
RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA) was
used to lyse cells with Complete Protease Inhibitor Cocktail
(Roche). Cell lysates at a density of 7×106 cells were
transferred to 1.5 ml tube and kept at −20°C before use. The
protein concentration was determined by BCA protein assay (Tiangen,
China). Twenty micrograms of protein in each sample was separated
by 12% SDS-PAGE, which was conducted to separate the cellular
proteins. Proteins were separated by 5% stacking gel and 10%
running gel. After blocking for 1 h, the membranes were incubated
with specific primary antibodies overnight at 4°C followed by
secondary antibodies for 2 h at room temperature. The molecular
weight of candidate proteins was referred to the Pre-stained
SeeBlue rainbow marker (Invitrogen) loaded in parallel. The
following antibodies were used: anti-Ki-67, anti-PCNA,
anti-caspase-3, anti-Bcl-2, anti-MMP-9 (active), anti-MMP-14,
anti-VEGF, anti-Notch2, anti-NICD, anti-CSL (Abcam, Cambridge, UK),
anti-GAPDH (Sigma, St. Louis, MO, USA). Blots were detected using a
Kodak film developer (Fujifilm, Japan).
Cell apoptosis analysis
Cell apoptosis was analyzed after appropriate
plasmids transfection using staining with Annexin V and PI (BD
Bioscience, San Jose, CA) according to the manufacturer's
instructions. After incubated for 15 min at room temperature in the
dark, the cells were analyzed by using a flow cytometry. Annexin
V-positive and PI-negative/positive staining cells represent
apoptotic cells.
Transwell migration assay
After transfection for 48 h, the cells were seeded
onto the upper part of a Transwell chamber (Corning Costar,
Rochester, NY, USA) containing a gelatin-coated polycarbonate
membrane filter (pore size, 8 μm). Culture medium with 20%
FBS was added to the lower chamber to stimulate cell migration.
After 24-h incubation at 37°C with 5% CO2, the cells
were stained with crystal violet (Sigma-Aldrich, Germany). Cells on
the underside of the filters were observed under a microscope
(Olympus IX71; Olympus Corp., Tokyo, Japan) at a magnification of
×100 and counted.
Wound healing assay
After transfection for 48 h, the subconfluent cell
monolayers were scraped in three parallel lines with a P-200
pipette tip. The detached cells were washed off twice gently, and
the medium was then replaced with 1% FBS complete medium. To
visualize wound healing, images were taken at 0 and 24 h. The
percentage of wound closure (original width-width after cell
migration/original width) was calculated.
Zymography
The activities of MMP-2 and MMP-9 were measured by
gelatin zymography according to a previous report (25). Briefly, proteins were separated by
electrophoresis on a 10% polyacrylamide gel (Sigma-Aldrich, USA).
Gels were washed for 1 h and incubated in 50 mM Tris-HCl buffer, pH
7.2, containing 10 mM CaCl2, 0.02% NaN3, and
2.5% Triton X-100 for 20 h at 37°C. Gels were stained with 0.1%
Coomassie Blue R-250 in 20% methanol, and 10% acetic acid, and
de-stained in 20% methanol and 10% acetic acid. MMP-2 and MMP-9
activities were detected as clear bands on a blue background.
Zymograms were captured by gel visualization system and quantified
with gene tools software (Syngene, Cambridge, UK). The activities
of gelatinases were expressed as the optical density of the
substrate lysis zone.
Luciferase reporter assay
The Luc-BANCR-WT and Luc-BANCR-MUT were constructed
as follows. The wild-type BANCR and mutant BANCR were amplified by
chemical synthesis and were inserted into a luciferase reporter
vector (pGL4.74) to generate Luc-BANCR-WT and Luc-BANCR-MUT
constructs, respectively. M14 cells cultured in 24-well plates were
co-transfected with Luc-BANCR-WT/Luc-BANCR-MUT, together with
miR-204 mock or miR-204 mimic for 24 h. Luciferase activities were
detected by a dual-luciferase reporter system according to the
manufacturer (E2920; Promega). The data are measured using the
ELISA plate reader (Bio-Rad, USA) at the wavelength of 490 nm.
Bioinformatic study
In silico prediction of the interaction
between BANCR/miR-204 or miR-204/Notch2, were performed using Diana
Tools. In addition, Mut-BANCR transcript was prepared according to
the binding sites of miR-204 within BANCR transcript.
Animal study
Male BALB/c mice (18–20 g, 4–5 weeks old) were
purchased from Beijing HFK Bioscience Co. Ltd. and kept under
sterile specific pathogen-free conditions. All experiments were
approved by the Animal Care and Ethics Committee of the First
Affiliated Hospital of Zhengzhou University and were in accordance
with NIH animal use guidelines. M14 cells were transfected with
BANCR shRNA or shRNA scramble and harvested from 6-well cell
culture plates. Approximately 2×106 cells in 50%
Matrigel (Becton-Dickinson, San Diego, CA, USA) were injected
subcutaneously into the right flanks of the mice. Tumor growth was
measured every 5 days, and tumor volume was calculated using the
formula, volume = 1/2 (length × width2). Twenty-five
days after injection, the mice were sacrificed and tumor weights
were measured.
Statistical analysis
Experimental values were obtained from at least
three independent experiments. Data are expressed as means ± SD.
Statistical analysis was performed by using the Student's t-test or
one-way analysis of variance (GraphPad Prism; GraphPad Software
Inc, La Jolla, CA, USA), where appropriate. The Bonferroni post hoc
test was used to determine the source of observed differences.
P-values of <0.05 were considered statistically significant.
Results
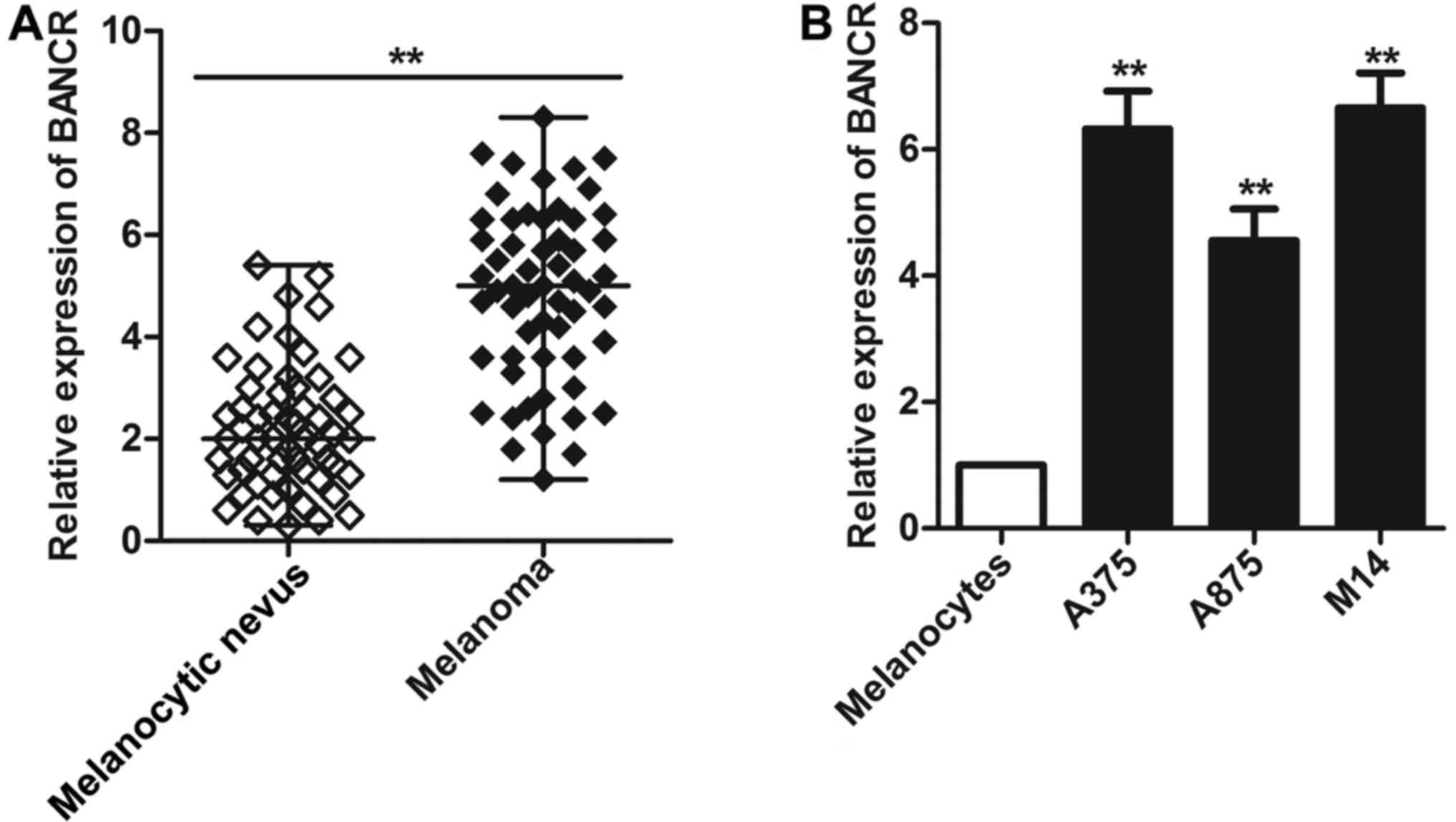
BANCR is frequently upregulated in
malignant melanoma tissues and cell lines
The expression of BANCR in melanoma tissues and cell
lines was evaluated using SYBR green quantitative PCR analysis by
real-time PCR. As shown in Fig.
1A, a significantly increased level of BANCR was seen in
patients with malignant melanoma compared with the levels detected
in age/gender-matched controls with melanocytic nevus (P<0.01).
Then we extended our test to three human melanoma cell lines (A375,
A875 and M14). As expected, high-level expression of BANCR was
observed in all three melanoma cell lines compared with human
epidermal melanocytes (P<0.01, Fig.
1B). These results suggest that BANCR may play an important
role in the development and progression of melanoma.
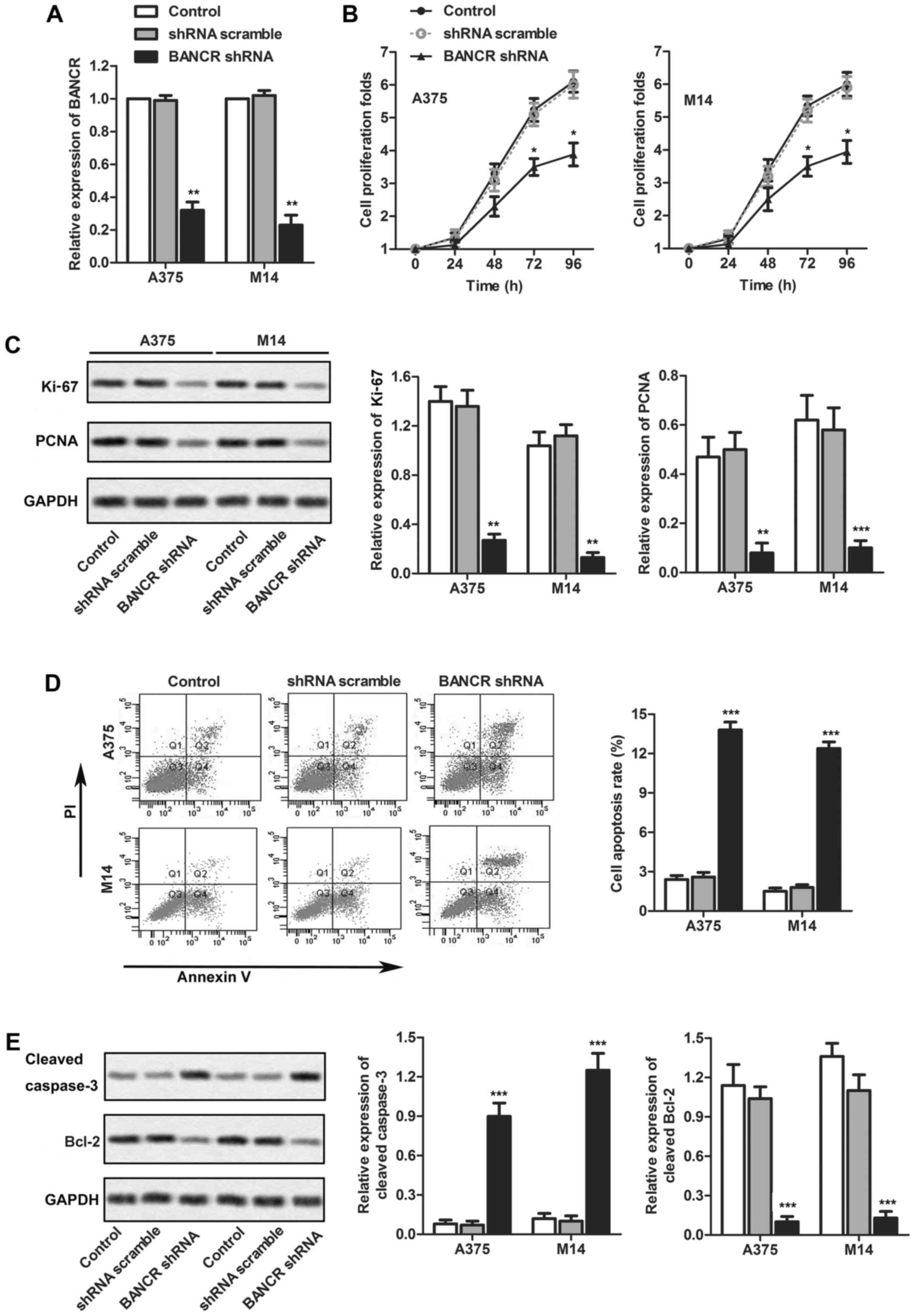
Knockdown of BANCR inhibits melanoma cell
proliferation in vitro
According to previous results, the overexpression of
BANCR was observed in all three cell lines. The expression level of
BANCR in A875 is a little lower compared with cell lines A375 and
M14, so we chose A375 and M14 for further investigation. To
determine the association of BANCR expression with melanoma cell
proliferation and apoptosis, BANCR shRNA or LV-NC (negative
control) were transfected into two human melanoma cell lines: A375
and M14. Compared with the control group, BANCR expression was
decreased in cells transfected with BANCR shRNA as measured by
real-time qPCR (P<0.01, Fig.
2A). Then, the effects of BANCR knockdown on the growth of
melanoma cells in vitro was measured by CCK-8 assays. When
compared with the control group, BANCR knockdown restrained the
proliferation folds of both A375 and M14 cell lines (P<0.05,
Fig. 2B). To better understand the
influence of BANCR on the melanoma cell proliferation, the
expression of Ki-67 and PCNA in the melanoma cell lines A375 and
M14 was assayed by western blotting (P<0.01 and P<0.05,
respectively, Fig. 2C). The
expression level of Ki-67 and PCNA were significantly reduced after
transfection with BANCR shRNA compared with control group. These
data strongly support a requirement for BANCR in the proliferation
of melanoma cells. Moreover, to explore potential mechanisms
underlying the growth-inhibitory effects of BANCR knockdown, we
assessed cell apoptosis in A375 and M14 cells by flow cytometry
analysis. As shown in Fig. 2D, the
proportion of apoptotic cells in the BANCR shRNA group was
significantly increased in comparison with that in the control
group. To further understand the influence of BANCR on the melanoma
cell apoptosis, the expression of cleaved caspase-3 and Bcl-2 in
the melanoma cell lines A375 and M14 were assayed by western
blotting (P<0.01, Fig. 2E). The
expression cleaved caspase-3 in BANCR shRNA group obviously
increased compared with the control group, while the expression of
Blc-2 reduced.
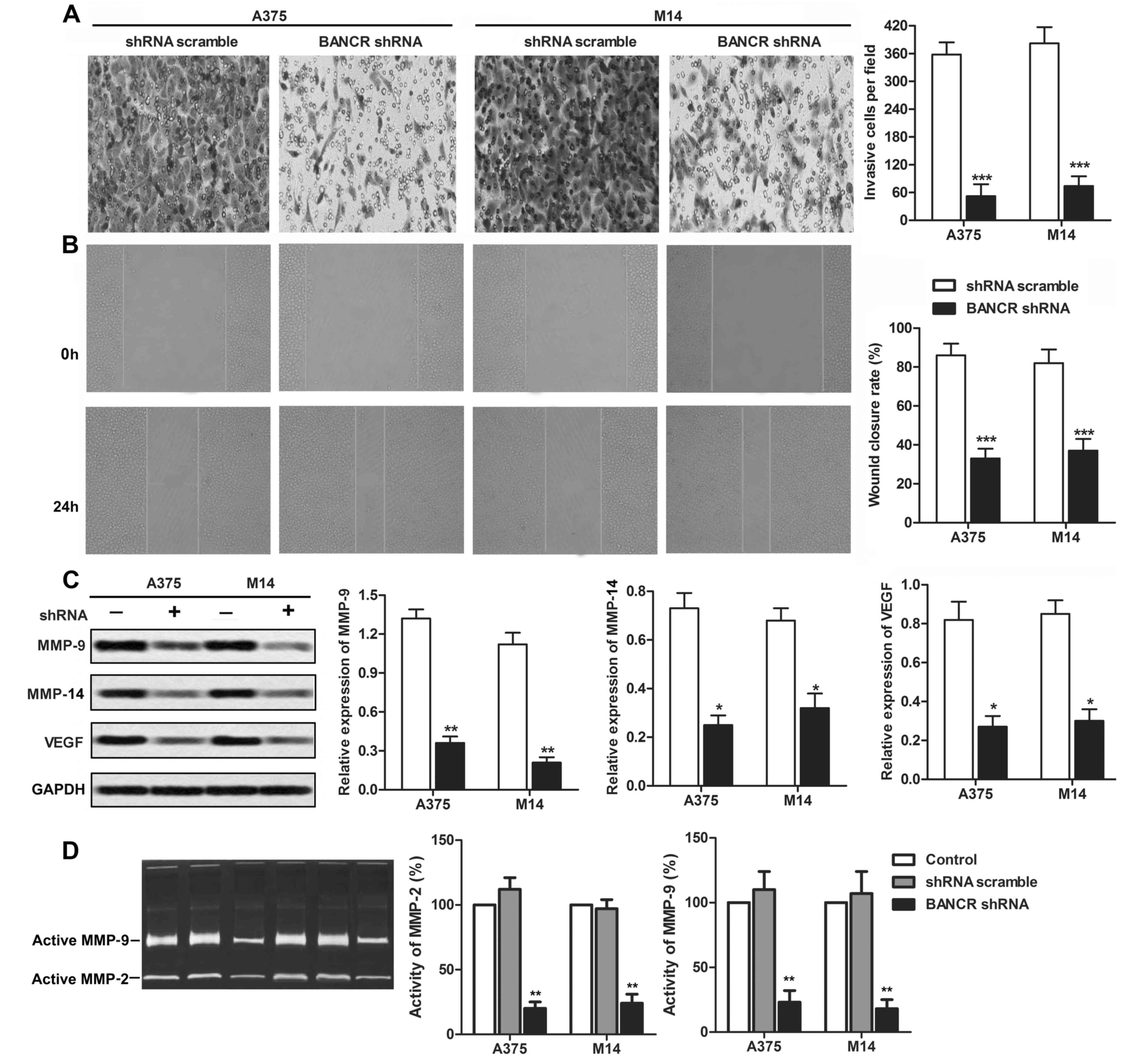
Knockdown of BANCR suppresses melanoma
cell migration and invasion in vitro
The effect of BANCR on the migratory capability was
assessed by Transwell chamber assay and wound healing assay in A375
and M14 cells. As illustrated in Fig.
3A, the Transwell assay showed that the number of migratory
cells in the BANCR shRNA group was significantly decreased compared
with that in the control group (P<0.001). The wound healing
assay also exhibited that the closing rate of scratch wounds was
significantly decreased by BANCR knockdown compared with the
control group (P<0.001, Fig.
3B). To better understand the influence of BANCR on the
melanoma cell migration and invasion, the expression of MMP-9,
MMP-14 and VEGF in the melanoma cell lines A375 and M14 were
assayed by western blotting (P<0.01, Fig. 2C). In addition, the activity of
MMP2 and MMP-9 was significantly decreased under BANCR shRNA
treatment by zymography assay (P<0.01, Fig. 3D). These results imply that BANCR
is involved in the promotion of melanoma cell motility.
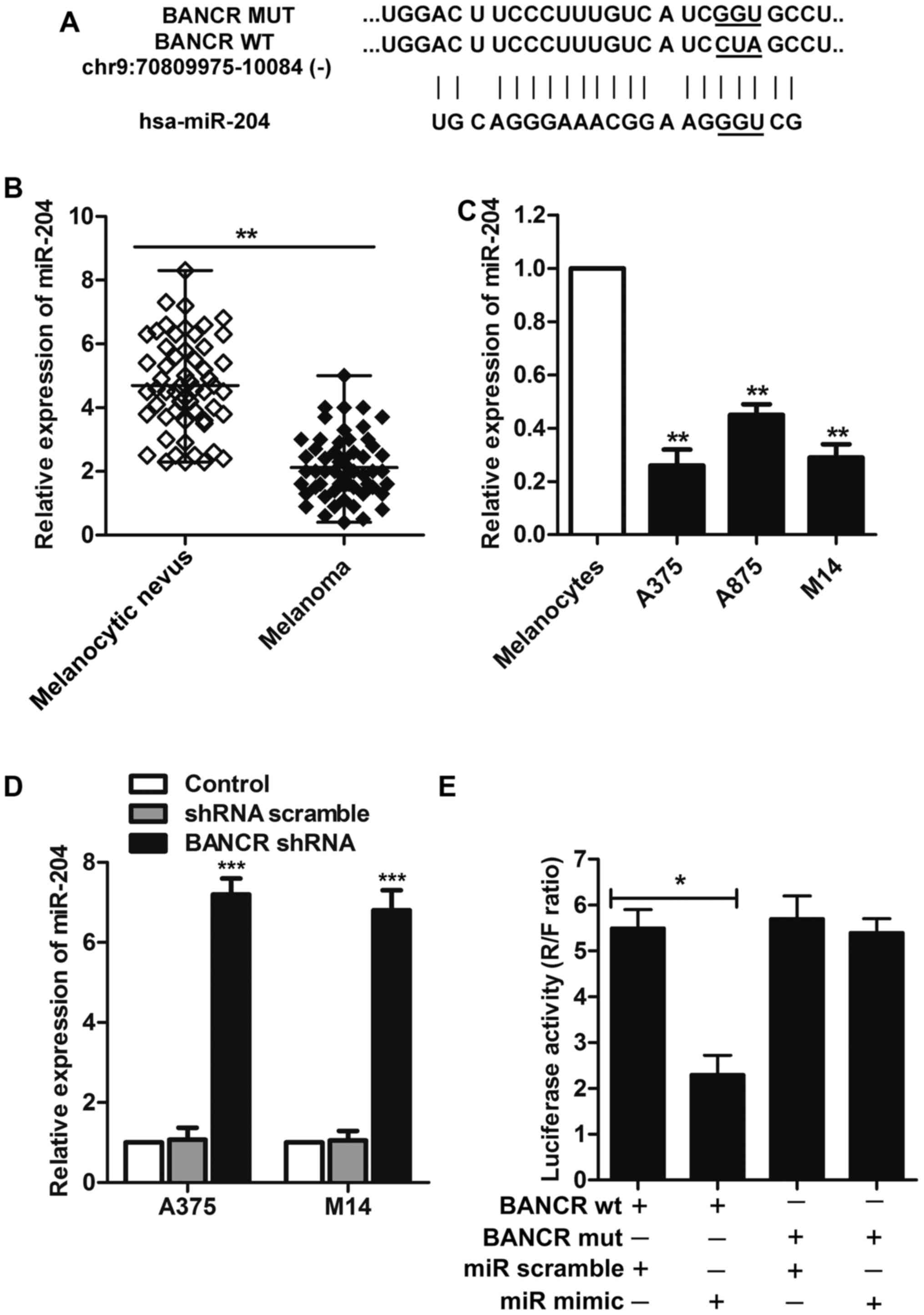
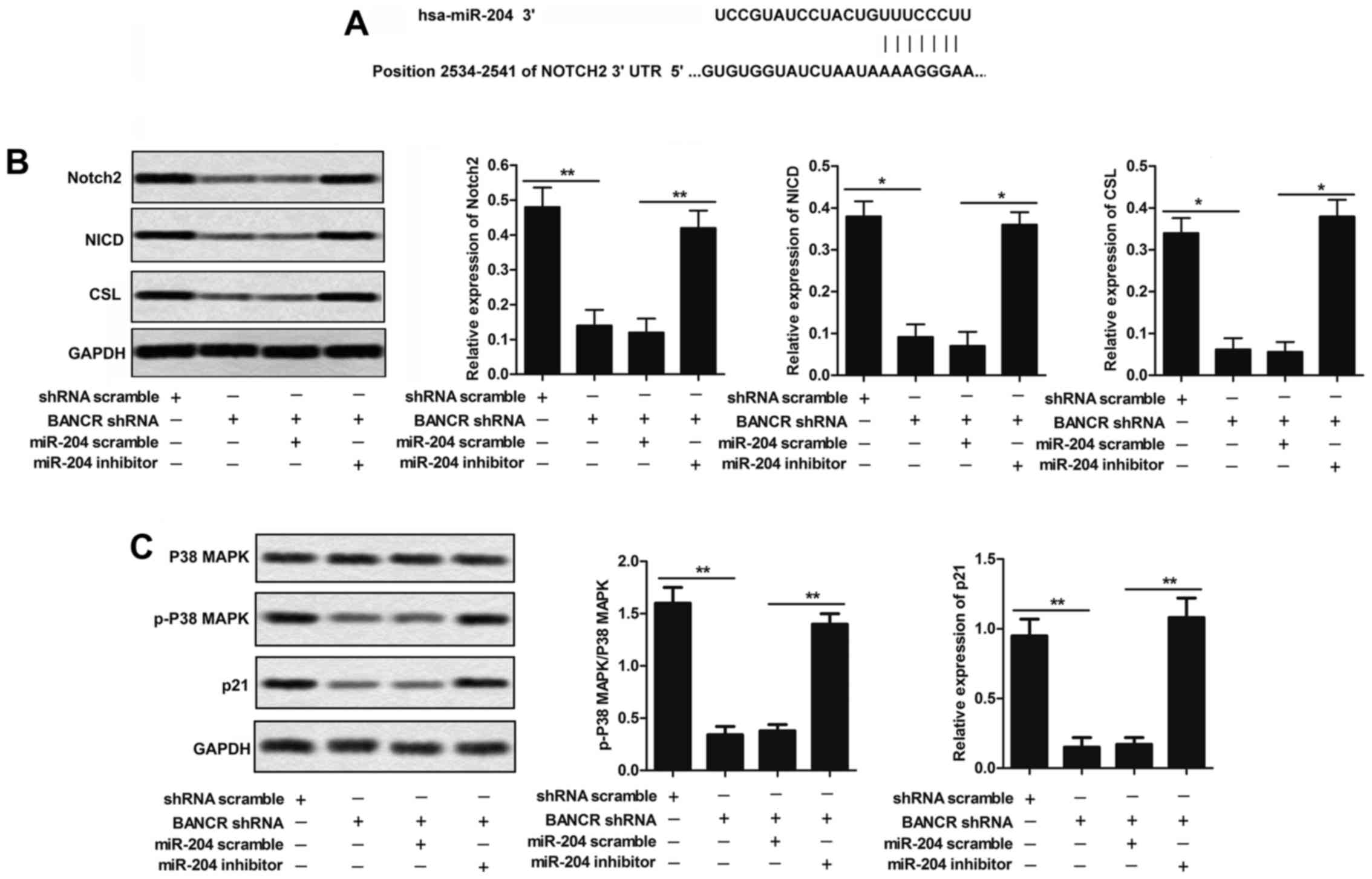
miR-204 is a direct target of BANCR
Growing evidence supports that miR-204 may functions
as a tumor suppressor and plays a prominent role in the development
of several cancers under some specific conditions (26–28).
Bioinformatics analyses predicted that BANCR might be a putative
target gene of miR-204, suggesting a potential interaction between
miR-204 and BANCR (Fig. 4A). Then,
we examined expression level of miR-204 in melanoma tissue samples
and melanoma cell lines (A375, A875 and M14). As illustrated in
Fig. 4B and C, miR-204 was
significantly downregulated in melanoma tissues compared with the
melanocytic nevus (P<0.01), while low-level expression of
miR-204 was also observed in melanoma cell lines rather than the
human epidermal melanocytes (P<0.01).
Considering the expression of miR-204 was lower in
A375 and M14 cells compared with cell lines A875. A375 and M14 were
chosen for the following experiments. To investigate whether
miR-204 is a direct target of BANCR, A375 and M14 cells were
transfected with BANCR shRNA or shRNA scramble. The results showed
that BANCR knockdown significantly caused upregulation of miR-204
compared with control group (P<0.001, Fig. 4D). Then, we generated two
luciferase reporter constructs: a wt-BANCR and a mut-BANCR. The
mut-BANCR contained a 3 bp mutation in the putative miR-204 binding
site (Fig. 4A). We use wt BANCR
transfected cells for convincing the targeting relationship between
BANCR and miR-204. The wild-type BANCR and mutant BANCR were
inserted into a luciferase reporter vector (pGL4.74) for luciferase
reporter assay. Considering the high levels of BANCR, these
wt-BANCR and mut-BANCR vectors and miR-204 scramble or miR-204
mimic were co-transfected into M14 cells, respectively. When
compared with the control group, luciferase activity of the
wt-BANCR vector deceased in cells transfected with miR-204 mimic
(P<0.05, Fig. 4E). The
repression of luciferase activity by miR-204 was not seen in cells
transfected with mut-BANCR (Fig.
4E). These results suggested a direct interaction between
miR-204 and BANCR via the 3-bp putative miR-204 binding site.
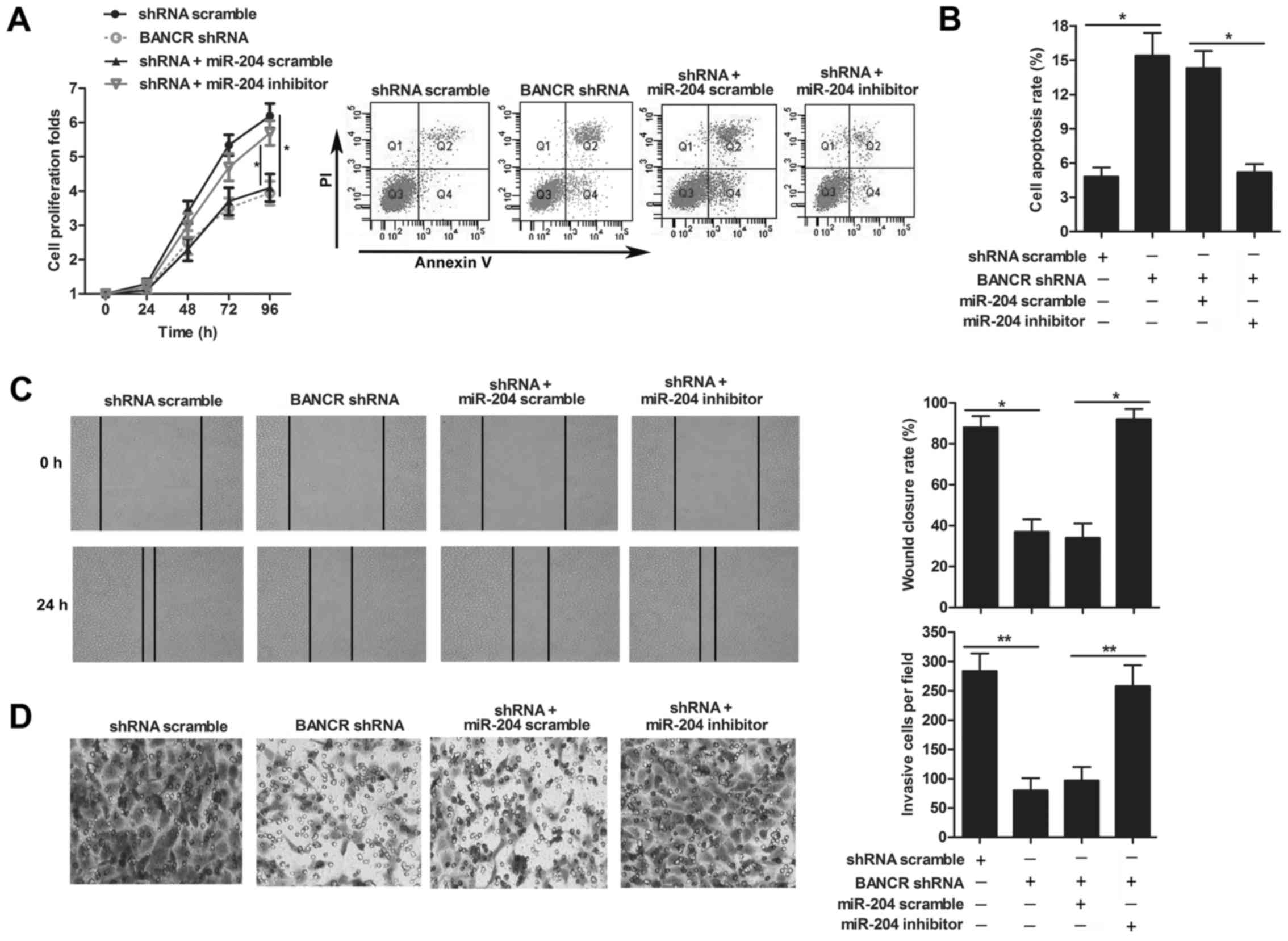
miR-204 is involved in the growth and
migration of melanoma
We further investigated the role of miR-204 in
melanoma, CCK8 assay displayed that the restrained proliferation of
M14 cells was enhanced adding miR-204 inhibitor in BANCR shRNA
group (P<0.05, Fig. 5A).
Moreover, we assessed cell apoptosis in M14 cells by flow cytometry
analysis. As shown in Fig. 5B, the
proportion of apoptotic cells in the miR-204 inhibitor group
co-transfected with BANCR shRNA was significantly lower than that
in BANCR shRNA group (P<0.001). In addition, to assess the
effect of miR-204 on the melanoma cell migratory capability, wound
healing assay and Transwell chamber assay were performed in M14
cells. As illustrated in Fig. 5C,
the wound healing assay showed that miR-204 inhibitor group
co-transfected with BANCR shRNA promoted the ability of melanoma
cells to close a gap compared with BANCR shRNA group (P<0.05).
Also, for the migratory cells in the miR-204 inhibitor group
co-transfected with BANCR shRNA was obviously increased compared
with that in BANCR shRNA detected by the Transwell assay
(P<0.01, Fig. 5D). Together,
these data demonstrated that miR-204 inhibited melanoma cell
proliferation and migration.
BANCR knockdown restrains the activation
of Notch 2 pathway
Bioinformatics analyses predicted that miR-204 might
be a putative target gene of Notch2 (Fig. 6A). To convince their target
relationship, we measured the expression of Notch2, NICD and CSL in
response to BANCR shRNA or shRNA scramble co-transfected with
miR-204 scramble or miR-204 inhibitor in human melanoma cell line
M14. The expression level of Notch2, NICD and CSL were
down-regulated by knockdown of BANCR compared with shRNA scramble
group as demonstrated by western blotting. miR-204 inhibitor
co-transfected with BANCR shRNA relatively upregulated Notch2, NICD
and CSL compared with BANCR shRNA group in M14 cells (P<0.05,
Fig. 6B). These results support a
direct correlation of BANCR, miR-204, and Notch2 expression in the
regulation of melanoma cell growth.
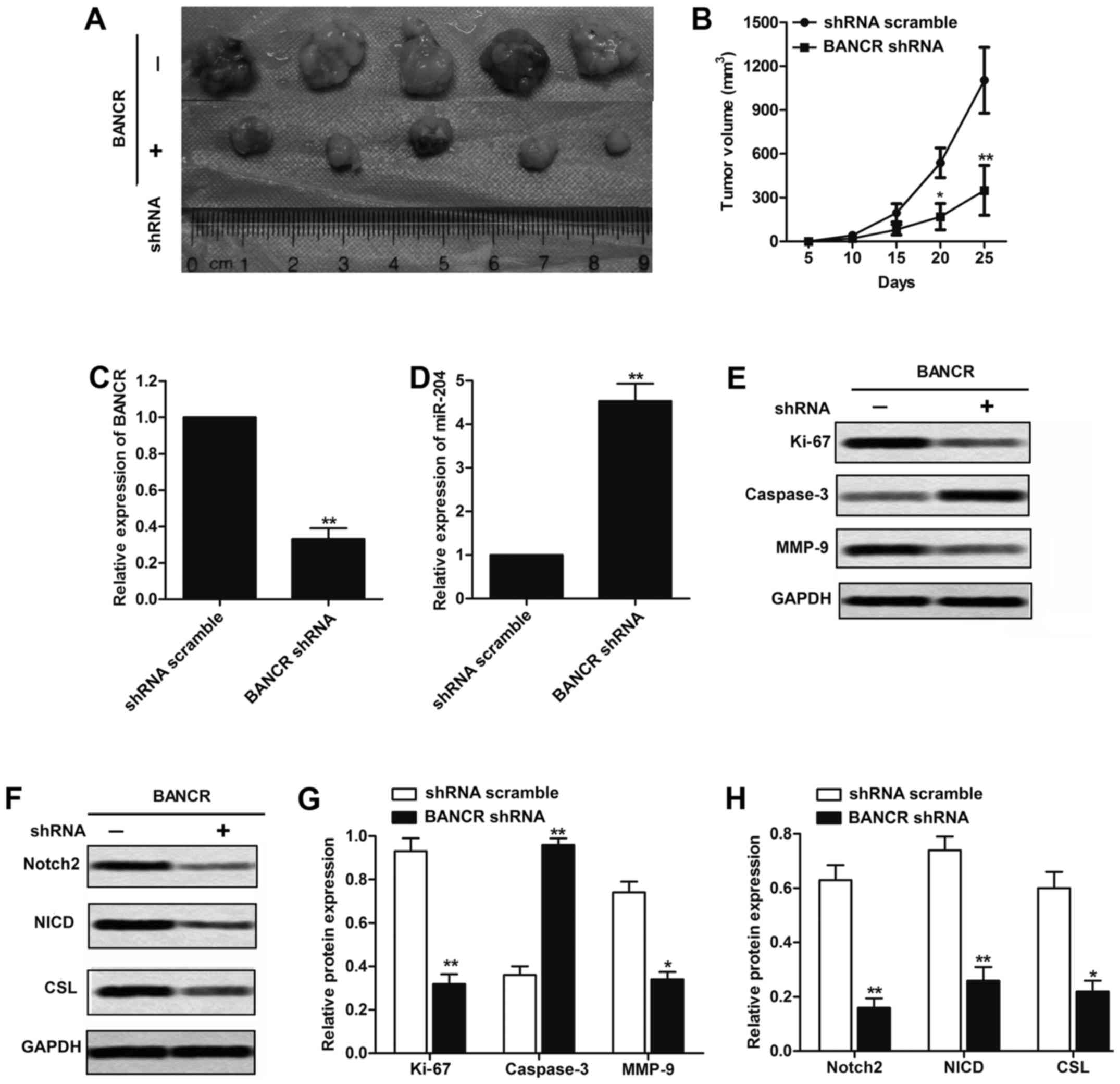
Downregulation of BANCR inhibits
tumorigenesis of melanoma cells in vivo
To confirm whether the expression level of BANCR
affects tumorigenesis in vivo, M14 cells transfected with
BANCR shRNA or shRNA scramble were subcutaneously inoculated into
nude mice. After injection, tumor growth in the BANCR shRNA group
was obviously slower than that in the shRNA scramble group,
especially after 20 days of observation (P<0.05, Fig. 7A and B). To confirm the mechanism
through which BANCR exerts its oncogenic effects in melanoma,
real-time qPCR was performed to analyze the mRNA expression of
BANCR, miR-204 in melanoma tissues. As shown in Fig. 7C and D, BANCR shRNA obviously
decreased the expression of BANCR and increased the expression of
miR-204 compared with the shRNA scramble group (P<0.01).
Furthermore, we also measured the expression of Ki-67, caspase-3
and MMP-9 in melanoma tissues. BANCR downregulation significantly
increased the protein level of caspase-3, while reduced the Ki-67
and MMP-9 compared with shRNA scramble group (P<0.05, Fig. 7E and F). In addition, BANCR shRNA
significantly decreased the protein levels of Notch2, NICD and CSL
compared with control group, further indicating that Notch2 pathway
indeed participates in the BANCR-mediated tumorigenesis of melanoma
(P<0.05, Fig. 7G and H). These
results demonstrate that BANCR/miR-204/Notch2 pathway plays a
crucial role in melanoma cell growth in vivo.
Discussion
Genome-wide studies have identified thousands of
lncRNAs lacking protein-coding capacity. Although lncRNAs only make
up a small proportion of the entire genome, recent studies suggest
that they facilitate normal growth and development and underpin
disease when dysfunctional (29,30).
For example, lncRNA HOTAIR correlates with disease progression in
bladder cancer (31).
Particularly, elevated level of lncRNA is often associated with the
progression of melanoma. For instance, the overexpressed lncRNA
SLNCR1 promotes melanoma invasion through a conserved SRA1-like
region (32). Upregulated lncRNA
SPEY4-IT1 induces apoptosis and promotes invasion of melanoma
(33). Although these studies are
beginning to unravel the importance of lncRNAs in melanoma, their
functions and mechanisms are largely unknown.
BANCR was found overexpressed in numerous cancer
tissues and cell lines and may play an important role in the
development of many malignancies. A report indicated that
overexpressed BANCR is associated with poor prognosis for non-small
cell lung cancer and promotes metastasis by affecting
epithelial-mesenchymal transition (EMT) (15); BANCR promotes proliferation and
invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway
(16). Besides, BANCR promotes
colorectal cancer migration by inducing epithelial-mesenchymal
transition (14). Other reports
have demonstrated that increased level of BANCR played a potential
functional role in melanoma cell proliferation and migration
through regulating MAPK pathway activation (17). Given the above reports, we applied
BANCR in the context of melanoma in this study. In accordance with
these reports, our study found that BANCR was highly expressed in
melanoma and BANCR konckdown could significantly inhibit melanoma
growth and migration, suggesting that BANCR functions as an
oncogene in the melanoma progression.
MicroRNAs regulate a variety of normal physiologic
processes, such as development, cell differentiation, and
regulation of cell cycle and apoptosis, and are involved in
pathogenesis of multiple malignancies, including melanoma (19,34).
Reports suggest that miR-204 plays a dual regulatory role in
cancer. Li et al, identified miR-204 an oncomiR through
targeting prostate-derived Ets factor (PDEF) and inhibiting the
PDEF tumor-suppressive function in PCa (35). miR-204 was also indicated to play a
tumor suppressive function in PAC cells, but acts as an oncomiR in
NEPC cells (36). However,
accumulated evidence suggests miR-204 plays a prominent role in
inhibiting the development of multiple types of cancer. For
example, VHL-regulated miR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell carcinoma
(26). Besides, MALAT1 interacted
with miR-204 to modulates human hilar cholangiocarcinoma
proliferation, migration and invasion by targeting CXCR4 (37). These results suggest that an
lncRNA-miRNA interaction might be important in the process of
tumorigenesis. Consistent with the above studies, our results
showed that an interaction between miR-204 and BANCR plays a role
in the regulation of melanoma tumorigenesis.
The Notch pathway controls diverse processes, such
as regulation of lymphoid development, differentiation and function
(38) enhancing retinal pigment
epithelial cell proliferation, while aberrant notch signaling has
been identified in numerous tumor types, such as activation of the
Notch pathway in head and neck cancer and in melanoma tissues and
cell lines (39). Inhibition of
Notch expression can suppress proliferation and invasion of
melanoma (40). BANCR contributes
to the progression of malignant melanoma by regulating MAPK pathway
activation. Studies also showed that the activation of Notch
pathway can be controlled by microRNAs in melanoma. For example,
miR-146a promotes the initiation and progression of melanoma by
activating Notch signaling (41).
Given these reports together with our previous results, we
speculated a BANCR/miR-204/Notch axis in regulating melanoma. The
results showed that BANCR knockdown induced suppressed growth,
metastasis and inactivation of Notch pathway were counteracted by
miR-204 inhibition. In addition, a melanoma xenograft animal model
was carried out to evaluate the effects of BANCR on miR-204 and
Notch2 expression in vivo, further indicating that miR-204
and Notch2 pathway indeed participate in the BANCR-mediated
tumorigenesis of melanoma.
In conclusion, these data support the
BANCR/miR-204/Notch2 axis in melanoma tumor progression whereby
BANCR promotes melanoma cell growth and migration through
activating Notch2 pathway via targeting miR-204. This is the first
time that BANCR, miR-204, and Notch2 have been linked in melanoma.
A better understanding of the microRNA-lncRNA interaction and their
regulation will provide new insight into mechanisms underlying
various aspects of tumorigenesis including tumor growth and
tumor-drug resistance, providing a new aspect for development of
novel treatment strategies.
Glossary
Abbreviations
Abbreviations:
|
BANCR
|
BRAF-activated non-coding RNA
|
|
lncRNAs
|
long non-coding RNAs
|
|
miR
|
microRNA
|
|
Notch2
|
neurogenic locus notch homolog protein
2
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Linares MA, Zakaria A and Nizran P: Skin
cancer. Prim Care. 42:645–659. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh S, Zafar A, Khan S and Naseem I:
Towards therapeutic advances in melanoma management: An overview.
Life Sci. 174:50–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gros A, Parkhurst MR, Tran E, Pasetto A,
Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts
IM, et al: Prospective identification of neoantigen-specific
lymphocytes in the peripheral blood of melanoma patients. Nat Med.
22:433–438. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slominski AT and Carlson JA: Melanoma
resistance: A bright future for academicians and a challenge for
patient advocates. Mayo Clin Proc. 89:429–433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shah DJ and Dronca RS: Latest advances in
chemotherapeutic, targeted, and immune approaches in the treatment
of metastatic melanoma. Mayo Clin Proc. 89:504–519. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Smith MA and Mattick JS: Structural and
functional annotation of long noncoding RNAs. Methods Mol Biol.
1526:65–85. 2017. View Article : Google Scholar
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
12
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang
S and Liu X: Long noncoding RNA MALAT1 as a potential therapeutic
target in osteosarcoma. J Orthop Res. 34:932–941. 2016. View Article : Google Scholar
|
|
14
|
Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang
D and Sun Y: BRAF-activated long non-coding RNA contributes to
colorectal cancer migration by inducing epithelial-mesenchymal
transition. Oncol Lett. 8:869–875. 2014.PubMed/NCBI
|
|
15
|
Sun M, Liu XH, Wang KM, Nie FQ, Kong R,
Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al: Downregulation of
BRAF activated non-coding RNA is associated with poor prognosis for
non-small cell lung cancer and promotes metastasis by affecting
epithelial-mesenchymal transition. Mol Cancer. 13:682014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu
J and Sun Y: BRAF-activated long non-coding RNA contributes to cell
proliferation and activates autophagy in papillary thyroid
carcinoma. Oncol Lett. 8:1947–1952. 2014.PubMed/NCBI
|
|
17
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L
and Sha N: Long non-coding RNA BANCR promotes proliferation in
malignant melanoma by regulating MAPK pathway activation. PLoS One.
9:e1008932014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Leucci E, Coe EA, Marine JC and Vance KW:
The emerging role of long non-coding RNAs in cutaneous melanoma.
Pigment Cell Melanoma Res. 29:619–626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Philippidou D, Schmitt M, Moser D, Margue
C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I and Kreis
S: Signatures of microRNAs and selected microRNA target genes in
human melanoma. Cancer Res. 70:4163–4173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang D, Wang D, Wang N, Long Z and Ren X:
Long non-coding RNA BANCR promotes endometrial cancer cell
proliferation and Invasion by regulating MMP2 and MMP1 via ERK/MAPK
signaling pathway. Cell Physiol Biochem. 40:644–656. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lv L, Jia JQ and Chen J: LncRNA CCAT1
upregulates proliferation and invasion in melanoma cells via
suppressing miR-33a. Oncol Res. Apr 12–2017.Epub ahead of print.
View Article : Google Scholar : 2017.PubMed/NCBI
|
|
22
|
Sun Y, Cheng H, Wang G, Yu G, Zhang D,
Wang Y, Fan W and Yang W: Deregulation of miR-183 promotes melanoma
development via lncRNA MALAT1 regulation and ITGB1 signal
activation. Oncotarget. 8:3509–3518. 2017.
|
|
23
|
Massi D, Tarantini F, Franchi A,
Paglierani M, Di Serio C, Pellerito S, Leoncini G, Cirino G,
Geppetti P and Santucci M: Evidence for differential expression of
Notch receptors and their ligands in melanocytic nevi and cutaneous
malignant melanoma. Modern Pathol. 19:246–254. 2006. View Article : Google Scholar
|
|
24
|
Suliman MA, Zhang Z, Na H, Ribeiro AL,
Zhang Y, Niang B, Hamid AS, Zhang H, Xu L and Zuo Y: Niclosamide
inhibits colon cancer progression through downregulation of the
Notch pathway and upregulation of the tumor suppressor miR-200
family. Int J Mol Med. 38:776–784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chegeni S, Khaki Z, Shirani D, Vajhi A,
Taheri M, Tamrchi Y and Rostami A: Investigation of MMP-2 and MMP-9
activities in canine sera with dilated cardiomyopathy. Iran J Vet
Res. 16:182–187. 2015.
|
|
26
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et
al: VHL-regulated miR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bao W, Wang HH, Tian FJ, He XY, Qiu MT,
Wang JY, Zhang HJ, Wang LH and Wan XP: A TrkB-STAT3-miR-204-5p
regulatory circuitry controls proliferation and invasion of
endometrial carcinoma cells. Mol Cancer. 12:1552013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
30
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berrondo C, Flax J, Kucherov V, Siebert A,
Osinski T, Rosenberg A, Fucile C, Richheimer S and Beckham CJ:
Expression of the long non-coding RNA HOTAIR correlates with
disease progression in bladder cancer and is contained in bladder
cancer patient urinary exosomes. PLoS One. 11:e01472362016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schmidt K, Joyce CE, Buquicchio F, Brown
A, Ritz J, Distel RJ, Yoon CH and Novina CD: The lncRNA SLNCR1
mediates melanoma invasion through a conserved SRA1-like region.
Cell Rep. 15:2025–2037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li T, Pan H and Li R: The dual regulatory
role of miR-204 in cancer. Tumour Biol. 37:11667–11677. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ding M, Lin B, Li T, Liu Y, Li Y, Zhou X,
Miao M, Gu J, Pan H, Yang F, et al: A dual yet opposite
growth-regulating function of miR-204 and its target XRN1 in
prostate adenocarcinoma cells and neuroendocrine-like prostate
cancer cells. Oncotarget. 6:7686–7700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tan X, Huang Z and Li X: Long Non-coding
RNA MALAT1 interacts with miR-204 to modulate human hilar
cholangiocarcinoma proliferation, migration, and invasion by
targeting CXCR4. J Cell Biochem. 118:3643–3653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Maillard I, Fang T and Pear WS: Regulation
of lymphoid development, differentiation, and function by the Notch
pathway. Annu Rev Immunol. 23:945–974. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schouwey K, Aydin IT, Radtke F and
Beermann F: RBP-Jκ-dependent Notch signaling enhances retinal
pigment epithelial cell proliferation in transgenic mice. Oncogene.
30:313–322. 2011. View Article : Google Scholar
|
|
40
|
Asnaghi L, Ebrahimi KB, Schreck KC, Bar
EE, Coonfield ML, Bell WR, Handa J, Merbs SL, Harbour JW and
Eberhart CG: Notch signaling promotes growth and invasion in uveal
melanoma. Clin Cancer Res. 18:654–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Forloni M, Dogra SK, Dong Y, Conte D Jr,
Ou J, Zhu LJ, Deng A, Mahalingam M, Green MR and Wajapeyee N:
miR-146a promotes the initiation and progression of melanoma by
activating Notch signaling. eLife. 3:e014602014. View Article : Google Scholar : PubMed/NCBI
|