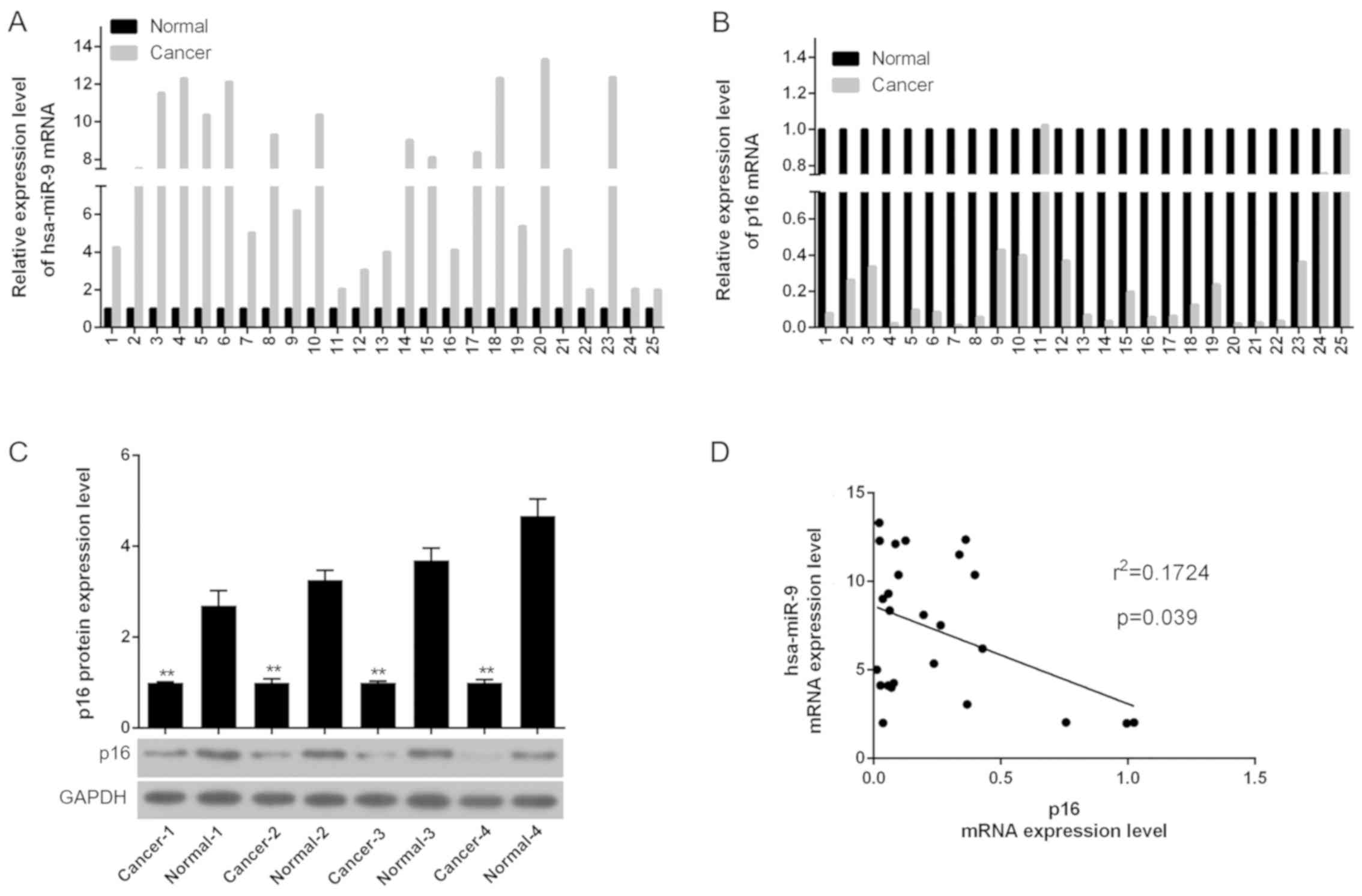
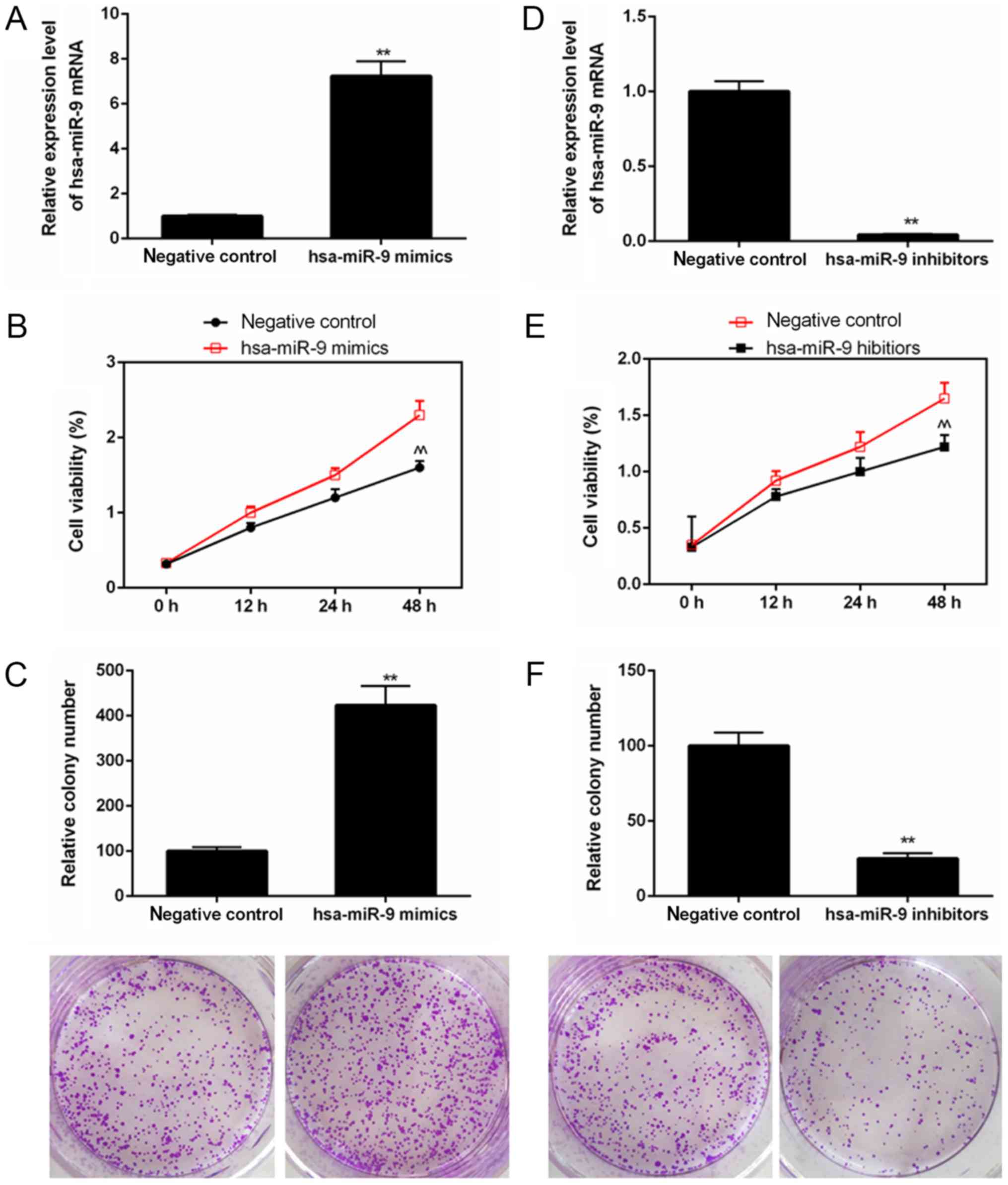
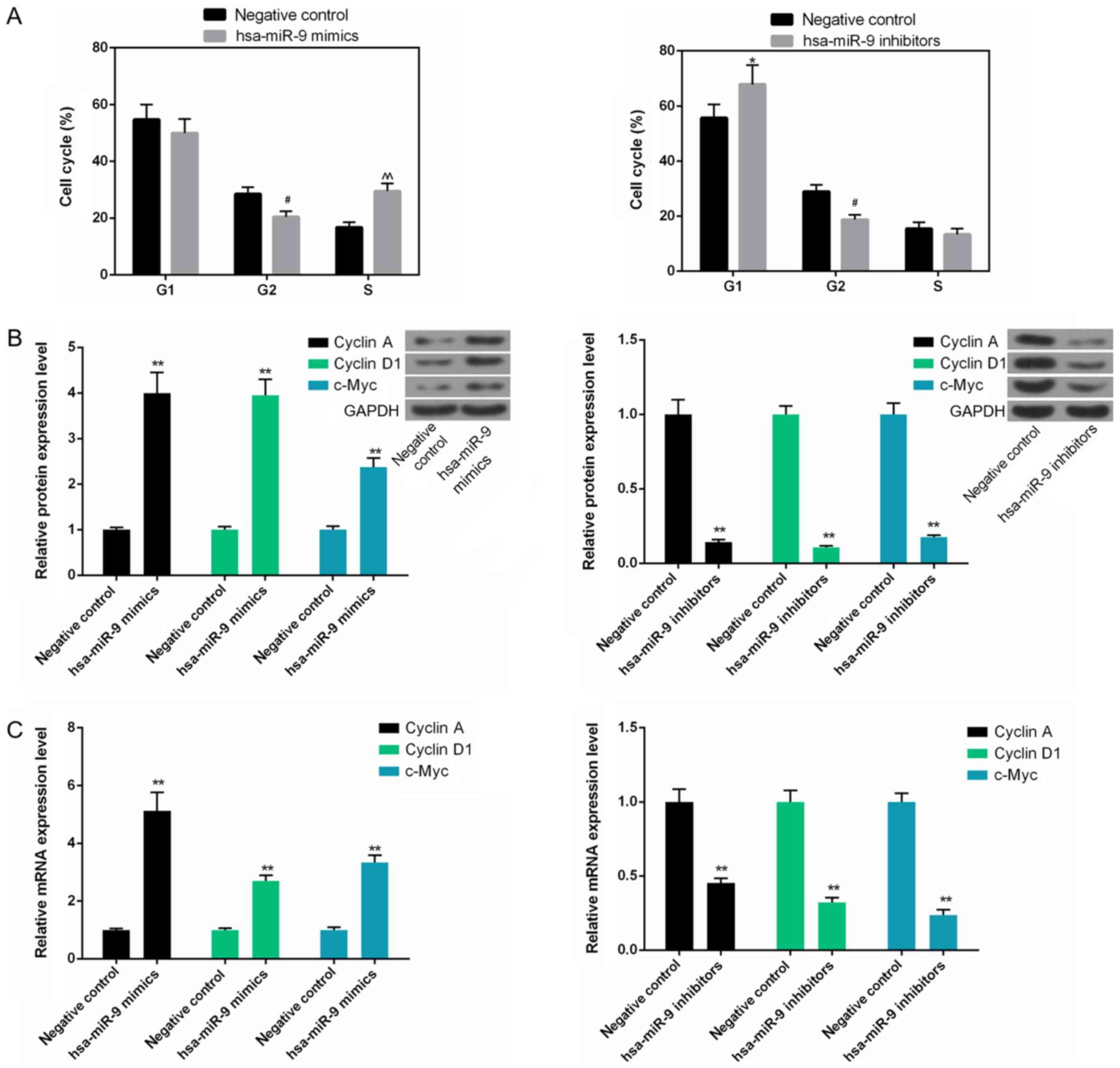
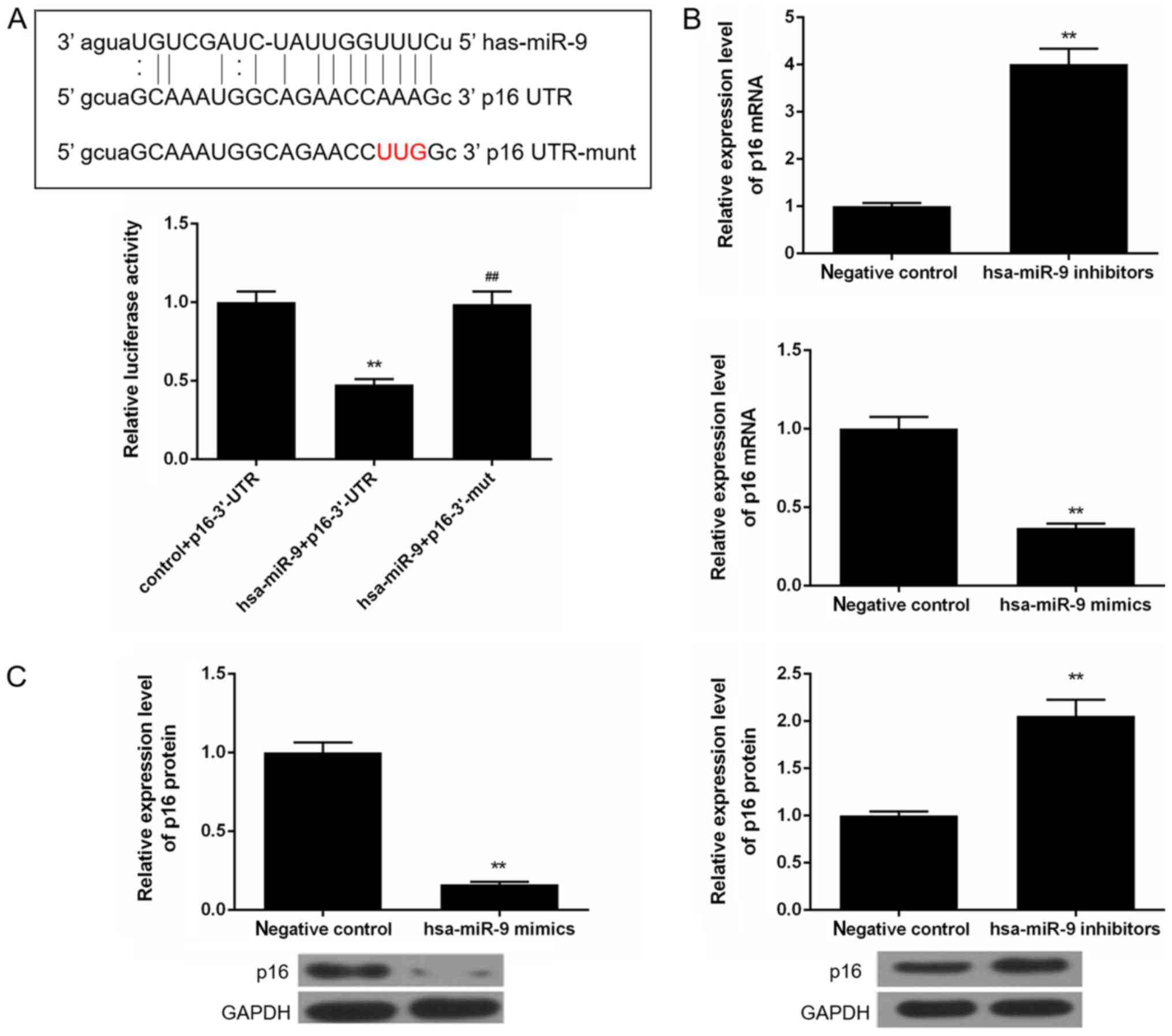
|
1
|
Garcia-Moure M, Martinez-Vélez N,
Patiño-García A and Alonso MM: Oncolytic adenoviruses as a
therapeutic approach for osteosarcoma: A new hope. J Bone Oncol.
9:41–47. 2016. View Article : Google Scholar
|
|
2
|
Misaghi A, Goldin A, Awad M and Kulidjian
AA: Osteosarcoma: A comprehensive review. SICOT J. 4:122018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guenther LM, Rowe RG, Acharya PT, Swenson
DW, Meyer SC, Clinton CM, Guo D, Sridharan M, London WB, Grier HE,
et al: Response Evaluation Criteria in Solid Tumors (RECIST)
following neoadjuvant chemotherapy in osteosarcoma. Pediatr Blood
Cancer. 65:652018. View Article : Google Scholar
|
|
4
|
Huang Z and Lou C: Application of the
alteration uptake ratio of 99mTc-MIBI scintigraphy for evaluating
the efficacy of neoadjuvant chemotherapy in osteosarcoma patients.
Hell J Nucl Med. 21:55–59. 2018.PubMed/NCBI
|
|
5
|
Cavit A, Özcanli H, Sançmiş M, Ocak GA and
Gürer EI: Tumorous conditions of the hand: A retrospective review
of 402 cases. Turk Patoloji Derg. 34:66–72. 2018.
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee J, Park H, Eom J and Kang SG:
MicroRNA-mediated regulation of the development and functions of
follicular helper T cells. Immune Netw. 18:e72018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation iC elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Z, Han Y, Song C, Wei H, Chen Y,
Huang K, Li S, Ma D, Wang S, Wang J, et al: Systematic review and
meta-analysis of the prognostic significance of microRNAs in
cervical cancer. Oncotarget. 9:17141–17148. 2017.
|
|
11
|
Hershkovitz-Rokah O, Geva P, Salmon-Divon
M, Shpilberg O and Liberman-Aronov S: Network analysis of
microRNAs, genes and their regulation in diffuse and follicular
B-cell lymphomas. Oncotarget. 9:7928–7941. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ichii O and Horino T: MicroRNAs associated
with the development of kidney diseases in humans and animals. J
Toxicol Pathol. 31:23–34. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iguchi T, Sakurai K, Tamai S and Mori K:
Circulating liver-specific microRNAs in cynomolgus monkeys. J
Toxicol Pathol. 31:3–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Peng R, Wang J, Qin Z and Xue L:
Circulating microRNAs as potential cancer biomarkers: The advantage
and disadvantage. Clin Epigenetics. 10:592018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen M, Liu YY, Zheng MQ, Wang XL, Gao XH,
Chen L and Zhang GM: microRNA-544 promoted human osteosarcoma cell
proliferation by downregulating AXIN2 expression. Oncol Lett.
15:7076–7082. 2018.PubMed/NCBI
|
|
16
|
Ding J, Sha L, Shen P, Huang M, Cai Q and
Li J: MicroRNA-18a inhibits cell growth and induces apoptosis in
osteosarcoma by targeting MED27. Int J Oncol. 53:329–338.
2018.PubMed/NCBI
|
|
17
|
Tang W, Wang W, Zhao Y and Zhao Z:
MicroRNA-874 inhibits cell proliferation and invasion by targeting
cyclin-dependent kinase 9 in osteosarcoma. Oncol Lett.
15:7649–7654. 2018.PubMed/NCBI
|
|
18
|
Fenger JM, Roberts RD, Iwenofu OH, Bear
MD, Zhang X, Couto JI, Modiano JF, Kisseberth WC and London CA:
MiR-9 is overexpressed in spontaneous canine osteosarcoma and
promotes a metastatic phenotype including invasion and migration in
osteoblasts and osteosarcoma cell lines. BMC Cancer. 16:7842016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jang MH, Kim HJ, Gwak JM, Chung YR and
Park SY: Prognostic value of microRNA-9 and microRNA-155 expression
in triple-negative breast cancer. Hum Pathol. 68:69–78. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Snezhkina AV, Krasnov GS, Zhikrivetskaya
SO, Karpova IY, Fedorova MS, Nyushko KM, Belyakov MM, Gnuchev NV,
Sidorov DV, Alekseev BY, et al: Overexpression of microRNAs miR-9,
-98, and -199 correlates with the downregulation of HK2 expression
in colorectal cancer. Mol Biol (Mosk). 52:220–230. 2018.In Russian.
View Article : Google Scholar
|
|
21
|
Wang H, Wu Q, Zhang Y, Zhang HN, Wang YB
and Wang W: TGF-β1-induced epithelial-mesenchymal transition in
lung cancer cells involves upregulation of miR-9 and downregulation
of its target, E-cadherin. Cell Mol Biol Lett. 22:222017.
View Article : Google Scholar
|
|
22
|
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang
X, Huang K and Tong Q: microRNA-9 suppresses the proliferation,
invasion and metastasis of gastric cancer cells through targeting
cyclin D1 and Ets1. PLoS One. 8:e557192013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
She H, He Y, Zhao Y and Mao Z: Autophagy
in inflammation: The p38α MAPK-ULK1 axis. Macrophage (Houst).
5:e16292018.
|
|
24
|
Tang Q, Tong M, Zheng G, Shen L, Shang P
and Liu H: Masquelet’s induced membrane promotes the osteogenic
differentiation of bone marrow mesenchymal stem cells by activating
the Smad and MAPK pathways. Am J Transl Res. 10:1211–1219.
2018.
|
|
25
|
Li S, Ma YM, Zheng PS and Zhang P: GDF15
promotes the proliferation of cervical cancer cells by
phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp
Clin Cancer Res. 37:802018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pinal N, Martín M, Medina I and Morata G:
Short-term activation of the Jun N-terminal kinase pathway in
apoptosis-deficient cells of Drosophila induces tumorigenesis. Nat
Commun. 9:15412018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang S, Guo L, Su Y, Wen J, Du J, Li X,
Liu Y, Feng J, Xie Y, Bai Y, et al: Nitric oxide balances
osteoblast and adipocyte lineage differentiation via the JNK/MAPK
signaling pathway in periodontal ligament stem cells. Stem Cell Res
Ther. 9:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aroui S, Aouey B, Chtourou Y, Meunier AC,
Fetoui H and Kenani A: Naringin suppresses cell metastasis and the
expression of matrix metalloproteinases (MMP-2 and MMP-9) via the
inhibition of ERK-P38-JNK signaling pathway in human glioblastoma.
Chem Biol Interact. 244:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Speth Z, Islam T, Banerjee K and Resat H:
EGFR signaling pathways are wired differently in normal 184A1L5
human mammary epithelial and MDA-MB-231 breast cancer cells. J Cell
Commun Signal. 11:341–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen HJ, Lin CM, Lee CY, Shih NC, Peng SF,
Tsuzuki M, Amagaya S, Huang WW and Yang JS: Kaempferol suppresses
cell metastasis via inhibition of the ERK-p38-JNK and AP-1
signaling pathways in U-2 OS human osteosarcoma cells. Oncol Rep.
30:925–932. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Joo SS and Yoo YM: Melatonin induces
apoptotic death in LNCaP cells via p38 and JNK pathways:
Therapeutic implications for prostate cancer. J Pineal Res.
47:8–14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao Li T, Ge J, Yang J, Song J, Wang X,
Mao C, Zhang J, Zou Y, Liu YY, et al: Particulate matter
facilitates C6 glioma cells activation and the release of
inflammatory factors through MAPK and JAK2/STAT3 pathways.
Neurochem Res. 41:1969–1981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Faustino-Rocha A, Oliveira PA,
Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, da Costa RG,
Colaço B, Pires MJ, Colaço J, Ferreira R, et al: Estimation of rat
mammary tumor volume using caliper and ultrasonography
measurements. Lab Anim (NY). 42:217–224. 2013. View Article : Google Scholar
|
|
35
|
Drakaki A, Hatziapostolou M, Polytarchou
C, Vorvis C, Poultsides GA, Souglakos J, Georgoulias V and
Iliopoulos D: Functional microRNA high throughput screening reveals
miR-9 as a central regulator of liver oncogenesis by affecting the
PPARA-CDH1 pathway. BMC Cancer. 15:5422015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu T, Liu X, Han L, Shen H, Liu L and Shu
Y: Up-regulation of miR-9 expression as a poor prognostic biomarker
in patients with non-small cell lung cancer. Clin Transl Oncol.
16:469–475. 2014. View Article : Google Scholar
|
|
37
|
Zhu SW, Li JP, Ma XL, Ma JX, Yang Y, Chen
Y and Liu W: miR-9 modulates osteosarcoma cell growth by targeting
the GCIP tumor suppressor. Asian Pac J Cancer Prev. 16:4509–4513.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Laios A, O’Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Lu J, Liu X, Peng X, Zhang Y, Wang B,
Luo L, Peng H, Li X, Tian GW, et al: Predictive value of miR-9 as a
potential biomarker for nasopharyngeal carcinoma metastasis. Br J
Cancer. 110:392–398. 2014. View Article : Google Scholar :
|
|
40
|
Jia GQ, Zhang MM, Wang K, Zhao GP, Pang MH
and Chen ZY: Long non-coding RNA PlncRNA-1 promotes cell
proliferation and hepatic metastasis in colorectal cancer. J Cell
Biochem. 119:7091–7104. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou Q, You C, Zheng C, Gu Y, Gu H, Zhang
R, Wu H and Sun B: 3-Nitroacridine derivatives arrest cell cycle at
G0/G1 phase and induce apoptosis in human breast cancer cells may
act as DNA-target anticancer agents. Life Sci. 206:1–9. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu W, Liu C, Yin B and Peng XZ: Functions
of miR-9 and miR-9* during aging in SAMP8 mice and their possible
mechanisms. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 37:253–258.
2015.PubMed/NCBI
|
|
43
|
Di Sante G, Di Rocco A, Pupo C, Casimiro
MC and Pestell RG: Hormone-induced DNA damage response and repair
mediated by cyclin D1 in breast and prostate cancer. Oncotarget.
8:81803–81812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Y, Zhang J, Gao W, Zhang L, Pan Y,
Zhang S and Wang Y: Insights on structural characteristics and
ligand binding mechanisms of CDK2. Int J Mol Sci. 16:9314–9340.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Roskoski R Jr: Cyclin-dependent protein
kinase inhibitors including palbociclib as anticancer drugs.
Pharmacol Res. 107:249–275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goetzman ES and Prochownik EV: The role
for Myc in coordinating glycolysis, oxidative phosphorylation,
glutaminolysis, and fatty acid metabolism in normal and neoplastic
tissues. Front Endocrinol (Lausanne). 9:1292018. View Article : Google Scholar
|
|
47
|
Wang H, Zhang W, Zuo Y, Ding M, Ke C, Yan
R, Zhan H, Liu J and Wang J: miR-9 promotes cell proliferation and
inhibits apoptosis by targeting LASS2 in bladder cancer. Tumour
Biol. 36:9631–9640. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou ZX, Zhang ZP, Tao ZZ and Tan TZ:
MiR-632 promotes laryngeal carcinoma cell proliferation, migration
and invasion through negative regulation of GSK3β. Oncol Res: Mar.
21:2018Epub ahead of print. View Article : Google Scholar
|
|
49
|
Lee JK, Lee KH, Kim SA, Kweon SS, Cho SH,
Shim HJ, Bae WK, Chung IJ, Chung WK, Yoon TM, et al: p16 as a
prognostic factor for the response to induction chemotherapy in
advanced hypo-pharyngeal squamous cell carcinoma. Oncol Lett.
15:6571–6577. 2018.PubMed/NCBI
|
|
50
|
Ottria L, Candotto V, Cura F, Baggi L,
Arcuri C, Nardone M, Gaudio RM, Gatto R, Spadari F and Carinci F:
HPV acting on E-cadherin, p53 and p16: Literature review. J Biol
Regul Homeost Agents. 32(Suppl 1): 73–79. 2018.PubMed/NCBI
|
|
51
|
Nobori T, Miura K, Wu DJ, Lois A,
Takabayashi K and Carson DA: Deletions of the cyclin-dependent
kinase-4 inhibitor gene in multiple human cancers. Nature.
368:753–756. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hirama T and Koeffler HP: Role of the
cyclin-dependent kinase inhibitors in the development of cancer.
Blood. 86:841–854. 1995.PubMed/NCBI
|
|
53
|
Serrano M, Hannon GJ and Beach D: A new
regulatory motif in cell-cycle control causing specific inhibition
of cyclin D/CDK4. Nature. 366:704–707. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu Pu X, Fu L, Fan Y, Zheng Z, Zhang J,
Yang B, Guan J, Wu W, Ye HQ, et al: Companied P16 genetic and
protein status together providing useful information on the
clinical outcome of urinary bladder cancer. Medicine (Baltimore).
97:e03532018. View Article : Google Scholar :
|
|
55
|
Goody D and Pfeifer A: MicroRNAs in brown
and beige fat. Biochim Biophys Acta Mol Cell Biol Lipids.
1864:29–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guo J and Cheng Y: MicroRNA-1247 inhibits
lipopolysac-charides-induced acute pneumonia in A549 cells via
targeting CC chemokine ligand 16. Biomed Pharmacother. 104:60–68.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Song Z, Cooper DKC, Cai Z and Mou L:
Expression and Regulation profile of mature MicroRNA in the pig:
Relevance to xenotransplantation. BioMed Res Int. 2018:29839082018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
O’Loghlen A, Brookes S, Martin N,
Rapisarda V, Peters G and Gil J: CBX7 and miR-9 are part of an
autoregulatory loop controlling p16(INK) (4a). Aging Cell.
14:1113–1121. 2015. View Article : Google Scholar
|
|
59
|
Wang C, Zhang X, Zhang C, Zhai F, Li Y and
Huang Z: MicroRNA-155 targets MAP3K10 and regulates osteosarcoma
cell growth. Pathol Res Pract. 213:389–393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gui ZL, Wu TL, Zhao GC, Lin ZX and Xu HG:
MicroRNA-497 suppress osteosarcoma by targeting MAPK/Erk pathway.
Bratisl Lek Listy. 118:449–452. 2017.PubMed/NCBI
|
|
61
|
Zhang J, Cheng J, Zeng Z, Wang Y, Li X,
Xie Q, Jia J, Yan Y, Guo Z, Gao J, et al: Comprehensive profiling
of novel microRNA-9 targets and a tumor suppressor role of
microRNA-9 via targeting IGF2BP1 in hepatocellular carcinoma.
Oncotarget. 6:42040–42052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ben-Hamo R and Efroni S: Correction: Gene
expression and network-based analysis reveals a novel role for
hsa-miR-9 and drug control over the p38 network in glioblastoma
multiforme progression. Genome Med. 4:872012. View Article : Google Scholar : PubMed/NCBI
|