Introduction
The ability of tumor cells to move through tissues
is required for local invasion and dissemination from the primary
tumor (1,2). Cellular movement depends on the
establishment of physical forces by means of protrusive forces and
traction forces, leading to membrane extensions and contractions
(3). These deformations are driven
by the spatially and temporally controlled assembly of the
cytoskeleton (4,5). Cell biological analyses have revealed
that the actin filament is mainly involved in the generation of the
forces responsible for cell motility by elongating or shortening at
specific sites on the membrane (5). This type of actin cytoskeleton is
regulated by a variety of actin-binding proteins; among these,
cofilin 1 plays a critical role in actin filament turnover by
interacting with monomeric actin, decreasing the availability of
polymerization-competent actin subunits (6,7). The
activity of cofilin 1 is controlled by the phosphorylation and
deposphorylation at Ser-3 by LIMK family protein kinases (8,9).
LIMK family is composed of two members, LIMK1 and LIMK2. Both of
these are protein kinases regulating the polymerization of actin
and decomposition of microtubules (10,11).
Recent findings have strongly indicated that LIMKs play important
roles in tumor cell invasion and metastasis (12-14).
Therefore, the understanding of the functions of LIMK as regards
the fine regulation of the balance between phosphorylated and
non-phosphorylated cofilin 1 may aid and accelerate the development
of novel therapeutic agents.
Sesquiterpenoids are interesting natural products
which have diverse structural frameworks and exhibit a series of
activities including anti-inflammatory, anti-fungal and anticancer
effects (15). Plants from
Chloranthaceae (genus Chloranthus) are important sources of
this type of compounds and their dimmers (16). Chloranthushenryl Hemsl. is a
perennial herb in this genus native to China, which is distributed
in the southern part of mainland China. It is widely used as a
‘folk remedy’ for lumbocrural pain, bone fractures, pruritus and
other ailments (16). In our
previous phytochemical and pharmacological investigations into the
genus Chloranthus, we found that there were several
sesquiterpenoid lactones which exhibited anti-metastatic
properties, most of which can inhibit the migration of tumor cells
(17-19). Based on these concepts, we focused
our research on the finding of chemicals from this genus with the
ability to suppress the motility of cancer cells. By
migration-inhibition screening, >70 chemicals from 4 plants from
the genus Chloranthus, including Sarcandra glabra
(Thunb.) Nakai, Chloranthus henryi Hemsl., Chloranthus
fortunei and Chloranthus multistachys, were
investigated. In the current study, we reported that curcolonol
(CCL), a furan type sesquiterpene isolated from
Chloranthushenryl Hemsl., suppressed the migration of breast
cancer cells by downregulating LIMK1 activity.
Materials and methods
Materials
HPLC grade CCL was purchased from Chroma
Biotechnology Co. Ltd. (Chengdu, China; cat. no. 217817-09-9;
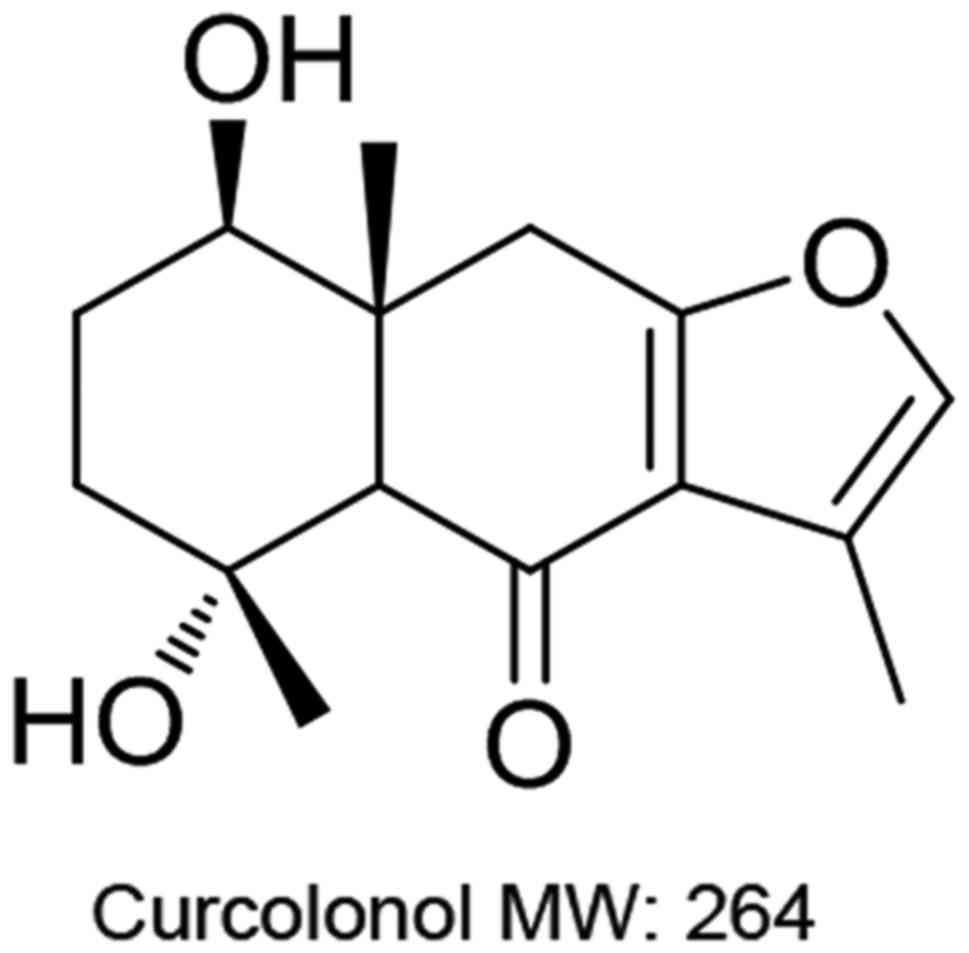
chemical structure shown in Fig.
1). A stock solution (1 mM) was prepared by dissolving CCL in
dimethyl sulfoxide (DMSO). Recombinant human transforming growth
factor (TGF)-β1 (PHG9211) was a product of Life Technologies/Thermo
Fisher Scientific (Waltham, MA, USA), and recombinant cofilin 1 was
purchased from Abcam (Cambridge, MA, USA; cat. no. ab85154). BB-94
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA; cat. no.
SC-203833), a broad spectrum matrix metalloproteinase (MMP)
inhibitor, served as a positive control in the cell migration
assay. Alexa Fluor™ 488 Phalloidin was purchased from Thermo Fisher
Scientific (cat. no. A12379). The TGF-βRI kinase inhibitor, SD-208
(Sigma-Aldrich, Beijing, China; cat. no. 627536-09-8), was used as
a positive control in the F-actin dying assay. Recombinant human
cofilin 1 (ab95396) protein was purchased from Abcam.
Antibodies and plasmids
Rabbit polyclonal antibodies against cofilin 1
(ab42824), p-cofilin 1 (ab12866), LIMK1 (ab81046) and rabbit
polyclonal to LIM kinase 1 (phospho T508, ab38508) were purchased
from Abcam. Mouse monoclonal antibody against β-actin (sc-130301)
is a product of Santa Cruz Biotechnology. Mouse monoclonal antibody
against Vimentin (sc-73258) was also obtained from Santa Cruz
Biotechnology. The pIRES2-LIMK1 eukaryotic expression plasmid and
the pIRES2-enhanced green fluorescent protein empty vector were
kindly provided by Professor Hongbo Wang from Yan Tai University
(Yan Tai, China). Small interfering RNA (siRNA) against cofilin 1
(sc-35078) and LIMK1 (sc-35810) are products of Santa Cruz
Biotechnology. Control siRNA-1 (sc37007) was used as a negative
control in the RNA interference (RNAi) experiments.
Cell culture
The human breast adenocarcinoma cell lines,
MDA-MB-231 and MDA-MB-468, purchased from the Cell Resource Center,
Institute of Basic Medical Sciences, Chinese Academy of Medical
Sciences (Beijing, China), and were maintained in DMEM
(Sigma-Aldrich) with 2 mM glutamine and 15% FCS at 37°C in 1%
CO2.
Vertical migration of breast cancer
cells
Firstly, the migratory capability of the cells was
assessed using an AP 48 chamber (Neuro Probe, Inc., Gaithersburg,
MD, USA). Briefly, the rough (lower) surface of the polycarbonate
membrane(8-µm pore size) was coated with 10 µg of
fibronectin in a volume of 50 µl overnight at 4°C.
Subsequently, the breast cancer cells (2×105) in 100
µl serum-free DMEM, which contained the vehicle (DMSO),
positive control or indicated concentrations of CCL were plated in
the upper chambers, while the lower part contained 30 µl
DMEM, including 10% serum and 10% collagen I. The chambers were
incubated for 18 h at 37°C. Following incubation, the cells which
had not migrated through the membrane pores were discarded by
wiping with a cotton swab. After being fixed and stained with
crystal violet (Sigma-Aldrich; final concentration, 0.5 g/ml).
Images of the migrating cells were captured under a microscope
(Leica DM 3000B; Leica Microsystems, Wetzlar, Germany) and the
migrating cells were then quantified with Image Pro plus software
5.0 (Media Cybernetics Inc.). The representative results are
illustrated in the figures. Each assay was performed in
triplicate.
Horizontal migration of breast cancer
cells
Subsequently, the migration of the breast cancer
cells was monitored in real-time using the ORIS™ cell migration
assay system (Platypus Technologies, Madison, WI, USA). Log-phase
cells were harvested, and resuspended to a final concentration of
1×106/ml in FBS-free DMEM. A total of 100 µl of
suspended cells was plated into each well through one of the side
ports of the Oris™ Cell Seeding Stopper. The plate containing
Stoppers was incubated in a humidified chamber (37°C, 5%
CO2) for 12 h to permit cell attachment. The stoppers
and media were removed, respectively. Subsequently, 100 µl
of FBS-free medium containing Calcein AM (Sigma-Aldrich, final
concentration 0.5 µg/ml) was added, and incubated in a
humidified chamber (37°C, 0.5% CO2) for 40 min. Images
were captured under a fluorescence microscope (Leica DM 3000B;
Leica Microsystems, Wetzlar, Germany), and these data served as an
initial control. Following fluorescence intensity examination, the
media were removed gently, and washed with PBS twice. Subsequently,
200 µl of full medium containing the vehicle (DMSO), 10 nM
BB-94 or indicated concentrations of CCL was added, and the cells
were incubated for 24 h at 37°C. At the end point of treatment,
data were obtained with the same methods mentioned above.
Phalloidin dying of F-actin
Log-phase MDA-MB-231 cells were harvested and
resuspended. A total of 300 µl of suspended cells was then
pipetted into each well of Lab-Tek® 16-well Chamber
Slides (Electron Microscopy Sciences, Hatfield, PA, USA) at a
density of 1×105/ml. After being subjected to the
treatments with the vehicle (DMSO), TGF-β (10 ng/ml), SD-208 (50
nM) or CCL (5 µM) for 24 h, the cells were fixed with 4%
paraformaldehyde (Sigma-Aldrich) in cytoskeleton buffer with
sucrose (CBS) [10 mM MES, pH 6.1, 138 mM KCl, 3 mM
MgCl2, 10 mM EGTA and 0.32 mM sucrose] for 30 min at
room temperature. The cells were permeabilized with 0.1% Triton
X-100 in PBS for 7 min and blocked with 1% BSA in PBS for 1 h at
37°C. Between each step described above, the cells were washed 3
times with PBS 5 min each. To visualize the actin cytoskeleton,
F-actin was stained with Alexa Fluor™ 488 Phalloidin in CBS for 1
h. The cells were then counterstained with 4 mg/ml
4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) and the samples
were mounted for fluorescence microscopy (Olymbus BX-63; Olympus,
Tokyo, Japan) examination.
Overexpression of LIM kinase1and
siRNA-mediated gene silencing
For the overexpression of LIMK1, the MDA-MB-231
cells were transiently transfected with the pIRES2-LIMK1 eukaryotic
expression plasmid and pIRES2-enhanced green fluorescent protein
empty vector using Plasmid Transfection Reagent (sc-108061; Santa
Cruz Biotechnology) according to the manufacturer’s instructions.
The cells were then maintained for 24 h at 37°C before being
harvested for further analyses. For gene silencing, at 24 h prior
to transfection, the MDA-MB-231 cells were seeded in a 6-well plate
in triplicate at a concentration so that the following day the
cells reached 70-80% confluency. Transfection was performed at a
final concentration of 200 nM using siRNA Transfection Reagent
(sc-29528; Santa Cruz Biotechnology) following the manufacturer’s
instructions. The cells were used at 48 h following transfection in
the further experiments.
Immunoprecipitation and kinase assay
The MDA-MB-231 and MDA-MB-468 cells were treated
with the indicated concentrations of CCL and stimulated with 10
ng/ml TGF-β1 for 24 h. The cells were then harvested and lysed in
radioimmunoprecipitation (RIPA) lysis buffer (50 mM Tris-HCl pH
7.5, 150 mM NaCl, 50 mM NaF, 1% NP-40, 0.1% sodium deoxycholate and
1 mM sodium pyrophosphate) with protease and phosphatase inhibitors
for 30 min on ice. Following centrifugation (10,000 × g for 15 min
at 4°C) to remove the debris, the supernatants were incubated with
the anti-LIMK1 antibody and protein G-Agarose beads (Santa Cruz
Biotechnology) for 4 h at 4°C. The immunoprecipitates were washed 3
times with lysis/kinase buffer and subjected to an in vitro
kinase reaction. In vitro kinase reactions were performed in
20 µl of the kinase buffer containing 15 µM ATP, 5μCi
of [γ-32P]-ATP (5,000 Ci/mmol; Amersham Biosciences,
Little Chalfont, UK) supplemented with 2 µg of recombinant
cofilin 1 at 30°C for 30 min. The reactions were terminated by the
addition of SDS sample buffer. Proteins were electrophoresed by 10%
SDS-PAGE and transferred onto nitrocellulose membranes, and
analyzed by autoradiography using a BAS1000 Bio-image analyzer
(Fuji Film, Tokyo, Japan), and by western blot analysis with
anti-cofilin 1, anti-LIMK1 and anti-p-LIMK1 antibodies.
Kinase-Glo® luminescent kinase
assay
LIM kinase1 activity was also determined with the
Kinase-Glo Plus luminescent kinase assay (Promega, Madison, WI,
USA). The principle of this assay is to evaluate kinase activity by
quantifying the amount of ATP remaining after a kinase reaction,
which is determined by a luciferase-catalyzed reaction. The LIMK1
reaction and the following luciferase reaction were performed in a
96-well plate. The MDA-MB-231 cells were treated with vehicle
(DMSO) or 2.5, 5 or 10 µM CCL and stimulated with 10 ng/ml
TGF-β1 for 24 h. The cells were then harvested and lysed in
lysis/kinase buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.5% NP-40,
5% glycerol, 1 mM MgCl2, 1 mM MnCl2, 10 mM
NaF, 1 mM Na3VO4, 1 mM dithiothreitol, 1 mM
phenyl-methylsulfonyl fluoride and 10 µg/ml leupeptin) for
30 min on ice. Following centrifugation (10,000 × g for 15 min at
4°C) to remove the debris, 50 µl supernatant were added to
each well. Subsequently, 50 µl of recombinant cofilin 1 (25
nM, final concentration) in kinase buffer (50 mmol/l HEPES, pH 7.3,
10 mmol/l MgCl2, 0.1% BSA, 2 mmol/l DTT) were added to
each well. A total of 10 µl ATP (1 µM, final
concentration) was then added to initiate the kinase reaction. The
reaction mixture was maintained at 30°C for 2 h in a water bath.
The mixture was then placed at room temperature for 10 min.
Subsequently, 50 µl of Kinase-Glo Assay Plus reagent were
added to initiate the luciferase reaction. Luminescence was
detected with the VICTOR3 multilabel counter (PerkinElmer, Waltham,
MA, USA) following a 10-min incubation.
Western blot analysis
The breast cancer cells were treated with the
indicated concentrations of CCL for 24 h. The cells were then
harvested and lysed in lysis buffer (Beyotime Biotech, Shanghai,
China). The concentration of the protein in the lysates was then
determined using a BCA kit (Beyotime Biotech). Aliquots of each
lysate containing equal quantities of protein (ranging between 500
and 1,000 µg between experiments) were added to SDS-PAGE
gels (ranging between 8 and 12%), and then transferred onto hybond
nitro blotting membranes and subjected to western blot analysis.
The membranes were blocked with 5% non-fat dried milk for 1 h at
room temperature and subsequently incubated with primary antibody
overnight at 4°C. Following washing 3 times with TTBS, the
membranes were incubated with the secondary antibody for 2 h at
room temperature. The immunoreactive bands were detected using an
enhanced chemiluminescence kit (Beyotime Biotech). β-actin
(1:1,000) served as an internal control. The signal intensities of
the bands of interest were quantified and normalized to β-actin
using the Image-Pro Plus software version 6.0 (Media Cybernetics,
Inc.). The primary antibodies used in the present study were as
follows: Rabbit polyclonal antibodies against cofilin 1 (1:1,000;
ab42824), p-cofilin 1 (1:1,000; ab12866), LIMK1 (1:1,000; ab81046),
rabbit polyclonal to LIM Kinase 1 (1:1,000; phospho T508, ab38508)
(all from Abcam) and mouse monoclonal antibody against Vimentin
(1:1,000; sc-73258; Santa Cruz Biotechnology, Inc.). The secondary
antibodies used in the present study were as follows: Goat
polyclonal secondary antibody to mouse IgG (1:5,000; ab6789) and
goat anti-rabbit IgG (1:5,000; ab6721) (both from Abcam).
Immunofluorescence analysis
Vimentin immunofluorescence in the breast cancer
cells was analyzed using an immunofluorescence staining kit
(Beyotime Biotech). Briefly, the cells were plated into wells of
8-well-Chamber Slide System (Nunc®; Thermo Fisher
Scientific), and treated with 5 µM CCL and/or the vehicle
(DMSO) for 24 h. At the end of the treatment, the cells cultured on
fibronectin-coated slides were fixed with 3.7%
paraformaldehyde-phosphate-buffered saline followed by incubation
with 0.5% Triton X-100 for 5 min at room temperature. After being
washed with PBS and blocked, the slides were triple-stained with
vimentin primary antibody, anti-mouse IgG conjugated with FITC
(4413; 1:500; Cell Signaling Technology, Danvers, MA, USA) and
ProLong Gold anti-fade reagent with DAPI (Thermo Fisher
Scientific). The samples were examined under a fluorescent
microscope (OlympusBX63; Olympus).
Statistical analysis
The data are presented as the means ± SD and were
analyzed using the SPSS for Windows (13.0) software program (SPSS
Inc., Chicago, IL, USA). Comparisons among different groups were
carried out by one-way analysis of variance (one-way ANOVA) and the
Fisher’s LSD test was used as a post hoc test following one-way
ANOVA. P-values 0.05 and 0.01 were assumed as the level of
significance for the statistic tests carried out.
Results
CCL attenuates the migratory capacity of
breast cancer cells
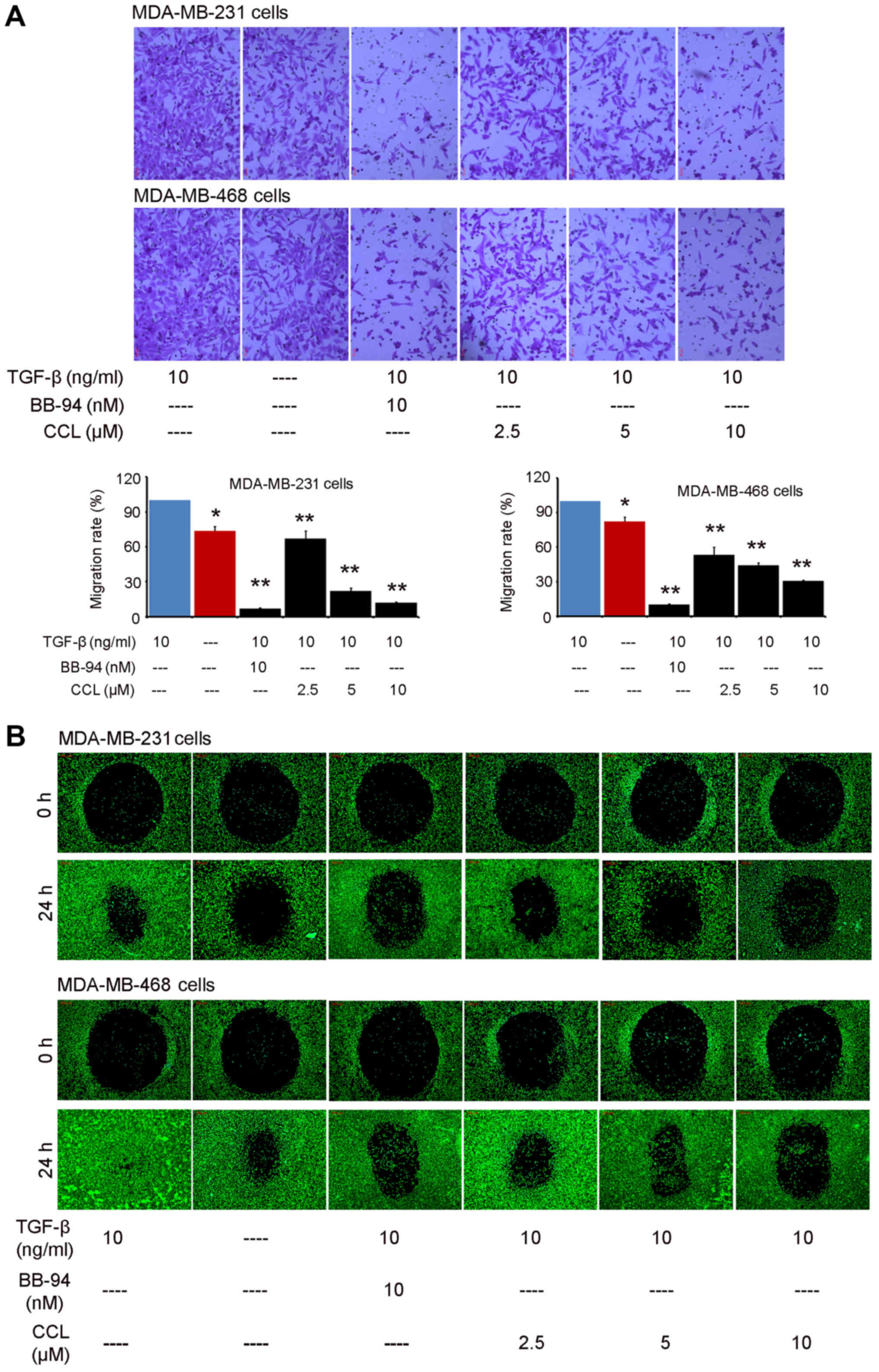
First, we investigated the effects of CCL (chemical
structure shown in Fig. 1) on cell
migration using an AP 48 chamber system. TGF-β1 (10 ng/ml) was used
to mimic the growth environment of cancer cells in vivo. It
was shown that CCL supplementation had a significant inhibitory
effect on the migration of the breast cancer cells (Fig. 2A). Compared with vehicle-treated
cells, the addition of TGF-β1 led to an obvious enhancement the
migration of both the MDA-MB-231 and MDA-MB-468 cells. When the
cells were cultured in presence of BB-94 for 18 h, both of the
breast cancer cell lines exhibited an obvious decrease in cell
migration compared with the TGF-β1-treated group. Following culture
in the presence of CCL for 18 h, both breast cancer cells exhibited
a weaker motility. The migration rates of the MDA-MB-231 cells were
66.94, 22.08 and 12.00% at concentrations of 2.5, 5 and 10
µM of CCL, respectively. The migration of the MDA-MB-468
cells was also significantly blocked by CCL (Fig. 2A). Consistent with these findings,
the real-time migration monitoring data from the ORIS™ cell
migration assay system indicated a dose-dependent decrease in cell
migration following the supplementation of CCL in both the
MDA-MB-231 and MDA-MB-468 cells (Fig.
2B). Thus, these results suggested that exposure to CCL
decreased the migratory potential of the breast cancer cells.
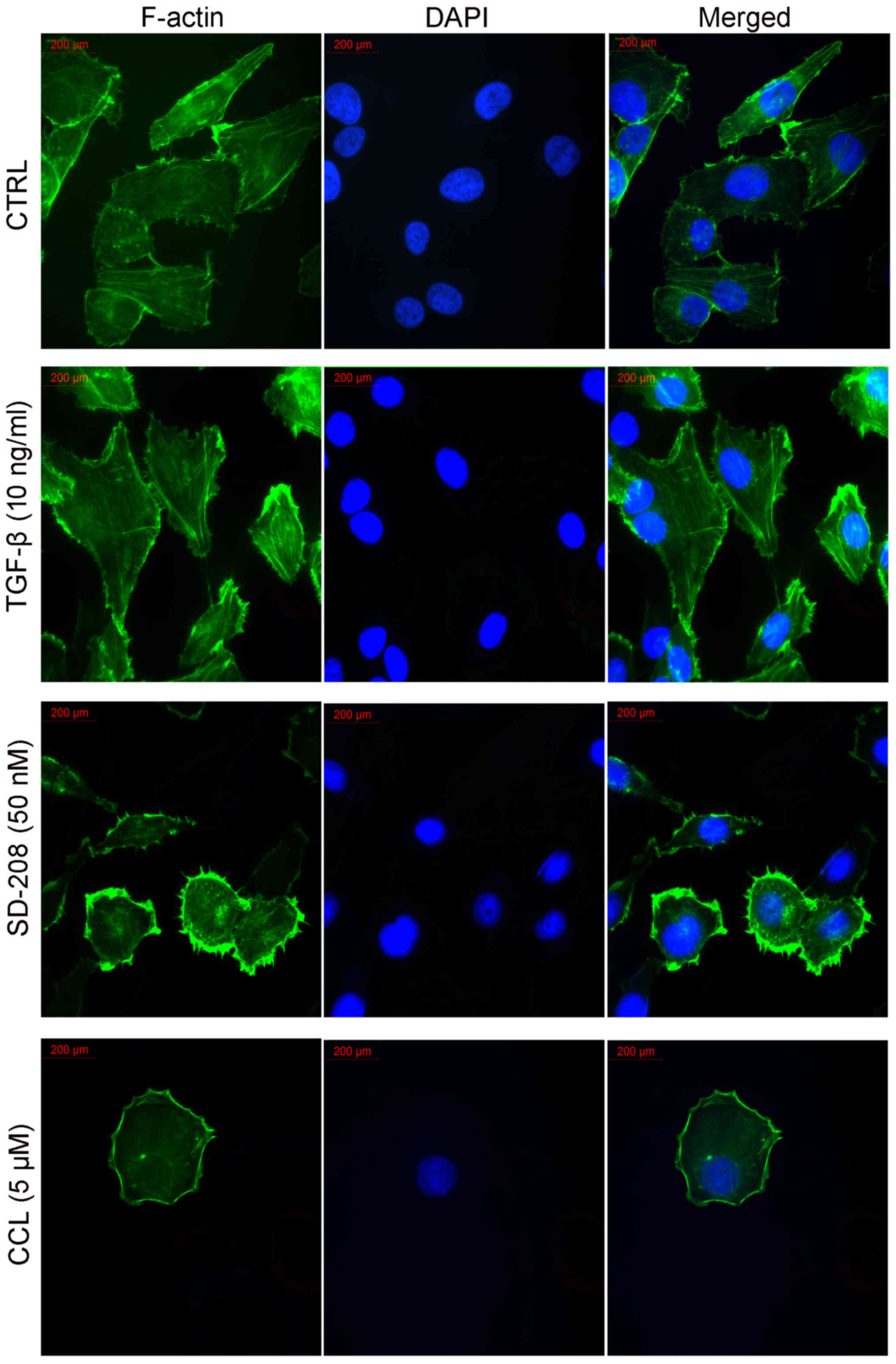
CCL impairs actin cytoskeleton
organization in MDA-MB-231 cells
We then investigated the effects of CCL on F-actin
microfilaments in the MDA-MB-231 cells. FITC-labeled phalloidin
clearly labeled the F-actin in the fixed and permeabilized cells,
as described in a range of earlier reports (4,5). As
shown in Fig. 3, the
vehicle-treated cells exhibited a regular aggregation of F-actin
present along the cells. When the cells were induced with TGF-β1,
this phenomenon became more pronounced. The addition of TGF-β1
resulted in the increased expression of F-actin and in the
formation of stress fibers at the cell perimeter. Furthermore,
there was an additional appearance of F-actin-rich microspikes
protruding from the cell periphery, which formed lamellipodia. When
the cells were treated with 50 nM SD-208, a TGF-βR inhibitor, there
were a significant reduction in F-actin fiber expression and a
disruption of F-actin arrangement inside the cells. However, there
were no obvious effects on the formation of lamellipodia at the
cell perimeter. When the cells were treated with CCL, we observed
not only a decrease in F-actin fiber expression and the disruption
of its arrangement, but also the disappearance of lamellipodia
around the cells. The cells became smooth at the cell perimeter.
Therefore, it seems clear that the inhibitory effects of CCL on
cell migration have a very close association with the
disorganization of F-actin.
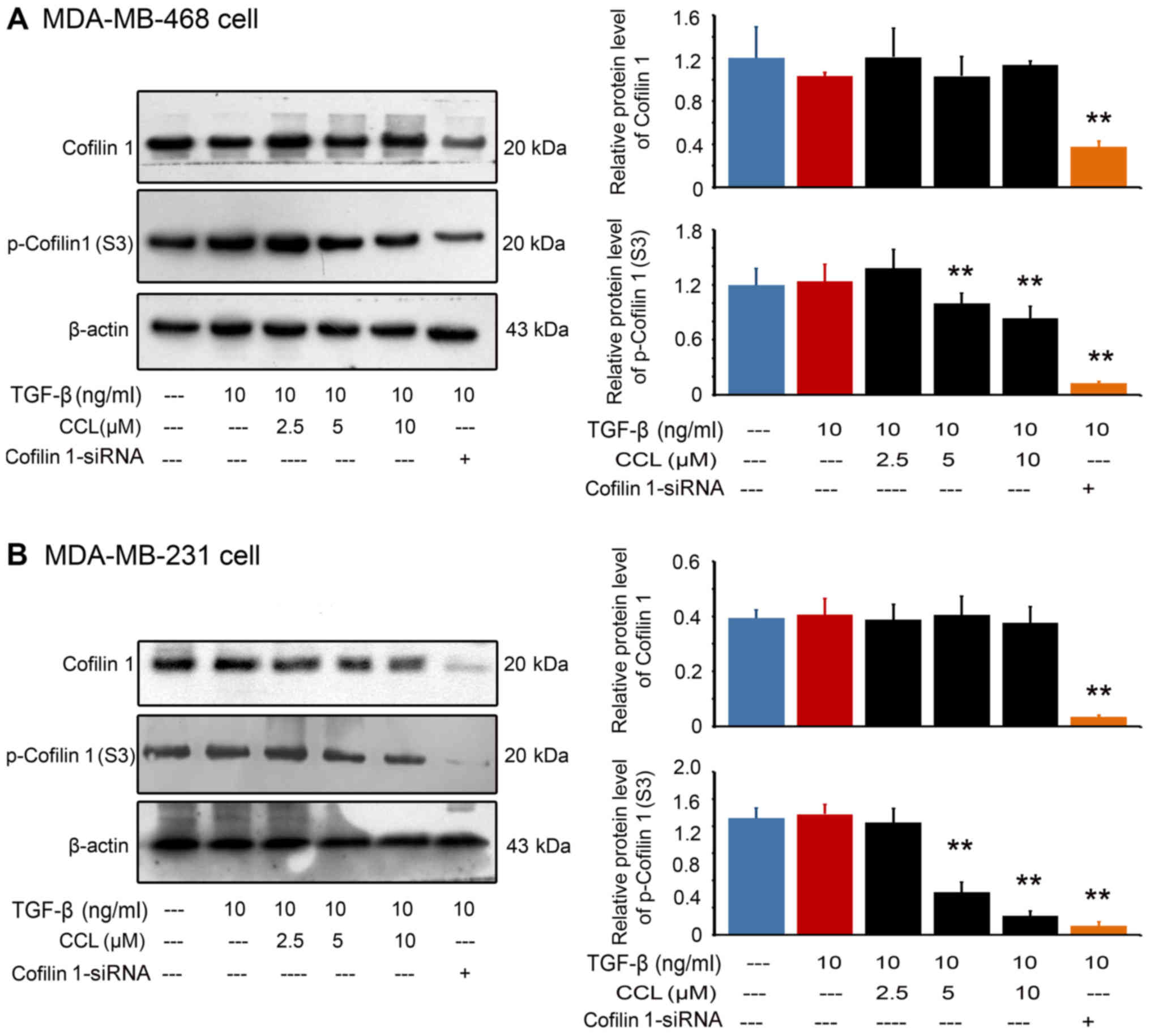
CCL disrupts actin cytoskeletal proteins
in breast cancer cells
We then analyzed the expression and phosphorylation
of cofilin 1, a well-known actin depolymerizing factor (ADF) by
western blot analysis (Fig. 4).
Our results revealed that, compared with the TGF-β1-stimulated
cells, CCL treatment, as well as cofilin 1 knockout, decreased the
expression of p-cofilin 1 in a dose-dependent manner in both the
MDA-MB-468 cells (Fig. 4A) and
MDA-MB-231 cells (Fig. 4B).
However, only were weak inhibitions of the expression of cofilin 1
in both breast cancer lines were observed.
CCL suppresses LIMK1 activity in breast
cancer cells
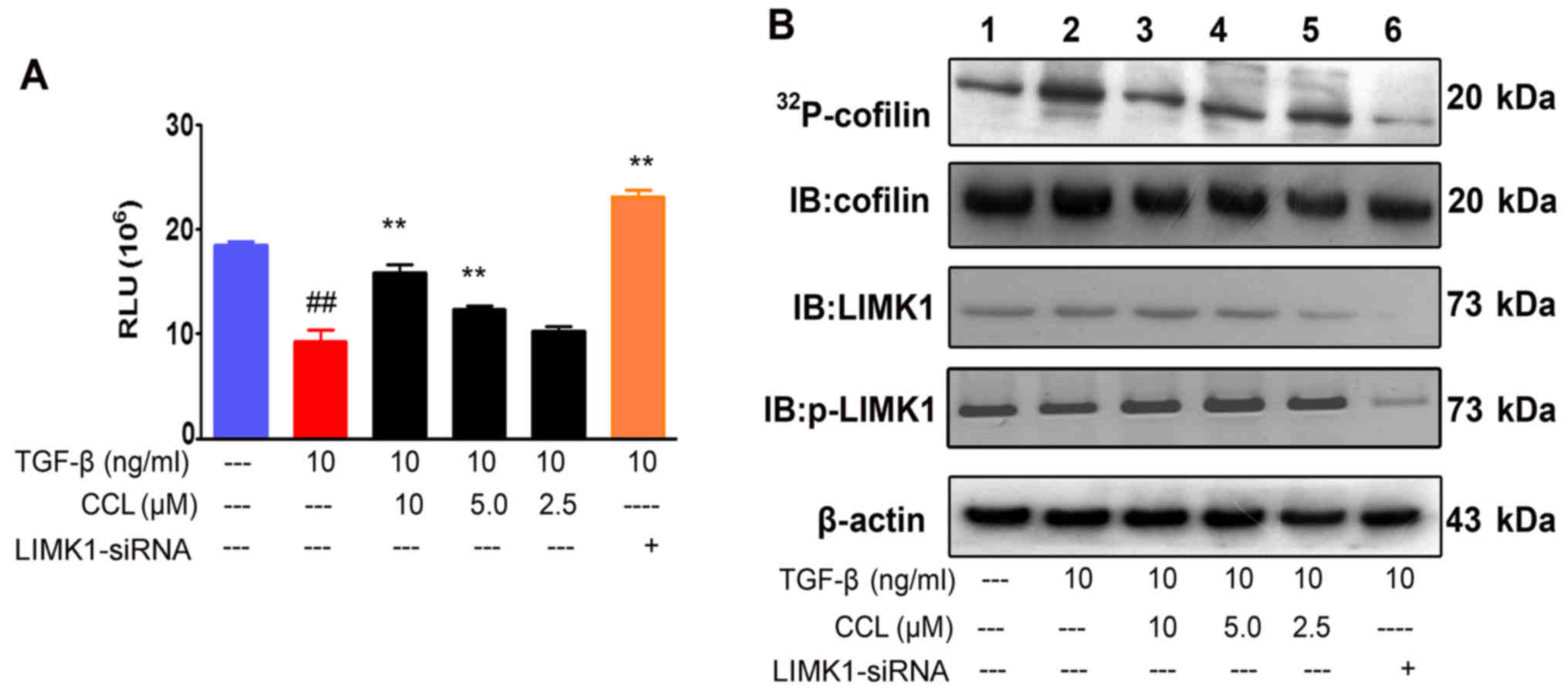
By kinase-Glo® luminescent kinase assay,
we then examined the effects of CCL on LIMK1. We demonstrated that
the consumption of ATP in this assay system was significantly
decreased after the supplementation of CCL in the MDA-MB-231 cells.
These data indicated that the inhibitory effects of CCL on cofilin
1 may be related to changes inLIMK1activity (Fig. 5A). To further distinguish these
findings, we analyzed LIMK1 activity by co-immunoprecipitation and
autoradiography-based assay. LIMK1 was purified by
immunoprecipitation and subjected to kinase assays in the presence
of [γ-32P]-ATP, cofilin 1 and indicated concentrations
of CCL. CCL exerted an inhibitory effect on cofilin 1
phosphorylation, but a weak inhibitory effect on the physical
interaction and expression of cofilin 1 and LIMK1. In addition,
there was no obvious inhibition on the phosphorylation of LIMK1.
Taken together, our data strongly suggested that CCL exerted marked
inhibitory effects on the catalytic activity of LIMK1, but not on
LIMK1 expression and phosphorylation.
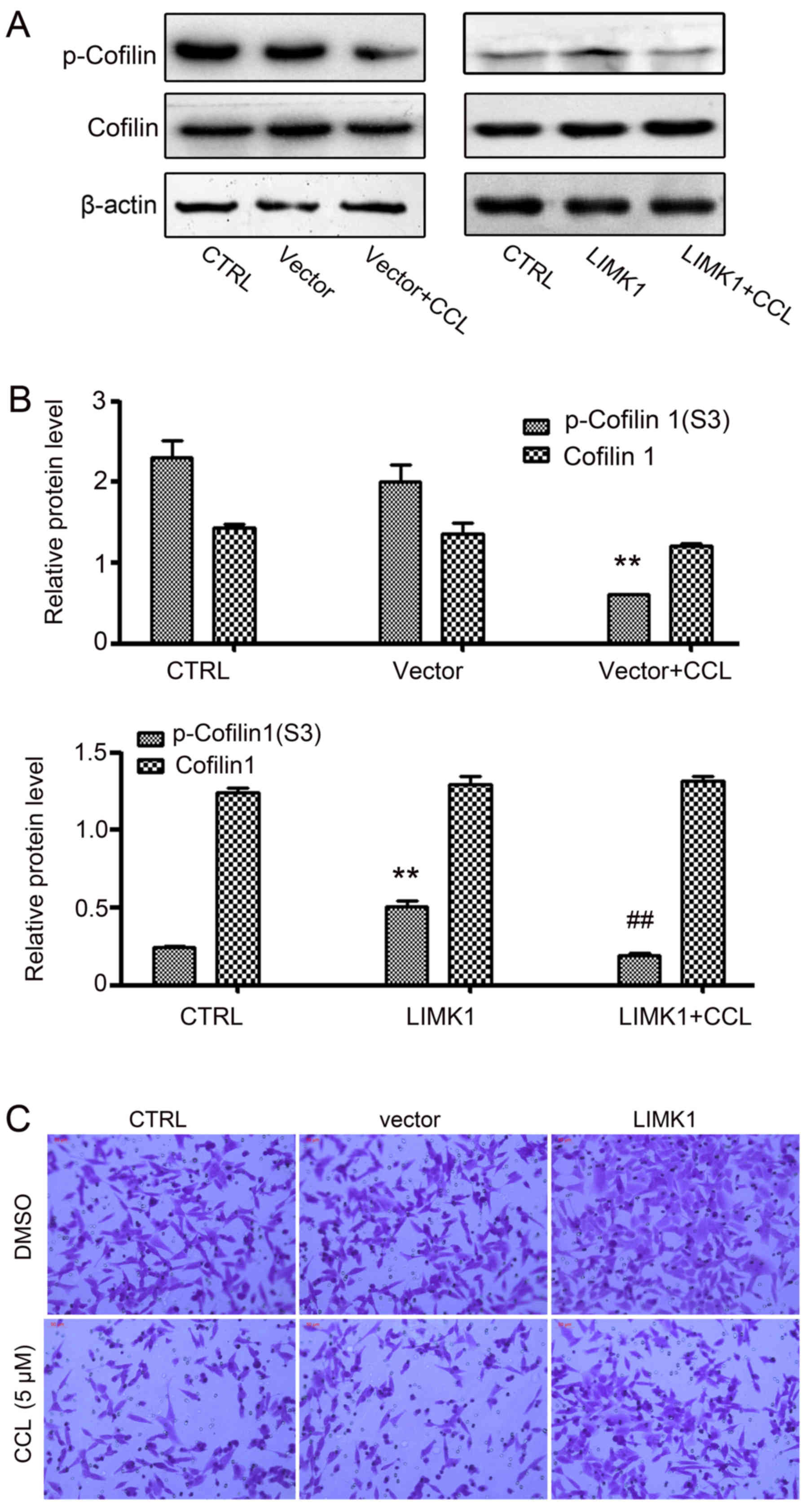
Overexpression of LIMK1 weakens the
effects of CCL on MDA-MB-231 cells
To further investigate the effects of CCL on LIMK1
activity, we constructed a transient LIMK1-overexpressing
MDA-MB-231 cell line and examined the impact of CCL on cofilin 1
expression and cell migration (Fig.
6). In the empty vector group and LIMK1 overexpression group,
no obvious changes were observed in the expression of total cofilin
1 compared to the control group. However, a significant increase in
the phosphorylation level of cofilin 1 was observed when the cells
were transiently transfected with the LIMK1 overexpression plasmid.
Following treatment with CCL for 24 h, we found that the expression
of p-cofilin 1 in the LIMK1-overexpressing cells was blocked by
CCL, although there was no clear change in cofilin 1 expression
(Fig. 6A and B). We then
investigated the effects of CCL on the migration of
LIMK1-overexpressing cells. It was found that the migratory
capacity of the cells was markedly increased in the
LIMK1-overexpressing group compared with the empty vector group
(Fig. 6C), when the cells were
treated with CCL for 24 h. This indicates that LIMK1 can neutralize
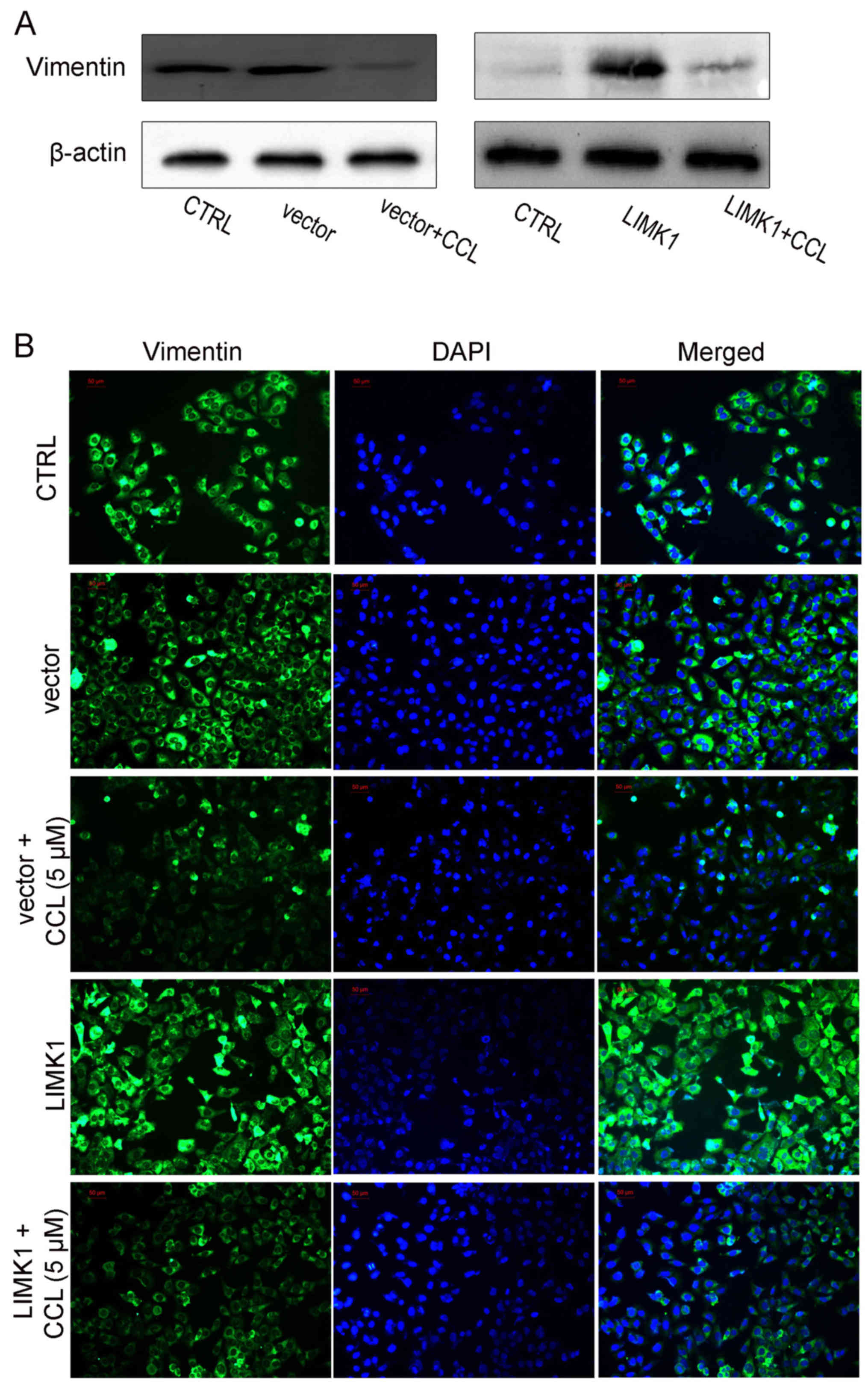
the inhibitory effects of CCL on cell migration. We also analyzed
the expression of vimentin, a mesenchymal-specific molecular mark
by immunofluorescence analysis, when the LIMK1-overexpressing cells
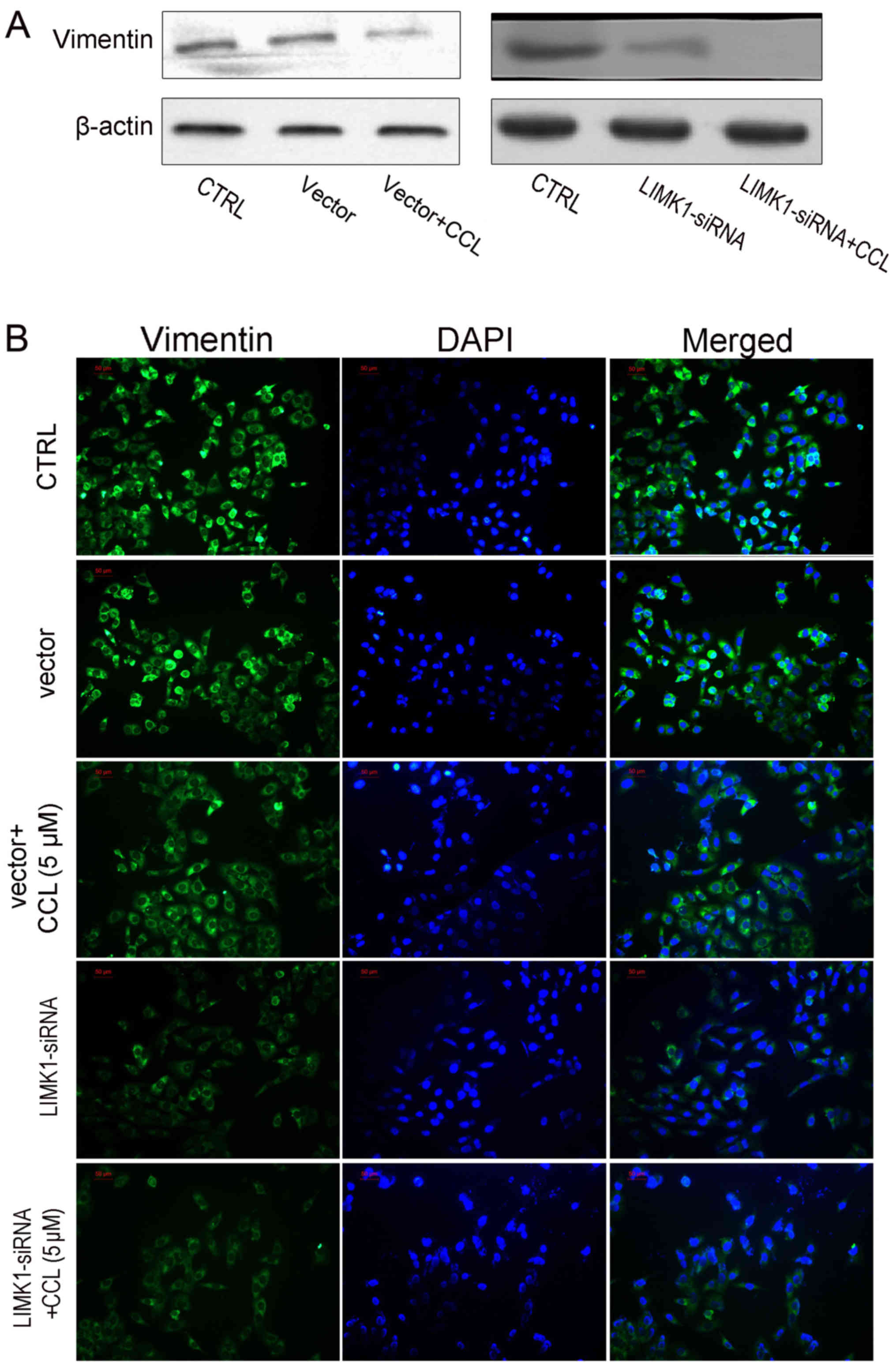
were treated with CCL. Similarly, LIMK1overexpression enhanced
vimentin expression and antagonized the effects of CCL on vimentin
expression in the MDA-MB-231 cells (Fig. 7).
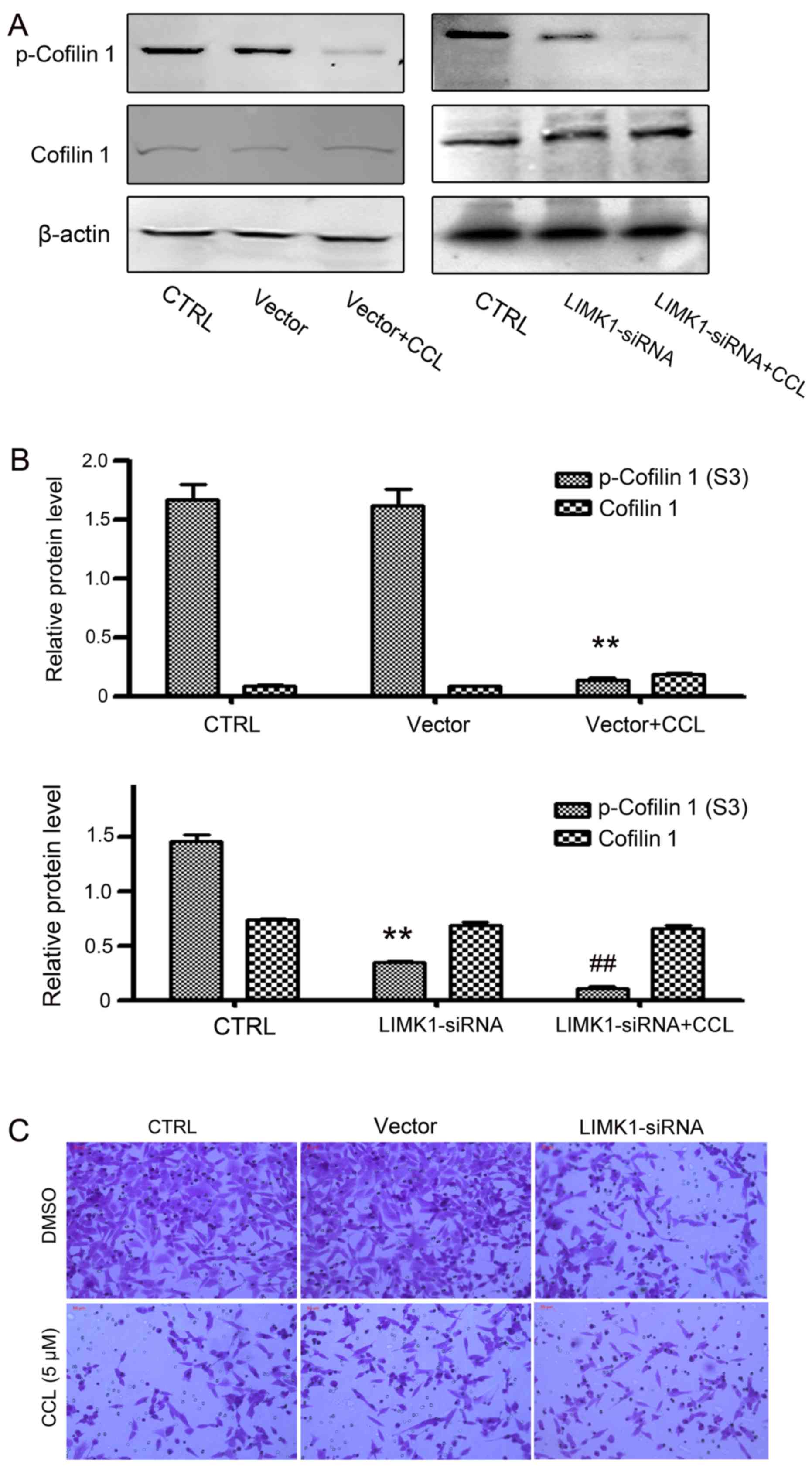
Downregulation of LIMK1 enhances the
effects of CCL on MDA-MB-231 cells
To confirm that LIMK1 was the critical target of
CCL, LIMK1 was knocked down by means of siRNA. Based on the results
of western blot analysis, the knockdown of LIMK1 significantly
inhibited the phosphorylation of cofilin 1, but did not affect
total cofilin 1 expression. The supplementation of CCL augmented
the inhibitory effects of LIMK1 on cofilin 1 phosphorylation
(Fig. 8A and B). Moreover, CCL
enhanced the effects of LIMK1-siRNA on cell activities. As shown in
Figs. 8C and 9, cell migration and vimentin expression
were significantly inhibited by CCL in the MDA-MB-231 cells and the
knockdown of LIMK1 by siRNA exerted more potent suppressive
effects. Taken together, these data indicated that the effects of
CCL on cell migration and cofilin 1 phosphorylation, are associated
with the suppression of LIMK1 activity.
Discussion
Chloranthus henryl is a perennial herb, which is
mainly distributed in the southern part of mainland China. It is
widely used as a ‘folk remedy’ for lumbocrural pain, bone
fractures, pruritus and other ailments (16). In our previous studies, it was
found that codonolactone, one of the sesquiterpenes extracted from
this herb, exhibited anti-metastatic properties. We confirmed that
codonolactone significantly suppressed the lung metastatic foci
formation of breast cancer in vivo and inhibited the f
invasive and migratory abilities of metastatic breast cancer cells.
Furthermore, it was also proven that this natural compound impaired
TGF-β1-induced EMT and the motility of breast cancer cells
(17-19). Therefore, in this study, we focused
our interests on the anti-metastatic activity and the probable
mechanisms of action of sesquiterpenes from this traditional
medical herb. Based on the previous data of anti-migratory activity
screening (20), we investigated
the effects of CCL on cancer cell motility.
By an AP 48 chamber system and ORIS™ cell migration
assay system, we first confirmed that CCL attenuated the migratory
capacity of breast cancer cells. Cell migration is governed by
multiple coordinated mechanisms which can influence the metastatic
potential of breast cancer cells. These processes involve the
reorganization of cell adhesion complexes and components of the
cytoskeleton. One of the rate-limiting steps in this mechanism is
F-actin microfilament organization. Actin is a key component of the
cytoskeleton and plays an important role in multitude cellular
functions, such as cell membrane dynamics, cell shape control,
movement and polarity (5). There
are two types of actin in cells, monomeric form (G-actin) and
filamentous form (F-actin) (21).
One of the properties of actin protein is the highly dynamics
turnover between G-actin and F-actin, and lead to rapid filament
polymerization and depolymerization, which is the rate-limiting
step of cell movement (21). In
light of the effects of CCL on breast cancer cell migration, in
this study, we investigated the effects of CCL on F-actin
microfilaments in MDA-MB-231 cells. We certified that CCL
significantly impaired the arrangement of F-actin in the MDA-MB-231
cells.
Studies have confirmed that the process of F-actin
micro-filaments organization is tightly controlled by a serial of
actin-binding proteins, including actin depolymerizing factor
(ADF)/cofilin family, which are essential for eukaryotes, and
important in actin filament dynamics in cells (22,23).
Cofilin 1, an important member of the ADF/cofilin 1 family, is
widely recognized for its ability to regulate actin polymerization
by severing filaments and enhancing their depolymerization. Studies
have suggested that cofilin 1 contributes to cancer development,
tumor progression, invasion and metastasis. The activity of cofilin
1 is predominantly regulated by phosphorylation on Ser3 by LIMKs,
which can block cofilin 1 activity of severing F-actin and results
in an increase in cell motility and invasion. Conversely, the
dephosphorylation of Ser3 results in the activation of cofilin 1
(24,25). Lee et al (26) proved that the overexpression of
cofilin 1 led to a decrease in the invasive abilities of human lung
cancerH1299 cells, which was also confirmed by other groups
(27,28). In the present study, we found that
CCL decreased p-cofilin 1 and increased non-p-cofilin 1 expression.
Taken together, our data strongly indicate that the inhibition of
CCL on cell migration may be associated with its effects on cofilin
1phosphorylation.
LIM kinase 1 is a serine protein kinase influencing
actin cytoskeletal dynamics. There are 39,499 base pairs with 16
exons in the LIMK1 gene which is located on human chromosome
7q11.23. The LIMK1 protein contains two amino-terminal LIM domains,
adjacent PDZ, proline/serine-rich regions, and followed by a
carboxyl-terminal kinase domain in tandem (10,11,29).
The LIM domains and PDZ domain are not only involved in mediating
protein-to-protein interactions, but are also clearly associated
with regulating LIMK activity. Studies on LIMK1 have indicated that
this kinase plays a central role in regulating the architecture of
the actin cytoskeleton through the phosphorylation and inactivation
of cofilin family members, which leads to the reorganization of the
actin cytoskeleton (10,11). In addition, this kinase has been
proven to be associated with the high invasion, migration and EMT
of cancer cells. Studies have found that the inhibition of LIMK1 by
pharmacological inhibitors and antisense RNAs targeting LIMK1
suppresses EMT, and the migration and invasion of cancer cells
(13,30). In this study, in order to further
determine the effects of CCL on the inhibition of cofilin 1
phosphorylation, we then investigated the effects of CCL on LIMK1
expression and activation in breast cancer cells. We demonstrated
that CCL significantly inhibited ATP consumption and cofilin 1
phosphorylation, but there were almost no effects on LIMK1
expression and phosphorylation. Furthermore, it was found that the
inhibitory effects of CCL were attenuated when LIMK1 was
overexpressed in the breast cancer cells. However, the
siRNA-mediated knockdown of LIMK1 enhanced the inhibitory effects
of CCL on cofilin 1 phosphorylation, cell migration and the EMT
phenotype. These data suggest that targeting the catalytic activity
of LIMK1 may be a mechanism through which CCL suppresses the
migration of breast cancer cells, and that there may be no effects
on the upstream signal transduction of LIMK1.
In conclusion, in this study, we demonstrated the
anti-motility properties of CCL on breast cancer cells and that
these effects are due to its potential to decrease of the
phosphorylation of cofilin 1, which is a protein controlling the
dynamics of actin filaments and the predominant substrate of LIMK1.
These effects may be associated with the inhibition of LIMK1
activity. Although our data strongly indicated that the
CCL-mediated suppression of cell migration was related to the
inhibition of the catalytic activity of LIMK1, confirmation that
LIMK1 is the direct target of CCL is still required. In future
studies, we aim to focus on the molecular interaction between
p-LIMK1 and CCL, and the effects of CCL on the upstream signal
transduction of LIMK1.
Funding
This study was supported by Grants from the National
Natural Science Foundation of China (Grant nos. 81560639, 81660680
and 81873043) and the Natural Science Foundation of Jiangxi
Province (Grant no. 20171BAB205097).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
HL performed the in vitro migration assay and
the Phalloidin dying of F-actin. JC contributed to experiments
involving the overexpression of LIM kinase 1 and siRNA-mediated
gene silencing. YL was responsible for the immunoprecipitation and
Kinase-Glo® luminescent kinase assay. HX participated in
the western blot analysis. LX performed the immunofluorescence
analysis. JF was responsible for the design of this study, and the
analysis and interpretation of the data. All authors read, edited
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Vanharanta S and Massagué J: Origins of
metastatic traits. Cancer Cell. 24:410–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Binamé F, Pawlak G, Roux P and Hibner U:
What makes cells move: Requirements and obstacles for spontaneous
cell motility. Mol Biosyst. 6:648–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carlier MF, Pernier J, Montaville P,
Shekhar S, Kühn S and Cytoskeleton Dynamics; Motility group:
Control of polarized assembly of actin filaments in cell motility.
Cell Mol Life Sci. 72:3051–3067. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shekhar S, Pernier J and Carlier MF:
Regulators of actin filament barbed ends at a glance. J Cell Sci.
129:1085–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanellos G and Frame MC: Cellular
functions of the ADF/cofilin family at a glance. J Cell Sci.
129:3211–3218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ostrowska Z and Moraczewska J: Cofilin - a
protein controlling dynamics of actin filaments. Postepy Hig Med
Dosw. 71:339–351. 2017. View Article : Google Scholar
|
|
8
|
He L, Seitz SP, Trainor GL, Tortolani D,
Vaccaro W, Poss M, Tarby CM, Tokarski JS, Penhallow B, Hung CY, et
al: Modulation of cofilin phosphorylation by inhibition of the Lim
family kinases. Bioorg Med Chem Lett. 22:5995–5998. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prunier C, Prudent R, Kapur R, Sadoul K
and Lafanechère L: LIM kinases: Cofilin and beyond. Oncotarget.
8:41749–41763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bernard O: Lim kinases, regulators of
actin dynamics. Int J Biochem Cell Biol. 39:1071–1076. 2007.
View Article : Google Scholar
|
|
11
|
Scott RW and Olson MF: LIM kinases:
Function, regulation and association with human disease. J Mol Med
(Berl). 85:555–568. 2007. View Article : Google Scholar
|
|
12
|
Park GB and Kim D: PI3K catalytic isoform
alteration promotes the LIMK1-related metastasis through the PAK1
or ROCK1/2 activation in cigarette smoke-exposed ovarian cancer
cells. Anticancer Res. 37:1805–1818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wan L, Zhang L, Fan K and Wang J: MiR-27b
targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol
Cell Biochem. 390:85–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liao Q, Li R, Zhou R, Pan Z, Xu L, Ding Y
and Zhao L: LIM kinase 1 interacts with myosin-9 and
alpha-actinin-4 and promotes colorectal cancer progression. Br J
Cancer. 117:563–571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu YH, Wu PQ, Hu QL, Pei YJ, Qi FM, Zhang
ZX and Fei DQ: Cytotoxic and antibacterial activities of iridoids
and sesquiterpenoids from Valeriana jatamansi. Fitoterapia.
123:73–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Editorial Committee of the Administration
Bureau of Traditional Chinese Medicine: Genus Chloranthaceae.
Chinese Materia Medica (Zhonghua Bencao). Shanghai Science &
Technology Press; Shanghai: pp. 2051–2061. 1998
|
|
17
|
Fu J, Ke X, Tan S, Liu T, Wang S, Ma J and
Lu H: The natural compound codonolactone attenuates TGF-β1-mediated
epithelial-to-mesenchymal transition and motility of breast cancer
cells. Oncol Rep. 35:117–126. 2016. View Article : Google Scholar
|
|
18
|
Wang W, Chen B, Zou R, Tu X, Tan S, Lu H,
Liu Z and Fu J: Codonolactone, a sesquiterpene lactone isolated
from Chloranthus henryi Hemsl, inhibits breast cancer cell
invasion, migration and metastasis by downregulating the
transcriptional activity of Runx2. Int J Oncol. 45:1891–1900. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu J, Wang S, Lu H, Ma J, Ke X, Liu T and
Luo Y: In vitro inhibitory effects of terpenoids from Chloranthus
multistachys on epithelial-mesenchymal transition via
down-regulation of Runx2 activation in human breast cancer.
Phytomedicine. 22:165–172. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang SS, Fu JJ, Chen HY, Tu LF, Xiao CR,
Zhang RZ, Liu DP and Luo YM: Sesquiterpenes with anti-metastasis
breast cancer activity fro Chloranthus henryi. Zhongguo Zhong Yao
Za Zhi. 42:3938–3944. 2017.In Chinese. PubMed/NCBI
|
|
21
|
Iwasa JH and Mullins RD: Spatial and
temporal relationships between actin-filament nucleation, capping,
and disassembly. Curr Biol. 17:395–406. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meberg PJ: Signal-regulated ADF/cofilin
activity and growth cone motility. Mol Neurobiol. 21:97–107. 2000.
View Article : Google Scholar
|
|
23
|
Mizuno K: Signaling mechanisms and
functional roles of cofilin phosphorylation and dephosphorylation.
Cell Signal. 25:457–469. 2013. View Article : Google Scholar
|
|
24
|
Shishkin S, Eremina L, Pashintseva N,
Kovalev L and Kovaleva M: Cofilin-1 and other ADF/cofilin
superfamily members in human malignant cells. Int J Mol Sci.
18:E102016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang W, Eddy R and Condeelis J: The
cofilin pathway in breast cancer invasion and metastasis. Nat Rev
Cancer. 7:429–440. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee YJ, Mazzatti DJ, Yun Z and Keng PC:
Inhibition of invasiveness of human lung cancer cell line H1299 by
overexpression of cofilin. Cell Biol Int. 29:877–883. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yap CT, Simpson TI, Pratt T, Price DJ and
Maciver SK: The motility of glioblastoma tumour cells is modulated
by intracellular cofilin expression in a concentration-dependent
manner. Cell Motil Cytoskeleton. 60:153–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hotulainen P, Paunola E, Vartiainen MK and
Lappalainen P: Actin-depolymerizing factor and cofilin-1 play
overlapping roles in promoting rapid F-actin depolymerization in
mammalian nonmuscle cells. Mol Biol Cell. 16:649–664. 2005.
View Article : Google Scholar :
|
|
29
|
Manetti F: LIM kinases are attractive
targets with many macromolecular partners and only a few small
molecule regulators. Med Res Rev. 32:968–998. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su B, Su J, Zeng Y, Liu F, Xia H, Ma YH,
Zhou ZG, Zhang S, Yang BM, Wu YH, et al: Diallyl disulfide
suppresses epithelial-mesenchymal transition, invasion and
proliferation by downregulation of LIMK1 in gastric cancer.
Oncotarget. 7:10498–10512. 2016.PubMed/NCBI
|