Introduction
Pancreatic carcinoma is one of the most lethal
diseases, with an extremely poor prognosis. Current cancer
statistics indicate that pancreatic carcinoma is the fourth leading
cause of cancer-associated mortality in the USA, with an incidence
of 53,000 new cases and a high mortality rate of 42,000 in 2016
(1). For patients with
unresectable or recurrent pancreatic carcinoma, the standard
chemotherapy is gemcitabine in combination with other
chemo-therapeutic agents (2).
Despite accumulated knowledge regarding pancreatic carcinoma
etiology, the prognosis has not significantly improved in the last
decade (3). The development of
gemcitabine resistance during chemotherapy serves an important role
in the prognosis of pancreatic carcinoma and has become an
increasingly common phenomenon (4). However, the molecular mechanisms
underlying gemcitabine resistance remain unclear. Therefore, to
improve the prognosis of patients with pancreatic carcinoma, it is
important to identify innovative biomarkers that are able to
predict the risk of recurrence and chemoresistance in patients who
are receiving gemcitabine-based chemotherapy.
MicroRNAs (miRNAs) represent novel single-stranded,
small non-coding RNA molecules. Mature miRNAs bind directly to
specific targets within mRNA 3′-untranslated regions of target RNAs
and negatively regulate translation or mRNA cleavage through
partial sequence homology at the post-transcriptional level
(5). Previous data have
demonstrated frequent deregulation of miRNAs in the majority of
malignant human tumors (6-9). Deregulated miRNAs are associated with
behavior as either oncogenes or tumor suppressor genes. Certain
miRNAs have been implicated in cellular processes involving
proliferation, invasiveness, apoptosis, and chemoresistance
(10-13). Moreover, current evidence has
demonstrated that miRNAs are critically involved in regulating drug
resistance-mediated epithelial to mesenchymal transition (EMT)
(14-16).
Among these miRNAs, there is evidence that miRNA
(miR)-200b is downregulated in numerous types of cancer, including
pancreatic, colorectal, gastric and lung cancer (17-22).
Additionally, miR-200b inhibition induces EMT through upregulated
zinc finger E-box-binding homeobox 1 (ZEB1) (23) and induces chemoresistance (24,25).
By contrast, miR-301 has been reported to be
associated with cell invasion, migration and drug-resistance in
breast cancer (26). More
recently, data have demonstrated that miR-301 is associated with
gemcitabine resistance through EMT and enhanced cell proliferation
in pancreatic carcinoma (13).
Based on these reports, the purpose of the present study was to
investigate the roles of miR-200b and miR-301 as potential
biomarkers for chemosensitivity or acquired chemoresistance in
pancreatic carcinoma. A total of six different pancreatic carcinoma
cell lines were used to investigate the mechanisms of miR-200b and
miR-301 expression in pancreatic carcinoma. It was observed that
the miR-200b expression level correlated with cadherin 1 (CDH1)
expression and chemosensitivity in the six cell lines. In addition,
it was demonstrated that transfection with miR-200b upregulated
gemcitabine sensitivity. Conversely, it was observed that stable
gemcitabine-resistant cell lines, Capan-2 and Panc-1, exhibited
increased miR-301 expression and inhibited CDH1 expression. Unlike
miR-200b, the overexpression of miR-301 induced chemoresistance and
reduced apoptosis. These findings indicated that the miR-200b/CDH1
and miR-301/CDH1 signaling axes serve important roles in mediating
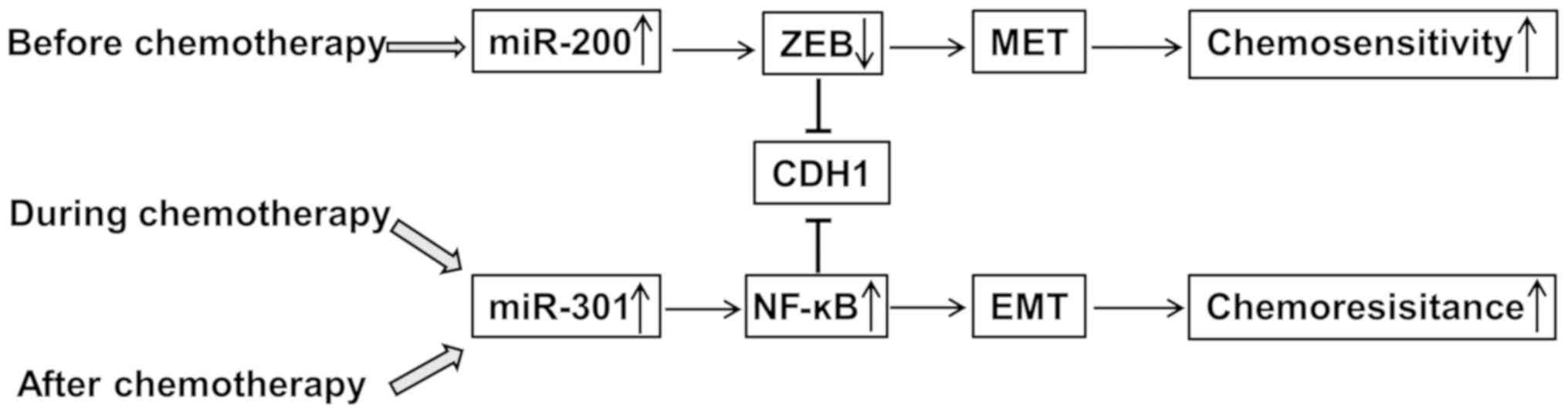
the response to chemotherapy in pancreatic carcinoma (Fig. 1). Moreover, these results implied
that miR-200b and miR-301 may be potential therapeutic targets in
pancreatic carcinoma.
Materials and methods
Oligonucleotides
Pre-miR-200b (cat. no. PM10492), pre-miR-301 (cat.
no. PM12929), negative control (cat. no. AM17110), miR-200b
inhibitor (cat. no. AM10492), miR-301 inhibitor (cat. no. AM12929)
and their negative controls (cat. no. AM17010) were purchased from
Ambion (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell culture conditions
Human pancreatic carcinoma cell lines (Capan-1,
Capan-2, Panc-1, MIAPaCa-2, BxPC-3 and PL45) were obtained from the
American Type Culture Collection (Manassas, VA, USA). Panc-1,
MIAPaCa-2 and PL45 were maintained in Dulbecco's modified Eagle's
medium (DMEM; Thermo Fisher Scientific, Inc.; cat. no. 10566-016)
supplemented with 10% fetal bovine serum (FBS). Capan-1, Capan-2
and BxPC-3 cells were grown in RPMI-1640 (cat. no. 61870-036) with
10% FBS (both from Thermo Fisher Scientific, Inc.; cat. no.
10437028). The two media contained antibiotics (100 U/ml penicillin
and 100 µg/ml strep tomycin). All cell lines were routinely
passaged as monolayer cultures at 37°C in a humidified atmosphere
of 95% air and 5% CO2.
Establishment and characterization of
gemcitabine-resistant Panc-1 and Capan-2 cells
In order to further identify the association between
chemoresistance and miRNA expression, gemcitabine-resistant cells
were generated. The gemcitabine-resistant cell lines (Capan-2 GEM-R
and Panc-1 GEM-R) were generated in RPMI-1640 or DMEM medium with
15% FBS and continuous exposure to the respective half-maximal
inhibitory concentration (IC50) of gemcitabine
(µM) (Tocris Bioscience, Bristol, UK) for >5 months
(27). To assess the gemcitabine
resistance of Capan-2 GEM-R and Panc-1 GEM-R cells, colorimetric
assays were performed following treatment with gemcitabine.
Secondly, in the two generated gemcitabine resistant cell lines,
miR-301 was measured by reverse transcription-quantitative
polymerase chain reaction.
RNA preparation and RT-qPCR analysis
RT-qPCR was performed as described previously
(28). Total RNA was extracted
from cultured cells using a standard TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) protocol. Cell pellets
were suspended in an aliquot of 1 ml/well of TRIzol in a 6-well
plate. Isolated RNA was reverse-transcribed using a High-Capacity
cDNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. cDNA was diluted and
stored at −20°C prior to use. Gene expression levels were measured
with a custom-designed, TaqMan qPCR (Applied Biosystems; Thermo
Fisher Scientific, Inc.) containing probes for six genes: CDH1 (ID:
Hs00156401_m1), ZEB1 (ID: Hs00232783_m1), ZEB2 (ID: Hs00207691_m1),
Vimentin (ID: Hs00185584_m1), interleukin (IL)-6 (ID:
Hs00174131_m1), miR-200b (ID: 002251) and miR-301 (ID: 002392),
with GAPDH (ID: Hs99999901_s1) for mRNA or RNAU6 (ID: 001002) for
miRNA as an internal control. The relative expression levels of
genes, miR-200b and miR-301 relative to GAPDH or RNAU6 were
calculated using the rela tive quantification ΔΔCq method (29). RT-qPCR reactions were performed
using TaqMan Gene Expression Assays on an ABI prism 7900HT Sequence
Detection instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.). qPCR was performed as follows: 95°C for 10 min, followed by
40 cycles of 95°C for 15 sec and 60°C for 60 sec, according to the
manufacturer's protocol. Each sample was assayed in triplicate.
Pre-miR-200b, miR-200b and miR-301
inhibitor transfection experiments
miRNA precursor molecules corresponding to miR-200b
or miR-200b inhibitor, and miR-301 inhibitor, were transfected
using the RNAiMAX Transfection Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) into Capan-2 and Panc-1 cells, and the effects on
the respective oligonucleotide were measured by RT-qPCR. Capan-2
and Panc-1 cells at a density of 1×105 cells/well were
transfected with 50 nM microRNA in a 6-well plate for RNA
extraction or a 10-cm dish for gemcitabine sensitivity and
proliferation assays, following the manufacturer's protocol. Cells
in the 6-well plate were collected 48 h post-transfection to
extract RNA and measured for miR-200b or miR-301 expression. After
12 h of transfection, transfected cells in the 10-cm dish were
seeded into 96-well plates for proliferation. These transfection
experiments were repeated independently three times.
Gemcitabine sensitivity assay with
transfection of pre-miR-200b or miR-200b and miR-301
inhibitors
The colorimetric assay was performed essentially as
described in a previous method (30). Briefly, cells were seeded in a 6-cm
dish at 70% confluency. After 12 h, pre-miR-200b or miR-200b/301
inhibitors and respective controls were transfected in each dish
overnight. Transfected cells were seeded in 96-well plates at 4,000
cells/well in triplicate. After incubating for 12 h, cell viability
was determined by treating the cells with stepwise 4-fold serial
dilutions of gemcitabine (from 100 µM) and incubated at 37°C
for 96 h. To evaluate cell survival, the cells were fixed with 25%
glutaraldehyde for 30 min at room temperature and stained with 200
µl 0.05% methylene blue for 20 min at room temperature. The
dye was eluted with 0.33 M HCl for 20 min with agitation.
Absorbance was measured using a microplate reader (model no. 3550;
Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 598 nm. The
IC50 for cell growth was calculated. The morphology of
Panc-1 and Capan-2 cells was assessed using light microscopy (×20
magnification).
Cell proliferation assay
The cell growth of pancreatic carcinoma cell lines
was studied using the colorimetric methylene blue assay, as
described previously (31,32). To test cell growth, cells were
transfected with miR-200b or 200b inhibitor or negative control in
a 10-cm dish, counting the first 12 h as Day 0. Transfected cells
at 4,000 cells/well were plated in a 96-well plate for 24 h. Mean
values were calculated from three different wells in triplicate for
4 days.
Apoptosis assay
To evaluate whether miR-200b or acquired gemcitabine
resistance contributes to a decrease in caspase-3/7, Capan-2 or
Panc-1 cells were cultured for 12 h in 96-well plates in
triplicate, and treated with 50 nM pre-miR-200b or miR-200b
inhibitor, or their respective controls. The assay was analyzed
using a caspase-3/7 assay kit (Promega Corporation, Madison, WI,
USA), according to the manufacturer's protocol.
Cell invasion assay
The invasion assay was performed in 24-well Biocoat
Matrigel invasion chambers (BD Biosciences, San Jose, CA, USA),
according to a previous protocol (25,33).
Briefly, cells were transfected with pre-miR-200b or pre-miR-301
and the negative control in a 10-cm dish. After 12 h transfection,
cells were harvested and plated in the Matrigel-coated wells
(4×104 cells/well) and control insert wells
(4×104 cells/well) using Capan-2 and Panc-1 cells,
respectively. After 20 h incuba tion, seeded cells on the membrane
were removed by wiping with a cotton swab, and the invasive cells
through the membrane were fixed with methanol for 5 min and stained
with crystal violet for 5 min. Under a light microscope (×20
magnification), invasive cells were counted in three random fields.
All assays were performed in triplicate.
Immunofluorescence imaging
Immunofluorescence was performed as previously
described (13). Briefly, cells
were seeded into a chamber slide at 40% confluence. Following
incubation overnight, the cells were fixed with 4% paraformaldehyde
for 20 min at room temperature and permeabilized with 0.15% Triton
X-100 in PBS for 20 min. Subsequently, the cells were blocked with
5% goat serum (cat. no. ab7481; Abcam, Cambridge, MA, USA) in PBS
for 1 h at room temperature. CDH1 and nuclear factor (NF)-κB
protein expression was detected using anti-CDH1 (cat. no. ab15148;
Abcam; 1:1,000) and anti-NF-κB (cat. no. 3033; Cell Signaling
Technology Inc., Danvers, MA, USA; 1:500) antibodies at 4°C
overnight, according to the manufacturer's protocol. An Alexa
Fluor-conjugated antibody (cat. no. ab1500083; 1:2,000) was used as
a secondary antibody at 37°C for 2 h. The cell nucleus was
counterstained with DAPI for 20 min at room temperature. The
coverslips were mounted on slides prior to viewing using a
fluorescence microscope (magnification, ×20).
Statistical analysis
All experiments were performed in triplicate and
conducted at least twice. Data are presented as the mean ± standard
deviation where applicable. GraphPad Prism version 5.0 (GraphPad
Software Inc., La Jolla, CA, USA) was used for all statistical
analysis. Levels of significance for comparisons between cell lines
were determined by the Student's t-test distribution. To assess the
correlation between miR-200b expression, CDH1 expression and
IC50, Pearson's correlation analysis was performed in
six different pancreatic carcinoma cell lines. To analyze multiple
comparisons, one-way analysis of variance and the Bonferroni test
as a post hoc test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-200b expression correlates negatively
with the IC50 of gemcitabine in pancreatic carcinoma
cell lines
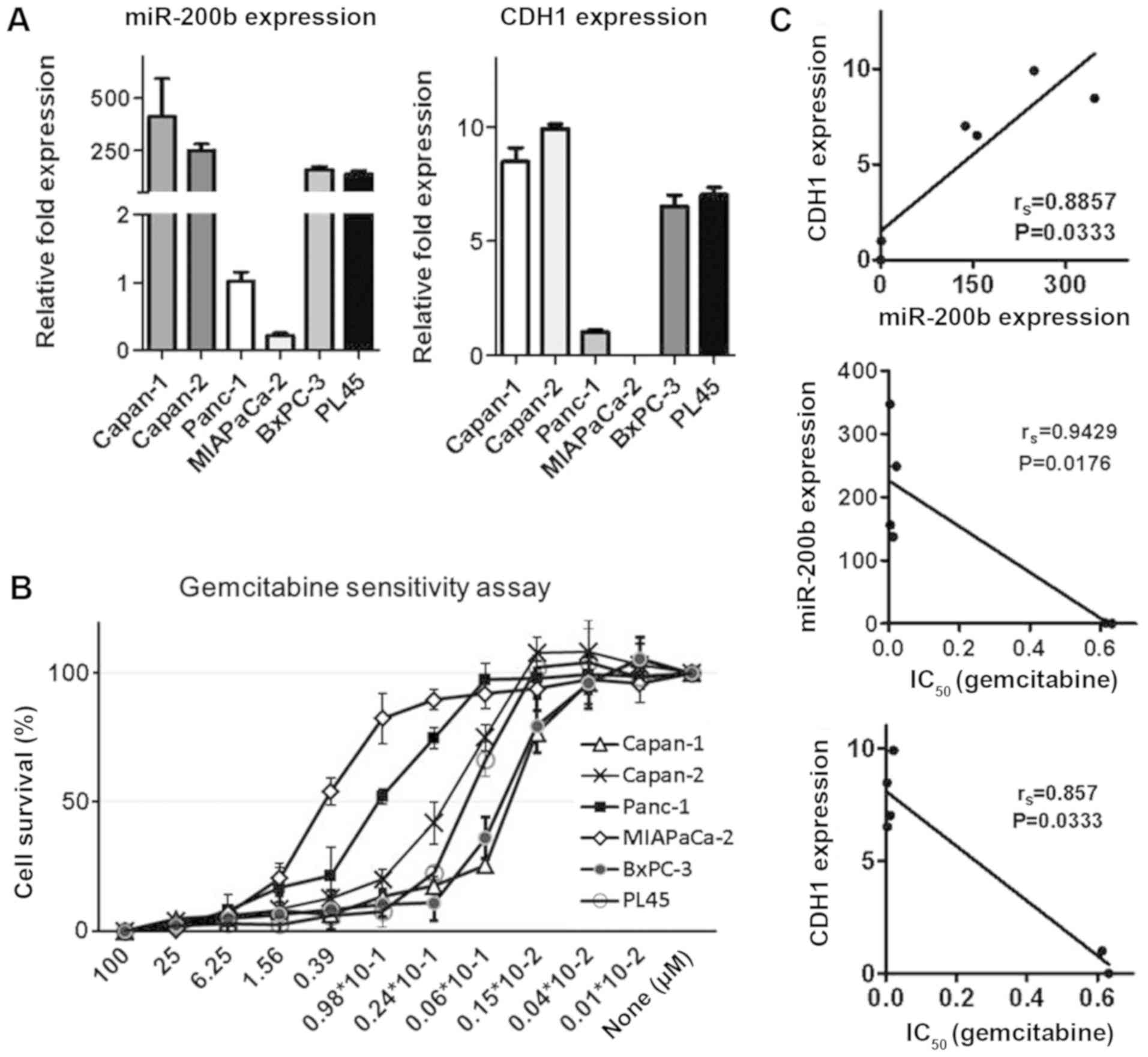
To examine the correlation between miR-200b and
IC50, the present study focused on CDH1 expression,
which is known to be a target for miR-200b and the EMT markers. To
identify the correlation, miR-200b and CDH1 expression was
initially investigated using six different cell lines (Capan-1,
Capan-2, Panc-1, MIAPaCa-2, BxPC-3 and PL45) using RT-qPCR
(Fig. 2A). Next, the
IC50 of gemcitabine was measured using a colorimetric
assay in the six pancreatic carcinoma cell lines (Fig. 2B). As expected, the results
demonstrated a clear positive correlation between miR-200b and CDH1
expression according to the Pearson data (r2=0.8165;
Fig. 2C). By contrast, an inverse
correlation between IC50 and miR-200b or CDH1 was
observed (r2=0.7042 and r2=0.9030,
respectively; Fig. 2C). These data
indicated that miR-200b is a putative biomarker for
chemosensitivity in pancreatic carcinoma.
Forced expression of miR-200b induces
CDH1 expression and promotes gemcitabine sensitivity in Capan-2 and
Panc-1 cells
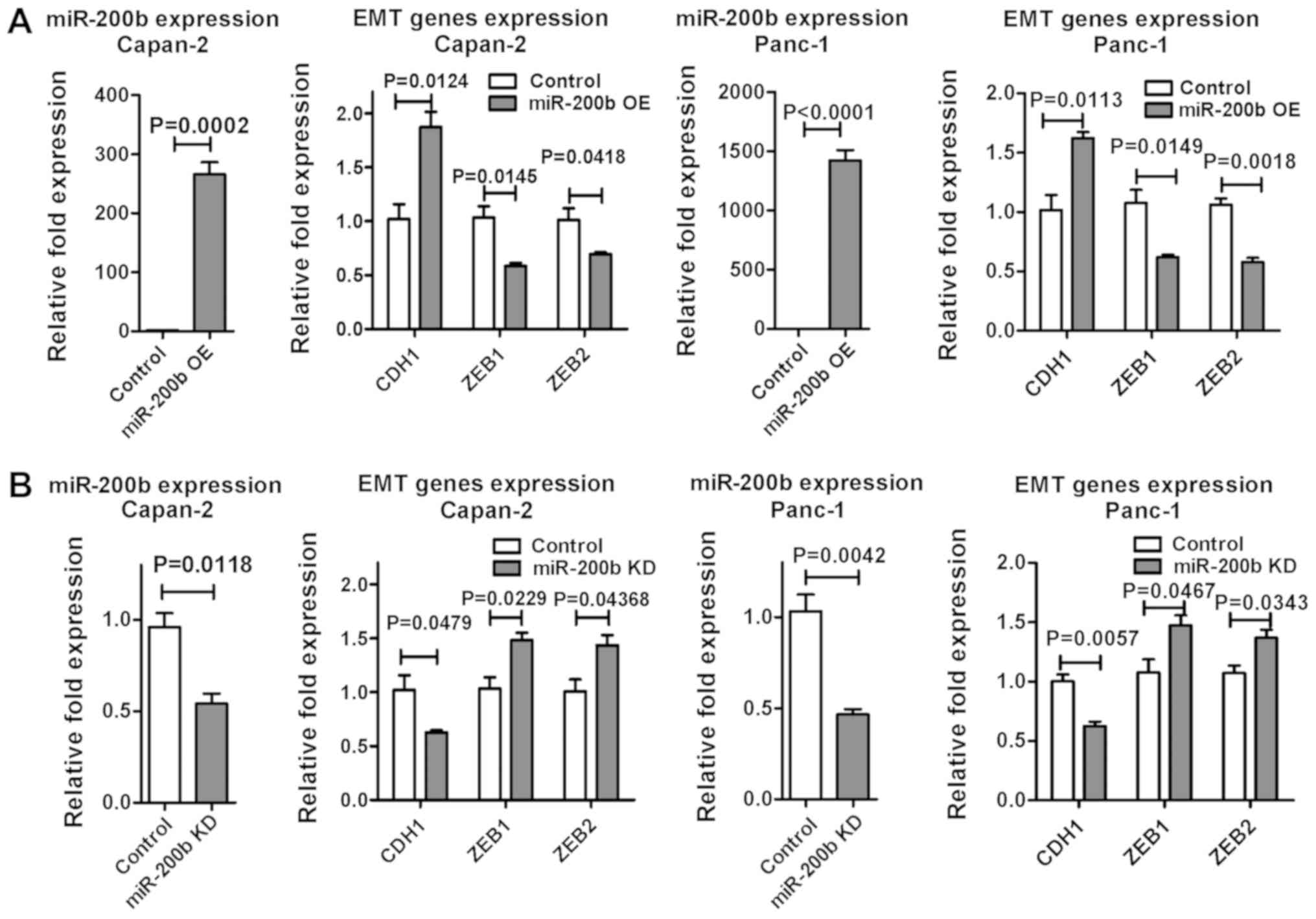
To evaluate the functional role of miR-200b in
pancreatic carcinoma, pre-miR-200b or miR-200b inhibitor were
transfected into Capan-2 and Panc-1 cells using RNAiMAX
Transfection Reagent. The efficacy of the transfection was
confirmed using RT-qPCR. miR-200b overexpression upregulated CDH1,
and suppressed ZEB1 and ZEB2 (Fig.
3A). Conversely, miR-200b inhibition reduced CDH1 expression in
the two cell lines (Fig. 3B).
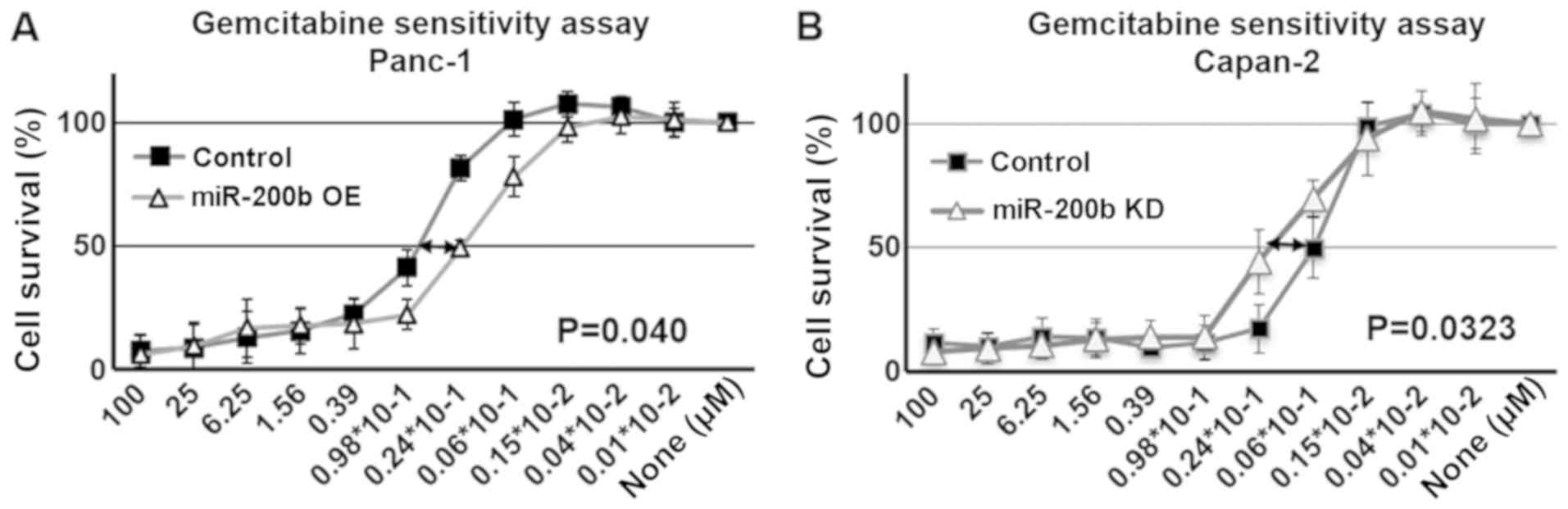
Moreover, miR-200b overexpression improved sensitivity to
gemcitabine, and miR-200b inhibitor significantly affected
gemcitabine sensitivity in the two cell lines (Fig. 4). Furthermore, the capacity for
cell growth was evaluated using a proliferation assay in the two
cell types. Forced expression of miR-200b reduced cell growth,
while miR-200b inhibition did not affect cell growth (Fig. 5A). These results were consistent
across the two cell lines were in accordance with a previous study
(25).
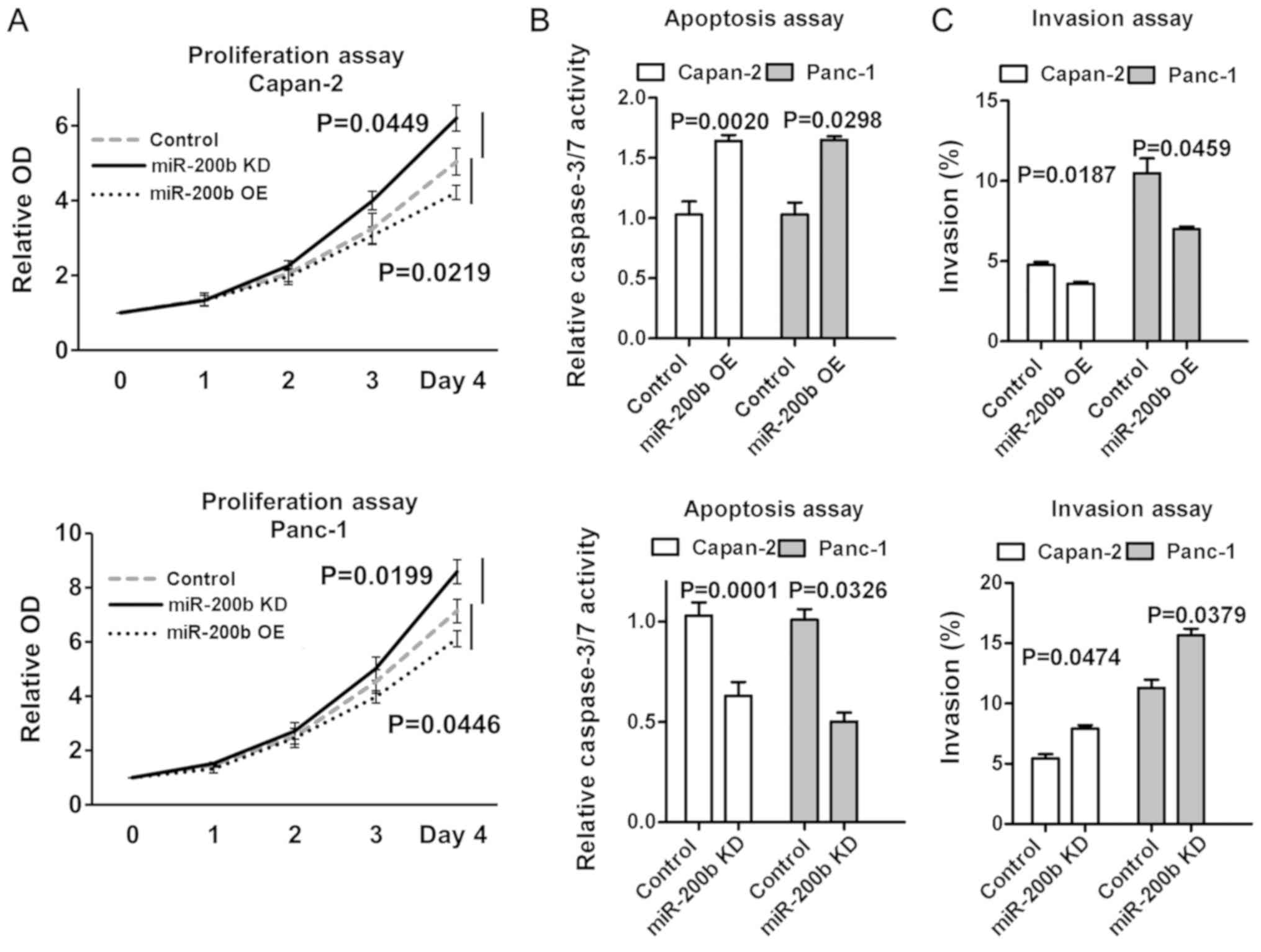 | Figure 5Further analysis of the effect of
miR-200b on cell proliferation, apoptosis and invasion. (A)
According to the colorimetric assay, miR-200b significantly reduced
cell proliferation in Capan-2 and Panc-1 cell lines compared with
the control. By contrast, miR-200b inhibition induced cell growth
in the two cell lines. Values are the mean of three independent
experiments performed in triplicate. Error bars indicate the
standard deviation. Data from Day 4 were used for the statistical
analysis. (B) Capan-2 or Panc-1 cells were seeded in 96-well plates
having been transfected overnight with 50 nM miR-200b (miR-200b OE)
or miR-200b inhibitor (miR-200b KD), or their controls. miR-200b OE
contributed to the induction of apoptosis. Successful induction of
apoptosis was assessed by measuring caspase-3/7 activity. Data
represent the mean of three replicates ± standard deviation. (C)
Following transfection with 50 nM miR-200b (miR-200b OE) or
miR-200b inhibitor (miR-200b KD), or their controls, cells were
seeded in Matrigel-coated chambers for the invasion assays.
miR-200b OE significantly reduced cell invasiveness in the two cell
lines. Invasive cells were fixed with methanol prior to being
stained with crystal violet and photographed. Stained cells were
counted in three separate microscopic fields per well. The values
were averaged, and the mean ± standard deviation was calculated
from triplicate samples. miR, microRNA; OE, overexpression; KD,
knockdown; OD, optical density. |
miR-200b and miR-301 expression affects
apoptosis and cell invasiveness
Overexpressed miR-200b enhanced apoptosis and
inhibited cell invasiveness in the two cell lines (Fig. 5B and C). By contrast, downregulated
miR-200b reduced apoptosis and enhanced cell invasiveness (Fig. 5B and C).
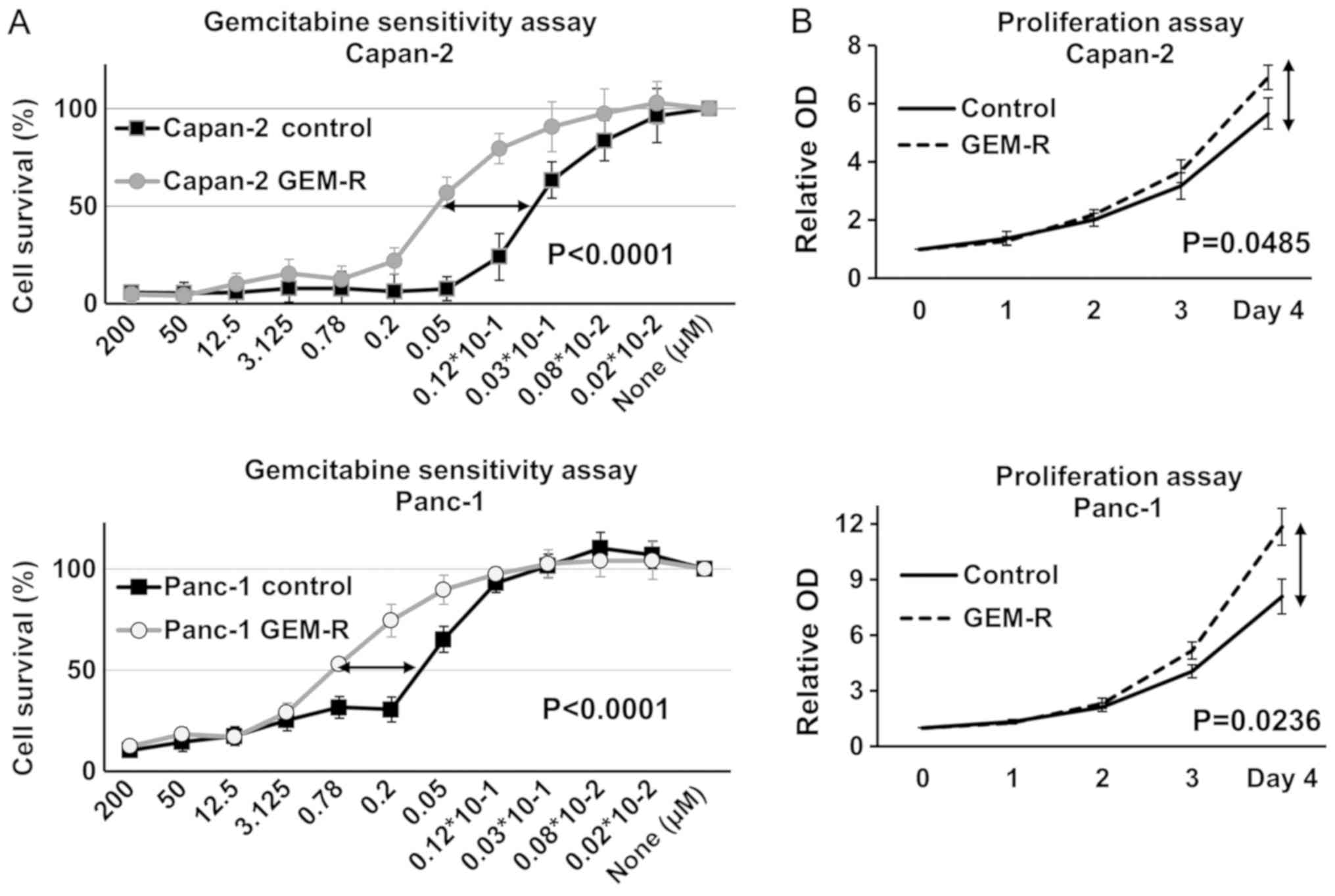
Gemcitabine-resistant cells were
generated from Capan-2 and Panc-1 cells
To examine the mechanisms underlying gemcitabine
resistance, two sub-cell lines were generated, which were derived
from Capan-2 and Panc-1 cells. These were cultured in the presence
of high concentrations of gemcitabine (IC50), by
gradually increasing the concentration of gemcitabine over 5
months. Cells in which the IC50 values were over four
times higher compared with their controls were defined as Capan-2
GEM-R and Panc-1 GEM-R. The IC50 of these cells was
evaluated using a colorimetric assay, and they exhibited strong
gemcitabine resistance compared with each control cell line
(Fig. 6A). Moreover, the two GEM-R
cell lines exhibited increased cell growth (Fig. 6B).
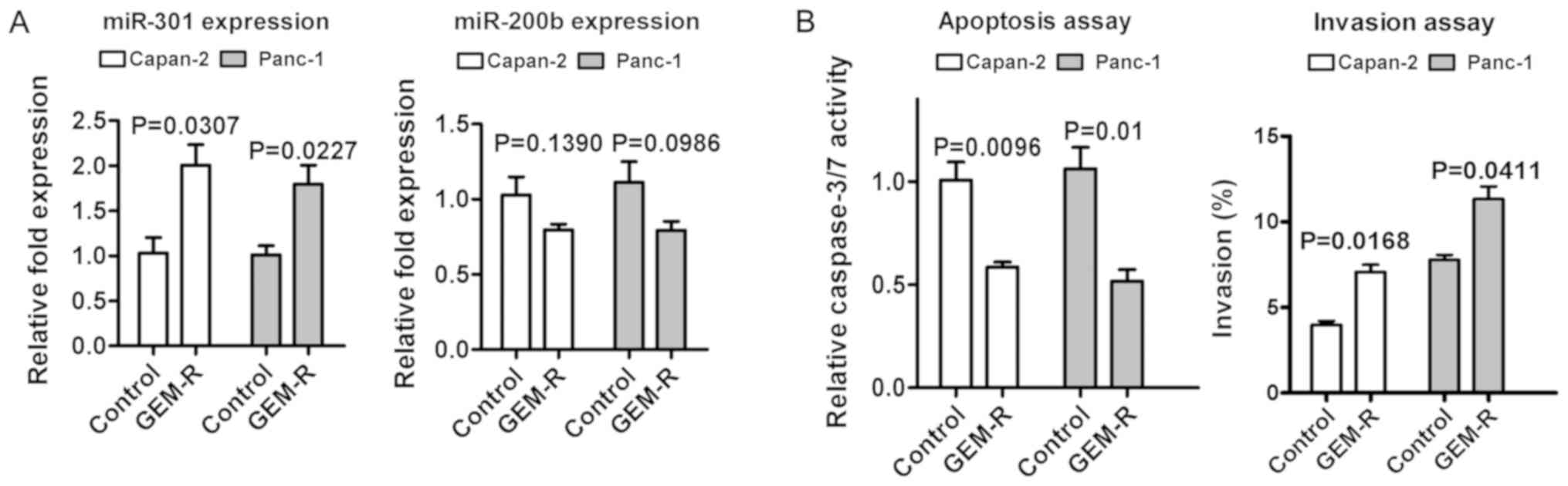
Gemcitabine-resistant Capan-2 and Panc-1
cells exhibited increased miR-301 expression
The present study also examined whether acquired
gemcitabine resistance in cells was associated with miR-301
expression. Gemcitabine-resistant cells exhibited increased miR-301
and reduced miR-200b expression (Fig.
7A). No explanation became clear as to why miR-200b was reduced
in gemcitabine-resistant cells. However, Wang et al
(34) recently reported that
gemcitabine treatment induced reduced miR-200b in pancreatic
carcinoma cell lines. In addition, resistant cells exhibited an
anti-apoptotic effect and increased cell invasiveness compared with
control cells (Fig. 7B).
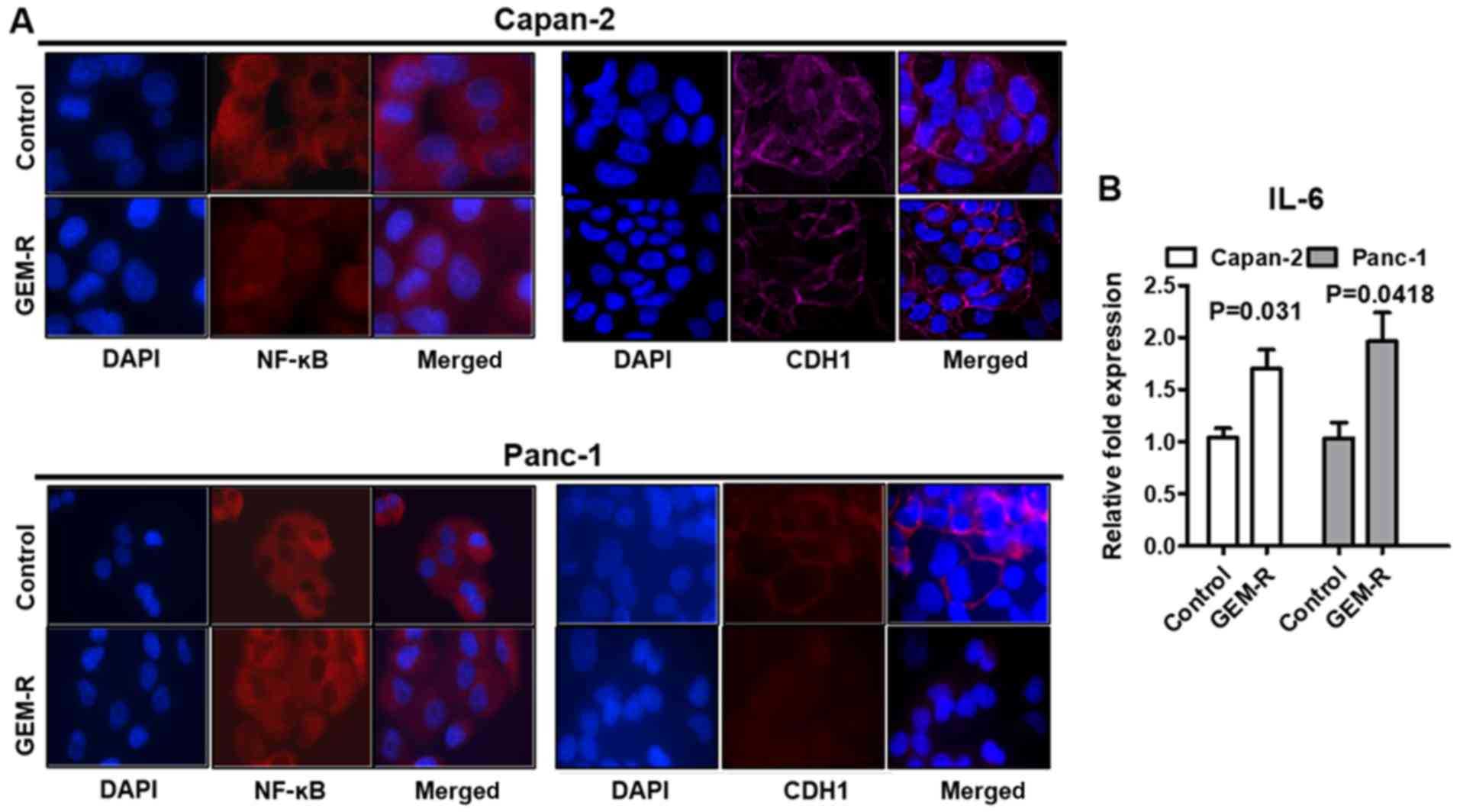
Gemcitabine-resistant cells revealed
activated NF-κB and decreased CDH1 expression at the protein
level
In the two gemcitabine-resistant cell lines,
immunofluorescence was performed for NF-κB and CDH1.
Gemcitabine-resistant cells exhibited NF-κB activation and
decreased CDH1 expression, as expected (Fig. 8A). The difference was less marked
in Panc-1 cells due to the lower expression levels of CHD1 and
higher activation levels of NF-κB in the control cells. It was not
possible within the scope of this investigation to perform western
blot analysis using a fractionated sample (Cyto/Nuc) and
quantification of the expression of target proteins of NF-κB. As an
alternative, IL-6 expression was measured to verify the NF-κB
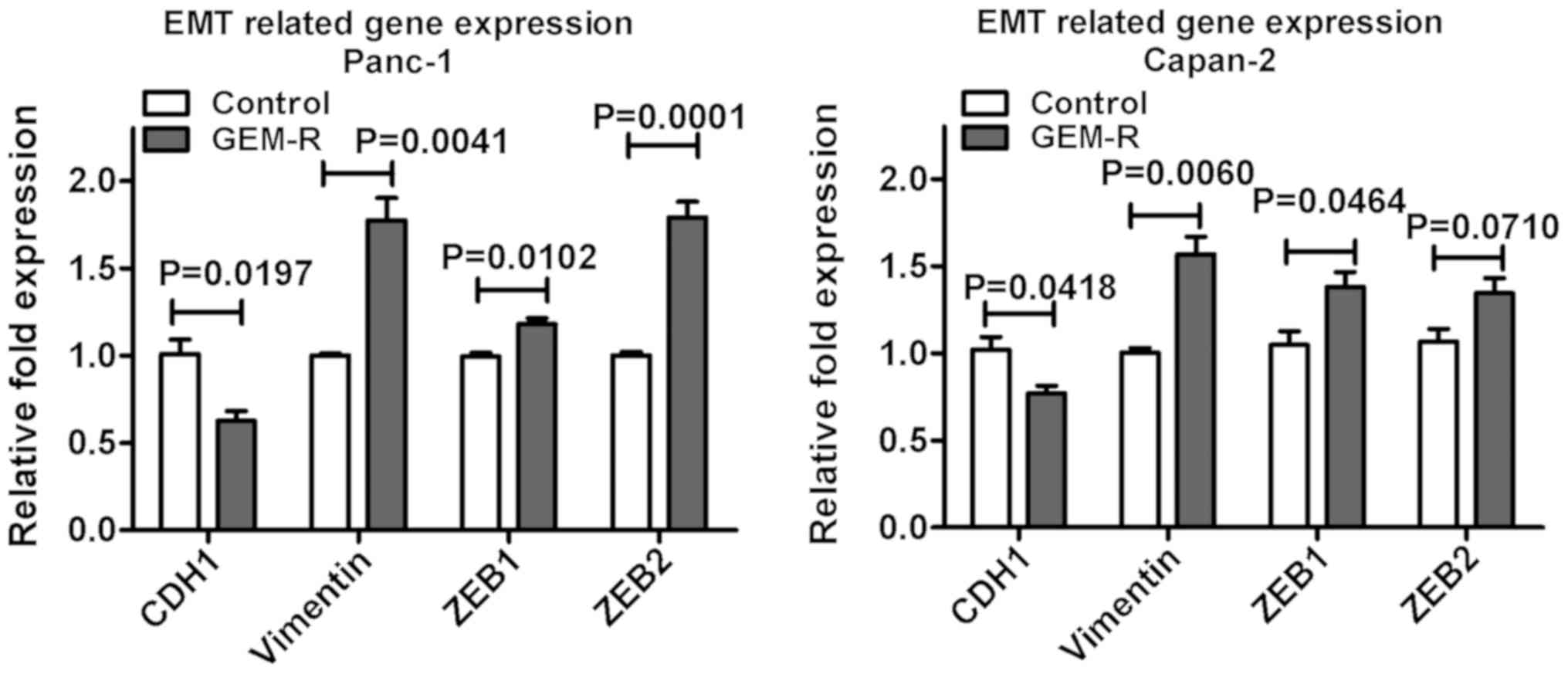
activation in gemcitabine-resistant cells (Fig. 8B). In addition, RT-qPCR analysis
demonstrated a decrease in CDH1 expression. The expression of
EMT-associated genes, including vimentin, ZEB1 and ZEB2, was
measured using RT-qPCR. The results did not exhibit a consistent
pattern of gene expression to explain how gemcitabine resistance
led to EMT in the cell lines (Fig.
9), although western blot analysis was not presented. In
addition, morphology remained unaltered in gemcitabine-resistant
cells (data not shown). In the same manner, miR-301 overexpression
revealed decreased CDH1 expression levels, an enhanced
anti-apoptotic effect, an increased IC50 of gemcitabine
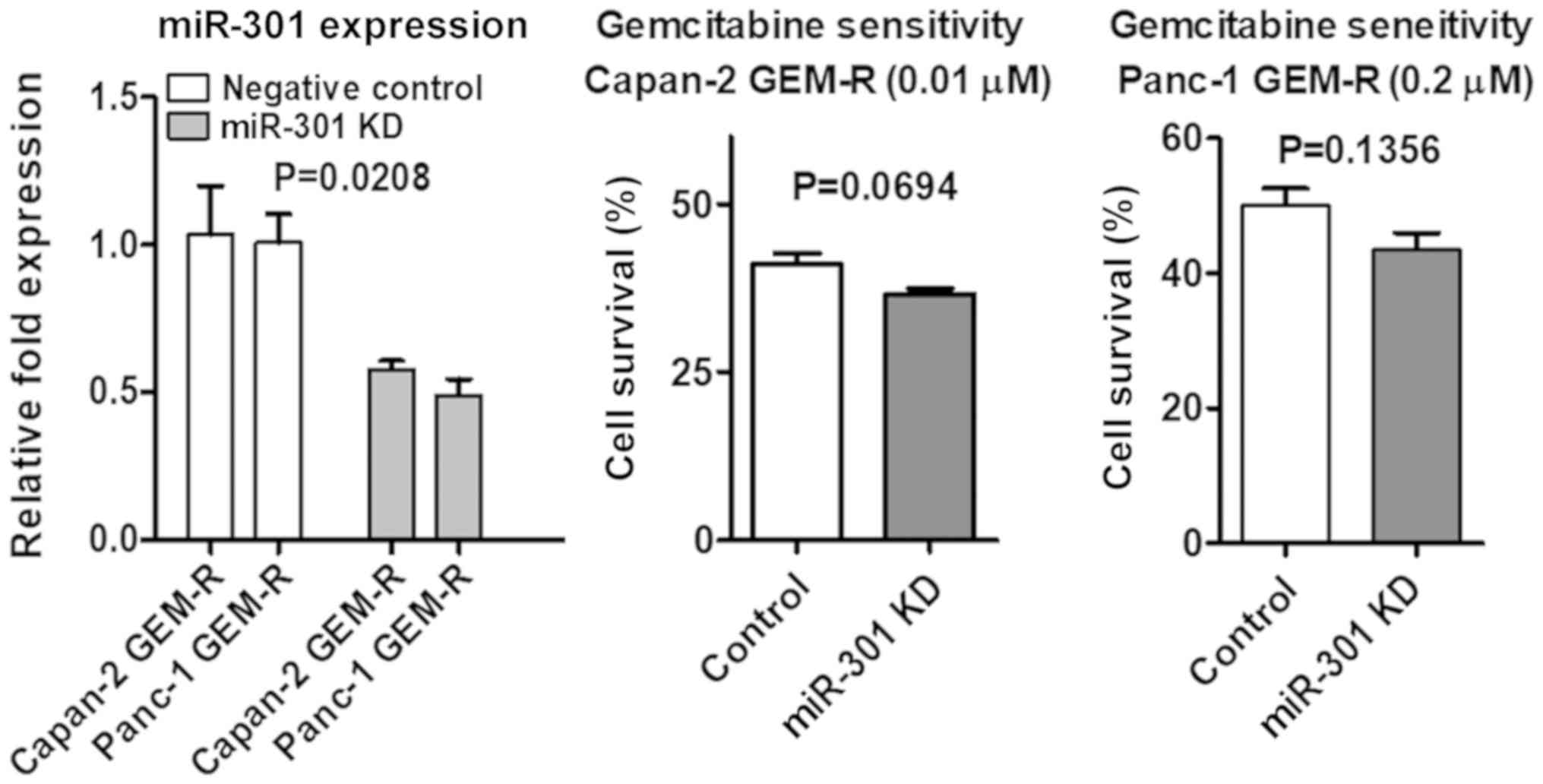
and increased cell invasiveness, as previously reported (13). Contrary to expectations, miR-301
inhibition did not induce the remission of gemcitabine resistance
(Fig. 10). These data are
consistent with a previous report (13). Based on the present data, elevated
miR-301 expression may affect acquired gemcitabine resistance, and
an increase in miR-301 may be a predictive biomarker for acquired
gemcitabine resistance in patients with pancreatic carcinoma.
Discussion
Pancreatic carcinoma has an extremely poor
prognosis; early diagnosis is difficult to achieve, and it exhibits
aggressive invasion, early distant metastasis, and resistance to
anticancer drugs (1,35). However, gemcitabine-based
chemotherapy remains the first-line chemotherapy regimen (36). Gemcitabine became the most widely
used anticancer drug for the initial treatment of advanced and
recurrent pancreatic carcinoma when Burris et al (37) reported in 1997 that gemcitabine had
better clinical outcomes compared with 5-fluorouracil. Gemcitabine
contributed only 6.2 months to the median survival time for locally
advanced or metastatic pancreatic carcinoma (36). Recently, a number of trials have
been performed to compare gemcitabine alone with combinations with
alternative drugs; however, the combined regimens, including
oxaliplatin, erlotinib and nab-paclitaxel, have not produced
markedly different results in terms of overall survival (38-40).
Therefore, it is necessary to identify effective biomarkers for
chemosensitivity to gemcitabine and to elucidate the mechanism
underlying the development of gemcitabine resistance. Recently,
evidence has suggested that a number of miRNAs may affect
chemosensitivity to gemcitabine for the treatment of malignant
tumors, including miR-301, miR-200b, miR-29a and miR-145 (13,21,41,42).
The present study focused on miR-200b and miR-301, as they are
known to be involved in the progression of several types of cancers
(13,17,18).
Since the molecular mechanisms of miR-200b and miR-301 in
pancreatic carcinoma in relation to gemcitabine treatment remain
unclear, it was hypothesized that the expression levels of miR-200b
and miR-301 may serve as surrogate predictors of chemosensitivity
and chemoresistance through EMT in pancreatic carcinoma treated
with gemcitabine. miR-200b is known to be a tumor suppressor, and
serves an important role in the development and progression of
malignant tumors. A number of studies have demonstrated that
miR-200b expression is reduced in various types of cancer,
including pancreatic carcinoma (18,22).
Additionally, Gui et al (43) reported that overexpressed miR-200b
significantly inhibited cell migration by targeting ZEB1 in
pancreatic carcinoma cells. Moreover, it was demonstrated that
overexpressed miR-200b increased chemosensitivity to gemcitabine
(43). Notably, a recent report
revealed that docetaxel chemoresistance is closely associated with
the downregulation of miR-200b and the corresponding upregulation
of autophagy-associated gene 12 in lung cancer (44). Furthermore, Asakura et al
(45) reported that miR-200b is
associated with proteasome inhibitor resistance by targeting
E-cadherin suppression. Consistently, our previous study also
illustrated the same phenomenon of gemcitabine resistance through
EMT in pancreatic carcinoma cells (25). Therefore, miR-200b may be
associated with the regulation of gemcitabine sensitivity as a
tumor suppressor gene. Secondarily, accumulated evidence has
revealed that miR-301 expression is upregulated in a number of
malignant tumors (46).
Furthermore, miR-301 reportedly functions as an oncogene (47). In addition, Shi et al
(26) reported that reduced
miR-301 expression increased tamoxifen sensitivity via targeting of
forkhead box F2, BCL2 biding component 3, phosphatase and tensin
homolog and collagen type II α1 chain. Moreover, a recent study
revealed that miR-301 contributes to the activation of NF-κB in
pancreatic carcinoma (48). In
previous work, it was demonstrated that the miR-301/NF-κB axis
critically promotes the upregulation of gemcitabine resistance in
pancreatic carcinoma cells (13).
Therefore, the present findings indicated that miR-301 may serve an
important role as an oncogene.
In the current study, it was identified that
miR-200b expression correlated positively with IC50 in
six pancreatic carcinoma cell lines, and overexpressed miR-200b
affected gemcitabine sensitivity, cell invasiveness, apoptosis and
cell proliferation. These results suggested that miR-200b
expression may predict gemcitabine sensitivity prior to the
introduction of initial chemotherapy. By contrast, it was observed
that miR-301 promoted gemcitabine resistance in Capan-2 and Panc-1
cells. Furthermore, it was demonstrated that gemcitabine-resistant
cells exhibited increased miR-301 expression in the Capan-2 and
Panc-1 cell lines. Capan-2-GEM-R and Panc-1-GEM-R cells exhibited
increased IC50 values for gemcitabine compared with
their respective control cell lines. By contrast, exogenous miR-301
inhibited apoptosis and enhanced cell invasion, consistent with
previous data (13). However, a
limitation of the present study is the lack of flow cytometry or
data on poly-ADP-ribose polymerase to reinforce the association
between miR-301 and apoptosis. Notably, miR-200b and miR-301
affected EMT through CDH1 expression (13,25).
However, no morphological alterations were observed in either cell
line despite the alteration of CDH1 expression. Based on these
results and the evidence above, the present data suggested that
miR-200b and miR-301 may be predictive markers for response to
gemcitabine in patients with pancreatic carcinoma. However, the
exact molecular mechanisms underlying the ability of miR-200b and
miR-301 to regulate chemosensitivity and acquired chemoresistance
are unknown, and these mechanisms require further investigation and
validation in additional studies in vivo.
In conclusion, the present study provided evidence
that miR-200b and elevated miR-301 may be useful biomarkers to
predict the chemosensitivity and acquired chemoresistance of
pancreatic carcinoma to gemcitabine. The present data also
indicated that tailoring treatments according to miR-200b and
miR-301 expression in such patients may be considered for the
treatment of pancreatic carcinoma in routine clinical practice.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NF performed the experimental studies, and drafted
and completed the manuscript. CRL and MK participated in the design
of the study. KY and YM conceived the project and supervised the
research. NF conceived of the study and performed the statistical
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Noriko Funamizu
(Department of Internal Medicine, Hirose Hospital, Ehime, Japan)
for helpful discussions throughout this work.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oettle H: Progress in the knowledge and
treatment of advanced pancreatic cancer: From benchside to bedside.
Cancer Treat Rev. 40:1039–1047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paulson AS, Tran Cao HS, Tempero MA and
Lowy AM: Therapeutic advances in pancreatic cancer.
Gastroenterology. 144:1316–1326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li D, Xie K, Wolff R and Abbruzzese JL:
Pancreatic cancer. Lancet. 363:1049–1057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nikitina EG, Urazova LN and Stegny VN:
MicroRNAs and human cancer. Exp Oncol. 34:2–8. 2012.PubMed/NCBI
|
|
8
|
Piepoli A, Tavano F, Copetti M, Mazza T,
Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia
G, et al: Mirna expression profiles identify drivers in colorectal
and pancreatic cancers. PLoS One. 7:e336632012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Subramani R, Gangwani L, Nandy SB,
Arumugam A, Chattopadhyay M and Lakshmanaswamy R: Emerging roles of
microRNAs in pancreatic cancer diagnosis, therapy and prognosis
(Review). Int J Oncol. 47:1203–1210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li C, Hashimi SM, Good DA, Cao S, Duan W,
Plummer PN, Mellick AS and Wei MQ: Apoptosis and microRNA
aberrations in cancer. Clin Exp Pharmacol Physiol. 39:739–746.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou
C and Zhou J: A miR-200b/200c/429-binding site polymorphism in the
3′ untranslated region of the AP-2α gene is associated with
cisplatin resistance. PLoS One. 6:e290432011. View Article : Google Scholar
|
|
12
|
Garofalo M, Romano G, Di Leva G, Nuovo G,
Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al:
EGFR and MET receptor tyrosine kinase-altered microRNA expression
induces tumorigenesis and gefitinib resistance in lung cancers. Nat
Med. 18:74–82. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Funamizu N, Lacy CR, Parpart ST, Takai A,
Hiyoshi Y and Yanaga K: MicroRNA-301b promotes cell invasiveness
through targeting TP63 in pancreatic carcinoma cells. Int J Oncol.
44:725–734. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Z, Li Y, Ahmad A, Azmi AS, Kong D,
Banerjee S and Sarkar FH: Targeting miRNAs involved in cancer stem
cell and EMT regulation: An emerging concept in overcoming drug
resistance. Drug Resist Updat. 13:109–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang WL, Zhang JH, Wu XZ, Yan T and Lv W:
miR-15b promotes epithelial-mesenchymal transition by inhibiting
SMURF2 in pancreatic cancer. Int J Oncol. 47:1043–1053. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bai Z, Sun J, Wang X, Wang H, Pei H and
Zhang Z: MicroRNA-153 is a prognostic marker and inhibits cell
migration and invasion by targeting SNAI1 in human pancreatic
ductal adenocarcinoma. Oncol Rep. 34:595–602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pacurari M, Addison JB, Bondalapati N, Wan
YW, Luo D, Qian Y, Castranova V, Ivanov AV and Guo NL: The
microRNA-200 family targets multiple non-small cell lung cancer
prognostic markers in H1299 cells and BEAS-2B cells. Int J Oncol.
43:548–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J,
Yang SH, Song H, Lim BJ and Kim SH: MicroRNA-200 family members and
ZEB2 are associated with brain metastasis in gastric
adenocarcinoma. Int J Oncol. 45:2403–2410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen A, Lin W, Chen Y, Liu L, Chen H,
Zhuang Q, Lin J, Sferra TJ and Peng J: Pien Tze Huang inhibits
metastasis of human colorectal carcinoma cells via modulation of
TGF-β1/ZEB/ miR-200 signaling network. Int J Oncol. 46:685–690.
2015. View Article : Google Scholar
|
|
20
|
Li L, Li B, Chen D, Liu L, Huang C, Lu Z,
Lun L and Wan X: miR-139 and miR-200c regulate pancreatic cancer
endothelial cell migration and angiogenesis. Oncol Rep. 34:51–58.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang L, Guo F, Huo B, Lv Y, Wang Y and
Liu W: Expression and clinical significance of the microRNA-200
family in gastric cancer. Oncol Lett. 9:2317–2324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M,
Zhang W, Chen W, Pan C, Liu Q, et al: MiR-200b and miR-15b regulate
chemotherapy-induced epithelial-mesenchymal transition in human
tongue cancer cells by targeting BMI1. Oncogene. 31:432–445. 2012.
View Article : Google Scholar
|
|
25
|
Funamizu N, Hu C, Lacy C, Schetter A,
Zhang G, He P, Gaedcke J, Ghadimi MB, Ried T, Yfantis HG, et al:
Macrophage migration inhibitory factor induces epithelial to
mesenchymal transition, enhances tumor aggressiveness and predicts
clinical outcome in resected pancreatic ductal adenocarcinoma. Int
J Cancer. 132:785–794. 2013. View Article : Google Scholar
|
|
26
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duxbury MS, Ito H, Zinner MJ, Ashley SW
and Whang EE: Inhibition of SRC tyrosine kinase impairs inherent
and acquired gemcitabine resistance in human pancreatic
adenocarcinoma cells. Clin Cancer Res. 10:2307–2318. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Funamizu N, Kamata Y, Misawa T, Uwagawa T,
Lacy CR, Yanaga K and Manome Y: Hydroxyurea decreases gemcitabine
resistance in pancreatic carcinoma cells with highly expressed
ribonucleotide reductase. Pancreas. 41:107–113. 2012. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
30
|
Funamizu N, Okamoto A, Kamata Y, Misawa T,
Uwagawa T, Gocho T, Yanaga K and Manome Y: Is the resistance of
gemcitabine for pancreatic cancer settled only by overexpression of
deoxycytidine kinase? Oncol Rep. 23:471–475. 2010.PubMed/NCBI
|
|
31
|
Funamizu N, Lacy CR, Fujita K, Furukawa K,
Misawa T, Yanaga K and Manome Y: Tetrahydrouridine inhibits cell
proliferation through cell cycle regulation regardless of cytidine
deaminase expression levels. PLoS One. 7:e374242012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kamada M, Akiyoshi K, Akiyama N, Funamizu
N, Watanabe M, Fujioka K, Ikeda K and Manome Y: Cholangiocarcinoma
cell line TK may be useful for the pharmacokinetic study of the
chemotherapeutic agent gemcitabine. Oncol Rep. 32:829–834. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Funamizu N, Lacy CR, Kamada M, Yanaga K
and Manome Y: MicroRNA-203 induces apoptosis by upregulating Puma
expression in colon and lung cancer cells. Int J Oncol.
47:1981–1988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Z, Chen Y, Lin Y, Wang X, Cui X,
Zhang Z, Xian G and Qin C: Novel crosstalk between KLF4 and ZEB1
regulates gemcitabine resistance in pancreatic ductal
adenocarcinoma. Int J Oncol. 51:1239–1248. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gillen S, Schuster T, Meyer Zum
Büschenfelde C, Friess H and Kleeff J: Preoperative/neoadjuvant
therapy in pancreatic cancer: A systematic review and meta-analysis
of response and resection percentages. PLoS Med. 7:e10002672010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Abou-Alfa GK, Letourneau R, Harker G,
Modiano M, Hurwitz H, Tchekmedyian NS, Feit K, Ackerman J, De Jager
RL, Eckhardt SG, et al: Randomized phase III study of exatecan and
gemcitabine compared with gemcitabine alone in untreated advanced
pancreatic cancer. J Clin Oncol. 24:4441–4447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Burris HA III, Moore MJ, Andersen J, Green
MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo
AM, Tarassoff P, et al: Improvements in survival and clinical
benefit with gemcitabine as first-line therapy for patients with
advanced pancreas cancer: A randomized trial. J Clin Oncol.
15:2403–2413. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Louvet C, Labianca R, Hammel P, Lledo G,
Zampino MG, André T, Zaniboni A, Ducreux M, Aitini E, Taïeb J, et
al GERCOR; GISCAD: Gemcitabine in combination with oxaliplatin
compared with gemcitabine alone in locally advanced or metastatic
pancreatic cancer: Results of a GERCOR and GISCAD phase III trial.
J Clin Oncol. 23:3509–3516. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vickers MM, Powell ED, Asmis TR, Jonker
DJ, Hilton JF, O'Callaghan CJ, Tu D, Parulekar W and Moore MJ:
Comorbidity, age and overall survival in patients with advanced
pancreatic cancer - results from NCIC CTG PA.3: A phase III trial
of gemcitabine plus erlotinib or placebo. Eur J Cancer.
48:1434–1442. 2012. View Article : Google Scholar
|
|
40
|
Von Hoff DD, Ramanathan RK, Borad MJ,
Laheru DA, Smith LS, Wood TE, Korn RL, Desai N, Trieu V, Iglesias
JL, et al: Gemcitabine plus nab-paclitaxel is an active regimen in
patients with advanced pancreatic cancer: A phase I/II trial. J
Clin Oncol. 29:4548–4554. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nagano H, Tomimaru Y, Eguchi H, Hama N,
Wada H, Kawamoto K, Kobayashi S, Mori M and Doki Y: MicroRNA-29a
induces resistance to gemcitabine through the Wnt/β-catenin
signaling pathway in pancreatic cancer cells. Int J Oncol.
43:1066–1072. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lin Y, Ge X, Wen Y, Shi ZM, Chen QD, Wang
M, Liu LZ, Jiang BH and Lu Y: MiRNA-145 increases therapeutic
sensibility to gemcitabine treatment of pancreatic adenocarcinoma
cells. Oncotarget. 7:70857–70868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gui Z, Luo F, Yang Y, Shen C, Li S and Xu
J: Oridonin inhibition and miR 200b 3p/ZEB1 axis in human
pancreatic cancer. Int J Oncol. 50:111–120. 2017. View Article : Google Scholar
|
|
44
|
Pan B, Feng B, Chen Y, Huang G, Wang R,
Chen L and Song H: MiR-200b regulates autophagy associated with
chemoresistance in human lung adenocarcinoma. Oncotarget.
6:32805–32820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Asakura T, Yamaguchi N, Ohkawa K and
Yoshida K: Proteasome inhibitor-resistant cells cause EMT-induction
via suppression of E-cadherin by miR-200 and ZEB1. Int J Oncol.
46:2251–2260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar
|
|
47
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu Z and Li Y, Takwi A, Li B, Zhang J,
Conklin DJ, Young KH, Martin R and Li Y: miR-301a as an NF-κB
activator in pancreatic cancer cells. EMBO J. 30:57–67. 2011.
View Article : Google Scholar
|