Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed malignant tumors in the digestive system, with an
age-standardized incidence rate of 36.3 per 100,000 individuals in
more developed countries and 13.7 per 100,000 individuals in less
developed countries (1). According
to the estimated data from the National Cancer Institute,
colorectal cancer accounts for approximately 8% of all patients
suffering from any type of tumor in 2017, regardless of sex
(2). The proximal segments of the
colon, cecum and ascending colon are considered similar to the
right-sided colon, and the morbidity and mortality of carcinomas of
these segments are included within right-sided colon carcinomas
(RCCs).
Due to its different embryological origins, the
colon is divided into two distinct parts: the proximal colonic
segments, which are developed from the midgut, and the distal
colonic segments, which originate from the hindgut (3). Considering the distal transverse
colon as the boundary, the proximal colon (right-sided) contains
the cecum, ascending colon and proximal two-thirds of the
transverse colon, whereas the distal colon (left-sided) includes
the distal one-third of the transverse colon, descending colon and
sigmoid colon (4,5). Based on the location of the primary
tumor, CRCs can be defined as either RCCs or left-sided colon
carcinomas (LCCs), each of which are associated with their
respective embryological origins (6,7).
Recent studies have demonstrated several differences in cancer
morbidity, clinical characteristics, overall survival, molecular
profiling and responses to various therapeutics between RCCs and
LCCs, indicating that RCCs and LCCs may be distinct diseases
(5,7,8).
Adenocarcinomas of the cecum and ascending colon are
both considered RCCs, which exhibit exophytic pathological behavior
and a poorer overall survival than LCCs (8,9).
Developing rapidly from the cecal diverticulum in the 5-week-old
embryo, the cecum differs from the ascending colon, which is
instead developed from the caudal limb of the midgut loop. Although
the cecum and ascending colon are both considered to originate from
the midgut, there may be differences between adenocarcinomas of the
cecum and ascending colon due to their different locations of
origin and developmental processes. The cecum, a junctional
structure separated from the ileum by the ileocecal valve, receives
chyme from the ileum and connects to the ascending colon. While the
cecum is usually intraperitoneal, the ascending colon is
retroperitoneal.
Different molecular carcinogenesis pathways are
considered another reason for the heterogeneous clinical behaviors
of CRCs, and elucidating these pathways may enhance our
understanding of CRCs, and may thus aid in making more appropriate
choices for therapeutic strategies (5,10).
In CRCs, three classical molecular carcinogenesis pathways have
been identified: the chromosomal instability (CIN), microsatellite
instability (MSI) and CpG island methylator phenotype (CIMP)
pathways (10,11). Additionally, several genes
associated with stem cell initiation are positively expressed in
RCCs, such as octamer-binding transcription factor 4 (Oct4)A
and ATP-binding cassette sub-family G member 2 (ABCG2) (5). Within RCCs, variances in carcinogenic
pathways and gene expression levels are still controversial and
require further investigation.
Therefore, the aim of this study was to analyze the
different clinicopathological characteristics of patients with
adenocarcinomas of the cecum and ascending colon and to further
investigate the potential genes with differential expression levels
in RCCs. These findings may explain the reasons for the differences
between carcinomas of the cecum and ascending colon.
Patients and methods
Patients
Data collected included the demographic and
pathological characteristics and survival (overall survival and
cancer-specific survival) of patients with adenocarcinoma of the
cecum and ascending colon. All patients were reported between 2004
and 2013 in the Surveillance, Epidemiology, and End Results (SEER)
database. The inclusion criteria were as follows: i) Patients with
pathologically diagnosed adenocarcinoma of the cecum and ascending
colon; and ii) Patients who underwent surgery and for whom exact
pathological details were available. Another dataset of patients
who fulfilled the inclusion criteria was collected from 2007 to
2011 at the First Affiliated Hospital of Xi'an Jiaotong University
in order to validate the functions of genes analyzed below. To
avoid the bias caused by the limitation of the retrospective
studies, the propensity score method was employed to solve the
problem of imbalance in baseline characteristics between the two
subgroups of carcinoma of the cecum and that of the ascending
colon. For our study, a signed SEER research data agreement form
was provided to the SEER program, and approval was granted to us to
access and analyze the SEER data. This study was also approved by
the Ethics Committee of the First Affiliated Hospital of Xi'an
Jiaotong University, Xi'an, China and all experiments were
performed in accordance with relevant guidelines and regulations.
All patients signed informed consent forms.
X-tile analysis
X-tile (Rimm Laboratory, Yale School of Medicine,
New Haven, CT, USA) was used to determine the optimal cut-off point
for predicting cancer-specific survival according to the number of
lymph nodes examined cases of carcinoma of the cecum and ascending
colon. X-tile creates separate training and validation cohorts by
first making separate lists of 'censored' and 'uncensored'
observations, which are ordered by the follow-up time. Patients are
alternately assigned to training and validation sets by reading
down the list and selecting every other patient. This technique
normalizes the base survival curve for both sets.
Gene Expression Omnibus (GEO) dataset
collection
All gene expression datasets were downloaded from
the NCBI GEO Database (http://www.ncbi.nlm.nih.gov). The exact location of
the colon segments must be determined for primary tumor analysis.
The final datasets (GEO microarray ID: GSE41258) included 32 cases
of carcinoma of the ascending colon, 29 cases of carcinoma of the
cecum, 47 cases of hepatic metastatic colon cancer and 17 normal
liver samples. The datasets were downloaded in the .CEL format.
Data normalization
Normalization of expression data helps adjust
individual hybridization intensities and balances data
appropriately so that meaningful biological comparisons can be
made. Moreover, this process clusters data so that the points are
less scattered. Data normalization was performed using
Bioconductor's RMA package.
Differential expression analysis and
clustering analysis
Differential expression analysis was performed using
the R 'limma' package. Based on the limma output for the most
differentially expressed genes, unsupervised hierarchical
clustering analysis was used to discover the gene expression
patterns of groups sharing common characteristics. Heatmap and
Volcano plots were constructed using R software.
TCGA based Kaplan-Meier plot
analysis
The prognostic value of queried genes in hepatic
metastatic colon cancer and non-metastatic colon cancer was
analyzed using PPISURV (http://www.bioprofiling.de). The overall survival of
patients with high and low levels of the queried gene was shown
using a Kaplan-Meier survival plot.
Oncomine database validation
analysis
Single-gene mRNA expression levels in the cases of
carcinoma of the cecum, colon cancer and metastatic sites were
compared with their matched normal tissues using TCGA and other
datasets in the Oncomine database (http://www.oncomine.org). The threshold to obtain the
most significant probes of the queried gene for each microarray
data included a 2-fold difference in the expression between cancer
tissues and normal tissues with a P-value <0.0001. Genes
co-expressed with a queried gene were also analyzed, and the map
was generated using Cytoscape 3.4.0.
Human Protein Atlas (HPA)
Immunohistochemical images were downloaded from the
publicly available HPA(http://www.proteinatlas.org). HPA version 8.0 is a
database of tissue microarray (TMA) images labeled with antibodies
against 11,250 human proteins. The tissue microarrays consist of
sections from 46 normal human tissues and 20 different human cancer
types.
RNA extraction and reverse
transcription-quantitative (real-time) PCR (RT-qPCR)
Total cellular RNA from all tissues was extracted
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified
by spectrophotometry. For RT-qPCR analysis, reverse transcription
was performed using the RevertAid First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Waltham, MA, USA) according to the
manufacturer's instructions. The forward primer sequence of
SLCO1B3 was 5′-ACAGCAGAGTCAGCATCTTCAG-3′ and the reverse
primer sequence of SLCO1B3 was 5′-ATCACAAGCAAATTTCCAATTT-3′. The
two-step SYBR-Green I (Takara Bio, Dalian, China) real-time PCR
conditions were as follows: Step 1: 95°C for 30 sec; step 2: 95°C
for 5 sec and 60°C for 30 sec for 40 cycles. The relative mRNA
level was expressed as the fold change relative to that of GAPDH
(forward, 5′-TCGACAGTCAGCCGCATCTTCTTT-3′ and reverse,
5′-ACCAAATCCGTTGACTCCGACCTT-3′). The 2−ΔΔCq method was
utilized to calculate the relative mRNA expression quantitatively
(12).
Statistical analysis
Continuous data are presented as the means ±
standard deviation. Categorical variables were grouped and compared
using the χ2 test. Continuous variables were compared
using the Student's t-test. Univariate and multivariate Cox
proportional hazard regression models were constructed to explore
the associations between clinicopathological factors and
cancer-specific survival. All parameters that were statistically
significant in the univariate analysis were included in the
multivariate Cox model. Cancer-specific survival was estimated
using the Kaplan-Meier method, and differences in survival were
examined using the log-rank test. All statistical tests were
two-sided, and P-values <0.05 were considered to indicate
statistically significant differences. Statistical analyses were
performed using SPSS 13.0 and R software version 3.3.0 (http://www.r-project.org) with the 'SEERaBomb',
'affy', 'affyPLM', 'ape', 'CBPS', 'RMA', 'limma', 'MatchIt' and
'PSAgraphics' packages.
Results
Patient characteristics
A total of 59,035 cases of carcinoma of the cecum
and ascending colon without distant metastasis were identified from
the SEER database. Of these, 31,362 were carcinomas of the cecum
cancer and 27,673 were carcinomas of the ascending colon. Carcinoma
of the cecum was more commonly observed in female patients (56.3%),
and it had a significantly higher prevalence than that of carcinoma
of the ascending colon. Patients older than 60 years accounted for
77.9% of the cases of carcinoma of the cecum and 78.3% of the cases
of carcinoma of the ascending colon. The depth of invasion and
regional lymph node metastasis differed significantly between the
two subgroups. However, the histological grade and mucous
expression did not exhibit obvious differences (the detailed
demographic and pathological information of the patients is
presented in Table I).
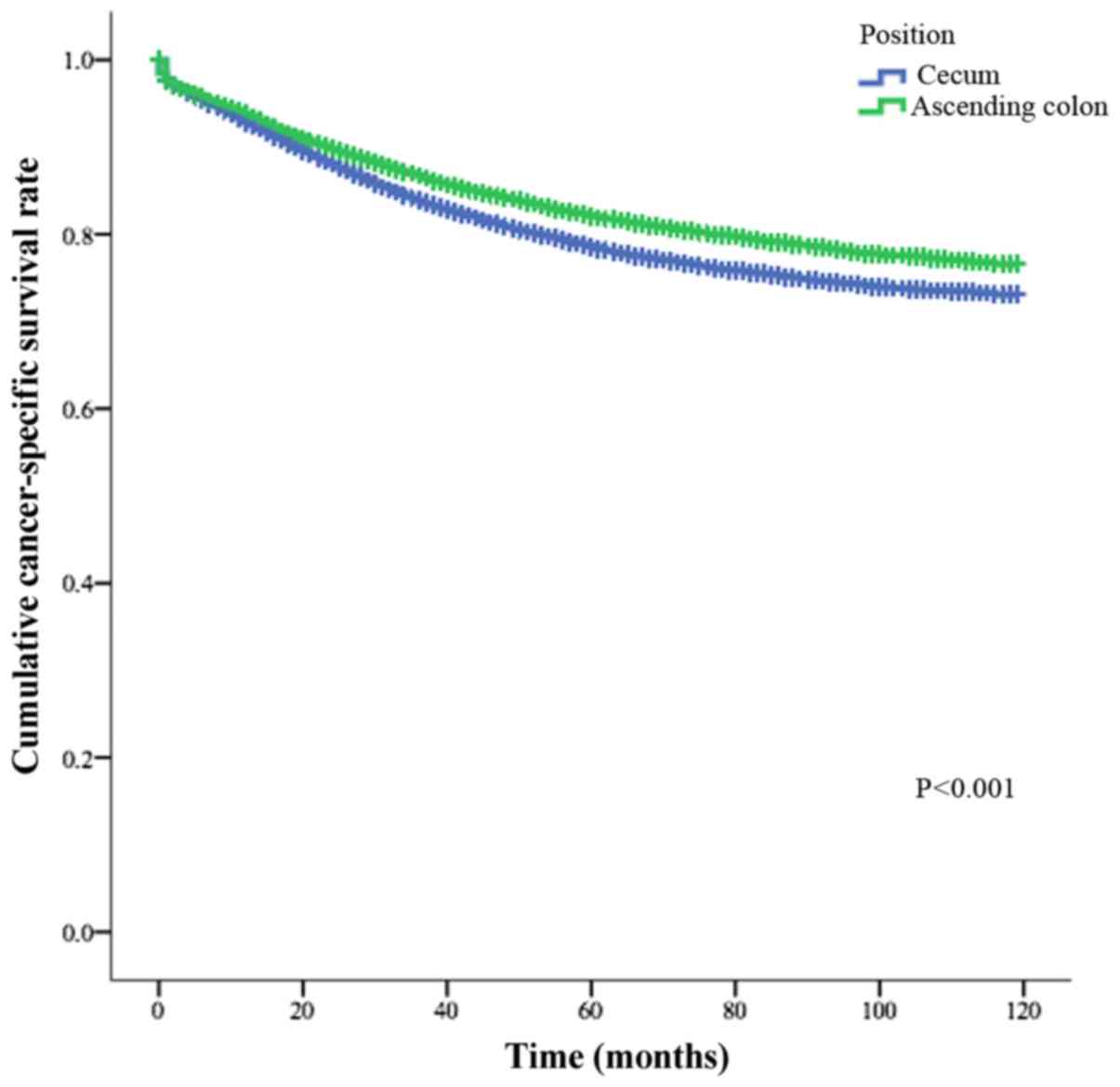
Furthermore, the cancer-specific survival of the patients with
carcinoma of the ascending colon was significantly greater than
that of patients with carcinoma of the cecum (Fig. 1, P<0.001).
 | Table IThe demographic and pathological
information of patients with carcinomas of the cecum and ascending
colon. |
Table I
The demographic and pathological
information of patients with carcinomas of the cecum and ascending
colon.
| Characteristic | Cecum
| Ascending colon
| P-valuea |
|---|
| Number | Percentage | Number | Percentage |
|---|
| Sex | | | | | |
| Male | 13,719 | 43.7 | 12,664 | 45.8 | |
| Female | 17,643 | 56.3 | 15,009 | 54.2 | <0.001 |
| Age (years) | | | | | |
| <40 | 535 | 1.7 | 502 | 1.8 | |
| 40–60 | 6,388 | 20.4 | 5,500 | 19.9 | |
| >60 | 24,439 | 77.9 | 21,671 | 78.3 | 0.219 |
| Race | | | | | |
| Caucasian | 25,640 | 81.8 | 22,251 | 80.4 | |
| Of African
descent | 3,827 | 12.2 | 3,371 | 12.2 | |
| Other | 1,895 | 6 | 2,051 | 7.4 | <0.001 |
| Depth of
invasion | | | | | |
| T1 | 4,430 | 14.1 | 4,398 | 15.9 | |
| T2 | 5,873 | 18.7 | 4,596 | 16.6 | |
| T3 | 16,601 | 52.9 | 16,219 | 58.6 | |
| T4 | 4,458 | 14.2 | 2,460 | 8.9 | <0.001 |
| Metastatic lymph
nodes | | | | | |
| N0 | 20,078 | 64 | 18,777 | 67.9 | |
| N1 | 6,906 | 22 | 5,904 | 21.3 | |
| N2 | 4,378 | 14 | 2,992 | 10.8 | <0.001 |
| Grade | | | | | |
| G1 | 3,203 | 10.2 | 2,897 | 10.5 | |
| G2 | 21,470 | 68.5 | 18,860 | 68.2 | |
| G3 | 5,914 | 18.9 | 5,271 | 19 | |
| G4 | 775 | 2.5 | 645 | 2.3 | 0.455 |
| Mucous | | | | | |
| With | 3,799 | 12.1 | 3,384 | 12.2 | |
| Without | 27,563 | 87.9 | 24,289 | 87.8 | 0.669 |
In order to validate the differences in the
cancer-specific survival of the two subgroups in the retrospective
study, propensity score analysis (PSA, matching method) (13) was used. After the cases of
carcinoma of the cecum and ascending colon were matched by
histological characteristics, metastatic lymph nodes, depth of
invasion, race, sex and surgical approach, 27,670 paired patients
were included in the validation set. The jittered plot of the
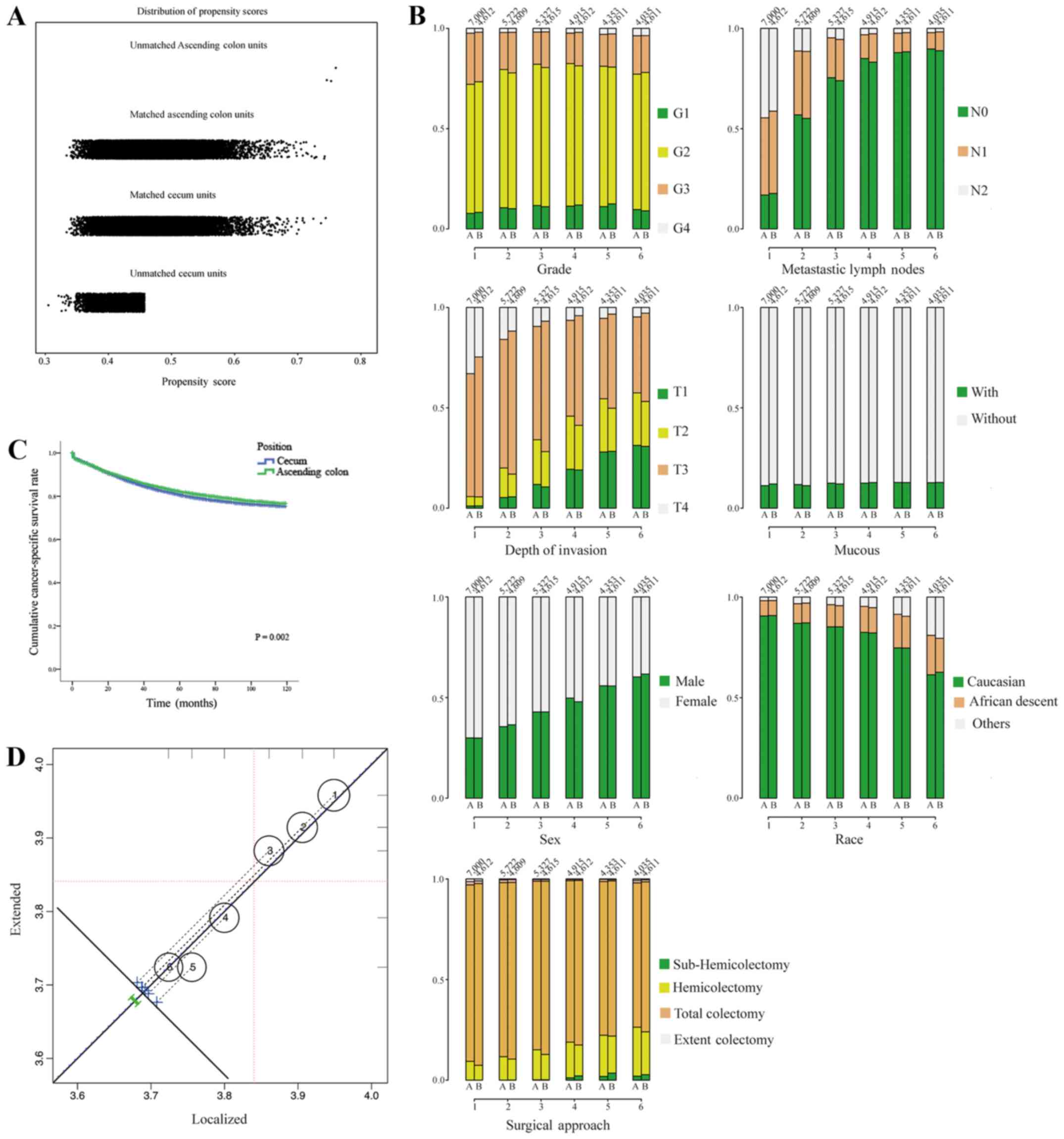
matched and unmatched observations, as well as the distributions of
their propensity score values are presented in Fig. 2A. In addition, side-by-side
barplots comparing the proportion of cases in each category for a
variety of risk factors are presented in Fig. 2B. After PSA matching, the results
also indicated that patients with carcinoma of the ascending colon
had a better cancer-specific survival than those with carcinoma of
the cecum (Fig. 2C). Furthermore,
surgical approaches, as the factors influenced by human beings,
were analyzed in the PSA analysis. The contributions of surgical
approaches (localized surgery or extended surgery) to the overall
effect with the weighting of contributions of individual strata
according to the relative sizes of the respective strata are shown
in Fig. 2D. The overall effect is
plotted as a heavy dashed diagonal line that runs parallel to the
identity diagonal. The influence of the surgical approach was
relatively limited in the PSA matching.
The risk factors of the cancer-specific survival of
the two subgroups were then analyzed. Table II depicts the prognostic value of
9 patient characteristics on the adjusted cancer-specific survival.
For patients with carcinoma of the cecum, age, race, depth of
invasion, regional lymph node metastasis, the number of resected
lymph nodes and histological characteristics were significant
factors affecting cancer-specific survival. In addition to the 6
factors listed above, the male sex was associated with a
significantly increased risk of cancer-specific mortality [hazard
ratio (HR) = 0.899; 95% CI, 0.862 to 0.938; P<0.001] of patients
with carcinoma of the ascending colon. Extended surgery for the
treatment of carcinoma of the ascending colon had a better
cancer-specific survival in the univariate analysis. However, this
result did not occur in patients with carcinoma of the cecum
(Table II).
 | Table IIUnivariate and multivariate analyses
of patients (after matching) with carcinomas of the cecum and
ascending colon. |
Table II
Univariate and multivariate analyses
of patients (after matching) with carcinomas of the cecum and
ascending colon.
| Characteristic | Cecum cancer
| Ascending colon
cancer
|
|---|
Univariate analysis
| Multivariate
analysis
| Univariate analysis
| Multivariate
analysis
|
|---|
| P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value |
|---|
| Sex | | | | | | |
| Male | | | | | | |
| Female | 0.151 | | | 0.02 | 0.899
(0.862–0.938) | <0.001 |
| Age (years) | | | | | | |
| <40 | | | | | | |
| 40–60 | | | | | | |
| >60 | <0.001 | 2.419
(2.284–2.562) | <0.001 | <0.001 | 2.498
(2.341–2.666) | <0.001 |
| Race | | | | | | |
| Caucasian | | | | | | |
| Of African
descent | | | | | | |
| Other | <0.001 | 0.926
(0.892–0.961) | <0.001 | <0.001 | 0.893
(0.859–0.929) | <0.001 |
| Depth of
invasion | | | | | | |
| T1 | | | | | | |
| T2 | | | | | | |
| T3 | | | | | | |
| T4 | <0.001 | 1.034
(1.032–1.037) | <0.001 | <0.001 | 1.029
(1.026–1.033) | <0.001 |
| Metastatic lymph
nodes | | | | | | |
| N0 | | | | | | |
| N1 | | | | | | |
| N2 | <0.001 | 1.040
(1.037–1.042) | <0.001 | <0.001 | 1.041
(1.038–1.044) | <0.001 |
| Examined lymph
nodes | | | | | | |
| <12 | | | | | | |
| ≥12 | <0.001 | 0.652
(0.626–0.679) | <0.001 | <0.001 | 0.662
(0.632–0.694) | <0.001 |
| Grade | | | | | | |
| G1 | | | | | | |
| G2 | | | | | | |
| G3 | | | | | | |
| G4 | <0.001 | 1.090
(1.055–1.126) | <0.001 | <0.001 | 1.129
(1.089–1.170) | <0.001 |
| Mucous | | | | | | |
| With | | | | | | |
| Without | 0.056 | | | <0.001 | 0.961
(0.904–1.021) | 0.199 |
| Surgical
approacha | | | | | | |
| Sub-hemi | | | | | | |
| Hemi | | | | | | |
| Total | | | | | | |
| Extent | 0.159 | | | <0.001 | 1.017
(0.962–1.075) | 0.552 |
Identification of optimal cut-off points
for regional lymph nodes examined in cases of carcinoma of the
cecum and ascending colon
Patients with carcinoma of the cecum had a median of
18.51 lymph nodes examined, and patients with carcinoma of the
ascending colon had 19.04 lymph nodes examined. The proportion of
patients with carcinoma of the ascending colon with node positivity
was 32.15%, almost the same as that for patients with carcinoma of
the cecum (31.32%). The depth of invasion was associated with the
rate of node positivity. Furthermore, compared with the patients
with carcinoma of the cecum, patients with carcinoma of the
ascending colon were more frequently examined, with >12 lymph
nodes (data not shown).
Compared with carcinoma of the ascending colon,
carcinoma of the cecum had a special anatomic location, which was
in the pelvis and near the rectum (data not shown). For patients
with carcinoma of the cecum, the extended surgical approach did not
provide cancer-specific survival benefits, irrespective of the
depth of tumor invasion. However, this phenomenon was not observed
in patients with carcinoma of the ascending colon, which might
indicate that the 'right colon' is not a single organ. Thus,
different surgical strategies should be adopted for the two parts
of the right colon. For example, localized surgery may be
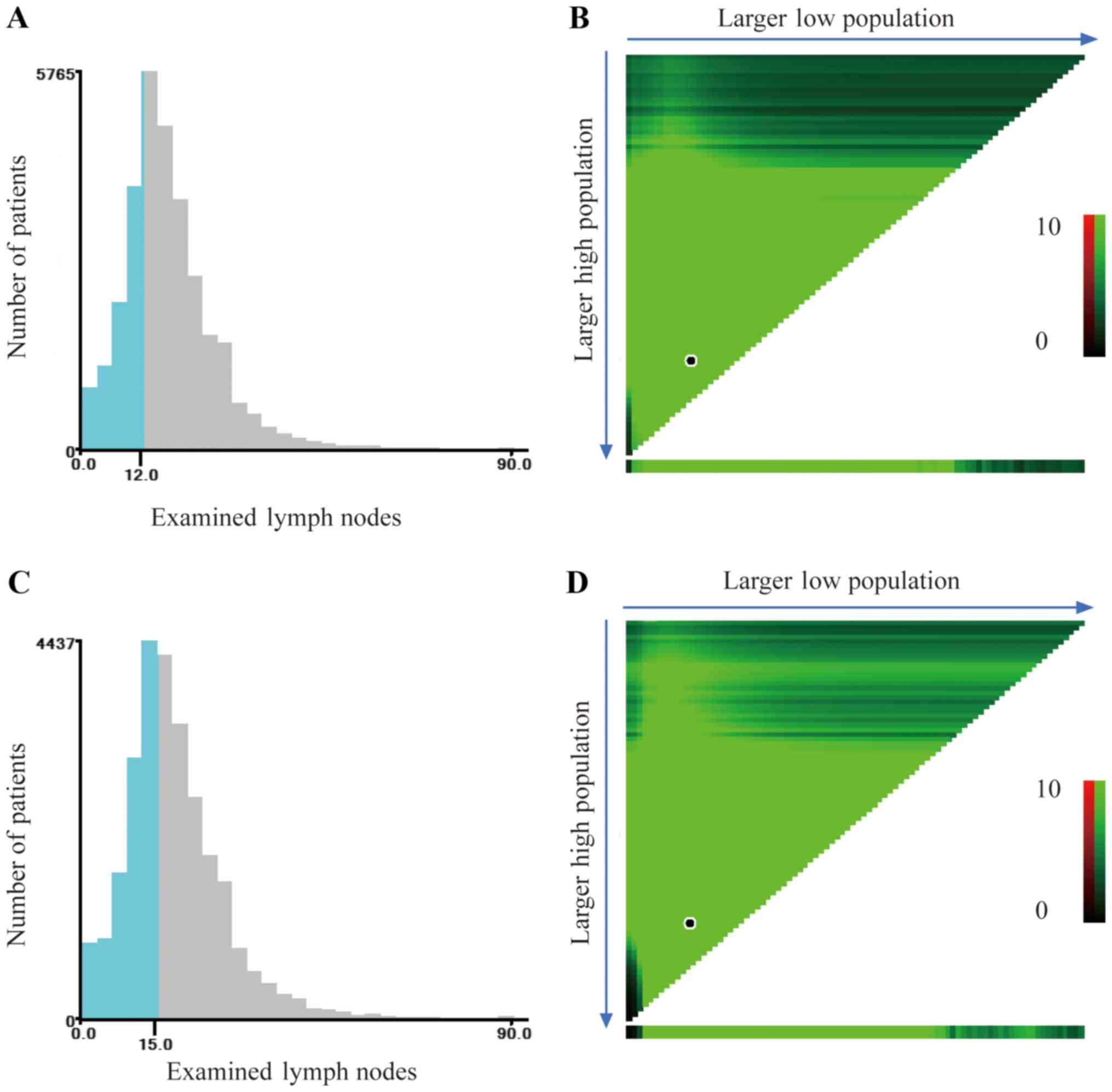
sufficient for carcinoma of the cecum. X-tile was used to determine
the optimal cut-off point for predicting cancer-specific survival
according to the number of examined lymph nodes. X-tile analysis
indicated that the maximum χ2 log-rank value was
produced with 12 as the cut-off value to identify patients with
carcinoma of the cecum with the strongest discriminatory capacity
(P<0.001) (Fig. 3A and B). With
the same method, we identified the optimal cut-off value of 15 for
patients with carcinoma of the ascending colon, corresponding to
the maximum of the χ2 log-rank value (P<0.001)
(Fig. 3C and D).
Patients with carcinoma of the cecum, as described
above, had a significantly lower cancer-specific survival than
those with carcinoma of the ascending colon. However, the results
indicated that the localized surgical strategy (sub-hemi and
hemicolectomy) may be sufficient, possibly as carcinoma of the
cecum is more likely to form distant metastasis, the strongest
prognostic factor for malignant tumors (Table II). The results indicated that
15.3% of the cases of carcinoma of the cecum had distant
metastasis, which was significantly higher than the number of
patients with carcinoma of the ascending colon with distant
metastasis (11.7%) (data not shown).
The mRNA expression patterns in carcinoma
of the cecum and ascending colon
We searched the GEO database and found 29 cases of
carcinoma of the cecum, 32 cases of carcinoma of the ascending
colon, 5 normal cecum tissues and 12 normal ascending colon tissues
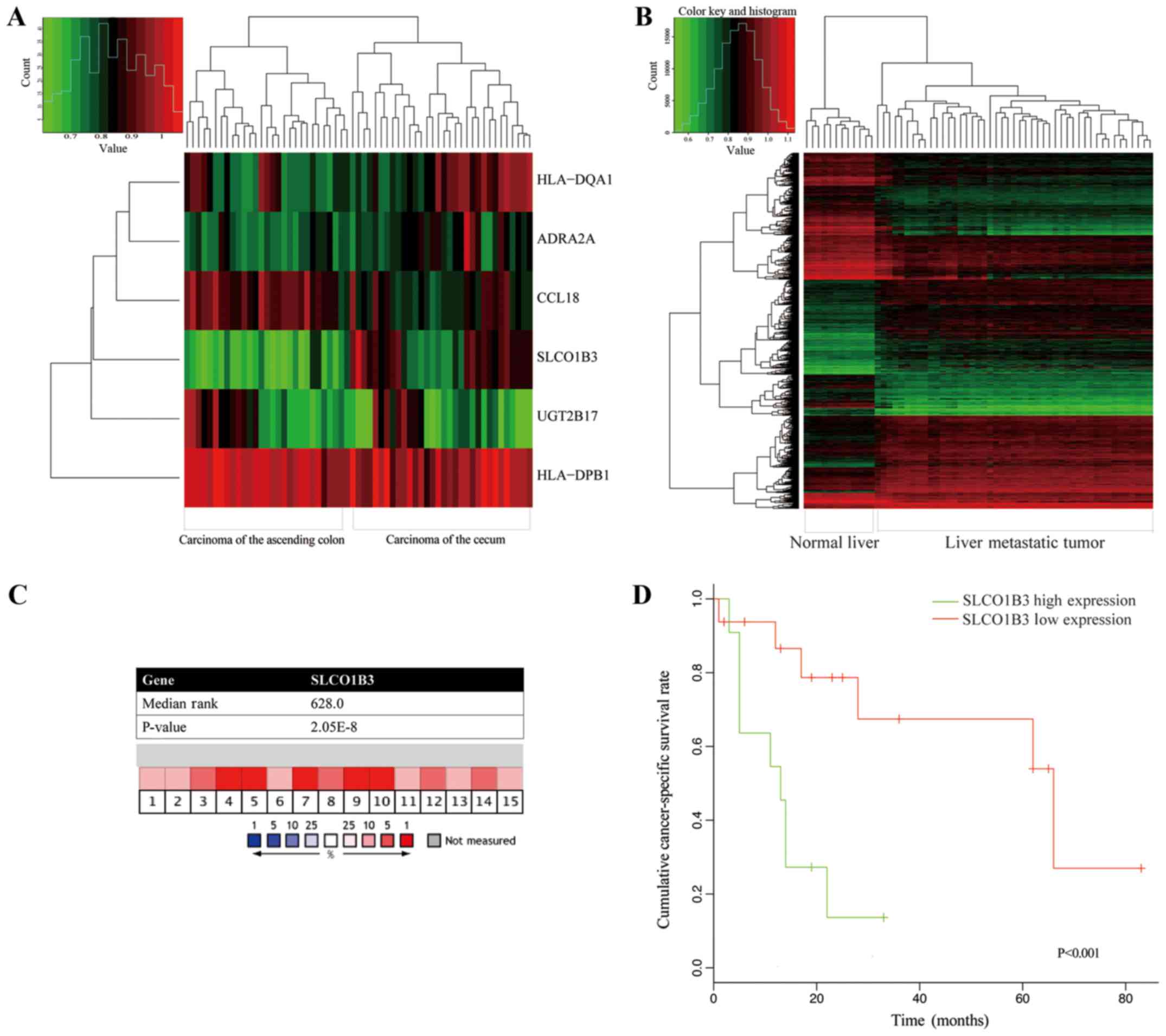
that met the inclusion criteria. A total of 6 genes (Fig. 4A) identified, the expression of
which differed significantly between the cases of carcinoma of the
cecum and those of carcinoma of the ascending colon. All 6 of these
genes were different between the cancer tissues and normal mucosa
(P<0.001, data not shown).
Furthermore, in the same GEO database, 47 liver
metastatic tumors (all from colon cancer) and 12 normal hepatic
tissues were used to analyze the differences in gene expression. In
total, 1,921 genes (Fig. 4B) were
identified with a significant difference between metastatic hepatic
tumors and normal liver tissues. Among these, 4 genes (ADRA2A,
SLCO1B3, HLA-DQA1 and UGT2B17) of carcinoma of the
ascending colon and cecum were also shown to have significant
differences in the hepatic tissues.
Of the 4 matched genes, SLCO1B3 encodes a
liver-specific member of the organic anion transporter, and it may
play an important role in hepatic metastasis. Validating the gene
expression differences between carcinoma of the cecum and ascending
colon, Oncomine analysis of cancer vs. normal tissue indicated that
SLCO1B3 mRNA expression was significantly high in colon
adenocarcinoma (Fig. 4C).
Kaplan-Meier analysis revealed that a high SLCO1B3 mRNA
expression was associated with the poor survival of patients with
colon cancer with distant metastasis (Fig. 4D). In the Human Protein Atlas (HPA)
database, SLCO1B3 protein expression was detected in 27% of
patients with colorectal cancer, while none was detected in normal
colon tissues.
SLCO1B3 is associated with the liver
metastasis rate
We evaluated the expression of SLCO1B3 in 32
cases of carcinoma of the ascending colon and cecum (stages I and
III) who underwent resection of the primary lesion. We performed
RT-qPCR using the RNA from these cancer tissues. SLCO1B3
expression was higher in the tissues obtained from patients with
carcinoma of the ascending colon or cecum that ultimately had liver
metastasis (11 patients) than in the tissues obtained from cases
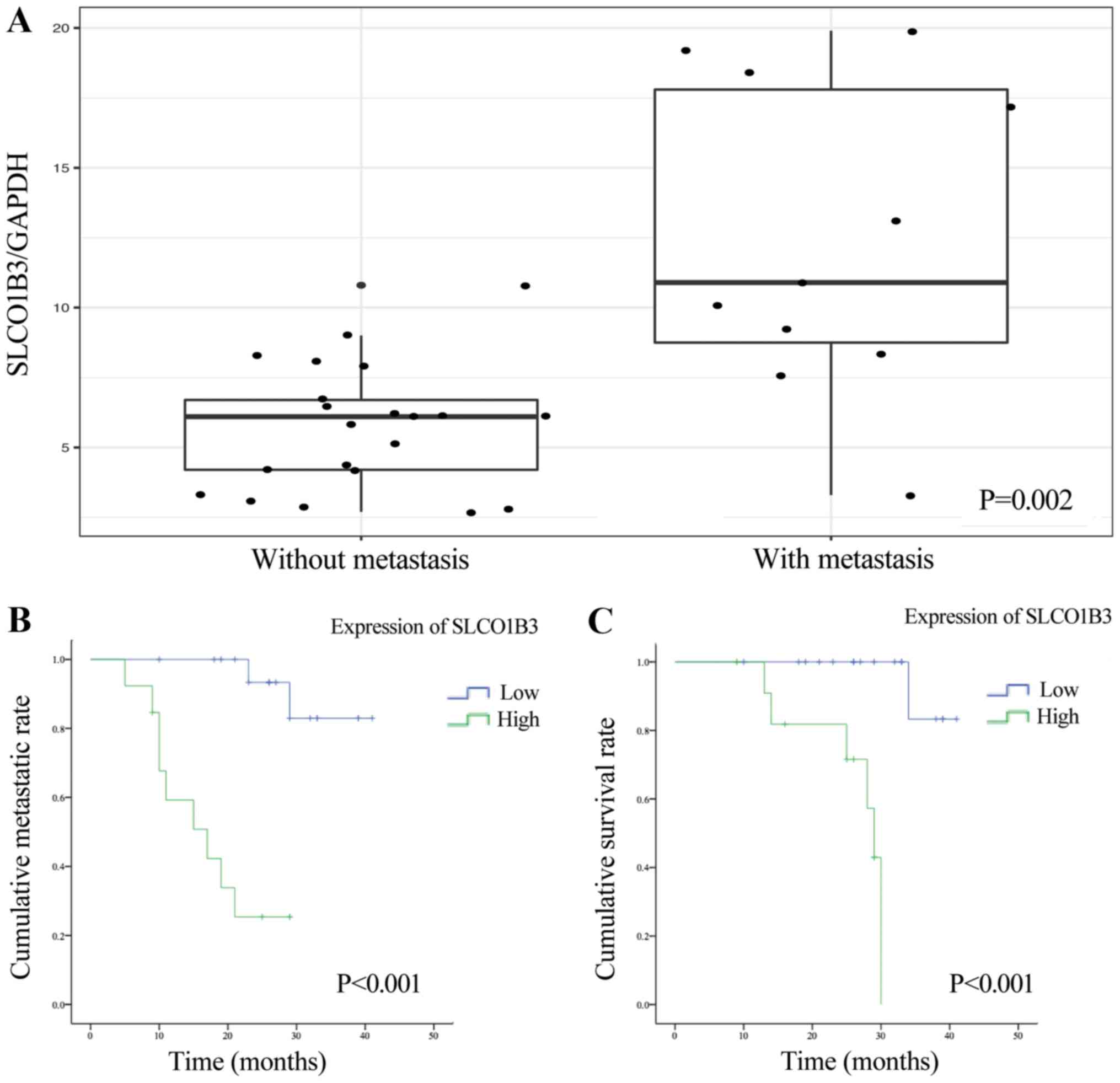
without metastasis (21 patients) (Fig.
5A, P=0.002). Tumors that metastasized to the liver had a
median relative SLCO1B3 expression of 12.47±5.50
(SLCO1B3 mRNA expression/GAPDH mRNA expression/%
calibrator) compared to a median relative SLCO1B3 expression
of 5.73±2.25 (SLCO1B3 mRNA expression/GAPDH mRNA
expression/% calibrator) in cases without metastases.
Furthermore, we subdivided the patients into 2
groups according to the median expression of SLCO1B3. After
performing log-rank analysis of the 2 groups, we found that
patients with a high SLCO1B3 expression were more likely to
have liver metastasis and a shorter time to distant metastasis than
those with a low SLCO1B3 expression (Fig. 5B, P<0.001). Finally, we
evaluated SLCO1B3 as a potential prognostic marker for
patient survival. We also performed Kaplan-Meier survival analysis
based on SLCO1B3 expression in primary tumors. Patients with
a high SLCO1B3 expression in their primary tumors had a
significantly shorter cancer-specific survival time than those with
a low SLCO1B3 expression (Fig.
5C, P<0.001).
Discussion
Due to the similar embryonic origin and adjacent
anatomic location in the gastrointestinal tract, the cecum and
ascending colon are considered the proximal and initial parts of
the right-sided colon, respectively (3,4).
Adenocarcinomas of the cecum or ascending colon are also classified
into the category of RCCs, with identical therapeutic strategies
and prognostic management for both adenocarcinomas, although there
are potential differences between these two 'similar'
adenocarcinomas (14,15). In this study, based on the analysis
of clinicopathological data from the SEER database for 59,035
patients with adenocarcinomas of the cecum or ascending colon, the
differences and associations between the tumor location, age at
diagnosis, race, sex, TNM stage, histological grade, numbers of
lymph node examined intraoperative and cancer-specific survival are
demonstrated. Furthermore, the SLCO1B3 gene, a novel
prognostic risk factor, was preliminarily identified and validated
in our study.
Developed from the caudal limb of the midgut loop,
the cecum is one of the most important junctional parts in the
gastrointestinal system. Anatomically connecting the ileum and
ascending colon via the ileocecal valve, the cecum receives
digested chyme with intestinal fluid and begins to absorb water and
other nutrition physiologically (16–18).
Therefore, it is anatomically and physiologically a transition site
for digestive function. Additionally, located at the initial part
of cecum, the appendix is a blind tube of the cecum and is
considered part of the immune system (19). The ascending colon, in addition to
its similar physiological function, is a simple digestive tract
connecting the cecum and transverse colon (16).
Including adenocarcinomas of the cecum or ascending
colon, RCCs have a worse prognosis than LCCs (5,9,20).
Consistent with the prognostic differences between RCCs and LCCs,
it has been found that tumor location is a significant prognostic
factor in patients with adenocarcinomas of the cecum or ascending
colon (21–23). In this study, compared to
adenocarcinomas of the ascending colon, patients with
adenocarcinomas of the cecum had a worse cancer-specific survival
(Fig. 1), indicating the
heterogeneity within different subtypes of RCC. This variability
may be attributed to the different tumor biological behavior or
potential gene expressive differences between these two subtypes.
We further explored the GEO database for potential genes that may
cause the differences between carcinomas of the cecum and ascending
colon or may be associated with the prognosis. Notably, in this
study, significant differences were observed in the depth of tumor
invasion and regional lymph node metastasis between adenocarcinomas
of the cecum and ascending colon (Table II), which indicated that
intraoperative local lymph node examination and regional
lymphadenectomy are essential in surgical therapy for
adenocarcinomas of the cecum and ascending colon.
The proximal-most portion of the colon is called the
cecum. As a junctional organ, the cecum is generally covered in
visceral peritoneum, and there is greater variability near the
transition to the ascending colon (24). In addition, the ascending colon is
'freed up' with the mobilization of the lateral retroperitoneal
attachments from the cecum to the hepatic flexure. For some
carcinomas of the cecum with serosal invasion, the surgical
strategy may differ for carcinoma of the ascending colon, perhaps
due to the higher likelihood that adenocarcinomas of the cecum will
invade the rectum or other pelvic organs. During the surgical
process, surgeons have to choose total pelvic exenteration for
locally advanced carcinomas of the cecum. In addition, while the
prevalence of interval colorectal cancer in a recent study was
3.28%, over half of interval colorectal cancers are distributed
over the cecum and rectum (25).
Although the number of lymph nodes evaluated for
colon cancer has markedly increased in recent decades (26), differences in nodal evaluation
among different colon or rectum segments continue to exist. It has
been reported that lymph node examination should be able to
discriminate between RCCs and LCCs, and cut-off values of 15 and 11
may be appropriate for patients with RCC and LCC, respectively
(27). In this study, the
resection of 12 lymph nodes was sufficient for carcinomas of the
cecum, while the resection of 15 nodes was required for carcinomas
of the ascending colon. The potential reasons for these differences
in the examined lymph node requirement were multifactorial.
Firstly, the surgeons may play an important role in the variability
of nodal evaluation between carcinomas of the cecum and ascending
colon. The extent of localized resection by surgeons directly
affects the number of examined lymph nodes; however, extended
resection often indicates extended invasion and extended
complications. Secondly, the differences in the immune response and
the molecular characteristics between the cecum and ascending colon
should be considered potential factors influencing the number of
examined lymph nodes. The cecum and ascending colon have different
features related to the immune response, anatomical, physiological
and molecular characteristics (28). For example, the cecum base is the
appendix, which is embryologically derived from the colon and has
been proposed to play a role in immune function. However, the SEER
database lacks chemotherapy information, and the potential
confounding effect of chemotherapy may not be separately assessed
for cecum and ascending colon cancer. Furthermore, adjuvant
chemotherapy plays an important role in the treatment of colon
cancer (29). Despite these
limitations, SEER remains a valuable resource to analyze trends and
patterns in patient characteristics, tumor features, cancer
treatments and survival outcomes (30).
Hepatic metastasis is common in CRC. Konopke et
al revealed that the location of hepatic metastasis could
reflect the location of primary CRC (31). RCCs mainly involve the right
hemi-liver due to the 'streaming' effect, which is influenced by
the hydrodynamic differences between the superior mesenteric and
splenic blood flow (31,32). Despite this blood flow, potential
gene expression differences may cause different characteristics in
hepatic metastasis.
As a potential prognostic risk factor, the
SLCO1B3 gene, which belongs to the solute carrier organic
anion transporter family, encodes the human organic
anion-transporting polypeptide 1B3 (OATP1B3) (33,34).
OATP1B3 has only a hepatic expression, and it transports organic
anions and drugs into the liver, such as digoxin, amanitin,
docetaxel and paclitaxel (34–36).
Expressed in tissues of primary breast cancer and prostate cancer,
SLCO1B3/OATP1B3 is considered a novel risk factor and
potential variable indicating tumor sensitivity to methotrexate
treatment (37–39). As regards CRC, several studies have
demonstrated that SLCO1B3/OATP1B3 is cytoplasmically
detected, which differs from its expression pattern in the normal
liver (34,38). Although the pathobiological
association between CRC and SLCO1B3, a liver-specific member
of the organic anion transporter family, has yet not been
determined, this study suggested that it may be a potential hepatic
metastasis risk factor for adenocarcinoma of the cecum and
ascending colon. Based on our analysis of the GEO database, the
expression of SLCO1B3 was significantly increased in
patients with hepatic metastasis from adenocarcinoma of the cecum
and ascending colon (Fig. 4C,
P<0.001), which indicates its potential utility in hepatic
metastasis prediction. Furthermore, a high SLCO1B3
expression was associated with a poor prognosis of patients with
colon cancer with distant metastasis (Fig. 4D, P<0.001 for Kaplan-Meier
analysis).
Due to the poor prognosis of patients with CRC with
hepatic metastasis (40), we
explored whether SLCO1B3 may be a prognostic risk factor for
patients with adenocarcinomas of the cecum and ascending colon.
Compared with patients with carcinomas of the cecum or ascending
colon without hepatic metastasis, patients with hepatic metastasis
had a higher expression of SLCO1B3 and a poorer
cancer-specific survival (Fig. 5).
Consistent with the findings of previous studies on different
tumors, SLCO1B3 was superior in predicting prognosis
(34,41). Furthermore, in this study, for
patients with adenocarcinomas of the cecum or ascending colon,
SLCO1B3 was suggested to be a novel risk factor for both
prognostic and hepatic metastasis prediction.
Our study has several limitations. Firstly, based on
the SEER database, the chemotherapy treatment details of the
patient were limited, which may influence the analysis of survival
estimation. As cancer is a complex disease requiring
multidisciplinary therapies, systematic chemotherapy strategies may
influence the prognosis of cancer patients. Additionally, although
our retrospective study was based on a large sample size of almost
60,000 patients for over a 10-year period, any subjective
diagnostic criterion, such as the histological grade, can cause
potential biases in the overall analysis. Furthermore, high-quality
prognostic factors should be more clinically homogeneous.
Therefore, our novel SLCO1B3 finding requires further
investigation in other tumors and pathobiological mechanisms.
In conclusion, the results of the present study
revealed the potential prognostic differences between
adenocarcinomas of the cecum and ascending colon, which may be
caused by the differential expression of the SLCO1B3 gene.
Including the expression level of SLCO1B3 in
intraoperatively examined lymph nodes, eight factors could predict
the prognosis for patients with cancers of the cecum and ascending
colon. Regarding surgical therapeutic strategies, resection of more
than 15 local lymph nodes is appropriate for improving
prognosis.
Acknowledgments
The authors of the present study would like to thank
all the authors of the studies cited herein. We also acknowledge
the Surveillance, Epidemiology, and End Results Program (SEER)
database, which provided the data, and the website (http://www.r-project.org), which provided the R
software.
Funding
This study was supported by the National Natural
Scientific Foundation of China (grant no. 81502442).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
All authors participated in the conception and
design of the study. XX, ZZ, YS and WW obtained and analyzed the
data. XX and ZZ drafted the manuscript. CD and HZ revised the
manuscript prior to submission. All authors have read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
A signed SEER research data agreement form was
provided to the SEER program, and approval was granted to us to
access and analyze the SEER data. This study was also approved by
the Ethics Committee of the First Affiliated Hospital of Xi'an
Jiaotong University, Xi'an, China and all experiments were
performed in accordance with relevant guidelines and regulations.
All patients signed informed consent forms.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gervaz P, Bucher P and Morel P: Two
colons-two cancers: Paradigm shift and clinical implications. J
Surg Oncol. 88:261–266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mik M, Berut M, Dziki L, Trzcinski R and
Dziki A: Right- and left-sided colon cancer - clinical and
pathological differences of the disease entity in one organ. Arch
Med Sci. 13:157–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trosko JE and Lenz HJ: What roles do colon
stem cells and gap junctions play in the left and right location of
origin of colorectal cancers? J Cell Commun Signal. 11:79–87. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jess P, Hansen IO, Gamborg M and Jess T;
Danish Colorectal Cancer Group: A nationwide Danish cohort study
challenging the categorisation into right-sided and left-sided
colon cancer. BMJ Open. 3:e0026082013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benedix F, Schmidt U, Mroczkowski P,
Gastinger I, Lippert H and Kube R; Study Group 'Colon/Rectum
Carcinoma (Primary Tumor)': Colon carcinoma - classification into
right and left sided cancer or according to colonic subsite? -
Analysis of 29,568 patients. Eur J Surg Oncol. 37:134–139. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yahagi M, Okabayashi K, Hasegawa H,
Tsuruta M and Kitagawa Y: The worse prognosis of right-sided
compared with left-sided colon cancers: A systematic review and
meta-analysis. J Gastrointest Surg. 20:648–655. 2016. View Article : Google Scholar
|
|
9
|
Wong HL, Lee B, Field K, Lomax A, Tacey M,
Shapiro J, McKendrick J, Zimet A, Yip D, Nott L, et al: Impact of
primary tumor site on bevacizumab efficacy in metastatic colorectal
cancer. Clin Colorectal Cancer. 15:e9–e15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bae JM, Kim JH and Kang GH: Molecular
subtypes of colorectal cancer and their clinicopathologic features,
with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab
Med. 140:406–412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamauchi M, Morikawa T, Kuchiba A, Imamura
Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower
C, et al: Assessment of colorectal cancer molecular features along
bowel subsites challenges the conception of distinct dichotomy of
proximal versus distal colorectum. Gut. 61:847–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
13
|
Zhang Z: Propensity score method: A
non-parametric technique to reduce model dependence. Ann Transl
Med. 5:72017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Network NCC: (NCCN)Clinical Practice
Guidelines in Oncology. Colon Cancer, Version 2. 2017, https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf.
Accessed March 13, 2017.
|
|
15
|
Messersmith WA: Systemic management of
colorectal cancer. J Natl Compr Canc Netw. 15(5S): 699–702. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thompson JS: Core Textbook of Anatomy. 1st
edition. Lippincott Williams & Wilkins; Riverwoods, IL:
1977
|
|
17
|
Guarner F and Malagelada JR: Gut flora in
health and disease. Lancet. 361:512–519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dabareiner RM and White NA II: Diseases
and surgery of the cecum. Vet Clin North Am Equine Pract.
13:303–315. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruoff C, Hanna L, Zhi W, Shahzad G,
Gotlieb V and Saif MW: Cancers of the appendix: Review of the
literatures. ISRN Oncol. 2011:7285792011.PubMed/NCBI
|
|
20
|
Price TJ, Beeke C, Ullah S, Padbury R,
Maddern G, Roder D, Townsend AR, Moore J, Roy A, Tomita Y, et al:
Does the primary site of colorectal cancer impact outcomes for
patients with metastatic disease? Cancer. 121:830–835. 2015.
View Article : Google Scholar
|
|
21
|
Loupakis F, Yang D, Yau L, Feng S,
Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ,
et al: Primary tumor location as a prognostic factor in metastatic
colorectal cancer. J Natl Cancer Inst. 107:dju4272015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He WZ, Liao FX, Jiang C, Kong PF, Yin CX,
Yang Q, Qiu HJ, Zhang B and Xia LP: Primary tumor location as a
predictive factor for first-line bevacizumab effectiveness in
metastatic colorectal cancer patients. J Cancer. 8:388–394. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tejpar S, Stintzing S, Ciardiello F,
Tabernero J, Van Cutsem E, Beier F, Esser R, Lenz HJ and Heinemann
V: Prognostic and predictive relevance of primary tumor location in
patients with RAS wild-type metastatic colorectal cancer:
Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA
Oncol. Oct 10–2016.Epub ahead of print. PubMed/NCBI
|
|
24
|
Timmcke AE: The ASCRS Textbook of Colon
and Rectal Surgery. Ochsner J. 7:482007.
|
|
25
|
Tsai CE, Wu KL, Chiu YC, Chuah SK, Tai WC,
Hu ML and Liang CM: The incidence and clinical associated factors
of interval colorectal cancers in Southern Taiwan. J Formos Med
Assoc. 117:185–190. 2018. View Article : Google Scholar
|
|
26
|
Parsons HM, Tuttle TM, Kuntz KM, Begun JW,
McGovern PM and Virnig BA: Association between lymph node
evaluation for colon cancer and node positivity over the past 20
years. JAMA. 306:1089–1097. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guan X, Chen W, Liu Z, Jiang Z, Hu H, Zhao
Z, Wang S, Chen Y, Wang G and Wang X: Whether regional lymph nodes
evaluation should be equally required for both right and left colon
cancer. Oncotarget. 7:59945–59956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen H, Yang J, Huang Q, Jiang MJ, Tan YN,
Fu JF, Zhu LZ, Fang XF and Yuan Y: Different treatment strategies
and molecular features between right-sided and left-sided colon
cancers. World J Gastroenterol. 21:6470–6478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Connor ES, Greenblatt DY, LoConte NK,
Gangnon RE, Liou JI, Heise CP and Smith MA: Adjuvant chemotherapy
for stage II colon cancer with poor prognostic features. J Clin
Oncol. 29:3381–3388. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guan X, Chen W, Jiang Z, Liu Z, Miao D, Hu
H, Zhao Z, Yang R and Wang X: Exploration of the optimal minimum
lymph node count after colon cancer resection for patients aged 80
years and older. Sci Rep. 6:389012016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Konopke R, Distler M, Ludwig S and
Kersting S: Location of liver metastases reflects the site of the
primary colorectal carcinoma. Scand J Gastroenterol. 43:192–195.
2008. View Article : Google Scholar
|
|
32
|
Desai AG, Park CH and Schilling JF:
'Streaming' in portal vein. Its effect on the spread of metastases
to the liver. Clin Nucl Med. 10:556–559. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
König J, Cui Y, Nies AT and Keppler D:
Localization and genomic organization of a new hepatocellular
organic anion transporting polypeptide. J Biol Chem.
275:23161–23168. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Evangeli L, Ioannis S, Valentinos K,
Antigony M, Elli I, Eleftheria H, Vasiliki G and Evangelos B:
SLCO1B3 screening in colorectal cancer patients using
High-Resolution Melting Analysis method and immunohistochemistry.
Tumour Biol. 39:1010428317691176. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kullak-Ublick GA, Ismair MG, Stieger B,
Landmann L, Huber R, Pizzagalli F, Fattinger K, Meier PJ and
Hagenbuch B: Organic anion-transporting polypeptide B (OATP-B) and
its functional comparison with three other OATPs of human liver.
Gastroenterology. 120:525–533. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Letschert K, Faulstich H, Keller D and
Keppler D: Molecular characterization and inhibition of amanitin
uptake into human hepatocytes. Toxicol Sci. 91:140–149. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kounnis V, Ioachim E, Svoboda M, Tzakos A,
Sainis I, Thalhammer T, Steiner G and Briasoulis E: Expression of
organic anion-transporting polypeptides 1B3, 1B1, and 1A2 in human
pancreatic cancer reveals a new class of potential therapeutic
targets. Onco Targets Ther. 4:27–32. 2011.PubMed/NCBI
|
|
38
|
Lee W, Belkhiri A, Lockhart AC, Merchant
N, Glaeser H, Harris EI, Washington MK, Brunt EM, Zaika A, Kim RB,
et al: Overexpression of OATP1B3 confers apoptotic resistance in
colon cancer. Cancer Res. 68:10315–10323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lockhart AC, Harris E, Lafleur BJ,
Merchant NB, Washington MK, Resnick MB, Yeatman TJ and Lee W:
Organic anion transporting polypeptide 1B3 (OATP1B3) is
overexpressed in colorectal tumors and is a predictor of clinical
outcome. Clin Exp Gastroenterol. 1:1–7. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zarour LR, Anand S, Billingsley KG, Bisson
WH, Cercek A, Clarke MF, Coussens LM, Gast CE, Geltzeiler CB,
Hansen L, et al: Colorectal cancer liver metastasis: Evolving
paradigms and future directions. Cell Mol Gastroenterol Hepatol.
3:163–173. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brenner S, Riha J, Giessrigl B, Thalhammer
T, Grusch M, Krupitza G, Stieger B and Jäger W: The effect of
organic anion-transporting polypeptides 1B1, 1B3 and 2B1 on the
antitumor activity of flavopiridol in breast cancer cells. Int J
Oncol. 46:324–332. 2015. View Article : Google Scholar
|