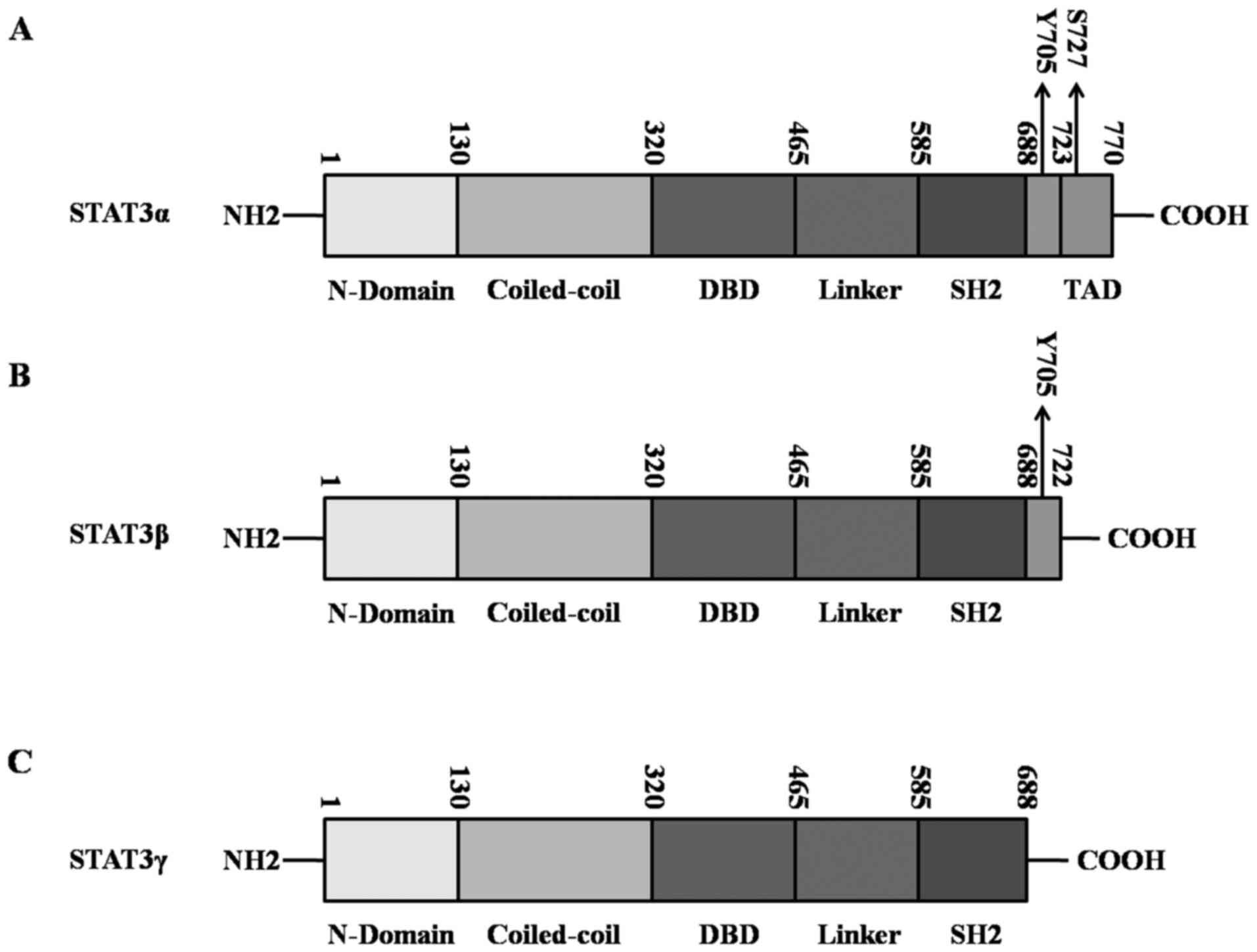
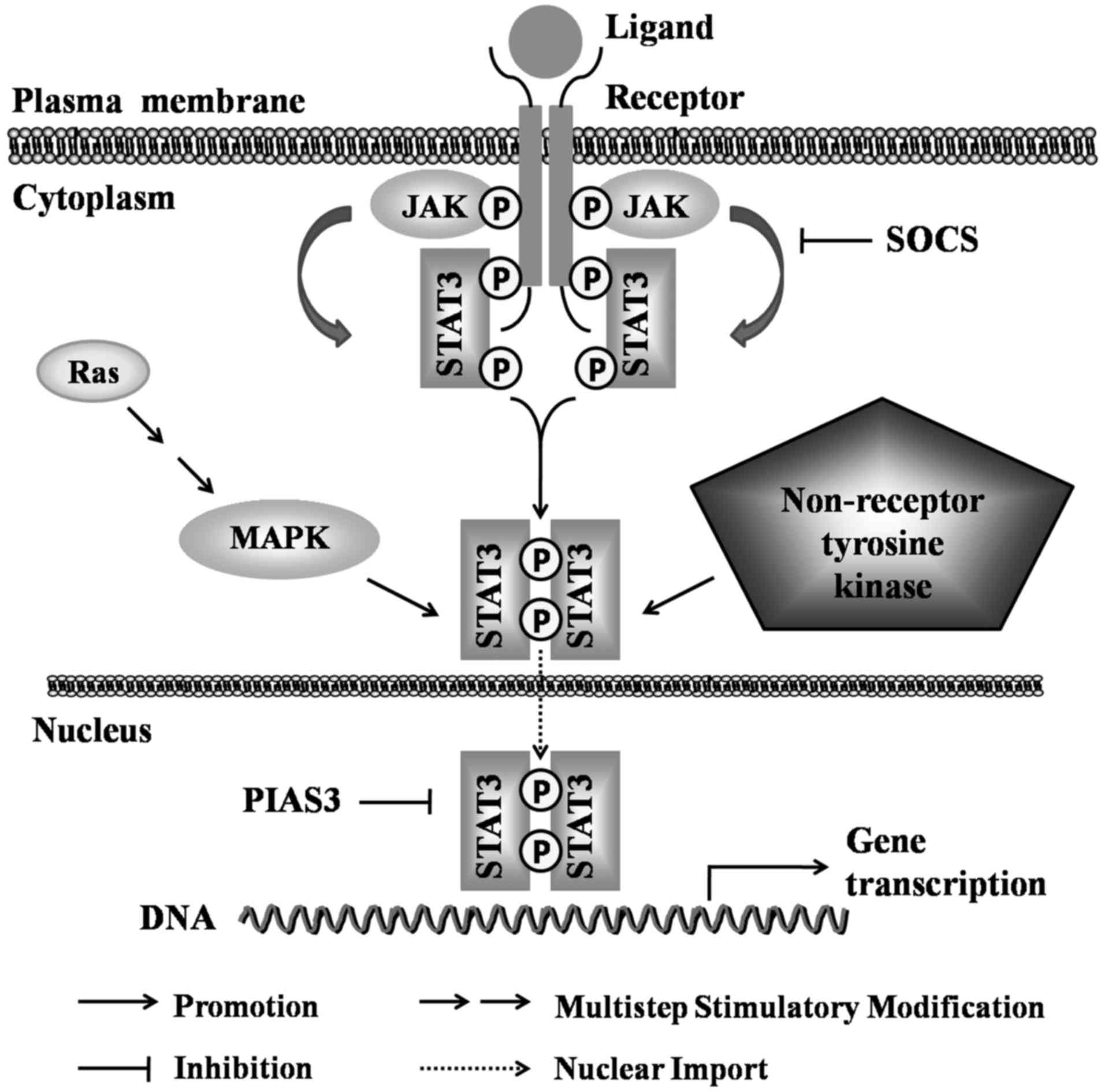
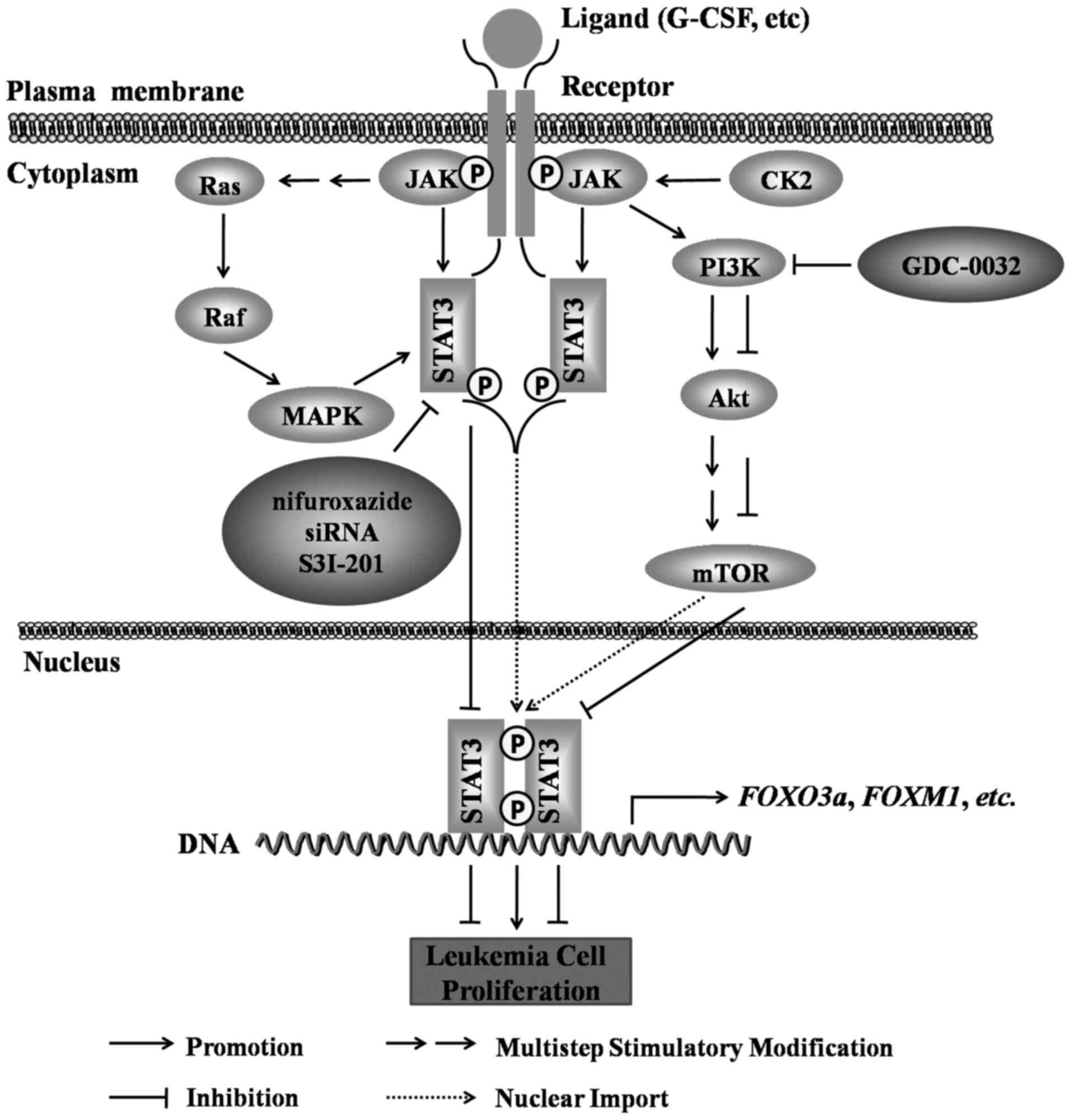
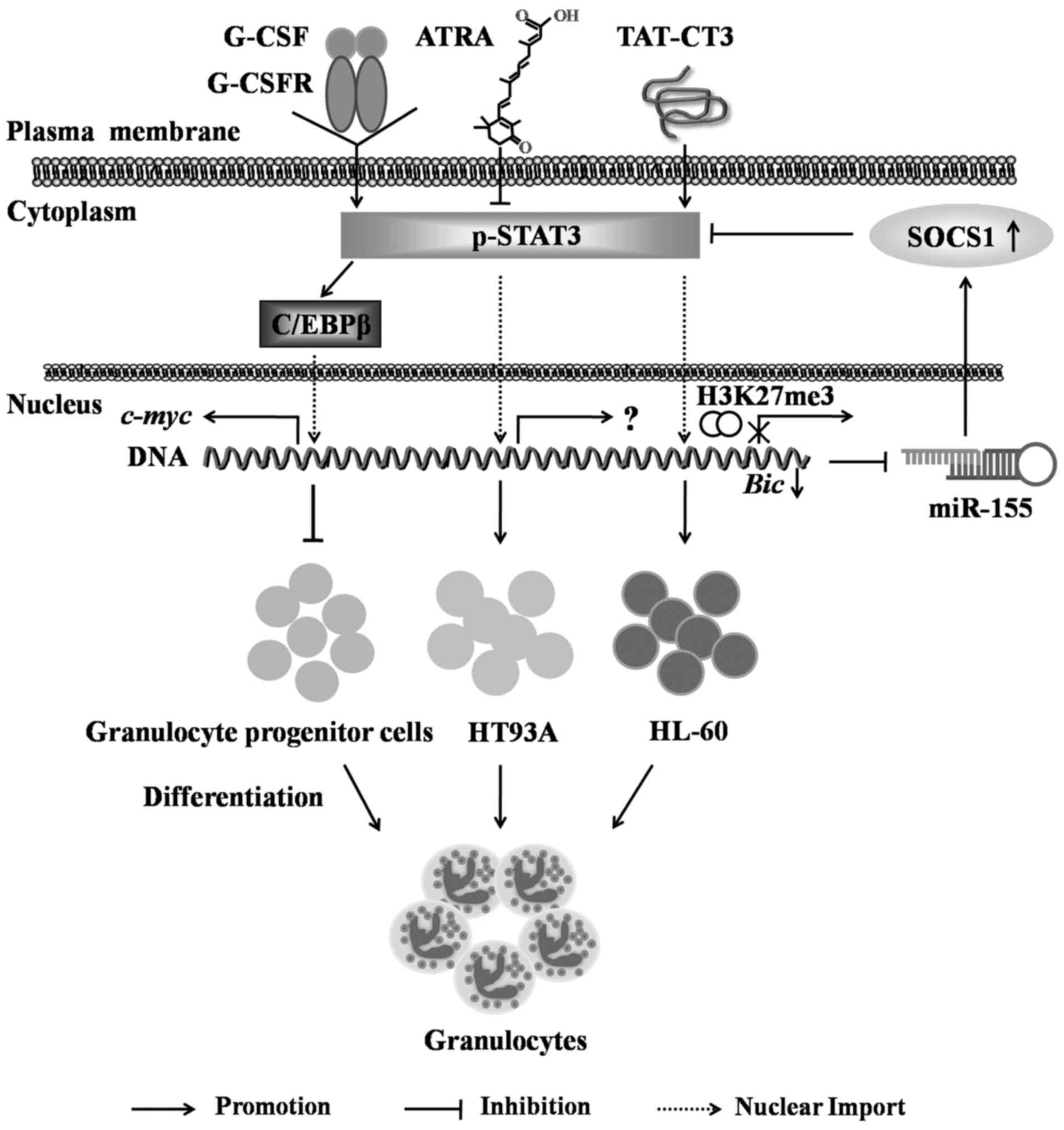
|
1
|
Chen WQ, Shan BE, Zheng SR, Lin GZ, Chen
JZ, Chen JG and He YT: Analysis of incidence and mortality of
leukemia in registration areas of China from 2003 to 2007. Tumor.
32:251–255. 2012.In Chinese.
|
|
2
|
Amitay EL and Keinan-Boker L:
Breastfeeding and childhood leukemia incidence: Ameta-analysis and
systematic review. JAMA Pediatr. 169:e1510252015. View Article : Google Scholar
|
|
3
|
Redaelli A, Stephens JM, Laskin BL, Pashos
CL and Botteman MF: The burden and outcomes associated with four
leukemias: AML, ALL, CLL and CML. Expert Rev Anticancer Ther.
3:311–329. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Namanja AT, Wang J, Buettner R, Colson L
and Chen Y: Allosteric communication across STAT3 domains
associated with STAT3 function and disease-causing mutation. J Mol
Biol. 428:579–589. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: Role of STAT3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar
|
|
6
|
Yu H and Jove R: The STATs of cancer - new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Villarino AV, Kanno Y, Ferdinand JR and
O'Shea JJ: Mechanisms of Jak/STAT signaling in immunity and
disease. J Immunol. 194:21–27. 2015. View Article : Google Scholar :
|
|
8
|
Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB
and Tweardy DJ: Stat3 signaling in acute myeloid leukemia:
Ligand-dependent and -independent activation and induction of
apoptosis by a novel small-molecule Stat3 inhibitor. Blood.
117:5701–5709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Benekli M, Xia Z, Donohue KA, Ford LA,
Pixley LA, Baer MR, Baumann H and Wetzler M: Constitutive activity
of signal transducer and activator of transcription 3 protein in
acute myeloid leukemia blasts is associated with short disease-free
survival. Blood. 99:252–257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rezvani K and Barrett J: STAT3: The
'Achilles' heel for AML? Blood. 123:1–2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Redell MS: A STAT3 decoy lures AML out of
hiding. Blood. 127:1628–1629. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yue P and Turkson J: Targeting STAT3 in
cancer: How successful are we? Expert Opin Investig Drugs.
18:45–56. 2009. View Article : Google Scholar :
|
|
13
|
Zhang T, Kee WH, Seow KT, Fung W and Cao
X: The coiled-coil domain of Stat3 is essential for its SH2
domain-mediated receptor binding and subsequent activation induced
by epidermal growth factor and interleukin-6. Mol Cell Biol.
20:7132–7139. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seidel HM, Milocco LH, Lamb P, Darnell JE
Jr, Stein RB and Rosen J: Spacing of palindromic half sites as a
determinant of selective STAT (signal transducers and activators of
transcription) DNA binding and transcriptional activity. Proc Natl
Acad Sci USA. 92:3041–3045. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song L and Shen BF: New development on
Jak/STAT sigal transduction pathway. Immunol J. 16:68–71. 2000.In
Chinese.
|
|
16
|
Dewilde S, Vercelli A, Chiarle R and Poli
V: Of alphas and betas: Distinct and overlapping functions of STAT3
isoforms. Front Biosci. 13:6501–6514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim BH, Yi EH and Ye SK: Signal transducer
and activator of transcription 3 as a therapeutic target for cancer
and the tumor microenvironment. Arch Pharm Res. 39:1085–1099. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ren Z, Mao X, Mertens C, Krishnaraj R, Qin
J, Mandal PK, Romanowski MJ, McMurray JS and Chen X: Crystal
structure of unphosphorylated STAT3 core fragment. Biochem Biophys
Res Commun. 374:1–5. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chakraborty A and Tweardy DJ: Stat3 and
G-CSF-induced myeloid differentiation. Leuk Lymphoma. 30:433–442.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ilaria RL Jr: STAT isoforms: Mediators of
STAT specificity or leukemogenesis? Leuk Res. 25:483–484. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Becker S, Groner B and Müller CW:
Three-dimensional structure of the Stat3beta homodimer bound to
DNA. Nature. 394:145–151. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benekli M, Baer MR, Baumann H and Wetzler
M: Signal transducer and activator of transcription proteins in
leukemias. Blood. 101:2940–2954. 2003. View Article : Google Scholar
|
|
23
|
Coffer PJ, Koenderman L and de Groot RP:
The role of STATs in myeloid differentiation and leukemia.
Oncogene. 19:2511–2522. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chakraborty A and Tweardy DJ: Granulocyte
colony-stimulating factor activates a 72-kDa isoform of STAT3 in
human neutrophils. J Leukoc Biol. 64:675–680. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hevehan DL, Miller WM and Papoutsakis ET:
Differential expression and phosphorylation of distinct STAT3
proteins during granulocytic differentiation. Blood. 99:1627–1637.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pilati C and Zucman-Rossi J: Mutations
leading to constitutive active gp130/JAK1/STAT3 pathway. Cytokine
Growth Factor Rev. 26:499–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar
|
|
28
|
Groner B, Lucks P and Borghouts C: The
function of Stat3 in tumor cells and their microenvironment. Semin
Cell Dev Biol. 19:341–350. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takeda K, Kaisho T, Yoshida N, Takeda J,
Kishimoto T and Akira S: Correction: Stat3 activation is
responsible for IL-6-dependent T cell proliferation through
preventing apoptosis: generation and characterization of T
cell-specific Stat3-deficient mice. J Immunol. 194:35262015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Levy DE and Lee CK: What does Stat3 do? J
Clin Invest. 109:1143–1148. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bournazou E and Bromberg J: Targeting the
tumor microenvironment: JAK-STAT3 signaling. JAK-STAT.
2:e238282013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo H, Zhou H, Lu J, Qu Y, Yu D and Tong
Y: Vascular endothelial growth factor: An attractive target in the
treatment of hypoxic/ischemic brain injury. Neural Regen Res.
11:174–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leonard WJ and O'Shea JJ: Jaks and STATs:
Biological implications. Annu Rev Immunol. 16:293–322. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schindler C and Strehlow I: Cytokines and
STAT signaling. Adv Pharmacol. 47:113–174. 2000. View Article : Google Scholar
|
|
35
|
Kisseleva T, Bhattacharya S, Braunstein J
and Schindler CW: Signaling through the JAK/STAT pathway, recent
advances and future challenges. Gene. 285:1–24. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang SK, Kang KS, Jee MK and Kim BS:
Vascular endothelial growth factor/kinase insult domain receptor
(KDR)/fetal liver kinase 1 (FLK1)-mediated skin-epithelial
progenitor cells reprogramming. Tissue Eng Part A. 16:2687–2697.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pastuschek J, Poetzsch J, Morales-Prieto
DM, Schleussner E, Markert UR and Georgiev G: Stimulation of the
JAK/STAT pathway by LIF and OSM in the human granulosa cell line
COV434. J Reprod Immunol. 108:48–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Marino VJ and Roguin LP: The granulocyte
colony stimulating factor (G-CSF) activates Jak/STAT and MAPK
pathways in a trophoblastic cell line. J Cell Biochem.
103:1512–1523. 2008. View Article : Google Scholar
|
|
39
|
Platanias LC: Mechanisms of type-I- and
type-II-interferon-mediated signalling. Nat Rev Immunol. 5:375–386.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kijima T, Niwa H, Steinman RA, Drenning
SD, Gooding WE, Wentzel AL, Xi S and Grandis JR: STAT3 activation
abrogates growth factor dependence and contributes to head and neck
squamous cell carcinoma tumor growth in vivo. Cell Growth Differ.
13:355–362. 2002.PubMed/NCBI
|
|
41
|
Ram PT and Iyengar R: G protein coupled
receptor signaling through the Src and Stat3 pathway: Role in
proliferation and transformation. Oncogene. 20:1601–1606. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ahmed ST and Ivashkiv LB: Inhibition of
IL-6 and IL-10 signaling and Stat activation by inflammatory and
stress pathways. J Immunol. 165:5227–5237. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Trengove MC and Ward AC: SOCS proteins in
development and disease. Am J Clin Exp Immunol. 2:1–29.
2013.PubMed/NCBI
|
|
44
|
Piessevaux J, Lavens D, Peelman F and
Tavernier J: The many faces of the SOCS box. Cytokine Growth Factor
Rev. 19:371–381. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shuai K: Regulation of cytokine signaling
pathways by PIAS proteins. Cell Res. 16:196–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma LD, Zhou M, Wen SH, Ni C, Jiang LJ, Fan
J and Xia L: Effects of STAT3 silencing on fate of chronic
myelogenous leukemia K562 cells. Leuk Lymphoma. 51:1326–1336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chakraborty A, White SM, Schaefer TS, Ball
ED, Dyer KF and Tweardy DJ: Granulocyte colony-stimulating factor
activation of Stat3 alpha and Stat3 beta in immature normal and
leukemic human myeloid cells. Blood. 88:2442–2449. 1996.PubMed/NCBI
|
|
48
|
Quotti Tubi L, Canovas Nunes S, Brancalion
A, Doriguzzi Breatta E, Manni S, Mandato E, Zaffino F, Macaccaro P,
Carrino M, Gianesin K, et al: Protein kinase CK2 regulates AKT,
NF-κB and STAT3 activation, stem cell viability and proliferation
in acute myeloid leukemia. Leukemia. 31:292–300. 2017. View Article : Google Scholar
|
|
49
|
Mencalha AL, Binato R, Ferreira GM, Du
Rocher B and Abdelhay E: Forkhead box M1 (FoxM1) gene is a new
STAT3 transcriptional factor target and is essential for
proliferation, survival and DNA repair of K562 cell line. PLoS One.
7:e481602012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuzugullu H, Von T, Thorpe LM, Walker SR,
Roberts TM, Frank DA and Zhao JJ: NTRK2 activation cooperates with
PTEN deficiency in T-ALL through activation of both the PI3K-AKT
and JAK-STAT3 pathways. Cell Discov. 2:160302016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cook AM, Li L, Ho Y, Lin A, Li L, Stein A,
Forman S, Perrotti D, Jove R and Bhatia R: Role of altered growth
factor receptor-mediated JAK2 signaling in growth and maintenance
of human acute myeloid leukemia stem cells. Blood. 123:2826–2837.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sasaki R, Ito S, Asahi M and Ishida Y:
YM155 suppresses cell proliferation and induces cell death in human
adult T-cell leukemia/lymphoma cells. Leuk Res. 39:1473–1479. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Avalos BR: Molecular analysis of the
granulocyte colony-stimulating factor receptor. Blood. 88:761–777.
1996.PubMed/NCBI
|
|
54
|
Hillmer EJ, Zhang H, Li HS and Watowich
SS: STAT3 signaling in immunity. Cytokine Growth Factor Rev.
31:1–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Panopoulos AD, Zhang L, Snow JW, Jones DM,
Smith AM, El Kasmi KC, Liu F, Goldsmith MA, Link DC, Murray PJ, et
al: STAT3 governs distinct pathways in emergency granulopoiesis and
mature neutrophils. Blood. 108:3682–3690. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Krönke M, Schlüter C and Pfizenmaier K:
Tumor necrosis factor inhibits MYC expression in HL-60 cells at the
level of mRNA transcription. Proc Natl Acad Sci USA. 84:469–473.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Johansen LM, Iwama A, Lodie TA, Sasaki K,
Felsher DW, Golub TR and Tenen DG: c-Myc is a critical target for
c/EBPalpha in granulopoiesis. Mol Cell Biol. 21:3789–3806. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu S, Xu Z, Liu B, Sun Q, Yang L, Wang J,
Wang Y and Liu H: LIFRα-CT3 induces differentiation of a human
acute myelogenous leukemia cell line HL-60 by suppressing miR-155
expression through the JAK/STAT pathway. Leuk Res. 38:1237–1244.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Uchino Y, Iriyama N, Hatta Y and Takei M:
Granulocyte colony-stimulating factor potentiates all-trans
retinoic acid-induced granulocytic differentiation in acute
promyelocytic leukemia cell line HT93A. Cancer Cell Int. 15:302015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu N, Tang XH, Pan W, Xie ZM, Zhang GF, Ji
MH, Yang JJ, Zhou MT and Zhou ZQ: Spared nerve injury increases the
expression of microglia M1 markers in the prefrontal cortex of rats
and provokes depression-like behaviors. Front Neurosci. 11:2092017.
View Article : Google Scholar :
|
|
61
|
Allman WR, Dey R, Liu L, Siddiqui S,
Coleman AS, Bhattacharya P, Yano M, Uslu K, Takeda K, Nakhasi HL,
et al: TACI deficiency leads to alternatively activated macrophage
phenotype and susceptibility to Leishmania infection. Proc Natl
Acad Sci USA. 112:E4094–E4103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou L, Zhuo H, Ouyang H, Liu Y, Yuan F,
Sun L, Liu F and Liu H: Glycoprotein non-metastatic melanoma
protein b (Gpnmb) is highly expressed in macrophages of acute
injured kidney and promotes M2 macrophages polarization. Cell
Immunol. 316:53–60. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Binnemars-Postma K, Storm G and Prakash J:
Nanomedicine strategies to target tumor-associated macrophages. Int
J Mol Sci. 18:9792017. View Article : Google Scholar :
|
|
64
|
Fu XL, Duan W, Su CY, Shen XQ, Zhuang Y,
Yu PW and Zhao YL: IL-6 cooperated with M-CSF induce
CD14+ monocytes differentiation into M2-like phenotype
macrophages in vitro. Immunol J. 32:1013–1018. 2016.In Chinese.
|
|
65
|
Komohara Y, Horlad H, Ohnishi K, Ohta K,
Makino K, Hondo H, Yamanaka R, Kajiwara K, Saito T, Kuratsu J, et
al: M2 macrophage/microglial cells induce activation of Stat3 in
primary central nervous system lymphoma. J Clin Exp Hematop.
51:93–99. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mangan JK, Rane SG, Kang AD, Amanullah A,
Wong BC and Reddy EP: Mechanisms associated with IL-6-induced
up-regulation of Jak3 and its role in monocytic differentiation.
Blood. 103:4093–4101. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Iwamoto T, Senga T, Adachi K and Hamaguchi
M: Stat3-dependent induction of interleukin-3 receptor expression
in leukemia inhibitory factor-stimulated M1 mouse leukemia cells.
Cytokine. 25:136–139. 2004. View Article : Google Scholar
|
|
68
|
Tanuma N, Nakamura K, Shima H and Kikuchi
K: Protein-tyrosine phosphatase PTPepsilon C inhibits Jak-STAT
signaling and differentiation induced by interleukin-6 and leukemia
inhibitory factor in M1 leukemia cells. J Biol Chem.
275:28216–28221. 2000.PubMed/NCBI
|
|
69
|
Yao Y, Zhou Q and Ericson SG: Vanadate
stimulates monocytic differentiation activity of IL-6 by enhancing
actin filament polymerization in HL-60 cells. J Biomed Sci.
11:940–949. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Woetmann A, Nielsen M, Christensen ST,
Brockdorff J, Kaltoft K, Engel AM, Skov S, Brender C, Geisler C,
Svejgaard A, et al: Inhibition of protein phosphatase 2A induces
serine/threonine phosphorylation, subcellular redistribution, and
functional inhibition of STAT3. Proc Natl Acad Sci USA.
96:10620–10625. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sun Q, Wang J, Xiong J, Yang L and Liu H:
Free LIF receptor α-chain distal cytoplasmic motifs enhance
Jak2-independent STAT3 phosphorylation and induce differentiation
in HL-60 cells. Oncol Rep. 26:399–404. 2011.PubMed/NCBI
|
|
72
|
Zhou J, Chong PS, Lu X, Cheong LL, Bi C,
Liu SC, Zhou Y, Tan TZ, Yang H, Chung TH, et al: Phosphatase of
regenerating liver-3 is regulated by signal transducer and
activator of transcription 3 in acute myeloid leukemia. Exp
Hematol. 42:1041–1052. e1–2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Banchereau J, Briere F, Caux C, Davoust J,
Lebecque S, Liu YJ, Pulendran B and Palucka K: Immunobiology of
dendritic cells. Annu Rev Immunol. 18:767–811. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu YJ, Kanzler H, Soumelis V and Gilliet
M: Dendritic cell lineage, plasticity and cross-regulation. Nat
Immunol. 2:585–589. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
75
|
Mohty M, Jarrossay D, Lafage-Pochitaloff
M, Zandotti C, Brière F, de Lamballeri XN, Isnardon D, Sainty D,
Olive D and Gaugler B: Circulating blood dendritic cells from
myeloid leukemia patients display quantitative and cytogenetic
abnormalities as well as functional impairment. Blood.
98:3750–3756. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Claxton DF, McMannis J, Champlin R and
Choudhury A: Therapeutic potential of leukemia-derived dendritic
cells: Preclinical and clinical progress. Crit Rev Immunol.
21:147–155. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu L and Liu YJ: Development of
dendritic-cell lineages. Immunity. 26:741–750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
McKenna HJ, Stocking KL, Miller RE, Brasel
K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J,
Pulendran B, et al: Mice lacking flt3 ligand have deficient
hematopoiesis affecting hematopoietic progenitor cells, dendritic
cells, and natural killer cells. Blood. 95:3489–3497.
2000.PubMed/NCBI
|
|
79
|
Waskow C, Liu K, Darrasse-Jèze G,
Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K and
Nussenzweig M: The receptor tyrosine kinase Flt3 is required for
dendritic cell development in peripheral lymphoid tissues. Nat
Immunol. 9:676–683. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li HS, Yang CY, Nallaparaju KC, Zhang H,
Liu YJ, Goldrath AW and Watowich SS: The signal transducers STAT5
and STAT3 control expression of Id2 and E2-2 during dendritic cell
development. Blood. 120:4363–4373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Brady MT, Miller A, Sait SN, Ford LA,
Minderman H, Wang ES, Lee KP, Baumann H and Wetzler M:
Down-regulation of signal transducer and activator of transcription
3 improves human acute myeloid leukemia-derived dendritic cell
function. Leuk Res. 37:822–828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhou J, Bi C, Janakakumara JV, Liu SC,
Chng WJ, Tay KG, Poon LF, Xie Z, Palaniyandi S, Yu H, et al:
Enhanced activation of STAT pathways and overexpression of survivin
confer resistance to FLT3 inhibitors and could be therapeutic
targets in AML. Blood. 113:4052–4062. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Nair RR, Tolentino JH and Hazlehurst LA:
Role of STAT3 in transformation and drug resistance in CML. Front
Oncol. 2:302012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li P, Harris D, Liu Z, Rozovski U,
Ferrajoli A, Wang Y, Bueso-Ramos C, Hazan-Halevy I, Grgurevic S,
Wierda W, et al: STAT3-activated GM-CSFRα translocates to the
nucleus and protects CLL cells from apoptosis. Mol Cancer Res.
12:1267–1282. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mencalha AL, Du Rocher B, Salles D, Binato
R and Abdelhay E: LLL-3, a STAT3 inhibitor, represses
BCR-ABL-positive cell proliferation, activates apoptosis and
improves the effects of Imatinib mesylate. Cancer Chemother
Pharmacol. 65:1039–1046. 2010. View Article : Google Scholar
|
|
86
|
Mangolini M, de Boer J,
Walf-Vorderwülbecke V, Pieters R, den Boer ML and Williams O: STAT3
mediates oncogenic addiction to TEL-AML1 in t(12;21) acute
lymphoblastic leukemia. Blood. 122:542–549. 2013. View Article : Google Scholar
|
|
87
|
Pathania AS, Kumar S, Guru SK, Bhushan S,
Sharma PR, Aithagani SK, Singh PP, Vishwakarma RA, Kumar A and
Malik F: The synthetic tryptanthrin analogue suppresses STAT3
signaling and induces caspase dependent apoptosis via ERK up
regulation in human leukemia HL-60 cells. PLoS One. 9:e1104112014.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pathania AS, Guru SK, Ul Ashraf N,
Riyaz-Ul-Hassan S, Ali A, Abdullah Tasduq S, Malik F and Bhushan S:
A novel stereo bioactive metabolite isolated from an endophytic
fungus induces caspase dependent apoptosis and STAT-3 inhibition in
human leukemia cells. Eur J Pharmacol. 765:75–85. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Rozovski U, Harris DM, Li P, Liu Z, Wu JY,
Grgurevic S, Faderl S, Ferrajoli A, Wierda WG, Martinez M, et al:
At high levels, constitutively activated STAT3 induces apoptosis of
chronic lymphocytic leukemia cells. J Immunol. 196:4400–4409. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Xia Z, Sait SN, Baer MR, Barcos M, Donohue
KA, Lawrence D, Ford LA, Block AM, Baumann H and Wetzler M:
Truncated STAT proteins are prevalent at relapse of acute myeloid
leukemia. Leuk Res. 25:473–482. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Stevens AM, Ruiz MJ, Gerbing RB, Alonzo
TA, Gamis AS and Redell MS: Ligand-induced STAT3 signaling
increases at relapse and is associated with outcome in pediatric
acute myeloid leukemia: A report from the Children's Oncology
Group. Haematologica. 100:e496–e500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Benekli M1, Xia Z, Donohue KA, Ford LA,
Pixley LA, Baer MR, Baumann H and Wetzler M: Constitutive activity
of signal transducer and activator of transcription 3 protein in
acute myeloid leukemia blasts is associated with short disease-free
survival. Blood. 2002:252–257. 2002. View Article : Google Scholar
|
|
93
|
Kristensen T, Larsen M, Rewes A,
Frederiksen H, Thomassen M and Møller MB: Clinical relevance of
sensitive and quantitative STAT3 mutation analysis using
next-generation sequencing in T-cell large granular lymphocytic
leukemia. J Mol Diagn. 16:382–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jerez A, Clemente MJ, Makishima H, Koskela
H, Leblanc F, Peng Ng K, Olson T, Przychodzen B, Afable M,
Gomez-Segui I, et al: STAT3 mutations unify the pathogenesis of
chronic lymphoproliferative disorders of NK cells and T-cell large
granular lymphocyte leukemia. Blood. 120:3048–3057. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Rajala HL, Olson T, Clemente MJ, Lagström
S, Ellonen P, Lundan T, Hamm DE, Zaman SA, Lopez Marti JM,
Andersson EI, et al: The analysis of clonal diversity and therapy
responses using STAT3 mutations as a molecular marker in large
granular lymphocytic leukemia. Haematologica. 100:91–99. 2015.
View Article : Google Scholar :
|
|
96
|
Lamy T and Loughran TP Jr: How I treat LGL
leukemia. Blood. 117:2764–2774. 2011. View Article : Google Scholar :
|
|
97
|
Lamy T, Moignet A and Loughran TP Jr: LGL
leukemia: From pathogenesis to treatment. Blood. 129:1082–1094.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Matutes E: Large granular lymphocytic
leukemia. Current diagnostic and therapeutic approaches and novel
treatment options. Expert Rev Hematol. 10:251–258. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ohgami RS, Ma L, Merker JD, Martinez B,
Zehnder JL and Arber DA: STAT3 mutations are frequent in
CD30+ T-cell lymphomas and T-cell large granular
lymphocytic leukemia. Leukemia. 27:2244–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhong Y, Wu J, Chen B, Ma R, Cao H, Wang
Z, Cheng L, Ding J and Feng J: Investigation and analysis of single
nucleotide polymorphisms in Janus kinase/signal transducer and
activator of transcription genes with leukemia. Leuk Lymphoma.
53:1216–1221. 2012. View Article : Google Scholar
|
|
101
|
Lautner-Csorba O, Gézsi A, Semsei AF,
Antal P, Erdélyi DJ, Schermann G, Kutszegi N, Csordás K, Hegyi M,
Kovács G, et al: Candidate gene association study in pediatric
acute lymphoblastic leukemia evaluated by Bayesian network based
Bayesian multilevel analysis of relevance. BMC Med Genomics.
5:422012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Sakamoto KM, Grant S, Saleiro D, Crispino
JD, Hijiya N, Giles F, Platanias L and Eklund EA: Targeting novel
signaling pathways for resistant acute myeloid leukemia. Mol Genet
Metab. 114:397–402. 2015. View Article : Google Scholar :
|
|
103
|
Bruserud Ø, Nepstad I, Hauge M, Hatfield
KJ and Reikvam H: STAT3 as a possible therapeutic target in human
malignancies: Lessons from acute myeloid leukemia. Expert Rev
Hematol. 8:29–41. 2015. View Article : Google Scholar
|
|
104
|
Munoz J, Dhillon N, Janku F, Watowich SS
and Hong DS: STAT3 inhibitors: Finding a home in lymphoma and
leukemia. Oncologist. 19:536–544. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Hayakawa F, Sugimoto K, Harada Y,
Hashimoto N, Ohi N, Kurahashi S and Naoe T: A novel STAT inhibitor,
OPB-31121, has a significant antitumor effect on leukemia with
STAT-addictive oncokinases. Blood Cancer J. 3:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhu Z, Lu X, Jiang L, Sun X, Zhou H, Jia
Z, Zhang X and Ma L: STAT3 signaling pathway is involved in
decitabine induced biological phenotype regulation of acute myeloid
leukemia cells. Am J Transl Res. 7:1896–1907. 2015.PubMed/NCBI
|
|
107
|
Selvi N, Kaymaz BT, Gündüz C, Aktan C,
Kiper HD, Sahin F, Cömert M, Selvi AF, Kosova B and Saydam G:
Bortezomib induces apoptosis by interacting with JAK/STAT pathway
in K562 leukemic cells. Tumour Biol. 35:7861–7870. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kiper HD, Tezcanli Kaymaz B, Gokbulut AA,
Selvi N, Avci CB, Kosova B, Iskender G, Yandim MK, Gunduz C, Sahin
F, et al: STAT pathway in the regulation of zoledronic acid-induced
apoptosis in chronic myeloid leukemia cells. Biomed Pharmacother.
67:527–532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhao S, Guo J, Zhao Y, Fei C, Zheng Q, Li
X and Chang C: Chidamide, a novel histone deacetylase inhibitor,
inhibits the viability of MDS and AML cells by suppressing
JAK2/STAT3 signaling. Am J Transl Res. 8:3169–3178. 2016.PubMed/NCBI
|
|
110
|
Chan KT, Li K, Liu SL, Chu KH, Toh M and
Xie WD: Cucurbitacin B inhibits STAT3 and the Raf/MEK/ERK pathway
in leukemia cell line K562. Cancer Lett. 289:46–52. 2010.
View Article : Google Scholar
|
|
111
|
Cai H, Qin X and Yang C: Dehydrocostus
lactone suppresses proliferation of human chronic myeloid leukemia
cells through Bcr/Abl-JAK/STAT signaling pathways. J Cell Biochem.
118:3381–3390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Jung JH, Kwon TR, Jeong SJ, Kim EO, Sohn
EJ, Yun M and Kim SH: Apoptosis induced by tanshinone IIA and
cryptotanshinone is mediated by distinct JAK/STAT3/5 and SHP1/2
signaling in chronic myeloid leukemia K562 cells. Evid Based
Complement Alternat Med. 2013:8056392013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ma L, Zhu Z, Jiang L, Sun X, Lu X, Zhou M,
Qian S and Jianyong L: Matrine suppresses cell growth of human
chronic myeloid leukemia cells via its inhibition of the
interleukin-6/Janus activated kinase/signal transducer and
activator of transcription 3 signaling cohort. Leuk Lymphoma.
56:2923–2930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Jia X, Yang W, Han J and Xiong H: Effects
of lentivirus mediated STAT3 silencing on human chronic myeloid
leukemia cells and leukemia mice. Int J Clin Exp Med. 7:4031–4037.
2014.
|
|
115
|
Stella S, Tirrò E, Conte E, Stagno F, Di
Raimondo F, Manzella L and Vigneri P: Suppression of survivin
induced by a BCR-ABL/JAK2/STAT3 pathway sensitizes
imatinib-resistant CML cells to different cytotoxic drugs. Mol
Cancer Ther. 12:1085–1098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Allen JC, Talab F, Zuzel M, Lin K and
Slupsky JR: c-Abl regulates Mcl-1 gene expression in chronic
lymphocytic leukemia cells. Blood. 117:2414–2422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Folgiero V, Goffredo BM, Filippini P,
Masetti R, Bonanno G, Caruso R, Bertaina V, Mastronuzzi A, Gaspari
S, Zecca M, et al: Indoleamine 2,3-dioxygenase 1 (IDO1) activity in
leukemia blasts correlates with poor outcome in childhood acute
myeloid leukemia. Oncotarget. 5:2052–2064. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Adamaki M, Tsotra M, Vlahopoulos S,
Zampogiannis A, Papavassiliou AG and Moschovi M: STAT transcript
levels in childhood acute lymphoblastic leukemia: STAT1 and STAT3
transcript correlations. Leuk Res. 39:1285–1291. 2015. View Article : Google Scholar
|
|
119
|
Zhong Y, Feng J, Chen B, Cheng L, Li Y,
Qian J, Ding J, Gao F and Xia G: Signal transducer and activator of
transcription 3 (STAT3) gene polymorphisms are associated with
treatment outcomes in acute myeloid leukemia. Int J Lab Hematol.
34:383–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Redell MS, Ruiz MJ, Gerbing RB, Alonzo TA,
Lange BJ, Tweardy DJ and Meshinchi S; Children's Oncology Group:
FACS analysis of Stat3/5 signaling reveals sensitivity to G-CSF and
IL-6 as a significant prognostic factor in pediatric AML: A
Children's Oncology Group report. Blood. 121:1083–1093. 2013.
View Article : Google Scholar :
|
|
121
|
Levidou G, Sachanas S, Pangalis GA,
Kalpadakis C, Yiakoumis X, Moschogiannis M, Sepsa A, Lakiotaki E,
Milionis V, Kyrtsonis MC, et al: Immunohistochemical analysis of
IL-6, IL-8/CXCR2 axis, Tyr p-STAT-3, and SOCS-3 in lymph nodes from
patients with chronic lymphocytic leukemia: Correlation between
microvascular characteristics and prognostic significance. BioMed
Res Int. 2014:2514792014. View Article : Google Scholar
|
|
122
|
Danis E, Yamauchi T, Echanique K, Zhang X,
Haladyna JN, Riedel SS, Zhu N, Xie H, Orkin SH, Armstrong SA, et
al: Ezh2 controls an early hematopoietic program and growth and
survival signaling in early T cell precursor acute lymphoblastic
leukemia. Cell Rep. 14:1953–1965. 2016. View Article : Google Scholar : PubMed/NCBI
|