Introduction
Gastric cancer is the 4th most common type of cancer
and is ranked 2nd among the causes of cancer-related mortality
worldwide (1-3). Despite significant progress in
surgical and chemotherapeutic treatment regimens, the prognosis for
patients with gastric cancer remains poor (4). Therefore, there is an urgent need for
a better understanding of the molecular mechanisms that underly
gastric cancer carcinogenesis, including oncogenes, tumor
progression and development, in the hope of discovering novel
prognostic markers and therapeutic targets.
The chemokine-like factor (CKLF)-like MARVEL
transmembrane domain-containing family (CMTM) is a novel family of
proteins that links classical chemokines and the transmembrane-4
superfamily (5). In humans, CMTM
comprises 9 genes, which are CKLF and CMTM1-8 (6). The CMTM family members play diverse
role in multiple types of cancer. Among these members, CMTM3, 5, 7
and 8 exhibit tumor suppressive properties in cancers, such as
clear cell renal cell carcinoma, oral squamous cell carcinoma,
non-small cell lung cancer and hepatocellular carcinoma (7-10).
In gastric cancer, the decreased expression of CMTM3 has been
observed, and the downregulation of CMTM3 promotes the metastasis
of gastric cancer cells. Mechanistic analyses have demonstrated
that the knockdown of CMTM3 promotes cancer cell migration and
invasion via the epithelial-mesenchymal transition (EMT) process
caused by the signal transducer and activator of transcription
(STAT)3/Twist1/EMT signaling pathway (11). However, the contribution of CMTM3
to gastric cancer cell proliferation has seldom been discussed, at
least to the best of our knowledge. Moreover, although some studies
have reported that the promoter hypermethylation inhibits CMTM3
expression in gastric cancer (12,13),
the regulatory mechanisms of CMTM3 in gastric cancer require
further comprehensive interpretation.
Over the past decades, the contribution of microRNAs
(miRNAs or miRs) to oncogenesis and cancer progression has been
widely discussed. miRNAs are small, endogenous, non-coding RNAs,
approximately 20–25 nucleotides in length, that regulate target
gene expression at the post-transcriptional level by inhibiting
translation or/and cleaving the targeted mRNA by binding to the
3′-untranslated regions (3′-UTRs) of target mRNAs (14). In gastric cancer, investigations on
miRNAs and their functions have provided novel targets that may be
used for the prediction of the prognosis and for the development of
novel therapeutic strategies (15). miR-135b is a well characterized
miRNA and has been reported to play a tumor-promoting role in
several types of cancer, such as non-small cell lung cancer and
breast cancer (16,17). Conversely, other studies have found
opposing results, demonstrating that miR-135b inhibits metastasis
in prostate cancer and reverses the chemoresistance of non-small
cell lung cancer cells by targeting STAT6 and Frizzled-1 (FZD1),
respectively (18,19). Moreover, miR-135b-5p has been shown
to inhibit lipopolysaccharide (LPS)-induced tumor necrosis factor
(TNF)-α production by silencing the AMP-activated protein kinase
(AMPK) phosphatase, Ppm1e (20).
These above-mentioned studies have illustrated the existence of
heterogeneity in the function of miR-135b due to its multiple
targets in various types of tumors. However, its role in gastric
cancer and the relative targets has seldom been discussed
previously, at least to the best of our knowledge.
In this study, we first identified that CMTM3 was
significantly decreased in gastric cancer tissues, whereas
miR-135b-5p was markedly upregulated. We then further investigated
the function of CMTM3 and miR-135b-5p in the progression of gastric
cancer. Furthermore, we demonstrate that miR-135b-5p promotes
gastric cancer progression by targeting CMTM3.
Materials and methods
Specimens
Gastric cancer tissues and normal adjacent tissues
were collected from patients who underwent curative resection at
the Department of Surgery, the Second Affiliated Hospital of
Wenzhou Medical University, Wenzhou, China between 2013 and 2015.
All samples were collected after obtaining written informed
consent. The tissues were immediately transported into liquid
nitrogen in operating theatres and then stored at −80°C. The study
protocol was approved by the Ethics Committee of the Second
Affiliated Hospital of Wenzhou Medical Univesity, and was carried
out according to the Declaration of Helsinki.
Cells and cell culture
The human normal gastric epithelial cell line,
GES-1, and the gastric cancer cell lines, SGC-7901, HGC-27, BGC-823
and MKN45, were purchased from the Cell Bank of the Chinese Academy
of Sciences. The 293T cells were a kind gift from Wenzhou Medical
University. The cells were maintained in DMEM (GES-1, SGC-7901 and
293T), RPMI-1640 medium (BGC-823 and MKN45), or MEM (HGC-27)
supplemented with 10% fetal bovine serum (all from HyClone, Salt
Lake City, UT, USA), 100 units/ml penicillin and 100 units/ml
streptomycin (Sigma, St. Louis, MO, USA). All these cells were
cultured in a humidified environment containing 5% CO2
and maintained at a constant temperature of 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA for CMTM3 and β-actin was extracted from
the clinical samples using TRIzol reagent (Invitrogen, Carlsbad,
CA, USA). The concentration of the RNA was measured using a
NanoDrop spectrophotometer at 260/280 nm (Thermo Fisher Scientific,
Waltham, MA, USA) and the RNA quality was determined via
electrophoresis. First-strand cDNA was synthesized PrimeScript RT
Reagent kit (Takara, Mountain View, CA, USA) according to the
manufacturer's instructions. Briefly, 4 µl of isolated RNA
(30 µg) was first mixed with 1 µl of random hexamer
primer and 7 µl of RNAse-free H2O and then
incubated at 65°C for 5 min. Subsequently, the microtubes were
cooled on ice followed by the addition of 4 µl of reaction
buffer, 1 µl of RNase inhibitor, 2 µl of dNTP mix,
and 1 µl of reverse transcriptase to each sample. The
samples were immediately incubated at 25°C for 5 min and then at
42°C for 60 min. Finally, the reaction was terminated by heating
the samples at 70°C for 5 min. The reverse transcription reaction
was performed with the final volume of 20 µl per tube.
Quantitative PCR was performed using SYBR-Green Ex Taq™ master mix
(Takara). The quantitative analysis was carried out using iQ 5
Real-Time PCR System (Bio-Rad, Hercules, CA, USA). The real-time
PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min,
then 40 cycles at 95°C for 15 sec, and 60°C for 1 min. The primers
used for β-actin and CMTM3 were as follows: β-actin forward,
5′-GGCACTCTTCCAGCCTTCC-3′; and reverse, 5′-GAGCCGCCGATCCACAC-3′;
and CMTM3 forward, 5′-TCTTGCGTGTGAATCTCTTACC-3′; and reverse,
5′-CAGGATCCACATTGGTGTTACC-3′. Total RNA for miR-135b-5p and U6 was
extracted from the clinical samples and cells using the miRNeasy
mini kit (Qiagen, Hilden, Germany). The RT-qPCR reactions of
miR-135b-5p and U6 were performed according to the manufacturer's
instructions of the All-in-One™ miRNA qRT-PCR Detection kit
(GeneCopoeia, Rockville, MD, USA). iQ-5 (Bio-Rad) was used to
monitor all these RT-qPCR reactions. RNA expression was relative
quantified using 2-ΔΔCt method.
Western blot analysis
Total protein was extracted using RIPA protein lysis
buffer (Beyotime, Shanghai, China) with 1% protease inhibitor
cocktail and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell
fractions were prepared using a Nuclear and Cytoplasmic Protein
Extraction kit (Beyotime) according to the manufacturer's
instructions. Generally, 50 µg of protein were used for
western blotting. Samples were separated by SDS-PAGE and
transferred onto PVDF membranes (Thermo Fisher Scientific, Waltham,
MA, USA). After blocking in 5% skim milk, the PVDF membranes were
incubated with primary antibodies in blocking buffer overnight at
4°C and then with HRP-conjugated secondary antibody for 2 h. The
primary antibodies used were as follows: anti-β-tubulin (1:5,000
dilution, sc-23949; Santa Cruz Biotechnology, Santa Cruz, CA, USA),
anti-CMTM3 (1:1,000 dilution, ab198016; Abcam, Cambridge, UK),
anti-caspase 3 [1:1,000 dilution, 9662; Cell Signaling Technology
(CST), Danvers, MA, USA], anti-poly(ADP-ribose) polymerase (PARP;
1:1,000 dilution, 9532; CST), anti-caspase 9 (1:1,000 dilution,
ab32539; Abcam), anti-cyclin B1 (1:1,000 dilution, ab32053; Abcam),
anti-p21 (1:1,000 dilution, ab109520; Abcam). The membranes were
then incubated with HRP-conjugated goat anti-mouse IgG (sc-2005;
Santa Cruz Biotechnology) or HRP-conjugated goat anti-rabbit IgG
(sc-2004; Santa Cruz Biotechnology). Reactive bands were visualized
with ECL reagent (Pierce, Rockford, IL, USA) and analyzed. Protein
expression was quantified using ImageJ software (National
Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry (IHC)
The frozen tumor tissues and adjacent tissues thawed
in 4°C prior to fixation, and washed with PBS twice. The tissues
were then fixed in 3.7% formalin and embedded in paraffin, and were
then cut into 4-µm-thick serial sections. The
paraffin-embedded tissue sections were dewaxed, rehydrated and
placed in 10 mmol/l citrate buffer (pH 6.0), and heated twice in a
microwave oven for 5 min each. The sections were incubated with 3%
H2O2 for 10 min, washed with PBS, blocked
with 10% normal goat serum for 30 min, and then incubated with 4
mg/l purified anti-CMTM3 (ab198016; Abcam) or normal rabbit IgG
(ab172730; Abcam) as a control at 4°C overnight. After washing, the
sections were stained with the catalyzed signed amplification
system kit (Agilent Technologies, Santa Clara, CA, USA) and
visualized with a Nikon E800 microscope (Nikon, Tokyo, Japan) and
images were acquired.
Oligonucleotide transfection and
Luciferase reporter assay
miR-135b-5p mimics and scramble control mimics were
purchased from GeneCopoeia Inc. The target gene of miR-135b-5p, was
firstly predicted by TargetScan (http://www.targetscan.org/mamm_31/), miRDB (http://www.mirdb.org/) and miRanda (http://www.microrna.org/), respectively. CMTM3 was
then filtered out from the inter-section of above 3 prediction
results. Wild-type CMTM3 3′UTR (CMTM3-3′UTR-wt) and miR-135B-5p
target site deletion mutation CMTM3 3′UTR (CMTM3-3′UTR-mu) were
constructed into the psiCHECK2 plasmid (Promega, Madison, WI, USA).
The 293T cells (105 cells) were seeded in 24-well plates
prior to transfection. According to the instructions of the
manufacturer, mimics and CMTM3 3′UTR reporter plasmids
(psiCHECK-CMTM3-3′UTR-wt or psiCHECK-CMTM3-3′UTR-mu) were
co-transfected using Lipofectamine® RNAiMAX (Thermo
Fisher Scientific, MA, USA) with a final concentration of 50 nM
(mimics) or 200 ng (PrLZ 3′UTR reporter plasmid). After 48 h, the
cells were collected and Luciferase activity was detected using the
Dual-Glo luciferase assay system (Promega) according to the
manufacturer's instructions.
Lentiviral infection
miR-135b-5p inhibitor lentivirus and CMTM3
overexpression lentivirus were purchased from Shanghai R&S
Biotechnology Co., Ltd. The SGC-7901 cells were planted into 10 cm
dishes (106 cells/dish) 24 h prior to infection.
Lentiviral infection was performed at a multiplicity of infection
(MOI) of 50. The infection efficiency was deter-mined by counting
the number of GFP-positive cells which should be guaranteed to be
>90%.
Cell proliferation assay
The cells were seeded in 96-well plates in
triplicate at densities of 3,000/well. Cell viability was evaluated
at the desired time points using CCK8 kits (Dojindo Molecular
Technologies, Kumamoto, Japan) according to the manufacturer's
instructions. Light absorbance of the solution was measured at 450
nm using a microplate reader (PR4100; Bio-Rad).
Cell invasion assay
Transwell chambers coated with Matrigel (BD
Biosciences, San Jose, CA, USA) were used for the analysis of cell
invasion. A total of 3×105 SGC-7901 cells in 100
µl serum-free DMEM were seeded on the upper chambers and
DMEM with 10% FBS was added to the lower chambers. After 24 h of
incubation, the invaded cells in the lower side of the membranes
were fixed with methanol and stained with crystal violet
(Beyotime). Images were acquired using an inverted microscope.
Invaded cells were counted from three different fields. The
experiment was repeated three times.
Apoptosis assay
For the analysis of cell apoptosis, the transfected
cells were harvested, washed and resuspended in 1 ml of binding
buffer. The cells were then stained with 5 µl of Annexin
V-APC (BD Biosciences) and 10 µl of propidium iodide
(Sigma-Aldrich) in the dark for 15 min at room temperature and
analyzed using a flow cytometer (BD Biosciences) equipped with
CellQuest software.
Cell cycle analysis
Propidium iodide (PI) staining with flow cytometry
was used to assess cell cycle distribution. Briefly, the
lentivirus-infected SGC-7901 cells were released by trypsinization,
collected, washed with cold PBS, and then fixed in 70% ethanol at
4°C overnight. The fixed cells were then suspended in 250 µl
of RNase A buffer (100 ng/ml), and labeled with a 2X solution of PI
(100 ng/ml) for 30 min at 4°C. Finally, the stained cells were
analyzed using a flow cytometer (BD Biosciences) equipped with
CellQuest software.
Xenograft mouse model
A total of 30 female BALB/c nude mice (6 weeks old,
weighing 18-22 g) were purchased from Nanjing Biomedical Research
Institute of Nanjing University (Nanjing, China). These BALB/c nude
mice were injected with the SGC-7901 cells subcutaneously (5×106
cells/mouse) into the right posterior shoulder area. For the
lentiviral infection groups, 10 µl lentivirus were
respectively injected into the tumors every 3 days when the tumor
volume up had reached up to 50 mm3. The tumor sizes were
measured every 3 days using micrometer calipers, and tumor volumes
were calculated as follows: Tumor volume = d2×D/2, where
d and D represented shortest and the longest diameters,
respectively. At 22 days after the first injection of the
lentivirus, the mice were euthanized and tissue was collected. The
tissues were immediately transported into liquid nitrogen and then
stored at −80°C for subsequent pathology and gene expression
detection. All animal experiment protocols were approved by the
Institutional Animal Care and Use Committee of Wenzhou Medical
University.
Statistical analysis
The results are presented as the means ± standard
deviations (SD) of 3 independent experiments. Significant
differences in mean values were evaluated by an unpaired t-test.
One-way ANOVA with Tukey's post hoc test was used to compare
continuous variables among 2 or more groups. Tests of association
were conducted using Pearson's χ2 test. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of CMTM3 and miR-135b-5p in
gastric cancer tissues and cells
RT-qPCR and IHC were used to examine the expression
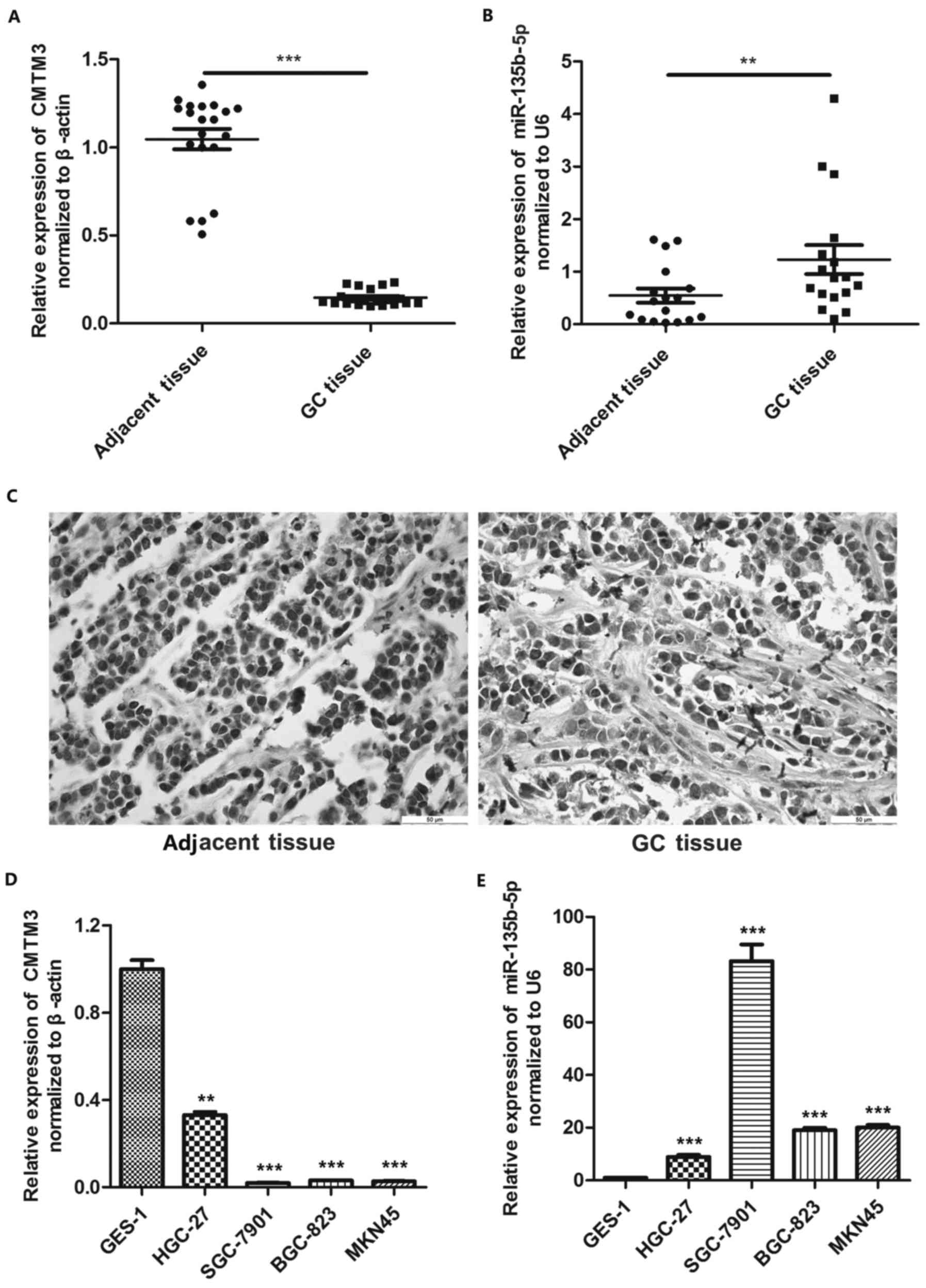
pattern of CMTM3 and miR-135b-5p in gastric cancer. The results of
RT-qPCR revealed that CMTM3 exhibited a lower expression pattern in
gastric cancer tissues compared with the adjacent tissues, while
the expression of miR-135b-5p was significantly higher in gastric
cancer tissues compared with the adjacent tissues (Fig. 1A and B). Moreover, IHC yielded
similar results, also showing that CMTM3 was markedly downregulated
in gastric cancer tissues (Fig.
1C). We then performed RT-qPCR to verify the differences in the
expression of CMTM3 and miR-135b-5p between the normal gastric
epithelial cell line, GES-1, and gastric cancer cell lines (HGC-27,
SGC-7901, BGC823 and MKN45). Our results indicated that CMTM3
expression was higher in the GES-1 cells than that in the HGC-27,
SGC-7901, BGC823 and MKN45 cells (Fig.
1D). By contrast, the miR-135b-5p expression level was
significantly lower in the GES-1 cells compared with the HGC-27,
SGC-7901, BGC823 and MKN45 cells (Fig.
1E).
miR-135b-5p targets CMTM3 expression
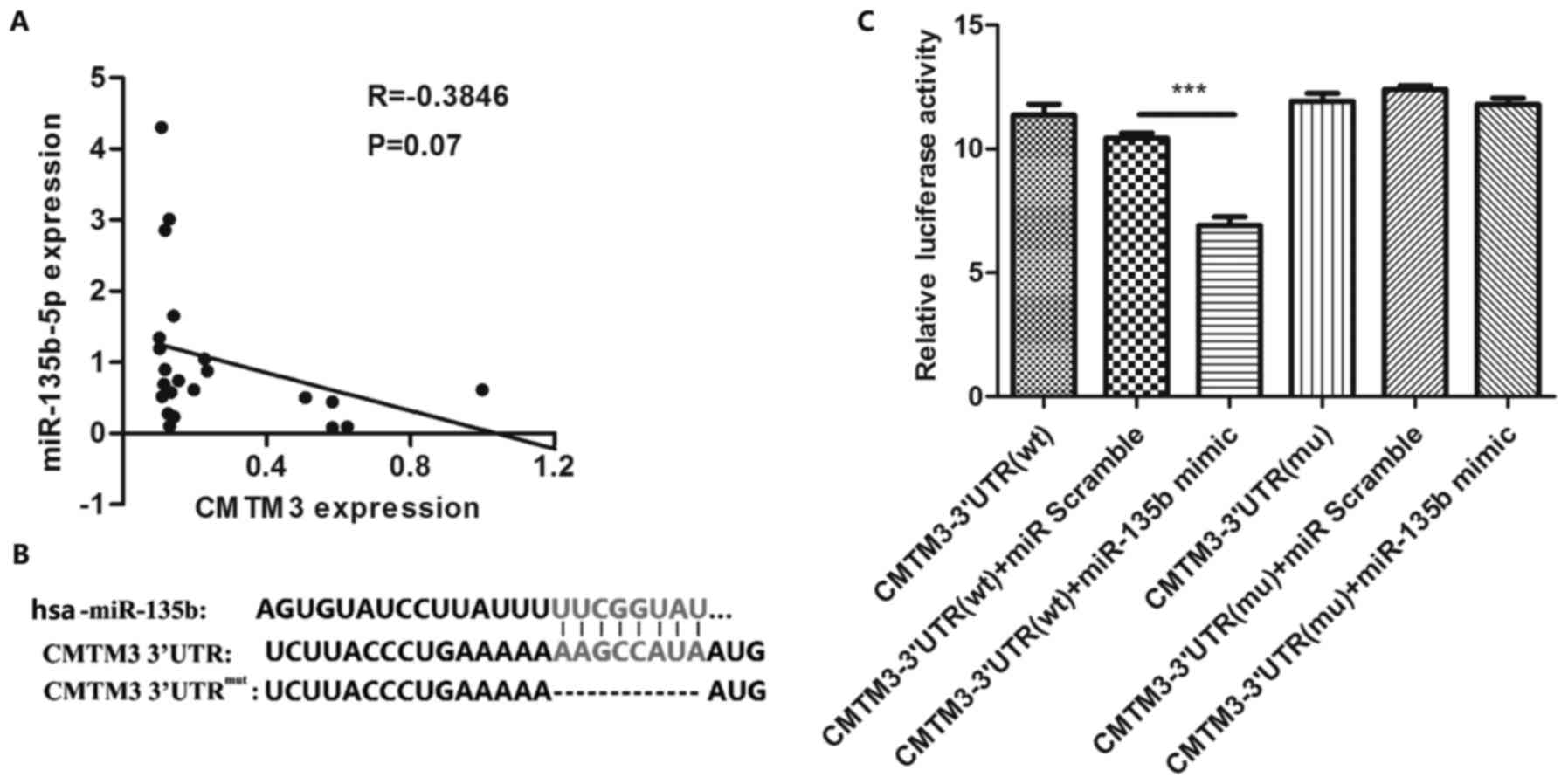
To verify the regulatory association between
miR-135b-5p and CMTM3, we first analyzed their expression
correlation in gastric cancer tissues. Pearson's correlation
analysis revealed that CMTM3 expression negatively correlated with
miR-135b-5p expression (Fig. 2A).
To identify the potential target site of miR449a, we used a
combination of three algorithms, TargetScan, miRDB and miRanda. The
target site of miR-135b-5p in CMTM3 3′-UTR predicted by using
TargetScan, miRDB and miRanda is shown in Fig. 2B. We further confirmed the target
association between miR-135b-5p and CMTM3 by Luciferase assay
(Fig. 2C). Our results indicated
that CMTM3 was specifically targeted by miR-135b-5p.
CMTM3 inhibits gastric cancer
progression
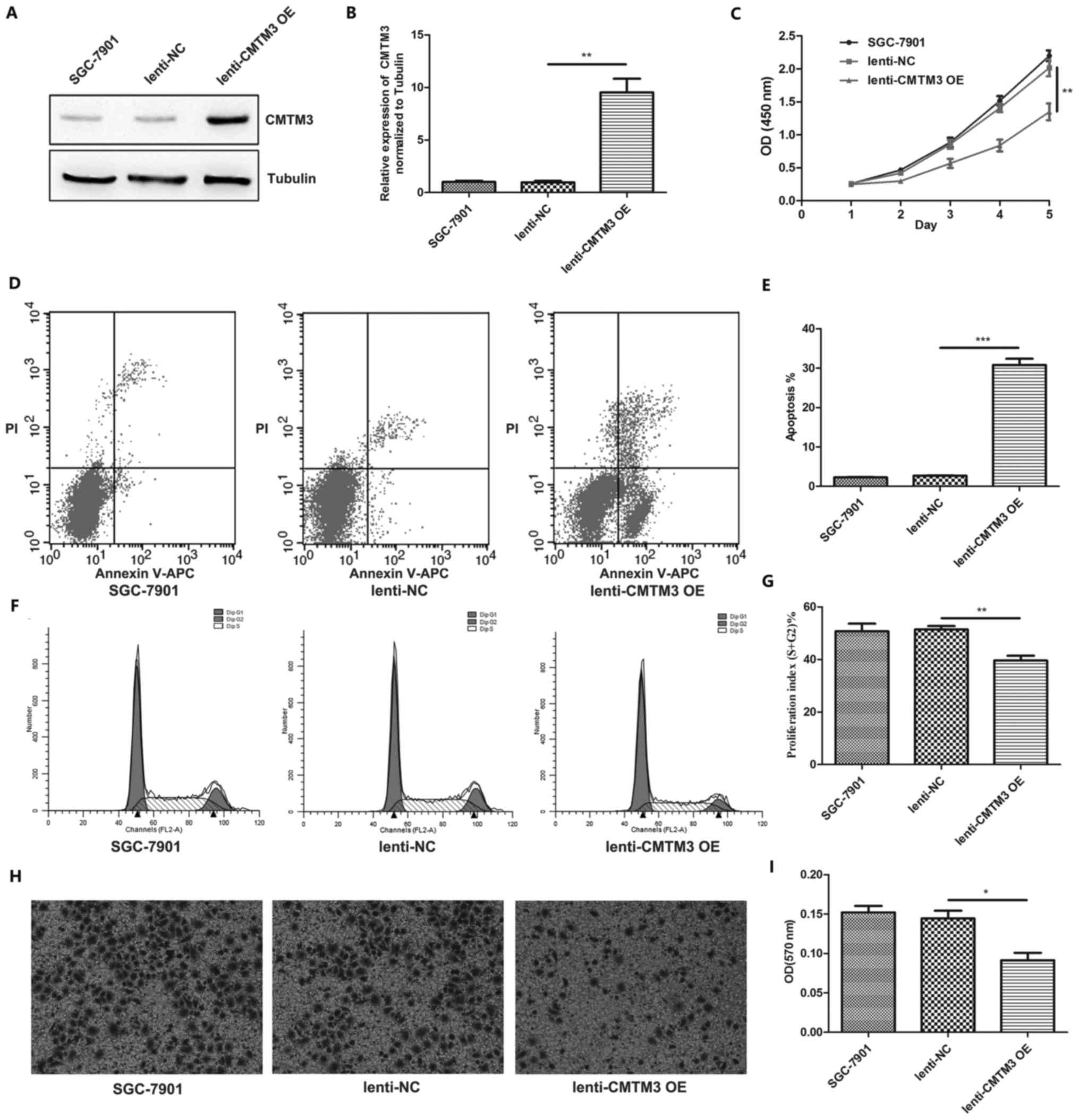
To verify the effect of CMTM3 on gastric cancer
cells in vitro, we needed to select a representative cell
line. As shown in Fig. 1D and E,
CMTM3 expression decreased most significantly in the SGC-7901 cells
compared with the GES-1 cells and other gastric cancer cell lines,
and miR-135b-5p expression increased most significantly in the
SGC-7901 cells. Thus, the SGC-7901 cells were selected for use in
the subsequent experiments. CMTM3 overexpression lentivirus was
used to introduce the exogenous expression of CMTM3 in the SGC-7901
cells. The effect of CMTM3 overexpression was validated by western
blot analysis (Fig. 3A and B). We
then performed cell proliferation assay using the CCK-8
proliferation kit. Our results suggested that CMTM3 inhibited
SGC-7901 cell proliferation (Fig.
3C). In addition, flow cytometry was used to assess the
function of CMTM3 in SGC-7901 cell apoptosis and cell cycle
progression, and the results indicated that CMTM3 overexpression
significantly promoted SGC-7901 cell apoptosis (Fig. 3D and E). Furthermore, CMTM3
overexpression markedly suppressed SGC-7901 cell cycle progression
(Fig. 3F and G). Transwell
invasion assays also indicated that CMTM3 overexpression inhibited
the invasiveness of the SGC-7901 cells (Fig. 3H and I).
miR-135b-5p inhibitor upregulates CMTM3
and suppresses gastric cancer progression
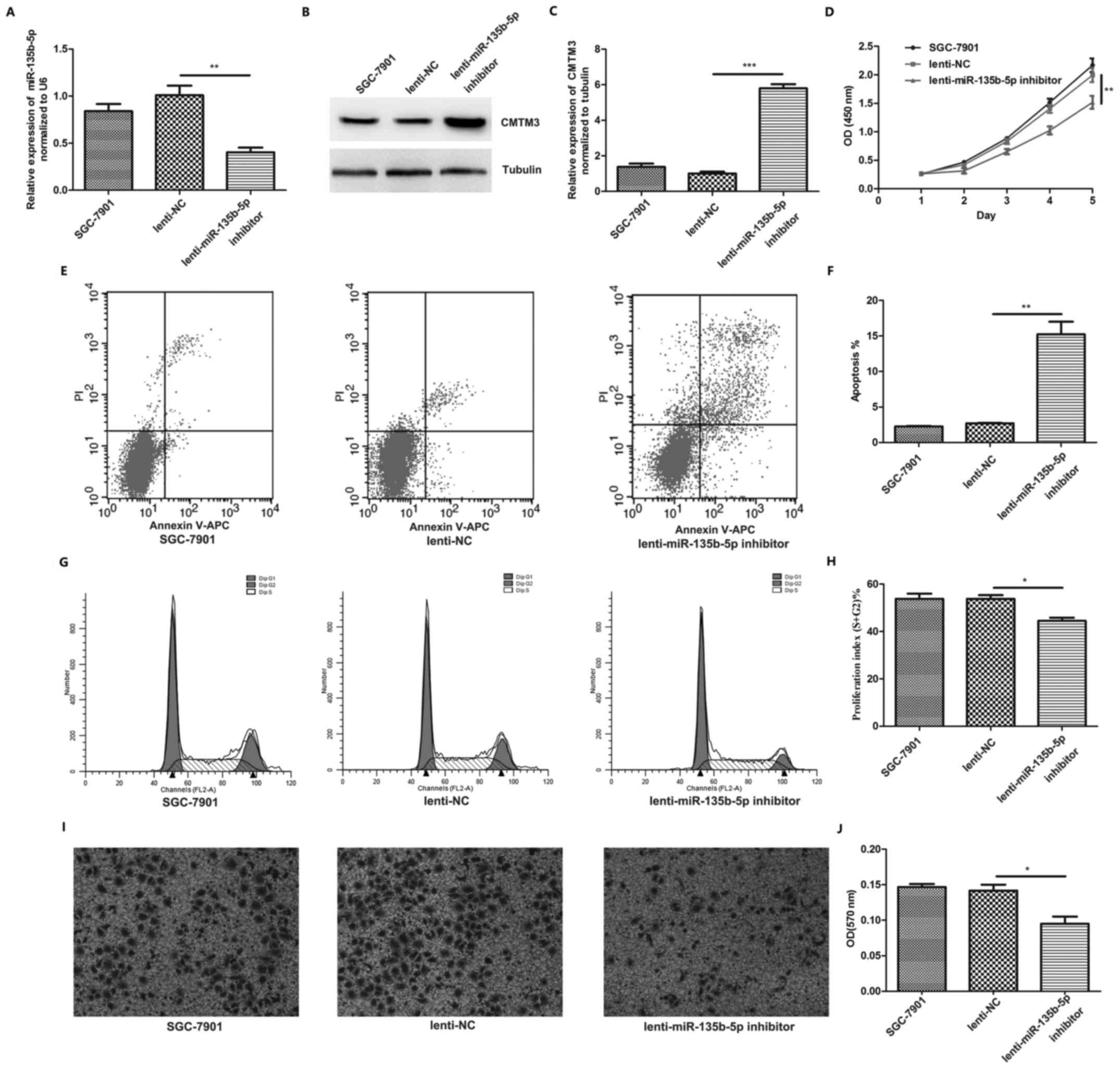
In order to investigate the function of miR-135b-5p
in gastric cancer progression, miR-135b-5p inhibitor lentivirus was
used to neutralize miR-135b-5p in the SGC-7901 cells. We found that
miR-135b-5p expression was decreased in the SGC-7901 cells
following infection with miR-135b-5p inhibitor lentivirus (Fig. 4A), while CMTM3 was significantly
upregulated (Fig. 4B and C).
Proliferation assay revealed that infection with miR-135b-5p
inhibitor inhibited SGC-7901 cell proliferation (Fig. 4D). Flow cytometry was also used to
assess the effects of miR-135b-5p inhibitor on SGC-7901 cell
apoptosis and cell cycle progression. The results indicated that
infection with miR-135b-5p inhibitor significantly promoted
SGC-7901 cell apoptosis (Fig. 4E and
F). Moreover, infection with miR-135b-5p inhibitor markedly
suppressed SGC-7901 cell cycle progression (Fig. 4G and H). Transwell invasion assays
also indicated that infection with miR-135b-5p inhibitor suppressed
the invasiveness of the SGC-7901 cells (Fig. 4I and J). The above-mentioned
results indicated that miR-135b-5p promoted gastric cancer
progression by downregulating CMTM3 expression.
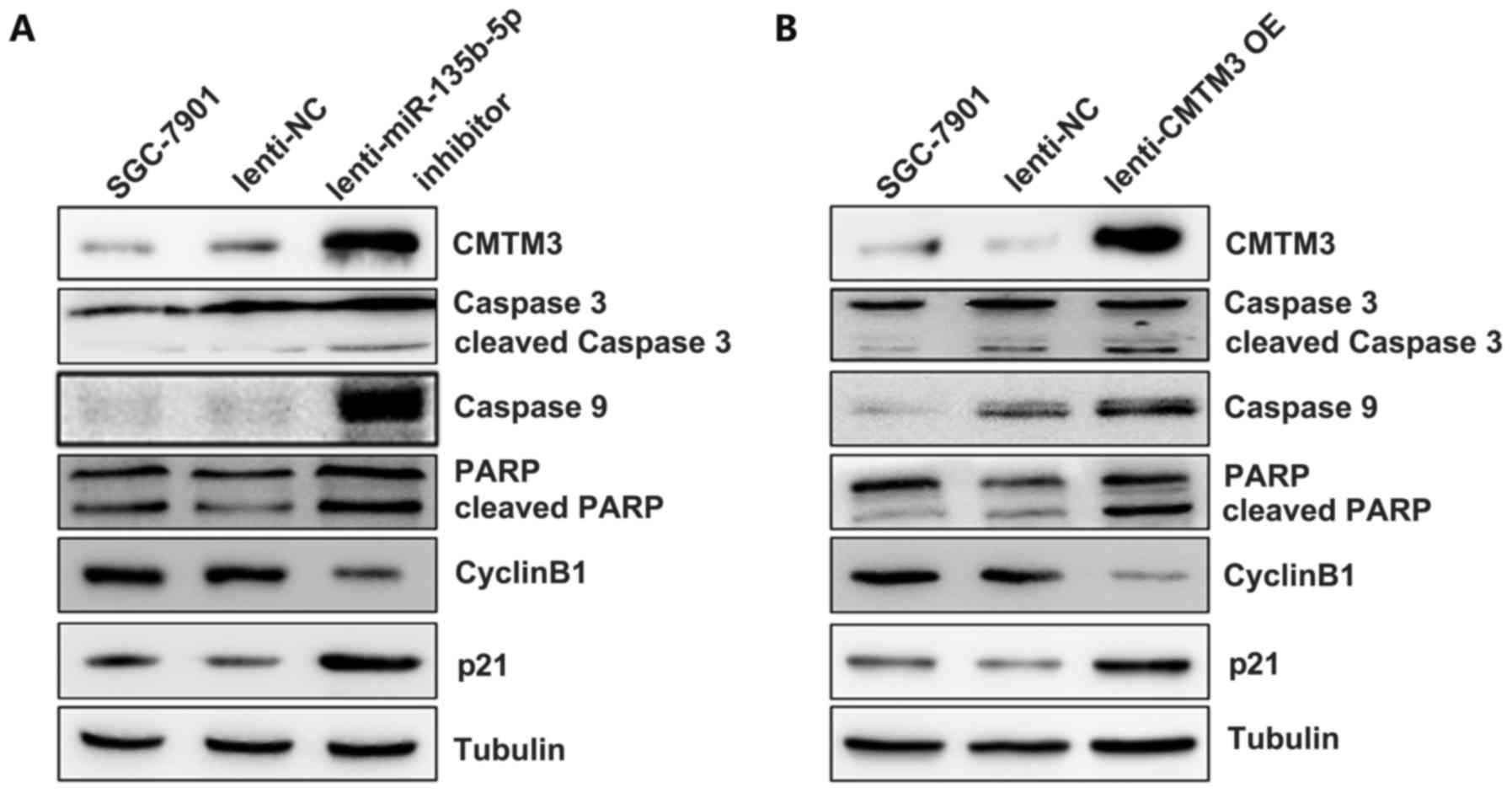
The miR-135b-5p/CMTM3 axis regulates
indicators of proliferation and apoptosis in SGC-7901 cells
In order to confirm the effects of the
miR-135b-5p/CMTM3 axis on cell proliferation, apoptosis and
invasion, we further examined the indicators of proliferation,
apoptosis and invasion by western blot analysis. The results
revealed that CMTM3 overexpression increased the level of cleaved
caspase 3, caspase 9 and cleaved PARP in the SGC-7901 cells, which
indicated that CMTM3 promoted gastric cancer cell apoptosis
(Fig. 5B). The expression of p21,
which has been reported to be an indicator of cell cycle arrest
(21), was significantly increased
after CMTM3 was overexpressed in the SGC-7901 cells. However, the
expression of cyclin B1, an important regulatory protein involved
in mitosis (22), was markedly
decreased in the SGC-7901 cells following infection with CMTM3
overexpression lentivirus (Fig.
5B). Moreover, infection with miR-135b-5p inhibitor, which
upregulated CMTM3 expression in the SGC-7901 cells, led to a
similar effect on the expression of these markers as observed with
CMTM3 overexpressoin (Fig. 5A).
These results indicated that miR-135b-5p promoted SGC-7901 cell
malignant phenotype via targeting CMTM3.
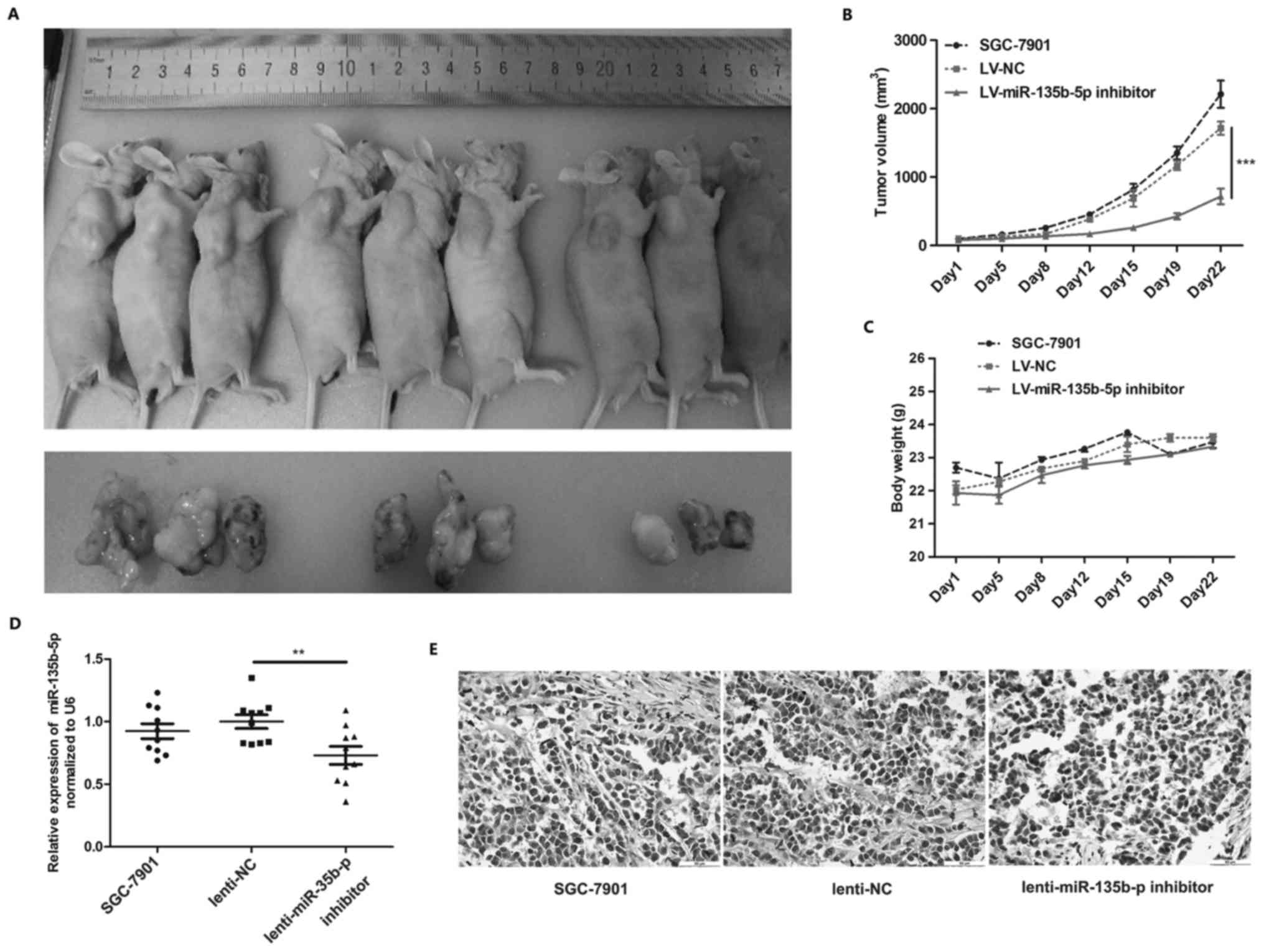
miR-135b-5p inhibitor suppresses SGC-7901
cell tumorigenesis in vivo
Nude mice with SGC-7901 subcutaneously transplanted
tumors were used to verify the function of miR-135b-5p in
vivo. miR-135b-5p inhibitor lentivirus was used to neutralize
miR-135b-5p in the tumors, and we found that the injection of the
miR-135b-5p inhibitor lentivirus markedly suppressed tumor
formation in vivo (Fig.
6A–C). We further examined miR-135b-5p and CMTM3 in the
transplanted tumors from the nude mice, and the results of RT-qPCR
revealed that miR-135b-5p expression was decreased in tumors after
the injection of miR-135b-5p inhibitor lentivirus (Fig. 6D). Moreover, IHC revealed that
CMTM3 expression was markedly increased in the tumors injected with
miR-135b-5p inhibitor lentivirus (Fig.
6E). These results indicated that miR-135b-5p promoted gastric
cancer progression in vivo.
Discussion
Despite significant progress being made in surgical
techniques and chemotherapeutic regimens for gastric cancer, the
death rate remains high. Thus, there is an urgent need for the
discovery of novel predictive strategies and therapeutic targets
for gastric cancer. In this study, we examined the expression
status of CMTM3 and miR-135b-5p in gastric cancer tissues and cell
lines, and analyzed the regulatory association between miR-135b-5p
and CMTM3. Furthermore, we examined the contribution of
miR-135b-5p/CMTM3 axis in gastric cancer progression; our results
revealed novel targets for gastric cancer.
CMTMs was shown to play a crucial role as a tumor
suppressor in gastric cancer, which is consistent with the findings
of previous studies reporting its role in other types of cancer.
For example, in prostate cancer, interleukin (IL)-30 treatment has
been shown to suppress the expression of CMTM3, leading to the
progression of prostate cancer (23). In testicular cancer cells, CMTM3
has been shown to suppress the proliferation and migration
capacities in vitro (24),
which is consistent with our observations on gastric cancer cells.
A recent study stated the role of CMTM3 in gastric cancer, and
stated that the silencing of CMTM3 expression promoted migration
and the EMT phenotype of gastric cancer cells. Mechanistically,
CMTM3 regulated the STAT3/Twist1/EMT signaling pathway (13). Although the positive role of CMTM3
in cancer progression has been demonstrated (25), it is well accepted to be a tumor
suppressor. Previous studies have suggested that promoter
hypermethylation is involved in the decreased expression of CMTM3
in various types of cancer, including gastric, colorectal and
breast cancer (11,12,26).
Currently, miRNA-mediated gene regulation is also considered to be
an important aspect of epigenetic regulation in tumorigenesis
(27,28). The present study has provided a
novel regulatory mechanism of CMTM3 regulation. This promted us to
carry out further investigations by the combination of miRNA
expression with CMTM3 expression in the diagnosis and treatment of
gastric cancer.
miR-135b plays contradictory roles in different
types of cancer. For example, in prostate cancer and glioblastoma,
miR-135b has been reported to act as a tumor suppressor (18,29).
However, in cancers such as breast cancer, hepatocellular carcinoma
and colorectal cancer, miR-135b has been reported to be a
tumor-promoting factor and to promote cancer cell proliferation and
migration (16,30,31).
A previous study indicated that the upregulated expression of
hsa-miR-135b is found in gastric lesions compared to normal gastric
mucosa and intestinal-type gastric adenocarcinoma samples,
suggesting the positive role of miR-135b on cancer progression
(32). This is consistent with our
observations, supporting the positive role of miR-135b in cancer
progression. Furthermore, the present study identified a novel
target of miR-135b, and broadened the significance of miR-135b in
cancers. We verified that miR-135b-5p functioned as an oncogene by
targeting CMTM3. Infectoin with miR-135b-5p inhibitor lentivirus
induced the upregulation of CMTM3 and increased the apoptotic rate
of the SGC-7901 cells, and suppressed SGC-7901 cell proliferation,
invasion and cell cycle progression. The results of corresponding
western blot analysis for indicator genes, including cleaved
caspase 3, caspase 9, cleaved PARP, p21 and cyclin B1 (33-36),
further confirmed our findings.
In conclusion, the present study uncovered a novel
miR-135b/CMTM3 axis in the progression of gastric cancer, and
provided a novel target for the prediction of the prognosis and for
the development of novel treatment strategies and therapeutic
targets for gastric cancer.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu X, Qiu H, Kong P, Zhou Z and Sun X:
Gastric cancer, nutritional status, and outcome. Onco Targets Ther.
10:2107–2114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maleki SS and Röcken C: Chromosomal
instability in gastric cancer biology. Neoplasia. 19:412–420. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han W, Lou Y, Tang J, Zhang Y, Chen Y, Li
Y, Gu W, Huang J, Gui L, Tang Y, et al: Molecular cloning and
characterization of chemokine-like factor 1 (CKLF1), a novel human
cytokine with unique structure and potential chemotactic activity.
Biochem J. 357:127–135. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han W, Ding P, Xu M, Wang L, Rui M, Shi S,
Liu Y, Zheng Y, Chen Y, Yang T, et al: Identification of eight
genes encoding chemokine-like factor superfamily members 1-8
(CKLFSF1-8) by in silico cloning and experimental validation.
Genomics. 81:609–617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie J, Yuan Y, Liu Z, Xiao Y, Zhang X, Qin
C, Sheng Z, Xu T and Wang X: CMTM3 is frequently reduced in clear
cell renal cell carcinoma and exhibits tumor suppressor activities.
Clin Transl Oncol. 16:402–409. 2014. View Article : Google Scholar
|
|
8
|
Shao L, Guo X, Plate M, Li T, Wang Y, Ma D
and Han W: CMTM5-v1 induces apoptosis in cervical carcinoma cells.
Biochem Biophys Res Commun. 379:866–871. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu B, Su Y, Li T, Yuan W, Mo X, Li H, He
Q, Ma D and Han W: CMTM7 knockdown increases tumorigenicity of
human non-small cell lung cancer cells and EGFR-AKT signaling by
reducing Rab5 activation. Oncotarget. 6:41092–41107. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang W, Mendoza MC, Pei X, Ilter D,
Mahoney SJ, Zhang Y, Ma D, Blenis J and Wang Y: Down-regulation of
CMTM8 induces epithelial-to-mesenchymal transition-like changes via
c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol
Chem. 287:11850–11858. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan W, Li T, Mo X, Wang X, Liu B, Wang W,
Su Y, Xu L and Han W: Knockdown of CMTM3 promotes metastasis of
gastric cancer via the STAT3/Twist1/EMT signaling pathway.
Oncotarget. 7:29507–29519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H,
Li H, Shu XS, Li H, Liu W, et al: CMTM3, located at the critical
tumor suppressor locus 16q22.1, is silenced by CpG methylation in
carcinomas and inhibits tumor cell growth through inducing
apoptosis. Cancer Res. 69:5194–5201. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su Y, Lin Y, Zhang L, Liu B, Yuan W, Mo X,
Wang X, Li H, Xing X, Cheng X, et al: CMTM3 inhibits cell migration
and invasion and correlates with favorable prognosis in gastric
cancer. Cancer Sci. 105:26–34. 2014. View Article : Google Scholar
|
|
14
|
Catela Ivkovic T, Voss G, Cornella H and
Ceder Y: microRNAs as cancer therapeutics: A step closer to
clinical application. Cancer Lett. 407:113–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng Q, Chen C, Guan H, Kang W and Yu C:
Prognostic role of microRNAs in human gastrointestinal cancer: A
systematic review and meta-analysis. Oncotarget. 8:46611–46623.
2017.PubMed/NCBI
|
|
16
|
Hua K, Jin J, Zhao J, Song J, Song H, Li
D, Maskey N, Zhao B, Wu C, Xu H, et al: miR-135b, upregulated in
breast cancer, promotes cell growth and disrupts the cell cycle by
regulating LATS2. Int J Oncol. 48:1997–2006. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xue Y, Ni T, Jiang Y and Li Y: Long
noncoding RNA GAS5 inhibits tumorigenesis and enhances
radiosensitivity by suppressing miR-135b expression in non-small
cell lung cancer. Oncol Res. 25:1305–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang N, Tao L, Zhong H, Zhao S, Yu Y, Yu
B, Chen X, Gao J and Wang R: miR-135b inhibits tumour metastasis in
prostate cancer by targeting STAT6. Oncol Lett. 11:543–550.
2016.PubMed/NCBI
|
|
19
|
Su W, Mo Y, Wu F, Guo K, Li J, Luo Y, Ye
H, Guo H, Li D and Yang Z: miR-135b reverses chemoresistance of
non-small cell lung cancer cells by downregulation of FZD1. Biomed
Pharmacother. 84:123–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li P, Fan JB, Gao Y, Zhang M, Zhang L,
Yang N and Zhao X: miR-135b-5p inhibits LPS-induced TNFα production
via silencing AMPK phosphatase Ppm1e. Oncotarget. 7:77978–77986.
2016.PubMed/NCBI
|
|
21
|
Li Z, Li X, Xu L, Tao Y, Yang C, Chen X,
Fang F, Wu Y, Ding X, Zhao H, et al: Inhibition of neuroblastoma
proliferation by PF-3758309, a small-molecule inhibitor that
targets p21-activated kinase 4. Oncol Rep. 38:2705–2716.
2017.PubMed/NCBI
|
|
22
|
Miles DC, van den Bergen JA, Sinclair AH
and Western PS: Regulation of the female mouse germ cell cycle
during entry into meiosis. Cell Cycle. 9:408–418. 2010. View Article : Google Scholar
|
|
23
|
Di Meo S, Airoldi I, Sorrentino C, Zorzoli
A, Esposito S and Di Carlo E: Interleukin-30 expression in prostate
cancer and its draining lymph nodes correlates with advanced grade
and stage. Clin Cancer Res. 20:585–594. 2014. View Article : Google Scholar
|
|
24
|
Li Z, Xie J, Wu J, Li W, Nie L, Sun X,
Tang A, Li X, Liu R, Mei H, et al: CMTM3 inhibits human testicular
cancer cell growth through inducing cell-cycle arrest and
apoptosis. PLoS One. 9:e889652014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Delic S, Thuy A, Schulze M, Proescholdt
MA, Dietrich P, Bosserhoff AK and Riemenschneider MJ: Systematic
investigation of CMTM family genes suggests relevance to
glioblastoma pathogenesis and CMTM1 and CMTM3 as priority targets.
Genes Chromosomes Cancer. 54:433–443. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Chen C, Bi X, Zhou C, Huang T, Ni C,
Yang P, Chen S, Ye M and Duan S: DNA methylation of CMTM3, SSTR2,
and MDFI genes in colorectal cancer. Gene. 630:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cho CJ, Myung SJ and Chang S: ADAR1 and
microRNA; A hidden crosstalk in cancer. Int J Mol Sci. 18:E7992017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Forrest ME and Khalil AM: Review:
Regulation of the cancer epigenome by long non-coding RNAs. Cancer
Lett. 407:106–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lulli V, Buccarelli M, Martini M, Signore
M, Biffoni M, Giannetti S, Morgante L, Marziali G, Ilari R,
Pagliuca A, et al: miR-135b suppresses tumorigenesis in
glioblastoma stem-like cells impairing proliferation, migration and
self-renewal. Oncotarget. 6:37241–37256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Liang H, Bai M, Ning T, Wang C, Fan
Q, Wang Y, Fu Z, Wang N, Liu R, et al: miR-135b promotes receptor
II (TGFBR2) in colorectal cancer. PLoS One. 10:e01301942015.
View Article : Google Scholar
|
|
31
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015. View Article : Google Scholar :
|
|
32
|
Vidal AF, Cruz AM, Magalhães L, Pereira
AL, Anaissi AK, Alves NC, Albuquerque PJ, Burbano RM, Demachki S
and Ribeiro-dos-Santos Â: hsa-miR-29c and hsa-miR-135b differential
expression as potential biomarker of gastric carcinogenesis. World
J Gastroenterol. 22:2060–2070. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: Epigenetic alteration and microRNA dysregulation in cancer.
Front Genet. 4:2582013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ando T, Yoshida T, Enomoto S, Asada K,
Tatematsu M, Ichinose M, Sugiyama T and Ushijima T: DNA methylation
of microRNA genes in gastric mucosae of gastric cancer patients:
Its possible involvement in the formation of epigenetic field
defect. Int J Cancer. 124:2367–2374. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Zhao X, Fiskus W, Lin J, Lwin T,
Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, et al: Coordinated
silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic
target of histone modification in aggressive B-Cell lymphomas.
Cancer Cell. 22:506–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan H, Choi AJ, Lee BH and Ting AH:
Identification and functional analysis of epigenetically silenced
microRNAs in colorectal cancer cells. PLoS One. 6:e206282011.
View Article : Google Scholar : PubMed/NCBI
|