1. Introduction
Circular RNAs (circRNAs) are a group of non-coding
RNAs that in recent years have emerged as potential regulators of
various cell processes. It is proposed that they serve a range of
functions, including regulation of RNA-binding proteins (RBPs),
promotion of mRNA cis-transcription and post-transcriptional
regulation through sponging of microRNAs (miRNAs/miRs) (1). They may also interact with splicing
proteins or RNA polymerase II (RNA pol II) to aid in
transcriptional regulation (2).
Due to their numerous and varied regulatory roles, circRNAs have
been postulated to serve key roles in cancer, acting in both
oncogenic and tumour-suppressive manners. Although research
focusing on circRNAs in cancer remains in its infancy, circRNAs
have already been discovered to be dysregulated in a number of
different cancers, as well as other types of diseases.
RNA was first discovered to be able to form natural
circles >40 years ago in both plant viroids and in hepatitis δ
virus (3). In 1991, circRNAs were
found in human cells when circular isoforms with out-of-order exons
from the gene DCC (encoding 'deleted in colorectal cancer'
protein) were discovered, however, they were dismissed as a
by-product from errors in RNA processing and splicing (4). Throughout the 1990s and early 2000s,
increasing evidence for the existence of circRNAs in mammalian
cells, such as transcripts with out-of-order exons, were
discovered, although they were still considered to be low-abundance
and non-functional RNAs (5).
Improvements in RNA sequencing techniques have since revealed that
circRNAs are expressed at far higher levels than initially
considered, whereas the discovery in 2013 of a circRNA that was
capable of sponging miRNAs led to the increased interest in their
roles (6). This key finding
provided evidence that circRNAs serve a functional role as master
regulators of gene expression and are not simple by-products of
splicing (7).
2. Biogenesis
The biogenesis of circRNAs is yet to be fully
elucidated; however, most are formed through backsplicing. Unlike
canonical splicing, backsplicing results in the 3′ splice donor
site being joined to the upstream 5′ splice acceptor site forming a
closed circle (8). These circRNAs
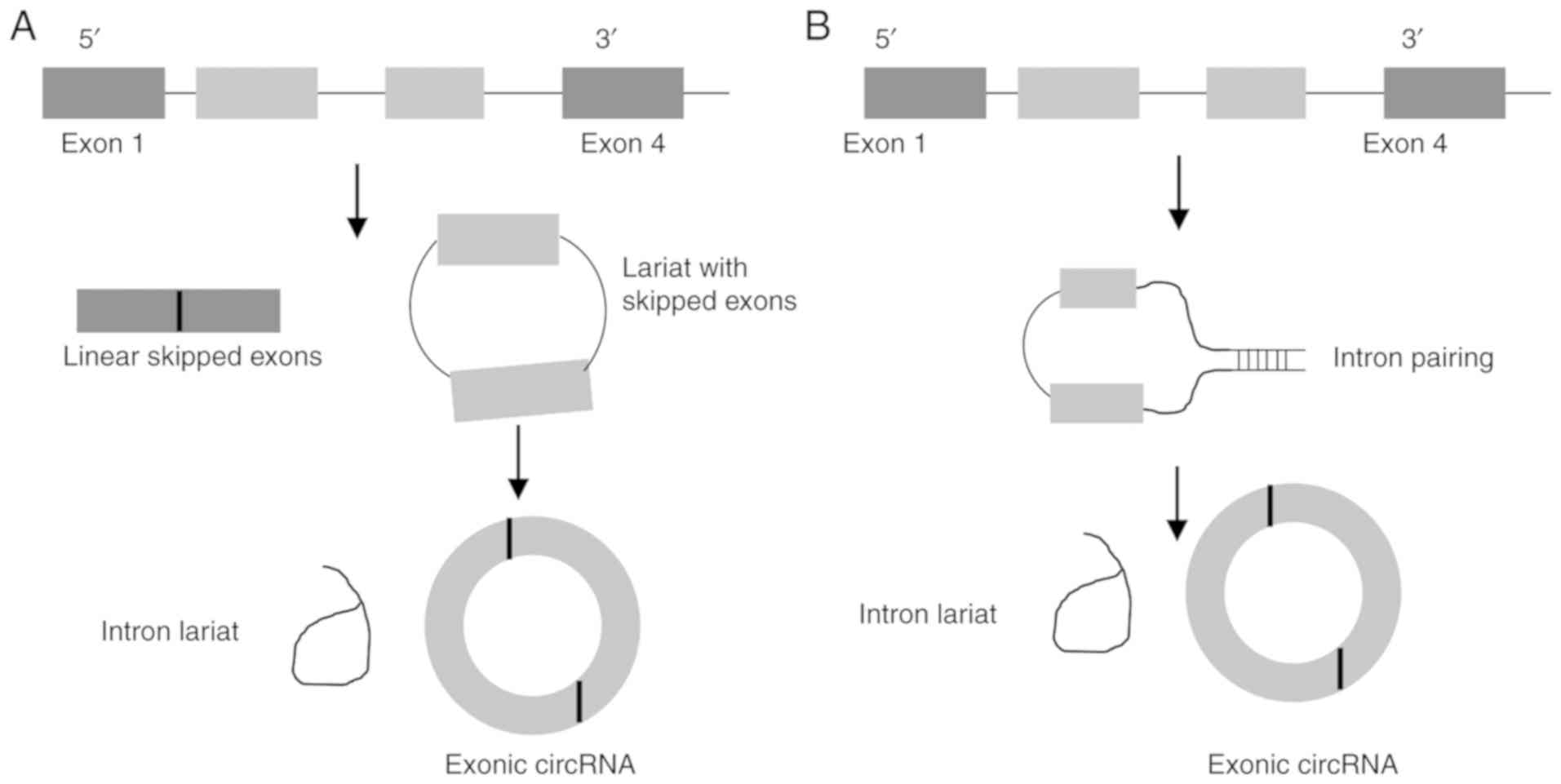
are characterised as either exonic circRNAs (Ecircs),
exonic-intronic circRNAs (EIcircs) or circular intronic circRNAs
(CIcircs) (9). The vast majority
of circRNAs discovered are Ecircs, with 2 main models for
formation: Intron-pairing or lariat-driven (Fig. 1) (10).
In intron-pairing, circularisation is driven by
flanking inverted repeats in the introns either side of the exons.
These repeats, particularly ALU elements, promote circRNA formation
through base pairing of the introns bringing backsplicing sites
closer together, enabling trans factors to facilitate
circularisation (11).
Lariat-driven formation occurs through exon skipping; this is a
form of alternative splicing where exons are spliced from the main
transcript, allowing greater diversity (12). These skipped exons can form a
lariat with the removed introns; under certain conditions, before
this lariat is degraded, it can undergo further splicing to remove
the introns, forming a circRNA from the spliced exons (13). Similarly to intron-pairing,
formation of a lariat drives circularisation through bringing the
backsplice sites closer together, although the specific cis
and trans factors that aid circularisation are not
known.
Similar to exon-containing circRNAs, intron
only-containing circRNA formation is promoted through cis
factors bringing splice sites into close proximity (14). With intron-containing circRNAs,
these cis factors contain two consensus motifs. These motifs
are a 7-nucleotide GU-rich element at the 5′ end and an
11-nucleotide C-rich element at the branch site; together, these
motifs bind and promote the formation of an intermediate circular
lariat. These lariats escape degradation before further processing
removes any exons or overhangs on the lariat, leading to the
formation of a stable intron-only circRNA (15).
3. Regulation and dysregulation of
circRNAs
The regulation of circRNAs during and after
transcription remains to be fully elucidated, with various complex
factors controlling their transcription and splicing. Specific RBPs
can promote circRNA formation; for example, the mannose-binding
lectin (Mbl) protein can bind to the flanking introns of circRNAs,
acting as a bridge to bring the splice sites closer and promote
circularisation (16). Another RBP
termed Quaking (QKI) has also been demonstrated to regulate circRNA
formation (17). QKI is
hypothesised to specifically regulate multiple circRNAs involved in
the epithelial-mesenchymal transition (EMT). QKI regulation,
similar to Mbl, requires binding motifs within the flanking introns
(17). These motifs are sufficient
to promote circularisation, with the addition of these motifs to
linear transcripts leading to circRNA formation. QKI protein binds
to these motifs, forming a dimer structure, bringing the splice
sites closer together to promote circularisation (17).
The nuclear export of Ecircs is also regulated,
although the mechanism is yet to be fully determined. A recent
study has suggested that the nuclear export of circRNAs is
size-dependent (18). Using an RNA
interference-mediated approach targeting known nuclear
export-related proteins, the impact on circRNA localization was
evaluated in Drosophila DL1 cells. Results showed that
depletion of the DExH/D-Box helicase, helicase at 25E (Hel25E),
resulted in nuclear accumulation of circRNAs which were >800
nucleotides in length. Of note, it was further demonstrated that
homologues of Hel25E, namely UAP56/DDX39B and URH49/DDX39A, have a
similar role in mammalian cells. Longer circRNAs are exported
through UAP56, whilst shorter circRNAs are exported through URH49
in humans (18). At present, the
protein which exports shorter circRNAs in Drosophila remains
to be determined. The regulation of circRNA nuclear export by these
RNA helicases may provide a novel therapeutic approach to prevent
circRNA dysregulation, as small molecule inhibitors are being
developed which inhibit UAP56 export ability (19).
Negative regulators of circRNA formation have also
been identified; these include the double-strand RNA editing enzyme
adenosine deaminase acting on RNA 1 (ADAR1) and the enzyme RNA
Helicase A (DHX9). Overexpression of ADAR leads to significantly
decreased levels of circRNAs; this mechanism is proposed to occur
due to disruption of base pairing of the flanking introns (20). DHX9 is hypothesised to have roles
in the regulation of ALU elements by binding and preventing
secondary structure formation (21). As ALU secondary structures aid in
promoting circularisation by bringing flanking introns in closer
proximity, through this binding, DHX9 can also suppress circRNA
formation (11). Furthermore,
other ALU element regulators, such as heterogeneous nuclear
riboprotein (hnRNP) C and hnRNP L, have also been shown to affect
circRNA expression (7).
Studies have found circRNAs are frequently
dysregulated in cancer, although exact mechanisms are not fully
elucidated (22-25). However, one known mechanism is
through mutations in flanking introns; certain cancers, such as
gastric cancer, often have flanking intron mutations; these can
lead to mismatches or shortenings of introns, leading to reduced
circRNA biogenesis (26).
Dysregulation of proteins that have functions in regulating circRNA
biogenesis has also been discovered; for example, ADAR is commonly
found dysregulated in cancers, from hepatocellular carcinoma (HCC)
and lung cancer to melanoma (27).
Through ADAR, base changes in flanking introns could lead to
dysregulated biogenesis. Both hnRNPs and DHX9 have also found to be
dysregulated in cancer, with both dysregulated in breast and lung
cancers (28,29).
There is also the potential for aberrant expression
of transcription factors or splicing factors in cancer affecting
circRNA levels. Although the role of splicing and transcription in
relation to circRNAs is not fully elucidated, key mutations could
lead to changes. For instance, the transcription factor C-myc
frequently acts an oncoprotein and is upregulated in cancer; it is
able to regulate specific circRNAs through myc-binding sites in
circRNA promoters (30).
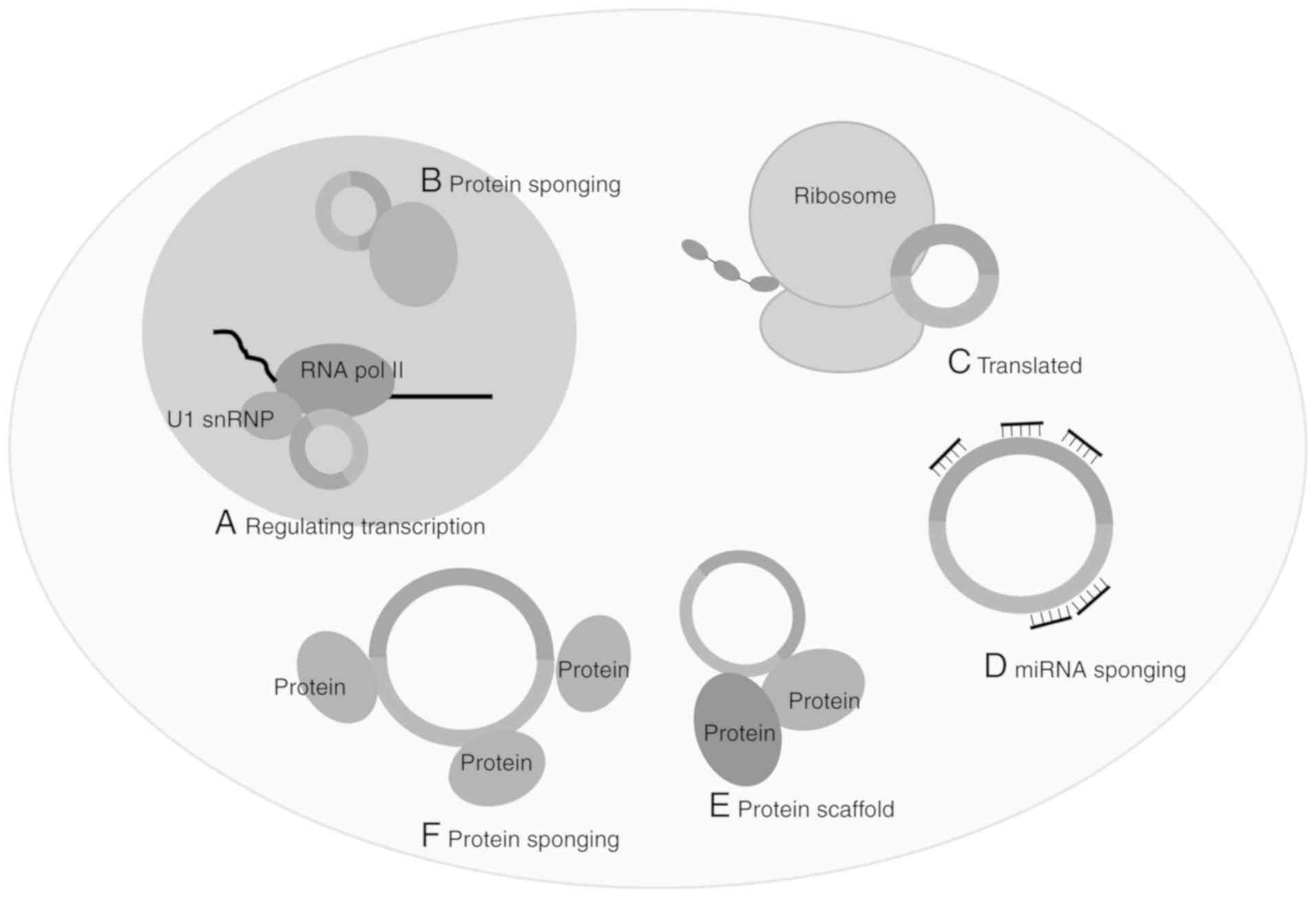
4. Functions
The proposed functions of circRNAs in regulating
gene expression include miRNA sponging, interaction with RBPs and
regulation of gene transcription (Fig.
2). Ecircs are proposed to carry out the majority of miRNA
sponging and interaction with RBPs, whereas CIcircs and EIcircs are
hypothesised to be involved in transcriptional control. Whether
this division of function is due to the composition of the circRNAs
or (more likely) their subcellular localisation, is unknown, with
Ecircs generally found in the cytoplasm and CI/EIcircs located in
the nucleus.
5. miRNA sponges
It is hypothesised that one of the main functions of
circRNAs is to act as miRNA sponges, acting as endogenous
regulators of miRNA activity (31). miRNAs are involved in
post-transcriptional regulation of gene expression through binding
to complementary mRNAs and instigating translational inhibition by
degradation or P-body storage of the mRNA (32). Through sponging of the miRNAs it is
hypothesised that circRNAs play a key role in regulating gene
expression and are often found to be dysregulated in diseases
associated with aberrant gene expression (33). Through sponging of miRNAs, circRNAs
act as crucial post-transcriptional regulators and play a key role
in a regulatory network involving circRNAs, miRNAs and mRNAs. As
they regulate through competition, circRNAs represent a novel class
of competing endogenous RNA (ceRNA) (34). Compared with other known ceRNAs,
circRNAs are more abundant and stable, and can contain a number of
binding sites for miRNAs, allowing efficient regulation (35). Due to this regulation, circRNAs may
play a key regulatory role in diseases associated with dysregulated
miRNAs, including cancer, Parkinson's disease, hypertension and
viral infection (36-38). This is supported by the fact that
numerous circRNAs have already been reported as dysregulated in
cancer; however, it must be noted that numerous circRNAs lack miRNA
binding sites, with one study finding only 12% of circRNAs studied
contained miRNA binding sites and an argonaute (AGO) footprint
(39). Furthermore, a number of
the circRNAs that do contain miRNA binding sites have far fewer
than the notable examples, such as ciRS-7 (also known as CDR1as)
and sex-determining region Y circRNA (40,41).
The first instance of circRNAs sponging miRNAs was
reported in 2013, when the circRNA derived from the CDR1
gene, ciRS-7, was shown to sponge miR-7 (6). ciRS-7 contains >70 binding sites
for miR-7, with 90% of its sequence composed of hexamer repeats
that comprise the miR-7 binding site (42). The use of
AGO-crosslinking/immunoprecipitation (IP) has been integral for the
confirmation of circRNAs as miRNA sponges. AGO is a key protein
member of the RNA-induced silencing complex (RISC) involved in
miRNA function (43).
miRNA-sponging circRNAs precipitate strongly with AGO2.
Furthermore, circRNAs are enriched in AGO-IPs upon over-expression
of target miRNAs (44). As such,
the target miRNA facilitates complex formation between the circRNA
and AGO2, enabling circRNA sponging ability and high occupancy of
AGO sites (6). ciRS-7 interacts
with miR-7, sequestering it from its target mRNA. Originally,
ciRS-7 was found to act in the neuronal cells of zebrafish embryos
and, by sponging miR-7, led to brain development defects via
dysregulation of the mTOR pathway (45). Outside the brain, other miR-7
target mRNAs include epidermal growth factor receptor (EGFR) and
X-linked inhibitor of apoptosis protein, two proteins frequently
dysregulated in cancer (46).
Furthermore, ciRS-7 has been found to be upregulated in pancreatic
ductal adeno-carcinoma (PDAC), where it aids in promoting
proliferation through the sponging of miR-7, leading to increased
activity of the EGFR/STAT3 pathway (47). By sponging miR-7, ciRS-7 acts as an
oncogenic circRNA, allowing increased levels of several proteins
that promote cell proliferation and negatively regulate apoptosis
(45).
One of the most extensively studied dysregulated
circRNAs is homeodomain interacting protein kinase 3 circRNA
(circHIPK3), a 1099-base pair circRNA formed from exon 2 of the
HIPK3 gene (48). Through
sequence analysis, it has been identified that circHIPK3 may
contain 18 binding sites for 12 different miRNAs, including binding
sites for miR-7, miR-29b and miR-124 that have been confirmed
(48-50). Through the sponging of miR-7,
circHIPK3 leads to an increase in the expression of focal adhesion
kinase (FAK), EGFR, YY1 transcription factor and insulin-like
growth factor 1 receptor (49).
These oncogenes can promote metastasis and tumour growth by
activating the PI3K/AKT and mitogen-activated protein kinase
kinase/ERK pathways, increasing cell proliferation and leading to
an increase in expression of pro-metastatic proteins, including
vascular endothelial growth factor, and metalloproteinase (MMP)2
and MMP9 (49). circHIPK3 also
sponges the tumour suppressor miR-124, leading to an increase in
the expression of miR-124 target mRNAs, including STAT3 and
cyclin-dependent kinase 4 (CDK4), both commonly found to be
upregulated in cancer (51). STAT3
is a transcription factor promoting cell growth through the
activation of pro-cell cycle progression proteins and
anti-apoptotic proteins, while CDK4 promotes cell cycle progression
(51,52). circHIPK3 has now been found to be
upregulated in a range of cancers, including HCC, and colorectal,
gastric, prostate and lung cancers, and may represent a novel
biomarker or therapeutic target (22,50,53).
Studies have shown some promise with silencing circHIPK3, reducing
the proliferation of cancer cells and inducing apoptosis, whereas
overexpression constructs promoted cell survival (49,51).
circRNAs can also act as tumour suppressors via the
sponging of oncomiRs, thereby decreasing the expression of
oncoproteins. La-related protein 4 circRNA (circLARP4) sponges
miR-424, leading to an increase in the expression of
serine/threonine-protein kinase LATS1, which in turn down-regulates
the pro-proliferative yes-associated protein (YAP) pathway
(Fig. 3) (54). Decreased expression of circLARP4
was found to be associated with poorer prognosis and survival in
gastric cancer (54). A second
study also reported that low circLARP4 expression was associated
with a poorer prognosis in HCC, with overexpression of circLARP4
leading to an increase in the levels of p53 and p21 (55). The study suggested that circLARP4
sponges miR-761, leading to upregulation of its target runt-related
transcription factor 3 (RUNX3), which in turn activates downstream
p53 and p21 signalling. RUNX3 is a transcription factor that has
previously been reported to function as a tumour suppressor, with
loss of expression associated with gastric cancer, although there
is a degree of controversy as to whether RUNX3 possesses
tumour-suppressive or oncogenic properties (56).
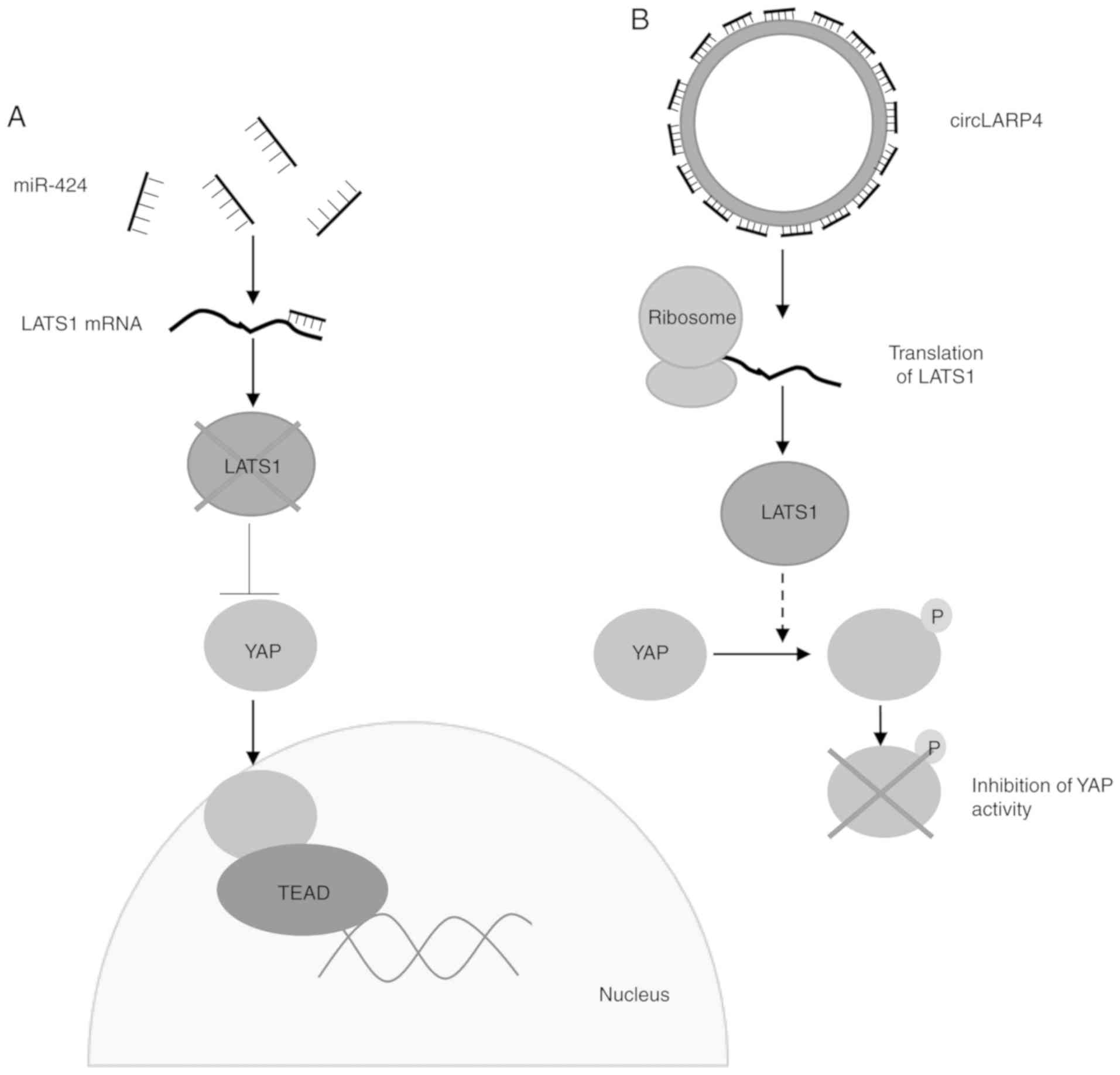 | Figure 3Proposed mechanisms for an
miRNA-sponging circRNA, circLARP4. (A) Low levels of circLARP4 lead
to increased miR-424, which binds to its target mRNA LATS1, leading
to reduced levels. Low levels of LATS1 allow YAP to translocate to
the nucleus, where it acts as a transcriptional coactivator through
associations with TEAD transcription factors, leading to cell
proliferation. (B) Increased circLARP4 sponges miR-424, which leads
to increased translation of LATS1. LATS1 phosphorylates YAP,
preventing its translocation to the nucleus and inhibiting its
activity. circRNA, circular RNA; LARP4, La-related protein 4;
miRNA/miR, microRNA; TEAD1, TEA domain family member 1; YAP,
yes-associated protein. |
An unusual example of post-transcriptional
regulation via circRNA sponging of miRNAs is itchy E3 ubiquitin
protein ligase circRNA (circITCH), as circITCH regulates the
expression of the linear transcript it is derived from: The
ITCH gene (57). circITCH
contains binding sites for miR-7, miR-17 and miR-214 (42). These miRNA seed sequences are found
in the 3′ untranslated region (UTR) of the linear ITCH
transcript, suggesting the miRNA may suppress linear ITCH's
expression. circITCH sponges these miRNAs, leading to an increase
in the mRNA expression of its linear form. ITCH functions as an E3
ubiquitin ligase, targeting proteins for degradation; one such
protein is dishevelled segment polarity protein 2 (Dvl2). Targeting
Dvl2 leads to a downregulation of the Wnt/β-catenin pathway
(57). By upregulating the levels
of ITCH mRNA levels (and therefore its protein levels) via miRNA
sponging, circITCH may exhibit tumour suppressive properties
(57,58). Further examples of miRNAs sponged
by circRNAs are presented in Table
I (24,25,59-65).
 | Table IExamples of known circRNA:miRNA:mRNA
regulatory networks. |
Table I
Examples of known circRNA:miRNA:mRNA
regulatory networks.
| circRNA | miRNA sponged | miRNA targets | Function
regulated | Associated
cancer |
|---|
| circAKT3 | miR-198 | PIK3R1 | Apoptosis, DNA
repair | Gastric (59) |
| circSRY | miR-138 | RhoC, TWIST2, H2AX,
Bcl-2, SOX4 | Migration and
invasion | CRC,
cholangiocarcinoma (60) |
| circPVT1 | miR-125a, miR-125b,
let-7 | HMGA2, KRAS,
E2F2 | Cell proliferation,
metastasis | Lung (61,62) |
| circCEP128 | miR-145 | SOX11 | Cell
proliferation | Bladder (63) |
| circUBAP2 | miR-143 | Bcl-2 | Apoptosis | Osteosarcoma
(64) |
| circLAMP1 | miR-615 | DDR2 | Cell growth and
apoptosis | T-cell lymphoma
(24) |
| circDLST | miR-502 |
NRAS/MEK1/ERK1/2 | Cell invasion, DNA
synthesis | Gastric (65) |
| circBIRC6 | miR-3918 | Bcl-2 | Apoptosis | HCC (25) |
6. Transcriptional regulators
A lesser-understood role of circRNAs involves acting
as transcriptional regulators. Intron-containing circRNAs are
retained in the nucleus, and are hypothesised to regulate the
transcription of genes. It is proposed that the circRNAs upregulate
their parental gene expression through the formation of a
circRNA-U1 small nuclear (sn)ribonucleoprotein complex that
interacts with RNA pol II (66).
Nuclear circRNAs can also interact with additional transcriptional
and splicing regulators, including U1A and U1C. It has been shown
that these circRNAs localise to the loci of their parental genes
and co-immunoprecipitate with their promoters, suggesting that they
may regulate their expression in cis (9). Conversely, they can also localise to
other areas, suggesting some trans regulatory mechanisms
(15,67). One circRNA proposed to regulate
transcription and splicing within the nucleus is E3
ubiquitin-protein ligase UBR5 circRNA (circUBR5). circUBR5 can bind
to QKI, Nova-1 and U1 snRNA, and is therefore hypothesised to serve
a regulatory role in splicing, although exact mechanisms remain to
be elucidated (68). Notably,
circUBR5 has been reported as downregulated in non-small cell lung
cancer.
7. Interactions with proteins
circRNAs also interact with proteins, acting as
scaffolds or sponges (also termed decoys); through this mechanism,
they can regulate crucial cellular processes, including ribosome
biogenesis and cell cycle progression (69,70).
Several of these protein-interacting circRNAs have also been
implicated in tumourigenesis. One such circRNA is poly(A) binding
protein nuclear 1 circRNA (circPABPN1); this can form complexes
with the RBP human antigen R (HuR), thereby competitively
preventing HuR from binding to PABPN1 mRNA (69). This suggests competition between
the linear and circular form of PABPN1 serves a role in regulating
their expression. PABN1 is required for efficient 3′ poly(A) tail
formation, and therefore mRNA nuclear export and mRNA stability
(71). circPABN1, through binding
and sponging of the HuR protein, reduces 3′ poly(A) tail formation,
leading to a reduction in mRNA export and stability.
Other circRNAs also interact with HuR, including
circAGO2; this binding enhances export of HuR to the cytoplasm,
where it binds to 3′ UTRs of mRNA, leading to reduced AGO2/RISC
complex binding and miRNA-mediated translational repression
(23). Through this mechanism,
circAGO2 promotes cancer metastasis and promotes tumourigenesis
both in vitro and in vivo. It has been found
upregulated in neuroblastoma, and gastric, colon and prostate
cancers, and is associated with a poor prognosis (23).
Protein-circRNA interactions can also aid in
suppressing tumourigenesis. Antisense non-coding RNA in the INK4
locus circRNA (circANRIL) binds to the c-terminal of pescadillo
ribosomal biogenesis factor 1 (PES1) (72). PES1 is an RBP that mediates the
processing of pre-mRNA through exonuclease activity and is
essential for 60S ribosomal assembly. Through binding to PES1,
circANRIL prevents its interactions with ribosomal RNA and impairs
biogenesis of the ribosomes, leading in turn to an increase in p53
expression and apoptosis (70).
One circRNA that has been thoroughly studied in
regards to protein binding is forkhead box O3 circRNA (circFOXO3),
which binds to numerous proteins aiding in reducing cell growth and
survival. In the cytoplasm, circFOXO3 binds to proteins including
FAK, hypoxia-inducible factor 1α, inhibitor of DNA binding 1 and
E2F transcription factor 1, preventing their translocation to the
nucleus and reducing their anti-cellular senescence functions
(73). It can also bind to CDK2
and p21, forming a tertiary complex and acting as a sponge for
CDK2, preventing its normal functions, which include enhancement of
cyclin A expression and promotion of G1/S transition (74). Therefore, increased expression of
circFOXO3 leads to decreased cell cycle progression.
circFOXO3 may also serve roles in cancer through its
regulation of FOXO3, with circFOXO3 increasing protein levels of
FOXO3. FOXO3 is considered to be a tumour suppressor, reducing cell
motility and invasion; reduced levels of FOXO3 lead to increases in
PTEN and increased AKT signalling, and FOXO3 is often reported as
downregulated in breast cancer (75). circFOXO3 increases FOXO3 levels by
binding to it and preventing its ubiquitination by mouse double
minute 2 homolog (MDM2). This in turn leads to increases in its
downstream target p53-upregulated modulator of apoptosis (PUMA),
promoting cell apoptosis (76).
Notably, circFOXO3 is also capable of acting as a scaffold for MDM2
and p53, promoting the ubiquitination-mediated degradation of p53
via MDM2, thereby repressing apoptosis. It is possible that by
promoting MDM2 and p53 interactions, circFOXO3 reduces the
interaction between MDM2 and FOXO3, leading to increases in PUMA
(7). Overexpression of circFOXO3
leads to cell apoptosis in vitro and reduces tumour growth
in vivo (76).
8. Translated circRNAs
Although defined as non-coding RNAs, there is some
evidence suggesting that certain circRNAs may be translated.
Originally, studies reported in principle and artificial systems,
that circRNAs may be able to be translated using an internal
ribosome entry site (IRES) (77,78).
In subsequent years, experimental evidence has further supported
this view. For example, it has been found that specific circRNAs
that contain the same start codon as the gene they are derived from
can associate with ribosomes (79). Secondly, a protein product derived
from the Muscleblind (Mbl) transcript was also discovered that
could only have originated from its circular form, translated using
an IRES in the 3′ UTR (77).
Another circRNA proposed be translated is zinc finger protein 609
circRNA (circZNF609). Similar to the translated Mbl circRNA,
circZNF609 contains the same start codon as its linear transcript
and an in-frame stop codon; the transcript was reported to
associate with ribosomes and be translated in a cap-independent
manner using an IRES (79). Of
note, Transcript Isoform in Polysomes sequencing showed 22% of all
circRNAs investigated were associated with ribosomal fractions,
suggesting that circRNAs may exhibit translatable activity
(80). This polysome profiling
also found that circRNAs in the ribosomal fractions mostly
accumulated in the lighter fractions, suggesting circRNAs generally
associate with fewer ribosomes along their length than linear mRNAs
(39,80). However, it must be noted that the
vast majority of circRNAs appear to not be translated. These two
translated circRNAs (Mb1 and circZNF609) are also found in brain
and muscle cells, respectively, where circRNAs are expressed at
higher levels; therefore, abundance of circRNA may contribute to
transcript expression (77,79).
In addition, the RNA modification N6-methyladenosine
(m6A) was found to be enriched in these circRNAs
(81). This modification aids in
translation through recruitment of translation initiation factors,
including eukaryotic translation initiation factor 4 γ 2 and YTH
domain-containing family protein 3 (82).
9. Oncogenic viruses and circRNAs
Viruses are estimated to be responsible for ~20% of
all global cancer cases, with 7 viruses currently classified as
human oncogenic viruses: Hepatitis B virus (HBV); hepatitis C
virus; Merkel cell polyomavirus; Epstein-Barr virus (EBV); Kaposi's
sarcoma (KS)-associated herpesvirus (KSHV); human papilloma virus;
and human T-lymphotrophic virus 1 (83). Of these viruses, circRNAs have been
reported to be encoded by EBV and KSHV (84,85).
In all viruses, only DNA viruses have been discovered to encode
circRNAs, with members of the herpesviridae family thus far
dominating. This is not unexpected, due to their larger genome size
and complexity; herpesvirus genomes are known to undergo the
complex splicing necessary for the biogenesis of circRNAs. Both EBV
and KSHV also encode their own miRNAs; EBV encodes at least 44
miRNAs, whereas KSHV encodes 18 (86,87).
This suggests that both EBV and KSHV, by encoding their own miRNAs
and circRNAs, seek to dysregulate the cellular non-coding RNA
network.
EBV expresses circRNAs from its BART locus; these
circRNAs include both Ecircs and EIcircs. These transcripts were
found to be expressed in cells latently infected with EBV,
including Burkitt's lymphoma and nasopharyngeal cancer cell lines
(84), suggesting a potential role
in EBV oncogenesis. Other circRNAs encoded by EBV may be expressed
both in the latent and lytic lifecycles. For example, circBHLF1 was
found to be expressed at low levels during latency with a large
upregulation during reactivation, suggesting a potential role in
regulating the lytic/latent switch, whereas circRPMS1_E4_ E3a and
circRPMS1_E4_E2 were both found to be expressed in EBV-positive
stomach cancer tissues (88).
KSHV has been found to constitutively express a
circRNA from its vIRF4 locus, as well as hundreds of low-copy
circRNAs from its PAN locus (84).
The circRNAs originating from the PAN locus were highly variable,
and the authors suggested that they may be merely artefacts from
processing PAN RNA due to the high copy number.
Notably, when searching for circRNAs encoded by
KSHV, the miRNAs these circRNA may sponge were found to be enriched
in pathways involved with the regulation of apoptosis and cancer,
suggesting the potential for regulating oncogenic processes
(89). These circRNAs were found
to be expressed during lytic replication and played roles in
altering cell replication. Using seed sequence analysis, the KSHV
circRNAs also exhibited the potential to sponge virally-encoded
miRNAs in addition to cellular miRNAs. These circRNAs were found in
KSHV-infected endothelial cells and from primary effusion lymphoma
(PEL) cells. Furthermore, these circRNAs were also discovered in
the lymph nodes of patients suffering from KSHV-associated
malignancies, including KS, PEL and multi-centric Castleman's
disease (89). Finally, there is
the potential for other herpesviruses to encode circRNAs, with the
monkey homologue for EBV, rhesus macaque lymphocryptovirus, and
another γ-herpesvirus, murine γ-herpesvirus 68, both reported to
encode circRNAs (85).
As well as encoding their own circRNAs, viruses have
been shown to dysregulate cellular circRNAs to aid their survival
and replication. The oncogenic virus, simian virus 40 (SV40), was
reported to dysregulate 134 cellular circRNAs during infection, of
which 103 were upregulated (90).
These circRNAs were found to be involved in regulation pathways
crucial to oncogenesis, including the p53 and Wnt/β-catenin
pathways. In addition, a potential circRNA:miRNA: mRNA regulatory
network was identified, suggesting that SV40 is capable of
dysregulating non-coding RNAs to aid viral manipulation of cellular
gene expression, supporting viral replication (90).
As well as oncogenic DNA viruses dysregulating
cellular circRNAs to aid in tumourigenesis, the retrovirus avian
sarcoma leukosis virus (ASLV) has been found to dysregulate
circVav3 (91). ASLV causes cancer
in various types of bird, including chickens, pheasants and quails.
circVav3 was found to be upregulated in liver tumours from
ASLV-infected birds, where it acted as a sponge for gga-miR-375,
leading to an increase in the expression of downstream mRNA
targets. Among these targets is the transcription factor YAP1,
which promotes cell proliferation and suppresses apoptosis. This
sponging also led to increases in EMT activity and several
EMT-associated markers, including N-cadherin and MMP2 (91). Furthermore, studies have shown
differences in circRNA expression in ASLV-resistant and
ASLV-susceptible birds, suggesting that circRNAs may either aid in
suppressing oncogenesis or in tumour induction, depending on
expression profiles (92).
Cellular circRNAs have also been reported as
dysregulated by non-oncogenic viruses, suggesting that the
mechanisms underlying virus-mediated dysregulation of circRNAs are
varied and understudied, and that dysregulation of circRNAs is
important in a range of diseases. For example, in early HIV
infection, >1,300 circRNAs have been shown to be dysregulated,
with 67 circRNAs proposed to serve a role in HIV replication
(93). An RNA regulatory network
of >500 dysregulated circRNAs, 900 dysregulated mRNAs and 21
miRNAs was also discovered. Similarly, a circRNA:miRNA network has
also been identified during Ebola virus infection (94). Finally, herpes simplex virus 1
dysregulates >500 circRNAs and >207 miRNAs, once again
emphasising the potential for dysregulation of the circRNA:miRNA
axis to attenuate cellular gene transcription (95).
10. circRNAs and exosomes
Exosomes are nanosized extracellular vesicles that
transport a wide variety of molecules, from nucleic acids and
lipids to proteins and hormones, with crucial roles in cell
communication. Exosomes have also been implicated in tumourigenesis
through roles in mediating the tumour microenvironment, modulating
the immune response and aiding metastasis (96). Mechanisms behind these roles
including transporting extracellular matrix remodelling enzymes and
growth promoting factors such as EGFR (97). Exosomes have also been shown to
deliver miRNAs that promote oncogenesis, including miR-9 and
miR-105 (98).
Recent studies have provided evidence that exosomes
may also transport and deliver circRNAs extracellularly; these so
called exocircRNAs may also have roles in both tumourigenesis and
tumour suppression through delivery of circRNAs. CDR1as, the first
functional circRNA discovered, has been shown to be delivered to
distant cells in exosomes and once delivered to new cells, reduces
the cellular levels of miR-7, inhibiting cell growth.
Furthermore, these exo-circRNAs may also act as
functional biomarkers in a range of cancers. circRNAs were first
shown to exist in exosomes in 2015, with >1,000 discovered in
human serum exosomes alone, with the ratio of circRNAs to their
respective linear transcripts found to be enriched in a number of
exosomes (99). These exo-circRNA
populations only mildly correlate with cellular populations,
suggesting a separate mechanism for circRNA regulation or
transportation into exosomes from the main cellular environment.
Studies have shown that exo-circRNAs can be excreted by tumours
into serum and fluid throughout the body; these exo-circRNAs have
been isolated, and significant differences were found between their
circRNA compositions compared with normal healthy cells (100,101). Tumour exosomes from colorectal
cancer had 257 novel circRNAs, and were missing 67 compared to
exosomes from healthy tissues (99).
Specific exo-circRNA functions in cancer have also
been investigated. Metastatic PDAC cells have been found to excrete
a circRNA in exosomes, phosphodiesterase 8A circRNA (circPDE8A)
(102). Increased levels of this
circRNA are associated with poor prognosis, increased metastasis
and increased lymphatic invasion. Once the exosome delivers
circPDE8A to new cells, it can sponge miR-338. Downregulation of
miR-338 leads to an increased in expression of
metastasis-associated in colon cancer protein 1, in turn leading to
activation of the MET/ERK and AKT signalling pathways, promoting
cell growth and cell survival (102). This circRNA, once secreted into
blood plasma inside of exosomes, was able to be detected, and may
not only be a biomarker for PDAC cancer, but suggest the severity
of the cancer. Specific exo-circRNAs have also been discovered to
be associated with HCC, gastric and urothelial cancer (101).
11. Biomarkers
Although research into circRNAs remains in its
infancy, these molecules possess the potential to become novel
biomarkers for a range of diseases, from cancer to viral
infections, due to their inherent properties. circRNAs are highly
stable due to their resistance to exonucleases, and have a
half-life of >48 h compared with mRNA's half-life of ~10 h
(103). They are often disease-,
tissue- and developmental stage-specific, allowing a precise
diagnosis and even a prognosis. Furthermore, due to the
identification of circRNAs in exosomes, this may enable the
detection of disease-specific circRNAs in the blood, thereby
allowing the detection of tumours early on through blood testing
without the invasiveness associated with solid biopsies. As well as
blood, circRNAs have been found enriched in a variety of body
fluids including saliva, urine and cerebrospinal fluid (99). circRNAs have also been touted to be
biomarkers in cardiovascular disease, neurological disease and
diabetes (103). Infections also
induce changes in cellular circRNAs, which could be used as a
biomarkers; chronic HBV infection leads to the dysregulation of at
least 99 circRNAs (104), whereas
Mycobacterium Tuberculosis has been reported to dysregulate
at least 61 cellular circRNAs during active infection (105). Of these, hsa_circ_0001953 and
hsa_circ_0009024 have been suggested to be viable biomarkers, both
displaying strong associations with active TB infection.
12. Fusion circRNAs (f-circRNAs)
The phenomenon of chromosomal translocation is a
well-studied area in cancer biology, with numerous examples of
translocations leading to the aberrant expression of oncogenic
fusion proteins, most notably Bcr-Abl and promyelocytic
leukaemia-retinoic acid receptor α (PML-RARα) (106). However, the effects chromosomal
translocations have on the expression of circRNAs is yet to be
determined. Recently, these f-circRNAs have been implicated in
several cancers and onco-genic processes. These fusion circRNAs
have two junctions, the junction where two genes far apart in the
genome have joined together and the backsplice junction. It has
been shown that in tumours caused by chromosomal translocations,
50% expressed f-circRNAs, including PML-RARα and nucleo-phosmin
1-anaplastic lymphoma kinase (NPM1-ALK) (107). NPM1-ALK-associated cancers have
been shown to express f-circRNAs; these f-circRNAs are uniquely
expressed only in the transformed cells and were not detected in
wild-type cells (108). An
f-circRNA, f-circAF9, was not only expressed in haematopoietic stem
cells, but also led to a proliferative advantage and contributed to
transformation of these cells, suggesting these f-circRNAs may
serve active roles during oncogenic processes (109).
13. Future prospects
Long dismissed as simple by-products and errors in
splicing, research into the roles and functions of circRNAs has
shown that not only do they serve key roles in regulating gene
expression in normal cells, but that their dysregulation can
contribute to tumour development. Through their roles in miRNA
sponging, protein sponging and gene expression, it is clear that
circRNAs play an important role and further investigation is
required. However, despite recent breakthroughs, a number of
questions remain concerning circRNAs, with gaps in knowledge in all
areas of circRNA research, particularly biogenesis, regulation and
function. Regarding circRNA biogenesis, factors that aid
circularisation, including longer flanking introns, nucleotide
repeats and cellular RBPs have been identified; however, more
unidentified factors undoubtedly remain and require further
characterisation. Furthermore, the specific process of biogenesis
remains poorly understood, with conflicting evidence suggesting
circRNAs are synthesised both co- and post-transcriptionally. For
example, circRNAs are often transcribed at a faster rate than
linear RNAs, implying that regulation of RNA pol II and therefore
its transcriptional speed may also play a role in their biogenesis
(39). The export of Ecircs from
the nucleus is also not fully elucidated. Initial research suggests
specific cellular export factors, such as UAP56, regulate this
process, but again which specific factors need to be identified.
Finally, the stability of circRNAs is a crucial area that must be
further investigated, as it contributes substantially to their
function. Initial studies suggest that they are resistant to
various RNases and capable of being exosomal markers (14). As such, several questions remain,
namely how cells regulate and control their degradation. This is
particularly relevant in respect to diseases such as cancer that
can lead to the downregulation of specific circRNAs. As further
research is conducted, the impact of circRNAs in cell regulation
and in diseases, such as cancer, should be revealed. In particular,
the potential for circRNAs to act as biomarkers or targets of novel
therapeutic approaches merits further investigation.
Acknowledgments
The authors would like to thank all members of the
Whitehouse laboratory for useful discussions.
Funding
This work was supported in part by grants from the
White Rose BBSRC Doctoral Training Partnership in Mechanistic
Biology (grant no. 95519935) and MRC DiMeN Doctoral Training
Partnership (grant no. 95505183).
Availability of data and materials
Not applicable.
Authors' contributions
All authors contributed to designing and drafting
this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar :
|
|
3
|
Taylor JM: Host RNA circles and the origin
of hepatitis delta virus. World J Gastroenterol. 20:2971–2978.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
8
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar :
|
|
10
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Zhou L, Ponnusamy M, Zhang L, Dong
Y, Zhang Y, Wang Q, Liu J and Wang K: A comprehensive review of
circRNA: From purification and identification to disease marker
potential. PeerJ. 6:e55032018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Zhu M, Pan J, Chen C, Xia S and
Song Y: Circular RNAs: A rising star in respiratory diseases.
Respir Res. 20:32019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S, Yang B, Chen BJ, Bliim N,
Ueberham U, Arendt T and Janitz M: The emerging role of circular
RNAs in transcriptome regulation. Genomics. 109:401–407. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schumann S, Jackson BR, Yule I, Whitehead
SK, Revill C, Foster R and Whitehouse A: Targeting the
ATP-dependent formation of herpesvirus ribonucleoprotein particle
assembly as an antiviral approach. Nat Microbiol. 2:162012016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aktaş T, Avşar Ilık İ, Maticzka D,
Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R and
Akhtar A: DHX9 suppresses RNA processing defects originating from
the Alu invasion of the human genome. Nature. 544:115–119. 2017.
View Article : Google Scholar
|
|
22
|
Chen D, Lu X, Yang F and Xing N: Circular
RNA circHIPK3 promotes cell proliferation and invasion of prostate
cancer by sponging miR-193a-3p and regulating MCL1 expression.
Cancer Manag Res. 11:1415–1423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar
|
|
24
|
Deng L, Liu G, Zheng C, Zhang L, Kang Y
and Yang F: Circ-LAMP1 promotes T-cell lymphoblastic lymphoma
progression via acting as a ceRNA for miR-615-5p to regulate DDR2
expression. Gene. 701:146–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P,
Liu Z and Li J: circ-BIRC6, a circular RNA, promotes hepatocellular
carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell
Cycle. 18:976–989. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Q, Li P, Wu M and Liu Q: Deregulation
of circular RNAs in cancer from the perspectives of aberrant
biogenesis, transport and removal. Front Genet. 10:162019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Zou J, Ma X, Wang E and Peng G:
Mechanisms and implications of ADAR-mediated RNA editing in cancer.
Cancer Lett. 411:27–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee T and Pelletier J: The biology of DHX9
and its potential as a therapeutic target. Oncotarget.
7:42716–42739. 2016.PubMed/NCBI
|
|
29
|
Goehe RW, Shultz JC, Murudkar C, Usanovic
S, Lamour NF, Massey DH, Zhang L, Camidge DR, Shay JW, Minna JD and
Chalfant CE: hnRNP L regulates the tumorigenic capacity of lung
cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin
Invest. 120:3923–3939. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gou Q, Wu K, Zhou JK, Xie Y, Liu L and
Peng Y: Profiling and bioinformatic analysis of circular RNA
expression regulated by c-Myc. Oncotarget. 8:71587–71596. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar
|
|
33
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mitra A, Pfeifer K and Park KS: Circular
RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer
Res. 7(Suppl 5): S624–S628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu L, Wang J, Khanabdali R, Kalionis B,
Tai X and Xia S: Circular RNAs: Isolation, characterization and
their potential role in diseases. RNA Biol. 14:1715–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leggio L, Vivarelli S, L'Episcopo F,
Tirolo C, Caniglia S, Testa N, Marchetti B and Iraci N: microRNAs
in Parkinson's disease: From pathogenesis to novel diagnostic and
therapeutic approaches. Int J Mol Sci. 18:E26982017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rothman A, Restrepo H, Sarukhanov V, Evans
WN, Wiencek RG Jr, Williams R, Hamburger N, Anderson K, Balsara J
and Mann D: Assessment of microRNA and gene dysregulation in
pulmonary hypertension by endoarterial biopsy. Pulm Circ.
7:455–464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Girardi E, López P and Pfeffer S: On the
importance of Host MicroRNAs during viral infection. Front Genet.
9:4392018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar
|
|
41
|
Quan G and Li J: Circular RNAs:
Biogenesis, expression and their potential roles in reproduction. J
Ovarian Res. 11:92018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pratt AJ and MacRae IJ: The RNA-induced
silencing complex: A versatile gene-silencing machine. J Biol Chem.
284:17897–17901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kluiver J, Gibcus JH, Hettinga C, Adema A,
Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ and
van den Berg A: Rapid generation of microRNA sponges for microRNA
inhibition. PLoS One. 7:e292752012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peng L, Yuan XQ and Li GC: The emerging
landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep.
33:2669–2674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu
CH, Shen MJ and Huang Q: Circular RNA ciRS-7 promotes the
proliferation and metastasis of pancreatic cancer by regulating
miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat
Dis Int. Mar 9;S1499-3872(19)30039-6. 2019.Epub ahead of print.
View Article : Google Scholar
|
|
48
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen G, Shi Y, Liu M and Sun J: circHIPK3
regulates cell proliferation and migration by sponging miR-124 and
regulating AQP3 expression in hepatocellular carcinoma. Cell Death
Dis. 9:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu H, Chen Y and Jiang P: Circular RNA
HIPK3 exerts oncogenic properties through suppression of miR-124 in
lung cancer. Biochem Biophys Res Commun. 506:455–462. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morgan EL, Wasson CW, Hanson L, Kealy D,
Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish
JL, et al: STAT3 activation by E6 is essential for the
differentiation-dependent HPV18 life cycle. PLoS Pathog.
14:e10069752018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang
L, Wu J and Wang X: circLARP4 induces cellular senescence through
regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular
carcinoma. Cancer Sci. 110:568–581. 2019. View Article : Google Scholar :
|
|
56
|
Levanon D, Bernstein Y, Negreanu V, Bone
KR, Pozner A, Eilam R, Lotem J, Brenner O and Groner Y: Absence of
Runx3 expression in normal gastrointestinal epithelium calls into
question its tumour suppressor function. EMBO Mol Med. 3:593–604.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar
|
|
58
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar :
|
|
61
|
Panda AC, Grammatikakis I, Kim KM, De S,
Martindale JL, Munk R, Yang X, Abdelmohsen K and Gorospe M:
Identification of senescence-associated circular RNAs (SAC-RNAs)
reveals senescence suppressor CircPVT1. Nucleic Acids Res.
45:4021–4035. 2017. View Article : Google Scholar :
|
|
62
|
Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan
H, Niu Y, Liu F, Gu H, Fan X and Gao J: Circular RNA circPVT1
promotes proliferation and invasion through sponging miR-125b and
activating E2F2 signaling in non-small cell lung cancer. Cell
Physiol Biochem. 51:2324–2340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu Z, Huang W, Wang X, Wang T, Chen Y,
Chen B, Liu R, Bai P and Xing J: Circular RNA CEP128 acts as a
sponge of miR-145-5p in promoting the bladder cancer progression
via regulating SOX11. Mol Med. 24:402018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017.PubMed/NCBI
|
|
65
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 18:802019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular functions and specific roles of circRNAs in the
cardiovascular system. Noncoding RNA Res. 3:75–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qin M, Wei G and Sun X: Circ-UBR5: An
exonic circular RNA and novel small nuclear RNA involved in RNA
splicing. Biochem Biophys Res Commun. 503:1027–1034. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Banerjee A, Apponi LH, Pavlath GK and
Corbett AH: PABPN1: Molecular function and muscle disease. FEBS J.
280:4230–4250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lapik YR, Fernandes CJ, Lau LF and Pestov
DG: Physical and functional interaction between Pes1 and Bop1 in
mammalian ribosome biogenesis. Mol Cell. 15:17–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
74
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu WY: Roles of the circular RNA
circ-Foxo3 in breast cancer progression. Cell Cycle. 16:589–590.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar :
|
|
77
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wesselhoeft RA, Kowalski PS and Anderson
DG: Engineering circular RNA for potent and stable translation in
eukaryotic cells. Nat Commun. 9:26292018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Floor SN and Doudna JA: Tunable protein
synthesis by transcript isoforms in human cells. Elife.
5:e109212016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol.
S1044-579X(18)30099-3. 2018.PubMed/NCBI
|
|
83
|
Luo GG and Ou JH: Oncogenic viruses and
cancer. Virol Sin. 30:83–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Toptan T, Abere B, Nalesnik MA, Swerdlow
SH, Ranganathan S, Lee N, Shair KH, Moore PS and Chang Y: Circular
DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA.
115:E8737–E8745. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ungerleider NA, Jain V, Wang Y, Maness NJ,
Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C, et al:
Comparative analysis of gammaherpesvirus circular RNA repertoires:
Conserved and unique viral circular RNAs. J Virol. 93:e01952–18.
2019.
|
|
86
|
Wang M, Yu F, Wu W, Wang Y, Ding H and
Qian L: Epstein-Barr virus-encoded microRNAs as regulators in host
immune responses. Int J Biol Sci. 14:565–576. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Qin Z, Jakymiw A, Findlay V and Parsons C:
KSHV-encoded MicroRNAs: Lessons for viral cancer pathogenesis and
emerging concepts. Int J Cell Biol. 2012:6039612012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ungerleider N, Concha M, Lin Z, Roberts C,
Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, et al: The
Epstein Barr virus circRNAome. PLoS Pathog. 14:e10072062018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tagawa T, Gao S, Koparde VN, Gonzalez M,
Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan
R and Ziegelbauer JM: Discovery of Kaposi's sarcoma
herpesvirus-encoded circular RNAs and a human antiviral circular
RNA. Proc Natl Acad Sci USA. 115:12805–12810. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shi J, Hu N, Li J, Zeng Z, Mo L, Sun J, Wu
M and Hu Y: Unique expression signatures of circular RNAs in
response to DNA tumor virus SV40 infection. Oncotarget.
8:98609–98622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Yan Y, Lin W, Li A, Zhang H, Lei
X, Dai Z, Li X, Li H, Chen W, et al: Circular RNA Vav3 sponges
gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol.
16:118–132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai
Z, Li X, Chen W, Lin W, Chen F, et al: Circular RNA alterations are
involved in resistance to avian leukosis virus subgroup-J-induced
tumor formation in chickens. Oncotarget. 8:34961–34970.
2017.PubMed/NCBI
|
|
93
|
Zhang Y, Zhang H, An M, Zhao B, Ding H,
Zhang Z, He Y, Shang H and Han X: Crosstalk in competing endogenous
RNA networks reveals new circular RNAs involved in the pathogenesis
of early HIV infection. J Transl Med. 16:3322018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang ZY, Guo ZD, Li JM, Zhao ZZ, Fu YY,
Zhang CM, Zhang Y and Liu LN, Qian J and Liu LN: Genome-wide search
for competing endogenous RNAs responsible for the effects induced
by Ebola virus replication and transcription using a trVLP system.
Front Cell Infect Microbiol. 7:4792017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shi J, Hu N, Mo L, Zeng Z, Sun J and Hu Y:
Deep RNA sequencing reveals a repertoire of human fibroblast
circular RNAs associated with cellular responses to Herpes simplex
virus 1 infection. Cell Physiol Biochem. 47:2031–2045. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tung KH, Ernstoff MS, Allen C and Shu S: A
review of exosomes and their role in the tumor microenvironment and
host-tumor 'Macroenvironment'. J Immunol Sci. 3:4–8. 2019.
View Article : Google Scholar :
|
|
97
|
Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng
J and Fan S: The roles of tumor-derived exosomes in non-small cell
lung cancer and their clinical implications. J Exp Clin Cancer Res.
37:2262018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tai YL, Chen KC, Hsieh JT and Shen TL:
Exosomes in cancer development and clinical applications. Cancer
Sci. 109:2364–2374. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang J, Zhang Q, Zhou S, Xu H, Wang D,
Feng J, Zhao J and Zhong S: Circular RNA expression in exosomes
derived from breast cancer cells and patients. Epigenomics.
11:411–421. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Bai H, Lei K, Huang F, Jiang Z and Zhou X:
Exo-circRNAs: A new paradigm for anticancer therapy. Mol Cancer.
18:562019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang Z, Yang T and Xiao J: Circular RNAs:
Promising biomarkers for human diseases. EBioMedicine. 34:267–274.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang M, Yu F and Li P: Circular RNAs:
Characteristics, function and clinical significance in
hepatocellular carcinoma. Cancers (Basel). 10:E2582018. View Article : Google Scholar
|
|
105
|
Huang Z, Su R, Qing C, Peng Y, Luo Q and
Li J: Plasma circular RNAs hsa_circ_0001953 and hsa_circ_0009024 as
diagnostic biomarkers for active tuberculosis. Front Microbiol.
9:20102018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
De Braekeleer E, Douet-Guilbert N and De
Braekeleer M: RARA fusion genes in acute promyelocytic leukemia: A
review. Expert Rev Hematol. 7:347–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Dal Molin A, Bresolin S, Gaffo E, Tretti
C, Boldrin E, Meyer LH, Guglielmelli P, Vannucchi AM, Te Kronnie G
and Bortoluzzi S: CircRNAs are here to stay: A perspective on the
MLL recombinome. Front Genet. 10:882019. View Article : Google Scholar :
|
|
108
|
Babin L, Piganeau M, Renouf B, Lamribet K,
Thirant C, Deriano L, Mercher T, Giovannangeli C and Brunet EC:
Chromosomal translocation formation is sufficient to produce fusion
circular RNAs specific to patient tumor cells. iScience. 5:19–29.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of Fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 166:1055–1056. 2016. View Article : Google Scholar : PubMed/NCBI
|