Introduction
Breast cancer is one of the leading causes of
cancer-associated mortalities worldwide and the most common cancer
among women (1). The majority of
patients are at advanced stages at the time of diagnosis, and the
prognosis of these patients remains poor (2). Currently, adjuvant chemotherapy and
radiotherapy following surgical resection is the most commonly used
treatment strategy, however, the 5-year survival rate of breast
cancer remains low, and the improvement of prognosis of breast
cancer patients has reached a plateau (3,4).
Therefore, finding promising therapeutic and prognostic targets is
essential for developing effective therapies for breast cancer
patients.
Drugs that target the mitotic spindle are among the
most effective cancer therapeutics currently in use. In addition,
there is hope that such drugs would not produce debilitating
neuropathies such as those caused by treatment of cancer with
taxanes (5). Monastrol, the
prototype anti-kinesin drug, is known to inhibit the mitotic motor
Eg5 (6-8), which is a member of a family of
kinesins crucial for maintaining separation of the half-spindles
(9,10). Treatment of dividing cells with
monastrol results in the collapse of the bipolar spindle into a
non-functional monastral spindle. However, the underlying
functional mechanism and interaction between Eg5 and monastrol is
not well known.
High-throughput RNA sequencing (RNA-Seq), an
emerging method to study the RNA regulation mechanism in the whole
genome, has been able to detect circular RNA (circRNAs) (11). Circular RNAs (circRNAs) are a large
class of endogenous RNAs that are formed by exon skipping or
back-splicing events with neither 5′ to 3′ polarity nor a
polyadenylated tail; however, they attracted little attention until
their function in post-transcriptional regulation of gene
expression was discovered. circRNAs are conserved and stable, and
numerous circRNAs seem to be specifically expressed in a cell type
or developmental stage (12,13).
The cell type- or developmental stage-specific expression patterns
indicate that circRNAs may be important regulators in various
diseases, including heart cerebrovascular disease, hematological
disease and malignant tumors (14).
circRNAs participate in several different biological
processes in cancer cells, including cell growth, metastasis, cell
cycle control, nuclear and cytoplasmic trafficking, cell
differentiation, RNA decay, transcription and translation (15). In the cytoplasm, circRNAs may
function as competing endogenous RNAs (ceRNAs), thus inducing the
suppression of genes that are targeted by specific miRNAs (16). Previously, it has been reported
that they can act as scaffolds to directly bind to specific
proteins and to hire gene-modifying bodies to silence or activate
targeted genes (17). To date,
verifying the deregulated circRNAs and further investigating the
underlying regulatory mechanism in breast cancer cells is still an
ongoing process.
The present study investigated monastrol-induced
circRNA regulation in breast cancer by performing a genome-wide
microarray in cells that were treated with monastrol. In addition,
it was identified that, circRNA MTO1 (hsa-circRNA-007874), inhibits
Eg5-mediated cell viability and promotes chemosensitivity through
sequestering TRAF4 from binding to Eg5 protein.
Materials and methods
Cell culture and reagents
The human breast cancer cell lines MDA-MB-231,
MCF-7, MDA-MB-453, SKBR-3, T47D and MDA-MB-468 were purchased from
Chinese Academy of Sciences (Shanghai, China). All breast cancer
cell lines were cultured with high glucose-Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at
37°C in a humidified incubator with 5% CO2.
Cycloheximide (CHX) was purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany) and used for treatment of breast cancer cells
for 80 min followed by western blot analysis to determine Eg5
protein level.
Development of monastrol-resistant breast
cancer cell lines
MCF-7-R and MDA-MB-231R cell lines were developed by
exposing parental MCF-7 and MDA-MB-231 parental cells to an initial
dose of 10 µM monastrol (Sigma-Aldrich; Merck KGaA) in DMEM
supplemented with 10% FBS for 6 weeks. Cells were cultured for
three passages to reach a confluence of 70%. The survival cells
were then cultured in 20 µM monas-trol for 8 weeks and 50
µM monastrol for a further 8 weeks to obtain the resistant
population. The monastrol-resistant cell lines were eventually
established by culturing the cells in monastrol at a concentration
of 100 µM. During the experiments, both monastrol-resistant
cell lines were cultured to no higher than 10 passages.
Expression profile analysis of
circRNAs
Total RNAs were extracted from breast cancer cells
and quantified using the NanoDrop ND-2000 (Nanodrop Technologies;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). circRNA
expression analysis was performed according to the Arraystar
protocol (18). Following
hybridization and washing of samples, six breast cancer cell lines
(MCF-7, MDA-MB-231, MDA-MB-468, MDA-MB-453, SKBR-3 and T47D) and
two monastrol resistant cell lines (MCF-7R and MDA-MB-231R) were
amplified and labeled using the RNeasy Mini kit (Qiagen GmbH,
Hilden, Germany). A hybridization solution was then prepared and
added to the circRNA expression microarray slide and incubated at
65°C for 16 h. Subsequently, the slides were scanned by the Axon
GenePix 4000B microarray scanner and imported into GenePix Pro 6.0
software (Molecular Devices, LLC, Sunnyvale, CA, USA). The
microarray analysis was conducted by Beijing Genomics
Institute/HuaDa-Shenzhen (Shenzhen, China).
RNA oligoribonucleotides and cell
transfection
The circRNA-MTO1 overexpression plasmid was
synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China) and
an empty vector used as a control. The Eg5 overexpression vector
was purchased from OriGene Technologies, Inc. (Rockville, MD, USA)
and an empty vector used as a control. The breast cancer cells were
plated at 5×104 cells/well in 24-well plates
approximately 24 h prior to transfection. After the cells reached
30–50% confluence, transfection was carried out using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) following the manufacturer's
protocol. The final concentration of circRNA-MTO1 overexpression
plasmid, Eg5 overexpression vector, and the respective empty
control vectors were 100 nM. Transfection efficiency was evaluated
by labeling vectors with green fluorescence protein (GFP) to ensure
that cells were successfully transfected. Functional experiments
were then performed 48 h following sufficient transfection.
Cell viability assay
The altered cell viability following transfection or
other treatments was assayed using a Cell Counting Kit (CCK)-8
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). In
brief, breast cancer cell lines were seeded into a 96-well plate in
triplicate and then treated with circ-MTO1 and (or) Eg5
overexpression vector for different time periods. Following this,
cell cultures were treated with the CCK8 reagent (10 ml) and
further cultured for 2 h. The optical density at a wavelength of
450 nm was measured with a spectrophotometer (Thermo Electron
Corporation, Waltham, MA, USA). The percentage of the control
samples of each cell line was calculated thereafter.
Nuclear fractionation
Nuclear fractionation was performed with a PARIS™
Kit (Ambion; Thermo Fisher Scientific, Inc.). For nuclear
fractionation, 1×107 cells were collected and
resuspended in the cell fraction buffer and incubated on ice for 10
min. Following centrifugation (4°C and 500 × g for 3 min), the
supernatant and nuclear pellet were preserved for RNA extraction
using a cell disruption buffer according to the manufacturer's
protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from breast cancer cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.,). RT-qPCR of individual RNAs was performed by
using a TaqMan RNA Reverse Transcription Kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and TaqMan Human RNA Assay kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. The comparative cycle
threshold (Cq) method was used to calculate the relative abundance
of RNA compared with GAPDH expression (19). The primer sequences used were as
follows: Forward, 5′-GGGTGTTTACGTAGACCAGAACC-3′, reverse,
5′-CTTCCAAAAGCCTTCTGCCTTAG-3′ for circRNA-MTO1; forward,
5′-GAACAATCATTAGCAGC AGAATRAF4-3′, reverse,
5′-TCAGTATAGACACCACAGTTG-3′ for Eg5 and forward,
5′-GCACCGTCAAGGCTGAGAAC-3′, reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′ for
GAPDH. Each experiment was performed in triplicate.
circRNAs immunoprecipitation
(circRIP)
Biotin-labeled circRNA-MTO1 probe
(5′-AAAGGAAGGATTACATGACATCTGACCCAAAACAACCCCACTGACA-3′-biotin) was
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and the
circRIP assay was performed as previously described with minor
modifications (20). circRIP was
conducted by using MCF-7 and MCF-7R cells with Magna RIP™
RNA-binding protein immunoprecipitation kit (EMD Millipore,
Billerica, MA, USA) according to the manufacturer's protocol. A
total of 1×107 cells were lysed in complete RNA lysis
buffer, then cell lysates were incubated with RIP
immunoprecipitation buffer containing magnetic beads conjugated
with human anti-TRAF4 antibody (EMD Millipore, cat. no. #MABC985)
or negative control IgG (EMD Millipore, cat. no. #PP64). Samples
were incubated with Proteinase K and then immunoprecipitated RNA
was isolated. Extracted RNAs were analyzed by RT-qPCR to identify
the interaction.
RNA pulldown and mass spectrometry
Cells were lysed and incubated with biotin-labeled
MTO1 (Sangon Biotech Co., Ltd.,) overnight. The proteins associated
with biotin-labeled RNA were then pulled down with Streptavidin
Magnetic Beads (Thermo Fisher Scientific, Inc.) after 1-h
incubation. The proteins were then washed and used for mass
spectrometry (MS) analysis. The MS analysis included a biotinylated
MTO1 pulldown group and streptavidin beads only pulldown groups as
negative controls. Briefly, both groups of proteins were eluted and
resolved by gel electrophoresis followed by staining with the
SilverQuest™ Silver Staining Kit (Thermo Fisher Scientific, Inc.).
Then, proteins were excised, de-stained, and digested prior to
analysis by using high-performance liquid chromatography with a
Thermo Electron LTQ OrbiTrap XL mass spectrometer at Central
Laboratory of Shanxi Province People's Hospital. Proteins in MTO1
pulldown group were filtered out with a spectral count less than
three, and a cutoff was set up for at least 3-fold peptide
enrichment of MTO1 group, as compared with beads only group.
The detailed information for mass spectrometry was
as follows: In-gel tryptic digests were fractionated by CapHPLC
using a Shimadzu Prominence HPLC system (Shimadzu Corp., Kyoto,
Japan) and were introduced directly into the LTQ-Orbitrap XL hybrid
MS (Thermo Fisher Scientific, Inc.) equipped with a dynamic
nanoelectrospray ion source (Proxeon, Odense, Denmark) and distal
coated silica emitters (30 µm i.d., 20 µm tip i.d;
New Objective, Woburn, MA, USA). Acidified samples were loaded onto
a 120 Å, 3 µm particle size, 300 µm by 10 mm C18-AQ
Reprosil-Pur trap column (SGE Australia Pty., Ltd., Clyde,
Australia) at 30 µl/min in 98% solvent A [0.1% (v/v) aqueous
formic acid] and 2% solvent B [80% (v/v) acetonitrile containing
0.1% (v/v) formic acid] for 3 min at 40°C, and were subsequently
gradient eluted onto a pre-equilibrated self-packed analytical
column (Dr Maisch GmbH Reprosil-Pur C18-AQ, 120 Å, 150 µm by
200 mm) using a flow rate of 900 nl/min. The LTQ-Orbitrap was
controlled using Xcalibur 2.0 SR1 (Thermo Fisher Scientific, Inc.).
Analyses were carried out in data-dependent acquisition mode,
whereby the survey full scan mass spectra (m/z 300-2000) were
acquired in the Orbitrap FT mass analyser at a resolution of 60,000
(at 400 m/z) after accumulating ions to an automatic gain control
target value of 5.0×105 charges in the LTQ mass
analyser. MS/MS mass spectra were concurrently acquired on the
eight most intense ions in the full scan mass spectra in the LTQ
mass analyser to an automatic gain control target value of
3.0×104 charges. Charge state filtering, where
unassigned precursor ions were not selected for fragmentation, and
dynamic exclusion (repeat count, 1; repeat duration, 30 sec;
exclusion list size, 500; and exclusion duration, 90 sec) were
used. Fragmentation conditions in the LTQ were: 35% normalised
collision energy, activation q of 0.25, isolation width of 3.0 Da,
30 ms activation time, and minimum ion selection intensity 500
counts. Maximum ion injection times were 500 ms for survey full
scans and 100 ms for MS/MS.
Fluorescence in situ hybridization
analysis (FISH)
Nuclear and cytosolic fraction separation was
performed using a PARIS kit (Thermo Fisher Scientific, Inc.), and
RNA FISH probes were designed and synthesized by Bogu according to
the manufacturer's protocol. Briefly, cells were fixed in 4%
formaldehyde for 15 min at room temperature and then washed with
PBS. The fixed cells were treated with pepsin and dehydrated
through ethanol. The air-dried cells were incubated further with 40
nM FISH probe in hybridization buffer. After hybridization, the
slide was washed, dehydrated and mounted with Prolong Gold Antifade
Reagent with DAPI for detection. The slides were visualized for
immunofluorescence with an Olympus fluorescence microscope with an
attached CCD camera (Olympus Corporation, Tokyo, Japan).
Immunohistochemistry (IHC)
IHC staining was performed on 4 µm-thick TMA
slides. Briefly, the slides were deparaffinized and antigen
retrieval was then performed in a steam cooker for 1.5 min in 1 mM
EDTA. Endogenous peroxidase was quenched by incubating the slides
in Peroxidazed I reagent (Biocare Medical, Pacheco, CA, USA) for 5
min and background staining was blocked by incubation in Background
Sniper reagent (Biocare Medical) for another 5 min at room
temperature. Rabbit anti-Eg5 polyclonal antibody (cat. no. ab61199;
Abcam, Cambridge, MA, USA) at 1:150 dilution was used for culture
overnight at 4°C. Universal secondary antibody (cat. no. E043201-6;
Goat Anti-Rabbit IgG, 1:5,000; Dako, Santa Clara, CA, USA) was
applied for 15 min at room temperature. Diaminobenzidine or
3-amino-9-ethylcarbazole was used as a chromogen and slides were
counterstained for 1 min at room temperature with hematoxylin
before mounting. The slides were visualized with an Olympus
microscope (Olympus Corporation; magnification, ×40). ROS1 IHC was
scored using a previously described scoring system (21).
Western blotting
Cell lysates were prepared with
radioimmunoprecipation buffer containing protease inhibitors
(Sigma-Aldrich; Merck KGaA). Protein concentrations were measured
with the BCA Protein Assay kit according to the manufacturer's
protocol (Beyotime Institute of Biotechnology, Shanghai, China).
Equal amounts of protein (25 µg) were separated by 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred onto polyvinylidene fluoride membranes (EMD Millipore).
Then, the membrane was blocked with 5% (5 g/100 ml) non-fat dry
milk in Tris-buffered saline plus Tween (TBS-T) buffer for 2 h at
room temperature. The membranes were incubated overnight at 4°C
with a 1:1,000 solution of antibodies: anti-Eg5 (Abcam; cat. no.
ab61199) and anti-β-actin (Abcam; cat. no. ab8227). The horseradish
peroxidase-conjugated anti-rabbit antibody (cat. no. #7074;
1:5,000; Cell Signaling Technology, Danvers, MA, USA) was used as a
secondary antibody for immunostaining for 1 h at room temperature.
Immunoblots were visualized using Immobilon™ Western
Chemiluminescent HRP Substrate (EMD Millipore). Densitometry was
performed by using ImageJ software version 1.51r (National
Institutes of Health, Bethesda, MD, USA) (22).
Cell invasion assay
A Transwell invasion assay was performed to evaluate
the invasive ability of breast cancer cells. Briefly, 100 µl
matrigel (Ambion; Thermo Fisher Scientific, Inc.) was firstly added
onto the bottom of the transwell chamber (24-well insert; 8-mm pore
size; Corning Costar Corp., Corning, NY, USA), then
1×105 cells in reduced serum medium (Opti-MEM; Gibco;
Thermo Fisher Scientific, Inc.) were placed on the coated membrane
in the chamber. DMEM supplemented with 10% FBS was placed in the
bottom wells as chemoattractant. After 24 h, cells that did not
migrate were removed from the top side of the inserts with a cotton
swab. Cells that migrated through the permeable membrane were fixed
in 4% paraformaldehyde for 15 min at room temperature, stained with
crystal violet for 5 min at room temperature, and counted under a
microscope (Olympus Corporation) at magnification, ×20 in 5 random
fields in each well.
Statistical analysis
The Mann-Whitney U test or Kruskal-Wallis test (post
hoc Mann-Whitney U test with Bonferroni's correction) as used for
evaluating the difference among different cell groups. The survival
curves of breast cancer cells were estimated via the Kaplan-Meier
method, and the difference in survival rate was analyzed using the
log-rank testing. All statistical analyses were performed with SPSS
software, version 17.0 (SPSS Inc., Chicago, IL, USA). The package
plots and function heatmap in R software (http://www.R-project.org/) were used for mapping.
Error bars in figures represent standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Acquisition of monastrol resistance
induces elevated cell viability in breast cancer cells
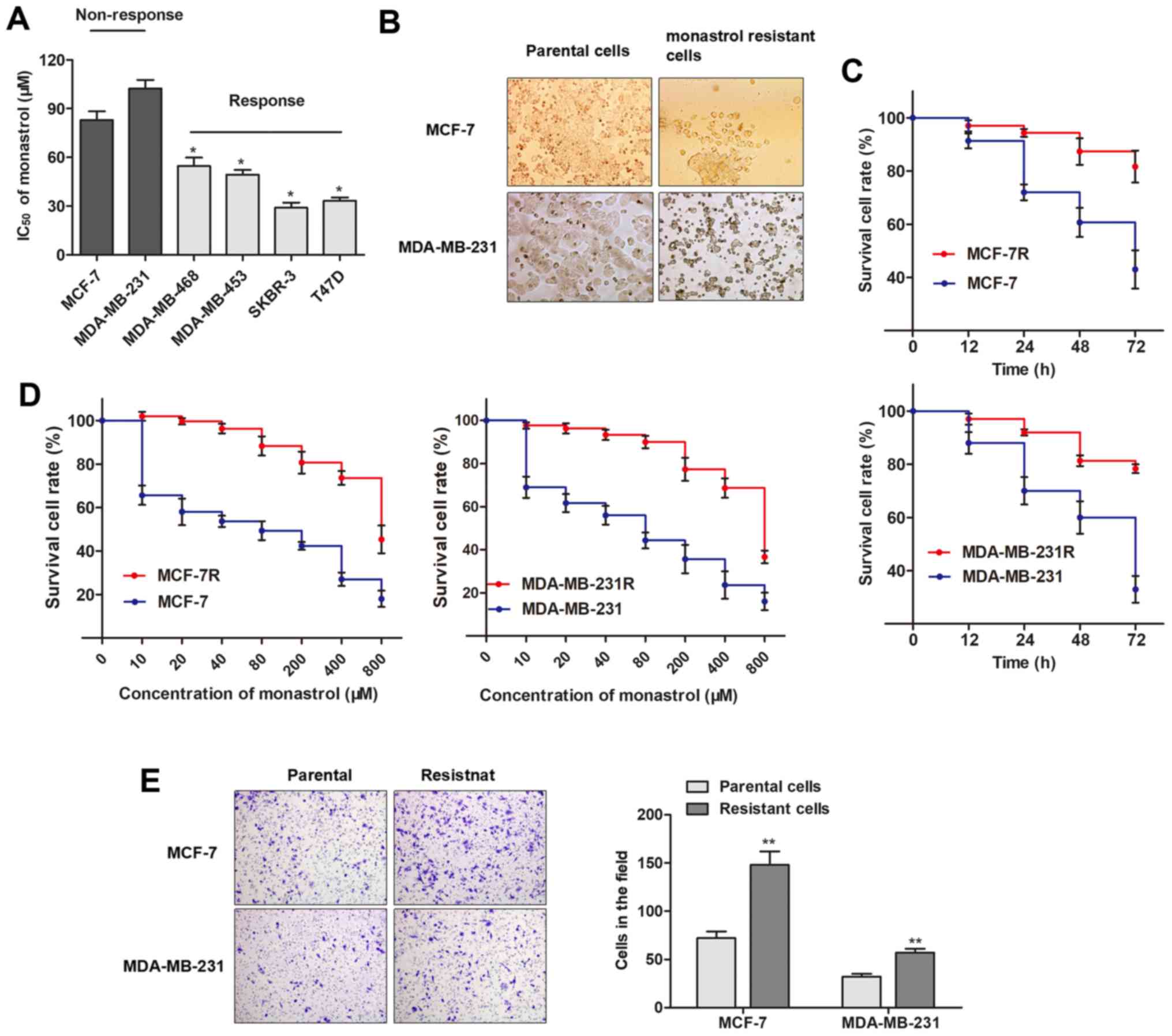
A panel of breast cancer cell lines was divided into
two groups according to their response to monastrol (Fig. 1A). The two cell lines that
exhibited little response to monastrol treatment, MCF-7 and
MDA-MB-231, were constantly exposed to a high concentration of
monas-trol (100 µM) to establish the monastrol-resistant
cell lines, MCF-7R and MDA-MB-231R. As presented in Fig. 1B, the establishment of monastrol
resistant cells induced specific morphological changes, including
loss of cell polarity, increased intercellular separation, and
increased formation of pseudopodia. Furthermore, MCF-7R and
MDA-MB-231R cells exhibited elevated cell viability compared with
the parental cells when incubated with culture medium containing
100 µM concentration of monastrol for 48 h (Fig. 1C). The concentration-effect curve
indicated that the IC50 of monastrol (48 h) on MCF-7R
was 726.3 µM, while the IC50 of monastrol on
MCF-7 was 81.5 µM, which indicated that monastrol resistance
for MCF-7R cells was 8.91 times higher compared with MCF-7 cells.
Similarly, the MDA-MB-231R was 8.49 times the ability of monastrol
resistance of MDA-MB-231 (607.9/71.6 µM, Fig. 1D). At 48 h, a significantly
increased number of chemo-resistant cells were observed to migrate
through the collagen membrane compared with parental cells
(Fig. 1E), indicating an increased
cell migratory ability. To conclude, the monastrol resistant cell
lines were successfully established.
Monastrol resistance induces
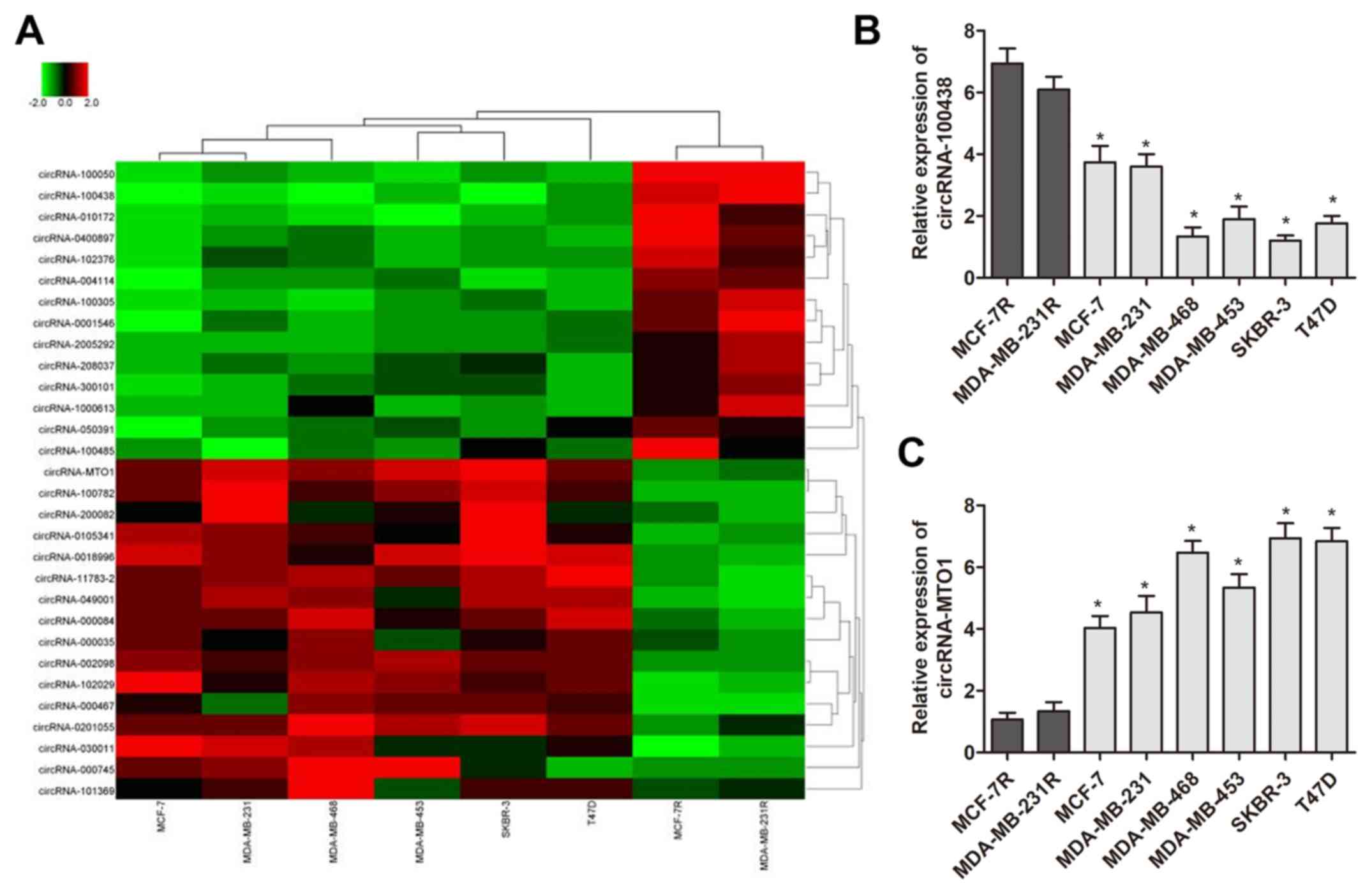
downregulation of circRNA-MTO1 in breast cancer cells
To identify specific circRNAs that have important
roles of monastrol resistance, circRNA microarray analysis was
performed using two monastrol resistant cell lines and six
non-resistant breast cancer cell lines. This assay identified 398
circRNAs which were deregulated between the two cell types. To
further identify the specific circRNAs, the results were narrowed
to 30 circRNAs (15 upregulated and 15 downregulated) which
exhibited the most altered expression levels (presented in heat
map, Fig. 2A). RT-qPCR was then
performed to verify the expression pattern of these 30 circRNAs in
monastrol resistant and parental cells. As presented in Fig. 2B and C, two circRNAs were
identified with a consistent expression gap between monastrol
resistant cells and parental cells as compared with microarray data
(P<0.05), including circRNA-MTO1 (hsa-circRNA-007874) and
circRNA-100438. In addition, the preliminary results demonstrated
that silencing of circRNA-100438 had no significant effect on
breast cancer cell viability (data not shown), while silencing of
circRNA-MTO1 significantly increased cell viability. Therefore, the
high percentage of circRNA-MTO1 decrease in monastrol-resistant
cells led to further investigation of the functional role of
circRNA-MTO1 in breast cancer resistance.
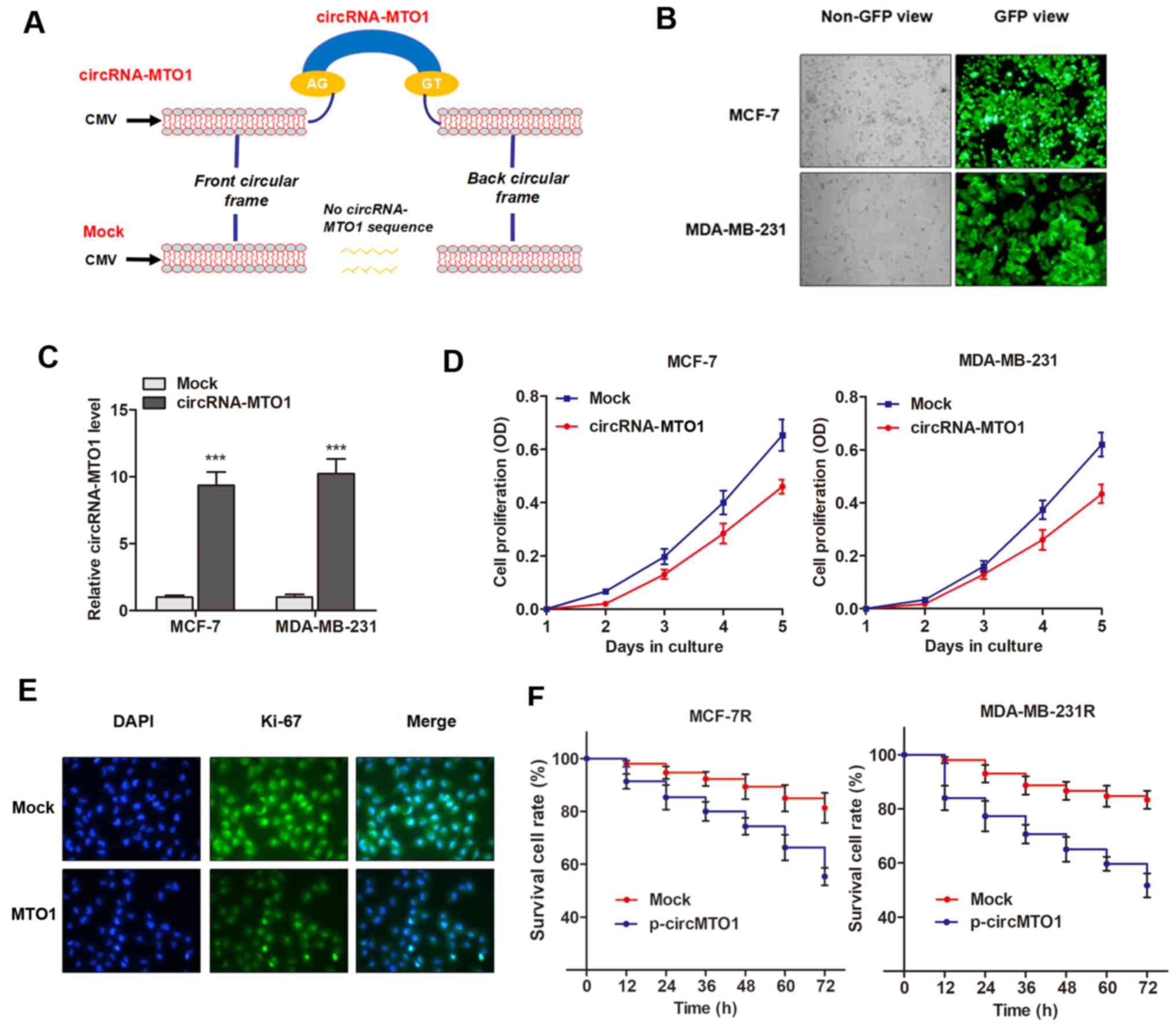
Overexpression of circRNA-MTO1 inhibits
cell viability and reverses monastrol resistance
The present study investigated the functional role
of MTO1 in cell viability and chemoresistance. To determine whether
the upregulation of MTO1 could inhibit cell viability and reverse
monastrol resistance of breast cancer cells, the specific plasmid
of circRNA-MTO1 was designed from MCF-7 cells and cloned into the
specific vector (Fig. 3A), then
MTO1 was upregulated via the transfection of pcDNA3.1-MTO1 vectors
(Fig. 3B and C). CCK-8 assay
revealed that enhanced MTO1 expression inhibited the viability of
breast cancer cells compared with control (Fig. 3D). This data was further validated
by the detection of Ki-67 expression, a known biomarker of cell
viability, in MCF-7 cells (Fig.
3E). However, MTO1 had little effect on cell apoptosis (data
not shown), indicating that MTO1 may regulate monastrol resistance
by influencing cell viability. Notably, monastrol resistant cells
with enhanced expression of MTO1 were more sensitive to the
treatment of monastrol at the concentration of 100 nM (Fig. 3F), indicating that MTO1 partially
reversed monastrol resistance.
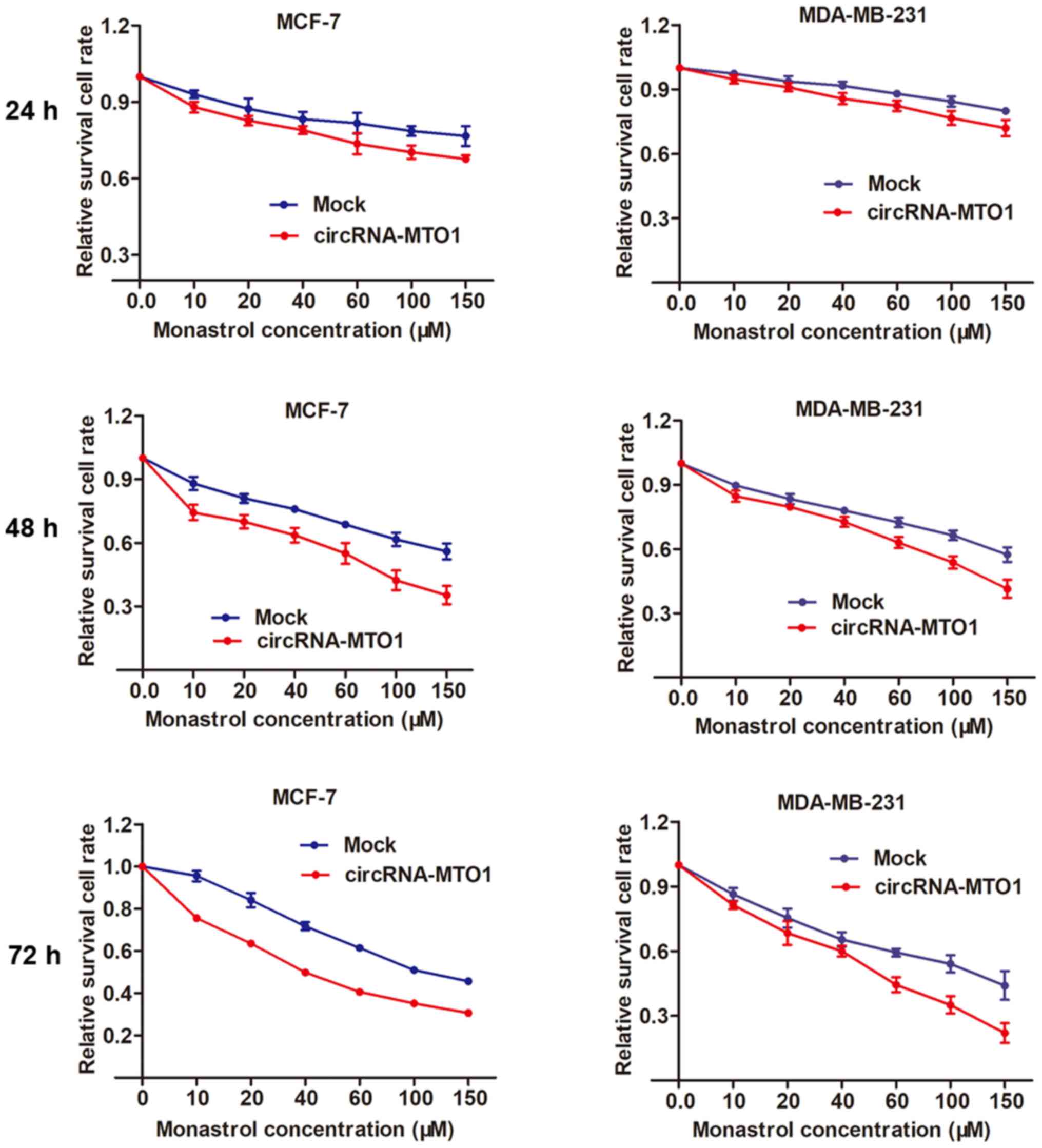
circRNA-MTO1 promotes monastrol-induced
cytotoxicity in breast cancer cells
By using the MCF-7 and MDA-MB-231 parental cells,
the present study then investigated whether MTO1 enhances monastrol
cytotoxicity. After being transfected with MTO1 overexpression
vector or negative control, MCF-7 and MDA-MB-231 cells were
incubated without monastrol or with a concentration gradient of 10,
20, 40, 60, 100, 150 µM monastrol for 24, 48 and 72 h. Then,
a dose-effect curve was constructed based on the viability of
cells. The graph showed that enhanced expression of MTO1 was
followed by increased cell death compared with the negative control
in monastrol treated cells at different concentrations (Fig. 4).
circRNA-MTO1 suppresses cell viability
and reverses monastrol resistance through targeting Eg5
protein
It is well known that monastrol exerts the
tumor-suppressive function mainly through blockage of mitotic
kinesin Eg5, which is required for the formation of a bipolar
spindle (8). Inhibition of Eg5 has
gained primary attention as an alternative strategy to interfere
with spindle function. As the synergistic effect of circRNA-MTO1
with monastrol treatment was verified, the present study then
investigated whether circRNA-MTO1 targets Eg5 protein, thereby
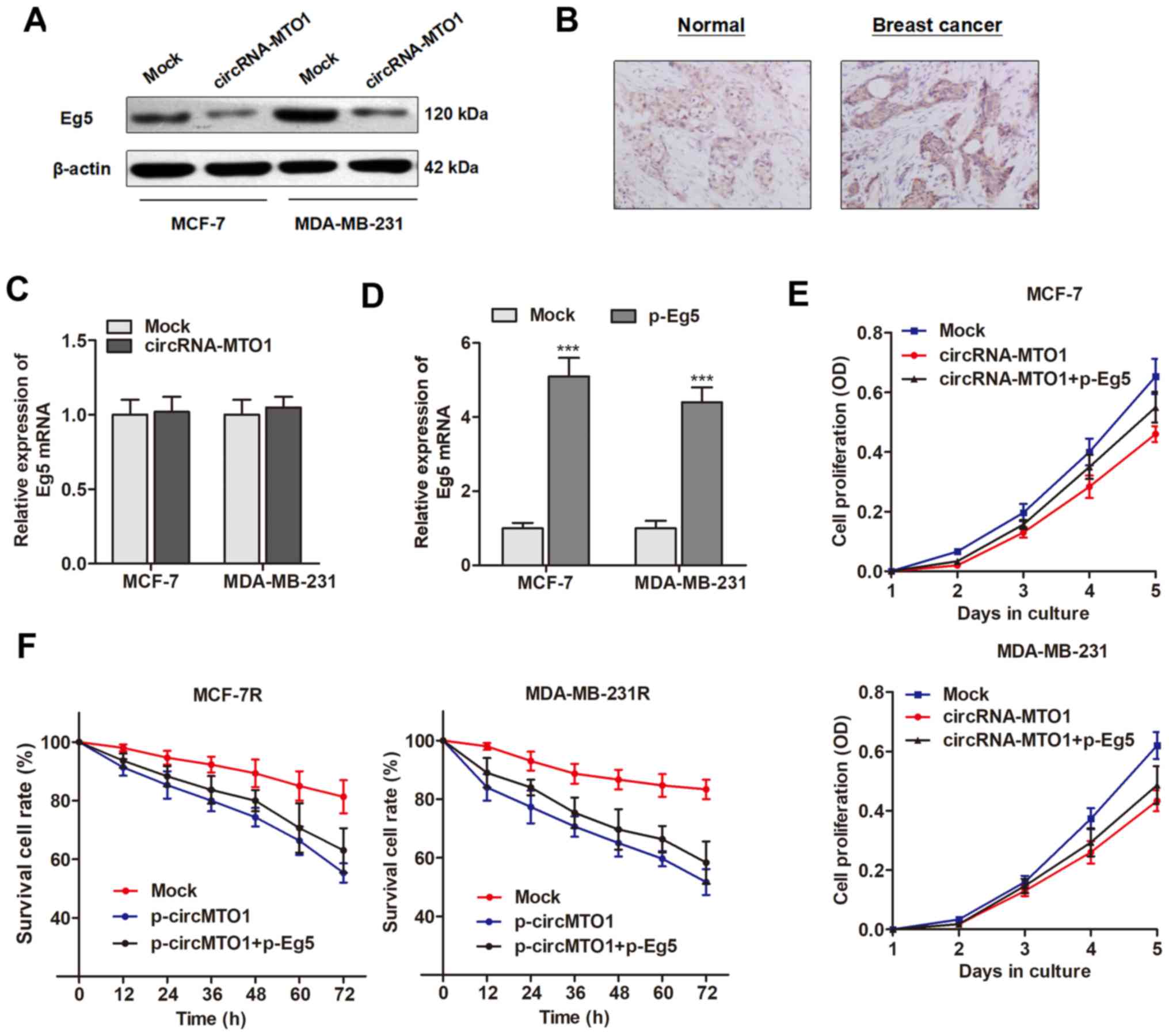
inducing a tumor suppressive effect. Western blotting revealed that
Eg5 protein was inhibited by overexpression of MTO1 in the two
breast cancer cell lines (Fig.
5A). Notably, MTO1 did not affect the Eg5 mRNA level,
suggesting that MTO1 may regulate Eg5 at a post transcriptional
level (Fig. 5B).
Next, it was determined whether Eg5 plays a causal
role during the MTO1-regulated cell viability and monastrol
resistance. Eg5 was upregulated by transfection of specific
Eg5-containing plasmid (Fig. 5C).
Enhanced expression of Eg5 significantly abrogated the decreased
cell growth and reversed the chemo response to monastrol induced by
MTO1 overexpression in MCF-7R and MDA-MB-231R cells (Fig. 5D and E). Collectively, the results
suggested that MTO1 regulates cell viability and monastrol
resistance through inhibiting Eg5 levels at the
post-transcriptional level.
circRNA-MTO1 sequesters TRAF4 from
binding to Eg5 gene
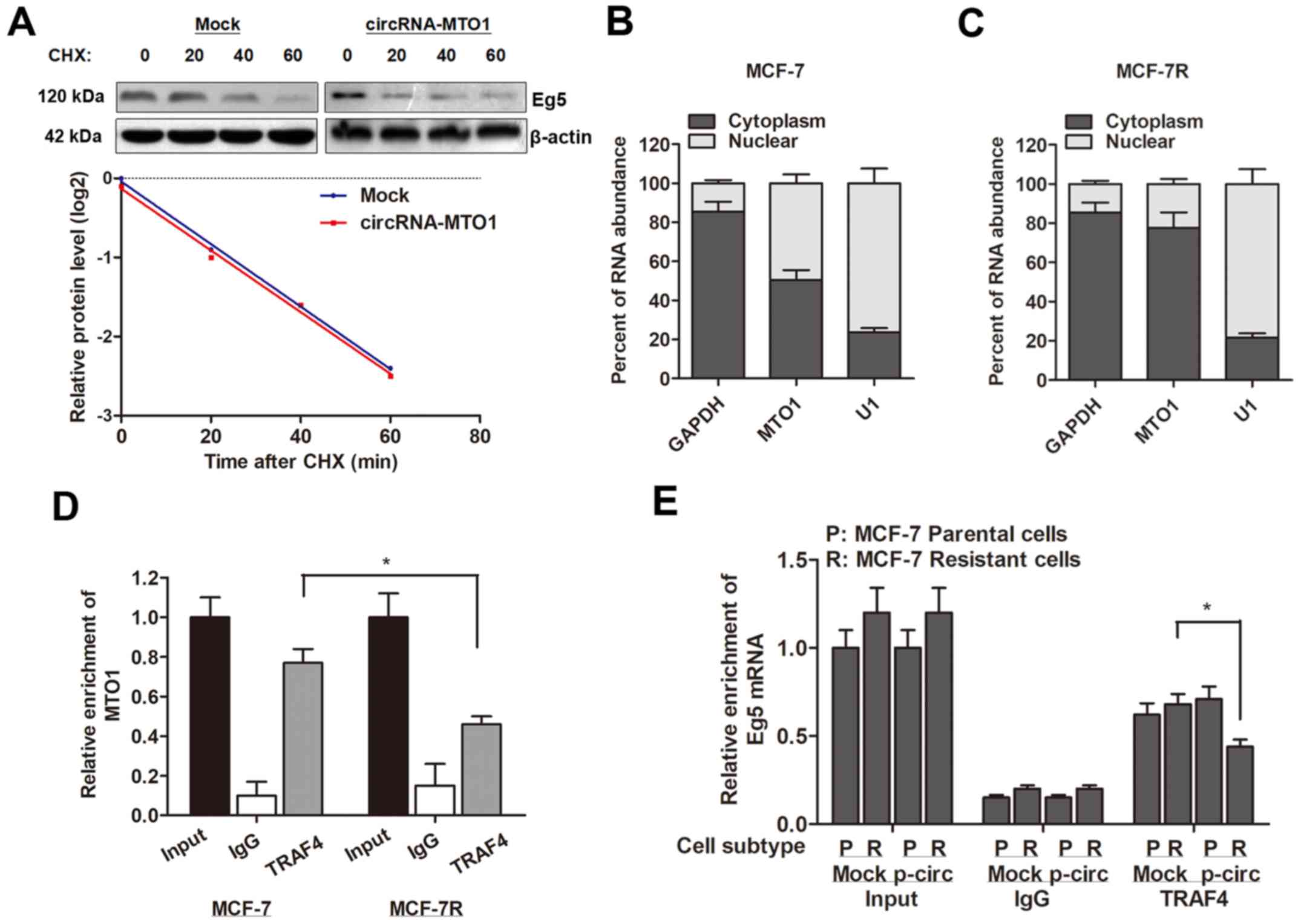
Furthermore, the present study investigated how
circRNA-MTO1 targets and inhibits the expression of Eg5 gene. By
treating with cycloheximide (CHX), a protein synthesis inhibitor,
it was revealed that MTO1 did not affect the protein stability of
Eg5 (Fig. 6A), indicating that
MTO1 may affect the protein level of Eg5 at post-transcriptional
level. Subsequently, RNA-pull down experiments were performed
followed by mass spectrometry to search for the MTO1-associated
proteins. As presented in Table I,
a list of correlative MTO1-associated proteins were identified.
Then, these potential regulator genes were evaluated to identify
the specific protein that may activate Eg5 at the
post-transcriptional level. The assays validated TRAF4 as a
potential interacting protein of MTO1. It has previously been
reported that TRAF4 is an (A+U)-rich elements (AREs)-binding
protein, and can interact with ARE areas within the 3′ UTR of
target gene and activates translation without influencing the mRNA
level (23). The present study
then localized the expression pattern of MTO1 and observed that
MTO1 was expressed in both nucleus and cytoplasm of MCF-7 parental
cells (Fig. 6B), however,
monastrol resistance significantly increased the proportion of MTO1
localization in the nucleus (Fig.
6C). In addition, the RIP assay revealed an interaction between
MTO1 and TRAF4 protein in MCF-7 cells; however, the enrichment
between MTO1 and TRAF4 protein was suppressed in MCF-7R cells
(Fig. 6D). MTO1 overexpression
decreased the interaction between TRAF4 and Eg5 gene in MCF-7R
cells (Fig. 6E). Taken together,
these data indicated that MTO1 serves as a competing endogenous RNA
(ceRNA) binding to TRAF4, and therefore inhibits Eg5 protein level
and reverses monastrol resistance.
 | Table IIdentification of circRNA-MTO1
binding proteins by mass spectrometry. |
Table I
Identification of circRNA-MTO1
binding proteins by mass spectrometry.
| Protein | Beads | MTO1 | Ratio
(MTO1/Beads) |
|---|
| TRAF4 | 0 | 3 | NA |
| U2AF1 | 1 | 3 | 3 |
| NKRF | 0 | 3 | NA |
| EF1D | 0 | 3 | NA |
| AIMP2 | 0 | 3 | NA |
| ROA0 | 0 | 3 | NA |
| RO60 | 0 | 3 | NA |
| ARP2 | 0 | 3 | NA |
| STT3B | 0 | 3 | NA |
| PCH2 | 1 | 3 | 3 |
| MRP1 | 0 | 3 | NA |
| LAS1L | 0 | 3 | NA |
| ARF6 | 1 | 3 | 3 |
| PLST | 0 | 3 | NA |
| PSAL | 0 | 3 | NA |
| TTL12 | 0 | 3 | NA |
| ERLN1 | 0 | 3 | NA |
| NSF | 0 | 3 | NA |
| AKAP8 | 0 | 3 | NA |
| GSTO1 | 0 | 3 | NA |
| AP1B1 | 0 | 3 | NA |
| DPM1 | 0 | 3 | NA |
| PSDE | 1 | 3 | 3 |
| KTN1 | 1 | 3 | 3 |
Discussion
Extensive efforts in the past have contributed to
the understanding of both molecular and cellular mechanisms of
action of chemoresistance, one of the major causes for the failure
of treatment in advanced cancer types. However, little progress has
been regarding this issue (24).
Thus, novel molecular signatures seem to hold great promise in
tumor characterization and could be used as potential prognostic
markers and treatment targets. The present study established
monastrol resistant cell lines, and sought to find potential
interactions between monastrol resistance and a novel group of gene
regulators, circRNAs. First, the downregulation of circRNA-MTO1 in
monastrol resistant cells was identified by utilizing microarray
analysis. Then, in vitro investigations suggested that
circRNA-MTO1 inhibited cell viability, and reversed monastrol
resistance of breast cancer cells by inhibiting Eg5 protein via
TRAF4.
Monastrol is a reversible, cell-permeable, small
molecule that selectively inhibits the plus-end-directed Kinesin-5
family member, Eg5 (25,26), a microtubule-based motor protein
that is required for the formation and maintenance of the bipolar
spindle (27). Monastrol treatment
of dividing cells results in spindle collapse and cell cycle arrest
with a monastrol spindle, which is similar to the phenotype
observed when Eg5 is inhibited by anti-Eg5 antibodies. The
anticancer effect of monastrol in breast cancer patients has been
frequently reported, and a chemoresistance has also been revealed
(28). The existence of circular
form RNAs in body fluid was firstly reported by Sanger et al
in 1976 (29), demonstrating that
this type of single-stranded closed circular RNA is stably
expressed from viroid to certain higher species, such as human
beings. With the development of gene investigations, it is
recognized that circRNAs are widely expressed in human cells, and
their expression levels can be 10-fold or higher compared to their
linear isomers. The two most important properties of circRNAs are
highly conserved sequences and a high degree of stability in
mammalian cells (30). Compared
with other noncoding RNA, such as microRNA (miRNA) and long
noncoding RNA (lncRNA), these properties provide circRNAs with the
potential to become ideal biomarkers in the diagnosis and prognosis
of cancers (31,32).
To date, only a few circRNAs have been explored. The
present study identified a circular RNA termed circRNA-MTO1 that
was significantly downregulated in monastrol resistant cells and
was associated with chemosensitivity. MTO1, a gene conserved in all
eukaryotes, encodes one of the two subunits of the enzyme that
catalyzes the 5-carboxymethylamino-methylation (mnm5s2U34) of the
wobble uridine base in the mitochondrial tRNA specific to Gln, Glu,
Lys, Leu (UUR), and possibly Trp (33,34).
The biological role of its circular form, circRNA-MTO1 was reported
only once. Han et al (20)
demonstrated that circRNA-MTO1 may serve as a competing endogenous
RNA (ceRNA) through binding to miR-9, thereby inducing the
suppression of hepatocellular progression. The study identified
circRNA-MTO1 as one circRNA significantly downregulated in HCC
tissues, and HCC patients with low circRNA-MTO1 expression had
shortened survival. In addition, circRNA-MTO1 suppresses HCC
progression by acting as the sponge of oncogenic miR-9 to promote
downstream p21 gene expression levels. The results of the present
study indicated that MTO1 may also serve as a tumor suppressor in
breast cancer. However, another identified circRNA, circRNA-100438,
demonstrated no effect on monastrol resistance, and whether it
participates in the oncogenic process remains unknown. Thus, the
functional role of circRNA-100438 requires further investigation in
future studies to verify whether it regulates progression of breast
cancer as well as other cancer types.
The functional role of MTO1 in breast cancer
progression and chemoresistance was then evaluated. To
comprehensively investigate the effect of MTO1 in chemoresistance,
the present study established two monastrol resistant cell lines,
MCF-7R and MDA-MB-231R. Consistent with the study by Han et
al, it was validated that MTO1 plays a tumor-suppressor role,
including the inhibition of cell growth, and the promotion of
monastrol chemosensitivity. Furthermore, the present study
identified the mechanism by which MTO1 serves a tumor-suppressive
role. Eg5 protein is well recognized as a direct target of
monastrol and MTO1 has a synergistic function with monastrol,
therefore whether MTO1 reverses monastrol resistance through
targeting Eg5 protein was investigated. The expression of Eg5 is
closely associated with cell viability and cancer. Overexpression
of Eg5 has been observed in bladder cancer (35) and pancreatic cancer (36). Furthermore, transgenic mice
overexpressing Eg5 are prone to developing a variety of tumors
(37). These and other
observations favor Eg5 as an attractive target for chemotherapy. In
addition, the in vitro experiments verified that MTO1
inhibited Eg5 protein level, however, MTO1 had little influence on
the mRNA level of Eg5 gene. In addition, the data obtained
following treatment of CHX indicated that MTO1 did not affect the
Eg5 protein stability.
To further explore how MTO1 regulates the protein
level of Eg5, an RNA pulldown assay and mass spectrometry were
performed, and it was observed that MTO1 physically interacted with
the TRAF4 gene. Furthermore, the proportion of MTO1 localized in
the cytoplasm was enhanced in cells resistant to monastrol
treatment. In addition, monastrol resistance elevated the physical
interaction between MTO1 and the TRAF4 gene. As one of the
important members of the TRAF family of proteins, TRAF4 was
initially isolated from breast carcinomas and identified as the
first member of the TRAFfamily to be upregulated in human
carcinomas (38). TRAF family
members mainly function in the immune system, where they mediate
signaling via tumor necrosis factor receptors and
interleukin-1/Toll-like receptors (39,40).
TRAF4 has been considered as an oncogene as it is overexpressed in
a wide range of human malignancies, including breast cancer, lung
cancer, colon adenocarcinomas, melanomas, neurogenic tumors and
lymphomas (41). Accumulating
evidence indicates that TRAF4 plays a critical role in breast
cancer, such as antiapoptosis, and promotes cell migration
(42). The data revealed a circRNA
network that that revealed that under conditions of monastrol
resistance, circRNA-MTO1 interacts with TRAF4, and may serve as a
ceRNA to repress TRAF4 from binding to the Eg5 gene, leading to
suppression of Eg5 protein levels, prevention of cell viability and
reversal of monastrol resistance.
In conclusion, the results of the present study
indicated a circRNA-involved regulatory pattern to mediate Eg5
protein expression during monastrol resistance. First, monastrol
resistance induced downregulated circRNA-MTO1 expression in breast
cancer cells. Second, overexpression of MTO1 repressed viability
and reversed monastrol resistance through inhibiting Eg5 protein
level and associating with TRAF4. Therefore, circRNA-MTO1 may be a
functional regulatory factor of breast cancer, and restoration of
MTO1 levels could be a future direction to overcome breast cancer
chemoresistance.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Basic
Research Projects of Shandong Province (grant no.
201601D202100).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YL and YD designed this work and created a draft of
the manuscript; YL, LZ and LS prepared the reagents and kits and
performed the in vitro experiments; YL and JL analyzed and
interpreted the original results, and performed statistical
analysis; YL and YD revised and approved the final version of the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of Shanxi Province People's Hospital (Taiyuan,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang B, Beeghly-Fadiel A, Long J and
Zheng W: Genetic variants associated with breast-cancer risk:
Comprehensive research synopsis, meta-analysis, and epidemiological
evidence. Lancet Oncol. 12:477–488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calaf GM, Zepeda AB, Castillo RL, Figueroa
CA, Arias C, Figueroa E and Farías JG: Molecular aspects of breast
cancer resistance to drugs (review). Int J Oncol. 47:437–445. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Zeng H and Zhang S: The
updated incidences and mortalities of major cancers in China, 2011.
Chin J Cancer. 34:502–507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quasthoff S and Hartung HP:
Chemotherapy-induced peripheral neuropathy. J Neurol. 249:9–17.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mayer TU, Kapoor TM, Haggarty SJ, King RW,
Schreiber SL and Mitchison TJ: Small molecule inhibitor of mitotic
spindle bipolarity identified in a phenotype-based screen. Science.
286:971–974. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapoor TM, Mayer TU, Coughlin ML and
Mitchison TJ: Probing spindle assembly mechanisms with monastrol, a
small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol.
150:975–988. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maliga Z, Kapoor TM and Mitchison TJ:
Evidence that monastrol is an allosteric inhibitor of the mitotic
kinesin Eg5. Chem Biol. 9:989–996. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blangy A, Lane HA, d'Hérin P, Harper M,
Kress M and Nigg EA: Phosphorylation by p34cdc2 regulates spindle
association of human Eg5, a kinesin-related motor essential for
bipolar spindle formation in vivo. Cell. 83:1159–1169. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharp DJ, Yu KR, Sisson JC, Sullivan W and
Scholey JM: Antagonistic microtubule-sliding motors position
mitotic centrosomes in Drosophila early embryos. Nat Cell Biol.
1:51–54. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sorek R and Cossart P: Prokaryotic
transcriptomics: A new view on regulation, physiology and
pathogenicity. Nat Rev Genet. 11:9–16. 2010. View Article : Google Scholar
|
|
12
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
14
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42D:D92–D97. 2014. View Article : Google Scholar
|
|
15
|
Huang G, Li S, Yang N, Zou Y, Zheng D and
Xiao T: Recent progress in circular RNAs in human cancers. Cancer
Lett. 404:8–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu SY, Huang X and Cheong KL: Recent
advances in marine algae polysaccharides: Isolation, structure, and
activities. Mar Drugs. 15:152017. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Remmele W and Stegner HE: Recommendation
for uniform definition of an immunoreactive score (IRS) for
immunohistochemical estrogen receptor detection (ER-ICA) in breast
cancer tissue. Pathologe. 8:138–140. 1987.PubMed/NCBI
|
|
22
|
Schindelin J, Rueden CT, Hiner MC and
Eliceiri KW: The ImageJ ecosystem: An open platform for biomedical
image analysis. Mol Reprod Dev. 82:518–529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Zhou F, García de Vinuesa A, de
Kruijf EM, Mesker WE, Hui L, Drabsch Y, Li Y, Bauer A, Rousseau A,
et al: TRAF4 promotes TGF-β receptor signaling and drives breast
cancer metastasis. Mol Cell. 51:559–572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J, et al: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
DeBonis S, Simorre JP, Crevel I, Lebeau L,
Skoufias DA, Blangy A, Ebel C, Gans P, Cross R, Hackney DD, et al:
Interaction of the mitotic inhibitor monastrol with human kinesin
Eg5. Biochemistry. 42:338–349. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan Y, Sardana V, Xu B, Homnick C,
Halczenko W, Buser CA, Schaber M, Hartman GD, Huber HE and Kuo LC:
Inhibition of a mitotic motor protein: Where, how, and
conformational consequences. J Mol Biol. 335:547–554. 2004.
View Article : Google Scholar
|
|
27
|
Sawin KE, LeGuellec K, Philippe M and
Mitchison TJ: Mitotic spindle organization by a plus-end-directed
microtubule motor. Nature. 359:540–543. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sashidhara KV, Avula SR, Sharma K, Palnati
GR and Bathula SR: Discovery of coumarin-monastrol hybrid as
potential antibreast tumor-specific agent. Eur J Med Chem.
60:120–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang YS, Jie N, Zou KJ and Weng Y:
Expression profile of circular RNAs in human gastric cancer
tissues. Mol Med Rep. 16:2469–2476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Suzuki T, Nagao A and Suzuki T: Human
mitochondrial tRNAs: Biogenesis, function, structural aspects, and
diseases. Annu Rev Genet. 45:299–329. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang X, Yan Q and Guan MX: Combination of
the loss of cmnm5U34 with the lack of s2U34 modifications of
tRNALys, tRNAGlu, and tRNAGln altered mitochondrial biogenesis and
respiration. J Mol Biol. 395:1038–1048. 2010. View Article : Google Scholar
|
|
35
|
Ding S, Xing N, Lu J, Zhang H, Nishizawa
K, Liu S, Yuan X, Qin Y, Liu Y, Ogawa O, et al: Overexpression of
Eg5 predicts unfavorable prognosis in non-muscle invasive bladder
urothelial carcinoma. Int J Urol. 18:432–438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu M, Wang X, Yang Y, Li D, Ren H, Zhu Q,
Chen Q, Han S, Hao J and Zhou J: Ectopic expression of the
microtubule-dependent motor protein Eg5 promotes pancreatic
tumourigenesis. J Pathol. 221:221–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Wu X, Du M, Chen X, Ning X, Chen
H, Wang S, Liu J, Liu Z, Li R, et al: Eg5 inhibitor YL001 induces
mitotic arrest and inhibits tumor proliferation. Oncotarget.
8:42510–42524. 2017.PubMed/NCBI
|
|
38
|
Zhang X, Wen Z, Sun L, Wang J, Song M,
Wang E and Mi X: TRAF2 regulates the cytoplasmic/nuclear
distribution of TRAF4 and its biological function in breast cancer
cells. Biochem Biophys Res Commun. 436:344–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chung JY, Park YC, Ye H and Wu H: All
TRAFs are not created equal: Common and distinct molecular
mechanisms of TRAF-mediated signal transduction. J Cell Sci.
115:679–688. 2002.PubMed/NCBI
|
|
40
|
Kedinger V and Rio MC: TRAF4, the unique
family member. Adv Exp Med Biol. 597:60–71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Camilleri-Broët S, Cremer I, Marmey B,
Comperat E, Viguié F, Audouin J, Rio MC, Fridman WH, Sautès-Fridman
C and Régnier CH: TRAF4 overexpression is a common characteristic
of human carcinomas. Oncogene. 26:142–147. 2007. View Article : Google Scholar
|
|
42
|
Zhang X, Wen Z and Mi X: Expression and
anti-apoptotic function of TRAF4 in human breast cancer MCF-7
cells. Oncol Lett. 7:411–414. 2014. View Article : Google Scholar : PubMed/NCBI
|