Introduction
Clock genes exist in almost all cells of the human
body (1,2). They not only perform important roles
in controlling the biological rhythm of the human body, but also
affect cellular functions by regulating numerous genes. The
downstream genes regulated by clock genes are called
clock-controlled genes (CCGs) (3).
CCGs are involved in various physiological activities, including
cell proliferation, apoptosis, cell cycling, metabolism, endocrine
signaling and immunity (4–8). As a result, abnormal expression of
clock genes can lead to a variety of diseases, including endocrine
disorders, cardiovascular diseases and tumors (9–12).
PER2 gene is an important clock gene. Changes in its
expression are closely associated with the occurrence and
development of cancers (13–17).
Studies have demonstrated that PER2 expression levels are
downregulated in multiple types of tumor, including gastric
carcinoma, liver cancer, colon cancer, and head and neck squamous
cell carcinomas (18–21). Furthermore, in vitro
investigations of PER2 overexpression in lung cancer LLC cells,
breast cancer EMT6 cells, osteosarcoma MG63 cells, and pancreatic
cancer Panc1 and Aspc1 cells have demonstrated significant
increases in tumor cell apoptosis and marked decreases in cell
proliferation (22–24). Our previous studies have also shown
that knockdown of PER2 in OSCC cells can greatly enhance cell
proliferation and reduce apoptosis (25,26).
The aforementioned studies thus implicate PER2 as an important
tumor-suppressor gene.
OSCC accounts for 90% of oral cancers, and is a
common malignant tumor (27). At
present, there is no clear opinion regarding the associations
between PER2 expression changes and the occurrence, development and
survival outcomes of OSCC. It has been shown that the development
of OSCC is closely related to expression changes of several
important tumor-related genes, such as PIK3CA, PTEN, P53, P14ARF
and caspase-8 (28–34). However, it is unclear whether there
is any correlation between PER2 expression and the aberrant
expression of these tumor-related genes.
In the present study, the mRNA and protein
expression levels of PIK3CA, PTEN, P53, P14ARF and caspase-8 were
assessed and compared between OSCC and cancer-adjacent oral mucosa.
In addition, the correlations between PER2 expression and
clinicopathological parameters, survival time and expression levels
of PIK3CA, PTEN, P53, P14ARF and caspase-8 were analyzed, so as to
provide a basis for further study into the function and mechanism
of PER2 in cancer development.
Materials and methods
Antibodies
All primary antibodies used in the study were rabbit
anti-human antibodies, and details are provided in Table I. Horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG secondary antibodies were
purchased from CWBio Biotechnology (Beijing, China).
 | Table IPrimary antibodies for detecting all
target proteins. |
Table I
Primary antibodies for detecting all
target proteins.
| Primary
antibodies | Brand | Catalog no. | Host | Applications and
dilutions |
|---|
| PER2 | Novus | NBP2-24596 | Rabbit
polyclonal | IHC-P:1:100 |
| PER2 | Abcam | ab208163 | Rabbit
polyclonal | WB:1:250 |
| PIK3CA | Bioworld | BS6052 | Rabbit
polyclonal | IHC-P:1:200
WB:1:1,000 |
| PTEN | CST | 9559T | Rabbit
monoclonal | IHC-P:1:160
WB:1:1,000 |
| P53 | CST | #2527 | Rabbit
monoclonal | IHC-P:1:160
WB:1:1,000 |
| P14ARF | Novus | NB200-111 | Rabbit
polyclonal | IHC-P:1:250
WB:1:1,000 |
| Caspase-8 | GeneTex | GTX110723 | Rabbit
polyclonal | IHC-P:1:500
WB:1:2,000 |
| GAPDH | CWBio | CW101M | Rabbit
polyclonal | WB:1:3,000 |
Patients and specimens
Fresh OSCC tissues and cancer-adjacent tissues were
obtained from 8 patients with OSCC who underwent surgery at the
First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between September 2016 and November 2016. The
clinicopathological data of these 8 OSCC patients are shown in
Table II. In addition,
paraffin-embedded tissue sections of 40 OSCC patients were obtained
from the Pathology Department of the First Affiliated Hospital of
Chongqing Medical University for immunohistochemical staining. The
40 patients were in-patients treated at the Department of Oral and
Maxillofacial Surgery in the First Affiliated Hospital of Chongqing
Medical University between 2007 and 2011; the clinicopathological
data of these 40 patients are presented in Table III. From the 40 paraffin-embedded
sections, 14 cancer-adjacent oral mucosa specimens were selected
and used as the control group. None of the patients underwent
radiotherapy or chemotherapy prior to surgery. The study was
approved by the Biomedical Ethics Committee of the First Affiliated
Hospital of Chongqing Medical University (approval no. 2016-124)
and written informed consent was obtained from all patients who
participated.
 | Table IIClinicopathological features of 8
patients with OSCC. |
Table II
Clinicopathological features of 8
patients with OSCC.
| Parameters | Cases |
|---|
| Tissue type | |
| ANT | 8 |
| OSCC | 8 |
| Sex | |
| Male | 6 |
| Female | 2 |
| Age | |
| ≥60 | 7 |
| <60 | 1 |
| Tumor
differentiation | |
| Poor | 5 |
| Moderate and
well | 3 |
| TNM staging | |
| T1+T2 | 2 |
| T3+T4 | 6 |
| Lymph node
metastases | |
| Yes | 2 |
| No | 6 |
| Clinical stage | |
| I+II | 2 |
| III+IV | 6 |
| Site | |
| Gingiva | 1 |
| Tongue | 3 |
| Buccal | 3 |
| The floor of the
oral | 1 |
 | Table IIIClinicopathological features of 40
patients with OSCC. |
Table III
Clinicopathological features of 40
patients with OSCC.
| Parameters | Cases |
|---|
| Tissue type | |
| ANT | 14 |
| OSCC | 40 |
| Sex | |
| Male | 25 |
| Female | 15 |
| Age | |
| ≥60 | 21 |
| 45–60 | 11 |
| <45 | 8 |
| Tumor
differentiation | |
| Poor | 17 |
| Moderate | 10 |
| Well | 13 |
| TNM stage | |
| T1+T2 | 21 |
| T3+T4 | 19 |
| Lymph node
metastases | |
| Yes | 12 |
| No | 28 |
| Clinical stage | |
| I+II | 17 |
| III+IV | 23 |
| Site | |
| Gingiva | 2 |
| Tongue | 18 |
| Buccal | 8 |
| The floor of the
oral | 2 |
| Palate | 2 |
Reverse transcription-quantitative PCR
(RT-qPCR)
A liquid nitrogen grinding method was used to
prepare the fresh OSCC and cancer-adjacent oral mucosa tissues from
8 patients. Total RNA was extracted using TRIzol reagent (Takara,
Shiga, Japan) according to the manufacturer's instructions. A
PrimeScript™ reagent kit with gDNA Eraser (Takara) was used to
synthesize cDNA by reverse transcription. The primers for the PER2,
PIK3CA, PTEN, P53, P14ARF and caspase-8 genes, as well as GAPDH,
were designed with Oligo 7.0. The forward and reverse primer
sequences for each gene are shown in Table IV. The reaction system included
12.5 µl 2X SYBR Premix Ex Taq™ II, 1 µl 0.4 µM
forward primer and reverse primer, 2 µl 50 ng/µl cDNA
and 8.5 µl dH2O, in a total volume of 25
µl. A C-1000™ Thermal Cycler (Bio-Rad, CA, USA) was used for
qPCR. The PCR steps included 95°C predegeneration for 90 sec,
followed by 40 amplification cycles of denaturation at 95°C for 10
sec and annealing/elongation at 60°C for 30 sec. The transcript
level of each gene was normalized to the expression of GAPDH. The
comparative threshold cycle method (2−ΔΔCt) was used to
calculate relative changes in expression. For every sample, the
experiment was repeated three times.
 | Table IVPrimer sequences and product size
used for reverse transcription-quantitative PCR. |
Table IV
Primer sequences and product size
used for reverse transcription-quantitative PCR.
| Gene | Primer
sequence | Product size
(bp) |
|---|
| PER2 | F:
5′-GCGTGTTCCACAGTTTCACC-3′ | 146 |
| R:
5′-GCGGATTTCATTCTCGTGGC-3′ | |
| PIK3CA | F:
5′-GGTTTTGCTGTTCGGTGCTT-3′ | 220 |
| R:
5′-GGCCAAACCTCTGGCTAACT-3′ | |
| PTEN | F:
5′-CTCAGCCGTTACCTGTGTGT-3′ | 129 |
| R:
5′-AGGTTTCCTCTGGTCCTGGT-3′ | |
| P53 | F:
5′-ACCTATGGAAACTACTTCCTGAAA-3′ | 257 |
| R:
5′-GCTGCCCTGGTAGGTTTTCT-3′ | |
| P14ARF | F:
5′-GTTTTCGTGGTTCACATCCCG-3′ | 101 |
| R:
5′-AGACGCTGGCTCCTCAGTA-3′ | |
| Caspase-8 | F:
5′-CGCAAAGGAAGCAAGAACCC-3′ | 209 |
| R:
5′-GGCAGAAATTTGAGCCCTGC-3′ | |
| GAPDH | F:
5′-GGATTTGGTCGTATTGGGCG-3′ | 171 |
| R:
5′-CTTCCCGTTCTCAGCCTTGA-3′ | |
Western blot analysis
OSCC and cancer-adjacent oral mucosa tissues were
weighed and ground into tissue powder in liquid nitrogen. Total
protein from the OSCC and cancer-adjacent tissues was extracted
using RIPA lysate (strong) (CWBio) containing a protease inhibitor
cocktail (CWBio). Protein concentrations were measured using a BCA
protein assay kit (CWBio). Subsequently, equal amounts of protein
(50 µg) were separated by SDS-PAGE and electrophoretically
transferred onto PVDF membranes (Solarbio). The PVDF membranes were
then blocked with 5% skim milk for 1 h at room temperature, prior
to being incubated with primary antibodies at 4°C overnight.
Affinity-purified HRP-conjugated IgG secondary antibodies (1:4,500,
CWBio) were incubated with the membranes in a 37°C incubator for 1
h. An ECL-Advance Western Blot Detection system (Bio-Rad) was used
for detection of the protein bands. Each experiment was repeated
three times.
Immunohistochemistry
The 'two-step method' immunohistochemistry method
was used to analyze protein expression in the paraffin-embedded
sections (thickness, 4 µm). The paraffin-embedded sections
were placed in incubators at 60°C for 1 h, then dewaxed and
rehydrated by conventional protocols. The sections were then placed
in citric acid solution (pH 6.0), heated in an oven until boiling,
then heated for 10 min at 95°C for antigen retrieval. The sections
were then naturally cooled to the ambient room temperature. To
eliminate endogenous peroxidase activity, the sections were
incubated in 3% H2O2 deionized water for 10
min. PBS was then used to wash the sections (3×3 min). Blocking was
performed with 5% normal goat serum for 20 min, and the sections
were subsequently incubated with primary antibodies (Table I) overnight at 4°C. Biotin-labeled
secondary antibodies (SP test kit; SP-9000; Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd., Beijing, China) were
incubated with the sections at room temperature for 10 min.
Following 3×3-min washes in PBS, Streptavidin-HRP (SP test kit;
SP-9000; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.)
was added to the sections and incubated at room temperature for 15
min. Further 3×3-min washes in PBS were performed. For microscopic
examination, DAB developer was added and incubated at room
temperature for 3 min. The sections were then stained with
hematoxylin, dehydrated, cleared, and observed under a light
microscope after mounting with neutral resin. Slides with PBS added
instead of primary antibody were used as the negative control. The
positive slide provided by the company was used as the positive
control. Five high-power fields were examined randomly in each
section.
Results were evaluated using semiquantitative
method. Intensity of positive staining (no staining, 0; light
yellow, 1; brown, 2; dark brown, 3) and the percentage of positive
cells (number of positive cells ≤25%, 0; 26–50%, 1; 51–75%, 2;
≥76%, 3) were scored. The sum of the intensity of positive staining
scores and the percentage of positive cells scores was used finally
to evaluate PER2 expression as follows: 0–1-negative (−),
2–3-weakly positive (+), 4–5-positive (++), and ≥6-strongly
positive (+++). Weakly positive (+), positive (++) and strongly
positive (+++) were judged as positive expression.
Statistical analysis
All data were analyzed using SPSS 19.0 software
(SPSS Inc., Chicago, IL, USA). The experimental data from the
RT-qPCR and western blot analyses were expressed as the mean ±
standard deviation (mean ± SD). A Student's t-test was used to make
comparisons between two groups. Immunohistochemical data were
expressed as the proportion of specimens with positive expression,
and a χ2 test was used to analyze the differences in
protein expression between OSCC and cancer-adjacent oral mucosa. A
Spearman's rank correlation analysis was used to analyze the
correlation between the level of PER2 and of other proteins.
Survival curves were drawn using the Kaplan-Meier method, and
log-rank was used to compare survival between PER2-positive and
PER2-negative cases (α=0.05, two-sided). P<0.05 was considered
to indicate a statistically significant difference.
Results
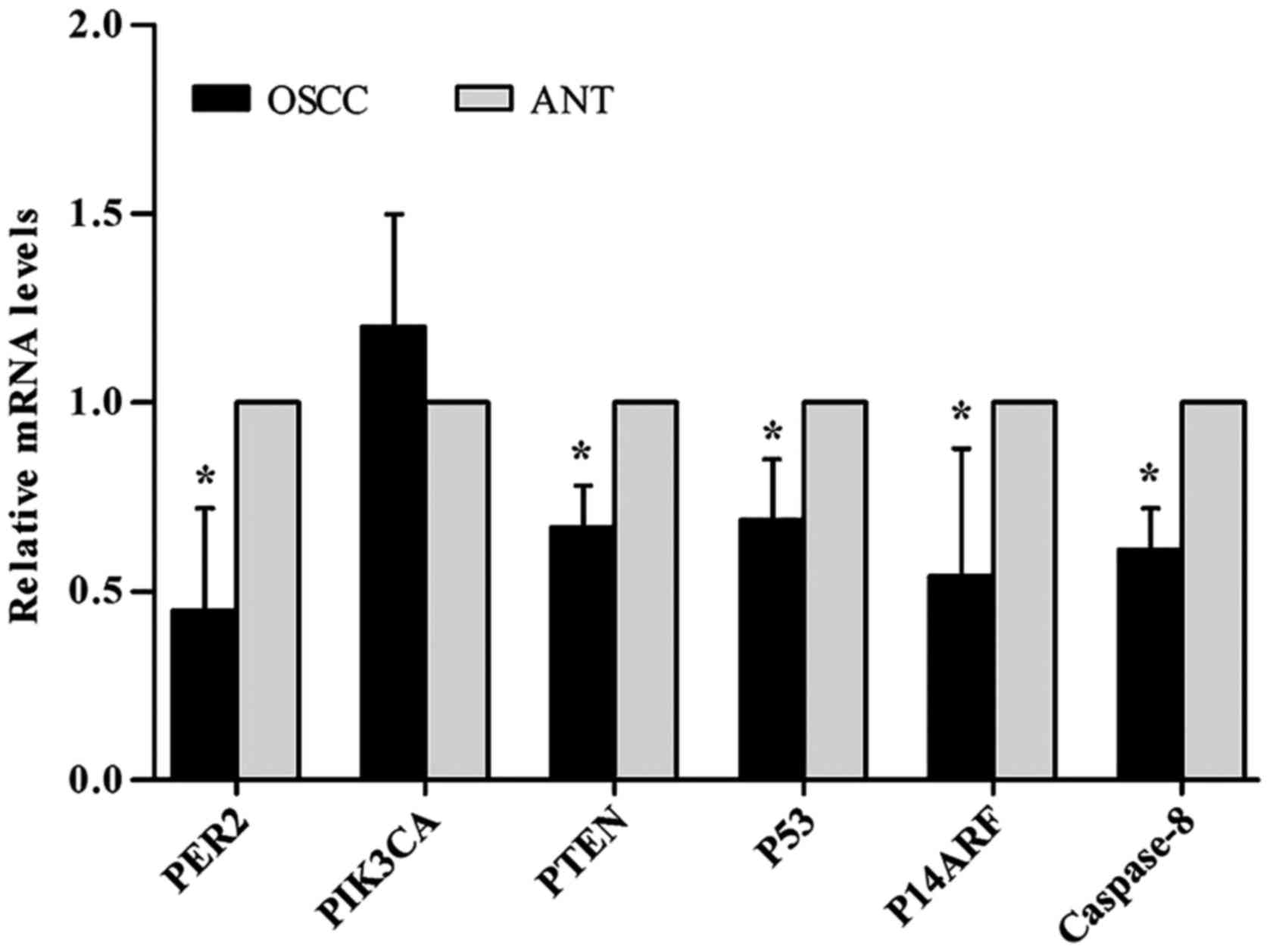
RT-qPCR analysis
To determine the PER2, PIK3CA, PTEN, P53, P14ARF and
caspase-8 mRNA expression levels in human OSCC, RT-qPCR was
performed on paired OSCC tissues and adjacent non-cancerous tissues
from 8 patients. The results revealed that the mRNA expression
levels of PTEN, P53, P14ARF and caspase-8 were reduced
significantly in OSCC compared with adjacent non-cancerous tissues
(P<0.05), whereas there were no significant differences in the
PIK3CA mRNA expression levels, as shown in Table V and Fig. 1.
 | Table VLevels of mRNA expression of each
gene in OSCC tissue and cancer-adjacent oral mucosa tissue. |
Table V
Levels of mRNA expression of each
gene in OSCC tissue and cancer-adjacent oral mucosa tissue.
| Gene | OSCC | ANT | F | t | P-value |
|---|
| PER2 | 0.45±0.27 | 1.00 | 7.28 | −4.00 | 0.007 |
| PIK3CA | 1.20±0.30 | 1.00 | 4.214 | −1.35 | 0.310 |
| PTEN | 0.67±0.11 | 1.00 | 4.47 | 6.18 | 0.025 |
| P53 | 0.69±0.16 | 1.00 | 5.46 | 3.92 | 0.030 |
| P14ARF | 0.54±0.34 | 1.00 | 7.38 | 2.67 | 0.039 |
| Caspase-8 | 0.61±0.11 | 1.00 | 16.00 | 6.31 | 0.390 |
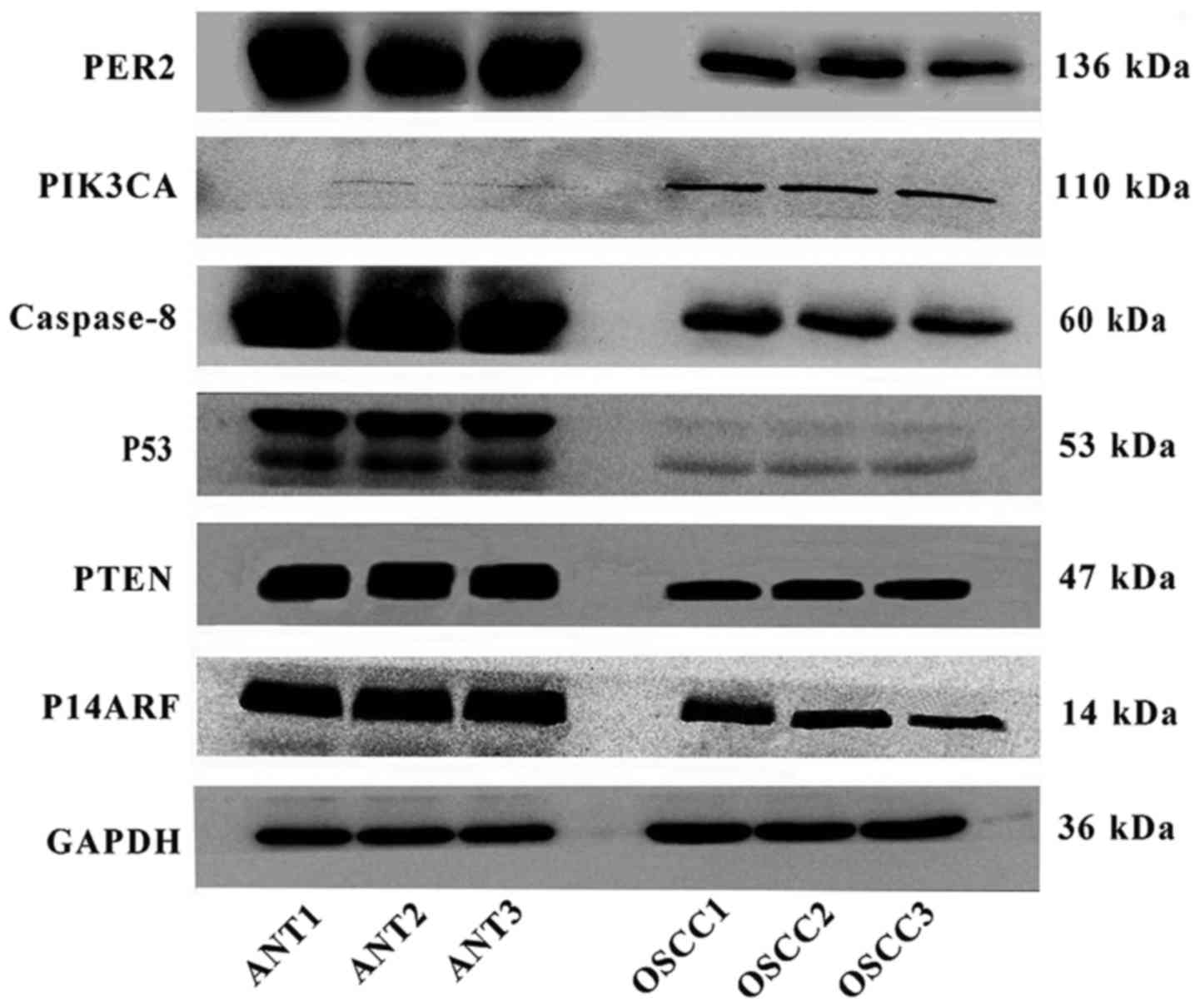
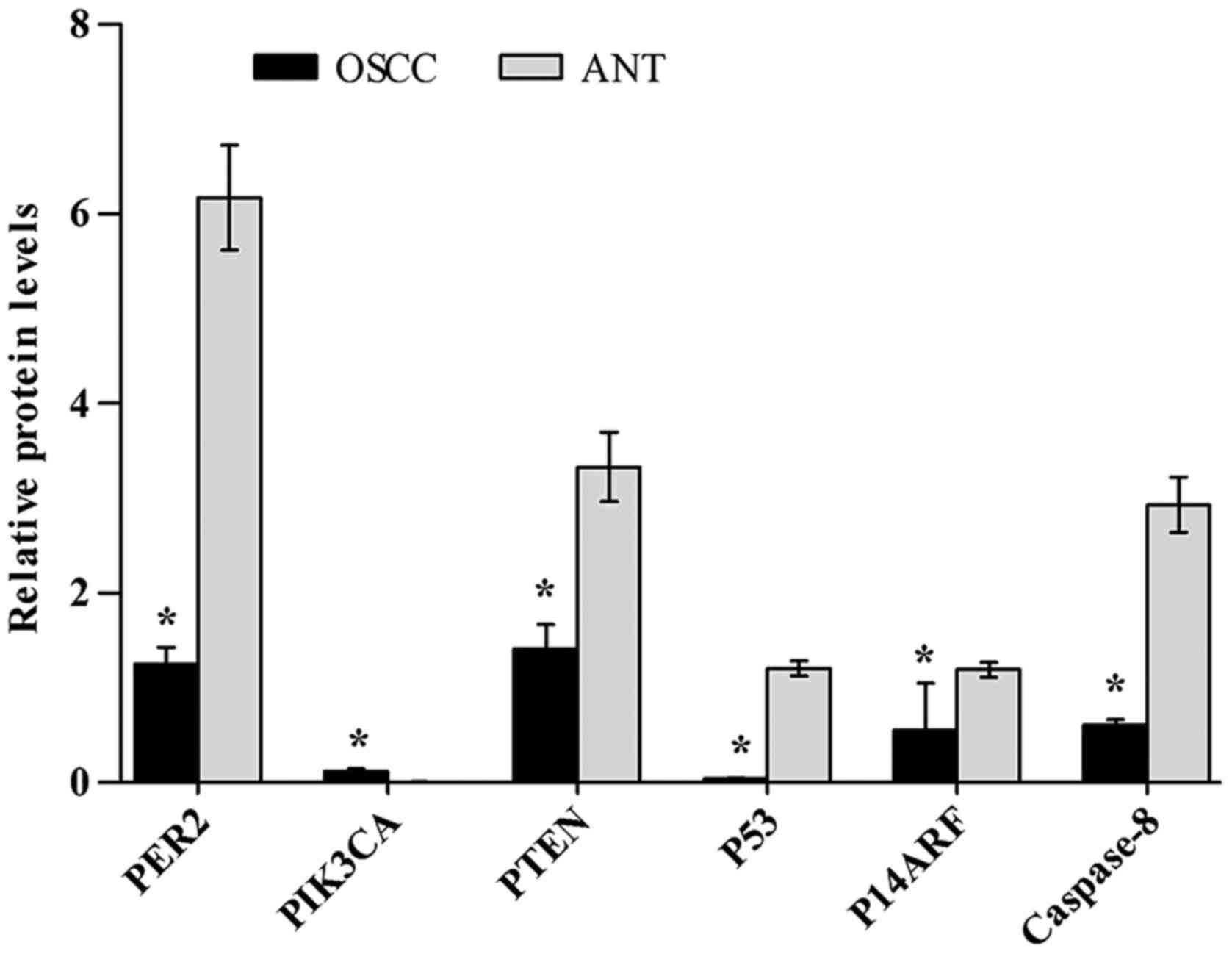
Western blot analysis
The PER2, PIK3CA, PTEN, P53, P14ARF and caspase-8
protein expression levels were determined by western blotting in
paired OSCC and adjacent non-cancerous tissues from 8 patients. The
western blot results showed that the protein expression levels of
PTEN, P53, P14ARF and caspase-8 in OSCC were significantly reduced
compared with those in non-cancerous tissues (P<0.05), while the
PIK3CA protein level was significantly increased (P<0.05), as
shown in Table VI, Figs. 2 and 3.
 | Table VIResults of protein average grey value
in paired OSCC tissues and adjacent non-cancerous tissues. |
Table VI
Results of protein average grey value
in paired OSCC tissues and adjacent non-cancerous tissues.
| Proteins | OSCC | ANT | t-test | P-value |
|---|
| PER2 | 1.24±0.18 | 6.17±0.55 | 14.77 | 0.000 |
| PIK3CA | 0.12±0.021 | 0.006±0.004 | −9.42 | 0.001 |
| PTEN | 1.41±0.26 | 3.33±0.37 | 7.34 | 0.003 |
| P53 | 0.04±0.007 | 1.20±0.08 | 25.16 | 0.001 |
| P14ARF | 0.55±0.50 | 1.19±0.08 | 5.73 | 0.005 |
| Caspase-8 | 0.60±0.06 | 2.93±0.29 | 13.51 | 0.000 |
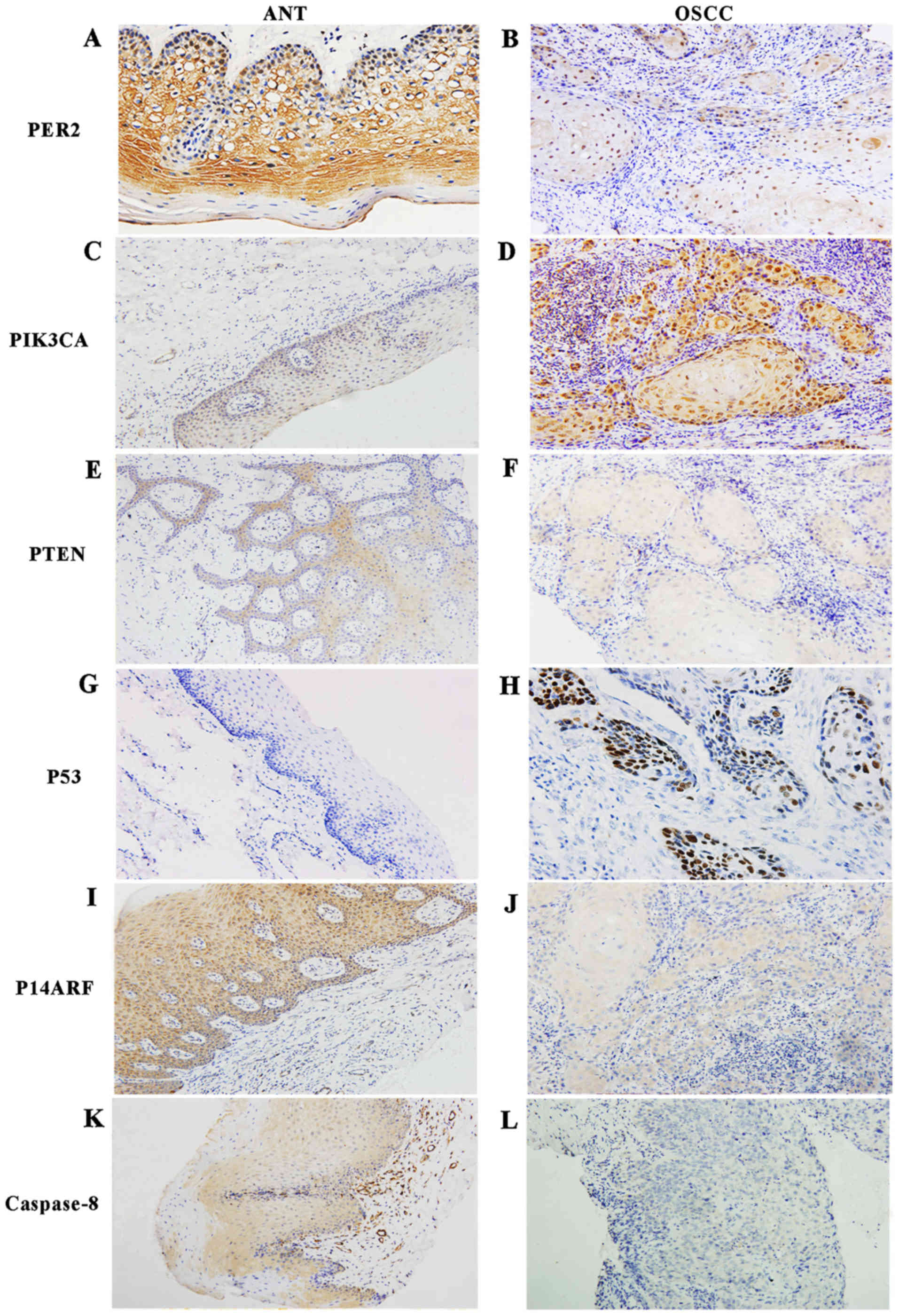
Immunohistochemical analysis
The protein expression levels of PER2, PIK3CA, PTEN,
P53, P14ARF and caspase-8 were analyzed in 40 OSCC tissues and 14
adjacent noncancerous tissues by immunohistochemical staining. The
semi-quantified results of the immunohistochemical staining are
presented in Table VII. The
rates of positive expression of PER2, PTEN, P53, P14ARF and
caspase-8 proteins in OSCC were significantly lower than those in
the adjacent non-cancerous tissues (P<0.05), while the rates of
positive expression of PIK3CA and P53 proteins in OSCC were
markedly higher than those in adjacent non-cancerous tissues
(P<0.05). Representative immunohistochemical staining of PIK3CA,
PTEN, P53, P14ARF and caspase-8 proteins is shown in Fig. 4. Additionally, PER2 protein was
mainly located in the cytoplasm in adjacent non-cancerous tissues,
while it appeared to be expressed in both the cytoplasm and the
cell nucleus in OSCC tissues according to subcellular localization
analysis. The staining of PER2 in the nuclei of the basal layer of
epithelial tissue was more intense than that in the subcutaneous
layer of cells, as shown in Fig.
4.
 | Table VIIResult of immunohistochemical
staining of each protein in OSCC tissues and cancer-adjacent
tissues. |
Table VII
Result of immunohistochemical
staining of each protein in OSCC tissues and cancer-adjacent
tissues.
| Group | Cases | Positive
(percentage)
|
|---|
| PER2 | PIK3CA | PTEN | P53 | P14ARF | Caspase-8 |
|---|
| ANT | 14 | 13 (92.9%) | 8 (57.1%) | 12 (85.7%) | 1 (7.1%) | 14 (100%) | 11 (78.6%) |
| OSCC | 40 | 16 (40%) | 37 (92.5%) | 19 (47.5%) | 17 (42.5%) | 4 (35%) | 10 (25.0%) |
| OSCC:ANT | χ2 | 9.62a | 6.96a | 4.73a | 8.71a | 15.04a | 10.37a |
Low expression of PER2 is associated with
the clinicopathological parameters of patients with OSCC
There was a clear association between PER2
expression and the clinical stage of OSCC: positive PER2 expression
was markedly more frequent in OSCC of stages I/II than in OSCC of
stages III/IV (P=0.000). Furthermore, PER2 expression was increased
in OSCC of stages T1/T2 compared with that of stages T3/T4
(P=0.020), and was significantly more frequent in patients without
lymphatic metastasis than in patients with lymphatic metastasis
(P=0.049). There were no significant associations between PER2 and
patient age, sex or pathological grade in OSCC (P<0.05), as
shown in Table VIII.
 | Table VIIIThe expression of PER2 protein and
its relationship with clinicopathological features of patients with
OSCC. |
Table VIII
The expression of PER2 protein and
its relationship with clinicopathological features of patients with
OSCC.
| Parameters | Case | PER2 expression
| χ2 | P-value |
|---|
| Negative | Positive |
|---|
| Tissue type | | | | | |
| OSCC | 40 | 24 | 16 | 11.653 | 0.001 |
| ANT | 14 | 1 | 13 | | |
| Age | | | | | |
| ≥60 | 21 | 14 | 7 | 0.047 | 0.972 |
| 45–60 | 11 | 7 | 4 | | |
| <45 | 8 | 5 | 3 | | |
| Sex | | | | | |
| Male | 25 | 16 | 9 | 0.029 | 0.864 |
| Female | 15 | 10 | 5 | | |
| Tumor
differentiation | | | | | |
| Well | 13 | 7 | 8 | 3.022 | 0.221 |
| Moderate | 10 | 6 | 4 | | |
| Poor | 17 | 13 | 4 | | |
| T staging | | | | | |
| T1+T2 | 21 | 9 | 12 | 5.414 | 0.020a |
| T3+T4 | 19 | 15 | 4 | 3.889 | 0.049a |
| Lymph node
metastasis | | | | | |
| No | 28 | 14 | 14 | 3.889 | 0.049a |
| Yes | 12 | 10 | 2 | | |
| Clinical stage | | | | | |
| I+II | 17 | 2 | 15 | 28.62 | 0.000a |
| III+IV | 23 | 22 | 2 | | |
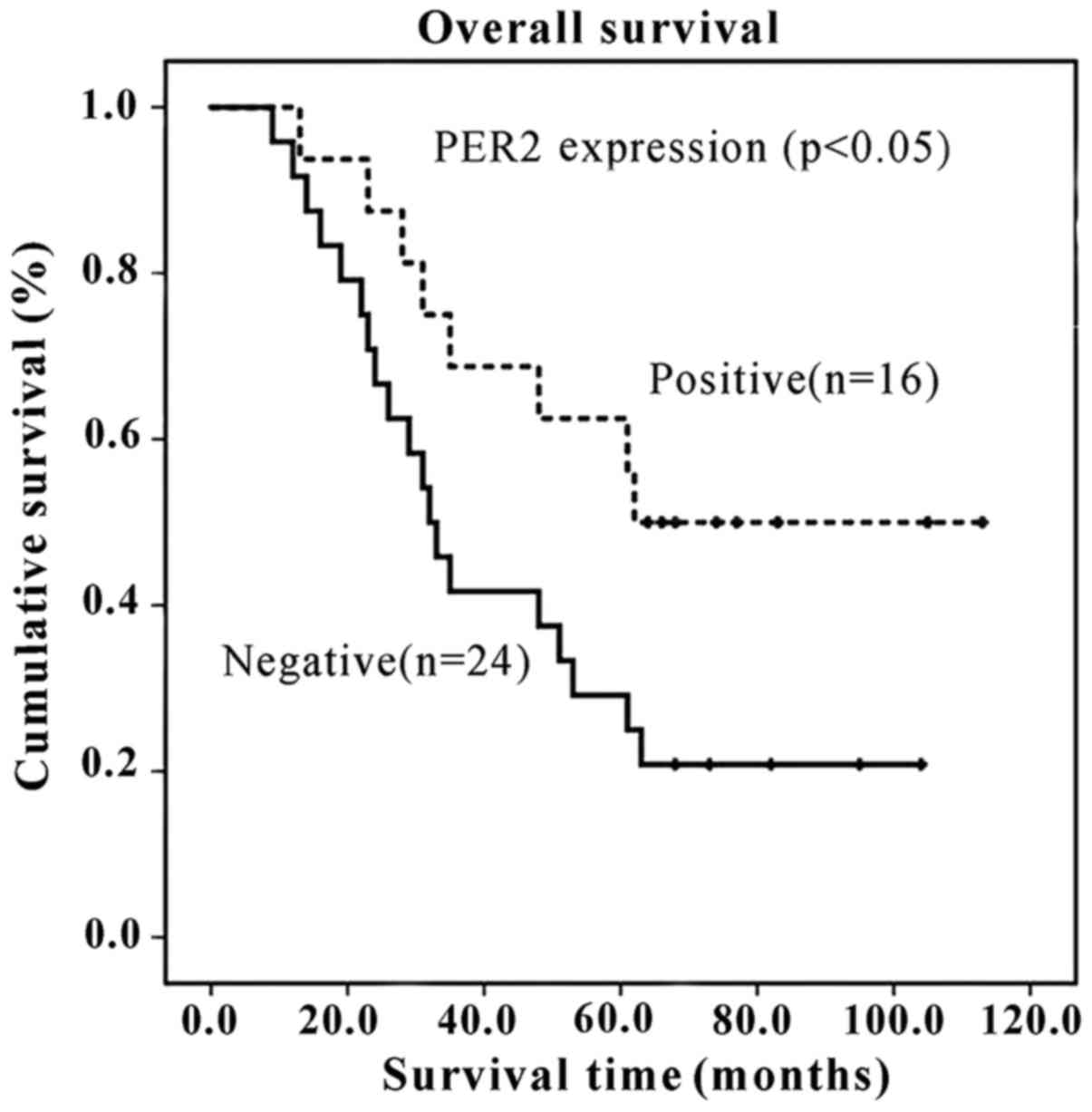
Low expression of PER2 is associated with
poor prognosis of patients with OSCC
Kaplan-Meier survival curves and log-rank tests
demonstrated that OSCC patients with PER2-negative expression had a
shorter overall survival time than those with PER2-positive
expression (P=0.044 and P<0.05). The median post-operative
survival times of patients with positive and negative PER2
expression, respectively, were 62 and 32 months. Furthermore, the
3- and 5-year survival rates of patients with PER2-positive
expression were 68.8% and 62.5%, respectively, whereas the 3-year
and 5-year survival rates of patients with PER2-negative expression
were 41.7% and 29.2%, respectively, as shown in Fig. 5.
PER2 expression is associated with the
expression of the tumor-related genes PIK3CA, PTEN, P53, P14ARF and
caspase-8
PER2 expression was negatively correlated with the
expression of PIK3CA and P53 (P<0.05), and was positively
correlated with the expression of PTEN, P14ARF and caspase-8
(P<0.05), as shown in Table
IX.
 | Table IXSpearman's rank correlation analysis
between PER2 protein and tumor related protein in patients with
OSCC. |
Table IX
Spearman's rank correlation analysis
between PER2 protein and tumor related protein in patients with
OSCC.
| Protein | N | PER2
| r | P-value |
|---|
| − | + | ++ | +++ |
|---|
| PIK3CA | | | | | | −0.603 | <0.01 |
| − | 3 | 0 | 1 | 0 | 2 | | |
| + | 10 | 5 | 5 | 4 | 0 | | |
| ++ | 10 | 9 | 1 | 0 | 0 | | |
| +++ | 17 | 10 | 2 | 5 | 0 | | |
| PTEN | | | | | | 0.442 | 0.004 |
| − | 21 | 15 | 6 | 0 | 0 | | |
| + | 8 | 6 | 1 | 0 | 1 | | |
| ++ | 7 | 2 | 2 | 2 | 1 | | |
| +++ | 4 | 1 | 0 | 2 | 1 | | |
| P53 | | | | | | −0.397 | 0.011 |
| − | 23 | 10 | 11 | 2 | 0 | | |
| + | 7 | 5 | 1 | 1 | 0 | | |
| ++ | 7 | 6 | 1 | 0 | 0 | | |
| +++ | 3 | 3 | 0 | 0 | 0 | | |
| P14ARF | | | | | | 0.317 | 0.047 |
| − | 26 | 17 | 8 | 0 | 1 | | |
| + | 7 | 5 | 1 | 1 | 0 | | |
| ++ | 4 | 1 | 0 | 1 | 2 | | |
| +++ | 3 | 1 | 0 | 2 | 0 | | |
| Caspase-8 | | | | | | 0.620 | <0.01 |
| − | 30 | 23 | 5 | 1 | 1 | | |
| + | 6 | 1 | 2 | 2 | 1 | | |
| ++ | 2 | 0 | 1 | 1 | 0 | | |
| +++ | 2 | 0 | 1 | 0 | 1 | | |
Discussion
Previous studies have demonstrated that PER2
expression was decreased in gastric carcinoma, liver cancer, colon
cancer and HNSCC (18–21). Similarly, this study also revealed
that PER2 expression was reduced in OSCC compared with adjacent
non-cancerous tissue. Further analysis found that the expression of
PER2 was associated with the clinical stage of OSCC, as well as the
lymphatic metastasis status and the patient survival time: reduced
expression of PER2 could promote the occurrence and metastasis of
OSCC and shorten survival time. This seems to be consistent with
research results of Zhao et al in gastric carcinoma
(18) and of Wang et al in
colon cancer (20).
The specific molecular mechanisms by which an
abnormal expression level of PER2 promotes the occurrence and
development of OSCC has remained unclear until now. P53 and P14ARF
are known to play important roles in the occurrence and development
of OSCC (32,33). P14ARF can restrain MDM2, thereby
relieving the ubiquitination and degradation of P53 and increasing
the intracellular level of functional P53 (35). P14ARF can thus positively regulate
P53, and can also form a feedback loop (35). Loss of p53 in OSCC may be partly
attributed to reduced P14ARF expression. Previous evidence has
shown that wild-type P53 (wt-P53) functions as a tumor suppressor,
whereas mutant type P53 (mt-P53) does not (36). Gröbe et al and Friedrich
et al reported that mt-P53 accounted for 40–60% of all P53
in OSCC (37,38). wt-p53 protein's half-life is very
short, its detection is difficult by general experimental methods.
However mt-p53 protein is more stable. mt-p53 protein can be
detected by immunohistochemistry. Although weaker staining of p53
protein might be detected due to increased stability when cells are
in a state of oxidative cellular stress, it is generally believed
that oxidative cellular stress had very little influence on the
positive rate of p53 when detected by immunohistochemistry. Judging
mutation status of p53 according to the result of positive
expression detected by immunohistochemistry with highly specificity
and sensitivity (39). Our results
of immunohistochemistry staining show that the p53-positive
expression rate in OSCC was 42.5%, which could be considered
roughly representing its mutation status in OSCC. Further analysis
show that PER2 was negatively correlated with p53 positive
expression, and positively correlated with P14ARF in OSCC.
Studies have revealed that PIK3CA and PTEN have an
important role in the occurrence and development of OSCC (23–31).
PIK3CA is the key molecule in the PI3K/AKT signaling pathway
(40), while PTEN is the most
important negative regulatory component in the PI3K/AKT signaling
pathway (41). Abnormal activation
of PI3K/AKT signaling may be associated with the functional loss of
PTEN (42). According to the study
by Chen et al, the downregulation of PER2 in
cisplatin-resistant lung cancer A549/DDP cells resulted in
increased PI3K/AKT/mTOR pathway activity. Conversely, after
upregulating PER2, PI3K/AKT/mTOR pathway activity was reduced,
thereby promoting apoptosis (43).
Wang et al injected a recombinant PER2 plasmid to induce
PER2 overexpression in an allograft ovarian cancer nude mouse
model, revealing that PIK3CA and AKT protein expression levels
could be reduced as a result (44). The present study also indicated
that PER2 expression in clinical specimens of OSCC was negatively
correlated with PIK3CA expression, but positively correlated with
PTEN expression. Therefore, there may be an interaction between
PER2 and the PI3K/AKT signaling pathway. PER2 could restrain the
activity of the PI3K signaling pathway through two modes: firstly,
PER2 might restrain PTEN expression, so as to weaken the negative
regulatory effect of PTEN on PI3K/AKT signaling, thus indirectly
activating PI3K/AKT signaling; secondly, PER2 might interact with
the P110 subunits, encoded by PIK3CA, in the PI3K/AKT signaling
pathway, so as to restrain the intracellular conduction of
extracellular signals. The molecular mechanism by which PER2
regulates PI3K/AKT should be further studied.
Our previous studies found that downregulating PER2
in OSCC cells could markedly reduce cell apoptosis (25,26).
Caspase-8 is an important apoptosis-related gene, and its abnormal
expression is closely associated with carcinogenesis (45). Previous studies have already
demonstrated decreased caspase-8 expression in various types of
cancers (34,46). Consistently, the results of the
current study also found that caspase-8 mRNA and protein expression
levels in OSCC were reduced, and further demonstrated a positive
correlation between PER2 and caspase-8. These findings suggest that
the abnormally decreased expression of PER2 may reduce apoptosis by
regulating caspase-8, thereby leading to carcinogenesis.
In the present study, a subcellular localization
analysis was conducted. We found that PER2 staining was mainly
present in the cytoplasm in cancer-adjacent oral mucosa cells,
while OSCC cells exhibited both cytoplasmic and nuclear PER2
staining. We also found that PER2 staining in the nuclei of stratum
basale cells was more intense than that in the nuclei of stratum
spinosum and granular layer cells. Ectopic expression of PER2 in
cancer cells may promote cell proliferation, and this may be one of
the causes of tumor occurrence and development.
In conclusion, the expression of PER2 is reduced in
OSCC. Reduced expression of PER2 is closely associated with the
occurrence and development of OSCC, and may therefore be an
important biological marker to predict OSCC prognosis. PER2 may
exert its antitumor effect via the P53/P14ARF, PIK3CA/AKT and
caspase-8 pathways. These findings suggest that PER2 may be an
important target for cancer therapy; however, the exact role of
PER2 must be further verified. Future studies should explore the
key molecules that interact with PER2 in the aforementioned
pathways, and investigate their molecular mechanisms.
Acknowledgments
This study was supported by the Program for Graduate
Student Research Innovation in Chongqing (CYS16168).
References
|
1
|
Dibner C, Schibler U and Albrecht U: The
mammalian circadian timing system: Organization and coordination of
central and peripheral clocks. Annu Rev Physiol. 72:517–549. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Welsh DK, Yoo SH, Liu AC, Takahashi JS and
Kay SA: Bioluminescence imaging of individual fibroblasts reveals
persistent, independently phased circadian rhythms of clock gene
expression. Curr Biol. 14:2289–2295. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi JS: Transcriptional architecture
of the mammalian circadian clock. Nat Rev Genet. 18:164–179. 2017.
View Article : Google Scholar
|
|
4
|
Miller BH, McDearmon EL, Panda S, Hayes
KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch
JB, et al: Circadian and CLOCK-controlled regulation of the mouse
transcriptome and cell proliferation. Proc Natl Acad Sci USA.
104:3342–3347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matsuo T, Yamaguchi S, Mitsui S, Emi A,
Shimoda F and Okamura H: Control mechanism of the circadian clock
for timing of cell division in vivo. Science. 302:255–259. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu Z, Hua B, Shang Z, Yuan G, Xu L, Li E,
Li X, Sun N, Yan Z, Qian R, et al: Altered clock and lipid
metabolism-related genes in atherosclerotic mice kept with abnormal
lighting condition. BioMed Res Int. 2016:54385892016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Cara F and King-Jones K: The circadian
clock is a key driver of steroid hormone production in Drosophila.
Curr Biol. 26:2469–2477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Terry EE, Fejer E, Gamba D, Hartmann
N, Logsdon J, Michalski D, Rois LE, Scuderi MJ, Kunst M, et al:
Achilles is a circadian clock-controlled gene that regulates immune
function in Drosophila. Brain Behav Immun. 61:127–136. 2017.
View Article : Google Scholar
|
|
9
|
Tao H, Li X, Qiu JF, Cui WZ, Sima YH and
Xu SQ: Inhibition of expression of the circadian clock gene period
causes metabolic abnormalities including repression of
glycometabolism in Bombyx mori cells. Sci Rep. 7:462582017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vieira E, Ruano E, Figueroa AL, Aranda G,
Momblan D, Carmona F, Gomis R, Vidal J and Hanzu FA: Altered clock
gene expression in obese visceral adipose tissue is associated with
metabolic syndrome. PLoS One. 9:e1116782014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akashi M, Matsumura R, Matsuo T, Kubo Y,
Komoda H and Node K: Hypercholesterolemia causes circadian
dysfunction: A potential risk factor for cardiovascular disease.
EBioMedicine. 20:127–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wood PA, Yang X and Hrushesky WJ: Clock
genes and cancer. Integr Cancer Ther. 8:303–308. 2009. View Article : Google Scholar
|
|
13
|
Zheng B, Larkin DW, Albrecht U, Sun ZS,
Sage M, Eichele G, Lee CC and Bradley A: The mPer2 gene encodes a
functional component of the mammalian circadian clock. Nature.
400:169–173. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Albrecht U, Bordon A, Schmutz I and
Ripperger J: The multiple facets of Per2. Cold Spring Harb Symp
Quant Biol. 72:95–104. 2007. View Article : Google Scholar
|
|
15
|
Toh KL, Jones CR, He Y, Eide EJ, Hinz WA,
Virshup DM, Ptácek LJ and Fu YH: An hPer2 phosphorylation site
mutation in familial advanced sleep phase syndrome. Science.
291:1040–1043. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu L, Pelicano H, Liu J, Huang P and Lee
C: The circadian gene Period2 plays an important role in tumor
suppression and DNA damage response in vivo. Cell. 111:41–50. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wood PA, Yang X, Taber A, Oh EY, Ansell C,
Ayers SE, Al-Assaad Z, Carnevale K, Berger FG, Peña MM, et al:
Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer
Res. 6:1786–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao H, Zeng ZL, Yang J, Jin Y, Qiu MZ, Hu
XY, Han J, Liu KY, Liao JW, Xu RH, et al: Prognostic relevance of
Period1 (Per1) and Period2 (Per2) expression in human gastric
cancer. Int J Clin Exp Pathol. 7:619–630. 2014.PubMed/NCBI
|
|
19
|
Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC,
Lin SF, Su WW and Chang JG: Disturbance of circadian gene
expression in hepatocellular carcinoma. Mol Carcinog. 47:925–933.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Hua L, Lu C and Chen Z: Expression
of circadian clock gene human Period2 (hPer2) in human colorectal
carcinoma. World J Surg Oncol. 9:1662011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu CM, Lin SF, Lu CT, Lin PM and Yang MY:
Altered expression of circadian clock genes in head and neck
squamous cell carcinoma. Tumour Biol. 33:149–155. 2012. View Article : Google Scholar
|
|
22
|
Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang
C, Wang X, Wang Z, Cornelissen-Guillaume G and Halberg F: Circadian
gene mPer2 overexpression induces cancer cell apoptosis. Cancer
Sci. 97:589–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng AY, Zhang Y, Mei HJ, Fang S, Ji P,
Yang J, Yu L and Guo WC: Construction of a plasmid for
overexpression of human circadian gene period2 and its biological
activity in osteosarcoma cells. Tumour Biol. 36:3735–3743. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oda A, Katayose Y, Yabuuchi S, Yamamoto K,
Mizuma M, Shirasou S, Onogawa T, Ohtsuka H, Yoshida H, Hayashi H,
et al: Clock gene mouse period2 overexpression inhibits growth of
human pancreatic cancer cells and has synergistic effect with
cisplatin. Anticancer Res. 29:1201–1209. 2009.PubMed/NCBI
|
|
25
|
Wang Q, Ao Y, Yang K, Tang H and Chen D:
Circadian clock gene Per2 plays an important role in cell
proliferation, apoptosis and cell cycle progression in human oral
squamous cell carcinoma. Oncol Rep. 35:3387–3394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Su X, Chen D, Yang K, Zhao Q, Zhao D, Lv X
and Ao Y: The circadian clock gene PER2 plays an important role in
tumor suppression through regulating tumor-associated genes in
human oral squamous cell carcinoma. Oncol Rep. 38:472–480. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma - an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wan X, Li X, Yang J, Lv W, Wang Q, Chen Y
and Li Y: Genetic association between PIK3CA gene and oral squamous
cell carcinoma: A case control study conducted in Chongqing, China.
Int J Clin Exp Pathol. 8:13360–13366. 2015.
|
|
29
|
Chen Y, Hou Q, Yan W, Luo J, Chen D, Liu
Z, He S and Ding X: PIK3CA is critical for the proliferation,
invasiveness, and drug resistance of human tongue carcinoma cells.
Oncol Res. 19:563–571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jasphin SS, Desai D, Pandit S, Gonsalves
NM, Nayak PB and Iype A: Immunohistochemical expression of
phosphatase and tensin homolog in histologic gradings of oral
squamous cell carcinoma. Contemp Clin Dent. 7:524–528. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rahmani A, Alzohairy M, Babiker AY, Rizvi
MA and Elkarimahmad HG: Clinicopathological significance of PTEN
and bcl2 expressions in oral squamous cell carcinoma. Int J Clin
Exp Pathol. 5:965–971. 2012.PubMed/NCBI
|
|
32
|
Mao C, Lu Y, Lai Q, Xia Y and Yang C:
Expression of p53 gene in oral squamous cell carcinoma and its
relation with clinical and pathological parameters and prognosis of
patients. Chin Med Sci J. 10:199–203. 1995.PubMed/NCBI
|
|
33
|
Shintani S, Nakahara Y, Mihara M, Ueyama Y
and Matsumura T: Inactivation of the p14(ARF), p15(INK4B) and
p16(INK4A) genes is a frequent event in human oral squamous cell
carcinomas. Oral Oncol. 37:498–504. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elrod HA, Fan S, Muller S, Chen GZ, Pan L,
Tighiouart M, Shin DM, Khuri FR and Sun SY: Analysis of death
receptor 5 and caspase-8 expression in primary and metastatic head
and neck squamous cell carcinoma and their prognostic impact. PLoS
One. 5:e121782010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J and Zhu MH: Interactive pathway of
ARF-mdm2-p53. Ai Zheng. 22:328–330. 2003.In Chinese. PubMed/NCBI
|
|
36
|
Liu J, Zhang C and Feng Z: Tumor
suppressor p53 and its gain-of-function mutants in cancer. Acta
Biochim Biophys Sin (Shanghai). 46:170–179. 2014. View Article : Google Scholar
|
|
37
|
Gröbe A, Hanken H, Al-Dam A, Cachovan G,
Smeets R, Krohn A, Clauditz T, Grob T, Simon R, Sauter G, et al:
P53 immunohistochemical expression does not correlate with clinical
features in 207 carcinomas of the oral cavity and in the head and
neck region. Clin Oral Investig. 18:211–217. 2014. View Article : Google Scholar
|
|
38
|
Friedrich RE, Giese M, Riethdorf S and
Loning T: P53-mutation in smears of oral squamous cell carcinoma.
Anticancer Res. 20D:4927–4930. 2000.
|
|
39
|
Takami H, Yoshida A, Fukushima S, Arita H,
Matsushita Y, Nakamura T, Ohno M, Miyakita Y, Shibui S, Narita Y,
et al: Revisiting TP53 mutations and immunohistochemistry - A
comparative study in 157 diffuse gliomas. Brain Pathol. 25:256–265.
2015. View Article : Google Scholar
|
|
40
|
Lai K, Killingsworth MC and Lee CS: Gene
of month: PIK3CA. J Clin Pathol. 68:253–257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee JO, Yang H, Georgescu MM, Di
Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P and Pavletich
NP: Crystal structure of the PTEN tumor suppressor: Implications
for its phosphoinositide phosphatase activity and membrane
association. Cell. 99:323–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Di Cristofano A and Pandolfi PP: The
multiple roles of PTEN in tumor suppression. Cell. 100:387–390.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen B, Tan Y, Liang Y, Li Y, Chen L, Wu
S, Xu W, Wang Y, Zhao W and Wu J: Per2 participates in AKT-mediated
drug resistance in A549/DDP lung adenocarcinoma cells. Oncol Lett.
13:423–428. 2017.PubMed/NCBI
|
|
44
|
Wang Z, Li L and Wang Y: Effects of Per2
overexpression on growth inhibition and metastasis, and on MTA1,
nm23-H1 and the autophagy-associated PI3K/PKB signaling pathway in
nude mice xenograft models of ovarian cancer. Mol Med Rep.
13:4561–4568. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fulda S: Caspase-8 in cancer biology and
therapy. Cancer Lett. 281:128–133. 2009. View Article : Google Scholar
|
|
46
|
Wang X, Fu Z, Chen Y and Liu L: Fas
expression is downregulated in gastric cancer. Mol Med Rep.
15:627–634. 2017. View Article : Google Scholar :
|