Introduction
Renal cell carcinoma (RCC) is one of the most common
malignancy affecting adults, and the incidence of RCC has increased
over the past two decades (1). The
mortality rate of patients with RCC appears to be increasing each
year (2). In addition, patients
with RCC respond poorly to conventional chemotherapy and
radiotherapy treatment (3). Thus,
further understanding of the molecular mechanisms responsible for
the development and progression of RCC is of particular
importance.
It is well known that protein-coding genes account
for <2% of the total genome DNA, whereas a large number of the
human genome can be transcribed into non-coding RNAs (ncRNAs)
(4-6). Recently, new members of the family of
ncRNAs (>200 nucleotides) with limited or no protein-coding
potential, long ncRNAs (lncRNAs) have been extensively studied in
both physiological and pathological processes (7,8).
Accumulating evidence has indicated that lncRNAs are emerging as
key molecules in human malignancies, disease progression and
metastasis (5,9,10).
However, the role of lncRNAs in RCC remains unclear.
The HOXA transcript at the distal tip (HOTTIP)
lncRNA, located at the 5′ end of the HOXA cluster, has recently
been functionally characterized (11). HOTTIP primarily targets WDR5/MLL
complexes across HOXA by directly binding the adaptor protein, WD
repeat-containing protein 5 (WDR5), leading to histone H3 lysine 4
trimethylation and the gene transcription of several 5′ HOXA genes
(12). It has been demonstrated
that HOTTIP plays a major role in tumor progression (13). An increased HOTTIP expression has
been reported in various types of cancer, such as non-small cell
lung cancer (14) gastric cancer
(15), colorectal cancer (16) and prostate cancer (17). In these tumors, HOTTIP may serve as
a potential oncogene and may be a poor prognostic factor for
patients. However, the functions of HOTTIP and its potential
mechanisms of action in RCC have not yet been fully elucidated.
Recently, a class of lncRNAs, referred to as
competing endogenous RNAs (ceRNAs), has been characterized,
including lncARSR and Linc00152 (18). For instance, lnc-MD1, the first
identified ceRNA involved in myogenesis, has been shown to control
muscle cell differentiation by competing for the binding of miR-133
and miR-135 (18). The function of
ceRNAs has been described as a novel regulatory mechanism of RNAs
(19), and this is a newly
proposed mechanism that describes a crosstalk among lncRNAs, mRNAs
and their shared microRNAs (miRNAs or miRs). ceRNAs protect mRNAs
by acting as molecular sponges for miRNAs that specifically repress
the target mRNAs. This hypothesis suggests that lncRNAs, mRNAs and
pseudogenes can communicate with each other by competing for common
miRNA response elements.
In this study, we demonstrate that HOTTIP expression
was significantly upregulated in RCC and was associated with a
larger tumor size and a higher clinical stage, lymph node
metastasis vascular invasion, and a shorter overall survival (OS)
and disease-free survival (DFS). The inhibition of HOTTIP
suppressed cell proliferation, migration and invasion in RCC. In
addition, IFG-2 was found to be a direct target gene of HOTTIP.
HOTTIP directly bound to miR-615 and effectively acted as a sponge
for miR-615 to modulate the suppression of IFG-2. To the best of
our knowledge, this study provides the first evidence that the
HOTTIP/miR-615/IFG-2 axis plays an important role in RCC
tumorigenesis and progression and may thus have diagnostic and
therapeutic potential for use as a target in RCC.
Materials and methods
Ethics statement
The use of tissues for this study was approved by
the Medical Ethics Committee of Dalian Medical University. Due to
the retrospective nature of the study, the Ethics Committee waived
the need of written informed consent by the patients. All the
samples were anonymous.
Clinical samples
Tumor specimens were collected from patients with
primary RCC, who underwent surgery at the Department of Urology,
the First Affiliated Hospital of Dalian Medical University (Dalian,
China) from August, 2009 to January, 2016. The cohort consisted of
57 patients, from whom fresh tumor samples coupled with adjacent
non-tumorous renal tissues 5-10 cm away from the tumor edge were
obtained and subjected to HOTTIP and insulin-like growth factor-2
(IGF-2) mRNA and protein expression analysis. All the fresh
specimens were stored at −80°C until use.
TCGA data analysis
The lncRNA expression profiles and corresponding
clinical information of the patients with RCC were obtained from
The Cancer Genome Atlas (TCGA) data portal (up to May 27, 2016;
https://cancergenome.nih.gov/). After
excluding the data without complete survival information, a total
of 529 patients with RCC and 72 paired non-tumor renal samples were
enrolled in this study. We also downloaded the detailed clinical
information of the patients with RCC, including age, sex, tumor
grade, AJCC cancer stage, etc.
Cell culture and transfection
The human RCC cell lines (A-498, 786-O, Caki-1,
Caki-2 and ACHN), the normal renal epithelial cells HK-2, 293T cell
line were obtained from the American Tissue Culture Collection
(ATCC, Manassas, VA, USA). The cells were cultured in RPMI-1640 or
DMEM (Gibco/Thermo Fisher Scientific, Waltham, MA, USA)
supplemented with 10% FBS (Gibco-BRL, Gaithersburg, MD, USA), 100
U/ml penicillin and 100 mg/ml streptomycin (Gibco/Thermo Fisher
Scientific) in humidified air of 5% CO2 at 37°C.
Small interfering RNAs (siRNAs) specifically
targeting human HOTTIP, IGF-2 and negative control siRNA (siNC)
were synthesized by Life Technologies/Thermo Fisher Scientific. The
target HOTTIP siRNA sequence was 5′-UGGGAACCCG CUAUUUCACUCUAUU-3′.
The target IGF-2 siRNA sequence was 5′-GGGUUUUCUUUUGACUUAUTT-3′.
miR-615-3p mimics (5′-UCCGAGCCUGGGUCUCCCUCUU-3′), miR-615-3p
inhibitor (5′-ACCGAGUCAGGGAUACCCA CAA-3′), miR-615-3p mimic
negative control (5′-UUCUCGA ACGUGUCACGUUUU-3′) and miR-615-3p
inhibitor negative control (5′-CAGUACUUUUGUGUAGUACAA-3′) were
chemically synthesized by RiboBio (Guangzhou, China). The cells
were grown in 6-well plates to 70% confluence and transfected with
the siRNAs (50 nM), mimics (20 nM) or inhibitors (20 nM) for 48 h
using Lipofectamine 2000 (Invitrogen/Thermo Fisher Scientific)
according to the manufacturer’s instructions. The transfection
efficiency was examined by reverse transcription-quantitative PCR
(RT-qPCR).
Cell proliferation assay
Cell proliferation was measured by MTT assay.
Briefly, the cells were seeded into 96-well plates at 1,000
cells/well and incubated for 1, 2, 3 and 4 days. Subsequently, 20
µl of MTT (5 mg/ml; Sigma, St. Louis, MO, USA) were added to
each well and followed by incubation at 37°C for 4 h. Viable cells
were evaluated by absorbance measurements at 490 nm using a
microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Flow cytometric analysis
To analyze cell cycle progression, the cells fixed
and incubated with RNase A (0.25 mg/ml; Sigma) followed by
treatment with propidium iodide (PI; Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China). The cell cycle was analyzed using a
FACSCalibur flow cytometer (Beckman Coulter, Brea, CA, USA). For
apoptosis assay, the cells were seeded at a density of
1×106 cells/well and cultured in 6-well plates at 37°C
and transfected with HOTTIP siRNA. After 48 h, the cells,
including, the adhesive and floating cells were harvested and
washed twice with cold PBS and re-suspended in 1X binding buffer
(Invitrogen/Thermo Fisher Scientific). Subsequently, 10 µl
of Annexin V-FITC (Invitrogen/Thermo Fisher Scientific) and 5
µl of PI were added to each cell suspension. The
fluorescence of the stained cells was then analyzed by flow
cytometry.
Matrigel invasion assay
A Matrigel invasion chamber (BD Biosciences,
Bedford, MA, USA) was used for the Matrigel invasion assay. In
brief, the cells were seeded into the top chamber with serum-free
medium. The lower chamber was filled with 10% FBS. The cells were
incubated for 48 h and the cells that did not invade through the
pores were removed using a cotton swab. The cells on the lower
surface of the membrane were stained with crystal violet at room
temperature for 20 min (Sigma) and counted under a dissecting
microscope (Olympus Optical, Tokyo, Japan). The experiment was
repeated 3 times.
RT-qPCR
RNA was extracted from cells or clinical samples
using TRIzol reagent (Invitrogen/Thermo Fisher Scientific).
Subsequently, 1 µg RNA was reverse transcribed into cDNA
using PrimeScript RT-polymerase (Takara, Dalian, China).
Quantitative PCR (qPCR) was performed using the SYBR Premix Ex Taq™
kit (Takara). qPCR for mRNA expression was conducted as follows:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles of
annealing at 95°C for 15 sec and extension at 60°C for 30 sec. qPCR
for miRNA expression was conducted as follows: Initial denaturation
at 95°C for 3 min, followed by 40 cycles of annealing at 95°C for
12 sec and extension at 62°C for 60 sec. GAPDH was used as an
internal control. The results were quantified using the
2−ΔΔCq method (20).
The primers were synthesized by Ribobio Co. The corresponding
primers are listed in Table I.
 | Table ISequences of primers used for
RT-qPCR. |
Table I
Sequences of primers used for
RT-qPCR.
| Name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| HOTTIP |
CCTAAAGCCACGCTTCTTTG |
TGCAGGCTGGAGATCCTACT-3 |
| IGF-2 |
AGACCCTTTGCGGTGGAGA |
GGAAACATCTCGCTCGGACT |
| pri-miR-615 |
GCAAGTCGAGCATTTTACCTGC |
GCCATGTGTCCACTGAAATGTG |
| pre-miR-615 |
ACACTCCAGCTGGGTCCGAGCCTGGGTCTC |
TGGTGTCGTGGAGTCG |
| GAPDH |
CAGCCTCAAGATCATCAGCA |
TGTGGTCATGAGTCCTTCCA |
| U6 |
ATTGGAACGATACAGAGAAGATT |
GGAACGCTTCACGAATTTG |
Western blot analysis
Western blot analyses were performed as previously
described (21). In brief, the
cells were lysed by a radio immunoprecipitation assay (RIPA) buffer
and extracted using Mammalian Protein Extraction Reagent (both from
Thermo Fisher Scientific). The concentration of the protein was
determined using the Bradford Protein Assay kit (Beyotime Institute
of Biotechnology). Protein lysates (30-50 µg) were separated
on a 10% SDS/PAGE gel and transferred onto polyvinylidene fluoride
membranes (PVDF; Millipore, Billerica, MA, USA). The membranes were
then blocked with 5% non-fat milk at room temperature for 1.5 h,
and the membranes were then incubated with primary antibody at 4°C
overnight, followed by 1 h of incubation with anti-mouse
IgG/horseradish peroxidase-conjugated secondary antibody (sc-2380;
Santa Cruz Biotechnology, Dallas, TX, USA) at room temperature.
Immunocomplexes were visualized by chemiluminescence using ECL
(Santa Cruz Biotechnology). The relative band intensity was
calculated using ImageJ software (version 1.50; National Institutes
of Health, Bethesda, MD, USA). The following antibodies and
dilutions were used: IGF-2 (1:2,000; cat. no. sc-293176, mouse
mAb), GAPDH (1:2,000; cat. no. sc-47724, mouse mAb) (both from
Santa Cruz Biotechnology).
Luciferase reporter assays
To determine whether IGF-2 is a direct target of
miR-615, Luciferase reporter vectors (control), or vectors
containing wild-type (pGL3-IGF-2-3′-UTR-Full or mutated
(pGL3-IGF-2-3′-UTR-Mut) 3′-UTRs of IGF-2 mRNA (Shanghai GenePharma
Co., Ltd.) were co-transfected into 293T cells with miR-615 mimics,
using Lipofectamine 2000. To confirm whether HOTTIP can interact
with miR-615 HOTTIP-WT/ HOTTIP-MUT were examined through PCR and a
mutagenesis kit. The fragments, including the predicted binding
sites, were cloned into a pmirGLO vector (Promega, Madison, WI,
USA) as pmirGLO-HOTTIP-WT/pmirGLO-HOTTIP-MUT plasmids. All the
plasmids were constructed by GeneChem (Shanghai, China).
pmirGLO-HOTTIP-WT/pmirGLO-HOTTIP-MUT were co-transfected with
miR-615 mimics into 293T cells. At 48 h post-transfection, the
cells were harvested and then assayed for reporter gene activities
using the Dual-Luciferase Reporter Assay System (Promega) according
to the manufacturer’s instructions.
miRNA target gene prediction
To identify the possible molecular mechanisms of
action of the miRNA and the miRNA target, we retrieved putative
miRNA-mRNA interactions by prediction algorithms from TargetScan
(http://www.targetscan.org/vert_71/).
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 software package (SPSS, Chicago, IL, USA). The
significance of differences between groups was estimated by one-way
ANOVA, followed by LSD post-hoc tests. The results are reported as
the means ± SD. A Chi-square test were used to compare the
associations between HOTTIP expression and the patient
clinicopathological parameters. Wilcoxon signed-rank test was used
to compare the differences of HOTTIP expression between normal and
cancer from TCGA database. Survival analyses were performed using
the Kaplan-Meier method and survival differences were estimated
using the log-rank test. The correlations of different genes were
assessed by Pearson’s correlation analysis. Statistical
significance was assigned at P<0.05. All experiments were
performed at least 3 times with triplicate samples.
Results
Association between HOTTIP expression and
patient clinicopathological characteristics
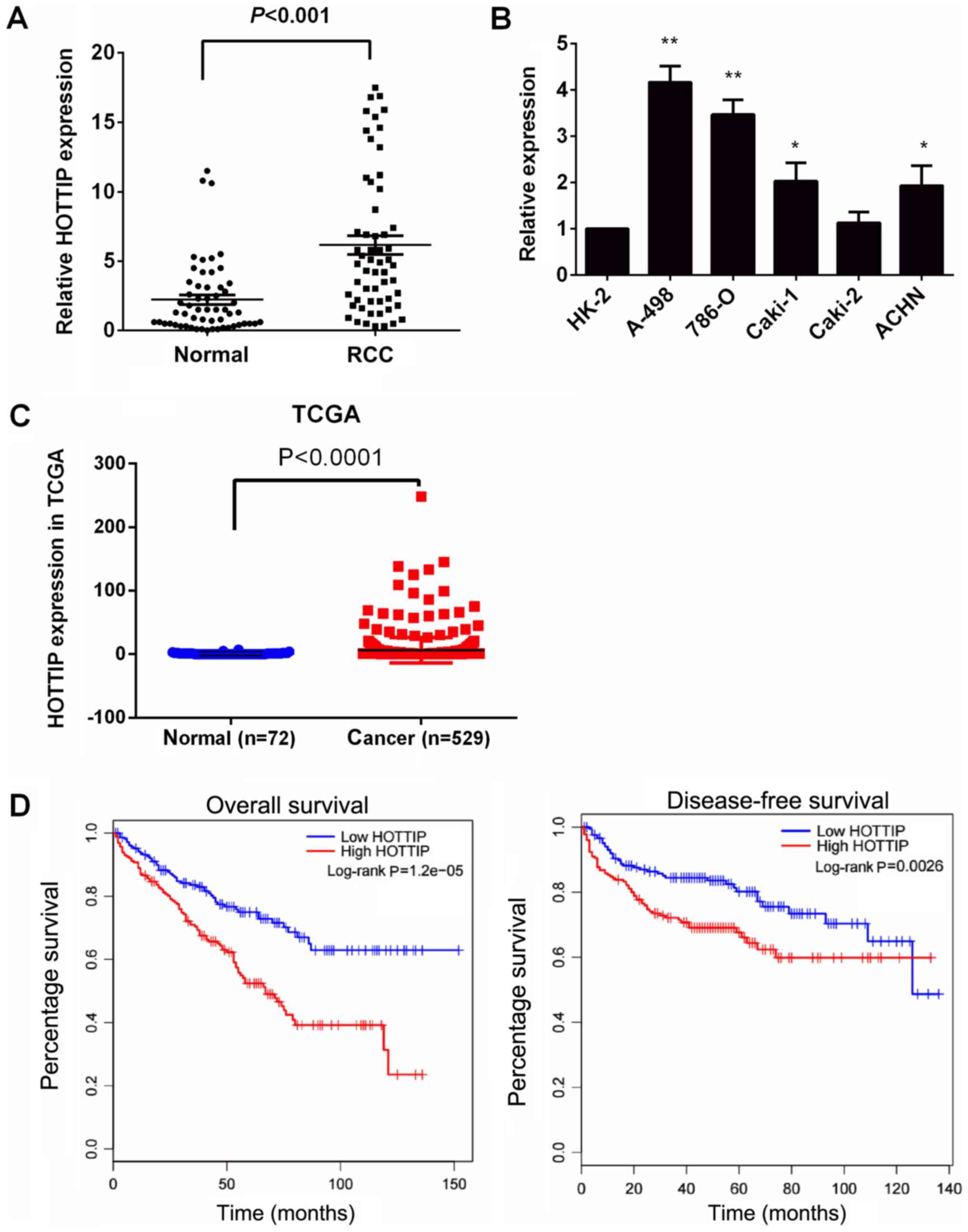
Initially, RT-qPCR was used to examine HOTTIP
expression in the 57 matched tissues and a panel of RCC cell lines
(A-498, 786-O, Caki-1, Caki-2 and ACHN). The results revealed that
HOTTIP expression was significantly increased in all renal cancer
tissues compared with the matched normal kidney tissues (Fig. 1A). HOTTIP expression was also
significantly increased in four RCC cell lines (A-498, 786-O,
Caki-1 and ACHN), but not in the Caki-2 cell lines, compared with
that in the normal prostate epithelial cell line, HK-2 (Fig. 1B, P<0.05). The A-498 and 786-O
cells exhibited the highest HOTTIP expression. Thus, the A-498 and
786-O cells were used as a model to perform the following
experiments to examine the effects of HOTTIP on the proliferation,
apoptosis, migration and invasion of RCC cells in vitro.
We then investigated the association between HOTTIP
expression with the clinicopathological characteristics of patients
with RCC from the TCGA database. Consistently, the quantification
of the HOTTIP levels demonstrated significantly higher expression
levels of HOTTIP in the tumors compared with the matched adjacent
normal tissues (Fig. 1C,
P<0.05). According to the median of the HOTTIP expression value
in TCGA, the samples were divided into the high and low expression
groups. As presented in Table II,
an increased HOTTIP expression was significantly associated with a
high TNM stage (P =0.003), tumor size (P=0.001), and pathological
grade (P=0.005). However, HOTTIP protein expression was not
associated with other clinicopathological characteristics such as
sex, lymph node involvement and distant metastasis. In addition,
patients with RCC with high HOTTIP expression levels had a shorter
OS and DFS than those with low HOTTIP expression levels (Fig. 1D), as shown by Kaplan-Meier
survival analysis. These results demonstrated that a high
expression level of HOTTIP was associated with a poor prognosis in
RCC. The upregulation of HOTTIP may thus play an important role in
RCC development and progression.
 | Table IIAssociation of HOTTIP expression with
clinicopathological characteristics of patients with renal cell
carcinoma from the TCGA database. |
Table II
Association of HOTTIP expression with
clinicopathological characteristics of patients with renal cell
carcinoma from the TCGA database.
| Covariate | Classification | HOTTIP
|
|---|
| Low expression, n
(%) | High expression, n
(%) | P-value |
|---|
| Age, years | <61 | 102 (19.28) | 179 (33.84) | 0.775 |
| ≥61 | 93 (17.58) | 155 (29.30) | |
| Sex | Female | 71 (13.42) | 114 (21.55) | 0.596 |
| Male | 124 (23.44) | 220 (41.59) | |
| Pathological
grade | I-II | 104 (19.77) | 137 (26.05) | 0.005 |
| III-IV | 89 (16.92) | 196 (37.26) | |
| Lymph node
involvement | NO-N1 | 88 (16.64) | 151 (28.54) | 0.986 |
| N2-NX | 107 (20.23) | 183 (34.59) | |
| Distant
metastasis | M0 | 162 (30.74) | 258 (48.96) | 0.139 |
| M1-MX | 33 (6.26) | 74 (14.04) | |
| Tumor T (size) | T1-T2 | 145 (27.41) | 200 (37.81) | 0.001 |
| T3-T4 | 50 (9.45) | 134 (25.33) | |
| TNM stage | I-II | 135 (25.67) | 187 (35.55) | 0.003 |
| III-IV | 59 (11.22) | 145 (27.57) | |
Effect of HOTTIP on the viability, cell
cycle progression, apoptosis and invasion of renal cancer
cells
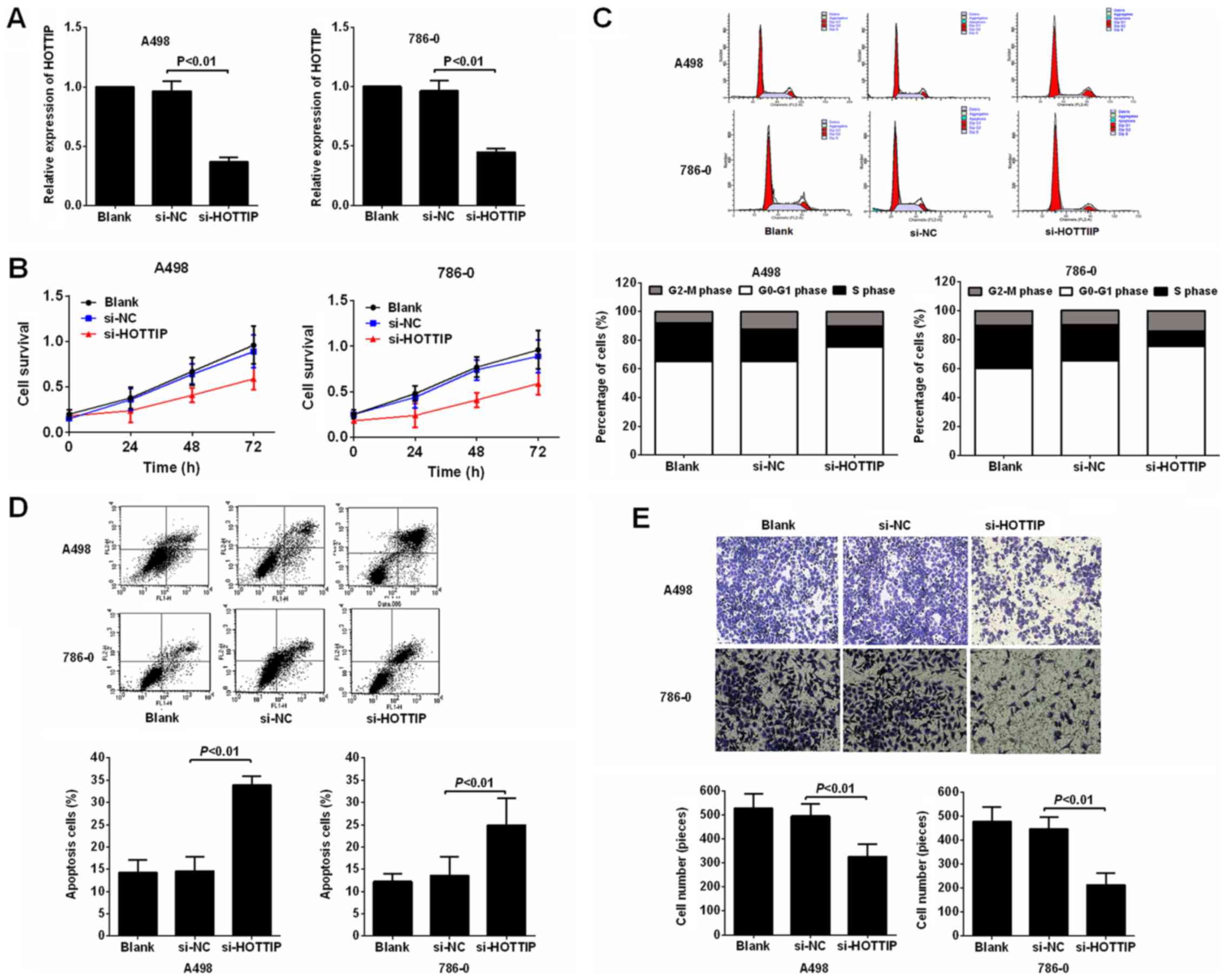
Subsequently, the knockdown of HOTTIP using siRNA in
the A-498 and 786-O cells was performed to evaluated the biological
role of HOTTIP in RCC cell proliferation, migration and invasion.
The results of RT-qPCR performed at 48 h post-transfection
confirmed the silencing efficiency. Following transfection with
HOTTIP siRNA (si-HOTTIP), HOTTIP expression was downregulated in
the A-498 and 786-O cells (Fig.
2A). The results of MTT assay then revealed that cell viability
was suppressed following transfection with si-HOTTIP compared with
the negative control in both the A-498 and 786-O cell lines
(Fig. 2B). To further investigate
the observed inhibition of proliferation following HOTTIP
knockdown, we compared the cell cycle profiles of the cells in
which HOTTIP was knocked down by flow cytometry. The results
revealed that the inhibition of HOTTIP led to a decrease in the
number of cells in the S phase and an increase in the percentage of
cells in the G0/G1 phase (Fig.
2C).
Some lncRNAs have been reported to play important
roles in tumor cell apoptosis (22,23).
Thus, in this study, to examine the effects of HOTTIP on RCC cell
apoptosis, the percentage of apoptotic cells was measured by flow
cytometry with PI and Annexin V double staining. The results
revealed that the apoptotic population was notably increased in the
A-498 and 786-O cells in which HOTTIP was knocked down compared
with the controls (Fig. 2D).
Subsequently, we examined the effects of HOTTIP knockdown on cell
invasion. We performed a Matrigel assay, as shown in Fig. 2E. The knockdown of HOTTIP induced a
significant decrease in invasiveness compared with the controls.
Collectively, these results clearly indicated that the
downregulation of HOTTIP markedly suppressed the proliferation and
invasion of RCC cells in vitro, and promoted cell
apoptosis.
Reciprocal repression of HOTTIP and
miR-615
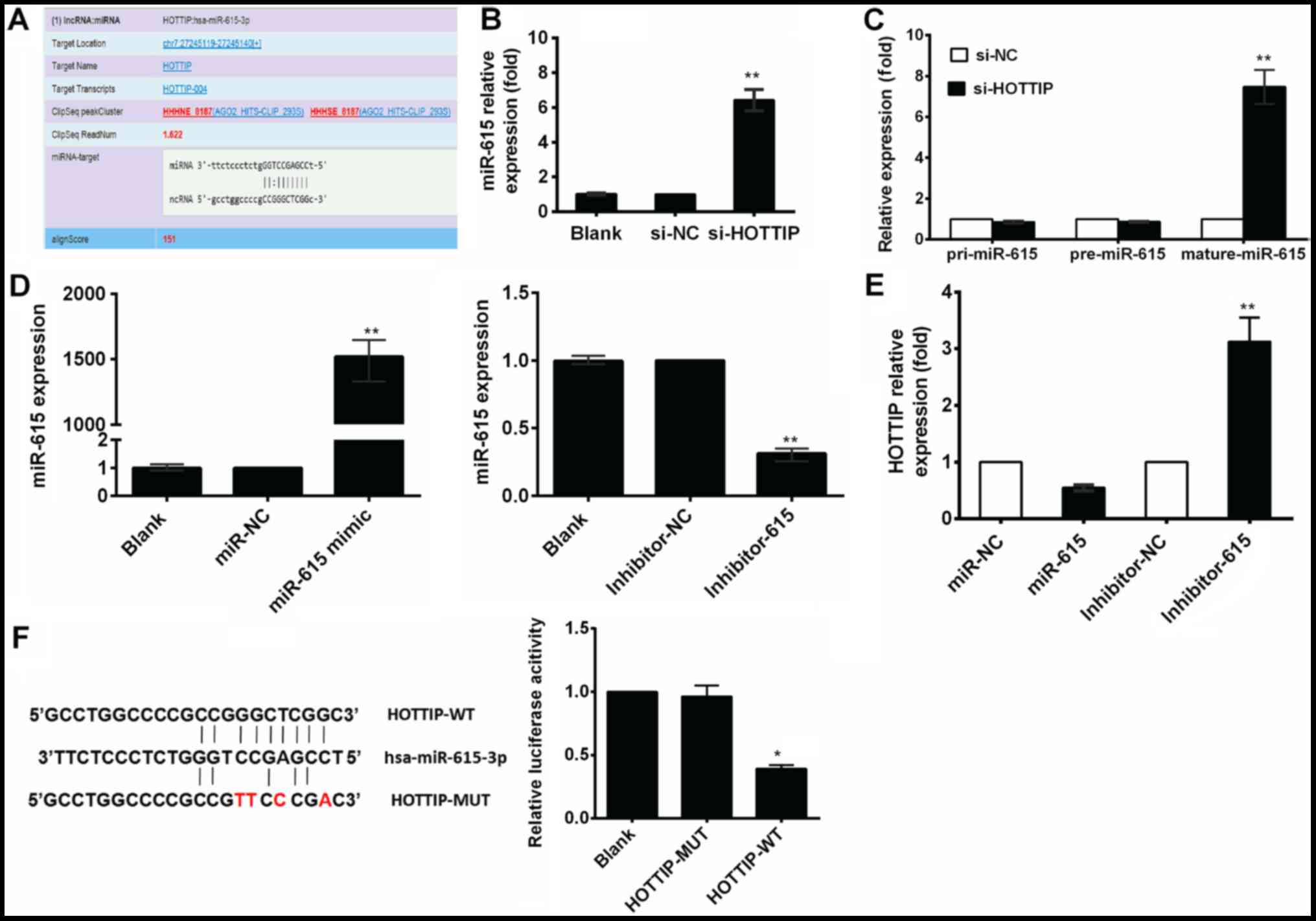
As stated in the Introduction, increasing numbers of
studies have suggested that lncRNAs may act as ceRNAs in regulating
the biological functions of miRNAs. In this study, to further
explore the underlying mechanisms of action of HOTTIP and its role
in the development of RCC, we first examined a set of miRNAs that
were predicted to bind HOTTIP using TargetScan. Bioinformatics
analysis revealed that the HOTTIP transcript contained putative
binding sites for miR-615-3p (Fig.
3A). To further examine this hypothesis, we performed RT-qPCR
assay to detect the changes in miR-615 expression in the A-498 and
786-O cells transfected with si-HOTTIP, and found that the
expression of miR-615 was increased following transfection with
si-HOTTIP (Fig. 3B). To explore
the underlying mechanisms of the regulation of miR-615 expression
by HOTTIP, we examined the effects of HOTTIP on the expression
levels of pri-miR-615 and pre-miR-615 by RT-qPCR. The expression
levels of pri-miR-615 or pre-miR-615 were not affected by si-HOTTIP
(Fig. 3C), suggesting that the
HOTTIP-mediated regulation of miR-615 was likely achieved through a
post-transcriptional mechanism. To determine whether miR-615
negatively and reciprocally regulated HOTTIP, miR-615 mimic or
inhibitor were transfected into the A-498 cells (Fig. 3D), and a significant inhibition of
HOTTIP expression by miR-615 mimic was observed. Conversely, the
miR-615 inhibitor markedly increased HOTTIP expression (Fig. 3E).
Moreover, we also performed a
dual-luciferase reporter assay to investigate the potential
interaction of HOTTIP with miR-615
We obtained HOTTIP-WT/HOTTIP-MUT through PCR and a
mutagenesis kit. The fragments, including the predicted binding
sites, were cloned into a pmirGLO vector as
pmirGLO-HOTTIP-WT/pmirGLO-HOTTIP-MUT. The recombinant plasmids were
transiently co-transfected with a miR-615 mimic and scrambled
oligonucleotides into 293T cells. Our results demonstrated miR-615
overexpression reduced the luciferase activity of the
pmirGLO-HOTTIP-WT reporter vector, but not that of the empty vector
or pmirGLO-HOTTIP-MUT reporter vector (Fig. 3F). These results suggested that the
interaction between HOTTIP and miR-615 had reciprocal effects.
The oncogenic functions of HOTTIP are
partially mediated through the negative regulation of miR-615
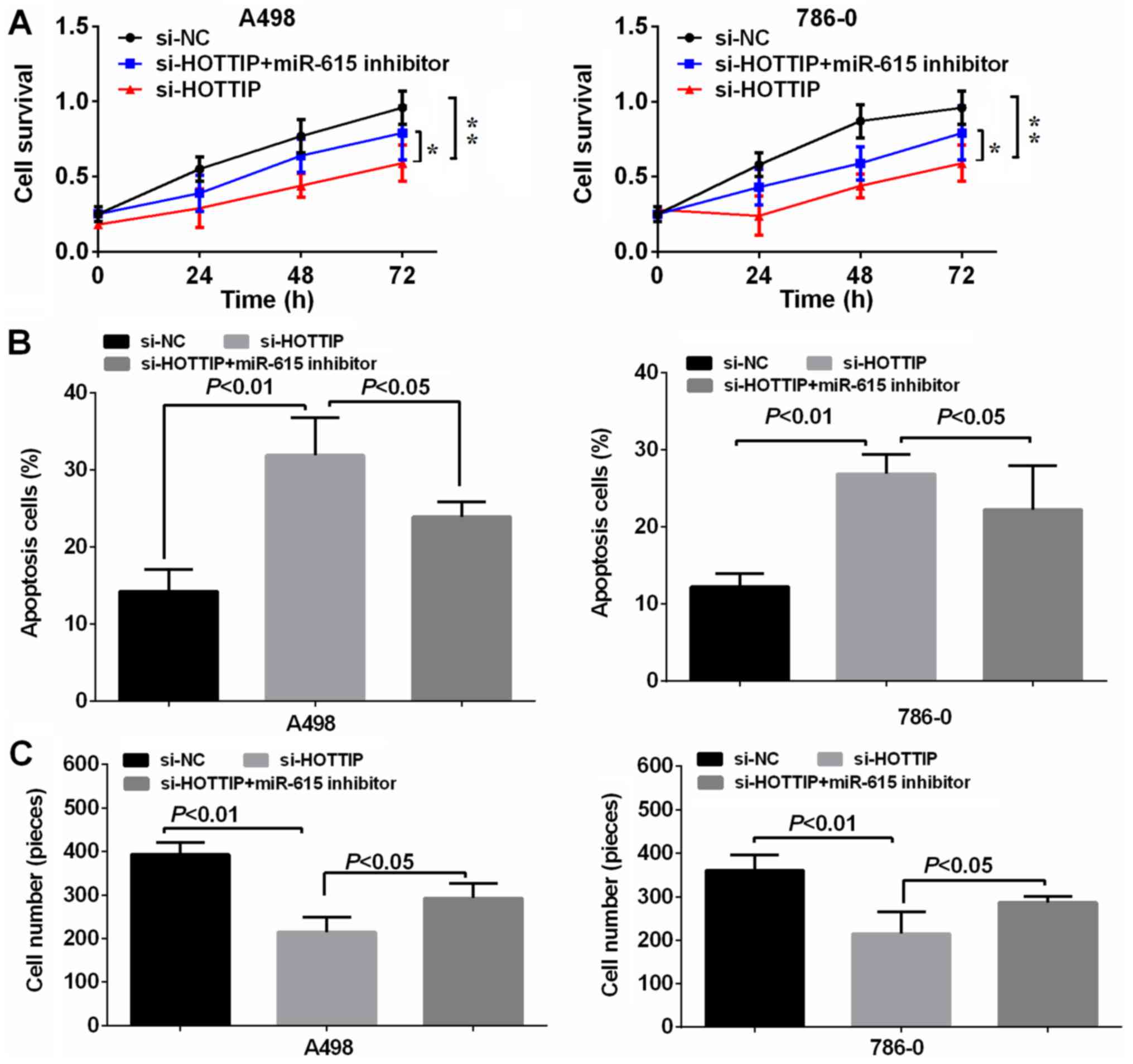
In order to verify the reciprocal repression of
HOTTIP and miR-615, and to determine whether HOTTIP exerts its
biological functions through miR-615, we performed a rescue
experiment. We co-transfected si-HOTTIP and miR-615 inhibitor into
the A-498 and 786-O cell lines. The results revealed that the
miR-615 inhibitor significantly reversed the suppressive effects
exerted by the knockdown of HOTTIP on cell proliferation and
invasion, and the promoting effects on the apoptosis of A-498 and
786-O cells (Fig. 4), suggesting
that HOTTIP plays its oncogenic role in RCC cells through
miR-615.
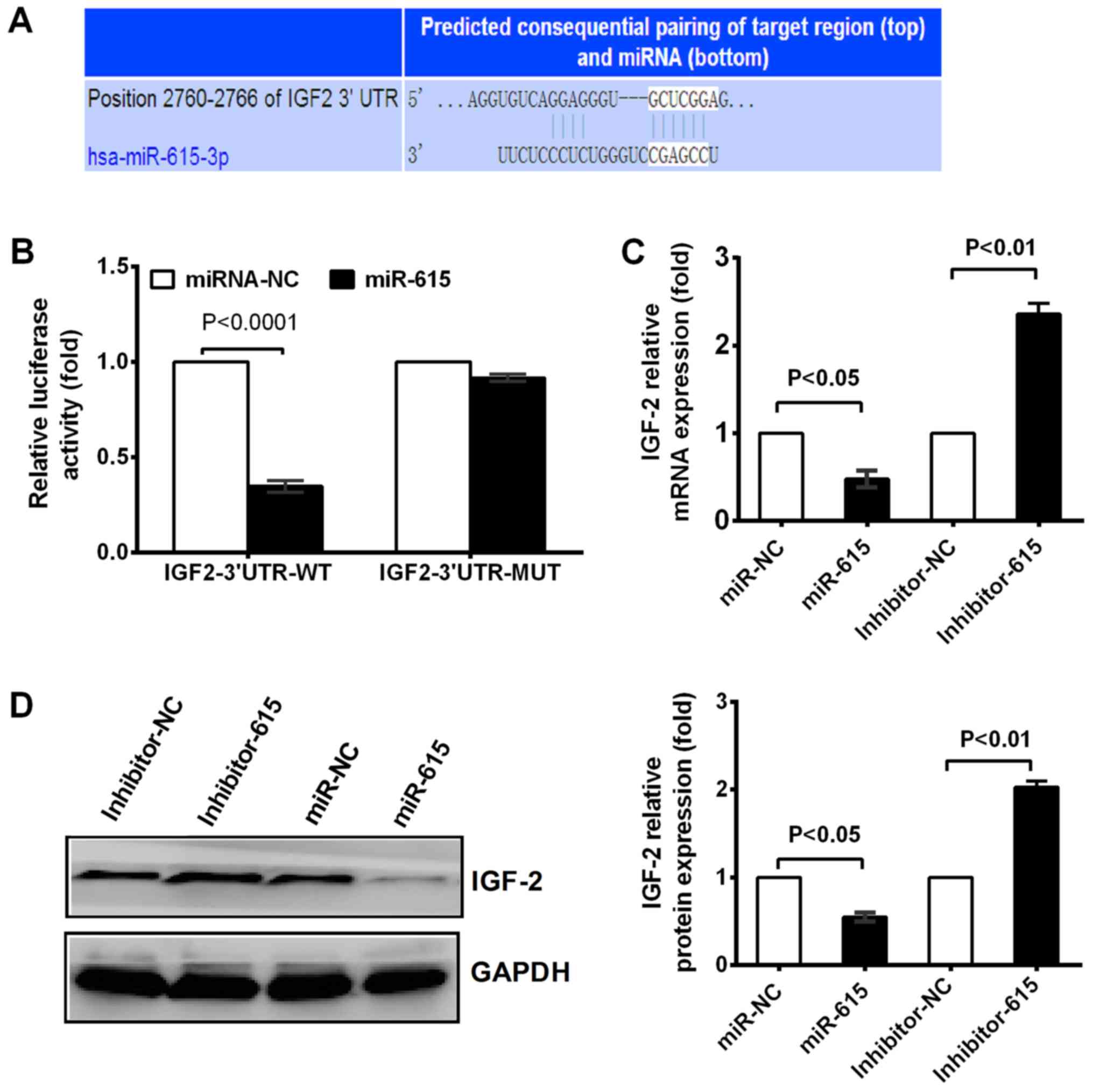
IGF-2 is a direct target of miR-615
Based on the above-mentioned results, we aimed to
identify the main target genes of miR-615, which were predicted
using the TargetScan website. IGF-2 was predicted to be one of the
targets of miR-615 (Fig. 5A). As
it has been reported that IGF-2 is involved in cancer progression
(24,25)], in this study, we thus selected
IGF-2 as our target gene to study the mechanisms responsible for
the HOTTIP-induced progression of RCC mediated by miR-615. The
results of luciferase reporter assay revealed that miR-615 mimics
significantly reduced luciferase activity compared to miR-615 NC in
the wild-type IGF-2 construct, whereas the luciferase activity in
the IGF-2-MUT construct exhibited no change in the cells
transfected with miR-615 mimics and miR-615 NC group (Fig. 5B), suggesting that miR-615 directly
binds to IGF-2-3′-UTR. Consistent with the results of reporter
assay, the IGF-2 mRNA and protein expression levels were decreased
in the presence of miR-615 mimic in the A-498 cells. Conversely,
when miR-615 expression was inhibited, the opposite effects were
observed (Fig. 5C and D). These
results indicated that IGF-2 was a direct target of miR-615.
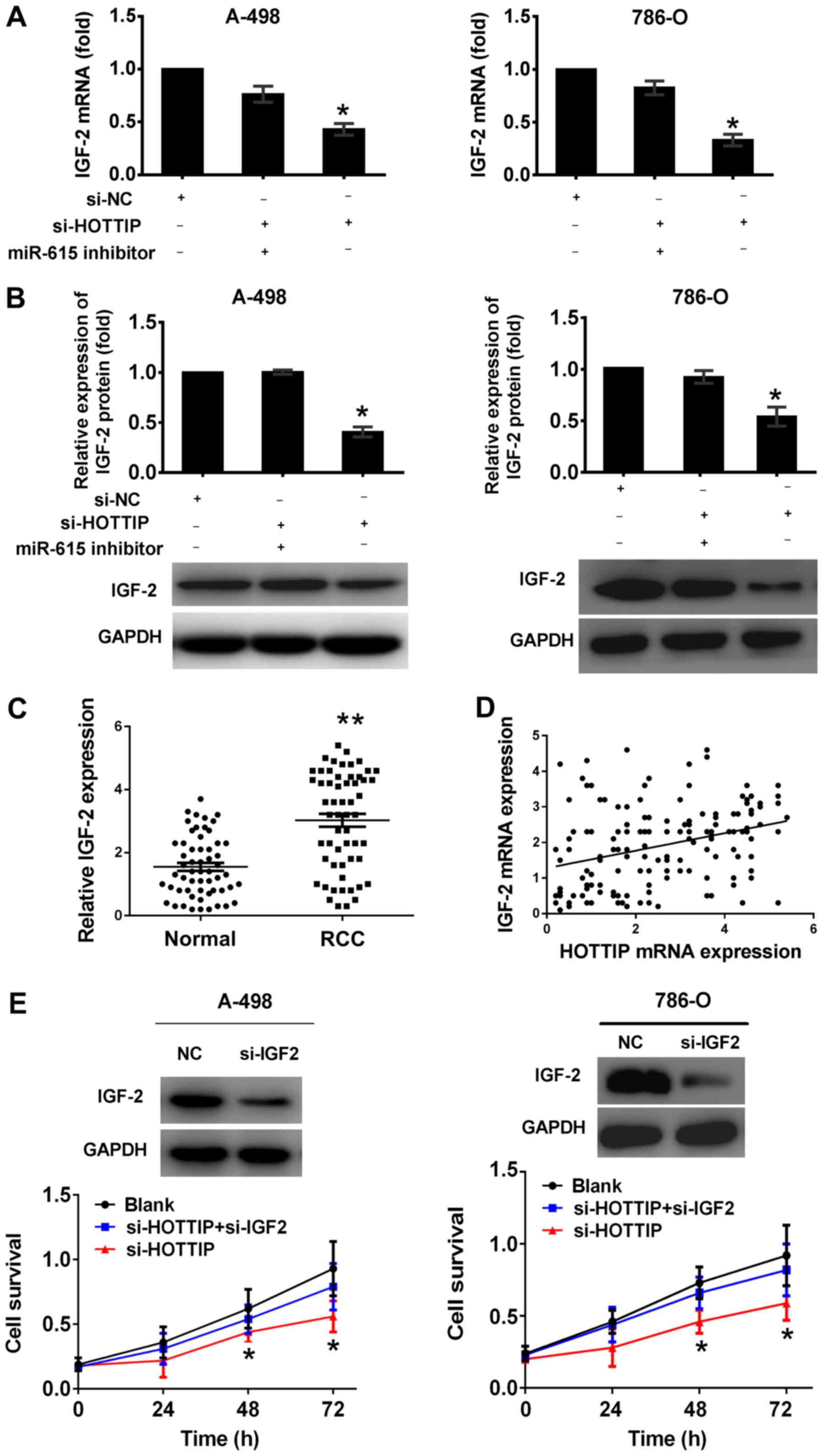
HOTTIP regulates IGF-2 expression in an
miR-615-dependent manner in RCC cells
According to the ceRNA concept, lncRNAs can act as
ceRNAs to carry out their regulatory functions (26). In this study, we found that HOTTIP
shared regulatory miR-615 binding sites with IGF-2 (Figs. 3A and 5A). To determine whether HOTTIP interacts
with miR-615 to regulate IGF-2 expression, we examined the mRNA and
protein levels of IGF-2. The inhibition of HOTTIP significantly
decreased the expression of IGF-2 at the mRNA and protein level,
and this effect was significantly attenuated by miR-615 inhibitor
(Fig. 6A and B), indicating that
HOTTIP regulated IGF-2 expression in a miR-615-dependent
manner.
Furthermore, we assessed the correlation between
HOTTIP mRNA and IGF-2 expression in RCC tissues. Compared with
normal kidney tissues, the mRNA levels of IGF-2 were significantly
higher in renal cancer tissues (Fig.
6C). In addition, we found that HOTTIP expression significantly
and positively correlated with IGF-2 expression (two-sided Pearson’
s correlation, r=0.314, P<0.0001; Fig. 6D).
To further examine whether HOTTIP carries out its
biological functions through IGF-2, we performed rescue experiments
by inhibiting IGF-2 expression in the cells in which HOTTIP was
knocked down (Fig. 6E). MTT assay
revealed that cell growth was reduced in the RCC cells in which
HOTTIP was knocked down, whereas si-IGF-2 partially reversed the
reduction of the cell proliferative ability (Fig. 6E).
Discussion
lncRNAs have been found to be involved in many
biological processes, including carcinogenesis (27,28).
In RCC, several lncRNAs have been reported to play important roles
in tumorigenesis (29-32). For instance, Linc00152 is a
positive prognostic factor in RCC. Hirata et al reported
that MALAT1 promoted aggressive RCC through Ezh2 and interacted
with miR-205 (32). In this study,
we investigated the functional role and clinical significance of
lncRNA HOTTIP in RCC. In several types of cancer, HOTTIP has been
reported to be a potent oncogene (14,17,33-35).
In the present study, our results confirmed that HOTTIP expression
was significantly higher in RCC tissues and cell lines. A high
HOTTIP expression was also associated with a poor clinical outcome
in patients with RCC. The downregulation of HOTTIP inhibited cell
proliferation and invasion, and promoted the apoptosis of the RCC
cells. In addition, we found that HOTTIP had a dual effect on cell
proliferation and invasive ability. However, we did not determine
whether the reduced invasive ability of the si-HOTTIP-transfected
cells was due to reduced viability and/ or increased apoptosis. On
the whole, however, our results suggest that HOTTIP functions as an
oncogene and may thus be a potential therapeutic target for
RCC.
Although HOTTIP has been suggested to act as an
oncogene in different cancers, little is known about the underlying
molecular mechanism. ceRNA theory is the most important theory
regarding lncRNA, and lncRNA has been shown to be a sponge for
regulating the expression and function of miRNA.
It has reported been that lncRNAs play a crucial
role in multiple processes in cells by acting as ceRNAs to regulate
miRNAs (9). In this study, we
discovered that HOTTIP and miR-615 negatively regulated each other
and were involved in the ceRNA regulatory network. HOTTIP knockdown
increased miR-615 expression due to HOTTIP functioning as an
endogenous miRNA sponge to bind to miR-615; the inhibition of
HOTTIP abrogated this sponge, leading to an increase in miR-615
expression. Recent studies have indicated that miR-615 plays a
tumor suppressive role in hepatoma cells (36), breast cancer cells (37) and pancreatic ductal adenocarcinoma
(38). However, its role on RCC
has not yet been investigated, at least to the best of our
knowledge. In this study, we provide evidence that miR-615 targets
not only protein-coding genes, but also the lncRNA HOTTIP. Hence,
the identification of HOTTIP as an miR-615 target expands the
repertoire of miR-615 targets. Furthermore, HOTTIP is able to
repress miR-615, forming a reciprocal negative regulatory loop.
These results provide further supporting evidence of the ceRNA
regulatory network. Moreover, we found that HOTTIP carried out is
oncogenic function through miR-615 in RCC cells. Taken together,
these data strongly suggest that HOTTIP directly targets miR-615
and affects the biological characteristics of RCC cells by
negatively regulating miR-615.
The IGF pathway is frequently activated in a variety
of cancers (39). IGF-2 is
maternally imprinted in the majority of normal tissues, with only
the paternal allele expressed (40). In many tumors, however, this
imprinting is lost, leading to the biallelic expression of the
gene. The overproduction of the growth factor promotes the
malignant behavior of tumor cells (41,42).
The ceRNA hypothesis indicates that lncRNAs may functions as ceRNAs
by acting as endogenous decoys for miRNAs, which in turn affects
the binding of miRNAs on their targets. In this study, the results
of reporter assays indicated that IGF-2 was a direct target of
miR-61 and that miR-615 downregulated the activity of the
IGF-2-3′-UTR in RCC cells. Furthermore, the overexpression of
miR-615 decreased IGF-2 expression in A-498 cells. In agreement
with HOTTIP being a decoy for miR-615, we proved that the
inhibition of HOTTIP significantly decreased IGF-2 expression,
which was partially reversed in the presence of miR-615 inhibitor.
Furthermore, we found that si-IGF-2 partially reversed the
reduction of cell proliferation ability induced by si-HOTTIP.
Therefore, the HOTTIP/miR-615/IGF-2 axis may play an important role
in RCC progression.
In conclusion, in this study we demonstrated that
HOTTIP contributed to carcinogenesis and functioned as a miRNA
sponge to attenuate the endogenous function of miR-615, which
negatively modulated IGF-2 expression in RCC. Due to this crucial
role which HOTTIP plays in the progression of RCC, it may thus
serve as a therapeutic target, as well as prognostic biomarker for
RCC.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
QL and XC conceived and designed the research; QW,
GW, ZZ and QT organized, analyzed and interpreted the data; QW, GW,
ZZ, QT, WZ, XC and FC performed the experiments. QW and GW drafted
and revised the manuscript; QL and XC supervised the study. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The use of tissues for this study was approved by
the Medical Ethics Committee of Dalian Medical University. Due to
the retrospective nature of the study, the Ethics Committee waived
the need of written informed consent by the patients. All the
samples were anonymous.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Linehan WM: Genetic basis of kidney
cancer: Role of genomics for the development of disease-based
therapeutics. Genome Res. 22:2089–2100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Porta C, Giglione P and Paglino C:
Targeted therapy for renal cell carcinoma: Focus on 2nd and 3rd
line. Expert Opin Pharmacother. 17:643–655. 2016. View Article : Google Scholar
|
|
4
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
8
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ntziachristos P, Abdel-Wahab O and
Aifantis I: Emerging concepts of epigenetic dysregulation in
hematological malignancies. Nat Immunol. 17:1016–1024. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ong CT and Corces VG: Enhancer function:
New insights into the regulation of tissue-specific gene
expression. Nat Rev Genet. 12:283–293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie H, Zhu D, Xu C, Zhu H, Chen P, Li H,
Liu X, Xia Y and Tang W: Long none coding RNA HOTTIP/HOXA13 act as
synergistic role by decreasing cell migration and proliferation in
Hirschsprung disease. Biochem Biophys Res Commun. 463:569–574.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lian Y, Cai Z, Gong H, Xue S, Wu D and
Wang K: HOTTIP: A critical oncogenic long non-coding RNA in human
cancers. Mol Biosyst. 12:3247–3253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao
Z, Li Q and Zhang W: Up-regulation of long non-coding HOTTIP
functions as an oncogene by regulating HOXA13 in non-small cell
lung cancer. Am J Transl Res. 8:2022–2032. 2016.PubMed/NCBI
|
|
15
|
Wang SS, Wuputra K, Liu CJ, Lin YC, Chen
YT, Chai CY, Lin CS, Kuo KK, Tsai MH, Wang SW, et al: Oncogenic
function of the homeobox A13-long noncoding RNA HOTTIP-insulin
growth factor-binding protein 3 axis in human gastric cancer.
Oncotarget. 7:36049–36064. 2016.PubMed/NCBI
|
|
16
|
Ren YK, Xiao Y, Wan XB, Zhao YZ, Li J, Li
Y, Han GS, Chen XB, Zou QY, Wang GC, et al: Association of long
non-coding RNA HOTTIP with progression and prognosis in colorectal
cancer. Int J Clin Exp Pathol. 8:11458–11463. 2015.PubMed/NCBI
|
|
17
|
Zhang SR, Yang JK, Xie JK and Zhao LC:
Long noncoding RNA HOTTIP contributes to the progression of
prostate cancer by regulating HOXA13. Cell Mol Biol
(Noisy-le-grand). 62:84–88. 2016.
|
|
18
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ergun S and Oztuzcu S: Oncocers:
ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways.
Tumour Biol. 36:3129–3136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
21
|
Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L
and Wang Z: Epigenetic silencing of miR-130b in ovarian cancer
promotes the development of multidrug resistance by targeting
colony-stimulating factor 1. Gynecol Oncol. 124:325–334. 2012.
View Article : Google Scholar
|
|
22
|
Wang Y, Zhang L, Zheng X, Zhong W, Tian X,
Yin B, Tian K and Zhang W: Long non-coding RNA LINC00161 sensitises
osteosarcoma cells to cisplatin-induced apoptosis by regulating the
miR-645-IFIT2 axis. Cancer Lett. 382:137–146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He A, Liu Y, Chen Z, Li J, Chen M, Liu L,
Liao X, Lv Z, Zhan Y, Zhuang C, et al: Over-expression of long
noncoding RNA BANCR inhibits malignant phenotypes of human bladder
cancer. J Exp Clin Cancer Res. 35:1252016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Y, Chen Y, Chen L, Liu Z, Ieong ML, Gao
F and Huang W: Promotion of Insulin-like growth factor II in cell
proliferation and epithelial-mesenchymal transition in
hepatocellular carcinoma. J Cancer Res Ther. 14:844–850. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kessler SM, Haybaeck J and Kiemer AK:
Insulin-like growth factor 2 - The oncogene and its accomplices.
Curr Pharm Des. 22:5948–5961. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J,
et al: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie W, Yuan S, Sun Z and Li Y: Long
noncoding and circular RNAs in lung cancer: Advances and
perspectives. Epigenomics. 8:1275–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shukla S, Zhang X, Niknafs YS, Xiao L,
Mehra R, Cieślik M, Ross A, Schaeffer E, Malik B, Guo S, et al:
Identification and Validation of PCAT14 as Prognostic Biomarker in
Prostate Cancer. Neoplasia. 18:489–499. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su H, Sun T, Wang H, Shi G, Zhang H, Sun F
and Ye D: Decreased TCL6 expression is associated with poor
prognosis in patients with clear cell renal cell carcinoma.
Oncotarget. 8:5789–5799. 2017.
|
|
30
|
Wu Y, Tan C, Weng WW, Deng Y, Zhang QY,
Yang XQ, Gan HL, Wang T, Zhang PP, Xu MD, et al: Long non-coding
RNA Linc00152 is a positive prognostic factor for and demonstrates
malignant biological behavior in clear cell renal cell carcinoma.
Am J Cancer Res. 6:285–299. 2016.PubMed/NCBI
|
|
31
|
Ellinger J, Alam J, Rothenburg J, Deng M,
Schmidt D, Syring I, Miersch H, Perner S and Müller SC: The long
non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients
with clear cell renal cell carcinoma. Am J Cancer Res. 5:2799–2807.
2015.PubMed/NCBI
|
|
32
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ye H, Liu K and Qian K: Overexpression of
long noncoding RNA HOTTIP promotes tumor invasion and predicts poor
prognosis in gastric cancer. Onco Targets Ther. 9:2081–2088.
2016.PubMed/NCBI
|
|
34
|
Li F, Cao L, Hang D, Wang F and Wang Q:
Long non-coding RNA HOTTIP is up-regulated and associated with poor
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
8:11414–11420. 2015.PubMed/NCBI
|
|
35
|
Chang S, Liu J, Guo S, He S, Qiu G, Lu J,
Wang J, Fan L, Zhao W and Che X: HOTTIP and HOXA13 are oncogenes
associated with gastric cancer progression. Oncol Rep.
35:3577–3585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Z, Wang X, Liu R, Chen L, Yi J, Qi B,
Shuang Z, Liu M, Li X, Li S, et al: KDM4B-mediated epigenetic
silencing of miRNA-615-5p augments RAB24 to facilitate malignancy
of hepatoma cells. Oncotarget. 8:17712–17725. 2017.
|
|
37
|
Bai Y, Li J, Li J, Liu Y and Zhang B:
MiR-615 inhibited cell proliferation and cell cycle of human breast
cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med.
8:3801–3808. 2015.PubMed/NCBI
|
|
38
|
Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M,
Kondo Y, Shinjo K, Zhu Y, Zhang J, et al: miR-615-5p is
epigenetically inactivated and functions as a tumor suppressor in
pancreatic ductal adenocarcinoma. Oncogene. 34:1629–1640. 2015.
View Article : Google Scholar
|
|
39
|
Heidegger I, Pircher A, Klocker H and
Massoner P: Targeting the insulin-like growth factor network in
cancer therapy. Cancer Biol Ther. 11:701–707. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rainier S, Johnson LA, Dobry CJ, Ping AJ,
Grundy PE and Feinberg AP: Relaxation of imprinted genes in human
cancer. Nature. 362:747–749. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen W, Feng Y, Zhao Q, Zhu Z and Dimitrov
DS: Human monoclonal antibodies targeting nonoverlapping epitopes
on insulin-like growth factor II as a novel type of candidate
cancer therapeutics. Mol Cancer Ther. 11:1400–1410. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ji Y, Wang Z, Li Z, Huang N, Chen H, Li B
and Hui B: Silencing IGF-II impairs C-myc and N-ras expressions of
SMMC-7721 cells via suppressing FAK/PI3K/Akt signaling pathway.
Cytokine. 90:44–53. 2017. View Article : Google Scholar
|