Introduction
Colorectal cancer (CRC) is one of the most common
causes of malignancy-associated mortality, and tumor metastasis is
the predominant component that leads to disease recurrence and
mortality in patients with CRC (1,2).
Previous studies have demonstrated that chronic intestinal
inflammation such as inflammatory bowel disease is associated with
the risk of CRC, and the inflammatory microenvironment around the
tumor serves a central function in the development and progression
of CRC (3–5). Various pro-inflammatory cytokines
released by infiltrating immune cells and other cells in the
microenvironment have been identified to induce CRC metastasis
(6). In particular, among these
cytokines, including interleukin (IL)-6, IL-8, IL-13 and IL-17 are
involved in the pathogenesis of CRC metastasis (7–10).
In addition, previous studies have focused on the overactivation of
IL-4 signaling in association with CRC cell invasiveness and
metastasis. In CRC cells expressing the IL-4 receptor (IL-4R), IL-4
stimulation downregulates the expression of the cell adhesion
molecule epithelial (E-)cadherin, suggesting that the cytokine may
be involved in the epithelial-mesenchymal transition (EMT) of CRC
(11). Additionally, IL-4Rα
appears to promote CRC growth, and blockade of the IL-4Rα-IL-4
interaction decreases proliferation and increases apoptosis
(12).
IL-4 initiates the signaling pathway by binding to
IL-4Rα, triggering a cascade, including the activation of Janus
kinase (JAK)1 and JAK3, which phosphorylates and dimerizes signal
transducer and activator of transcription 6 (STAT6). Finally, STAT6
homodimers translocate to the nucleus and bind to the promoters of
IL-4-responsive genes to induce various biological functions
(13). Therefore, STAT6 mediates
signal transduction and determines the cellular response to IL-4
stimulation. However, abnormal STAT6 activation induced by IL-4
promotes pro-metastatic processes including migration and invasion,
proliferation, and survival in cancer cells. For example, in
colorectal cancer stem cells (CR-CSCs), the IL-4/STAT6 signaling
pathway helps cells to escape cell death by increasing the nuclear
survivin pool (14). In addition,
Liu et al (15) identified
that the IL-4/STAT6 signaling pathway accelerates CRC cell
proliferation through enhancing the expression of
nicotinamide-adenine dinucleotide phosphate oxidase 1.
Although E2F1 is recognized as a strong
apoptosis-driver following chemotherapy-induced DNA damage in all
types of human cancer, evidence suggests that E2F1 is associated
with cancer progression. Increased abundance of E2F1 in various
cancer cells has been identified to trigger invasion and metastasis
by activating cytokine receptor signaling pathways. In malignant
melanoma, E2F1-dependent progression is mediated via upregulation
of epidermal growth factor receptor (EGFR) and activation of the
mitogen-activated protein kinase/extracellular-signal-regulated
kinase and phosphoinositide 3-kinase/protein kinase B signaling
cascades (16). Other evidence
indicates that E2F1 transactivates integrin-linked kinase, which
drives the IL-6/nuclear factor-κB (NF-κB) signaling loop in
triple-negative breast cancer (17). In our previous study, different
expression levels of E2F1 were detected in several CRC cell lines,
and knockdown of E2F1 was identified to impair the aggressive
phenotypes of CRC cells with highly expressed E2F1 (18). Therefore, whether E2F1 contributes
to CRC development promoted by inflammatory cytokines requires
further investigation.
In the present study, a previously unknown function
of E2F1 as an enhancer in the IL-4/STAT6 signaling pathway was
identified. The results supported the hypothesis that E2F1 is able
to induce STAT6 expression by upregulating specificity protein 3
(SP3), which was identified as a transcription activator of the
STAT6 gene in CRC cells. An increase in total STAT6 protein led to
a strong response to IL-4 stimulation, as indicated by a high level
of STAT6 phosphorylation. Finally, as targets of the activated
STAT6, zinc finger E-box-binding homeobox (Zeb)1 and Zeb2 boosted
EMT and aggressiveness of CRC cells. Thus, the existence of an
aberrant E2F1/SP3/STAT6 axis may amplify the IL-4 signaling which
facilitates CRC metastasis.
Materials and methods
Cell lines and culture
Human CRC cell lines SW480, HCT116, RKO, HT-29 and
DLD1 were purchased from the American Type Culture Collection
(Manassas, VA, USA). The cells were cultured and stored according
to the supplier's protocol. In detail, all the cells were cultured
in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in
a humidified atmosphere containing 5% CO2. Cell passage
was performed every 2 days. Short hairpin RNA targeting E2F1
(shE2F1, 5′-CUGCAGAGCAGAUGGUUAU-3′; GenePharma, Shanghai,
China)-stably transfected HCT116 cells were derived from the
parental cells using G418 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) selection. Human recombinant IL-4 (R&D Systems,
Minneapolis, MN, USA) at 20 ng/ml was used to treat CRC cells. Cell
morphology was visualized by phase-contrast microscopy
(magnification, ×400).
Reagents and antibodies
Recombinant human IL-4 was purchased from PeproTech,
Inc. (Rocky Hill, NJ, USA). Antibodies against E-cadherin (cat. no.
sc-7870; 1:1,000), vimentin (cat. no. sc-6260; 1:1,000), E2F1 (cat.
no. sc-193; 1:1,000) and GAPDH (cat. no. sc-365062; 1:5,000) were
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Antibodies against zonula occludens-1 (ZO-1; cat. no. ab59720;
1:500), fibronectin (cat. no. ab2413; 1:1,000) and SP3 (cat. no.
ab129099; 1:1,000) were purchased from Abcam (Cambridge, UK).
Antibodies against phosphorylated (p-)JAK1 (cat. no. 3331; 1:500),
JAK1 (cat. no. 3332; 1:1,000), p-STAT6 (cat. no. 9361; 1:500) and
STAT6 (cat. no. 9362; 1:1,000) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the CRC cell lines
(HCT116, RKO, HT-29 or DLD1) with RNAiso Plus (Takara Bio, Inc.,
Otsu, Japan), according to the manufacturer's protocol. RNA quality
and concentration was evaluated using a NanoDrop ND-1000
spectrophotometer. A total of 0.5 μg RNA was
reverse-transcribed using a cDNA Reverse Transcription kit (Takara
Bio, Inc.), according to the manufacturer's protocol. The resulting
cDNA was analyzed in triplicate using qPCR (95°C for 5 min,
followed by 40 cycles of 95°C for 45 sec, annealing at 55°C for 45
sec and extension at 72°C for 1 min) on a Step One Plus Real-Time
PCR system (Thermo Fisher Scientific, Inc.) with SYBR-Green Master
Mix (Roche Diagnostics, Basel, Switzerland), according to the
manufacturer's protocol. Target gene expression was normalized to
GAPDH levels in respective samples as an internal control and
calculated using the 2−ΔΔCq method (19). The sequences of the qPCR primers
are listed in Table I.
 | Table ISequences of qPCR primer pairs. |
Table I
Sequences of qPCR primer pairs.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| STAT6 |
AGGTGTACCCACCACACTCT |
GGTCACATCTGAGCAGAGCA |
| Slug |
CAGTATGTGCCTTGGGGGAG |
AGGCACTTGGAAGGGGTATTG |
| Zeb1 |
AGAGCGCTAGCTGCCAATAA |
GGGCGGTGTAGAATCAGAGT |
| Zeb2 |
GGCCTACACCTACCCAACTG |
ACAGGAGTCGGAGTCTGTCA |
| Snail1 |
CCTGTCTGCGTGGGTTTTTG |
ACCTGGGGGTGGATTATTGC |
| Twist1 |
GCATTCTCAAGAGGTCGTGC |
GGTTTTGCAGGCCAGTTTGAT |
| GAPDH |
ATGGGGAAGGTGAAGGTCGGAGT |
TGACAAGCTTCCCGTTCTCAGCC |
Western blotting
Cells were harvested and lysed using
radioimmunoprecipitation lysis buffer [50 mM Tris/HCl, pH 7.4, 100
mM 2-mercaptoethanol, 2% (w/v) SDS and 10% glycerol]. The protein
concentration was determined using the Bradford method (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The proteins (40
μg/lane) were separated by SDS-PAGE (10 gels) and
transferred onto nitrocellulose membranes (Whatman, Maidstone, UK).
The membrane was blocked with 5% non-fat milk powder (diluted in
Tris-buffered saline with Tween-20) for 2 h at room temperature and
then incubated with diluted primary antibodies followed by
IRDye® 800CW- or IRDye 680RD-conjugated secondary
antibodies (LI-COR Biosciences, Lincoln, NE, USA): IRDye 680RD goat
anti-rabbit immunoglobulin G (IgG) [heavy and light chains (H + L)]
(cat. no. 926-68071), IRDye 800CW goat anti-rabbit IgG (H + L)
(cat. no. 926-32211), IRDye 680RD goat anti-mouse IgG (H + L) (cat.
no. 926-68070) and IRDye 800CW goat anti-mouse IgG (H + L) (cat.
no. 926-32210). All the secondary antibodies were diluted at
1:20,000, and detection was performed using the Odyssey Infrared
Imaging System (LI-COR Biosciences, Lincoln, NE, USA). The obtained
signals were converted into grayscale images (Application Software,
version 2.1.12; LI-COR Biosciences).
Luciferase assay
In total, 1×105 HCT116 cells were plated
onto 24-well plates. The next day, cells were co-transfected with
firefly luciferase reporter constructs and pRL-SV40 Renilla
luciferase reporter plasmids (Promega Corporation, Madison, WI,
USA). The pRL-SV40 plasmid was used to normalize the transfection
efficiency. The luciferase activities were measured using a
dual-luciferase reporter assay system (Promega Corporation) and a
luminometer (LB 9507; Berthold Technologies GmbH and Co. KG, Bad
Wildbad, Germany). All experiments were performed in
triplicate.
Chromatin immunoprecipitation (ChIP)
Chromatin was cross-linked with 1% formaldehyde and
ChIP assays were performed using a SimpleChIP® Enzymatic
Chromatin IP kit (Cell Signaling Technology, Inc.; cat. no. 9003),
according to the manufacturer's protocol. The immunoprecipitated
DNA fragments were quantified using RT-qPCR using the following
primer pairs for the STAT6 promoter: −1,300 bp,
5′-TGGTTAGCAGCATTAGC AGG-3′ (forward) and
5′-CAGCTCCTCCTCCTCCATCC-3′ (reverse); −400 bp,
5′-TCAGTCCAAGGGACTCCTAG-3′ (forward) and 5′-GCTCGATGCCAGCCAACTCC-3′
(reverse); +500 bp, 5′-GGTTTCAGCTAGTGTTGGTG-3′ (forward) and
5′-GAGGGGCACTCCTTCACCTC-3′ (reverse).
Wound healing assay
Cells were seeded in 6-well plates. Following
transfection for 24 h, the monolayer was gently and slowly
scratched with a pipette tip across the center of the well.
Following scratching, the well was gently washed several times with
PBS to remove the detached cells. The wells were replenished with
fresh medium without serum, and the cells were allowed to grow for
an additional 48 h, and the images of the stained monolayer were
captured at ×40 magnification under a light microscope (Eclipse Ti,
Nikon, Kyoto, Japan). The wound was evaluated using ImageJ (version
1.4.3.67; National Institutes of Health, Bethesda, MD, USA).
Cell invasion assays
Cell invasion was assessed in Boyden chambers with
Matrigel (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. First, an 8-mm-porosity polycarbonate
membrane was covered with 200 μl serum-free medium
containing 1×105 cells/well. The plates were then
incubated with 10% FBS for 48 h at 37°C in a humidified atmosphere
containing 5% CO2. The invading cells on the lower
surface of the filter were fixed, stained and counted under a light
microscope (Eclipse Ti) at ×100 magnification.
Xenografts
Animal experiments were performed with the approval
of the Ethics Committee of Sanmen People's Hospital (Sanmen, China)
and the Animal Care and Use Committee of Zhejiang University
(Hangzhou, China). A total of 28 female BALB/c nude mice
(5-week-old; weight, 18 g; housed at 25°C with filtered air by
efficiency particulate air filters with 40–60% humidity on a 10-h
light/14-h dark cycle, with; free access to food and water) were
purchased from the Laboratory Animal Center of Zhejiang University
and randomly divided into HCT116/shCtrl, IL-4-HCT116/shCtrl,
HCT116/shE2F1 and IL-4-HCT116/shE2F1 groups (n=7 per group), where
shCtrl is control short hairpin RNA (5′-UUC UCCGAACGUGUCACGU-3′;
GenePharma, Shanghai, China), and equal amounts of indicated cells
(5×106) were injected subcutaneously into each mouse.
IL-4 was administered by intraperitoneal injection at a dose of 10
μg/kg three times weekly for 2 weeks. Treatment began on day
7, when the tumor sizes were measurable. The tumors were examined
every 2 days, the length and width measurements were obtained with
calipers, and the tumor volumes were calculated. On day 21, the
animals were euthanized using decapitation under Midazolam
anesthesia (5 mg/kg), and the tumors were excised and weighed. The
IL-4/STAT6 signaling in the tumor tissues was analyzed by
immunohistochemistry.
Immunohistochemistry
Experiments using human tissues were performed with
the approval of the Ethics Committee of Sanmen People's Hospital.
In total, 218 human CRC samples were collected at Sanmen People's
Hospital, and written informed consent was obtained from all
patients. Immunohistochemistry was performed and analyzed as
described in our previous study (20).
Statistical analysis
Statistical analysis was performed with SPSS
software (version 22.0; IBM Corp., Armonk, NY, USA) and GraphPad
Prism (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA).
Analysis of differences was performed using a two-tailed Student's
t-test and analysis of variance (one-way with Tukey's post hoc
test; two-way with Sidak's post hoc test). The results are
presented as the mean ± standard deviation of three separate
experiments. χ2 test or Fisher's exact probability test
was used to compare the clinicopathological features of the
patients with protein expression. A Spearman's rank correlation
test was used for analyzing the correlation. Kaplan-Meier plots and
log-rank tests were used for survival analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
IL-4-induced EMT is more evident in the
CRC cell line HCT116 compared with the RKO
Although expression of IL-4 and IL-4R is involved in
the process of local metastases in colorectal cancer (21), the underlying molecular mechanisms
have not been clearly elucidated. In the present study, IL-4
immunoreactivity was observed in clinical CRC tissues, but not in
adjacent normal tissues, and a higher IL-4 level was detected in
invasive CRC tissues compared with in non-invasive ones (Fig. 1A). Additionally, IL-4 was
identified to induce an EMT-like morphological change in CRC cells
(Fig. 1B). The potential function
of IL-4 in EMT induction in CRC cells was investigated. Following
exogenous IL-4 stimulation, the migration of HCT116 and RKO cells
was identified to be increasing, and the CRC cells exposed to IL-4
also acquired the capacity for migration and invasion (Fig. 1C and D). Furthermore, IL-4
treatment decreased the expression of the membranous epithelial
marker E-cadherin and increased the expression of the cytoplasmic
mesenchymal marker vimentin at the mRNA and protein levels in
HCT116 and RKO cells (Fig. 1E and
F). In addition, repression of ZO-1 accompanied by the
induction of fibronectin was observed in IL-4-treated HCT116 and
RKO cells (Fig. 1E and F).
However, it is notable that IL-4 exposure led to an increased EMT
change in HCT116 cells compared with in RKO cells.
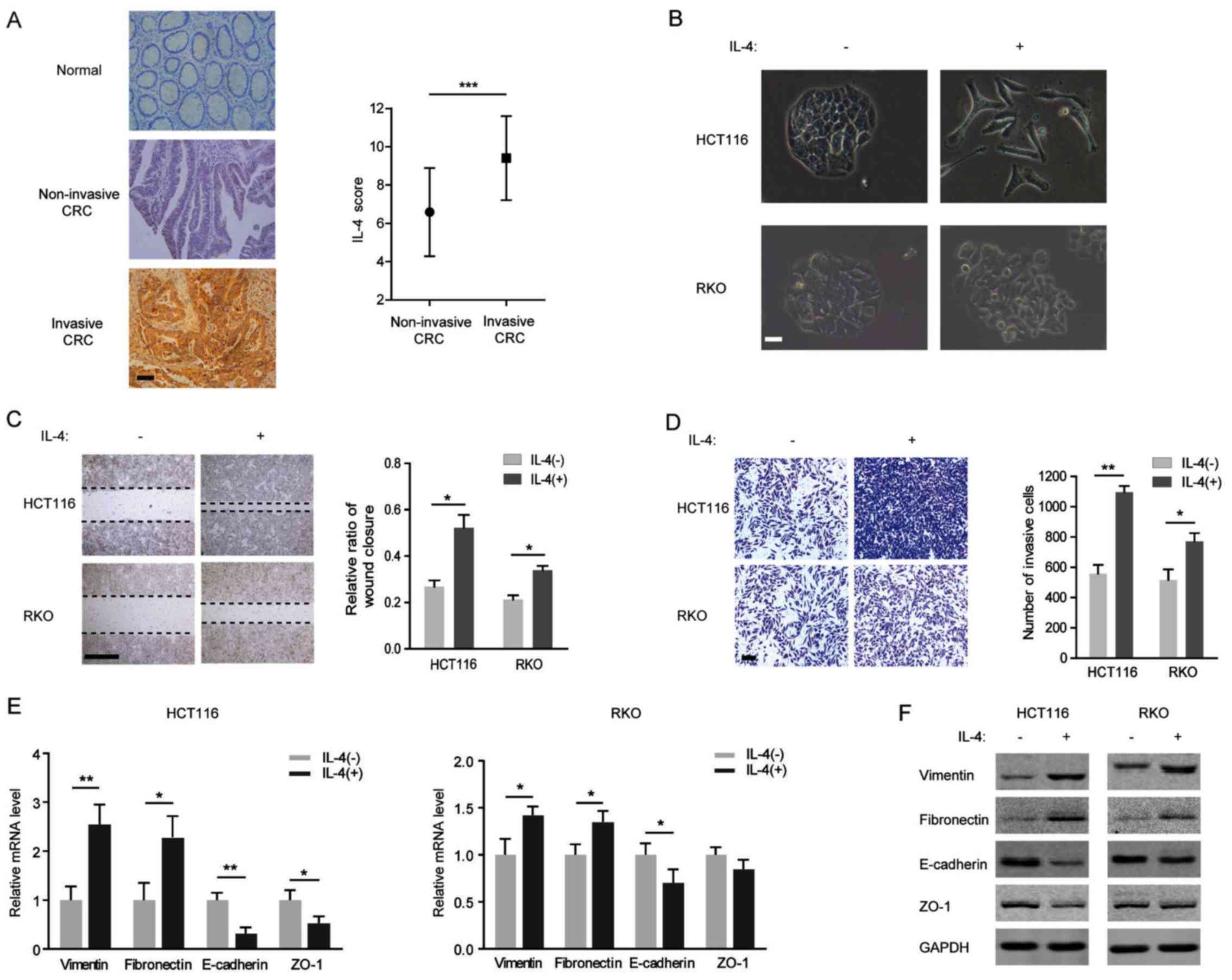 | Figure 1IL-4-induced epithelial-mesenchymal
transition program is more evident in HCT116 cells compared with in
RKO cells. (A) Left: Representative images of IL-4 immunostaining
in human colorectal cancer tissue and para-tumor tissue. Scale bar,
200 μm. Right: Immunostaining scores of IL-4 in invasive or
non-invasive CRC tissues. (B) Morphology of HCT116 and RKO cells
treated with or without IL-4 (20 ng/ml) for 72 h visualized using
phase-contrast microscopy. Scale bar, 500 μm. (C) Left:
Representative images from wound healing assays with HCT116 and RKO
cells treated with 20 ng/ml IL-4. Scale bar, 100 μm. Right:
Percentage wound closure 48 h after addition of IL-4. (D) Left:
Representative images of HCT116 and RKO cells penetrating the
Matrigel in invasion assays following treatment with 20 ng/ml IL-4.
Scale bar, 100 μm. Right: numbers of invasive cells treated
with IL-4 for 48 h. (E) Relative mRNA levels of vimentin,
fibronectin, E-cadherin and ZO-1 in HCT116 (left) and RKO (right)
cells treated with 20 ng/ml IL-4 for 48 h. (F) Western blots of
vimentin, fibronectin, E-cadherin and ZO-1 with specific antibodies
in HCT116 (left) and RKO (right) cells treated with 20 ng/ml IL-4
for 48 h. *P<0.05; **P<0.01. IL,
interleukin; CRC, colorectal cancer; E-cadherin, epithelial
cadherin; ZO-1, zonula occludens-1. |
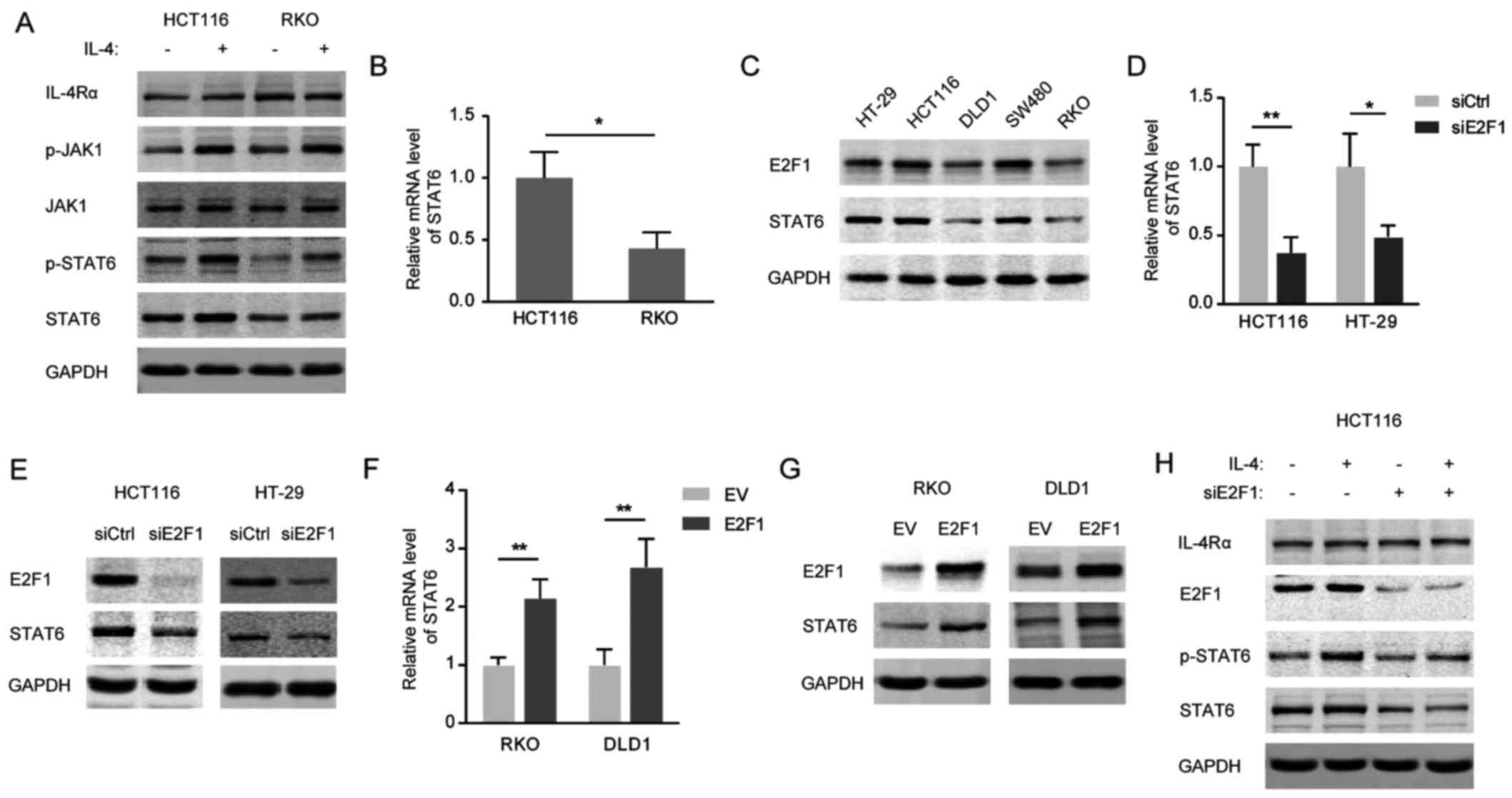
Constitutive expression of STAT6 is
dependent on E2F1 in CRC cells
To understand the different responses to IL-4 in
HCT116 and RKO cells, the IL-4/STAT6 signaling pathway was
investigated in the two CRC cell lines. As presented in Fig. 2A, exogenous IL-4 resulted in a
marked increase in p-STAT6 in HCT116, but not in RKO, cells due to
a high level of STAT6 protein in HCT116 cells. Consistently, the
mRNA level of STAT6 was significantly increased in HCT116 cells
compared with in RKO cells (Fig.
2B). Furthermore, the expression levels of STAT6 were
paralleled with those of E2F1 in different CRC cell lines (Fig. 2C). Small interfering
(siRNA)-mediated E2F1 knockdown markedly decreased STAT6 expression
at the mRNA (Fig. 2D) and protein
(Fig. 2E) levels in HCT116 and
HT-29 cells, and STAT6 was identified to be markedly upregulated
following overexpression of E2F1 in RKO and DLD1 cells (Fig. 2F and G). As expected, the induced
activation of IL-4/STAT6 signaling was blunted in the E2F1-silenced
HCT116 cells, which exhibited attenuated STAT6 expression (Fig. 2H). From these results, it may be
concluded that ectopically expressed E2F1 is required for the
abnormal activation of IL-4/STAT6 signaling in CRC cells.
E2F1-induced SP3 transactivates STAT6 in
CRC cells
Considering E2F1 is a well-known transcriptional
activator, serial deletion constructs of the STAT6 gene promoter
were examined using luciferase reporter assays to identify the
transcriptional regulatory region responsive to E2F1 in HCT116
cells. It was identified that the STAT6 promoter without the region
between −500 and −350 lost the ability to respond to E2F1 silencing
(Fig. 3A). However, ChIP-PCR
analysis revealed no enrichment of E2F1 protein into the STAT6
promoter (Fig. 3B). The promoter
sequence (−500/−350) was analyzed and two potential GT-box elements
predicted to be bound by SP3 were identified (Fig. 3C). Mutations in either the GT#1 or
GT#2 sites decreased the reporter activity of the STAT6 gene
promoter in HCT116 cells, and knockdown of SP3 mimicked the effect
of these mutations (Fig. 3D and
E). In turn, SP3 overexpression induced the reporter activity
of the wild-type STAT6 promoter, but failed to induce the
GT#1/2-mutated STAT6 promoter (Fig.
3F). Consistently, the mRNA level of STAT6 was decreased
following knockdown of SP3 in HCT116 cells (Fig. 3G). ChIP-PCR analysis also confirmed
that SP3 directly bound to the STAT6 promoter at ~−400 bp, upstream
of the transcription start site (Fig.
3H). Furthermore, E2F1 knockdown diminished the expression of
SP3 and overexpression of SP3 rescued the attenuated STAT6
expression that resulted from E2F1 deprivation, indicating that SP3
mediates the E2F1-dependent increased level of expression of STAT6
in CRC cells (Fig. 3I).
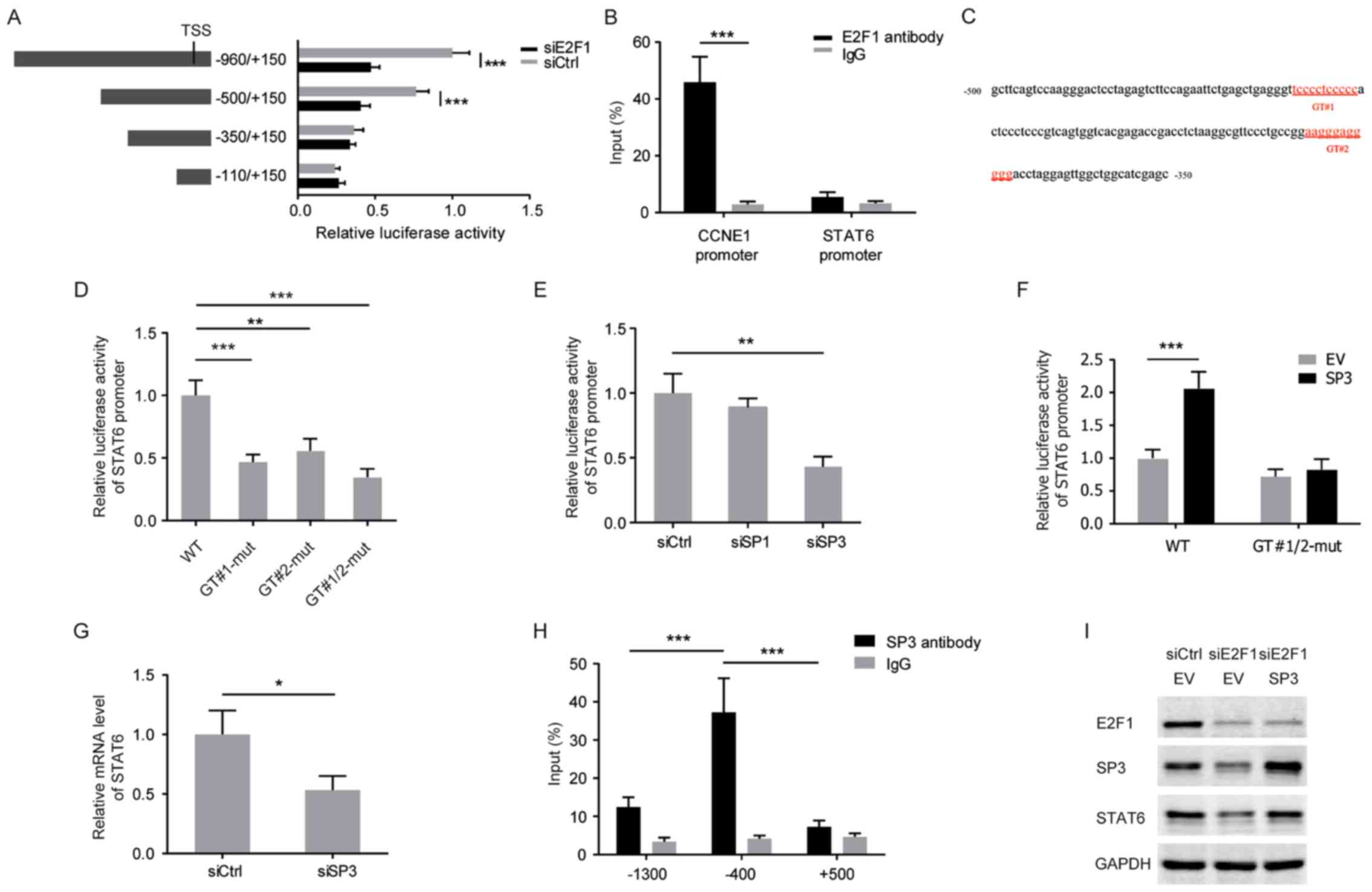 | Figure 3E2F1-induced SP3 transactivates STAT6
in CRC cells. (A) Relative luciferase activities were measured with
a series of truncated constructs of the STAT6 promoter in HCT116
cells with E2F1 knockdown (internal control, pRL-SV40). (B) HCT116
cells were analyzed by ChIP assays using anti-E2F1 antibody and
IgG. qPCR was performed with the immunoprecipitated DNA or soluble
chromatin using the specific primer pairs for STAT6 promoter (~−400
bp). CCNE1 served as a positive control. (C) The STAT6 promoter
region (-500/-350) with two potential GT-box elements. (D) Relative
luciferase activities in HCT116 cells transfected with the
wild-type or GT-box-mutated STAT6 promoter reporter constructs. (E)
Relative luciferase activity of STAT6 promoter in HCT116 cells with
knockdown of SP1 or SP3. (F) Relative luciferase activity of the
wild-type or GT-box-mutated STAT6 promoter in HCT116 cells with
overexpression of SP3. (G) mRNA level of STAT6 in HCT116 cells with
SP3 knockdown. (H) HCT116 cells were analyzed by ChIP assays using
anti-SP3 antibody and IgG. qPCR was performed with the
immunoprecipitated DNA or soluble chromatin using the specific
primer pairs for STAT6 promoter regions (~−1300, −400 and +500 bp).
(I) Western blots of the indicated proteins with specific
antibodies in HCT116 cells with E2F1 knockdown and SP3
overexpression. *P<0.05; **P<0.01;
***P<0.001. SP3, specificity protein 3; STAT6, signal
transducer and activator of transcription 6; CRC, colorectal
cancer; ChIP, chromatin immunoprecipitation; IgG, immunoglobulin G;
qPCR, quantitative polymerase chain reaction; TSS, transcription
start site; si, small interfering RNA; Ctrl, control; CCNE1, cyclin
E1; WT, wild-type; mut, mutant; EV, empty vector. |
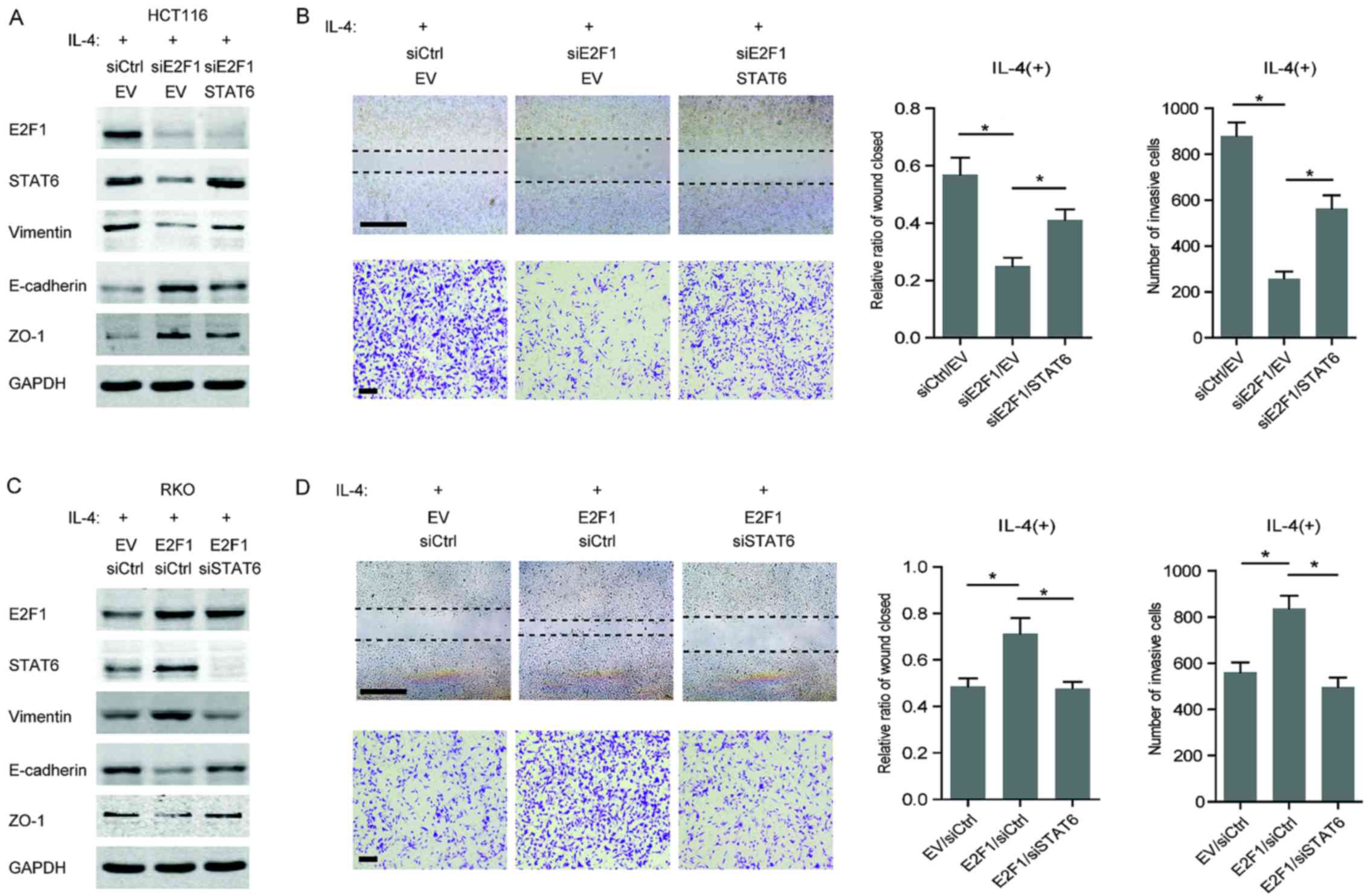
Highly expressed E2F1 promotes the
IL-4-induced aggressiveness by upregulating STAT6
On the basis of the aforementioned results, the
function of E2F1 in the IL-4/STAT6 signaling-induced EMT process
was next investigated. In IL-4-stimulated HCT116 cells, a decrease
in a mesenchymal marker (vimentin) and an increase in epithelial
markers (E-cadherin and ZO-1) were observed following knockdown of
E2F1, whereas overexpression of STAT6 partially recovered the
mesenchymal phenotype (Fig. 4A).
In addition, the acquisition of migratory and invasive capabilities
in HCT116 cells exposed to IL-4 was attenuated by E2F1 silencing,
but restored when STAT6 was overexpressed (Fig. 4B). Similarly, overexpression of
E2F1 enhanced the promoting effect of IL-4 on the mesenchymal
phenotype in RKO cells; however, knockdown of STAT6 impaired the
E2F1-dependent EMT properties (Fig.
4C). As expected, E2F1 overexpression strengthened the
IL-4-induced migration and invasion of RKO cells, but failed to
disrupt STAT6 (Fig. 4D). These
results demonstrate that the regulation of STAT6 by E2F1 is
essential for IL-4-induced malignancy in CRC cells.
As a target gene of STAT6, Zeb1 is
important in IL-4 triggered-EMT
Cao et al (22) identified that the IL-13/STAT6/Zeb1
pathway serves a critical function in promoting EMT and
aggressiveness of CRC cells. The alterations in EMT-associated
transcription factors including Snail1, Slug, Zeb1, Zeb2 and Twist1
in response to IL-4/STAT6 signaling were investigated. As presented
in Fig. 5A and B, the mRNA and
protein levels of Zeb1 and Zeb2 were markedly increased in HCT116
cells following IL-4 stimulation. However, in HCT116 cells with
STAT6 knockdown, IL-4 was not able to induce Zeb1 and Zeb2
(Fig. 5C and D). Furthermore, it
was identified that knockdown of STAT6 prevented IL-4 from inducing
Zeb1 and EMT markers, whereas ectopic expression of Zeb1 recovered
the cell's EMT properties (Fig.
5E). Therefore, consistent with a previous study (22), Zeb1 functions as a critical
downstream effector of IL-4/STAT6 signaling during EMT process.
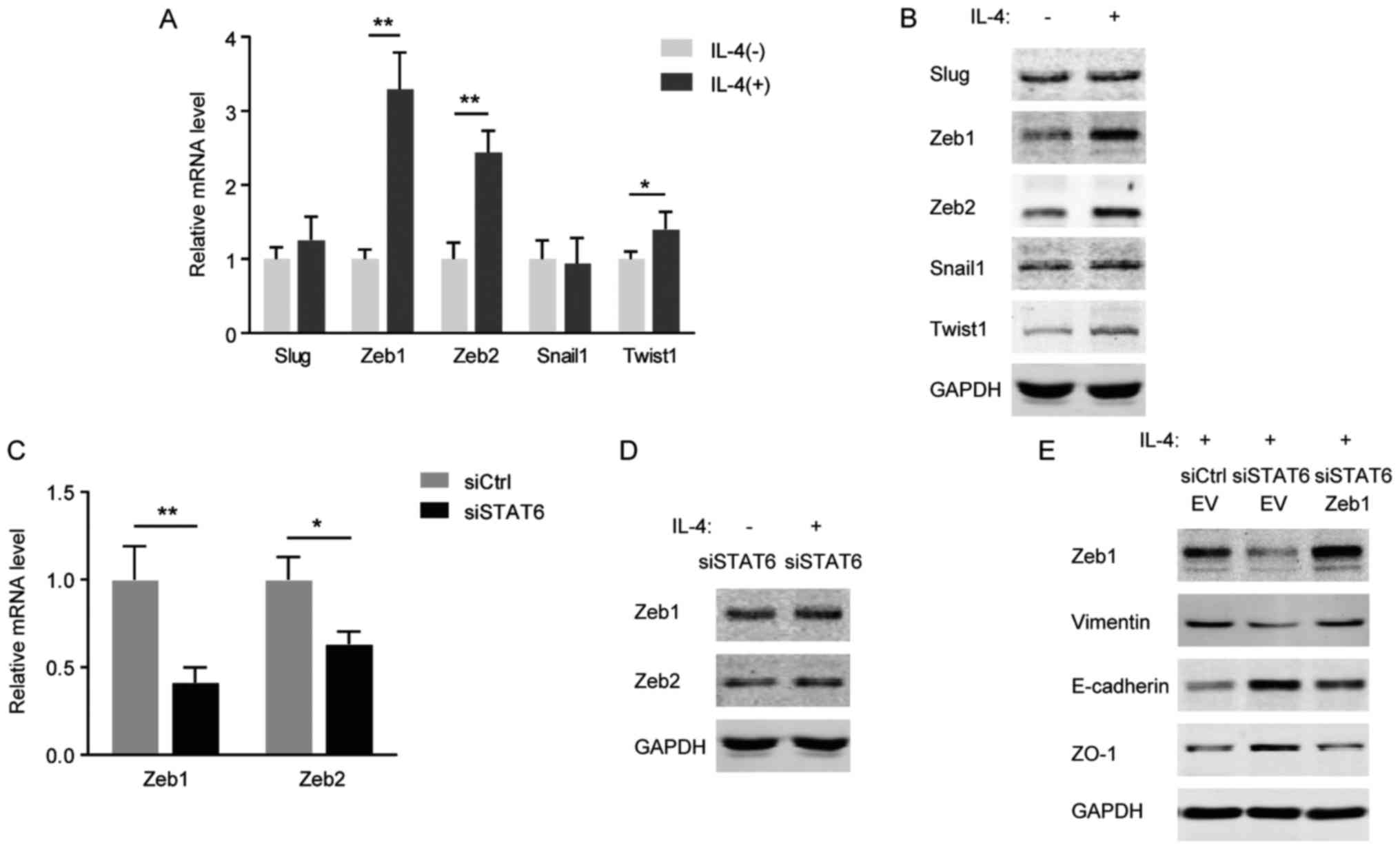 | Figure 5As a target gene of STAT6, Zeb1 is
important in EMT triggered by IL-4. (A) mRNA levels of EMT
transcription factors in HCT116 cells treated with 20 ng/ml IL-4
for 24 h. (B) Western blots of the indicated proteins with specific
antibodies in HCT116 cells treated with 20 ng/ml IL-4 for 24 h. (C)
mRNA levels of Zeb1/2 in HCT116 cells with STAT6-knockdown. (D)
Western blots of the indicated proteins with specific antibodies in
HCT116 cells transfected with STAT6 siRNA and exposed to 20 ng/ml
IL-4 for 24 h. (E) Western blots of the indicated proteins with
specific antibodies in HCT116 cells co-transfected with STAT6
siRNA/Zeb1 overexpression plasmid and exposed to 20 ng/ml IL-4 for
48 h. *P<0.05; **P<0.01. STAT6, signal
transducer and activator of transcription 6; EMT,
epithelial-mesenchymal transition; IL, interleukin; Zeb, zinc
finger E-box-binding homeobox; si, small interfering; Ctrl,
control; E-cadherin, epithelial cadherin; ZO-1, zonula occludens-1;
EV, empty vector. |
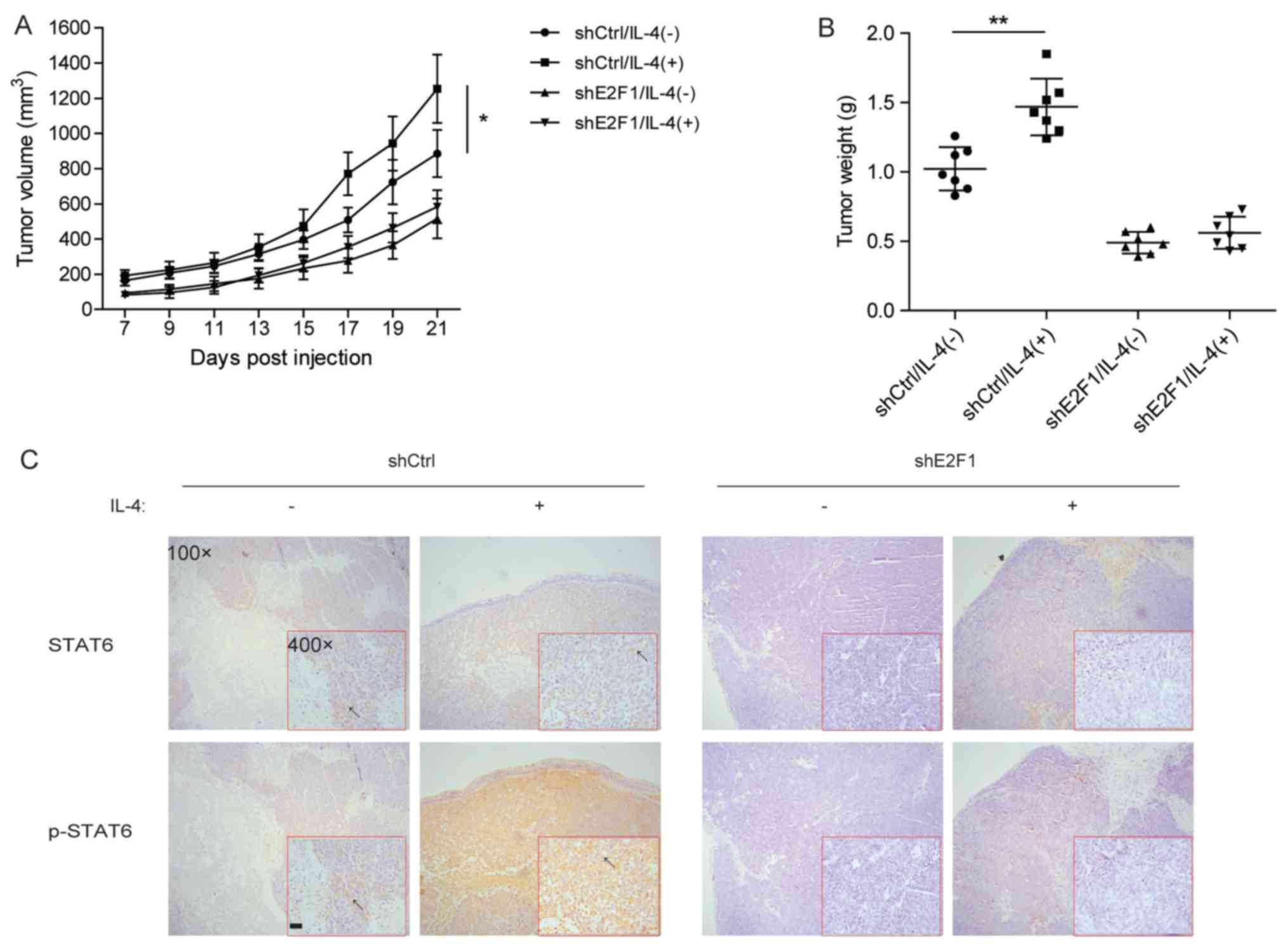
E2F1 is required for IL-4-induced CRC
tumorigenesis in vivo
To further determine the involvement of E2F1 in
IL-4-induced tumorigenesis, HCT116 cells with stable knockdown of
E2F1 were constructed and subcutaneously injected into nude mice.
Tumor formation was evaluated during the 21 days after injection.
It was identified that IL-4 significantly promoted the
tumorigenesis of HCT116 cells, but not HCT116 cells with stable
knockdown of E2F1 (Fig. 6A and B).
Consistent with these results, immunostaining for the xenograft
indicated that silencing of E2F1 inhibited IL-4-induced
accumulation of p-STAT6 through decreasing the total STAT6 level
(Fig. 6C). Thus, in vivo
experiments suggest that E2F1 may exert a crucial function in
IL-4-induced CRC tumorigenesis by regulating STAT6 signaling.
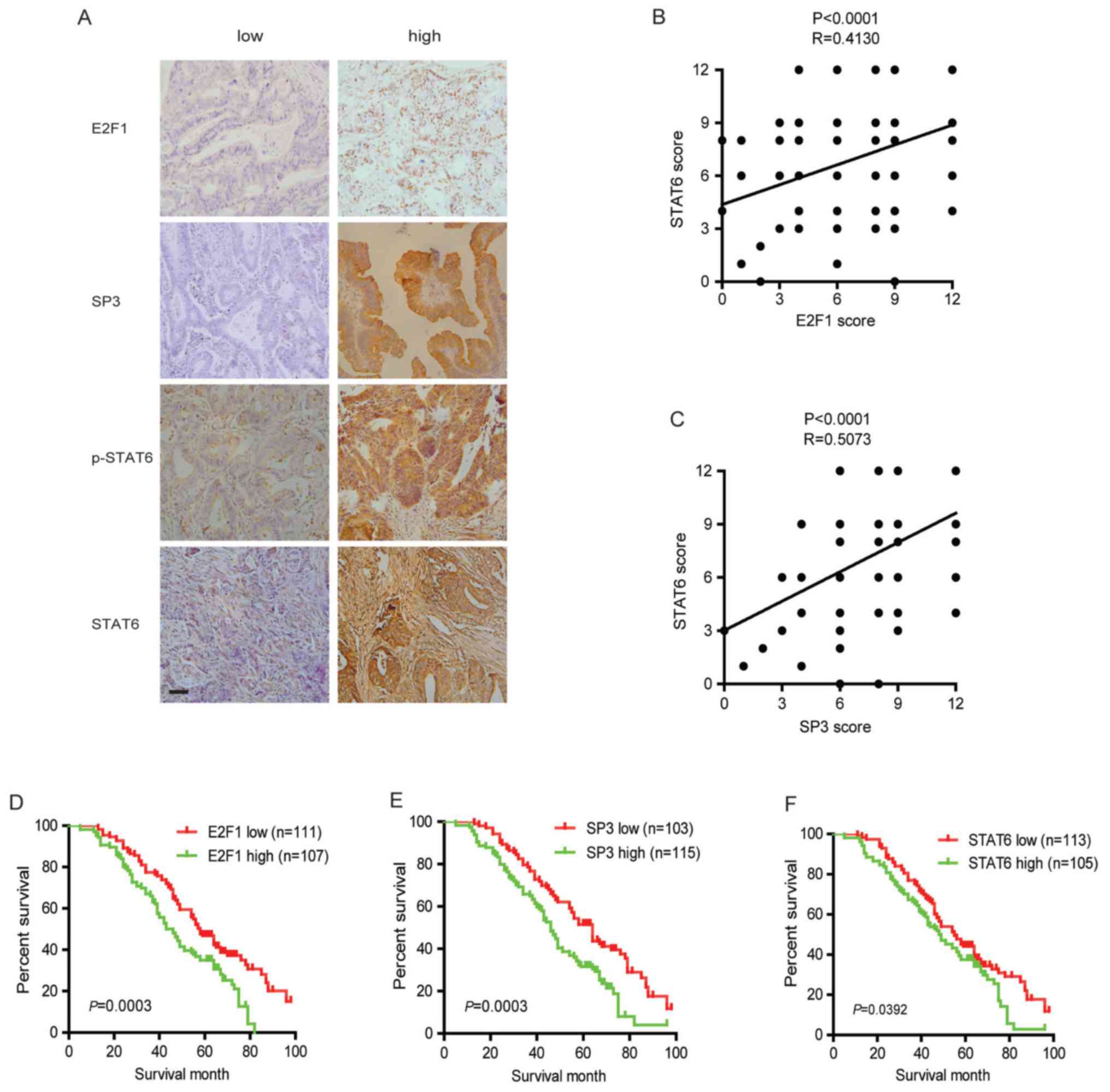
Abnormal activation of E2F1/SP3/STAT6
axis is associated with cancer aggressiveness in CRC samples
Immunohistochemical staining was performed to
evaluate the protein levels of E2F1, SP3, STAT6 and p-STAT6 in
human CRC specimens. As presented in Fig. 7A, different immunoreactivity
intensities of E2F1, SP3, STAT6 and p-STAT6 were observed in CRC
specimens. Further analysis revealed that all three factors were
significantly correlated with the TNM stage and distant metastasis,
but not with other clinical characteristics (Table II). In addition, immunostaining
using consecutive sections revealed a positively correlation of the
expression levels of STAT6 with the levels of E2F1 and SP3
(Fig. 7B and C). The potential
associations between immunostaining and overall survival were
retrospectively evaluated in 218 patients with CRC. A Kaplan-Meier
analysis identified that increased expression of E2F1, SP3 or STAT6
significantly indicated poor survival (Fig. 7D–F). These results suggested that
the E2F1/SP3/STAT6 axis is involved in CRC progression and
metastasis.
 | Table IICorrelation of the expression of SP3,
E2F1 and STAT6 with clinicopathological features in CRC. |
Table II
Correlation of the expression of SP3,
E2F1 and STAT6 with clinicopathological features in CRC.
| Characteristic | Total | SP3 expression
| E2F1 expression
| STAT6 expression
|
|---|
| Low | High | P-value | Low | High | P-value | Low | High | P-value |
|---|
| N | 218 | 103 | 115 | | 111 | 107 | | 113 | 105 | |
| Tumor location | | | | 0.5832 | | | 0.7549 | | | 0.6933 |
| Colon | 90 | 41 | 49 | | 44 | 46 | | 46 | 44 | |
| Rectum | 128 | 62 | 66 | | 67 | 61 | | 67 | 61 | |
| Sex | | | | 0.9837 | | | 0.3220 | | | 0.1288 |
| Male | 116 | 56 | 60 | | 54 | 62 | | 54 | 62 | |
| Female | 102 | 47 | 55 | | 57 | 45 | | 59 | 43 | |
| Age, years | | | | 0.8908 | | | 0.7756 | | | 0.6932 |
| ≤65 | 88 | 43 | 45 | | 44 | 44 | | 46 | 42 | |
| >65 | 130 | 60 | 70 | | 67 | 63 | | 67 | 63 | |
| Differentiation
status | | | | 0.9279 | | | 0.1521 | | | 0.6771 |
| Well | 46 | 24 | 22 | | 22 | 24 | | 25 | 21 | |
| Moderate | 147 | 68 | 79 | | 82 | 65 | | 78 | 69 | |
| Poor | 25 | 11 | 14 | | 7 | 18 | | 10 | 15 | |
| Tumor size, cm | | | | 0.2307 | | | 0.3672 | | | 0.3444 |
| <5 | 105 | 46 | 59 | | 55 | 50 | | 50 | 55 | |
| ≥5 | 113 | 57 | 56 | | 56 | 57 | | 63 | 50 | |
| Lymph node
metastasis | | | | 0.0194a | | | 0.0957 | | | 0.0079a |
| N0 | 125 | 67 | 58 | | 70 | 55 | | 71 | 54 | |
| N1 | 69 | 28 | 41 | | 33 | 36 | | 35 | 34 | |
| N2 | 24 | 8 | 16 | | 8 | 16 | | 7 | 17 | |
| TNM | | | | 0.0055a | | | 0.0126a | | | 0.0439a |
| I | 35 | 20 | 15 | | 19 | 16 | | 17 | 18 | |
| II | 83 | 39 | 44 | | 43 | 40 | | 50 | 33 | |
| III | 83 | 41 | 42 | | 45 | 38 | | 41 | 42 | |
| IV | 17 | 3 | 14 | | 4 | 13 | | 5 | 12 | |
| Distant
metastasis | | | | 0.0005a | | | 0.0010a | | | 0.0089a |
| M0 | 201 | 102 | 99 | | 107 | 94 | | 114 | 87 | |
| M1 | 17 | 3 | 14 | | 4 | 13 | | 5 | 12 | |
Discussion
It is well-accepted that cytokines including ILs
produced by tumor cells or, more often, by the B- or T-lymphocytes
recruited to the tumor microenvironment promote the proliferation
of tumor cells, perturb their differentiation and support the
metastasis of cancer cells (23).
Several lines of evidence suggest that IL-4 may suppress
cancer-directed immunosurveillance and enhance tumor metastasis.
For example, IL-4 has the ability to exert an autocrine
proliferation stimulation effect in pancreatic cancer cells and
furthermore serves paracrine functions on the surrounding
infiltrating immune cells, repressing immunoresponses (24). In addition, it has been observed
that secretion of IL-4, for instance, by malignant cells from the
bladder, lung and colon, confers resistance to chemotherapy-induced
cell death (25,26). Particularly for CR-CSC cells,
resistance to oxaliplatin is dependent on autocrine production of
IL-4 (27). In the present study,
the acquired EMT-like characteristics of several CRC cell lines
stimulated by IL-4 were identified. However, it was observed that
different CRC cell lines (HCT116 and RKO) with different expression
levels of STAT6 exhibited entirely different responses to IL-4. As
a key signaling transducer in the IL-4 pathway, STAT6 may determine
the IL-4-induced aggressiveness of CRC cells. Consistent with this,
it was identified previously that a lack of STAT6 attenuates
inflammation and prevents early steps of colitis-associated CRC
(28). Furthermore, HT-29 cells
with an active STAT6 phenotype exhibit more aggressive metastasis
compared with Caco-2 cells with defective STAT6 (29). Additionally, knockdown of STAT6
inhibits proliferation and induces apoptosis in HT-29 cells
(30). Therefore, the STAT6 level
may determine the CRC development facilitated by inflammatory
cytokines.
Our previous study focused on E2F1, a critical
transcription factor for CRC development, which promoted the
migration and invasion of CRC cells (18). In the present study, it was
identified that the levels of STAT6 protein were paralleled with
E2F1 in several CRC cell lines. Further in vitro experiments
confirmed an E2F1-dependent transcription of STAT6. However, no
evident enrichment of E2F1 into the E2F1-response region located at
the STAT6 promoter was identified in HCT116 with increased
expression of STAT6, indicating an indirect regulation for STAT6 by
E2F1. In addition, SP3, a member of the SP transcription factor
family, was verified to mediate the transcription of the STAT6
gene. SP transcription factors including SP3 are have been
identified to be overexpressed in tumors, whereas SP levels are
relatively low in non-tumor tissues (31). Functional studies have identified
that SP transcription factors have a function in cancer cell
proliferation, survival, angiogenesis, migration and invasion by
inducing oncogenes including survivin, B-cell lymphoma 2, p65
(NF-κB), c-MET, EGFR and other receptor tyrosine kinases (32). The overexpressed SP members in CRC
cells, SP1 and SP3, are required for migration and invasion
(33). Even though SP1 and SP3
share >90% DNA sequence homology in the DNA-binding domain and
bind to the same DNA element with similar affinity (34), it was identified in the present
study that SP3 rather than SP1 contributed to the transcription
activation of the STAT6 gene in CRC cells. More importantly,
increased expression of SP3 appeared to depend on E2F1, suggesting
the E2F1/SP3/STAT6 axis as a potential regulatory pathway for STAT6
signaling in CRC. However, the reasons for SP3, but not SP1, being
the major regulatory activator of the transcription of the STAT6
gene remain to be determined, and the details of how E2F1 regulates
SP3 remain unknown.
On the surface of many solid tumors, IL-4R consists
of the IL-4Rα and IL-13Rα1 subunits. Thus, IL-4R may be bound and
activated by IL-13, although IL-4 binds with higher affinity
(35). Previous studies
demonstrate that IL-4 and IL-13 share signaling events by
initiating a signaling cascade that activates the JAK/STAT pathway
(particularly STAT6) in human CRC cells (36). Recent study by Cao et al
(22) identified that IL-13/STAT6
signaling induces the aggressive properties of HT29 and SW480 cells
through transactivating Zeb1, which is a well-known EMT core
regulator and triggers malignant transformation. Therefore, in the
present study, the expression patterns of EMT-associated
transcription factors, including Snail1, Slug, Zeb1, Zeb2 and
Twist1, were also investigated. However, the results of the present
study were not completely consistent with those of Cao et al
(22), as demonstrated by the
critical function of Zeb2 in the EMT progress driven by IL-4/STAT6
signaling. Despite STAT6 functioning as the common transducer in
IL-4 and IL-13 signaling, activated STAT6 induces different
expression profiles of downstream EMT-drivers following IL-4 and
IL-13 stimulation.
In summary, we propose a model in which the high
expression of STAT6 arose from abnormal activation of the
E2F1/SP3/STAT6 axis and is phosphorylated by IL-4 and then promotes
EMT progression by upregulating Zeb1/2, therefore enhancing the
migration and invasion of CRC cells (Fig. 8). The discovery of such a
regulatory pathway for IL-4 signaling suggests the potential of the
E2F1/SP3/STAT6 axis to serve as a biomarker and represents a
promising target for therapeutic intervention against CRC.
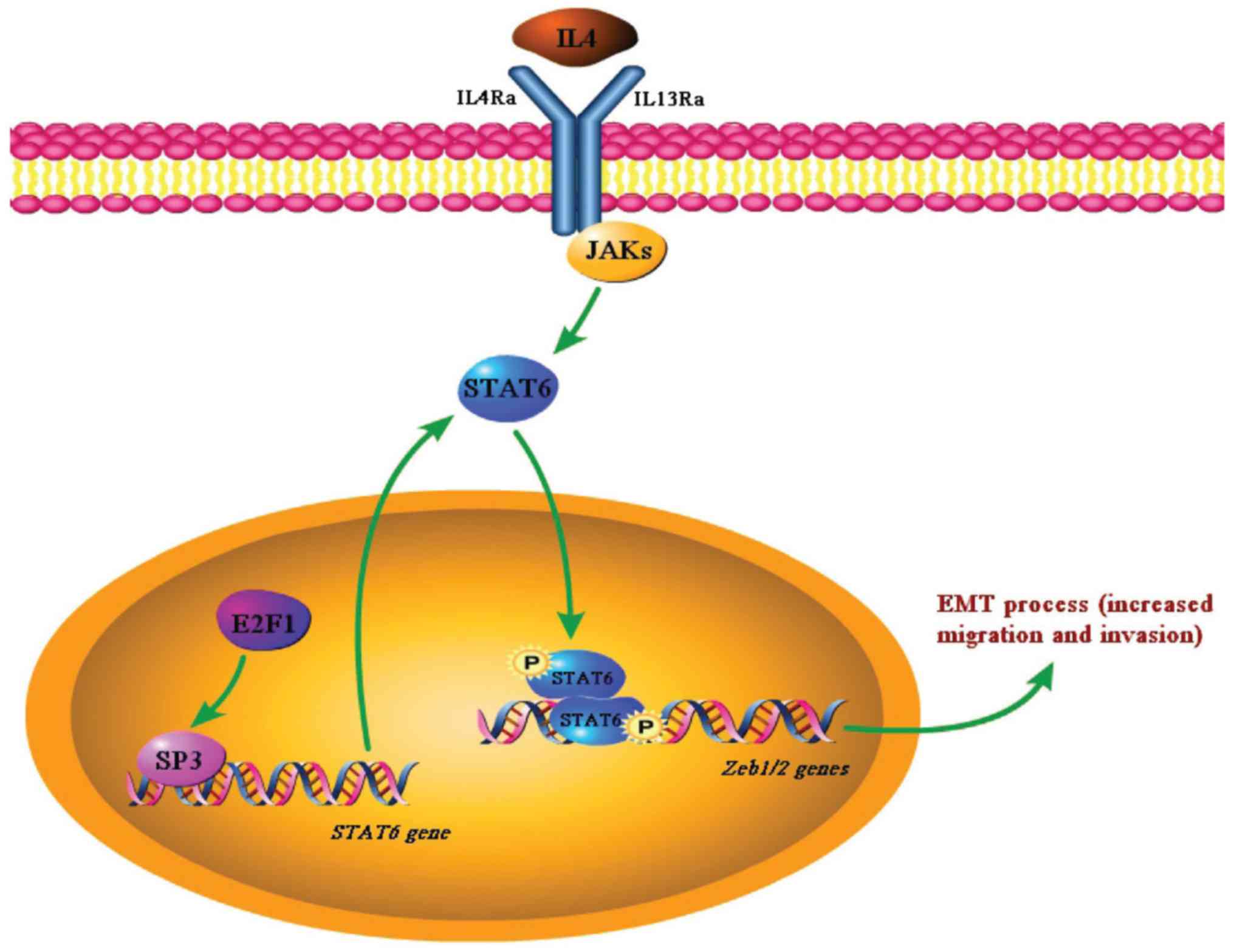 | Figure 8Schema indicating the crucial
function of the E2F1/SP3/STAT6 axis in IL-4-induced EMT. In
IL-4-responsive CRC cells such as HCT116, highly expressed E2F1
induces the expression of SP3, which transactivates STAT6. At the
same time, IL-4 signaling leads to STAT6 phosphorylation, which
then promotes the EMT of CRC cells by activating the transcription
of Zeb1/2. SP3, specificity protein 3; STAT6, signal transducer and
activator of transcription 6; IL, interleukin; EMT,
epithelial-mesenchymal transition; CRC, colorectal cancer; Zeb,
zinc finger E-box-binding homeobox; IL4Ra, IL-4 receptor α; IL13Rα,
IL-13 receptor α; JAK, Janus kinase. |
Acknowledgments
Not applicable.
Funding
The present study was supported by the Zhejiang
Medical and Health Science and Technology Plan (grant no.
2018KY920) and the Taizhou Science and Technology Plan (1601KY61).
It was also supported by the Zhejiang Medical and Health Science
and Technology Plan (grant nos. 2018KY184, 2016KYB330 and
2017KY722) and the Science and Technology Program of Sanmen County
Public Technology Social Development Project (grant nos. 16302,
16304 and 16312).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HL and XC conceived and designed the study. JC, CG,
HM, ZL, ZF, QC, ML, XJ, YH, WW, XZ, XC and HL performed the
experiments and wrote the manuscript. JC, CG, HM, ZL and ZF
analyzed and interpreted data. JC, HL, XC, ZF and WW contributed
reagents and materials. All authors helped to draft the manuscript
and read and approved the final manuscript.
Ethics approval and consent to
participate
Experiments using human tissues were performed with
the approval of the Ethics Committee of Sanmen People's Hospital
(Sanmen, China). Animal experiments were performed with the
approval of the Ethics Committee of Sanmen People's Hospital and
the Animal Care and Use Committee of Zhejiang University (Hangzhou,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sears CL and Garrett WS: Microbes,
microbiota, and colon cancer. Cell Host Microbe. 15:317–328. 2014.
View Article : Google Scholar
|
|
2
|
Todaro M, Gaggianesi M, Catalano V,
Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S,
Cocorullo G, et al: CD44v6 is a marker of constitutive and
reprogrammed cancer stem cells driving colon cancer metastasis.
Cell Stem Cell. 14:342–356. 2014. View Article : Google Scholar
|
|
3
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology.
138:2101–2114.e5. 2010. View Article : Google Scholar
|
|
4
|
Shawki S, Ashburn J, Signs SA and Huang E:
Colon cancer: Inflammation-associated cancer. Surg Oncol Clin N Am.
27:269–287. 2018. View Article : Google Scholar
|
|
5
|
Wang K and Karin M: Tumor-elicited
inflammation and colorectal cancer. Adv Cancer Res. 128:173–196.
2015. View Article : Google Scholar
|
|
6
|
Ullman TA and Itzkowitz SH: Intestinal
inflammation and cancer. Gastroenterology. 140:1807–1816. 2011.
View Article : Google Scholar
|
|
7
|
Waldner MJ, Foersch S and Neurath MF:
Interleukin-6 - a key regulator of colorectal cancer development.
Int J Biol Sci. 8:1248–1253. 2012. View Article : Google Scholar
|
|
8
|
Lee YS, Choi I, Ning Y, Kim NY,
Khatchadourian V, Yang D, Chung HK, Choi D, LaBonte MJ, Ladner RD,
et al: Interleukin-8 and its receptor CXCR2 in the tumour
microenvironment promote colon cancer growth, progression and
metastasis. Br J Cancer. 106:1833–1841. 2012. View Article : Google Scholar
|
|
9
|
Barderas R, Bartolomé RA,
Fernandez-Aceñero MJ, Torres S and Casal JI: High expression of
IL-13 receptor α2 in colorectal cancer is associated with invasion,
liver metastasis, and poor prognosis. Cancer Res. 72:2780–2790.
2012. View Article : Google Scholar
|
|
10
|
Hyun YS, Han DS, Lee AR, Eun CS, Youn J
and Kim HY: Role of IL-17A in the development of colitis-associated
cancer. Carcinogenesis. 33:931–936. 2012. View Article : Google Scholar
|
|
11
|
Kanai T, Watanabe M, Hayashi A, Nakazawa
A, Yajima T, Okazawa A, Yamazaki M, Ishii H and Hibi T: Regulatory
effect of interleukin-4 and interleukin-13 on colon cancer cell
adhesion. Br J Cancer. 82:1717–1723. 2000.
|
|
12
|
Koller FL, Hwang DG, Dozier EA and
Fingleton B: Epithelial interleukin-4 receptor expression promotes
colon tumor growth. Carcinogenesis. 31:1010–1017. 2010. View Article : Google Scholar
|
|
13
|
Bankaitis KV and Fingleton B: Targeting
IL4/IL4R for the treatment of epithelial cancer metastasis. Clin
Exp Metastasis. 32:847–856. 2015. View Article : Google Scholar
|
|
14
|
Di Stefano AB, Iovino F, Lombardo Y,
Eterno V, Höger T, Dieli F, Stassi G and Todaro M: Survivin is
regulated by interleukin-4 in colon cancer stem cells. J Cell
Physiol. 225:555–561. 2010. View Article : Google Scholar
|
|
15
|
Liu H, Antony S, Roy K, Juhasz A, Wu Y, Lu
J, Meitzler JL, Jiang G, Polley E and Doroshow JH: Interleukin-4
and interleukin-13 increase NADPH oxidase 1-related proliferation
of human colon cancer cells. Oncotarget. 8:38113–38135. 2017.
|
|
16
|
Alla V, Engelmann D, Niemetz A, Pahnke J,
Schmidt A, Kunz M, Emmrich S, Steder M, Koczan D and Pützer BM:
E2F1 in melanoma progression and metastasis. J Natl Cancer Inst.
102:127–133. 2010. View Article : Google Scholar
|
|
17
|
Hsu EC, Kulp SK, Huang HL, Tu HJ, Chao MW,
Tseng YC, Yang MC, Salunke SB, Sullivan NJ, Chen WC, et al:
Integrin-linked kinase as a novel molecular switch of the
IL-6-NF-κB signaling loop in breast cancer. Carcinogenesis.
37:430–442. 2016. View Article : Google Scholar
|
|
18
|
Fang Z, Gong C, Liu H, Zhang X, Mei L,
Song M, Qiu L, Luo S, Zhu Z, Zhang R, et al: E2F1 promote the
aggressiveness of human colorectal cancer by activating the
ribonucleotide reductase small subunit M2. Biochem Biophys Res
Commun. 464:407–415. 2015. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Fang Z, Gong C, Yu S, Zhou W, Hassan W, Li
H, Wang X, Hu Y, Gu K, Chen X, et al: NFYB-induced high expression
of E2F1 contributes to oxaliplatin resistance in colorectal cancer
via the enhancement of CHK1 signaling. Cancer Lett. 415:58–72.
2018. View Article : Google Scholar
|
|
21
|
Formentini A, Braun P, Fricke H, Link KH,
Henne-Bruns D and Kornmann M: Expression of interleukin-4 and
interleukin-13 and their receptors in colorectal cancer. Int J
Colorectal Dis. 27:1369–1376. 2012. View Article : Google Scholar
|
|
22
|
Cao H, Zhang J, Liu H, Wan L, Zhang H,
Huang Q, Xu E and Lai M: IL-13/STAT6 signaling plays a critical
role in the epithelial-mesenchymal transition of colorectal cancer
cells. Oncotarget. 7:61183–61198. 2016. View Article : Google Scholar
|
|
23
|
Cavallo F, De Giovanni C, Nanni P, Forni G
and Lollini PL: 2011: The immune hallmarks of cancer. Cancer
Immunol Immunother. 60:319–326. 2011. View Article : Google Scholar
|
|
24
|
Prokopchuk O, Liu Y, Henne-Bruns D and
Kornmann M: Interleukin-4 enhances proliferation of human
pancreatic cancer cells: Evidence for autocrine and paracrine
actions. Br J Cancer. 92:921–928. 2005. View Article : Google Scholar
|
|
25
|
Conticello C, Pedini F, Zeuner A, Patti M,
Zerilli M, Stassi G, Messina A, Peschle C and De Maria R: IL-4
protects tumor cells from anti-CD95 and chemotherapeutic agents via
up-regulation of antiapoptotic proteins. J Immunol. 172:5467–5477.
2004. View Article : Google Scholar
|
|
26
|
Todaro M, Zerilli M, Ricci-Vitiani L, Bini
M, Perez Alea M, Maria Florena A, Miceli L, Condorelli G, Bonventre
S, Di Gesù G, et al: Autocrine production of interleukin-4 and
interleukin-10 is required for survival and growth of thyroid
cancer cells. Cancer Res. 66:1491–1499. 2006. View Article : Google Scholar
|
|
27
|
Todaro M, Alea MP, Di Stefano AB,
Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G,
Medema JP, et al: Colon cancer stem cells dictate tumor growth and
resist cell death by production of interleukin-4. Cell Stem Cell.
1:389–402. 2007. View Article : Google Scholar
|
|
28
|
Leon-Cabrera SA, Molina-Guzman E,
Delgado-Ramirez YG, Vázquez-Sandoval A, Ledesma-Soto Y,
Pérez-Plasencia CG, Chirino YI, Delgado-Buenrostro L,
Rodríguez-Sosa M, Vaca-Paniagua F, et al: Lack of STAT6 attenuates
inflammation and drives protection against early steps of
colitis-associated colon cancer. Cancer Immunol Res. 5:385–396.
2017. View Article : Google Scholar
|
|
29
|
Li BH, Yang XZ, Li PD, Yuan Q, Liu XH,
Yuan J and Zhang WJ: IL-4/Stat6 activities correlate with apoptosis
and metastasis in colon cancer cells. Biochem Biophys Res Commun.
369:554–560. 2008. View Article : Google Scholar
|
|
30
|
Zhang M, Zhou Y, Xie C, Zhou F, Chen Y,
Han G and Zhang WJ: STAT6 specific shRNA inhibits proliferation and
induces apoptosis in colon cancer HT-29 cells. Cancer Lett.
243:38–46. 2006. View Article : Google Scholar
|
|
31
|
Vizcaíno C, Mansilla S and Portugal J: Sp1
transcription factor: A long-standing target in cancer
chemotherapy. Pharmacol Ther. 152:111–124. 2015. View Article : Google Scholar
|
|
32
|
Safe S, Imanirad P, Sreevalsan S, Nair V
and Jutooru I: Transcription factor Sp1, also known as specificity
protein 1 as a therapeutic target. Expert Opin Ther Targets.
18:759–769. 2014. View Article : Google Scholar
|
|
33
|
Hedrick E, Cheng Y, Jin UH, Kim K and Safe
S: Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4
are non-oncogene addiction genes in cancer cells. Oncotarget.
7:22245–22256. 2016. View Article : Google Scholar
|
|
34
|
Wierstra I: Sp1: emerging roles - beyond
constitutive activation of TATA-less housekeeping genes. Biochem
Biophys Res Commun. 372:1–13. 2008. View Article : Google Scholar
|
|
35
|
LaPorte SL, Juo ZS, Vaclavikova J, Colf
LA, Qi X, Heller NM, Keegan AD and Garcia KC: Molecular and
structural basis of cytokine receptor pleiotropy in the
interleukin-4/13 system. Cell. 132:259–272. 2008. View Article : Google Scholar
|
|
36
|
Murata T, Noguchi PD and Puri RK: IL-13
induces phosphorylation and activation of JAK2 Janus kinase in
human colon carcinoma cell lines: similarities between IL-4 and
IL-13 signaling. J Immunol. 156:2972–2978. 1996.
|