Introduction
Worldwide, there were ~18.1 million new cancer cases
and 9.6 million cancer-associated mortalities in 2018 (1). Among all types of cancer, lung cancer
has the highest incidence and mortality rates, with 2.1 million new
lung cancer cases and 1.8 million mortalities predicted in 2018,
which accounts for ~18.4% of all cancer-associated mortalities
(1). Non-small cell lung carcinoma
(NSCLC) is the most common histological type of lung malignancy.
Lung adenocarcinoma (LUAD) accounts for ~50% of all lung cancer
cases (2). In the past few
decades, clinical trials for the treatment and examination of lung
adenocarcinoma have increased, including studies involving
chemotherapy, molecular targeted therapy, stereotactic radiotherapy
and immunotherapy (3-6). Although NSCLC has been investigated
in numerous molecular studies aimed at developing new treatment
strategies, the 5-year overall survival rate remains at 4-17%, and
65% of patients with NSCLC are classified as stage III or IV at
diagnosis (4,7). Therefore, improved understanding
regarding the regulatory mechanisms of NSCLC development and
progression is urgently required for the improvement of cancer
treatment.
Centrosome protein E (CENPE) is a kinesin-like motor
protein that accumulates at the G2 phase of the cell cycle
(8). Unlike other
centrosome-associated proteins, it is not present during interphase
and first appears at the centromere region of chromosomes during
prometaphase (9). Therefore, cells
with rapid proliferation have higher levels of CENPE expression and
CENPE is upregulated in numerous types of solid cancer (10). Certain studies have identified that
reducing the expression of CENPE can inhibit cancer cell
proliferation. In prostate cancer, genetic deletion or
pharmacological inhibition of CENPE was demonstrated to
significantly decrease tumor growth (11). In high-grade glioma cells,
knockdown of CENPE, kinesin family member 14 or non-SMC condensin I
complex subunit G combined with temozolomide-treatment resulted in
a combined suppressive effect on cell proliferation (12). In addition, small interfering RNA
(siRNA)-induced knockdown of CENPE in human neuroblastoma cell
lines can inhibit cellular proliferation (13). Targeting CENPE with the small
molecular inhibitor GSK923295 inhibited the in vitro proliferation
of 19 neuroblastoma cell lines and delayed tumor growth in three
xenograft models (13).
Furthermore, one study identified that CENPE can interact with FA
complementation group A, which may serve an important role in cell
cycle control and the pathogenesis of tumor (14).
In summary, previous studies have demonstrated that
CENPE is associated with the proliferation of certain cancer cells.
However, to the best of our knowledge, the role and regulatory
mechanisms of CENPE in NSCLC have not been studied. The present
study identified that the expression level of CENPE is higher in
LUAD tissues compared with normal tissues. Cell proliferation in
the A549 and PC9 cell lines was significantly inhibited following
knockdown of CENPE. In addition, it was identified that the
expression of CENPE in LUAD is directly regulated by FOXM1.
Materials and methods
Sample collection and cell culture
Between March 2018 and June 2018, seven LUAD tumor
samples and seven paired normal lung tissue samples were collected
from patients at The First Affiliated Hospital of Jinzhou Medical
University (Jinzhou, China). The tumor and normal tissue samples
were obtained by lobectomy, and the distance between the tumor
tissue and normal lung tissue was ~6 mm. The patients included five
males and two females, with a mean age of 56 years (range, 41-78
years). The present study was approved by the Ethics Committee of
the First Affiliated Hospital of Jinzhou Medical University and
written informed consent was obtained from each individual. The
A549 and PC9 human LUAD cell lines were purchased from the American
Type Culture Collection (Manassas) and cultured in DMEM high
glucose (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere containing 5%
CO2.
siRNA and plasmid transfection
A549 or PC9 cells (5×105/well) were
seeded in 6-well plates, cultured overnight and then transfected
with 100 nmol/l CENPE siRNA, 100 nmol/l FOXM1 siRNA or 100 nmol/l
negative control siRNA using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The sequences of the siRNAs were as
follows: CENPE, 5′-GGCUGUAAUAUAA AUCGAA-3′ (11); FOXM1 5′-GCUCAUACCUGGUACC UAU-3
(15); and negative control,
5′-UUCUCCGAACGUG UCACGU-3′ (Shanghai GenePharma, Co., Ltd.).
The FOXM1 expression plasmid and control plasmid
were purchased from Guangzhou FulenGen Co., Ltd. (Guangzhou, China)
and transfected into A549 cells due to the lower expression of
FOXM1 in these cells compared with PC9 cells (16). Transfections were performed in
6-well plates using Lipofectamine® 3000 transfection
reagent (Gibco; Thermo Fisher Scientific, Inc.,), according to the
manufacturer's protocol. In total, the cells were transfection with
2 µg plasmid/well. Cells were harvested and subjected to
analysis 48 h after transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA was reverse transcribed into cDNA using the
RT reagent kit with gDNA Eraser (Takara Bio, Inc.). The reaction
conditions were as follows: 42°C for 2 min, 37°C for 15 min and
87°C for 5 sec. qPCR was performed in a reaction mix with SYBR
Green (Takara Bio, Inc.) and was performed in triplicate with an
ABI 7500 Prism Sequence Detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following conditions were
used: 95°C for 30 sec for initial denaturation followed by 40
cycles of 95°C for 5 sec and 60 °C for 34 sec. All qPCR primers
were purchased from Invitrogen (Invitrogen; Thermo Fisher
Scientific, Inc.) and the sequences were as follows: CENPE forward,
5′-GATTCTGCCATACAAGGCTACAA-3′ and reverse,
5′-TGCCCTGGGTATAACTCCCAA-3′; FOXM1 forward,
5′-GGAGCAGCGACAGGTTAAGG-3′ and reverse,
5′-GTTGATGGCGAATTGTATCATGG-3′; and GAPDH forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′. Relative expression levels were
calculated using the 2−∆∆Cq method (17).
Western blot analysis
Total protein was extracted from the cells or tissue
samples using RIPA lysis buffer (Beyotime Institute of
Biotechnology) with 1% protease inhibitor phenylmethanesulfonyl
fluoride (Beyotime Institute of Biotechnology). The lysates were
centrifuged at 1,3000 × g for 15 min at 4°C and protein
concentration was measured using a BCA kit (Beyotime Institute of
Biotechnology). Subsequently, 40 µg protein/lane was
separated by 10% SDS-PAGE and the proteins were then transferred to
a PVDF membrane (Roche Applied Science). The membrane was blocked
with 5% BSA (Wuhan Boster Biological Technology, Ltd.) for 1 h at
room temperature and then incubated at 4°C overnight with primary
antibodies. Following incubation with appropriate HRP-conjugated
secondary antibodies, including goat anti-mouse IgG (cat. no.
BA1050; 1:5,000; Wuhan Boster Biological Technology, Ltd.) and goat
anti-rabbit IgG (cat. no. BA1054; 1:5,000; Wuhan Boster Biological
Technology, Ltd.), for 1 h at room temperature, the membranes were
visualized with an ECL kit (Beyotime Institute of Biotechnology)
The primary antibodies for western blot analysis included mouse
monoclonal anti-GAPDH (1:2,000; cat. no. BM1985; Wuhan Boster
Biological Technology, Ltd.), rabbit anti-FOXM1 (1:1,000; cat. no.
ab184637; Abcam) and rabbit anti-CENPE (1:1,000; cat. no. ab133583;
Abcam).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) and 5-ethynyl-2'-deoxyuridine (EdU;
GeneCopoeia, Inc.) assays were used to detect proliferation. In the
CCK-8 assay, 24 h post-transfection, cells were seeded in a 96-well
plate at a density of 3,000 cells/well and cultured overnight at
37°C in a humidified atmosphere containing 5% CO2. CCK-8
reagent was then added to the wells and the absorbance at 450 nm
was detected following culture for 24, 48 and 72 h at 37°C.
The iClick™ EdU Andy Fluor™ 647 Flow Cytometry assay
kit (GeneCopoeia, Inc.) was used to evaluate cell proliferation in
the EdU assay. A total of 24 h post-transfection, A549 and PC9
cells, cultured in DMEM high glucose supplemented with 10% FBS,
were treated with EdU (10 µM) for 6 h and the assays were
performed according to manufacturer's protocol. The cells were then
analyzed using a flow cytometer and analysis was performed with
FlowJo v10 software (FlowJo LLC).
Chromatin immunoprecipitation (ChIP)
assay
The FOXM1 binding site on the CENPE promoter region
was analyzed using the Cistrome Data Browser (http://cistrome.org/db/#/) (18), with the following parameters:
Biological sources; Neuroblastoma cell, Brain; and Factors; FOXM1,
SK-N-SH, CENPE (19). The CENPE
promoter sequence was obtained from the Eukaryotic Promoter
Database (https://epd.epfl.ch/) (20). Subsequently, the binding site was
analyzed using the AnimalTFDB3.0 database (http://bioinfo.life.hust.edu.cn/AnimalTFDB/)
(21). The following two pairs of
primers were used in the present study: Primer 1 (114 bp) forward,
5′-GACGGCAATTCTGTTTGGGT-3′ and reverse, 5′-CTGCCAAAAGCTAGCGAACG-3′;
and primer 2 (218 bp) forward, 5′-CCCTCTCCTGTTTAGCAGTG-3′ and
reverse, 5′-CCGCCATCCTATCAGGCTG-3′.
The ChIP assay was performed using the SimpleChIP™
Enzymatic Chromatin IP kit (cat. no. 9003; Cell Signaling
Technology, Inc.), according to the manufacturer's protocol. In
brief, 4×106 A549 cells were fixed with formaldehyde at
room temperature for 15 min and lysed on ice for 5 min using the
lysate in the kit, and then chromatin was harvested and fragmented
using sonication. Antibodies specific to FOXM1 (1:100; cat. no.
ab184637; Abcam) were used to recruit the target DNA overnight at
4°C and the complex was precipitated by Protein G magnetic beads
for 2 h at 4°C. Normal rabbit IgG (1:300; cat. no. 2729; Cell
Signaling Technology, Inc.) was used as the negative control and
incubated with the sample overnight at 4°C, followed by
precipitation of the complex with Protein G magnetic beads for 2 h
at 4°C. Following immunoprecipitation, the protein-DNA complex was
reversed and the DNA was purified. The enriched DNA was subjected
to PCR analysis. PCR was performed with PCR Master mix (cat. no.
KT201; Tiangen Biotech, Co., Ltd.), 2 µl DNA, 0.5 µl
of each forward and reverse primers, and 7 µl
ddH2O. The reaction conditions were as follows: 95°C for
4 min, 35 cycles of 95°C for 1 min, 60°C for 1 min and 72°C for 1
min, and a final step of 72°C for 5 min. The PCR products were
screened on 2% agarose gel and signals were detected using enhanced
chemiluminescence reagent (Beyotime Institute of Biotechnology),
followed by scanning using a FluorChem HD imaging system
(ProteinSimple). The following primers were used to detect the
co-immunoprecipitated CENPE promoter region by PCR: P r imer 1 (114
bp) for wa rd, 5′-GACGGCAATTCTGTTTGGGT-3′ and reverse,
5′-CTGCCAAAAGCTAGCGAACG-3′; and primer 2 (218bp) forward,
5′-CCCTCTCCTGTTTAGCAGTG-3′ and reverse,
5′-CCGCCATCCTATCAGGCTG-3′.
The Cancer Genome Atlas (TCGA) data
analyses
To analyze the differential CENPE/FOXM1 expression
levels in normal and LUAD tissues, and the survival of patients
with NSCLC the Gene Expression Profiling Interactive Analysis
server (http://gepia.cancer-pku.cn/)
(22) was used to performed
Kaplan-Meier analysis followed by a log-rank test. The CENPE and
FOXM1 RNA-sequencing data were downloaded from the cBioPortal
(http://www.cbioportal.org/index.do)
(23). The expression difference
analysis was performed with an unpaired Student's t-test using
UALCAN (http://ualcan.path.uab.edu/index.html) (24) and Pearson's correlation analysis
was performed using GraphPad Prism 6 software (GraphPad Software,
Inc.).
Statistical analysis
Each experiment was independently repeated three
times. Statistical analysis was performed with GraphPad Prism 6
software (GraphPad Software, Inc.). All data are presented as the
mean ± standard deviation. Data were analyzed using Student's
t-test for two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
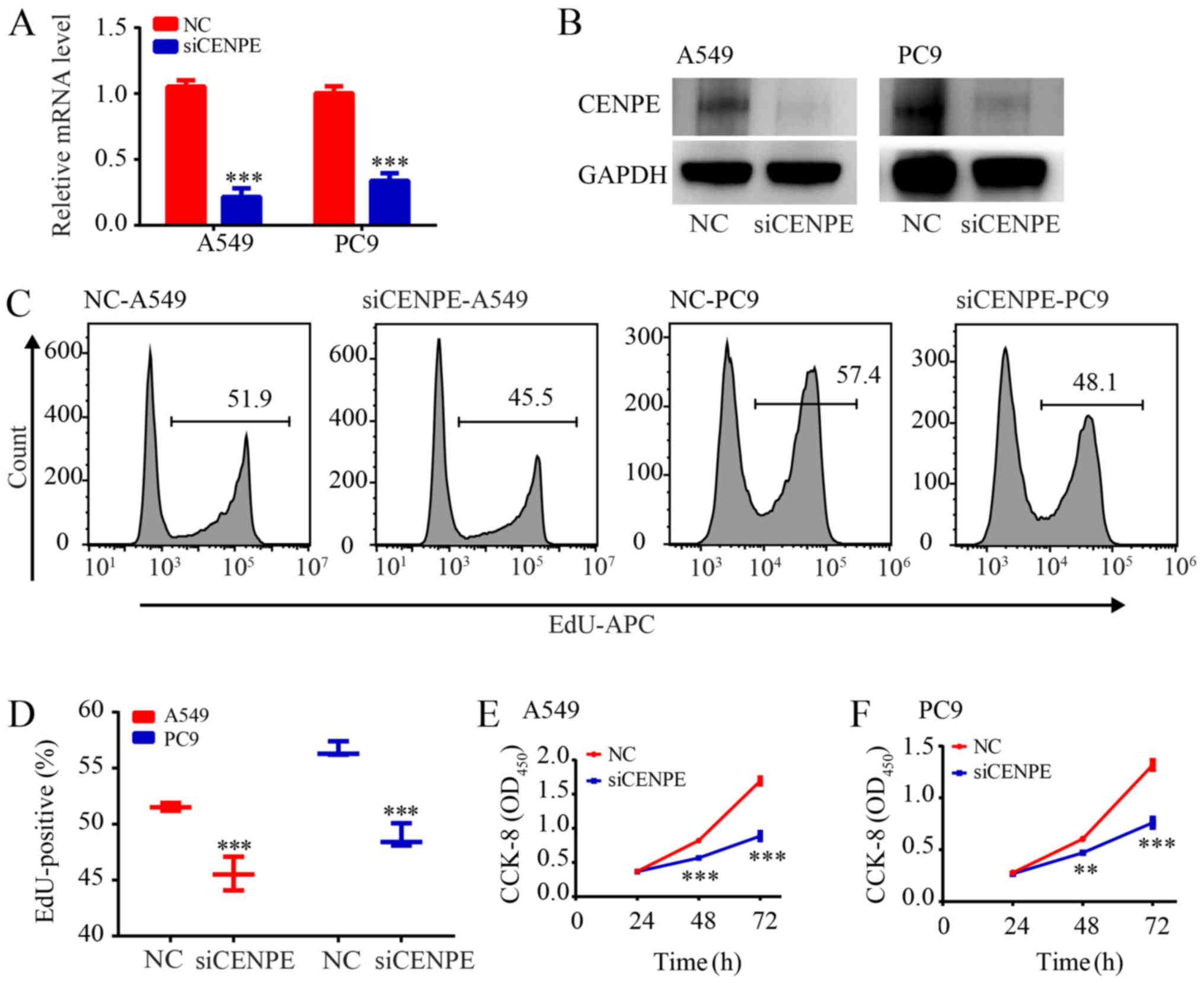
CENPE enhances the proliferation rate of
LUAD cells
CCK-8 and EdU assays were used to examine the effect
of CENPE on LUAD cell proliferation. The siRNA was designed to
knockdown CENPE in A549 and PC9 cells. CENPE mRNA and protein
levels were significantly reduced following RNA interference
(P<0.001; Fig. 1A and B). The
EdU assay demonstrated that the proliferation of LUAD cells was
inhibited following CENPE-knockdown (P<0.001; Fig. 1C and D). The CCK-8 assay
demonstrated the same phenomenon (P<0.01; Fig. 1E and F).
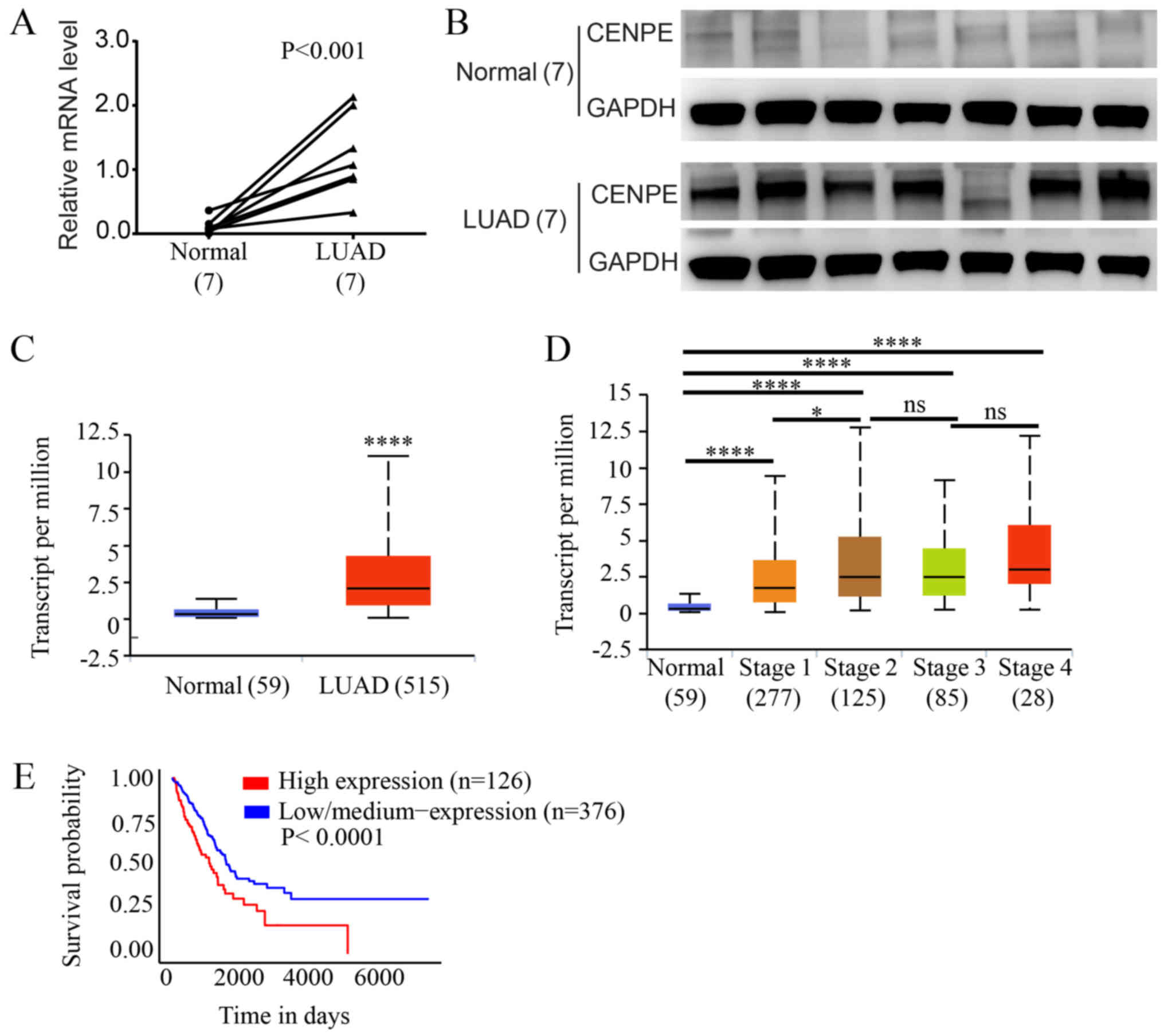
Expression of CENPE in human LUAD
tissues
The expression of CENPE in lung adenocarcinoma and
adjacent normal tissues was evaluated by RT-qPCR and western blot
analysis. The results demonstrated that the mRNA levels of CENPE in
the LUAD tissues were significantly upregulated compared with those
in the paired normal tissues (P<0.01, Fig. 2A), and similar results were
observed following western blot analysis of protein levels
(Fig. 2B). Subsequently, the
expression of CENPE in TCGA data was analyzed, and the expression
level of CENPE was significantly higher in the LUAD samples
compared with the normal tissue samples (P<0.0001; Fig. 2C). In addition, all tumor grades
expressed higher levels of CENPE compared with normal lung tissue
and the expression of CENPE was significantly higher in stage II
tumors compared with stage I tumors (Fig. 2D). Furthermore, Kaplan-Meier
analysis of TCGA data revealed that a lower CENPE expression was
associated with improved patient outcomes (P<0.0001; Fig. 2E).
CENPE levels are associated with the
expression of FOXM1
To further understand the regulatory mechanisms of
CENPE, genes associated with CENPE expression were analyzed using
TCGA database. It was identified that CENPE expression was highly
correlated with transcription factor FOXM1 expression (r=0.8509;
P<0.0001; Fig. 3A).
Furthermore, in the tissue samples collected in the present study,
the expression levels of FOXM1 and CENPE were significantly
correlated (r=0.7057; P<0.0001; Fig. 3B). In addition, it was identified
that FOXM1 expression was significantly higher in LUAD tissues
compared with normal tissues (P<0.05; Fig. 3C). Analysis of the data from TCGA
demonstrated the same result (P<0.001; Fig. 3D). All grades of tumors expressed
higher levels of FOXM1 compared with normal lung tissue and the
expression of CENPE was significantly higher in stage II tumors
compared with stage I tumors (Fig.
3E). Additionally, Kaplan-Meier analysis of TCGA data
demonstrated that lower FOXM1 expression was associated with an
improved patient outcome (P=0.0015; Fig. 3F).
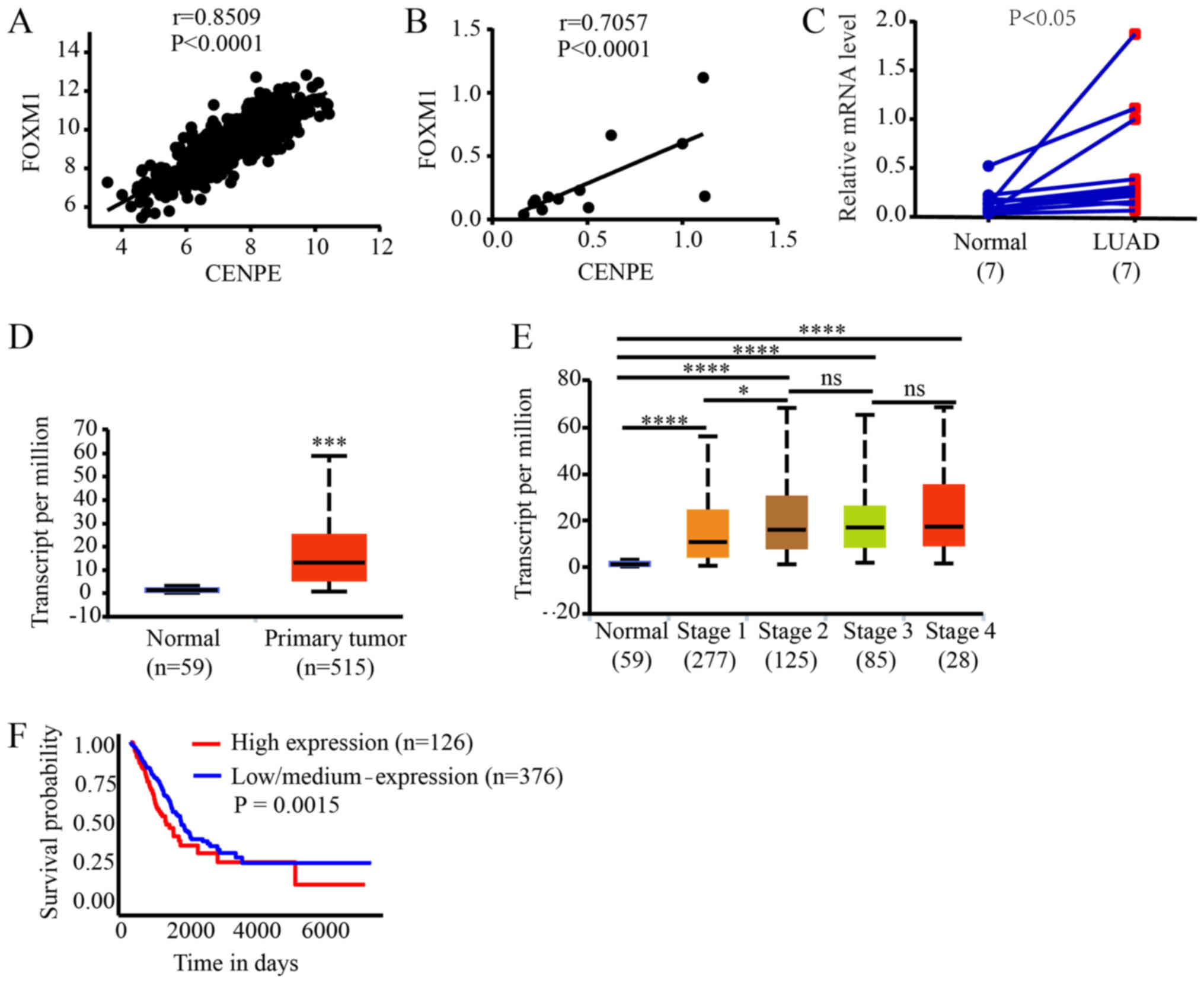 | Figure 3CENPE expression is associated with
the expression of FOXM1. (A) Correlation analysis between CENPE and
FOXM1 expression in LUAD, according to data from TCGA database. (B)
Correlation analysis between CENPE and FOXM1 expression in LUAD,
according to data from the patient samples. (C) The mRNA levels of
FOXM1 in LUAD and normal tissues, according to data from the
patient samples. (D) The mRNA levels of FOXM1 in LUAD and normal
tissues, according to data from TCGA database.
***P<0.001. (E) CENPE expression based on individual
cancer stages, according to data from TCGA database.
*P<0.05, ****P<0.0001. (F) Kaplan-Meier
analysis of FOXM1 in the LUAD RNA-sequencing data from TCGA
database. CENPE, centromere protein E; LUAD, lung adenocarcinoma;
TCGA, The Cancer Genome Atlas; FOXM1, forkhead box M1; ns, not
significant. |
Silencing of FOXM1 inhibits proliferation
of LUAD cells
To verify that FOXM1 regulates the expression of
CENPE, siRNAs were designed to knockdown FOXM1 expression in A549
and PC9 cells. It was first demonstrated that FOXM1 was
significantly knocked down in both cell lines and CENPE expression
was also significantly reduced (P<0.01; Fig. 4A and B). The EdU assay revealed
that silencing FOXM1 significantly inhibited the proliferation of
LUAD cells (Fig. 4C and D;
P<0.01). The CCK-8 assay further demonstrated that knockdown of
FOXM1 significantly inhibited the proliferation of LUAD cells
(P<0.05; Fig. 4E and F).
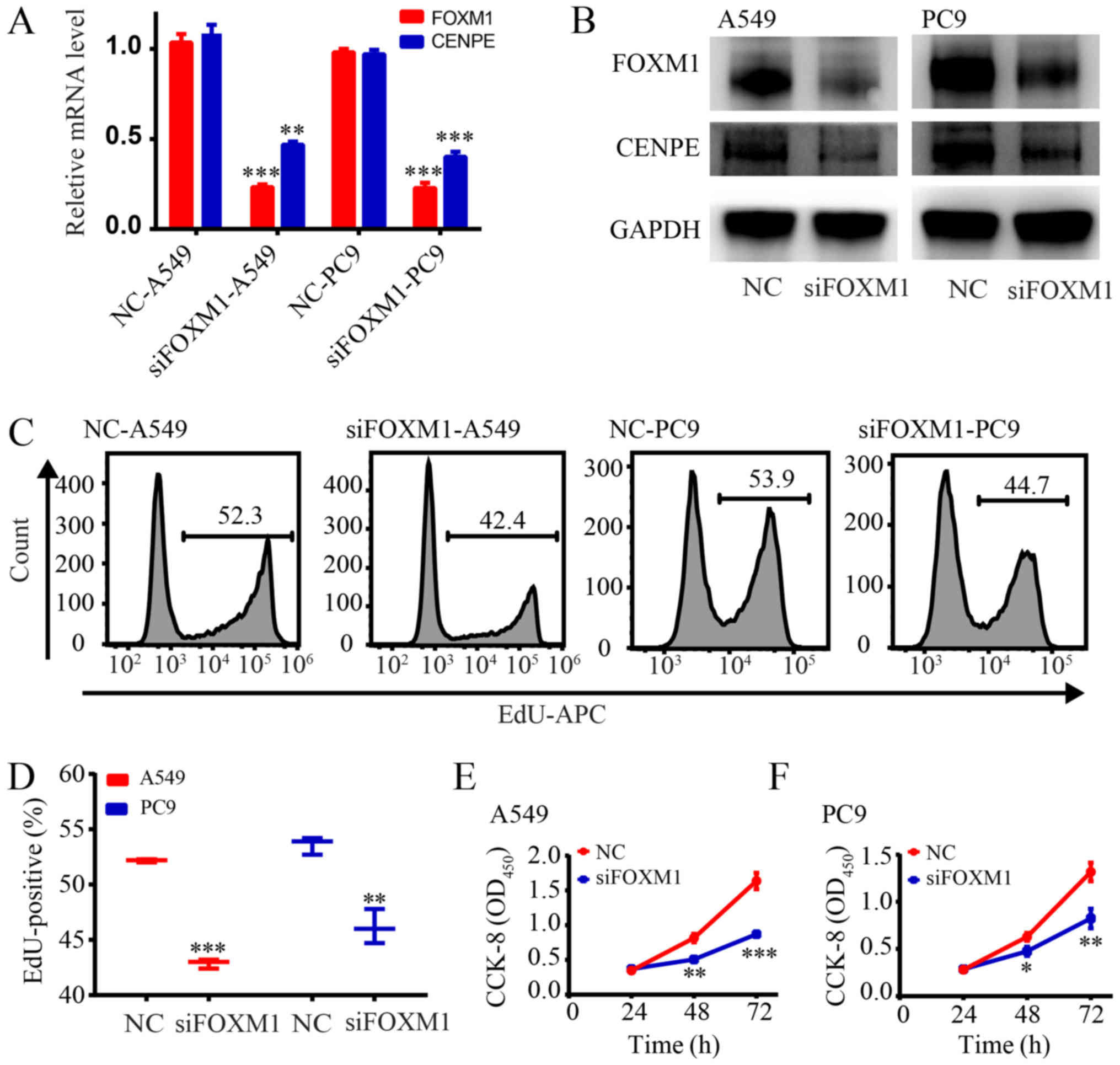 | Figure 4Silencing of FOXM1 inhibits the
proliferation of LUAD cells. (A) Reverse transcription-quantitative
polymerase chain reaction analysis of CENPE in A549 and PC9 cells
transfected with control siRNA or siFOXM1. (B) Western blot
analysis of FOXM1 and CENPE in A549 and PC9 cells transfected with
control siRNA or siFOXM1. (C) Cell proliferation rates as
determined by EdU assays with A549 and PC9 cells. (D)
Quantification of EdU-positive A549 and PC9 cells transfected with
control siRNA or siFOXM1. (E) CCK-8 results for the A549 group. (F)
CCK-8 results for the PC9 group. *P<0.05,
**P<0.01, ***P<0.001 vs NC. FOXM1,
forkhead box M1; LUAD, lung adenocarcinoma; CENPE, centromere
protein E; si, small interfering; NC, negative control; CCK-8, Cell
Counting Kit-8; OD, optical density; APC, allophycocyanin; EdU,
5-ethynyl-2'-deoxyuridine. |
Overexpression of FOXM1 promotes
proliferation of LUAD cells
To further confirm that FOXM1 regulates CENPE and
promotes proliferation of LUAD cells, FOXM1 was overexpressed in
A549 cells. RT-qPCR and western blot analysis demonstrated that
FOXM1 was significantly overexpressed, and CENPE levels were
significantly higher in cells overexpressing FOXM1 (P<0.001;
Fig. 5A and B). After FOXM1 was
overexpressed, the CCK-8 results revealed a significantly increased
proliferation rate (P<0.05; Fig.
5C). Furthermore, the EdU assay demonstrated that
overexpression of FOXM1 significantly promoted proliferation of
A549 cells (P<0.001; Fig. 5D and
E).
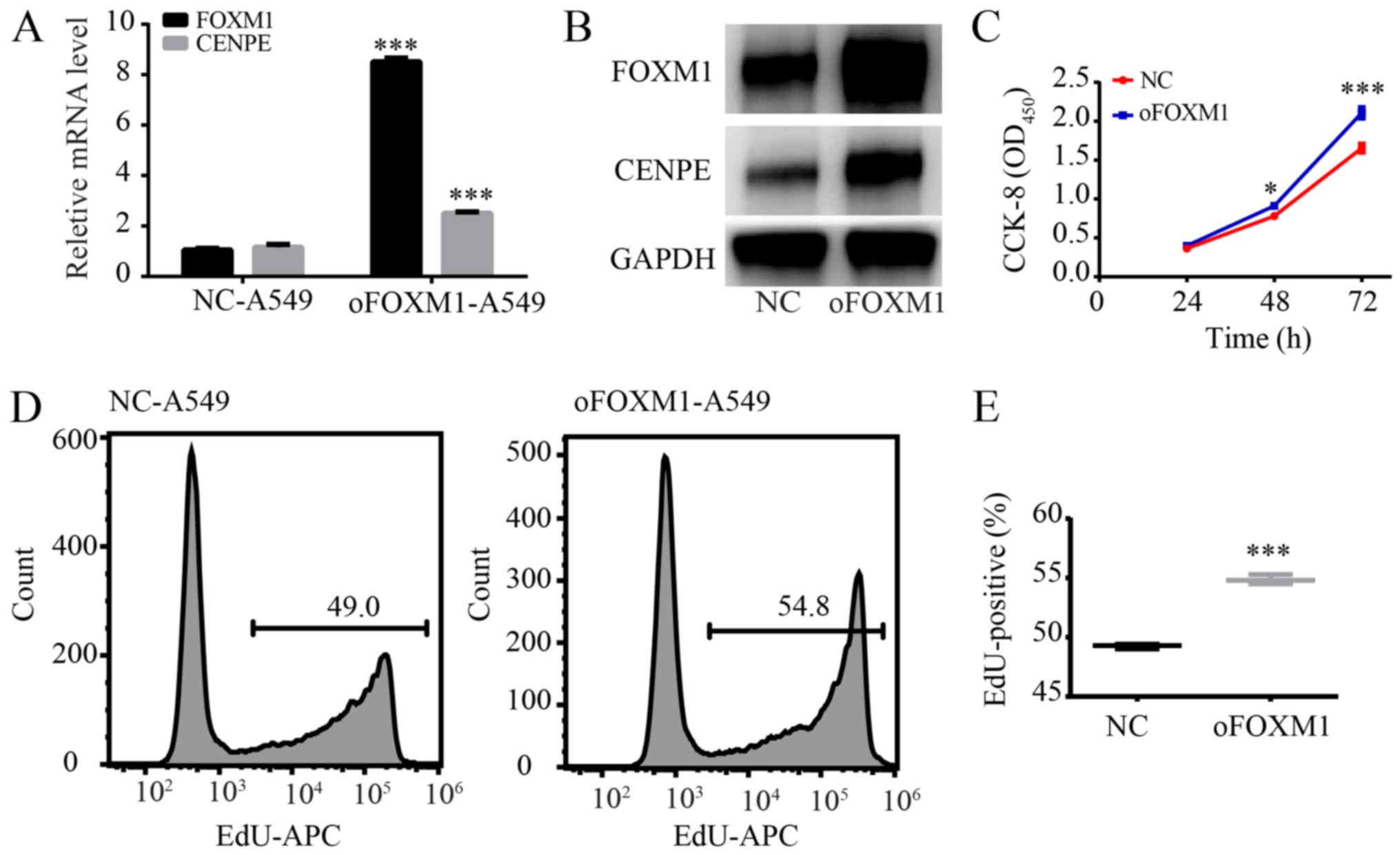 | Figure 5Overexpression of FOXM1 promotes
proliferation of A549 cells. (A) Reverse transcription-quantitative
polymerase chain reaction analysis of CENPE and FOXM1 in A549 cells
transfected with control vector or oFOXM1 vector. (B) Western blot
analysis of CENPE and FOXM1 in A549 cells transfected with control
vector or oFOXM1 vector. (C) CCK-8 assay results demonstrating that
overexpression of FOXM1 promoted proliferation of A549 cells. (D)
Cell proliferation rates as determined by EdU assays with A549
cells. (E) Quantification of EdU-positive A549 cells transfected
with control vector or oFOXM1 vector. *P<0.05,
***P<0.001 vs. NC. FOXM1, forkhead box M1; CENPE,
centromere protein E; si, small interfering; NC, negative control;
CCK-8, Cell Counting Kit-8; OD, optical density; APC,
allophycocyanin; oFOXM1, forkhead box M1-overexpression; EdU, EdU,
5-ethynyl-2'-deoxyuridine. |
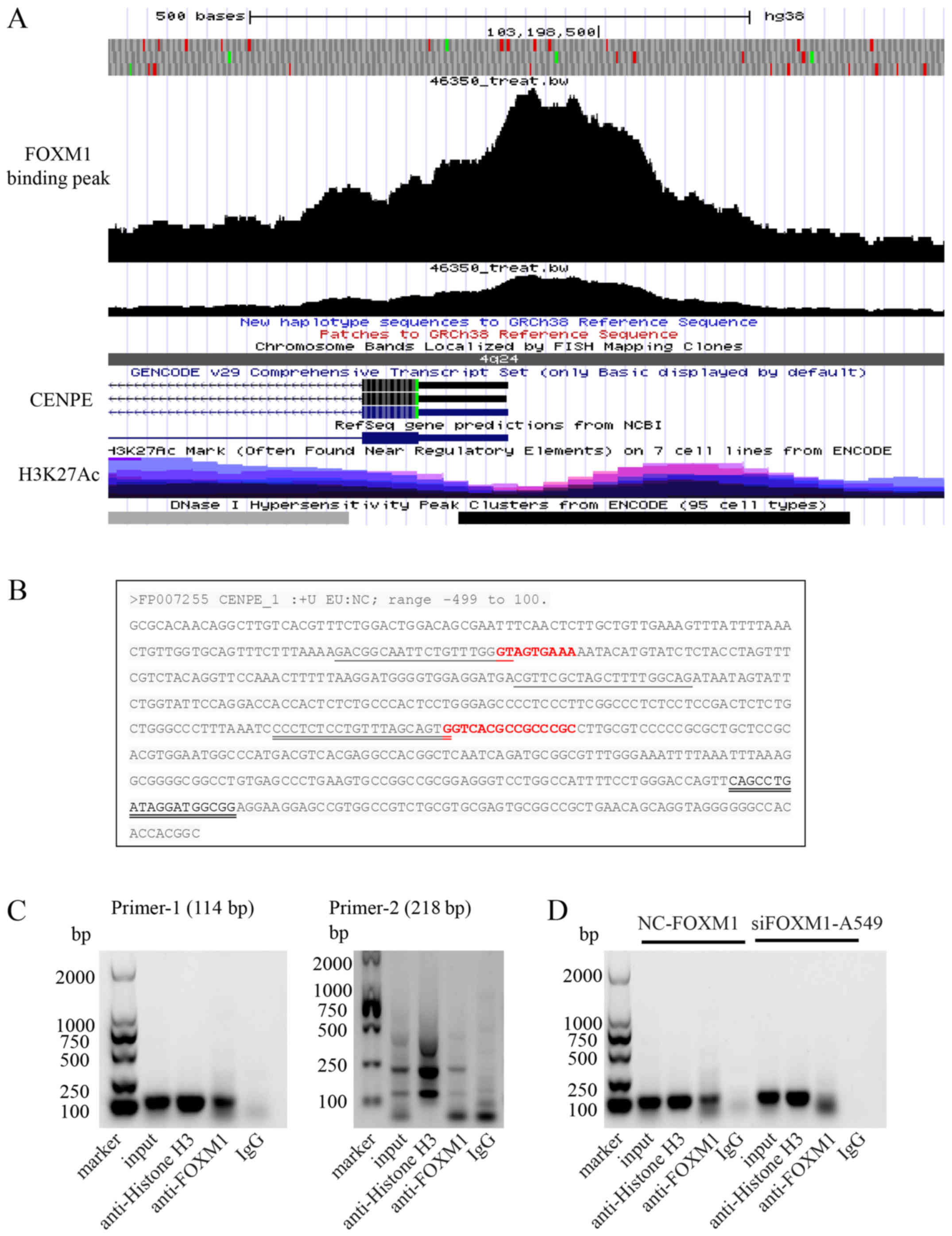
CENPE expression is directly regulated by
FOXM1
From a previous study, it is understood that
transcription factors can bind to the promoter region of genes and
directly regulate their transcription (25). Therefore, the present study further
analyzed the regulatory mechanism of FOXM1 on CENPE. First, the
ChIP-sequencing data of FOXM1 was analyzed using the Cistrome
database. The results demonstrated that FOXM1 has a binding peak in
the promoter region of CENPE (Fig.
6A). The FOXM1 binding site on the CENPE promoter region (-499
to 100) was then analyzed. It was identified that there are two
FOXM1 binding sites in the region of CENPE (Fig. 6B). Primers were then designed using
DNA sequences that bind to the peak positions to further validate
the conclusions of the binding. The ChIP assay of A549 cells
confirmed that the CENPE promoter region contains a binding site
for FOXM1 (Fig. 6C). To further
confirm this conclusion, the band strengths obtained by CHIP from
the control and FOXM1-interference groups were compared, and a
lower intensity was observed for the FOXM1-interference group
(Fig. 6D). Therefore, the current
results indicate that FOXM1 can directly regulate the expression of
CENPE in LUAD.
Discussion
Mitosis is a key event of the cell cycle. The
ability to maintain proliferation is a hallmark of cancer and
microtubules serve an important part in maintaining a sustained
cell cycle (9). At the same time,
numerous microtubule-associated proteins are highly expressed in
tumor tissues (9), and targeting
microtubule-associated proteins is one way to treat tumors
(26). Therefore, further
understanding of the regulatory mechanisms of
microtubule-associated proteins in lung cancer may improve
understanding regarding the cell cycle and also provide new targets
for the treatment of tumors. CENPE is a microtubule-associated
protein that is highly expressed in numerous types of cancer,
including prostate cancer (11)
and glioma (27). The present
study identified a high expression of CENPE in LUAD tissues and
CENPE-knockdown was demonstrated to inhibit the proliferation of
LUAD cell lines.
Firstly, the current study identified that CENPE is
highly expressed in lung adenocarcinoma using TCGA database, and in
grade I and II lung adenocarcinoma the expression was revealed to
increase with increasing grade. Subsequently, it was demonstrated
that the proliferation of A549 and PC9 cells was inhibited
following CENPE-knockdown, indicating that CENPE is associated with
the proliferation rate of lung adenocarcinoma. Therefore, CENPE may
be a marker of the proliferation rate of a tumor. In other tumor
types, there are reports that CENPE can be used as a marker for
tumor proliferation. In prolactin pituitary tumors, it was
identified that seven genes, including ADAM metallopeptidase with
thrombospondin type 1 motif 6 (ADAMTS6), collapsing response
mediator protein 1 (CRMP1), pituitary tumor transforming gene,
aspartokinase (ASK), cyclin B1 (CCNB1), aurora kinase B and CENPE,
were associated with tumor recurrence or progression, and five of
these genes (ADAMTS6, CRMP1, ASK, CCNB1 and CENPE) were also
associated with the pathological classification (28). In glioma, RNA levels of eight major
mitotic spindle assembly checkpoint genes, including CENPE, have
been reported to be significantly associated with glioma grade and
survival time (27). In invasive
ductal carcinoma, CENPE was identified to be one of the top ten
potential crucial genes (29). In
addition, studies have identified that the expression of CENPE is
associated with drug tolerance (30-32).
In paclitaxel-resistant ovarian cell lines, it was revealed that
CENPE expression is lower (31).
Following bioinformatics analysis, the present study
identified that CENPE has a significant correlation with FOXM1
expression, and FOXM1 is also highly expressed in lung
adenocarcinoma. In addition, the overall survival rate of patients
with high expression of FOXM1 was worse; however, there was a
difference between the survival trend of FOXM1 and the survival
trend of CENPE. FOXM1 is a transcription factor that participates
in all stages of tumor development, predominantly through control
of the cell cycle and proliferation, regulating the expression of
genes involved in the G1/S and G2/M transitions and M phase
progression (33). The forkhead
box O3 and FOXM1 transcription factors, functioning downstream of
the essential PI3K-Akt, Ras-ERK and JNK/p38MAPK signaling cascades,
are crucial for cell proliferation, differentiation, cell survival,
senescence, DNA damage repair and cell cycle control (34). Therefore, numerous studies have
identified that FOXM1 can promote the proliferation of a number of
types of tumor. It has been reported that DEP domain containing 1,
negatively regulated by microRNA (miR)-26b, promotes cell
proliferation and tumor growth by upregulating FOXM1 expression,
indicating an important underlying mechanism of regulating the
progression of triple negative breast cancer (35). G2 and S phase-expressed-1
contributes to cell proliferation, migration and invasion by
regulating the p53/forkhead box 1 (FOXM1)/CCNB1 pathway and
predicts poor prognosis in bladder cancer (36). In U2OS cells, enforced expression
of FOXM1 suppressed diallyl disulfide-induced antiproliferation and
anti-invasion (37). In
glioblastoma cells, miR-876-5p may inhibit the development of
glioblastoma by directly targeting FOXM1 (38). In ovarian cancer, hsa_circ_0061140
has been demonstrated to promote cell growth and metastasis through
regulation of the miR-370/FOXM1 pathway, mediating the
epithelial-mesenchymal transition (39). In ovarian cancer cells, FOXM1
transcriptionally activated PCNA-associated factor to drive cell
proliferation (40). In
meningioma, the FOXM1/Wnt signaling axis is associated with a
mitotic gene expression program, poor clinical outcomes and
proliferation (41). In colorectal
cancer, one study reported that FOXM1 is critical for the
proliferation and growth of colorectal cancer (42). In summary, the aforementioned
studies suggest that the expression of FOXM1 is associated with
proliferation and mainly regulates the G2/M phase. Therefore, the
present study hypothesized that the expression of CENPE may be
regulated by FOXM1. Through interference, overexpression and ChIP
experiments, it was demonstrated that CENPE can be directly
regulated by FOXM1.
In conclusion, the current study revealed that CENPE
is highly expressed in LUAD tissues. In addition, FOXM1 is
correlated with the expression of CENE and is associated with the
proliferation of lung cancer cells. The ChIP assay demonstrated
that FOXM1 binds directly to the promoter region of CENPE.
Therefore, the current data indicate that CENPE can promote the
proliferation of LUAD cells and is directly regulated by FOXM1. In
the future it will be important to further understand the signaling
pathways that regulate CENPE expression, and the mechanism by which
CENPE regulates the cell cycle. In addition, more studies should
investigate centromere-associated proteins that are abnormally
expressed in tumors. The study of more centromere-associated
proteins may provide new targets and a theoretical basis for
targeted therapy of cell cycle-associated proteins.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81700050).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XS and WC contributed to the experiment design,
drafted the manuscript and performed data analysis. LS conceived or
designed the experiments, performed the experiments, wrote the
manuscript and performed data analysis. MZ designed the
experiments. YL, HN and YS performed the experiments. SY analyzed
the data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experiments were approved by the Ethics
Committee of The First Affiliated Hospital of Jinzhou Medical
University (Jinzhou, China) and written informed consent was
obtained from each individual.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Gang Zhang (State
Key Laboratory of Bioactive Substances and Function of Natural
Medicine, Peking Union Medical College and Chinese Academy of
Medical Science, Beijing, China) for editing a draft of the
manuscript.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
5
|
Mathew M, Enzler T, Shu CA and Rizvi NA:
Combining chemotherapy with PD-1 blockade in NSCLC. Pharmacol Ther.
186:130–137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rotow J and Bivona TG: Understanding and
targeting resistance mechanisms in NSCLC. Nat Rev Cancer.
17:637–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bradbury P, Sivajohanathan D, Chan A,
Kulkarni S, Ung Y and Ellis PM: Postoperative adjuvant systemic
therapy in completely resected non-small-cell lung cancer: A
systematic review. Clin Lung Cancer. 18:259–273.e258. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Testa JR, Zhou JY, Bell DW and Yen TJ:
Chromosomal localization of the genes encoding the kinetochore
proteins CENPE and CENPF to human chromosomes 4q24-->q25 and
1q32-->q41, respectively, by fluorescence in situ hybridization.
Genomics. 23:691–693. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rath O and Kozielski F: Kinesins and
cancer. Nat Rev Cancer. 12:527–539. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hitti E, Bakheet T, Al-Souhibani N,
Moghrabi W, Al-Yahya S, Al-Ghamdi M, Al-Saif M, Shoukri MM, Lánczky
A, Grépin R, et al: Systematic analysis of AU-rich element
expression in cancer reveals common functional clusters regulated
by key RNA-binding proteins. Cancer Res. 76:4068–4080. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang Y, Ahmed M, Guo H, Soares F, Hua JT,
Gao S, Lu C, Poon C, Han W, Langstein J, et al: LSD1-mediated
epigenetic reprogramming drives CENPE expression and prostate
cancer progression. Cancer Res. 77:5479–5490. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai
CF, Chao ME, Liu DJ, Chu SS, Chen W, Liu YR, et al: Downregulation
of miR-137 and miR-6500-3p promotes cell proliferation in pediatric
high-grade gliomas. Oncotarget. 7:19723–19737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Balamuth NJ, Wood A, Wang Q, Jagannathan
J, Mayes P, Zhang Z, Chen Z, Rappaport E, Courtright J, Pawel B, et
al: Serial transcriptome analysis and cross-species integration
identifies centromere-associated protein E as a novel neuroblastoma
target. Cancer Res. 70:2749–2758. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du J, Chen L and Shen J: Identification of
FANCA as a protein interacting with centromere-associated protein
E. Acta Biochim Biophys Sin (Shanghai). 41:816–821. 2009.
View Article : Google Scholar
|
|
15
|
Hasson SA, Kane LA, Yamano K, Huang CH,
Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, et
al: High-content genome-wide RNAi screens identify regulators of
parkin upstream of mitophagy. Nature. 504:291–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cancer Cell Line Encyclopedia Consortium;
Genomics of Drug Sensitivity in Cancer Consortium: Pharmacogenomic
agreement between two cancer cell line data sets. Nature.
528:84–87. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H,
Chen CH, Brown M, Zhang X, Meyer CA, et al: Cistrome Data Browser:
Expanded datasets and new tools for gene regulatory analysis.
Nucleic Acids Res. 47(D1): D729–D735. 2019. View Article : Google Scholar :
|
|
19
|
Gertz J, Savic D, Varley KE, Partridge EC,
Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE and Myers RM:
Distinct properties of cell-type-specific and shared transcription
factor binding sites. Mol Cell. 52:25–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dreos R, Ambrosini G, Groux R, Cavin
Périer R and Bucher P: The eukaryotic promoter database in its 30th
year: Focus on non-vertebrate organisms. Nucleic Acids Res. 45(D1):
D51–D55. 2017. View Article : Google Scholar :
|
|
21
|
Hu H, Miao YR, Jia LH, Yu QY, Zhang Q and
Guo AY: AnimalTFDB 3.0: A comprehensive resource for annotation and
prediction of animal transcription factors. Nucleic Acids Res.
47(D1): D33–D38. 2019. View Article : Google Scholar :
|
|
22
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:pl12013. View Article : Google Scholar
|
|
24
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 172:650–665. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Casaluce F, Sgambato A, Maione P,
Ciardiello F and Gridelli C: Emerging mitotic inhibitors for
non-small cell carcinoma. Expert Opin Emerg Drugs. 18:97–107. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bie L, Zhao G, Cheng P, Rondeau G,
Porwollik S, Ju Y, Xia XQ and McClelland M: The accuracy of
survival time prediction for patients with glioma is improved by
measuring mitotic spindle checkpoint gene expression. PLoS One.
6:e256312011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He M, Agbu S and Anderson KV: Microtubule
motors drive hedgehog signaling in primary cilia. Trends Cell Biol.
27:110–125. 2017. View Article : Google Scholar :
|
|
29
|
Li C, Luo L, Wei S and Wang X:
Identification of the potential crucial genes in invasive ductal
carcinoma using bioinformatics analysis. Oncotarget. 9:6800–6813.
2017.
|
|
30
|
Nara M, Teshima K, Watanabe A, Ito M,
Iwamoto K, Kitabayashi A, Kume M, Hatano Y, Takahashi N, Iida S, et
al: Bortezomib reduces the tumorigenicity of multiple myeloma via
downregulation of upregulated targets in clonogenic side population
cells. PLoS One. 8:e569542013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chong T, Sarac A, Yao CQ, Liao L, Lyttle
N, Boutros PC, Bartlett JMS and Spears M: Deregulation of the
spindle assembly checkpoint is associated with paclitaxel
resistance in ovarian cancer. J Ovarian Res. 11:272018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Horning AM, Wang Y, Lin CK, Louie AD,
Jadhav RR, Hung CN, Wang CM, Lin CL, Kirma NB, Liss MA, et al:
Single-cell RNA-seq reveals a subpopulation of prostate cancer
cells with enhanced cell-cycle-related transcription and attenuated
androgen response. Cancer Res. 78:853–864. 2018. View Article : Google Scholar
|
|
33
|
Liao GB, Li XZ, Zeng S, Liu C, Yang SM,
Yang L, Hu CJ and Bai JY: Regulation of the master regulator FOXM1
in cancer. Cell Commun Signal. 16:572018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yao S, Fan LY and Lam EW: The FOXO3-FOXM1
axis: A key cancer drug target and a modulator of cancer drug
resistance. Semin Cancer Biol. 50:77–89. 2018. View Article : Google Scholar
|
|
35
|
Zhang L, Du Y, Xu S, Jiang Y, Yuan C, Zhou
L, Ma X, Bai Y, Lu J and Ma J: DEPDC1, negatively regulated by
miR-26b, facilitates cell proliferation via the up-regulation of
FOXM1 expression in TNBC. Cancer Lett. 442:242–251. 2019.
View Article : Google Scholar
|
|
36
|
Liu A, Zeng S, Lu X, Xiong Q, Xue Y, Tong
L, Xu W, Sun Y, Zhang Z and Xu C: Overexpression of G2 and S
phase-expressed-1 contributes to cell proliferation, migration, and
invasion via regulating p53/FoxM1/CCNB1 pathway and predicts poor
prognosis in bladder cancer. Int J Biol Macromol. 123:322–334.
2019. View Article : Google Scholar
|
|
37
|
Li Y, Wang Z, Li J and Sang X: Diallyl
disulfide suppresses FOXM1-mediated proliferation and invasion in
osteosarcoma by upregulating miR-134. J Cell Biochem. Nov
1–2018.Epub ahead of print. View Article : Google Scholar
|
|
38
|
Wang L, Lu J, Zhang H, Lyu X and Sun Z:
MicroRNA-876-5p inhibits the progression of glioblastoma multiforme
by directly targeting Forkhead box M1. Oncol Rep. 41:702–710.
2019.
|
|
39
|
Chen Q, Zhang J, He Y and Wang Y:
hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and
metastasis in ovarian cancer through miR-370 sponge activity. Mol
Ther Nucleic Acids. 13:55–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin C, Liu Z, Li Y, Bu H, Wang Y, Xu Y,
Qiu C, Yan S, Yuan C, Li R, et al: PCNA-associated factor P15PAF,
targeted by FOXM1, predicts poor prognosis in high-grade serous
ovarian cancer patients. Int J Cancer. 143:2973–2984. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vasudevan HN, Braunstein SE, Phillips JJ,
Pekmezci M, Tomlin BA, Wu A, Reis GF, Magill ST, Zhang J, Feng FY,
et al: Comprehensive molecular profiling identifies FOXM1 as a key
transcription factor for meningioma proliferation. Cell Rep.
22:3672–3683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida Y, Wang IC, Yoder HM, Davidson NO
and Costa RH: The forkhead box M1 transcription factor contributes
to the development and growth of mouse colorectal cancer.
Gastroenterology. 132:1420–1431. 2007. View Article : Google Scholar : PubMed/NCBI
|