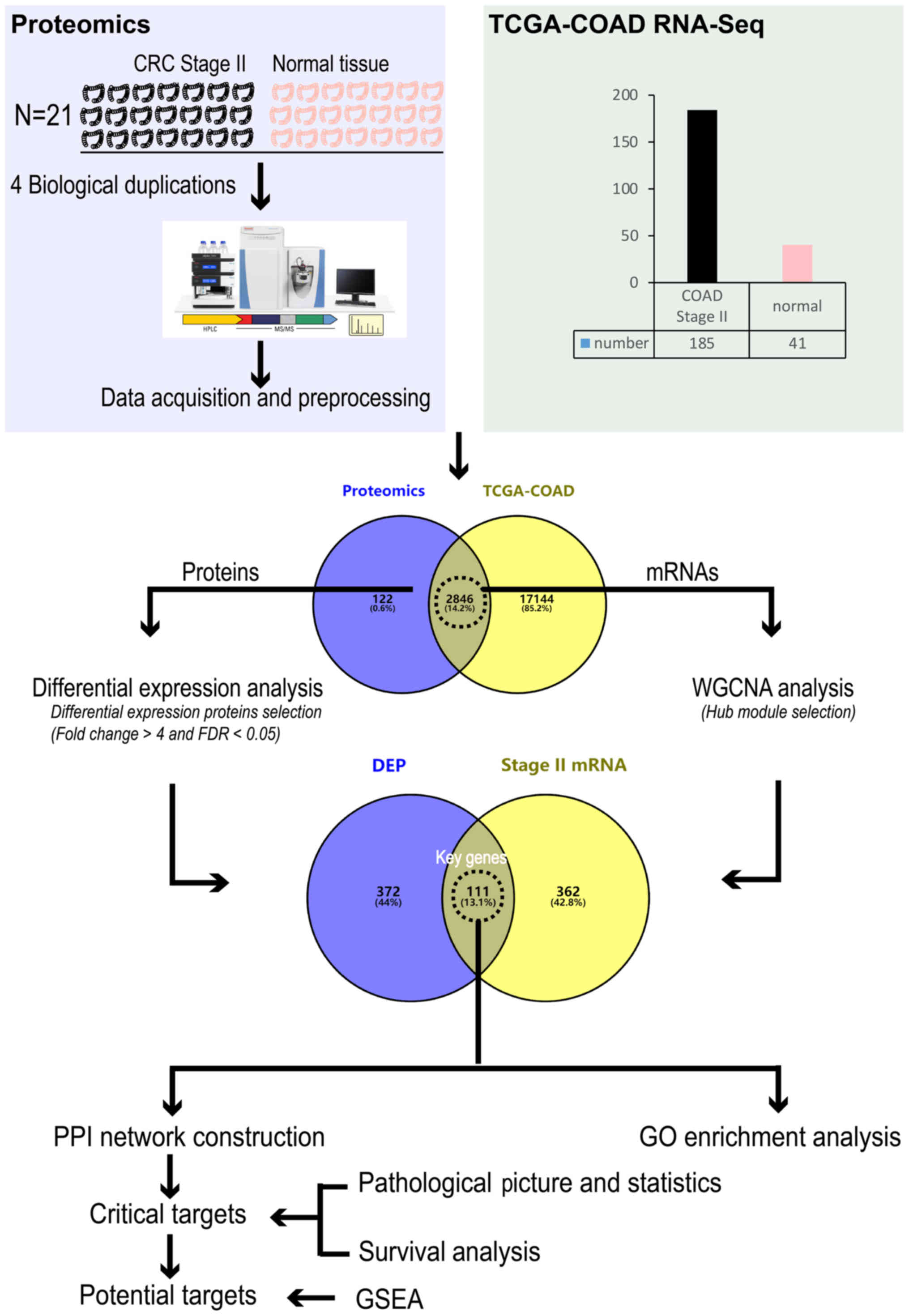
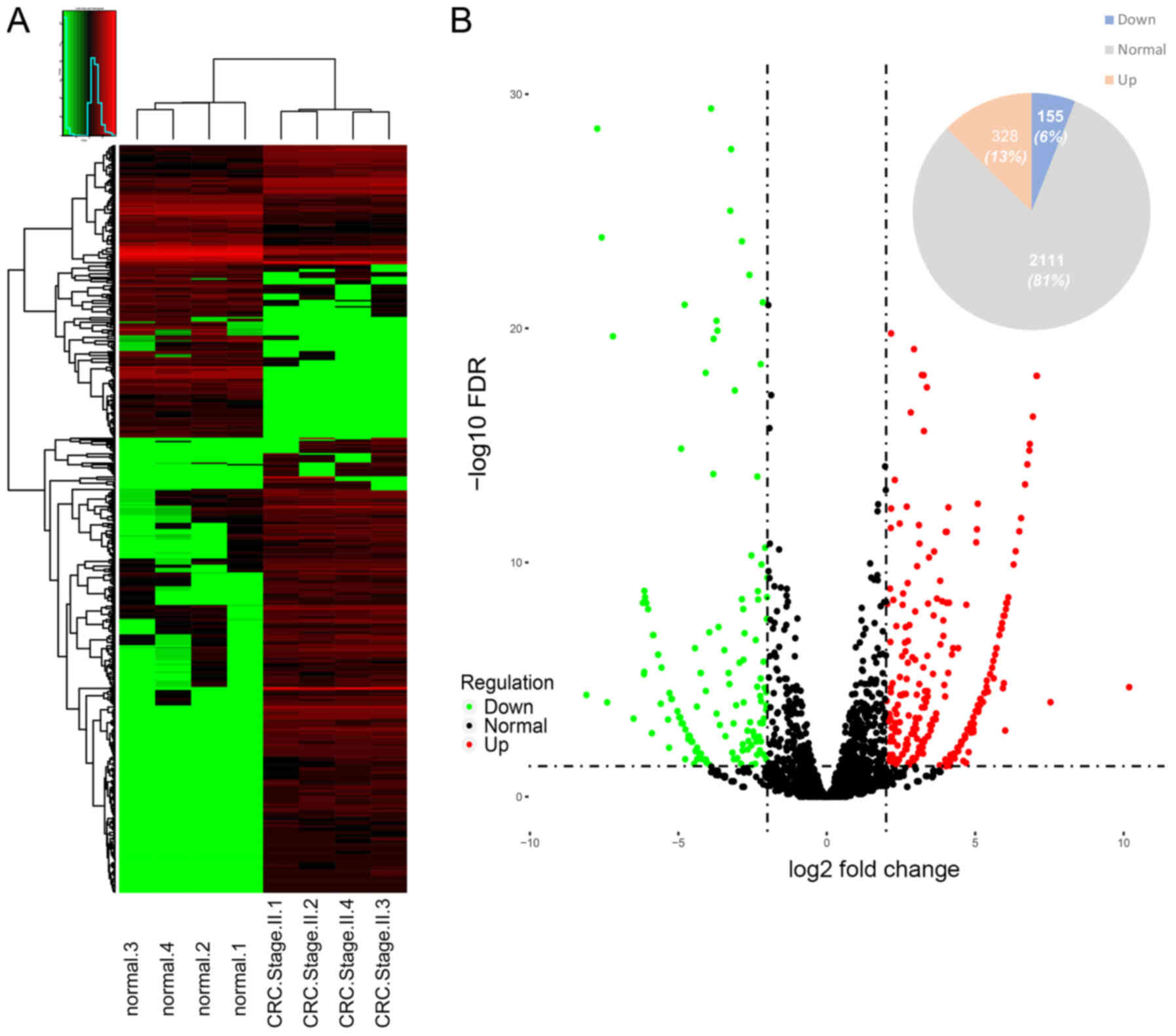
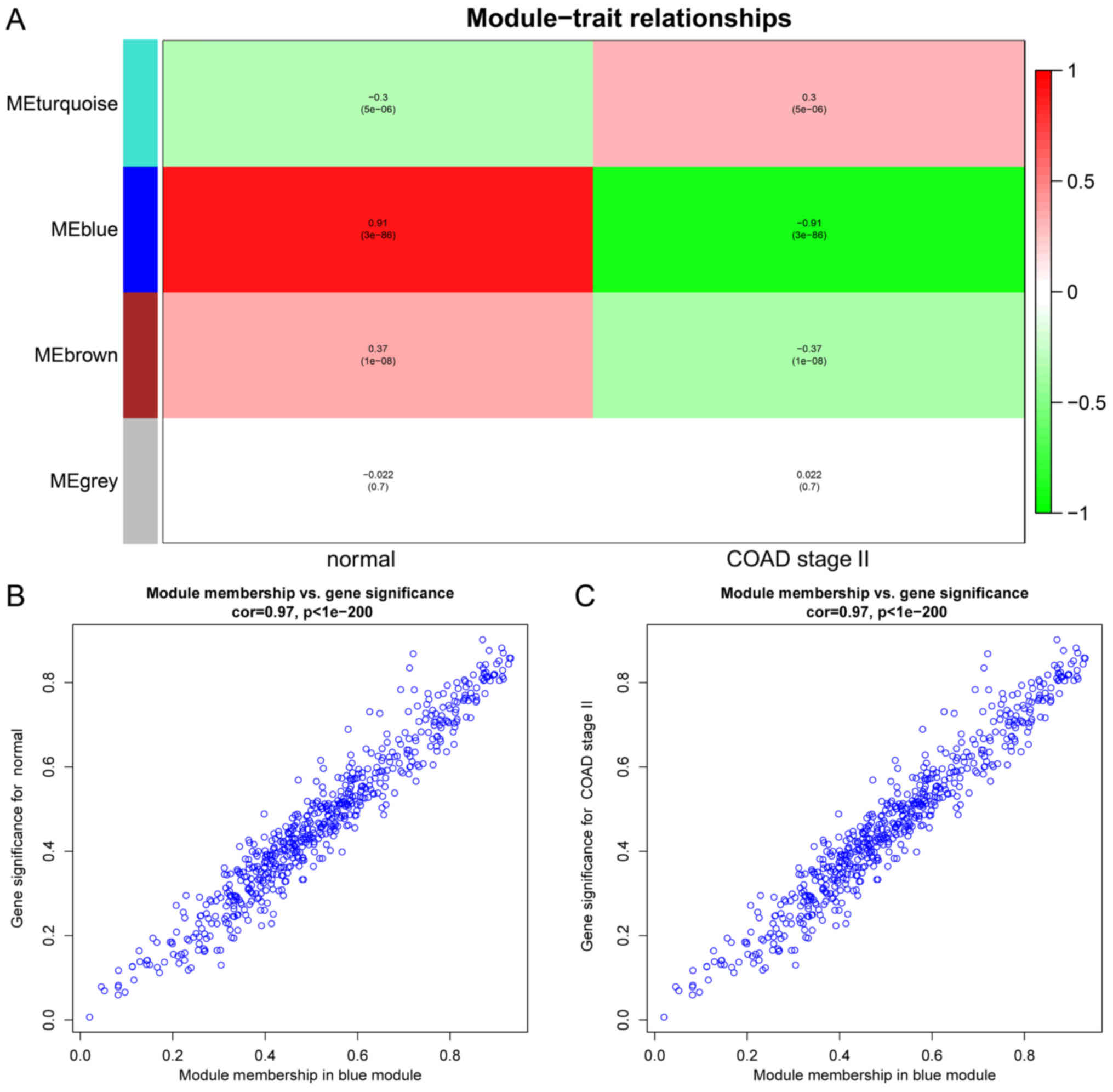
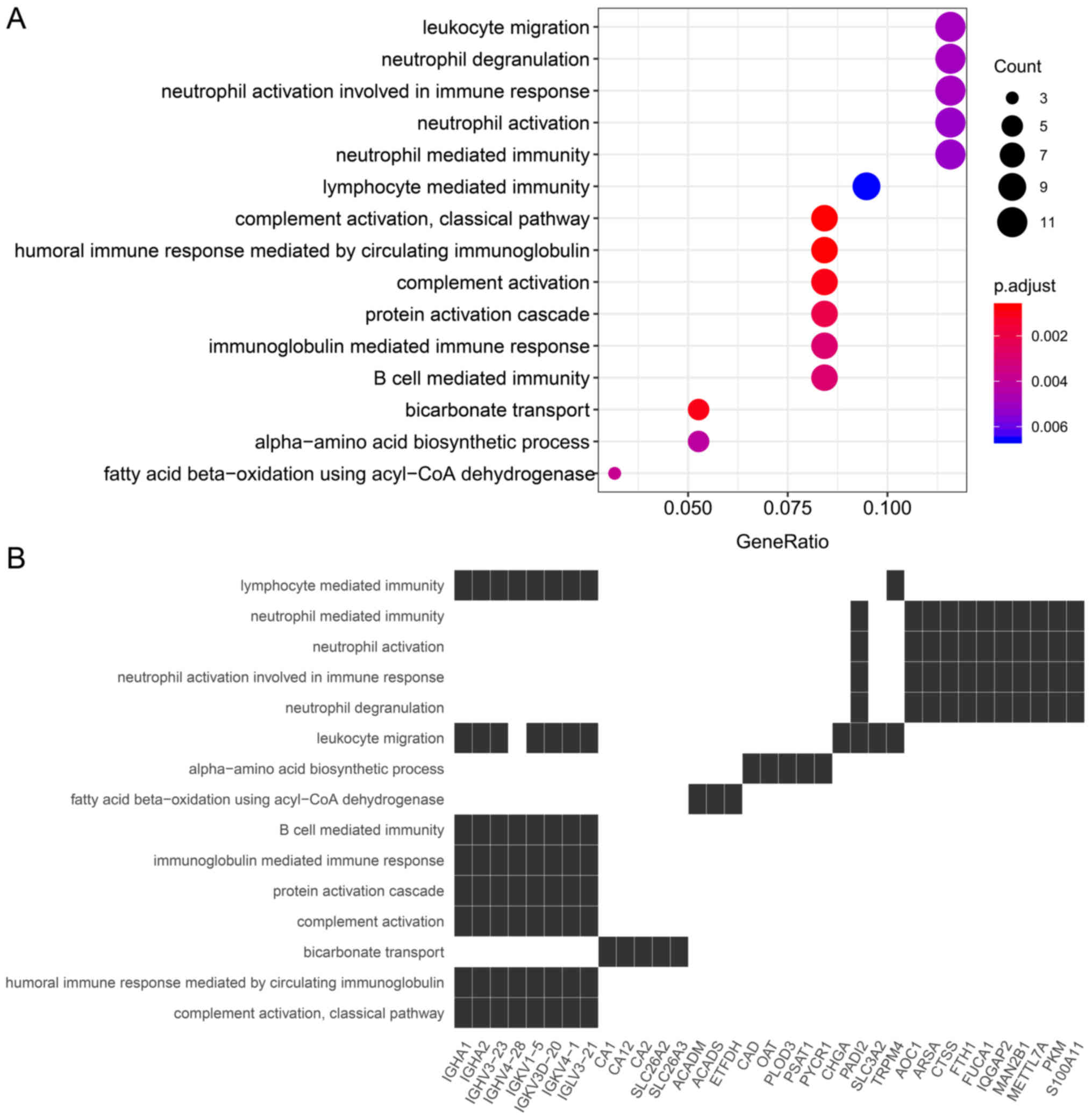
|
1
|
Brody H: Colorectal cancer. Nature.
521:S12015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar
|
|
4
|
Weinberg BA, Marshall JL and Salem ME: The
Growing Challenge of Young Adults With Colorectal Cancer. Oncology
(Williston Park). 31:381–389. 2017.
|
|
5
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Osterman E and Glimelius B: Recurrence
risk after up- to- date colon cancer staging, surgery, and
pathology: Analysis of the entire Swedish population. Dis Colon
Rectum. 61:1016–1025. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roncucci L and Mariani F: Prevention of
colorectal cancer: How many tools do we have in our basket? Eur J
Intern Med. 26:752–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aran V, Victorino AP, Thuler LC and
Ferreira CG: Colorectal cancer: Epidemiology, disease mechanisms
and interventions to reduce onset and mortality. Clin Colorectal
Cancer. 15:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stoffel EM and Boland CR: Genetics and
genetic testing in hereditary colorectal cancer. Gastroenterology.
149:1191–1203.e2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naldini L: Gene therapy returns to centre
stage. Nature. 526:351–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu L, Johnson M and Sato M:
Transcriptionally targeted gene therapy to detect and treat cancer.
Trends Mol Med. 9:421–429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mair B, Kubicek S and Nijman SM:
Exploiting epigenetic vulnerabilities for cancer therapeutics.
Trends Pharmacol Sci. 35:136–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dalerba P, Sahoo D, Paik S, Guo X, Yothers
G, Song N, Wilcox- Fogel N, Forgó E, Rajendran PS, Miranda SP, et
al: CDX2 as a prognostic biomarker in stage II and stage III colon
cancer. N Engl J Med. 374:211–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mo A, Jackson S, Varma K, Carpino A,
Giardina C, Devers TJ and Rosenberg DW: Distinct transcriptional
changes and epithelial-stromal interactions are altered in
early-stage colon cancer development. Mol Cancer Res. 14:795–804.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lucena- Cacace A, Otero-Albiol D,
Jiménez-García MP, Muñoz-Galvan S and Carnero A: NAMPT is a potent
oncogene in colon cancer progression that modulates cancer stem
cell properties and resistance to therapy through Sirt1 and PARP.
Clin Cancer Res. 24:1202–1215. 2018. View Article : Google Scholar
|
|
17
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.
|
|
18
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å Kampf C, Sjöstedt
E, Asplund A, et al: Proteomics. Tissue-based map of the human
proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:3562017. View Article : Google Scholar
|
|
20
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:3572017. View Article : Google Scholar
|
|
21
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun N, Sun W, Li S, Yang J, Yang L, Quan
G, Gao X, Wang Z, Cheng X, Li Z, et al: Proteomics analysis of
cellular proteins co- immunoprecipitated with nucleoprotein of
influenza A virus (H7N9). Int J Mol Sci. 16:25982–25998. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Davis CA, Hitz BC, Sloan CA, Chan ET,
Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan
AK, et al: The Encyclopedia of DNA elements (ENCODE): Data portal
update. Nucleic Acids Res. 46:D794–D801. 2018. View Article : Google Scholar :
|
|
24
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar :
|
|
25
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
26
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA- Seq
experiments with respect to biological variation. Nucleic Acids
Res. 40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ito K and Murphy D: Application of ggplot2
to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol.
2:e792013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gene Ontology Consortium: Gene Ontology
Consortium: Going forward. Nucleic Acids Res. 43:D1049–D1056. 2015.
View Article : Google Scholar :
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality- controlled protein- protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar
|
|
31
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
33
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A knowledge-
based approach for interpreting genome- wide expression profiles.
Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar
|
|
35
|
Xu H, Wang C, Song H, Xu Y and Ji G: RNA-
Seq profiling of circular RNAs in human colorectal Cancer liver
metastasis and the potential biomarkers. Mol Cancer. 18:82019.
View Article : Google Scholar
|
|
36
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL and
Yang Y: RNA sequencing reveals the expression profiles of circRNA
and indicates that circDDX17 acts as a tumor suppressor in
colorectal cancer. J Exp Clin Cancer Res. 37:3252018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jones MF, Hara T, Francis P, Li XL, Bilke
S, Zhu Y, Pineda M, Subramanian M, Bodmer WF and Lal A: The CDX1-
microRNA- 215 axis regulates colorectal cancer stem cell
differentiation. Proc Natl Acad Sci USA. 112:E1550–E1558. 2015.
View Article : Google Scholar
|
|
38
|
Triner D, Devenport SN, Ramakrishnan SK,
Ma X, Frieler RA, Greenson JK, Inohara N, Nunez G, Colacino JA,
Mortensen RM, et al: Neutrophils restrict tumor-associated
microbiota to reduce growth and invasion of colon tumors in mice.
Gastroenterology. 156:1467–1482. 2019. View Article : Google Scholar
|
|
39
|
Wu L, Saxena S, Awaji M and Singh RK:
Tumor- associated neutrophils in cancer: Going Pro. Cancers
(Basel). 11:112019. View Article : Google Scholar
|
|
40
|
Galdiero MR, Bianchi P, Grizzi F, Di Caro
G, Basso G, Ponzetta A, Bonavita E, Barbagallo M, Tartari S,
Polentarutti N, et al: Occurrence and significance of tumor-
associated neutrophils in patients with colorectal cancer. Int J
Cancer. 139:446–456. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ning C, Li YY, Wang Y, Han GC, Wang RX,
Xiao H, Li XY, Hou CM, Ma YF, Sheng DS, et al: Complement
activation promotes colitis-associated carcinogenesis through
activating intestinal IL-1β/IL- 17A axis. Mucosal Immunol.
8:1275–1284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Piao C, Zhang WM, Li TT, Zhang CC, Qiu S,
Liu Y, Liu S, Jin M, Jia LX, Song WC, et al: Complement 5a
stimulates macrophage polarization and contributes to tumor
metastases of colon cancer. Exp Cell Res. 366:127–138. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Piao C, Cai L, Qiu S, Jia L, Song W and Du
J: Complement 5a enhances hepatic metastases of colon cancer via
monocyte chemoattractant protein-1-mediated inflammatory cell
infiltration. J Biol Chem. 290:10667–10676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wikberg ML, Ling A, Li X, Öberg Å, Edin S
and Palmqvist R: Neutrophil infiltration is a favorable prognostic
factor in early stages of colon cancer. Hum Pathol. 68:193–202.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Governa V, Trella E, Mele V, Tornillo L,
Amicarella F, Cremonesi E, Muraro MG, Xu H, Droeser R, Däster SR,
et al: The interplay between neutrophils and CD8+ T
cells improves survival in human colorectal cancer. Clin Cancer
Res. 23:3847–3858. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou G, Peng K, Song Y, Yang W, Shu W, Yu
T, Yu L, Lin M, Wei Q, Chen C, et al: CD177+ neutrophils
suppress epithelial cell tumourigenesis in colitis-associated
cancer and predict good prognosis in colorectal cancer.
Carcinogenesis. 39:272–282. 2018. View Article : Google Scholar
|
|
47
|
Guo W, Zheng Y, Xu B, Ma F, Li C, Zhang X,
Wang Y and Chang X: Investigating the expression, effect and
tumorigenic pathway of PADI2 in tumors. OncoTargets Ther.
10:1475–1485. 2017. View Article : Google Scholar
|
|
48
|
Funayama R, Taniguchi H, Mizuma M,
Fujishima F, Kobayashi M, Ohnuma S, Unno M and Nakayama K: Protein-
arginine deiminase 2 suppresses proliferation of colon cancer cells
through protein citrullination. Cancer Sci. 108:713–718. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cantariño N, Musulén E, Valero V, Peinado
MA, Perucho M, Moreno V, Forcales SV, Douet J and Buschbeck M:
Downregulation of the deiminase PADI2 is an early event in
colorectal carcinogenesis and indicates poor prognosis. Mol Cancer
Res. 14:841–848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shangkuan WC, Lin HC, Chang YT, Jian CE,
Fan HC, Chen KH, Liu YF, Hsu HM, Chou HL, Yao CT, et al: Risk
analysis of colorectal cancer incidence by gene expression
analysis. PeerJ. 5:e30032017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qu Y, Olsen JR, Yuan X, Cheng PF, Levesque
MP, Brokstad KA, Hoffman PS, Oyan AM, Zhang W, Kalland KH, et al:
Small molecule promotes β-catenin citrullination and inhibits Wnt
signaling in cancer. Nat Chem Biol. 14:94–101. 2018. View Article : Google Scholar
|
|
52
|
Pelaseyed T, Bergström JH, Gustafsson JK,
Ermund A, Birchenough GM, Schütte A, van der Post S, Svensson F,
Rodríguez-Piñeiro AM, Nyström EE, et al: The mucus and mucins of
the goblet cells and enterocytes provide the first defense line of
the gastrointestinal tract and interact with the immune system.
Immunol Rev. 260:8–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Johansson ME, Thomsson KA and Hansson GC:
Proteomic analyses of the two mucus layers of the colon barrier
reveal that their main component, the Muc2 mucin, is strongly bound
to the Fcgbp protein. J Proteome Res. 8:3549–3557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee S, Bang S, Song K and Lee I:
Differential expression in normal- adenoma- carcinoma sequence
suggests complex molecular carcinogenesis in colon. Oncol Rep.
16:747–754. 2006.PubMed/NCBI
|
|
55
|
Xiong L, Wen Y, Miao X and Yang Z: NT5E
and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal
transition (EMT) are associated with tumor progression and survival
of patients with gallbladder cancer. Cell Tissue Res. 355:365–374.
2014. View Article : Google Scholar
|
|
56
|
Wang Y, Liu Y, Liu H, Zhang Q, Song H,
Tang J, Fu J and Wang X: FcGBP was upregulated by HPV infection and
correlated to longer survival time of HNSCC patients. Oncotarget.
8:86503–86514. 2017.PubMed/NCBI
|
|
57
|
Zoni E, van der Pluijm G, Gray PC and
Kruithof- de Julio M: Epithelial plasticity in cancer: Unmasking a
MicroRNA Network for TGF-β-, Notch-, and Wnt-Mediated EMT. J Oncol.
2015:1989672015. View Article : Google Scholar
|
|
58
|
Chiu HC, Li CJ, Yiang GT, Tsai AP and Wu
MY: Epithelial to mesenchymal transition and cell biology of
molecular regulation in endometrial carcinogenesis. J Clin Med.
8:82019. View Article : Google Scholar
|
|
59
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Vié N, Copois V, Bascoul-Mollevi C, Denis
V, Bec N, Robert B, Fraslon C, Conseiller E, Molina F, Larroque C,
et al: Overexpression of phosphoserine aminotransferase PSAT1
stimulates cell growth and increases chemoresistance of colon
cancer cells. Mol Cancer. 7:142008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ma L, Tao Y, Duran A, Llado V, Galvez A,
Barger JF, Castilla EA, Chen J, Yajima T, Porollo A, et al: Control
of nutrient stress- induced metabolic reprogramming by PKCζ in
tumorigenesis. Cell. 152:599–611. 2013. View Article : Google Scholar : PubMed/NCBI
|