Introduction
New gastric cancer (GC) cases have declined by 40%
worldwide over the past 50 years (1-3).
However, GC continues to be a significant public health concern
(1-3). GC is the third most common type of
cancer and the second leading cause of cancer-associated mortality
worldwide (4,5). It has been estimated that globally,
GC accounts for 989,600 new cases and 738,000 deaths annually,
rendering its case-fatality ratio higher compared with other common
malignancies, including colon, breast and prostate cancer (3).
Extracellular vesicles (EVs) are small membrane sacs
that are released by a variety of cell types under normal or
pathological conditions (6).
Previously known as 'cellular dust', EVs are now recognized as
important mediators during cell-to-cell communication (6). It has been demonstrated that EVs
transport bioactive cargo between cells in the form of
transmembrane receptors, mRNAs, microRNAs (miRNAs or miRs) and
signaling molecules, which are able to modify the extracellular
microenvironment (6-10). For example, cancer EVs carrying
transforming growth factor β (TGF-β) have been indicated to induce
fibroblast differentiation into cancer associated fibroblasts with
increased levels of α-smooth muscle actin (11). A previous study also demonstrated
that exosomes containing extracellular matrix metalloprotease (MMP)
inducer, which is released by lung carcinoma cells, enhanced MMPs
in fibroblasts and led to tumor progression and metastasis
(12). Wieckowski et al
(13) demonstrated that EVs
isolated from head and neck squamous cell carcinoma cells expressed
melanoma-associated antigen 3/6 and Fas ligand, and were able to
induce apoptosis in CD8+ T lymphocytes. The
aforementioned studies indicated that cancer cells may release EVs
to escape immunological surveillance by exerting a direct effect on
normal cells. There is increasing evidence that cancer cells may
also communicate via autologous EV release to impair an effective
anti-tumor response from the immune system. Gamperl et al
(14) revealed that microparticles
containing tissue factor that were isolated from pancreatic
adenocarcinoma cells were able to induce cell migration in tumor
cells by activating the protease activated receptor 2 (PAR2)
signaling pathway. An additional study demonstrated that EVs
derived from pancreatic cancer cells led to the down-regulation of
Hes-1 transcription factor, an intracellular target of the Notch-1
signaling pathway, which was indicated to induce the apoptosis of
these cells (15). Al-Nedawi et
al (16) reported that
glioblastoma (GMB)-derived EVs containing epithelial growth factor
receptor variant III (EGFRvIII) were transferred from
EGFRvIII-positive GMB cells to EGFRvIII-negative GMB cells, which
led to the activation of the mitogen-activated protein kinase
(MAPK) and Akt signaling pathways, and resulted in the expression
of vascular endothelial growth factor (VEGF).
The subject of the current study was the human GC
cell line, GC1415 (patent pending), of Caucasian origin that was
previously established and characterized in our laboratory
(17). The aim of the present
study was to investigate whether the autologous tumor-derived
microvesicles (auto-TMVs) released by GC1415 cells are capable of
altering the following: i) Gene expression; ii) chemotactic
potential; iii) signal transduction; and iv) cellular respiration
in GC1415 cells that are exposed to them.
Materials and methods
Isolation of TMVs
Cells of the previously characterized GC1415 cell
line (17) were cultured using
bi-weekly passages in DMEM (GE Healthcare) supplemented with 5% FBS
(Biowest), which was centrifuged at 50,000 × g for 1 h at room
temperature to remove bovine EVs. The cells were regularly tested
for Mycoplasma sp. contamination using a PCR-ELISA kit
(Roche Diagnostics GmbH) and for endotoxin contamination using a
Limulus test (Charles River Laboratories, Inc.), according to the
manufacturers' protocols. Supernatants from cell cultures were
collected at ~90% confluency and centrifuged using a modified
protocol that was previously described (18). Briefly, the collected supernatants
were spun at 3,000 × g for 10 min at room temperature to remove
cellular debris and transferred to a new Eppendorf-type test tube
and spun again for 30 min at 13,000 × g at room temperature.
Supernatants were subsequently transferred to new Eppendorf-type
test tube and centrifuged for 2 h at 100,000 × g at 4°C.
Supernatants were then discarded and the remaining pellets were
re-suspended in 100 µl filtered (0.22 µm) PBS. The
auto-TMV protein concentration was obtained according to the
Bradford method's protocol using Quick Start Bradford Dye reagent
(Bio-Rad Laboratories, Inc.). Isolated auto-TMVs were tested for
endotoxin contamination using a Limus test and stored at -20°C
until further use. Based on our previous studies, the quantity of
auto-TMVs used in the subsequent experiments was determined to be 3
µg (18,19).
Nanoparticle tracking analysis (NTA)
Average size, modal size and size distribution of
auto-TMVs were obtained using the NANOSIGHT LM10-HS488FT14
Nanoparticle Characterization system (Malvern Instruments, Ltd.) as
previously described (18). A
total of 1 µl auto-TMV suspension was then diluted to
1:1,000 in filtered (0.22 µm) PBS to obtain the total sample
volume of 1 ml. Subsequently, ~700 µl of the sample was
loaded manually into the measuring chamber using a 1-ml
insulin-type syringe (Terumo Corp.) and the syringe was mounted
onto the pump to deliver the sample at a constant flow rate of 80
units. Subsequently, three 1-min videos were recorded using the
sCMOS camera for each sample and analyzed using NanoSight NTA 3.1
analytical software (Malvern Instruments, Ltd.).
Scanning electron microscopy (SEM)
For morphological and size analyses, auto-TMVs were
fixed using 2.5% glutar-aldehyde at room temperature for 10 min and
particles were washed twice in PBS using an ultracentrifugation
step for 30 min at 100,000 × g and 4°C. The auto-TMV pellet was
then re-suspended in 100 µl PBS, and a 20-µl
suspension was applied to the double-sided adhesive conductive
carbon tape (Agar Scientific, Ltd.) and allowed to air-dry for ~1 h
at room temperature. The air-dried auto-TMVs were then subjected to
dehydration using gentle, stepwise washing with ethanol at 50, 70,
90 and 100%. Imaging was performed using a Tescan Vega 3 scanning
electron microscope (magnification, 80.1 kx) equipped with a
tungsten cathode (TESCAN), which was set at the acceleration
voltage of 10 kV, and a secondary electron detector.
RT-qPCR arrays
For mRNA gene expression, the GC1415 cells
(1×106) were suspended in 500 µl of the DMEM (GE
Healthcare) cell medium supplemented with 5% FBS (Biowest) and
exposed to auto-TMVs (3 µg) for 2 h (37°C; 5%
CO2). The same number of GC1415 cells cultured under the
same culturing conditions but without the exposure to auto-TMVs was
used a control. Total RNA isolation and first-strand cDNA synthesis
were subsequently performed as previously described (20). The generated cDNA was used for the
analyses of mRNA gene expression (in duplicate) using the
pathway-focused (angiogenesis, tumor metastasis, oncogene and tumor
suppressor genes) RT2 Profiler PCR Arrays (Qiagen GmbH)
according to the manufacturer's protocol. A total of 25 µl
prepared PCR component mix, which included 5 µl cDNA, was
added to each well of a 96-well PCR array plate and capped with cap
strips. A PCR reaction was then performed (initial denaturation for
10 min at 95°C, then 15 sec at 95°C and 1 min at 60°C for 40
cycles) using the 7300 Real Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Following each PCR run, a melting
curve analysis was performed using 7300 System SDS software version
1.2.3 (Applied Biosystems; Thermo Fisher Scientific, Inc.) to
verify the amplified PCR product. The obtained PCR array data were
analyzed using the on-line Data Analysis Center (Qiagen GmbH).
Chemotaxis assay
The chemotaxis assay was performed using the μ-slide
chemotaxis set (ibidi GMbH) according to the manufacturer's
protocol. At 1 day prior to the experiment, all components of the
μ-slide chemotaxis set, including the slide, caps, plugs and cell
medium (DMEM + 5% FBS) were placed in a CO2 incubator
(37°C; 5% CO2) overnight for gas equilibration.
Additionally, the microscope component of the HoloMonitor M4 system
(Phase Holographic Imaging AB) was switched on and left overnight
inside the CO2 incubator to achieve parameter stability.
The following day, a total of 6 µl GC1415 cell suspension
(~3×106 cells/ml) was applied to the μ-slide seeding
channel via its filling port. The μ-slide was then placed into a
Petri dish with a sterile wet tissue surrounding the slide and was
left in the CO2 incubator for 3 h until cell seeding was
achieved. Following cell seeding, a total of 65 µl cell
medium was applied to each reservoir located on the sides of the
μ-slide seeding channel, according to the manufacturer's protocol.
A total of 30 µl cell medium containing auto-TMVs (3
µg) was introduced into one of the reservoirs to achieve a
uniform auto-TMVs gradient. All filling ports were closed with
plugs apart from those of the cell seeding channel, which were
covered with caps. The μ-slide was then mounted onto the microscope
stage inside the CO2 incubator with the cells in the
cell seeding channel focused under a X10 objective, and a 24-h
observation interval was set to capture images every 5 min.
Following a period of 24 h, the recordings were analyzed using the
M4 Studio tracking software version 2.6.2. (Phase Holographic
Imaging AB).
Western blot analysis
For protein expression analyses, GC1415 cells
(1×106 cells) alone or with auto-TMVs (3 µg) were
incubated for 0.5, 2 and 24 h (37°C; 5% CO2) in a
CO2 incubator. Cell lysates were then prepared and
electrophoresis was performed, as previously described (21). A total of 20 µg lysed
protein of each sample was heated (70°C; 10 min) and
electrophoresis was performed in 14% polyacrylamide gel. The
proteins were then transferred onto PVDF membranes, blocked (room
temperature; 1 h) using Tris-Buffered Saline with 0.1% Tween and 1%
bovine serum albumin (TTBS) and incubated (overnight; 4°C) with the
following monoclonal antibodies (mAbs; all, 1:1,000): Rabbit
anti-phospho-AKT (cat. no. 4060S), anti-phospho-p44/42 MAP kinase
(cat. no. 9106S) and anti-GAPDH (cat. no. 2118; all, Cell Signaling
Technology, Inc.). The membranes were subsequently washed in TTBS
and incubated with secondary goat anti-rabbit antibody (cat. no.
7074S; Cell Signaling Technology, Inc.; 1:4,000) conjugated with
horseradish peroxidase (HRP) at room temperature for 1 h. The
protein bands were visualized using SuperSignal West Pico PLUS
Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.) and
analyzed with the Kodak Gel Logic 1500 imaging system (Kodak).
Where applicable, densitometric analysis was performed using the
Kodak Molecular Imaging software version 4.5.0 (Kodak).
Additionally, lysates obtained from auto-TMVs alone (20 µg)
were tested for the presence of exosomal markers, including Alix,
CD9 and CD63, using the same protocol as aforementioned. The
following mAbs (all, 1:1,000) were used: Mouse anti-Alix (cat. no.
2171; Cell Signaling Technology, Inc.), rabbit anti-CD9 (cat. no.
13174; Cell Signaling Technology, Inc.) and rabbit anti-CD63 (cat.
no. sc5275; Santa-Cruz Biotechnology, Inc.). The following
secondary antibodies were used (all HRP-conjugated, 1:4,000): for
Alix detection- horse anti-mouse (cat. no. 7076P2, Cell Signaling
Technology, Inc.) and for CD9 and CD63 detection-goat anti-rabbit
(cat. no. 7074S, Cell Signaling Technology, Inc.). Visualization of
the respective proteins was performed as aforementioned.
Cellular respiration
For the assessment of mitochondrial respiration, the
oxygen consumption rate (Ocr) was measured using the Seahorse XF
analyzer and the Seahorse XF Cell Mito Stress Test, according to
the manufacturer's protocol (Agilent Technologies, Inc.). The
GC1415 cells (2×104) were suspended in the DMEM cell
culture medium supplemented with 5% FBS and transferred into wells
of Cell Culture Miniplates (Agilent Technologies, Inc.) to allow
overnight cell seeding under standard conditions (37°C; 5%
CO2). To achieve the required stability, the chambers of
the Extracellular Flux Cartridge (Agilent Technologies, Inc.) were
filled with the XF Calibrant (Agilent Technologies, Inc.) and
placed overnight in an incubator (37°C) without CO2.
After a period of 24 h, the culture medium was carefully aspirated
and discarded, and the GC1415 cells were washed twice with 200
µl pre-warmed (37°C) stabilized Seahorse Medium (Agilent
Technologies, Inc.) that was prepared according to the
manufacturer's protocol. A total of 180 µl Seahorse Medium
was added to each well containing the washed GC1415 cells, and the
Cell Culture Miniplate was then placed in an incubator (37°C)
without CO2 for 1 h. Using the Seahorse Medium,
concentrations of auto-TMVs (3 µg), oligomycin,
p-triflouro-methoxyphenylhydrazone and rotenone/antimycin A
(all from Agilent Technologies, Inc.) mix were prepared and loaded
into the appropriate chambers of the calibrated Extracellular Flux
Cartridge, which was then placed into the Seahorse XF analyzer. The
stabilized Cell Culture Miniplate containing the GC1415 tumor cells
was then placed into the Seahorse XF analyzer and the Acute
Injection Seahorse XF Cell Mito Stress Test protocol was started
with the initial injection of auto-TMVs. The GC1415 cells from each
of the wells were subsequently lysed using the M-PER reagent
(Thermo Fisher Scientific, Inc.) and their protein concentration
was assessed (Bradford assay) for normalization. All the results
were analyzed using Wave software version 2.6.0.31 (Agilent
Technologies, Inc.).
Statistical analysis
All the data in the present study were analyzed
using GraphPad software version 5.0 (GraphPad Software, Inc.) and
are presented as the means ± standard error of the mean. An
unpaired Student's t-test (western blot densitometric analysis) and
one-way ANOVA followed by Dunnett's post hoc test (Ocr analysis)
were used to assess the differences among groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characterization of auto-TMVs
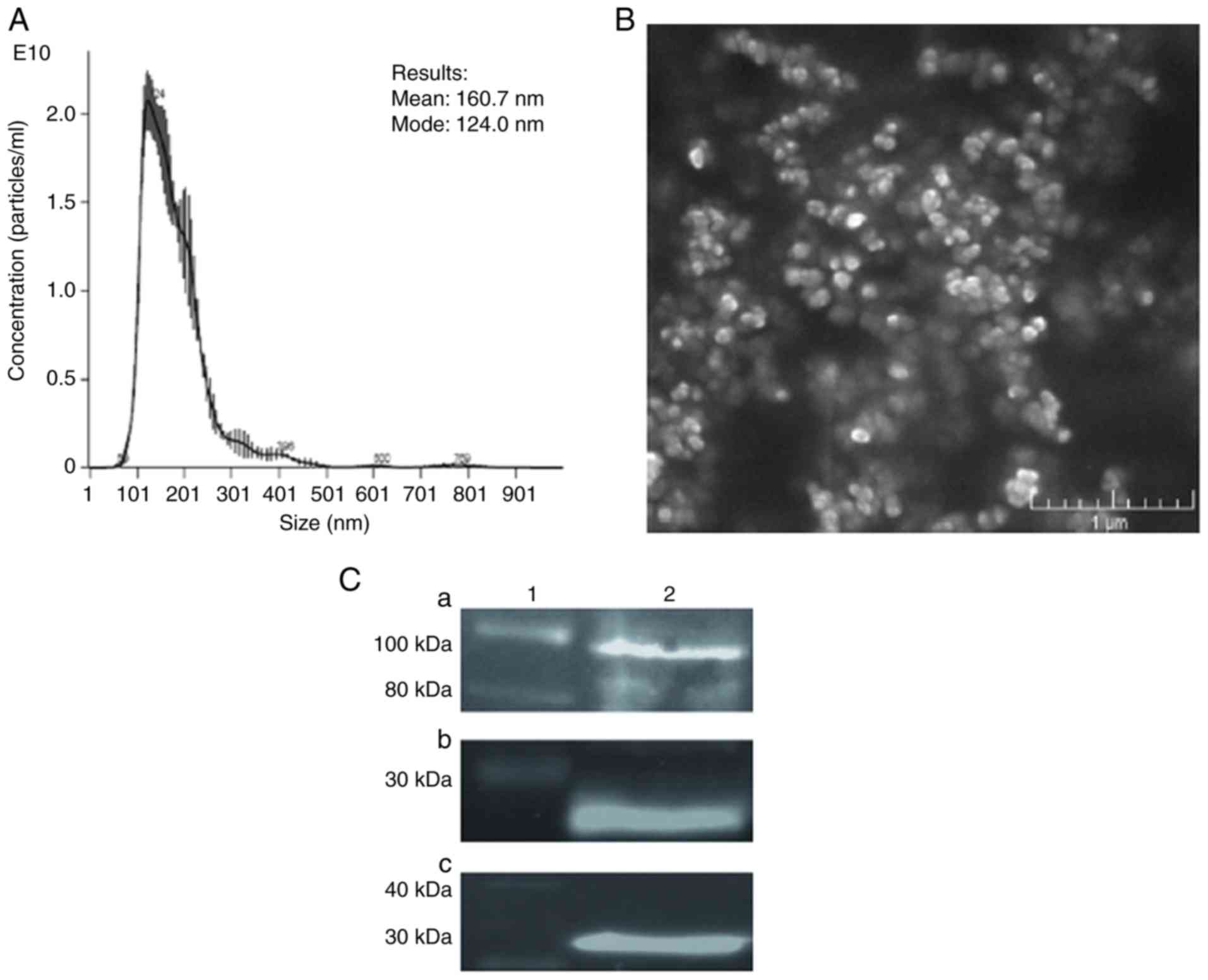
The NTA indicated the size range of auto-TMVs to be
90-800 nm, demonstrating that the predominant population of studied
vesicles was composed of microvesicles (Fig. 1A), which, according to the position
statement of the International Society for Extracellular Vesicles
on minimal information for studies of extracellular vesicles 2018
guidelines, are defined as medium/large EVs (22). The average size of the analyzed
auto-TMVs was 160.7 nm, and their modal size was 124 nm. An image
of auto-TMVs obtained using SEM confirmed these results (Fig. 1B). Western blot analysis revealed
the presence of endosomal/cytosolic markers (Alix and CD9; Fig. 1C) in auto-TMVs, indicating that
they are composed, in part, of a population that exhibits an
intracellular origin.
PCR arrays
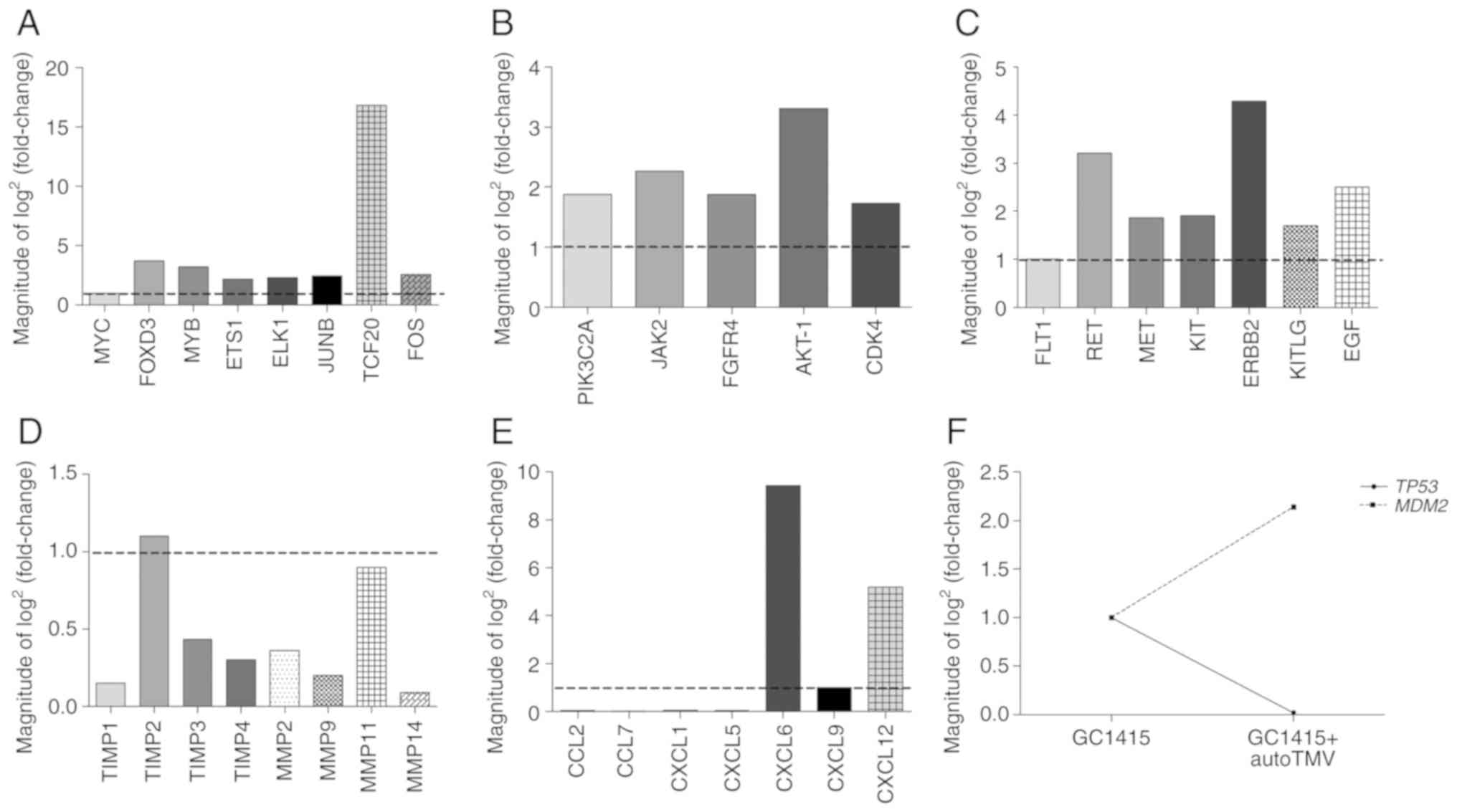
The gene expression data identified the
overexpression of 75 genes and the underexpression of 96 genes in
GC1415 cells following interaction with auto-TMVs. These genes
included oncogenes, tumor suppressor genes and genes associated
with angiogenesis and metastasis. A number of the overexpressed
genes included genes that code for transcription factors or their
co-activators [including MYC, forkhead box D3
(FOXD3), MYB, ETS Proto-Oncogene 1 (ETS1), ETS
Transcription Factor ELK1 (ELK1), JUNB,
transcription factor protein 20 (TCF20) and FOS;
Fig. 2A], with TCF20
exhibiting the highest fold increase (16.85). Another set of
overexpressed genes included genes that code for proteins with
kinase activity [including phosphatidylinositol-4-phosphate
3-kinase catalytic subunit type 2 alpha (PIK3C2A), Janus
kinase 2 (JAK-2), fibroblast growth factor receptor 4
(FGFR4), AKT-1 and CDK4; Fig. 2B], receptor tyrosine kinases
(RTKs), including [Fms related tyrosine kinase 1 (FLT1),
RET, MET, KIT and Erb-B2 receptor tyrosine kinase 2
(ERBB2); Fig. 2C] and their
potential ligands [including KIT ligand (KITLG) and
epidermal growth factor (EGF); Fig. 2C]. In the protein kinase group, the
AKT-1 gene exhibited the highest fold increase (3.31),
ERBB2 indicated the highest fold increase in the RTK group
(4.29) and EGF exhibited the highest fold increase in the
RTKs ligands group (2.5) compared to the control. A large number of
affected transcripts comprised of tissue inhibitors of
metalloproteinases (TIMPs; including TIMP-1, -2, -3 and
-4; Fig. 2D) and matrix
metalloproteinases (MMPs; including MMP-2, -9, -11 and
-14; Fig. 2D). In the
current study, all the transcripts were downregulated apart from
TIMP-2, which was slightly elevated with a fold increase at
1.10. The mRNA expression of chemokine ligands presented a more
challenging scenario. In this case, a number of genes were
underexpressed [including C-C motif chemokine ligand (CCL)2,
CCL7, C-X-C motif chemokine ligand (CXCL)1 and
CXCL5; Fig. 2E], while
other genes were overexpressed (including CXCL6, CXCL9 and
CXCL12; Fig. 3E). The obtained
data also indicated an association between the tumor suppressor
protein p53 (TP53), which was downregulated (6.79) and mouse
double minute 2 homolog (MDM2), which was upregulated (3.35;
Fig. 2F). Other mRNA transcripts
that were substantially overexpressed in the GC1415 cells following
contact with auto-TMV, including methionine aminopeptidase 2
[(METAP2); 7.33-fold increase], interleukin-1 β
[(IL1-β); 7.95-fold increase] and nitric oxide synthase 3
[(NOS3); 6.88-fold increase; Fig. S1]. The expression heatmaps of all
tested genes are presented in Fig.
S1.
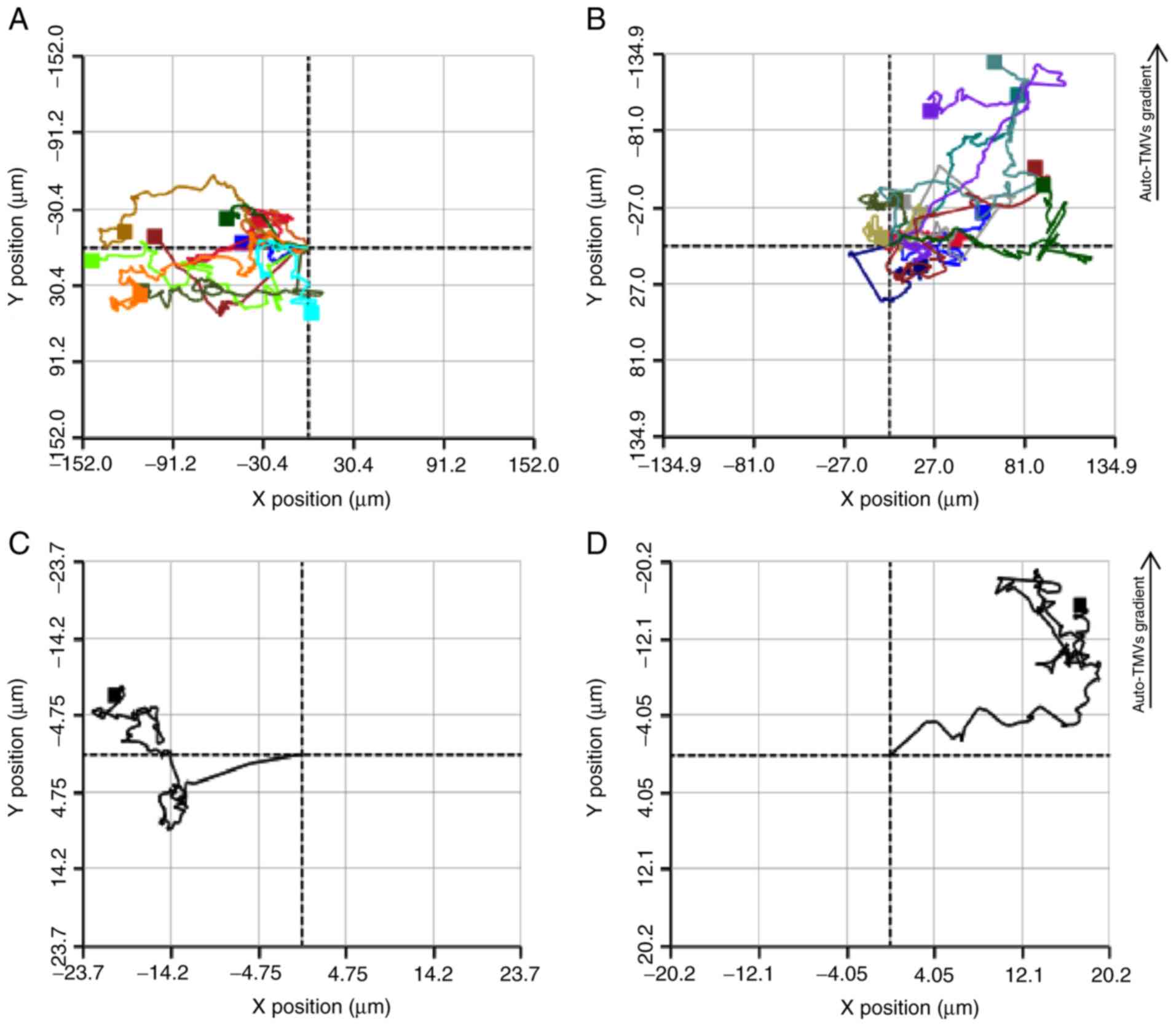
Chemotaxis
The average migration distance by the control GC1415
cells was 71.6 µm over a period of 12 h. The average
motility speed over a period of 12 h was 31.15 µm/h with the
highest speed being recorded at 11 h (46.7 µm/h). The
average distance migrated by the GC1415 cells towards their
auto-TMVs gradient was 80.5 µm over a period of 12 h. The
average motility speed for the same time period was 27.20
µm/h with the highest being 59.3 µm/h, which was
observed at the 10 min mark of the experiment. However, the average
motility speed was lower for the GC1415 cells exposed to the
auto-TMVs gradient, and they covered a higher distance compared
with control GC1415 cells. Additionally, the migration tracking
graph of GC1415 cells exposed to the auto-TMVs gradient, as
presented in Fig. 3, exhibited a
more orderly pattern compared with the control GC1415 cells,
implying the chemoattractant capabilities of auto-TMVs. The highest
motility speed for the gradient-exposed GC1415 cells was observed
almost immediately (10 min) after the cell tracking was initiated
when compared with the control GC1415 cells, which was recorded at
the 11-h time point.
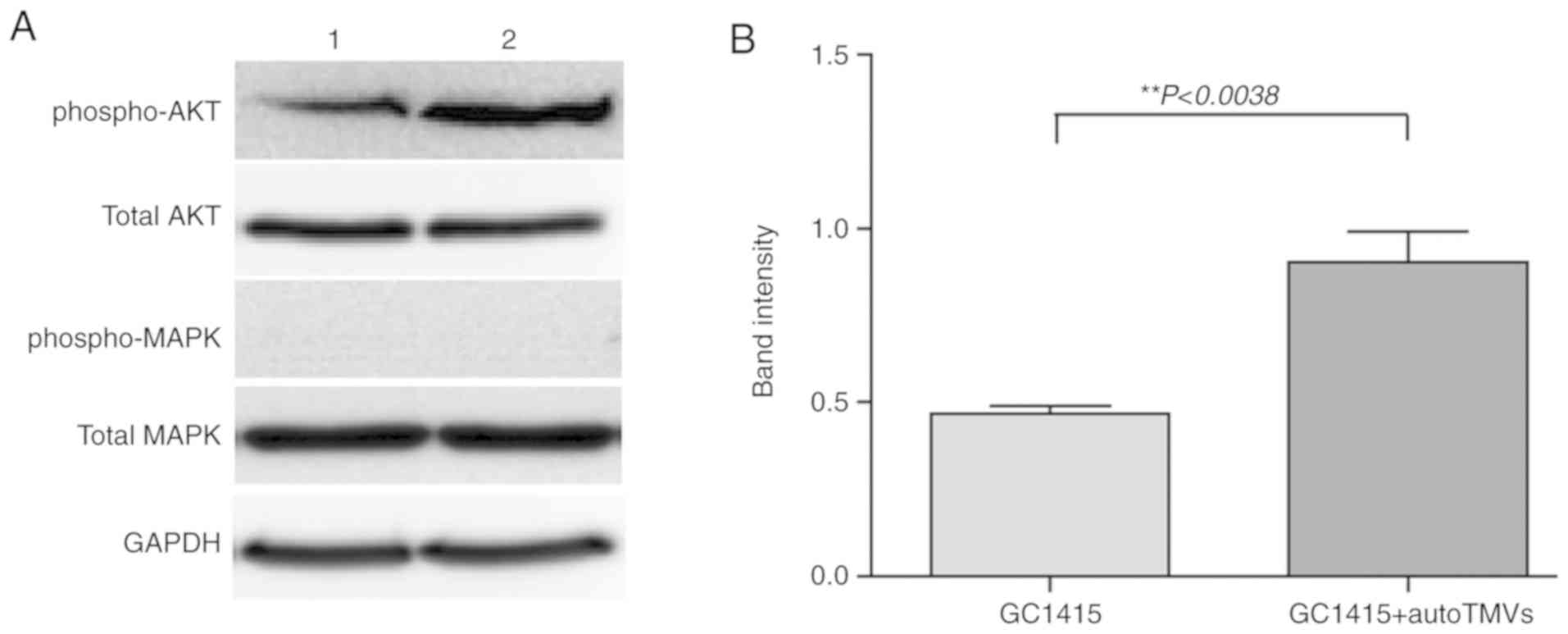
Western blot analysis
The PI3K/AKT and MAPK/ERK signaling pathways were
analyzed in the GC1415 cells and in the GC1415 cells treated with
auto-TMVs at time intervals of 0.5, 2 and 24 h. The MAPK/ERK
signaling pathway constituents (p44/p42 MAP kinase) were not
indicated to be influenced by auto-TMVs at any of the tested time
points (Fig. 4A). However, an
increase in the phosphorylated form of AKT kinase (phospho-AKT;
Fig. 4A and B) was observed at the
0.5 h time point, indicating that auto-TMVs-stimulated signal
transduction may occur via the PI3K/AKT pathway.
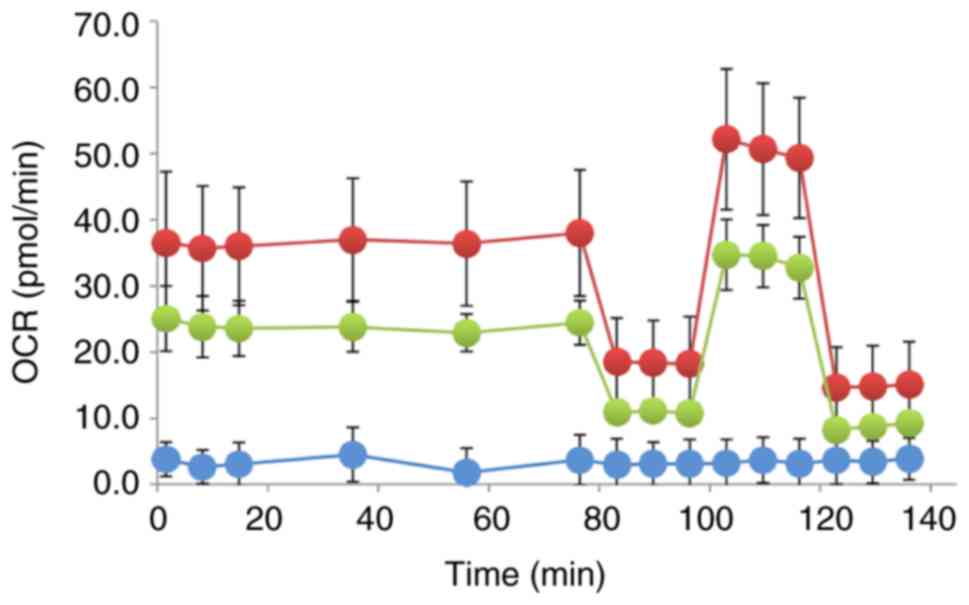
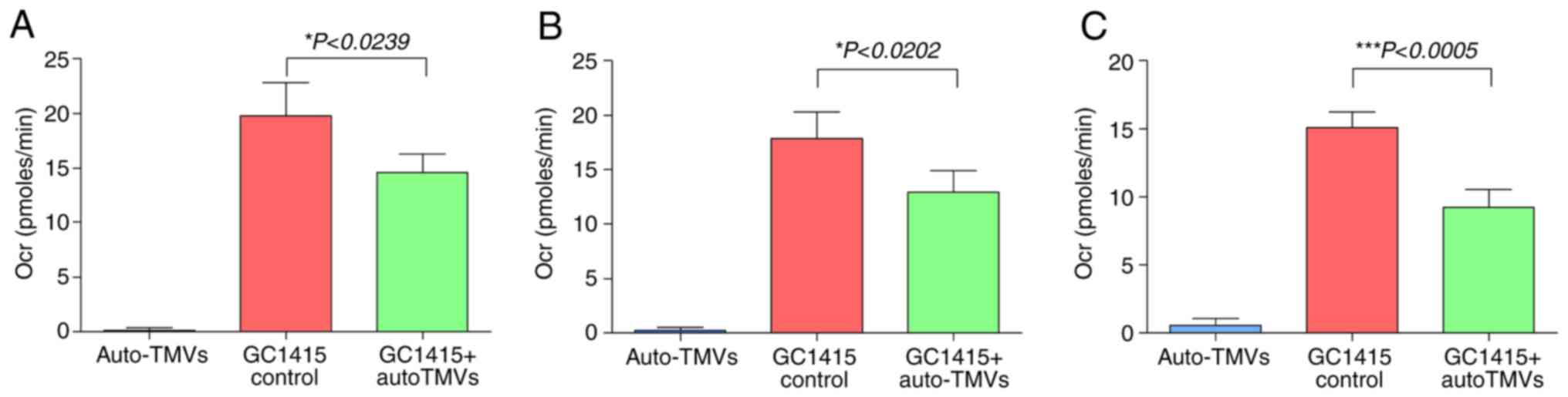
Cellular respiration
Treatment of the GC1415 cells with auto-TMVs
resulted in a decrease in the Ocr compared with the control GC1415
cells, as presented in Fig. 5,
indicating a decrease in mitochondrial respiration. The Ocr values
measured during the basal respiration phase were decreased in
treated GC1415 cells, which suggested that auto-TMVs decreased
initial cellular energy requirements (Fig. 6A). This decrease in cellular energy
demand was also evident during the ATP production phase of the
experiment and in the Ocr values obtained for the spare respiratory
capacity measurements (Fig. 6B and
C).
Discussion
Cancer develops as a consequence of the continual
and uncontrolled proliferation of cells, the outcome of which is an
unfavorable transformation, leading to the invasion of normal
tissue/organs and throughout the body (23). The loss of responsiveness of cancer
cells to signals controlling normal cell behavior is a result of
abnormalities that develop in multiple cell regulatory systems. The
current study demonstrated that a number of genes, protein
expression and cell behavior/metabolism are affected in
auto-TMV-exposed GC1415 cells and this may suggest that, auto-TMVs
play a role in facilitating cancer proliferation and
metastasis.
An abnormal gene expression profile in cancer cells
is associated with their potential to proliferate and spread
(23). The data obtained in the
current study indicated a variety of genes that were affected in
GC1415 cells following contact with auto-TMVs. For example, TCF20
mRNA, which was highly overexpressed (Fig. 2A), is a transcriptional coactivator
that enhances the activity of Jun and Sp1 transcription factors
(24). With the Fos subunit, Jun
can form AP-1 transcription factor, which can bind to the promoter
gene regions of a number of genes (including IL-2, CD95L,
MMP-1 and TGF-β) that are associated with cell
proliferation, differentiation and apoptosis (22-25).
Sp1 is also a transcription factor that binds GC/GT-rich promoter
elements through its zinc fingers and has been indicated to play a
role in tumor growth and metastasis by regulating cell cycle genes
and VEGF (24). Genes encoding
TIMPs, which are glycoproteins that are natural inhibitors of
MMPs-peptides and are associated with the degradation of the
extracellular matrix (26,27), were also affected (underexpressed)
in auto-TMVs-exposed GC1415 cells (Fig. 2D). This indicated their potential
involvement in metastasis. A simultaneous underexpression of MMP
(including, MMP-2, -9, -11 and -14; Fig. 2D) mRNA transcripts was also
observed. However, the downregulation of TIMP mRNA transcripts was
demonstrated, MMP mRNA underexpression and its significance
requires future investigation. The results of the current study
indicated an association between TP53 and MDM2
transcript expression (Fig. 2F),
which may also imply that auto-TMVs may be initiating mechanisms in
GC1415 cells that increase tumor proliferation. The p53 protein is
a known tumor suppressor that is capable of initiating DNA damage
repair, apoptosis and/or cell cycle arrest in affected cells, which
are important mechanisms in tumorigenesis (28,29).
The MDM2 protein, selectively binds to the p53 protein, and causes
its inactivation, which limits its anticancer potential (30,31).
The interaction between antigens and surface
receptors initiates an intracellular signaling cascade that
generates an appropriate cellular response (23). However, the mechanisms through
which auto-TMVs initiate the PI3K/AKT signaling pathway in GC1415
cells has not yet been determined. However, this may be associated
with one of the receptors exhibiting tyrosine kinase activity
(namely RTKs) (30-32). The expression of HER-2/neu
receptor, a member of the RTK family, on GC1415 cells, has been
previously reported (18). In the
current study, the upregulation of HER/2 neu (ERBB2)
mRNA gene expression (Fig. 2C) was
observed in GC1415 cells treated with auto-TMVs, and this result
may demonstrate its involvement in signal transduction. The
overexpression of HER/2 neu (ERBB2) protein and its
mRNA has been reported in a variety of tumor types, which may lead
to the activation of downstream protein kinases and transcription
factors that can propagate tumor progression and metastasis
(29-33). The simultaneous upregulation of
AKT-1 mRNA (Fig. 2A) and
the phosphorylated form of AKT 1 kinase protein (Fig. 4A), in auto-TMV-exposed GC1415
cells, support this hypothesis. Additionally, it has been reported
that AKT-1, through the activation of mTOR, induces
hypoxia-inducible factor 1α levels, which is a complex that is
responsible for binding to hypoxia-responsible elements of the
promoters of genes coding for glycolytic enzymes (34). This is in concurrence with the
results of the current study, which indicated a decrease in the
Ocrs (Figs. 5 and 6), suggesting that contact with auto-TMVs
may cause a metabolic switch from oxidative phosphorylation to
glycolysis in GC1415 cells, a phenomenon frequently observed in
tumor metabolism and referred to as the Warburg effect (35). It should be pointed out, however,
that the observed Ocrs were relatively low (~40 pmol/min), which,
in part, may be explained by the low auto-TMVs concentration (3
µg) used in the experiments. During growth, tumor cells
require increasing amounts of energy to meet their cellular demands
(34,36,37).
Glycolysis, although not as efficient as oxidative phosphorylation,
metabolizes glucose more rapidly and delivers ATP to growing tumors
at a faster rate (38).
Additionally, intermediates generated during glycolysis have been
indicated to be shunted into subsidiary pathways causing the de
novo generation of nucleotides, lipids and amino acids, which
can be utilized by tumor cells to propagate further proliferation
(39).
Metastasis is the final stage of cancer and requires
a migratory ability of cancer cells that is facilitated by surface
chemokine receptors that are present at the cell surface (35,38,39).
A higher expression of CXCR4, CXCR7 and their ligand, CXCL12/SDF-1,
has been observed in GC cells, particularly in the intestinal-type,
where it has been associated with lymph node and liver metastasis
(40,41). Previous phenotyping has revealed
that GC1415 cells express a number of chemokine receptors,
including CXCR4 (17). However, in
this aforementioned study, CXCR4 expression was relatively low
(<10%) (17). Other researchers
have revealed that exposure of the GC1415 cells to auto-TMVs
induces an enhancement of tumor growth and cancer-cell induced
angiogenesis in NOD-SCID mice (18). It has also been previously reported
that surface receptors and mRNA transcripts can be transferred to
target cells by EVs (40-42). The gene expression data acquired in
the current study demonstrated that CXCR4 and its ligand's
(CXCL12/SDF-1) mRNA were upregulated in GC1415 cells treated with
auto-TMVs and this may indicate their chemotactic abilities
(Figs. 2E and S1A). The CXCR4 receptor and its mRNA may
be transferred to GC1415 cells via auto-TMVs, which may lead to an
instant enhancement in the receptor's surface expression, or de
novo synthesis from the transferred mRNA transcript. However,
the CXCL12 mRNA translation in exposed GC1415 cells may lead to the
synthesis of the ligand and its potential release via
auto-TMVs, which may serve as a chemoattractant for
CXCR4+ cells.
The aim of the present study was to assess the
effects of auto-TMVs on GC1415 cells. The initial characterization
of auto-TMVs revealed their size range as 90-800 nm with an average
size of 160.7 nm, suggesting that the majority of auto-TMVs are
within the same size range as microvesicles. The obtained gene
expression data indicated an array of genes to be overexpressed and
underexpressed in GC1415 cells that were exposed to auto-TMVs.
These genes coding for proteins engaged in cellular processes,
including signal transduction, metabolism, chemotaxis, angiogenesis
and metastasis. Chemotaxis experiments performed in the present
study revealed that GC1415 cells migrated towards their auto-TMVs
gradient. Signal transduction experiments performed in the current
study indicated the PI3K/AKT signaling pathway to be affected in
auto-TMVs-exposed GC1415 cells. The obtained data on cellular
metabolism in GC1415 cells exposed to auto-TMVs have revealed a
metabolic shift from oxidative phosphorylation to glycolysis.
In conclusion, the data from the current study
demonstrate that EVs released by tumor cells may also interact and
induce behavioral changes under in vitro conditions in other
tumor cells. These changes may enhance pro-tumorigenic activity in
cancer cells interacting with auto-TMVs. Further studies focusing
on the individual aspects of the presented data (including signal
transduction, metabolism and chemotaxis) will provide increased
knowledge regarding the effect of autologous TMVs on tumor
cells.
Supplementary Data
Funding
This research was funded by National Science Centre,
grant no. 2012/07/B/NZ6/03499 and European Commission
H2020-MSCA-RISE-2017; grant no. 777682 'CANCER'.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
RS and MBK were involved in the conception and
design of the study. RS, KW, MBK, MSt, JB and MSi were involved in
the study methodology. MPW was involved in SEM imaging. RS, KW and
MBK were involved in data acquisition. RS was involved in data
analysis. RS was involved in the writing and preparation of the
original draft. RS, MBK, JB and MSi were involved in the writing,
reviewing and editing of the article. RS and MBK were involved in
study supervision. RS was also involved in project administration.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Joanna
Cichy and Dr Mateusz Kwitniewski (both, Department of Immunology,
Jagiellonian University) for disclosing the Seahorse XF Analyzer
and providing assistance with the cellular respiration
experiments.
Abbreviations:
|
TMVs
|
tumor-derived microvesicles
|
|
EVs
|
extracellular vesicles
|
|
GC
|
gastric cancer
|
|
NTA
|
nanoparticle tracking analysis
|
References
|
1
|
Yada T, Yokoi C and Uemura N: The current
state of diagnosis and treatment for early gastric cancer. Diagn
Ther Endosc. 2013:2413202013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture Eur J Cancer. 37(Suppl
8): S4–S66. 2001.
|
|
5
|
Parkin DM: International variation.
Oncogene. 23:6329–6340. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ratajczak J, Wysoczynski M, Hayek F,
Janowska-Wieczorek A and Ratajczak MZ: Membrane-derived
microvesicles: Important and underappreciated mediators of
cell-to-cell communication. Leukemia. 20:1487–1495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Pol E, Hoekstra AG, Sturk A, Otto
C, van Leeuwen TG and Nieuwland R: Optical and non-optical methods
for detection and characterization of microparticles and exosomes.
J Thromb Haemost. 8:2596–2607. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: Artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baj-Krzyworzeka M, Szatanek R, Weglarczyk
K, Baran J, Urbanowicz B, Brański P, Ratajczak MZ and Zembala M:
Tumour-derived microvesicles carry several surface determinants and
mRNA of tumour cells and transfer some of these determinants to
monocytes. Cancer Immunol Immunother. 55:808–818. 2006. View Article : Google Scholar
|
|
10
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Webber J, Steadman R, Mason MD, Tabi Z and
Clayton A: Cancer exosomes trigger fibroblast to myofibroblast
differentiation. Cancer Res. 70:9621–9630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sidhu SS, Mengistab AT, Tauscher AN,
LaVail J and Basbaum C: The microvesicle as a vehicle for EMMPRin
in tumor-stromal interactions. Oncogene. 23:956–963. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wieckowski EU, Visus C, Szajnik M,
Szczepanski MJ, Storkus WJ and Whiteside TL: Tumor-derived
microvesicles promote regulatory T cell expansion and induce
apoptosis in tumor-reactive activated CD8+ T lymphocytes. J
Immunol. 183:3720–3730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gamperl H, Plattfaut C, Freund A, Quecke
T, Theophil F and Gieseler F: Extracellular vesicles from malignant
effusions induce tumor cell migration: Inhibitory effect of LMWH
tinzaparin. Cell Biol Int. 40:1050–1061. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ristorcelli E, Beraud E, Mathieu S,
Lombardo D and Verine A: Essential role of Notch signaling in
apoptosis of human pancreatic tumoral cells mediated by exosomal
nanoparticles. Int J Cancer. 125:1016–1026. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumour cells. Nat
Cell Biol. 10:619–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mytar B, Stec M, Szatanek R, Węglarczyk K,
Szewczyk K, Szczepanik A, Drabik G, Baran J, Siedlar M and
Baj-Krzyworzeka M: Characterization of human gastric
adeno-carcinoma cell lines established from peritoneal ascites.
Oncol Lett. 15:4849–4858. 2018.PubMed/NCBI
|
|
18
|
Stec M, Szatanek R, Baj-Krzyworzeka M,
Baran J, Zembala M, Barbasz J, Waligórska A, Dobrucki JW, Mytar B,
Szczepanik A, et al: Interactions of tumour-derived micro(nano)
vesicles with human gastric cancer cells. J Transl Med. 13:3762015.
View Article : Google Scholar
|
|
19
|
Baj-Krzyworzeka M, Mytar B, Szatanek R,
Surmiak M, Węglarczyk K, Baran J and Siedlar M: Colorectal
cancer-derived microvesicles modulate differentiation of human
monocytes to macrophages. J Transl Med. 14:362016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szatanek R, Drabik G, Baran J,
Kolodziejczyk P, Kulig J, Stachura J and Zembala M: Detection of
isolated tumour cells in the blood and bone marrow of patients with
gastric cancer by combined sorting, isolation and determination of
MAGE-1, -2 mRNA expression. Oncol Rep. 19:1055–1060.
2008.PubMed/NCBI
|
|
21
|
Rutkowska-Zapała M, Suski M, Szatanek R,
Lenart M, Węglarczyk K, Olszanecki R, Grodzicki T, Strach M,
Gąsowski J and Siedlar M: Human monocyte subsets exhibit divergent
angiotensin I-converting activity. Clin Exp Immunol. 181:126–132.
2015. View Article : Google Scholar
|
|
22
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extracellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
23
|
Feitelson MA, Arzumanyan A, Kulathinal RJ,
Blain SW, Holcombe RF, Mahajna J, Marino M, Martinez-Chantar ML,
Nawroth R, Sanchez-Garcia I, et al: Sustained proliferation in
cancer: Mechanisms and novel therapeutic targets. Semin Cancer
Biol. 35(Suppl): S25–S54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Safe S and Abdelrahim M: Sp transcription
factor family and its role in cancer. Eur J Cancer. 41:2438–2448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McMurray RW, Ndebele K, Hardy KJ and
Jenkins JK: 17-beta-estradiol suppresses IL-2 and IL-2 receptor.
Cytokine. 14:324–333. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gialeli C, Theocharis AD and Karamanos NK:
Roles of matrix metalloproteinases in cancer progression and their
pharmacological targeting. FEBS J. 278:16–27. 2011. View Article : Google Scholar
|
|
27
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sionov RV and Haupt Y: The cellular
response to p53: The decision between life and death. Oncogene.
18:6145–6157. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prives C and Hall PA: The p53 pathway. J
Pathol. 187:112–26. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kubbutat MH, Jones SN and Vousden KH:
Regulation of p53 stability by Mdm2. Nature. 387:299–303. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haupt Y, Maya R, Kazaz A and Oren M: Mdm2
promotes the rapid degradation of p53. Nature. 387:296–299. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iyer P, Shrikhande SV, Ranjan M, Joshi A,
Gardi N, Prasad R, Dharavath B, Thorat R, Salunkhe S, Sahoo B, et
al: ERBB2 and KRAS alterations mediate response to EGFR inhibitors
in early stage gallbladder cancer. Int J Cancer. 144:2008–2019.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Slamon DJ, Godolphin W, Jones LA, Holt JA,
Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al:
Studies of the HER-2/neu proto-oncogene in human breast and ovarian
cancer. Science. 244:707–712. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shaw RJ: Glucose metabolism and cancer.
Curr Opin Cell Biol. 18:598–608. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
WARBURG O: Origin of cancer cells.
Oncologia. 9:75–83. 1956.In German. View Article : Google Scholar
|
|
36
|
Azarhoosh R, Ebneghasem R and Besharat S:
HER-2/neu gene amplification in gastric adenocarcinoma and its
relationship with clinical and pathological findings. J
Gastrointest Oncol. 8:1046–1050. 2017. View Article : Google Scholar
|
|
37
|
Tateishi M, Ishida T, Mitsudomi T, Kaneko
S and Sugimachi K: Prognostic value of c-erbB-2 protein expression
in human lung adenocarcinoma and squamous cell carcinoma. Eur J
Cancer. 27:1372–1375. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pfeiffer T, Schuster S and Bonhoeffer S:
Cooperation and competition in the evolution of ATP-producing
pathways. Science. 292:504–507. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hamanaka RB and Chandel NS: Targeting
glucose metabolism for cancer therapy. J Exp Med. 209:211–215.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iwasa S, Yanagawa T, Fan J and Katoh R:
Expression of CXCR4 and its ligand SDF-1 in intestinal-type gastric
cancer is associated with lymph node and liver metastasis.
Anticancer Res. 29:4751–4758. 2009.PubMed/NCBI
|
|
41
|
Ma DM, Luo DX and Zhang J: SDF-1/CXCR7
axis regulates the proliferation, invasion, adhesion, and
angiogenesis of gastric cancer cells. World J Surg Oncol.
14:2562016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Okuma A, Hanyu A, Watanabe S and Hara E:
p16Ink4a and p21Cip1/Waf1 promote tumour
growth by enhancing myeloid-derived suppressor cells chemotaxis.
Nat Commun. 8:20502017. View Article : Google Scholar
|