Introduction
Oct4, also known as Oct3 or POU class 5 homeobox 1
(POU5F1) gene, is a member of the POU transcription factor
family, the members of which have a structurally characteristic
POU-specific domain (POU-S) and a POU homeodomain (POU-H) that are
required for specific and efficient DNA binding (1,2).
Oct3/4 is specifically expressed in totipotent mouse and human
embryonic stem (ES) and germ cells (3,4) and
is considered to be a gatekeeper during the early steps of
mammalian embryogenesis due to its pivotal role in the regulation
and maintenance of ES cell pluripotency and self-renewal (4–6).
Loss of Oct3/4 alters the fate of inner blastocyst cells in the
early mouse embryo to prevent them from becoming the inner cell
mass or further differentiating into the trophectoderm lineage
(7). Oct3/4 expression is
downregulated when embryonic stem cells are triggered to
differentiate and does not occur in normal somatic cells in
differentiated tissues (7,8).
There is mounting evidence for an association
between Oct3/4 and carcinogenesis. Increased Oct3/4 expression (up
to 150% of normal expression) in ES cells promotes the potential of
these cells to form tumors in syngeneic hosts, from 4% to >80%,
whereas inactivation causes regression of a malignant phenotype
(9). Furthermore, Oct3/4 has been
defined as a diagnostic marker for human testicular germ cell tumor
precursor carcinoma, in situ/intratubular germ cell
neoplasia undifferentiated, seminoma and embryonal carcinoma
(10). In addition to its
expression in germ cell tumors, Oct3/4 has been detected in several
somatic tumors, including brain, bladder, gastric, pancreatic, lung
and rectal cancer (8,10–18).
A transgenic study demonstrated that in vivo activation of
Oct3/4 in adult mouse somatic tissues inhibits cell differentiation
in a manner similar to that observed in embryonic cells, resulting
in epithelial tissue dysplastic growth in the small intestine and
epidermis coupled with progenitor cell expansion (19); the animals die shortly after Oct3/4
expression is activated. Furthermore, chimeric mice with somatic
tissues composed of a mixture of wild type and Oct3/4-inducible
cells develop visible skin tumors 3 weeks after induction of Oct4
expression; however, these tumors completely disappear when Oct3/4
induction is withdrawn (19).
Oct3/4 expression has been observed in several
malignant human breast cancer cell lines, including MCF-7, SKBr3
and MDA-MB-453 cells, but not in normal human breast tissues and
cells (8,12,20–22).
In breast cancer cells, Oct3/4 expression is repressed by
all-trans-retinoic acid and is associated with decreased
cell proliferation (22). Oct3/4
expression in these cells upregulates expression of fibroblast
growth factor-4 (22), a gene that
stimulates MCF-7 cells to become more tumorigenic and metastatic in
ovariectomized and tamoxifen-treated nude mice (23). Furthermore, exogenous
overexpression of Oct3/4 in the 4T1 mouse mammary cancer cell line
increases its ability to form tumor spheres in vitro as well
as its tumorigenic potential in vivo (24).
Notably, computational analysis has revealed six
putative Oct3/4 pseudogenes in the human genome (25) and expression of these pseudogenes
has been detected in cancer cells and primary tumors (26). Thus, it is possible that certain
previous reports of Oct3/4 expression in cancer may have been the
result of Oct-3/4 pseudogene expression. Additionally, two
alternatively spliced Oct3/4 isoforms, originally named Oct3A and
Oct3B, in humans have been cloned (27), though it is not known which
isoform(s) is/are expressed in breast cancer cells.
In this study, we compared levels of Oct3/4
expression in 28 primary breast tumors and 9 normal breast tissues.
Importantly, we reveal, for the first time, differential expression
of Oct3/4 isoforms between breast tumor and normal breast
tissues.
Materials and methods
Breast tumor and normal breast tissue
specimen collection
Breast tumor and normal breast tissue specimens were
collected from 28 patients with primary breast cancer following
lumpectomy or mastectomy and from 9 women undergoing cosmetic
mammoplasties at the Department of Surgery, University of Vermont
Medical Center (Burlington, VT, USA) from March 2008 to February
2009. Tumor clinicopathological parameters for each patient were
also obtained in a blinded manner. Patient ages were unknown, and
all patient tissue samples from which RNA could be isolated were
used in this study. The study protocol was approved and the
requirement for informed patient consent was waived by the
Institutional Review Boards of the University of Vermont (no.
M08-186). The tissues were immediately snap-frozen in liquid
nitrogen and stored at −80°C until RNA and protein isolation. For
histological and immunofluorescence analyses, 1–2 small pieces (~1
mm3) of individual mammary tissues were fixed in freshly
prepared 4% (w/v) paraformaldehyde (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 4 h at 4°C, followed by overnight immersion
in 0.5 M sucrose in phosphate-buffered saline (PBS) at 4°C. The
fixed tissues were embedded in an optimum cutting temperature
compound (Sakura Finetek USA, Inc., Torrance, CA, USA), frozen in
liquid-nitrogen-chilled isopentane and stored at −80°C.
F9 cell lysate and culture of MCF-7 and
MCF-10A cells
F9 cell lysate was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). MCF-7 (cat. no. HTB-2) and
MCF-10A (cat. no. CRC-10317) cells were obtained from the American
Type Culture Collection (ATCC; Manassas, VA, USA). MCF-7 cells were
cultured in Eagle’s minimum essential medium (ATCC; cat. no.
30-2003) supplemented with 10% fetal bovine serum (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 0.01 mg/ml
bovine insulin (Sigma-Aldrich; Merck KGaA), and 50 mg/ml gentamycin
(Invitrogen; Thermo Fisher Scientific, Inc.). MCF-10A cells were
grown in Mammary Epithelial Growth Medium (cat. no. CC-3051;
Cambrex Corporation, East Rutherford, NJ, USA) supplemented with
100 ng/ml cholera toxin (cat. no. 227035; Calbiochem; Merck
KGaA).
RNA isolation and purification and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
polyA+ RNA was isolated from MCF-7 and
MCF-10A cells using FastTrack® 2.0 mRNA Isolation kit
(Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was
extracted from tumor and normal breast tissues using the
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
treated with 0.3 U DNase I at room temperature for 15 min, and
column-purified using the Qiagen RNAeasy Mini kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer’s instructions.
Total RNA (1 μg) was used to generate cDNA using the GeneAMP
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR
analysis was performed using an ABI PRISM 7900 with TaqMan Assays
On Demand Gene Expression kits (Applied Biosystems; Thermo Fisher
Scientific, Inc.) specific for human Oct3/4 (cat. no.
Hs00999632_g1) and for the housekeeping genes hypoxanthine
phosphoribosyltransferase 1 (HPRT1; cat. no. Hs99999909_m1)
and GAPDH (cat. no. Hs99999905_m1). Reactions were performed
in duplicate in a 20-μl volume containing 10 μl
Quanta PerfeCTa qPCR SuperMix (Quantabio, Beverly, MA, USA), 1.0
μl TaqMan assay and 1 μl diluted cDNA (corresponding
to 50 ng of reverse-transcribed total RNA). The following PCR
conditions were used: Initial denaturation at 95°C for 10 min and
40 cycles of 95°C for 15 sec and 60°C for 1 min. Relative
expression of Oct3/4 was calculated with the 2−∆∆CT
method (28) and normalized
against HPRT and GAPDH.
Rapid amplification of cDNA ends (RACE)
of human Oct 3/4 isoforms and cloning of full-length Oct3/4
transcript variant cDNAs
The 3′ and 5′ sequences of human Oct3/4 were
obtained by RACE using SMART RACE cDNA Amplification kit (Clontech
Laboratories, Inc., Mountainview, CA, USA). 3′- and 5′-RACE-ready
first-strand cDNAs were synthesized using polyA+ RNA
from MCF-7 or MCF-10A cells or total RNA pooled from all 28 breast
cancer tissues or all 9 normal breast tissues. The Oct3/4 5′
sequences were first amplified using the provided universal primer
(UPR; Clontech Laboratories, Inc.) and an Oct3/4-specific reverse
primer (159R; Table I) and then
reamplified using the provided nested universal primer (NUP;
Clontech Laboratories, Inc.) and a nested Oct3/4-specific reverse
primer (155R for cells or 161R for tissues; Table I). Similarly, the 3′ sequence of
Oct3/4 was first amplified using UPM and an Oct3/4-specific forward
primer (154F; Table I) and
reamplified using NUP and a nested Oct3/4-specific forward primer
(158F for cells or 160F for tissues; Table I). The resulting PCR products were
gel-purified, cloned into the pCR® 2.1 vector (Clontech
Laboratories, Inc.), and sequenced by the Vermont Cancer Center DNA
Core Facility using an ABI 3730 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The RACE product sequences were verified in at
least three independent clones.
 | Table ISequences of oligonucleotide primers
used for PCR and rapid amplification of 5′ and 3′ cDNA ends. |
Table I
Sequences of oligonucleotide primers
used for PCR and rapid amplification of 5′ and 3′ cDNA ends.
| Primer name | Sequence |
|---|
| 154 F |
5′-CCAAGCTCCTGAAGCAGAAG-3′ |
| 155 R |
5′-GCCGGTTACAGAACCACACT-3′ |
| 158 F |
5′-GGCTCTGCAGCTTAGCTTCA-3′ |
| 159 R |
5′-CCGAGGAGTACAGTGCAGTG-3′ |
| 160 F |
5′-CACTTCACTGCACTGTACTC-3′ |
| 161 R |
5′-CACATCGGCCTGTGTATATC-3′ |
| 164 R |
5′-TTACTGTGTCCCAGGCTTCT-3′ |
| 165 R |
5-TTCACCTTCCCTCCAACCAG-3′ |
| 166 F |
5′-GATCCAAGTGGGCAACTTGA-3′ |
| 168 F |
5′-CTGAGACATGATGCTCTTCC-3′ |
| 170 F |
5′-TGATGCTTCAGGCACTGTGT-3′ |
| 172 F |
5′-AATCTGGACCTGAGCGAGAA-3′ |
Full-length Oct3/4 transcript variant cDNAs were
first PCR-amplified from the 5′-RACE-ready first-strand cDNAs from
either MCF-7 or MCF-10A cells using Advantage 2 polymerase
(Clontech Laboratories, Inc.) with the UPM-A primer (Clontech
Laboratories, Inc.) and a gene-specific primer (164R or 165R,
Table I); the sequences were
reamplified with 165R and the following specific forward primer for
each individual transcript: i) 170F (Oct3; GenBank accession no.
DQ486513.1); ii) 166F (Oct4; GenBank accession no. DQ486514.1);
iii) 168F (GenBank accession nos. DQ486515.1 and DQ486516.1); and
iv) 172F (GenBank accession no. DQ486517.1; Table I). The primers were designed to
correspond with the far 5′ or 3′ region of each individual
transcript. The PCR products were gel-purified and cloned into the
pCR 2.1 vector. Each full-length cDNA was sequenced, verified in at
least three independent clones, and submitted to GenBank under
accession nos. DQ486513.1-DQ486517.1.
DNA sequence analysis
cDNA sequence analysis was conducted using Lasergene
(version 3; DNASTAR, Inc., Madison, WI, USA) computer program and
the National Center for Biotechnology Information (blast.ncbi.nlm.nih.gov/Blast.cgi) BLAST
site. Multiple sequence alignment was performed using the European
Molecular Biology Laboratory MAFFT site (ebi.ac.uk/Tools/msa/mafft) and viewed with the GeneDoc
program (psc.edu/biomed/genedoc).
In vitro transcription and
translation
Full-length Oct3/4 transcript cDNAs were transcribed
in vitro by T7 polymerase and translated in the presence of
L-[35S]methionine (GE Healthcare, Chicago, IL, USA) using the
TNT® Coupled Reticulocyte Lysate System (Promega
Corporation, Madison, WI, USA) according to the manufacturer’s
instructions. The translation products were resolved by 12% (w/v)
SDS-PAGE and the radioimaging was captured by a phosphor-capture
screen and analyzed using Quantity One software (version 4.2.1)
with a Molecular Imager FX (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Western blot analysis
Whole-tissue and cell lysates (25 μg total
protein) extracted using T-PER™ Tissue Protein Extraction Reagent
(Thermo Fisher Scientific, Inc.) were resolved by 12% (w/v)
SDS-PAGE using Mini-PROTEAN III Cell system (Bio-Rad Laboratories,
Inc.). Proteins were electrophoretically transferred at 4°C to a
polyvinylidene fluoride membrane (GE Healthcare). The blot was
blocked for 1 h at room temperature in Tris-buffered saline (TBS;
50 mM Tris-Cl, pH 7.5 and 150 mM NaCl) with 0.1% Tween-20 (TBS-T)
containing 5% non-fat dry milk (w/v) and incubated overnight at 4°C
in TBS-T containing 5% non-fat dry milk and 1 μg/ml
anti-Oct3/4 antibody (cat. no. ab19857; Abcam, Cambridge, MA, USA)
or 1 μg/ml anti-Oct3/4 antibody that had been pre-incubated
with a 5-fold excess (molar concentration) of blocking polypeptide
(cat. no. ab20650; Abcam) for 1 h at room temperature. The membrane
was washed twice at room temperature for 20 min with TBS-T and
incubated for 1 h at room temperature in TBS-T with 2% non-fat
dried milk and a 1:5,000 dilution of a goat anti-rabbit secondary
antibody conjugated with horseradish peroxidase (cat. no. NA9340;
GE Healthcare). Detection of the protein-primary antibody-secondary
antibody immune complex was performed using the SuperSignal West
Pico Chemiluminescent substrate (Pierce; Thermo Fisher Scientific,
Inc.).
Immunofluorescence staining
For Oct3/4 immunohistochemical staining, embedded
tissues were sectioned (10 μm), thaw-mounted onto the
surface of gelatin-coated slides, and incubated in 10% normal goat
serum (Thermo Fisher Scientific, Inc.) at room temperature for 30
min to block non-specific antibody binding. The sections were then
incubated with either diluted anti-Oct-3/4 (cat. no. ab19857;
Abcam; 2 μg/ml) in PBS with 1% bovine serum albumin (BSA) or
normal rabbit IgG for 1 h at 37°C. For an additional control, some
sections were incubated with an anti-Oct3/4 antibody that had been
pre-incubated with a 5-fold excess (molar concentration) of
blocking polypeptide (cat. no. ab20650; Abcam) for 1 h at room
temperature. After being washed twice in PBS with 1% BSA for 5 min,
the sections were incubated for 1 h at room temperature in the dark
with a 1:4,000 dilution of secondary antibody conjugated with Alexa
Fluor 647 (cat. no. A-21244; Molecular Probes; Thermo Fisher
Scientific, Inc.). After three washes in PBS with 1% BSA for 5 min,
the sections were stained with the nuclear stain DAPI (Molecular
Probes; Thermo Fisher Scientific, Inc.) diluted 1:10,000 in PBS for
5 min at room temperature. Finally, the sections were washed twice
in PBS with 1% BSA for 15 min, once in PBS, and once in distilled
water before being mounted onto glass microscope slides using
Aqua-Poly/Mount (Polysciences, Inc., Warrington, PA, USA). The
sections were then examined under a confocal microscope (LSM 510;
Zeiss AG, Thornwood, NY, USA).
Statistical analysis
Data are presented as the mean ± standard error.
Statistical analysis of mRNA expression was performed using the JMP
software package (SAS Institute, Inc., Cary, NC, USA). Differences
in the means of Oct3/4 mRNA expression between breast cancer and
normal breast tissues were compared using Student’s t-test.
Logistic regression of Oct3/4 mRNA expression and tumor types was
analyzed using the logistic regression package in the R language
program. P<0.05 was considered to indicate a statistically
significant difference.
Results
Strong Oct3/4 expression in primary
breast cancer tissues
Oct3/4 expression was previously detected in several
malignant human breast cancer cell lines (12,22).
To examine Oct3/4 expression in primary breast carcinomas, 28
primary breast tumor tissues were obtained from patients following
lumpectomy or mastectomy and 9 normal breast tissues from women
undergoing cosmetic surgery. RT-qPCR analysis revealed that,
compared with normal breast tissues, all tumor tissues,
irrespective of type or clinicopathological status (fibroadenoma,
ductal in situ carcinoma, differentiation level, TNM
classification, or estrogen, progesterone and receptor
tyrosine-protein kinase erbB-2 (Her2) receptor status), strongly
expressed Oct3/4 (Table II).
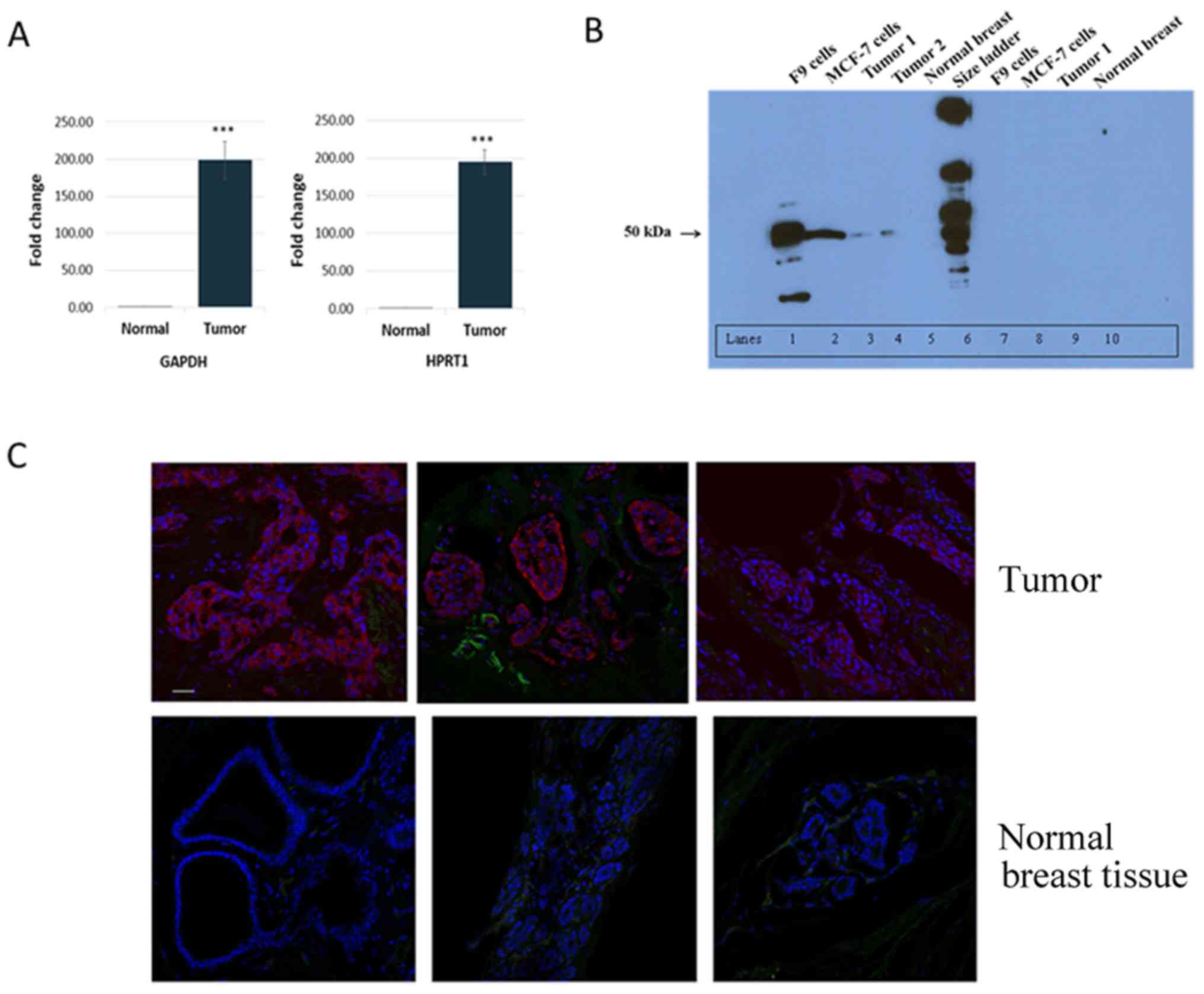
Specifically, Oct3/4 mRNA levels were 193-fold higher in tumors
than in normal breast tissues (P<0.001; Fig. 1A), ranging from 35- to 746-fold
based on normalization to the expression level of either
GAPDH or HPRT1 (Table
II). However, logistic regression analysis did not reveal any
significant association between Oct3/4 expression level and tumor
phenotype (data not shown).
 | Table IIOct3/4 mRNA expression in 28 primary
breast tumor tissues. |
Table II
Oct3/4 mRNA expression in 28 primary
breast tumor tissues.
| Patient no. | Tumor
histology | Level of
differentiation | Tumor sizea | Lymph node
spreada | Metastasisa | Receptor status
| Oct3/4 mRNAd
|
|---|
| ERb | PRb | Her2c | GAPDH | HPRT1 |
|---|
| 01 | IDCA | Poor | T2 | N0 | Mx |
0 |
0 | 0 |
92.31 | 172.42 |
| 04 | IDCA | Poor | T1c | N0 | +M |
0 |
0 | 0 |
74.96 | 121.76 |
| 05 | IDCA | Moderate | T1c | N0 | Mx | 90 | 90 | 2 NA | 170.26 | 228.6 |
| 07 | IDCA | Well | T1c | N0 | Mx | 90 | 90 | 0 | 214.66 | 139.69 |
| 08 | IDCA | Moderate | T1c | N0 | Mx |
0 |
0 | 1 | 71.5 |
92.76 |
| 10 | Fibroepthelial
fibroadenoma and phyllodes | | | | | | | | 311.56 | 339.49 |
| 11 | IDCA | Poor | T1c | N3 | Mx | >90 | <5 | 0 | 305.95 | 149.96 |
| 12 | IDCA | Poor | T2 | N0 | Mx | 80 | 90 | 0 |
160.2 | 148.65 |
| 14 | Lobular and
IDCA | Moderate | T2 | N1 | Mx | >90 | >90 | 0 |
98.87 | 63.84 |
| 15 | IDCA | Poor | T2 | N0 | Mx | 70 | 90 | 2 A | 157.96 | 228.54 |
| 16 | IDCA | Poor | T2 | N3 | Mx | >90 |
0 | 0 | 116.73 | 153.52 |
| 17 | IDCA | Moderate | T1c | N0 | Mx | 90 | 80 | 1 | 129.51 | 225.54 |
| 18 | IDCA | Poor | T1c | N0 | Mx |
0 | 60 | 3 |
34.59 | 101.30 |
| 19 | IDCA | Moderate | T1c | N0 | Mx | 90 | >90 | 0 | 330.59 | 208.13 |
| 21 | IDCA | Poor | T2 | N0 | Mx |
0 |
0 | 0 | 746.42 | 485.89 |
| 23 | IDCA | Poor | T2 | N1 | Mx | 90 |
0 | 0 | 170.83 | 171.86 |
| 24 | IDCA | Moderate | T1c | N0 | Mx | 90 | 30 | 1 | 149.59 |
82.77 |
| 25 | IDCA | Moderate | T2 | N0 | Mx | 90 | >90 | 0 | 176.65 | 284.85 |
| 27 | IDCA | Poor | T2 | N0 | Mx |
0 |
0 | 2 | 230.54 | 229.01 |
| 28 | IDCA | Mild | T2 | N | Mx | >90 | >90 | 0 | 136.86 | 191.85 |
| 30 | IDCA | Moderate | T3 | N | Mx | 90 |
0 | 0 | 128.65 | 125.84 |
| 31 | IDCA | Moderate | T2 | N0 | Mx | >90 |
0 | 0 | 265.90 | 237.90 |
| 34 | IDCA | Moderate | T1b | N0 | Mx | >90 | 80 | 0 | 360.97 | 240.54 |
| 35 | IDCA | Poor | T2 | N1 | Mx | 80 | 90 | 1 | 162.73 | 222.87 |
| 36 | IDCA | Lobular
features | T2 | N0 | Mx | >90 | 60 | 2 | 157.54 | 129.37 |
| 37 | IDCA and focal
medullary features | Poor | T1c | N0 | Mx |
0 |
0 | 2 | 188.84 | 236.58 |
| 41 | IDCA and medullary
features | Poor | T2 | N0 | Mx |
0 |
0 | 0 | 231.80 | 273.61 |
| 42 | IDCA | Poor | T2 | N0 | Mx | | | | 173.42 | 152.30 |
To examine Oct3/4 protein expression in tumor
tissues, western blot analysis was performed. As shown in Fig. 1B, Oct3/4 protein was detected in
two primary breast tumors examined, MCF-7 breast cancer cells, and
the positive control F9 cells, but not in normal breast tissues.
Preincubation of the anti-Oct3/4 antibody with a control peptide
essentially blocked recognition, supporting the specificity of the
antibody.
Immunofluorescence staining of Oct3/4 was performed
to further examine and determine the localization of Oct3/4
expression in breast tumors and normal tissues. Strong Oct3/4
staining was observed in all tumors examined, whereas no staining
was observed in normal breast tissues (Fig. 1C; staining of three tumors and
three normal tissues). The strong staining of Oct3/4 in tumors
appeared to mainly occur in cancer cells and not in the stromal
tissues surrounding cancer cells within ducts. Essentially, no
staining was observed in the tumor and normal tissue sections
incubated with either normal rabbit IgG or anti-Oct3/4 preincubated
with the control peptide (data not shown).
Expression of different Oct3/4
transcripts in breast tumor and normal breast tissues
Multiple Oct3/4 isoforms have been cloned (27), and six pseudogenes highly
homologous to Oct3/4 have been previously reported (25). To verify expression of the
functional Oct3/4 gene and identify Oct3/4 mRNA isoforms in breast
cancer cells and tumors, 5′- and 3′-RACE was performed using
polyA+ RNA isolated from MCF-7 breast cancer cells and
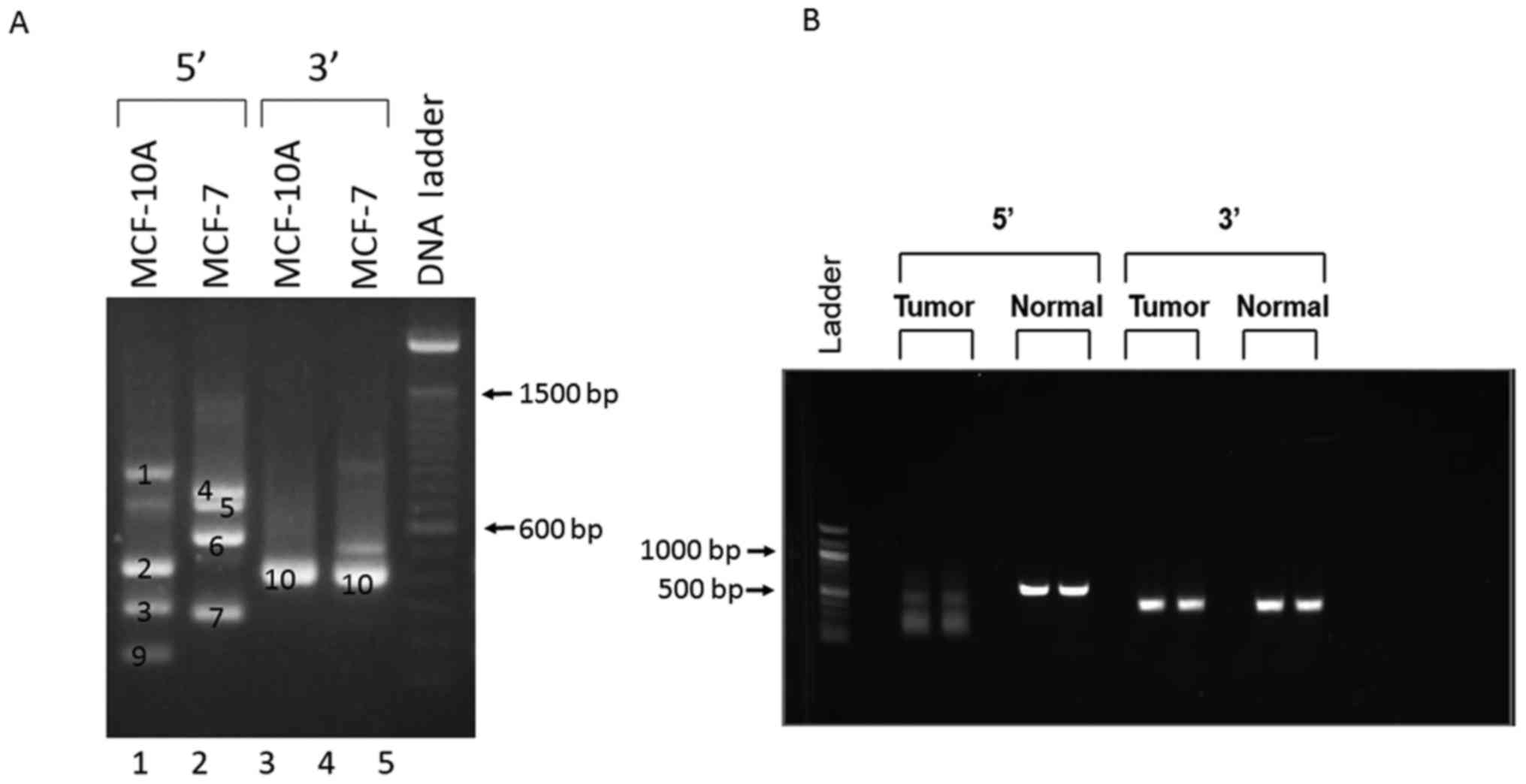
MCF-10A non-tumorigenic breast epithelial cells (Fig. 2). In 3′-RACE, major, similar-sized
products were obtained from both cells (band 10). Cloning and
sequencing of band 10 confirmed that it contained the 3′ sequences
of Oct3/4 [the last two exons are the same in all Oct4 transcript
variants (POU5F1 on chromosome 6) and 98% identical to the
3′ mRNA sequence of Oct3 (POU5F1B on chromosome 8), Fig. 3 and see below]. However, 5′-RACE
produced different products from the two cell lines (Fig. 2). Cloning and sequencing of the
5′-RACE products from MCF-10A cells showed that bands 1, 2, 3, and
9 were various Oct4 gene transcripts with different transcription
start sites at positions 31167170, 31166838, 31166211 and 31166081,
respectively, on chromosome 6 (GRCh38.p7 primary assembly sequence
ID: NC_000006.12) and/or exhibited different RNA splicing (Fig. 3). By contrast, the major 5′-RACE
products from MCF-7 cells (bands 4-6) consisted of various Oct3
transcripts (intronless) starting at different positions on
chromosome 8, and band 7 was revealed to be another Oct4 transcript
that starts at position 31180731 on chromosome 6 (Fig. 3). Thus, the RACE results indicated
that MCF-10A and MCF-7 cells express different Oct4 (POU5F1)
and Oct3 (POU5F1B) gene transcripts from different
chromosomes.
To examine the Oct3/4 transcripts expressed in
breast tumor and normal breast tissues, RACE was performed using
total RNA pooled from all 28 breast tumors or all 9 normal breast
tissues. As shown in Fig. 2 and
consistent with the RACE results obtained using MCF-7 and MCF-10A
cells, one similar-sized band was obtained from tumor and normal
tissues by 3′-RACE, whereas 5′-RACE resulted in different bands
from tumor and normal tissues (one single bright band from normal
tissues vs. weak, smaller and diffused bands from tumor tissues).
Verification by cloning and sequencing revealed that the 3′-RACE
products from tumor tissues comprised both Oct3 and Oct4
transcripts and a transcript from chromosome 1 (an Oct4
pseudogene), and the 3′ products from normal tissues contained the
Oct4 transcript and the same transcript from chromosome 1. The
5′-RACE products from tumor and normal tissues were all Oct4
transcripts. However, the transcription start sites of the Oct4
transcripts from breast tumors were between 31,186,202 and
31,186,092 on chromosome 6, whereas the start site of the Oct4
transcript from normal breast tissues was at 31,166,274 on
chromosome 6 (Fig. 3).
Cloning and in vitro transcription and
translation of Oct3/4 transcript variants, genomic organization and
sequence alignment of Oct3/4 isoforms
The full-length cDNAs of various Oct3/4 transcripts
were amplified from the MCF-7 or MCF-10A cell 5′-RACE libraries.
For the Oct3 transcript (the longest sequence corresponds to bands
4-6 in Fig. 2) and each Oct4
transcript (corresponding to bands 1-3, 7 and 9), the first PCR
reactions were performed using the UPM-A primer and a reverse
Oct3/4-specific primer close to the polyA+ site; the
second PCR was performed using a nested forward primer at the
farthest possible 5′ site in each individual transcript with either
the same reverse primer used in the first PCR or a nested reverse
primer very close to the first reverse primer. Using this approach,
the full-length cDNAs of Oct3 (DQ486513) and several Oct4
transcripts were successfully amplified, including DQ486514
(corresponding to the band 1 transcript, that also included bands 3
and 9), DQ486515 (transcript A of band 2), DQ489516 (transcript B
of band 2), and DQ486517 (band 7; Fig.
3 and Table III); these cDNA
sequences were submitted to GenBank under accession nos.
DQ486513.1-DQ486517.1, respectively. Sequence analysis of these
transcripts indicated that the Oct3 transcript contains an open
reading frame (ORF) that encodes the full-length Oct3 protein;
conversely, none of the Oct4 transcripts harbor an ORF that encodes
an Oct4 isoform with intact POU-S and POU-H domains.
 | Table IIIPutative human Oct3/4 isoforms. |
Table III
Putative human Oct3/4 isoforms.
| Namea | Gene
| Protein
|
|---|
| Symbol | Chromosome | Transcripts
(GenBank accession no.) | Exons | GenBan accession
no. | Amino acid
length | Size (Da) |
|---|
| Oct4(A) | POU5F1 (NCBI ID
5460) | 6 (6p21.33)c | Variant 1e (NM_002701.5, Z11898.1) | 5 | NP_002692.2 | 360 | 38,571 |
| Oct4B | POU5F1 | 6 (6p21.33)c | Variant 5e (NM_001285987.1, Z11899.1),
KY781166e,f | 4 | NP_001272916.1 | 265 | 29,954 |
| Oct4-190b | POU5F1 | 6 (6p21.33)c | Variant 2e (DQ486515.1f, NM_203289.5) | 4 | NP_976034.4,
NP_001167002.1 | 190 | 21,127 |
| Variant 3e (DQ486516.1f, NM_001173531.2) | 5 | | | |
| DQ486517.1e,f | 5 | | | |
| KY781137e,f | 5 | | | |
| Oct4-164 | POU5F1 | 6 (6p21.33)c | Variant 4
(DQ486514.1f,
NM_001285986.1) | 3 | NP_001272915 | 164 | 18,310 |
| Oct3 | POU5F1B (NCBI ID
5462) | 8 (8Q24.21)d | DQ486513.1f, NM_001159542.1, Z11901.1 | Intronless | NP_001153014.1 | 359 | 38,457 |
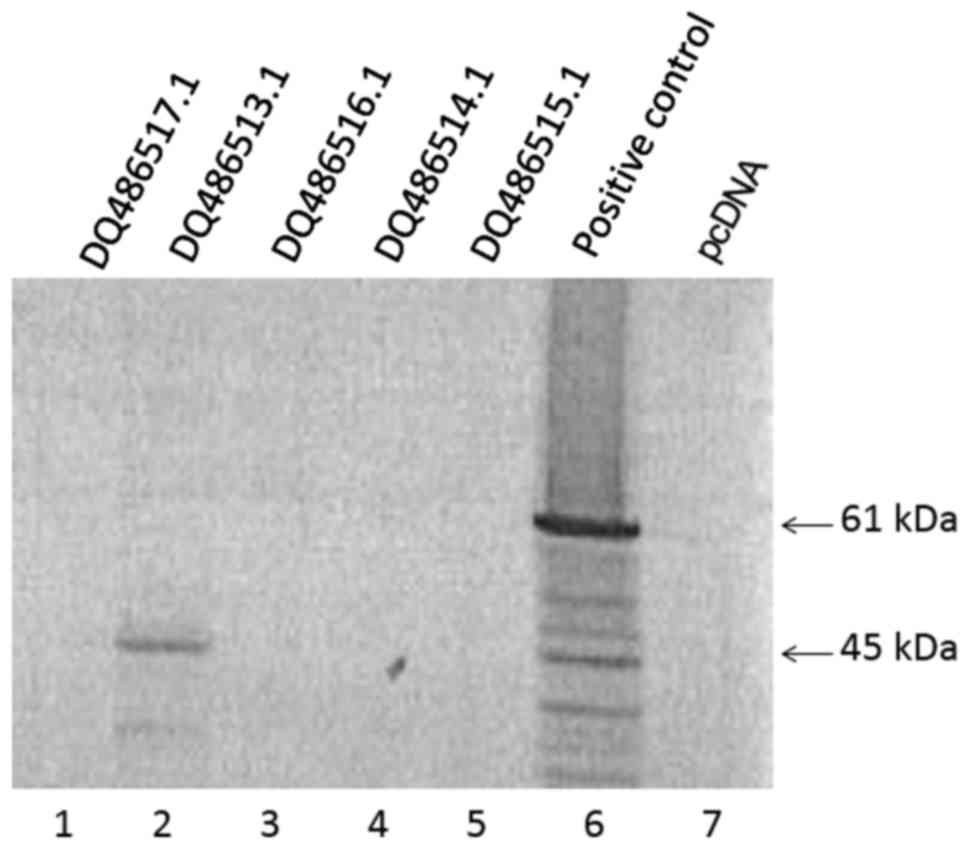
Subsequently, coupled in vitro transcription
and translation was performed to characterize the different Oct3/4
full-length cDNA sequences obtained (Fig. 4). Consistent with sequence
analysis, only DQ486513 (Oct3 transcript), but none of the Oct4
transcripts, produced translation products.
Furthermore, two new Oct4 transcripts were predicted
based on our 5′- and 3′-RACE results from normal breast and breast
tumor tissues (KY781166 and KY781167; Fig. 3). KY781167, which is expressed in
breast tumors, uses the farthest 5′ exon and is initiated on
chromosome 6 at position 31,186,202 in a sequence (LC050991.1)
called psoriasis susceptibility 1 candidate 3 (PSORS1C3); it does
not contain an ORF to encode an intact Oct4 isoform. KY781166,
which is expressed in normal breast tissues, is a shorter DQ486515
5′ region sequence with an important single-nucleotide polymorphism
(SNP) that creates an ‘ATG’ translation initiation codon to encode
a 265-aa Oct4B isoform (Table
III and Fig. 3). All sequenced
clones of the 5′-RACE product from normal breast tissues contain
this SNP.
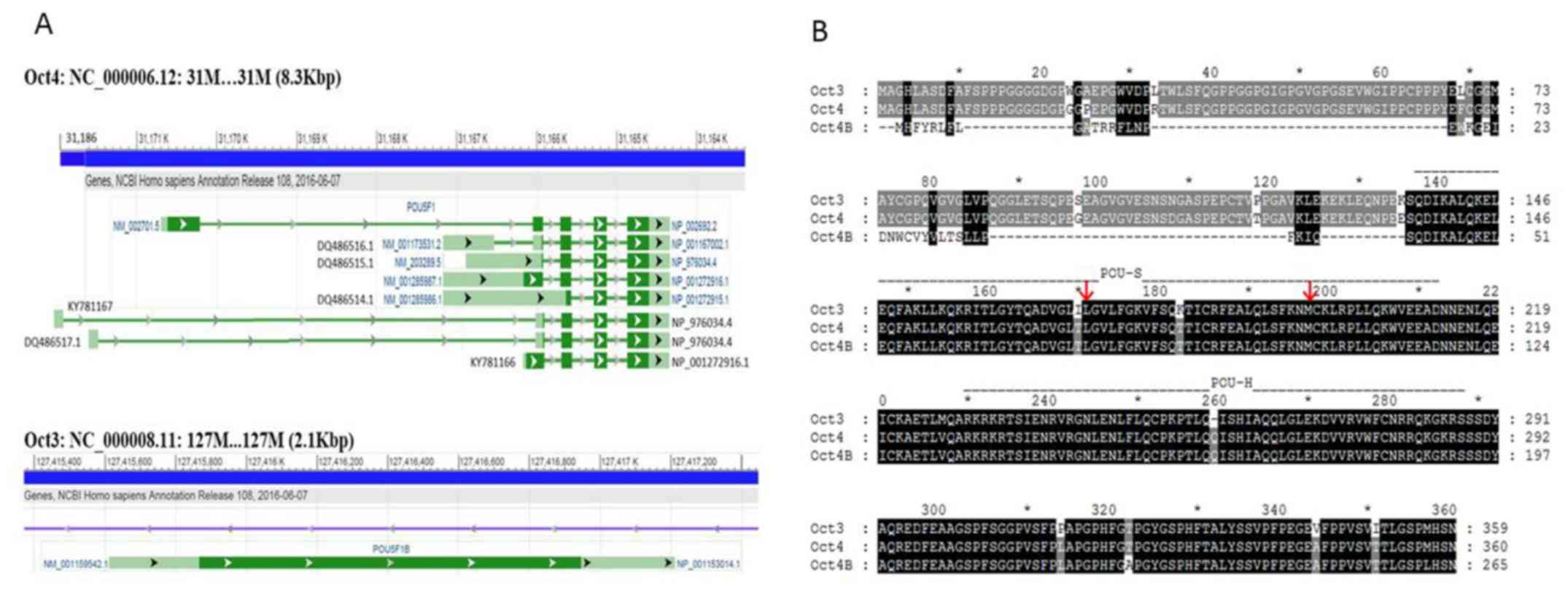
The genomic organization of the Oct3 and Oct4 genes
is shown in Fig. 3A. Oct3 is an
intronless gene, whereas the Oct4 gene has ≥8 exons. Specifically,
the Oct4 gene has multiple transcription initiation sites in the
exon ranging from sites 31,167,170 to 31,165,926. The last two
exons are retained in all Oct4 transcript variants. The amino acid
sequences of Oct3 (NP_001153014.1), Oct4 (NP_002692.2) and Oct4B
(NP_001272916.1) are aligned in Fig.
3B. Oct4 has 360 amino acids and is one amino acid longer and
96% identical to Oct3. Oct4B consists of 265 amino acids, with its
last 225 being the same as those of Oct4; it contains intact POU-S
and POU-H domains.
Discussion
In the present study, all breast tumors examined,
regardless of tumor type, expressed Oct3/4 mRNAs at levels 10- to
100-fold higher than those in normal breast tissues. Additionally,
Oct3/4 protein expression in breast tumors was confirmed by western
blot analysis and immunostaining. These observations are consistent
with previous findings for normal breast and breast cancer cell
lines (8,12,22)
as well as recent clinicopathological evaluations in which Oct3/4
expression was reported to be associated with poor clinical
outcomes (20,29–31).
However, Oct3/4 mRNA levels were not correlated with tumor type,
tumor size, level of differentiation or estrogen/progesterone/Her2
receptor status in this study, which is inconsistent with positive
correlations between Oct4 expression and breast cancer type (such
as lymph node metastasis and histological grade) reported in other
studies (29,31). This discrepancy was likely due to
the small number of tumor samples included in this study.
Notably and for the first time (to the best of our
knowledge), the findings of the current study provided strong
evidence that breast tumor and normal breast tissues express
different Oct3/4 genes and transcripts. The 5′-RACE results
revealed different Oct3/4 transcripts between breast tumors and
MCF-7 cells and normal breast tissues and MCF-10A cells. These
different RACE products are likely not RNA degradation products
because 3′-RACE using the same RNAs only produced a single major
product for each tissue and cell. Indeed, sequencing of the RACE
products indicated that the different products resulted from
expression of different genes, the use of different transcription
initiation sites, or variable gene splicing. MCF-10A cells and
normal breast tissues only expressed POU5F1 transcripts,
though MCF-7 cells and breast tumors expressed both POU5F1B
and POU5F1 transcripts, with predominant POU5F1B
expression in the former. The reason for the scant products and
lack of POU5F1B transcripts in the 5′-RACE experiments with
breast tumors was most likely due to the lower RNA amounts used in
RACE conducted using tissues compared with that using cells
(polyA+ vs. total RNA). A single-nucleotide mismatch in
the 159R primer used in the 5′-RACE for POU5F1B, which was
realized following the experiment, may have also contributed to
these results. Nevertheless, the 3′-RACE results demonstrated
expression of POU5F1B and POU5F1 in tumor
tissues.
The possible differential expression of
POU5F1 and POU5F1B in breast tumors and normal
tissues warrants clearly distinguishing between these two genes. We
previously proposed that the two sequences are two individual genes
and designated POU5F1 as Oct4 and POU5F1B as Oct3
(1,2), as was used in the present study.
These designations are also supported by the following: i) The two
sequences are located on different chromosomes, with POU5F1
on chromosome 6 and POU5F1B on chromosome 8; ii) POU5F1 has
multiple exons and belongs to the POU-V family, whereas POU5F1B is
intronless and belongs to the POU-III family (1); and iii) the Oct4 protein is one amino
acid longer than and 4% different from the Oct3 protein.
The present study identified multiple Oct4
transcripts expressed in breast tissues and cells, displaying
different transcription initiation sites and/or RNA splicing.
Surprisingly, the Oct4 transcript (NM_002701) that encodes the
major form of the Oct4 protein in ES cells (NP_002692, 360 aa) was
not detected. The full-length cDNAs of the majority of the Oct4
transcripts detected in this study were cloned (DQ486513-DQ486517).
However, computational analysis of these cDNAs failed to identify
an ORF that encodes an Oct4 protein with intact POU-S and POU-H
domains, and no products were obtained when using these cDNAs in an
in vitro transcription and translation assay, indicating
that these transcripts are untranslatable. The putative Oct4
transcript obtained from normal breast tissues (KY781166) contains
an SNP that creates an ‘ATG’ initiation codon from an ‘AGG’ and an
ORF that can encode the 265-amino acid Oct4B protein. This SNP
(reference no. 3130932; ncbi.nlm.nih.gov/snp/) is also found in the Celera and
GenBank human genome assemblies and may have important functional
significance. Oct4B was not detected in normal breast tissues by
western blot analysis (a limitation of our western blot analysis
was the lack of a loading control to confirm there was protein in
each lane) and immunohistological staining, which was likely to be
because the protein levels were too low to be detected. In
addition, there is evidence that Oct4 transcripts can be translated
using a downstream in-frame non-AUG (CUG) start codon to express a
190-amino acid isoform (Table
III) or a downstream AUG start codon to express a 164- amino
acid isoform (32). These isoforms
were not detected in the in vitro transcription and
translation assay in the present study. Nevertheless, even though
these short isoforms are produced, they may not have any of the
biological functions of Oct4 because they do not contain intact POU
domains, which are required to efficiently bind to target gene DNA
elements. Taken together, the data demonstrated that normal breast
tissues express low levels of Oct4B, which may be functionally
different from Oct4 due to the lack of a 5′ transactivation domain,
whereas breast tumors express both Oct3 and untranslated Oct4
transcripts.
In addition to the high homology between the Oct4
and Oct3 sequences [98% identity between Oct4 mRNA (NM_002701) and
Oct3 mRNA (NM_001159542)], the 3′ sequences of Oct3/4 cDNAs also
have high homology to the Oct3/4 pseudogenes (25,26,33)
on chromosomes 1, 3, 8, 10 and 12 (Table IV). Expression of these
pseudogenes has been observed in tumors and cells (26,33).
As a transcript from chromosome 1 (POU5F1P4) was amplified
by 3′-RACE in this study, the primers used for analyzing Oct3/4
expression should not be designed based on Oct3/4 3′ mRNA
sequences. Indeed, only short 5′ sequences should be used, but with
caution, as the 5′ sequence is both Oct3/4 and isoform specific. It
is important to note that because the majority of Oct4 transcripts
may not be translated or may not produce proteins with Oct4
biological functions, detection of Oct4 transcripts does not
necessarily indicate Oct4 protein expression in tissues and
cells.
 | Table IVOct3 mRNA homology to sequences in
the human genome.a |
Table IV
Oct3 mRNA homology to sequences in
the human genome.a
| Corresponding
gene | Chromosome
location | NCBI reference
sequence (accession no.) | Region of Oct3
cDNA | Sequence range | Length (bp) | Homology (%) |
|---|
| POU5F1P4 | 1q22 | NC_000001.11 | 216–1,561 |
155,433,140–155,434,489 | 1,349 | 96 |
| POU5F1P6 | 3q21.3 | NC_000003.12 | 211–780 |
128,677,051–128,676,528 |
523 | 80 |
| | | 768–949 |
128,675,568–128,675,388 |
180 | 92 |
| | | 1,048–1,583 |
128,674,928–128,674,421 |
507 | 86 |
| POU5F1 (Oct4) | 6p21.33 | NC_000006.12 | 214–660 |
31,170,662–31,170,216 |
446 | 97 |
| | | 659–782 |
31,166,049–31,165,929 |
120 | 99 |
| | | 781–915 |
31,165,702–31,165,568 |
134 | 98 |
| | | 911–1,071 |
31,165,288–31,165,125 |
163 | 98 |
| | | 1,069–1,583 |
31,164,867–31,164,352 |
515 | 98 |
| POU5F1B (Oct3) | 8q24.21 | NC_000008.11 | 1–1,583 |
127,415,612–127,417,194 | 1,582 | 100 |
| POU5F1P2 | 8q22.3 | | 516–1,579 |
102,621,247–102,622,271 | 1,024 | 80 |
| POU5F1P5 | 10q21.3 | NC_000010.11 | 659–1,583 |
68,010,879–68,009,954 |
925 | 88 |
| POU5F1P3 | 12p13.31 | NC_000012.12 | 234–1,583 |
8,133,523–8,134,873 | 1,350 | 96 |
Based on its aberrant expression in breast tumors,
Oct3/4 is a potential prognostic marker for breast cancer. These
data support the hypothesis that adult stem cells serve as targets
for carcinogenesis (34,35) due to the essential role of Oct4 in
the regulation and maintenance of stem cell pluripotency and
self-renewal (5). It has been
demonstrated that Oct3/4-expressing breast cancer stem cells
isolated from breast cancer lesions exhibit stem/progenitor cell
properties (36). Furthermore,
because Oct4 is the most important among the four embryonic
transcription factors that reprogram adult somatic cells into
undifferentiated, pluripotent, and self-renewable states (referred
to as induced pluripotent stem cells) (1,37),
aberrant expression of Oct3/4 in adult somatic cells may also
induce tumorigenesis. Furthermore, expression of Oct3/4 may be
required for the maintenance of transformed cancer cells. Thus,
Oct3/4 may represent an excellent target for gene-directed
therapeutic interventions for the prevention and treatment of
breast cancer. In summary, the results of the current study suggest
that Oct3, rather than Oct4, represents a potential prognostic
marker and an excellent target for gene-directed therapeutic
interventions for breast cancer.
Acknowledgments
We thank Dr Timothy Hunter, Mr. Scott Tighe and Ms.
Mary Lou Shane at the Vermont Cancer Center (Burlington, VT, USA)
for their help with the DNA sequencing analysis.
Funding
This research was funded by the Department of
Defense Breast Cancer Research Program Concept Award (grant no.
W81XWH-05-1-0432; to FQZ), the Vermont Cancer Center VCC/LCCRO
Pilot Award (grant no. 022820; to FQZ), and the Vermont Genetics
Network graduate assistantship (to YM).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
FQZ, YM, DBL, MPW, GZ, QT and KB performed the
experiments. YM, DK, DW and JT were involved in the breast tumor
and normal breast tissue collection. DWL performed the statistical
analysis. FQZ wrote the manuscript. All authors have read and
approved the manuscript.
Ethics approval and consent to
participate
The study protocol was approved and the requirement
for informed patient consent was waived by the Institutional Review
Boards of the University of Vermont (no. M08-186).
Consent for publication
Informed patient consent was waived by the
Institutional Review Boards of the University of Vermont (no.
M08-186).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhao FQ: Octamer-binding transcription
factors: Genomics and functions. Front Biosci. 18:1051–1071. 2013.
View Article : Google Scholar
|
|
2
|
Qian X and Zhao FQ: Regulatory roles of
Oct proteins in the mammary gland. Biochim Biophys Acta.
1859:812–819. 2016. View Article : Google Scholar
|
|
3
|
Schöler HR, Dressler GR, Balling R,
Rohdewohld H and Gruss P: Oct-4: A germline-specific transcription
factor mapping to the mouse t-complex. EMBO J. 9:2185–2195.
1990.
|
|
4
|
Niwa H, Miyazaki J and Smith AG:
Quantitative expression of Oct-3/4 defines differentiation,
dedifferentiation or self-renewal of ES cells. Nat Genet.
24:372–376. 2000. View
Article : Google Scholar
|
|
5
|
Babaie Y, Herwig R, Greber B, Brink TC,
Wruck W, Groth D, Lehrach H, Burdon T and Adjaye J: Analysis of
Oct4-dependent transcriptional networks regulating self-renewal and
pluripotency in human embryonic stem cells. Stem Cells. 25:500–510.
2007. View Article : Google Scholar
|
|
6
|
Pesce M and Schöler HR: Oct-4: Gatekeeper
in the beginnings of mammalian development. Stem Cells. 19:271–278.
2001. View Article : Google Scholar
|
|
7
|
Pesce M and Schöler HR: Oct-4: Control of
totipotency and germline determination. Mol Reprod Dev. 55:452–457.
2000. View Article : Google Scholar
|
|
8
|
Tai MH, Chang CC, Kiupel M, Webster JD,
Olson LK and Trosko JE: Oct4 expression in adult human stem cells:
Evidence in support of the stem cell theory of carcinogenesis.
Carcinogenesis. 26:495–502. 2005. View Article : Google Scholar
|
|
9
|
Gidekel S, Pizov G, Bergman Y and Pikarsky
E: Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer
Cell. 4:361–370. 2003. View Article : Google Scholar
|
|
10
|
de Jong J and Looijenga LH: Stem cell
marker OCT3/4 in tumor biology and germ cell tumor diagnostics:
History and future. Crit Rev Oncog. 12:171–203. 2006. View Article : Google Scholar
|
|
11
|
Monk M and Holding C: Human embryonic
genes re-expressed in cancer cells. Oncogene. 20:8085–8091. 2001.
View Article : Google Scholar
|
|
12
|
Jin T, Branch DR, Zhang X, Qi S, Youngson
B and Goss PE: Examination of POU homeobox gene expression in human
breast cancer cells. Int J Cancer. 81:104–112. 1999. View Article : Google Scholar
|
|
13
|
Chen Z, Xu WR, Qian H, Zhu W, Bu XF, Wang
S, Yan YM, Mao F, Gu HB, Cao HL, et al: Oct4, a novel marker for
human gastric cancer. J Surg Oncol. 99:414–419. 2009. View Article : Google Scholar
|
|
14
|
Iki K and Pour PM: Expression of Oct4, a
stem cell marker, in the hamster pancreatic cancer model.
Pancreatology. 6:406–413. 2006. View Article : Google Scholar
|
|
15
|
Saigusa S, Tanaka K, Toiyama Y, Yokoe T,
Okugawa Y, Ioue Y, Miki C and Kusunoki M: Correlation of CD133,
OCT4, and SOX2 in rectal cancer and their association with distant
recurrence after chemoradiotherapy. Ann Surg Oncol. 16:3488–3498.
2009. View Article : Google Scholar
|
|
16
|
Li B, Yao Z, Wan Y and Lin D:
Overexpression of OCT4 is associated with gefitinib resistance in
non-small cell lung cancer. Oncotarget. 7:77342–77347. 2016.
|
|
17
|
Zhou J, Dong D, Cheng R, Wang Y, Jiang S,
Zhu Y, Fan L, Mao X, Gui Y, Li Z, et al: Aberrant expression of
KPNA2 is associated with a poor prognosis and contributes to OCT4
nuclear transportation in bladder cancer. Oncotarget.
7:72767–72776. 2016.
|
|
18
|
Asadi MH, Khalifeh K and Mowla SJ: OCT4
spliced variants are highly expressed in brain cancer tissues and
inhibition of OCT4B1 causes G2/M arrest in brain cancer cells. J
Neurooncol. 130:455–463. 2016. View Article : Google Scholar
|
|
19
|
Hochedlinger K, Yamada Y, Beard C and
Jaenisch R: Ectopic expression of Oct-4 blocks progenitor-cell
differentiation and causes dysplasia in epithelial tissues. Cell.
121:465–477. 2005. View Article : Google Scholar
|
|
20
|
Cai S, Geng S, Jin F, Liu J, Qu C and Chen
B: POU5F1/Oct-4 expression in breast cancer tissue is significantly
associated with non-sentinel lymph node metastasis. BMC Cancer.
16:1752016. View Article : Google Scholar
|
|
21
|
Wang D, Lu P, Zhang H, Luo M, Zhang X, Wei
X, Gao J, Zhao Z and Liu C: Oct-4 and Nanog promote the
epithelial-mesenchymal transition of breast cancer stem cells and
are associated with poor prognosis in breast cancer patients.
Oncotarget. 5:10803–10815. 2014.
|
|
22
|
Wang P, Branch DR, Bali M, Schultz GA,
Goss PE and Jin T: The POU homeodomain protein OCT3 as a potential
transcriptional activator for fibroblast growth factor-4 (FGF-4) in
human breast cancer cells. Biochem J. 375:199–205. 2003. View Article : Google Scholar
|
|
23
|
McLeskey SW, Kurebayashi J, Honig SF,
Zwiebel J, Lippman ME, Dickson RB and Kern FG: Fibroblast growth
factor 4 transfection of MCF-7 cells produces cell lines that are
tumorigenic and metastatic in ovariectomized or tamoxifen-treated
athymic nude mice. Cancer Res. 53:2168–2177. 1993.
|
|
24
|
Kim RJ and Nam JS: OCT4 expression
enhances features of cancer stem cells in a mouse model of breast
cancer. Lab Anim Res. 27:147–152. 2011. View Article : Google Scholar
|
|
25
|
Pain D, Chirn GW, Strassel C and Kemp DM:
Multiple retropseudogenes from pluripotent cell-specific gene
expression indicates a potential signature for novel gene
identification. J Biol Chem. 280:6265–6268. 2005. View Article : Google Scholar
|
|
26
|
Suo G, Han J, Wang X, Zhang J, Zhao Y,
Zhao Y and Dai J: Oct4 pseudogenes are transcribed in cancers.
Biochem Biophys Res Commun. 337:1047–1051. 2005. View Article : Google Scholar
|
|
27
|
Takeda J, Seino S and Bell GI: Human Oct3
gene family: cDNA sequences, alternative splicing, gene
organization, chromosomal location, and expression at low levels in
adult tissues. Nucleic Acids Res. 20:4613–4620. 1992. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Gwak JM, Kim M, Kim HJ, Jang MH and Park
SY: Expression of embryonal stem cell transcription factors in
breast cancer: Oct4 as an indicator for poor clinical outcome and
tamoxifen resistance. Oncotarget. 8:36305–36318. 2017. View Article : Google Scholar
|
|
30
|
Kaufhold S, Garbán H and Bonavida B: Yin
Yang 1 is associated with cancer stem cell transcription factors
(SOX2, OCT4, BMI1) and clinical implication. J Exp Clin Cancer Res.
35:842016. View Article : Google Scholar
|
|
31
|
Liu T, Sun B, Zhao X, Li Y, Gu Q, Dong X
and Liu F: OCT4 expression and vasculogenic mimicry formation
positively correlate with poor prognosis in human breast cancer.
Int J Mol Sci. 15:19634–19649. 2014. View Article : Google Scholar
|
|
32
|
Wang X, Zhao Y, Xiao Z, Chen B, Wei Z,
Wang B, Zhang J, Han J, Gao Y, Li L, et al: Alternative translation
of OCT4 by an internal ribosome entry site and its novel function
in stress response. Stem Cells. 27:1265–1275. 2009. View Article : Google Scholar
|
|
33
|
Zhao S, Yuan Q, Hao H, Guo Y, Liu S, Zhang
Y, Wang J, Liu H, Wang F, Liu K, et al: Expression of OCT4
pseudogenes in human tumours: Lessons from glioma and breast
carcinoma. J Pathol. 223:672–682. 2011. View Article : Google Scholar
|
|
34
|
Cotsarelis G, Sun TT and Lavker RM:
Label-retaining cells reside in the bulge area of pilosebaceous
unit: Implications for follicular stem cells, hair cycle, and skin
carcinogenesis. Cell. 61:1329–1337. 1990. View Article : Google Scholar
|
|
35
|
Aguilar-Gallardo C and Simón C: Cells,
stem cells, and cancer stem cells. Semin Reprod Med. 31:5–13. 2013.
View Article : Google Scholar
|
|
36
|
Ponti D, Costa A, Zaffaroni N, Pratesi G,
Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar
|
|
37
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar
|