Introduction
Colorectal cancer (CRC) is a very common malignancy
worldwide and is the third leading cause of cancer-associated
mortality (1). The survival rate
of patients with CRC remains unsatisfactory, despite effective
therapeutic strategies including surgical resection and
chemo-radiotherapy. 5-Fluorouracil (5-FU), antimetabolite
medication, is a fluorinated analog active against a wide range of
solid tumors, including pancreatic cancer (2–4),
biliary tract cancer (5), breast
cancer (6,7) and hepatocellular cancer (8,9).
5-FU-based therapy is one of the treatment regimens as the adjuvant
or palliative chemotherapy for patients with CRC (10,11).
5-FU acts through various mechanisms, but principally as a
thymidylate synthase inhibitor, eliciting cytotoxicity by
interrupting the function of nucleotide synthetic enzyme
thymidylate synthase. Additionally, it incorporates
fluoronucleotides into RNA and DNA (12,13).
A subset of patients with CRC is primarily refractory or have
acquired chemoresistance to 5-FU, which has poses a major challenge
for clinical practice to improve therapeutic efficiency. Thus,
investigation of the mechanisms and discovery of potential
treatments targets for patients with CRC with 5-FU resistance is
urgently required.
Zinc finger and BTB domain containing 7A (ZBTB7), is
part of the POZ and Krüppel/zinc finger and BTB (POK) protein
family (14). It is involved in
pleiotropic functions and cellular differentiation. ZBTB7
physically interacts with other POK family members, such as B-cell
lymphoma 6 proteins (15). ZBTB7,
considered to be an oncogene, is overexpressed in a variety of
cancer types, and involved in the pathogenesis of cancer (16), including lung cancer (17), prostate cancer (18), liver cancer (19) and oral cancer (20). ZBTB7 drives tumorigenesis as a
master regulator and is required for the regulation of other tumor
suppressor genes, such as p53 (21). ZBTB7 has also been documented to be
an important regulator of CRC initiation and progression (22). ZBTB7 knockdown could inhibit cell
proliferation, and induce cell cycle arrest and promote apoptosis
independently of p14-E3 ubiquitin-protein ligase MDM2-p53 pathway
(23).
Currently, there limited knowledge of the role of
ZBTB7 in CRC, and the specific mechanisms the mediate the oncogenic
role of ZBTB7 in CRCs is unknown. To the best of our knowledge, no
previous studied have investigated the association between of ZBTB7
expression in CRC and 5-FU resistance. In the present study, it was
demonstrated that ZBTB7 knockdown sensitized the 5-FU response to
in CRC, potentially via the nuclear factor (NF)-κB signaling
pathway.
Materials and methods
Microarray data analysis
The genetic profiles of patients with CRC were
acquired from the Gene Expression Omnibus (GEO; ncbi.nlm.nih.gov/geo/) data repository (GSE39582,
GSE36133, GSE17538, GSE31595, GSE33113, GSE37892, GSE38832 and
GSE39084). The method used for quality control and raw data
processing was previously described (24).
Differences in ZBTB7 expression among multiple
cancer types were assessed using data also from GEO database
(GSE39582). The subtypes with high or low expression levels of
ZBTB7 mRNA were defined as: Low, 4.3352-6.1425; and high,
6.1433-7.1295. Cell line data were from GEO (GSE36133). Two
subtypes of cell lines were defined (C1 and C2) according to the
genetic signature, including epithelial-mesenchymal transition
(EMT)-associated genetic expression, NF-κB signaling pathway, ABCG
family and apoptosis-associated genes.
Unsupervised subtype identification
Unsupervised clustering was performed using the
Brunet algorithm from the R package non-negative matrix
factorization (NMF), based on most variant probe sets (n=1,000) of
chemotherapy resistance associated pathway genes in Kyoto
Encyclopedia of Genes and Genomes (KEGG; genome.jp/kegg/) in 53
CRCs cell lines. After determining the optimal number of subtypes
corresponding to high cophenetic and dispersion coefficients, the
final subtype assignment was regenerated for this number of
subtypes, using 200 runs.
Gene set enrichment analysis
Gene set enrichment analysis was performed using
java GSEA Desktop Application (Broad Institute, Cambridge, MA, USA)
with the hallmark gene sets (n=50) and KEGG gene sets (n=186)
implemented in Molecular Signatures Database (v 5.1), expression
data and phenotype data were formatted following user guide,
samples were permutated with NMF clustering subtype or ZBTB7
expression level for 1,000 times.
Cell lines and reagents
Human SW620 (CCL-227), SW480 (CCL-228), LoVo
(CCL-229), HCT-116 (CCL-247), and HT-29 (HTB-38) CRC cell lines
were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). FHC
colon epithelial cell was cultured in DMEM:F12 medium supplemented
with 10% fetal bovine serum. All the cell lines were purchased from
the American Type Culture Collection (Manassas, VA, USA), and were
cultured at 37°C in 5% CO2 atmosphere. Spheroids of
cells were formed using a 3D culture technique (25, 26). As described previously (27), cells were seeded in 24-well plates
coated with 2% SeaPlaque agarose (BioWhittaker™; Lonza Group, Ltd.,
Basel, Switzerland) in DMEM with 5×104 cells/well in 500
μl DMEM. 5-FU was purchased from Selleck Chemicals (Houston,
TX, USA). NF-κB inhibitor SN50 was purchased from MedChemExpress
(Shanghai, China) and used at the concentration at 18 μmol/l
for the indicated times.
Construction of overexpression vector and
short hairpin RNA (shRNA) treatment
A ZBTB7 overexpression vector and shRNA system was
constructed to explore the function and mechanism. pcDNA3.1-ZBTB7A
was generated by inserting the coding region (116-1,870 bp) from
SW620 cells into pcDNA3.1 vector, using HindIII restriction
enzyme digestion and ligation (Invitrogen; Thermo Fisher
Scientific, Inc.) The procedure was performed according to previous
research (19). The resulting
plasmid was used to express ZBTB7. Plasmid (4 μg) was
transfected into cells at 50–70% confluence using Lipofectamine
2000®. ZBTB7 primer pairs were
5′-CTTAAGCTTGCCACCATGGCCGGCGGCGTGG-3′ and
5′-GTCAAGCTTTTAGGCGAGTCCGGCTGTGAAGTTAC-3′. The shRNA sequences used
were as follows: Control shRNA, forward
CGCGAATTCACCATGGCCGGCGGCGTGG, reverse
TGGCTCGAGTTAGGCGAGTCCGGCTGTGAAGT TAC; ZBTB7 shRNA, forward
GATCCCGCCCACAACTACGACCTGAATTGATATCCGTTCAGGTCGTAGTTGTGGGTTTTTTCCAAA,
reverse
AGCTTTTGGAAAAAACCCACAACTACGACCTGAACGGATATCAATTCAGGTCGTAGTTGTGGGCGG.
The subsequent experiments were performed 48 h after the
transfection.
Transwell assay
The chambers (BD Biosciences, San Jose, CA, USA)
were precoated with Matrigel (BD Biosciences, San Jose, CA, USA).
HCT116 cells (5×106) and ZBTB7-shRNA cells were counted
and suspended in 100 μl FBS-free medium at the upper
chamber. Medium containing 10% FBS was added in the lower chamber,
and incubated for 24 h at the temperature of 37°C. Cells were fixed
with 90% ethanol for 1 h at 4°C. Migrant cells on membranes were
visualized with 0.1% crystal violet staining for 15 min at room
temperature. Following drying, the migrant cells were counted in
five ×200 microscopic fields.
In vitro chemosensitivity assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) to determine the
IC50 values of cells. Following trypsinization, the
transfected cell suspensions (control shRNA, ZBTB7-shRNA and
ZBTB7-OE) were transferred and dispersed into 96-well plates. The
density was ~5,000 cells/well. 5-FU was added to treatment groups.
After 72 h, cells were incubated with 10 μl CCK-8 reagents
for another 2 h, then detected using a microplate reader at 450 nm
absorbance (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Mouse xenografts models
Control HCT116 cells (~5×106) and
ZBTB7-shRNA cells were subcutaneously implanted into the posterior
flank of BALB/c nude mice (male, 6 weeks old, purchased from
Beijing Vital River Laboratory Animal Technology Co., Ltd.
(Beijing, China). A total of 12 nude mice were used with 3 mice in
each group. Tumor size was measured every 3 days and recorded by
using the following equation: Volume (mm3) = (length ×
width2)/2. When the tumors reached 100 mm3, 5-FU was
administered to the mice. Intraperitoneal injection of 5-FU was
administered at a concentration of 25 mg/kg every 3 days. After ~1
month, the mice were sacrificed. The tumor tissues were extracted
and the volume and weight were calculated. The maximum tumor volume
was 4,200 mm3. Ethics approval for the animal
experiments was provided by The Institutional Animal Care and Use
Committee of Chongqing Fuling Central Hospital (Chongqing, China;
ethics approval no. 2014015).
Western blotting
The cells were lysed in radioimmuno-precipitation
assay lysis buffer (Thermo Fisher Scientific, Inc.) and cell
lysates were collected. Then bicinchoninic acid method was used to
detect the protein concentration. Equal amounts of protein (20
μg) was separated by 10% SDS-PAGE (Thermo Fisher Scientific,
Inc.) and then the proteins were transferred onto polyvinylidene
difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA).
Subsequently, the membranes were blocked at room temperature with
5% bovine serum albumin (Thermo Fisher Scientific, Inc.) [BSA; in
TBS-Tween (TBST)] for 1 h and incubated at 4°C with the indicated
antibodies overnight. TBST was used to wash the PVDF membranes
three times and incubated with secondary antibodies, anti-mouse IgG
(cat. no. 7076; Cell Signaling Technology, Inc., Danvers, MA, USA),
anti-rabbit IgG (cat. no. 7074; Cell Signaling Technology, Inc.;
both diluted at 1:1,000) for 1 h at room temperature. TBST was used
to wash again three times. Finally, proteins were observed with a
chemiluminescence kit (Thermo Fisher Scientific, Inc.) by using
ImageQuant LAS 4000 (GE Healthcare Life Sciences, Little Chalfont,
UK). The following primary antibodies were used: GAPDH (1:3,000;
Cell Signaling Technology, Inc. A; cat. no. 5174), ZBTB7 (1:1,000;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no.
sc-33683), E-cadherin (1:1,000; Cell Signaling Technology, Inc.;
cat. no. 14472), E-selectin (1:1,000; Santa Cruz Biotechnology,
Inc.; cat. no. sc-18852), integrin β1 (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 34971), integrin αV (1:500; Santa Cruz
Biotechnology, Inc.; cat. no. 4711) and fibronectin (1:1,000; Santa
Cruz Biotechnology, Inc.; cat. no. sc-69681).
Immunofluorescence assay
HCT116 control or HCT116-shRNA cells were cultured
overnight with or without 5-FU (50 μM) to reach 80–90%
confluence. Then, the cells were cultured in complete medium for
another 48 h, and fixed in 4% paraformaldehyde for 10 min at room
temperature. The cells were permeabilized in PBS containing 0.2 %
Triton X-100 for 5 min at room temperature, washed three times in
TBST and blocked with 5% BSA for 30 min at room temperature. The
cells were incubated with antibody against cleaved caspase-3
(1:400; Cell Signaling Technology, Inc.; cat. no. 9661), ZBTB7
(1:100; Santa Cruz Biotechnology, Inc.; cat. no. sc-33683)
overnight at 4°C. After washing, the cells were labeled with 5
μg/ml Alexa Fluor 488-conjugated secondary antibody at 4°C
for 30 min (cat. no. A-10631; Thermo Fisher Scientific, Inc.),
followed by examination under a fluorescence microscope (Nikon
Corporation, Tokyo, Japan). DAPI was purchased from Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany) and was incubated at room
temperature for 10 min at a concentration of 1 μg/ml. Five
randomly selected fields at a magnification of ×200 were counted in
each slide. The experiment was repeated for three times.
High-performance liquid chromatography
(HPLC) analysis of bases released from the DNA of FU-treated
cells
HPLC assay was performed to measure the
intracellular 5-FU level in the cells. The methods for HPLC assay
were performed according to a standard procedure (28). Exponentially growing cells
(×107) were treated with FU-2-14C (53 mCi
mmol−1; Sigma-Aldrich; Merck KGaA), treated with
non-radiolabeled FU and labeled with uracil-2-14C (52 mCi
mmol-1; Sigma-Aldrich; Merck KGaA), or treated with
non-radiolabeled FU. The concentration of FU (and uracil) used was
10 μmol/l and cells were exposed to FU for 48 h (less than
two cell divisions). Genomic DNA was isolated from the cells
(Wizard Genomic DNA purification kit; Promega Corporation, Madison,
WI, USA), and was precipitated with ethanol and the supernatant
analyzed by high-performance liquid chromatography (HPLC).
Fractions were collected at 0.5-min intervals, and released DNA
bases detected by scintillation counting or UV absorbance at 254
nm; reference compounds were detected by UV absorbance.
Statistical analysis
The statistics analyses were performed using SPSS
20.0 software package (IBM Corp., Armonk, NY, USA). At least three
independent experiments were performed to calculate the results
presented as the mean ± standard deviation. For comparison between
two groups, two-tailed Student’s t-test was performed. Multiple
group comparisons were performed with one-way analysis of variance.
Least significant difference was used as a post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
ZBTB7 was upregulated in CRC and promoted
CRC progression
In order to explore genetic initiation involved in
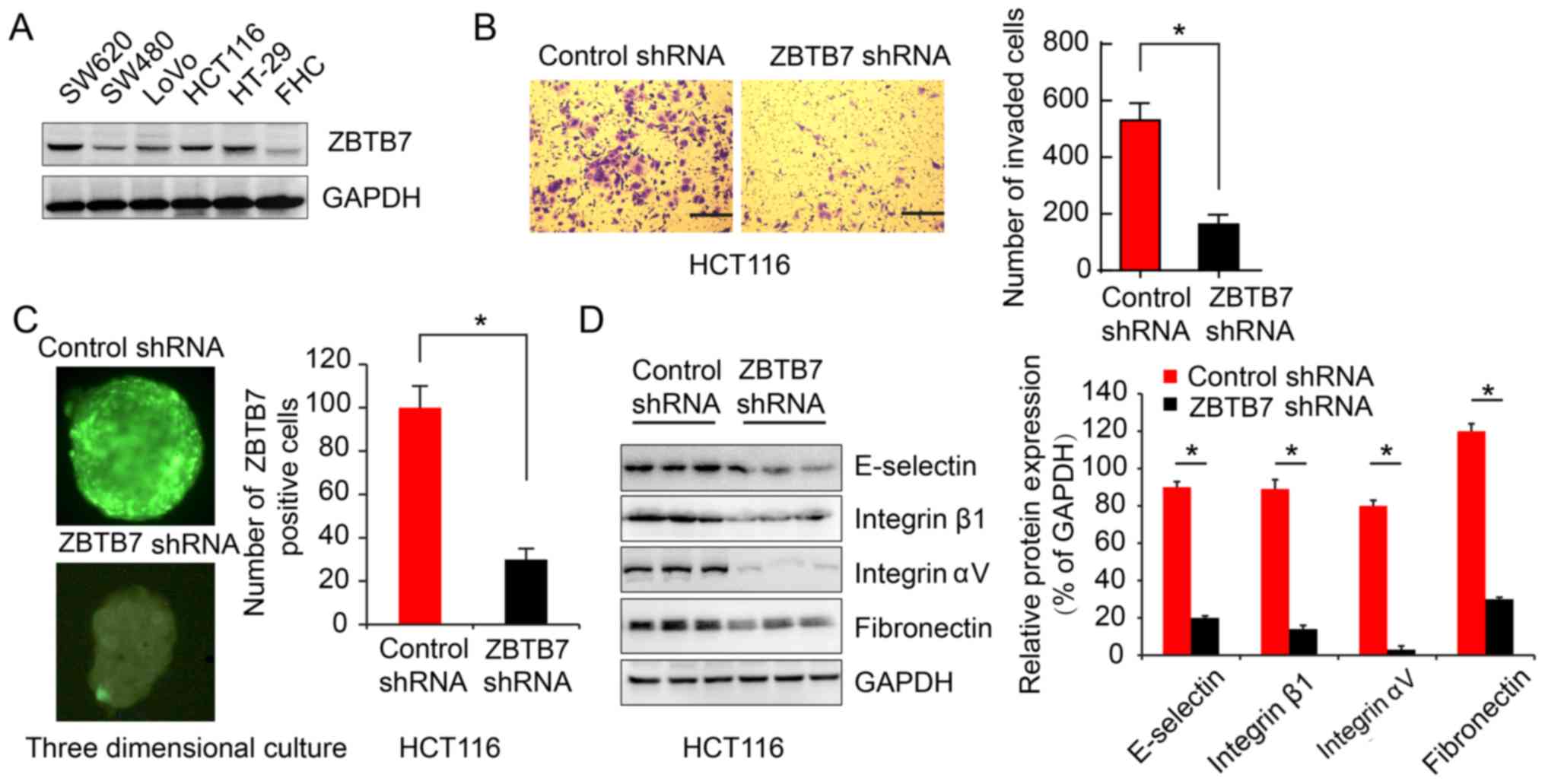
CRC, ZBTB7 expression was analyzed in CRC cell lines and tissue.
ZBTB7 was upregulated in CRC cells lines compared with a normal
colon epithelial cell line, and HCT116 cell had a relatively high
ZBTB7 level (Fig. 1A). ZBTB7
function was evaluated in HCT116 cell lines. Transwell assay
demonstrated that ZBTB7 shRNA reduced the invasion of HCT116 cells
(162.7±34.37) compared with the control group (530.3±60.73;
P<0.05; Fig. 1B). Furthermore,
three-dimensional cultures of cells were performed to mimic the
complex in vivo tumor microenvironment. Immunofluorescence
showed control RNA cells and ZBTB7 shRNA cells (Fig. 1C). The results showed the same
trends (Fig. 1D).
Silencing ZBTB7 increases the 5-FU
sensitivity in CRC
To explore the functional role of ZBTB7 in 5-FU
resistance, IC50 values were determined in ZBTB7-shRNA
HCT116 cells and control groups cells by detecting cell viability
in both cell lines at different concentration of 5-FU and over
time. Compared with the control group, the IC50 values
were significantly reduced in the ZBTB7-shRNA group. As the 5-FU
concentration and time increased, the cell viability was decreased,
with ZBTB7-shRNA group decreased more than the control (P<0.05,
Fig. 2A–C). By contrast,
overexpression of ZBTB7 in SW480 cells (relatively low ZBTB7 level
compared with other CRC cell lines; Fig. 1A) exhibited the opposite effects
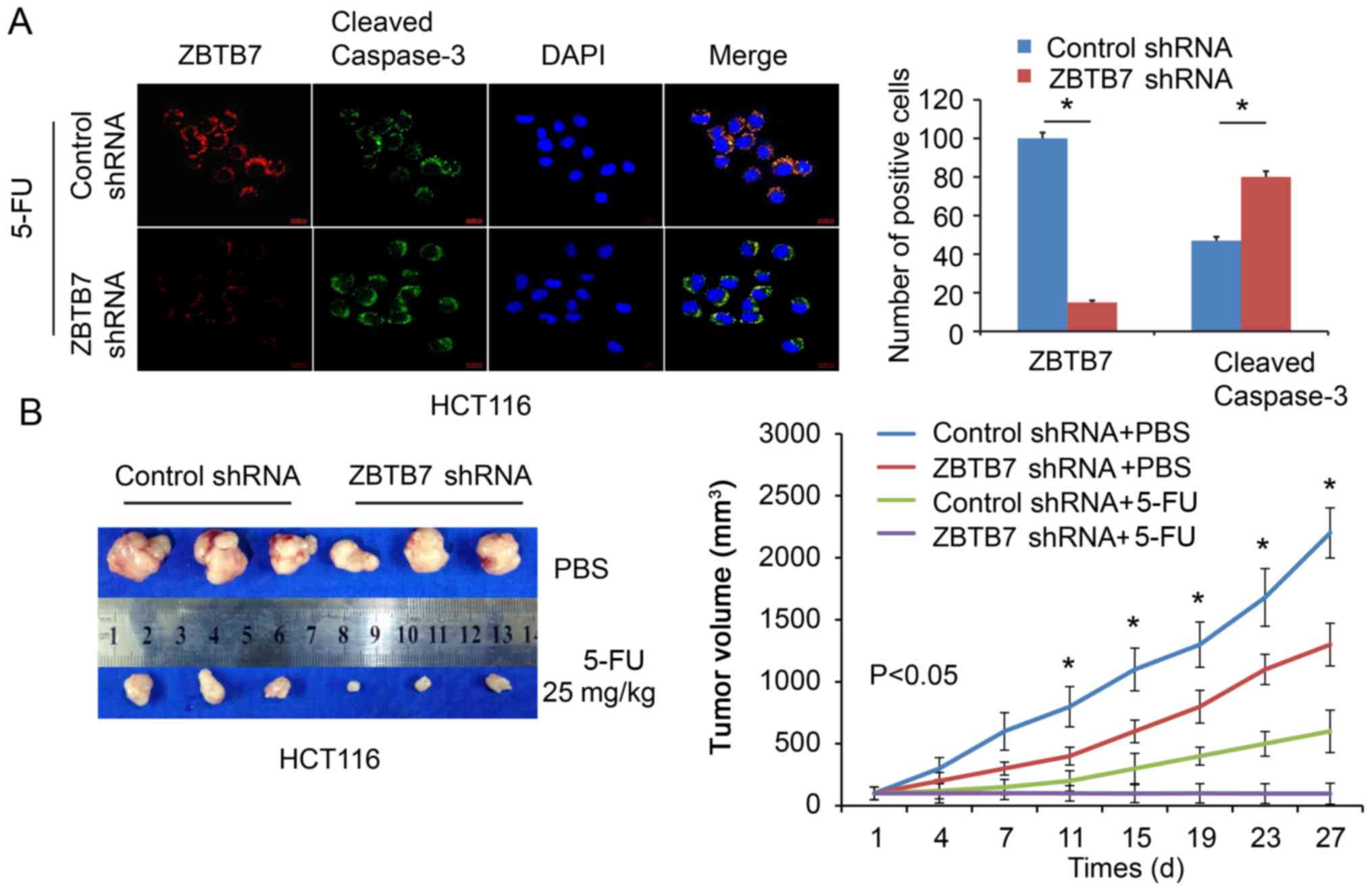
(P<0.05; Fig. 2D–F). Cleaved
caspase-3 is an apoptotic protein marker, and immunofluorescence
demonstrated that 5-FU-induced apoptosis was increased in
ZBTB7-shRNA cells compared with the control group (P<0.05;
Fig. 3A). Furthermore, a xenograft
mouse model demonstrated that the size of tumors was reduced in the
ZBTB7-shRNA group following treatment with 5-FU (Fig. 3B). The tumors volume were
calculated and recorded. ZBTB7 shRNA cell-derived tumors and 5-FU
were the slowest growing among all the groups (P<0.05; Fig. 3B).
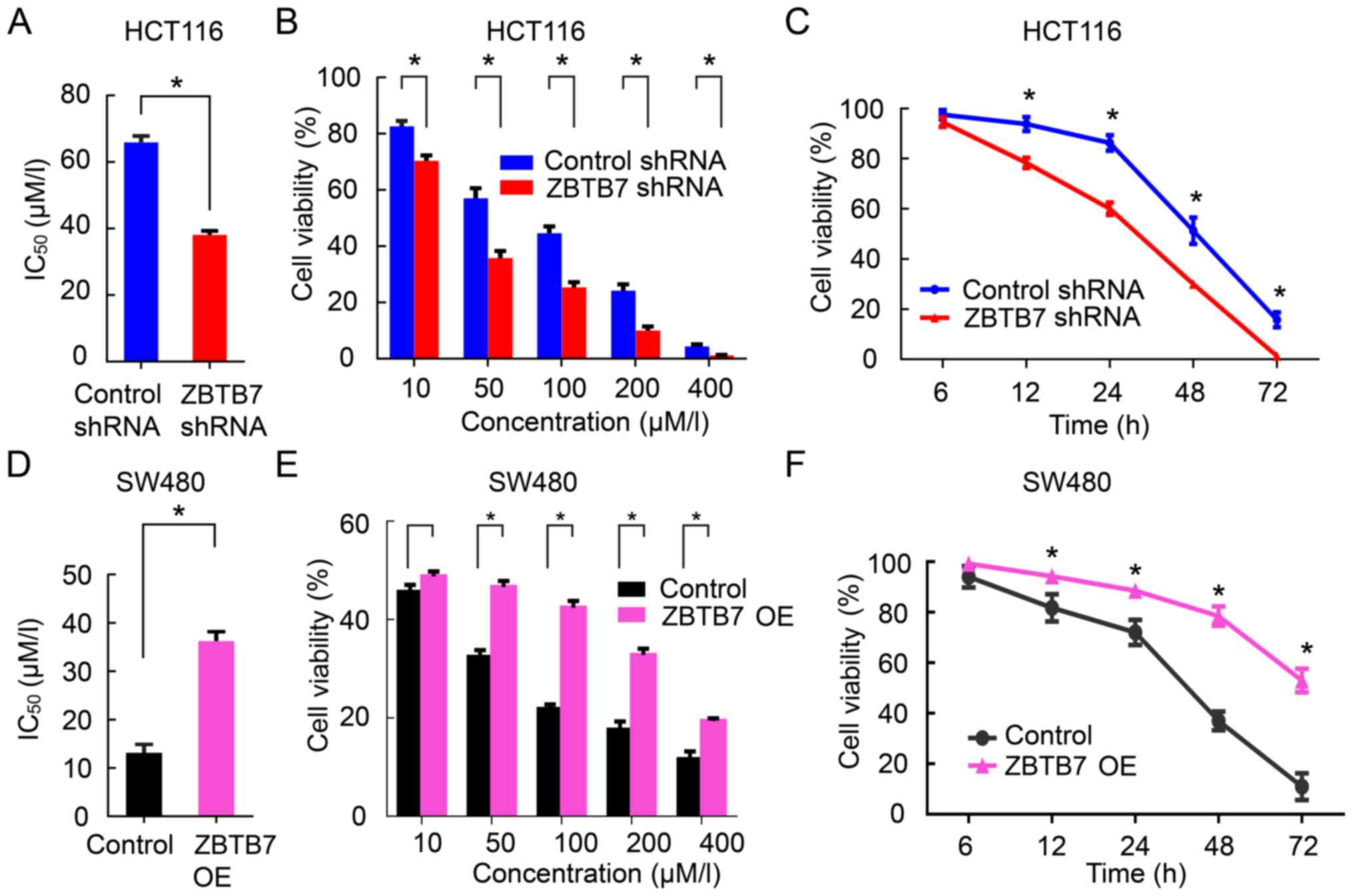 | Figure 2Silencing ZBTB7 restores the
sensitivity of colorectal cancer to 5-FU. (A) CCK-8 method was
measured IC50 values in control or ZBTB7-shRNA groups
exposed to 5-FU. (B) Cell viability in control or ZBTB7-shRNA
groups exposed to different 5-FU concentration (10, 50, 100, 200
and 400 μM). (C) Cell viability in control or ZBTB7-shRNA
groups exposed to 5-FU at different time-points (6, 12, 24, 48 and
72 h). (D) IC50 values in control or ZBTB7-OE groups
(SW480) exposed to 5-FU. (E) Cell vitality in control or ZBTB7-OE
groups (SW480) exposed to different 5-FU concentration (10, 50,
100, 200 and 400 μM). (F) Cell viability in control or
ZBTB7-OE groups (SW480) exposed to 5-FU at different time-points
(6, 12, 24, 48, and 72 h). *P<0.05 vs. control. 5-FU,
5-fluorouracil; shRNA, short hairpin RNA; ZBTB7, zinc finger and
BTB domain containing A; OE, overexpression. |
ZBTB7 is associated with NF-κB signaling
pathways
Subsequently, it was aimed to uncover the potential
mechanism of ZBTB7-driven 5-FU resistance. The original genetic
profiles of patients with CRC were acquired from the GEO data
repository. Differences in ZBTB7 expression among multiple cancer
types were assessed using data from GEO. To explore the potential
role and the mechanism of ZBTB7 in regulating chemotherapeutic
resistance, data from cell lines (GSE36133) derived from naturally
occurring tumors were analyzed because they recapitulate various
aspects of the tissue type and genomic context of cancer. NMF, a
recently established approach for consensus clustering, was
performed onto 53 CRC cell lines according to their transcriptional
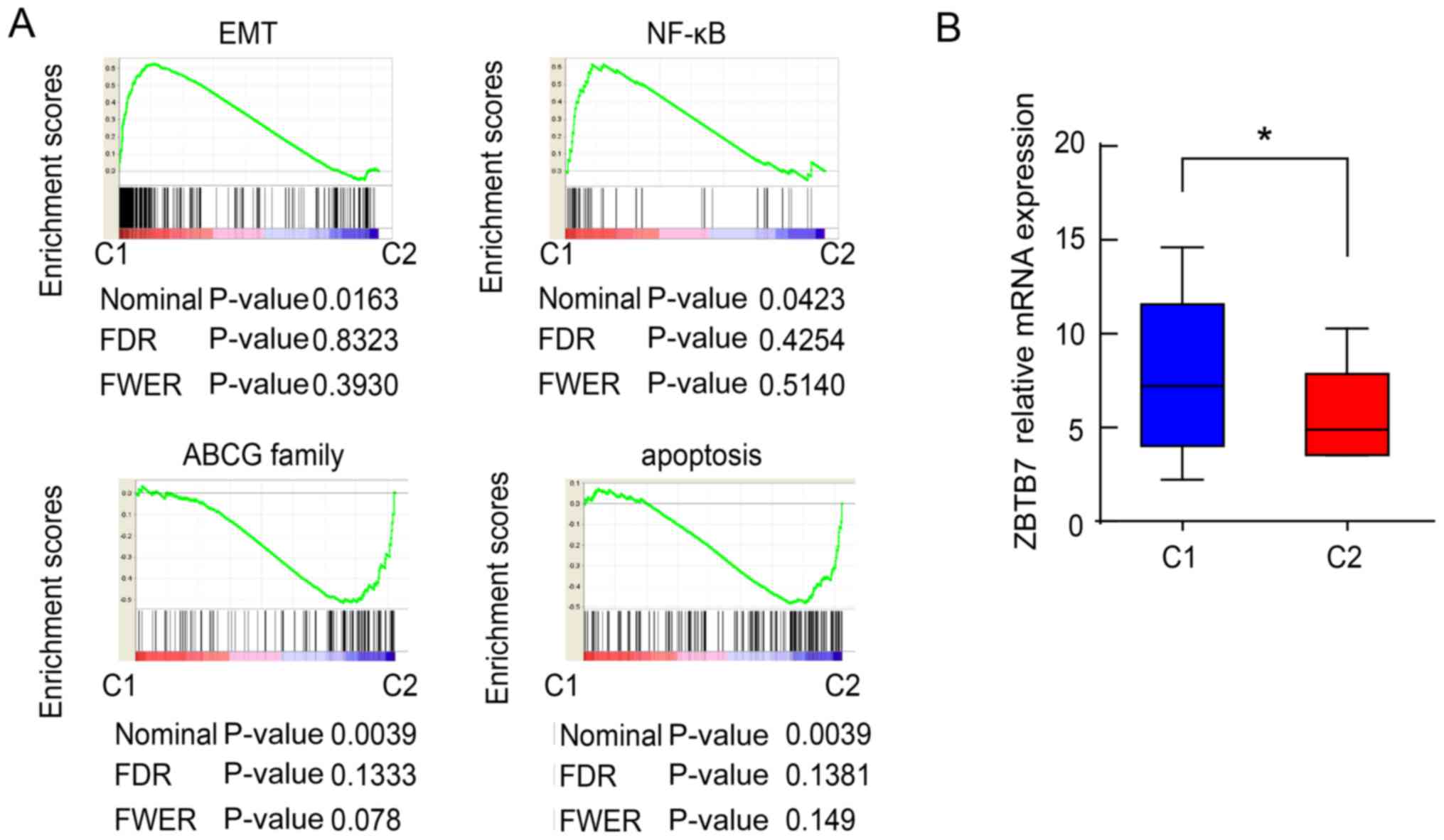
features. This analysis revealed two subtypes of CRC cell lines
with adequate data coherence. Bioinformatics analysis split cell
lines into two subtypes according to the EMT-associated genetic
expression, the NF-κB signaling pathway, ABCG family and
apoptosis-associated genes. One subtype (39.6% of all lines, n=21)
was especially high in epithelial- mesenchymal transition and NF-κB
(Fig. 4A), and was termed C1
subtype. The other subtype (56.2% of all lines, n=27) especially
high in ABCG family and apoptosis (Fig. 4B) were named C2 subtype. Notably,
the relative ZBTB7 level in group C1 was significantly higher than
that in group C2 (Fig. 4B). This
evidence indicates that ZBTB7 may be involved in chemotherapeutic
resistance of CRCs via regulation of the NF-κB pathway.
NF-κB inhibitor SN50 reverses
ZBTB7-induced resistance in CRC
The role of NF-κB in ZBTB7-driven 5-FU resistance in
CRC was investigated. 5-FU reduced cell viability, and
overexpression of ZBTB7 depleted the inhibitory effects. Treatment
with NF-κB inhibitor SN50 reversed the proliferation enhancing
effects of ZBTB7 (P<0.05; Fig.
5A). These results were verified in vivo. The tumor
volume (P<0.05; Fig. 5B) and
weight (P<0.05; Fig. 5C)
exhibited the same trends. Finally, ZBTB7 overexpression increased
the 5-FU level while SN50 significantly increased the intracellular
5-FU level (P<0.05; Fig.
5D).
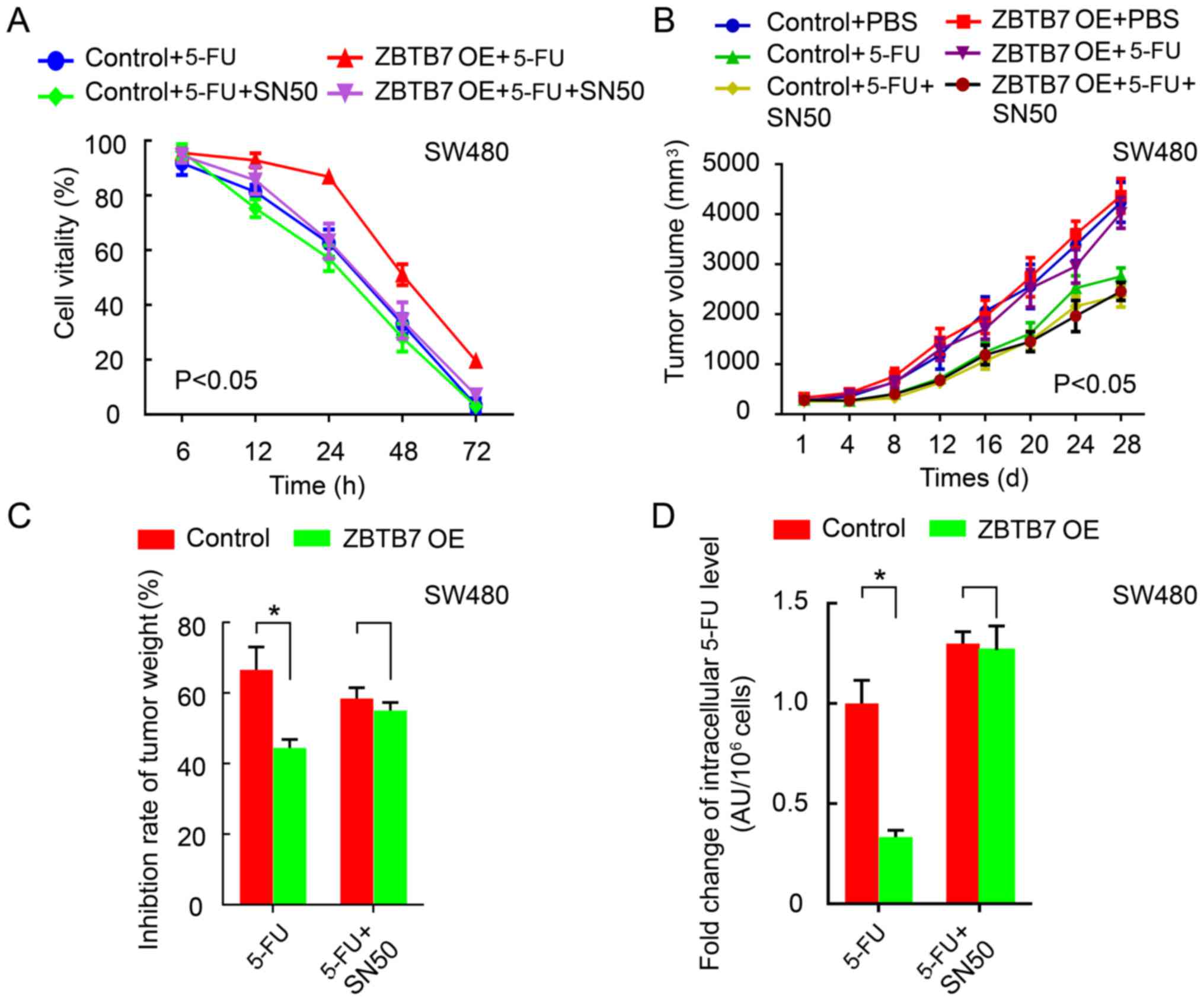 | Figure 5NF-κB inhibitor SN50 reverses
ZBTB7-induced resistance in colorectal cancer cells. (A) CCK8
method was used to detect cell viability in control+5-FU with or
without SN50 group, in OE+5-FU with or without SN50 group. (B)
Tumor volume in control+PBS, control+5-FU, control+5-FU+SN50, in
OE+PBS, OE+5-FU, OE+5-FU+SN50 group. (C) Inhibition rate of tumor
weight in control+5-FU with or without SN50 group, in OE+5-FU with
or without SN50 group. (D) HPLC assay was performed to measure
intracellular 5-FU concentration in control+5-FU with or without
SN50 group, in OE+5-FU with or without SN50 group in control and
ZBTB7-OE cells with or without 5-FU and SN50.
*P<0.05. NF-κB, nuclear factor-κB; 5-FU,
5-fluorouracil; OE, overexpression. |
ZBTB7 mRNA level is associated with 5-FU
sensitivity and prognosis
The above results had demonstrated that higher ZBTB7
expression promoted tumorigenesis and susceptibility to 5-FU
resistance through the NF-κB pathway. The clinical significance was
investigated using GSE39582 from the GEO database. The patients
received 5-FU alone or 5-FU-based adjuvant chemotherapy; the
subgroup with ZBTB7-low tumors substantially benefited from
adjuvant chemotherapy and exhibited a significantly increased
probability of relapse-free survival and overall survival relative
to those in the subgroup with ZBTB7-high tumors. (P<0.05;
Fig. 6; Table I).
 | Table IEffect of ZBTB7 expression on overall
survival and relapse-free survival of patients with stage II/III
colorectal cancer according to Gene Expression Omnibus (GSE39582)
clinical annotations. |
Table I
Effect of ZBTB7 expression on overall
survival and relapse-free survival of patients with stage II/III
colorectal cancer according to Gene Expression Omnibus (GSE39582)
clinical annotations.
A, Overall survival
|
|---|
| Treatment | Mean months
(standard error)
| P-valueb |
|---|
| Total | ZBTB7 Lowa | ZBTB7 Higha |
| 5-FU | 52.28 (1.84) | 53.99 (1.98) | 47.46 (4.07) | 0.041 |
| CCc | 53.19 (1.70) | 55.60 (1.84) | 48.81 (3.43) | 0.043 |
B, Relapse-free
survival
|
|---|
| Treatment | Mean months
(standard error)
| P-valueb |
|---|
| Total | ZBTB7 Lowa | ZBTB7 Higha |
|---|
| 5-FU | 43.16 (2.61) | 46.60 (2.80) | 32.93 (5.58) | 0.019 |
| CCc | 47.36 (2.66) | 51.53 (2.79) | 39.77 (5.16) | 0.103 |
Discussion
5-FU-based chemotherapy is the basic classical
treatment for patients with CRC (29). However, chemoresistance, either
primary or acquired, is as a major challenge for clinical
prac-tice. 5-FU primary chemoresistance is predominantly due to
increased thymidylate synthetase mRNA or protein level. Acquired
chemoresistance mechanisms include the depletion of certain
enzymes, such as thymidilate synthase, which is the target of 5-FU,
or certain genetic mutations (30). Up to 40% of patients with stage II
and III CRC receiving 5-FU-based adjuvant chemotherapy experience
recurrence or mortality within 8 years of follow-up (31). Of patients with metastatic CRC, 50%
are resistant to 5-FU-based chemotherapy (32,33).
It is important to investigate the 5-FU resistance mechanisms and
set up therapeutic strategies to reverse the chemoresistance to
improve prognosis. In the current study, ZBTB7 was overexpressed in
CRC cells compared with normal colon epithelial cells.
Additionally, ZBTB7 knockdown increased 5-FU sensitivity.
Mechanistically, ZBTB7 potentially promoted 5-FU resistance through
the NF-κB signaling pathway. Targeting NF-κB signaling using SN50
reversed ZBTB7-mediated 5-FU resistance. Therefore, a ZBTB7/NF-κB
axis is involved in mediating 5-FU resistance in patients with
CRC.
Though ZBTB7 is aberrantly overexpressed in human
cancers, little is known about the mechanism that regulates this. A
previous study demonstrated that the ZBTB7 gene is at the genomic
locus chromosome 19p13.3, which is frequently mutated. The
t(14;19)(q32;p13.3) translocation is common in B-cell non-Hodgkin’s
lymphoma (34). ZBTB7
overexpres-sion may aberrantly activate regulatory pathways, such
as fibronectin-mediated h1-integrin ligation in precursor B
leukemia cells (35). However, its
function in 5-FU resistance is unknown. In the current study, ZBTB7
was upregulated in CRC cell lines compared with normal colon
epithelial cells. A Transwell assay demonstrated that ZBTB7-shRNA
cells were less invasive than the control group. E-selectin,
integrin β1, integrin αV and fibronectin are adhesion molecules.
Typically, cancer cells interactions initially require a
selectin-mediated initial attachment and then the circulating
cancer cells roll along the endothelium. Locally released
chemokines activate the rolling cancer cells. This triggers
integrin activation, making a firmer adhesion to cell adhesion
molecules, initiating and driving the trans-endothelial migration
and extravasation processes (36).
In the current study, three-dimensional cultures were used to mimic
the tumor microenvironment. The results revealed that integrin β1,
integrin αV and fibronectin expression were reduced in ZBTB7-shRNA
cells compared with the control group. These results indicated that
ZBTB7 could regulate adhesion molecules expression in vitro
and in vivo, leading to tumor progression. Furthermore,
ZBTB7 knockdown and 5-FU treatment could increase apoptosis. EMT
was previously reported to be associated with 5-FU resistance in
pancreatic cancer (37) and lung
cancer (38). We hypothesized that
ZBTB7 may be involved in 5-FU resistance in CRC. Previous research
demonstrated that ZBTB7 promoted the migration and invasion of
hepatocellular carcinoma by increasing myocyte enhancer factor 2D
expression (19). Mak et al
(39) reported that ZBTB7 promoted
cell migration and invasion via phosphoinositide 3-kinase/Akt
signaling. Taken together, the results suggest that ZBTB7 may
regulate EMT- and apoptosis-associated proteins to promote cell
invasion and decrease cell apoptosis, which may mediate cell
chemoresistance.
ZBTB7 was reported to be associated with efficacy of
paclitaxel and cisplatin combination chemotherapy and overall
survival in patients with NSCLC (40). The function of ZBTB7 in 5-FU
resistance was examined in the present study. IC50
values were determined using ZBTB7-shRNA cells and control groups
by measuring cell viability over different concentrations and
time-points of 5-FU. The results demonstrated that the 5-FU
IC50 values were reduced upon ZBTB7 depletion compared
with control group, in dose- and time-dependent manners.
Additionally, overexpression of ZBTB7 exerted the opposite effects.
The results were also validated in animal models. The volume and
the weights of the tumor were lower in the ZBTB7 shRNA group
following exposure to 5-FU.
GEO data was analyzed to identify genes that may be
involved in the 5-FU resistance of CRC. The results demonstrated
that the 5-FU resistance may be associated with the NF-κB signaling
pathways. NF-κB signaling is reported to mediate chemoresistance in
various ways (41,42). Kwon et al (43) reported that gastric cancer cell
resistance to 5-FU was mediated via activation of NF-κB. ZBTB7, as
an oncogene, participated in NF-κB signaling pathway. For instance,
ZBTB7 reduces Bcl-2 expression through NF-κB in hepatocellular
carcinoma (44). In the current
study, NF-κB altered in ZBTB7-driven 5-FU resistance in CRC. 5-FU
inhibited cell viability and overexpression of ZBTB7 depleted the
inhibitory effects. NF-κB inhibitor SN50 reversed the proliferation
enhancing effects of ZBTB7 in vitro and in vivo.
Finally, clinical data was analyzed and indicated that patients
with high expression of ZBTB7 have worse prognosis under 5-FU
adjuvant therapy.
In conclusion, the finding indicated that the
ZBTB7/NF-κB axis contributes to 5-FU resistance of patients with
CRC, and may be serve as potential therapeutic targets to overcome
5-FU resistance.
Funding
This study was supported by The Health Bureau of
Chongqing (grant no. 2011-2-437).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
ZW and XZ performed the cellular and animal studies,
the statistical analysis and drafted the manuscript. WW and MZ
analyzed the GEO database. YiL, YaL, JG and GX performed the
cellular studies. CW and RL participated in designing the study. QZ
was involved in designing the study and drafting the manuscript and
revising it critically for important intellectual content and given
final approval of the version to be published. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval for the animal experiments was
provided by The Institutional Animal Care and Use Committee of
Chongqing Fuling Central Hospital (ethics approval no.
2014015).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Okusaka T, Funakoshi A, Furuse J, Boku N,
Yamao K, Ohkawa S and Saito H: A late phase II study of S-1 for
metastatic pancreatic cancer. Cancer Chemother Pharmacol.
61:615–621. 2008. View Article : Google Scholar
|
|
3
|
Morizane C, Okusaka T, Furuse J, Ishii H,
Ueno H, Ikeda M, Nakachi K, Najima M, Ogura T and Suzuki E: A phase
II study of S-1 in gemcitabine-refractory metastatic pancreatic
cancer. Cancer Chemother Pharmacol. 63:313–319. 2009. View Article : Google Scholar
|
|
4
|
Sudo K, Yamaguchi T, Nakamura K, Denda T,
Hara T, Ishihara T and Yokosuka O: Phase II study of S-1 in
patients with gemcitabine-resistant advanced pancreatic cancer.
Cancer Chemother Pharmacol. 67:249–254. 2011. View Article : Google Scholar
|
|
5
|
Sasaki T, Isayama H, Yashima Y, Yagioka H,
Kogure H, Arizumi T, Togawa O, Matsubara S, Ito Y, Nakai Y, et al:
S-1 monotherapy in patients with advanced biliary tract cancer.
Oncology. 77:71–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsuji W, Ishiguro H, Tanaka S, Takeuchi M,
Ueno T and Toi M: Orally administered S-1 suppresses circulating
endothelial cell counts in metastatic breast cancer patients. Int J
Clin Oncol. 19:452–459. 2014. View Article : Google Scholar
|
|
7
|
Saek T, Takashima S, Sano M, Horikoshi N,
Miura S, Shimizu S, Morimoto K, Kimura M, Aoyama H, Ota J, et al: A
phase II study of S-1 in patients with metastatic breast cancer - a
Japanese trial by the S-1 Cooperative Study Group, Breast Cancer
Working Group. Breast Cancer. 11:194–202. 2004. View Article : Google Scholar
|
|
8
|
Lee SJ, Lee J, Park SH, Park JO, Park YS,
Kang WK, Lee J, Yim DS and Lim HY: Phase 1 trial of S-1 in
combination with sorafenib for patients with advanced
hepatocellular carcinoma. Invest New Drugs. 30:1540–1547. 2012.
View Article : Google Scholar
|
|
9
|
Furuse J, Okusaka T, Kaneko S, Kudo M,
Nakachi K, Ueno H, Yamashita T and Ueshima K: Phase I/II study of
the pharma-cokinetics, safety and efficacy of S-1 in patients with
advanced hepatocellular carcinoma. Cancer Sci. 101:2606–2611. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pectasides D, Karavasilis V, Papaxoinis G,
Gourgioti G, Makatsoris T, Raptou G, Vrettou E, Sgouros J, Samantas
E, Basdanis G, et al: Randomized phase III clinical trial comparing
the combination of capecitabine and oxaliplatin (CAPOX) with the
combination of 5-fluorouracil, leucovorin and oxaliplatin (modified
FOLFOX6) as adjuvant therapy in patients with operated high-risk
stage II or stage III colorectal cancer. BMC Cancer. 15:3842015.
View Article : Google Scholar
|
|
11
|
Argilés G, Saunders MP, Rivera F, Sobrero
A, Benson A III, Guillén Ponce C, Cascinu S, Van Cutsem E,
Macpherson IR, Strumberg D, et al: Regorafenib plus modified
FOLFOX6 as first-line treatment of metastatic colorectal cancer: A
phase II trial. Eur J Cancer. 51:942–949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Álvarez P, Marchal JA, Boulaiz H, Carrillo
E, Vélez C, Rodríguez-Serrano F, Melguizo C, Prados J, Madeddu R
and Aranega A: 5-Fluorouracil derivatives: A patent review. Expert
Opin Ther Pat. 22:107–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maeda T, Hobbs RM and Pandolfi PP: The
transcription factor Pokemon: A new key player in cancer
pathogenesis. Cancer Res. 65:8575–8578. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Davies JM, Hawe N, Kabarowski J, Huang QH,
Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G, et
al: Novel BTB/POZ domain zinc-finger protein, LRF, is a potential
target of the LAZ-3/BCL-6 oncogene. Oncogene. 18:365–375. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CC, Zhou JP, Liu YP, Liu JJ, Yang XN,
Jazag A, Zhang ZP, Guleng B and Ren JL: The silencing of Pokemon
attenuates the proliferation of hepatocellular carcinoma cells in
vitro and in vivo by inhibiting the PI3K/Akt pathway. PLoS One.
7:e519162012. View Article : Google Scholar
|
|
17
|
Zhao Z, Wang J, Wang S, Chang H, Zhang T
and Qu J: LncRNA CCAT2 promotes tumorigenesis by over-expressed
Pokemon in non-small cell lung cancer. Biomed Pharmacother.
87:692–697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aggarwal H, Aggarwal A and Agrawal DK:
Corrigendum to ‘Epidermal growth factor increases LRF/Pokemon
expression in human prostate cancer cells’ [Exp. Mol. Pathol. 91
(2011) 496-501]. Exp Mol Pathol. 100:3612016. View Article : Google Scholar
|
|
19
|
Kong J, Liu X, Li X, Wu J, Wu N, Chen J
and Fang F: Pokemon promotes the invasiveness of hepatocellular
carcinoma by enhancing MEF2D transcription. Hepatol Int.
10:493–500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sartini D, Lo Muzio L, Morganti S, Pozzi
V, Di Ruscio G, Rocchetti R, Rubini C, Santarelli A and Emanuelli
M: Pokemon proto-oncogene in oral cancer: Potential role in the
early phase of tumorigenesis. Oral Dis. 21:462–469. 2015.
View Article : Google Scholar
|
|
21
|
He S, Liu F, Xie Z, Zu X, Xu W and Jiang
Y: P-Glycoprotein/MDR1 regulates pokemon gene transcription through
p53 expression in human breast cancer cells. Int J Mol Sci.
11:3309–051. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao GT, Yang LJ, Li XX, Cui HL and Guo R:
Expression of the proto-oncogene Pokemon in colorectal cancer -
inhibitory effects of an siRNA. Asian Pac J Cancer Prev.
14:4999–5005. 2013. View Article : Google Scholar
|
|
23
|
Zhao Y, Yao YH, Li L, An WF, Chen HZ, Sun
LP, Kang HX, Wang S and Hu XR: Pokemon enhances proliferation, cell
cycle progression and anti-apoptosis activity of colorectal cancer
independently of p14ARF-MDM2-p53 pathway. Med Oncol. 31:2882014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Reyniès A, Assié G, Rickman DS, Tissier
F, Groussin L, René-Corail F, Dousset B, Bertagna X, Clauser E and
Bertherat J: Gene expression profiling reveals a new classification
of adreno-cortical tumors and identifies molecular predictors of
malignancy and survival. J Clin Oncol. 27:1108–1115. 2009.
View Article : Google Scholar
|
|
25
|
Desoize B and Jardillier J: Multicellular
resistance: A paradigm for clinical resistance? Crit Rev Oncol
Hematol. 36:193–207. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Green SK, Francia G, Isidoro C and Kerbel
RS: Antiadhesive antibodies targeting E-cadherin sensitize
multicellular tumor spheroids to chemotherapy in vitro. Mol Cancer
Ther. 3:149–159. 2004.PubMed/NCBI
|
|
27
|
Zheng C, Zhou Q, Wu F, Peng Q, Tang A,
Liang H and Zeng Y: Semaphorin3F down-regulates the expression of
integrin alpha(v) beta3 and sensitizes multicellular tumor
spheroids to chemotherapy via the neuropilin-2 receptor in vitro.
Chemotherapy. 55:344–352. 2009. View Article : Google Scholar
|
|
28
|
Butler WR and Guthertz LS: Mycolic acid
analysis by high-performance liquid chromatography for
identification of Mycobacterium species. Clin Microbiol Rev.
14:704–726. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan BR, Thomas F, Myerson RJ, Zehnbauer B,
Trinkaus K, Malyapa RS, Mutch MG, Abbey EE, Alyasiry A, Fleshman
JW, et al: Thymidylate synthase genotype-directed neoadjuvant
chemoradiation for patients with rectal adenocarcinoma. J Clin
Oncol. 29:875–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Temraz S, Mukherji D, Alameddine R and
Shamseddine A: Methods of overcoming treatment resistance in
colorectal cancer. Crit Rev Oncol Hematol. 89:217–230. 2014.
View Article : Google Scholar
|
|
31
|
Sargent D, Sobrero A, Grothey A, O’Connell
MJ, Buyse M, Andre T, Zheng Y, Green E, Labianca R, O’Callaghan C,
et al: Evidence for cure by adjuvant therapy in colon cancer:
Observations based on individual patient data from 20,898 patients
on 18 randomized trials. J Clin Oncol. 27:872–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Douillard JY, Cunningham D, Roth AD,
Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J,
Alakl M, et al: Irinotecan combined with fluorouracil compared with
fluo-rouracil alone as first-line treatment for metastatic
colorectal cancer: A multicentre randomised trial. Lancet.
355:1041–1047. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Giacchetti S, Perpoint B, Zidani R, Le
Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y,
Coudert B, et al: Phase III multicenter randomized trial of
oxaliplatin added to chronomodulated fluorouracil-leucovorin as
first-line treatment of metastatic colorectal cancer. J Clin Oncol.
18:136–147. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gozzetti A, Davis EM, Espinosa R III,
Fernald AA, Anastasi J and Le Beau MM: Identification of novel
cryptic translocations involving IGH in B-cell non-Hodgkin’s
lymphomas. Cancer Res. 62:5523–5527. 2002.PubMed/NCBI
|
|
35
|
Astier AL, Xu R, Svoboda M, Hinds E, Munoz
O, de Beaumont R, Crean CD, Gabig T and Freedman AS: Temporal gene
expression profile of human precursor B leukemia cells induced by
adhesion receptor: Identification of pathways regulating B-cell
survival. Blood. 101:1118–1127. 2003. View Article : Google Scholar
|
|
36
|
Walzog B and Gaehtgens P: Adhesion
molecules: The path to a new understanding of acute inflammation.
News Physiol Sci. 15:107–113. 2000.
|
|
37
|
Arumugam T, Ramachandran V, Fournier KF,
Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey
DJ and Choi W: Epithelial to mesenchymal transition contributes to
drug resistance in pancreatic cancer. Cancer Res. 69:5820–5828.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu D, Zhao B, Qi X, Peng F, Fu H, Chi X,
Miao QR and Shao S: Nogo-B receptor promotes epithelial-mesenchymal
transition in non-small cell lung cancer cells through the
Ras/ERK/Snail1 pathway. Cancer Lett. 418:135–146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mak VC, Wong OG, Siu MK, Wong ES, Ng WY,
Wong RW, Chan KK, Ngan HY and Cheung AN: FBI-1 is overexpressed in
gestational trophoblastic disease and promotes tumor growth and
cell aggressiveness of choriocarcinoma via PI3K/Akt signaling. Am J
Pathol. 185:2038–2048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang QL, Xing XZ, Li FY, Xing YJ and Li
J: Pretreatment Pokemon level as a predictor of response to
cisplatin and Paclitaxel in Patients with Unresectable Non-Small
Cell Lung Cancer. Oncol Res Treat. 38:496–502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim JK, Kim KD, Lee E, Lim JS, Cho HJ,
Yoon HK, Cho MY, Baek KE, Park YP, Paik SG, et al: Up-regulation of
Bfl-1/A1 via NF-kappaB activation in cisplatin-resistant human
bladder cancer cell line. Cancer Lett. 212:61–70. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang CY, Guttridge DC, Mayo MW and Baldwin
AS Jr: NF-kappaB induces expression of the Bcl-2 homologue A1/Bfl-1
to preferentially suppress chemotherapy-induced apoptosis. Mol Cell
Biol. 19:5923–5929. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kwon OH, Kim JH, Kim SY and Kim YS:
TWEAK/Fn14 signaling mediates gastric cancer cell resistance to
5-fluorouracil via NF-κB activation. Int J Oncol. 44:583–590. 2014.
View Article : Google Scholar
|
|
44
|
Zhao X, Ning Q, Sun X and Tian D: Pokemon
reduces Bcl-2 expression through NF-κ Bp65: A possible mechanism of
hepa-tocellular carcinoma. Asian Pac J Trop Med. 4:492–497. 2011.
View Article : Google Scholar : PubMed/NCBI
|