Introduction
Melanoma antigen family A4 (MAGEA4), a cancer/testis
antigen, belongs to a family of genes, MAGEA1 to A12, located on
human X-chromosome q28 (1). MAGEA4
is expressed in various malignant tumors, including non-small cell
lung carcinoma, but not in adult normal tissues, excluding germ
cells (2). Accordingly, MAGEA4 is
widely used as a target for cancer vaccine therapy (3,4). The
biological function of MAGEA4, however, remains controversial, with
some studies suggesting that MAGEA proteins are oncoproteins that
promote tumor cell survival (5–7), and
others indicating that MAGEA proteins are tumor suppressors that
elicit apoptosis (8–10). The fact that different methods are
used to assess MAGEA4 expression and subcellular localization in
non-small cell lung cancers further complicates this issue
(Table I) (6,8,11–14).
For example, several reports have suggested that MAGEA
overexpression is associated with a poorer overall survival
(13,14); however, this association is
strongly variable among cohorts, presumably as a result of assay
cross-reactivity among MAGEA proteins (Table II) (15,16),
or the lack of standard criteria to interpret such assays. In this
study, we have now obtained a specific antibody to MAGEA4, and
established specific conditions with which to distinguish
MAGEA4 from other MAGEA genes by reverse
transcriptase-quantitative polymerase chain reaction (RT-qPCR),
western blot analysis and immunohistochemistry.
 | Table IPrevious reports on MAGEA4 expression
in non-small cell lung cancer. |
Table I
Previous reports on MAGEA4 expression
in non-small cell lung cancer.
| Author (Ref.) | Year | Expression rates
| Intracellular
localization | Association with
prognosis |
|---|
| RT-PCR | WB | IHC |
|---|
| Gure et al
(11) | 2005 | 35% | NA | NA | NA | NA |
| Groeper et
al (12) | 2006 | 18% | NA | 74% [57B] |
Nucleus/cytoplasm | NA |
| Peikert et
al (8) | 2006 | 60% | 38% [6C1] | [6C1]a | Nucleus | Apoptosis
promotion |
| Yoshida et
al (13) | 2006 | 28% | NA | NA | NA | Poor survival in
advanced stage cancers |
| Shigematsu et
al (14) | 2010 | 20% | NA | NA | NA | Poor overall
survival |
 | Table IISequence homological ratios between
MAGEA4 and the other MAGEA family genes. |
Table II
Sequence homological ratios between
MAGEA4 and the other MAGEA family genes.
| MAGEA1 | MAGEA2B | MAGEA3 | MAGEA12 |
|---|
| Homological ratio
with MAGEA4 in: | | | | |
| Nucleotide
sequence | 87% | 82% | 83% | 83% |
| Amino acid
sequence | 75% | 68% | 69% | 69% |
Based on this assay, we found that the prognostic
significance of MAGEA4 is dependent on its subcellular localization
and on the p53 status. These data enhance our understanding of the
function of MAGEA4, and help clarify the seemingly contradictory
effects of MAGEA4 on apoptosis.
Materials and methods
Patients
Using tissue microarray, we investigated MAGEA4
expression in 240 patients who received surgery for non-small cell
lung cancer between 1996 and 2004 at the Department of Surgical
Oncology, Hokkaido University Hospital, Sapporo, Japan. Patients
were classified according to the TNM Classification of Malignant
Tumours 7th edition (17). This
study was approved by the Ethics Committee of Hokkaido University.
Informed consent for the use of tissue samples which were stored
prior to the establishment of ethics approval, were originally
obtained from the patients at the time of surgery. The Hokkaido
University Institutional Review Board confirmed that this study was
fully ethically compliant and the second informed consent was
waived by the ethics committee of our institution.
Cells and cell culture
The 293F and 293FT cells were obtained from
Invitrogen (Carlsbad, CA, USA), while the human lung squamous cell
carcinoma cell lines, PC10, H226, LK2 and LC-1, were obtained from
the Japanese Cancer Research Resources Bank (Ibaraki, Japan), along
with lung adenocarcinoma cell lines, A549, REPF-LC-MS, VMRC-LCD and
ABC-1. The cells were grown at 37°C in a humidified incubator with
5% CO2, and in culture medium with 10% fetal bovine
serum and 1% penicillin/streptomycin.
Cloning
MAGEA genes were amplified from human testis
cDNA (Takara Bio, Inc., Tokyo, Japan) using suitable PCR primers
and high-fidelity KOD-Plus-polymerase (Toyobo, Tokyo, Japan).
Amplicons were then inserted into pcDNA-IRES-GFP vector by
restriction enzyme cloning using T4 ligase (Promega, Madison, WI,
USA). The vector is a pcDNA3.1(+) plasmid (Invitrogen) that
contains an internal ribosomal entry site and green fluorescent
protein (GFP) originally synthesized in our previous study
(18). Following plasmid
amplification in JM-109 competent cells (Takara Bio, Inc.), inserts
were confirmed by full sequencing (Hokkaido System Science Co.,
Ltd., Sapporo, Japan).
Transfection
The 293F or 293FT cells were transfected with the
MAGEA expression plasmids using Lipofectamine LTX
(Invitrogen), following the manufacturer' s instructions.
Transfection was confirmed by GFP fluorescence. Transfectants were
then used to optimize MAGEA detection by RT-qPCR, western
blot analysis and immunohistochemistry. Untransformed cells were
used as negative controls, along with cells transfected with the
empty vector. Transfected cells were exposed for 24–48 h to 1
μM cisplatin (Bristol-Myers Squibb, New York, NY, USA) when
investigating subcellular localization under cytotoxic
conditions.
MAGEA detection by RT-qPCR
Total RNA was extracted from the transfected cells
or non-small cell lung cancer cells using TRI reagent
(Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's
instructions, and digested with RQ1 RNase-Free DNase (Promega).
cDNA was then reverse transcribed from 2 μg RNA using the
Superscript II or Superscript VILO cDNA Synthesis kit (Invitrogen),
and digested with Ribonuclease H (Takara Bio, Inc.). MAGEA4
was then amplified with GoTaq® DNA Polymerase (Promega),
using the primers MAGEA4_forward, 5′-ATGTCTTCTGAGCAGAAG AGTCAGC-3′
and MAGEA4_reverse, 5′-TCAGACTCCCTCT TCCTCCTCT-3′. The reaction
consisted of pre-heating at 94°C for 5 min and 30 cycles of
denaturation at 94°C for 30 sec, annealing at 60.4°C for 30 sec,
extension at 72°C for 1 min, followed by final extension for 5 min
at 72°C. Subsequently, MAGEA4 expression was assessed by
quantitative RT-PCR on an ABI PRISM 7000, using Power
SYBR®-Green PCR Master Mix (both from Life
Technologies/Applied Biosystems, Carlsbad, CA, USA), the internal
probe 5′-CTGGAGCATGT GGTCAGGGTCAAT-3′ and the reverse primer used
in the initial PCR reaction. Expression was normalized to β-actin,
which was amplified with the internal probe, 5′-CCAGGC
TGTGCTATCCCTGTACGC-3′ and reverse primer, 5′-ACC
GGAGTCCATCACGATGC-3′.
Monoclonal antibody to MAGEA4
CB6F1 mice were immunized with human recombinant
MAGEA4, and clone designated as MCV-1 was affinity-purified with
protein G. MCV-1 was selected based on epitope specificities of
hybridoma supernatants, as assessed by ELISA. This process was
conducted at Mie University, Tsu, Japan, and purified antibody to
MCV-1 was supplied by the University.
Mice and tumor xenograft models
CB17/SCID mice were purchased from Charles River
Laboratories Japan (Yokohama, Japan). All mice were female, 4–6
weeks of age, and were maintained under specific pathogen-free
conditions and were treated under the guidelines of the Hokkaido
University Institutional Animal Care and Use Committee. PC10, H226,
LK2, LC-1, A549, REPF-LC-MS, VMRC-LCD and ABC-1 cells
(5×106) were subcutaneously injected in a volume of 100
μl of phosphate-buffered saline into the left flank region
of each mouse. The mice were monitored once every 2 or 3 days after
the injection. When the tumor diameter exceeded 10 mm, the mice
were sacrificed and the tumors were separated into 2 blocks: one
block was frozen using liquid nitrogen to extract proteins for
western blot analysis, and the other was immersed in formalin for
immunohistochemical analysis. If multiple tumors were developed in
a mouse, the largest one was used as a sample. All animal
experiments were conducted with the Institutional Animal Care and
Use Committee approval at that time.
Western blot analysis
Lysates from cell lines and from SCID mouse
xenografts were prepared in SDS buffer containing 62.5 mm Tris-HCL
(pH 6.8), 2% w/v SDS, 10% glycerol, 50 mm DTT, 0.1% w/v bromphenol
blue and 1 mm PMSF. Protein concentration was measured by the
Bradford method using a commercial protein assay kit (Bio-Rad
Laboratories, Hercules, CA, USA). Heat-denatured cell lysate (10
μg) were electrophorased in 15% SDS-polyacrylamyde gels and
were blotted on nitrocellulose membranes. They were probed for 1 h
at room temperature with 1:1,000 dilutions of monoclonal antibodies
to MAGEA4 (MCV-1), β-actin (#MAB1501, clone C4; Millipore,
Temecula, CA, USA), a control for protein loading, and GFP
(#632375, Living Colors GFP Monoclonal Antibody; Clontech, Mountain
View, CA, USA), a marker of transfection. The blots were then
labeled for 1 h at room temperature with a 1:10,000 dilution of
peroxidase-conjugated goat anti-mouse IgG (#115-035-003; Jackson
ImmunoResearch Laboratories, West Grove, PA, USA) and visualized
with ECL Plus Western Blotting Detection Reagents (Amersham
Biosciences, Buckinghamshire, UK).
Immunohistochemistry
Formalin-fixed, paraffin-embedded cell lines,
non-small cell lung cancer xenografts and clinical specimens were
evaluated by immunohistochemistry based on streptavidin, biotin and
peroxidase. The samples were labeled overnight at 4°C with
monoclonal antibodies to MAGEA4 (MCV-1, 2.8 mg/ml) at a dilution of
1:2,000. To stain p53, the samples were labeled for 1 h at room
temperature with mouse monoclonal antibodies to human p53 (#M7001,
DO-7; Dako Japan, Tokyo, Japan). After washing, the sections were
incubated for 30 min at room temperature with peroxidase-labeled
goat anti-mouse and anti-rabbit polyclonal IgG (Fab'), and then
with Histofine Simple Stain MAXPO (MULTI) (Nichirei, Tokyo, Japan).
After further washing, specimens were visualized with freshly
prepared 3,3′-diaminobenzidine tetrahydrochloride supplied with the
Histofine SAB-PO (M) kit (Nichirei), counter-stained with
hematoxylin and mounted. Secimens probed with a mix of mouse
isotype IgG1 and IgG2a (Dako, Glostrup, Denmark) at a dilution of
1:20 were used as negative controls, whereas normal human testis
(#T2234260, Paraffin Tissue Section; BioChain, Newark, CA, USA) was
used as a positive control. Apoptosis was assessed by
immunostaining for cleaved caspase-3, using Autostainer Plus (Dako,
Carpinteria, CA, USA) and a rabbit monoclonal antibody to cleaved
caspase-3 (Asp175) (5A1E) (#9664; Cell Signaling Technology,
Danvers, MA, USA), which was diluted 1:100 in S2022 Antibody
Diluent (Dako). Specimens were then reacted for 60 min at room
temperature with the EnVision™ + Dual Link Detection kit
(K4063; Dako), a polymer-based detection system with
3′,3′-diaminobenzidine as the chromogen. Staining was quantified in
five fields per specimen using Aperio Image Scope software (Leica
Biosystems, Nussloch, Germany) and averaged.
Tissue microarray
We selected three cancer spots and two non-cancerous
spots from paraffin-embedded resected non-small cell lung cancer
tissues in each of the 240 patients described above. These
specimens were arrayed in a second paraffin block using Tissue
Arrayer (Beecher Instruments, Alphelys, Plaisir, France) with a
diameter of 0.6 mm. MAGEA4 and p53 staining in 1,200 samples from
240 different tissues was independently assessed by A.F.-K. and the
late Dr Masaki Miyamoto. The staining intensity was evaluated by
scoring ('−' for 0%; '+' for 1–100%) in both the nucleus and
cytoplasm, and we considered staining intensity as positive if any
of three cancerous spots indicated positive (+) expression for
MAGEA4 or p53. p53 staining was evaluated only with respect to
nuclear expression as all specimens expressing cytoplasmic p53 also
expressed nuclear p53. A specimen was considered positive if judged
by both investigators as positive.
Statistical analysis
The association between MAGEA4 and
clinicopathological variables was evaluated using the χ2
test with StatView J version 5.0 software (SAS Institute Inc.,
Cary, NC, USA). Apoptotic levels based on cleaved caspase-3 were
compared using the Student's t-test. The overall survival was
assessed by Kaplan-Meier analysis and the log-rank test. Uni- and
multivariate regression was performed using Cox's proportional
hazards model. P-values <0.05 were considered to indicate
statistically significant differences.
Results
Detection of MAGEA4 mRNA
MAGEA4 was detected by RT-qPCR in 293FT cells
transfected with MAGEA4 expression plasmid, but not in cells
transfected with other MAGEA genes (Fig. 1A). Endogenous MAGEA4
expression was also detected in four (PC10, LC-1, REPF-LCMS and
VMRC-LCD) of eight non-small cell lung cancer cell lines (50.0%,
Fig. 1B). The endogenous
expression of other MAGEA genes was also quantified
(Table III).
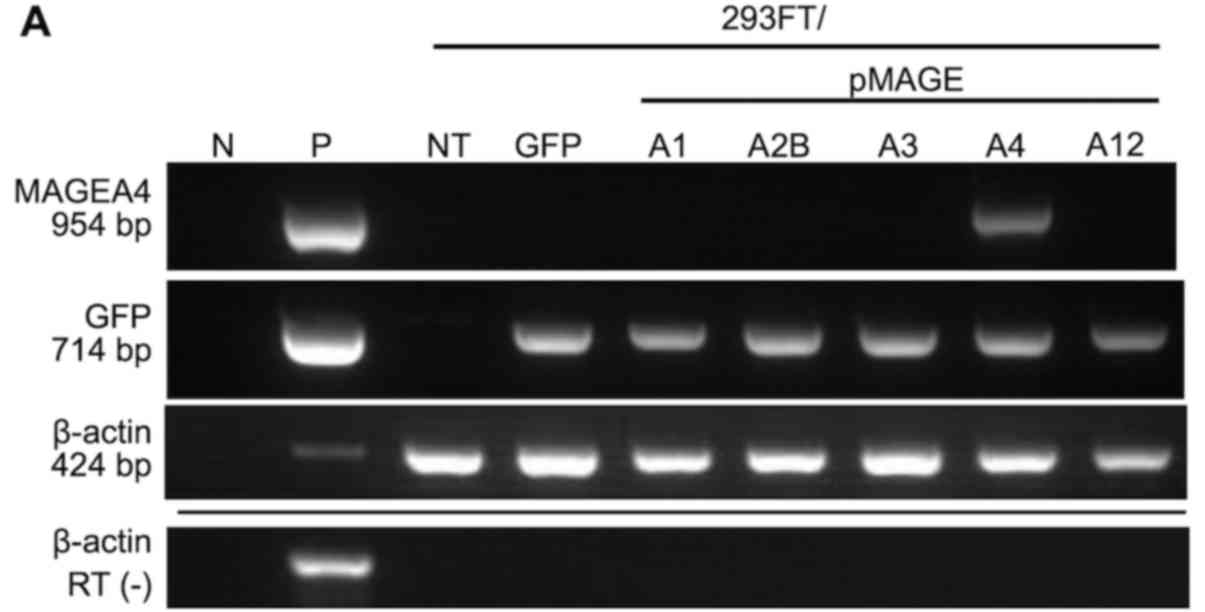 | Figure 1MAGEA4 expression in
transfected cells and non-small cell lung cancer cell lines. RT-PCR
of (A) transfected cells and (B) non-small cell lung cancer cells
using specific primers for MAGEA4, MAGEA2B, GFP and
β-actin (internal control). Transfection was assessed by GFP
expression. MAGEA4 was expressed in two of four (PC10 and
LC-1) squamous cell carcinoma cell lines, and in two of four
(REPF-LC-MS and VMRC-LCD) adenocarcinoma cell lines. (C) Western
blot analysis of transfected cells (upper panel) and non-small cell
lung cancer cells (lower panel) using MCV-1, a monoclonal antibody
to MAGEA4 that crossreacts with MAGEA2B and MAGEA12. Endogenous
MAGEA4 was detected in PC10 and LC-1 squamous cell carcinoma cell
lines, and in REPF-LCMS and VMRC-LCD adenocarcinoma cell lines. An
invalid cell line, that had been reported to be contaminated with
an astrocytoma cell line, was originally applied in the empty lane.
N, negative control; P, positive control; NT, no treatment; RT (−),
reverse transcription without RNA templates; 293FT/pMAGEA,
MAGEA-transfected cells; IB, immunoblot; MW, molecular weight
markers. |
 | Table IIIExpression of MAGEA family genes in
non-small cell lung cancer cell lines by RT-qPCR. |
Table III
Expression of MAGEA family genes in
non-small cell lung cancer cell lines by RT-qPCR.
| Squamous cell
carcinoma cells
| Adenocarcinoma
cells
|
|---|
| PC10 | H226 | LK2 | LC-1 | A549 | REPF-LCMS | VMRC-LCD | ABC-1 |
|---|
| MAGEA1 | − | − | − | − | − | − | − | − |
| MAGEA2B | + | − | + | − | − | + | + | − |
| MAGEA3 | + | − | + | − | − | + | + | − |
| MAGEA4 | + | − | − | + | − | + | + | − |
| MAGEA12 | + | − | − | − | − | + | + | − |
Evaluation of the anti-MCV-1
antibody
In western blots of 293FT cells transfected with
various MAGEA genes, the anti-MAGEA4 antibody, MCV-1,
crossreacted with MAGEA2B and MAGEA12 (Fig. 1C, upper panel). However, MCV-1
detected only the 37 kDa MAGEA4 protein in the cell lines, as
determined by RT-qPCR. The cells endogenously expressed the
MAGEA4 gene, but not MAGEA2B (at 35 kDa) (Fig. 1C, lower panel). Although MCV-1 also
crossreacted with MAGEA2B and MAGEA12 when used for
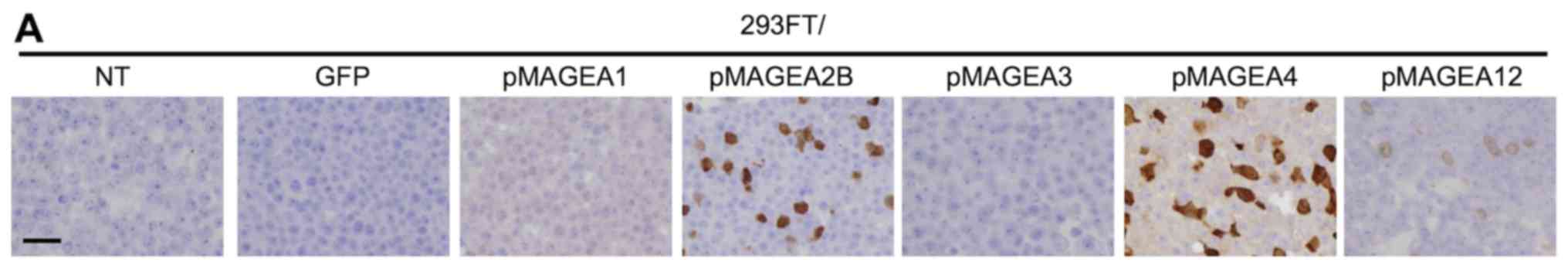
immunohistochemistry of the transfected cells (Fig. 2A), the antibody detected only
MAGEA4 in MAGEA4-positive cell lines (Fig. 2B). For example, MCV-1 did not stain
xenografted LK2 tissue, which was shown to express MAGEA2B, but not
MAGEA4 by RT-qPCR (Fig. 1B and
Table III). Collectively, these
results indicate that MCV-1 specifically detects endogenous MAGEA4
(Table IV).
 | Table IVOverall results of MAGEA4 expression
analyses in non-small cell lung cancer cell lines and
MAGEA-specific controls based on the same condition. |
Table IV
Overall results of MAGEA4 expression
analyses in non-small cell lung cancer cell lines and
MAGEA-specific controls based on the same condition.
| Non-small cell lung
cancer cell lines
| Testis | 293FT/pMAGE
|
|---|
| PC10 | H226 | LK2 | LC-1 | A549 | REPF-LC-MS | VMRC-LCD | ABC-1 | A1 | A2B | A3 | A4 | A12 |
|---|
| RT-PCR | ++ | − | − | ++ | − | ++ | + | − | | − | − | − | ++ | − |
| RT-qPCR |
10−2 | − | − |
10−2 | − |
10−2 |
10−3 | − | | | | | | |
| WB | | | | | | | | | | | | | | |
| MCV-1 | ++ | − | − | ++ | − | ++ | + | − | | − | ++ | − | ++ | + |
| 57B | ++ | − | − | + | − | ++ | + | − | | − | + | + | + | + |
| 6C1 | +− | − | − | + | − | + | − | − | | + | + | + | ++ | − |
| IHC | | | | | | | | | | | | | | |
| MCV-1 | ++ | − | − | ++ | − | ++ | + | − | ++ | − | + | − | + | + |
| 57B | + | − | − | + | − | + | + | − | ++ | + | ++ | ++ | ++ | ++ |
| 6C1 | − | − | − | +− | − | +− | − | − | + | ++ | +− | − | + | +− |
Subcellular localization of MAGEA4
MAGEA4 was shown to be accumulated in the cytoplasm
of transfected 293FT cells, as assessed by fluorescence from the
genetically fused GFP (Fig. 2C and
D). However, endogenous MAGEA4 was detected not only in the
cytoplasm, but also in the nuclei of xenografts derived from
MAGEA4-positive cell lines (Fig.
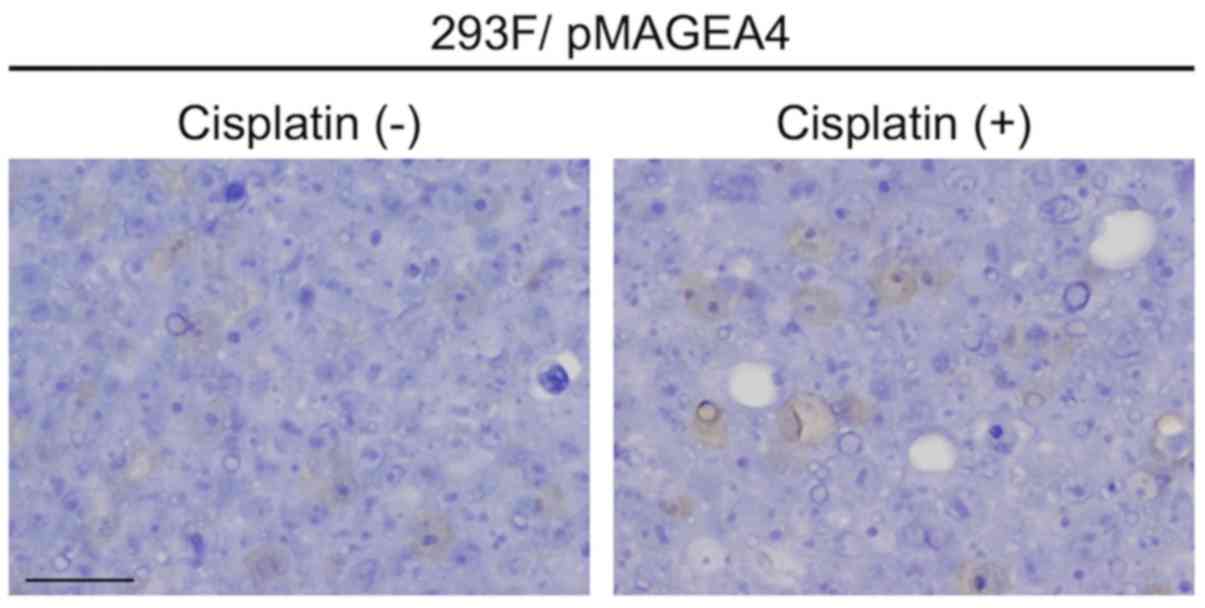
2B). Notably, exposure for 24–48 h to 1 μM cisplatin, a
cytotoxic anticancer agent, did not alter the intracellular
localization of MAGEA4 in transfected 293F cells (Fig. 3).
Cytoplasmic MAGEA4 inhibits
apoptosis
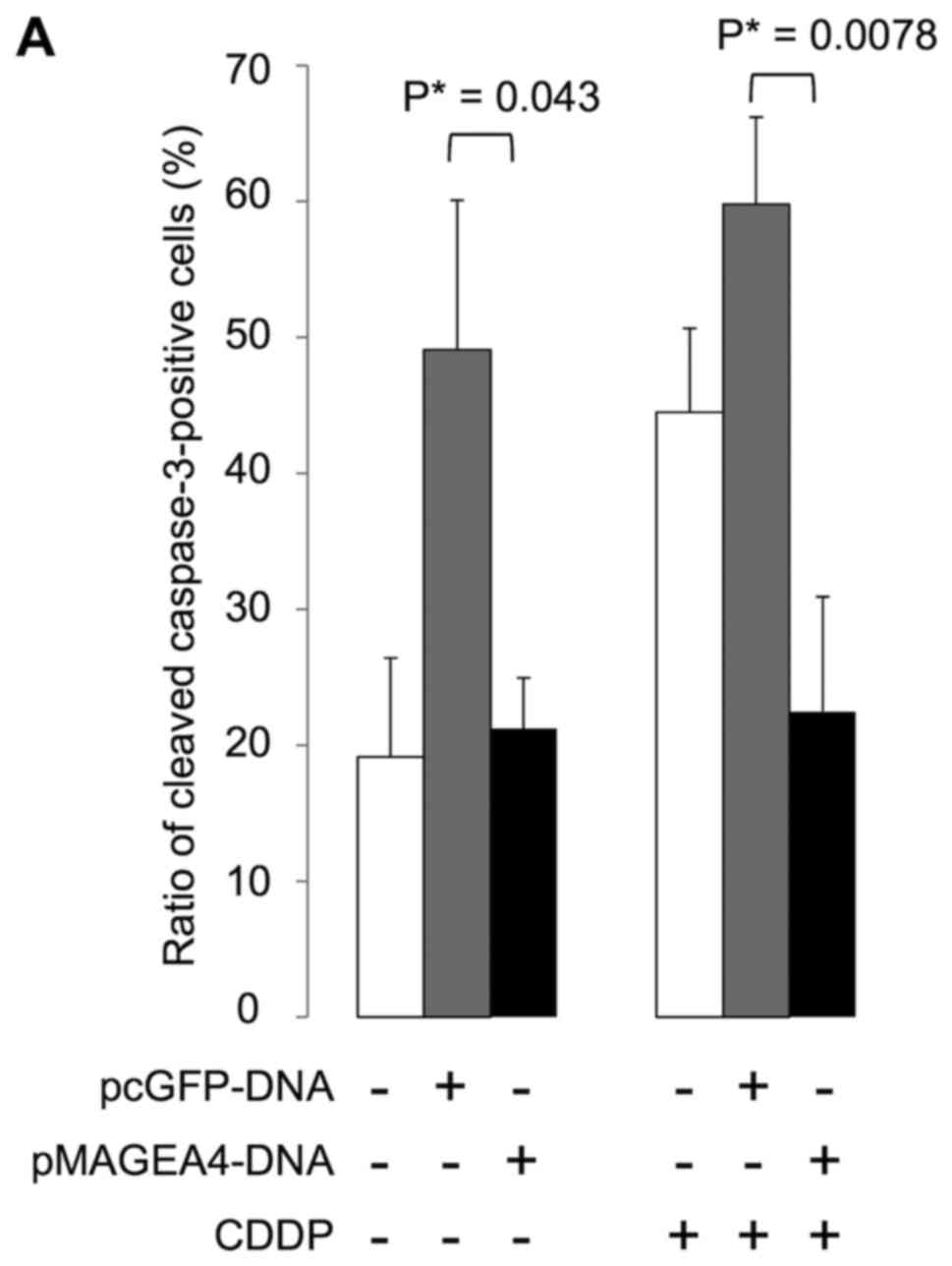
Using immunohistochemistry, we examined caspase-3
activation in stably transfected 293F cells to investigate whether
MAGEA4 expression induces or inhibits apoptosis in response to
genotoxic stress. We found that cells expressing cleaved caspase-3
were significantly fewer in number in the cultures transfected with
MAGEA4 than in the cultures transfected with the empty vector, with
(P=0.0078) or without (P=0.043) exposure to cisplatin for 24 h
(Fig. 4). These results suggest
that MAGEA4 inhibits apoptosis via caspase.
MAGEA4 subcellular localization and
prognostic value in clinical non-small cell lung cancers
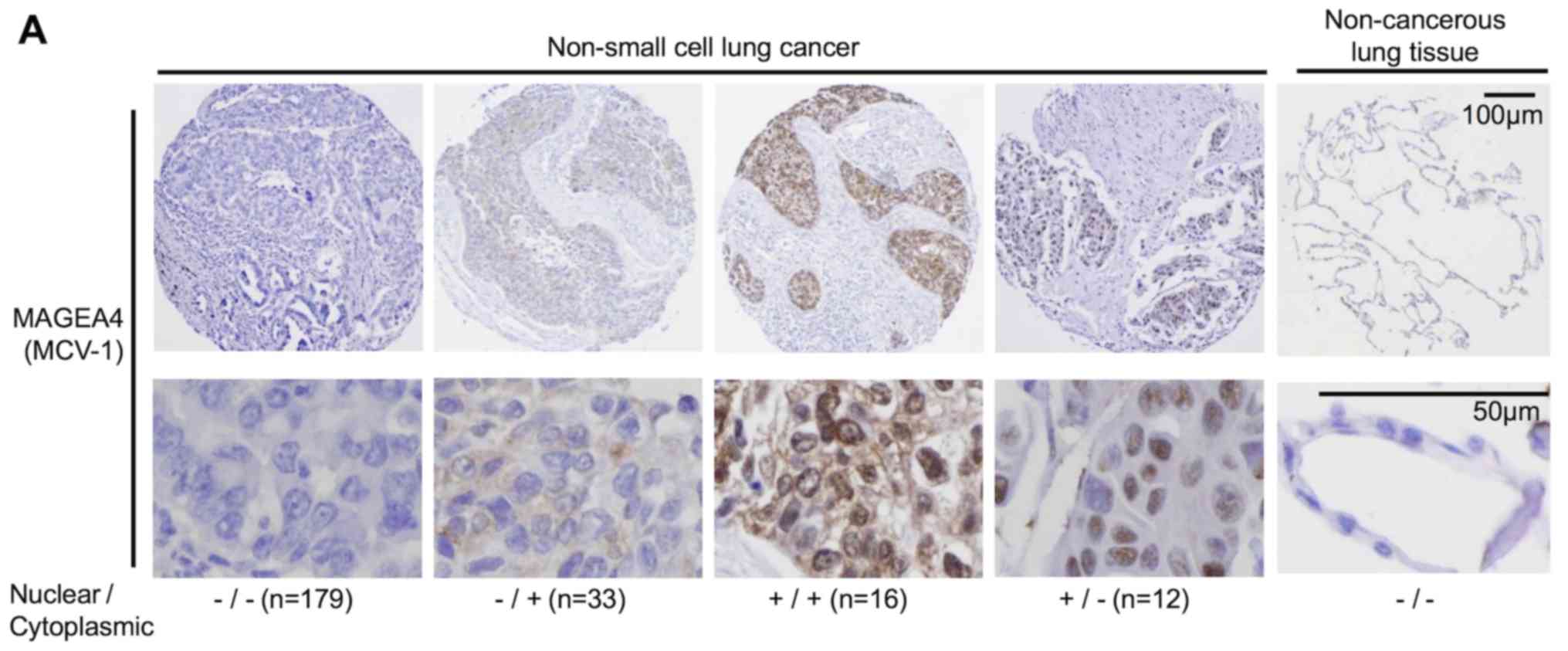
MAGEA4 was detected in 61 of 240 patients (25.4%);
33 cases in the cytoplasm only, 12 cases in the nucleus only and 16
cases in both the cytoplasm and nucleus (Table V). MAGEA4 expression was
significantly associated with male patients, of whom 31% were
positive for MAGEA4, although only 18% of female patients exhibited
positivity (P=0.0251). MAGEA4 expression was also associated with
squamous cell carcinomas, with MAGEA4 detected in 47% of cases, but
in only 16% of adenocarcinomas (P<0.0001). Finally, the positive
expression rate was significantly higher in tissues with an
advanced pathological stage, being detected in 21, 32 and 34% of
patients in stage I, II and III/IV disease (P=0.0309), respectively
(Table VI). Of note, cytoplasmic
MAGEA4 was also associated with the male sex (P=0.0049) and
squamous cell carcinomas (P<0.0001). However, no significant
association was found between nuclear MAGEA4 expression and
clinicopathological variables (Table
VI). A representative immunohistochemical analysis of
cytoplasmic and nuclear MAGEA4 expression is shown in Fig. 5A. Of the 240 tumor specimens, 12
(5.0%) exhibited nuclear, but not cytoplasmic MAGEA4 expression,
whereas 33 specimens (13.8%) exhibited cytoplasmic, but not nuclear
MAGEA4 expression. Both the nuclear and cytoplasmic forms were
detected in 16 cases (6.7%, Table
V). Patients expressing only nuclear or only cytoplasmic MAGEA4
exhibited a significantly poorer overall survival (P=0.0424 and
P=0.0340, respectively) (Fig. 5B),
than those with neither. Intriguingly, the accumulation of both the
nuclear and cytoplasmic forms was not prognostic (P=0.9101,
Fig. 5B). Therefore, we
hypothesized that the intracellular localization of MAGEA4 was
functionally significant, and was thus associated with
prognosis.
 | Table VMAGEA4 expression pattern according
to its subcellular localization. |
Table V
MAGEA4 expression pattern according
to its subcellular localization.
| MAGEA4 in cytoplasm
| Total (%) |
|---|
| − | + |
|---|
| MAGEA4 in nucleus
(%) | − | 179 (74.6) | 33 (13.8) | 212 (88.3) |
| + | 12 (5.0) | 16 (6.7) | 28 (11.7) |
| Total (%) | | 191 (79.6) | 49 (20.4) | 240 (100.0) |
 | Table VIClinicopathological characteristics
for the tissue microarray analysis according to MAGEA4
expression. |
Table VI
Clinicopathological characteristics
for the tissue microarray analysis according to MAGEA4
expression.
| Total
n=240 | MAGEA4 expression
| Cytoplasmic MAGEA4
| Nuclear MAGEA4
|
|---|
+
n (%) | −
n (%) | P-value | +
n (%) | −
n (%) | P-value | +
n (%) | −
n (%) | P-value |
|---|
| Sex | | | | | | | | | | |
| Male | 144 | 44 (31) | 100 (69) | 0.0251 | 38 (26) | 106 (74) | 0.0049 | 18 (12) | 126 (88) | 0.6223 |
| Female | 96 | 17 (18) | 79 (82) | | 11 (11) | 85 (89) | | 10 (10) | 86 (90) | |
| Age (years) | | | | | | | | | | |
| ≥65 | 138 | 30 (22) | 108 (78) | 0.1280 | 23 (17) | 115 (83) | 0.0937 | 17 (12) | 121 (88) | 0.7143 |
| <65 | 102 | 31 (30) | 71 (70) | | 26 (25) | 76 (75) | | 11 (11) | 91 (89) | |
| Histological
type | | | | | | | | | | |
| ADC | 158 (66) | 25 (16) | 133 (84) |
<0.0001a | 16 (10) | 142 (90) |
<0.0001a | 13 (8) | 145 (92) | 0.0509a |
| SqCC | 68 (28) | 32 (47) | 36 (53) | | 30 (44) | 38 (56) | | 13 (19) | 55 (81) | |
| Other | 14 (6) | 4 (29) | 10 (71) | | 3 (21) | 11 (79) | | 2 (14) | 12 (86) | |
| pStage | | | | | | | | | | |
| I | 146 | 30 (21) | 116 (79) |
0.0309b | 24 (16) | 122 (84) | 0.0567b | 15 (10) | 131 (90) | 0.4023b |
| II | 50 | 16 (32) | 34 (68) | | 14 (28) | 36 (72) | | 4 (8) | 46 (92) | |
| III, IV | 44 | 15 (34) | 29 (66) | | 11 (25) | 33 (75) | | 9 (20) | 35 (80) | |
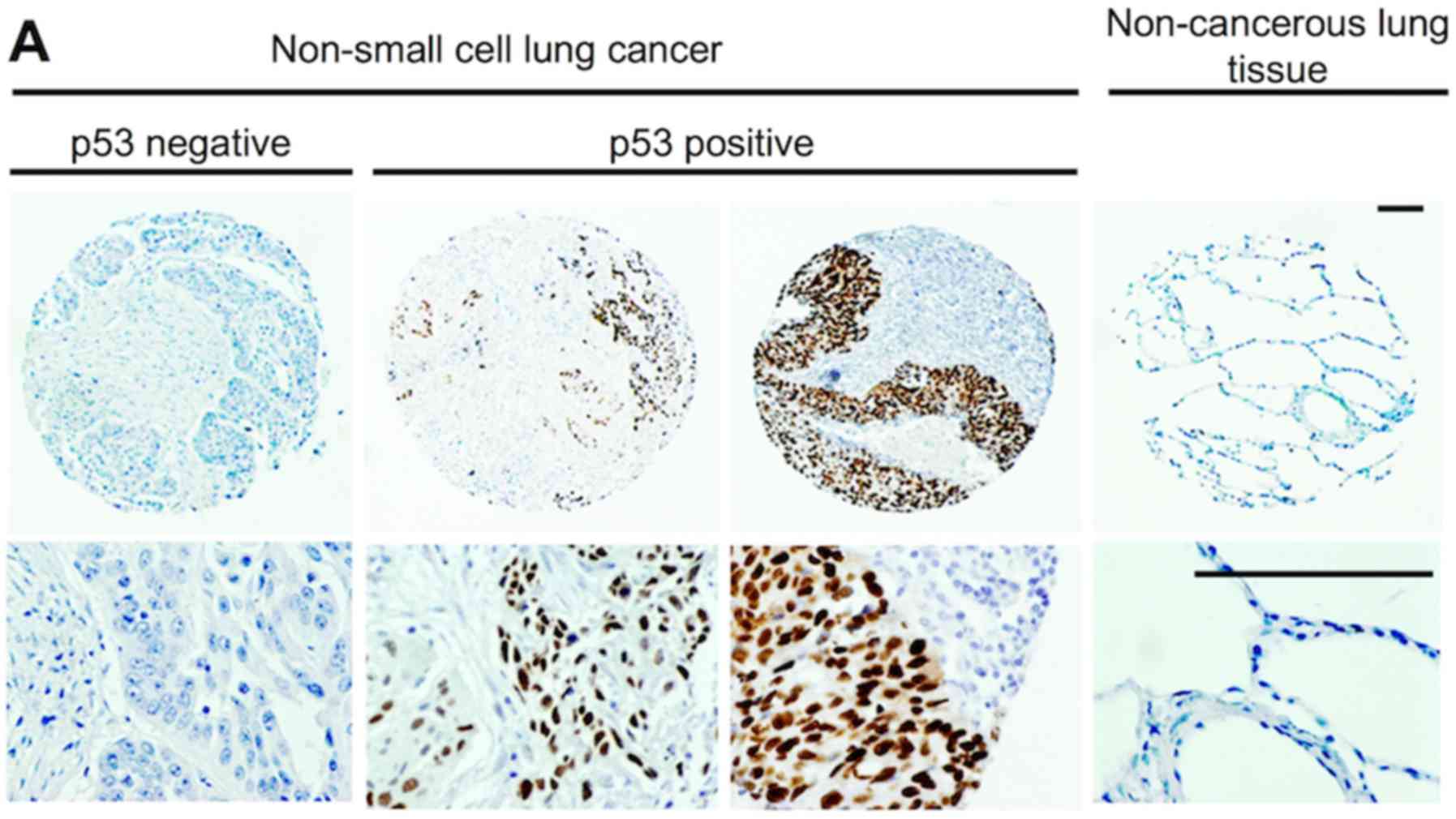
Association with the p53 status
In light of the link between the MAGEA4 subcellular
localization and prognosis, we also examined the association
between p53 and MAGEA4, as MAGEA proteins have been reported to be
in complex with p53 at p53 cognate sites in chromatin (5). The p53 status was
immunohistochemically surveyed in clinical specimens (Fig. 6A). p53 expression itself did not
have significant prognostic value (Fig. 6B). In addition, there was no
difference in survival among patients without nuclear MAGEA4,
regardless of p53 expression. However, patients with nuclear MAGEA4
expression, but not p53 expression exhibited a significantly poorer
survival than others (P=0.0017; Fig.
6C, left panel). Conversely, the survival rate was 100% in
patients with nuclear MAGEA4 and p53 expression (Fig. 6C, right panel). Moreover, the
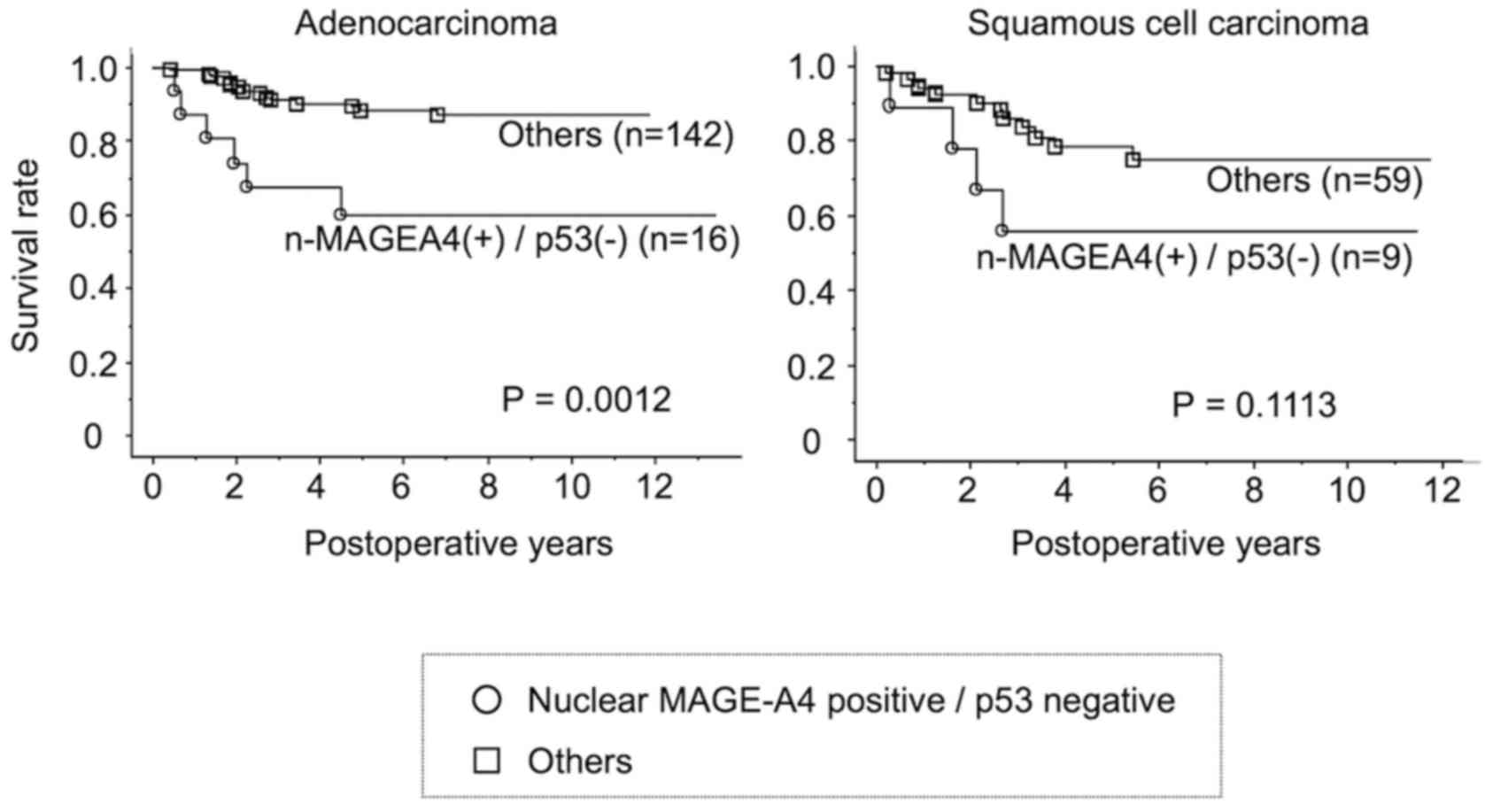
patients with lung adenocarcinoma with nuclear MAGEA4 expression,
but not p53 expression exhibited a shorter overall five-year
survival than patients with other forms of adenocarcinoma
(P=0.0012, Fig. 7). A similar
trend was observed in patients with squamous cell carcinomas,
although the difference was not statistically significant
(P=0.1113).
Uni- and multivariate analysis implied that an
advanced pT status (P= 0.0001), advanced pN status (P<0.0001),
non-adenocarcinoma histology (P=0.0436) and the accumulation of
nuclear MAGEA4, but not p53 expression (P=0.0022) were
significantly associated with a poor prognosis (Table VII). Multivariate analysis also
showed that expression of nuclear MAGEA4 but not p53 was an
independent prognostic factor in patients with non-small cell lung
cancers (P=0.0116), as were pT (P=0.0066) and pN status (P=0.0035,
Table VII).
 | Table VIIPrognostic factors in Cox's
proportional hazards model in non-small cell lung cancer
(n=231a). |
Table VII
Prognostic factors in Cox's
proportional hazards model in non-small cell lung cancer
(n=231a).
| Variables | Risk ratio |
Univariate
95% CI | P-value | Risk ratio |
Multivariate
95% CI | P-value |
|---|
| Age (years) | | | | | | |
| ≥65/<65 | 1.650 | 0.829-3.286 | 0.1540 | | | |
| Sex | | | | | | |
| Male/female | 1.927 | 0.951-3.906 | 0.0687 | | | |
| pT | | | | | | |
| pT2-4/pT1 | 5.025 | 2.203-11.494 | 0.0001b | 3.333 | 1.399-7.937 | 0.0066b |
| pN | | | | | | |
| pN1-2/pN0 | 4.274 | 2.217-8.197 | <0.0001b | 2.755 | 1.395-5.435 | 0.0035b |
| Histology | | | | | | |
| ADC/non-ADC | 1.949 | 1.019-3.717 | 0.0436b | 1.497 | 0.773-2.899 | 0.2318 |
| Nuclear MAGEA4 (+)
p53 (−) | | | | | | |
| Yes/no | 3.953 | 1.642-9.524 | 0.0022b | 3.155 | 1.292-7.692 | 0.0116b |
Discussion
Immunohistochemical staining is a standard analysis
used to detect specific proteins in tissues, but is limited by
issues of sensitivity and specificity. In particular, it is not
rare for an antibody used in western blot analysis or
immunohistochemistry to crossreact with other proteins. For
example, as previously demonstrated, the monoclonal antibody, 57B,
which was raised against MAGEA3, stains cells transfected with
plasmids encoding multiple MAGEA genes, and preferentially reacts
with MAGEA4 in tissue sections (16). In addition, variable results were
previously obtained when MAGEA4 was surveyed by RT-PCR, western
blot analysis and immunohistochemistry (8,12).
These inconsistencies are likely due to the lack of standard
criteria to evaluate results and to inadequate assay validation.
Accordingly, we have previously highlighted the importance of
optimizing immunohistochemical staining conditions based on
suitable controls (19). Such
staining conditions are already established for widely studied
proteins, such as p53, but not for many cancer-related proteins.
Therefore, we optimized the staining conditions for MAGEA4 in the
present study, with a view toward enabling patient selection for
clinical trials of cancer vaccines against MAGEA4 (4,20,21).
This was achieved by the analysis of cells transfected with MAGEA
genes and of xenografted non-small cell lung cancer cell lines.
The function of MAGEA4 remains controversial. For
example, Peikert et al (8)
reported that exogenous MAGEA4 accumulated in the nucleus, induced
caspase-mediated apoptosis and sensitized non-small cell lung
cancers to chemotherapeutic agents, implying that MAGEA4 was a
tumor suppressor. Consistent with this supposition, MAGEA4 was
processed by the proteasome to generate a pro-apoptotic C-terminal
fragment that ultimately boosted p53, thereby eliciting apoptosis
(10). This process is triggered
by low doses of adriamycin, either to maintain cellular homeostasis
or initiate apoptosis depending on MAGEA4 expression (22). By contrast, we found that exogenous
MAGEA4 expression inhibited apoptosis, particularly under genotoxic
stress, as assessed by immunohistochemistry for active (cleaved)
caspase-3. This result implies that MAGEA4 favors tumor cell
survival, and thus functions as an oncoprotein. Of note, we found
that exogenous MAGEA4 expression was accumulated exclusively in the
cytoplasm, and was insensitive to cisplatin. We hypothesized that
this difference in subcellular localization may explain why our
results are contradictory those of Peikert et al (8) and others (10,22).
Furthermore, the antibody we raised against MAGEA4, MCV-1,
recognizes amino acids 71–87 of 318 amino acids, and thus may not
react with the pro-apoptotic C-terminal fragment nor show its
distribution. Hence, antibodies specific to this fragment may help
clarify the function of MAGEA4.
Notably, endogenous MAGEA4 was accumulated in the
cytoplasm and nucleus of xenografted non-small cell lung cancer
cell lines and of resected clinical specimens. Importantly,
exclusively nuclear or exclusively cytoplasmic MAGEA4 was
indicative of a poor prognosis, whereas MAGEA4 accumulation in both
compartments was associated with a favorable prognosis similar to
that of MAGEA4-negative patients. Collectively, these findings
indicate that to better understand the function and prognostic
value of MAGEA4, it is critical to examine not only mRNA expression
but also subcellular localization.
Some studies have shown that endogenous MAGEA
proteins inhibit the apoptosis of cancer cells in association with
wild-type p53, and contribute to tumor aggressiveness (5,23).
For example, suppression of class-1 MAGE (A, B and C) induces
apoptosis in p53 wt/wt HCT116 cancer cells, but not in
p53−/− cells, implying that the anti-apoptotic effects
of MAGEA proteins depend on p53 (23). MAGEA proteins also block p53
binding to cognate sites in chromatin, suppressing cell death
(5). Accordingly, we found that
the accumulation of nuclear MAGEA4 in the absence of p53 resulted
in a significantly shorter survival, and was an independent
indicator of poor prognosis in non-small cell lung cancers.
Therefore, nuclear MAGEA4 may inhibit the apoptosis of cancer cells
by suppressing wild-type p53, or by enhancing malignant progression
via p53-independent pathways. Finally, our data clearly indicate
that the accumulation of nuclear MAGEA4 together with p53 are
associated with the apoptosis of cancer cells and with a better
prognosis.
We noted that the DO-7 antibody we used to detect
p53 reacts with both mutant p53 and overexpressed wild-type p53
(24), and hence we could not
confirm whether p53-positive cells were expressing mutant p53 or
overexpressing wild-type p53. p53 was detected in 108 of 212 cases
without nuclear MAGEA4 (50.9%), a rate consistent with previous
surveys investigating mutant p53 as a prognostic factor in lung
cancer (25). Nevertheless, we
found that p53 itself may be of little prognostic value in
non-small cell lung cancer (Fig.
6B), differently from previous surveys (25,26).
Although we speculate that cells without nuclear MAGEA4 expression
probably express mutant p53, whereas cells with nuclear MAGEA4
expression probably overexpress wild-type p53, a more comprehensive
study is warranted in order to fully evaluate the association
between MAGEA4 and p53.
It should also be noted that the major purpose of
this study was to enable accurate patient selection for cancer
vaccine therapy against MAGEA4, not to show the potential for
MAGEA4 as a marker in patients with non-small cell carcinoma. We
demonstrated that 25.4% of patients with lung carcinoma, and up to
47% of those with lung squamous cell lung carcinoma have the
potential to benefit from vaccine therapy.
In conclusion, whereas MAGEA4 expression by itself
may be indicative of a poor prognosis, the prognostic value
entirely depends on its subcellular localization and on the p53
status. Indeed, the accumulation of nuclear MAGEA4 expression
without p53 expression is significantly associated with a poor
survival, implying that MAGEA4 inhibits apoptosis and increases
tumorigenesis. However, the accumulation of both p53 and nuclear
MAGEA4 is indicative of favorable prognosis, suggesting the
induction of apoptosis via a MAGEA4/p53 pathway. Although the
mechanistic basis of the association between p53 and MAGEA4 remains
unknown, our data may help to resolve the controversy over whether
MAGEA4 promotes or inhibits apoptosis. Our observations may also
help clarify the role of MAGEA4 in non-small cell lung
carcinogenesis.
Acknowledgments
The authors would like to specifically thank Mr.
Hiraku Shida (Tonan Hospital, Sapporo, Hokkaido, Japan), who
performed the p53 immunohistochemistry and to Mr. Takatomo Funayama
(Morphotechnology Co., Ltd., Sapporo, Japan), who performed
immunohistochemistry for cleaved caspase-3. The authors would also
thank Mr. Daiyoon Lee (Virginia Commonwealth University, Richmond,
VA, USA) for correcting the manuscript and Editage (www.editage.jp) for English language editing.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
AFK performed the experimental design, most of the
experiments and analysis, drafted manuscript, and was involved in
the conception and design of the study. TK analyzed the data,
performed the statistical analysis, and contributed to the writing
of manuscript. TA contributed to the preparation for the tissue
microarray. NK, MI, KT and TT conducted some supporting
experiments. TN and SH supervised the study and were involved in
the conception and design of the study. YH, KK, YM were involved in
collecting tissue samples and accessing clinical databases. HI, SK
and HS participated in the planning/design of the experiments and
supplied the antibodies. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Hokkaido University. Informed consent for the use of tissue samples
which were stored prior to the establishment of ethics approval,
were originally obtained from the patients at the time of surgery.
The Hokkaido University Institutional Review Board confirmed that
this study was fully ethically compliant and the second informed
consent was waived by the ethics committee of our institution. All
animal experiments were conducted according to the guidelines of
the Hokkaido University Institutional Animal Care and Use Committee
with the Institutional Animal Care and Use Committee approval.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
De Plaen E, Arden K, Traversari C, Gaforio
JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethé B,
Lurquin C, et al: Structure, chromosomal localization, and
expression of 12 genes of the MAGE family. Immunogenetics.
40:360–369. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Caballero OL and Chen YT: Cancer/testis
(CT) antigens: Potential targets for immunotherapy. Cancer Sci.
100:2014–2021. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi N, Ohkuri T, Homma S, Ohtake J,
Wakita D, Togashi Y, Kitamura H, Todo S and Nishimura T: First
clinical trial of cancer vaccine therapy with artificially
synthesized helper/killer-hybrid epitope long peptide of MAGE-A4
cancer antigen. Cancer Sci. 103:150–153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saito T, Wada H, Yamasaki M, Miyata H,
Nishikawa H, Sato E, Kageyama S, Shiku H, Mori M and Doki Y: High
expression of MAGE-A4 and MHC class I antigens in tumor cells and
induction of MAGE-A4 immune responses are prognostic markers of
CHP-MAGE-A4 cancer vaccine. Vaccine. 32:5901–5907. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marcar L, Maclaine NJ, Hupp TR and Meek
DW: Mage-A cancer/testis antigens inhibit p53 function by blocking
its interaction with chromatin. Cancer Res. 70:10362–10370. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhan S, Chuang A, Negi SS, Glazer CA and
Califano JA: MAGEA4 induces growth in normal oral keratinocytes by
inhibiting growth arrest and apoptosis. Oncol Rep. 28:1498–1502.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duan Z, Duan Y, Lamendola DE, Yusuf RZ,
Naeem R, Penson RT and Seiden MV: Overexpression of MAGE/GAGE genes
in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin
Cancer Res. 9:2778–2785. 2003.PubMed/NCBI
|
|
8
|
Peikert T, Specks U, Farver C, Erzurum SC
and Comhair SA: Melanoma antigen A4 is expressed in non-small cell
lung cancers and promotes apoptosis. Cancer Res. 66:4693–4700.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagao T, Higashitsuji H, Nonoguchi K,
Sakurai T, Dawson S, Mayer RJ, Itoh K and Fujita J: MAGE-A4
interacts with the liver oncoprotein gankyrin and suppresses its
tumorigenic activity. J Biol Chem. 278:10668–10674. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sakurai T, Itoh K, Higashitsuji H, Nagao
T, Nonoguchi K, Chiba T and Fujita J: A cleaved form of MAGE-A4
binds to Miz-1 and induces apoptosis in human cells. J Biol Chem.
279:15505–15514. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gure AO, Chua R, Williamson B, Gonen M,
Ferrera CA, Gnjatic S, Ritter G, Simpson AJ, Chen YT, Old LJ, et
al: Cancer-testis genes are coordinately expressed and are markers
of poor outcome in non-small cell lung cancer. Clin Cancer Res.
11:8055–8062. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Groeper C, Gambazzi F, Zajac P, Bubendorf
L, Adamina M, Rosenthal R, Zerkowski HR, Heberer M and Spagnoli GC:
Cancer/testis antigen expression and specific cytotoxic T
lymphocyte responses in non small cell lung cancer. Int J Cancer.
120:337–343. 2007. View Article : Google Scholar
|
|
13
|
Yoshida N, Abe H, Ohkuri T, Wakita D, Sato
M, Noguchi D, Miyamoto M, Morikawa T, Kondo S, Ikeda H, et al:
Expression of the MAGE-A4 and NY-ESO-1 cancer-testis antigens and T
cell infiltration in non-small cell lung carcinoma and their
prognostic significance. Int J Oncol. 28:1089–1098. 2006.PubMed/NCBI
|
|
14
|
Shigematsu Y, Hanagiri T, Shiota H, Kuroda
K, Baba T, Mizukami M, So T, Ichiki Y, Yasuda M, So T, et al:
Clinical significance of cancer/testis antigens expression in
patients with non-small cell lung cancer. Lung Cancer. 68:105–110.
2010. View Article : Google Scholar
|
|
15
|
Rimoldi D, Salvi S, Schultz-Thater E,
Spagnoli GC and Cerottini JC: Anti-MAGE-3 antibody 57B and
anti-MAGE-1 antibody 6C1 can be used to study different proteins of
the MAGE-A family. Int J Cancer. 86:749–751. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landry C, Brasseur F, Spagnoli GC, Marbaix
E, Boon T, Coulie P and Godelaine D: Monoclonal antibody 57B stains
tumor tissues that express gene MAGE-A4. Int J Cancer. 86:835–841.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin L, Gospodarowics M and Wittekind C:
International Union Against Cancer: TNM Classification of Malignant
Tumours. Wiley-Blackwell; Chichester, West Sussex: 2009
|
|
18
|
Ichinokawa M, Miyamoto M, Tanaka K,
Kyogoku N, Kuroda A, Maki T, Yamamura Y and Hirano S: Sequence
confirmation and characterization of the mouse Ssxa gene: Ssxa
protein is cleaved and the N-terminal cleaved fragment translocates
into the nucleus. Int J Mol Med. 28:705–710. 2011.PubMed/NCBI
|
|
19
|
Ishikawa K, Miyamoto M, Yoshioka T, Kadoya
M, Li L, Mishra R, Ichinokawa K, Shoji Y, Matsumura Y, Hida Y, et
al: Method for the validation of immunohistochemical staining using
SCID mouse xenografts: Expression of CD40 and CD154 in human
non-small cell lung cancer. Oncol Rep. 29:1315–1321. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kyogoku N, Ikeda H, Tsuchikawa T, Abiko T,
Fujiwara A, Maki T, Yamamura Y, Ichinokawa M, Tanaka K, Imai N, et
al: Time-dependent transition of the immunoglobulin G subclass and
immunoglobulin E response in cancer patients vaccinated with
cholesteryl pullulan-melanoma antigen gene-A4 nanogel. Oncol Lett.
12:4493–4504. 2016. View Article : Google Scholar
|
|
21
|
Miyauchi K, Tsuchikawa T, Wada M, Abiko T,
Kyogoku N, Shichinohe T, Miyahara Y, Kageyama S, Ikeda H, Shiku H,
et al: Clinical relevance of antigen spreading pattern induced by
CHP-MAGE-A4 cancer vaccination. Immunotherapy. 8:527–540. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakurai T, Kudo M, Itoh K, Ryu U,
Higashitsuji H and Fujita J: Adriamycin enhances
proteasome-mediated generation of the proapoptotic processed form
of MAGE-A4 in hepatoma cells. Oncology. 81(Suppl 1): 30–35. 2011.
View Article : Google Scholar
|
|
23
|
Yang B, O' Herrin SM, Wu J, Reagan-Shaw S,
Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et
al: MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1
and suppress p53-dependent apoptosis in MAGE-positive cell lines.
Cancer Res. 67:9954–9962. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nielsen S: Flex Ready-to-Use Atlas of
Stains. Dako; Santa Clara, CA: 2012
|
|
25
|
Steels E, Paesmans M, Berghmans T, Branle
F, Lemaitre F, Mascaux C, Meert AP, Vallot F, Lafitte JJ and
Sculier JP: Role of p53 as a prognostic factor for survival in lung
cancer: A systematic review of the literature with a meta-analysis.
Eur Respir J. 18:705–719. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mitsudomi T, Hamajima N, Ogawa M and
Takahashi T: Prognostic significance of p53 alterations in patients
with non-small cell lung cancer: A meta-analysis. Clin Cancer Res.
6:4055–4063. 2000.PubMed/NCBI
|