|
1
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar :
|
|
3
|
Taylor JM: Host RNA circles and the origin
of hepatitis delta virus. World J Gastroenterol. 20:2971–2978.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
8
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar :
|
|
10
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Zhou L, Ponnusamy M, Zhang L, Dong
Y, Zhang Y, Wang Q, Liu J and Wang K: A comprehensive review of
circRNA: From purification and identification to disease marker
potential. PeerJ. 6:e55032018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Zhu M, Pan J, Chen C, Xia S and
Song Y: Circular RNAs: A rising star in respiratory diseases.
Respir Res. 20:32019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S, Yang B, Chen BJ, Bliim N,
Ueberham U, Arendt T and Janitz M: The emerging role of circular
RNAs in transcriptome regulation. Genomics. 109:401–407. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schumann S, Jackson BR, Yule I, Whitehead
SK, Revill C, Foster R and Whitehouse A: Targeting the
ATP-dependent formation of herpesvirus ribonucleoprotein particle
assembly as an antiviral approach. Nat Microbiol. 2:162012016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aktaş T, Avşar Ilık İ, Maticzka D,
Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R and
Akhtar A: DHX9 suppresses RNA processing defects originating from
the Alu invasion of the human genome. Nature. 544:115–119. 2017.
View Article : Google Scholar
|
|
22
|
Chen D, Lu X, Yang F and Xing N: Circular
RNA circHIPK3 promotes cell proliferation and invasion of prostate
cancer by sponging miR-193a-3p and regulating MCL1 expression.
Cancer Manag Res. 11:1415–1423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar
|
|
24
|
Deng L, Liu G, Zheng C, Zhang L, Kang Y
and Yang F: Circ-LAMP1 promotes T-cell lymphoblastic lymphoma
progression via acting as a ceRNA for miR-615-5p to regulate DDR2
expression. Gene. 701:146–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P,
Liu Z and Li J: circ-BIRC6, a circular RNA, promotes hepatocellular
carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell
Cycle. 18:976–989. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Q, Li P, Wu M and Liu Q: Deregulation
of circular RNAs in cancer from the perspectives of aberrant
biogenesis, transport and removal. Front Genet. 10:162019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Zou J, Ma X, Wang E and Peng G:
Mechanisms and implications of ADAR-mediated RNA editing in cancer.
Cancer Lett. 411:27–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee T and Pelletier J: The biology of DHX9
and its potential as a therapeutic target. Oncotarget.
7:42716–42739. 2016.PubMed/NCBI
|
|
29
|
Goehe RW, Shultz JC, Murudkar C, Usanovic
S, Lamour NF, Massey DH, Zhang L, Camidge DR, Shay JW, Minna JD and
Chalfant CE: hnRNP L regulates the tumorigenic capacity of lung
cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin
Invest. 120:3923–3939. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gou Q, Wu K, Zhou JK, Xie Y, Liu L and
Peng Y: Profiling and bioinformatic analysis of circular RNA
expression regulated by c-Myc. Oncotarget. 8:71587–71596. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar
|
|
33
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mitra A, Pfeifer K and Park KS: Circular
RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer
Res. 7(Suppl 5): S624–S628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu L, Wang J, Khanabdali R, Kalionis B,
Tai X and Xia S: Circular RNAs: Isolation, characterization and
their potential role in diseases. RNA Biol. 14:1715–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leggio L, Vivarelli S, L'Episcopo F,
Tirolo C, Caniglia S, Testa N, Marchetti B and Iraci N: microRNAs
in Parkinson's disease: From pathogenesis to novel diagnostic and
therapeutic approaches. Int J Mol Sci. 18:E26982017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rothman A, Restrepo H, Sarukhanov V, Evans
WN, Wiencek RG Jr, Williams R, Hamburger N, Anderson K, Balsara J
and Mann D: Assessment of microRNA and gene dysregulation in
pulmonary hypertension by endoarterial biopsy. Pulm Circ.
7:455–464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Girardi E, López P and Pfeffer S: On the
importance of Host MicroRNAs during viral infection. Front Genet.
9:4392018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar
|
|
41
|
Quan G and Li J: Circular RNAs:
Biogenesis, expression and their potential roles in reproduction. J
Ovarian Res. 11:92018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pratt AJ and MacRae IJ: The RNA-induced
silencing complex: A versatile gene-silencing machine. J Biol Chem.
284:17897–17901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kluiver J, Gibcus JH, Hettinga C, Adema A,
Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ and
van den Berg A: Rapid generation of microRNA sponges for microRNA
inhibition. PLoS One. 7:e292752012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peng L, Yuan XQ and Li GC: The emerging
landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep.
33:2669–2674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu
CH, Shen MJ and Huang Q: Circular RNA ciRS-7 promotes the
proliferation and metastasis of pancreatic cancer by regulating
miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat
Dis Int. Mar 9;S1499-3872(19)30039-6. 2019.Epub ahead of print.
View Article : Google Scholar
|
|
48
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen G, Shi Y, Liu M and Sun J: circHIPK3
regulates cell proliferation and migration by sponging miR-124 and
regulating AQP3 expression in hepatocellular carcinoma. Cell Death
Dis. 9:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu H, Chen Y and Jiang P: Circular RNA
HIPK3 exerts oncogenic properties through suppression of miR-124 in
lung cancer. Biochem Biophys Res Commun. 506:455–462. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morgan EL, Wasson CW, Hanson L, Kealy D,
Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish
JL, et al: STAT3 activation by E6 is essential for the
differentiation-dependent HPV18 life cycle. PLoS Pathog.
14:e10069752018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
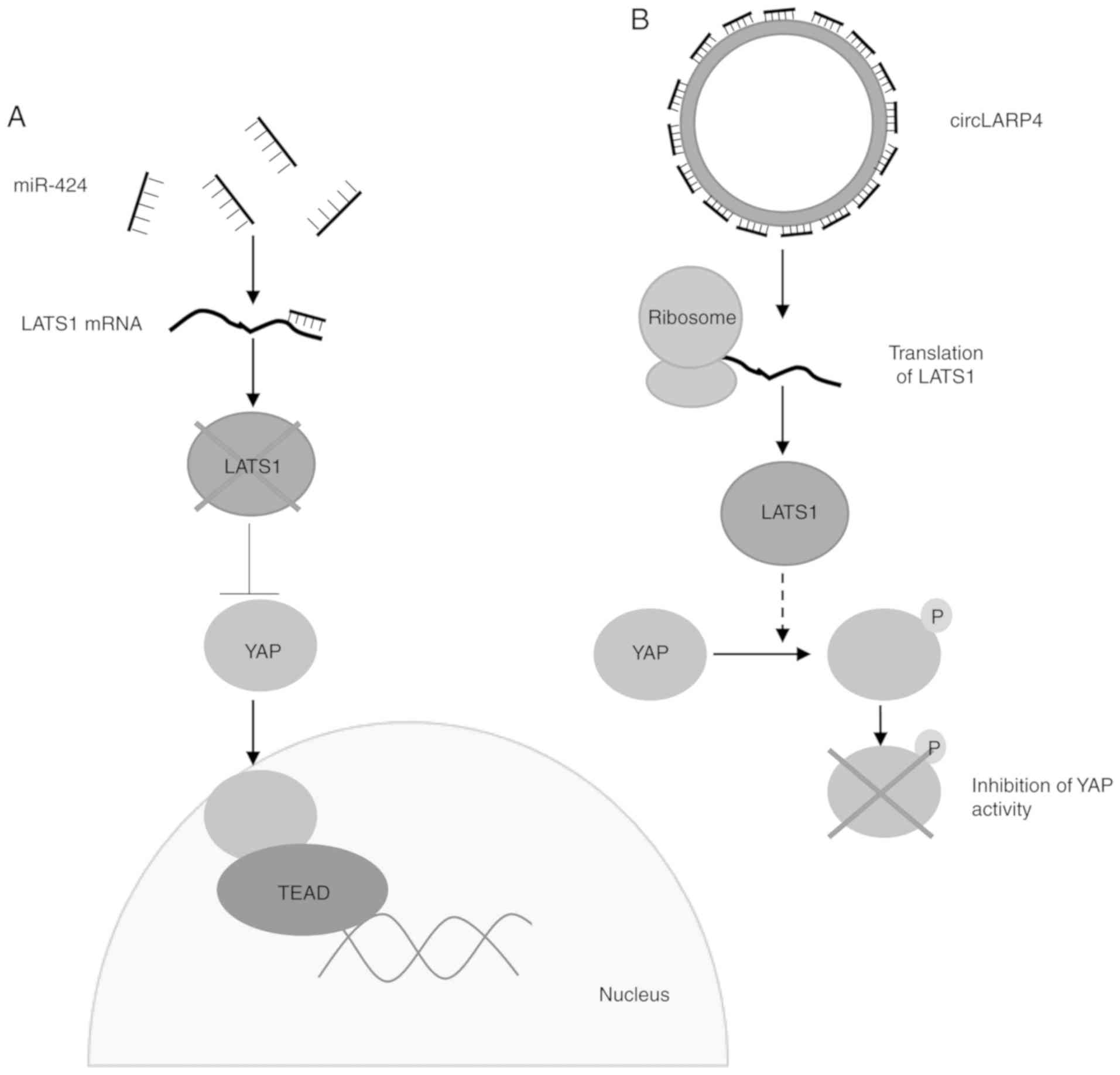
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang
L, Wu J and Wang X: circLARP4 induces cellular senescence through
regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular
carcinoma. Cancer Sci. 110:568–581. 2019. View Article : Google Scholar :
|
|
56
|
Levanon D, Bernstein Y, Negreanu V, Bone
KR, Pozner A, Eilam R, Lotem J, Brenner O and Groner Y: Absence of
Runx3 expression in normal gastrointestinal epithelium calls into
question its tumour suppressor function. EMBO Mol Med. 3:593–604.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar
|
|
58
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar :
|
|
61
|
Panda AC, Grammatikakis I, Kim KM, De S,
Martindale JL, Munk R, Yang X, Abdelmohsen K and Gorospe M:
Identification of senescence-associated circular RNAs (SAC-RNAs)
reveals senescence suppressor CircPVT1. Nucleic Acids Res.
45:4021–4035. 2017. View Article : Google Scholar :
|
|
62
|
Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan
H, Niu Y, Liu F, Gu H, Fan X and Gao J: Circular RNA circPVT1
promotes proliferation and invasion through sponging miR-125b and
activating E2F2 signaling in non-small cell lung cancer. Cell
Physiol Biochem. 51:2324–2340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu Z, Huang W, Wang X, Wang T, Chen Y,
Chen B, Liu R, Bai P and Xing J: Circular RNA CEP128 acts as a
sponge of miR-145-5p in promoting the bladder cancer progression
via regulating SOX11. Mol Med. 24:402018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017.PubMed/NCBI
|
|
65
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 18:802019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular functions and specific roles of circRNAs in the
cardiovascular system. Noncoding RNA Res. 3:75–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qin M, Wei G and Sun X: Circ-UBR5: An
exonic circular RNA and novel small nuclear RNA involved in RNA
splicing. Biochem Biophys Res Commun. 503:1027–1034. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Banerjee A, Apponi LH, Pavlath GK and
Corbett AH: PABPN1: Molecular function and muscle disease. FEBS J.
280:4230–4250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lapik YR, Fernandes CJ, Lau LF and Pestov
DG: Physical and functional interaction between Pes1 and Bop1 in
mammalian ribosome biogenesis. Mol Cell. 15:17–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
74
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu WY: Roles of the circular RNA
circ-Foxo3 in breast cancer progression. Cell Cycle. 16:589–590.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar :
|
|
77
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wesselhoeft RA, Kowalski PS and Anderson
DG: Engineering circular RNA for potent and stable translation in
eukaryotic cells. Nat Commun. 9:26292018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Floor SN and Doudna JA: Tunable protein
synthesis by transcript isoforms in human cells. Elife.
5:e109212016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol.
S1044-579X(18)30099-3. 2018.PubMed/NCBI
|
|
83
|
Luo GG and Ou JH: Oncogenic viruses and
cancer. Virol Sin. 30:83–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Toptan T, Abere B, Nalesnik MA, Swerdlow
SH, Ranganathan S, Lee N, Shair KH, Moore PS and Chang Y: Circular
DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA.
115:E8737–E8745. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ungerleider NA, Jain V, Wang Y, Maness NJ,
Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C, et al:
Comparative analysis of gammaherpesvirus circular RNA repertoires:
Conserved and unique viral circular RNAs. J Virol. 93:e01952–18.
2019.
|
|
86
|
Wang M, Yu F, Wu W, Wang Y, Ding H and
Qian L: Epstein-Barr virus-encoded microRNAs as regulators in host
immune responses. Int J Biol Sci. 14:565–576. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Qin Z, Jakymiw A, Findlay V and Parsons C:
KSHV-encoded MicroRNAs: Lessons for viral cancer pathogenesis and
emerging concepts. Int J Cell Biol. 2012:6039612012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ungerleider N, Concha M, Lin Z, Roberts C,
Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, et al: The
Epstein Barr virus circRNAome. PLoS Pathog. 14:e10072062018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tagawa T, Gao S, Koparde VN, Gonzalez M,
Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan
R and Ziegelbauer JM: Discovery of Kaposi's sarcoma
herpesvirus-encoded circular RNAs and a human antiviral circular
RNA. Proc Natl Acad Sci USA. 115:12805–12810. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shi J, Hu N, Li J, Zeng Z, Mo L, Sun J, Wu
M and Hu Y: Unique expression signatures of circular RNAs in
response to DNA tumor virus SV40 infection. Oncotarget.
8:98609–98622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Yan Y, Lin W, Li A, Zhang H, Lei
X, Dai Z, Li X, Li H, Chen W, et al: Circular RNA Vav3 sponges
gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol.
16:118–132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai
Z, Li X, Chen W, Lin W, Chen F, et al: Circular RNA alterations are
involved in resistance to avian leukosis virus subgroup-J-induced
tumor formation in chickens. Oncotarget. 8:34961–34970.
2017.PubMed/NCBI
|
|
93
|
Zhang Y, Zhang H, An M, Zhao B, Ding H,
Zhang Z, He Y, Shang H and Han X: Crosstalk in competing endogenous
RNA networks reveals new circular RNAs involved in the pathogenesis
of early HIV infection. J Transl Med. 16:3322018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang ZY, Guo ZD, Li JM, Zhao ZZ, Fu YY,
Zhang CM, Zhang Y and Liu LN, Qian J and Liu LN: Genome-wide search
for competing endogenous RNAs responsible for the effects induced
by Ebola virus replication and transcription using a trVLP system.
Front Cell Infect Microbiol. 7:4792017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shi J, Hu N, Mo L, Zeng Z, Sun J and Hu Y:
Deep RNA sequencing reveals a repertoire of human fibroblast
circular RNAs associated with cellular responses to Herpes simplex
virus 1 infection. Cell Physiol Biochem. 47:2031–2045. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tung KH, Ernstoff MS, Allen C and Shu S: A
review of exosomes and their role in the tumor microenvironment and
host-tumor 'Macroenvironment'. J Immunol Sci. 3:4–8. 2019.
View Article : Google Scholar :
|
|
97
|
Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng
J and Fan S: The roles of tumor-derived exosomes in non-small cell
lung cancer and their clinical implications. J Exp Clin Cancer Res.
37:2262018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tai YL, Chen KC, Hsieh JT and Shen TL:
Exosomes in cancer development and clinical applications. Cancer
Sci. 109:2364–2374. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang J, Zhang Q, Zhou S, Xu H, Wang D,
Feng J, Zhao J and Zhong S: Circular RNA expression in exosomes
derived from breast cancer cells and patients. Epigenomics.
11:411–421. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Bai H, Lei K, Huang F, Jiang Z and Zhou X:
Exo-circRNAs: A new paradigm for anticancer therapy. Mol Cancer.
18:562019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang Z, Yang T and Xiao J: Circular RNAs:
Promising biomarkers for human diseases. EBioMedicine. 34:267–274.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang M, Yu F and Li P: Circular RNAs:
Characteristics, function and clinical significance in
hepatocellular carcinoma. Cancers (Basel). 10:E2582018. View Article : Google Scholar
|
|
105
|
Huang Z, Su R, Qing C, Peng Y, Luo Q and
Li J: Plasma circular RNAs hsa_circ_0001953 and hsa_circ_0009024 as
diagnostic biomarkers for active tuberculosis. Front Microbiol.
9:20102018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
De Braekeleer E, Douet-Guilbert N and De
Braekeleer M: RARA fusion genes in acute promyelocytic leukemia: A
review. Expert Rev Hematol. 7:347–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Dal Molin A, Bresolin S, Gaffo E, Tretti
C, Boldrin E, Meyer LH, Guglielmelli P, Vannucchi AM, Te Kronnie G
and Bortoluzzi S: CircRNAs are here to stay: A perspective on the
MLL recombinome. Front Genet. 10:882019. View Article : Google Scholar :
|
|
108
|
Babin L, Piganeau M, Renouf B, Lamribet K,
Thirant C, Deriano L, Mercher T, Giovannangeli C and Brunet EC:
Chromosomal translocation formation is sufficient to produce fusion
circular RNAs specific to patient tumor cells. iScience. 5:19–29.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of Fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 166:1055–1056. 2016. View Article : Google Scholar : PubMed/NCBI
|