Introduction
Colorectal cancer (CRC) is the third most common
type of cancer in men and the second most common in women world
wide; every year, ~1.4 million new cases of CRC are diagnosed and
it is responsible for ~700,000 cases of mortality (1,2).
Although screening and multidisciplinary treatment has improved
therapeutic outcomes in several countries, due to its high
incidence, CRC remains a major healthcare challenge worldwide.
Therefore, specific biomarkers for predicting treatment outcomes,
as well as key target molecules responsible for cancer progression,
need to be identified.
Transcriptional activity is frequently dysregulated
in cancer, due to genomic and epigenetic alterations. Polymerase
(Pol) II is responsible for the transcription of mRNAs, including
oncogenes, and other Pol enzymes, including Pol I and III, are also
involved in cancer progression via ribosomal RNA (rRNA) and
transfer RNA (tRNA) biosynthesis (3,4).
Although transcription of protein coding genes is Pol II dependent,
oncogenes and tumor suppressors, including MYC, phosphoinositide 3
kinase and phosphatase and tensin homolog directly or indirectly
activate or inhibit transcription by Pol I and III, thus resulting
in altered rRNA production, which is required for rapid cell growth
(5).
The present study focused on MAF1 homolog, negative
regulator of RNA Pol III (MAF1). Although MAF1 has been
demonstrated to inhibit Pol III dependent transcription, a certain
number of Pol II dependent genes are also considered to be
important targets of this gene (6,7). For
example, previous studies demonstrated a conserved function of MAF1
in the maintenance of intracellular lipid pools, through regulation
of fatty acid synthase and acetyl coA carboxylase expression
(8,9). These findings suggest that MAF1 may
not only be involved in a cell growth through rRNA and tRNA
mediated post transcriptional regulation, but may also affect
various biological and pathological processes, including malignant
potential of cancer.
Previous studies have reported the tumor suppressive
effect of MAF1 in certain solid malignancies (7,8,10).
However, the clinical significance has not been investigated in
CRC, which has a different genetic background and immune
environment. The present study used surgical specimens and a large
scale, multi layered open database to investigate the clinical
significance of MAF1 in CRC. Furthermore, the association between
MAF1 expression in cancer cells and tumor immunity, which is an
important factor for predicting prognosis, was explored.
Materials and methods
Clinical samples
Primary CRC specimens were collected from 146
patients who underwent surgery at the Department of
Gastroenterological Surgery, Osaka University (Suita, Japan)
between January 2011 and December 2012. All patients were clearly
diagnosed with CRC, based on the clinicopathological criteria
described by the Japanese Society for Cancer of the Colon and
Rectum (11). None of the patients
received preoperative chemotherapy or radiotherapy. Specimens were
fixed in 10% buffered formalin overnight at room temperature,
processed through graded ethanol solutions and embedded in
paraffin. The follow-up periods ranged between 1 month and 7 years,
with a mean of 4 years. All data, including age, sex, tumor size
and depth, lymphatic invasion, lymph node metastasis, vascular
invasion, liver metastasis, peritoneal dissemination, distant
metastasis and histological grade were obtained from clinical and
pathological records. The present study was approved by the
Research Ethics Committee of Osaka University (approval ID: 08226)
and written informed consent was obtained from all patients
included in this study.
Cell lines and cell culture
The human CRC cell lines RKO, HT29, HCT 116,
Colo205, SW480 and DLD 1 were purchased from the American Type
Culture Collection (Manassas, VA, USA). These cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) containing 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Waltham, MA,
USA) at 37˚C in a humidified incubator containing 5%
CO2.
RNA interference
Two types of MAF1-specific small inter fering RNA
(siRNA; Sigma Aldrich; Merck KGaA) were used to knockdown MAF1
mRNA. MAF1 siRNAs, or the negative control siRNA, were transfected
into RKO and HCT 116 cells, which were seeded at 2×105
cells/well in a 2 ml volume in 6-well flat-bottomed microtiter
plates, at a final concentration of 50 nM using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). These cells were maintained at 37˚C in a
humidified incubator containing 5% CO2 for 48 h,
according to the manufacturer's protocol. The sequences of siRNAs
against MAF1 were as follows: #1, CCACGCUCAAUGAGU CCUUTT; #2,
GGCUCAAGCGAAUCGUCUUTT. The MISSION® siRNA Universal
Negative Control (cat. no. SIC001; Sigma Aldrich; Merck KGaA) was
used as a negative control siRNA.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using
TRIzol® RNA Isolation Reagents (Invitrogen; Thermo
Fisher Scientific, Inc.) 48 h post-transfection as previously
described (12). RNA quality was
confirmed (RNA concentra tion >0.5 µg/µl and
optical density 260/280=1.8 2.0). cDNA was synthesized from 10 ng
total RNA using the ReverTra® Ace qPCR RT Master Mix
(Toyobo Life Science, Osaka, Japan), according to the
manufacturer's protocol. qPCR was performed using a
LightCycler® 2.0 system (Roche Applied Science,
Penzberg, Germany) and LightCycler® FastStart DNA Master
SYBR Green I (Roche Applied Science). The amplification conditions
were as follows: Initial denaturation at 95°C for 10 min, followed
by 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C
for 10 sec and extension at 72°C for 10 sec. Data were normalized
to the expression of GAPDH, which was used as an internal control
for each experiment. The following primers were used: MAF1, sense
5' ctcacagctgactgtggagact 3', antisense 5' aacatgtgtttgtcgtctc ctg
3'; and GAPDH, sense 5' agccacatcgctcagacac 3' and anti sense 5'
gcccaatacgaccaaatcc 3'.
Immunohistochemical staining
Protein expression levels of MAF1 were assessed by
immunohistochemical staining of the 146 CRC specimens. The anti
MAF1 rabbit antibody (cat. no. HPA058548; Atlas Antibodies AB,
Bromma, Sweden) and VECTASTAIN® Elite ABC Rabbit
Immunoglobulin G kit (cat. no. PK 6101; Vector Laboratories, Inc.,
Burlingame, CA, USA) were used for immunohistochemical staining,
according to the manufacturer's protocol. Specimens were fixed in
10% buffered formalin overnight at room temperature, processed
through graded ethanol solutions and embedded in paraffin. Tissue
sections (3.5 µm) were prepared from paraffin-embedded
blocks. Following antigen retrieval in 10 mM citrate buffer (pH
6.0) at 115°C for 15 min using Decloaking Chamber™ NxGen (Biocare
Medical, Pacheco, CA, USA), and the slides were incubated overnight
at 4°C with the primary antibody at 1:300 dilution, followed by
incubation at room temperature for 30 min with the secondary
antibody at 1:200 dilution. With reference to the Human Protein
Atlas (image available from v18. proteinatlas.org/ENSG00000179632MAF1/tissue/esophagus#img),
an intensity score of 2 was assigned to nuclei stained as intensely
as normal esophageal mucosa, whereas unstained nuclei were assigned
a score of 0. Nuclei that exhibited weaker staining than normal
esophageal mucosa were assigned a score of 1. In the subsequent
analysis, a score of 0 was defined as the MAF1-negative group, and
a score of 1 or 2 was defined as the MAF1 positive group. Staining
was reviewed by three independent pathologists without the
knowledge of patient outcomes. The specimens were visualized on the
light field of a confocal microscope BZ X710 (Keyence Corporation,
Osaka, Japan) and were analyzed using a BZ X analyzer (v. 1.3.0.3;
Keyence Corporation).
Scratch wound healing assay
Cells were seeded at a density of 5×105
cells/well in 6 well plates and were grown to confluence under
standard conditions. Briefly, a scratch was generated in the cell
layer using a 200 µl pipette tip, and the cells were
cultured under standard conditions in DMEM supplemented with only
1% FBS to prevent proliferation. Plates were washed with DMEM
supplemented with 1% FBS to remove non adherent cells prior to
image capture. Images were captured at 0, 24 and 48 h after scratch
generation using a confocal microscope BZ X710 (Keyence
Corporation) and were analyzed using a BZ X analyzer (v. 1.3.0.3;
Keyence Corporation). Cell migration was evaluated by measuring the
average distance between the wound edges at 10 random areas.
Chemosensitivity assay
Cells were seeded at a density of 4×103
cells/well in 96 well plates and were precultured for 24 h.
Subsequently, the cells were exposed to various concentrations of 5
fluouracil (5 FU; Tokyo Chemical Industry Co., Ltd., Tokyo, Japan)
and oxaliplatin (L OHP; Yakult Honsha Co., Ltd., Tokyo, Japan) at
37°C in a humidified incubator containing 5% CO2 for 72
h. The in vitro cytotoxic effects of 5 FU and L OHP were
evaluated using the Cell Counting kit 8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan), according to the
manufacturer's protocol. The half maximal inhibi tory concentration
values were calculated from the viability data of CRC cells treated
with each concentration of L OHP. The results were analyzed using
Bioconductor package 'drc' (https://cran.rproject.org/web/packages/drc/index.html)
with default settings; this package uses non linear regression
models.
Western blot analysis
Total proteins were extracted from cultured cells
using radioimmunoprecipitation assay buffer containing protease and
phosphatase inhibitors (Thermo Fisher Scientific, Inc.) and
concentration was determined using the bicinchoninic acid method.
Briefly, 15 µg proteins were separated by 10% SDS-PAGE and
were electrob lotted onto polyvinylidene fluoride membranes (Merck
KGaA, Darmstadt, Germany) at 300 mA for 60 min. After blocking with
3% skim milk at room temperature for 1 h, these membranes were
incubated with primary antibodies against MAF1 (cat. no. HPA058548;
Atlas Antibodies AB), poly (ADP ribose) polymerase (PARP), cleaved
PARP, caspase 3, cleaved caspase 3 (apoptosis antibody sampler kit;
cat. no. 9915; Cell Signaling Technology, Inc., Danvers, MA, USA)
and actin (cat. no. A2066; Sigma Aldrich; Merck KGaA) at 4°C
overnight. After incubating with secondary antibodies (1:100,000;
cat. no. NA934; GE Healthcare, Little Chalfont, UK), the protein
bands were detected using the Amersham Enhanced Chemiluminescence
Prime Western Blotting Detection Reagent (GE Healthcare).
Data processing
Experiments were conducted in triplicate, and data
are presented as the means ± standard deviation. The Cancer Genome
Atlas (TCGA) mRNA expression data and clinical information were
downloaded from the GDAC Firehose website (http://gdac.broadinstitute.org); the colorectal
adenocarcinoma data set (COADREAD) (n=615) was used. Regulatory T
cell (Treg) infiltration in each sample was calculated using
CIBERSORT (http://cibersort.stanford.edu) (13,14).
R programming language v3.5.0, JMP® Pro 13.2.1 (SAS
Institute Inc., Cary, NC, USA) and Microsoft®
Excel® Version 14.7.1 (Microsoft Corporation, Redmond,
WA, USA) were used for analysis. Overall survival (OS) rates and
relapse free survival (RFS) rates were calculated according to the
Kaplan Meier method and were measured from the day of surgery.
Differences between groups were estimated using χ2 test,
Student's t test, one way analysis of variance followed by post hoc
Dunnett test, or the log rank test. For univariate and multivariate
analyses of clinicopathological factors, variables with P<0.05
in the univariate analysis were used in a subsequent multivariate
analysis based on the Cox proportional hazards regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
High MAF1 expression is associated with
advanced clinicopathological factors and poor prognosis
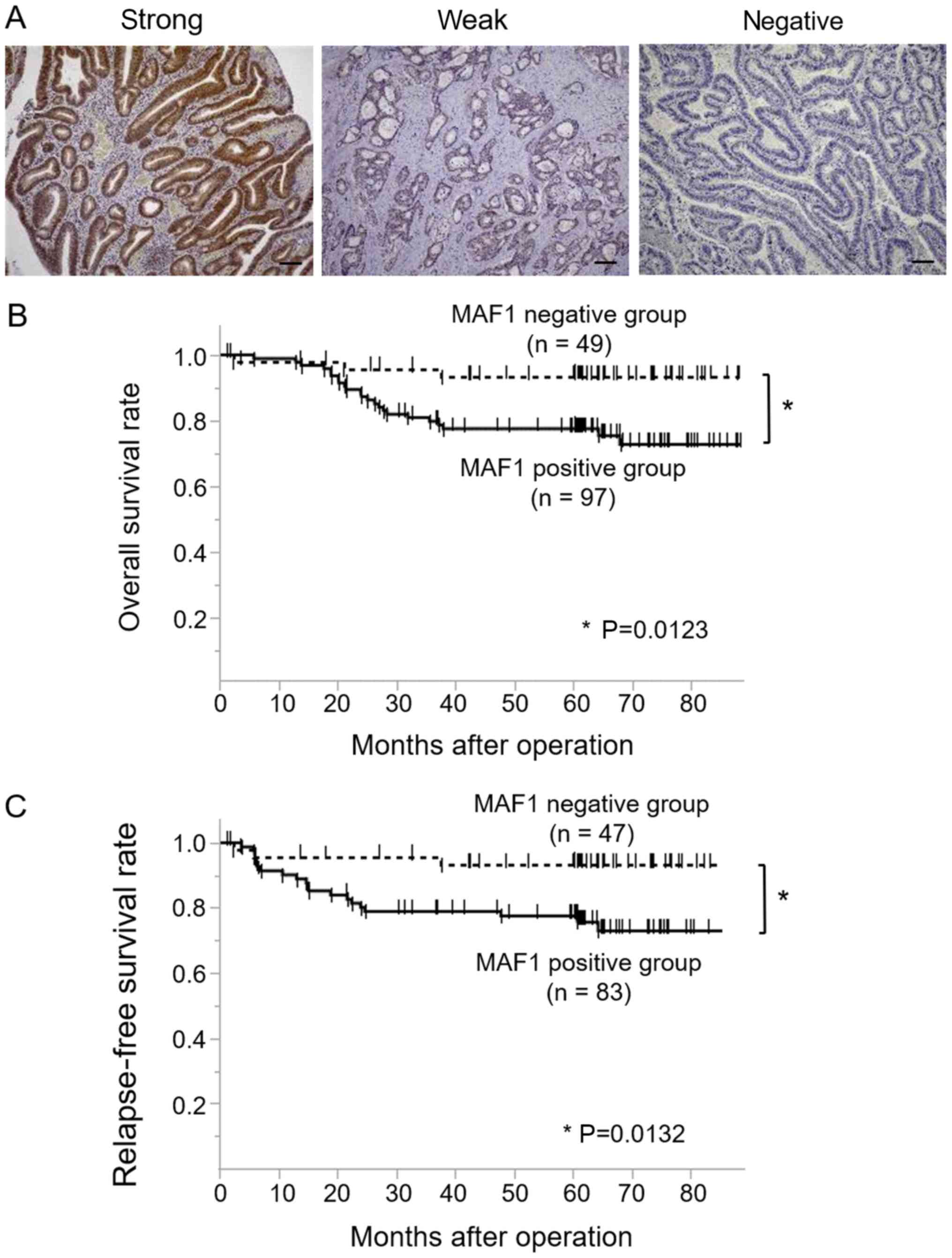
Of the 146 CRC cases, 97 cases were classified as
MAF1-positive (52 cases with strong staining and 45 cases with weak
staining), whereas 49 cases were classified as the MAF1 negative
(Fig. 1A). A clinicopathological
analysis demonstrated that high MAF1 expression was associated with
tumor malignancy, including tumor depth, lymph node metastasis,
distant metastasis and a poorer cancer stage (Table I). Kaplan Meier curves for OS
(n=146) and RFS (n=130) revealed that the MAF1 positive group
exhibited a significantly poorer prognosis (P=0.0123) and higher
relapse rate (P=0.0132) compared with the MAF1 negative group
(Fig. 1B and C). Univariate
analysis indicated that tumor depth, lymph node metastasis,
lymphatic invasion, venous invasion and MAF1 expression were
prognostic factors for overall survival, whereas multivariate
analysis revealed that tumor depth, lymph node metastasis and
venous invasion were independent prognostic factors (Table II).
 | Table IMAF1 expression and
clinicopathological factors of patients with colorecetal
cancer. |
Table I
MAF1 expression and
clinicopathological factors of patients with colorecetal
cancer.
| Factor | Positive (n=97)
Number (%) | Negative (n=49)
Number (%) | P-value |
|---|
| Age (mean ±
SD) | 63.8±1.35 | 67.8±1.56 | 0.0544 |
| Sex | |
| Male | 60 (61.9) | 32 (65.3) | 0.6827 |
| Female | 37 (38.1) | 17 (34.7) | |
| Histological
grade | |
| tub1, tub2,
pap | 87 (89.7) | 45 (91.8) | 0.6738 |
| por, muc | 10 (10.3) | 4 (8.2) | |
| Depth of tumor
invasion | |
| Tis, T1, T2 | 36 (37.1) | 31 (63.3) | 0.0027a |
| T3, T4 | 61 (62.9) | 18 (36.7) | |
| Lymph node
metastasis | |
| Absent | 61 (62.9) | 44 (89.8) | 0.0003a |
| Present | 36 (37.1) | 5 (10.2) | |
| Lymphatic
invasion | |
| Absent | 32 (33.0) | 23 (47.0) | 0.2139 |
| Present | 64 (66.0) | 25 (51.0) | |
| No data | 1 (1.0) | 1 (2.0) | |
| Venous
invasion | |
| Absent | 67 (69.1) | 39 (79.6) | 0.2890 |
| Present | 29 (29.9) | 9 (18.4) | |
| No data | 1 (1.0) | 1 (2.0) | |
| Distant
metastasis | |
| Absent | 83 (85.6) | 47 (95.9) | 0.0417a |
| Present | 14 (14.4) | 2 (4.1) | |
| Stage | |
| 0, I, II | 57 (58.8) | 44 (89.8) | 0.0011a |
| IIIa, IIIb,
IV | 40 (41.2) | 5 (10.2) | |
 | Table IIUnivariate and multivariate analyses
of overall survival in patients with colorectal cancer (Cox
regression model). |
Table II
Univariate and multivariate analyses
of overall survival in patients with colorectal cancer (Cox
regression model).
| Factors | Univariate analysis
| Multivariate
analysis
|
|---|
| RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age (≤65/>65
years) | 0.59 | 0.26-1.28 | 0.1855 | - | - | - |
| Sex
(male/female) | 1.35 | 0.60-3.28 | 0.4769 | - | - | - |
| Histology grade
(muc, por/tub1, tub2, pap) | 2.65 | 0.88-6.54 | 0.0778 | - | - | - |
| Depth of tumor
invasion (T3, T4/Tis, T1, T2) | 11.8 | 3.50-73.4 | <0.0001a | 7.76 | 1.95 52.7 | 0.0021a |
| Lymph node
metastasis (positive/negative) | 5.28 | 2.42-12.1 | <0.0001a | 2.84 | 1.10 8.30 | 0.0303a |
| Lymphatic invasion
(positive/negative) | 3.34 | 1.27-11.5 | 0.0123a | 0.49 | 0.13 2.10 | 0.3138 |
| Venous invasion
(positive/negative) | 5.13 | 2.32-11.8 | <0.0001a | 2.46 | 1.05 6.09 | 0.0389a |
| MAF1 expression
(positive/negative) | 4.12 | 1.43-17.4 | 0.0062a | 1.94 | 0.64 8.43 | 0.2666 |
MAF1 expression is associated with the
malignant potential of CRC cells
To elucidate how MAF1 contributes to the malig nant
potential of CRC, in vitro loss of function assays were
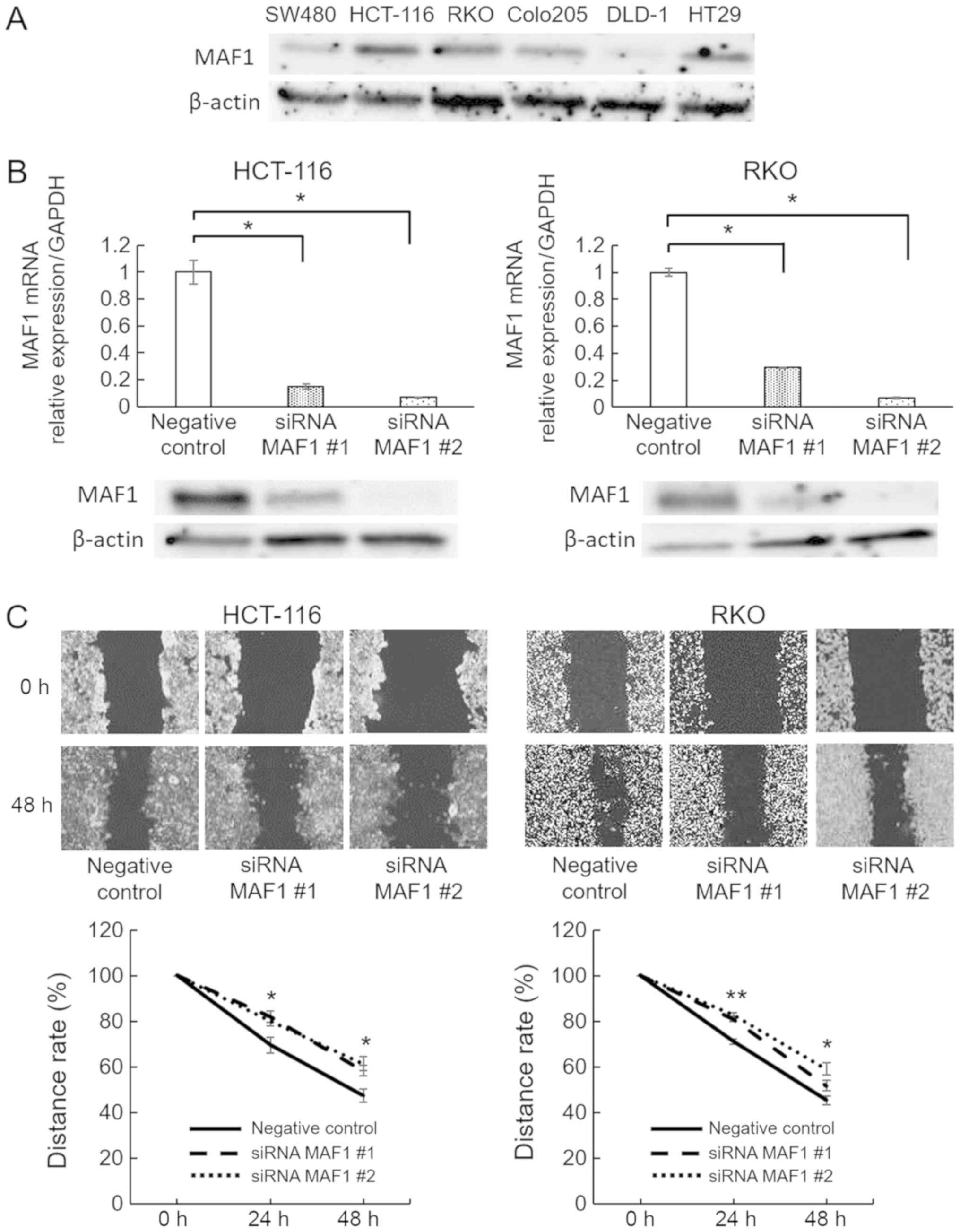
performed using RNA interference. HCT 116 and RKO CRC cell lines
were used for MAF1knockdown experiments, since they exhibited
relatively high expression levels of MAF1 in the CRC cell lines
investigated (Fig. 2A). Knockdown
efficiency was confirmed using RT-qPCR and western blotting
(Fig. 2B). Subsequently, MAF1
knockdown significantly inhibited the migratory ability of HCT 116
and RKO cells (Fig. 2C), whereas
proliferation rate was not affected (data not shown). To further
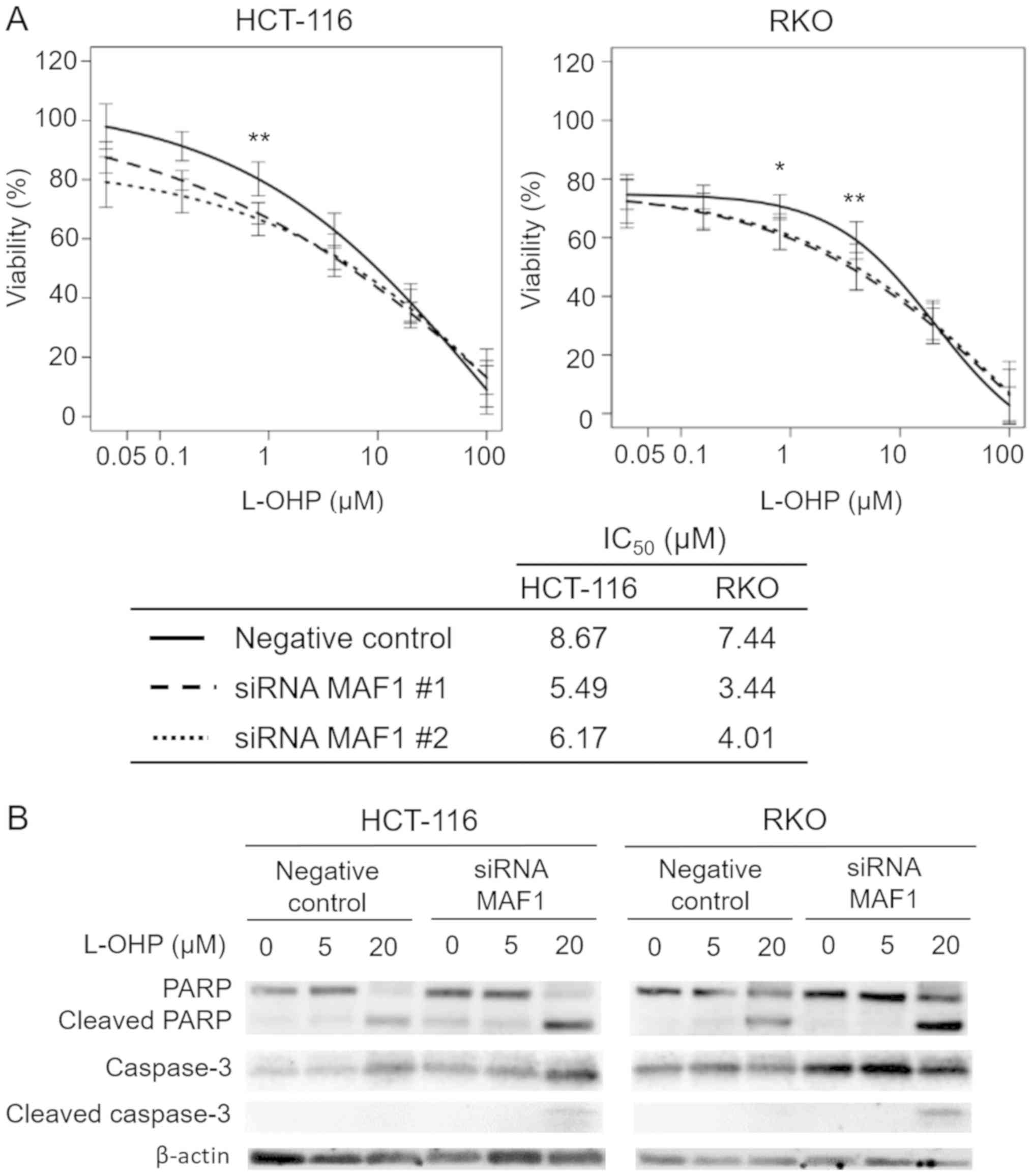
investigate the involvement of MAF1 in CRC progression, the
association between drug resistance of cancer cells and MAF1
expression was explored. The results from the chemosensitivity
assay revealed that knockdown of MAF1 significantly improved
chemosensitivity to L-OHP, which is one of the standard
chemotherapy drugs used to treat CRC (Fig. 3A), whereas chemosensitivity to 5 FU
was not affected (data not shown). Western blotting was performed
to confirm the enhanced induction of apoptosis in MAF1 knockdown
cells treated with L OHP. Knockdown of MAF1 markedly enhanced the
expression of cleaved PARP and caspase 3 in L OHP treated cells,
thus demonstrating that MAF1 may be critically involved in the
avoidance of CRC cell apoptosis following exposure to cytotoxic
agents (Fig. 3B).
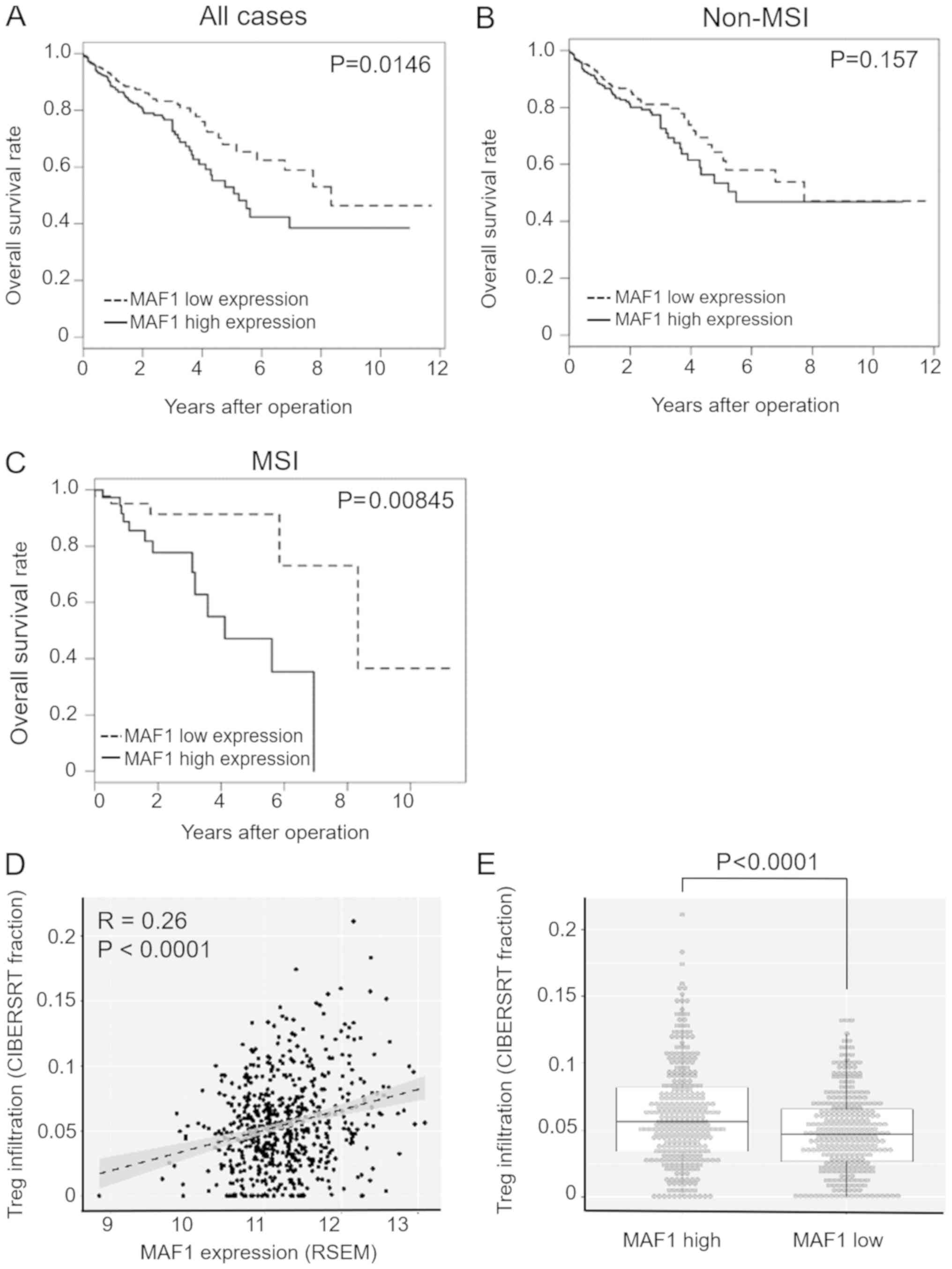
High MAF1 expression is associated with
poor prognosis in CRC with microsatellite instability (MSI)
To further evaluate the clinical significance and
prognostic impact of MAF1 expression in CRC, the present study
analyzed a large scale CRC dataset from TCGA (n=615), which
consisted of 87 MSI positive cases and 528 non MSI cases. Since Pol
III activity is closely associated with immune response in various
cell types (15,16), this study focused on MAF1
expression and MSI status, which strongly affects activation of
tumor immunity. 'MSI high' samples were considered MSI positive,
whereas all other samples were categorized in the non MSI group;
this is because of the differences in clinical and immunological
behaviors between MSI high tumors and other tumors (17,18).
In the subgroup analysis using MSI status information, the
prognostic impact of MAF1 was significantly higher in MSI positive
cases (P=0.00845) compared with all cases (P=0.0146) or non MSI
cases (P=0.157) (Fig. 4A C).
Multivariate analysis of MSI positive cases (n=87) indicated that
high MAF1 expression was an independent prognostic factor for
overall survival (Table III). To
further investigate the association between MAF1 expression and the
immune microenvironment, bioinformatics analysis was performed
using CIBERSORT (13,14). CIBERSORT is a computational program
used to predict the relative contribution of each immune cell type
in a mixed cell population using transcriptome data (13,14).
A mild correlation between MAF1 expression and Treg infiltration
was detected (Fig. 4D), and the
extent of Treg infiltration was significantly higher in the MAF1
high expression group in the TCGA dataset (P<0.0001; Fig. 4E).
 | Table IIIUnivariate and multivariate analyses
of overall survival in MSI cases using The Cancer Genome Atlas
dataset (Cox regression model). |
Table III
Univariate and multivariate analyses
of overall survival in MSI cases using The Cancer Genome Atlas
dataset (Cox regression model).
| Factor | Univariate analysis
| Multivariate
analysis
|
|---|
| RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age (≤65/>65
years) | 0.33 | 0.05-1.19 | 0.0962 | - | - | - |
| Sex
(male/female) | 0.24 | 0.06-0.76 | 0.0131a | 0.21 | 0.05 0.69 | 0.0079a |
| Depth of tumor
invasion (T3, T4/T1, T2) | 3.21 | 0.65-58.2 | 0.1797 | - | - | - |
| Lymph node
metastasis (negative/positive) | 0.82 | 0.30-2.61 | 0.7124 | - | - | - |
| MAF1 expression
(low/high) | 0.25 | 0.07-0.70 | 0.0075a | 0.22 | 0.06 0.64 | 0.0046a |
Discussion
The present used two independent datasets and
demonstrated that high MAF1 expression may be closely associated
with cancer progression in patients with CRC. The results of
clinicopathological analysis revealed that high MAF1 expression was
highly associated not only with tumor depth, but also with lymph
node and distant metastasis, thus suggesting that MAF1 may
contribute to metastatic ability. In addition, in vitro
analyses revealed that MAF1 inhibition significantly suppressed the
migratory ability of CRC cells. MAF1 knockdown also rendered cancer
cells more sensitive to a chemotherapeutic agent. In the present
analysis, MAF1 was significantly highly expressed in patients with
stage III and IV cancer; these patients have been reported to
benefit from standard chemotherapy (19). Therefore, the MAF1 gene may be a
useful biomarker, as well as therapeutic target, for patients
receiving chemotherapy. However, further in vivo
investigation is required to fully confirm the improvement of
chemoresistance to L OHP by knockdown of MAF1 expression.
High MAF1 expression was not considered an
independent prognostic indicator in all patients with CRC, whereas
analysis of an MSI positive population clearly demonstrated that
MAF1 was a significant independent prognostic indicator. These find
ings suggested that the prognostic value of MAF1 expression may be
particularly important in MSI positive patients.
Although MAF1 expression has been reported to
suppress cancer proliferation through negative regulation of Pol
III mediated transcription in some cell lines, the malignant
potential of MAF1, including its effects on chemoresistance, have
not been sufficiently explored. Cellular stress, which can cause
growth arrest, also induces cell dormancy, leading to stress
tolerance and cell survival, depending on cellular context
(20,21). Notably, MAF1 has been reported to
be required for maintaining a cell dormancy like state and
subsequent cell survival under nutrient starvation in Plasmodium
falciparum (22). Furthermore,
previous reports have revealed that MAF1 can directly or indirectly
regulate Pol II dependent genes (7,8,23),
indicating that MAF1 may control various cellular functions,
including the malignant potential of cancer cells. However, since
this study did not identify direct targets of the MAF1 gene that
regulate cell mobility and survival, further exploration is
required to determine the direct involvement of MAF1 in the
metastatic process of CRC.
The present results obtained from clinical samples
indicated an oncogenic role for the MAF1 gene in CRC; however,
other reports have demonstrated a tumor suppressive role of MAF1 in
other malignancies, with the exception of CRC (8,10).
To explain this discrepancy, this study focused on the immune
microenvironment, which differs markedly among tissues. Growing
evidence has suggested that both tumor antigenicity and the immune
environment have a marked influence on the prognosis of patients
with cancer (24,25). In addition, because Pol III
activity is known to be closely associated with the immune response
in various cell types (15,16),
the present study considered the possibility that MAF1 may serve an
important role in the immune microenvironment. Immune checkpoint
inhibitors are increasingly being used in the clinic, and have been
reported to be very effective for the treatment of MSI high tumors,
regardless of histology (18).
This is because MSI tumors activate the immune system through
producing a large number of neoantigens (18,26
29). Notably, the present
subgroup anal ysis using a CRC TCGA database revealed that the
prognostic impact of MAF1 was more evident in MSI high CRC, thus
suggesting that MAF1 may exert its malignant potential more
powerfully under immune activated conditions. In this context,
transcriptome based gene enrichment analysis demonstrated that Treg
infiltration was more abundant in tumors with high expression
levels of MAF1. Tregs are known to suppress the immune response
against tumor antigens in several types of tissue (30), thus indicating a close association
between MAF1 expression in cancer and immune suppression in CRC
tissues. However, further investigation is required to fully
understand the effects of MAF1 expression on tumor immunity.
In conclusion, the present study revealed that MAF1
expression may be considered a useful prognostic indicator in
patients with CRC, particularly in MSI positive cases. These
results may help to understand the complex gene regulatory network,
including both Pol II and III dependent transcription, and may
provide novel medical information that may lead to breakthroughs in
the treatment of CRC.
Funding
This study was supported in part by a Grant-in-Aid
for Scientific Research (C), JSPS KAKENHI (grant nos. 18K07970,
17K106310, 17K106320 and 16K086210) from the Ministry of Education,
Culture, Sports, Science and Technology; and partial support was
received from Takeda Science Foundation and Senri Life Science
Foundation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contribution
KH initiated this project. KH, NN, NM, HT, NH, TH,
CM and TM designed experiments and wrote the manuscript. KH and NN
performed in vitro experiments and bioinformatics analysis.
CM, TM, YD and MM provided clinical samples and designed the study.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Osaka University (approval ID: 08226) and
written informed consent was obtained from all patients included in
this study.
Patient consent for publication
Consent for publication was obtained from all
patients included in this study.
Competing interests
NN received research grants from Chugai Co. Ltd,
Yakult Honsha Co. Ltd. and Ono Pharmaceutical Co., Ltd. These
funders had no role in supplying expenses, study design, data
analysis, decision to publish or preparation of the article.
Acknowledgments
The authors would like to thank Ms. Asuka Yasueda
and Dr Satoshi Ishikawa (Departments of Gastroenterological
Surgery, Osaka University Graduate School of Medicine) for helpful
arrangement of the experiments, as well as Dr Hirofumi Yamamoto
(Division of Health Sciences, Osaka University) for fruitful
discussion.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM and Forman D: Bray F. Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Colvin H, Mizushima T, Eguchi H, Takiguchi
S, Doki Y and Mori M: Gastroenterological surgery in Japan: The
past, the present and the future. Ann Gastroenterol Surg. 1:5–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pradhan A, Hammerquist AM, Khanna A and
Curran SP: The C box rgion of MAF1 regulates transcriptional
activity and protein stability. J Mol Biol. 429:192–207. 2017.
View Article : Google Scholar
|
|
4
|
Vannini A and Cramer P: Conservation
between the RNA poly merase I, II, and III transcription initiation
machineries. Mol Cell. 45:439–446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
White RJ: RNA polymerases I and III, non
coding RNAs and cancer. Trends Genet. 24:622–629. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khanna A, Pradhan A and Curran SP:
Emerging roles for MAF1 beyond the regulation of RNA polymerase III
activity HHS Public Access. J Mol Biol. 427:2577–2585. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson SS, Zhang C, Fromm J, Willis IM
and Johnson DL: Mammalian Maf1 is a negative regulator of
transcription by all three nuclear RNA polymerases. Mol Cell.
26:3673792007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Palian BM, Rohira AD, Johnson SAS, He L,
Zheng N, Dubeau L, Stiles BL and Johnson DL: Maf1 is a novel target
of PTEN and PI3K signaling that negatively regulates oncogenesis
and lipid metabolism. PLoS Genet. 10:e10047892014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khanna A, Pradhan A and Curran SP:
Emerging roles for Maf1 beyond the regulation of RNA polymerase III
activity. J Mol Biol. 427:2577–2585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Tsang CK, Wang S, Li XX, Yang Y, Fu
L, Huang W, Li M, Wang HY and Zheng XF: MAF1 suppresses AKT mTOR
signaling and liver cancer through activation of PTEN tran
scription. Hepatology. 63:1928–1942. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watanabe T, Itabashi M, Shimada Y, Tanaka
S, Ito Y, Ajioka Y, Hamaguchi T, Hyodo I, Igarashi M, Ishida H, et
al: Japanese Society for Cancer of the Colon and Rectum: Japanese
Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2010
for the treatment of colorectal cancer. Int J Clin Oncol. 17:1–29.
2012. View Article : Google Scholar
|
|
12
|
Sugimura K, Fujiwara Y, Omori T, Motoori
M, Miyoshi N, Akita H, Gotoh K, Kobayashi S, Takahashi H, Noura S,
et al: Clinical importance of a transcription reverse transcription
concerted (TRC) diagnosis using peritoneal lavage fluids obtained
pre and post lymphadenectomy from gastric cancer patients. Surg
Today. 46:654–660. 2016. View Article : Google Scholar
|
|
13
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graczyk D, White RJ and Ryan KM:
Involvement of RNA poly merase III in immune responses. Mol Cell
Biol. 35:1848–1859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chiu YH, Macmillan JB and Chen ZJ: RNA
polymerase III detects cytosolic DNA and induces type I interferons
through the RIG I pathway. Cell. 138:576–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim CG, Ahn JB, Jung M, Beom SH, Kim C,
Kim JH, Heo SJ, Park HS, Kim JH, Kim NK, et al: Effects of
microsatellite insta bility on recurrence patterns and outcomes in
colorectal cancers. Br J Cancer. 115:25–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD 1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson
A, Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giancotti FG: Mechanisms governing
metastatic dormancy and reactivation. Cell. 155:750–764. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Senft D and Ronai ZA: Adaptive stress
responses during tumor metastasis and dormancy. Trends Cancer.
2:429–442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McLean KJ and Jacobs Lorena M: Plasmodium
falciparum Maf1 confers survival upon amino acid starvation. MBio.
8:e02317 e162017. View Article : Google Scholar
|
|
23
|
Orioli A, Praz V, Lhôte P and Hernandez N:
Human MAF1 targets and represses active RNA polymerase III genes by
preventing recruitment rather than inducing long term
transcriptional arrest. Genome Res. 26:624–635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Whiteside TL: Immune responses to cancer:
Are they potential biomarkers of prognosis? Front Oncol. 3:1072013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Galon J, Costes A, Sanchez Cabo F,
Kirilovsky A, Mlecnik B, Lagorce Pagès C, Tosolini M, Camus M,
Berger A, Wind P, et al: Type, density, and location of immune
cells within human colorectal tumors predictclinical outcome.
Science. 313:1960–1964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ribic CM, Sargent DJ, Moore MJ, Thibodeau
SN, French AJ, Goldberg RM, Hamilton SR, Laurent Puig P, Gryfe R,
Shepherd LE, et al: Tumor microsatellite instability status as a
predictor of benefit from fluorouracil-based adjuvant chemo therapy
for colon cancer. N Engl J Med. 349:247–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weisenberger DJ, Siegmund KD, Campan M,
Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D,
Buchanan D, et al: CpG island methylator phenotype underlies
sporadic microsatellite instability and is tightly associated with
BRAF mutation in colorectal cancer. Nat Genet. 38:787–793. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
ZhangBWangJWangXZhuJLiuQShiZChambersMCZimmermanLJShaddoxKFKimSet
al NCI CPTAC: Proteogenomic characterization of human colon and
rectal cancer. Nature. 513:382–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saito T, Nishikawa H, Wada H, Nagano Y,
Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et
al: Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the
prognosis of colorectal cancers. Nat Med. 22:679–684. 2016.
View Article : Google Scholar : PubMed/NCBI
|