Introduction
Breast cancer is one of the most common
heterogeneous malignant tumors in women. It affects developed and
developing countries, and its incidence continues to increase by
~3% each year (1). Despite
substantial progress in improving diagnostic and therapeutic
methods in recent years, breast cancer remains the second leading
cause of cancer-associated mortality in women; and there were
>500,000 mortalities in 2011 according to the World Health
Organization statistics (2).
Previous research suggests that recurrent metastasis in patients
with breast cancer remains incurable and only 3% of cases can
achieve complete remission for >5 years (3). The malignant biological behavior of
breast cancer is regulated by multiple genes. The occurrence and
increased tumorigenesis of breast cancer are associated with the
loss of tumor suppressor genes (4).
Previous studies have confirmed that the
transfection of tumor suppressor genes can significantly reduce the
growth and metastatic ability of breast cancer cells (5). Breast cancer suppressor candidate-1
(BCSC-1; also termed von Willebrand factor A domain containing 5A
and LOH11CR2A) is a newly identified tumor suppressor gene
localized at chromosome 11q23–q24. It encodes a protein with 786
amino acids and a molecular mass of 86 kDa. BCSC-1 contains two
conserved domains: An inter-α-trypsin inhibitor domain at the
N-terminus and a von Willebrand factor type A domain at the
C-terminus. Northern blot analysis of BCSC-1 gene expression
indicated a lack of expression in 33/41 (80%) tumor cell lines, and
ectopic expression of BCSC-1 led to the suppression of
tumorigenicity in vitro and in vivo (6). Other studies have also demonstrated
that BCSC-1 is lost or expressed at low levels in different
carcinomas, including lung adenocarcinoma, nasopharyngeal
carcinoma, esophageal squamous carcinoma and melanoma (7–9). Our
previous study confirmed that BCSC-1 can suppress the malignant
biological behavior of various types of tumor cells including human
CNE-2L2 nasopharyngeal carcinoma cells and human NCI-H446 small
cell lung cancer cells. The metastatic ability of these tumor cells
was significantly reduced following transfection with the BCSC-1
gene (8). These data suggest that
BCSC-1 may exert tumor suppressor activity, and that low expression
of BCSC-1 may lead to increased tumorigenesis. In the present
study, the expression level of BCSC-1 was detected in breast cancer
using tissue microarray with immunohistochemical methods. Tissue
microarrays, also termed tissue chips, are an important branch of
biochip technology. Many specimens of different individuals are
arranged in a regular array of the same carrier in a tissue
microarray. Tissue microarrays are used to study the expression of
the same gene or protein molecule in different cells or tissues.
Additionally, a stably transfected MDA-MB-231 cell line with high
BCSC-1 expression was established, and the effects of BCSC-1 on the
biological behavior of breast cancer were investigated.
Previous studies have suggested that matrix
metalloproteinases (MMPs) exert specific proteolytic activity on
components of the ECM, and the overexpression of MMPs has been
linked to the invasiveness of breast cancer cells, a process that
may be regulated by the nuclear factor-κB (NF-κB) pathway (10,11).
Osteopontin (OPN) is a multifunctional, secretory, phosphorylated
glycoprotein that can promote cell adhesion and migration. In
recent years, OPN has been identified as a critical protein
participating in the malignant transformation of tumor cells
(12,13). Thus, changes in the expression of
MMPs (particularly, MMP-2, MMP-7 and MMP-9), OPN and the NF-κB
signaling pathway were evaluated in the current study.
Lentivirus-mediated RNA interference (shRNA) methods were used to
knockdown BCSC-1 gene expression in MCF-7 breast cancer cells.
Stably silenced BCSC-1 in MCF-7 cells resulted in a higher capacity
for metastasis. These results suggest that BCSC-1 may be a
potential anticancer target in breast cancer.
Materials and methods
Tissue microarrays and clinical specimens
for immunohistochemistry
A tissue microarray containing 69 cases of breast
cancer tissue and 3 cases of normal breast tissue was purchased
from Alenabio (Xian, China; cat. no. BC08013a). Additionally, 40
pathologically and clinically confirmed patients with breast cancer
were recruited from the Breast Surgery Centre of Weifang People’s
Hospital (Weifang, China) from February to May 2016 in order to
obtain cancer and adjacent non-tumor tissue samples for further
investigation of BCSC-1. All patients were required to provide
written informed consent and the experiments were approved by the
Institutional Ethics Committee of Weifang Medical University. Thus,
a total of 109 cases of breast cancer tissues and 43 cases of
adjacent non-tumor tissues were included in the present study. All
of the patients were women, and ranged from 28–76 years old. All
patients were pathologically and clinically confirmed with breast
cancer, patients with incomplete information were excluded. The
fresh tissue samples were stored at −80°C until use for
immunohistochemistry. Prior to immunohistochemistry, samples were
fixed in 10% formalin solution and embedded in paraffin. The
paraffin sections were cut at 5 µm thickness and were
deparaffinized in xylene for 20 min, rehydrated in 100, 95, 80 and
70% ethanol for 10 min each step at room temperature and incubated
in 3% H2O2 for 10 min at room temperature to
block endogenous peroxidase. All slides were placed into 10%
citrate buffer and boiled at 90°C for 30 min for antigen retrieval,
then were incubated in blocking solution containing 10% goat serum
(Bioss, Beijing, China) at 37°C for 15 min. Then incubated with a
mouse anti-human BCSC-1 antibody (cat. no. ab64977; 1:1,000; Abcam,
Cambridge, MA, USA) at 4°C overnight. The corresponding
biotin-labeled goat anti-mouse IgG (ready-to-use reagents) were
added at 37°C for 30 min and reacted with 100 µl horseradish
peroxidase (HRP)-labeled avidin (ready-to-use reagents) at 37°C for
30 min, and 100 µl diaminobenzidine chromogenic substrate
(ready-to-use reagents) were added at 37°C for 10 min in a dark
box, according to the manufacturer’s instructions of
Streptavidin-Peroxidase Immunohistochemical staining kit (cat. no.
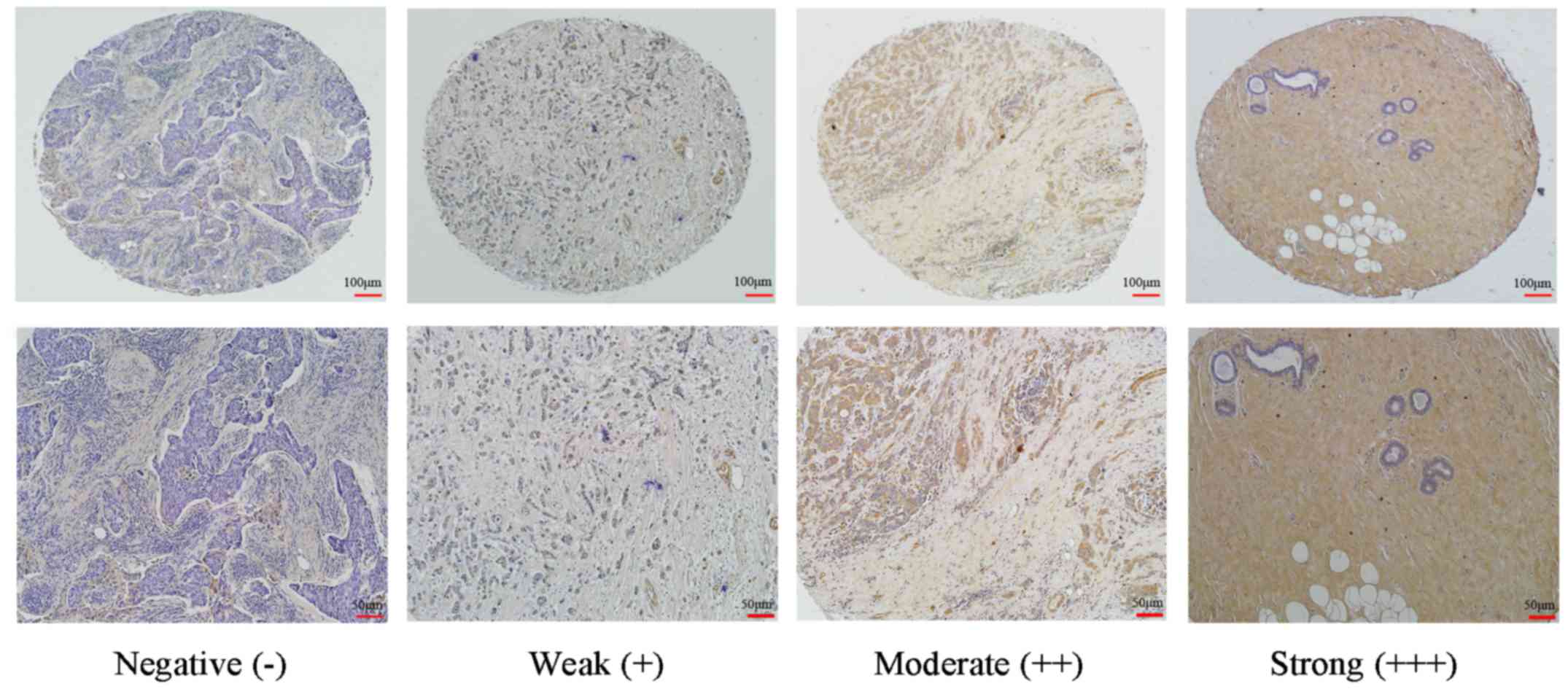
SP-0022; Bioss). Cytoplasmic and membranous staining intensity were
categorized as follows: Absent, 0; weak, 1; moderate, 2; and
strong, 3. The percentage of stained cells were categorized as
follows: No staining, 0; 1–10%, 1; 11–50%, 2; 51–80%, 3; and
81–100%, 4. The final score for each tissue was calculated by
multiplying the staining and the percentage score. The final scores
were categorized as: <2, negative (−); 3–4, weak (+); 6–8,
moderate (++); and 9–12, strong (+++).
Cells and cell culture
Human MDA-MB-231 and MCF-7 breast cancer cell lines,
and the 293T cell line were purchased from the American Type
Culture Collection (Manassas, VA, USA). All cell lines were
routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with
10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and
incubated at 37°C and 5% CO2.
Generation of an MDA-MB-231 cell line
stably overexpressing BCSC-1
MDA-MB-231 cells were routinely cultured in DMEM;
1×105 cells/well were seeded into a 6-well plate for
transfection with BCSC-1 plasmid. Transfection was conducted when
~80% cell confluence was reached. The full length of the human
BCSC-1 gene (NM_014622) was amplified from plasmid pcDNA4/myc-His
A-BCSC-1 (8) and cloned into the
eukaryotic expression vector pcDNA3.1, and the plasmid containing
BCSC-1 gene was stored routinely in our laboratory. The following
primer pairs were used in this study: BCSC-1, forward,
5′-GTGGAATTCTATGGTGCACTTCTGTGGCCTAC-3′ and reverse,
5′-ATTCTCGAGCGAAAGGCAAAGATAGCAGGA-3′. pcDNA3.1/BCSC-1 (4 µg)
and the empty pcDNA3.1 vector (negative control) were transfected
into MDA-MB-231 cells with Lipofectamine™ 2000. The cells were
cultured in DMEM containing 600 µg/ml G418 (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) for selection, and a stable BCSC-1
expression cell line was obtained 4 weeks later. Quantitative
polymerase chain reaction (qPCR), western blot analysis and
immunocytochemistry methods were performed in order to detect
changes in BCSC-1 expression in the MDA-MB-231 cells following
transfection.
Immunocytochemistry
Cells were seeded into 6-well plates (150,000/well)
at 37°C for 24 h and fixed in 4% polyformaldehyde at room
temperature for 15 min, then incubated with 0.5% Triton X-100/PBS
solution at room temperature for 20 min and followed with 3%
H2O2 at room temperature for 15 min, then
incubated in blocking solution containing 10% goat serum at 37°C
for 15 min. Cells were incubated with mouse anti-human BCSC-1
antibody (1:1,000; cat. no. ab64977; Abcam) at 4°C overnight, then
incubated with 100 µl biotin-labeled goat anti-mouse IgG
(ready-to-use reagents) and reacted with 100 µl HRP-labeled
avidin (ready-to-use reagents) and stained with 100 µl
diaminobenzidine (ready-to-use reagents), according to the
manufacturer’s instructions of Streptavidin-Peroxidase
Immunohistochemical staining kit (cat. no. SP-0022; Bioss). The
slides stained with hematoxylin at room temperature for 30 sec,
then dehydrated in 70, 80, 95 and 100% ethanol, and xylene at room
temperature for 5 min each step. The results were observed and
imaged by using an electron microscope (Olympus BX43; Olympus
Corporation, Tokyo, Japan).
RNA extraction and reverse transcription
(RT)-qPCR
Total cellular RNA from MDA-MB-231 cells was
extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer’s protocol.
Subsequently, 0.5 µg of total RNA was reverse transcribed
into cDNA using an RT reaction kit (cat. no. DRR036s; Takara
Biotechnology Co., Ltd., Dalian, China) with incubation at 37°C for
15 min, 85°C for 5 sec and 4°C for 10 min. qPCR was performed using
SYBR™ PremixEX Taq (cat. no. DRR820s; Takara Biotechnology Co.,
Ltd.) in a LightCycler 480 instrument (Roche Diagnostics, Basel,
Switzerland). The reaction conditions were as follows:
Predegeneration at 95°C for 1 min, then 35 cycles of denaturation
at 94°C for 30 sec, renaturation at 55°C for 30 sec and extension
at 72°C for 45 sec. The primer sequences are listed in Table I. The relative mRNA expression
levels were determined using the 2−ΔΔCq formula. Cq was
the cycle quantitation; ΔCq = Cq (target gene) - Cq (β-actin)
(14).
 | Table IPrimer sequences used in the present
study. |
Table I
Primer sequences used in the present
study.
| Gene | Accession | Forward | Reverse |
|---|
| BCSC-1 | NM_014622 |
5′-TGCTTCTGCCCCATTGAAGA-3′ |
5′-CTGTGCTGGTCCTTGTGAC-3′ |
| β-actin | NM_001101.4 |
5′-CCTAGAAGCATTTGCGGTGG-3′ |
5′-GAGCTACGAGCTGCCTGACG-3′ |
| MMP-2 | NM_001302510.1 |
5′-GATACCCCTTTGACGGTAAGGA-3′ |
5′-CCTTCTCCCAAGGTCCATAGC-3′ |
| MMP-7 | NM_002423.4 |
5′-AGATGTGGAGTGCCAGATGT-3′ |
5′-TAGACTGCTACCATCCGTCC-3′ |
| MMP-9 | NM_004994.2 |
5′-TCTATGGTCCTCGCCCTGAA-3 |
5′-CATCGTCCACCGGACTCAAA-3′ |
| OPN | NM_001251830.1 |
5′-CTCCATTGACTCGAACGACTC-3′ |
5′-CAGGTCTGCGAAACTTCTTAGAT-3′ |
Western blot analysis
Cells (106 cells/ml) were harvested by
scraping from the wells and washed twice with PBS. Proteins were
extracted using a protein extraction solution (cat. no. P0013;
Beyotime Institute of Biotechnology, Haimen, China) and the
concentrations were measured using a bicinchoninic acid assay
protein assay kit (cat. no. P0010; Beyotime Institute of
Biotechnology), then 40 µg of protein per lane was separated
by 10% SDS-PAGE and electrotransferred onto nitrocellulose
membranes (cat. no. FFN05; Beyotime Institute of Biotechnology).
The membranes were blocked with 5% skim milk at 37°C for 1 h and
incubated with primary antibodies against BCSC-1 (cat. no.
sc-137568; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) and mouse anti-human β-actin (cat. no. AA128; 1:500; Beyotime
Institute of Biotechnology) at 4°C overnight, then incubated with
the corresponding secondary antibody conjugated to horseradish
peroxidase (cat. nos. A0181 and A0216; 1:1,000; Beyotime Institute
of Biotechnology) for 2 h. The detection of specific proteins was
performed with an enhanced chemiluminescence kit (Sangon Biotech
Co., Ltd., Shanghai, China), according to the recommended
procedure. The visualized bands were imaged and analyzed using
ImageJ software (version 1.47; National Institutes of Health,
Bethesda, MD, USA), which measured the density of each band using
β-actin as the loading control.
Cell invasion and migration assays in
vitro
For invasion and migration assays, MDA-MB-231 cells
were serum-starved overnight. For the invasion assay,
5×105 cells were seeded into Matrigel (BD Biosciences,
San Jose, CA, USA) pre-coated chambers with 8.0-µm pores
(Corning, Inc., Corning, NY, USA) and cultured for 48 h. For the
migration assay, 1×105 cells were seeded onto uncoated
membranes and cultured for 12 h. Cells remaining on the upper
filter surfaces were removed with sterile swabs, and the invaded or
migrated cells were fixed in 70% precooled methanol for 1 h, then
stained with 1% crystal violet at 37°C for 10 min. The numbers of
invaded or migrated cells were counted in five randomly selected
fields under an inverted fluorescence microscope (Nikon
Corporation, Tokyo, Japan).
Scratch wound-healing assay
MDA-MB-231 cells were cultured in DMEM with 10% FBS.
Cells were seeded into 6-well plates at a density of
1×105 cells/ml to form a confluent monolayer. The
monolayer was scratched with a sterile 200 µl pipette tip
across the center of the well at the indicated time points (0 and
24 h). The extent of cell migration was imaged and measured using
an NIS electron microscope (Nikon Corporation).
Changes in MMPs, OPN and NF-κB in
MDA-MB-231 cells affected by BCSC-1 overexpression
MDA-MB-231 cells were cultured in DMEM and
harvested, following which changes in MMPs and OPN in MDA-MB-231
cells were detected by RT-qPCR and western blot analysis. The
primer sequences used for RT-qPCR are described in Table I, the antibodies used for western
blot analysis were as follows: MMP-2 (cat. no. ab92536; 1:1,000);
MMP-7 (cat. no. ab205525; 1:1,000) (both from Abcam); MMP-9 (cat.
no. 13667; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA); OPN (cat. no. ab69498; 5 µg/ml; Abcam); NF-κB/p65
(cat. no. 8242; 1:1,000); phospho-NF-κB p65 (Ser536; cat. no. 3033;
1:1,000) (both from Cell Signaling Technology, Inc.); mouse
anti-human β-actin (cat. no. AA128; 1:500; Beyotime Institute of
Biotechnology); the corresponding secondary antibody conjugated to
horseradish peroxidase (cat. nos. A0208 and A0216; 1:1,000;
Beyotime Institute of Biotechnology). The detailed operating
procedures of RT-qPCR and western blot were performed according to
the aforementioned protocols.
Lentivirus-mediated short hairpin RNA
(shRNA) knockdown of BCSC-1 gene expression and the effect on MCF-7
cell metastasis capacity
Lentiviral production and transductions were
performed. The plasmids psPAX2 and pMD2.G were purchased from
Addgene, Inc. (Cambridge, MA, USA). Briefly, an interference
sequences specific to the human BCSC gene (LV-BCSC-1) and a
negative control sequence (LV-NC) were designed and cloned into a
PLKO.1-SP6-PGK-GFP vector (Animal Core Facility of Nanjing Medical
University, Nanjing, China). A three-plasmid lentiviral packaging
cell system (vector plasmid + psPAX2 + pMD2.G) was used for the
co-transfection of 293T cells using Lipofectamine™ 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer’s instructions. The supernatants containing the
lentivirus were collected after 48 h and purified with a 0.4
µm filter, then the lentiviral titer was determined. MCF-7
cells were infected with lentiviral particles at a multiplicity of
infection of 5, followed by puromycin selection for 10 days. The
efficacy of transfection was evaluated by calculating the number of
fluorescent-positive cells with flow cytometry (BD FACSVerse™; BD
Bioscience) and FlowJo 7.6.1 software (FlowJo LLC, Ashland, OR,
USA). The changes in BCSC-1 expression were detected by qPCR and
western blotting methods. Changes to the invasive ability of MCF-7
cells were measured by an invasion assay, according to the
aforementioned procedures. The target sequences are described in
Table II.
 | Table IITarget sequences used in LV-mediated
short hairpin RNA interference. |
Table II
Target sequences used in LV-mediated
short hairpin RNA interference.
| Target name | Target
sequences | Target site |
|---|
| LV-BCSC-1 |
5′-GAGTTTACCTATAGGCTGTTA-3′ | 1,048-1,068 |
| LV-NC |
5′-CCTAAGGTTAAGTCGCCCTC-3′ | n/a |
Statistical analysis
Data are presented as the mean ± standard deviation
and were analyzed using SPSS 10.0 software (SPSS, Inc., Chicago,
IL, USA). A χ2 test was used to analyze the
immunohistochemical expression of BCSC-1. Differences between two
groups were analyzed using the Student’s t-test. Differences
between more than two groups were analyzed using one-way analysis
of variance. The least significant difference method was used to
conduct multiple comparisons in cases of homogeneity of variance,
whereas the Games-Howell method was applied in cases of
heterogeneity of variance. P<0.05 was considered to indicate a
statistically significant difference.
Results
BCSC-1 is expressed at low levels in
breast cancer tissues
To investigate the exact effect of BCSC-1 in breast
cancer, the expression of BCSC-1 in breast cancer tissue was
investigated using microarrays and clinical tissue specimens using
immunohistochemistry. Of the 109 breast cancer samples, 45 (41.28%)
were negative, 30 (27.53%) were weakly positive, 23 (21.1%) were
moderately positive and 11 (10.09%) were strongly positive. In
normal breast tissue samples or adjacent non-tumor tissues, 6
(13.95%) were negative, 7 (16.28%) were weakly positive, 14
(32.56%) were moderately positive and 16 (37.21%) were strongly
positive. BCSC-1 expression in breast cancer tissue was
significantly lower than in normal breast or adjacent non-tumor
tissues. These results suggest that BCSC-1 is expressed at low
levels in breast cancer and may be involved in the pathogenesis of
breast cancer (Fig. 1 and Table III).
 | Table IIIExpression of BCSC-1 in breast cancer
as evaluated by immunohistochemistry. |
Table III
Expression of BCSC-1 in breast cancer
as evaluated by immunohistochemistry.
| Samples | n | BCSC-1
immunostaining [n (%)]
| χ2 | P-value |
|---|
| Negative (−) | Weak (+) | Moderate (++) | Strong (+++) |
|---|
| Breast cancer
tissue | 109 | 45 (41.28) | 30 (27.53) | 23 (21.1) | 11 (10.09) | 22.89 | <0.001 |
| Adjacent non-tumor
tissue | 43 | 6 (13.95) | 7 (16.28) | 14 (32.56) | 16 (37.21) | | |
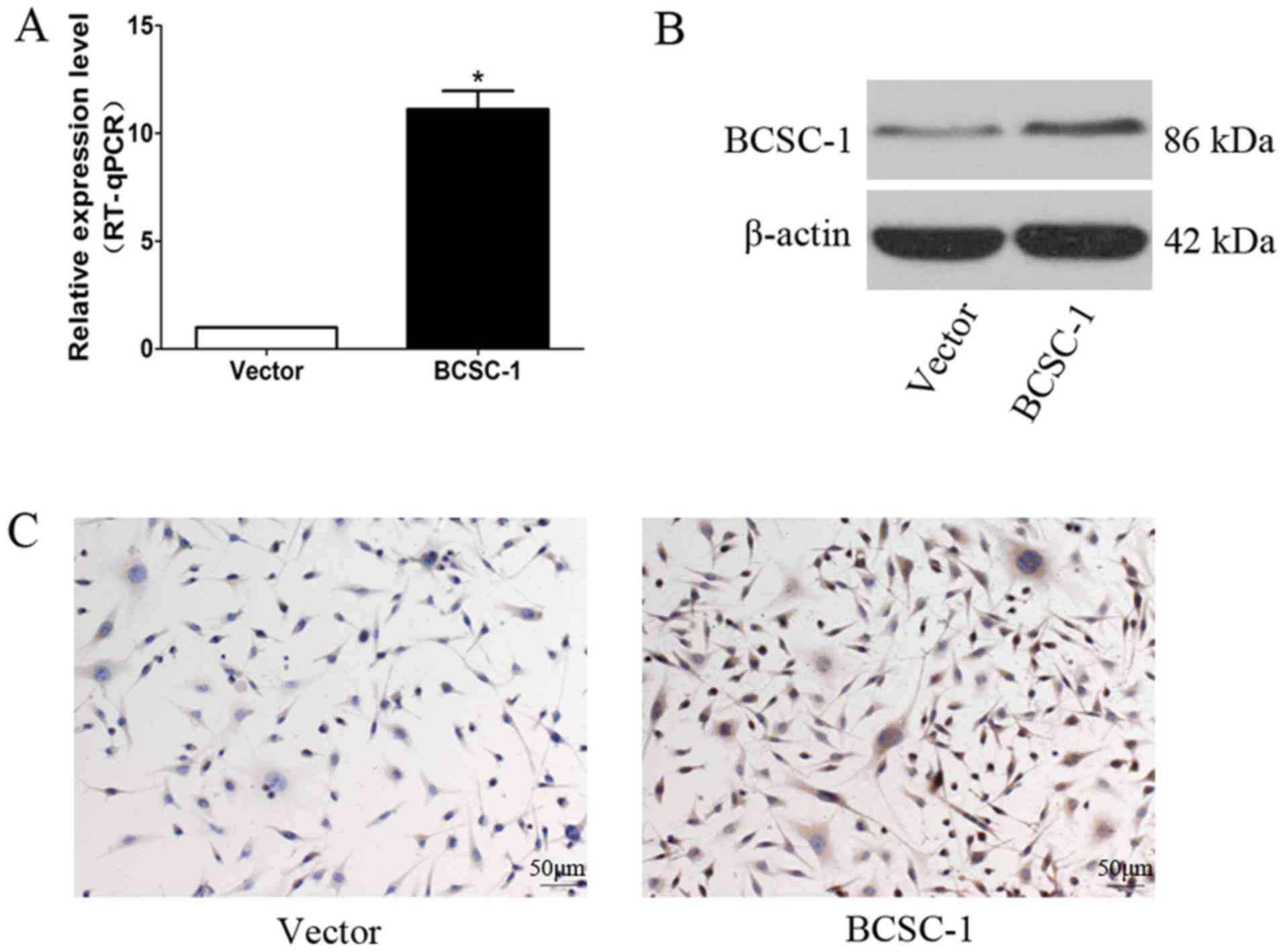
Establishment of an MDA-MB-231 stable
cell line overexpressing BCSC-1
An MDA-MB-231 cell line with stable high BCSC-1
expression, and an empty plasmid cell line, was successfully
established by transfection with the BCSC-1 plasmid and subsequent
screening with G418. The results of RT-qPCR and western blotting
demonstrated that the mRNA and protein expression levels of BCSC-1
in MDA-MB-231 cells were significantly increased in the BCSC-1
group compared with in the control group (Fig. 2A and B), and these results were
further confirmed by immunocytochemistry methods (Fig. 2C).
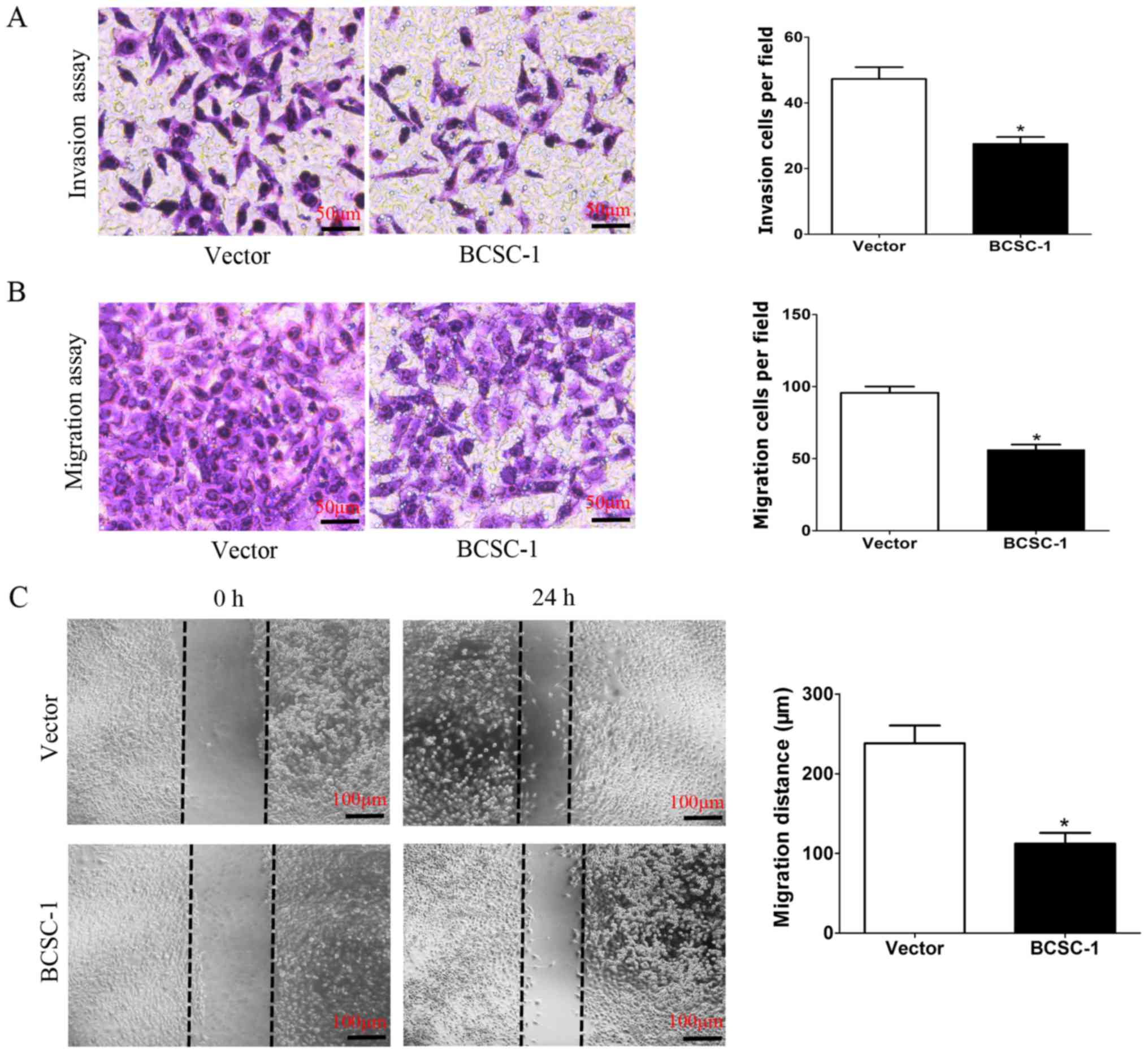
Overexpression of BCSC-1 can inhibit the
metastasis capacity of MDA-MB-231 cells in vitro
To clarify the association between BCSC-1 and the
metastatic capability of MDA-MB-231 cells, an MDA-MB-231 cell line
with stable high BCSC-1 expression and an MDA-MB-231 cell line with
empty plasmid were established as the control. It was subsequently
identified that overexpression of BCSC-1 inhibited the metastasis
capacity of MDA-MB-231 cells in vitro. In the invasion
assay, 27.5±2.08 cells/field in the BCSC-1 group invaded to the
lower chamber, while 47.25±3.59 cells/field invaded in the control
group (Fig. 3A). In the migration
assay, 56.25±3.5 cells/field migrated in the BCSC-1 group and
95.75±4.35 cells/field migrated in the control group (Fig. 3B). In the migration and invasion
assays, the number of BCSC-1-overexpressing cells that passed
through the membrane was significantly lower than the cells of the
control group. In the wound-healing assay, the average migration
distance recorded in the BCSC-1 group was 112.2±13.57 µm,
which was significantly shorter, compared with that of the control
group (238.2±22.16 µm) (Fig.
3C).
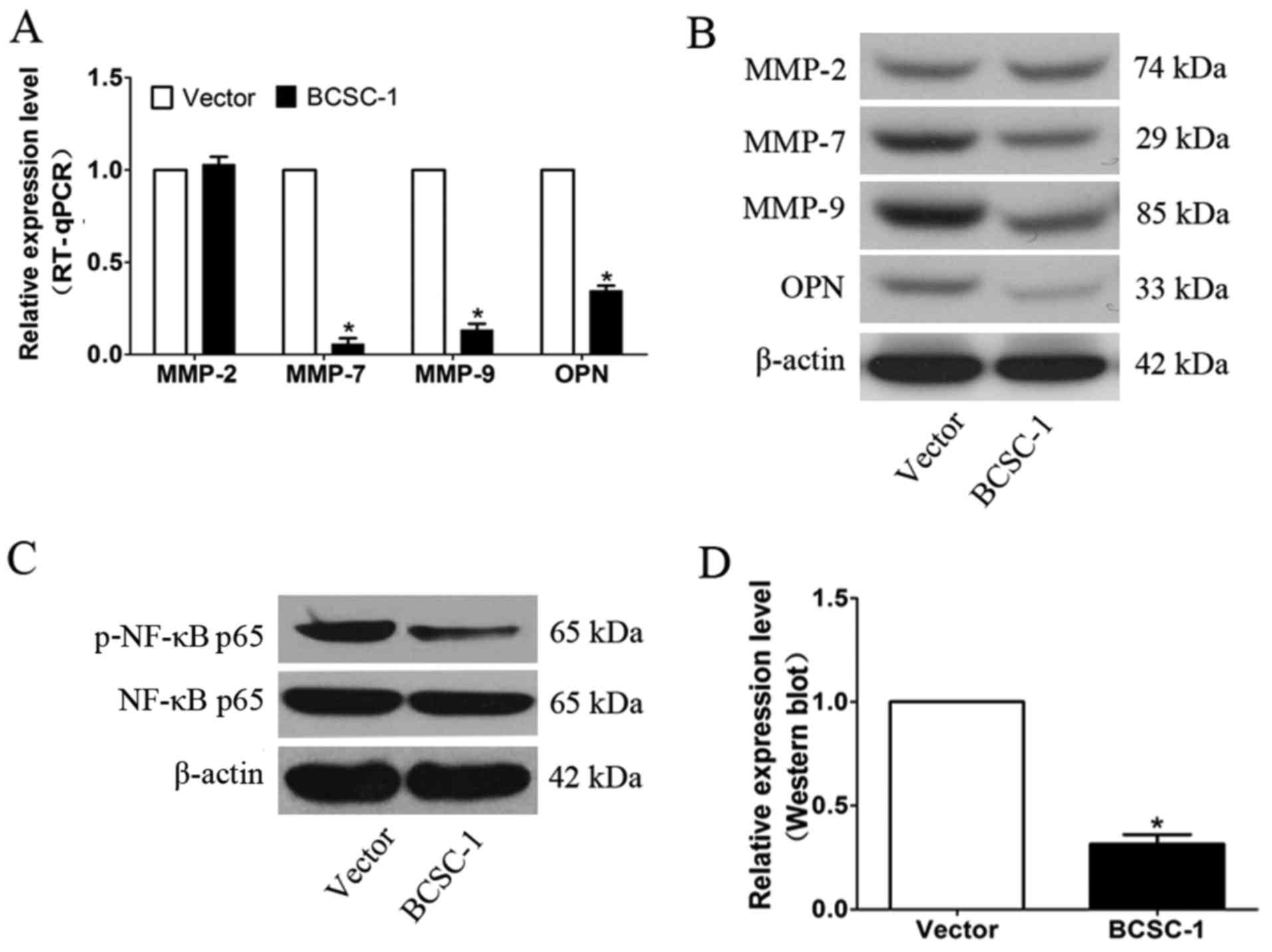
Expression of MMPs and OPN, and NF-κB
transcriptional activity in MDA-MB-231 cells is affected by BCSC-1
gene overexpression
The expression levels of MMPs (MMP-2, MMP-7 and
MMP-9) and OPN were detected by RT-qPCR and western blot. The
results demonstrated that the levels of MMP-7, MMP-9 and OPN were
reduced in cells over-expressing BCSC-1 compared with the vector
control cells (Fig. 4A and B).
NF-κB/p65 is a critical factor involved in tumor metastasis. The
effect of BCSC-1 gene overexpression on NF-κB/p65 activity was
determined via western blot analysis. The results revealed that
BCSC-1 expression decreased NF-κB/p65 activity, as compared with
the control group, indicating that BCSC-1 may mediate breast cancer
cell metastasis through the NF-κB pathway (Fig. 4C and D).
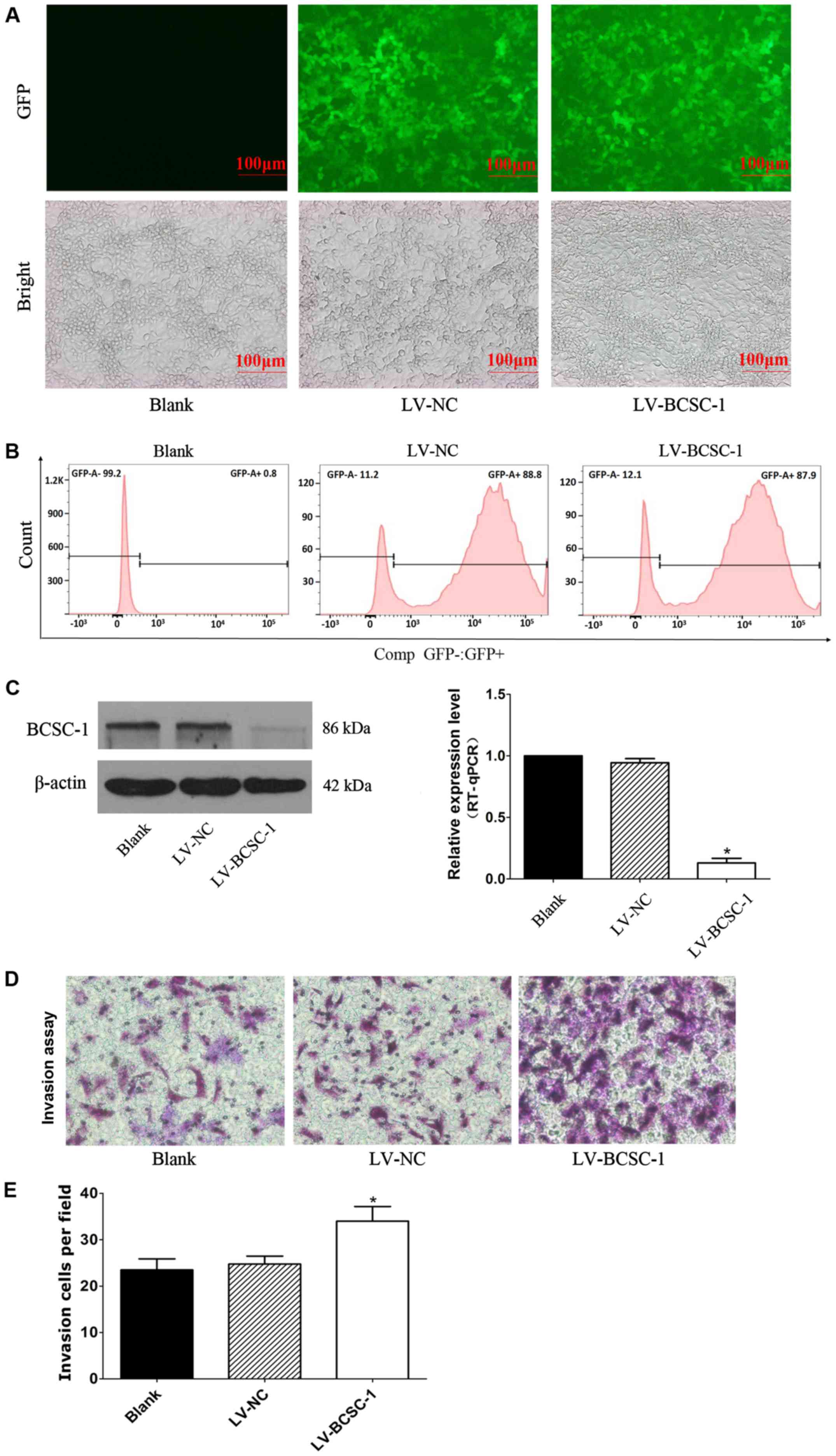
Efficient knockdown of BCSC-1 expression
by lentiviral-mediated RNA interference (RNAi) and its effect on
the invasion ability of MCF-7 cells
To investigate the function of BCSC-1 in breast
cancer cells, lentivirus-mediated RNAi technology was used to
knockdown BCSC-1 gene expression in MCF-7 cells that exhibited high
BCSC-1 expression. At 4 days after lentiviral infection, the cells
emitted bright fluorescence visible under a fluorescent microscope
(Fig. 5A). The number of positive
cells was determined using flow cytometry, and the results from
FlowJo software analysis indicated that the infection efficiency
was >80% (Fig. 5B). Results
from RT-qPCR and western blotting indicated that BCSC-1 expression
was downregulated in the LV-BCSC-1 group compared with in the LV-NC
and blank group (Fig. 5C and D).
Results from the invasion assay indicated that 23.5±2.39 and
24.75±1.71 cells/field invaded in the blank and LV-NC groups,
respectively, which was significantly higher than the LV-BCSC-1
group (34±3.16 cells/field; P<0.05). This indicated that the
downregulation of BCSC-1 increased MCF-7 cell invasion capacity
in vitro (Fig. 5E).
Discussion
Breast cancer is one of the most common type of
malignancy diagnosed among women worldwide (2). Metastasis is the main cause of
mortality in patients with breast cancer. The function and
mechanism of tumor suppressor genes in the invasion and metastasis
processes of tumor cells have increasingly received the attention
of researchers in recent years (15).
BCSC-1 is a novel tumor suppressor gene, recently
identified by Martin et al (6). The BCSC-1 gene is located at
chromosome 11q23 and encodes a predicted 786 amino acid protein
containing two conserved domains: A vault protein inter-α-trypsin
inhibitor domain at the N-terminus and a von Willebrand factor type
A domain at the C-terminus (6).
Previous study has demonstrated that BCSC-1 is either not expressed
or is expressed at low levels in a variety of tumors, and that the
ectopic expression of BCSC-1 can reduce the tumorigenicity of
carcinoma cells. BCSC-1 can inhibit melanoma tumor formation in
vivo and tumor cell proliferation in vitro through
downregulating melanogenesis associated transcription factor
expression, resulting in a switch of melanoma from a proliferative
phenotype to a migratory phenotype (9). Our prior study indicated that BCSC-1
has a key role in the tumorigenesis of nasopharyngeal carcinoma by
increasing E-cadherin and α-catenin expression through the Wnt
signaling pathway (8). We also
confirmed that BCSC-1 was expressed at low levels in human
esophageal squamous cell carcinoma, and that the downregulation of
BCSC-1 was associated with the grade of tumor differentiation
(7). However, the association
between BCSC-1 and breast cancer is remains poorly understood.
In the present study, the expression levels of
BCSC-1 in breast cancer tissues were determined via
immunohistochemistry analysis of a tissue microarray; the results
indicated that BCSC-1 was expressed at low levels in breast cancer,
compared with in normal breast tissues (P<0.05). Subsequently,
cancer and adjacent non-tumor tissue samples were obtained from 40
patients with breast cancer for further confirmation, and the
results were concordant. This indicated that BCSC may be involved
in the pathogenesis of breast cancer. Following this, the human
MDA-MB-231 breast cancer line, with low BCSC-1 expression, was
selected for use in the current study. It was observed that the
ectopic expression of BCSC-1 inhibited the metastatic ability of
cells in vitro compared with control cells.
MMPs are zinc-dependent endopeptidases that have an
important role in the invasion and metastasis of breast cancer by
degrading various extracellular matrix proteins; this may be
regulated by the NF-κB signaling pathway (10,11).
OPN is a multifunctional secretory phosphorylated glycoprotein that
can promote cell adhesion and migration. OPN is considered to be a
critical protein involved in the malignant transformation of tumor
cells. OPN has a close association with MMPs, and stimulates MMP
expression via the NF-κB signaling pathway (16). Thus, the expression of MMPs, OPN
and the NF-κB p65 were detected in the present study. The results
demonstrated that overexpression of BCSC-1 inhibited the expression
of MMP-7, MMP-9 and OPN, and that NF-κB/p65 activity was decreased
in MDA-MB-231 cells, indicating that the anti-metastatic effects of
BCSC-1 in human breast cancer may be mediated through inhibition of
the NF-κB signaling pathway.
As a transcription factor, NF-κB is present in the
cytoplasm of most cells and translocates to the nucleus when
activated. Activation of NF-κB can be induced by stress, cigarette
smoke, viruses, bacteria, inflammatory cytokines and tumors, among
other factors (17). Previous
studies have demonstrated that NF-κB can affect a wide range of
biological behaviors, including inflammation, cell cycle and
apoptosis (18). NF-κB also has
important roles in the occurrence and development of malignancy.
Aberrant constitutive activation of NF-κB has been observed in
large variety of cancer cells, including liver cancer, lung cancer
and stomach cancer, and the inhibition of NF-κB activity can reduce
chemoresistance (19–22). The angiogenesis of breast cancer
depends on inflammatory factors, such as chemokines (interleukin-8
and C-X-C motif chemokine ligand 8) or growth factors (vascular
endothelial growth factor) produced by neutrophils or macrophages
following activation of the NF-κB signaling pathway. In a previous
report, high levels of NF-κB expression were more common in
patients with breast cancer, and were associated with a larger
tumor size or higher grade, which are poor prognostic factors in
breast cancer. Various genes that are involved in breast cancer
invasion have been found to be regulated by NF-κB, including cell
adhesion molecules, inflammatory cytokines and MMPs (23–25).
Abnormal activation of NF-κB can promote breast cancer cell
proliferation, invasion and angiogenesis, and reduce cell
sensitivity to apoptotic stimuli through regulating downstream
target genes including MMPs or intercellular adhesion molecule 1,
therefore facilitating the survival of tumor cells (26). Studies have confirmed that small
interfering RNA against NF-κB can simultaneously inhibit the growth
and suppress the distant metastasis of MDA-MB-231 and MCF-7 cells
(27).
There have been multiple reports of increased levels
of MMPs in breast cancer, and high expression of MMP-7 and MMP-9
has been reported to be associated with an unfavorable prognosis
for patients with breast cancer (28–30).
A meta-analysis also demonstrated that polymorphisms in the
promoter regions of MMP-7 and MMP-9 may be associated with
metastasis in breast cancer (31).
Studies have demonstrated that MMP-7 is involved in the invasion
process of MDA-MB-231 cells, and the endogenous long non-coding RNA
urothelial cancer associated 1 (non-protein coding) can
significantly reduce the number of invading cells via inhibition of
MMP-7 (32). Other studies have
demonstrated that common MMP-7 genetic polymorphisms are
significant determinants of survival in Chinese patients with
breast cancer (33).
Overexpression of MMP-9 induced by the inflammatory cytokine
interleukin-1β can promote MCF-7 cell metastasis, and the
inhibition of MMP-9 can inhibit breast cancer cell metastasis
(34–37). Further studies indicated that MMP-9
is regulated by the NF-κB pathway, and that inhibition of MMP-9 via
the NF-κB signaling pathway can suppress the MCF-7 cell invasion
ability (38–41). In the present study, the expression
of MMPs in MDA-MB-231 cells was affected by BCSC-1 overexpression.
The results indicated that MMP-7 and MMP-9 were reduced
significantly by BCSC-1 overexpression (P<0.05). These data
suggest that BCSC-1-mediated inhibition of breast cancer cell
metastasis may be attributed to the down-regulation of MMP-7 and
MMP-9.
OPN is a type of phosphorylated glycoprotein present
in the extracellular matrix. It is synthesized and secreted by
various tissue cells, including tumor cells. OPN was first
identified by Senger et al (42) in malignant transformed epithelial
cells in 1979. An increasing number of studies have reported that
OPN is overexpressed in a variety of malignant tumor types, and
that it is closely associated with tumor cell metastasis and growth
(43,44). Overexpression of OPN can promote
tumor cell metastasis by stimulating proliferation, inducing the
formation of new blood vessels and promoting tumor cell metastasis
through binding CD44 or integrins (45–47).
Previous study has also confirmed that OPN is associated with
breast cancer. OPN is highly expressed in breast cancer tissues
compared with in normal breast tissues, and the overall survival
time of patients with breast cancer and high OPN expression is
significantly lower than for those with low OPN expression
(48).
A meta-analysis also indicated that OPN
overexpression is a positive candidate prognostic biomarker for
patients with breast cancer (49).
Certain experiments have demonstrated a high level of OPN secretion
in MDA-MB-435 cells, and a significant decrease in the metastasis
capacity of tumor cells following inhibition of OPN expression.
This indicates that OPN has an important role in the metastasis of
breast cancer cells (50).
In the present study, lentivirus-mediated RNAi
methods were used to knockdown BCSC-1 in MCF-7 cells that had high
BCSC-1 expression. The results indicated that stable silencing of
BCSC-1 in MCF-7 cells resulted in a higher capacity for metastasis.
These results further confirmed a tumor suppressor function for
BCSC-1 in breast cancer.
In conclusion, the results of the current study
suggested that BCSC-1 is expressed at low levels in breast cancer
tissues, and it can suppress human breast cancer cell metastasis by
changing the expression of MMP7, MMP9, OPN, and the activity of the
NF-κB pathway, indicating that BCSC-1 may serve as a biomarker for
the treatment of breast cancer in the future. The results of the
present study provided novel insights into the role of BCSC-1 in
breast cancer and improved the understanding of the mechanisms
underlying the progression of breast cancer.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81373185 and
81572578), the Natural Science Foundation of Shandong, China (grant
nos. ZR2009CM019, ZR2014HL058 and ZR2015HM028) and Shandong
Province Health Department (grant nos. 2013WS0287 and
2014WS0462).
[2] Availability
of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
[3] Authors’
contributions
JJ and CZ were responsible for experimental design
and manuscript writing. DD and LC performed the histological
examination of the breast cancer tissue and were major contributors
in writing the manuscript. YG and LW performed the western blot
analysis and cell experiments. All authors read and approved the
final manuscript.
[4] Ethics
approval and consent to participate
The experiments were approved by the Institutional
Ethics Committee of Weifang Medical University (Weifang, China).
All patients were required to provide written informed consent.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare no that they have no competing
interests.
References
|
1
|
Hong W and Dong E: The past, present and
future of breast cancer research in China. Cancer Lett. 351:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Organization WH: Breast cancer: Prevention
and control. World Health Stat Annu. 41:697–700. 2012.
|
|
3
|
Müller V, Fuxius S, Steffens CC,
Lerchenmüller C, Luhn B, Vehling-Kaiser U, Hurst U, Hahn LJ,
Soeling U, Wohlfarth T, et al: Quality of life under capecitabine
(Xeloda®) in patients with metastatic breast cancer:
Data from a german non-interventional surveillance study. Oncol Res
Treat. 37:748–755. 2014. View Article : Google Scholar
|
|
4
|
Lu J and Jin ML: Short-hairpin
RNA-mediated MTA2 silencing inhibits human breast cancer cell line
MDA-MB231 proliferation and metastasis. Asian Pac J Cancer Prev.
15:5577–5582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shanker M, Jin J, Branch CD, Miyamoto S,
Grimm EA, Roth JA and Ramesh R: Tumor suppressor gene-based
nanotherapy: From test tube to the clinic. J Drug Deliv.
2011:4658452011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin ES, Cesari R, Pentimalli F, Yoder
K, Fishel R, Himelstein AL, Martin SE, Godwin AK, Negrini M and
Croce CM: The BCSC-1 locus at chromosome 11q23–q24 is a candidate
tumor suppressor gene. Proc Natl Acad Sci USA. 100:11517–11522.
2003. View Article : Google Scholar
|
|
7
|
Zhao CL, Yu WJ, Gao ZQ, Li WT, Gao W, Yang
WW, Feng WG and Ju JY: Association of BCSC-1 with human esophageal
squamous cell carcinoma. Neoplasma. 62:765–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou YQ, Chen SL, Ju JY, Shen L, Liu Y,
Zhen S, Lv N, He ZG and Zhu LP: Tumor suppressor function of BCSC-1
in nasopharyngeal carcinoma. Cancer Sci. 100:1817–1822. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anghel SI, Correa-Rocha R, Budinska E,
Boligan KF, Abraham S, Colombetti S, Fontao L, Mariotti A, Rimoldi
D, Ghanem GE, et al: Breast cancer suppressor candidate-1 (BCSC-1)
is a melanoma tumor suppressor that down regulates MITF. Pigment
Cell Melanoma Res. 25:482–487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mali AV, Wagh UV, Hegde MV, Chandorkar SS,
Surve SV and Patole MV: In vitro anti-metastatic activity of
enterolactone, a mammalian lignan derived from flax lignan, and
down-regulation of matrix metalloproteinases in MCF-7 and MDA MB
231 cell lines. Indian J Cancer. 49:181–187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tong W, Wang Q, Sun D and Suo J: Curcumin
suppresses colon cancer cell invasion via AMPK-induced inhibition
of NF-κB, uPA activator and MMP9. Oncol Lett. 12:4139–4146. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hao C, Cui Y, Hu MU, Zhi X, Zhang L, Li W,
Wu W, Cheng S and Jiang WG: OPN-a splicing variant expression in
non-small cell lung cancer and its effects on the bone metastatic
abilities of lung cancer cells in vitro. Anticancer Res.
37:2245–2254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishigamori R, Komiya M, Takasu S, Mutoh M,
Imai T and Takahashi M: Osteopontin deficiency suppresses
intestinal tumor development in Apc-deficient min mice. Int J Mol
Sci. 18:182017. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Xu YM, Wang HJ, Chen F, Guo WH, Wang YY,
Li HY, Tang JH, Ding Y, Shen YC, Li M, et al: HRD1 suppresses the
growth and metastasis of breast cancer cells by promoting IGF-1R
degradation. Oncotarget. 6:42854–42867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding F, Wang J, Zhu G, Zhao H, Wu G and
Chen L: Osteopontin stimulates matrix metalloproteinase expression
through the nuclear factor-κB signaling pathway in rat
temporomandibular joint and condylar chondrocytes. Am J Transl Res.
9:316–329. 2017.
|
|
17
|
Ahn KS and Aggarwal BB: Transcription
factor NF-kappaB: A sensor for smoke and stress signals. Ann NY
Acad Sci. 1056:218–233. 2005. View Article : Google Scholar
|
|
18
|
Shishodia S and Aggarwal BB: Nuclear
factor-kappaB activation: A question of life or death. J Biochem
Mol Biol. 35:28–40. 2002.
|
|
19
|
Lin A and Karin M: NF-kappaB in cancer: A
marked target. Semin Cancer Biol. 13:107–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uetsuka H, Haisa M, Kimura M, Gunduz M,
Kaneda Y, Ohkawa T, Takaoka M, Murata T, Nobuhisa T, Yamatsuji T,
et al: Inhibition of inducible NF-kappaB activity reduces
chemoresistance to 5-fluorouracil in human stomach cancer cell
line. Exp Cell Res. 289:27–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang X, Liu D, Shishodia S, Ozburn N,
Behrens C, Lee JJ, Hong WK, Aggarwal BB and Wistuba II: Nuclear
factor-kappaB (NF-kappaB) is frequently expressed in lung cancer
and preneoplastic lesions. Cancer. 107:2637–2646. 2006. View Article : Google Scholar
|
|
22
|
Hu Y, Guo R, Wei J, Zhou Y, Ji W, Liu J,
Zhi X and Zhang J: Effects of PI3K inhibitor NVP-BKM120 on
overcoming drug resistance and eliminating cancer stem cells in
human breast cancer cells. Cell Death Dis. 6:e20202015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bharti AC and Aggarwal BB: Nuclear
factor-kappa B and cancer: Its role in prevention and therapy.
Biochem Pharmacol. 64:883–888. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashikawa K, Majumdar S, Banerjee S, Bharti
AC, Shishodia S and Aggarwal BB: Piceatannol inhibits TNF-induced
NF-kappaB activation and NF-kappaB-mediated gene expression through
suppression of IkappaBalpha kinase and p65 phosphorylation. J
Immunol. 169:6490–6497. 2002. View Article : Google Scholar
|
|
25
|
Sethi G, Sung B and Aggarwal BB: Nuclear
factor-kappaB activation: From bench to bedside. Exp Biol Med
(Maywood). 233:21–31. 2008. View Article : Google Scholar
|
|
26
|
Ahn KS, Sethi G and Aggarwal BB: Nuclear
factor-kappa B: From clone to clinic. Curr Mol Med. 7:619–637.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee CH, Jeon YT, Kim SH and Song YS:
NF-kappaB as a potential molecular target for cancer therapy.
Biofactors. 29:19–35. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abbas A, Aukrust P, Russell D,
Krohg-Sørensen K, Almås T, Bundgaard D, Bjerkeli V, Sagen EL,
Michelsen AE, Dahl TB, et al: Matrix metalloproteinase 7 is
associated with symptomatic lesions and adverse events in patients
with carotid atherosclerosis. PLoS One. 9:e849352014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cuadriello EF, Fernández-Guinea Ó, Eiró N,
González LO, Junquera S and Vizoso FJ: Relationship between
morphological features and kinetic patterns of enhancement of the
dynamic breast magnetic resonance imaging and tumor expression of
metalloproteases and their inhibitors in invasive breast cancer.
Magn Reson Imaging. 34:1107–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Makhoul I, Todorova VK, Siegel ER,
Erickson SW, Dhakal I, Raj VR, Lee JY, Orloff MS, Griffin RJ,
Henry-Tillman RS, et al: Germline genetic variants in TEK, ANGPT1,
ANGPT2, MMP9, FGF2 and VEGFA are associated with pathologic
complete response to bevacizumab in breast cancer patients. PLoS
One. 12:e01685502017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu D, Guo H, Li Y, Xu X, Yang K and Bai
Y: Association between polymorphisms in the promoter regions of
matrix metalloproteinases (MMPs) and risk of cancer metastasis: A
meta-analysis. PLoS One. 7:e312512012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao C, Wu CH and Hu HZ: LncRNA UCA1
promotes epithelial-mesenchymal transition (EMT) of breast cancer
cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med
Pharmacol Sci. 20:2819–2824. 2016.PubMed/NCBI
|
|
33
|
Beeghly-Fadiel A, Shu XO, Long J, Li C,
Cai Q, Cai H, Gao YT and Zheng W: Genetic polymorphisms in the
MMP-7 gene and breast cancer survival. Int J Cancer. 124:208–214.
2009. View Article : Google Scholar :
|
|
34
|
Mon NN, Senga T and Ito S: Interleukin-1β
activates focal adhesion kinase and Src to induce matrix
metalloproteinase-9 production and invasion of MCF-7 breast cancer
cells. Oncol Lett. 13:955–960. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hwang JK, Yu HN, Noh EM, Kim JM, Hong OY,
Youn HJ, Jung SH, Kwon KB, Kim JS and Lee YR: DHA blocks
TPA-induced cell invasion by inhibiting MMP-9 expression via
suppression of the PPAR-γ/NF-κB pathway in MCF-7 cells. Oncol Lett.
13:243–249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chung TW, Choi H, Lee JM, Ha SH, Kwak CH,
Abekura F, Park JY, Chang YC, Ha KT, Cho SH, et al: Oldenlandia
diffusa suppresses metastatic potential through inhibiting matrix
metal-loproteinase-9 and intercellular adhesion molecule-1
expression via p38 and ERK1/2 MAPK pathways and induces apoptosis
in human breast cancer MCF-7 cells. J Ethnopharmacol. 195:309–317.
2017. View Article : Google Scholar
|
|
37
|
Mohammadzadeh R, Saeid Harouyan M and Ale
Taha SM: Silencing of bach1 gene by small interfering RNA-mediation
regulates invasive and expression level of miR-203, miR-145, matrix
metalloproteinase-9, and CXCR4 receptor in MDA-MB-468 breast cancer
cells. Tumour Biol. 39:10104283176959252017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim JM, Noh EM, Kim HR, Kim MS, Song HK,
Lee M, Yang SH, Lee GS, Moon HC, Kwon KB, et al: Suppression of
TPA-induced cancer cell invasion by Peucedanum japonicum Thunb.
extract through the inhibition of PKCα/NF-κB-dependent MMP-9
expression in MCF-7 cells. Int J Mol Med. 37:108–114. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matsumoto G, Namekawa J, Muta M, Nakamura
T, Bando H, Tohyama K, Toi M and Umezawa K: Targeting of nuclear
factor kappaB Pathways by dehydroxymethylepoxyquinomicin, a novel
inhibitor of breast carcinomas: Antitumor and antiangiogenic
potential in vivo. Clin Cancer Res. 11:1287–1293. 2005.PubMed/NCBI
|
|
40
|
Yu H, Guo C, Feng B, Liu J, Chen X, Wang
D, Teng L, Li Y, Yin Q, Zhang Z, et al: Triple-layered
pH-responsive micelleplexes loaded with siRNA and cisplatin prodrug
for NF-kappa B targeted treatment of metastatic breast cancer.
Theranostics. 6:14–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gutsche K, Randi EB, Blank V, Fink D,
Wenger RH, Leo C and Scholz CC: Intermittent hypoxia confers
pro-metastatic gene expression selectively through NF-κB in
inflammatory breast cancer cells. Free Radic Biol Med. 101:129–142.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Senger DR, Wirth DF and Hynes RO:
Transformed mammalian cells secrete specific proteins and
phosphoproteins. Cell. 16:885–893. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bao LH, Sakaguchi H, Fujimoto J and Tamaya
T: Osteopontin in metastatic lesions as a prognostic marker in
ovarian cancers. J Biomed Sci. 14:373–381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Coppola D, Szabo M, Boulware D, Muraca P,
Alsarraj M, Chambers AF and Yeatman TJ: Correlation of osteopontin
protein expression and pathological stage across a wide variety of
tumor histologies. Clin Cancer Res. 10:184–190. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brown LF, Papadopoulos-Sergiou A, Berse B,
Manseau EJ, Tognazzi K, Perruzzi CA, Dvorak HF and Senger DR:
Osteopontin expression and distribution in human carcinomas. Am J
Pathol. 145:610–623. 1994.PubMed/NCBI
|
|
46
|
Denhardt DT, Mistretta D, Chambers AF,
Krishna S, Porter JF, Raghuram S and Rittling SR: Transcriptional
regulation of osteopontin and the metastatic phenotype: Evidence
for a Ras-activated enhancer in the human OPN promoter. Clin Exp
Metastasis. 20:77–84. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ito T, Hashimoto Y, Tanaka E, Kan T,
Tsunoda S, Sato F, Higashiyama M, Okumura T and Shimada Y: An
inducible short-hairpin RNA vector against osteopontin reduces
metastatic potential of human esophageal squamous cell carcinoma in
vitro and in vivo. Clin Cancer Res. 12:1308–1316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Psyrri A, Kalogeras KT, Wirtz RM,
Kouvatseas G, Karayannopoulou G, Goussia A, Zagouri F, Veltrup E,
Timotheadou E, Gogas H, et al: Association of osteopontin with
specific prognostic factors and survival in adjuvant breast cancer
trials of the Hellenic Cooperative Oncology Group. J Transl Med.
15:302017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu YY, Zhang YY, Lu WF, Mi YJ and Chen YQ:
Prognostic value of osteopontin expression in breast cancer: A
meta-analysis. Mol Clin Oncol. 3:357–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore
BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC and Welch
DR: Alterations of BRMS1-ARID4A interaction modify gene expression
but still suppress metastasis in human breast cancer cells. J Biol
Chem. 283:7438–7444. 2008. View Article : Google Scholar : PubMed/NCBI
|