Introduction
Metastasis, one of the hallmarks of cancer, is the
leading cause of mortality in patients with cancer (1). The first step of metastasis is the
invasion of cancer cells into the basement membrane and
extracellular matrix (ECM) (2). It
has been widely accepted that during the invasion process, cancer
cells require epithelial-mesenchymal transition (EMT) for
acquisition of mesenchymal phenotypes and in order to invade
(3). During EMT, tumor cells lose
their intercellular adhesion and detach from the primary tumor
mass. Subsequently, the cells invade into the ECM and further
metastasize to distant organs as single cells (4,5).
However, increasing evidence has demonstrated that a number of
human cancer cells retain intercellular adhesion and invade as
cohesive multicellular groups, which is a phenomenon termed
collective cell invasion or migration (4,6,7).
The multicellular units of collective cell invasion
are composed of heterogeneous populations of cancer cells that
differ in gene expression signatures, as well as in their
proliferative, invasive and metastatic abilities (8). Leader cells are the group of cancer
cells located in the invasive front of multicellular units, at the
interface of the tumor and ECM, whereas follower cells are located
in the rear and follow (9,10). Notably, leader cells steer and
maintain the migration of a cohesive cell group by exploring the
tissue environment, generating traction force and finding or
generating the path by ECM remodeling (11,12).
In order to investigate the roles of heterogeneous populations in
collective invasion, an increasing number of molecules have been
identified that define leader cells in different types of cancer.
In breast cancer, leader cells are defined by basal epithelial
genes, such as cytokeratin-14 and p63, whereas senescent tumor
cells have been demonstrated to lead collective invasion in thyroid
cancer (13,14). Although the collective invasion of
cancer cells has been extensively investigated, the contributions
of different heterogeneous populations of cancer cells to this
process remain unclear.
The degradation of basement membranes and connective
tissues by matrix proteases provides the pathways for cancer cell
invasion and metastasis (15,16).
Previous studies have demonstrated that collective cell invasion
requires proteolysis-mediated ECM remodeling to degrade the
physical barrier of the ECM and generate the path for invasion, and
leader cells play critical roles in ECM remodeling (15). Wolf et al (17) reported that in the collective cell
invasion of breast cancer, leader cells selectively realign fibers
via matrix metalloproteinase (MMP)-14 on the cell surface and
generate tube-like microtracks, which are further enlarged into
macrotracks to accommodate cell groups of collective movement. In
addition, MMP inhibitors and the knockdown of MMP-14 have been
demonstrated to inhibit collective cell invasion and induce
collective-to-amoeboid transition (17,18).
Cathepsin B (CTSB) is also an important matrix protease that is
involved in protein turnover in lysosomes (19,20).
The increased expression of CTSB has been reported in numerous
types of cancer, including breast, brain and colorectal cancer, and
is considered a marker of a poor prognosis (21-23).
It has been demonstrated that the overexpression of CTSB in breast
cancer cells is an indicator of higher ECM proteolysis and an
enhanced collective cell invasion (21). However, the contribution of CTSB to
the collective cell invasion of salivary adenoid cystic carcinoma
(SACC) and the underlying mechanisms remain unclear.
In the present study, the invasive pattern in human
SACC samples was examined and the expression of EMT markers and
CTSB in the invasive front of SACC was detected. Collective cell
invasion was commonly observed, accompanied by partial EMT, and
CTSB was overexpressed in the invasion front of SACC. Subsequently,
a 3D spheroid invasion assay was established to recapitulate the
collective cell invasion of SACC, and the role of CTSB in
collective invasion was investigated. The data demonstrated that
CTSB plays an important role in leader cells among migrating SACC
cell groups.
Materials and methods
Histological analysis
A total of 76 SACC specimens were obtained from the
Department of Oral Pathology, West China Hospital of Stomatology,
Sichuan University (Chengdu, China) between 2007 and 2008. The
human tissue samples and clinical data were obtained with written
informed consent, and the protocols were approved by the
Institutional Ethics Committee of the West China Medical Center,
Sichuan University (Chengdu, China; approval no.
WCHSIRB-D-2016-176). The collected SACC specimens were fixed with
10% buffered formalin and embedded in paraffin. The
4-µm-thick sections were stained with hematoxylin and eosin
for the examination of tumor invasion.
Cell culture and transfection
SACC-83 cells were obtained from the State Key
Laboratory of Oral Diseases of Sichuan University (Chengdu, China).
The cells were cultured in DMEM high-glucose (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (Gibco/Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and maintained in an incubator at 37°C and 5%
CO2. A negative control siRNA and two siRNAs against
CTSB were purchased from Shanghai Genechem Co., Ltd. (Shanghai,
China). The cells were transfected with siRNA (20 pM) using
Lipofectamine® 2000 reagent (Invitrogen/Thermo Fisher
Scientific, Inc.) according to the manufacturer’s instructions. The
sequences of the primers were as follows: Duplex-1,
GCCUCUAUGAAUCCCAUGUTTACAUGGGAUUCAUAG AGGCTT; duplex-2,
GUCCCACCAUCAAAGAGAUTTAUCUCUUUGAUGGUGGGACTT; duplex-3,
GUGGAAUCGAAUCAGAAGUTTACUUCUGAUUCGAUUCCA CTT; and control,
UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT. The transfected cells
were subjected to reverse transcription-quantitative polymerase
chain reaction (RT-qPCR), western blot analysis, immunofluorescence
staining, migration and invasion assays, and 3D spheroid culture at
48 h following transfection. To label the SACC-83 cells, cDNA
encoding mCherry or GFP (Shanghai Genechem Co., Ltd, Shanghai,
China) was cloned into the Ubi-MCS-SV40-Cherry-IRES-puro vector or
Ubi-MCS-SV40-GFP-IRES-puro vector (Shanghai Genechem Co., Ltd.,
Shanghai, China). The SACC-83 cells were infected with the virus
and subsequently selected with puromycin.
Immunohistochemistry (IHC)
The 4-µm-thick sections of SACC were
deparaffinized and then rehydrated in a graded ethanol series to
distilled water. The sections were autoclaved for antigen retrieval
and blocked with 3% hydrogen peroxide and goat serum albumin,
followed by routine immunohistochemical staining procedures, as
previously described (24,25). The primary antibodies used were as
follows: Rabbit anti-E-cadherin, 1:200 (cat. no. UM870076; OriGene
Technologies, Inc., Shanghai, China); mouse anti-N-cadherin, 1:200
(cat. no. UM500023; OriGene Technologies, Inc.); rabbit
anti-vimentin, 1:200 (cat. no. UM800054CF; OriGene Technologies,
Inc.); and rabbit anti-CTSB, 1:50 (cat. no. ab230401; Abcam,
Cambridge, MA, USA).
Within each tumor, the cancer cells that were
closest to the connective tissue or other normal tissue (such as
the salivary glands and nerves) were defined as the invasive tumor
front and the other tumor areas were regarded as the central tumor
areas (26). The
immunohistochemical staining was scored by two independent
researchers (JSW and SSW), and the invasive front and central tumor
areas were scored separately. The intensity of immunostaining was
scored as follows: 0, no staining; 1, mild staining; 2, moderate
staining; and 3, strong staining. The percentages of stained cells
were recorded as follows: 0, negative; 1, 0-10% positive; 2, 10-50%
positive; or 3, >50% positive. The H score was then calculated
by multiplying the two variables and the result was categorized as
negative (0-4) or positive (>4) expression.
RT-qPCR
Total RNA was extracted using an RNeasy Mini kit
(Qiagen Co. Ltd., Shanghai, China CA,) and reverse transcribed into
cDNA using the High-Capacity cDNA Reverse Transcription kit
(Applied Biosystems/Thermo Fisher Scientific, Inc.). The primer
sequences used were as follows: CTSB forward,
5′-CACTGACTGGGGTGACAATG-3′ and reverse, 5′-GCCACCACTTCTGATTCGAT-3′;
β-actin forward, 5′-GGCCTCCAA GGAGTAAGACC-3′ and reverse,
5′-AGGGGTCTACATGGCAACTG-3′; ROCK1 forward,
5′-CGAAGATGCCATGTTAAGTGC-3′ and reverse, 5′-ATC
TTGTAGAAAGCGTTCGAG-3′; ROCK2 forward, 5′-CAA CTGTGAGGCTTGTATGAAG-3′
and reverse, 5′-TGCAAG GTGCTATAATCTCCTC-3′; RhoA forward, 5′-TATCGA
GGTGGATGGAAAGC-3′ and reverse, 5′-TCTGGGGTCCA CTTTTCTG-3′; and
MMP-9 forward, 5′-CAACTGTCCCTG CCCGAGA-3′ and reverse,
5′-CAAAGGCGTCGTCAATCA CC-3′. The PCR reactions were performed using
SYBR-Green (Roche Diagnostics, Indianapolis, IN, USA) based on the
following protocol: Preincubation at 94°C for 10 min, followed by
45 cycles of 15 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C.
The sample values were normalized to the housekeeping gene,
β-actin.
Western blot analysis
The protein samples were harvested using a total
protein lysates kit (cat no. KGP250; Nanjing KeyGen Biotech Co.
Ltd., Nanjing, China). The protein concentrations were determined
using the BCA Protein Assay kit (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) according to the manufacturer’s instructions. A
total of 25 µg protein from each sample was subjected to
SDS-polyacrylamide gel electrophoresis (gel, 10%; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and transferred to a PVDF
membrane (Immobilon; EMD Millipore, Bedford, MA, USA).
Immunoprobing was performed by incubating the membrane with
anti-GAPDH antibody (1:1,000; cat. no. 5174; Cell Signaling
Technology, Inc., Danvers, MA, USA), anti-CTSB antibody (1:1,000;
cat. no. ab230401, Abcam), anti-FAK antibody (1:1,000; cat. no.
WL01696; Wanleibio Co., Ltd., Shanghai, China) and anti-MMP-9
antibody (1:1,000; cat. no. 10375-2-AP; Proteintech Group, Inc.,
Chicago, IL, USA) overnight at 4°C, followed by binding with a
HRP-conjugated secondary antibody (1:2,000; cat. no. 7074; Cell
Signaling Technology, Inc.) for 1 h at room temperature. The Pierce
ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) was
used for detection.
Immunofluorescence staining
The cells cultured in 3D gel were fixed with 4%
formaldehyde for 20 min. After washing with PBS for 10 min, the
cells were permeabilized with 0.2% Triton X-100 for 30 min. The
cells were then blocked with 1% bovine serum albumin in PBS for 30
min. F-actin was stained with Rhodamine phalloidin (1:400;
Invitrogen Molecular Probes/Thermo Fisher Scientific, Inc.) for 25
min in order to visualize the cell cytoskeleton. The cells were
incubated with primary antibodies against E-cadherin (1:400; cat.
no. 20874-1-AP; Proteintech Group, Inc.) and CTSB (1:50; cat. no.
ab230401; Abcam) at 4°C overnight. After washing with PBS, the
cells were incubated with TRITC goat anti-rabbit IgG (1:100;
catalog no. ZF-0316; ZSGB-Bio, Inc., Beijing, China) for 1 h at
room temperature, and the nuclei were stained DAPI. Fluorescence
was observed using a confocal laser microscope (Olympus FluoView
FV1000; Olympus Corp., Tokyo, Japan).
Migration and invasion assays
The cells were seeded in 6-well plates at a density
of 1.5×105 cell/ml. After 24 h, the cells were cultured
in serum-free medium, and wounds were created by scraping each
plate with a 200-µl pipette tip. Images of identical regions
of the scratch were captured at various time points using a phase
contrast microscope (Olympus IX 71; Olympus Corp., Tokyo, Japan).
Matrigel invasion assays were performed in Transwell chambers
(8-µm pore size; Costar; Corning Inc., NY, USA) coated with
100 µl Matrigel. A total of 5-6×104 cells were
seeded in the upper compartment in 200 ml serum-free medium and the
chambers were then placed in 24-well dishes containing 700
µl DMEM with 10% FBS. After 24 or 48 h, the membranes were
fixed with 4% formaldehyde and stained with 10% Giemsa staining
solution for 30 min at room temperature. After removing the cells
on the upper surface of the membrane, the cells on the lower
membrane surface were counted under a microscope (Olympus IX 71;
Olympus Corp.).
3D spheroid culture
Multicellular spheroids were generated using the
hanging drop technique or by plating the cells on 96-well ultra-low
attachment plates (Corning, Inc., NY, USA) or soft agar-coated
96-well plates. For the hanging drop method, the cells were
re-suspended in DMEM growth medium supplemented with 10%
methylcellulose at a density of 5×104 cells/ml, and
incubated in 25 µl droplets overnight as previously
described (27). For the latter
method, 1% agar was used to coat 96-well plates. The cells were
re-suspended in 200 µl DMEM growth medium at a density of
5×104 cells/ml and seeded in 96-well ultra-low
attachment plates or soft agar-coated 96-well plates. The cells
were cultured for 48-72 h to ensure multicellular aggregation.
Following cell aggregation, the spheroids were incorporated into
non-pepsinized rat tail collagen solution (BD Biosciences, San
Jose, CA, USA) prior to collagen polymerization. Subsequently, DMEM
growth medium was added and the plates were incubated at 37°C for
24-48 h. The definition of leader cells was based on their
positions in the cell cluster, with the leader and follower cells
located at the front and the back of the cluster, respectively
(10). Therefore, protrusive cells
at the front of the invasive strands were identified as leader
cells. To determine whether CTSB was required for leader cell
formation, mixed spheroid culture was performed by co-culturing the
control untreated GFP-expressing cells with siCTSB-transfected
mCherry-expressing cells, or by co-culturing control
mCherry-expressing cells with siCTSB-transfected GFP-expressing
cells. A confocal laser microscope (Olympus FluoView FV1000;
Olympus Corp.) was used to observe the tumor organoids and assess
the incidence of GFP- or mCherry-expressing cells leading
collective invasion strands according to the following formula:
100× (number of GFP- or mCherry-expressing leader cells/total
number of leader cells). In order to assess force-mediated ECM
remodeling, 2.5×104 cells were mixed with 1 ml collagen
I and Matrigel mixture, and the gel mix was plated in a 96-well
plate at a density of 100 µl per well as described
previously (28). After 4 days,
the plates were scanned, and the relative diameter of the well and
the gel were measured using ImageJ (National Institutes of Health,
Bethesda, MD, USA). The extent of contraction was assessed using
the following formula: 100× (well diameter – gel diameter)/well
diameter.
Scanning electron microscopy (SEM)
The samples were fixed with 4% glutaraldehyde for 15
min at room temperature, and after washing with PBS, the samples
were dehydrated with serially increasing concentrations of ethanol
at 30, 40, 50, 60, 70, 80, 90, 95, 99 and 100% for 10 min each. The
sample was critical point dried and sputter-coated with gold before
examination under a FEI Quanta FEG 250 scanning electron microscope
(FEI, Hillsboro, OR, USA).
Statistical analysis
Statistical analyses were performed using the
Statistical Package for Social Science software (version 22.0; IBM
Corp., Armonk, NY, USA) and GraphPad Prism (version 5.03; GraphPad
Software, Inc., La Jolla, CA, USA). The numerical data are
expressed as the means ± standard deviation. The data were analyzed
with the Student’s t-test (comparisons between 2 groups), or with
one-way analysis of variance and Student-Newman-Keuls test
(comparisons between >2 groups). The association between CTSB
expression and the clinicopathological characteristic of the
patients with SACC was analyzed using the Chi-square test. Survival
analysis was performed using the Kaplan-Meier method, and
significant differences were compared using the log-rank test. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
SACC cells invade as multicellular units
without a complete EMT signature
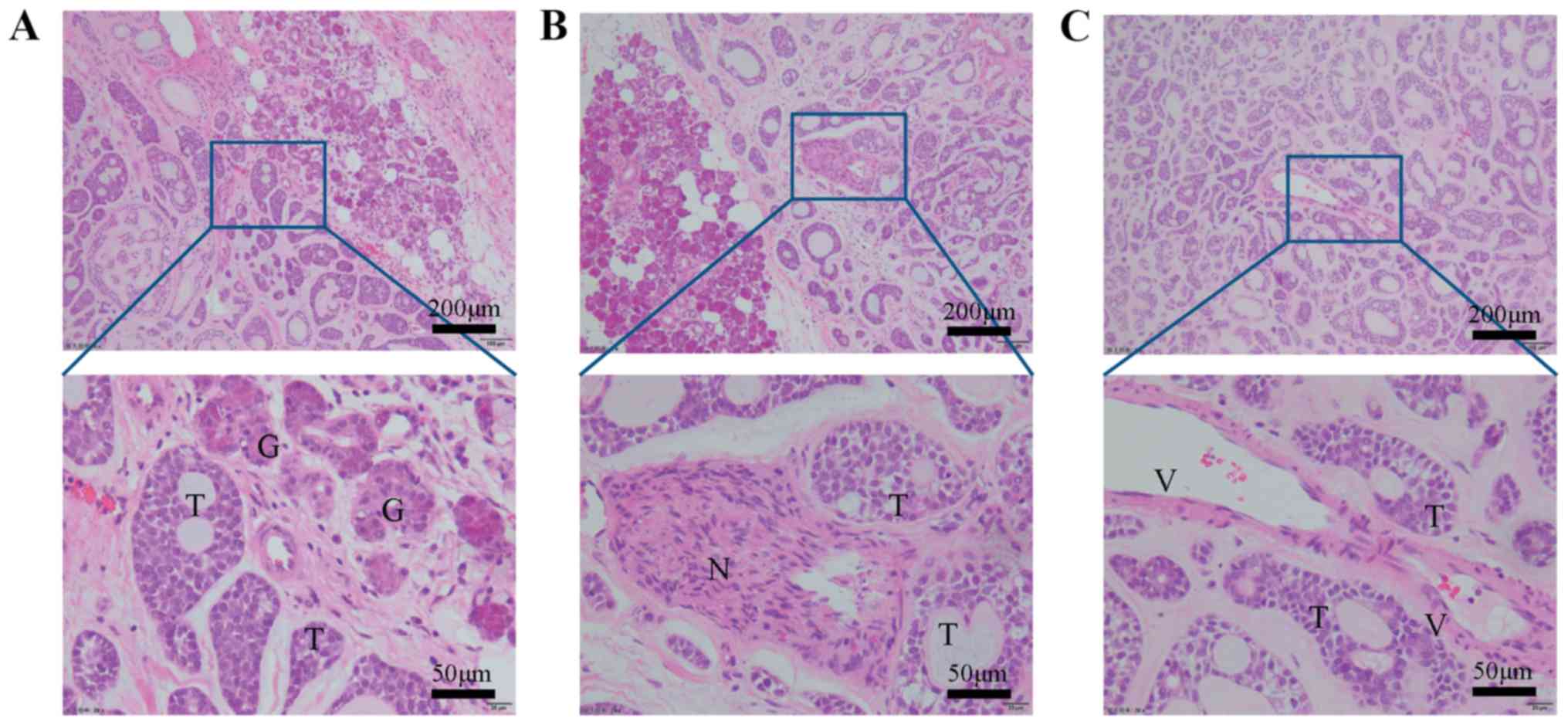
In the SACC invasive regions, the neoplastic
epithelial cells invaded into the adjacent gland, nerve and vessel
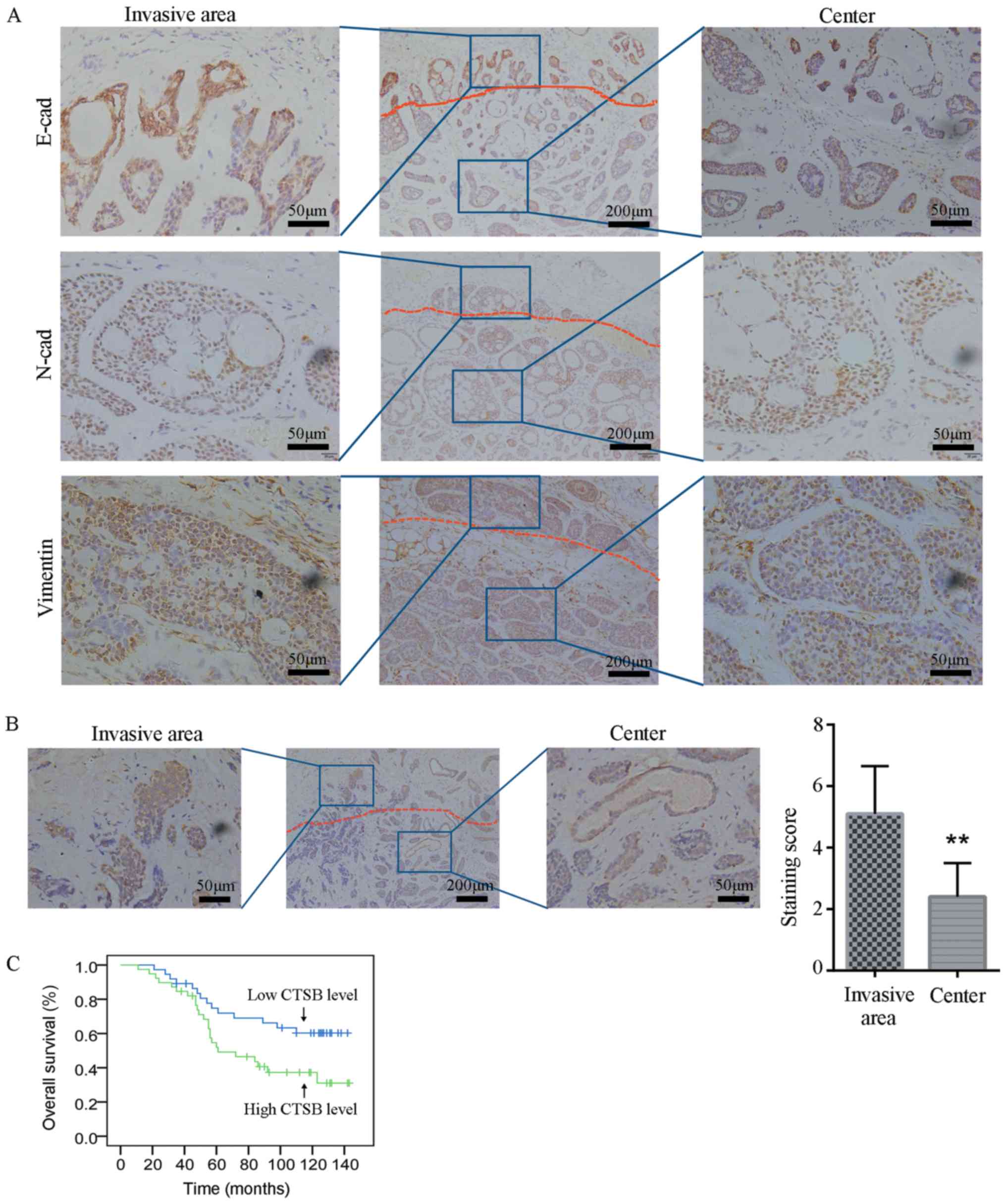
tissue and organized into multicellular units (Fig. 1). The IHC results revealed that the
invasion front preserved E-cadherin expression, indicating that
tumor cells of SACC invade as collective cell groups (Fig. 2A). To further assess whether the
EMT occurs in the invasion border of SACC, the expression of
mesenchymal markers was investigated by IHC. Notably, the results
demonstrated that N-cadherin and vimentin were expressed in the
entire cancer area of SACC without a loss of expression in the
invasive front (Fig. 2A). These
data indicated that invasive cancer cells of SACC lack a complete
EMT signature.
CTSB is overexpressed at the invasion
border and is associated with a poor prognosis of patients with
SACC
As CTSB promotes cancer invasion and metastasis, the
expression of CTSB was investigated at the invasion border of 76
SACC samples. CTSB staining was present predominantly in the
cytoplasm of the SACC cells. Notably, the cytoplasmic
immunoreaction for CTSB protein was higher at the invasion border
compared to the tumor center (Fig.
2B). The clinicopathological and prognostic implications of the
CTSB overexpression status at the invasion border of SACC were
subsequently examined. The increased CTSB expression at the
invasion border was closely associated with a poor pathological
type, nerve invasion, advanced clinical tumor-node-metastasis
stage, recurrence and metastasis of SACC (Table I). Kaplan-Meier survival curve
analysis also demonstrated that the CTSB level was associated with
the overall survival of patients with SACC and that the patients
with a a low CBST expression level lived longer than those with a
high CTSB level (P<0.05; Fig.
2C).
 | Table IAssociation between CTSB expression
and the clinicopathological characteristics of patients with
SACC. |
Table I
Association between CTSB expression
and the clinicopathological characteristics of patients with
SACC.
| Parameter | n | CTSB expression
| P-value |
|---|
| Low | High |
|---|
| Sex | | | | 0.484 |
| Male | 31 | 17 (54.84%) | 14 (45.16%) | |
| Female | 45 | 20 (44.4%) | 25 (55.56%) | |
| Age (years) | | | | 0.815 |
| <60 | 28 | 13 (46.43%) | 15 (53.57%) | |
| ≥60 | 48 | 24 (50.00%) | 24 (50.00%) | |
| Tumor location | | | | 0.262 |
| Major
salivary | 36 | 15 (41.67%) | 21 (58.33%) | |
| Minor
salivary | 40 | 22 (55.00%) | 18 (45.00%) | |
| Pathological
type | | | | 0.034a |
|
Cribriform/tubular | 57 | 32 (56.14%) | 25 (43.86%) | |
| Solid | 19 | 5 (26.32%) | 14 (73.68%) | |
| TNM stage | | | | 0.043a |
| I-II | 22 | 15 (68.18%) | 7 (31.82%) | |
| III-IV | 54 | 22 (40.74%) | 32 (59.26%) | |
| Perineural
invasion | | | | 0.022a |
| Yes | 24 | 10 (41.67%) | 14 (58.33%) | |
| No | 52 | 37 (71.15%) | 15 (28.85%) | |
| Recurrence | | | | 0.008a |
| Yes | 19 | 4 (21.05%) | 15 (78.95%) | |
| No | 57 | 33 (57.89%) | 24 (42.11%) | |
| Distant
metastasis | | | | 0.022a |
| Yes | 17 | 5 (29.41%) | 12 (70.59%) | |
| No | 49 | 32 (65.31%) | 17 (34.69%) | |
CTSB is expressed in leader cells of SACC
in 3D spheroid culture
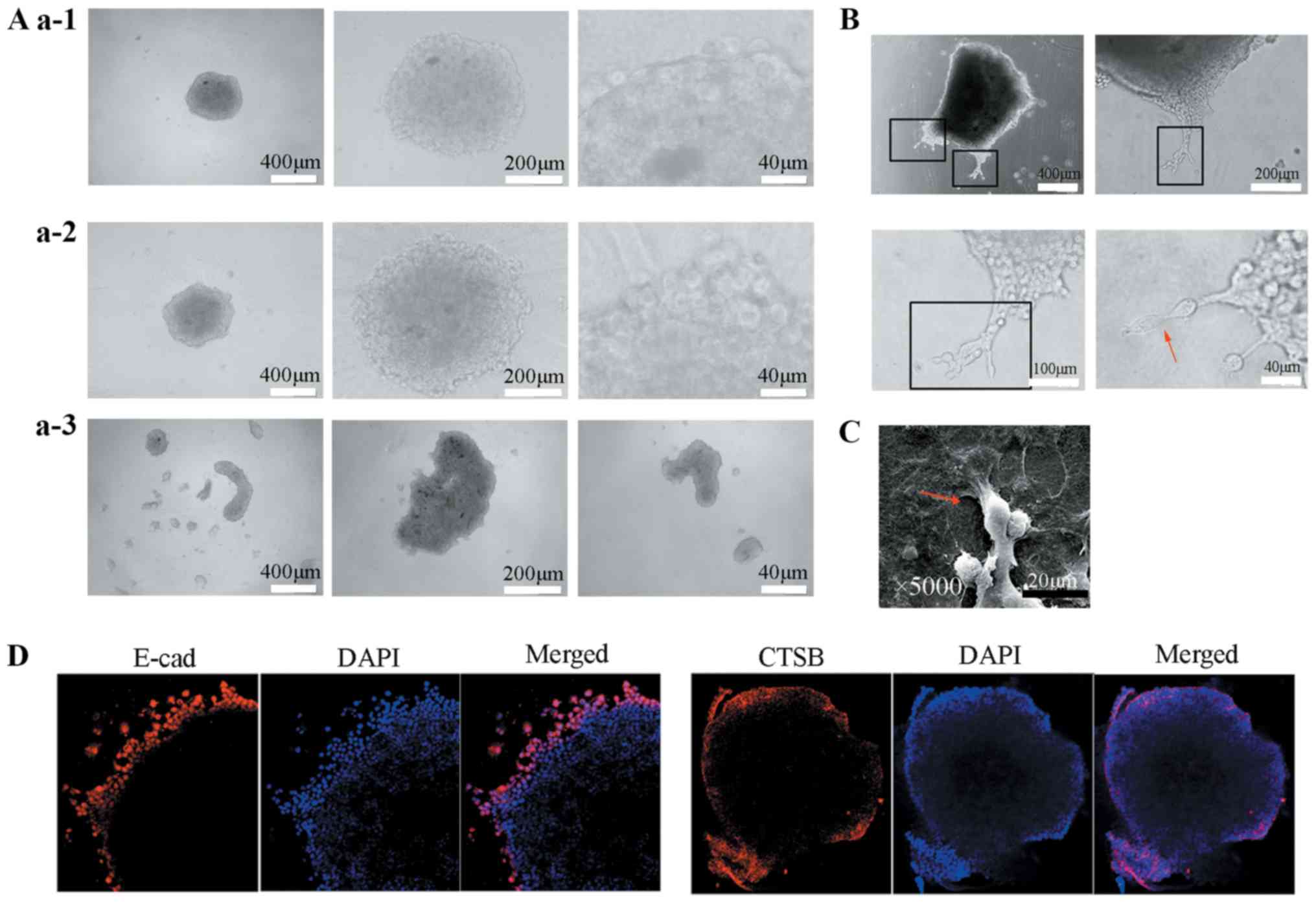
To recapitulate the collective cell invasion of SACC
in vitro, a 3D spheroid invasion assay was performed and the
hanging drop technique, 96-well ultra-low attachment plates and
soft agar-coated 96-well plates were used to generate multicellular
spheroids. To investigate the spheroid formation efficiency of
these three methods, the morphology of the spheroids was observed
using transmitted light microscopy. After 48 h, the multicellular
spheroids generated by the hanging drop technique were irregular
and loosely aggregated (Fig. 3A,
panels a1), while those produced by the 96-well ultra-low
attachment plates and soft agar-coated 96-well plates were regular
and densely compacted (Fig. 3A,
panels a2). Therefore, the multicellular spheroids generated by the
96-well ultra-low attachment plates and soft agar-coated 96-well
plates were embedded in type I collagen gels to perform a 3D
spheroid invasion assay.
After 16-24 h, the tumor organoids progressively
extended multicellular strands of cancer cells from the margin into
the collagen I and the leader cells were observed at the front of
invasive strands (Fig. 3B). The
close examination by light microscopy and SEM revealed that the
leader cells exhibited an elongated phenotype and extended
subcellular protrusions compared to the follower cells, which
exhibited an epithelial cell morphology and few protrusions
(Fig. 3B and C). In addition, the
immunofluorescence staining demonstrated that the invasive cells
retained E-cadherin expression, particularly the leader cells.
Subsequently, we determined whether CTSB was overexpressed in the
leader cells in the 3D spheroid invasion assay. Immunofluorescence
staining revealed that the leader cells expressed CTSB in the
cytoplasm, while the follower cells did not (Fig. 3D), indicating that CTSB may be a
marker of leader cells in SACC collective cell invasion.
CTSB is required for leader cell
formation in SACC collective cell invasion
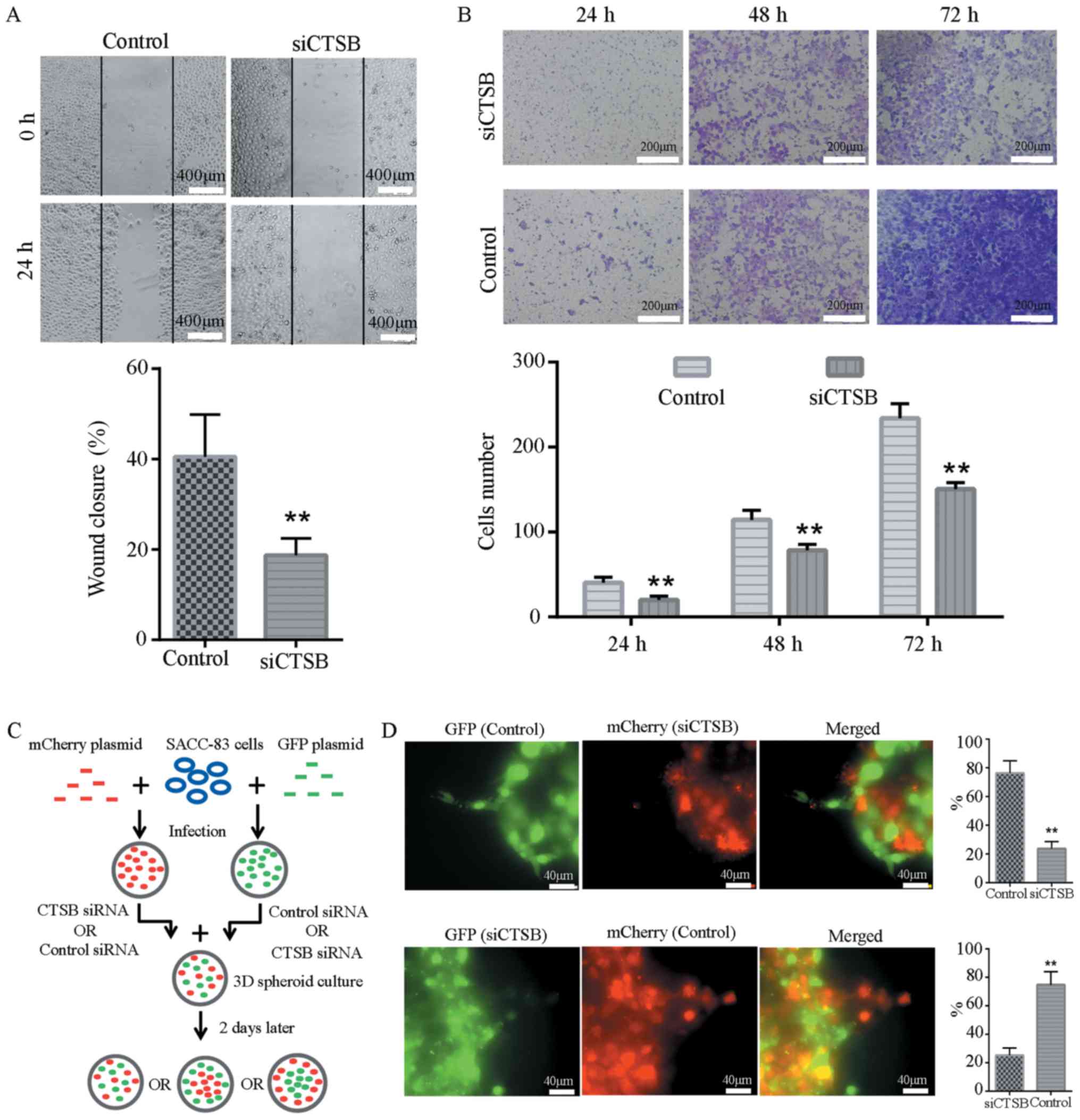
To determine the contribution of CTSB to SACC-83
cell invasion, three siRNA duplexes for CTSB were characterized.
All of them efficiently knocked down their target, as determined by
western blot analysis and RT-qPCR. Among the three siRNA duplexes,
siRNA 3 exhibited the highest efficiency and was used to perform
the subsequent experiments (Fig.
S1). In the wound healing and Transwell assays, siCTSB
transfection effectively reduced the migratory and invasive ability
of the SACC-83 cells compared to the control siRNA-transfected
cells (Fig. 4A and B). To
determine whether CTSB is required for leader cell formation, the
SACC-83 cells were labeled with either GFP or mCherry and mixing
experiments were performed by co-culturing the control
GFP-expressing cells with siCTSB-transfected mCherry-expressing
cells, or by co-culturing the control mCherry-expressing cells with
siCTSB-transfected GFP-expressing cells (Fig. 4C). The control GFP- and
mCherry-expressing cells were observed at the leading tip of the
invasive strands in 76.3 and 74.8% cases, respectively, while the
mCherry- and GFP-expressing cells with siCTSB treatment were only
observed at the front of the invasive strands in 23.7 and 25.2%
cases, respectively (Fig. 4D).
Knockdown of CTSB inhibits ECM
remodeling
In the present study, the effects of CTSB depletion
on protease- and force-mediated ECM remodeling were investigated.
The SACC-83 cells transfected with siCTSB expressed lower levels of
MMP-9 at the mRNA and protein level compared to those transfected
with control siRNA (Fig. 5A and
B). Force-mediated ECM remodeling was assessed by microscopic
analysis of the extent of matrix contraction. The results revealed
that there was a significant reduction in ECM contraction in
response to knockdown of CTSB (Fig.
5C). Phalloidin staining and SEM were used to examine the
effects of siCTSB transfection on cell cytoskeletal organization
and cell morphology (Fig. 5D and
E). The results of SEM analysis demonstrated that the SACC-83
cells transfected with siCTSB lost their elongated phenotype, but
exhibited an epithelial cell morphology and a reduced number of
subcellular protrusions. CTSB siRNA transfection disrupted the
normal cytoskeletal organization of the SACC-83 cells. It is widely
established that Rho-ROCK function and cytoskeletal organization
play a key role in force-mediated ECM remodeling (29,30).
The depletion of CTSB inhibited the expression of focal adhesion
kinase (FAK), an adaptor molecule involved in connecting integrins
to filamentous actin in the cytoskeleton. Furthermore, RT-qPCR
revealed that the expression levels of RhoA, ROCK1 and ROCK2 were
significantly downregulated following transfection with siCTSB
(Fig. 5A).
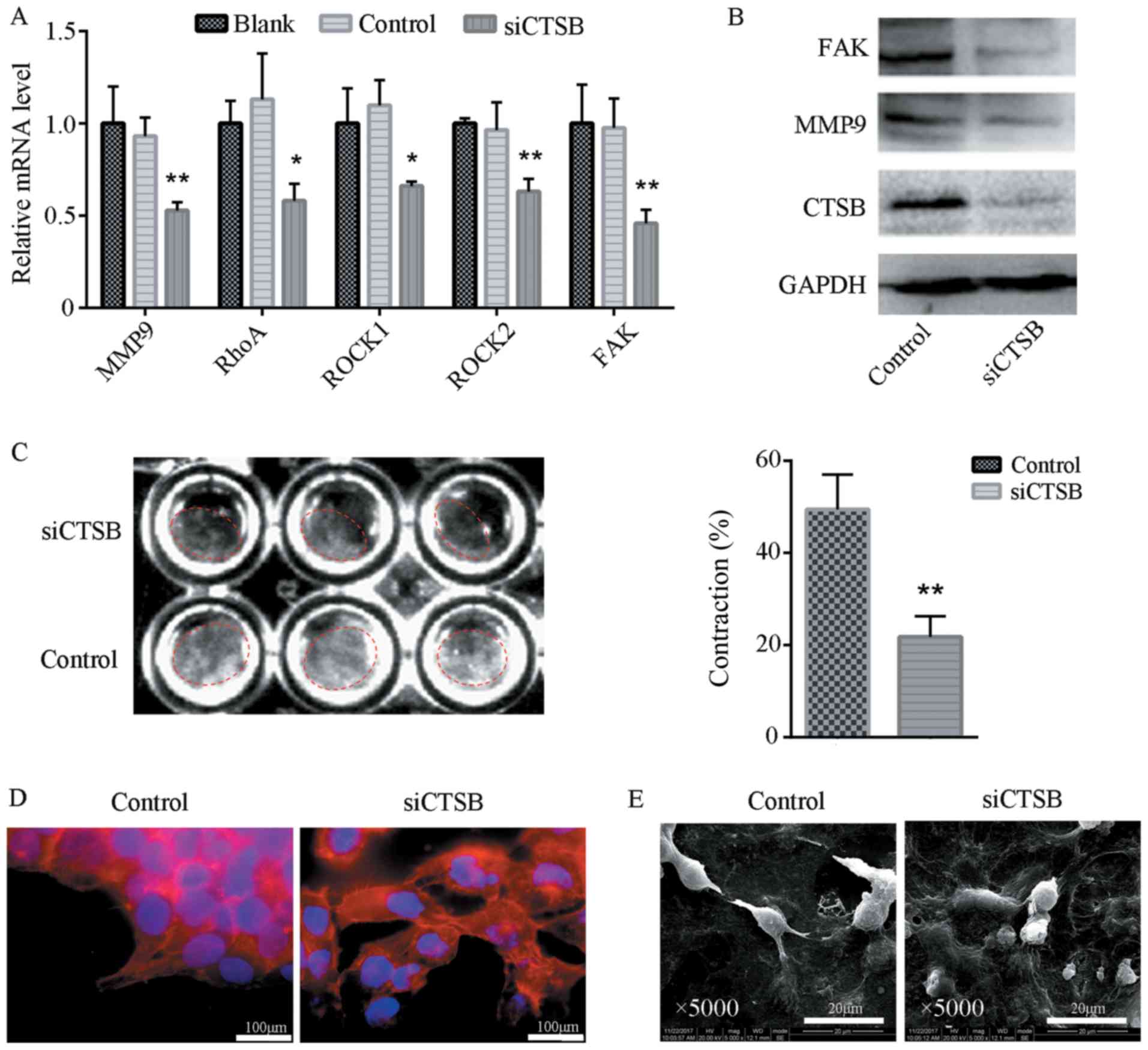 | Figure 5Knockdown of CTSB inhibits ECM
remodeling. (A) The expression levels of MMP-9, RhoA, ROCK1, ROCK2
and FAK mRNA in SACC-83 cells transfected with siCTSB or a blank
control. The knockdown of CTSB inhibited the mRNA levels of the ECM
remodeling-related genes. Data are presented as the means ±
standard deviation. *P<0.05, **P<0.001.
(B) Western blot analysis of CTSB, MMP-9 and FAK protein in SACC-83
cells transfected with siCTSB or a blank control. The knockdown of
CTSB inhibited the levels of ECM remodeling-related proteins. (C)
An ECM contraction assay of force-mediated ECM remodeling. The
percentage of contraction area per well revealed that contraction
was inhibited in response to knockdown of CTSB. Data are presented
as the means ± standard deviation. **P<0.001. (D)
CBST knockdown in SACC-83 cells cultured on glass coverslips
reduced cytoskeletal F-actin staining. (E) The morphologic
alteration of SACC-83 cells after CBST knockdown. siCTSB-treated
SACC-83 cells exhibited epithelial cell morphology and a reduced
number of subcellular protrusions compared to the control group.
CTSB, cathepsin B; ECM, extracellular matrix; MMP, matrix
metalloproteinase; FAK, focal adhesion kinase; SACC, salivary
adenoid cystic carcinoma. |
Discussion
The collective invasion of multicellular groups that
are comprised of leader cells and follower cells is critical for
cancer metastasis. Collective cell invasion requires the guidance
of leader cells; however, the molecular markers that define leader
cells and the mechanisms through which leader cells promote
collective cell invasion remain unknown. In the present study, CTSB
was revealed to be overexpressed in the invasive front compared to
the central area of SACC and this was associated with a poor
prognosis of patients with the disease. The knockdown of CTSB
impaired the formation of leader cells, disrupted cytoskeletal
organization, altered cell morphology, and inhibited ECM remodeling
through the downregulation of MMP-9, FAK and Rho/ROCK function.
These findings demonstrate that CTSB may be a useful marker of
leader cells in the collective invasion of SACC, and may be
involved in the regulation of protease- and force-mediated ECM
remodeling.
EMT is a key event in cancer invasion and is
characterized by the acquisition of mesenchymal phenotypes and the
loss of cell-to-cell adhesions. However, increasing evidence has
demonstrated that collective cell invasion is another model of
cancer invasion (14,31). In the present study, neoplastic
epithelial cells invaded into the adjacent tissue as multicellular
units in the SACC samples and retained E-cadherin expression.
Notably, the IHC results revealed that the cancer cells at the
invasive front lacked a complete EMT signature. In the 3D spheroid
invasion assay, the leader cells exhibited a more elongated
phenotype, but retained E-cadherin expression, indicating a partial
EMT phenotype. These results are consistent with those of some
previous studies, in which leader cells also lacked a complete EMT
signature and could not be identified by canonical mesenchymal
markers (8,32). It has been reported that EMT is
rarely equally pronounced in the entire tumor tissue, and partial
EMT is more frequent in cancer than complete EMT (7,33).
The cancer cells undergoing partial EMT acquire the expression of
mesenchymal markers and maintain certain epithelial characteristics
simultaneously (6). On the other
hand, previous studies have revealed that metastatic cancer cells
with mesenchymal features must undergo the reverse transition,
mesenchymal-epithelial transition (MET), at the secondary site
(34-36). MET has been demonstrated to
increase the survival of these cells and recapitulate the pathology
of the primary tumors in numerous types of cancer, including breast
cancer, small cell lung cancer and ovarian cancer (34,35,37).
However, few studies have focused on the role of MET in SACC
metastases and further studies are therefore required.
The invasive front of a tumor represents the tumor
cell-host organ interface, in which tumor cells invade into the
surrounding tissue by secreting proteases, such as MMPs and CTSB
(38,39). It has been reported that tumor
budding or dedifferentiation at the invasion front can be a
predictor of poor outcomes in colorectal cancers (38). Previous studies have confirmed that
CTSB is overexpressed in different types of cancer, including
breast cancer (21), non-small
cell lung carcinoma (40),
colorectal cancer (41) and oral
squamous cell carcinoma (42), and
it indicates poor outcomes for patients. The association between an
increased CTSB expression and the looseness of cancerous
interstitial tissue also indicates the pivotal roles of CTSB in
cancer invasion and metastasis (21). CTSB levels are frequently elevated
at the invasive edge of tumors and during tumor budding in
colorectal cancer (38,43). In the present study, the data
demonstrated that CTSB was overexpressed in the cytoplasm of SACC
cells at the invasive front compared to the tumor center and was
associated with a poor prognosis of patients with SACC.
Furthermore, CTSB overexpression was associated with perineural
invasion (PNI) in SACC. PNI, a hallmark of SACC, is an independent
indicator of tumor recurrence and a poor prognosis (44-46).
The role of cathepsins in PNI of SACC was also demonstrated in our
previous study, in which cathepsin D was overexpressed in the nerve
invasion frontier and associated with a poor prognosis of patients
with SACC (47).
In collective cell invasion, the multicellular units
are considered as phenotypically heterogeneous cancer cell
populations and the leader cells are protrusive at the front of the
invasive strands (32). The
present study demonstrated that leader cells of SACC were located
in the invasion edge and exhibit a more elongated phenotype with
subcellular protrusions compared to follower cells. Furthermore, in
the 3D spheroid invasion assay, CTSB was overexpressed in the
leader cells, which corresponds with the high expression of CTSB at
the invasive front in SACC. Previous studies have revealed that
cytokeratin-14, p63 and integrin β are overexpressed in leader
cells of breast cancer (14,
48). These results suggest that
leader cells are specialized cells that express particular markers,
including CTSB, cytokeratin-14, p63 and integrin β, and CTSB may be
a marker of leader cells in SACC. In addition, IHC and
immunofluorescence staining in the present study revealed that CTSB
was localized in the cytoplasm of leader cells. In previous
studies, CTSB has been identified in perinuclear lysosomes,
vesicles, mitochondria and the cell periphery (20). CTSB in the lysosomes can facilitate
autophagy, while in the nucleus it is involved in the nuclear
scaffold and cell division (49).
However, the multifaceted roles of CTSB in leader cells of SACC
have not yet been detected and therefore further studies are
required.
CTSB has been shown to mediate the dissemination and
invasion of cancer cells by degrading components of the ECM, and it
plays an active role in EMT and tumor angiogenesis (50-53).
Consistent with the findings of previous studies, the results of
the present study demonstrated that the knockdown of CTSB inhibited
the migratory and invasive abilities of SACC cells. The degradation
of basement membranes and connective tissues by various proteases
is considered as a prerequisite for the collective invasion of
cancer cells. In addition, CTSB has been implicated in the
initiation of the proteolytic cascade that involves uPA,
plasminogen and plasmin, which indicates that CTSB can activate
other ECM-degrading proteases (54). MMPs are key proteases involved in
proteolytically cleaving ECM components and reducing ECM barriers
during cancer cell invasion (55).
The expression levels of MMP-9 are elevated in various types of
cancer, including breast cancer, head and neck carcinoma and colon
cancer, and they indicate a poor prognosis in these patients
(38,56,57).
In a previous study, CTSB was revealed to destroy the inhibitors of
MMPs and maintain a high level of MMPs, thereby promoting ECM
degradation in human articular chondrocytes (53). Consistent with this study, the
present study demonstrated that CTSB knockdown inhibits mRNA and
protein expression of MMP-9, indicating that apart from its direct
proteolytic activity, CTSB may also serve an important role in
MMP-9-mediated proteolysis. Another MMP member, MMP-14, has been
identified as a key regulator in collective cell migration by
sterically generating tube-like microtracks and enlarging
microtracks into macrotracks for accommodation of cell groups
(17,58).
In addition to protease-mediated ECM remodeling,
force-mediated ECM remodeling is another major mechanism of ECM
remodeling (15). Tumor cells
generate contractile force to deform collagen fibers and move
through dense ECM without proteolysis (59). ROCK-dependent force-mediated ECM
remodeling is specifically required for collective cell invasion
(58,59). The function of Rho-ROCK has been
reported to regulate myosin activity, which is essential for the
formation of cell protrusions and ECM remodeling (17,29,30).
Previous studies have revealed that RhoA and ROCKs play an
important role in mediating collective cell invasion by controlling
leader-follower cell hierarchy, multicellular contractility and
mechanocoupling (60,61). In the present study, CTSB knockdown
inhibited the levels of Rho and ROCK, reduced the extension of
subcellular protrusions and disrupted the normal cytoskeletal
organization of SACC-83 cells, which indicates that CTSB is
involved in ROCK-dependent force-mediated ECM remodeling. FAK is an
intracellular protein tyrosine kinase and it functions as a key
regulator of integrin-dependent matrix adhesions (62). Previous studies have demonstrated
that FAK expression is elevated in numerous types of cancer and is
considered as an indicator of a poor prognosis for these patients
(63,64). Another study revealed that FAK may
regulate collective cell invasion by modulating cell-cell adhesion
through controlling E-cadherin internalization (65). The present study indicated that the
knockdown of CTSB in SACC-83 cells inhibited FAK expression. This
result is in agreement with the findings of a previous study, in
which the downregulation of CTSB induced cytoskeletal
disorganization by downregulating the expression of FAK (66). These findings suggest that FAK may
be involved in CTSB-mediated ECM remodeling.
In conclusion, the findings of this study
demonstrate that CTSB is overexpressed in the invasive front of
SACC and is associated with a poor prognosis of patients with SACC.
The knockdown of CTSB disrupts cytoskeletal organization, alters
cell morphology, inhibits ECM remodeling and impairs the formation
of leader cells. The present study provides evidence that CTSB may
define leader cells in SACC and is required for collective cell
invasion as a potential key regulator of ECM remodeling. However,
further studies are required in order to elucidate the underlying
mechanisms. In addition, further studies are warranted to explore
the effects of CTSB-specific inhibitors on tumor-bearing mice which
may shed light on SACC therapy by targeting CTSB.
Supplementary Materials
Funding
This study was supported by National Natural Science
Foundation of China grants (nos. 81572650, 81672672, 81772891,
81502357 and 81621062), the Natural Science Foundation of Zhejiang
Province (Q142114001), the Zhoushan Science and Technology Bureau
Project (2014C31068), the Fundamental Research Funds for the
Central Universities (2017), and by the State Key Laboratory of
Oral Diseases Special Funded Projects (2016).
Availability of data and materials
The datasets used during this study are available
from the corresponding author on reasonable request.
Authors’ contributions
JSW, MiZ and YLT conceived the study, participated
in its design and wrote the manuscript. JSW, ZFL, HFW, XP, JBW and
XHY performed the research. SSW, MeZ, XY, MXC, YJT, XHL interpreted
the results, analyzed data and reviewed the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The use of human tissue samples and clinical data
were obtained with written informed consent, and the protocols were
approved by the Institutional Ethics Committee of the West China
Medical Center, Sichuan University, China (WCHSIRB-D-2016-176).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
CTSB
|
cathepsin B
|
|
ECM
|
extracellular matrix
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FAK
|
focal adhesion kinase
|
|
GFP
|
green fluorescent protein
|
|
PNI
|
perineural invasion
|
|
SACC
|
salivary adenoid cystic carcinoma
|
|
SNK
|
Student-Newman-Keuls
|
|
IHC
|
immunohistochemistry
|
|
MMP
|
matrix metalloproteinase
|
|
SEM
|
scanning electron microscopy
|
|
TNM
|
tumor-node-metastasis
|
|
uPA
|
uridylyl phosphate adenosine
|
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sahai E: Illuminating the metastatic
process. Nat Rev Cancer. 7:737–749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong IY, Javaid S, Wong EA, Perk S, Haber
DA, Toner M and Irimia D: Collective and individual migration
following the epithelial-mesenchymal transition. Nat Mater.
13:1063–1071. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Revenu C and Gilmour D: EMT 2.0: Shaping
epithelia through collective migration. Curr Opin Genet Dev.
19:338–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krakhmal NV, Zavyalova MV, Denisov EV,
Vtorushin SV and Perelmuter VM: Cancer Invasion: Patterns and
Mechanisms. Acta Naturae. 7:17–28. 2015.PubMed/NCBI
|
|
8
|
Westcott JM, Prechtl AM, Maine EA, Dang
TT, Esparza MA, Sun H, Zhou Y, Xie Y and Pearson GW: An
epigenetically distinct breast cancer cell subpopulation promotes
collective invasion. J Clin Invest. 125:1927–1943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rørth P: Collective guidance of collective
cell migration. Trends Cell Biol. 17:575–579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mayor R and Etienne-Manneville S: The
front and rear of collective cell migration. Nat Rev Mol Cell Biol.
17:97–109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khalil AA and Friedl P: Determinants of
leader cells in collective cell migration. Integr Biol (Camb).
2:568–574. 2010. View Article : Google Scholar
|
|
12
|
Gov NS: Collective cell migration
patterns: Follow the leader. Proc Natl Acad Sci USA.
104:15970–15971. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim YH, Choi YW, Lee J, Soh EY, Kim JH and
Park TJ: Senescent tumor cells lead the collective invasion in
thyroid cancer. Nat Commun. 8:152082017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheung KJ, Gabrielson E, Werb Z and Ewald
AJ: Collective invasion in breast cancer requires a conserved basal
epithelial program. Cell. 155:1639–1651. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu JS, Sheng SR, Liang XH and Tang YL: The
role of tumor microenvironment in collective tumor cell invasion.
Future Oncol. 13:991–1002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar S, Kapoor A, Desai S, Inamdar MM and
Sen S: Proteolytic and non-proteolytic regulation of collective
cell invasion: Tuning by ECM density and organization. Sci Rep.
6:199052016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wolf K, Wu YI, Liu Y, Geiger J, Tam E,
Overall C, Stack MS and Friedl P: Multi-step pericellular
proteolysis controls the transition from individual to collective
cancer cell invasion. Nat Cell Biol. 9:893–904. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fisher KE, Sacharidou A, Stratman AN, Mayo
AM, Fisher SB, Mahan RD, Davis MJ and Davis GE: MT1-MMP- and
Cdc42-dependent signaling co-regulate cell invasion and tunnel
formation in 3D collagen matrices. J Cell Sci. 122:4558–4569. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Premzl A, Zavašnik-Bergant V, Turk V and
Kos J: Intracellular and extracellular cathepsin B facilitate
invasion of MCF-10A neoT cells through reconstituted extracellular
matrix in vitro. Exp Cell Res. 283:206–214. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aggarwal N and Sloane BF: Cathepsin B:
Multiple roles in cancer. Proteomics Clin Appl. 8:427–437. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yano M, Hirai K, Naito Z, Yokoyama M,
Ishiwata T, Shiraki Y, Inokuchi M and Asano G: Expression of
cathepsin B and cystatin C in human breast cancer. Surg Today.
31:385–389. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Strojnik T, Kos J, Zidanik B, Golouh R and
Lah T: Cathepsin B immunohistochemical staining in tumor and
endothelial cells is a new prognostic factor for survival in
patients with brain tumors. Clin Cancer Res. 5:559–567.
1999.PubMed/NCBI
|
|
23
|
Kos J, Nielsen HJ, Krasovec M, Christensen
IJ, Cimerman N, Stephens RW and Brünner N: Prognostic values of
cathepsin B and carcinoembryonic antigen in sera of patients with
colorectal cancer. Clin Cancer Res. 4:1511–1516. 1998.PubMed/NCBI
|
|
24
|
Tsukamoto H, Kato T, Enomoto A, Nakamura
N, Shimono Y, Jijiwa M, Asai N, Murakumo Y, Shibata K, Kikkawa F,
et al: Expression of Ret finger protein correlates with outcomes in
endometrial cancer. Cancer Sci. 100:1895–1901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iwakoshi A, Murakumo Y, Kato T, Kitamura
A, Mii S, Saito S, Yatabe Y and Takahashi M: RET finger protein
expression is associated with prognosis in lung cancer with
epidermal growth factor receptor mutations. Pathol Int. 62:324–330.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Søland TM, Brusevold IJ, Koppang HS,
Schenck K and Bryne M: Nerve growth factor receptor (p75 NTR) and
pattern of invasion predict poor prognosis in oral squamous cell
carcinoma. Histopathology. 53:62–72. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Korff T and Augustin HG: Integration of
endothelial cells in multicellular spheroids prevents apoptosis and
induces differentiation. J Cell Biol. 143:1341–1352. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hooper S, Gaggioli C and Sahai E: A
chemical biology screen reveals a role for Rab21-mediated control
of actomyosin contractility in fibroblast-driven cancer invasion.
Br J Cancer. 102:392–402. 2010. View Article : Google Scholar :
|
|
29
|
Meshel AS, Wei Q, Adelstein RS and Sheetz
MP: Basic mechanism of three-dimensional collagen fibre transport
by fibroblasts. Nat Cell Biol. 7:157–164. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Russo JM, Florian P, Shen L, Graham WV,
Tretiakova MS, Gitter AH, Mrsny RJ and Turner JR: Distinct
temporal-spatial roles for rho kinase and myosin light chain kinase
in epithelial purse-string wound closure. Gastroenterology.
128:987–1001. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clark AG and Vignjevic DM: Modes of cancer
cell invasion and the role of the microenvironment. Curr Opin Cell
Biol. 36:13–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Konen J, Summerbell E, Dwivedi B, Galior
K, Hou Y, Rusnak L, Chen A, Saltz J, Zhou W, Boise LH, et al:
Image-guided genomics of phenotypically heterogeneous populations
reveals vascular signalling during symbiotic collective cancer
invasion. Nat Commun. 8:150782017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krebs MG, Metcalf RL, Carter L, Brady G,
Blackhall FH and Dive C: Molecular analysis of circulating tumour
cells-biology and biomarkers. Nat Rev Clin Oncol. 11:129–144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hamilton G, Hochmair M, Rath B, Klameth L
and Zeillinger R: Small cell lung cancer: Circulating tumor cells
of extended stage patients express a mesenchymal-epithelial
transition phenotype. Cell Adhes Migr. 10:360–367. 2016. View Article : Google Scholar
|
|
35
|
Li Y, Gong W, Ma X, Sun X, Jiang H and
Chen T: Smad7 maintains epithelial phenotype of ovarian cancer
stem-like cells and supports tumor colonization by
mesenchymal-epithelial transition. Mol Med Rep. 11:309–316. 2015.
View Article : Google Scholar
|
|
36
|
Aokage K, Ishii G, Ohtaki Y, Yamaguchi Y,
Hishida T, Yoshida J, Nishimura M, Nagai K and Ochiai A: Dynamic
molecular changes associated with epithelial-mesenchymal transition
and subsequent mesenchymal-epithelial transition in the early phase
of metastatic tumor formation. Int J Cancer. 128:1585–1595. 2011.
View Article : Google Scholar
|
|
37
|
Souid S, Elsayed HE, Ebrahim HY, Mohyeldin
MM, Siddique AB, Karoui H, El Sayed KA and Essafi-Benkhadir K:
131 -Oxophorbine protopheophorbide A from Ziziphus lotus
as a novel mesenchymal-epithelial transition factor receptor
inhibitory lead for the control of breast tumor growth in vitro and
in vivo. Mol Carcinog. 57:1507–1524. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guzińska-Ustymowicz K: MMP-9 and cathepsin
B expression in tumor budding as an indicator of a more aggressive
phenotype of colorectal cancer (CRC). Anticancer Res. 26(2B):
1589–1594. 2006.
|
|
39
|
Ivanovska J, Zlobec I, Forster S,
Karamitopoulou E, Dawson H, Koelzer VH, Agaimy A, Garreis F, Söder
S, Laqua W, et al: DAPK loss in colon cancer tumor buds:
Implications for migration capacity of disseminating tumor cells.
Oncotarget. 6:36774–36788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Werle B, Kraft C, Lah TT, Kos J,
Schanzenbächer U, Kayser K, Ebert W and Spiess E: Cathepsin B in
infiltrated lymph nodes is of prognostic significance for patients
with nonsmall cell lung carcinoma. Cancer. 89:2282–2291. 2000.
View Article : Google Scholar
|
|
41
|
Zhang J, He P, Zhong Q, Li K, Chen D, Lin
Q and Liu W: Increasing Cystatin C and Cathepsin B in Serum of
Colorectal Cancer Patients. Clin Lab. 63:365–371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang WE, Ho CC, Yang SF, Lin SH, Yeh KT,
Lin CW and Chen MK: Cathepsin B Expression and the Correlation with
Clinical Aspects of Oral Squamous Cell Carcinoma. PLoS One.
11:e01521652016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guzińska-Ustymowicz K, Zalewski B, Kasacka
I, Piotrowski Z and Skrzydlewska E: Activity of cathepsin B and D
in colorectal cancer: Relationships with tumour budding. Anticancer
Res. 24(5A): 2847–2851. 2004.
|
|
44
|
Zhang M, Zhu ZL, Gao XL, Wu JS, Liang XH
and Tang YL: Functions of chemokines in the perineural invasion of
tumors (Review). Int J Oncol: Mar. 8:2018.(Epub ahead of
print).
|
|
45
|
Shen Z, Li T, Chen D, Jia S, Yang X, Liang
L, Chai J, Cheng X, Yang X and Sun M: The CCL5/CCR5 axis
contributes to the perineural invasion of human salivary adenoid
cystic carcinoma. Oncol Rep. 31:800–806. 2014. View Article : Google Scholar
|
|
46
|
Shan C, Wei J, Hou R, Wu B, Yang Z, Wang
L, Lei D and Yang X: Schwann cells promote EMT and the Schwann-like
differentiation of salivary adenoid cystic carcinoma cells via the
BDNF/TrkB axis. Oncol Rep. 35:427–435. 2016. View Article : Google Scholar
|
|
47
|
Zhang M, Wu JS, Yang X, Pang X, Li L, Wang
SS, Wu JB, Tang YJ, Liang XH, Zheng M, et al: Overexpression
Cathepsin D Contributes to Perineural Invasion of Salivary Adenoid
Cystic Carcinoma. Front Oncol. 8:4922018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kato T, Enomoto A, Watanabe T, Haga H,
Ishida S, Kondo Y, Furukawa K, Urano T, Mii S, Weng L, et al:
TRIM27/MRTF-B-dependent integrin β1 expression defines leading
cells in cancer cell collectives. Cell Rep. 7:1156–1167. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gondi CS and Rao JS: Cathepsin B as a
cancer target. Expert Opin Ther Targets. 17:281–291. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Podgorski I and Sloane BF: Cathepsin B and
its role(s) in cancer progression. Biochem Soc Symp. 70:263–276.
2003. View Article : Google Scholar
|
|
51
|
Poreba W, Gawlik K and Gutowicz J:
Cathepsin B and neoplasm invasiveness. Postepy Biochem. 48:111–120.
2002.In Polish.
|
|
52
|
Wellner U, Schubert J, Burk UC,
Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D,
zur Hausen A, et al: The EMT-activator ZEB1 promotes tumorigenicity
by repressing stemness-inhibiting microRNAs. Nat Cell Biol.
11:1487–1495. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kostoulas G, Lang A, Nagase H and Baici A:
Stimulation of angiogenesis through cathepsin B inactivation of the
tissue inhibitors of matrix metalloproteinases. FEBS Lett.
455:286–290. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Somanna A, Mundodi V and Gedamu L:
Functional analysis of cathepsin B-like cysteine proteases from
Leishmania donovani complex. Evidence for the activation of latent
transforming growth factor beta. J Biol Chem. 277:25305–25312.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tummalapalli P, Spomar D, Gondi CS,
Olivero WC, Gujrati M, Dinh DH and Rao JS: RNAi-mediated abrogation
of cathepsin B and MMP-9 gene expression in a malignant meningioma
cell line leads to decreased tumor growth, invasion and
angiogenesis. Int J Oncol. 31:1039–1050. 2007.PubMed/NCBI
|
|
56
|
Vento SI, Jouhi L, Mohamed H, Haglund C,
Mäkitie AA, Atula T, Hagström J and Mäkinen LK: MMP-7 expression
may influence the rate of distant recurrences and disease-specific
survival in HPV-positive oropharyngeal squamous cell carcinoma.
Virchows Arch. 472:975–981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Upmanyu N, Bulldan A, Papadopoulos D,
Dietze R, Malviya VN and Scheiner-Bobis G: Impairment of the
Gnα11-controlled expression of claudin-1 and MMP-9 and collective
migration of human breast cancer MCF-7 cells by DHEAS. J Steroid
Biochem Mol Biol. 182:50–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gaggioli C, Hooper S, Hidalgo-Carcedo C,
Grosse R, Marshall JF, Harrington K and Sahai E: Fibroblast-led
collective invasion of carcinoma cells with differing roles for
RhoGTPases in leading and following cells. Nat Cell Biol.
9:1392–1400. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wyckoff JB, Pinner SE, Gschmeissner S,
Condeelis JS and Sahai E: ROCK- and myosin-dependent matrix
deformation enables protease-independent tumor-cell invasion in
vivo. Curr Biol. 16:1515–1523. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tamir S, Rotem-Bamberger S, Katz C, Morcos
F, Hailey KL, Zuris JA, Wang C, Conlan AR, Lipper CH, Paddock ML,
et al: Integrated strategy reveals the protein interface between
cancer targets Bcl-2 and NAF-1. Proc Natl Acad Sci USA.
111:5177–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Murakami R, Matsumura N, Mandai M,
Yoshihara K, Tanabe H, Nakai H, Yamanoi K, Abiko K, Yoshioka Y,
Hamanishi J, et al: Establishment of a Novel Histopathological
Classification of High-Grade Serous Ovarian Carcinoma Correlated
with Prognostically Distinct Gene Expression Subtypes. Am J Pathol.
186:1103–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Laskin DM, Abubaker AO and Strauss RA:
Accuracy of predicting the duration of a surgical operation. J Oral
Maxillofac Surg. 71:446–447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gabarra-Niecko V, Schaller MD and Dunty
JM: FAK regulates biological processes important for the
pathogenesis of cancer. Cancer Metastasis Rev. 22:359–374. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sandilands E, Serrels B, McEwan DG, Morton
JP, Macagno JP, McLeod K, Stevens C, Brunton VG, Langdon WY, Vidal
M, et al: Autophagic targeting of Src promotes cancer cell survival
following reduced FAK signalling. Nat Cell Biol. 14:51–60. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Serrels A, Canel M, Brunton VG and Frame
MC: Src/FAK-mediated regulation of E-cadherin as a mechanism for
controlling collective cell movement: Insights from in vivo
imaging. Cell Adhes Migr. 5:360–365. 2011. View Article : Google Scholar
|
|
66
|
Alapati K, Gopinath S, Malla RR, Dasari VR
and Rao JS: uPAR and cathepsin B knockdown inhibits
radiation-induced PKC integrated integrin signaling to the
cytoskeleton of glioma-initiating cells. Int J Oncol. 41:599–610.
2012. View Article : Google Scholar : PubMed/NCBI
|