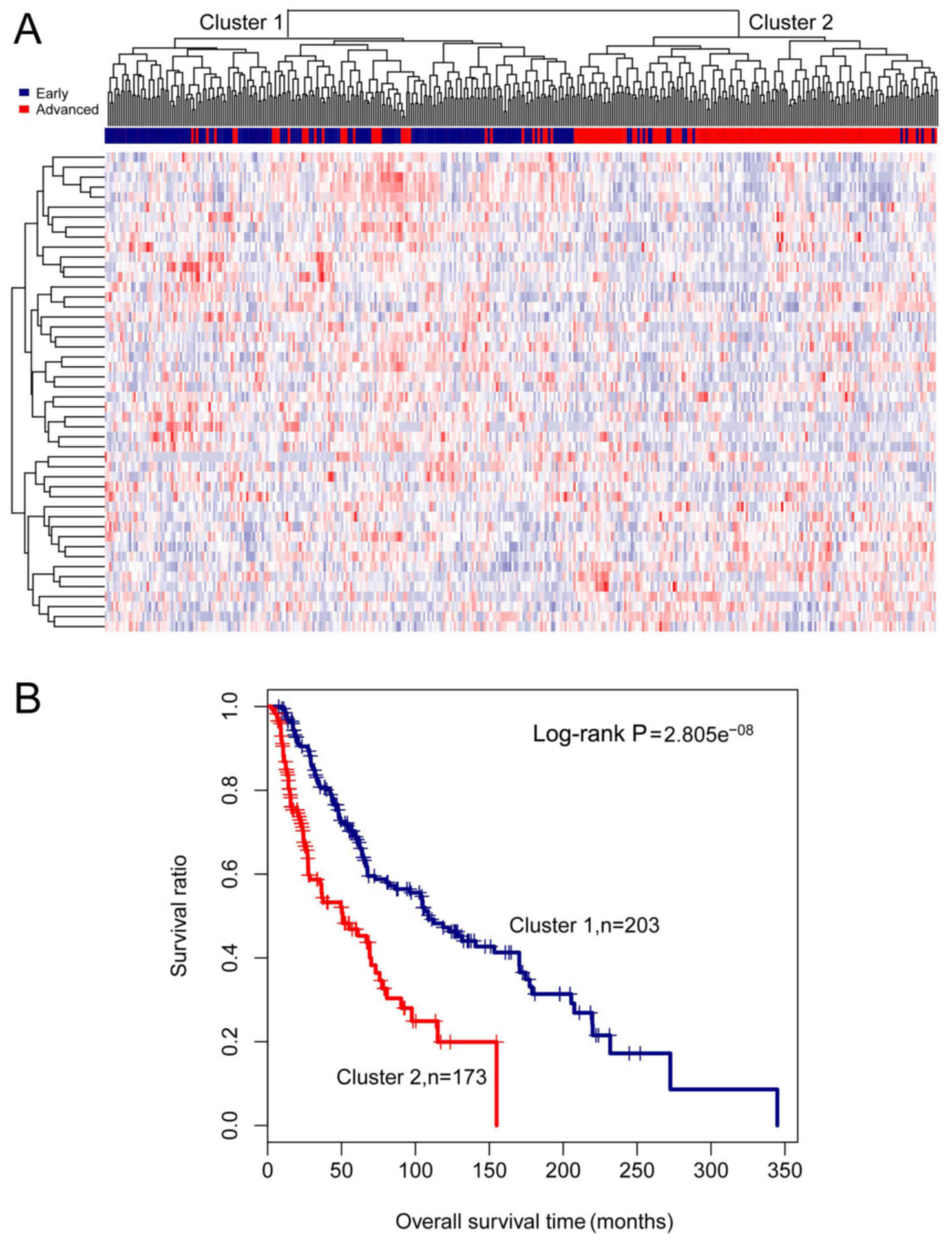
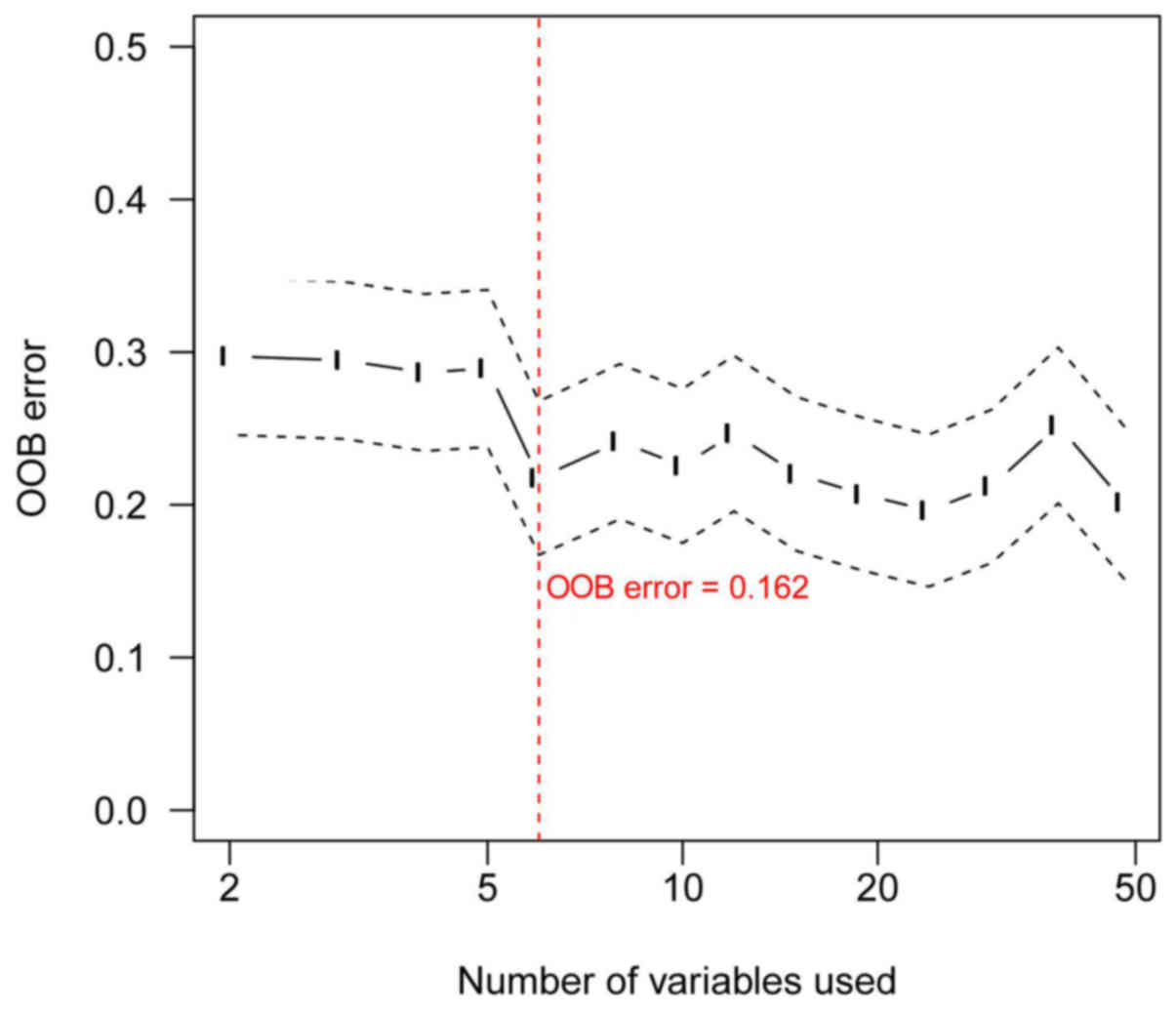
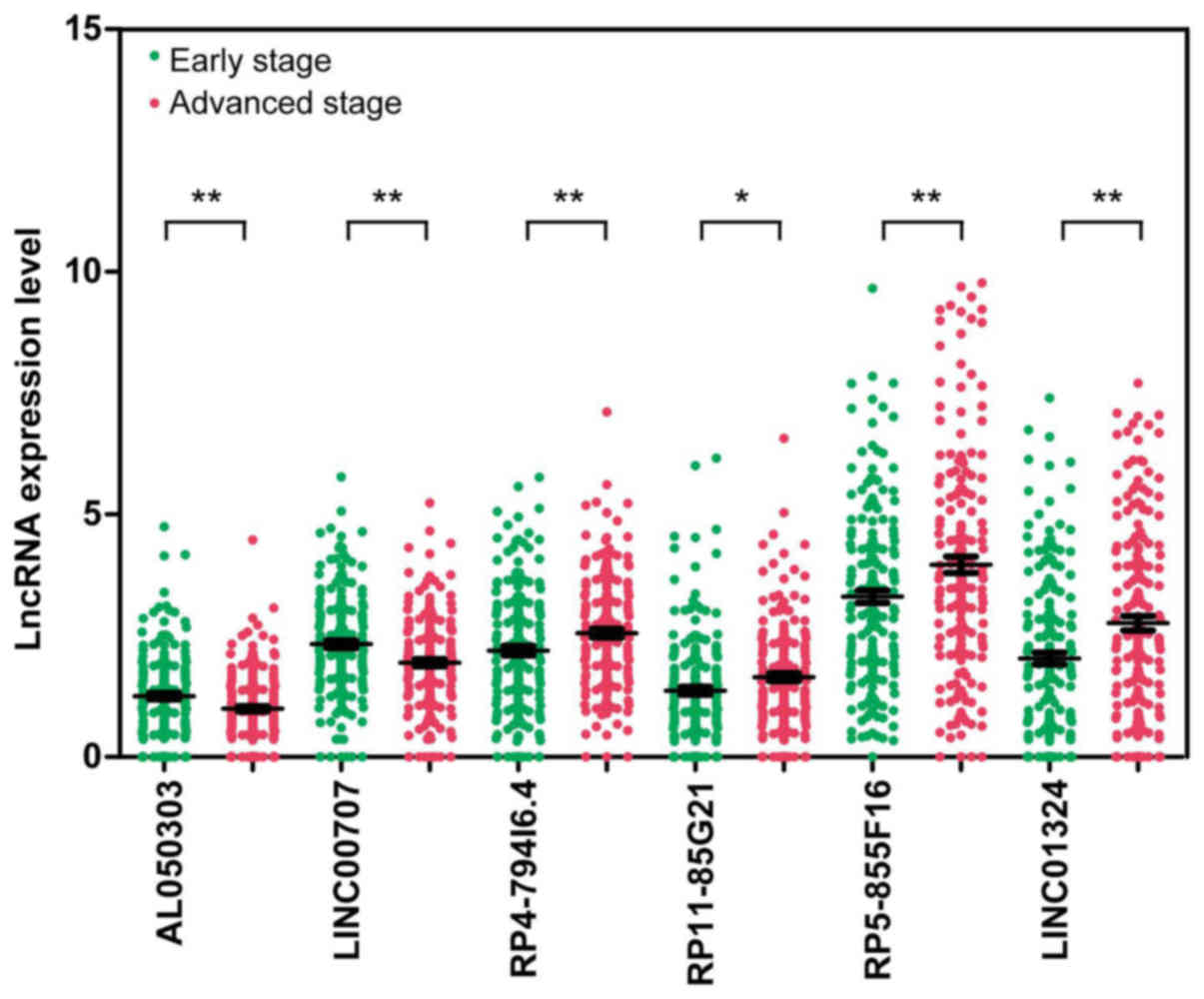
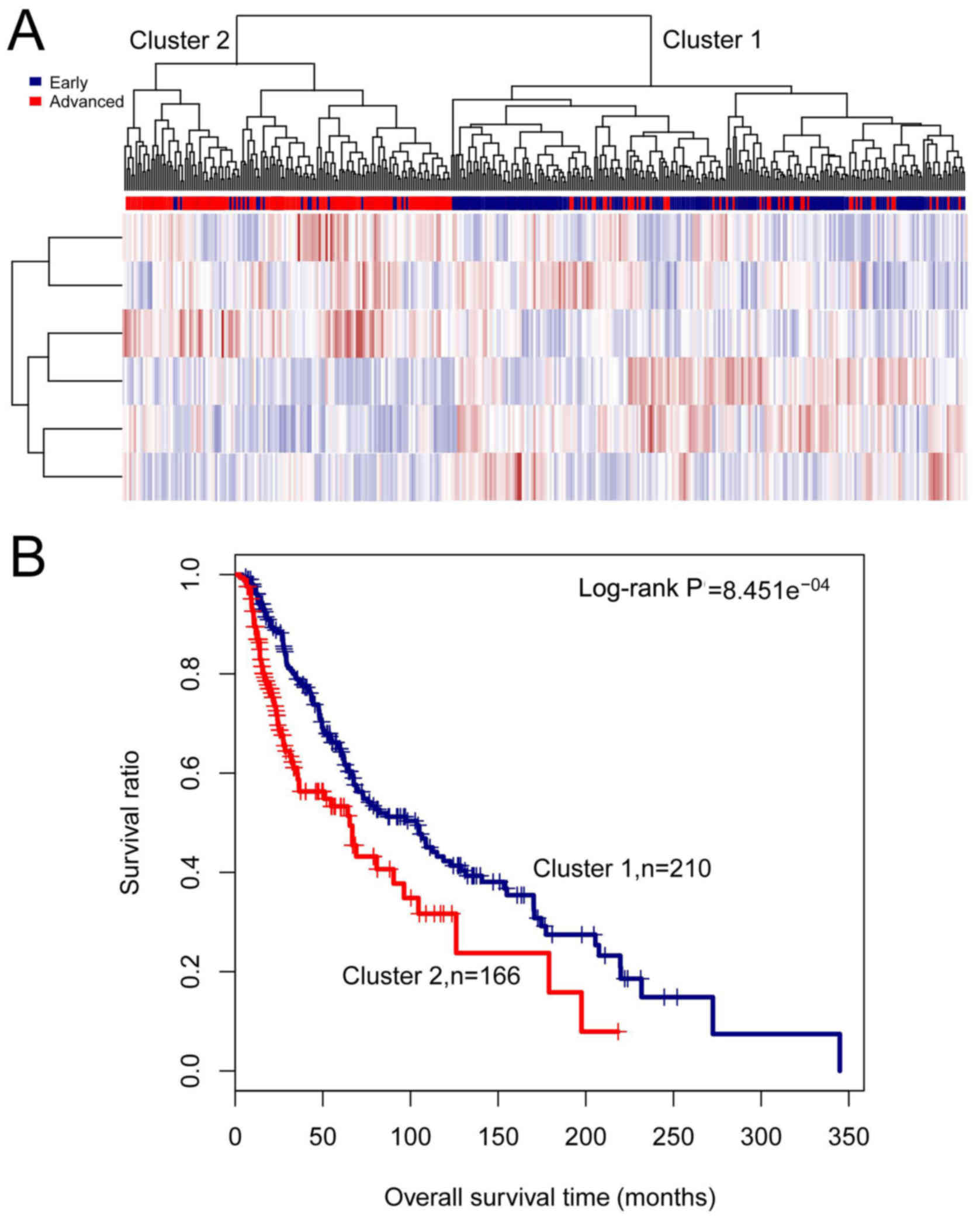
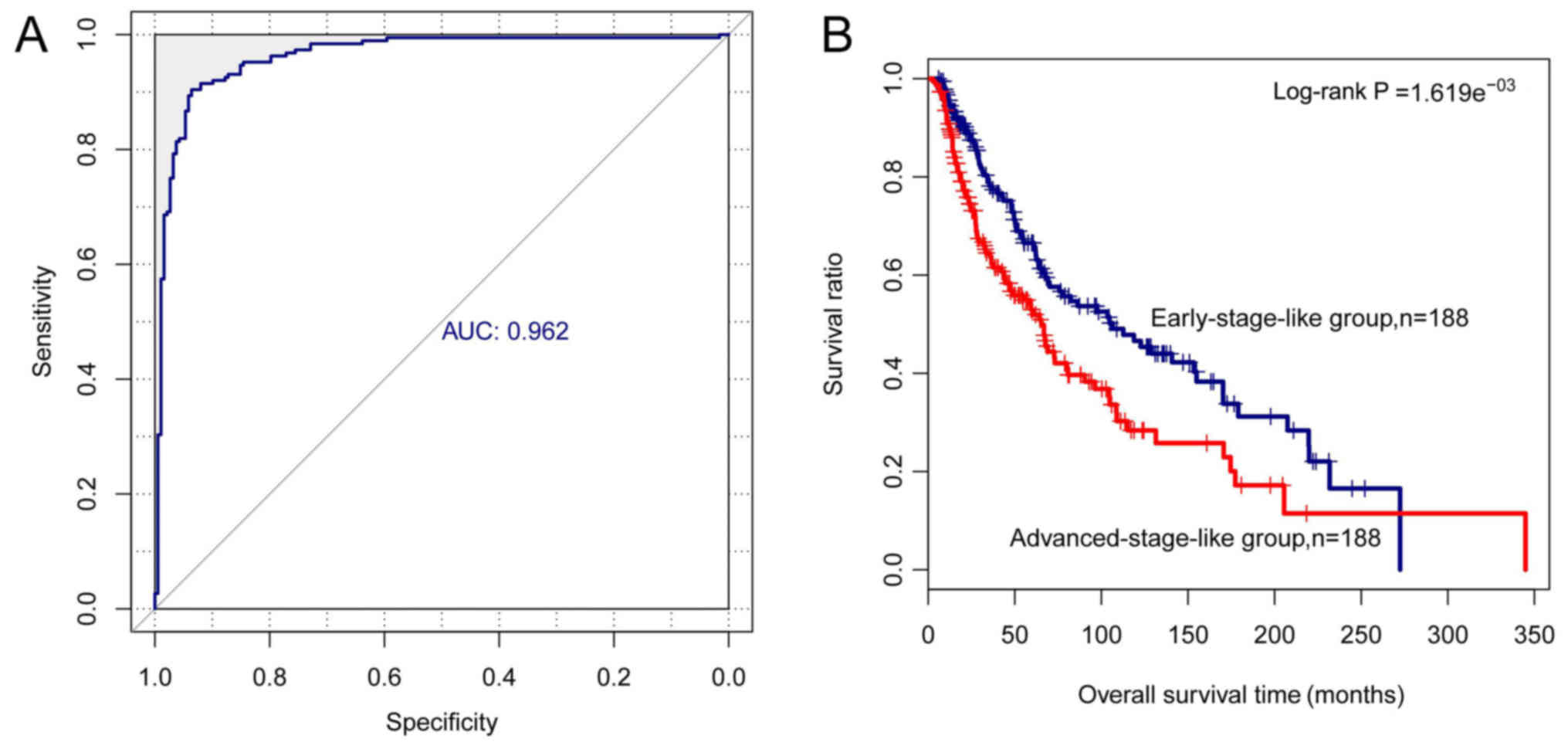
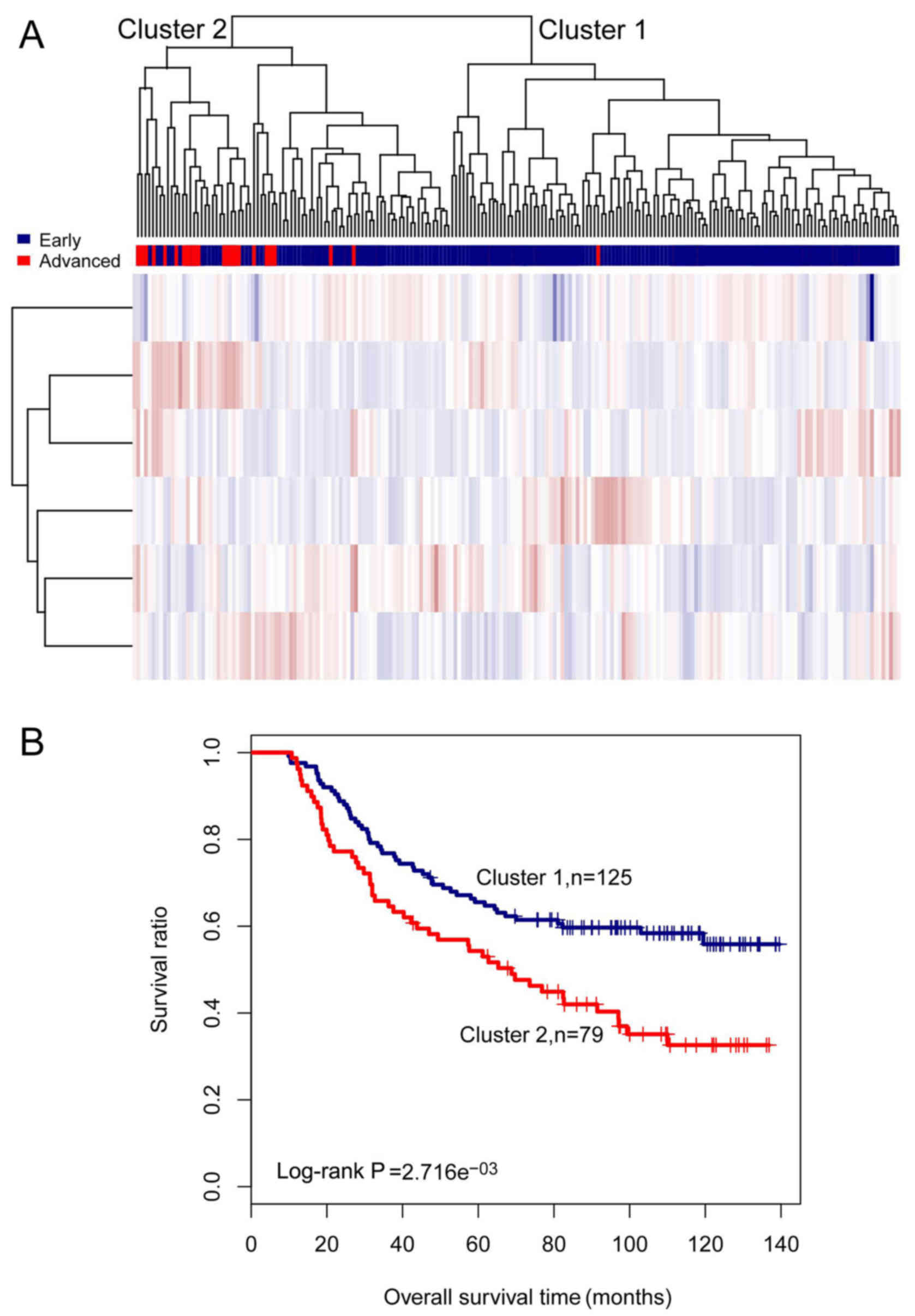
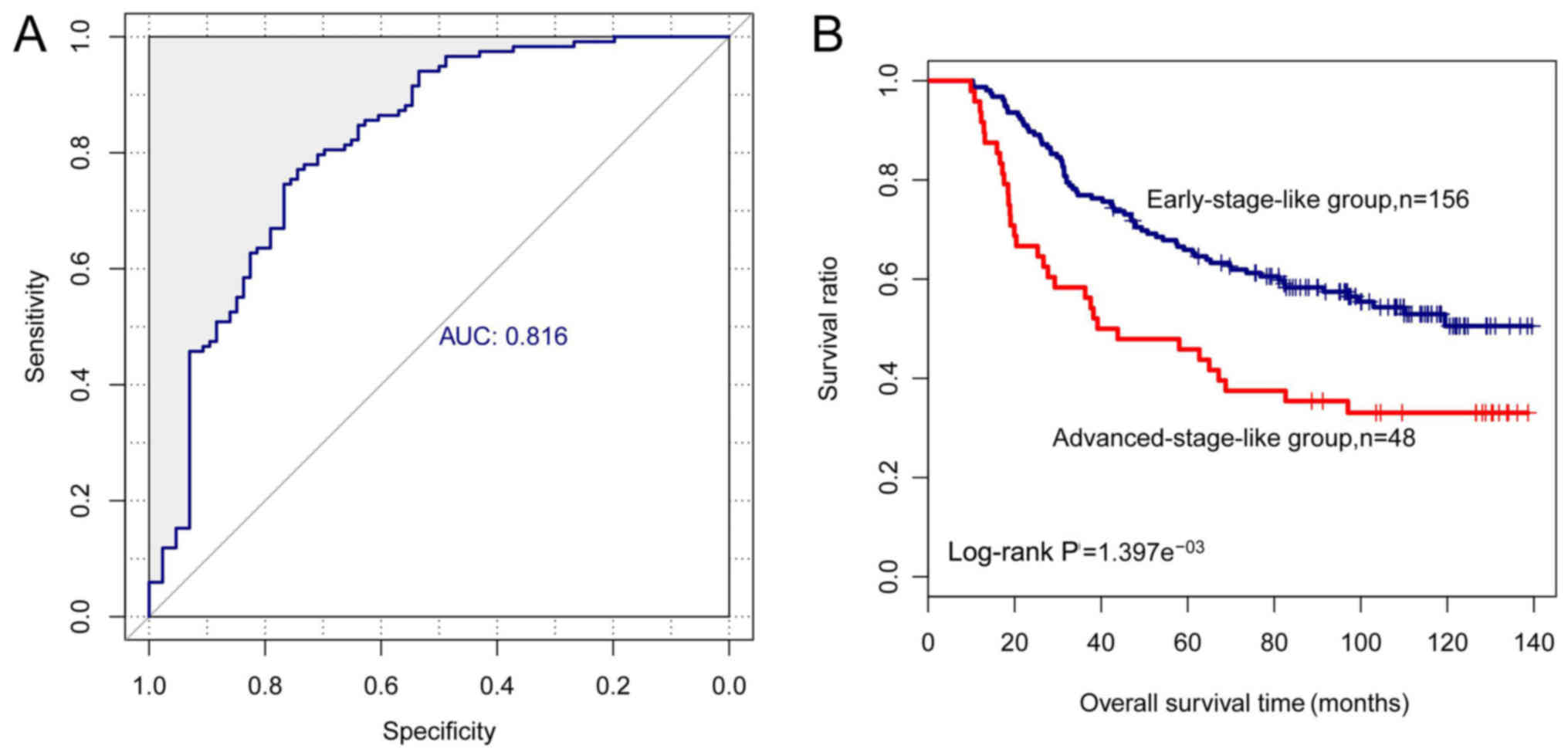
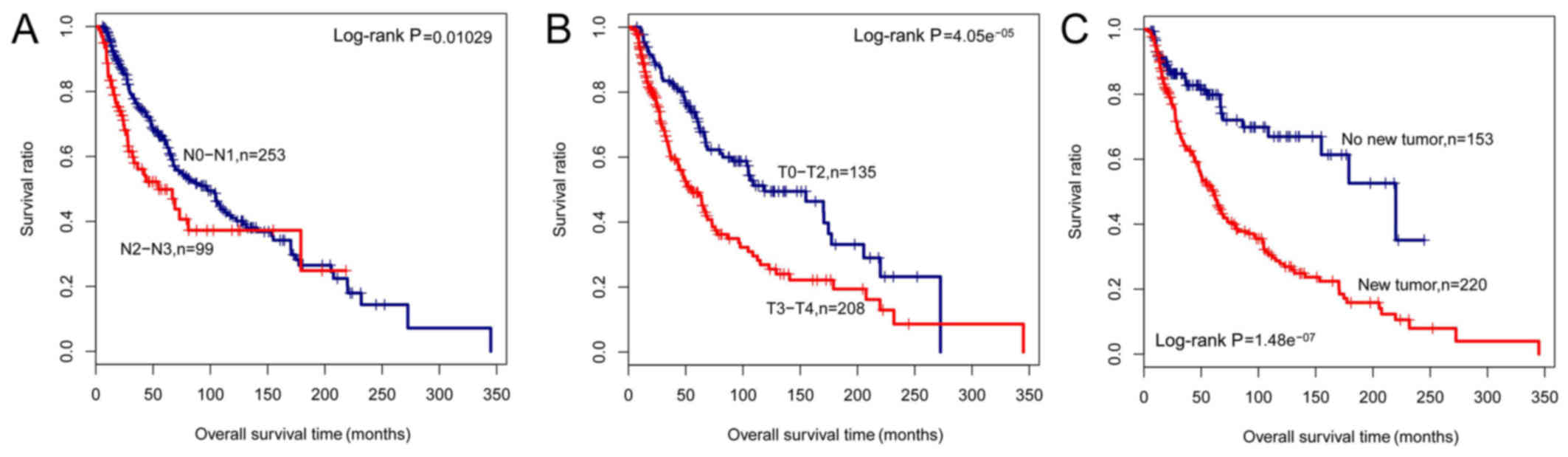
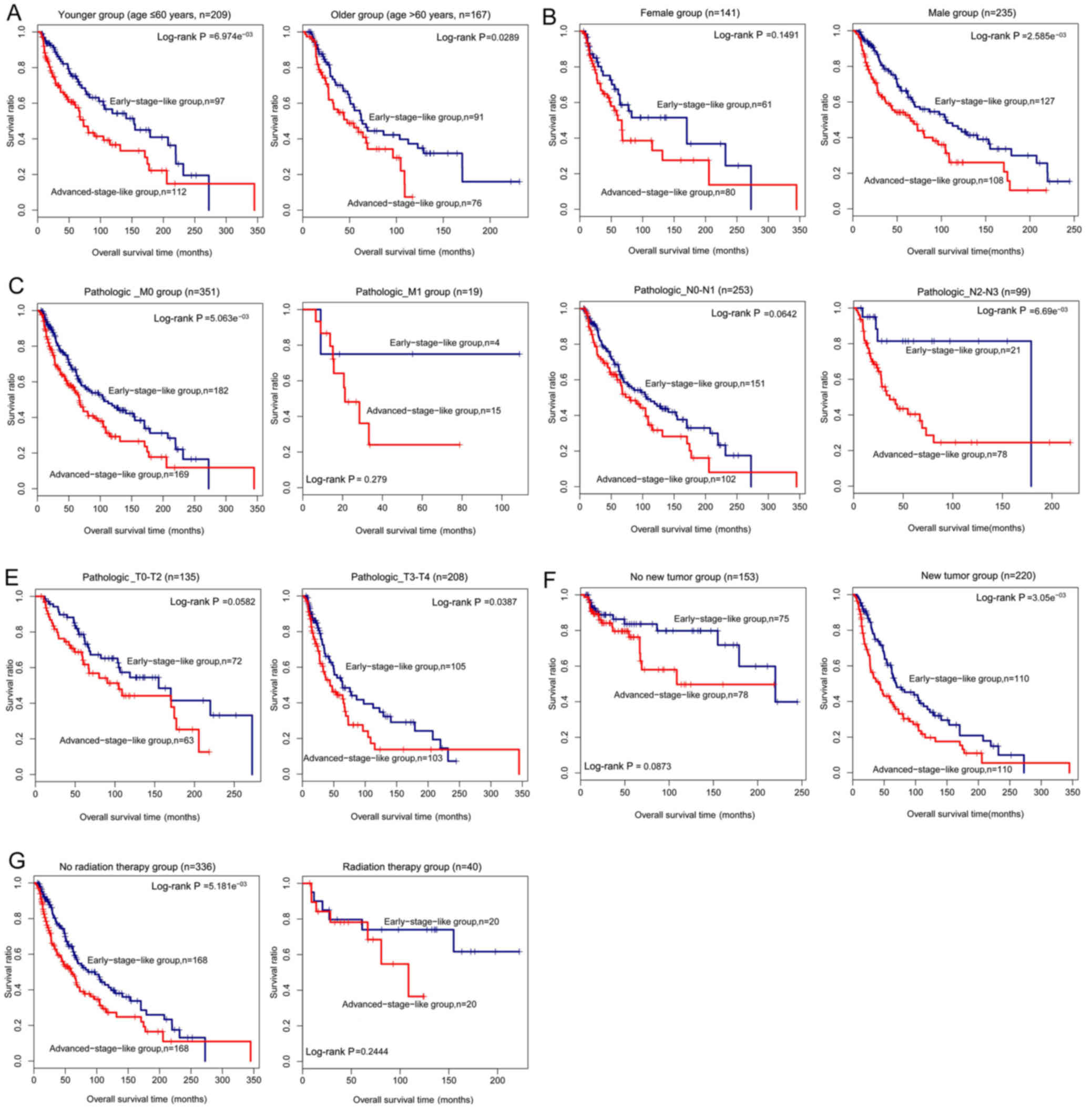
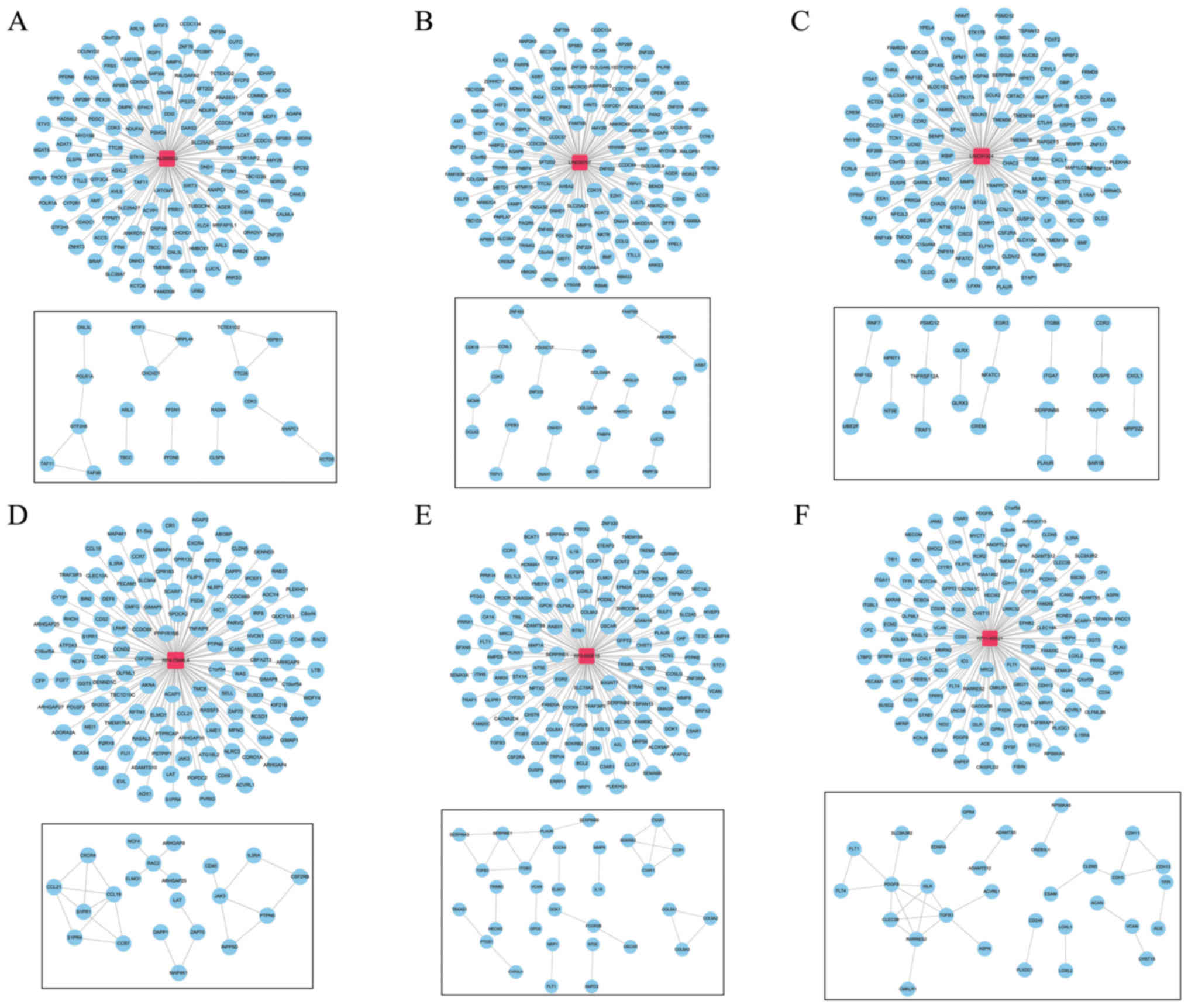
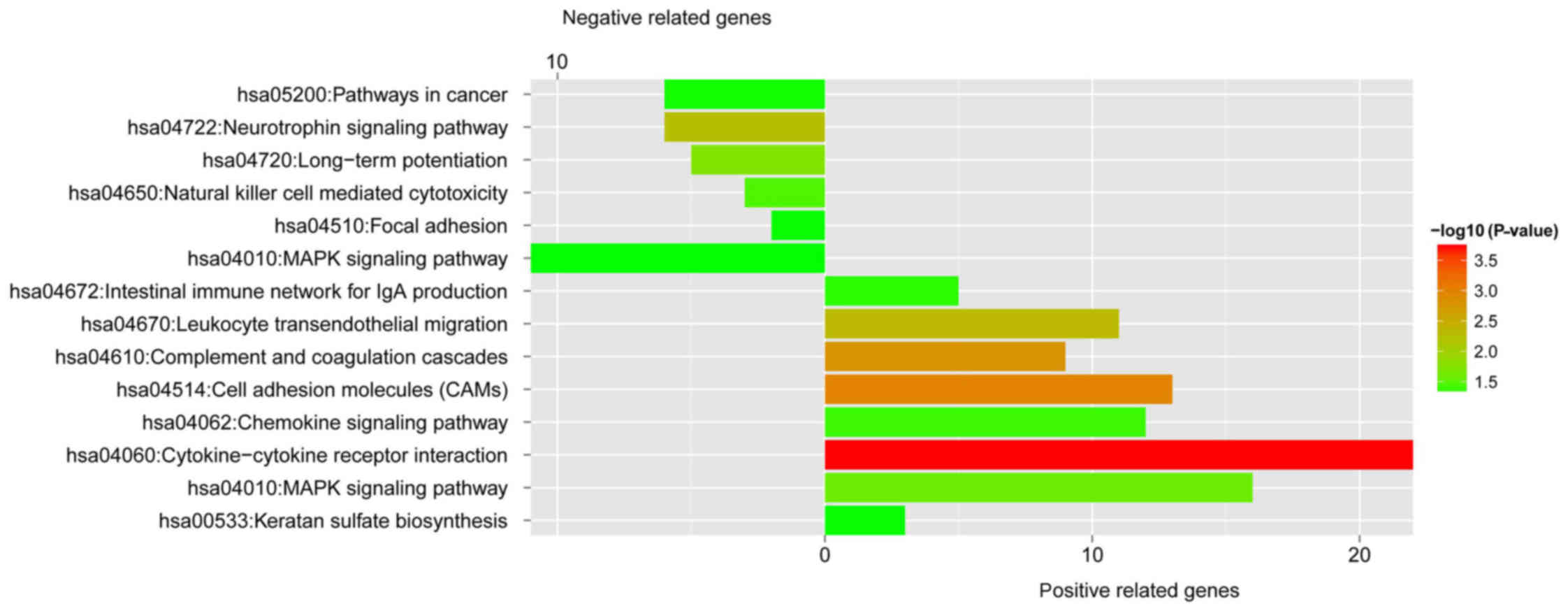
|
1
|
Wang H, Naghavi M, Allen C, Barber RM,
Bhutta ZA, Carter A, Casey DC, Charlson FJ, Chen AZ, Coates MM, et
al: GBD 2015 Mortality and Causes of Death Collaborators: Global,
regional, and national life expectancy, all-cause mortality, and
cause-specific mortality for 249 causes of death, 1980–2015: A
systematic analysis for the Global Burden of Disease Study.
2015.Lancet. 388:1459–1544. 2016. View Article : Google Scholar
|
|
2
|
MacKie RM, Hauschild A and Eggermont AMM:
Epidemiology of invasive cutaneous melanoma. Ann Oncol. 20(Suppl
6): vi1–vi7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piris A, Lobo AC and Duncan LM: Melanoma
staging: Where are we now. Dermatol Clin. 30:581–592. 2012.
View Article : Google Scholar
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hulstaert E, Brochez L, Volders PJ,
Vandesompele J and Mestdagh P: Long non-coding RNAs in cutaneous
melanoma: Clinical perspectives. Oncotarget. 8:43470–43480. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion, and
metastatic potential of metastatic melanoma. Biomed Res Int.
2013:2510982013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L
and Sha N: Long non-coding RNA BANCR promotes proliferation in
malignant melanoma by regulating MAPK pathway activation. PLoS One.
9:e1008932014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Guo W, Xu XJ, Su F, Wang Y, Zhang
Y, Wang Q and Zhu L: Melanoma long non-coding RNA signature
predicts prognostic survival and directs clinical risk-specific
treatments. J Dermatol Sci. 85:226–234. 2017. View Article : Google Scholar
|
|
11
|
Nsengimana J, Laye J, Filia A, Walker C,
Jewell R, Van den Oord JJ, Wolter P, Patel P, Sucker A, Schadendorf
D, et al: Independent replication of a melanoma subtype gene
signature and evaluation of its prognostic value and biological
correlates in a population cohort. Oncotarget. 6:11683–11693. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
14
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H, et al: RNA-seq analyses of multiple
meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
17
|
Tolosi L and Lengauer T: Classification
with correlated features: Unreliability of feature ranking and
solutions. Bioinformatics. 27:1986–1994. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zapf A, Brunner E and Konietschke F: A
wild bootstrap approach for the selection of biomarkers in early
diagnostic trials. BMC Med Res Methodol. 15:432015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Q and Liu X: Screening of feature
genes in distinguishing different types of breast cancer using
support vector machine. Onco Targets Ther. 8:2311–2317.
2015.PubMed/NCBI
|
|
20
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar :
|
|
21
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
22
|
Inamdar GS, Madhunapantula SV and
Robertson GP: Targeting the MAPK pathway in melanoma: Why some
approaches succeed and other fail. Biochem Pharmacol. 80:624–637.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grimaldi AM, Simeone E, Festino L, Vanella
V, Palla M and Ascierto PA: Novel mechanisms and therapeutic
approaches in melanoma: Targeting the MAPK pathway. Discov Med.
19:455–461. 2015.PubMed/NCBI
|
|
24
|
Wellbrock C and Arozarena I: The
Complexity of the ERK/MAP-kinase pathway and the treatment of
melanoma skin cancer. Front Cell Dev Biol. 4:332016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ito M, Yoshioka K, Akechi M, Yamashita S,
Takamatsu N, Sugiyama K, Hibi M, Nakabeppu Y, Shiba T and Yamamoto
KI: JSAP1, a novel jun N-terminal protein kinase (JNK)-binding
protein that functions as a Scaffold factor in the JNK signaling
pathway. Mol Cell Biol. 19:7539–7548. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meyer C, Sevko A, Ramacher M, Bazhin AV,
Falk CS, Osen W, Borrello I, Kato M, Schadendorf D, Baniyash M, et
al: Chronic inflammation promotes myeloid-derived suppressor cell
activation blocking antitumor immunity in transgenic mouse melanoma
model. Proc Natl Acad Sci USA. 108:17111–17116. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Keibel A, Singh V and Sharma MC:
Inflammation, microenvi-ronment, and the immune system in cancer
progression. Curr Pharm Des. 15:1949–1955. 2009. View Article : Google Scholar
|
|
28
|
Truzzi F, Marconi A, Lotti R, Dallaglio K,
French LE, Hempstead BL and Pincelli C: Neurotrophins and their
receptors stimulate melanoma cell proliferation and migration. J
Invest Dermatol. 128:2031–2040. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang R, Bi J, Ampah KK, Ba X, Liu W and
Zeng X: Lipid rafts control human melanoma cell migration by
regulating focal adhesion disassembly. Biochim Biophys Acta.
1833:3195–3205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hess AR, Postovit LM, Margaryan NV, Seftor
EA, Schneider GB, Seftor RE, Nickoloff BJ and Hendrix MJ: Focal
adhesion kinase promotes the aggressive melanoma phenotype. Cancer
Res. 65:9851–9860. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tímár J, Gyorffy B and Rásó E: Gene
signature of the metastatic potential of cutaneous melanoma: Too
much for too little. Clin Exp Metastasis. 27:371–387. 2010.
View Article : Google Scholar
|