Introduction
Breast cancer is the most prevalent type of cancer
among women worldwide and it is characterized by a high morbidity
(1). Metastasis is considered one
of the most crucial stages of tumor progression as it accounts for
>90% of all cancer-related deaths (2,3).
There have been notable improvements in survival over the past 3
decades for the majority of cancer types and the 5-year relative
survival rate for breast cancer patients has increased from 63% in
the early 1960s to 91% between 2007 and 2013. However, the survival
rate is low among women with more advanced stages of the disease at
the time of diagnosis. Thus, the relative survival is 99% for
localized disease, 85% for regional disease and 27% for
distant-stage disease (4).
According to the World Health Organization (WHO), the improvement
of the survival of breast cancer patients by early detection
remains the foundation of breast cancer regulations.
Epithelial-mesenchymal transition (EMT) is a
biological process through which a polarized epithelial cell, which
normally interacts with the basement membrane via its basal
surface, which endows the cell with the ability to undergo multiple
biochemical alterations. These alterations enable the cell to
acquire a mesenchymal phenotype; consequently, the cell also
acquires an enhanced migratory and invasive capacity, and increased
resistance to apoptosis (5).
AXL is a receptor of the TAM family of receptor
tyrosine kinases (Tyro3, Axl and Mer). It transduces signals from
the extracellular matrix into the cytoplasm by binding to its main
ligand, thus inducing dimerization and autophosphorylation and the
activation of signaling pathways involved in a wide variety of
cellular processes, including survival, proliferation, migration,
invasion and EMT (6). AXL is
considered to function as an oncogene and is known to be
overexpressed in breast, lung, ovarian, gastric, pancreatic and
prostate cancers. In addition, AXL is regulated by tumor suppressor
microRNAs (miRNAs or miRs), such as miR-34a and it is associated
with an enhanced metastatic potential (7), a poor prognosis (8) and resistance to therapy (9) in several types of cancer.
Slug is a C2H2-type zinc-finger transcription factor
and is a member of the Snail family. It is known to play a key role
in the development of cancer and in cancer-related EMT. The Slug
protein binds to E-box motifs in the promoter of downstream genes
and has been shown to suppress E-cadherin transcription in breast
cancer, gastric cancer and esophageal squamous cell carcinoma
(10,11). It has also been demonstrated that
Slug is negatively associated with estrogen receptor (ER)α in both
breast and lung cancer (12,13).
Cluster of differentiation 24 (CD24) is a small
GPI-linked membrane glycoprotein with glycosylation sites that bind
P-selectin (14). CD24 is an
adhesion molecule, and it has been shown to be widely expressed in
a number of types of cancer, including renal, ovarian, lung and
pancreatic cancer (15-17). A previous study also suggested that
CD24 expression is a candidate marker for the prognosis of patients
with breast cancer (18).
Ras homolog gene family member A (Rho-A) is a member
of the Ras-related C3 botulinum toxin substrate (Rac) subfamily of
the Rho family and is a small (~22 kDa) G protein/guanosine
triphosphatase (19). Rho-A can
induce the reorganization of the cell cytoskeleton and can regulate
cell migration by activating effector proteins, such as
Rho-associated coiled-coil kinase (ROCK) (20); these types of alterations are
associated with tumor invasion and migration in several types of
cancer cells (21,22). In breast cancer, an increased
expression of Rho-A has been shown to be associated with cancer
progression (23,24). In addition, an increased expression
of Rho-A seems to promote an enhanced cell invasion and metastasis
(24).
There is evidence to suggest that the expression of
miRNAs play a pivotal role in regulating the acquisition of the EMT
phenotype (25). miRNAs are
endogenous, non-coding RNAs approximately 21-23 nucleotides in
length and have been identified as key negative regulators of gene
expression through the endogenous RNA interference machinery.
miRNAs are able to regulate gene expression by binding to miRNA
recognition elements (MREs) located in the 3′ untranslated region
(3′-UTR) of target messenger RNAs (mRNAs), leading to their
translational repression or degradation (25,26).
It has been previously demonstrated either that miRNAs function as
oncogenes or tumor suppressor genes, depending on their target
genes of regulation, in order to control cell proliferation,
invasion, migration, differentiation and apoptosis (27). Recently, the abnormal expression of
miRNAs in several types of cancer has been confirmed, including
colon cancer, hepatocellular carcinoma, lung cancer and breast
cancer (28); in these types of
cancer, miRNAs have been shown to be associated with tumor
progression, invasion, metastasis and angiogenesis (28,29).
miRNAs as tumor suppressors have been found to be
over-expressed in a variety of human cancers, contributing to tumor
development. miRNA-34a (miR-34a) has been reported to function as
tumor suppressor miRNA and has been shown to be downregulated in
cancers (28,29); it also plays an important role in
tumorigenesis and the progression of cancer (30).
It is known that certain herbs have the ability to
regulate miRNAs associated with cancer (31). Among these, curcumin, a naturally
occurring compound derived from the rhizome of Curcuma
longa, and its analogs are known to exert potent
anti-carcinogenic effects (32)
through the regulation of multiple downstream cancer-related
signaling molecules (33). Studies
have demonstrated a protective role of curcumin in the oxidative
stress of breast cells (34,35).
It has been demonstrated that curcumin affects breast cancer cells
transformed by low concentrations of radiation and estrogen
(36). Among the genes involved in
such processes, CD44 expression has been shown to be associated
with oxidative stress in such cells (37). On the other hand, it has been
demonstrated that curcumin suppresses the invasive capabilities of
breast cancer cell lines through EMT (38).
MDA-MB-231 is a basal-like triple-negative breast
cancer cell line. Triple-negative breast cancer [negative for ER,
progesterone receptor (PgR) and human epidermal growth factor
receptor 2 (HER2)] is the most aggressive breast cancer subtype and
is associated with a poor prognosis. The aim of the present study
was to determine whether curcumin modulates miRNA expression,
whether it induces EMT-related changes, and thereof, whether it
affects the motility of breast cells.
Materials and methods
Breast cell lines
MCF-10F human breast and MDA-MB-231 human breast
cancer cell lines were obtained from the American Type Culture
Collection (ATCC). The MCF-10F cells were grown in Dulbecco’s
modified Eagle’s medium (DMEM)/F-12 (1:1) supplemented with 100
U/ml penicillin, 100 μg/ml streptomycin (all obtained from Life
Technologies, Thermo Fisher Scientific) and 5% equine serum
(Biofluids), 0.5 µg/ml hydrocortisone (Sigma) and 0.02
µg/ml epidermal growth factor (Collaborative Research). The
MDA-MB-231 cells were maintained in RPMI-1640 medium (Life
Technologies, Thermo Fisher Scientific) supplemented with
penicillin (100 U/ml), streptomycin (100 µg/ml), 0.1 mM
non-essential amino acids, 0.2 mM glutamine, 1 mM pyruvate and 10%
heat-inactivated fetal bovine serum, and incubated in a 5%
CO2 humidified atmosphere at 37°C. The cells were grown
to 80% confluence prior to the treatments.
Cell viability assay
The viability of the cells was determined in the
presence of curcumin (Sigma-Aldrich; Merck KGaA). Firstly, the
MCF-10F and MDA-MB-231 cell lines were seeded in 96-well
microplates (25×103 cells/well) in a final volume of 100
µl and incubated in culture medium for 48 h at 37°C and a 5%
concentration of CO2. Following overnight incubation at
37°C, the cells were treated with 0, 5, 10, 15, 20, 25, 30 and 35
µM of curcumin. Curcumin was dissolved in 0.1% of DMSO
solvent and used as a blank. Cells not treated curcumin functioned
as the control group. Following 48 h of incubation at 37ºC, the
metabolic activity of the living cells was determined by
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) assay (Sigma-Aldrich). The medium was removed, and the
insoluble formazan crystals were dissolved in 200 µl of
DMSO. Subsequently, 20 µl MTT (5 mg/ml) were added to each
well. The cells were then incubated at 37°C for a further 4 h. The
reduction of MTT was determined following the manufacturer’s
instructions. The absorbance was determined at 570 nm (Autobio
Labtec Instruments) in a microplate reader (Molecular Devices). The
results were expressed as the percentage of cell survival relative
to the control. The 50% inhibitory concentration (IC50)
was defined as the curcumin concentration that induced a 50%
reduction in the viability of the cells compared to control, as
previously described (34-39).
RNA extraction and cDNA synthesis
For RNA isolation, two separate purification
procedures were carried out. The measurement of small RNA
(containing miRNA) and larger RNA (containing mRNA) from the cells
was determined using the RNeasy Plus Mini kit (cat. no. 74204) and
the Rneasy MinElute Cleanup kit (cat. no. 74134), according to the
manufacturer’s instructions (Qiagen). For mRNAs, 2 µg of the
longer RNA fraction were reverse transcribed into cDNA using the
High Capacity cDNA Reverse Transcription kit (Applied Biosystems).
For miRNAs, reverse transcription was carried out using 2 to 10 ng
of the small RNA fraction, the TaqMan® MicroRNA Reverse
Transcription kit (cat. no. 4366596; Applied Biosystems) and
gene-specific RT-primers according to the TaqMan MicroRNA assay
protocol (cat. no. 4427975; Applied Biosystems).
RT-qPCR for the analysis of mRNA
expression
RNA was obtained using RNeasy Plus Mini kit and
RNeasy MinElute kit (Qiagen) following the manufacturer’s protocol
and for reverse transcription the High capacity cDNA Reverse
Transcription (Applied Biosystems). An aliquot of a dilution 1/50
of cDNA (2 µl) was used in a 20 µl qPCR reaction
containing SYBR-Green PCR Master Mix (Agilent Technologies) and 5
μM of each primer for the target genes, such as Axl,
Slug, CD24 and Rho-A. The primers for the
selected genes are presented in Table
IA. The reaction was carried out on a CFX 96 Touch Real-Time
PCR Detection System (Bio-Rad Laboratories) with the following
conditions: 95°C for 10 min and 40 cycles of a 2-step program of
95°C for 10 sec and 61°C for 45 sec when fluorescence-reading
occurs. The reactions were carried out in triplicate and the
threshold of the cycle was obtained using Bio-Rad CFX Manager 2.1
software and the average gene expression was normalized using the
reference housekeeping gene β-actin and the relative expression
level was calculated as previously described (36,38),
using the 2−ΔΔCq method (40).
 | Table ISequences of primers used for
RT-qPCR. |
Table I
Sequences of primers used for
RT-qPCR.
A. Sequence and
annealing temperature of the primers used in the RT-qPCR reactions
|
|---|
| Gene name | Primer
sequencea | Annealing
temperature |
|---|
| Axl | F:
GTTTGGAGCTGTGATGGAAGGC | 62.01 |
| R:
CGCTTCACTCAGGAAATCCTCC | 61 |
| Slug | F:
GACCCTGGTTGCTTCAAGGA | 59.89 |
| R:
TGTTGCAGTGAGGGCAAGAA | 60.11 |
| CD24 | F:
AACTAATGCCACCACCAGG | 58.1 |
| R:
GACGTTTCTTGGCCTGAGTC | 58.9 |
| Rho-A | F:
CCATCATCCTGGTGGGGAAT | 54.8 |
| R:
CATCCCAAAAGCGCCA | 56.1 |
|
B. Alignment of
mature miR-200a, miR-200c, let-7a, let-7c, let-7b, miR-21 and
miR-34a sequences
|
| Assay ID | miR | Mature
sequenceb |
| 000502 | hsa-miR-200a |
UAACACUGUCUGGUAACGAUGU |
| 000505 | hsa-miR-200c |
UAAUACUGCCGGGUAAUGAUGG |
| 000377 | hsa-let-7a |
UGAGGUAGUAGGUUGUAUAGUU |
| 000379 | hsa-let-7c |
UGAGGUAGUAGGUUGUAUGGUU |
| 000378 | hsa-let-7b |
UGAGGUAGUAGGUUGUGUGGUU |
| 000397 | hsa-miR-21 |
UAGCUUAUCAGACUGAUGUUGA |
| 000425 | hsa-miR-34a |
UGGCAGUGUCUUAGCUGGUUGU |
RT-qPCR for the analysis of miRNA
expression
It was done by using RNeasy Plus Mini kit and RNeasy
MinElute Cleanup kit (Qiagen) according to the manufacturer’s
protocol. An aliquot of cDNA (2 µl) was used in 15 µl
qPCR reaction containing TaqMan Small RNA Assays specific for each
miRNA (cat. no. 4427975) and TaqMan Universal PCR Master Mix II
(cat. no. 4440040; Applied Biosystems). The alignment of mature
miR-200a, miR-200c, let-7a, let-7c, let-7b, miR-21 and miR-34a
sequences is presented in Table
IB. The reaction was carried out on a CFX 96 Touch Real-Time
PCR Detection Systems (Bio-Rad Laboratories) with the following
conditions; 50°C for 2 min, 95°C for 10 min and 40 cycles of a
2-step program of 95°C for 15 sec and 60°C for 60 sec when
fluorescence-reading occurs. The reactions were carried out in
triplicate and the threshold of the cycle was obtained using
Bio-Rad CFX Manager 2.1 software and the average gene expression
was normalized using the reference snRNA U6. The relative
expression of each miRNA was normalized against snRNA U6 (ID no.
001973) expression using a Bio-Rad CFX Manager 2.1 software.
Oligonucleotide transfection
The MCF-10F and MDA-MB-231 cells were transfected
with 75 pmol anti-miR-34a (cat. no. AM11030) or anti-miR negative
control (cat. no. AM17010) (Life Technologies, Thermo Fisher
Scientific) using Lipofectamine® 2000 (Invitrogen,
Thermo Fisher Scientific) according to the manufacturer’s
instructions. The cells were assayed 24, 48 or 72 h
post-transfection.
Cell migration and invasion assays
Migration and invasion assays were performed using
modified Boyden’s chambers (Corning, Inc.) constructed with
multiwall cell culture plates and cell culture inserts as
previously described (36,38). For the invasion assay, the upper
chambers of Transwells with 8-µm membrane pores were
pre-coated with Matrigel matrix gel (60 µl) (BD Biosciences)
at least 1 h prior to the seeding of the tested cells. The cells
were pre-treated with curcumin (30 μM) and cultured for 24 h. A
total of 3×105 cells in 100 µl of medium without
fetal bovine serum was added to the upper chambers and 600
µl of medium with 10% FBS was placed in the lower chambers
as a chemoattractant. The Matrigel invasion chamber was incubated
for 16 h in a humidified tissue culture incubator. The upper
chambers were then removed from the lower chambers and then wiped
using cotton swabs. The invaded and migrated cells were fixed using
100% methanol at room temperature for 15 min, visualized and
quantified using DAPI. Ten fields of each chamber were photographed
using a fluorescence microscope (×40 magnification, Olympus Corp.).
Migration assay was carried out the same way as the invasion assay;
however, the Transwell insert was not coated with Matrigel. This
experiment was independently repeated 3 times.
Protein expression by immunoperoxidase
staining
Exponentially growing cells were plated on a glass
chamber slide (Nunc Inc.) at a density of 1×104 cells/ml
of medium and allowed to grow for 2-3 days until 70% confluent as
previously described (39). The
cells were then fixed with buffered paraformaldehyde at room
temperature, incubated with 1% H2O2 in
methanol to block endogenous peroxidase and washed again twice with
buffer solution. Subsequently, the cell cultures were then covered
with normal horse serum for 30 min at room temperature and
incubated with either anti-mouse or anti-goat monoclonal or
polyclonal antibodies: Anti-Axl (mouse, sc-166269), anti-Slug
(mouse, sc-166476), anti-CD24 (mouse, sc-65257) and anti-Rho-A
(mouse, sc-418), (all from Santa Cruz Biotechnology, Inc.) at a
1:500 dilution overnight at 4°C. The cells were subsequently
incubated for 45 min with diluted biotinylated secondary antibody
solution (Vector Laboratories Inc.) and Vectastin Elite ABC reagent
(Vector Laboratories Inc.) was used. The experiments were repeated
twice in cells with identical passages in vitro. A
semi-quantitative estimation based on the relative staining
intensity of protein expression both in the untreated control and
treated cells was determined. The number of immunoreactive cells
(30 cells/field) was counted in 5 randomly selected microscopy
fields per sample and the percentage of relative fold protein
expression was calculated.
Statistical analysis
Numerical data are expressed as the means ± standard
error of the mean (SEM). Comparisons between 2 groups were made
using the t-test and between several treatment groups and the
controls by ANOVA with Dunnett’s test. Data were analyzed using
release IBM SPSS 22.0 (SPSS, Inc.). A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Cell viability of the cells following
treatment with curcumin
Cell viability assay was performed using MTT assay
to examine the effects of curcumin at various concentrations
(fluctuating from 0 to 35 µM for 48 h) on the viability of
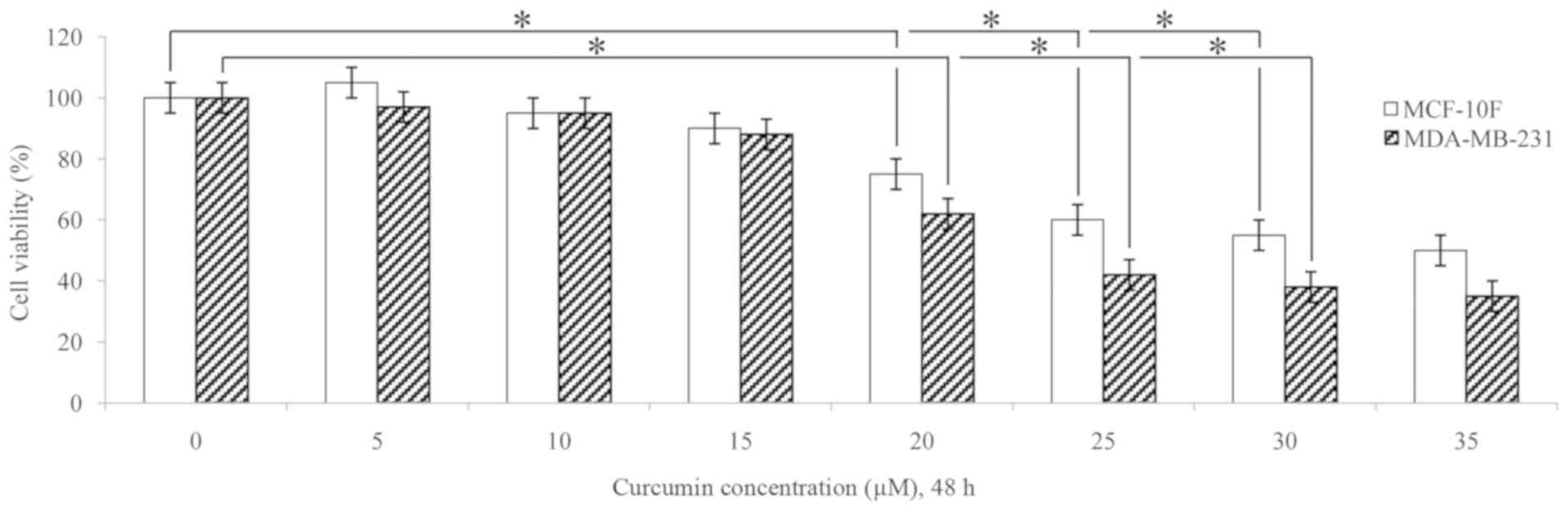
the MCF-10F and MDA-MB-231 cells. As shown in Fig. 1, a significant (P<0.05) decrease
in cell viability was observed in both cell lines following
treatment with 20 and 30 µM curcumin for 48 h in comparison
to the untreated cells (0 µM). The concentrations of 10 and
30 µM curcumin were used in several experiments.
Effects of curcumin on MCF-10F gene and
protein expression levels
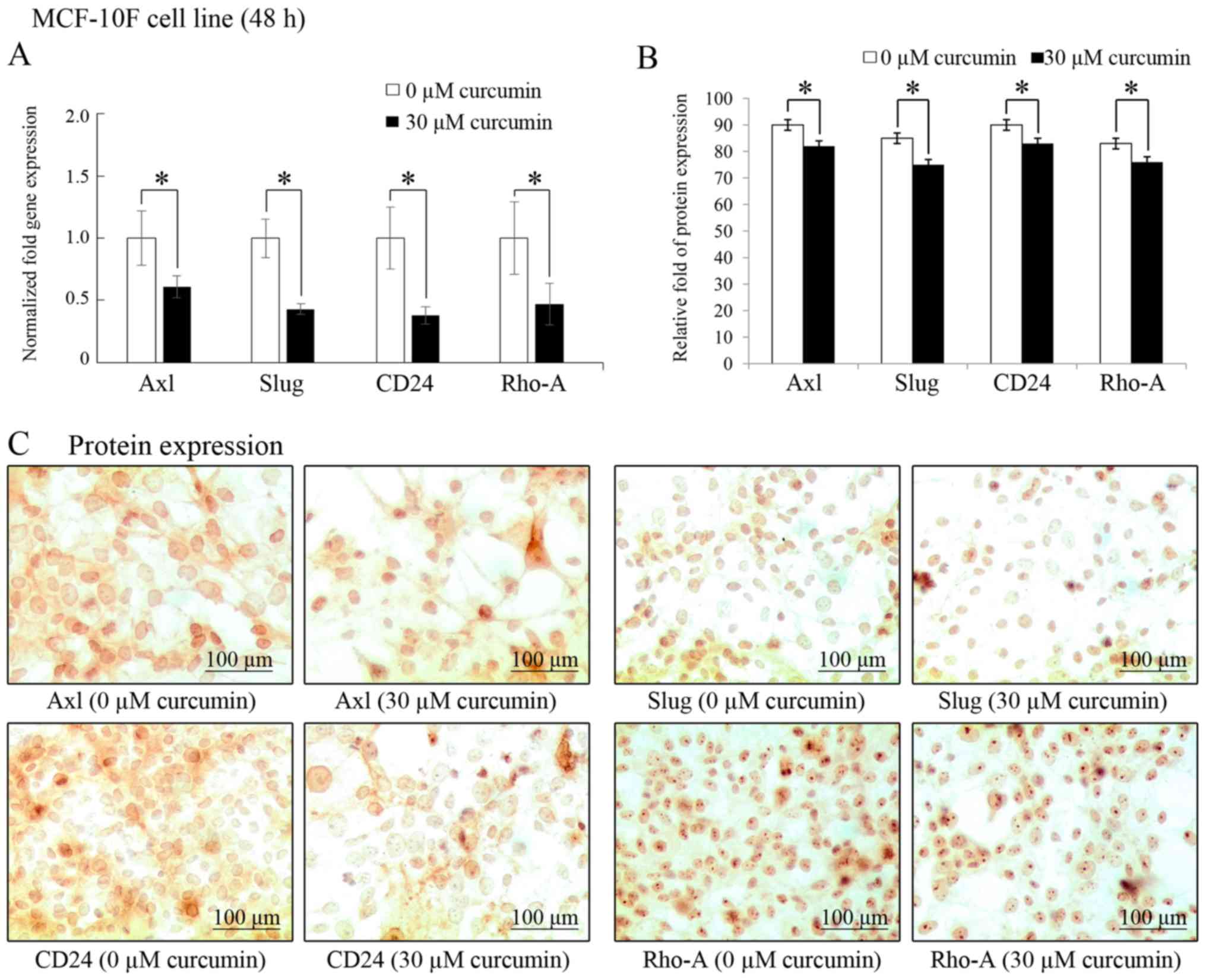
As shown in Fig.
2A, curcumin (30 µM for 48 h) significantly (P<0.05)
decreased the Axl (39%), Slug (57%), CD24
(62%) and Rho-A (53%) gene transcript levels. In addition,
protein expression was examined by immunocytochemistry in MCF-10F
cell line in comparison to its own controls (Fig. 2B). Similar to the mRNA levels,
curcumin also induced a decrease in the protein levels of Axl,
Slug, CD24 and Rho-A in the cells. Representative images of the
effects of curcumin on protein expression levels are presented in
Fig. 2C.
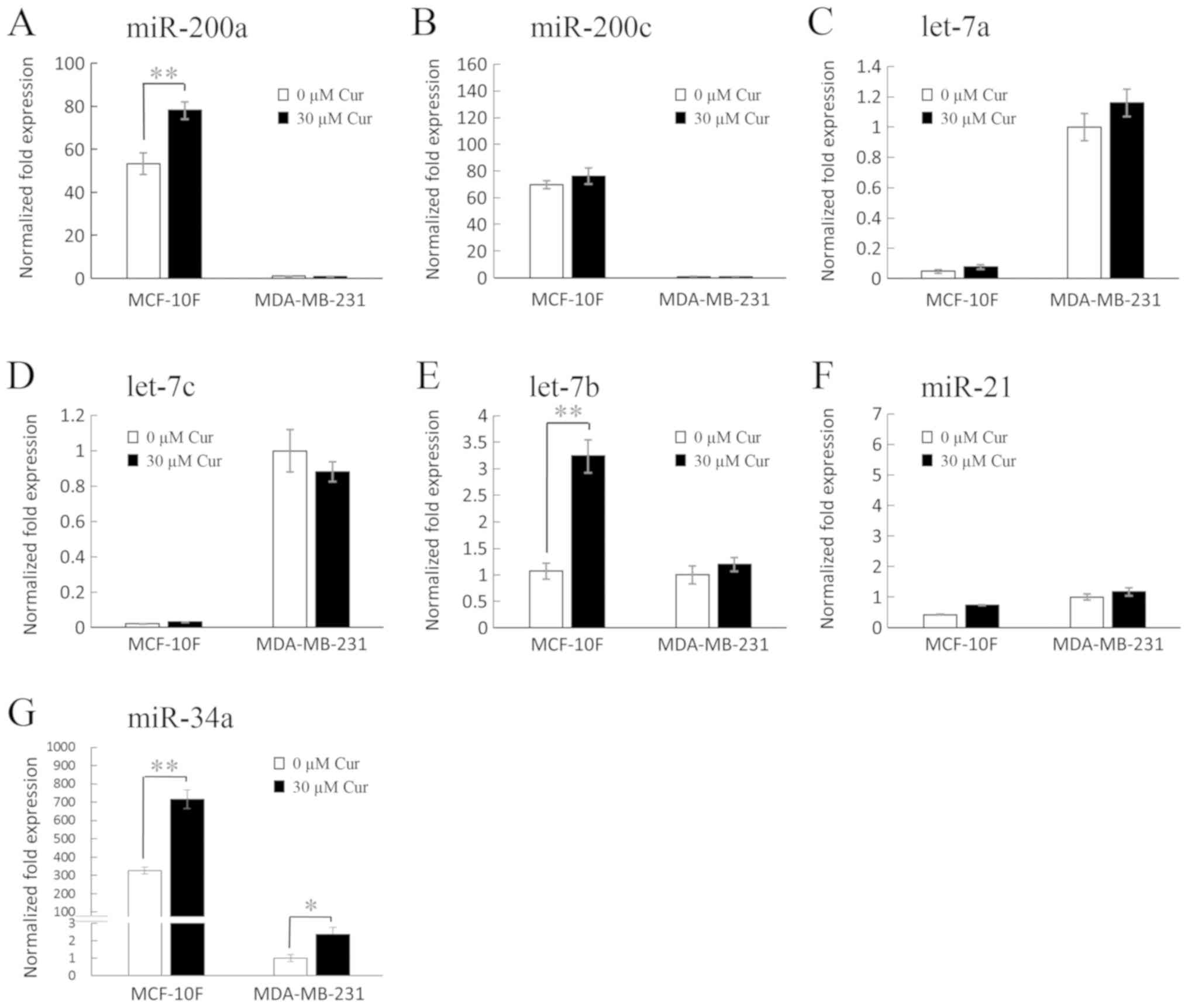
Effects of curcumin on miRNA levels
As shown in Fig. 3,
treatment with 30 µM curcumin significantly (46%, P<0.05)
increased miR-200a expression in the MCF-10F cell line in
comparison with its own control (Fig.
3A); however, it had no marked effect on the MDA-MB-231 cell
line. In addition, curcumin did not exert any notable effects on
the expression levels of miR-200c (Fig. 3B), let-7a (Fig. 3C), let-7c (Fig. 3D) and miR-21 (Fig. 3F) in the MCF-10F and MDA-MB-231
cell lines; however, it significantly (P<0.05) increased let-7b
expression (202%) in the MCF-10F cell line in comparison to its own
control (Fig. 3E), although no
notable effect was observed on the MDA-MB-231 cell line. On the
other hand, as shown in Fig. 3G,
curcumin significantly increased miR-34a expression in both the
MCF-10F and MDA-MB-231 cell lines (P<0.005, 120% and P<0.05,
138%, respectively), when compared to their own controls.
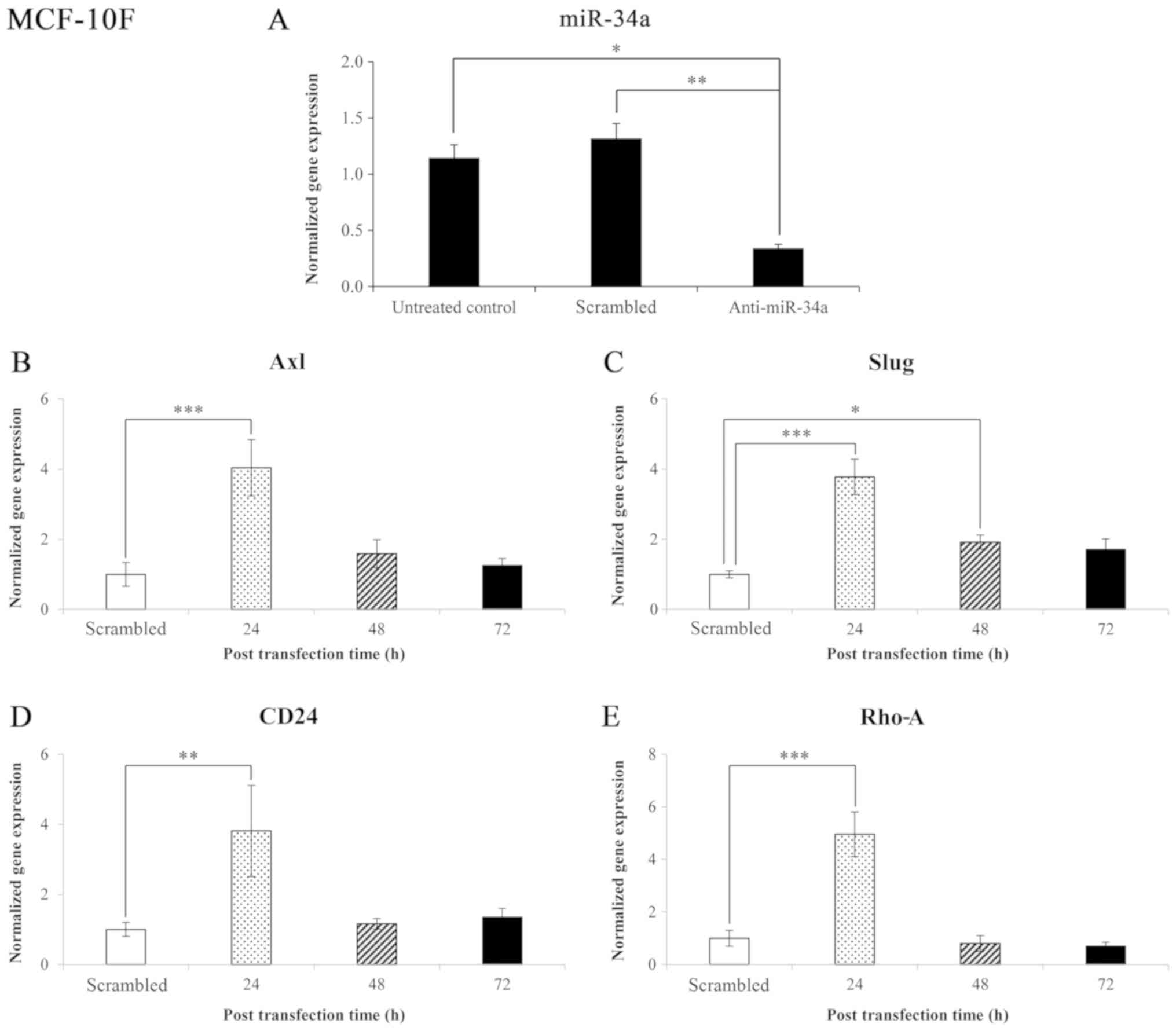
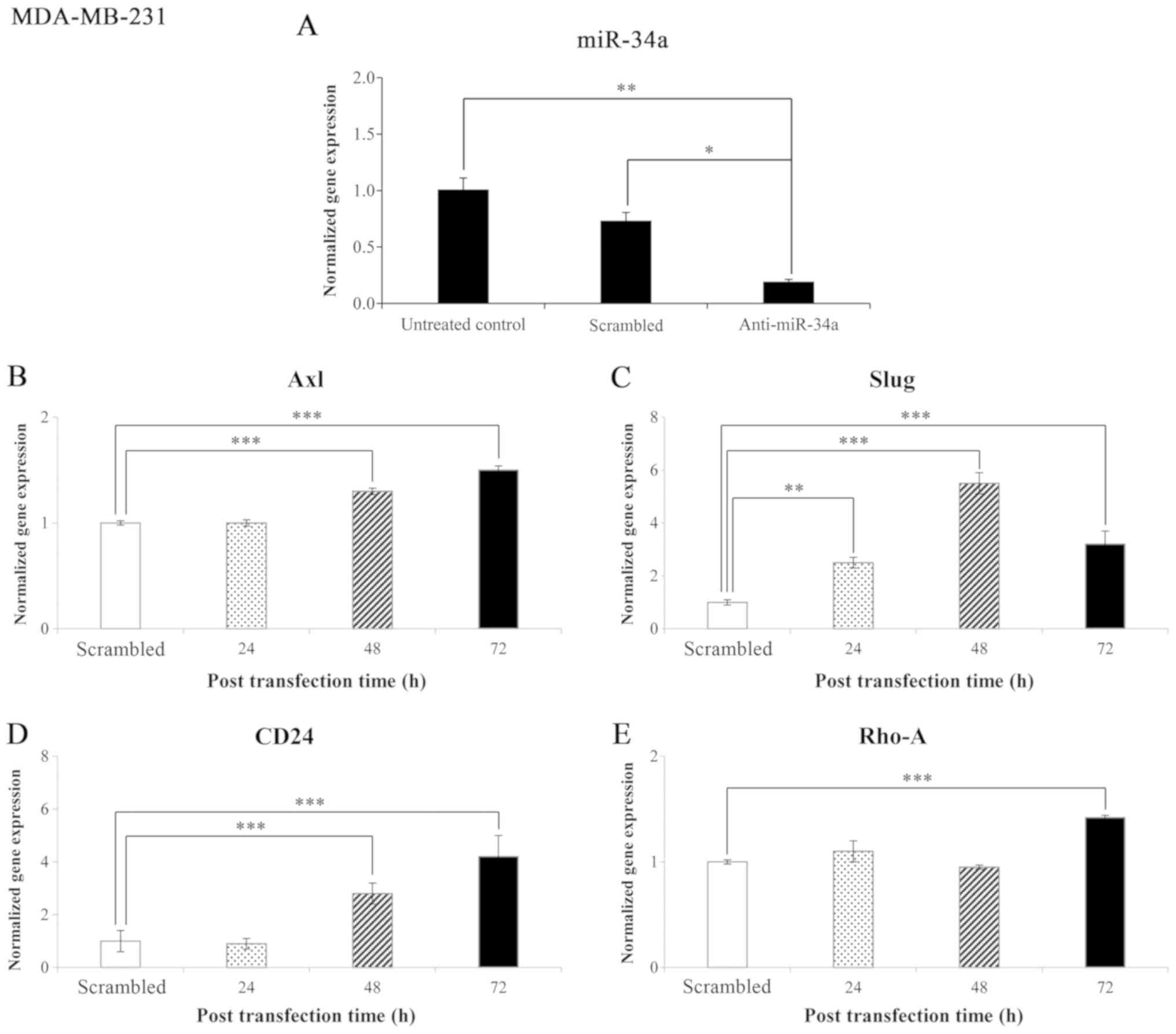
Effects of anti-miR-34a and the time of
post-transfection on gene expression levels in the MCF-10F cell
line
The results of the miR-34a expression level in the
MCF-10F cell line transfected with anti-miR-34a and negative
control (scrambled) are shown in Fig.
4A. The results revealed that miR-34a expression significantly
(P<0.005 and P<0.05) decreased in those cells transfected
with anti-miR-34a compared to the scrambled and untreated control,
respectively. The normalized gene expression levels following
transfection with anti-miR-34a of genes that code for proteins
involved in EMT in the MCF-10F cell line are shown in Fig. 4B-E. The effect of blocking miR-34a
on Axl, Slug, CD24, and Rho-A gene
expression was analyzed. The knockdown of miR-34a significantly
(P<0.001) increased Axl (304%) expression after 24 h; no
significant changes were observed after 48 and 72 h (Fig. 4B). The knockdown of miR-34a also
significantly increased Slug expression after 24 and 48 h
(278%, P<0.001; and 92%, P<0.05, respectively) (Fig. 4C). Following the knockdown of
miR-34a, an increase was also observed in the CD24 (281%,
P<0.005) (Fig. 4D) Rho-A
(395%, P<0.001) (Fig. 4E)
expression levels following 24 h of transfection.
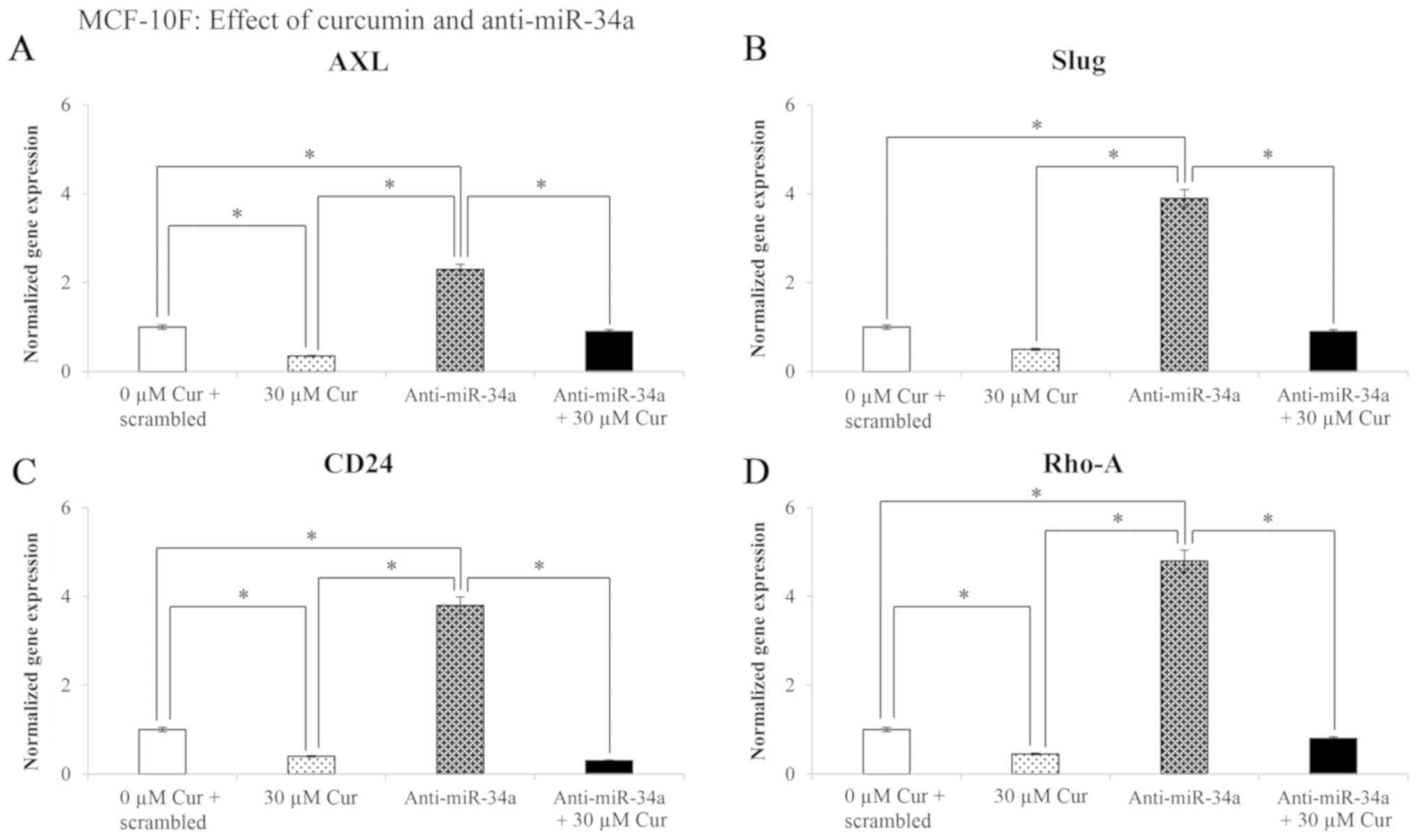
Effect of curcumin and anti-miR-34a on
gene expression levels in the MCF-10F cell line
The effects of treatment with 30 µM curcumin
for 48 h on the Axl, Slug, CD24 and
Rho-A gene expression levels in the MCF-10F cell line are
shown in Fig. 5. The results
revealed that curcumin significantly (P<0.05) decreased
Axl (65%), Slug (50%), CD24 (62%), and
Rho-A (55%) gene expression following transfection of the
cells with negative control (scrambled) in comparison with the
curcumin-untreated cells. Transfection with anti-miR-34a
significantly (P<0.05) increased the gene expression of
Axl (133%), Slug (290%), CD24 (282%) and
Rho-A (380%); however, treatment with curcumin plus
anti-miR-34a significantly (P<0.05) decreased the levels of the
examined genes in comparison to the cells transfected with
anti-miR-34a and not treated with curcumin (Fig. 5).
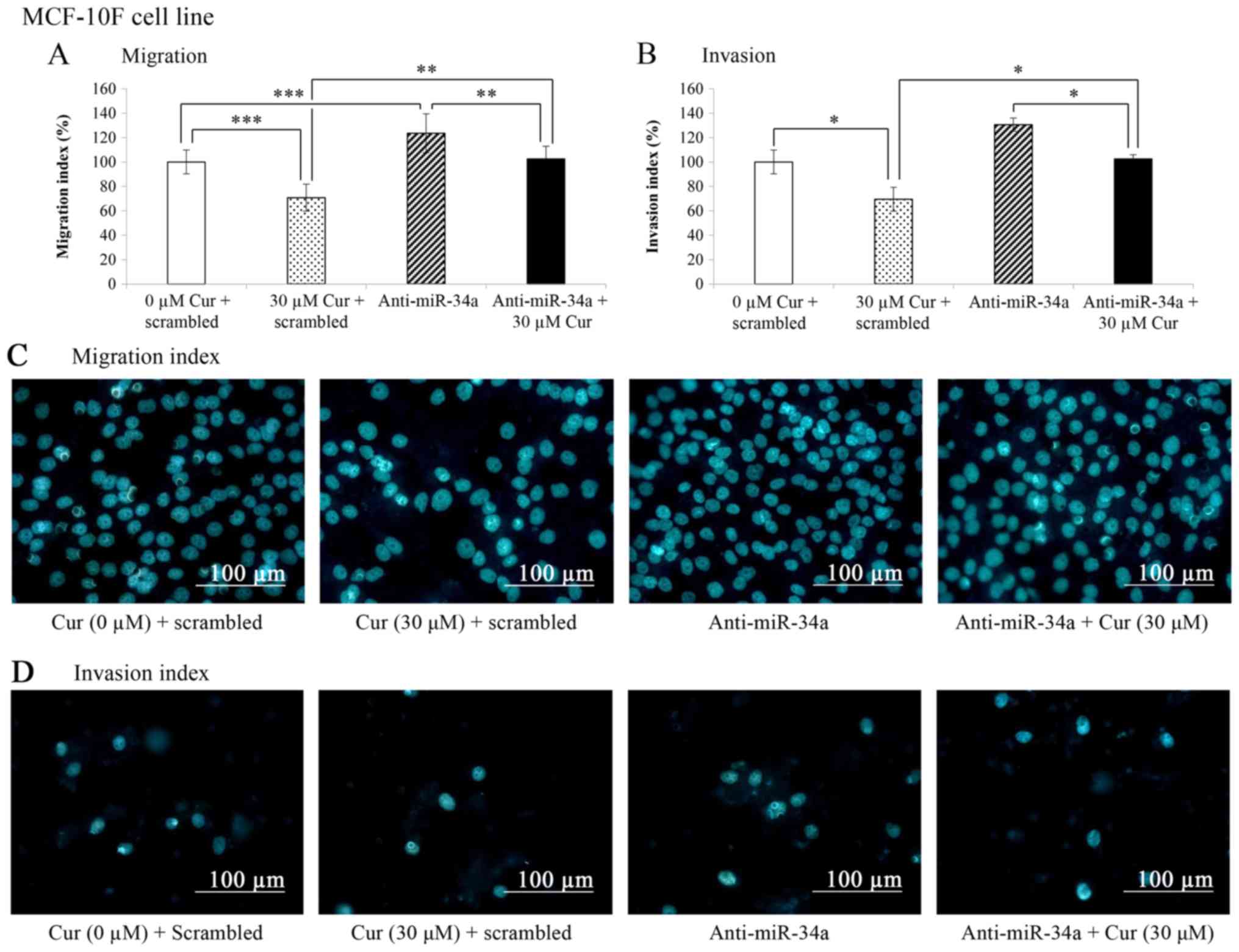
Effects of curcumin on the migratory and
invasive capabilities of the MCF-10F cell line
The migratory and invasive capabilities were
analyzed by migration and invasion assays carried out in a Boyden
chamber (Fig. 6A and B). The
results presented in Fig. 6A
revealed that curcumin significantly (P<0.001) decreased the
migration (29.3%) of the MCF-10F cells transfected with the
negative control (scrambled) in comparison with the untreated
control cells (scrambled + 0 µM Cur). In addition, a
significant increase (24%, P<0.001) in the migration of the
MCF-10F cells transfected with anti-miR-34a in comparison with the
untreated control cells (scrambled + 0 µM Cur) was observed.
Curcumin plus anti-miR-34a significantly (P<0.005) decreased the
migration of the MCF-10F cell line in comparison to anti-miR-34a
alone. The percentage of migrated cells treated with curcumin plus
anti-miR-34a was similar to the untreated control (scrambled + 0
µM Cur) following transfection. When miR-34a was blocked,
the migratory capability of the cells decreased.
The effects of the knockdown of miR-34a on the
invasive ability of the MCF-10F cells were also evaluated. As shown
in Fig. 6B, curcumin significantly
(P>0.05) decreased the invasion (30.5%) of the MCF-10F cells
transfected with the negative control (scrambled) in comparison
with the untreated control cells (scrambled + 0 µM Cur). The
invasive ability was significantly (P<0.05) decreased (39.1%) in
the MCF-10F cells transfected with anti-miR-34a and treated with
curcumin compared with anti-miR-34a alone. Cell invasion decreased
to the level of the untreated control (scrambled + 0 µM Cur)
when miR-34a was blocked and the cells were treated with curcumin
(30 µM for 24 h). Representative images of the migratory
capabilities following treatment with curcumin and miR-34a blockage
in the MCF-10F cell line are shown in Fig. 6C. In addition, representative
images of the invasive capabilities following treatment with
curcumin and miR-34a blockage are shown in Fig. 6D.
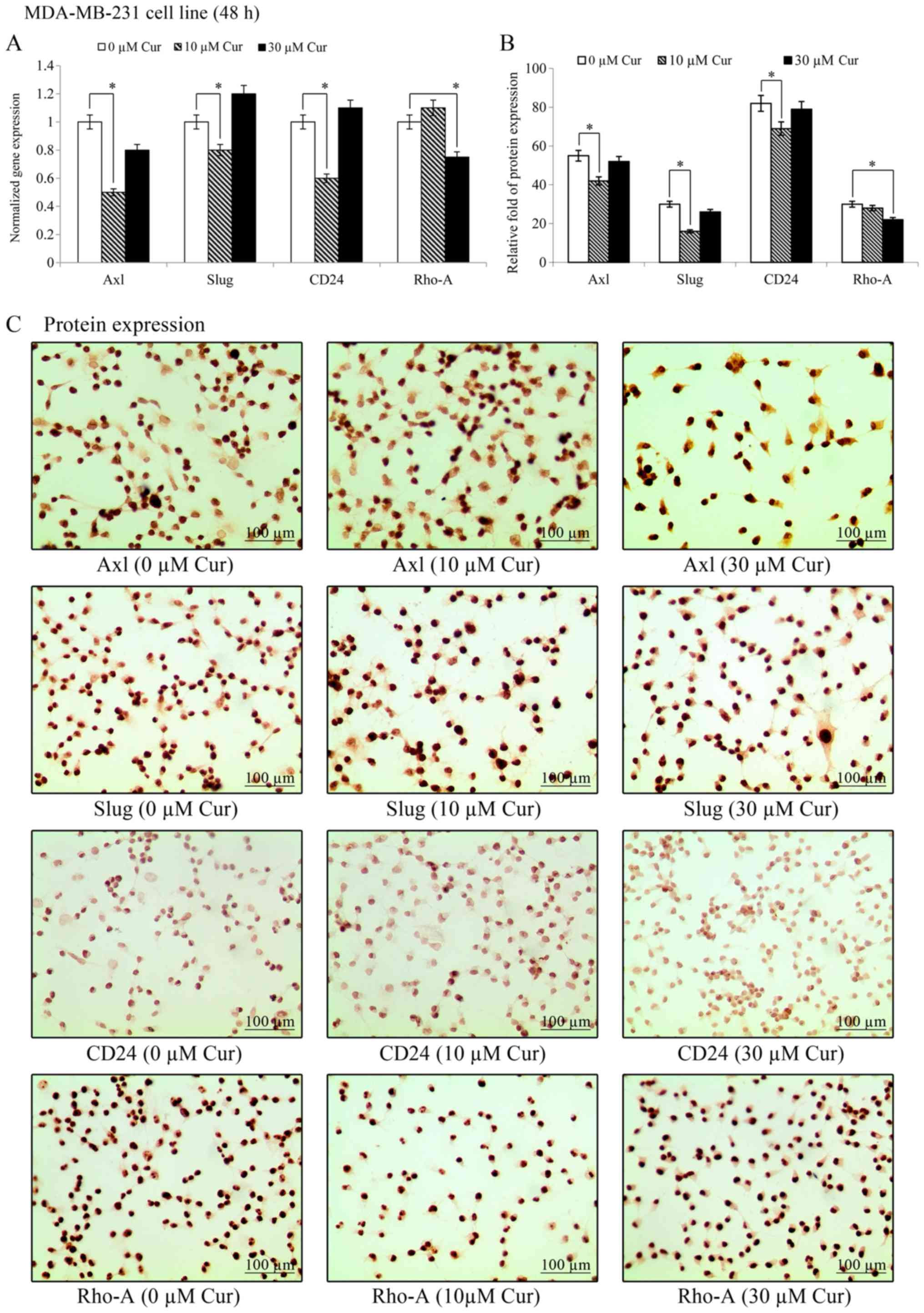
Effects of curcumin on gene and protein
expression levels in the MDA-MB-231 cell line
The effects of treatment with 10 and 30 µM
curcumin on Axl, Slug and CD24 gene expression
on the MDA-MB-231 cell line are presented in Fig. 7. The results revealed that
treatment with 10 µM curcumin significantly (P<0.05)
decreased the gene transcript levels of Axl, Slug and
CD24, while those of Rho-A remained unaffected.
However, treatment with 30 µM curcumin decreased the
Rho-A transcript levels, while those of the other genes were
unaffected. As shown in Fig. 7B,
treatment with 10 µM curcumin decreased the Axl, Slug, and
CD24 protein expression levels in the MDA-MB-231 cell line;
however, the levels of Rho-A were unaffected by this concentration.
However, treatment with 30 µM curcumin decreased only Rho-A
protein expression in comparison to its own control. Representative
images of the effects of curcumin on protein expression levels in
the MDA-MB-231 cell line are shown in Fig. 7C. The results revealed that
treatment with 10 µM curcumin decreased Axl, Slug and CD24
protein expression. However, 30 µM curcumin decreased only
Rho-A protein expression in comparison to its own control.
Effects of anti-miR-34a and the time of
post-transfection on gene expression levels in the MDA-MB-231 cell
line
The results of the miR-34a expression level in the
MDA-MB-231 cell line in comparison to the untreated control and
negative control (scrambled) are presented in Fig. 8A. The results indicated that
miR-34a expression significantly decreased (P<0.05) in the
anti-miR-34a-transfected cells compared to the cells transfected
with the scramble control and the untreated cells.
The normalized gene expression levels of genes that
code for proteins involved in EMT in the MDA-MB-231 cell line
following transfection with anti-miR-34a are shown in Fig. 8B-E. miR-34a knockdown significantly
(P<0.001) increased Axl expression after 48 and 72 h of
transfection (28 and 50%, respectively) (Fig. 8B). miR-34a knockdown also
significantly increased Slug gene expression after 24, 48
and 72 h of transfection (155, 452, 226%, respectively; 24 h,
P<0.005; 48 and 72 h, P<0.001) (Fig. 8C). Following miR-34a knockdown, a
significant (P<0.001) increase was also observed in CD24
gene expression after 48 h and after 72 h of transfection (179 and
325%, respectively) (Fig. 8D).
miR-34a knockdown also significantly (P<0.001) increased
Rho-A gene expression (43%) after 72 h of transfection
(Fig. 8E).
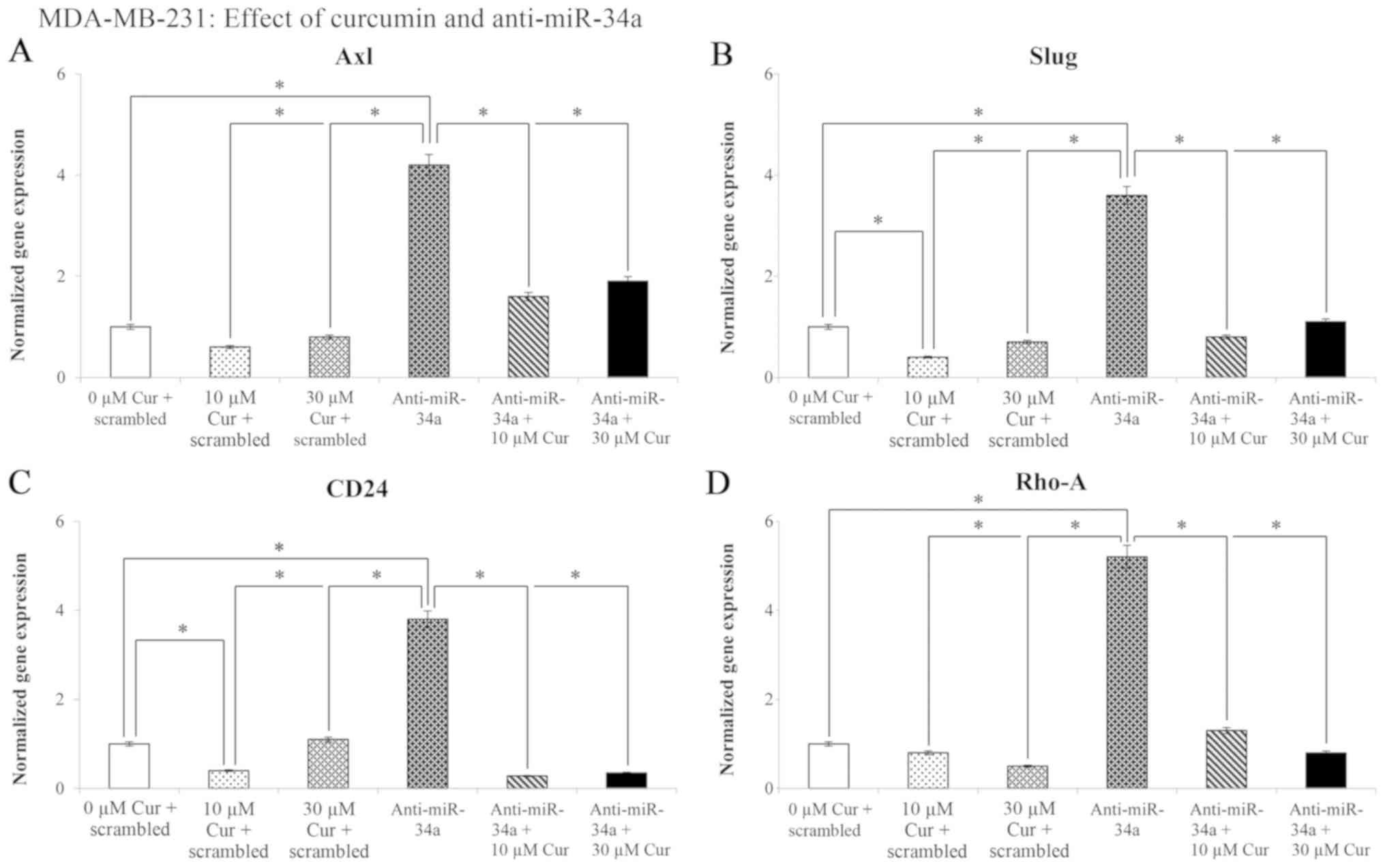
The effects of treatment with 10 and 30 µM
curcumin on Axl, Slug, CD24 and Rho-A
gene expression are presented in Fig.
9. The results revealed a significant (P<0.05) decrease in
Axl (39%), Slug (57%), CD24 (62%) and
Rho-A (53%) gene expression following transfection with the
negative control (scramble) in comparison with the untreated
control cells (0 µM cur + scrambled). Transfection with
anti-miR-34a increased the expression levels of the Axl,
Slug, CD24 and Rho-A genes (304, 260, 282 and
430%, respectively). However, following treatment with curcumin and
miR-34a knockdown, a significant (P<0.05) decrease was observed
in the levels of the examined genes.
Effects of the blockade of miR-34a on the
migratory and invasive capabilities of the MDA-MB-231 cell
line
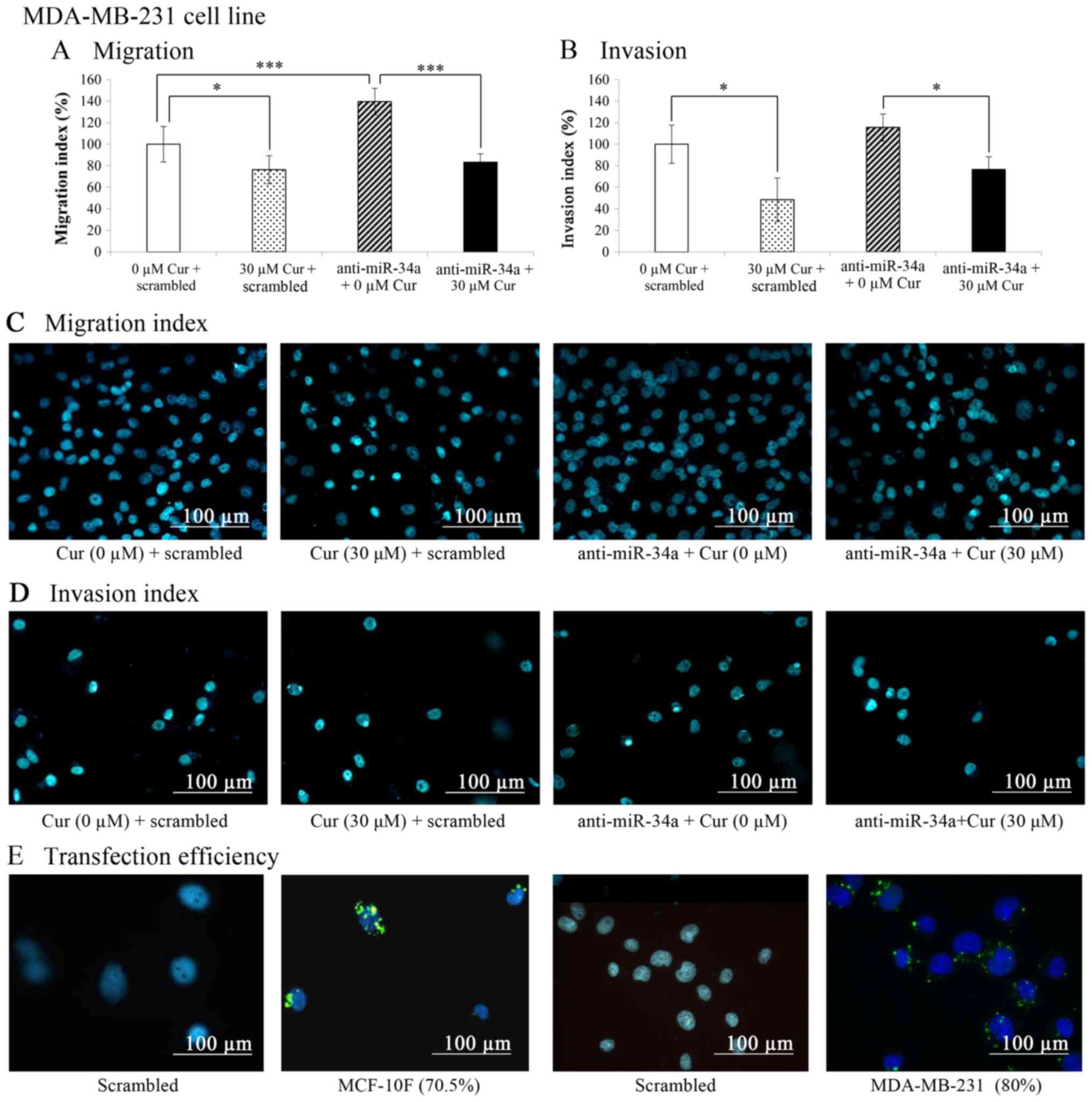
The results of the migration and invasion assays
carried out in a Boyden chamber, as indicated in the graphs
depicted in Fig. 10A and B
revealed that curcumin (30 µM for 48 h) significantly
(P<0.05) decreased the migration (23.7%) and invasion (51.6%) of
the MDA-MB-231 cell line in comparison with the untreated control
cells (0 µM cur + scrambled). On the other hand, when the
cells were transfected with anti-miR-34a (Fig. 10A), cell migration significantly
(P<0.001) increased in comparison with the untreated control (0
µM cur + scrambled). Curcumin plus anti-miR-34a
significantly (P<0.001) decreased the migration of the
MDA-MB-231 cell line in comparison with the cells treated with
anti-miR-34a alone. The percentage of migrated treated cells
treated with curcumin was lower than that of the untreated control
following transfection with anti-miR-34a. Representative images of
the migratory capabilities of the MDA-MB-231 cell line following
treatment with curcumin and miR-34a blockade are shown in Fig. 10C.
Curcumin also significantly (P<0.05) decreased
(39.1%) cell invasion. The invasive capabilities of the cells were
significantly (P<0.05) decreased in the cells transfected with
anti-miR-34a and treated with curcumin compared with the cells
treated with anti-miR-34a alone. Curcumin significantly (P<0.05)
decreased cell invasion to the level of the control (0 µM
cur + scrambled) when miR-34a was blocked and the cells were
treated with curcumin. Representative images of the invasive
capabilities of the MDA-MB-231 cell line following treatment with
curcumin and the miR-34a blockade are shown in Fig. 10D.
In addition, transfection conditions were
standardized by determining the transfection efficiency using a
negative control (scrambled) that was previously marked by using
fluorescence. As shown in Fig.
10E, the transfection efficiency was 70.5% in the MCF-10F and
80% in the MDA-MB-231 cell line in comparison with their respective
controls.
Discussion
Studies have reported that curcumin possesses
antioxidant, anti-proliferative, anti-migratory and apoptotic
effects in several types of human cancers (32,33,35).
We have previously demonstrated such effects in breast cell lines
and it has been corroborated by other authors in different cancers
(34,35,37,41).
The present study demonstrated that curcumin acted upon Axl,
Slug, CD24, genes implicated in EMT, as well as on
Rho-A in a non-malignant MCF-10F and a malignant MDA-MB-231
cell line. As regards the fact that the MDA-MB-231 cells were more
sensitive to low concentrations of curcumin, whereas at 30
µM the effect on gene expression was lost probably due to
the negative ER and ERB2 receptors that render the cells to behave
differently in comparison with a non-malignant cell line, as the
MCF-10F cell line.
Curcumin also affected miRNAs as a regulator of
genes implicated in EMT and Rho-A, affecting migration and
invasion independent of their ER, PgR and HER2 receptors. Curcumin
decreased Axl, Slug, CD24 and Rho-A
gene and protein expression in the MCF-10F cell line when compared
to the untreated control cells. It has been previously demonstrated
that Slug and Axl induce EMT and regulate several
aspects of this process (42,43).
These results are in agreement with the downregulation of
Axl reversing EMT in mesenchymal normal human mammary
epithelial cells that regulated breast cancer stem cell renewal
(44,45).
In a previous study, we examined the effects of
curcumin on breast carcinogenesis in an established in vitro
experimental model of breast cancer (termed the Alpha-model)
(46); we examined the effects of
curcumin on EMT in breast cancer cells transformed by low
concentrations of alpha particles and estrogen. This in
vitro experimental model was developed by the exposure of the
MCF-10F immortalized breast epithelial cell line to low radiation
concentrations of high linear energy transfer (LET) α-particles
(150 keV/µm) followed by culture in the presence of
17β-estradiol (estrogen). Three difference cell lines were used,
namely the MCF-10F (normal), the Alpha5 (pre-tumorigenic) and the
Tumor2 cells derived from Alpha5 injected into the nude mice. It
has been demonstrated (36) that
curcumin influences changes in the levels of genes associated with
EMT and which are situated at the core of several signaling
pathways known to mediate the transition and such results indicated
that curcumin decreased the protein expression levels of
N-cadherin, β-catenin, Slug, AXL, Twist1, Vimentin and Fibronectin,
independently of the positivity of the markers in these cell lines.
Curcumin was also shown to decrease the migratory and invasive
capabilities of the cells compared with their respective controls.
Thus, it was concluded that curcumin was able to prevent or
attenuate cancer progression through the disruption of the EMT
processes (36), i.e., by
affecting the gene expression E-cadherin and other genes in the
MDA-MB-231 cells. In another study, it was demonstrated that
curcumin decreased the expression of levels of genes (such as
E-cadherin, N-cadherin, ZEB2, Twist1, Slug, AXL, vimentin,
STAT-3, andfibronectin) involved in the EMT process in
the Tumor2 cells when compared to the respective control. In
addition, it was demonstrated that curcumin altered the levels of
the p53 and caveolin-1 genes, as well as those of the
apoptotic genes, caspase-3 andcaspase-8, as well
cyclin D1 andNF-κB. These changes in expression
levels led to a decrease in the migratory and invasive capabilities
of such a cell line (38). This
suggests that curcumin may affect the apoptosis and metastatic
properties of malignant cells, exerting antitumor effects on breast
cancer cells transformed by low concentrations of α-particles and
estrogen in vitro (38).
The miR-34 family was originally cloned and
character-ized in 2007 as a p53 target gene (47,48).
Soon thereafter, it became evident this miRNA played a main role as
a master regulator of tumor suppression. Notably, the
overexpression of miR-34 directly and indirectly suppresses several
oncogenes, resulting in an increase in cancer cell death (including
cancer stem cells), and in the inhibition of metastasis. Moreover,
the expression of miR-34 is known to be deregulated in several
types of human cancers (49). In
2013, a miR-34 mimic became the first miRNA to reach phase 1
clinical trials (50).
Others (31) have
reported that the regulation of miRNAs by natural agents is a novel
new strategy for the treatment of cancer. In the study by
Tarasovet al (2007), the differential regulation of miRNAs
by p53 revealed by massively parallel sequencing revealed miR-34a
as a target of p53 that induces apoptosis and G1-arrest. Thus,
miR-34a suppressed transcription factors implicated in EMT, thus
inhibiting metastasis (51). It
has also been demonstrated that miR-34 and SNAIL form a
double-negative feedback loop to regulate EMT (52). Axl targeted by miR-34a is well
known and its expression is significantly associated with
metastatic cancer of an advanced clinical stage. Of note, Axl is
associated with a poor prognosis in different types of cancer
(53).
In this study, the effects of curcumin on the
expression of EMT-associated genes in MCF-the 10F cell line were
examined following the knockdown miR-34a. The transfection
conditions were standardized by determining the transfection
efficiency using a negative control (scrambled) by using
fluorescence methods in the MCF-10F and in MDA-MB-231 cell lines in
comparison with the control. The results revealed that curcumin
decreased Axl, Slug, CD24, and Rho-A
gene expressions after transfection with negative control
(scrambled). Transfection with anti-miR-34a increased the
expression of these genes. However, the cells in which miR-34a was
knocked down were also treated with curcumin, a decrease in the
expression of these genes was observed in comparison to the
untreated cells. The knockdown of miR-34a expression in the
MDA-MB-231 cell line increased Axl, Slug, CD24
and Rho-A gene expression. However, when curcumin was added
to the cells in which miR-34a was knocked down, there was a
decrease in anti-miR-34a plus curcumin-treated group in comparison
to the untreated cells. The effects of anti-miR-34a transfection
were validated by using the Anti-miR™ negative control. This is a
random commercial molecule which has been extensively tested in
human cell lines by us and others. In addition, it has been widely
demonstrated that this control does not alter known functions of
miRNAs and mRNAs (54-57). In this study, the results suggested
that this scrambled sequence did not affect the expression of
miR-34a or that of EMT-related genes evaluated at 24 and 48 h
post-transfection in the MCF-10F and MDA-MB-231 cell lines.
Although other exposure times were analyzed on EMT-related genes
without using this scrambled group, the lack of this control did
not alter our general conclusions as the main focus of this study
was on the rapid-response phenotypic changes related to EMT, such
as cell migration and invasion. Therefore, only short-time
exposures to curcumin and anti-miR-34a were evaluated. However,
this rationale could be an important limitation if these results
are extrapolated to a different context, such as biomedical
applications. In this scenario, robust in vivo assays are
warranted to validate the anti-miR-34a administration mechanisms
and the integrity of curcumin over time. Currently, nanotechnology
offers novel materials that guarantee high efficiency in the supply
of therapeutic agents; for example, gold nanostructures, dextran
nanobubbles or nanoemulsions are emerging tools that improve
aqueous solubility and the supply of curcumin to the tissue of
interest (58-61). Similarly, RNA nanotechnology to
administer anti-miRNA is already being validated in cancer
(62). Therefore of this study may
provide a novel strategy with which to combat breast cancer if
nanomaterials are implemented in improving delivery over time and
the specificity of therapeutic agents.
Previously, we determined the potential effects of
curcumin on EMT in relation to migration and invasion and compared
the Tumor2 and MDA-MB-231 cell lines (36,38).
Curcumin inhibited the migration and invasive capabilities through
EMT in the breast cancer cells. It has also been found that
curcumin inhibits the proliferation and invasion of different types
of cancer (41). In the present
study, curcumin decreased cell migration and invasion in the
control-transfected MCF-10F and MDA-MB-231 cell lines in comparison
with the untreated cells. When these cells were transfected with
anti-miR-34a, cell migration increased.
miRNAs are known as important targets for curcumin
(63,64). These molecules can affect numerous
cellular and molecular events (65-70),
since it has been found that curcumin can affect the
chemoresistance of cancer cells by changing the expression of
certain miRNAs. Thus, curcumin functions as a master regulator of
the genome by modulating the expression of thousands of genes,
simultaneously controlling multiple signaling pathways (43,71).
The therapeutic use of miRNAs has been considered for all these
reasons. However, further studies are warranted using additional
cell lines.
The results of this study indicated that curcumin
decreased the migration of the MDA-MB-231 cell line. When these
cells were transfected with anti-miR-34a, cell migration increased.
However, curcumin decreased the migration of the MDA-MB-231 cell
line transfected with anti-miR-34a in comparison with the control
untreated cells. The percentage of migrated treated cells with
curcumin was lower than the control following transfection with
anti-miR-34a. This suggests that additional mechanisms may be
involved in mediating the observed effects of curcumin on the
invasive and migratory abilities of these cells.
An increasing number of preclinical studies support
the hypothesis that curcumin may be a promising anticancer drug.
In vitro studies require higher amounts of chemical
compounds to analyze different physiological effects on cells;
thus, the 10 and 30 µM concentrations of curcumin were used
in this study. However, the poor bioavailability has limited the
efficacy of curcumin in clinical trials, and plasma curcumin levels
remain low (ng/ml) despite patients taking gram doses of curcumin,
which is insufficient to yield the anticancer benefits of curcumin.
This issue has been resolved by the development of highly
bioavailable forms of curcumin (THERACURMIN®), and
higher plasma curcumin levels can now be achieved without increased
toxicity in patients. At the present time, some researchers are
testing this substance in nanoparticles to improve the
bioavailability (72).
Chemotherapy is known as one of the effective
therapeutic approaches for the treatment of a wide variety of
cancers (73,74). Recently, the regulation of miRNAs
by natural, nontoxic chemopreventive agents, including curcumin has
been described. Over the past decade, research on curcumin for its
chemoprophylactic and anti-inflammatory properties has been
increasing. Faduset al (2017) reviewed that trials on
curcumin trials are mainly focused on colorectal cancer,
hepatocellular carcinoma and several other tissues (75). The authors indicated that there
were at least 12 active clinical trials of curcumin in the USA,
Israel and Hong Kong. As of July 2012, there have been observations
from 67 clinical trials that have been published, with another 35
clinical trials which were in progress at that time (75).
In conclusion, the present study demonstrated that
the Axl, Slug and CD24 genes were implicated
in EMT. Rho-A was also found to be involved in the migration
and invasion of the MCF-10F and MDA-MB-231 cell lines. Curcumin
affected the expression levels of genes involved in EMT and
invasion by controlling miR-34a expression in the breast cell
lines. Curcumin acted upon miRNAs as a regulator of genes
implicated in EMT, as well as on Rho-A, affecting the
migration and invasion of the non-malignant MCF-10F and malignant
MDA-MB-231 breast cancer cell lines, independent of their ER, PgR
and HER2 receptor status; these cell lines are both negative for
such receptors.
Therefore, it is demonstrated that natural agents
have the ability to inhibit cancer progression, increase drug
sensitivity, reverse EMT and prevent metastasis through the
modulation of miRNAs. These findings may provide a novel
therapeutic approach for cancer treatment, particularly when used
in combination with conventional therapeutics (31). Furthermore, to the best of our
knowledge, this the first study to examine the effects of curcumin
on Rho-A and genes involved in EMT, such as Axl,
Slug and CD24. It is demonstrated that curcumin
prevents the migration and invasion of breast cells by targeting
miR-34a as a regulator of the above-mentioned genes.
Funding
The present study was supported by FONDECYT
#1120006 (to GMC), grant UTA-MINEDUC (UTA1117, to GMC), FONDECYT
#1161219 (to FA), FONDECYT #3190744 (to JPM) and CONICYT-FONDAP
#15130011 (to FA).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors’ contributions
MG and GMC conceived and designed the study. MG,
UK, FA, JPM, TCB and GMC performed the experiments. MG and GMC
wrote the manuscript. GMC, UK, and FA, JPM and TCB edited and
reviewed the manuscript. All authors have read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work is appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Consent for publication
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to acknowledge the technical
support provided by Mr. Leodán A. Crispin, Mr. Richard Ponce Cusi,
Ms. Guiliana Rojas and Mrs. Georgina Vargas Marchant from
Universidad de Tarapacá.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Sethi N and Kang Y: Unravelling the
complexity of metastasis - molecular understanding and targeted
therapies. Nat Rev Cancer. 11:735–748. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Howlader N, Noone AM, Krapcho M, Miller D,
Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, et al:
SEER Cancer Statistics Review, 1975-2014. National Cancer
Institute; Bethesda, MD: 2016, https://seer.cancer.gov/csr/1975_2014/.
Accessed April 2017.
|
|
5
|
Kalluri R and Neilson EG:
Epithelial-mesenchymal transition and its implications for
fibrosis. J Clin Invest. 112:1776–1784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graham DK, DeRyckere D, Davies KD and Earp
HS: The TAM family: Phosphatidylserine sensing receptor tyrosine
kinases gone awry in cancer. Nat Rev Cancer. 14:769–785. 2014.
View Article : Google Scholar
|
|
7
|
Kirane A, Ludwig KF, Sorrelle N, Haaland
G, Sandal T, Ranaweera R, Toombs JE, Wang M, Dineen SP, Micklem D,
et al: Warfarin Blocks Gas6-Mediated Axl Activation Required for
Pancreatic Cancer Epithelial Plasticity and Metastasis. Cancer Res.
75:3699–3705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Linger RM, Keating AK, Earp HS and Graham
DK: TAM receptor tyrosine kinases: Biologic functions, signaling,
and potential therapeutic targeting in human cancer. Adv Cancer
Res. 100:35–83. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dunne PD, McArt DG, Blayney JK, Kalimutho
M, Greer S, Wang T, Srivastava S, Ong CW, Arthur K, Loughrey M, et
al: AXL is a key regulator of inherent and chemotherapy-induced
invasion and predicts a poor clinical outcome in early-stage colon
cancer. Clin Cancer Res. 20:164–175. 2014. View Article : Google Scholar
|
|
10
|
Hajra KM, Chen DY and Fearon ER: The SLUG
zinc-finger protein represses E-cadherin in breast cancer. Cancer
Res. 62:1613–1618. 2002.PubMed/NCBI
|
|
11
|
Castro Alves C, Rosivatz E, Schott C,
Hollweck R, Becker I, Sarbia M, Carneiro F and Becker KF: Slug is
overexpressed in gastric carcinomas and may act synergistically
with SIP1 and Snail in the down-regulation of E-cadherin. J Pathol.
211:507–515. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX,
Shetuni B and Barsky SH: ERalpha signaling through slug regulates
E-cadherin and EMT. Oncogene. 29:1451–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Atmaca A, Wirtz RW, Werner D, Steinmetz K,
Claas S, Brueckl WM, Jäger E and Al-Batran SE: SNAI2/SLUG and
estrogen receptor mRNA expression are inversely correlated and
prognostic of patient outcome in metastatic non-small cell lung
cancer. BMC Cancer. 15:3002015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pirruccello SJ and LeBien TW: The human B
cell-associated antigen CD24 is a single chain sialoglycoprotein. J
Immunol. 136:3779–3784. 1986.PubMed/NCBI
|
|
15
|
Daniel L, Lechevallier E, Bouvier C,
Coulange C and Pellissier JF: Adult mesoblastic nephroma. Pathol
Res Pract. 196:135–139. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kristiansen G, Denkert C, Schlüns K, Dahl
E, Pilarsky C and Hauptmann S: CD24 is expressed in ovarian cancer
and is a new independent prognostic marker of patient survival. Am
J Pathol. 161:1215–1221. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jacob J, Bellach J, Grützmann R, Alldinger
I, Pilarsky C, Dietel M and Kristiansen G: Expression of CD24 in
adenocar-cinomas of the pancreas correlates with higher tumor
grades. Pancreatology. 4:454–460. 2004. View Article : Google Scholar
|
|
18
|
Kristiansen G, Winzer KJ, Mayordomo E,
Bellach J, Schlüns K, Denkert C, Dahl E, Pilarsky C, Altevogt P,
Guski H, et al: CD24 expression is a new prognostic marker in
breast cancer. Clin Cancer Res. 9:4906–4913. 2003.PubMed/NCBI
|
|
19
|
Yu LLG and Gu JY: Advances in the role of
Rho sub-family in tumor invasion. Fudan Univ J Med Sci. 37:617–619.
2010.
|
|
20
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
DerMardirossian C and Bokoch GM: GDIs:
Central regulatory molecules in Rho GTPase activation. Trends Cell
Biol. 15:356–363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Olofsson B: Rho guanine dissociation
inhibitors: Pivotal molecules in cellular signalling. Cell Signal.
11:545–554. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fritz G, Brachetti C, Bahlmann F, Schmidt
M and Kaina B: Rho GTPases in human breast tumours: Expression and
mutation analyses and correlation with clinical parameters. Br J
Cancer. 87:635–644. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burbelo P, Wellstein A and Pestell RG:
Altered Rho GTPase signaling pathways in breast cancer cells.
Breast Cancer Res Treat. 84:43–48. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zaravinos A: The Regulatory Role of
MicroRNAs in EMT and Cancer. J Oncol. 2015:8658162015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ng EK, Wong CL, Ma ES and Kwong A:
MicroRNAs as New Players for Diagnosis, Prognosis, and Therapeutic
Targets in Breast Cancer. J Oncol. 2009:3054202009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pichler M and Calin GA: MicroRNAs in
cancer: From developmental genes in worms to their clinical
application in patients. Br J Cancer. 113:569–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hermeking H: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar
|
|
30
|
Li Y, Kong D, Wang Z and Sarkar FH:
Regulation of microRNAs by natural agents: An emerging field in
chemoprevention and chemotherapy research. Pharm Res. 27:1027–1041.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sethi S, Li Y and Sarkar FH: Regulating
miRNA by natural agents as a new strategy for cancer treatment.
Curr Drug Targets. 14:1167–1174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mirzaei H, Shakeri A, Rashidi B, Jalili A,
Banikazemi Z and Sahebkar A: Phytosomal curcumin: A review of
pharmacokinetic, experimental and clinical studies. Biomed
Pharmacother. 85:102–112. 2017. View Article : Google Scholar
|
|
33
|
Kuttan R, Bhanumathy P, Nirmala K and
George MC: Potential anticancer activity of turmeric (Curcuma
longa). Cancer Lett. 29:197–202. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calaf GM, Echiburú-Chau C, Roy D, Chai Y,
Wen G and Balajee AS: Protective role of curcumin in oxidative
stress of breast cells. Oncol Rep. 26:1029–1035. 2011.PubMed/NCBI
|
|
35
|
Calaf GM: Curcumin, oxidative stress and
breast cancer. Oxidative stress and dietary antioxidants. Preedy
VR: Elsevier Inc; London: pp. 159–169. 2014
|
|
36
|
Gallardo M and Calaf GM: Curcumin and
epithelial-mesenchymal transition in breast cancer cells
transformed by low doses of radiation and estrogen. Int J Oncol.
48:2534–2542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Echiburú-Chau C, Roy D and Calaf GM:
Metastatic suppressor CD44 is related with oxidative stress in
breast cancer cell lines. Int J Oncol. 39:1481–1489.
2011.PubMed/NCBI
|
|
38
|
Gallardo M and Calaf GM: Curcumin inhibits
invasive capabilities through epithelial mesenchymal transition in
breast cancer cell lines. Int J Oncol. 49:1019–1027. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Calaf GM and Abarca-Quinones J: Ras
protein expression as a marker for breast cancer. Oncol Lett.
11:3637–3642. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
41
|
Kunnumakkara AB, Anand P and Aggarwal BB:
Curcumin inhibits proliferation, invasion, angiogenesis and
metastasis of different cancers through interaction with multiple
cell signaling proteins. Cancer Lett. 269:199–225. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alves CC, Carneiro F, Hoefler H and Becker
KF: Role of the epithelial-mesenchymal transition regulator Slug in
primary human cancers. Front Biosci. 14:3035–3050. 2009. View Article : Google Scholar
|
|
43
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao Y, Sun X, Jiang L, Yang F, Zhang Z
and Jia L: Differential expression of Axl and correlation with
invasion and multidrug resistance in cancer cells. Cancer Invest.
30:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vuoriluoto K, Haugen H, Kiviluoto S,
Mpindi JP, Nevo J, Gjerdrum C, Tiron C, Lorens JB and Ivaska J:
Vimentin regulates EMT induction by Slug and oncogenic H-Ras and
migration by governing Axl expression in breast cancer. Oncogene.
30:1436–1448. 2011. View Article : Google Scholar
|
|
46
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Raver-Shapira N, Marciano E, Meiri E,
Spector Y, Rosenfeld N, Moskovits N, Bentwich Z and Oren M:
Transcriptional activation of miR-34a contributes to p53-mediated
apoptosis. Mol Cell. 26:731–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Agostini M and Knight RA: miR-34: From
bench to bedside. Oncotarget. 5:872–881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Beg MS, Brenner AJ, Sachdev J, Borad M,
Kang YK, Stoudemire J, Smith S, Bader AG, Kim S and Hong DS: Phase
I study of MRX34, a liposomal miR-34a mimic, administered twice
weekly in patients with advanced solid tumors. Invest New Drugs.
35:180–188. 2017. View Article : Google Scholar
|
|
51
|
Tarasov V, Jung P, Verdoodt B, Lodygin D,
Epanchintsev A, Menssen A, Meister G and Hermeking H: Differential
regulation of microRNAs by p53 revealed by massively parallel
sequencing: miR-34a is a p53 target that induces apoptosis and
G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Siemens H, Jackstadt R, Hünten S, Kaller
M, Menssen A, Götz U and Hermeking H: miR-34 and SNAIL form a
double-negative feedback loop to regulate epithelial-mesenchymal
transitions. Cell Cycle. 10:4256–4271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mudduluru G, Ceppi P, Kumarswamy R,
Scagliotti GV, Papotti M and Allgayer H: Regulation of Axl receptor
tyrosine kinase expression by miR-34a and miR-199a/b in solid
cancer. Oncogene. 30:2888–2899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Komina A, Palkina N, Aksenenko M,
Tsyrenzhapova S and Ruksha T: Antiproliferative and Pro-Apoptotic
Effects of MiR-4286 Inhibition in Melanoma Cells. PLoS One.
11:e01682292016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang X, Zhang Y, Fu Y, Zhang J, Yin L, Pu
Y and Liang G: MicroRNA-125b may function as an oncogene in lung
cancer cells. Mol Med Rep. 11:3880–3887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mizuno R, Chatterji P, Andres S, Hamilton
K, Simon L, Foley SW, Jeganathan A, Gregory BD, Madison B and
Rustgi AK: Differential Regulation of LET-7 by LIN28B
Isoform-Specific Functions. Mol Cancer Res. 16:403–416. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lemecha M, Morino K, Imamura T, Iwasaki H,
Ohashi N, Ida S, Sato D, Sekine O, Ugi S and Maegawa H: MiR-494-3p
regulates mitochondrial biogenesis and thermogenesis through PGC1-α
signalling in beige adipocytes. Sci Rep. 8:150962018. View Article : Google Scholar
|
|
58
|
Inostroza-Riquelme M, Vivanco A, Lara P,
Guerrero S, Salas-Huenuleo E, Chamorro A, Leyton L, Bolaños K,
Araya E, Quest AFG, et al: Encapsulation of Gold Nanostructures and
Oil-in-Water Nanocarriers in Microgels with Biomedical Potential.
Molecules. 23:E12082018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bessone F, Argenziano M, Grillo G, Ferrara
B, Pizzimenti S, Barrera G, Cravotto G, Guiot C, Stura I, Cavalli
R, et al: Low-dose curcuminoid-loaded in dextran nanobubbles can
prevent metastatic spreading in prostate cancer cells.
Nanotechnology. 30:2140042019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Guerrero S, Inostroza-Riquelme M,
Contreras-Orellana P, Diaz-Garcia V, Lara P, Vivanco-Palma A,
Cárdenas A, Miranda V, Robert P, Leyton L, et al: Curcumin-loaded
nano-emulsion: A new safe and effective formulation to prevent
tumor reincidence and metastasis. Nanoscale. 10:22612–22622. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gera M, Sharma N, Ghosh M, Huynh DL, Lee
SJ, Min T, Kwon T and Jeong DK: Nanoformulations of curcumin: An
emerging paradigm for improved remedial application. Oncotarget.
8:66680–66698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P
and Shu D: Delivery of Anti-miRNA for Triple-Negative Breast Cancer
Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133.
Mol Ther. 27:1252–1261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Momtazi AA, Shahabipour F, Khatibi S,
Johnston TP, Pirro M and Sahebkar A: Curcumin as a MicroRNA
Regulator in Cancer: A Review. Rev Physiol Biochem Pharmacol.
171:1–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rabieian R, Boshtam M, Zareei M, Kouhpayeh
S, Masoudifar A and Mirzaei H: Plasminogen Activator Inhibitor
Type-1 as a Regulator of Fibrosis. J Cell Biochem. 119:17–27. 2018.
View Article : Google Scholar
|
|
65
|
Banikazemi Z, Haji HA, Mohammadi M,
Taheripak G, Iranifar E, Poursadeghiyan M, Moridikia A, Rashidi B,
Taghizadeh M and Mirzaei H: Diet and cancer prevention: Dietary
compounds, dietary MicroRNAs, and dietary exosomes. J Cell Biochem.
119:185–196. 2018. View Article : Google Scholar
|
|
66
|
Saeedi Borujeni MJ, Esfandiary E,
Taheripak G, Codoñer-Franch P, Alonso-Iglesias E and Mirzaei H:
Molecular aspects of diabetes mellitus: Resistin, microRNA, and
exosome. J Cell Biochem. 119:1257–1272. 2018. View Article : Google Scholar
|
|
67
|
Golabchi K, Soleimani-Jelodar R, Aghadoost
N, Momeni F, Moridikia A, Nahand JS, Masoudifar A, Razmjoo H and
Mirzaei H: MicroRNAs in retinoblastoma: Potential diagnostic and
therapeutic biomarkers. J Cell Physiol. 233:3016–3023. 2018.
View Article : Google Scholar
|
|
68
|
Mirzaei H, Khataminfar S, Mohammadparast
S, Sales SS, Maftouh M, Mohammadi M, Simonian M, Parizadeh SM,
Hassanian SM and Avan A: Circulating microRNAs as Potential
Diagnostic Biomarkers and Therapeutic Targets in Gastric Cancer:
Current Status and Future Perspectives. Curr Med Chem.
23:4135–4150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rashidi B, Hoseini Z, Sahebkar A and
Mirzaei H: Anti-Atherosclerotic Effects of Vitamins D and E in
Suppression of Atherogenesis. J Cell Physiol. 232:2968–2976. 2017.
View Article : Google Scholar
|
|
70
|
Mirzaei H, Masoudifar A, Sahebkar A, Zare
N, Sadri Nahand J, Rashidi B, Mehrabian E, Mohammadi M, Mirzaei HR
and Jaafari MR: MicroRNA: A novel target of curcumin in cancer
therapy. J Cell Physiol. 233:3004–3015. 2018. View Article : Google Scholar
|
|
71
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kanai M, Imaizumi A, Otsuka Y, Sasaki H,
Hashiguchi M, Tsujiko K, Matsumoto S, Ishiguro H and Chiba T:
Dose-escalation and pharmacokinetic study of nanoparticle curcumin,
a potential anticancer agent with improved bioavailability, in
healthy human volunteers. Cancer Chemother Pharmacol. 69:65–70.
2012. View Article : Google Scholar
|
|
73
|
Sinha D, Biswas J, Sung B, Aggarwal BB and
Bishayee A: Chemopreventive and chemotherapeutic potential of
curcumin in breast cancer. Curr Drug Targets. 13:1799–1819. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhou S, Zhang S, Shen H, Chen W, Xu H,
Chen X, Sun D, Zhong S, Zhao J and Tang J: Curcumin inhibits cancer
progression through regulating expression of microRNAs. Tumour
Biol. 39:10104283176916802017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fadus MC, Lau C, Bikhchandani J and Lynch
HT: Curcumin: An age-old anti-inflammatory and anti-neoplastic
agent. J Tradit Complement Med. 7:339–346. 2016. View Article : Google Scholar
|