Introduction
According to the World Health Organization (WHO)
classification, glioblastoma (GBM), a high grade primary glioma
(grade IV), ranks as the most fatal type of glioma in adults.
Previous studies have indicated that the present traditional
therapy for glioma produces limited curative effects and is
unsatisfactory (1,2). Only 10% of patients with GBM, who are
diagnosed with a favorable Karnofsky performance scale (KPS;
>70%) and treated with standard therapies live for >5 years
(3). Therefore, the identification
of novel and effective biomarkers may provide more effective
therapeutic targets.
In general, a cohort of untranslatable transcripts,
known as long non-coding RNAs (lncRNAs) when >200 nucleotides in
length, is not responsible for coding proteins. However, they have
been demonstrated to be involved in the regulation of gene
expression (4,5). An increasing number of studies have
demonstrated that lncRNAs are involved in several intracellular
biological functions (6,7). Additionally, the abnormal regulation
of lncRNAs has been demonstrated to be associated with the
deterioration of various types of tumor (8). Small nucleolar RNA host gene 12
(SNHG12), a lncRNA located at chromosome 1p35.3, has primarily been
shown to be markedly upregulated in endometrial tumors (9). Furthermore, interference with the
expression of SNHG12 in endometrial cancer cells has been shown to
inhibit cell proliferation and induce apoptosis (9). A recent study demonstrated that
SNHG12 was upregulated and may be an oncogene in various types of
human tumors, including osteosarcoma (10), nasopharyngeal carcinoma (11) and lung cancer (12). These studies suggest that SNHG12
plays an important role in the growth, viability, invasion and
metastasis of cancer cells. However, to the best of our knowledge,
the possible roles of SNHG12 in GBM remain unknown.
As an RNA-binding protein belonging to the ELAV
family, Hu antigen R (HuR) binds to mRNAs containing adenine
(AU)-or uridine (U)-rich sequence elements (ARE) located at the
3'UTR and plays important roles in the post-transcriptional
regulation of oncogenes involved in tumorigenesis (13). A previous study demonstrated that
the expression of HuR (ELAV1) was elevated in primary brain cancer
and maintained the RNA stability of certain growth factors,
including vascular endothelial growth factor (VEGF) and interleukin
(IL)-8 (14). Previous studies
have determined the expression of HuR in various types of cancer,
including breast (15), colorectal
(16), ovarian (17) and pancreatic (18) cancer. Previous studies have also
indicated that HuR may be selected as a promising biomarker of
certain disease activities (19–24).
Filippova et al (14)
demonstrated that the regulation of the expression of HuR by
genetic manipulation-induced alterations in the proliferation and
the apoptosis of glioma cells, which indicates the potential
application of HuR as a promising biomarker and target in glioma.
Thus, the present study aimed to investigate the possible roles of
SNHG12 involved in the oncogenic events of glioma and the
interaction between SNHG12 and HuR in glioma cells.
Materials and methods
Ethics approval
This study complied with the Declaration of Helsinki
and was approved by the Medical Ethics Committee of Biomedicine
Research, General Hospital of Shenyang Military Command (Shenyang,
China). Glioma tumor specimens were obtained from consenting
patients at the General Hospital of Shenyang Military Command
(Shenyang, China). The patients were informed and provided written
consent.
Sample collection and molecular
detection
A total of 79 patients consented to participate in
the present study. Between January, 2011 and December, 2012, the
patients (n=79) attended the clinic of the General Hospital of
Shenyang Military Command. All patients had received surgery at
this hospital and had not received any anticancer therapy,
including surgical resection, until they were admitted to that
hospital. The resected specimens were histopathologically verified
as primary glioma on the basis of the WHO Classification of Tumors
of the Central Nervous System by three independent senior
pathologists (25,26). Primary tumor samples and matched
adjacent non-carcinoma tissues were resected for further analysis.
The matched adjacent normal tissues were resected ≥3 cm away from
the primary carcinoma, as previously described (27).
Molecular feature detection was also performed.
Using a DNeasy Blood & Tissue kit (Qiagen, Tokyo, Japan), DNA
was extracted from the frozen cancer tissue according to the
manufacturer's instructions. The C228T and C250T mutation hotspots
in the TERT promoter, and the presence of isocitrate
dehydrogenase (IDH) gene 1 (R132) and 2 (R172) hotspot
mutations, were assessed by pyrosequencing and partly by Sanger
sequencing, as previously described (28,29).
The methylation status of the MGMT promoter was also
detectedusing a customized pyrosequencing assay, as previously
reported (30).
Cell culture and transfection
The normal human skin fibroblast HF cell line and
the glioma cell lines, U87, LN229, U373 and U251, were purchased
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). It should be noted that the cell line U373 has been
reported to be misidentified according to the Cellosaurus
(https://web.expasy.org/cellosaurus/CVCL_2219).
Therefore, the identity of the cell line used in the present study
was validated via an STR profile test by the Cell Bank of the
Chinese Academy of Sciences. The results revealed that the matching
ratio between the test sample and ATTC standard data was 88.8%,
which indicates that the cell line used in the present study is
identifiable as U373 according to the ASN-0002-2011 criterion. The
U87 cell line has also been reported to be
misidentified/contaminated and this cell line is considered
glioblastoma of unknown origin. The U87 cell line was also
authenticated using an STR profile test by the Cell Bank of the
Chinese Academy of Sciences. The results revealed that the matching
ratio between the test sample and ATTC standard data was 94.4% and
confirmed the identity of the cell line used in this study as U87.
Dulbecco's modified Eagle's medium (Gibco/Thermo Fisher Scientific,
Shanghai, China), supplemented with 10% fetal bovine serum as well
as 100 U/ml penicillin and 100 µg/ml streptomycin (Biowest,
Nuaillé, France) were employed for cell culture (31).
Specific small interfering RNAs (siRNA) targeting
human SNHG12 mRNA (si-SNHG12) were designated and purchased from
Sangon Biotech Co., Ltd. (Shanghai, China). The sequences were as
follows: Sense, 5′-GCAGUGUGCUAC UGAACUUTT-3′ and antisense,
5′-AAGUUCAGUAGCACA CUGCTT-3′ (32). Using Lipofectamine 2000 reagent
(Invitrogen, Carlsbad, CA, USA), 2×105 HF, U87 and U251
cells were prepared and transfected for 48 h, respectively. The
negative control duplex containing siRNA (si-NC; Sangon Biotech
Co., Ltd.) was employed as the control.
For SNHG12 overexpression, the full-length SNHG12
cDNA was synthesized by Biomarker Technologies (Beijing, China) and
bound with the pcDNA3.1(+) vector (Invitrogen), as previously
described (31,32). PcDNA3.1-SNHG12 (p-SNHG12) and
control blank vector (p-NC) were transfected into the U87 and U251
cells using Lipofectamine 2000 reagent. As the normal control, HF
cells were also prepared for trans-fection with p-SNHG12 or p-NC.
The cells were collected for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis at 48 h after
transfection.
The expression of HuR in the U87 and U251 cells was
upregu-lated or specifically downregulated in order to determine
whether SNHG12 is associated with HuR, as previously described
(33). Briefly, the pMSCV-PIG
plasmid ligated into HuRDNA fragments (CMV-HuR) and
pMDH-PGK-EGFP2.0 vector inserted with HuR siRNA (si-HuR) were
prepared. The HuR siRNA sequence was as follows:
5′-UUGUCAAACCGGAUAAACGCA-3′. The levels of SNHG12 in the U87 and
U251 cells, transfected with CMV-HuR/control vector or
si-HuR/negative control (si-NC) were detected by using RT-qPCR.
RT-qPCR
The expression of SNHG12 in the human glioma cells
or specimens from patients with glioma was evaluated by RT-qPCR as
previously described (32).
Briefly, TRIzol reagent was used for total RNA extraction. The
extracted RNA was then reverse transcribed into cDNA using the
ProtoScript® First Strand cDNA Synthesis kit (New
England Biolabs, Ipswich, MA, USA) according to the manufacturer's
instructions. iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA)
was employed for RT-qPCR analysis according to the manufacturer's
instructions. The 2−ΔΔCt or 2−ΔΔCq method
(33) was performed to analyze the
relative changes of SNHG12 after normalizing to GAPDH (endogenous
control). The primer sequences were as follows: GAPDH sense,
5′-CGAGATCCCTCCAAAATCAA-3′ and antisense,
5′-TTCACACCCATGACGAACAT-3′; SNHG12 sense,
5′-TCTGGTGATCGAGGACTTCC-3′ and antisense,
5′-ACCTCCTCAGTATCACACACT-3′ (32);
HuR sense, 5'-ATG AAGACCACATGGCCGAAGACT-3′ and antisense,
5′-TGTGGTCATGAGTCCTTCCACGAT-3′ (34).
Cell viability assay
Following various treatments, cell viability was
evaluated by MTT assay as previously described (31). Briefly, 1×105 cells/ml
were seeded into culture plates with 200 µl culture medium
per well. After 48 h, the cells were incubated with 20 µl of
5 mg/ml MTT solution at 37°C for 4 h, followed by the addition of
150 µl dimethyl sulfoxide. A microplate reader (Multiskan
Mk3; Thermo Fisher Scientific) was used to measure the absorbance
of each sample at 570 nm and the data were collected for
analysis.
Migration and invasion assays
Migration and invasion assays were performed as
previously described (35). Cells
at a density of 1×105/ml were prepared in serum-free
medium and plated in the upper chamber for the cell migration
assay. Achamber pre-coated with 500 ng/ml Matrigel solution (BD
Biosciences, Franklin Lakes, NJ, USA) was obtained for the cell
invasion assay. Cells that had migrated to the upper membrane
surface were physically removed after 48 h of incubation. Those
that had migrated or invaded to the lower side of the membrane were
collected for statistical analysis. The cells were then fixed,
stained and imaged under a microscope as previously described
(36).
Scratch wound assay
A scratch wound assay was performed to evaluate the
migration of glioma cells after the various treatments, as
previously described (37).
Briefly, at ~80% confluency, the cells transfected with various
chemicals were seeded onto 6-well plates and incubated at 37°C,
respectively. Using a 10-µl pipette tip, a vertical scratch
wound was made through the center of each well plate. After washing
3 times with PBS, fresh serum-free medium was added and the cells
were incubated for 48 h. Subsequently, a light microscope (Olympus,
Tokyo, Japan) was used to examine the mobility of the cell
monolayer at a magnification of ×200.
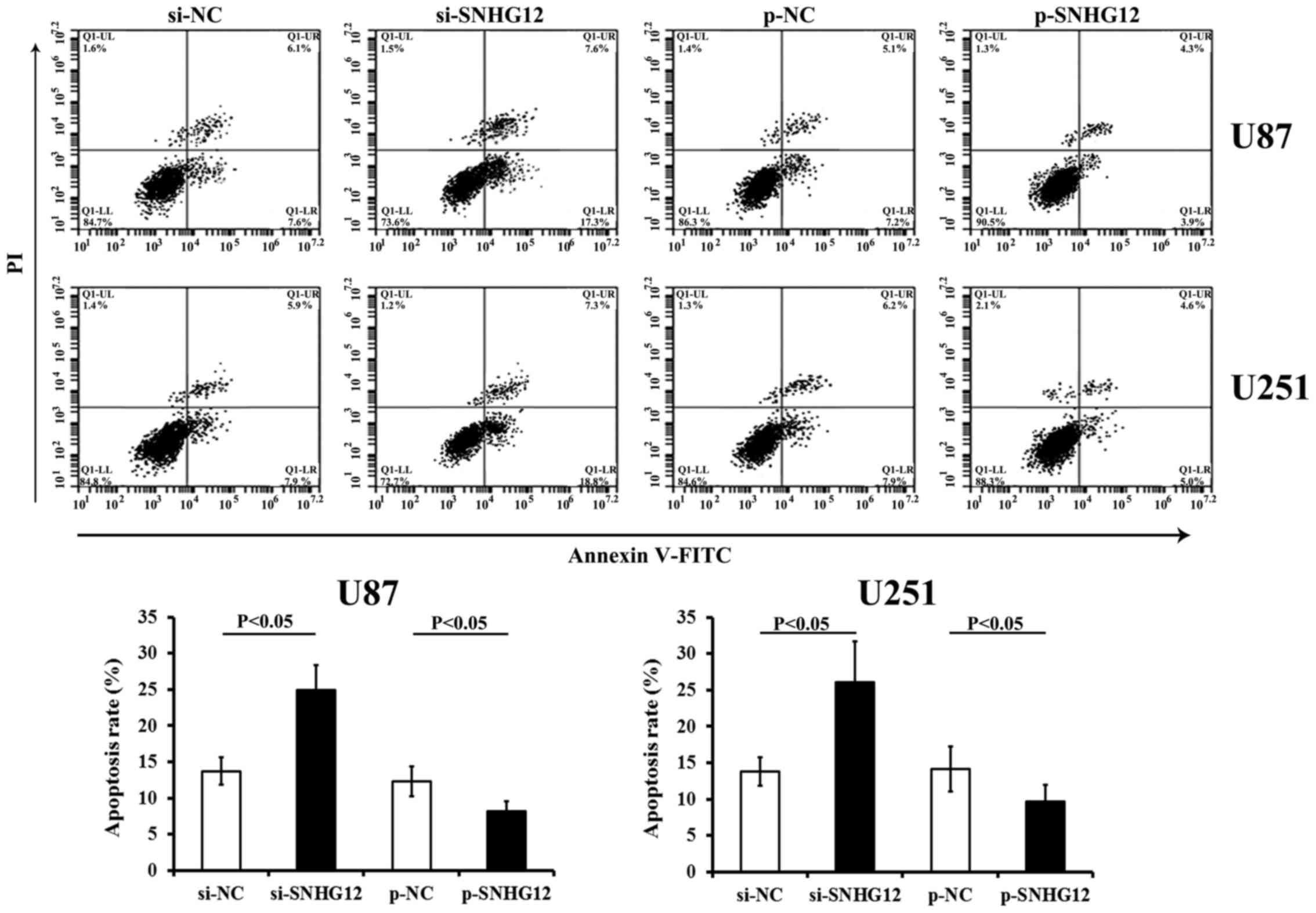
Apoptosis assay
Flow cytometric analysis was performed to evaluate
the effects of SNHG12 on the apoptosis of the cells, by
transfecting the U87 and U251 cells with either si-SNHG12 or
p-SNHG12. An Annexin V-fluorescein isothiocyanate (FITC), propidium
iodide (PI) assay kit (4A Biotech Co., Ltd.), FACScan flow
cytometer and Cell Quest software (both from BD Immunocytometry
Systems, San Jose, CA, USA) were employed to determine the rates of
cell apoptosis.
Bioinformatics analysis
A total of 118,777 transcripts were obtained from
LNCipedia (www.lncipedia.org)as previously
described (34). As HuR directly
binds to mRNA at 3'UTR containing ARE elements, transcripts
containing ARE elements were selected as the potential targets for
HuR.
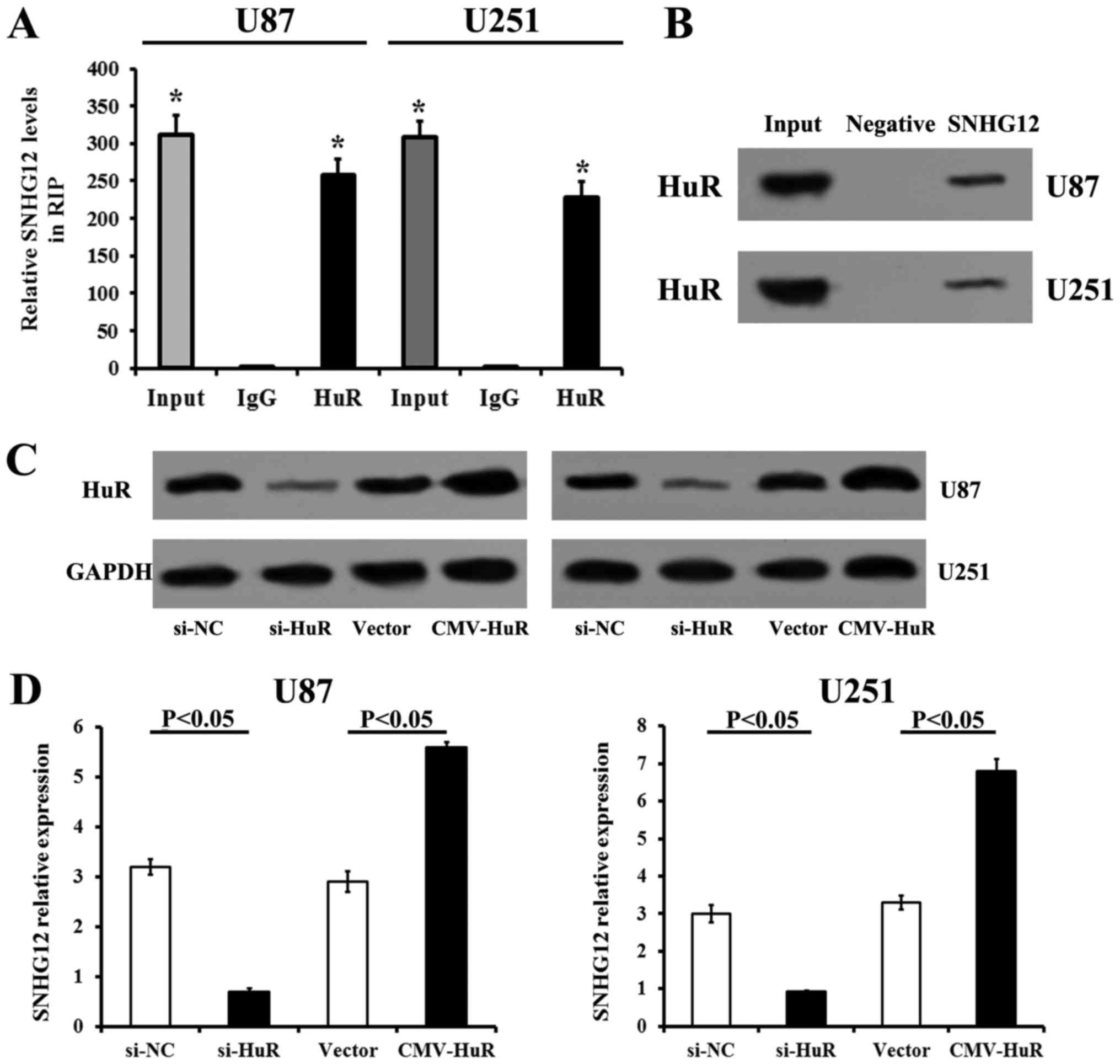
RNA immunoprecipitation (RIP) assay
Using the Magna RIP kit (Millipore, Billerica, MA,
USA), RIP assay was performed according to the manufacturer's
instructions. Briefly, at 80–90% confluency, the U87 and U251 cells
were collected and lysed in RIP lysis buffer. The cell extract (100
µl) was prepared and incubated with RIP buffer
pre-conjugated with HuR antibodies (1.5 µg per 500 µg
of total protein; cat. no. sc-5261; Santa Cruz Biotechnology, Santa
Cruz, CA, USA) or control mouse IgG (cat. no. HP6069, Millipore) at
4°C for 4 h. The complexes were treated with Proteinase K for 30
min with shaking at 55°C. Immunoprecipitated RNA in the
precipitates was purified using TRIzol RNA reagent (Invitrogen) and
analyzed for SNHG12 by RT-qPCR.
RNA pull-down assays
The interaction between SNHG12 and HuR was further
determined by an RNA pull-down assay as previously described
(31). The pCDNA3.1-SNHG12 vector
was cleaved by NruI and incubated with RNase-free DNaseI
(Takara, Dalian, China). The mMESSAGE mMACHINE T7® kit
(Ambion/Thermo Fisher Scientific) was used to transcribe SNHG12
from the pCDNA3.1-SNHG12 vector. The transcribed SNHG12 was then
purified using a RNeasy Mini kit (Qiagen, Valencia, CA, USA). A
Pierce RNA 3' End Desthiobiotinylation kit (Thermo Fisher
Scientific) was employed to label the 3' end of SNHG12 as per the
instructions provided with the kit. The non-specific IgG antibody
was used as the negative control. The binding protein isolated from
the RNA-protein complex was then determined by western blot
analysis.
Western blot analysis
Total proteins were extracted from the cells with
various treatments using cell lysis buffer for western blot
analysis (Beyotime Biotechnology, Shanghai, China). The
concentration of protein wasthen detected using the BCA Assay kit
(Beyotime Biotechnology) according to the manufacturer's
instructions. The expression of HuR protein was detected as
previously described (34).
Briefly, cell lysates were prepared with RIPA lysis buffer
containing protease inhibitor cocktail (both from Beyotime
Biotechnology). The nitrocellulose membranes on which the protein
samples were transferred to were blocked for 2 h at room
temperature and then incubated overnight at 4°C with the primary
antibodies, including HuR (dilution 1:800; cat. no. sc-5261) and
GAPDH (dilution 1:1500; cat. no. sc-47724) (both from Santa Cruz
Biotechnology). After washing, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibody (dilution
1:1,000; no. A0216; Beyotime Biotechnology) for 1 h. Protein blots
were determined using enhanced chemiluminescence luminol reagent
(Millipore) and quantified by densitometric analysis using Quantity
One software (Bio-Rad).
Statistical analysis
Data are presented as the means ± standard
deviation. SPSS software version 19.0 (SPSS Inc., Chicago, IL, USA)
was employed for analyses. The association between SNHG12 levels
(low or high level) and the clinicopathological or genetic
characteristics of the patients with glioma was evaluated using
multivariate logistic regression analysis. Odds ratio (OR) with 95%
confidence interval (CI) were employed to evaluate the strength of
association. Ordinal data were collected and analyzed using
logistic regression. Survival curves were esti mated with the
Kaplan-Meier method, and differences were compared using the
log-rank test. The statistical significance of SNHG12 expression,
MTT cell activity, migration, invasion and apoptosis rate among
groups with different treatments was analyzed by one-way ANOVA. A
least significant difference post hoc test by Student-Newman-Keuls
test was used to obtain individual P-values following ANOVA, as
previously described (31). P≤0.05
was considered to indicate a statistically significant
difference.
Results
SNHG12 expression is elevated in glioma
tissues and cells
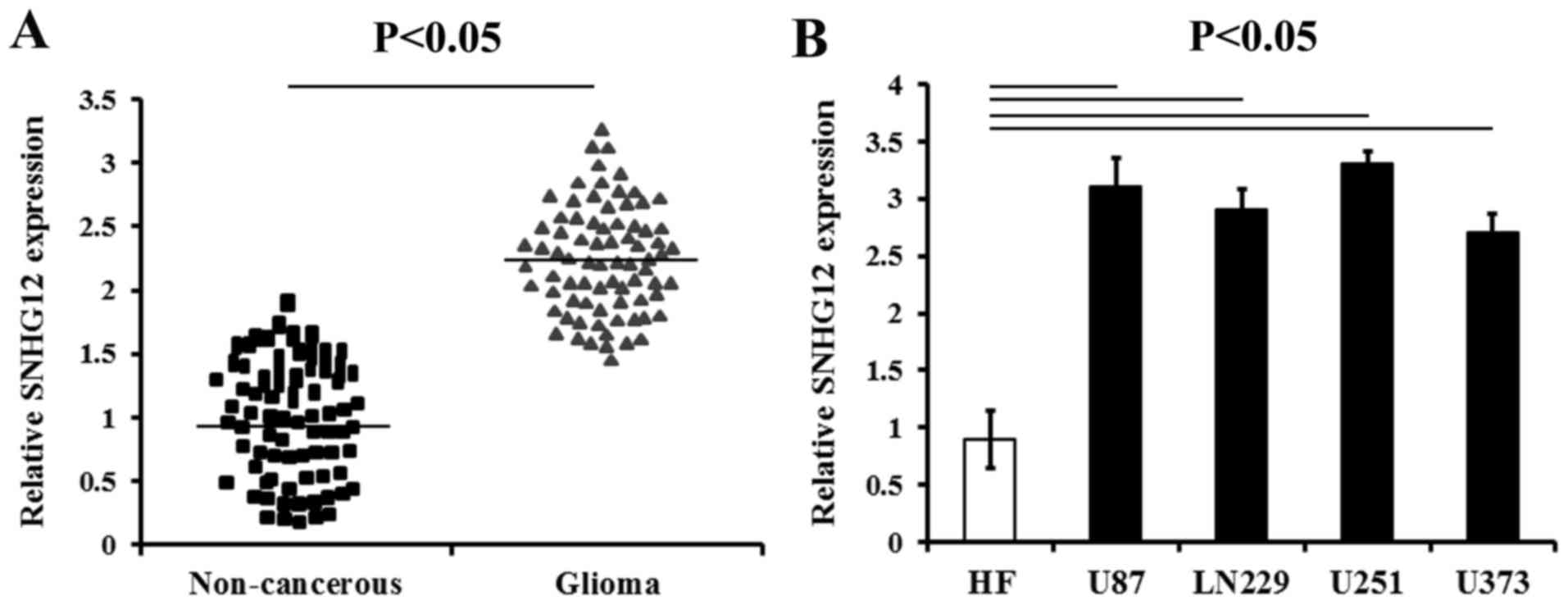
The results of RT-qPCR analysis revealed that the
expression levels of SNHG12 were prominently increased in the
glioma tissues compared with those of the matched non-cancerous
brain tissues (P<0.05) (Fig.
1A). The SNHG12 level were also increased in all the human
glioma cell lines (U87, LN229, U373 and U251) compared to the HF
cells, respectively (P<0.05) (Fig.
1B).
Increased SNHG12 expression is associated
with glioma progression
According to the SNHG12 level (determined by
RT-qPCR), the patients enrolled in the present study were divided
into 2 groups as follows: Those with less than or equal to the
median of the SNHG12 levels (low level) and those with higher than
the median of the SNHG12 levels (high level). The associations
between the clinicopathological characteristics of the patients
with glioma and the SNHG12 mRNA levels were analyzed. The data
demonstrated that high levels of SNHG12 were significantly
associated with age (r=0.176, P=0.021), WHO grade (r=0.349,
P=0.003) and KPS score (r=0.498, P=0.001) (Table I). Sex, resection status, tumor
size and tumor location were not found to be associated with SNHG12
expression (Table I).
 | Table IAssociation of SNHG12 expression is
with the clinicopathological characteristics of patients with
glioma. |
Table I
Association of SNHG12 expression is
with the clinicopathological characteristics of patients with
glioma.
| Parameters | SNHG12 expression,
n (%)
| rs | P-value | Adjusted OR (95%
CI) |
|---|
| Low level n=40 | High level
n=39 |
|---|
| Ages (years) | | | 0.176 | 0.021 | |
| <55 | 28 (35.4) | 14 (17.7) | | | 0.42
(0.23–0.83) |
| ≥55 | 12 (15.2) | 25 (31.7) | | | 5.21
(2.67–10.79) |
| Sex | | | 0.007 | 0.671 | |
| Male | 21 (26.5) | 19 (24.1) | | | 1.23
(0.46–2.17) |
| Female | 19 (24.05) | 20 (25.3) | | | 1.03
(0.52–2.95) |
| Resection
status | | | 0.009 | 0.572 | |
| Total | 17 (21.5) | 21 (26.6) | | | 1.34
(0.67–2.09) |
| Subtotal | 23 (29.1) | 18 (22.8) | | | 1.19
(0.54–4.13) |
| Tumor size
(cm) | | | 0.069 | 0.108 | |
| <5 | 22 (27.84) | 16 (20.3) | | | 1.21
(0.75–3.46) |
| ≥5 | 18 (22.8) | 23 (29.1) | | | 1.42
(0.63–1.69) |
| Tumor location | | | 0.014 | 0.595 | |
| Frontal | 15 (19) | 14 (17.7) | | | 1.12
(0.89–2.25) |
| Occipital | 5 (6.3) | 5 (6.3) | | | 1.02
(0.93–1.13) |
| Temporal | 7 (8.9) | 8 (10.1) | | | 1.24
(0.82–3.33) |
| Other | 13 (16.5) | 12 (15.2) | | | 1.08
(0.69–2.21) |
| WHO grade | | | 0.349 | 0.003 | |
| Low grade
(I–II) | 31 (39.2) | 10 (12.7) | | | 0.43
(0.24–0.88) |
| High grade
(III–IV) | 9 (11.4) | 29 (36.7) | | | 4.54
(2.09–9.97) |
| Number of tumor
nodules | | | 0.009 | 0.705 | |
| Multiple | 2 (2.5) | 3 (3. 8) | | | 1.04
(0.76–3.57) |
| Single | 38 (48.1) | 36 (45.6) | | | 1.21
(0.76–2.86) |
| KPS score | | | 0.498 | 0.001 | |
| <80 | 11 (13.9) | 31 (39.3) | | | 5.07
(2.93–8.02) |
| ≥80 | 29 (36.7) | 8 (10.1) | | | 0.67
(0.39–0.78) |
The assessment of the molecular characteristics of
these unselected patients with glioma was also performed. The data
revealed that SNHG12 expression was significantly associated with
TERT promoter mutation (r=0.448, P=0.001), IDH1
mutation (r=0.609, P=0.0007), 1p/19q status (r=0.712,
P=0.0005) and MGMT methylated status (r=0.401, P=0.002)
(Table II).
 | Table IIAssociation between SNHG12 expression
and molecular characteristics of patients with glioma. |
Table II
Association between SNHG12 expression
and molecular characteristics of patients with glioma.
| SNHG12 expression,
n (%)
| rs | P-value | Adjusted OR (95%
CI) |
|---|
| Low level n=40 | High level
n=39 |
|---|
| TERT
status | | | 0.448 | 0.001 | |
| WT | 29 (36.7) | 13 (16.5) | | | 0.31
(0.22–0.65) |
| mut | 11 (13.9) | 26 (32.9) | | | 5.23
(3.09–11.28) |
| IDH1/2
status | | | 0.609 | 0.0007 | |
| WT | 10 (12.7) | 28 (35.4) | | | 4.75
(6.98–15.76) |
| mut | 30 (38) | 11 (13.9) | | | 0.36
(0.18–0.79) |
| 1p/19q
status | | | 0.712 | 0.0005 | |
| Non-codel | 35 (44.3) | 7 (8.9) | | | 0.21
(0.13–0.58) |
| Codel | 5 (6.3) | 32 (40.5) | | | 6.73
(3.49–10.96) |
| MGMT
status | | | 0.401 | 0.002 | |
| Methylation | 12 (15.2) | 27 (34.2) | | | 5.54
(2.33–11.64) |
| Unmethylation | 28 (35.4) | 12 (15.2) | | | 0.44
(0.29–0.79) |
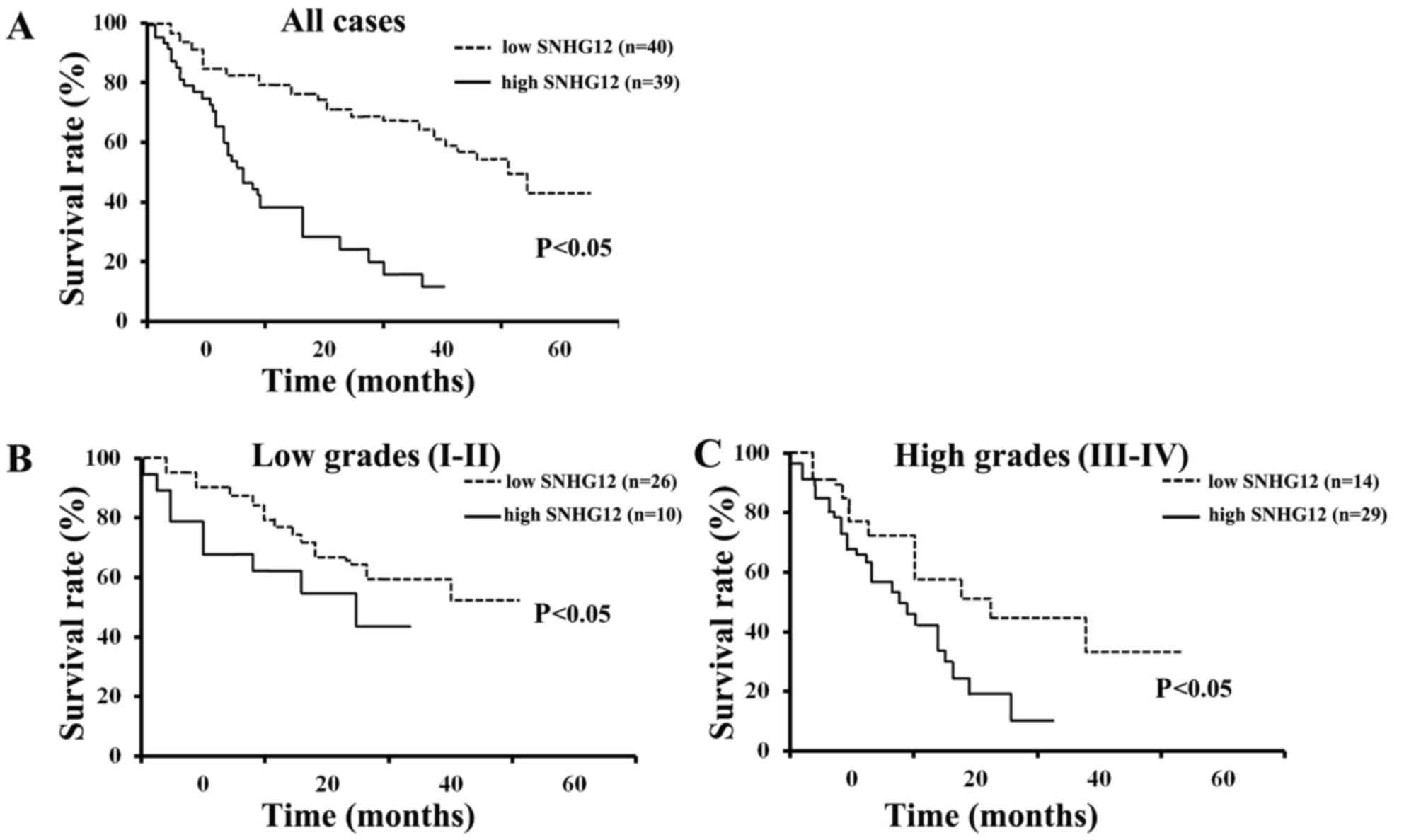
Kaplan-Meier survival analysis revealed that the
patients with low levels of SNHG12 had a significantly increased
overall survival (OS) compared to those with high levels of SNHG12
expression (P<0.05) (Fig. 2A).
The data also indicated that patients with either low grade (I–II)
or high grade (III–IV) glioma and low levels of SNHG12 expression
had an increased OS, compared to those with high levels of SNHG12
expression (P<0.05) (Fig. 2B and
C). These results indicate that the SNHG12 level is a promising
indicator for patients with glioma of all WHO grades, particularly
in those with grade III–IV glioma.
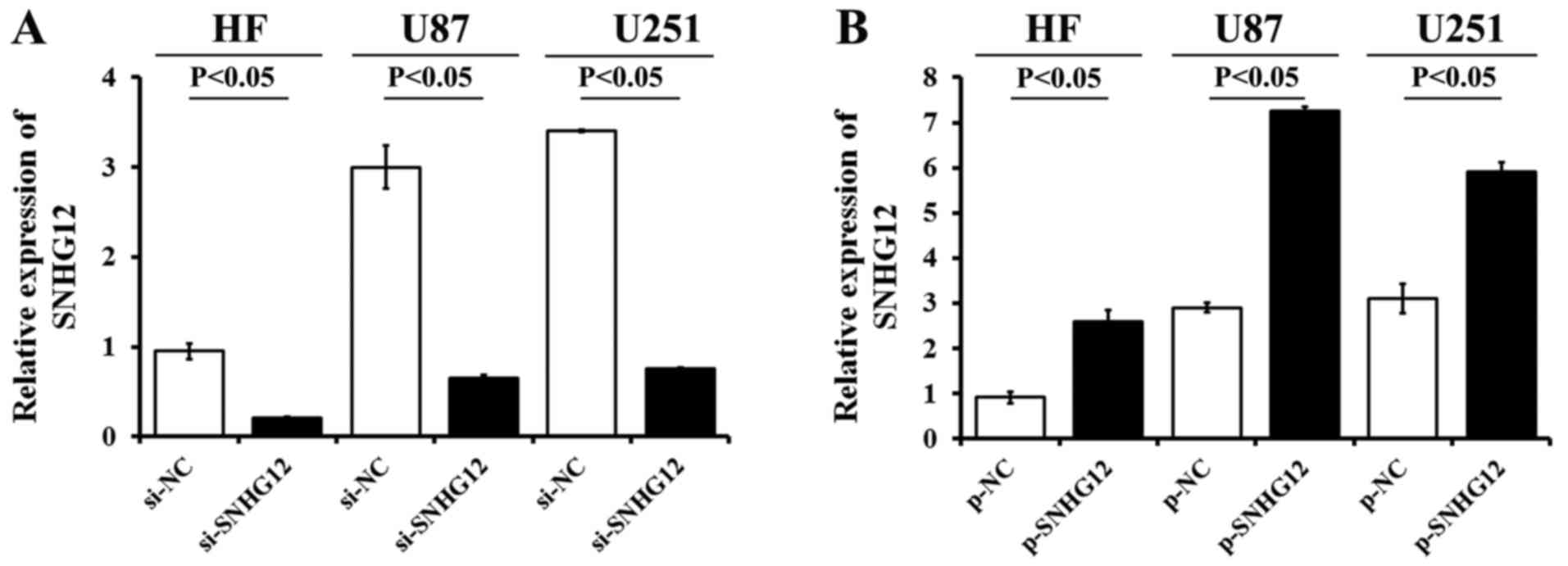
SNHG12 expression following transfection
with SNHG12 siRNA or overexpression plasmid
The expression of SNHG12 in the HF cells and glioma
cell lines transfected with si-SNHG12, p-SNHG12 or the matched
control si-NC, p-NC, was determined by RT-qPCR. The data revealed a
significant decrease (>77%) in SNHG12 mRNA expression following
stable transfection of the U87, U251 and HF cells with si-SNHG12 in
(Fig. 3A). Following p-SNHG12
transfection, the SNHG12 levels were markedly higher than those in
the matched p-NC-treated ones, respectively (P<0.05) (Fig. 3B). The expression levels of SNHG12
increased 2.8-fold in the HF cells, 2.5-fold in the U87 cells and
1.9-fold in the U251 cells, compared to the matched p-NC-treated
cells, respectively.
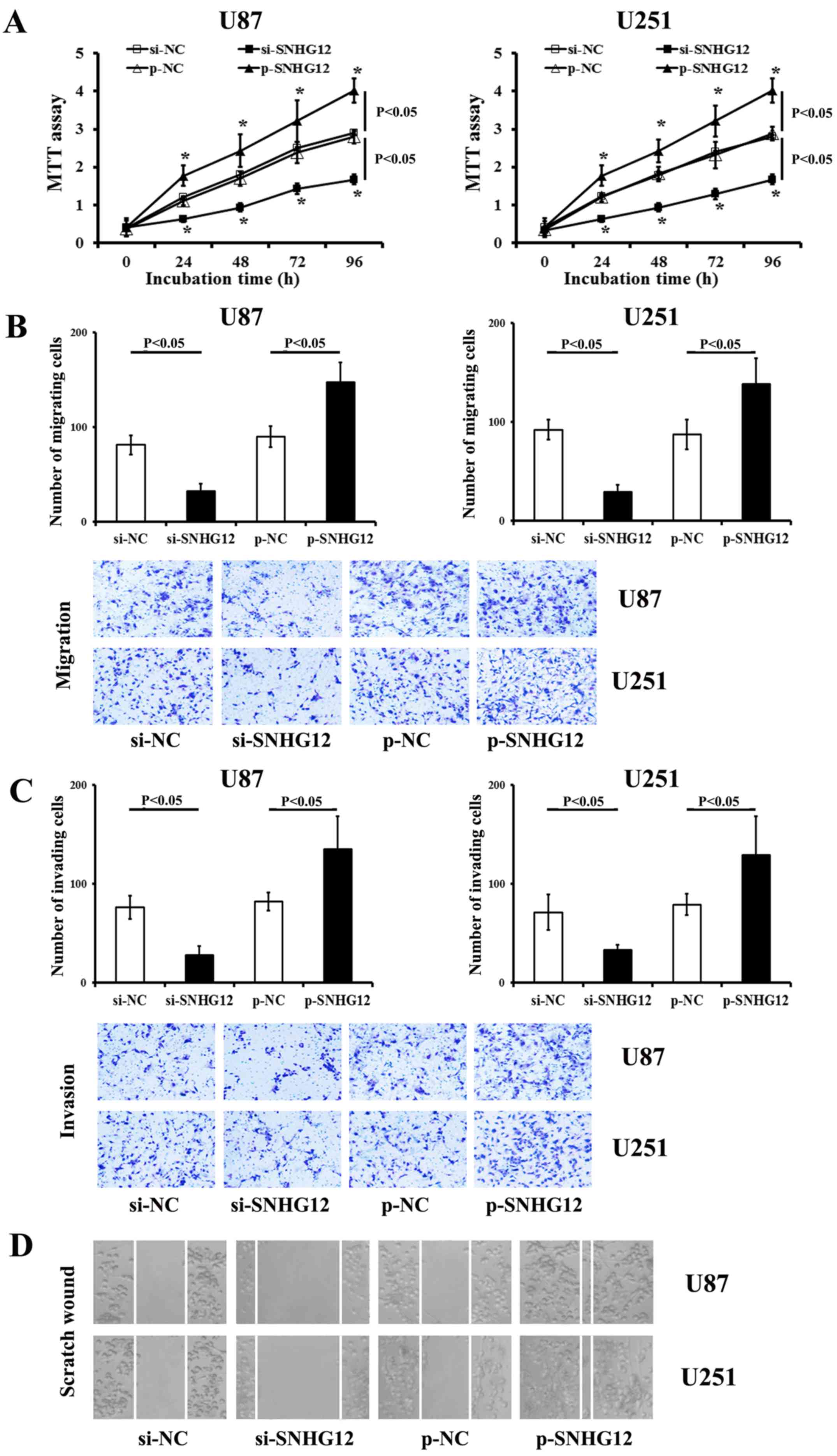
SNHG12 increases the viability and
mobility of glioma cells
The functional inhibition of endogenous SNHG12
significantly decreased the viability of both the U87 and U251
cells (compared to the matched si-NC-treated ones, respectively)
(P<0.05) (Fig. 4A). Conversely,
transfection with SNHG12 by specific p-SNHG12 induced a significant
promotion of the viability of both the U87 and U251 cells
(P<0.05) (Fig. 4A).
As shown by results of the migration and invasion
assays, there were less stained cells observed in the
si-SNHG12-transfected group than that in the matched si-NC control
groups (P<0.05) (Fig. 4B and
C). Transfection with p-SNHG12 increased the levels of
migration and invasion in both the glioma cell lines compared to
the p-NC-treated cells (P<0.05) (Fig. 4B and C). Scratch wound assay
revealed that transfection with si-SNHG12 significantly impaired
the invasiveness of the U87 and U251 cells (Fig. 4D), when compared to the control
si-NC-treated ones, respectively (P<0.05). Conversely, p-SNHG12
transfection significantly promoted the migration and invasion of
both glioma cells (compared to the matched p-NC control,
respectively) (P<0.05) (Fig.
4B–D).
SNHG12 inhibits the apoptosis of glioma
cells
Transfection with si-SNHG12 promoted the apoptosis
of either the U87 andU251 cells (compared to those transfected with
si-NC, P<0.05) (Fig. 5). The
overexpression of SNHG12 by transfection with p-SNHG12
significantly inhibited the apoptosis of both glioma cell lines
(compared to the matched p-NC control, respectively) (P<0.05)
(Fig. 5).
SNHG12 directly interacts with HuR in
glioma cells
The binding of SNHG12 with HuR was confirmed by
RIP-RT-qPCR in the U87 and U251 cells. RIP assays and subsequent
RT-qPCR revealed that SNHG12 was rich in RIP samples treated with
HuR antibodies, when compared to the non-specific IgG antibody
treated ones, which confirmed the specificity of RIP assays and
RT-qPCR (Fig. 6A). RNA pull-down
assays and western blot analysis also confirmed the interaction
between SNHG12 and HuR (Fig. 6B).
Transfection with si-HuR significantly decreased the HuR protein
levels in the U87 and U251 cells (compared to those transfected
with si-NC, P<0.05) (Fig. 6C).
The upregulation of HuR by CMV-HuR transfection also induced an
increase in HuR expression (compared to those transfected with the
vector) (P<0.05) (Fig. 6C).
Additionally, the RT-qPCR data demonstrated that the silencing of
HuR expression decreased the expression of SNHG12, while the
overexpression of HuR increased the level of SNHG12 in both glioma
cell lines (Fig. 6D). These
results revealed that SNHG12 directly bound to HuR in both U87 and
U251 cells.
Discussion
The findings of the present study are the following:
i) An abnormally elevated SNHG12 expression is associated with
certain clinicopathological and genetic characteristics of patients
with glioma; ii) patients with glioma and high levels of SNHG12
exhibit a reduced OS rate compared with those with lower levels;
iii) the silencing of SNHG12 using siRNA inhibits human glioma cell
proliferation, invasion and migration, and promotes apoptosis; and
iv) SNHG12 interacts with HuR in glioma cells and HuR modulates the
stabilization of SNHG12.
The dysregulation of lncRNAs has been demonstrated
to be involved in multiple intercellular and cellular processes of
numerous types of tumor (38). A
novel lncRNA, SNHG12, was recently reported to be elevated in
cancer and to play oncogenic roles in various types of tumor
(32,39,40).
A previous study demonstrated that the upregulation of SNHG12
(induced by c-MYC) increased cell viability and inhibited apoptosis
in triple-negative breast tumors (39). By regulating the levels of matrix
metalloproteinase (MMP)13, SNHG12 was reported to be involved in
cell migration in breast cancer (39). Furthermore, SNHG12 was proven to be
an oncogene that contributes to the promotion of tumorigenesis and
metastasis in hepatocellular carcinoma (40). However, to the best of our
knowledge, there is limited information on SNHG12 in the
progression of glioma. In the present study, it was found that the
endogenous SNHG12 expression was significantly increased in glioma
tissues and cells, while low levels of SNHG12 were associated with
certain clinicopathological characteristics and the OS rate of
patients with glioma. High levels of SNHG12 expression were
associated with the WHO grade (III–IV) and KPS score (<80),
which indicated the deterioration of patients with glioma. The
evaluation of the molecular genetic features of these unselected
glioma cohort revealed that SNHG12 expression was also associated
with the mutation of TERT and IDH1/2, as well as with
the 1p/19q status and MGMT methylated status.
The present study demonstrated that the TERT
promoter mutations and IDH-wild-type are the common
genotypes present in cohorts with high levels of SNHG12, which are
likely to become molecular indicators associated with a poor
prognosis (Table II). A previous
study demonstrated that TERT promoter mutations coinciding
with IDH1/2 mutations and 1p/19q co-deletion is the
most common genotypes detected in oligodendrogliomas, whereas a
combined genotype of IDH-wild-type present and TERT
promoter mutation has been observed in GBM (41). Therefore, considering the
expression of SNHG12, TERT promoter and IDH1/2
mutations may be used to distinguish the subclasses of glioma and
predict outcomes in high-grade glioma.
Studies have reported that the presence of
IDH1/2 mutation in patients diagnosed with astrocytoma is
associated with a favorable impact on survival rate (42,43).
These genotypes are barely observed in malignant glioma subclasses,
such as primary GBM and pilocytic astrocytomas, and are usually
accompanied by MGMT methylation, a well-established
prognostic marker for primary GBM (44–46).
The present study demonstrated that the presence of IDH1/2
mutation was positively associated with a relatively prolonged OS
in patients with glioma and lower levels of SNHG12, while the
presence of IDH-wild-type in those with high levels of
SNHG12 was associated with a poor OS. In the unselected cohorts,
the presence of MGMT methylation was detected in those with
high levels of SNHG12 expression, of which many more patients were
diagnosed with high grades (III–IV) (Table I). These data indicate that SNHG12
expression is highly associated with MGMT methylation and
the presence of IDH1/2 mutation. The present study suggests
that the combined assessment of SNHG12 expression and molecular
characteristics may be valuable as a prognostic indicator of
high-grade glioma. However, further investigations of the
association between SNHG12 and the well-established molecular
markers are required.
The present study demonstrated that SNHG12 may
interact with HuR in the tumorigenesis of glioma cells. HuR is an
ubiquitous, multi-faceted RNA-binding protein involved in the
stabilization, splicing and translational regulation of mRNAs
(24). HuR can enhance
tumorigenesis by interaction with or regulation of the target
oncogenes that encode proteins which regulate intracellular
inflammation, angiogenesis, cell cycle, migration, invasion and
metastasis, and chemotherapeutic sensitivity (14,24).
The number of potential HuR-regulated mRNAs active in these disease
pathways is potentially large, as up to 8% of the transcribed
genome may contain an AU-rich 3′UTR (47). In malignant GBM, HuR has been
demonstrated to amplify the expression of cancer-related genes,
including tumor necrosis factor (TNF)-α, IL-8, IL-6, VEGF,
cyclo-oxygenase (COX)2, transforming growth factor (TGF)-β and
other tumorigenic genes, which contributes to increased cell
proliferation, apoptosis evasion and angiogenesis, as well as to
enhanced invasion and metastasis (48–50).
The totality of this regulation results in the necessity of glioma
cells to sustain and increase growth (51). A recent study demonstrated that HuR
plays crucial roles in the growth of glioma and its activity and
viability were considered as potential therapeutic targets for
malignant glioma (14). In the
present study, RIP assays and RNA pull-down assay suggested that
HuR directly interacts with SNHG12 mRNA, which contains potential
AU-rich elements amenable for regulation by HuR. The overexpressing
of HuR increased the levels of SNHG12, while the suppression of HuR
expression reduced the SNHG12 level in glioma cell lines. SNHG12
was significantly elevated in cohorts with glioma, while SNHG12
expressional interference inhibited viability, invasion and
migration, and promoted the apoptosis of human glioma cell lines.
These results suggest that SNHG12 is associated with and is
stabilized by HuR in glioma cells. The HuR-SNHG12 interaction may
function as an oncogenic axis and may be considered as a novel
therapeutic target in glioma, particularly malignant glioma.
The present study found an elevated SNHG12
expression in tissues of patients with glioma and in glioma cells.
High levels of SNHG12 were associated with certain
clinicopathological characteristics and the 5-year survival rate of
glioma cohorts. Interference with SNHG12 expression using
transfection with specific siRNA inhibited the viability and
motility, and promoted the apoptosis of human glioma cells. The
data also indicated that SNHG12 was directly associated with and
was stabilized by HuR. These data demonstrate that the HuR-SNHG12
axis may play an oncogenic function and may therefore could be
considered as a novel therapeutic target in glioma.
Acknowledgments
The authors would like to thank Yunnan Labreal
Biotechnology Co., Ltd. (Kunming, China) for their technical
support.
References
|
1
|
Weller M, van den Bent M, Hopkins K, Tonn
JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D,
Henriksson R, Balana C, et al European Association for
Neuro-Oncology (EANO) Task Force on Malignant Glioma: EANO
guideline for the diagnosis and treatment of anaplastic gliomas and
glioblastoma. Lancet Oncol. 15:e395–e403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanai N, Polley MY, McDermott MW, Parsa AT
and Berger MS: An extent of resection threshold for newly diagnosed
glioblastomas. J Neurosurg. 115:3–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Diederichs S: The four dimensions of
noncoding RNA conservation. Trends Genet. 30:121–123. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang JL, Zheng L, Hu YW and Wang Q:
Characteristics of long non-coding RNA and its relation to
hepatocellular carcinoma. Carcinogenesis. 35:507–514. 2014.
View Article : Google Scholar
|
|
7
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
8
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhai W, Li X, Wu S, Zhang Y, Pang H and
Chen W: Microarray expression profile of lncRNAs and the
upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int J
Oncol. 46:2125–2137. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruan W, Wang P, Feng S, Xue Y and Li Y:
Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes cell proliferation and migration by upregulating
angiomotin gene expression in human osteosarcoma cells. Tumour
Biol. 37:4065–4073. 2016. View Article : Google Scholar
|
|
11
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brennan CM and Steitz JA: HuR and mRNA
stability. Cell Mol Life Sci. 58:266–277. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Filippova N, Yang X, Wang Y, Gillespie GY,
Langford C, King PH, Wheeler C and Nabors LB: The RNA-binding
protein HuR promotes glioma growth and treatment resistance. Mol
Cancer Res. 9:648–659. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heinonen M, Fagerholm R, Aaltonen K,
Kilpivaara O, Aittomäki K, Blomqvist C, Heikkilä P, Haglund C,
Nevanlinna H and Ristimäki A: Prognostic role of HuR in hereditary
breast cancer. Clin Cancer Res. 13:6959–6963. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Denkert C, Koch I, von Keyserlingk N,
Noske A, Niesporek S, Dietel M and Weichert W: Expression of the
ELAV-like protein HuR in human colon cancer: Association with tumor
stage and cyclooxygenase-2. Mod Pathol. 19:1261–1269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Erkinheimo TL, Lassus H, Sivula A,
Sengupta S, Furneaux H, Hla T, Haglund C, Butzow R and Ristimäki A:
Cytoplasmic HuR expression correlates with poor outcome and with
cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer
Res. 63:7591–7594. 2003.PubMed/NCBI
|
|
18
|
Costantino CL, Witkiewicz AK, Kuwano Y,
Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M
and Brody JR: The role of HuR in gemcitabine efficacy in pancreatic
cancer: HuR Up-regulates the expression of the gemcitabine
metabolizing enzyme deoxycytidine kinase. Cancer Res. 69:4567–4572.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
López de Silanes I, Fan J, Yang X,
Zonderman AB, Potapova O, Pizer ES and Gorospe M: Role of the
RNA-binding protein HuR in colon carcinogenesis. Oncogene.
22:7146–7154. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heinonen M, Bono P, Narko K, Chang SH,
Lundin J, Joensuu H, Furneaux H, Hla T, Haglund C and Ristimäki A:
Cytoplasmic HuR expression is a prognostic factor in invasive
ductal breast carcinoma. Cancer Res. 65:2157–2161. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cho NP, Han HS, Soh Y, Lee KY and Son HJ:
Cytoplasmic HuR over-expression is associated withincreased
cyclooxygenase-2 expression in laryngeal squamous cell carcinomas.
Pathology. 39:545–550. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim SJ, Kim HJ, Kim JY, Park K and Lee CM:
Expression of HuR is associated with increased cyclooxygenase-2
expression in uterine cervical carcinoma. Int J Gynecol Pathol.
26:229–234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ido K, Nakagawa T, Sakuma T, Takeuchi H,
Sato K and Kubota T: Expression of vascular endothelialgrowth
factor-A and mRNA stability factor HuR in human astrocytic tumors.
Neuropathology. 28:604–611. 2008.PubMed/NCBI
|
|
24
|
Ortega AD, Sala S and Espinosa E:
Gonzalez-Baron M and Cuezva JM: HuR and the bioenergetics signature
of breast cancer: a low tumor expression of the RNA-binding protein
predicts a higher risk of disease recurrence. Carcinogenesis.
29:2053–2061. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Ellison DW and Figarella-Branger D: World Health Organization
Histological Classification of Tumours of the Central Nervous
System. International Agency for Research on Cancer; France:
2016
|
|
26
|
Louis DN and Ohgaki H; Wiestler O and
Cavenee W: World Health Organization Histological Classification of
Tumours of the Central Nervous System. International Agency for
Research on Cancer; Lyon: 2007
|
|
27
|
Song S, Fajol A, Tu X, Ren B and Shi S:
miR-204 suppresses the development and progression of human
glioblastoma by targeting ATF2. Oncotarget. 7:70058–70065. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arita H, Narita Y, Matsushita Y, Fukushima
S, Yoshida A, Takami H, Miyakita Y, Ohno M, Shibui S and Ichimura
K: Development of a robust and sensitive pyrosequencing assay for
the detection of IDH1/2 mutations in gliomas. Brain Tumor Pathol.
32:22–30. 2015. View Article : Google Scholar
|
|
29
|
Arita H, Narita Y, Fukushima S, Tateishi
K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP,
Kawahara N, et al: Upregulating mutations in the TERT promoter
commonly occur in adult malignant gliomas and are strongly
associated with total 1p19q loss. Acta Neuropathol. 126:267–276.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mulholland S, Pearson DM, Hamoudi RA,
Malley DS, Smith CM, Weaver JM, Jones DT, Kocialkowski S, Bäcklund
LM, Collins VP, et al: MGMT CpG island is invariably methylated in
adult astrocytic and oligodendroglial tumors with IDH1 or IDH2
mutations. Int J Cancer. 131:1104–1113. 2012. View Article : Google Scholar
|
|
31
|
Wang XP, Shan C, Deng XL, Li LY and Ma W:
Long non-coding RNA PAR5 inhibits the proliferation and progression
of glioma through interaction with EZH2. Oncol Rep. 38:3177–3186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang P, Chen D, Ma H and Li Y: LncRNA
SNHG12 contributes to multidrug resistance through activating the
MAPK/Slug pathway by sponging miR-181a in non-small cell lung
cancer. Oncotarget. 8:84086–84101. 2017.PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
34
|
Wang L, Ye S, Wang J, Gu Z, Zhang Y, Zhang
C and Ma X: HuR Stabilizes lnc-Sox5 mRNA to Promote Tongue
Carcinogenesis. Biochemistry (Mosc). 82:438–445. 2017. View Article : Google Scholar
|
|
35
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar
|
|
36
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b
suppressesed human liver cancer cell proliferation and metastasis
by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He Z, Huang C, Lin G and Ye Y:
siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits
the invasion of human lung cancer SPC-A1 cells. Oncol Rep.
35:1933–1940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen QN, Wei CC, Wang ZX and Sun M: Long
non-coding RNAs in anti-cancer drug resistance. Oncotarget.
8:1925–1936. 2017.
|
|
39
|
Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C,
Wang J, Tan Q, Cheng Y, Xia E, et al: C-MYC-induced upregulation of
lncRNA SNHG12 regulates cell proliferation, apoptosis and migration
in triple-negative breast cancer. Am J Transl Res. 9:533–545.
2017.PubMed/NCBI
|
|
40
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arita H, Yamasaki K, Matsushita Y,
Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M,
Shimizu S, et al: A combination of TERT promoter mutation and MGMT
methylation status predicts clinically relevant subgroups of newly
diagnosed glioblastomas. Acta Neuropathol Commun. 4:792016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Turkalp Z, Karamchandani J and Das S: IDH
mutation in glioma: New insights and promises for the future. JAMA
Neurol. 71:1319–1325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang YA, Ma X, Sathe A, Fujimoto J,
Wistuba I, Lam S, Yatabe Y, Wang YW, Stastny V, Gao B, et al:
Validation of SCT methylation as a hallmark biomarker for lung
cancers. J Thorac Oncol. 11:346–360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Malmström A, Grønberg BH, Marosi C, Stupp
R, Frappaz D, Schultz H, Abacioglu U, Tavelin B, Lhermitte B, Hegi
ME, et al Nordic Clinical Brain Tumour Study Group (NCBTSG):
Temozolomide versus standard 6-week radiotherapy versus
hypofractionated radiotherapy in patients older than 60 years with
glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol.
13:916–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wick W, Platten M, Meisner C, Felsberg J,
Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M,
et al NOA-08 Study Group of Neuro-oncology Working Group (NOA) of
German Cancer Society: Temozolomide chemotherapy alone versus
radiotherapy alone for malignant astrocytoma in the elderly: The
NOA-08 randomised, phase 3 trial. Lancet Oncol. 13:707–715. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bakheet T, Williams BR and Khabar KS: ARED
3.0: The large and diverse AU-rich transcriptome. Nucleic Acids
Res. 34:D111–D114. 2006. View Article : Google Scholar :
|
|
48
|
Katsanou V, Milatos S, Yiakouvaki A,
Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger
M and Kontoyiannis DL: The RNA-binding protein Elavl1/HuR is
essential for placental branching morphogenesis and embryonic
development. Mol Cell Biol. 29:2762–2776. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Katsanou V, Papadaki O, Milatos S,
Blackshear PJ, Anderson P, Kollias G and Kontoyiannis DL: HuR as a
negative posttranscriptional modulator in inflammation. Mol Cell.
19:777–789. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Streffer JR, Rimner A, Rieger J, Naumann
U, Rodemann HP and Weller M: BCL-2 family proteins modulate
radiosensitivity in human malignant glioma cells. J Neurooncol.
56:43–49. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bolognani F, Gallani AI, Sokol L, Baskin
DS and Meisner-Kober N: mRNA stability alterations mediated by HuR
are necessary to sustain the fast growth of glioma cells. J
Neurooncol. 106:531–542. 2012. View Article : Google Scholar
|