Introduction
Anaplastic thyroid carcinoma (ATC) is an
undifferentiated and aggressive type of cancer, which accounts for
1-2% of all thyroid carcinomas. Despite its rareness, ATC is
associated with a high recurrence rate and is responsible for the
majority of deaths caused by thyroid cancer (1). Current treatments for ATC are
diverse, including surgery, radiotherapy and chemotherapy; however,
due to the resistance of ATC to almost all conventional treatments,
the prognosis of patients with ATC remains very poor (2). Therefore, a better understanding of
the molecular mechanisms underlying chemoresistance in ATC is
required for the treatment of patients with ATC.
Long non-coding RNAs (lncRNAs), which are defined as
non-coding transcripts >200 nucleotides in length, are important
regulators of a wide range of biological and cellular processes
(3). The abnormal expression of
lncRNAs is involved in the tumorigenesis, progression and
chemo-resistance of human cancer, including thyroid cancer
(4). The lncRNA nuclear
paraspeckle assembly transcript 1 (NEAT1) is a 4-kb lncRNA
localized to the nucleus, which has been demonstrated to act as an
oncogene involved in numerous types of human cancer (5-8).
Accumulating evidence has suggested that NEAT1 serves a critical
role in cancer chemoresistance, such as in osteosarcoma, ovarian
cancer and leukemia (9-11). Furthermore, upregulation of NEAT1
contributes to the progression of thyroid carcinoma by sponging
microRNA (miRNA/miR)-214 (12).
Conversely, NEAT1 knockdown suppresses the progression of papillary
thyroid cancer through the miR-129-5p/kallikrein-related peptidase
7 axis (13). However, whether
NEAT1 is associated with the chemoresistance of ATC remains
unclear.
miRNAs are an abundant class of small non-coding,
single-stranded RNAs, ~22 nucleotides long, which function as
negative gene regulators by binding to the 3′untranslated region
(3′-UTR) of their mRNA-target (14). The competing endogenous RNA (ceRNA)
hypothesis proposes that lncRNAs function as ‘miRNA sponges' to
protect target mRNAs from suppression by sequestering specific
miRNAs (15). The ceRNA concept is
involved in numerous physiopathological processes, including cell
differentiation, pluripotency, and tumorigenesis and
chemoresistance (16). The aim of
the present study was to investigate the functional role and
molecular mechanism of NEAT1 in DDP-resistant ATC. The present
study confirmed that NEAT1 silencing resulted in decreased
DDP-resistance in ATC in vitro and in vivo.
Furthermore, NEAT1 functioned as a ceRNA of miR-9-5p, and
sperm-associated antigen 9 (SPAG9) was a direct target of miR-9-5p.
These findings indicated that NEAT1 silencing sensitized ATC cell
lines to DDP by suppressing miR-9-5p sponging and regulating SPAG9
expression.
Materials and methods
Clinical specimens and cell culture
A total of 26 pairs of tumor tissues and adjacent
non-tumor thyroid tissues were collected from patients with ATC at
Henan Provincial People's Hospital between May 2015 and March 2016;
written informed consent was provided by all patients. The basic
characteristics of all volunteers are shown in Table I. No conventional treatments were
conducted prior to specimen collection. All specimens were stored
as −80°C until RNA extraction. The present study was approved by
the Institutional Ethics Review Board of Henan Provincial People's
Hospital. A human normal thyroid cell line (Nthy-ori 3-1) and two
ATC cell lines (SW1736 and 8505C) were purchased from BeNa Culture
Collection, and 293 cells were obtained from American Type Culture
Collection. The cells were maintained in DMEM supplemented with 10%
FBS and 1% penicillin/streptomycin (all Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified incubator containing 5%
CO2.
 | Table IAssociation of NEAT1 expression with
clinical characteristics of patients with anaplastic thyroid
carcinoma. |
Table I
Association of NEAT1 expression with
clinical characteristics of patients with anaplastic thyroid
carcinoma.
| Clinical
feature | n | NEAT1 expression
| P-value |
|---|
| High | Low |
|---|
| Age, years | | | | 0.899 |
| ≥60 | 16 | 10 | 6 | |
| <60 | 10 | 6 | 4 | |
| Sex | | | | 0.420 |
| Male | 8 | 4 | 4 | |
| Female | 18 | 12 | 6 | |
| TNM stage | | | | 0.008a |
| IVA-B | 15 | 6 | 9 | |
| IVC | 11 | 10 | 1 | |
| Smoking status | | | | 0.769 |
| Yes | 6 | 4 | 2 | |
| No | 20 | 12 | 8 | |
| Lymph node
metastasis | | | | 0.024a |
| N0 | 11 | 4 | 7 | |
| N1 | 15 | 12 | 3 | |
Cell transfection
NEAT1 and SPAG9 overexpression vectors (Vector-NEAT1
and Vector-SPAG9) were commercially synthesized by Guangzhou
Ribobio Co., Ltd., and an empty vector (pcDNA3.1; Guangzhou Ribobio
Co., Ltd.) was used as negative control. Briefly, ATC cells
(2×105 cells/well) were transfected with 20 mM small
interfering RNA (siRNA) targeting NEAT1 (si-NEAT1; Guangzhou
Ribobio Co., Ltd.) or non-targeting control siRNA (Scramble;
Guangzhou Ribobio Co., Ltd.); 50 mM mature miR-9-5p mimics
(Guangzhou Ribobio Co., Ltd.) or negative control (miR-NC mimics);
50 mM miR-9-5p inhibitor (anti-miR-9-5p; Guangzhou Ribobio Co.,
Ltd.) or control anti-NC; or 10 ng Vector-NEAT1/SPAG9 or empty
vector using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Transfection efficiency was confirmed by reverse
transcription-quantitative PCR (RT-qPCR). Successful NEAT1 vector,
SPAG9 vector and anti-miR-9-5p transfection is shown in Fig. S2. A total of 24 h
post-transfection, cells with positive transfection were harvested
for subsequent treatment and experiments. The following sequences
were used for transfection: si-NEAT1, 5′-GCC AUC AGC UUU GAA UAA
AUU-3′; Scramble siRNA, 5′-UUC UCC GAA CGU GUC ACG U-3′; miR-9-5p
mimic, 5′-GGU UAU CUA GCU GUA UGA-3′; miR-NC mimic, 5′-UCA CAA CCU
CCU AGA AAG AGU-3′; anti-miR-9-5p, 5′-AUA CAG CUA GAU AAC CAA AG-3′
and anti-NC, 5′-GUG UAA CAC GUC UAU ACG CCC A-3′.
RT-qPCR
RT-qPCR assays were performed to detect the
expression levels of NEAT1, miR-9-5p and SPAG9. Briefly, total RNA
was isolated from ATC tissues and cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and was
treated with RNase-free DNase I (Roche Diagnostics GmbH). The
quality and concentration of RNA extracts were assessed using a
NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.). For NEAT1 and SPAG9 mRNA detection, 500 ng RNA
was reverse transcribed to cDNA using the iScript cDNA Synthesis
kit (Bio-Rad Laboratories, Inc.), according to the manufacturer's
protocol, after which, qPCR was performed on an IQ5 Multi-color
RT-PCR Detection system (Bio-Rad Laboratories, Inc.) with SYBR
Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). GAPDH was used as an endogenous control. For miR-9-5p
detection, TaqMan miRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and Taqman MicroRNA
Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) were
used in line with the manufacturer's protocols, and miR-9-5p
expression was normalized to the expression of U6. The
amplification parameters for NEAT1, SPAG9 and miR-9-5p
quantification were: Denaturation at 95°C for 10 min, followed by
40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for
30 sec and extension at 72°C for 1 min. The relative expression
levels of NEAT1, SPAG9 and miR-9-5p were calculated using the
2−ΔΔ Cq method (17).
For RT-qPCR, the following primers were used: NEAT1, forward 5′-GTA
CGC GGG CAG ACT AAC AC-3′, reverse 5′-TGC GTC TAG ACA CCA CAA
CC-3′; SPAG9, forward 5′-ATG TCC ATA ATT ATA TGG AAC ATT TA-3′,
reverse 5′-TAA GTT GAT GAC CCA TTA TTA ACC A-3′; GAPDH, forward
5′-TTG CCA TCA ATG ACC CCT TCA-3′, reverse 5′-CGC CCC ACT TGA TTT
TGG A-3′; miR-9-5p, forward 5′-GTG CAG GGT CCG AGG T-3′, reverse
5′-GCG CTC TTT GGT TAT CTA GC-3′; and U6, forward 5′-GCT TCG GCA
GCA CAT ATA CTA AAA T-3′ and reverse 5′-CGC TTC ACG AAT TTG CGT GTC
AT-3′.
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was measured using the CCK-8
(Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. Transfected cells (2×103
cells/well) were treated with or without 30 µg/ml DDP
(Shanghai Aladdin Bio-Chem Technology Co., Ltd.) in 96-well plates.
At the indicated time (0, 24, 48 and 72 h), 10 µl CCK-8
solution was added to each well for 2 h at 37°C, after which,
absorbance at a wavelength of 450 nm was determined.
Flow cytometry
Cell apoptosis was assessed by flow cytometry using
the Annexin V-FITC Apoptosis Detection kit (BD Biosciences).
Briefly, transfected cells were harvested and washed with
pre-cooled PBS (Invitrogen; Thermo Fisher Scientific, Inc.), then
fixed with 70% cold ethanol on ice for 1 h at 4°C. Subsequently,
cells were resuspended in 500 µl Binding Buffer, and were
then labeled with 5 µl Annexin V-FITC and 10 µl
propidium iodide for 10 min in the dark at 37°C. Finally, the
samples were immediately analyzed using a FACScan flow cytometer
(BD Biosciences) with Cell Quest software v6.0 (BD
Biosciences).
Transwell assay
A total of 200 µl serum-free DMEM containing
transfected cells (5×104 cells/well) was added to the
upper chamber of 24-well Transwell plates with a Matrigel-coated
membrane (pore size, 8 µm; Corning, Inc.), and 600 µl
growth medium containing 10% FBS was added to the lower chamber of
the plates. After 48 h at 37°C, the invasive cells were stained
with 0.1% crystal violet at 37°C for 15 min. The images were
captured and the number of invaded cells was counted under a
microscope (×100 magnification; Leica DM 300; Leica Microsystems
GmbH).
Western blot analysis
Total protein was obtained from treated cells using
ice-cold lysis buffer (50 nM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 1 mM
EGTA, 1% Triton X-100, 0.1 mM phenylmethyl sulfonyl fluoride, 30 mM
sodium pyro-phosphate; pH 7.4) containing a protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA). Protein concentrations were
measured using a Bicinchoninic Acid Protein Assay kit (Novagen; EMD
Millipore). Equivalent amounts of protein (50 µg) were
separated by 10% SDS-PAGE and were then transferred onto
nitrocellulose membranes (EMD Millipore). After blocking in 5%
nonfat milk for 1 h at room temperature, the membranes were
incubated with primary antibodies for 1 h at room temperature,
followed by incubation with horseradish peroxidase-conjugated
anti-mouse or anti-rabbit immunoglobulin G (IgG; cat. no. ab6728 or
ab6721; 1:2,000; Abcam) as secondary antibodies. The protein bands
were visualized using an enhanced chemiluminescence (ECL) substrate
kit (Bio-Rad Laboratories, Inc.) with the ECL western blotting
system (Bio-Rad Laboratories, Inc.). The following primary
antibodies were used: Anti-Bax (cat. no. ab32503; 1:2,000; Abcam),
anti-Bcl-2 (cat. no. ab32124; 1:1,000; Abcam), anti-cleaved
(C)-caspase 3 (cat. no. ab49822; 1:1,000; Abcam), anti-p62 (cat.
no. ab56416; 1:1,000; Abcam), anti-microtubule-associated proteins
1A/1B light chain 3B (LC3B) (cat. no. ab168831; 1:500; Abcam),
anti-SPAG9 (cat. no. ab12331; 1:1,000; Abcam) and anti-GAPDH (cat.
no. ab8245; 1:1,000; Abcam).
Dual-luciferase reporter assay
Online software StarBase v.2.0 (starbase.sysu.edu.cn) was used to search for the
directly interacting miRNAs of NEAT1. TargetScan 6.2 online
software (www.targetscan.org/vert_71) was used
to predict the targets of miRNAs. NEAT1 wild-type or mutant-type
reporter vectors (NEAT1-wt or NEAT1-mut), and SPAG9 wild-type or
mutant-type reporter vectors (SPAG9-wt or SPAG9-mut) constructed by
Guangzhou Ribobio Co., Ltd., were cotransfected into cells
(5×104 cells/well) together with 20 nM miR-NC mimics or
miR-9-5p mimics for 48 h at room temperature. A total of 48 h
post-transfection, luciferase activity was analyzed using a
Dual-luciferase Reporter Assay system (Promega Corporation),
according to the manufacturer's protocol.
RNA immunoprecipitation (RIP) assay
RIP assay was performed to determine whether NEAT1
or SPAG9 was associated with miR-9-5p in RNA-induced silencing
complex (RISC) using the Magna RIP™ RNA-binding immunoprecipitation
kit (EMD Millipore), according to the manufacturer's protocol.
Briefly, cells were transfected with miR-9-5p mimics and lysed with
lysis buffer containing a protease inhibitor cocktail (EMD
Millipore). Argonaute2 (Ago2) is the core component of the RISC and
serves a crucial role in the mature process of miRNAs (14). Therefore, cell lysates were
incubated with magnetic beads and anti-Ago2 (cat. no. ab32381;
1:500; Abcam) or anti-IgG (ab172730; 1:100; Abcam) at 4°C for 24 h.
Finally, the beads were collected using a magnetic micro centrifuge
tube rack at 3,000 × g for 10 min at 4°C and digested with 1
µg/ml DNase (cat. no. M6101; Promega Corporation) and 50
µg/ml Proteinase K (cat. no. P6556; Sigma-Aldrich; Merck
KGaA) at 37°C for 10 min, and NEAT1 or SPAG9 mRNA enrichment was
measured by RT-qPCR.
RNA pull-down assay
To detect the endogenous interaction between NEAT1
and miR-9-5p in ATC cells, a RNA pull-down assay was performed.
Biotinylated RNA was prepared using the Biotin 3′End DNA Labeling
kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Biotinylated miR-9-5p mimics
(bio-miR-9-5p; 20 nM) were transfected into ATC cells
(3×105 cells/well) at 37°C for 48 h using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and biotinylated miR-NC mimic (bio-miR-NC; 20
nM) was used as a negative control. After 48 h, cells were lysed
using ice-cold lysis buffer and the supernatant was collected.
Subsequently, cell lysates were incubated with Dynabeads M-280
Streptavidin (BD Biosciences) to absorb bio-miR-9-5p at 4°C for 2
h, forming bio-miRNA-lncRNA complexes. Finally, the complexes were
collected by centrifugation at 3,000 x g for 10 min at 4°C and the
relative enrichment of NEAT1 was assessed in bio-miRNA-lncRNA
complexes by RT-qPCR.
Short hairpin RNA (shRNA)-mediated
silencing of NEAT1
Lentiviral vectors expressing NEAT1-targeted shRNA
(lenti-shNEAT1, 5′-GAC CGU GGU UUG UUA CUA U-3′) or non-target
control shRNA (lenti-Scramble, 5′-UUC UCC GAA CGU GUC ACG U-3′)
were constructed using BLOCK-iT™ Lentiviral RNAi Expression System
(cat. no. K494400; Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Lenti-shNEAT1 (10 nM) or
lenti-Scramble (10 nM) vectors were cotransfected into 293 cells
(1×106 cells; American Type Culture Collection) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) together with psPAX2 and pMD2.G (10 ng/ml;
Addgene) to generate lentiviral particles at 37°C. After 48 h,
shRNA-expressing lentivirus particles were collected and used to
infect ATC cell lines (5×105 cells/well) at a
multiplicity of infection of 100:1. After 24 h, cells were cultured
in 1 µg/ml puromycin to select stable NEAT1-silenced ATC
cells, and PT-qPCR was used to assess the expression of NEAT1.
In vivo assay
Male BALB/c mice (weight; 18-22 g; age, 6-8 weeks;
n=16) were purchased from Hubei Research Center of Laboratory
Animal. Mice were maintained under specific pathogen-free
conditions at a constant temperature of 22±2°C and 60% humidity,
under a 12-h light/dark, and were fed a standard chow diet ad
libitum for at least 1 week before experimentation.
Approximately 5.0×106 SW1736 or 8505C cells transfected
with lenti-Scramble or lenti-shNEAT1 were subcutaneously injected
into nude mice to develop xenografts (n=8). At 3 days
post-injection, PBS solution or DDP solution (3 mg/kg) was
intravenously administered into in each mouse every 4 days. After 4
weeks, the mice were sacrificed and tumor tissues were removed,
weighed and analyzed. All animal experiments were conducted
according to the national standard of the care and use of
laboratory animals, and the study was approved by the Committee of
Animal Research of Henan Provincial People's Hospital.
Statistical analysis
All data were analyzed using SPSS 18.0 software
(SPSS, Inc.) and are presented as the mean ± standard deviation.
Fold changes in tissue gene expression were analyzed using paired
Student's t-test, and differences between two other groups were
analyzed by unpaired Student's t-test. Multiple groups were
compared by one-way ANOVA with an honestly significant difference-q
test. Correlations between SPAG9 and NEAT1 or miR-9-5p were
analyzed by Spearman's test. χ2 test was used to
evaluate the association between NEAT1 expression and clinical
characteristics of patients with ATC. P<0.05 was considered to
indicate a statistically significant difference. Each assay was
performed independently at least three times.
Results
NEAT1 expression is upregulated in ATC
tissues and cell lines
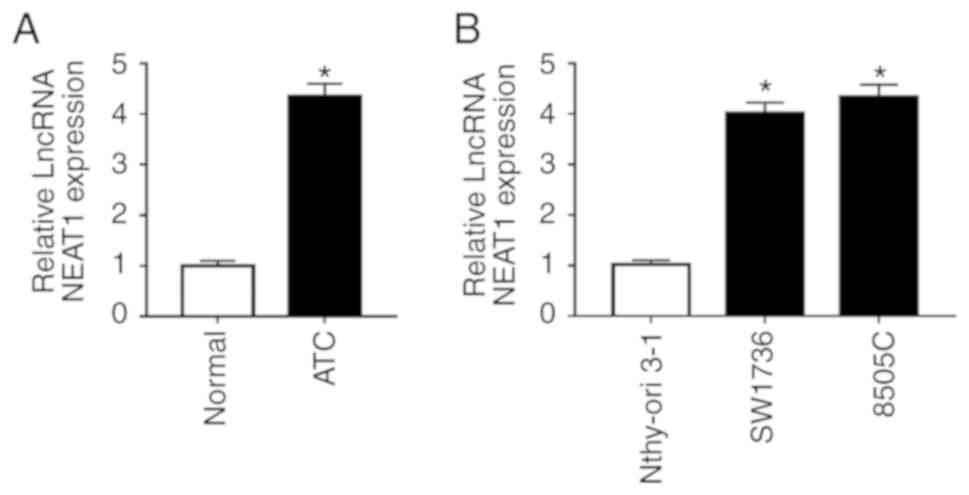
Initially, the present study analyzed NEAT1
expression in ATC tissues and adjacent normal thyroid tissues by
RT-qPCR. The results revealed that NEAT1 was significantly
upregulated in tumor tissues compared with in adjacent non-tumor
tissues (Fig. 1A). In addition,
the expression levels of NEAT1 were analyzed in ATC cell lines
(SW1736 and 8505C) and in a human normal thyroid cell line
(Nthy-ori 3-1). As shown in Fig.
1B, NEAT1 expression levels were highly elevated in ATC cell
lines compared with in the normal control.
Association between NEAT1 expression and
clinical characteristics
Subsequently, the association between NEAT1
expression and clinical characteristics was determined. As shown in
Table I, NEAT1 expression was
significantly associated with TNM stage (18) (P=0.008) and lymph node metastasis
(P=0.024). Conversely, other clinical characteristics were not
associated with NEAT1 expression.
NEAT1 silencing reduces DDP-resistance of
SW1736 and 8505C cells
To explore the function of NEAT1 on DDP-resistance
of ATC, loss-of-function experiments were performed by transfecting
SW1736 and 8505C cells with si-NEAT1, followed by treatment with or
without DDP. As shown in Fig. 2A,
compared with in the Scramble siRNA group, transfection with
si-NEAT1 resulted in a 57% reduction in NEAT1 expression in SW1736
cells, and a 62% reduction in 8505C cells. Subsequent functional
experiments revealed that NEAT1 silencing markedly suppressed cell
proliferation and invasion, and promoted cell apoptosis compared
with in the Scramble group (Fig. 2B
and C; Fig. S1). Furthermore,
western blot analysis revealed that NEAT1 silencing significantly
inhibited Bcl-2 expression, and increased Bax and C-caspase 3
levels, supporting the hypothesis that NEAT1 silencing may promote
cell apoptosis (Fig. 2D and E).
The expression levels of LC3, an autophagosome membrane protein,
and receptor protein p62 are widely acknowledged to reflect the
activity of autophagy (19).
Therefore, the expression levels of LC3-II/I and p62 were detected,
in order to explore the role of NEAT1 in cell autophagy. The
results demonstrated that NEAT1 silencing resulted in a decrease in
p62 expression, and an increase in LC3-II/I, thus indicating that
NEAT1 silencing may elevate autophagy in SW1736 and 8505C cells
(Fig. 2F and G).
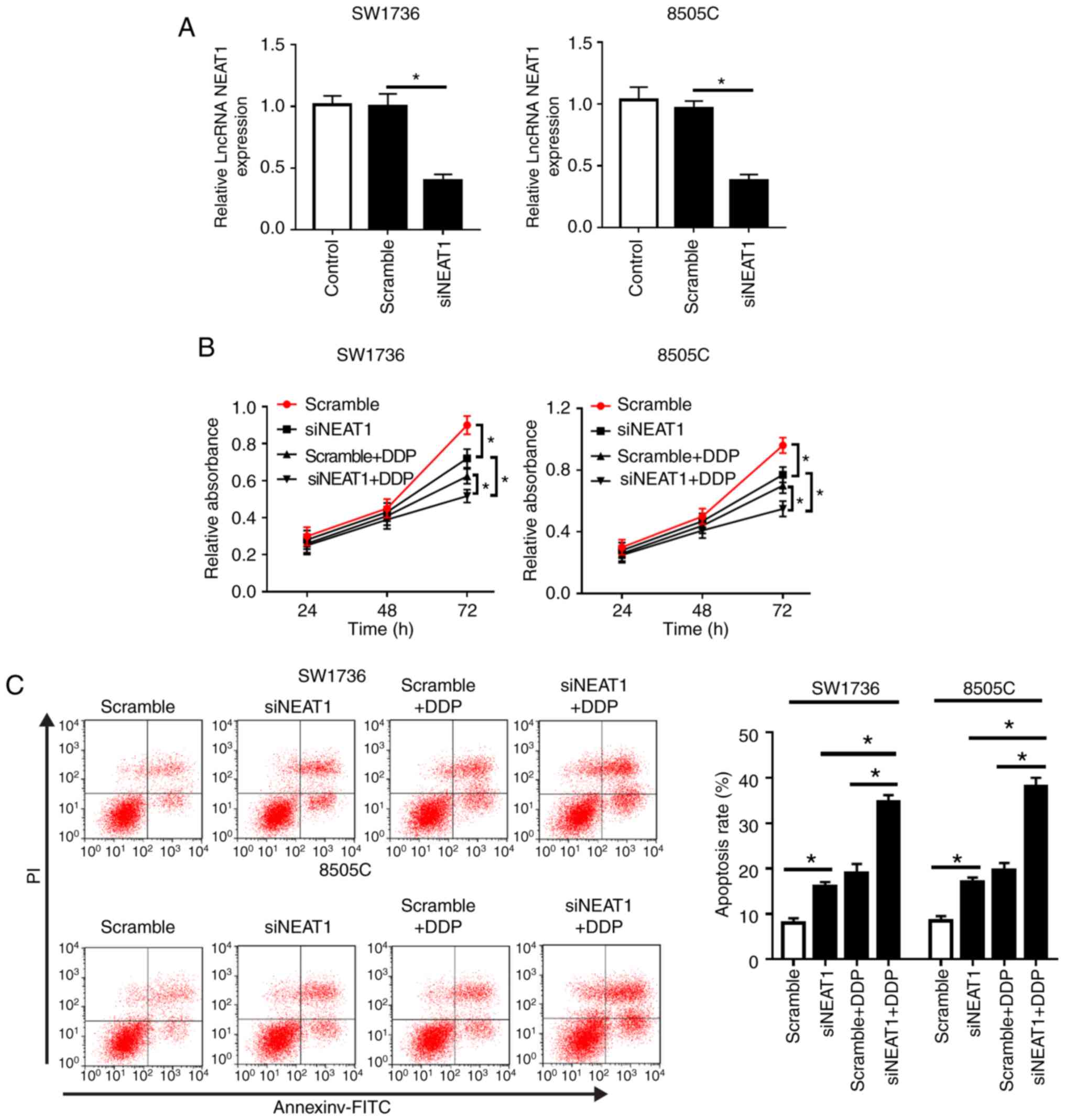 | Figure 2NEAT1 silencing decreases
DDP-resistance of SW1736 and 8505C cells. SW1736 and 8505C cells
were transfected with Scramble or siNEAT1, and were then stimulated
with or without 30 µg/ml DDP. (A) NEAT1 expression was
assessed by reverse transcription-quantitative PCR in transfected
cells. (B) At the indicated times, cell proliferation was detected
by Cell Counting kit-8 in treated cells. (C) After 48 h of DDP
treatment, cell apoptosis was measured by flow cytometry. (D and E)
After 48 h of treatment, the expression levels of Bax, Bcl-2 and
C-caspase3 were determined by western blotting.
*P<0.05 as indicated. NEAT1 silencing decreases
DDP-resistance of SW1736 and 8505C cells. (F and G) After 48 h of
treatment, the expression levels of p62, LC3-I and LC3-II were
assessed by western blotting in treated cells.
*P<0.05 as indicated. C-, cleaved; DDP, cisplatin;
LC3, microtubule-associated proteins 1A/1B light chain 3B; lncRNA,
long non-coding RNA; NEAT1, nuclear paraspeckle assembly transcript
1; PI, propidium iodide; si, small interfering RNA. |
Notably, compared with in the si-NEAT1 or Scramble +
DDP group, simultaneous NEAT1 silencing and DDP treatment in SW1736
and 8505C cells led to a more distinct inhibition of cell
proliferation and a more obvious promotion of cell apoptosis
(Fig. 2B-E). Furthermore, NEAT1
silencing in DDP-treated cells markedly promoted cell autophagy
(Fig. 2F and G). These data
indicated that NEAT1 silencing promoted ATC cell sensitivity to
DDP.
NEAT1 binds to miR-9-5p and suppresses
miR-9-5p expression
To further investigate the molecular mechanism by
which NEAT1 affected the DDP-resistance of ATC cells, StarBase
v.2.0 software was used to search for the directly interacting
miRNAs of NEAT1. Notably, the data revealed that miR-9-5p harbored
putative binding sites with NEAT1 (Fig. 3A). To verify this finding,
dual-luciferase reporter, RIP and RNA pull-down assays were
performed. For the dual-luciferase reporter assay, NEAT1-wt or
NEAT1-mut vectors were transfected into SW1736 and 8505C cells,
together with miR-9-5p or NC mimics. The results indicated that the
luciferase activity of NEAT1-wt was markedly reduced
post-transfection with miR-9-5p mimics; however, no change was
observed in the activity of cells transfected with NEAT1-mut in the
presence of miR-9-5p mimics (Fig.
3B). For the RIP assay, cells were transfected with miR-9-5p
mimics, and lysates were incubated with anti-Ago2. As presented in
Fig. 3C, NEAT1 was substantially
enriched in the presence of miR-9-5p mimics. Additionally, for the
RNA pull-down assay, cells were transfected with bio-miR-9-5p and
lysates were incubated with streptavidin-coupled beads to form
bio-miRNA-lncRNA complexes. As shown in Fig. 3D, NEAT1 enrichment was higher in
the bio-miR-9-5p group compared with in the control group.
Furthermore, this study aimed to determine whether NEAT1 regulated
miR-9-5p expression in SW1736 and 8505C cells by transfecting cells
with siNEAT1 or Vector-NEAT1. RT-qPCR assays demonstrated that
miR-9-5p expression was significantly elevated by NEAT1 silencing,
whereas it was reduced in the presence of Vector-NEAT1 (Fig. 3E). These results indicated that
NEAT1 may directly target miR-9-5p.
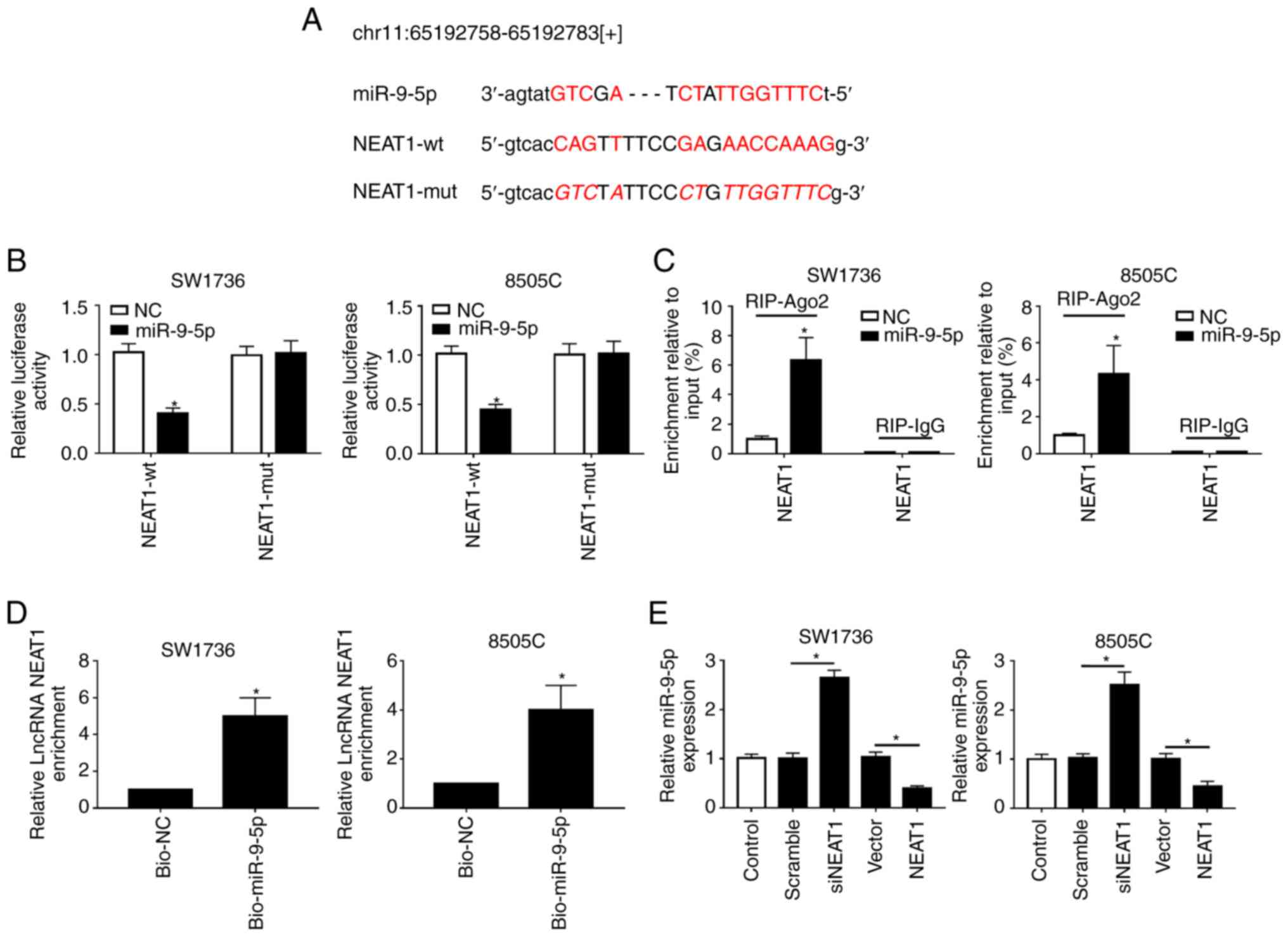 | Figure 3NEAT1 binds to miR-9-5p and
suppresses it expression. (A) Schematic diagram of the predicted
binding sites between NEAT1 and miR-9-5p, and the mutation in the
binding sequence of NEAT1-mut. (B) Relative luciferase activities
were assessed in SW1736 and 8505C cells cotransfected with NEAT1-wt
or NEAT1-mut and miR-NC mimics or miR-9-5p mimics. (C) SW1736 and
8505C cells were transfected with miR-9-5p mimics or miR-NC mimics,
and their lysates were incubated with anti-Ago2, followed by the
measurement of NEAT1 enrichment by RT-qPCR assay. (D) SW1736 and
8505C cells were transfected with bio-miR-9-5p mimics or bio-NC
mimics and their lysates were incubated with streptavidin-coupled
beads to form bio-miRNA-lncRNA complexes, followed by the detection
of NEAT1 enrichment. (E) RT-qPCR assay of miR-9-5p expression in
SW1736 and 8505C cells transfected with Scramble, siNEAT1, Vector
and Vector-NEAT1. *P<0.05 vs. NC, or as indicated.
Ago2, Argonaute2; bio, biotinylated; lncRNA, long non-coding RNA;
miR-9-5p, microRNA-9-5-p; mut, mutant; NC, negative control; NEAT1,
nuclear paraspeckle assembly transcript 1; RIP, RNA
immunoprecipiation; RT-qPCR, reverse transcription-quantitative
PCR; si, small interfering RNA; wt, wild type. |
miR-9-5p sensitizes SW1736 and 8505C
cells to DDP
The present study detected miR-9-5p expression in
ATC tissues and cell lines. The results revealed that miR-9-5p was
significantly downregulated in ATC tissues and cell lines compared
with in controls (Fig. 4A and B).
Subsequently, in order to explore the effects of miR-9-5p on
DDP-resistance in ATC, gain-of-function experiments were performed
by transfecting SW1736 and 8505C cells with miR-9-5p mimics,
followed by treatment with or without DDP. As presented in Fig. 4C, transfection with miR-9-5p mimics
resulted in a ~4.55-fold increase of miR-9-5p expression in SW1736
cells, and a ~4.53-fold increase in 8505C cells. Furthermore,
functional experiments demonstrated that miR-9-5p overexpression in
SW1736 and 8505C cells led to a decrease in cell proliferation
(Fig. 4D), an enhancement of cell
apoptosis (Fig. 4E and F), and
promotion of cell autophagy (Fig.
4G). Furthermore, simultaneous miR-9-5p overexpression and DDP
treatment led to a more distinct inhibition on cell proliferation,
and a more obvious promotion on cell apoptosis and autophagy
compared with miR-9-5p mimics alone or NC + DDP group (Fig. 4D-G). Taken together, these results
indicated that miR-9-5p may sensitize SW1736 and 8505C cells to
DDP.
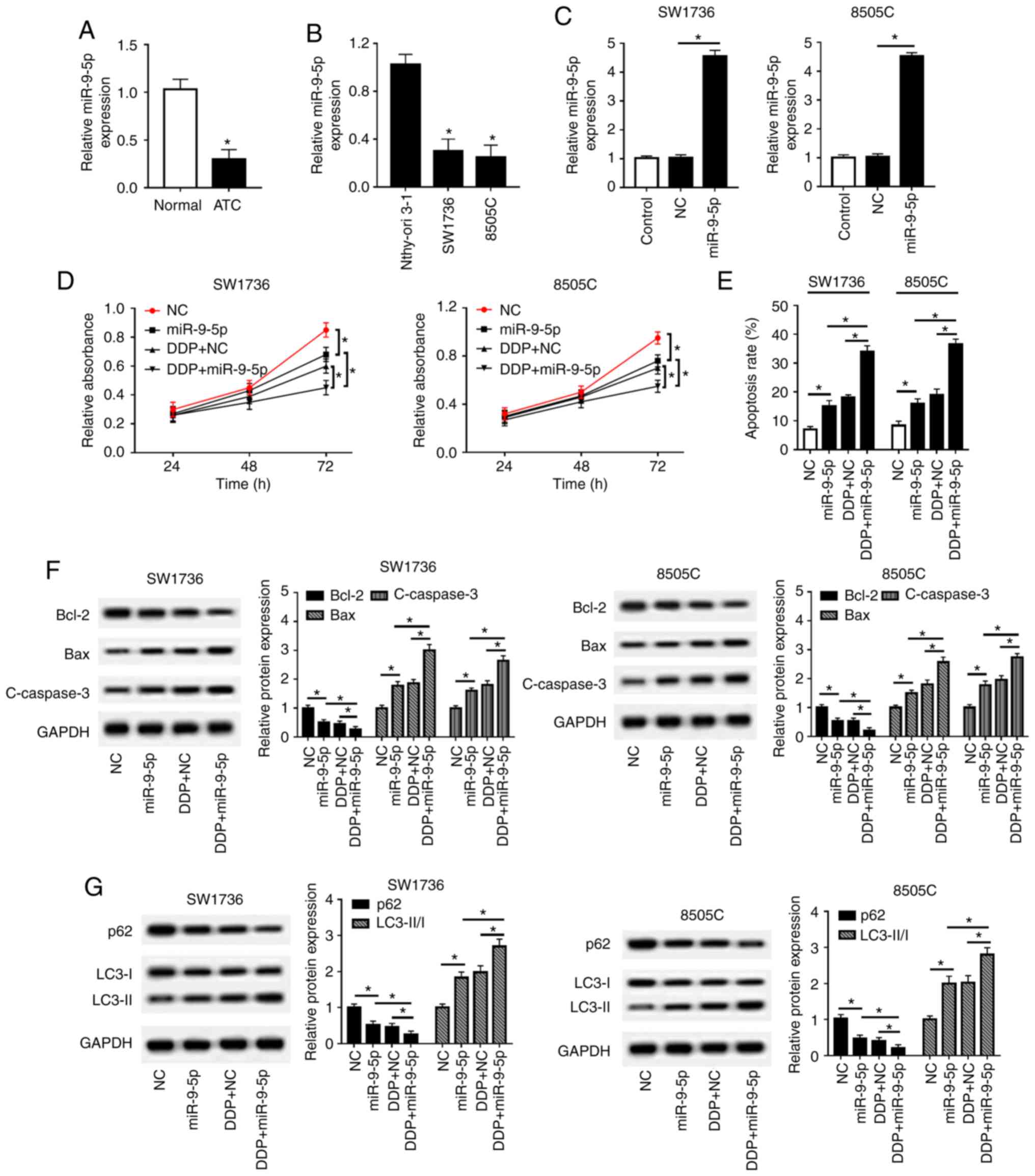 | Figure 4miR-9-5p sensitizes SW1736 and 8505C
cells to DDP. RT-qPCR assay of miR-9-5p expression in (A) ATC
tissues and adjacent normal thyroid tissues, and in (B) two ATC
cell lines (SW1736 and 8505C) and Nthy-ori 3-1 cells. SW1736 and
8505C cells were transfected with miR-9-5p mimics or miR-NC mimics,
and were then treated with or without DDP, followed by (C)
determination of miR-9-5p expression by RT-qPCR assay, (D) cell
proliferation by Cell Counting kit-8 assay, (E) cell apoptosis by
flow cytometry, and the expression levels of (F) Bax, Bcl-2 and
C-caspase3, and (G) p62, LC3-I, LC3-II and GAPDH by western
blotting. *P<0.05, as indicated. ATC, anaplastic
thyroid carcinoma; C-, cleaved; DDP, cisplatin; LC3,
microtubule-associated proteins 1A/1B light chain 3B; miR-9-5p,
microRNA-9-5-p; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR. |
Inhibitory role of NEAT1 silencing on
DDP-resistance of SW1736 and 8505C cells is mediated by
miR-9-5p
To provide further mechanistic insight into the
association between NEAT1 and miR-9-5p on DDP-resistance in ATC,
SW1736 and 8505C cells were cotransfected with siNEAT1 and
anti-miR-9-5p, followed by treatment with or without DDP. As shown
in Fig. 5A, cotransfection with
anti-miR-9-5p markedly antagonized the enhancing effects of NEAT1
silencing on miR-9-5p expression. CCK-8 assay revealed that
compared with NC cotransfection, anti-miR-9-5p cotransfection in
SW1736 and 8505C cells antagonized the inhibitory effects of NEAT1
silencing on cell proliferation upon DDP treatment (Fig. 5B). Further functional experiments
demonstrated that the effects of NEAT1 silencing on cell apoptosis
and autophagy were markedly reduced following cotransfection with
anti-miR-9-5p (Fig. 5C-E).
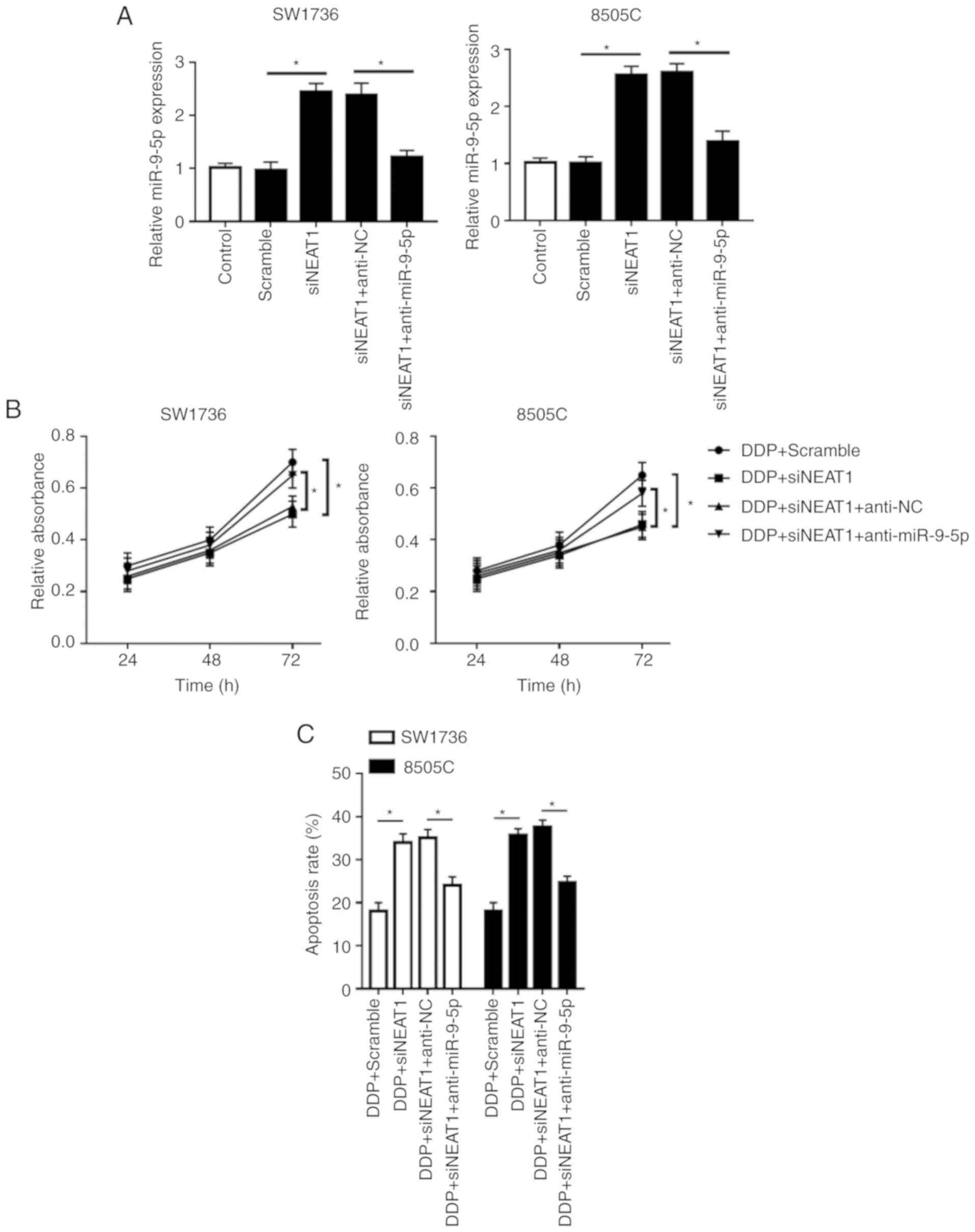 | Figure 5Inhibitory role of NEAT1 silencing on
DDP-resistance of SW1736 and 8505C cells is mediated by miR-9-5p.
SW1736 and 8505C cells were transfected with Scramble, siNEAT1,
siNEAT1 + anti-NC or siNEAT1 + anti-miR-9-5p, and were then treated
with or without DDP, followed by measurement of (A) miR-9-5p
expression by reverse transcription-quantitative PCR assay, (B)
cell proliferation by Cell Counting kit-8 assay, (C) cell apoptosis
by flow cytometry. *P<0.05, as indicated. Inhibitory
role of NEAT1 silencing on DDP-resistance of SW1736 and 8505C cells
is mediated by miR-9-5p. SW1736 and 8505C cells were transfected
with Scramble, siNEAT1, siNEAT1 + anti-NC or siNEAT1 +
anti-miR-9-5p, and were then treated with or without DDP, followed
by measurement of the expression levels (D) Bax, Bcl-2 and
C-caspase3 by western blotting. *P<0.05, as
indicated. Inhibitory role of NEAT1 silencing on DDP-resistance of
SW1736 and 8505C cells is mediated by miR-9-5p. SW1736 and 8505C
cells were transfected with Scramble, siNEAT1, siNEAT1 + anti-NC or
siNEAT1 + anti-miR-9-5p, and were then treated with or without DDP,
followed by measurement of the expression levels (E) p62, LC3-I and
LC3-II by western blotting. *P<0.05, as indicated.
C-, cleaved; DDP, cisplatin; LC3, microtubule-associated proteins
1A/1B light chain 3B; miR-9-5p, microRNA-9-5-p; NC, negative
control; NEAT1, nuclear paraspeckle assembly transcript 1; si,
small interfering RNA. |
SPAG9 is a direct target of miR-9-5p
TargetScan Human software was used to predict the
target genes of miR-9-5p. The results revealed that the 3′-UTR of
SPAG9 mRNA contained a putative complementary site of miR-9-5p
(Fig. 6A). To confirm whether
SPAG9 was a target of miR-9-5p, dual-luciferase reporter and RIP
assays were performed. Compared with in the NC group, the
luciferase activities of SPAG9-wt in SW1736 and 8505C cells were
significantly weakened by miR-9-5p overexpression (Fig. 6B). Conversely, there was little
change in the luciferase activities of SPAG9-mut in the presence of
miR-9-5p mimics (Fig. 6B).
Furthermore, miR-9-5p overexpression resulted in an abundant
enrichment of SPAG9 mRNA with anti-Ago2 in SW1736 and 8505C cells
(Fig. 6C). In addition, this study
verified whether miR-9-5p affected the expression of SPAG9. Western
blot analysis demonstrated that miR-9-5p negatively regulated SPAG9
expression in SW1736 and 8505C cells (Fig. 6D).
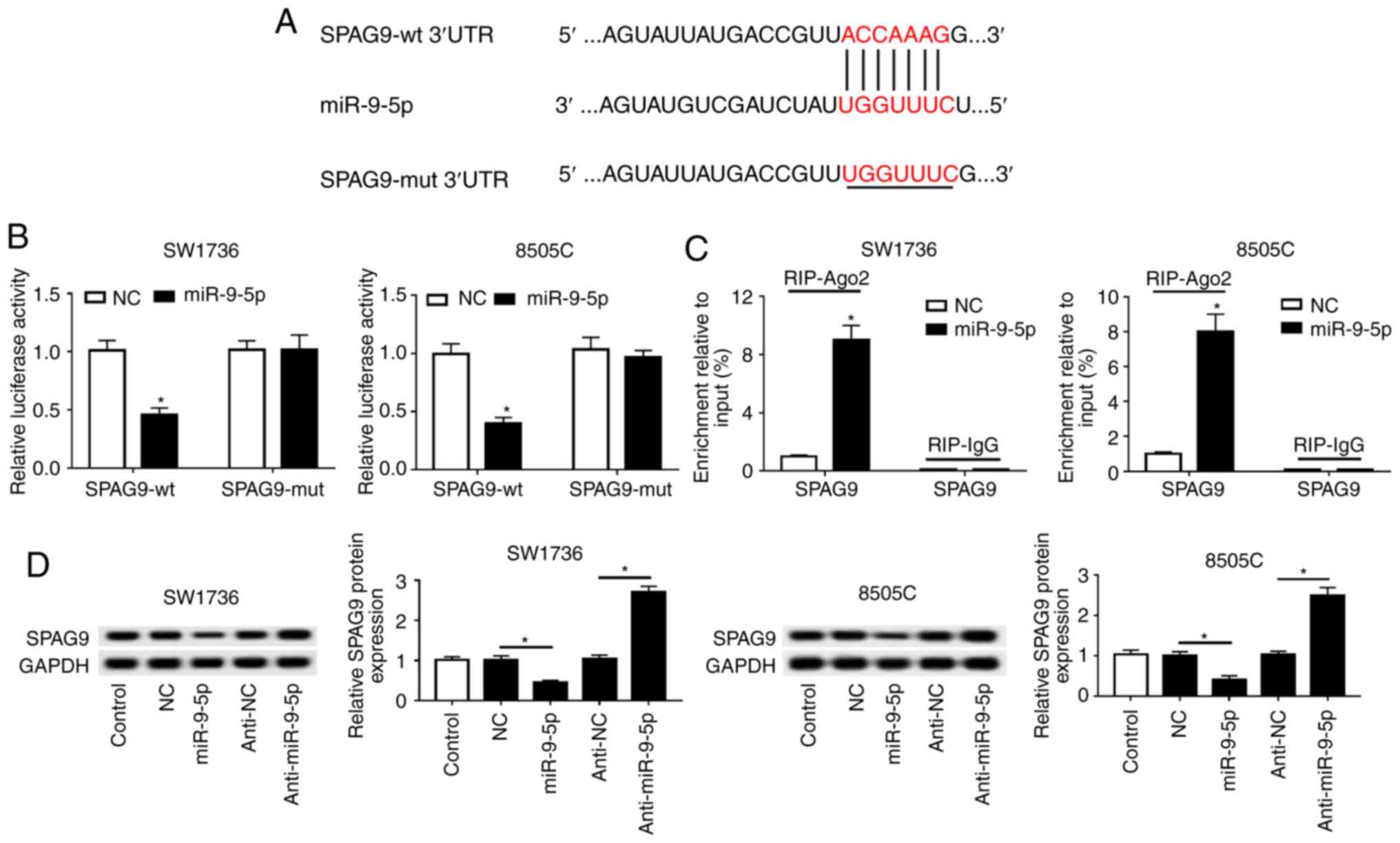 | Figure 6SPAG9 is a direct target gene of
miR-9-5p. (A) Schematic diagram of the predicted binding sites
between miR-9-5p and SPAG9, and the mutation in seeded region of
SPAG9-mut. (B) Relative luciferase activities were detected in
SW1736 and 8505C cells cotransfected with SPAG9-wt or SPAG-mut and
miR-NC mimics or miR-9-5p mimics. (C) SW1736 and 8505C cells were
transfected with miR-NC mimics or miR-9-5p mimics, and their
lysates were incubated with anti-Ago2 or anti-IgG, followed by the
measurement of SPAG9 mRNA enrichment by reverse
transcription-quantitative PCR assay. (D) Western blot analysis of
SPAG9 expression in SW1736 and 8505C cells transfected with miR-NC
mimics, miR-9-5p mimics, anti-miR-NC or anti-miR-9-5p.
*P<0.05 vs. NC, or as indicated. 3′-UTR,
3′-untranslated region; Ago2, Argonaute2; miR-9-5p, microRNA-9-5-p;
mut, mutant; NC, negative control; SPAG9, sperm-associated antigen
9; wt, wild type. |
SPAG9 antagonizes the inhibitory role of
NEAT1 silencing on DDP-resistance of SW1736 and 8505C cells
As expected, SPAG9 mRNA expression was highly
upregulated in ATC tissues compared with in normal control tissues
(Fig. 7A). Furthermore, SPAG9 mRNA
expression was positively correlated with NEAT1 expression, whereas
it was inversely correlated with miR-9-5p expression in ATC tissues
(Fig. 7B and C). In addition,
SPAG9 mRNA expression was also increased in ATC cell lines
(Fig. 7D).
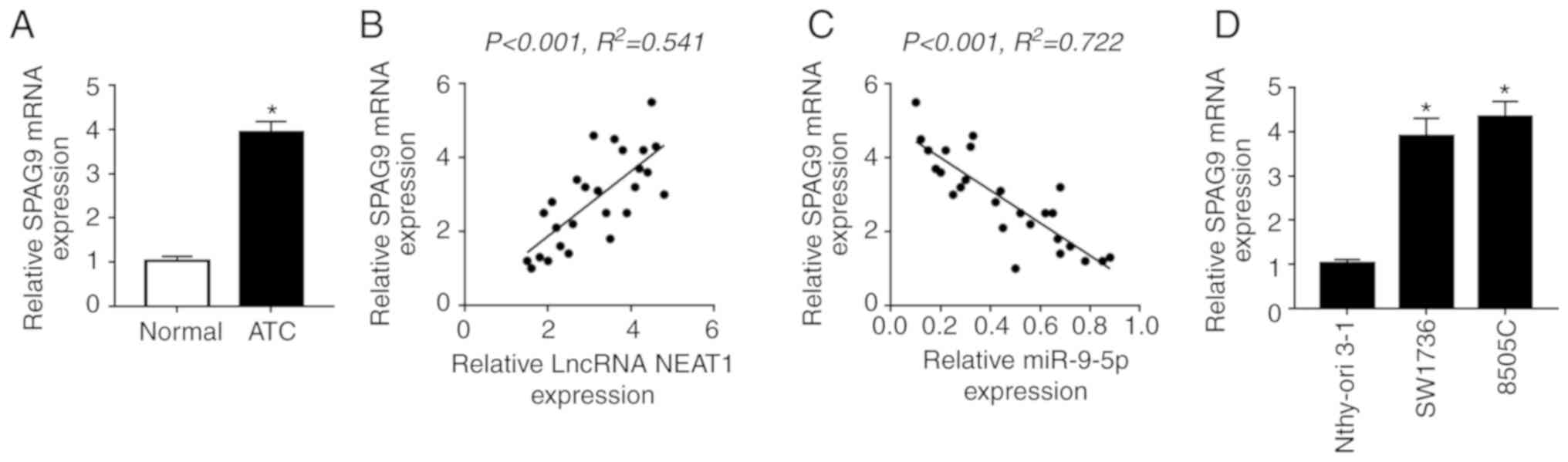 | Figure 7Inhibitory role of NEAT1 silencing on
DDP-resistance is antagonized by SPAG9 in SW1736 and 8505C cells.
(A) RT-qPCR assay of SPAG9 mRNA expression in ATC tissues and
adjacent normal thyroid tissues. Correlation between (B) SPAG9 mRNA
and NEAT1 expression or (C) SPAG9 mRNA and miR-9-5p expression was
determined. (D) RT-qPCR assay of SPAG9 mRNA expression in two ATC
cell lines (SW1736 and 8505C) and Nthy-ori 3-1 cells. SW1736 and
8505C cells were transfected with Scramble, siNEAT1, siNEAT1 +
Vector or siNEAT1 + Vector-SPAG9, followed by detection of (E)
SPAG9 expression by western blotting. SW1736 and 8505C cells were
transfected with Scramble, siNEAT1, siNEAT1 + Vector or siNEAT1 +
Vector-SPAG9, and were then treated with DDP, followed by the
measurement of (F) cell proliferation by Cell Counting kit-8 assay.
*P<0.05 vs. as indicated. Inhibitory role of NEAT1
silencing on DDP-resistance is antagonized by SPAG9 in SW1736 and
8505C cells. SW1736 and 8505C cells were transfected with Scramble,
siNEAT1, siNEAT1 + Vector or siNEAT1 + Vector-SPAG9, and were then
treated with DDP, followed by the measurement of (G) cell apoptosis
by flow cytometry, and the expression levels of (H and I) Bax,
Bcl-2 and C-caspase3, and (J) p62, LC3-I and LC3-II by western
blotting. *P<0.05 vs. as indicated. C-, cleaved; DDP,
cisplatin; LC3, microtubule-associated proteins 1A/1B light chain
3B; NC, negative control; NEAT1, nuclear paraspeckle assembly
transcript 1; RT-qPCR, reverse transcription-quantitative PCR; si,
small interfering RNA. |
The present study aimed to determine whether SPAG9
was involved in the regulatory effect of NEAT1 on DDP-resistance in
ATC. The results showed that NEAT1 silencing resulted in a
significant decrease in SPAG9 expression in SW1736 and 8505C cells,
whereas this effect was abrogated by cotransfection with
Vector-SPAG9 (Fig. 7E). Further
functional experiments demonstrated that the regulatory effect of
NEAT1 silencing on cell proliferation, apoptosis and autophagy upon
DDP treatment were markedly reversed by SPAG9 expression
restoration in SW1736 and 8505C cells (Fig. 7F-J).
NEAT1 silencing decreases DDP-resistance
in tumors in vivo
Given the present in vitro findings, this
study further evaluated the role of NEAT1 in DDP-resistance of
tumors in vivo. SW1736 and 8505C cells infected with
lenti-Scramble or lenti-shNEAT1 were subcutaneously injected into
nude mice to develop xenografts, followed by treatment with DDP or
PBS. At the end of the in vivo experiments, the average
diameter of the control tumors reached approximately 10 mm
(Fig. 8A and B). The present
results revealed that shNEAT1 transfection or DDP treatment highly
suppressed tumor growth compared with in the respective control
groups; this was presented as a decrease in tumor volume and weight
(Fig. 8A and B). Furthermore,
simultaneous shNEAT1 transfection and DDP treatment resulted in a
more distinct inhibition on tumor growth, suggesting the inhibitory
role of NEAT1 silencing on the DDP-resistance of ATC cells in
vivo (Fig. 8A and B). In
addition, NEAT1 and SPAG9 expression levels were downregulated,
whereas miR-9-5p expression was upregulated in tumors derived from
lenti-shNEAT1-infected cells with or without DDP treatment
(Fig. 8C-E). Additionally, western
blot analysis indicated that NEAT1 silencing led to a decrease in
p62 expression and an increase in LC3-II/I expression in xenograft
tissues with or without DDP treatment (Fig. 8F).
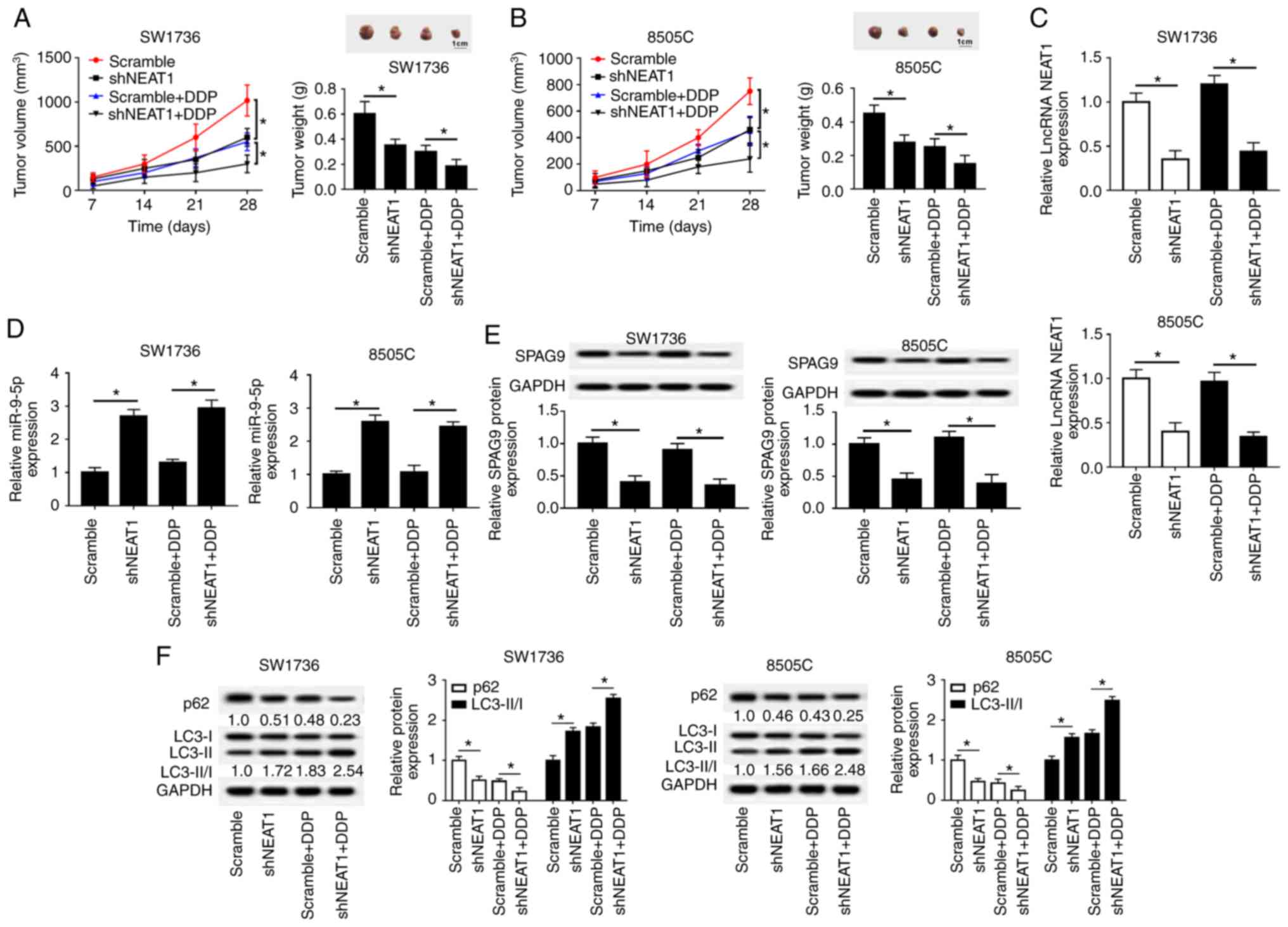 | Figure 8NEAT1 silencing decreases
DDP-resistance of tumors in vivo. Briefly,
5.0×106 SW1736 and 8505C cells stably transfected with
lenti-Scramble or lenti-shNEAT1 were subcutaneously injected into
nude mice to develop xenografts (n=8). At 3 days after injection,
PBS solution or DDP solution (3 mg/kg) were intravenously
administered into each mouse every 4 days. After 4 weeks, mice were
sacrificed. (A and B) Tumor volume, tumor images and average weight
were analyzed in excised tumor tissues. (C and D) Reverse
transcription-quantitative PCR assay of NEAT1 and miR-9-5p
expression in xenograft tissues. (E and F) Western blot analysis of
SPAG9, p62, LC3-I and LC3-II levels in xenograft tumor tissues.
*P<0.05 vs. as indicated. DDP, cisplatin; LC3,
microtu-bule-associated proteins 1A/1B light chain 3B; lncRNA, long
non-coding RNA; NEAT1, nuclear paraspeckle assembly transcript 1;
sh, short hairpin RNA. |
Discussion
Accumulating evidence has suggested that lncRNAs are
tightly linked to cancer chemoresistance, providing a possibility
that lncRNAs may be potential therapeutic targets for
chemoresistant cancer. For example, Han et al (20) reported that the lncRNA colorectal
neoplasia differentially expressed contributes to
5-fluoruracil-resistance in colorectal cancer cell through sponging
miR-181a-5p and regulating Wnt/β-catenin signaling. Gu et al
(21) demonstrated that HOXD
antisense growth-associated lncRNA knockdown ameliorates the
chemoresistance of castration-resistant prostate cancer cells by
recruiting WD repeat domain 5. Additionally, Yoshida et al
(22) reported that curcumin
sensitizes pancreatic cancer cells to gemcitabine through weakening
lncRNA PVT1 expression. Furthermore, Wang et al (23) verified that the lncRNA papillary
thyroid carcinoma susceptibility candidate 3 ameliorates drug
resistance of ATC cells to doxorubicin by regulating the
STAT3/INO80 pathway.
The present study demonstrated that NEAT1 was
upregulated in ATC tissues and cell lines, and NEAT1 silencing
sensitized ATC cell lines to DDP. Similar to these findings,
inhibition of NEAT1 has been reported to attenuate chemoresistance
in a series of cancer cells, including naso-pharyngeal carcinoma
cells (24), gastric cancer cells
(25) and lung cancer cells
(26). Furthermore, Adriaens et
al (27) demonstrated that
NEAT1 knockdown sensitizes MCF-7 cells to DNA-damaging agents, such
as doxorubicin or platinum compounds. Parasramka et al
(28) also verified that NEAT1
depletion leads to a reduction in tumor cell proliferation under
gemcitabine treatment, indicating it as a downstream effector of
gemcitabine sensitivity in cholan-giocarcinoma.
It is widely accepted that lncRNAs may act as ‘miRNA
sponges' to protect target mRNAs from suppression by sequestering
target miRNAs (15). Therefore,
StarBase v.2.0 software was used to search for the target miRNAs of
NEAT1. Among these candidates, miR-9-5p was selected for further
analysis, due to its crucial involvement in human cancer (29-33).
In addition, this study verified that NEAT1 suppressed miR-9-5p
expression by directly binding to miR-9-5p; this finding is
consistent with a recent study (29). miR-9-5p results from the
transcription of miR-9 genes located on chromosomes 1, 5 and 15,
and functions as an oncogene or a tumor suppressor involved in the
carcinogenesis and metastatic process of numerous types of cancer
(30), such as breast cancer
(31), colorectal cancer (32) and glioblastoma (33). Furthermore, miR-9 over-expression
is associated with decreased resistance of primary epithelial
ovarian cancer cells to DDP treatment (34). miR-9 also promotes DDP-sensitivity
of non-small lung cancer cells by targeting eukaryotic translation
inhibition factor 5A2 (35).
Conversely, miR-9 has been reported to enhance the resistance of
glioblastoma multiforme cells to temozolomide via a Sonic
Hedgehog-independent method by targeting patched 1 (36). In this study, it was reported that
miR-9-5p was down-regulated in ATC tissues and cell lines,
suggesting its role as a potential tumor suppressor in ATC, in
accordance with a recent study (37). Furthermore, the present data
indicated that miR-9-5p sensitized ATC cell lines to DDP, which is
similar to the findings of previous studies by Zhao et al
and Pan et al (34,35). Furthermore, this study validated
that the inhibitory role of NEAT1 silencing on DDP-resistance of
ATC cell lines was mediated by miR-9-5p.
TargetScan software was used to predict the target
genes of miR-9-5p. Among these candidates, SPAG9 was selected for
further study due to its well-recognized oncogenic role in human
cancer, including in endometrial carcinoma (38), lung cancer (39) and triple-negative breast cancer
(40). Additionally, SPAG9
expression is upregulated in the peripheral blood of patients with
hepatocellular carcinoma and lung cancer, highlighting its role as
a potential tumor-specific biomarker for tumor detection, therapy
and prognosis (41). SPAG9 has
also been demonstrated to be involved in the progression of thyroid
cancer (42). Furthermore, SPAG9
overexpression enhances drug resistance of breast cancer cells to
taxol (43). This study validated
that SPAG9 was a direct target of miR-9-5p, and confirmed that
SPAG9 mRNA expression levels were upregulated in ATC tissues and
cell lines. Furthermore, SPAG9 expression was positively correlated
with NEAT1 expression, whereas it was inversely correlated with
miR-9-5p expression in ATC tissues. The inhibitory role of NEAT1
silencing on DDP-resistance of ATC cell lines was antagonized by
SPAG9.
Finally, the present study demonstrated that NEAT1
silencing might decrease DDP-resistance of tumors in vivo
partly by regulating the miR-9-5p/SPAG9 axis. Since the number of
mice was relatively small and no ATC mouse model was generated, the
role and molecular mechanism of NEAT1 on DDP-resistance of ATC
in vivo cannot be confirmed. Therefore, further studies
regarding the relationship between NEAT1 and DDP-resistance of ATC
in vivo are required.
In conclusion, this study revealed that NEAT1
silencing ameliorated DDP-resistance of ATC cells, at least in part
by reducing miR-9-5p sponging and regulating SPAG9 expression.
Targeting NEAT1 may be a potential therapeutic strategy for the
treatment of DDP-resistant ATC.
Supplementary Data
Funding
The present study was supported by Henan Provincial
People's Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
This study was designed and conceived by PY and ZS.
The experimental procedures and data analysis were carried out by
PY, ZS, ZZ and TG. The manuscript was prepared by PY and ZZ. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Ethics
Review Board of Henan Provincial People's Hospital; all patients
provided prior written informed consent. All animal experiments
were carried out following the national standard of the care and
use of laboratory animals, and the study was approved by the
Committee of Animal Research of Henan Provincial People's
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Molinaro E, Romei C, Biagini A, Sabini E,
Agate L, Mazzeo S, Materazzi G, Sellari-Franceschini S, Ribechini
A, Torregrossa L, et al: Anaplastic thyroid carcinoma: From
clinicopathology to genetics and advanced therapies. Nat Rev
Endocrinol. 13:644–660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smallridge RC and Copland JA: Anaplastic
thyroid carcinoma: Pathogenesis and emerging therapies. Clin Oncol
(R Coll Radiol). 22:pp. 486–497. 2010, View Article : Google Scholar
|
|
3
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
4
|
Murugan AK, Munirajan AK and Alzahrani AS:
Long noncoding RNAs: Emerging players in thyroid cancer
pathogenesis. Endocr Relat Cancer. 25:R59–R82. 2018. View Article : Google Scholar
|
|
5
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen ZJ, Zhang Z, Xie BB and Zhang HY:
Clinical significance of up-regulated lncRNA NEAT1 in prognosis of
ovarian cancer. Eur Rev Med Pharmacol Sci. 20:3373–3377.
2016.PubMed/NCBI
|
|
7
|
Choudhry H, Albukhari A, Morotti M, Haider
S, Moralli D, Smythies J, Schödel J, Green CM, Camps C, Buffa F, et
al: Tumor hypoxia induces nuclear paraspeckle formation through
HIF-2a dependent transcriptional activation of NEAT1 leading to
cancer cell survival. Oncogene. 34:4482–4490. 2015. View Article : Google Scholar
|
|
8
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015.PubMed/NCBI
|
|
9
|
Hu Y, Yang Q, Wang L, Wang S, Sun F, Xu D
and Jiang J: Knockdown of the oncogene lncRNA NEAT1 restores the
availability of miR-34c and improves the sensitivity to cisplatin
in osteosarcoma. Biosci Rep. 38:pp. pii: BSR201803752018,
View Article : Google Scholar
|
|
10
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao C, Zhang J, Wang Q and Ren C:
Overexpression of lncRNA NEAT1 mitigates multidrug resistance by
inhibiting ABCG2 in leukemia. Oncol Lett. 12:1051–1057. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar
|
|
13
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: Long noncoding RNA NEAT1 regulate papillary thyroid
cancer progression by modulating miR-1295p/KLK7 expression. J Cell
Physiol. 233:6638–6648. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Shaha AR: TNM classification of thyroid
carcinoma. World J Surg. 31:879–887. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barth S, Glick D and Macleod KF:
Autophagy: Assays and artifacts. J Pthol. 221:117–124. 2010.
View Article : Google Scholar
|
|
20
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L and Cui BB: The lncRNA CRNDE
promotes colorectal cancer cell proliferation and chemoresistance
via miR-181a5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar
|
|
21
|
Gu P, Chen X, Xie R, Han J, Xie W, Wang B,
Dong W, Chen C, Yang M, Jiang J, et al: lncRNA HOXD-AS1 regulates
proliferation and chemo-resistance of castration-resistant prostate
cancer via recruiting WDR5. Mol Ther. 25:1959–1973. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshida K, Toden S, Ravindranathan P, Han
H and Goel A: Curcumin sensitizes pancreatic cancer cells to
gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1
expression. Carcinogenesis. 38:1036–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang XM, Liu Y, Fan YX, Liu Z, Yuan QL,
Jia M, Geng ZS, Gu L and Lu XB: LncRNA PTCSC3 affects drug
resistance of anaplastic thyroid cancer through STAT3/INO 80
pathway. Cancer Biol Ther. 19:590–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Tai Y and Ma J: LncRNA
NEAT1/let-7a5paxis regulates the cisplatin resistance in
nasopharyngeal carcinoma by targeting Rsf-1 and modulating the
Ras-MAPK pathway. Cancer Biol Ther. 19:534–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Zhao B, Chen X, Wang Z, Xu H and
Huang B: Silence of long noncoding RNA NEAT1 inhibits malignant
biological behaviors and chemotherapy resistance in gastric cancer.
Pathol Oncol Res. 24:109–113. 2018. View Article : Google Scholar
|
|
26
|
Jiang P, Wu X, Wang X, Huang W and Feng Q:
NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin
sensitivity in lung cancer cells. Oncotarget. 7:43337–43351.
2016.PubMed/NCBI
|
|
27
|
Adriaens C, Standaert L, Barra J, Latil M,
Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W,
et al: p53 induces formation of NEAT1 lncRNA-containing
paraspeckles that modulate replication stress response and
chemosensitivity. Nat Med. 22:861–868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Parasramka M, Yan IK, Wang X, Nguyen P,
Matsuda A, Maji S, Foye C, Asmann Y and Patel T: BAP1 dependent
expression of long non-coding RNA NEAT-1 contributes to sensitivity
to gemcitabine in cholangiocarcinoma. Mol Cancer. 16:222017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie Q, Lin S, Zheng M, Cai Q and Tu Y:
Long noncoding RNA NEAT1 promoted the growth of cervical cancer
cells via sponging miR-95p. Biochem Cell Biol. 97:100–108. 2018.
View Article : Google Scholar
|
|
30
|
Lujambio A, Calin GA, Villanueva A, Ropero
S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso
MS, Faller WJ, et al: A microRNA DNA methylation signature for
human cancer metastasis. Proc Natl Acad Sci USA. 105:13556–13561.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gwak JM, Kim HJ, Kim EJ, Chung YR, Yun S,
Seo AN, Lee HJ and Park SY: MicroRNA-9 is associated with
epithelial-mesenchymal transition, breast cancer stem cell
phenotype, and tumor progression in breast cancer. Breast Cancer
Res Treat. 147:39–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park YR, Lee ST, Kim SL, Liu YC, Lee MR,
Shin JH, Seo SY, Kim SH, Kim IH, Lee SO and Kim SW: MicroRNA-9
suppresses cell migration and invasion through downregulation of
TM4SF1 in colorectal cancer. Int J Oncol. 48:2135–2143. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gomez GG, Volinia S, Croce CM, Zanca C, Li
M, Emnett R, Gutmann DH, Brennan CW, Furnari FB and Cavenee WK:
Suppression of microRNA-9 by mutant EGFR signaling upregulates
FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res.
74:1429–1439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao HM, Wei W, Sun YH, Gao JH, Wang Q and
Zheng JH: MicroRNA-9 promotes tumorigenesis and mediates
sensitivity to cisplatin in primary epithelial ovarian cancer
cells. Tumour Biol. 36:6867–6873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pan Q, Sun L, Zheng D, Li N, Shi H, Song
J, Shao G and Xu G: MicroRNA-9 enhanced cisplatin sensitivity in
nonsmall cell lung cancer cells by regulating eukaryotic
translation initiation factor 5A2. Biomed Res Int.
2018:17690402018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Munoz JL, Rodriguez-Cruz V, Ramkissoon SH,
Ligon KL, Greco SJ and Rameshwar P: Temozolomide resistance in
glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic
hedgehog level. Oncotarget. 6:1190–1201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo F, Hou X and Sun Q: MicroRNA-9-5p
functions as a tumor suppressor in papillary thyroid cancer via
targeting BRAF. Oncol Lett. 16:6815–6821. 2018.PubMed/NCBI
|
|
38
|
Zhang L, Yan L, Cao M, Zhang H, Li C, Bai
Y, Yu P, Li M and Zhao X: SPAG9 promotes endometrial carcinoma cell
invasion through regulation of genes related to the
epithelial-mesenchymal transition. Eur J Gynaecol Oncol.
37:312–319. 2016.PubMed/NCBI
|
|
39
|
Ren B, Wei X, Zou G, He J, Xu G, Xu F,
Huang Y, Zhu H, Li Y, Ma G and Yu P: Cancer testis antigen SPAG9 is
a promising marker for the diagnosis and treatment of lung cancer.
Oncol Rep. 35:2599–2605. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jagadish N, Gupta N, Agarwal S, Parashar
D, Sharma A, Fatima R, Topno AP, Kumar V and Suri A:
Sperm-associated antigen 9 (SPAG9) promotes the survival and tumor
growth of triple-negative breast cancer cells. Tumour Biol.
37:13101–13110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ren B, Luo S, Xu F, Zou G, Xu G, He J,
Huang Y, Zhu H and Li Y: The expression of DAMP proteins HSP70 and
cancer-testis antigen SPAG9 in peripheral blood of patients with
HCC and lung cancer. Cell Stress Chaperones. 22:237–244. 2017.
View Article : Google Scholar :
|
|
42
|
Zhen Z, Dong F, Shen H, Wang QG, Yang L
and Hu J: MiR-524 inhibits cell proliferation and induces cell
apoptosis in thyroid cancer via targeting SPAG9. Eur Rev Med
Pharmacol Sci. 22:3812–3818. 2018.PubMed/NCBI
|
|
43
|
Yang C, Shen B, Zhang J and Zhang Q:
Sperm-associated antigen 9 overexpression correlates with poor
prognosis and insensitive to Taxol treatment in breast cancer.
Biomarkers. 21:62–67. 2016. View Article : Google Scholar
|