Introduction
Lung cancer is a common malignant tumour of the
respiratory system and the morbidity and mortality rates rank among
the highest worldwide. Lung cancer is a disease with multiple
complex molecular networks underlying its development and
progression (1). In recent years,
there have been notable advancements in the understanding of the
molecular mechanisms involved in lung adenocarcinoma, which has
resulted in the identification of numerous targeted drug therapies,
which exhibit notably improved survival and prognosis in patients
with lung cancer (2-4). Gefitinib, erlotinib and bevacizumab
are the most frequently used drugs for treatment of lung cancer
(5-7). However, patients may exhibit adverse
reactions, drug resistance and other complications when assigned
regimens containing these drugs (8-10).
Understanding the molecular mechanisms underlying the development
and progression of lung cancer may assist in the development of
treatment measures with enhanced efficacy and improved outcomes,
and improve early detection in patients with lung cancer.
The various stages of lung cancer are associated
with up- and downregulation of various genes. Wang et al
(11) demonstrated that microRNA
(miR)-513b regulates the effects of high mobility group box 3 on
cell proliferation, apoptosis, invasion and migration by regulating
the mTOR signalling pathway in non-small cell lung cancer (NSCLC).
Qiu et al (12)
demonstrated that circFGFR3 increases the expression of galetin-1,
phosphorylated (p)-AKT and p-ERK1/2 through competitive binding
with miR-22-3p, thus promoting the invasion and proliferation of
NSCLC. Upregulated expression of circFGFR3 is associated with a
poor prognosis in patients with lung cancer (13). However, studies based on individual
gene expression are insufficient for the investigation of the
mechanism of lung cancer. Interactions between genes influence gene
expression and a comprehensive understanding of the direct and
indirect interactions between genes will greatly assist in
developing a comprehensive description of cell mechanisms and
functions both in physiologically healthy cells and in cancerous
cells.
Advances in genomics, transcriptomics and sequencing
technology, and the used of gene co-expression networks has
developed and been expanded in biological research (14-16).
Gene co-expression networks are widely used in the analysis of
high-throughput chip data, RNA sequencing, DNA methylation and
other types of genome data analyses (17-19).
The most representative gene co-expression network is the weighted
gene co-expression network analysis (WGCNA) (20). WGCNA has provided meaningful
advances in our understanding of multi-species gene analysis, such
as in humans and mice, and has become a widely used network
analysis tool (21). In addition,
the core genes obtained by network screening can be supplemented
and verified by biological experiments to further explore and
verify the identified mechanisms. This strategy avoids a
potentially blind approach in experimental research and confirms
the validity or highlights potential flaws of network analyses. Sun
et al (22) identified CD36
as a core gene based on WGCNA screening. Differential expression
and increased methylation of CD36 in lung cancer were confirmed by
reverse transcription-quantitative PCR and western blotting,
confirming the inhibitory effect of CD36 in the development of lung
cancer (22). An et al
(23) used Gene Set Enrichment
Analysis (GSEA) and WGCNA to identify potential metabolic pathways
associated with the core gene KIBRA, which is involved in
regulation of lung cancer. KIBRA reduced proliferation and invasion
of lung cancer cells and induced apoptosis, and this was verified
in in vitro experiments (23).
The aim of the present study was to identify core
genes associated with lung cancer and construct a WGCNA network
based on data obtained from Gene Expression Omnibus (GEO) and
analyse the data in regards to the clinical information and
survival information of the patients. Additionally, the effects of
immunoglobulin superfamily member 10 (IGSF10) on proliferation of
lung cancer cells, cell-cell and cell-extracellular matrix
adhesions, and associated metabolic pathways were determined in
vitro. The mechanisms of the identified core genes were further
explored highlighting potential biomarkers for the diagnosis of
patients with lung cancer.
Materials and methods
Selection criteria and acquisition of the
data
The GSE19804 lung cancer dataset (24) was obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/). The dataset
contained information from non-smoking women with NSCLC. NSCLC
accounts for 85% of all lung cancer cases (25), and the majority of cases of lung
cancer in male patients are associated with smoking, whereas the
majority of lung cancer cases in females are not associated with
smoking (26,27). In the present study, biomarkers
associated NSCLC in non-smoking female patients were examined. A
total of 120 samples were analysed, and the information did not
include normal tissue from patients with pneumonia, but did contain
information from the normal adjacent lung tissue samples (60 tumour
tissues and 60 adjacent tissues). The Affymetrix Human Genome U133
Plus 2.0 Array (Affymetrix; Thermo Fisher Scientific, Inc.)
annotation platform was utilized to match probes with gene names.
Relevant clinical information was used for WGCNA.
Construction of a gene co-expression
network
The gene co-expression network was constructed using
the WGCNA package in R (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/).
The top 25% of genes showing the highest levels of variance were
screened for the construction of a weighted co-expression network.
The power value was calculated through the pickSoft-Threshold
function of WGCNA package. The dynamic tree cutting algorithm of
WGCNA package was used to segment the network module.
Identification of important clinical
modules
The correlation between modules and clinical
features was evaluated using Pearson's correlation coefficient
analysis. Clinical information included age and stage. The
correlation between the eigengenes of the module and the clinical
features were assessed to identify key modules. Gene significance
(GS) was defined as the linear relationship between gene expression
and clinical information. Module significance was defined as the
average GS, screening for all genes in each module to identify key
modules.
Gene Ontology (GO) and Kyoto
Encyclopaedia of Genes and Genomes (KEGG) enrichment analysis
GO enrichment analysis and KEGG enrichment analysis
were performed on key modules using the R package clusterProfiler
3.14.0 (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
P<0.05 was defined as a meaningful enrichment analysis
result.
Identification of hub genes
Genes exhibited high levels of connectivity to nodes
in a module were considered to have important functions. A key
module network diagram was created using Cytoscape 3.72 (https://cytoscape.org) to screen for the top 30 genes
with the highest levels of connectivity in the module network.
Survival analysis
GEPIA (http://gepia.cancer-pku.cn/) is a website used to
analyse RNA expression data of tumours and normal samples in The
Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). GEPIA was used to
perform survival analysis of the previously identified hub genes.
The gene expression was stratified into high and low expression
according to the median values, and the significance of expression
of these genes on survival was determined using a log-rank
test.
Dataset validation
GEPIA contains RNA sequencing expression data from
9,736 tumor samples and 8,587 normal samples from 33 malignant
tumors of TCGA and GTEx (28). In
the present study, lung adenocarcinoma and lung squamous cell
carcinoma data from TCGA database were used to validate gene
expression data for selected key genes, and the function of
'BoxPlots' was run to analyze whether the hub gene was
differentially expressed between lung adenocarcinoma and lung
squamous cell carcinoma and normal samples.
Oncomine analysis
Oncomine (https://www.oncomine.org/) is a database and
integrated data mining platform based on gene chip, in which the
data can be screened and mined according to determinable
requirements. In the present study, the following conditions were
set: i) Cancer Type, 'Lung Cancer'; ii) Gene, 'IGSF10'; iii)
Analysis Type, 'Cancer vs. Normal Analysis'; iv) critical value
setting conditions '(P value<1E-4, fold change>2, gene
rank=top 10%)'. Hou et al (29) and Okayama et al (30) lung datasets were determined to meet
the selection criterion.
Reagents
RPMI-1640 medium was purchased from Gibco; Thermo
Fisher Scientific, Inc. FBS was purchased from Biological
Industries. Integrin-β1 (catalog no. 9699S), p-FAK (catalog no.
8556S), FAK (catalog no. 71433S), p-AKT (catalog no. 4060S) and AKT
(catalog no. 4691S) primary antibodies were purchased from Cell
Signalling Technology, Inc. The IGSF10 antibody was purchased from
Novus Biologicals, Ltd. (catalog no. H00285313-A01). The β-actin
antibody (catalog no. sc-47778), secondary goat anti-rabbit
antibody (horseradish peroxidase-conjugated; catalog no. sc-2004)
and secondary goat anti-mouse antibody (horseradish
peroxidase-conjugated; catalog no. sc-2005) were purchased from
Santa Cruz Biotechnology, Inc.
Cell culture
Human lung adenocarcinoma cell lines H1299, HCC827,
A549 and PC9 were cultured in RPMI-1640 medium supplemented with
10% FBS, and incubated at 37°C with 5% CO2. Cells in the
logarithmic growth phase were used for subsequent experiments.
Reverse transcription-quantitative
(RT-q)PCR
Cell culture dishes were placed on ice, culture
medium was removed and cells were washed three times with PBS.
Total RNA was extracted using a Trizol® kit (Invitrogen;
Thermo Fisher Scientific, Inc.). The extracted RNA was reverse
transcribed into cDNA using a PrimeScript® RT Reagent
kit with DNA Eraser (Takara Bio Inc.) according to the
manufacturer's instructions. qPCR was performed using
SYBR® Premix EX TaqTM II (Tli RNaseH Plus, Takara Bio,
Inc.) on an Applied Biosystems® 7500 Real-Time PCR
System (Thermo Fisher Scientific, USA), and 18s was used as the
internal reference gene. The qPCR conditions were 10 min at 95°C
followed by 45 cycles at 95°C for 15 sec and 58°C for 34 sec. The
sequences of the primers used were: IGSF10 forward,
5'-CTGGGGAGTCCAATTGCTGT-3' and reverse, 5'-GCTGCCTTTGCTGACATC-3';
and 18S forward, 5'-GGTGAAGGTCGGAGTCAACGG-3' and reverse,
5'-GAGGTCAATGAAGGGGTCATTG-3'.
Western blotting
Protein samples were lysed at 4°C using RIPA lysis
buffer (1% Triton X-100, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 10 mM
EDTA, 100 mM NaF, 1 mM Na3VO4, 1 mM PMSF and 2 µg/ml
aprotinin) for 40 min. The samples were centrifuged at 18,620 × g
for 25 min at 4°C. The supernatants were obtained, and the protein
concentration was quantified using the Coomassie Brilliant Blue
method (31). Samples were mixed
with 3x sample buffer solution and boiled for 5 min. Protein
samples were loaded on a 12% SDS-gel (30-50 µg/lane) and
resolved using SDS-PAGE for 3 h. Resolved proteins were transferred
to a nitrocellulose membrane (voltage, 2 mV/cm2 for 120
min). Membranes were blocked with 5% skimmed milk for 1 h at room
temperature, and the membrane was cut according to the molecular
weight of the protein of interest based on a pre-stained protein
ladder. Subsequently, the membranes were incubated with the primary
antibody overnight at 4°C. The primary antibody dilutions were
prepared as follows: Integrin-β1, 1:1,000; p-FAK, 1:500; FAK,
1:1,000; p-AKT, 1:1,000; AKT, 1,000; IGSF10, 1:1000; and β-actin,
1:500. The following day, the membranes were washed four times with
TBST buffer (10 mM Tris-Cl pH 7.4, 150 mM NaCl, 0.1% Tween-20) and
incubated with the appropriate secondary antibody (1:2,000) for 30
min at room temperature. Membranes were washed again four times
again with TBST and signals were visualized using enhanced
chemiluminescence reagent (SuperSignal Western Pico
Chemiluminescent Substrate; Pierce; Thermo Fisher Scientific,
Inc.). Densitometry analysis was performed using ImageJ Pro Plus
6.0 (National Institutes of Health).
Liposome-mediated cell transfection
Healthy cells in the logarithmic growth phase were
trypsinized to a single cell suspension, plated in a 6-well plate
at a density of 2×105/well and incubated overnight. Once
the cells had adhered, they were transfected with 5 µl
IGSF10-small interfering (si)RNA or negative control (NC)-siRNA
from Santa Cruz Biotechnology, Inc. The sequences of the
IGSF10-specific siRNA and NC-siRNA were
5'-AGGUGUUUCCCAGAUUACCdTdt-3' and 5'-UUCUCCGAACGUGUCACGUTT-3',
respectively. Lipofectamine® 2000 (5 µl) was
mixed with RPMI-1640 medium and left to stand for 5 min at room
temperature. Subsequently, 10 µl siRNA was added to 240
µl RPMI-1640 medium and mixed with the
Lipofectamine® 2000 and RPMI-1640 mixture prepared
above. The 6-well plates containing the cells were incubated for 20
min, after which the medium was removed and 1.5 ml RPMI-1640 medium
was added, and the transfection solution prepared above was added.
Cells were incubated with the transfection mixture for 6-8 h, after
which the medium was replaced with supplemented RPMI-1640
medium.
Cell viability
To determine cell viability, cells were prepared and
transfected as described above. The absorbance values were measured
after transfection to evaluate the effect of IGSF10-knockdown on
cell viability. To measure viability, 20 µl MTT solution (5
mg/ml) was added to each well and incubated for another 4 h.
Subsequently, the supernatant was removed, 200 µl DMSO was
added to each well the plate was gently agitated until the formazan
crystals were completely dissolved. Absorbance was measured at 570
nm with a micro-plate reader at 0, 24, 48, 72 and 96 h after MTT
was added.
Colony formation assay
A total of 48 h after transfection, 500 cells were
plated in a 12-well culture plate and incubated. The growth status
of the cells was observed every 3 days. After 2 weeks, the colonies
were fixed with formaldehyde for 10 min at room temperature and
stained with 0.5% crystal violet solution for 40 min at room
temperature. Three fields were randomly counted under a light
microscope (magnification,×40). The number of colonies was
calculated.
Transwell migration and invasion
assays
Cells were trypsinized, and the samples were
centrifuged at 300 × g for 5 min at room temperature. After
discarding the supernatant, the samples were resuspended in
RPMI-1640 medium, centrifuged at 300 × g for 5 min at room
temperature to wash cells with PBS. Cells were resuspended in 200
µl RPMI-1640 medium and the density of cells was determined
by hemocytometer. For the invasion assays, Transwell membranes were
coated with Matrigel (BD Biosciences). A total of 1×104
cells were placed in the upper chamber of a microporous
(8-µm pores) Transwell insert. In the lower chamber, 500
µl RPMI-1640 supplemented with 10% FBS was added and the
cells were incubated. Migration and invasion was determined by
counting the number of cells that had successfully migrated through
the membrane (migration) or invaded through the Matrigel matrix
(invasion). After 24 h, the chamber was removed, cells which had
not migrated or invaded were removed using a cotton swab, and the
insert was dried at room temperature. Cells were subsequently fixed
with 4% paraformaldehyde for 10 min at room temperature and dyed
for 1 min using the Wright Stain Method (32) at room temperature. Cells were
incubated with diluted Giemsa and re-dyed for 40 min at room
temperature. The filter membrane was dried with a cotton swab, and
the sample was photographed.
Wound healing assay
The cells were selected, digested and counted, and
then inoculated into 6-well culture plates and incubated overnight.
The next day, IGSF10-siRNA and NC-siRNA were used for transfection.
A total of 48 h after transfection, when the confluence was close
to 100%, a monolayer of the cells was scratched with a
200-µl pipette tip and photographed using an inverted
microscope at ×200. The 6-well culture plate was placed in the
incubator and photographed again 24 h later. The areas of the
scratches in the two photos were compared.
Adhesion experiment
Cells were plated in 96-well plates, which were
precoated with 10 µg/ml Matrigel, overnight at 37°C, at a
density of 2×104 cells/well with serum-free RPMI-1640.
Following incubation at 37°C for 30 min, cells that did not adhere
to the plates were washed off with PBS. Adherent cells were fixed
in 4% paraformaldehyde for 10 min at room temperature, stained with
Wright-Giemsa for 40 min at room temperature, counted in five
random fields under a light microscope (magnification, ×200) and
analyzed statistically.
Flow cytometry
The effect of IGSF10-siRNA on apoptosis was
detected. The cells to be treated were digested with trypsin,
centrifuged at 300 × g for 5 min at room temperature, washed in PBS
and suspended in 200 µl buffer solution. Subsequently, 5
µl Annexin V-FITC (BD Biosciences) was added to the
195-µl cell suspension. After full mixing and incubation at
room temperature for 10 min, the cells were washed with 200
µl buffer solution and resuspended in 190 µl buffer
solution. Then, 10 µl propidium iodide (20 µg/ml) was
added for 30 min at 37°C. The samples were analysed using am Accuri
C6 flow cytometer with CFlow Plus analysis software version 1.5 (BD
Biosciences)
Statistical analysis
All data were the results of three independent
experiments, and expressed as the mean ± standard deviation. SPSS
22.0 (IBM Corp.) was used for statistical analysis. Multiple
comparisons of the means were performed using one-way analysis of
variance followed by Student-Newman-Keuls post hoc test. P<0.05
was considered to indicate a statically significant difference.
Results
Acquisition of microarray data
GSE19804 raw data were downloaded from the GEO
database. The Affymetrix Human Genome U133 Plus 2.0 Array platform
annotation information was used to match probes and gene names.
Ultimately, the present study obtained a total of 120 samples,
including the expression data of 60 normal samples and 60 lung
cancer samples, as well as their related clinical information
(Table SI).
WGCNA construction and gene module
recognition
The first 25% of variance genes in the GSE19804 chip
data were used for cluster analysis through the WGCNA package. To
ensure the reliability of the network structure, no outlier samples
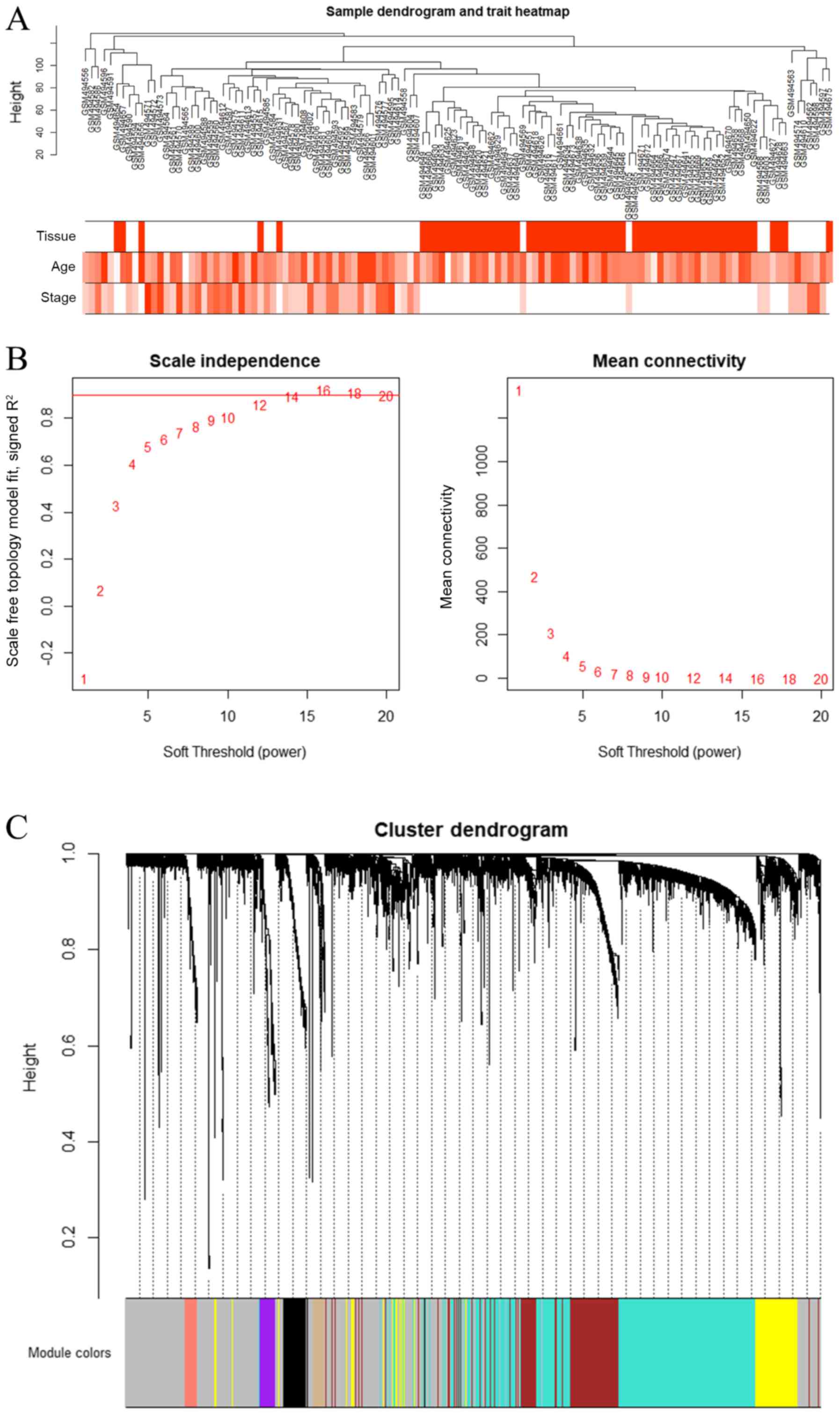
were included after calculation (Fig.
1A). The first 25% of the gene expression data were used to
construct a WGCNA. The power value of 14 was selected (Fig. 1B), and 14 modules were generated
(Fig. 1C), where the grey module
was a gene that was not co-expressed.
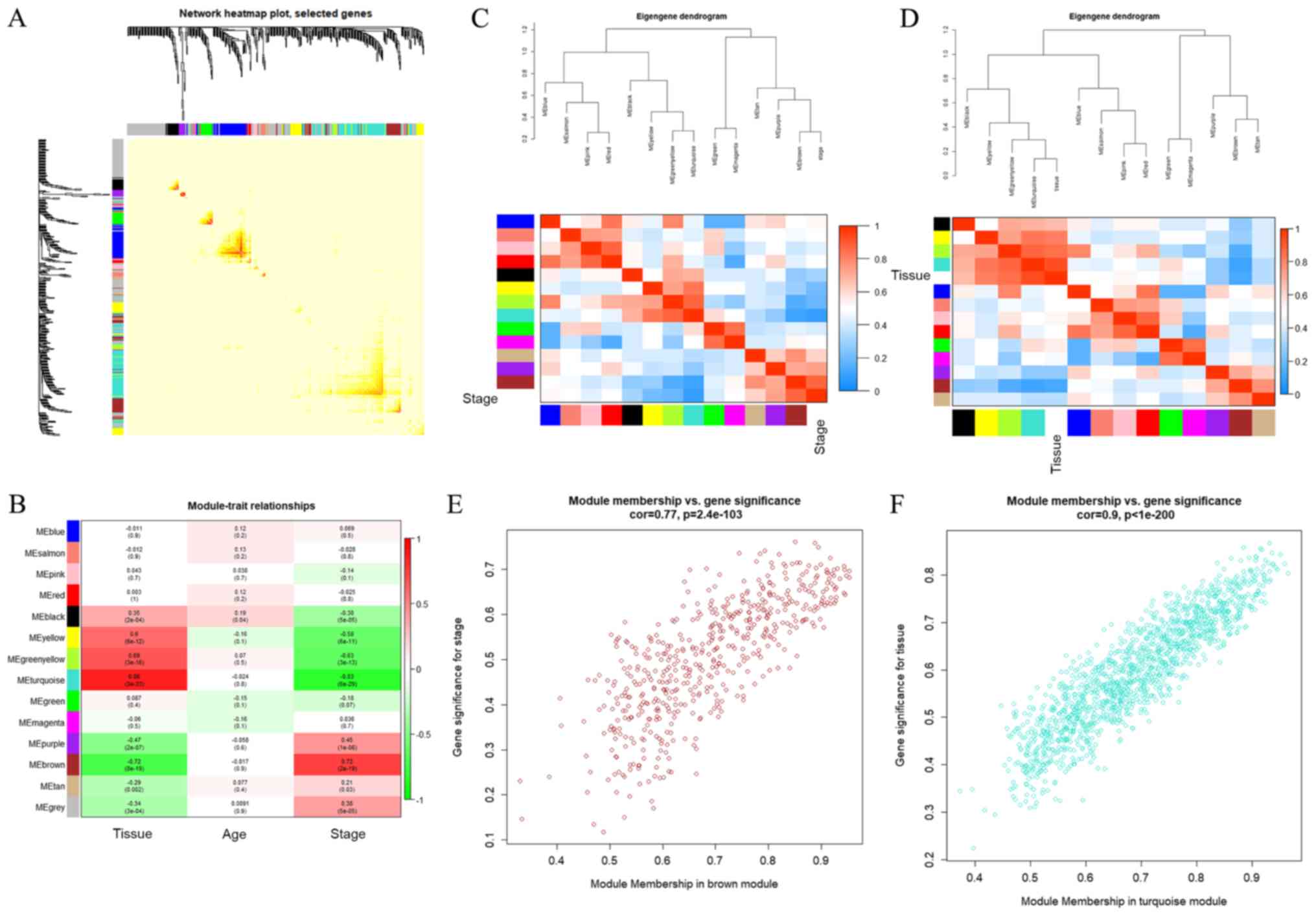
The interaction between the 14 modules was analysed
and a network heat map was generated, which demonstrated the
relative independence between the modules (Fig. 2A). As presented in Fig. 2B, compared with other modules, the
black, yellow, yellow-green and turquoise modules were positively
correlated with tissue (having cancer or not) and negatively
correlated with stage (cancer development stage). The purple and
brown modules were negatively correlated with tissue (having cancer
or not) and positively correlated with stage (stage of cancer
development). In addition, the present study calculated the
eigengenes of the module and clustered them according to their
correlation with tissue. Among them, the brown module was most
closely related to stage. Similar results were demonstrated by heat
maps based on adjacencies (Fig. 2C and
D). Therefore, it was determined that the turquoise module and
the brown module were the modules most relevant to lung cancer.
Fig. 2E and F illustrate the
associations between the brown and turquoise modules and the
genetic significance.
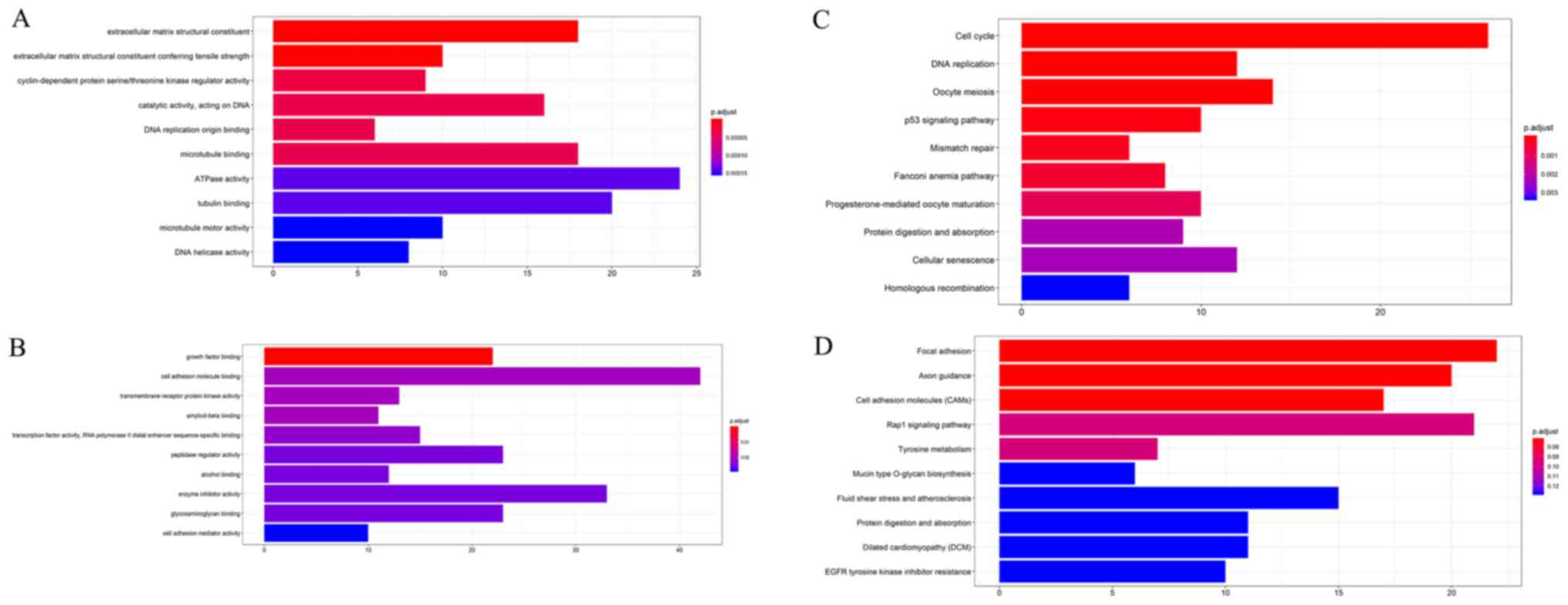
GO enrichment analysis and KEGG pathway
analysis
The GO enrichment analysis and KEGG enrichment
analysis of the brown module and turquoise module were performed
using the R package clusterProfiler. P<0.05 was defined as a
significant result of enrichment analysis. The results of the
enrichment analysis were closely associated with lung cancer, which
demonstrated the correctness of the present analysis results, as
presented in Fig. 3A-D and
Tables SII-V.
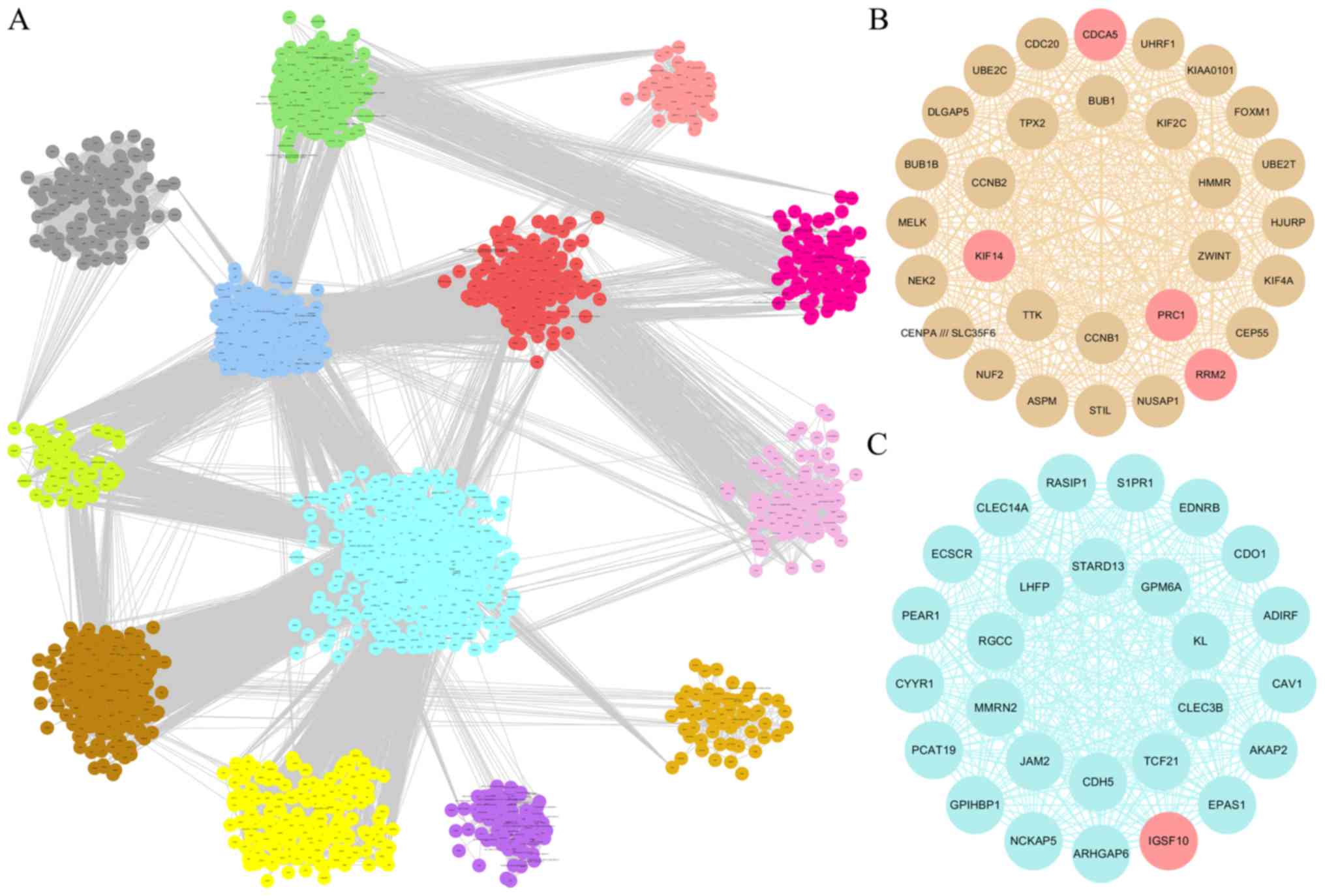
Network analysis identifies hub
nodes
The present study conducted a visualized analysis of
all the modules in Cytoscape, as presented in Fig. 4A, where the interrelationships
between the modules are shown. The turquoise module and brown
module were imported into Cytoscape for topology analysis. The
topological parameters of all nodes in the two module networks
(Tables SVI and SVII) were
calculated and the top 30 nodes of each module were screened
(33), which were selected to draw
the network diagram, as presented in Fig. 4B and C. A total of 60 nodes were
used as candidate key nodes for subsequent analysis.
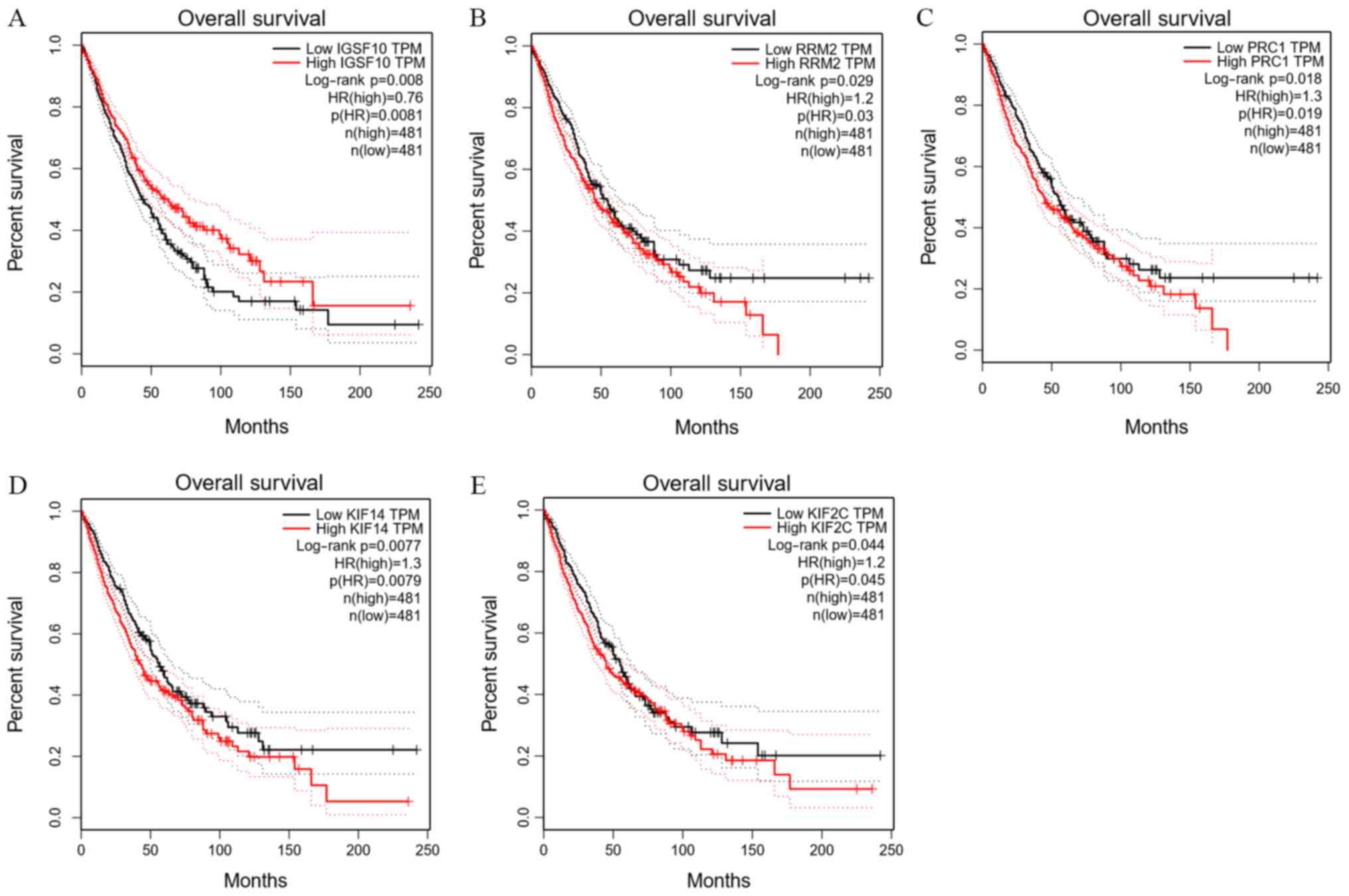
Survival analysis of the hub genes
GEPIA was used to analyse the overall survival and
P<0.05 was considered to be statistically significant. Further
survival analysis was performed on a total of 60 key genes selected
above. A total of 5 genes were significantly associated with the
prognosis of patients (P<0.05), including the IGSF10 gene of the
turquoise module, and the ribonucleotide reductase regulatory
subunit M2 (RRM2), protein regulator of cytokinesis 1 (PRC1),
kinesin family member (KIF)14 and KIF2C genes of the brown module
(Fig. 5). As the expression levels
of RRM2, PRC1, KIF14 and KIF2C in the brown module increased, the
total survival time was significantly reduced. By contrast, as the
expression of IGSF10 in the turquoise module decreased, the total
survival time was significantly decreased.
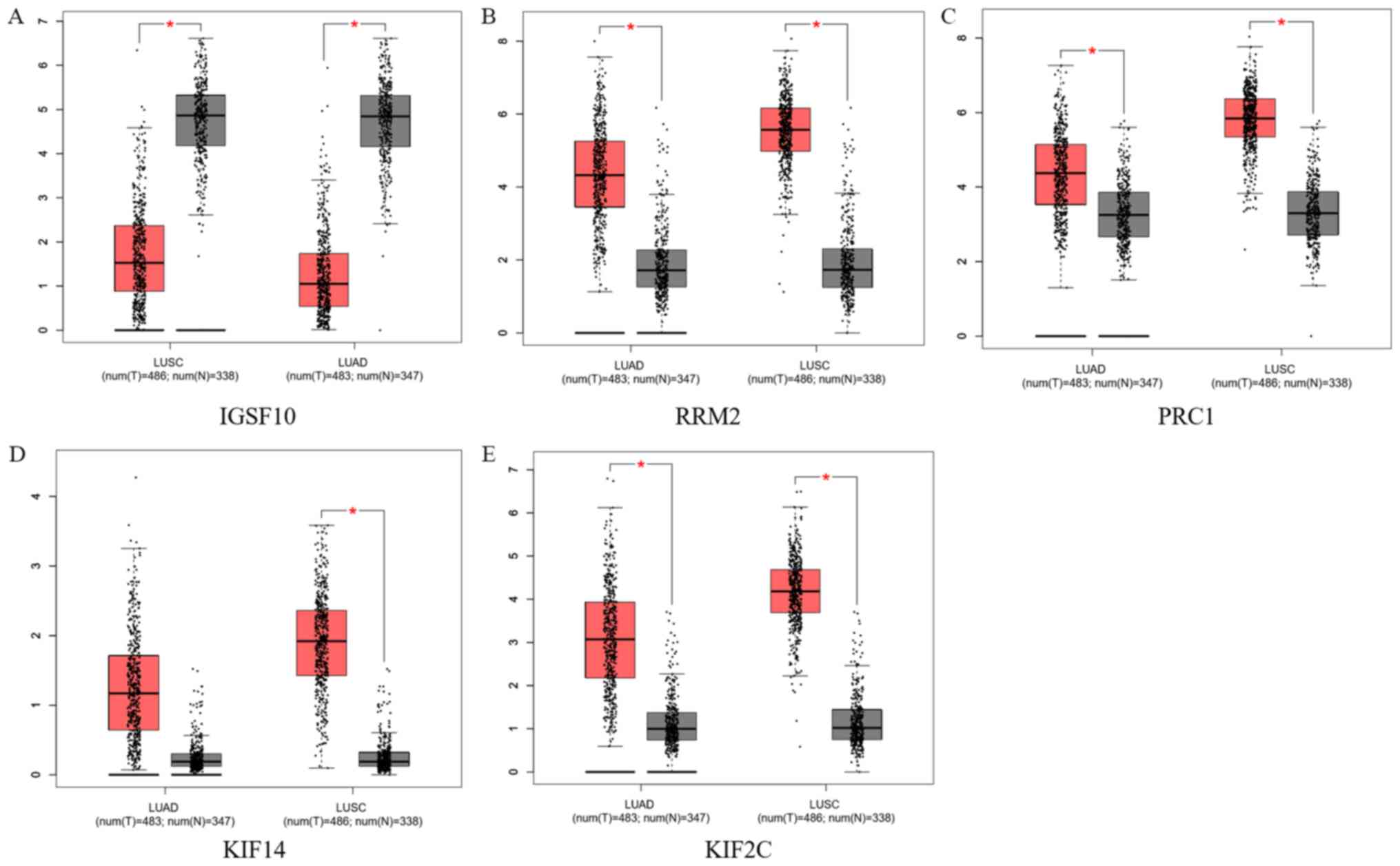
Dataset validation
Lung adenocarcinoma and lung squamous cell carcinoma
data from TCGA database were used to validate the screened key
genes, as presented in Fig. 6. The
expression of IGSF10 from the turquoise module was significantly
lower in patients compared with normal controls. The expression
levels of RRM2, PRC1, KIF14 and KIF2C from the brown module were
higher in patients compared with normal controls. These results
were consistent with those of the survival analysis, except that no
significant difference was identified in KIF14 expression between
patients with lung adenocarcinoma and normal controls.
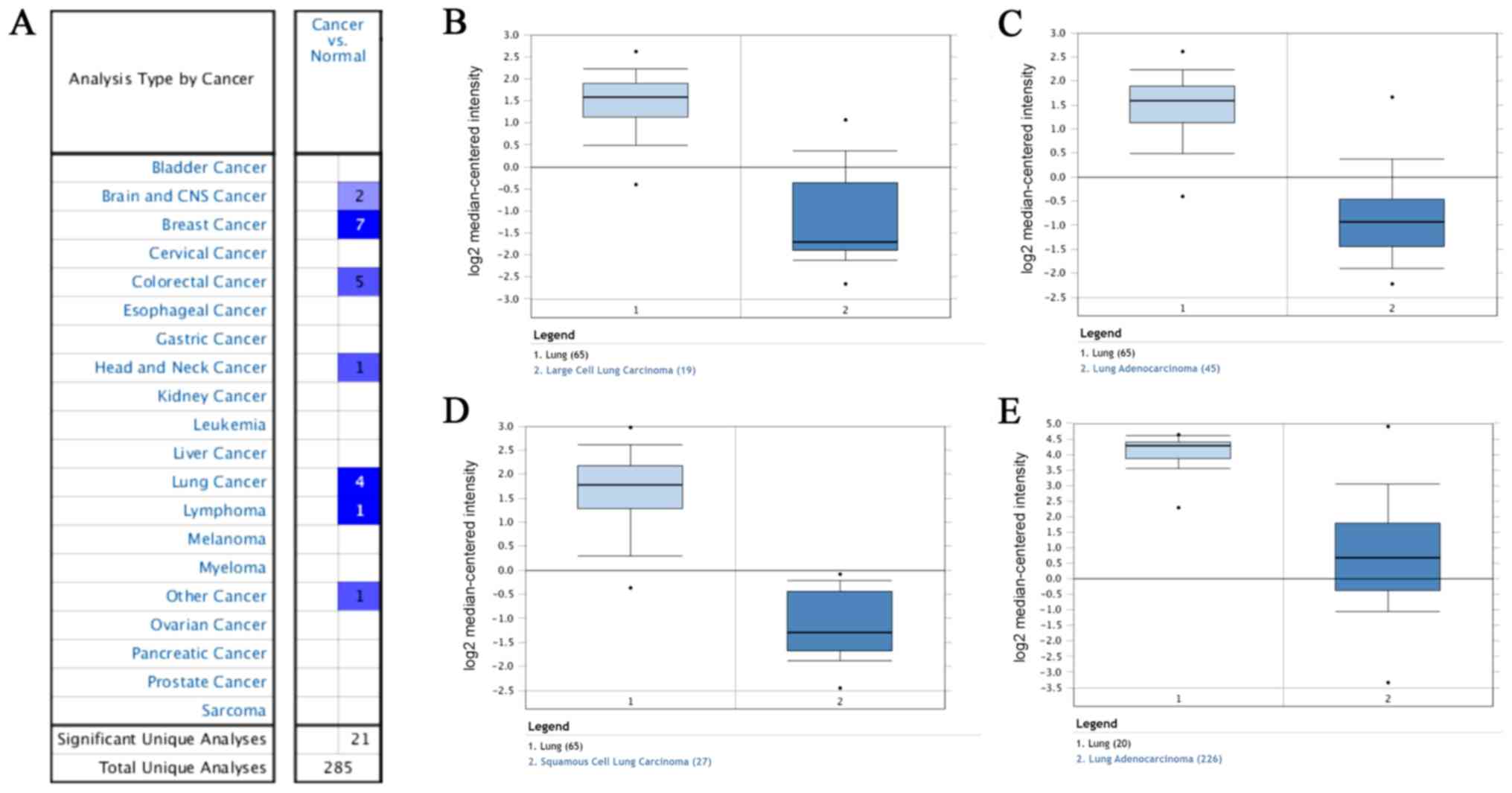
Expression of IGSF10 in different types
of cancer in the Oncomine database
The expression of IGSF10 in normal controls and in
different types of lung cancer was compared in the Oncomine
database. As presented in Fig. 7A,
the expression of IGSF10 was low in brain and CNS cancer, breast
cancer, colorectal cancer, head and neck cancer, lung cancer and
lymphoma, The expression of IGSF10 in the Hou Lung and Okayama Lung
datasets was downregulated, as presented in Fig. 7B-E.
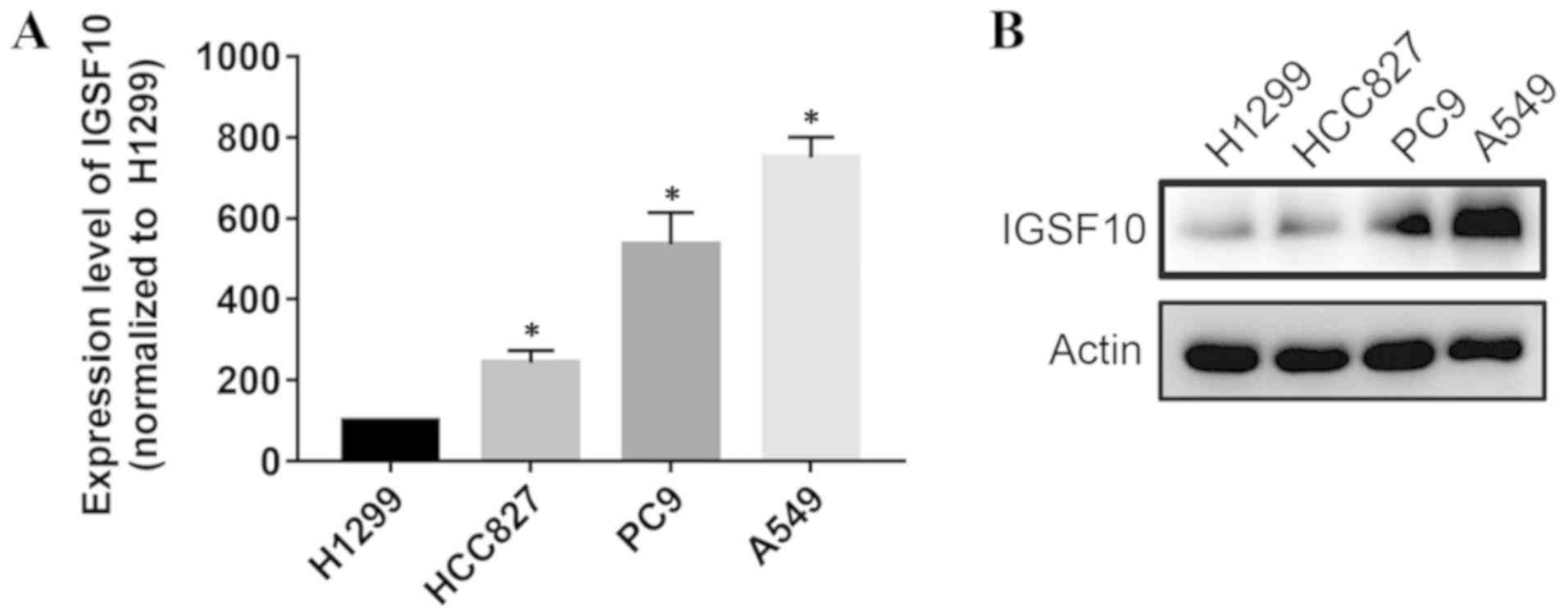
Knockdown of IGSF10 in vitro can promote
the proliferation of lung cancer cells
To select the appropriate cell model for the
following experiments, the present study first compared the
expression levels of IGSF10 in lung adenocarcinoma cells (A549,
H1299, HCC827 and PC9) (Fig. 8).
A549 and PC9 cell lines were selected for further analysis as the
expression levels of IGSF10 in these cells were the highest. The
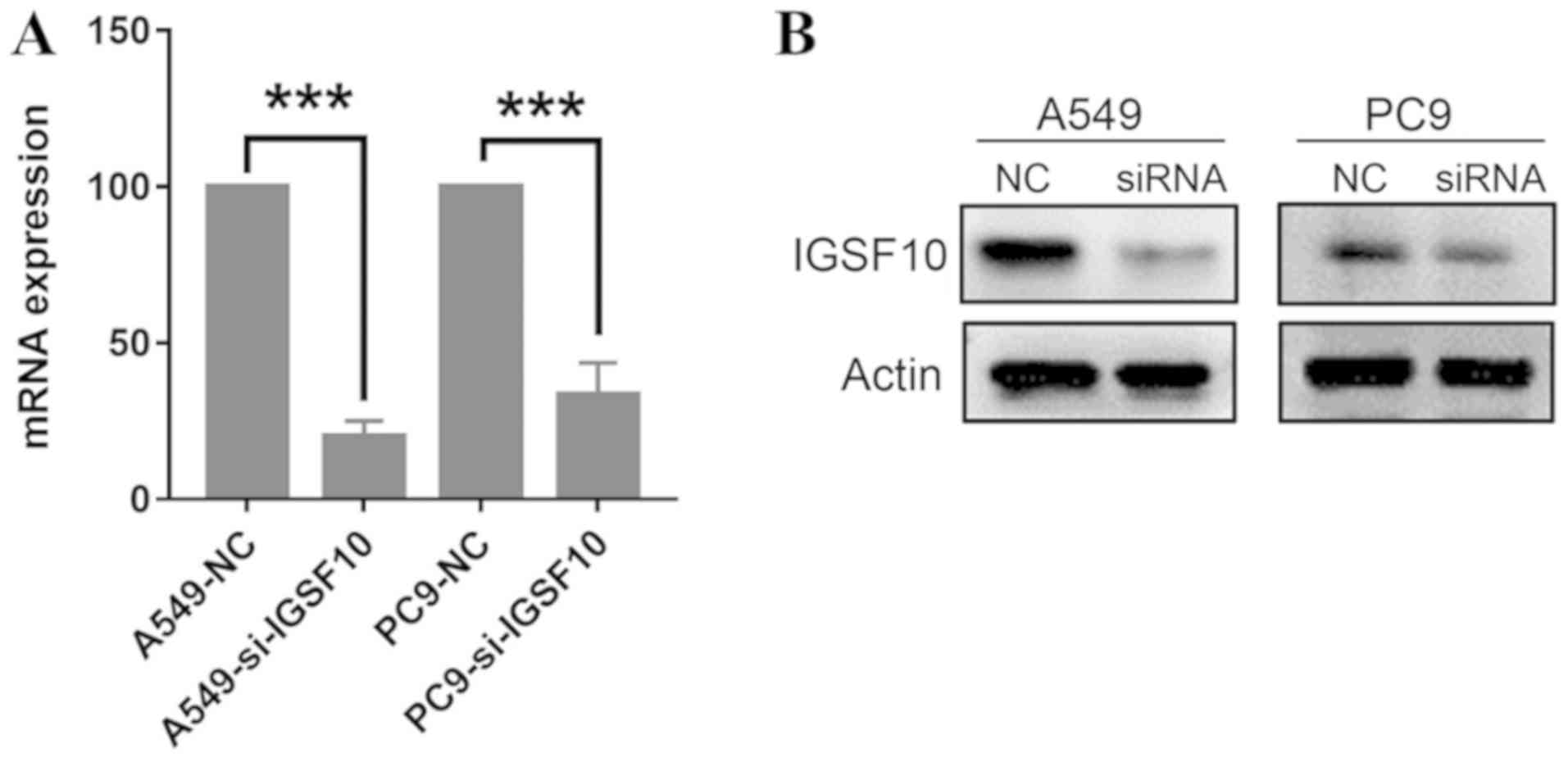
reduction efficiency was evaluated by siRNA knockdown of IGSF10,
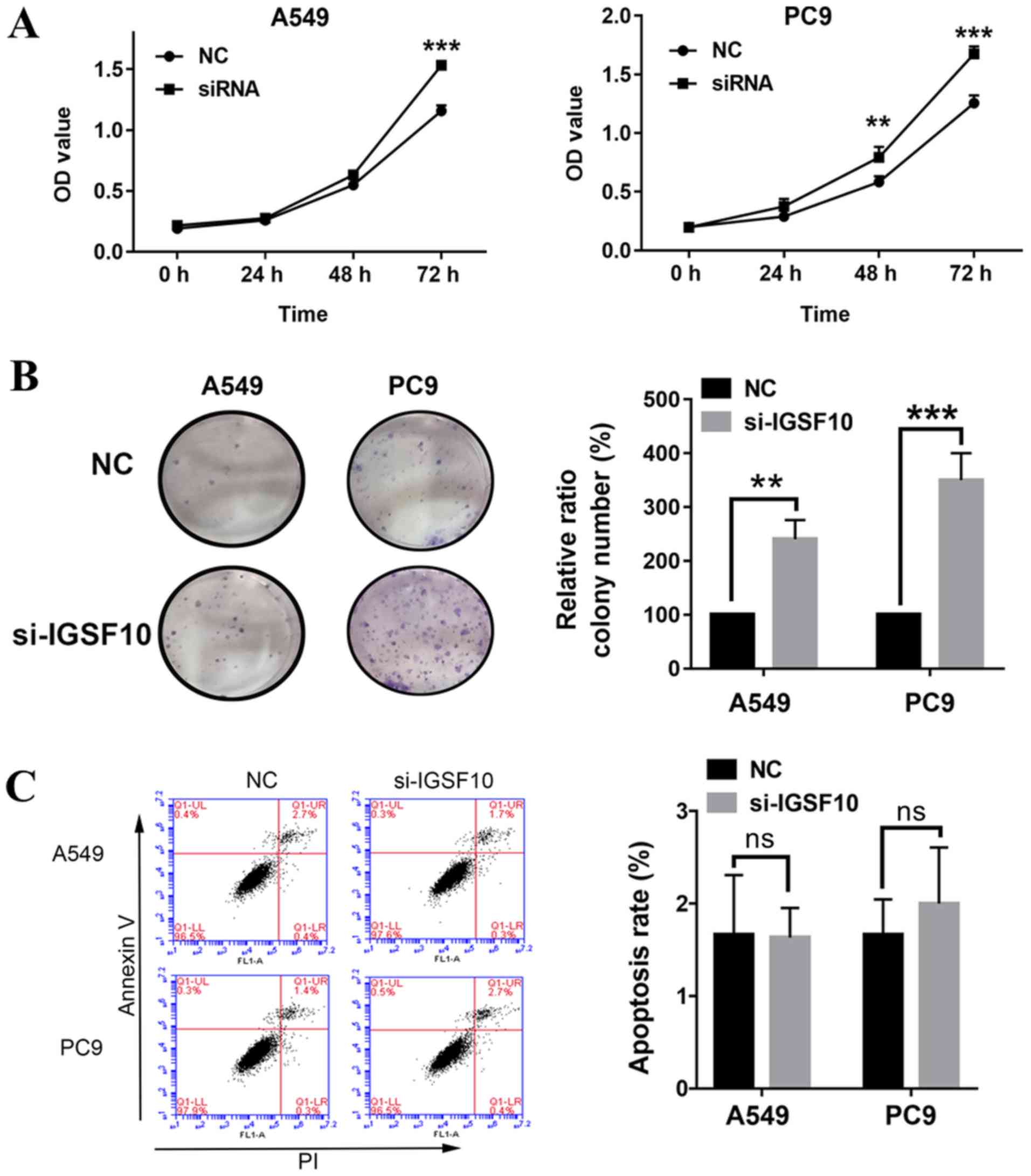
followed by RT-qPCR and western blotting (Fig. 9). MTT experiments revealed that the
proliferative ability of A549 and PC9 cells was significantly
increased following IGSF10-knockdown (Fig. 10A). The colony formation
experiments demonstrated that the number of colonies in the
IGSF10-siRNA transfection group was significantly higher compared
with that in the NC-siRNA group (Fig.
10B). Ultimately, the effect of IGSF10 on apoptosis was
evaluated by flow cytometry analysis. As shown in Fig. 10C, there was no significant
difference in apoptosis between the IGSF10-siRNA transfection group
and the NC-siRNA group. These results suggest that knockdown of
IGSF10 significantly promoted the proliferation of lung cancer
cells.
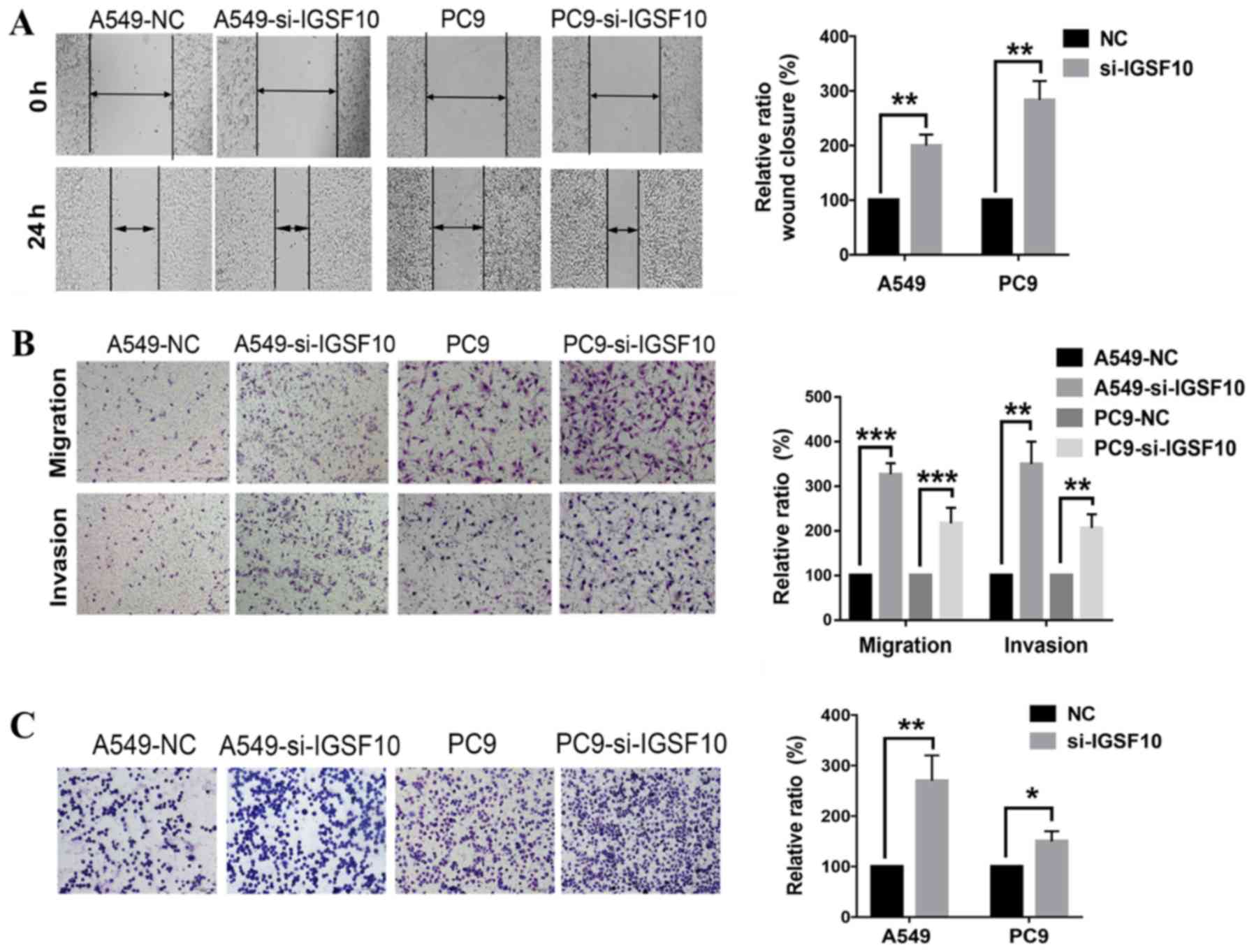
Knockdown of IGSF10 in vitro enhances the
invasion, migration and adhesion of lung cancer cells
In addition, it was investigated whether IGSF10 can
affect the invasion and migration of lung cancer cells. Wound
healing experiments revealed that the knockdown of IGSF10
significantly increased the migration ability of A549 and PC9 cells
(Fig. 11A). This result was
further validated by Transwell and Matrigel experiments in which
the migration and invasion ability of the IGSF10 siRNA transfection
group was significantly enhanced (Fig. 11B). In Fig. 11C, the adhesion experiment
confirmed that the adhesion between cells and the matrix was
significantly enhanced following IGSF10-knockdown.
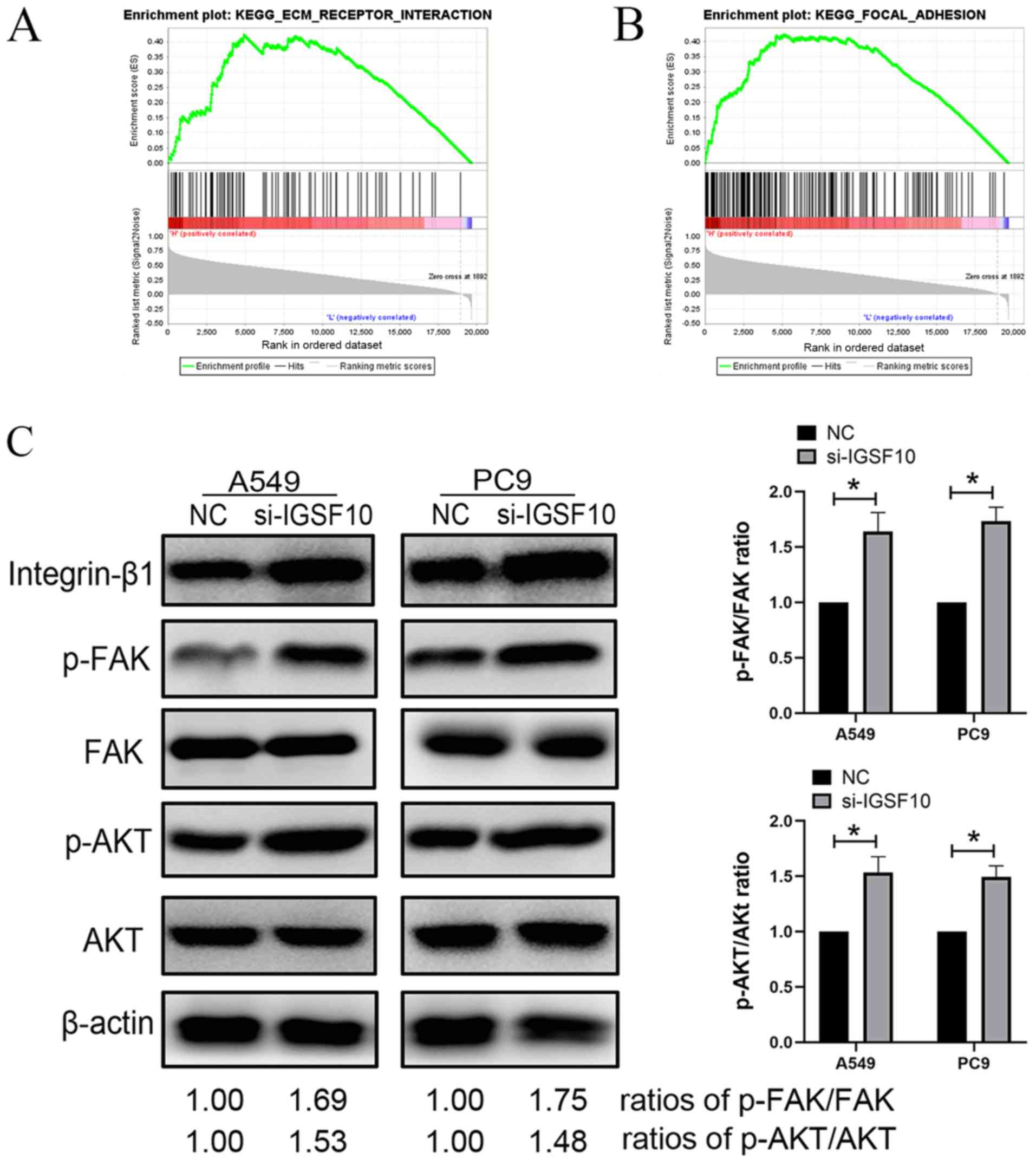
Integrin-β1/FAK pathway is activated
following knockout of IGSF10 in vitro
GSEA demonstrated that IGSF10 was significantly
associated with 'ECM receptor interaction' and 'Focal adhesion'.
Therefore, these results were verified by western blotting.
Integrins are located on the surface of the cell
membrane and are cross-membrane receptors that promote the adhesion
of the cell-extracellular matrix. The integrin family plays an
important role in cell adhesion (34,35).
The present study aimed to investigate the relationship between
IGSF10 and the integrin pathway in lung cancer. As presented in
Fig. 12, the protein expression
levels of integrin-β1, p-FAK and p-AKT were significantly
upregulated following the knockout of IGSF10 in A549 and PC9 cells.
The protein expression levels of FAK and AKT did not change
significantly. These results showed that after knocking out IGSF10,
the activation of the integrin-β1/FAK pathway in NSCLC cells may
promote the malignant phenotype of tumour cells (Fig. 12).
Discussion
The occurrence and development of lung cancer is
similar to that of most tumours, which is a multi-stage development
process with multiple genes and multiple factors (36,37).
An in-depth understanding of the molecular mechanism of the
occurrence and development of lung cancer is conducive to
developing effective targeted therapies, while also providing clues
for the early diagnosis of lung cancer (38). The present study used WGCNA to
investigate biomarkers associated with the pathogenesis of lung
cancer. A total of 14 gene modules were divided in the WGCNA
network based on the GSE19804 dataset, of which two modules were
significantly correlated with lung cancer (both P<0.001). Five
core genes, including IGSF10, RRM2, PRC1, KIF14 and KIF2C, were
obtained from these two modules. Among them, there have been
reports on the mechanisms of RRM2, PRC1, KIF14 and KIF2C in lung
cancer.
RRM2 is often highly active in lung cancer cells
(39), and its expression level is
associated with tumour cell invasion, tumour angiogenesis, tumour
metastasis and the prognosis of patients (40-43).
Therefore, RRM2 is closely related to the biological behaviour and
metastasis potential of malignant tumours.
The PRC1 gene causes disorders in the body in a
specific carcinogenic pattern that is associated with the
occurrence of a variety of human cancer types (44). The results of gene expression
analysis have demonstrated that the expression of PRC1 is
upregulated in numerous types of clinical cancer, including colon
cancer, non-small cell lung cancer, pancreatic cancer and breast
cancer (45-47). This suggests that PRC1 may be an
important tumour-promoting gene, playing an important role in the
occurrence and development of numerous malignant tumours. Chen
et al (48) found that the
high expression of PRC1 can promote the proliferation and
metastasis of hepatocellular carcinoma (HCC) cells through a mutual
regulation of the Wnt/β-catenin signalling pathway, thus promoting
the early recurrence and poor prognosis of HCC. Tang et al
(49) suggested that PRC1 may be
associated with poor prognosis in non-small cell lung cancer. At
the same time, the expression level of PRC1 in the cancer tissues
of patients with non-small cell lung cancer after chemotherapy was
lower than that before chemotherapy. This suggests that under
certain conditions, the mRNA level of PRC1 can not only predict the
prognosis of patients but may also be used as an important
reference index to evaluate the effect of tumour treatment.
Hung et al (50) studied the expression levels of
KIF14 in 122 cases of lung adenocarcinoma and found that ~30% of
patients with lung adenocarcinoma exhibited a downregulation of
KIF14 expression. In addition, the decreased expression of KIF14
was significantly correlated with the overall survival rate of
patients with lung cancer. Corson et al (51) found that the expression level of
KIF14 is significantly associated with the disease-free survival
rate and overall survival rate, and can be used as a prognostic
marker of lung cancer.
Bai et al (52) analysed the microarray data of
GSE31210 containing lung adenocarcinoma (n=226) and normal lung
tissue (n=20) samples and found that the high expression of KIF2C
was closely associated with the recurrence of lung adenocarcinoma
and tumour stage. The overall survival rate of patients with lung
adenocarcinoma with a high expression of KIF2C was significantly
decreased.
Song et al (53) also obtained similar conclusions
based on a differential gene expression analysis in GEO
datasets.
At present, there are few studies on IGSF10 in the
literature, and the mechanism related to lung cancer is unclear,
warranting further study (54,55).
The present study used the Oncomine database to investigate the
expression of IGSF10 in different cancer types, which revealed that
the expression of IGSF10 was low in numerous types of cancer. In
addition to low expression of IGSF10 in lung cancer, low expression
was found in breast cancer, colon cancer and head and neck cancer.
To clarify the mechanism of IGSF10 in lung cancer, the present
study further examined the effect of IGSF10 on the proliferation of
lung cancer cells, the adhesion between cells and the matrix, and
the related metabolic pathways through cell biology experiments.
The experimental results showed that knockout of IGSF10
significantly promoted the proliferation of lung cancer cells,
enhanced the adhesion between cells and the matrix, and activated
the integrin-β1/FAK pathway, as demonstrated by an increase in the
protein expression of integrin-β1, p-FAK and p-AKT. Integrin-β1 is
one of the subunits of integrin. The FAK-mediated signal
transduction activated by integrin-β1 plays an important role in
this process. It regulates a variety of cellular functions,
including apoptosis, cell proliferation, cell adhesion and
migration by mediating tumour and basal membranes, tumour and host
cell adhesion as well as signal transduction (56). Therefore, it plays an important
role in the occurrence and metastasis of tumour cells (57). FAK is the mediator connecting
integrin and downstream signal molecules in the integrin-β1/FAK
signalling pathway, which is at the intersection of multiple signal
pathways. Activated FAK can further activate the FAK-AKT pathway,
FAK-Ras-MAPK pathway, FAK-PI3K pathway, FAK-STAT pathway and other
signalling pathways, thus controlling transcription, translation,
the cell cycle, apoptosis and other biological effects (58,59).
Therefore, activation of the integrin-β1/FAK pathway following
IGSF10-knockout may be responsible for the promotion of cell
proliferation, enhancement of adhesion between cells and the
matrix, and the affect on the survival rate of patients.
In conclusion, the present study revealed that the
biomarker IGSF10 is closely associated with lung cancer based on
WGCNA. The possible mechanism and effects of IGSF10 on tumour cells
were preliminarily investigated through biological experiments.
Currently, there are a number or further analyses that are
required. For example, the use of mRNA data to analyse lung cancer
with high heterogeneity is inadequate, and combining these data
with genome, proteome, methylation data and other multi-omics data
for more in-depth research is necessary. In addition, with the
advent of the high-throughput era, the construction of a more
comprehensive bioinformatics database on lung cancer coupled with
the expansion of the sample size will help to improve the accuracy
of screening to explore potential gene biomarkers.
Supplementary Data
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81660549) and
Natural Science Foundation of Guangxi (grant no.
2016GXNSFAA380276).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL, YP and GQ designed the current study. BL, LL and
YJ performed the experiments. BL, XL and YH analyzed and
interpreted the data. BL wrote the manuscript. YP and GQ supervised
the study. All authors read and approved the final version of the
manuscript and agreed to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Yang D, Dai R, Zhang Q and Fang P:
Apatinib for heavily treated patients with non-small cell lung
cancer: Report of a case series and literature review. Saudi J Biol
Sci. 25:888–894. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gobbi G, Donati B, Do Valle IF, Reggiani
F, Torricelli F, Remondini D, Castellani G, Ambrosetti DC,
Ciarrocchi A and Sancisi V: The Hippo pathway modulates resistance
to BET proteins inhibitors in lung cancer cells. Oncogene.
38:6801–6817. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pan J: Dabrafenib Plus Trametinib for BRAF
V600E-Mutant Non-small Cell Lung Cancer: A Patient Case Report.
Clin Drug Investig. 39:1003–1007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu Y, Xiong ZC, Sun X, Sun L, Zhang SL,
Ma JT and Han CB: Impact of apatinib in combination with
osimertinib on EGFR T790M-positive lung adenocarcinoma. Transl
Cancer Res. 8:2151–2163. 2019. View Article : Google Scholar
|
|
5
|
Ito T, Kumagai Y, Itano K, Maruyama T,
Tamura K, Kawasaki S, Suzuki T and Murakami Y: Mathematical
analysis of gefitinib resistance of lung adenocarcinoma caused by
MET amplification. Biochem Biophys Res Commun. 511:544–550. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nelson AW, Schrock AB, Pavlick DC, Ali SM,
Atkinson EC and Chachoua A: Novel SPECC1L-MET Fusion Detected in
Circulating Tumor DNA in a Patient with Lung Adenocarcinoma
following Treatment with Erlotinib and Osimertinib. J Thorac Oncol.
14:e27–e29. 2019. View Article : Google Scholar
|
|
7
|
Nakasuka T, Ichihara E, Makimoto G, Maeda
Y and Kiura K: Primary Resistance to Alectinib Was Lost after
Bevacizumab Combined Chemotherapy in ALK-Rearranged Lung
Adenocarcinoma. J Thorac Oncol. 14:e168-e1692019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D
and Zhao S: SNHG14 confers gefitinib resistance in non-small cell
lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed
Pharmacother. 116:1089952019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu Y, Zhu X, Cao B, Wu X, Tong X, Shao YW
and Liang L: Transformation to small cell lung cancer and
activation of KRAS during long-term erlotinib maintenance in a
patient with non-small cell lung cancer: A case report. Oncol Lett.
17:5219–5223. 2019.PubMed/NCBI
|
|
10
|
Saito H, Fukuhara T, Furuya N, Watanabe K,
Sugawara S, Iwasawa S, Tsunezuka Y, Yamaguchi O, Okada M, Yoshimori
K, et al: Erlotinib plus bevacizumab versus erlotinib alone in
patients with EGFR-positive advanced non-squamous non-small-cell
lung cancer (NEJ026): Interim analysis of an open-label,
randomised, multicentre, phase 3 trial. Lancet Oncol. 20:625–635.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Sheng Z and Cai Y: Effects of
microRNA-513b on cell proliferation, apoptosis, invasion, and
migration by targeting HMGB3 through regulation of mTOR signaling
pathway in non-small-cell lung cancer. J Cell Physiol.
234:10934–10941. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiu BQ, Zhang PF, Xiong D, Xu JJ, Long X,
Zhu SQ, Ye XD, Wu Y, Pei X, Zhang XM and Wu YB: CircRNA fibroblast
growth factor receptor 3 promotes tumor progression in non-small
cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J
Cell Physiol. 234:11256–11264. 2019. View Article : Google Scholar
|
|
13
|
Braicu C, Zimta A-A, Harangus A, Iurca I,
Irimie A, Coza O and Berindan-Neagoe I: The function of non-coding
RNAs in lung cancer tumorigenesis. Cancers (Basel). 11:6052019.
View Article : Google Scholar
|
|
14
|
Lukas TJ, Mirzoeva S, Slomczynska U and
Watterson DM: Identification of novel classes of protein kinase
inhibitors using combinatorial peptide chemistry based on
functional genomics knowledge. J Med Chem. 42:910–919. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kellner U, Steinert R, Seibert V, Heim S,
Kellner A, Schulz HU, Roessner A, Krüger S and Reymond M:
Epithelial cell preparation for proteomic and transcriptomic
analysis in human pancreatic tissue. Pathol Res Pract. 200:155–163.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang C, Peng L, Zhang Y, Liu Z, Li W,
Chen S and Li G: The identification of key genes and pathways in
hepatocellular carcinoma by bioinformatics analysis of
high-throughput data. Med Oncol. 34:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu X, Lu J, Liao B, Li X, Qian X and Li K:
Driver pattern identification over the gene co-expression of drug
response in ovarian cancer by integrating high throughput genomics
data. Sci Rep. 7:161882017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Busch R, Qiu W, Lasky-Su J, Morrow J,
Criner G and DeMeo D: Differential DNA methylation marks and gene
comethylation of COPD in African-Americans with COPD exacerbations.
Respir Res. 17:1432016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pei G, Chen L and Zhang W: WGCNA
application to proteomic and metabolomic data analysis. In. Methods
in enzymology. 585. Elsevier; pp. 135–158. 2017, View Article : Google Scholar
|
|
21
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun Q, Zhang W, Wang L, Guo F, Song D,
Zhang Q, Zhang D, Fan Y and Wang J: Hypermethylated CD36 gene
affected the progression of lung cancer. Gene. 678:395–406. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An Y, Zhang Q, Li X, Wang Z, Li Y and Tang
X: Upregulated microRNA miR-21 promotes the progression of lung
adenocarcinoma through inhibition of KIBRA and the Hippo signaling
pathway. Biomed Pharmacother. 108:1845–1855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, et al: Identification
of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in
nonsmoking women. Cancer Epidemiol Biomarkers Prev. 19:2590–2597.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He J, Hu Y, Hu M and Li B: Development of
PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment
for Non-Small Cell Lung Cancer. Sci Rep. 5:131102015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakamura H, Ando K, Shinmyo T, Morita K,
Mochizuki A, Kurimoto N and Tatsunami S: Female gender is an
independent prognostic factor in non-small-cell lung cancer: A
meta-analysis. Ann Thorac Cardiovasc Surg. 17:469–480. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsuo K, Ito H, Yatabe Y, Hiraki A,
Hirose K, Wakai K, Kosaka T, Suzuki T, Tajima K and Mitsudomi T:
Risk factors differ for non-small-cell lung cancers with and
without EGFR mutation: Assessment of smoking and sex by a
case-control study in Japanese. Cancer Sci. 98:96–101. 2007.
View Article : Google Scholar
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar
|
|
31
|
Welinder C and Ekblad L: Coomassie
staining as loading control in Western blot analysis. J Proteome
Res. 10:1416–1419. 2011. View Article : Google Scholar
|
|
32
|
Dunning K and Safo AO: The ultimate
Wright-Giemsa stain: 60 years in the making. Biotech Histochem.
86:69–75. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu W, Chen X, Ning L and Jin K: Network
Analysis Reveals TNF as a Major Hub of Reactive Inflammation
Following Spinal Cord Injury. Sci Rep. 9:9282019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cooper J and Giancotti FG: Integrin
Signaling in Cancer: Mechanotransduction, Stemness, Epithelial
Plasticity, and Therapeutic Resistance. Cancer Cell. 35:347–367.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hamidi H and Ivaska J: Every step of the
way: Integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Scarpa A, Sikora K, Fassan M, Rachiglio
AM, Cappellesso R, Antonello D, Amato E, Mafficini A, Lambiase M,
Esposito C, et al: Molecular typing of lung adenocarcinoma on
cytological samples using a multigene next generation sequencing
panel. PLoS One. 8:e804782013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tessema M, Yingling CM, Liu Y, Tellez CS,
Van Neste L, Baylin SS and Belinsky SA: Genome-wide unmasking of
epigenetically silenced genes in lung adenocarcinoma from smokers
and never smokers. Carcinogenesis. 35:1248–1257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hamidaddin MA, AlRabiah H and Darwish IA:
Development and comparative evaluation of two immunoassay platforms
for bioanalysis of crizotinib: A potent drug used for the treatment
of non-small cell lung cancer. Talanta. 201:217–225. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Heidel JD, Liu JY, Yen Y, Zhou B, Heale
BS, Rossi JJ, Bartlett DW and Davis ME: Potent siRNA inhibitors of
ribonucleotide reductase subunit RRM2 reduce cell proliferation in
vitro and in vivo. Clin Cancer Res. 13:2207–2215. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Duxbury MS and Whang EE: RRM2 induces
NF-kappaB-dependent MMP-9 activation and enhances cellular
invasiveness. Biochem Biophys Res Commun. 354:190–196. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lundin D, Berggren G, Logan DT and Sjöberg
BM: The origin and evolution of ribonucleotide reduction. Life
(Basel). 5:604–636. 2015.
|
|
42
|
Li Y, Wang Z, Tang L, Hu R, Huang L and
Ding J: RRM2 overexpression in glioblastoma enhances the
proliferation and invasion of cancer cells. Int J Clin Exp Pathol.
9:11623–11630. 2016.
|
|
43
|
Chen WX, Yang LG, Xu LY, Cheng L, Qian Q,
Sun L and Zhu YL: Bioinformatics analysis revealing prognostic
significance of RRM2 gene in breast cancer. Biosci Rep.
39:392019.
|
|
44
|
Federico A, Sepe R, Cozzolino F, Piccolo
C, Iannone C, Iacobucci I, Pucci P, Monti M and Fusco A: The
complex CBX7-PRMT1 has a critical role in regulating E-cadherin
gene expression and cell migration. Biochim Biophys Acta Gene Regul
Mech. 1862:509–521. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu F, Zhou C, Zeng H, Liu Y and Li S: BMI1
activates WNT signaling in colon cancer by negatively regulating
the WNT antagonist IDAX. Biochem Biophys Res Commun. 496:468–474.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ramaiah MJ and Vaishnave S: BMI1 and PTEN
are key determinants of breast cancer therapy A plausible
therapeutic target in breast cancer. Gene. 678:302–311. 2018.
View Article : Google Scholar
|
|
47
|
Zhan P, Zhang B, Xi GM, Wu Y, Liu HB, Liu
YF, Xu WJ, Zhu QQ, Cai F, Zhou ZJ, et al: PRC1 contributes to
tumorigenesis of lung adenocarcinoma in association with the
Wnt/β-catenin signaling pathway. Mol Cancer. 16:1082017. View Article : Google Scholar
|
|
48
|
Chen J, Rajasekaran M, Xia H, Zhang X,
Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL,
et al: The microtubule-associated protein PRC1 promotes early
recurrence of hepatocellular carcinoma in association with the
Wnt/β-catenin signalling pathway. Gut. 65:1522–1534. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tang H, Xiao G, Behrens C, Schiller J,
Allen J, Chow CW, Suraokar M, Corvalan A, Mao J, White MA, et al: A
12-gene set predicts survival benefits from adjuvant chemotherapy
in non-small cell lung cancer patients. Clin Cancer Res.
19:1577–1586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hung PF, Hong TM, Hsu YC, Chen HY, Chang
YL, Wu CT, Chang GC, Jou YS, Pan SH and Yang PC: The motor protein
KIF14 inhibits tumor growth and cancer metastasis in lung
adenocarcinoma. PLoS One. 8:e616642013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Corson TW, Zhu CQ, Lau SK, Shepherd FA,
Tsao MS and Gallie BL: KIF14 messenger RNA expression is
independently prognostic for outcome in lung cancer. Clin Cancer
Res. 13:3229–3234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bai Y, Xiong L, Zhu M, Yang Z, Zhao J and
Tang H: Co-expression network analysis identified KIF2C in
association with progression and prognosis in lung adenocarcinoma.
Cancer Biomark. 24:371–382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Song YJ, Tan J, Gao XH and Wang LX:
Integrated analysis reveals key genes with prognostic value in lung
adenocarcinoma. Cancer Manag Res. 10:6097–6108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Daino K, Ugolin N, Altmeyer-Morel S,
Guilly MN and Chevillard S: Gene expression profiling of
alpha-radiation-induced rat osteosarcomas: Identification of
dysregulated genes involved in radiation-induced tumorigenesis of
bone. Int J Cancer. 125:612–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Thutkawkorapin J, Picelli S, Kontham V,
Liu T, Nilsson D and Lindblom A: Exome sequencing in one family
with gastric- and rectal cancer. BMC Genet. 17:412016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhao G, Gong L, Su D, Jin Y, Guo C, Yue M,
Yao S, Qin Z, Ye Y, Tang Y, et al: Cullin5 deficiency promotes
small-cell lung cancer metastasis by stabilizing integrin β1. J
Clin Invest. 129:972–987. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boudria A, Abou Faycal C, Jia T, Gout S,
Keramidas M, Didier C, Lemaître N, Manet S, Coll JL, Toffart AC, et
al: VEGF165b, a splice variant of VEGF-A, promotes lung tumor
progression and escape from anti-angiogenic therapies through a β1
integrin/VEGFR autocrine loop. Oncogene. 38:1050–1066. 2019.
View Article : Google Scholar
|
|
58
|
Mitra SK, Hanson DA and Schlaepfer DD:
Focal adhesion kinase: In command and control of cell motility. Nat
Rev Mol Cell Biol. 6:56–68. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hayashi I, Vuori K and Liddington RC: The
focal adhesion targeting (FAT) region of focal adhesion kinase is a
four-helix bundle that binds paxillin. Nat Struct Biol. 9:101–106.
2002. View
Article : Google Scholar : PubMed/NCBI
|