Introduction
Breast cancer is the most commonly diagnosed cancer
and the second leading cause of cancer-associated death among women
in the USA (1). With the
development of more effi-cient treatments and techniques for early
diagnosis, cancer mortality has been greatly reduced. Nevertheless,
the occurrence of cancer recurrence, metastasis and treatment
failure remains inescapable (2).
Therefore, it is critical to identify the underlying mechanisms of
breast cancer progression and metastasis. Currently, there is
increasing evidence that cancer stem cells (CSCs) are a major cause
of breast cancer recurrence, metastasis and treatment resistance
(3-5).
According to the CSC hypothesis, CSCs are a subset
of tumor cells that are capable of developing new tumors via
dysfunction of the self-renewal progress (6). Under normal conditions, the
self-renewal capacity of CSCs is governed by several critical
signaling pathways, including the Wnt, Notch and sonic hedgehog
signaling pathways (7). However,
should the normal regulatory mechanisms become disordered, they may
also contribute to sustaining the behaviors of CSCs (7-10).
Due to the control of the aforementioned signaling pathways in both
normal stem cells and CSCs, CSC-targeted therapies specific to
these pathways will inevitably result in substantial toxicity.
Therefore, novel targets for eliminating CSCs are urgently
required.
Long non-coding RNAs (lncRNAs) are characterized as
functional RNAs that are >200 nucleotides long and do not have a
protein-coding capacity (11,12).
In general, lncRNAs are linked to diverse biological processes,
such as cell proliferation, differentiation and apoptosis, which
allows them to serve as gene modulators (12-15).
In addition to biological processes, several studies have revealed
that dysregulated lncRNAs are closely associated with a number of
human diseases, including breast cancer (16-18).
For example, HOX transcript antisense RNA (HOTAIR), which is a
well-studied lncRNA, has been demonstrated to be aberrantly
expressed during breast cancer progression (19). In addition, the global level of
HOTAIR is enhanced in breast cancer metastasis and could serve as a
strong predictor of this event (19). Additionally, a previous study has
demonstrated that the lncRNA urothelial carcinoma-associated 1
could serve as an oncogene in breast cancer (20). Conversely, some lncRNAs seem to act
as tumor suppressors in breast cancer. Growth arrest-specific
transcript 5 serves a vital role in controlling apoptosis and cell
proliferation. Its transcript level is remarkably decreased in
breast cancer tissues relative to adjacent normal tissues (21). These reports collectively indicate
that lncRNAs may exert both positive and negative effects in breast
cancer. However, the expression profile, function and regulation of
lncRNAs in breast cancer stem cells (BCSCs) remain to be
identified. The present study analyzed the expression profile of
lncRNAs in BCSCs and matched non-BCSCs. Subsequently, the possible
functions and potential involved pathways of the identified lncRNAs
were analyzed using bioinformatics. Some stemness-related pathways
were closely associated with the lncRNAs. In addition, a
lncRNA-mRNA interaction network was constructed to further
determine the possible functions. lncRNA CUE domain containing 1
(lncCUEDC1; lncRNA id, ENST00000584746) was significantly
down-regulated and exhibited a negative regulatory effect in BCSCs.
Collectively, the present study provided a substantial foundation
for studying the roles of lncRNAs in BCSCs and evidence for novel
approaches to target BCSCs.
Materials and methods
Antibodies
Rabbit monoclonal anti-Sox2 (cat. no. 3579S),
anti-Nanog (cat. no. 4903S) and anti-GAPDH (cat. no. 5174S)
antibodies, and polyclonal anti-Oct4 (cat. no. 2750S) antibodies
used for western blotting were purchased from Cell Signaling
Technology, Inc. Goat anti-rabbit horseradish peroxidase-conjugated
immunoglobulin G (cat. no. sc-2004) was obtained from Santa Cruz
Biotechnology, Inc. PE-conjugated CD24 (cat. no. 130-095-953) and
FITC-conjugated CD44 (cat. no. 130-095-195) antibodies used for
fluorescence-activated cell sorting (FACS) were purchased from
Miltenyi Biotec, Inc.
Cell culture
Human MCF-7, MDA-MB-231 breast cancer cells and 293T
cells were obtained from the American Type Culture Collection and
cultured in the recommended media according to standard methods.
Briefly, MCF7 cells and 293T cells were cultured at 37°C in a
humidified atmosphere with 5% CO2 in DMEM (HyClone; GE
Healthcare Life Sciences), whereas MDA-MB-231 cells were cultured
under the same conditions in DMEM (HyClone; GE Healthcare Life
Sciences), but without CO2. In addition, all media were
supplemented with 10% FBS (Gibco: Thermo Fisher Scientific, Inc.),
100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) and 100
mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Cells
were passaged upon reaching 90% confluence.
Mammosphere formation assay
The mammosphere culture platform has been previously
established (22). Briefly,
adherent cells were digested with trypsin (Gibco; Thermo Fisher
Scientific, Inc.) into a single-cell suspension at a concentration
of 104 cells/ml. Subsequently, the cells were plated in
ultra-low attachment 6-well plates and maintained in DMEM/F12 (1:1;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 50X B27
(Invitrogen; Thermo Fisher Scientific, Inc.), 20 ng/ml human basic
fibroblast growth factor (Gibco; Thermo Fisher Scientific, Inc.),
20 ng/ml human epidermal growth factor (Gibco; Thermo Fisher
Scientific, Inc.) and 5 µg/ml human insulin (Sigma-Aldrich;
Merck KGaA). The medium was replaced every 3 days, and on day 7
significant mammospheres (diameter, ≥100 µm) were observed
under a light microscope at ×40 magnification.
Colony formation assay
Breast cancer cells were detached and resuspended in
a single-cell suspension. Subsequently, 400 cells were inoculated
into a 6-well plate and cultured in DMEM containing 10% FBS, 100
U/ml penicillin and 100 mg/ml streptomycin. On day 14, the colonies
were imaged under a light microscope.
Flow cytometry analysis
Mammosphere cells were harvested on day 7 and
resuspended in a single-cell suspension with flow cytometry buffer
(1X PBS containing 2.5% FBS) without blocking. The suspension was
incubated with anti-CD44 (dilution, 1:100) and anti-CD24 (dilution,
1:100) antibodies according to the manufacturer's protocols. The
stained cells were sorted with a FACSAria system (BD Biosciences)
and divided into two groups: CD44+CD24− cells
(BCSCs) and non-CD44+CD24− cells (non-BCSCs).
FlowJo software was used for data analysis (version 7.6; FlowJo
LLC) and 20,000 cells were included in the analysis.
Microarray analysis
The GeneChip® Human Exon 1.0 ST Array
(Affymetrix; Thermo Fisher Scientific, Inc.) is designed for
screening lncRNAs and protein coding transcripts in BCSCs. The
chips can be used to investigate 44,414 human lncRNAs and 22,012
human mRNAs. These lncRNAs were screened with several databases,
including RefSeq (https://www.ncbi.nlm.nih.gov/refseq/),
GENCODE/ENSEMBLE (http://ensembl.org/index.html), NONCODE (http://www.noncode.org/) and UCSC Genome Browser
(http://genome.ucsc.edu/). Briefly, total RNA was
transcribed into cDNA and labelled with GeneChip2 WT Terminal
Labelling kit and Controls kit (Affymetrix; Thermo Fisher
Scientific, Inc.). Subsequently, the samples were hybridized
overnight at 45°C and washed with washing buffer (Affymetrix;
Thermo Fisher Scientific, Inc.). The chips were scanned using the
Affymetrix GeneChip Command Console with default settings
(Affymetrix; Thermo Fisher Scientific, Inc.). Data were collected
with the Affymetrix Launcher (version 1.0; Affymetrix; Thermo
Fisher Scientific, Inc.).
Data analysis
Data summarization, normalization and quality
control were carried out using GeneSpring software v13.0 (Agilent
Technologies, Inc.). For hierarchical clustering, differentially
expressed genes were selected according to the following criteria:
Fold change ≥2 and P<0.05. Subsequently, the fold change was
log2-transformed, and the data were visualized by
generating a heatmap in Java TreeView (version 6; https://sourceforge.net/projects/jtreeview/). Finally,
two-way hierarchical clustering analysis (Cluster Profiler; V3.6;
http://www.bioconductor.org/packages/release/bioc/html/clus-terProfiler.html)
was applied to categorize the samples with similar gene expression
patterns and reveal associations between the samples.
Gene Ontology (GO) enrichment and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis
To the best of our knowledge, the majority of the
identified lncRNAs have not yet been functionally characterized.
Therefore, their functionalities can only be predicted by analyzing
their mRNA counterparts (23).
Accordingly, GO (http://geneontology.org/) and KEGG analyses
(https://www.genome.jp/kegg/pathway.
html) were performed to explore the corresponding mRNA functions
using R software (version 3.4.3; https://www.r-project. org/). Distinct GO categories
were considered statistically significant with P<0.05. The
P-values shown in the KEGG pathway analyses represent the
significance of the pathways.
lncRNA-mRNA network
Crosstalk between lncRNAs and mRNAs was established
as previously described (24).
Briefly, the interaction network was built based on the normalized
signal intensity of the expression of specific mRNAs and lncRNAs.
Correlations between the aberrantly expressed lncRNAs and their
reciprocal genes were calculated to select significantly correlated
pairs with which the network was constructed.
Reverse transcription-quantitative PCR
(RT-qPCR)
To validate the microarray results, six lncRNAs were
randomly selected for RT-qPCR, which was performed using the
FastStart Universal SYBR-Green Master kit (Roche Diagnostics)
according to the manufacturer's protocols on an ABI PRISM 7900HT
sequence detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Total RNA extraction was performed with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according the manufacturer's protocol, and reverse
transcription was performed using Super-Script II (Takara Bio,
Inc.) according to the manufacturer's protocol. The RT-qPCR
protocol was as follows: 2 min at 50°C, followed by 10 min at 95°C
and 40 cycles of PCR following standard conditions with 15 sec of
denaturation at 95°C, elongation at 60°C for 1 min, 95°C for 15 sec
and 1 min at 60°C, and extension at 72°C for 5 min. GAPDH was used
as an internal control, and the relative expression levels were
analyzed by the 2−ΔΔCq method (25). All primers were designed using
primer premier 5 (Premier Biosoft International) and are listed in
Table I.
 | Table IList of primers. |
Table I
List of primers.
| Gene | Primer sequences
(5′-3′) |
|---|
| RBM47 | F:
TATAGGCATGAGCCACCACA |
| R:
AGCGCCCACTATTTACAAGG |
| NONHSAT114469 | F:
TGCACCTCTACACCCAGCTA |
| R:
TCACACCTGTAATCCCAGCA |
| GOLPH | F:
TGCAGTTAGGTTTGCTAGGC |
| R:
TGGTTAGAGCACAATTCTAAGACC |
| CUEDC1 | F:
ACTTTCACCCAGTCCCTTCC |
| R:
GCCCTTCCAGTCCTTGTTTC |
| TMA16 | F:
CTGCATATTACAGAACCCATCTG |
| R:
TTCCTTTAAGCACCTCAATGTC |
| ATG12 | F:
TCCTGCTTCATTTGCCTGTA |
| R:
GCACACAGCCAAAAATCAAT |
| NANOG | F:
AAAGCCTCCCAATCCCAAACA |
| R:
GCGGGCTCAATTTATAGAAACCGGG |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
| R:
GGCTGTTGTCATACTTCTCATGG |
| SOX2 | F:
TACAGCATGTCCTACTCGCAG |
| R:
GAGGAAGAGGTAACCACAGGG |
| OCT4 | F:
GCCGCTGGCTTATAGAAGGT |
| R:
CTCTCCCCAGCTTGCTTTGA |
Sanger sequencing
PCR products were resolved on a 1.2% agarose gel
containing GelRed Nucleic Acid Gel stain (Yeasen). The products
were confirmed by Sanger sequencing (Sangon Biotech Co., Ltd.).
Western blotting
Total protein was extracted from mammo-sphere cells
and adherent cells, which were collected and lysed in RIPA buffer
(Invitrogen; Thermo Fisher Scientific, Inc.). The extracted protein
was subjected to BCA buffer (Invitrogen; Thermo Fisher Scientific,
Inc.) for protein quantity analysis. Equal amounts (20
µg/lane) of protein were separated by 10% SDS-PAGE and
transferred onto a PVDF membrane (EMD Millipore). Subsequently, the
membranes were blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) at
room temperature for 1 h. The membranes were incubated with primary
antibodies (dilution, 1:1,000) overnight at 4°C, and GAPDH served
as the loading control. After the membranes were incubated with
secondary antibodies (dilution, 1:5,000) at 37°C for 1 h, they were
subjected to ECL Prime Western Blotting Detection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The immunoreactive
bands were visualized using a LAS-3000 Imager (Fujifilm).
Silencing of lncCUEDC1
LncCUEDC1 small interfering RNA (siRNA) was
synthesized by Shanghai GenePharma Co., Ltd. MCF-7 and MDA-MB-231
cells were transfected with 20 pmol lncCUEDC1 siRNA or negative
control (NC) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Subsequent experiments were
performed after transfection for 48 h. The lncCUEDC1 and NC siRNA
sequences were as follows: lncCUEDC1 forward, 5′-CCA GAG GCU UCU
CAA CUU UTT and reverse, 5′-AAA GUU GAG AAG CCU CUG GTT; NC
forward, 5′-UUC UCC GAA CGU GUC ACG UTT and reverse, 5′-ACG UGA CAC
GUU CGG AGA ATT.
Fluorescence in situ hybridization
(FISH)
In situ hybridization was performed according
to the fluorescent in situ hybridization kit instructions
(cat. no. C1090; Guangzhou RiboBio Co., Ltd.). Briefly, breast
cancer cells were digested into a single-cell suspension and seeded
on glass coverslips. Subsequently, the cells were fixed with 4%
paraformaldehyde for 10 min at 4°C and permeabilized with 0.5%
Triton X-100 (Biosharp) for 5 min at 4°C. After blocking for 30 min
at 37°C, the cells were inoculated with a Cy3 labeled lncCUEDC1
FISH probe overnight at 37°C, all blocking and hybridization
buffers were included in the FISH kit. The images were captured
under a fluorescence microscope (Carl Zeiss AG) and were analyzed
with Photoshop (version 6.0; Adobe Systems, Inc.).
Luciferase reporter assay
Luciferase reporter assays were performed according
to the instructions of the Pierce™ Renilla-Firefly Luciferase Dual
Assay kit (cat. no. 16186; Thermo Fisher Scientific, Inc.).
Specifically, green Renilla luciferase acted as an
experimental reporter with constitutively active red firefly as a
normalization control. NANOG (1-512 bp) was amplified by PCR and
inserted into the psiCHECK2.0 vector (psiCHECK2.0 vector; Addgene,
Inc.). Full length lncCUEDC1 was cloned into the pcDNA 3.1 vector.
Subsequently, 293T cells were co-transfected with NANOG and
lncCUEDC1 using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for 48 h. Cells were collected and used to
perform the luciferase reporter assay according to the
manufacturer's protocols. Luciferase activity was measured using a
microplate reader (Tecan Group, Ltd.).
Statistical analysis
All data were analyzed using Prism v6 (GraphPad
Software, Inc.), unless otherwise specified. Independent-sample
t-testing was used to compare the global levels of lncRNAs between
the different groups. Fisher's exact test was applied in GO and
KEGG analysis. Data are presented as the mean ± SD of three
independent experiments. P<0.05 was considered to indicate a
statistically significant difference.
Results
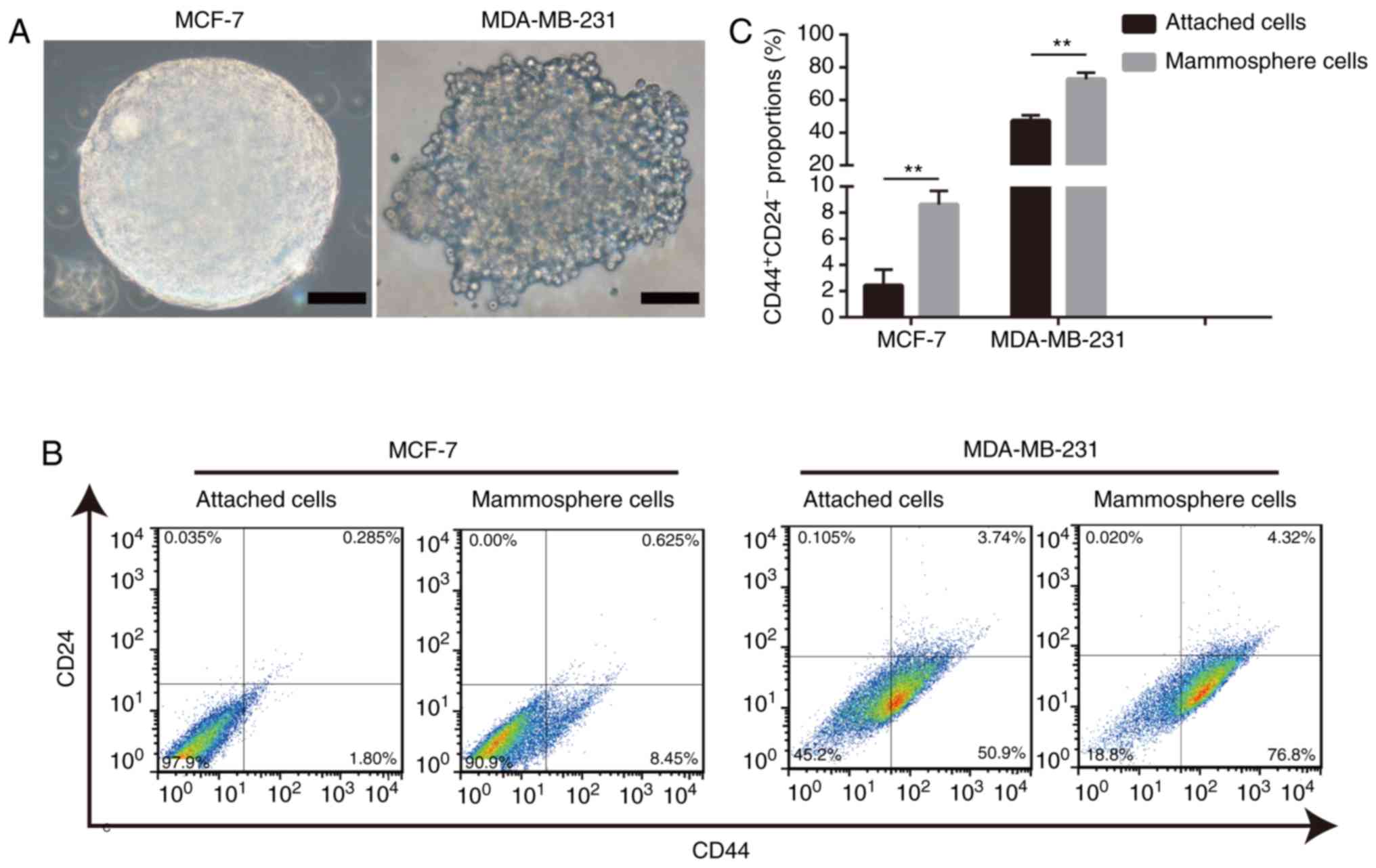
Mammosphere-derived cells exhibit a
relatively higher proportion of cells with the
CD44+CD24− phenotype
A commonly used method, the mammosphere formation
assay, was performed to enrich BCSCs (26). Mammospheres with diameters ≥100
µm were considered significant (Fig. 1A). The results of the present study
demonstrated that mammosphere-derived cells exhibited a higher
percentage of BCSCs with the CD44+CD24−
phenotype than adherent cells (Fig. 1B
and C). Overall, these data validated the system used in the
present study and supported the theory that the mammosphere culture
assay is an effective method for enriching BCSCs.
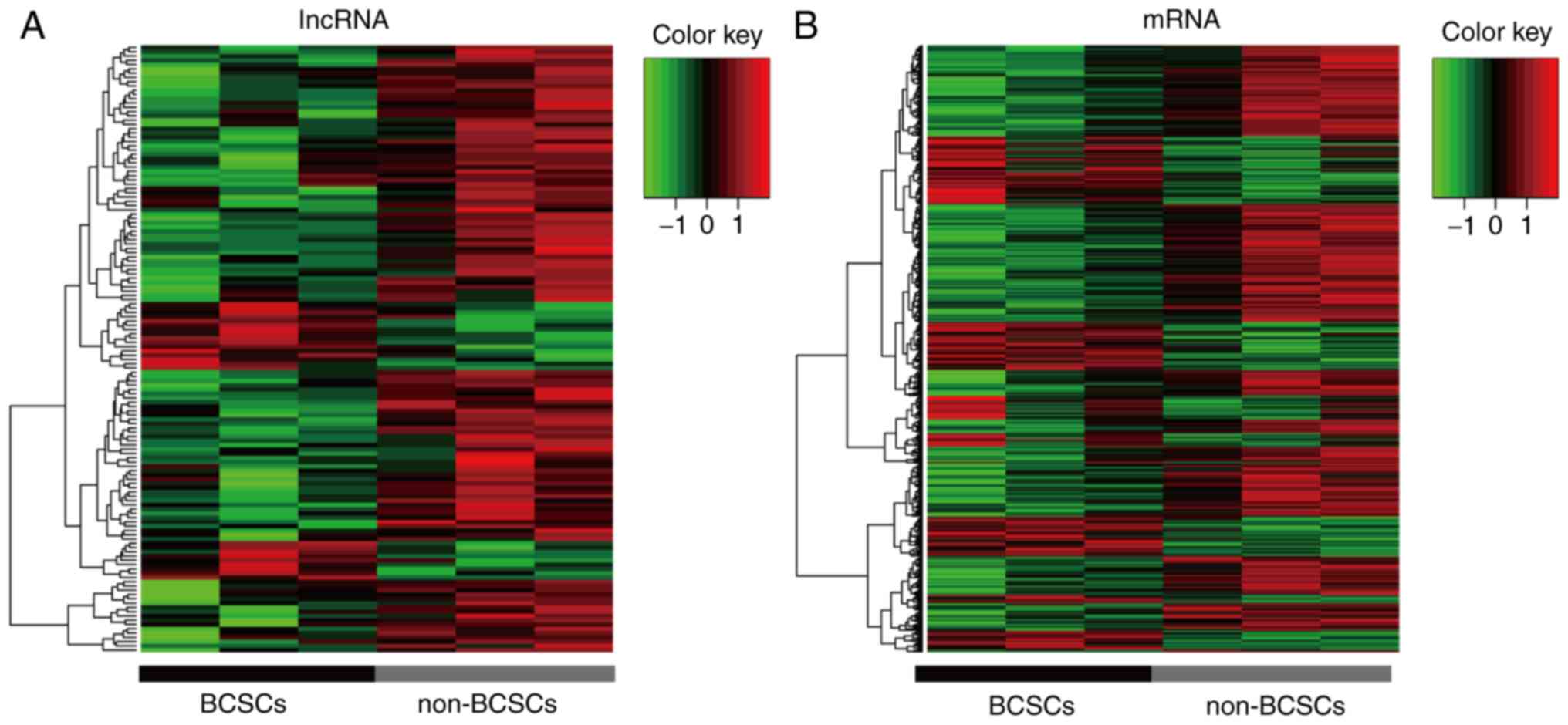
In the present study, a GeneChip® Human
Exon 1.0 ST Array was used to detect the lncRNA profile of BCSCs. A
total of 44,414 human lncRNAs and 22,012 human mRNA candidates were
identified in three pairs of samples. Additionally, 142
differentially expressed lncRNAs with a fold change >2.5 were
identified between BCSCs and non-BCSCs. Among these, 117 were
upregulated and 25 were downregulated (Fig. 2A). Additionally, the cluster of
corresponding mRNAs is presented in Fig. 2B. The information regarding the
differentially expressed lncRNA is listed in Table II by fold change.
 | Table IIDifferentially expressed lncRNAs in
breast cancer stem cells. |
Table II
Differentially expressed lncRNAs in
breast cancer stem cells.
| lncRNA_id | P-value | Fold change | Upregulation or
downregulation |
|---|
|
ENST00000584746 | 0.005225 | 0.33 | Down |
|
ENST00000531648 | 0.000236 | 0.34 | Down |
| NONHSAT118570 | 0.001455 | 0.35 | Down |
| NONHSAT060713 | 0.005342 | 0.35 | Down |
|
ENST00000565771 | 0.003509 | 0.36 | Down |
|
ENST00000332858 | 0.004131 | 0.36 | Down |
|
ENST00000553466 | 0.006265 | 0.36 | Down |
|
ENST00000450469 | 0.008614 | 0.36 | Down |
|
ENST00000590962 | 0.009127 | 0.36 | Down |
|
ENST00000466288 | 0.000708 | 0.37 | Down |
|
ENST00000583426 | 0.007507 | 0.37 | Down |
| NONHSAT077496 | 0.008404 | 0.37 | Down |
|
ENST00000568635 | 0.030779 | 0.37 | Down |
| NONHSAT032633 | 0.004160 | 0.38 | Down |
|
ENST00000467521 | 0.004415 | 0.38 | Down |
|
ENST00000426367 | 0.005238 | 0.38 | Down |
|
ENST00000528553 | 0.006575 | 0.38 | Down |
| NONHSAT078914 | 0.009508 | 0.38 | Down |
|
ENST00000559600 | 0.011918 | 0.38 | Down |
| NONHSAT030084 | 0.001486 | 0.39 | Down |
|
ENST00000505899 | 0.001553 | 0.39 | Down |
| NONHSAT127846 | 0.002345 | 0.39 | Down |
| NONHSAT022603 | 0.005557 | 0.39 | Down |
|
ENST00000496353 | 0.007553 | 0.39 | Down |
| NONHSAT107636 | 0.018664 | 0.39 | Down |
| NONHSAT032003 | 0.001439 | 2.51 | Up |
|
ENST00000553031 | 0.013427 | 2.51 | Up |
|
ENST00000480155 | 0.024014 | 2.51 | Up |
| NONHSAT143607 | 0.028454 | 2.51 | Up |
|
ENST00000481059 | 0.004178 | 2.52 | Up |
|
ENST00000413961 | 0.005341 | 2.53 | Up |
|
ENST00000589496 | 0.015149 | 2.53 | Up |
|
ENST00000600535 | 0.025085 | 2.53 | Up |
|
ENST00000602510 | 0.015045 | 2.54 | Up |
| NONHSAT074190 | 0.000795 | 2.55 | Up |
| NONHSAT102055 | 0.012780 | 2.55 | Up |
| NONHSAT027878 | 0.048534 | 2.55 | Up |
| NONHSAT112187 | 0.026605 | 2.56 | Up |
| n346329 | 0.002635 | 2.57 | Up |
|
ENST00000542306 | 0.001966 | 2.58 | Up |
| FR0049460 | 0.019297 | 2.58 | Up |
| NONHSAT003589 | 0.001326 | 2.59 | Up |
| n346329 | 0.006665 | 2.59 | Up |
|
ENST00000570137 | 0.018127 | 2.59 | Up |
| FR0011909 | 0.006207 | 2.6 | Up |
|
ENST00000519690 | 0.014859 | 2.6 | Up |
| NONHSAT138818 | 0.000899 | 2.61 | Up |
|
ENST00000492009 | 0.002426 | 2.61 | Up |
|
ENST00000429998 | 0.002882 | 2.61 | Up |
|
ENST00000558124 | 0.006331 | 2.62 | Up |
| NONHSAT073839 | 0.004252 | 2.63 | Up |
|
ENST00000551655 | 0.016965 | 2.63 | Up |
|
ENST00000586718 | 0.012420 | 2.64 | Up |
|
ENST00000470108 | 0.012616 | 2.64 | Up |
|
ENST00000471763 | 0.001250 | 2.65 | Up |
|
ENST00000422822 | 0.003282 | 2.65 | Up |
| NONHSAT115467 | 0.003438 | 2.66 | Up |
|
ENST00000515570 | 0.002535 | 2.67 | Up |
| NONHSAT002344 | 0.003232 | 2.67 | Up |
| NONHSAT114469 | 0.046724 | 2.68 | Up |
|
ENST00000478375 | 0.003936 | 2.69 | Up |
|
ENST00000487510 | 0.009022 | 2.69 | Up |
|
ENST00000589496 | 0.032779 | 2.69 | Up |
|
ENST00000484567 | 0.004827 | 2.7 | Up |
|
ENST00000529334 | 0.006399 | 2.7 | Up |
|
ENST00000462814 | 0.024536 | 2.71 | Up |
|
ENST00000512214 | 0.001719 | 2.72 | Up |
|
ENST00000597336 | 0.002131 | 2.72 | Up |
|
ENST00000513044 | 0.002454 | 2.73 | Up |
|
ENST00000462264 | 0.002692 | 2.73 | Up |
| FR0069759 | 0.006313 | 2.74 | Up |
|
ENST00000536823 | 0.031865 | 2.74 | Up |
| NONHSAT079829 | 0.004599 | 2.75 | Up |
| NONHSAT103651 | 0.011939 | 2.75 | Up |
|
ENST00000464026 | 0.002435 | 2.76 | Up |
|
ENST00000466287 | 0.012798 | 2.77 | Up |
|
ENST00000524936 | 0.040883 | 2.77 | Up |
| NONHSAT015408 | 0.001016 | 2.78 | Up |
|
ENST00000555596 | 0.020474 | 2.79 | Up |
| NONHSAT107508 | 0.040523 | 2.81 | Up |
|
ENST00000505252 | 0.003757 | 2.82 | Up |
|
ENST00000421480 | 0.004384 | 2.82 | Up |
| NONHSAT000825 | 0.007246 | 2.82 | Up |
|
ENST00000436137 | 0.013414 | 2.84 | Up |
|
ENST00000600056 | 0.037878 | 2.84 | Up |
|
ENST00000477648 | 0.042366 | 2.84 | Up |
| NONHSAT138537 | 0.021098 | 2.86 | Up |
|
ENST00000497638 | 0.002303 | 2.88 | Up |
|
ENST00000475697 | 0.011090 | 2.88 | Up |
|
ENST00000549100 | 0.013755 | 2.89 | Up |
|
ENST00000466522 | 0.001427 | 2.9 | Up |
|
ENST00000477648 | 0.001554 | 2.9 | Up |
|
ENST00000596267 | 0.000511 | 2.91 | Up |
|
ENST00000492497 | 0.005636 | 2.91 | Up |
|
ENST00000580010 | 0.016036 | 2.91 | Up |
|
ENST00000488850 | 0.000664 | 2.95 | Up |
|
ENST00000530163 | 0.001084 | 2.95 | Up |
|
ENST00000548283 | 0.002213 | 2.95 | Up |
|
ENST00000480707 | 0.003638 | 2.95 | Up |
|
ENST00000588390 | 0.003598 | 2.97 | Up |
|
ENST00000490265 | 0.004425 | 2.99 | Up |
| NONHSAT103765 | 0.000385 | 3.01 | Up |
|
ENST00000565390 | 0.011921 | 3.02 | Up |
|
ENST00000482284 | 0.001966 | 3.03 | Up |
| NONHSAT097185 | 0.014811 | 3.04 | Up |
| NONHSAT006994 | 0.009113 | 3.06 | Up |
|
ENST00000592636 | 0.021185 | 3.06 | Up |
| NONHSAT074169 | 0.003962 | 3.08 | Up |
|
ENST00000490258 | 0.013133 | 3.08 | Up |
| NONHSAT017862 | 0.000362 | 3.11 | Up |
| NONHSAT103002 | 0.008231 | 3.11 | Up |
|
ENST00000476635 | 0.000925 | 3.15 | Up |
|
ENST00000494618 | 0.011662 | 3.19 | Up |
|
ENST00000471892 | 0.019328 | 3.21 | Up |
|
ENST00000483207 | 0.024033 | 3.21 | Up |
|
ENST00000514842 | 0.000120 | 3.24 | Up |
|
ENST00000486261 | 0.001738 | 3.24 | Up |
|
ENST00000585864 | 0.035171 | 3.24 | Up |
|
ENST00000434942 | 0.008521 | 3.26 | Up |
|
ENST00000564690 | 0.001087 | 3.27 | Up |
| NONHSAT122723 | 0.000366 | 3.28 | Up |
|
ENST00000479971 | 0.017701 | 3.31 | Up |
|
ENST00000534296 | 0.001884 | 3.32 | Up |
|
ENST00000510321 | 0.009749 | 3.32 | Up |
| NONHSAT065985 | 0.002444 | 3.34 | Up |
|
ENST00000552938 | 0.021853 | 3.35 | Up |
| NONHSAT076663 | 0.001175 | 3.36 | Up |
|
ENST00000506675 | 0.002164 | 3.38 | Up |
| NONHSAT046699 | 0.002645 | 3.41 | Up |
|
ENST00000472297 | 0.021161 | 3.47 | Up |
|
ENST00000554812 | 0.000307 | 3.51 | Up |
|
ENST00000506067 | 0.000610 | 3.56 | Up |
| NONHSAT023366 | 0.002519 | 3.59 | Up |
|
ENST00000481407 | 0.004871 | 3.68 | Up |
|
ENST00000498815 | 0.000260 | 3.72 | Up |
| NONHSAT113228 | 0.032323 | 3.75 | Up |
|
ENST00000559075 | 0.000676 | 3.76 | Up |
| NONHSAT081217 | 0.002130 | 3.77 | Up |
|
ENST00000479757 | 0.001441 | 3.82 | Up |
|
ENST00000452181 | 0.001118 | 4.09 | Up |
| NONHSAT008483 | 0.022491 | 4.38 | Up |
|
ENST00000506214 | 0.000043 | 5.39 | Up |
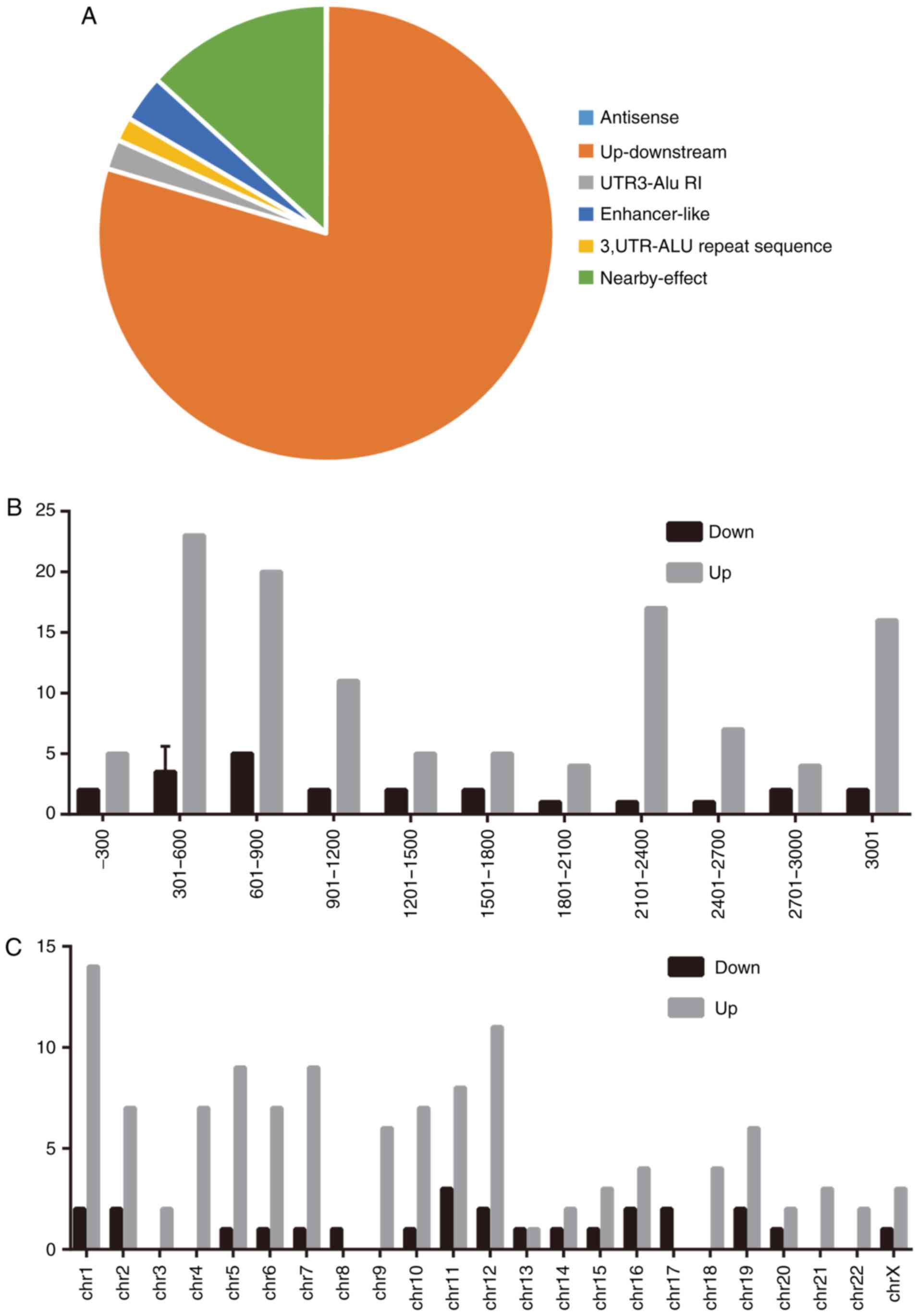
Characteristics of identified
lncRNAs
To better define the identified lncRNAs, lncRNAs
expression characteristics, including their classification,
chromosome distribution and length distribution, were analyzed. The
present study demonstrated that the identified lncRNAs could be
attributed to six classes, including natural 'antisense',
'up-downstream', 'UTR3-Alu RI' (UTR3 regulation), 'enhancer-like',
'3'UTR-Alu repeat sequence' and 'nearby-effect', based on their
position on the genome. The data revealed that most lncRNAs were
located up-downstream of their mRNA counterparts (Fig. 3A) and that their lengths were
mainly distributed between 301-600 and 601-900 nucleotides
(Fig. 3B). These findings,
including the length and chromosome location, partially agreed with
those of a previous study (26).
Furthermore, a chromosome distribution analysis revealed a
difference in the chromosome distribution between upregulated
lncRNAs and downregulated lncRNAs (Fig. 3C).
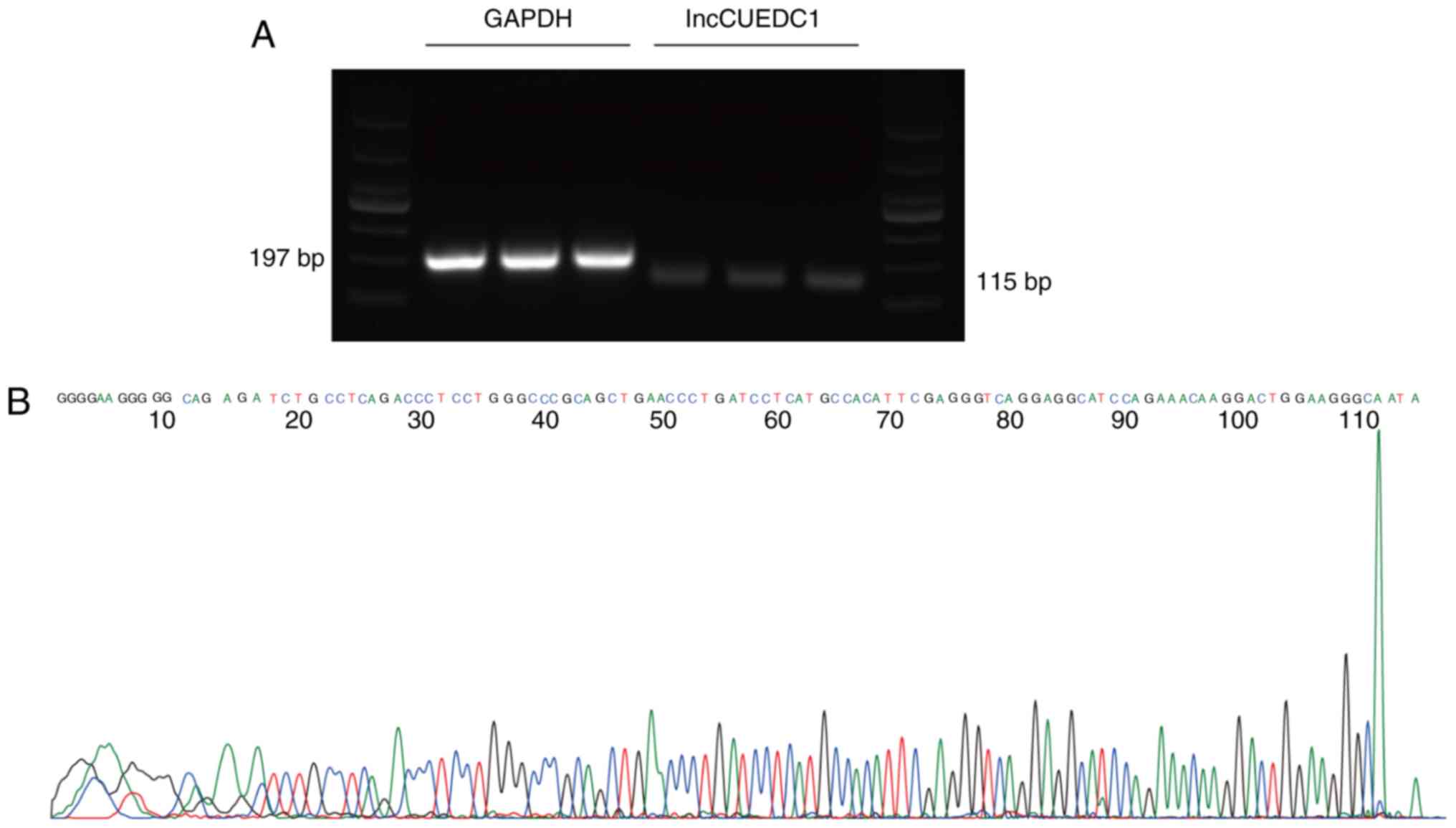
Validation of differentially expressed
lncRNAs
Six lncRNAs, including lncRBM47, lncNONHSAT114469,
lncGOPLH, lncCUEDC1, lncTMA16 and lncATG12, were randomly selected
to validate the microarray results by qPCR, which confirmed that
these lncRNAs were aberrantly expressed in mammosphere cells
compared with in adherent cells (Fig.
4A and B). The log2-fold changes were used for
analysing the microarray and qPCR results (Fig. 4C and D). The qPCR results were
consistent with the results of the microarray assay. In addition,
Sanger sequencing of lncCUEDC1, which was the most downregulated
lncRNA, further validated the microarray data (Fig. 5). These findings indicated that
these lncRNAs were differentially expressed between BCSCs and
non-BCSCs.
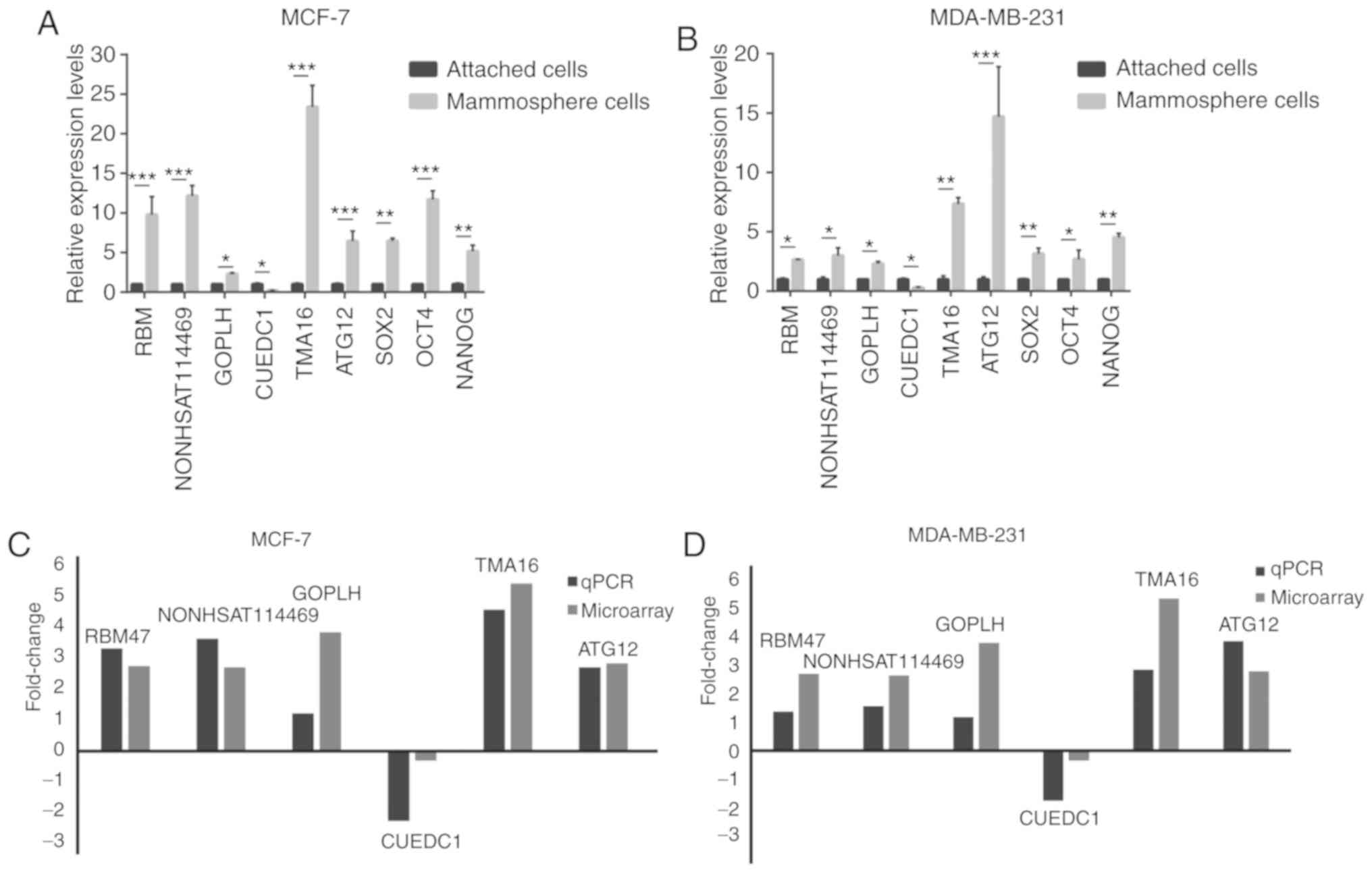 | Figure 4qPCR validation of the differences in
the expression of six lncRNAs, SOX2, NANOG and OCT4 between BCSCs
and non-BCSCs. (A) Expression levels of six lncRNAs, SOX2, NANOG
and OCT4 in adherent cells and floating mammospheres derived from
MCF7 cells. (B) Expression levels of six lncRNAs, SOX2, NANOG and
OCT4 in adherent cells and floating mammospheres derived from the
MDA-MB-231 cell line. (C) Comparison of the micro-array data and
qPCR results in MCF7 cells. (D) Comparison of the microarray data
and qPCR results in MDA-MB231 cells. The y-axis shows the average
fold change (log2 transformed) for each lncRNA measured by qPCR and
microarray analysis. *P<0.05, **P<0.01
and ***P<0.001, as indicated. qPCR, quantitative PCR;
BCSC, breast cancer stem cell; lncRNA, long non-coding RNA. |
Functional prediction analysis for
identified lncRNAs
Previous studies have revealed that lncRNAs are
generally transcribed along with their corresponding mRNAs and have
the potential to modulate the expression of their adjacent genes in
multiple manners (28,29). In addition, the functions of
lncRNAs are partially reflected by their counterpart mRNAs
(30). Therefore, GO enrichment
and KEGG pathway analyses were performed to predict the functions
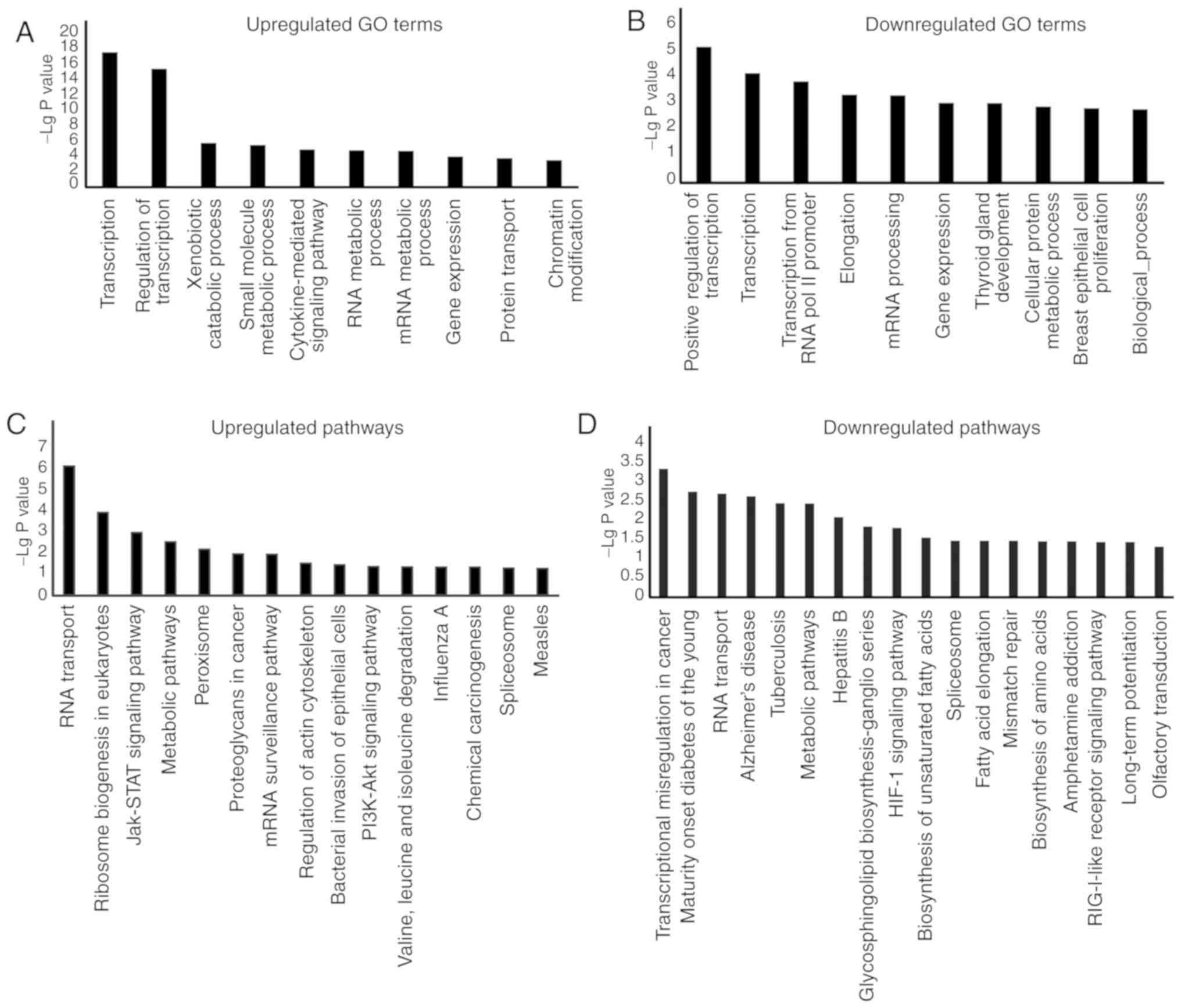
of identified lncRNAs. GO analysis of the upregulated and
downregulated lncRNAs revealed that the most frequently enriched
functions of lncRNAs were mainly associated with 'transcription',
'regulation of transcription', 'small molecule metabolic process'
and 'cytokine-mediated signalling pathway' (Fig. 6A and B). Intriguingly,
'transcription' and 'regulation of transcription' were the two most
significantly enriched terms for lncRNAs in BCSCs, which
demonstrated that lncRNAs may be involved in regulating expression
levels of stemness-related genes. In the KEGG pathway analysis, 142
aberrantly expressed lncRNAs were distributed among 305 KEGG
pathways (Fig. 6C and D). Of
these, the 'JAK-STAT signaling pathway', 'PI3K-Akt signaling
pathway' and 'HIF-1 signaling pathway', all of which are associated
with BCSC modulation, were found to be associated with the
identified lncRNAs. Several studies have indicated that these
signaling pathways serve critical roles in stem cell behavior
(31-33). These findings suggested that
lncRNAs could be implicated in BCSCs behaviors.
lncRNA-mRNA co-expression network
To further decipher the functionalities of the
lncRNAs identified here, a lncRNA-mRNA co-expression network was
constructed based on correlation analysis (34). The crosstalk between lncRNAs and
mRNAs is shown in Fig. S1. In
addition, the interaction network revealed that lncCUEDC1 could
interact with Musashi 2 (MSI2). Overall, these findings suggested
that lncRNAs may control BCSCs by modulating the expression of
their corresponding mRNAs.
lncCUEDC1 exhibits inhibitory effects on
BCSCs
Based on the results, it was postulated that
lncCUEDC1 may serve an inhibitory role in BCSCs. Hence, siRNAs
targeting lncCUEDC1 were designed to validate this hypothesis and
explore its effect on BCSCs behavior and stemness. After knocking
down the expression of lncCUEDC1 using siRNA, the expression levels
of stemness-related markers were greatly enhanced compared with
those in cells transfected with the NC siRNA at the mRNA and
protein levels (Fig. 7A-C).
Additionally, breast cancer cells transfected with lncCUEDC1 siRNA
exhibited a substantial increase in the proportion of
CD44+CD24− BCSCs compared with cells
transfected with the NC (Fig. 7D and
E). Additionally, mammosphere and colony formation capacities
were increased in cells transfected with lncCUEDC1 siRNA (Fig. 7F-I). Overall, these data suggested
that lncCUEDC1 could suppress BCSCs stemness, which supports the
potential of lncCUEDC1 as a target for eliminating BCSCs.
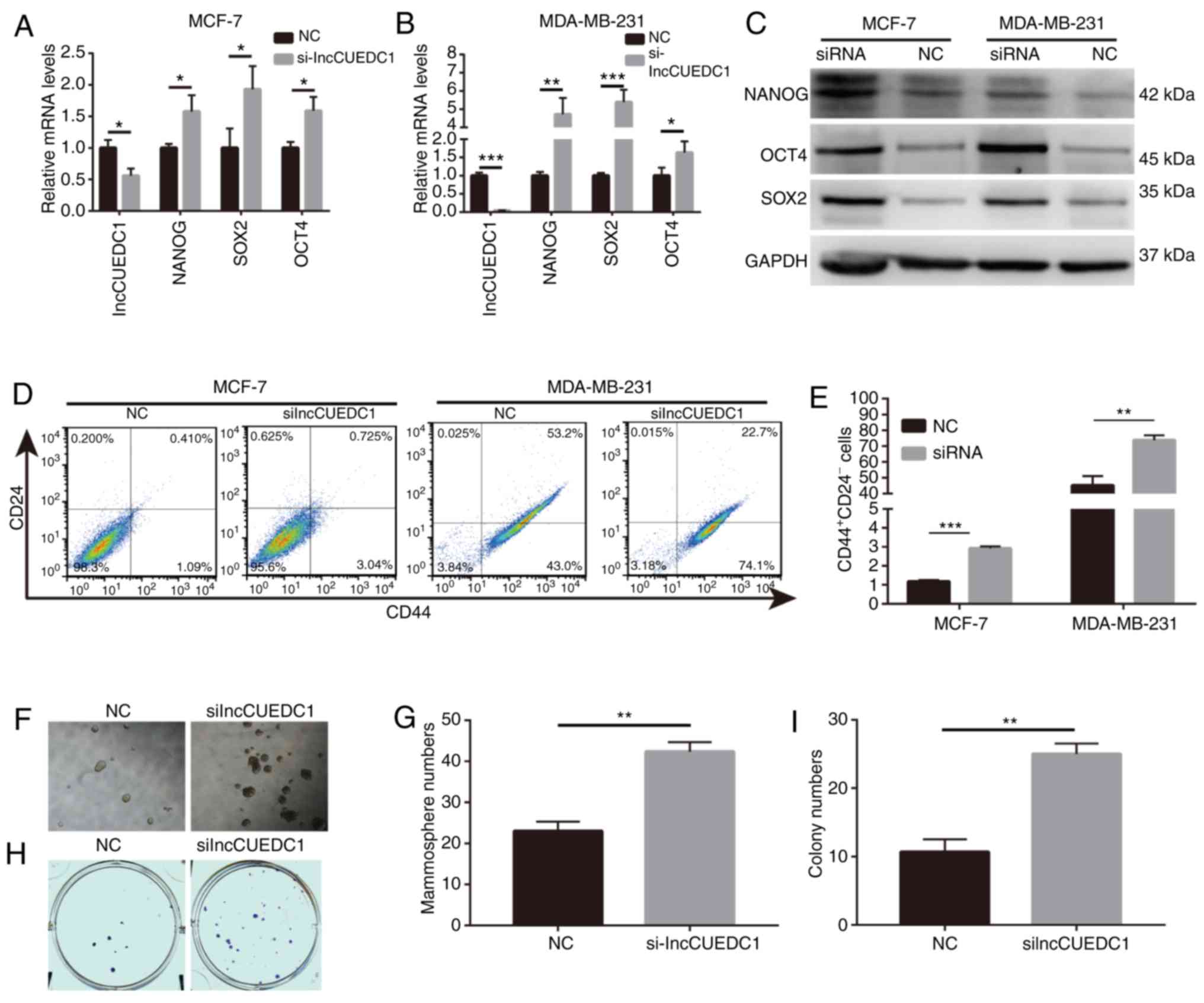 | Figure 7lncCUEDC1 negatively regulates BCSCs
in vitro. (A) mRNA expression levels of NANOG, SOX2 and OCT4
were markedly increased following loss of lncCUEDC1 in MCF-7 cells.
(B) mRNA expression levels of NANOG, SOX2 and OCT4 were markedly
increased following loss of lncCUEDC1 in MDA-MB-231 cells. (C)
Protein expression levels of NANOG, SOX2 and OCT4 were markedly
increased following loss of lncCUEDC1 in MCF-7 and MDA-MB-231
cells. (D) Flow cytometry revealed that
CD44+CD24− putative BCSCs were largely
expanded after loss of lncCUEDC1 in vitro. (E) When the
breast cancer cells were treated with lncCUEDC1 siRNA for 48 h, the
proportions of CD44+CD24− putative BCSCs were
largely increased compared with cells treated with the NC. (F) Size
of mammospheres was enhanced after loss of lncCUEDC1. (G) Numbers
of mammospheres were enhanced after loss of lncCUEDC1. (H)
lncCUEDC1 depletion increased the size of colonies. (I) lncCUEDC1
depletion increased the numbers of colonies. All results were
obtained from three independent experiments, and the data are
presented as the mean ± SD. *P<0.05,
**P<0.01 and ***P<0.001, as indicated.
lncCUEDC1, long non-coding RNA CUE domain containing 1; BCSC,
breast cancer stem cell; NC, negative control; siRNA, small
interfering RNA. |
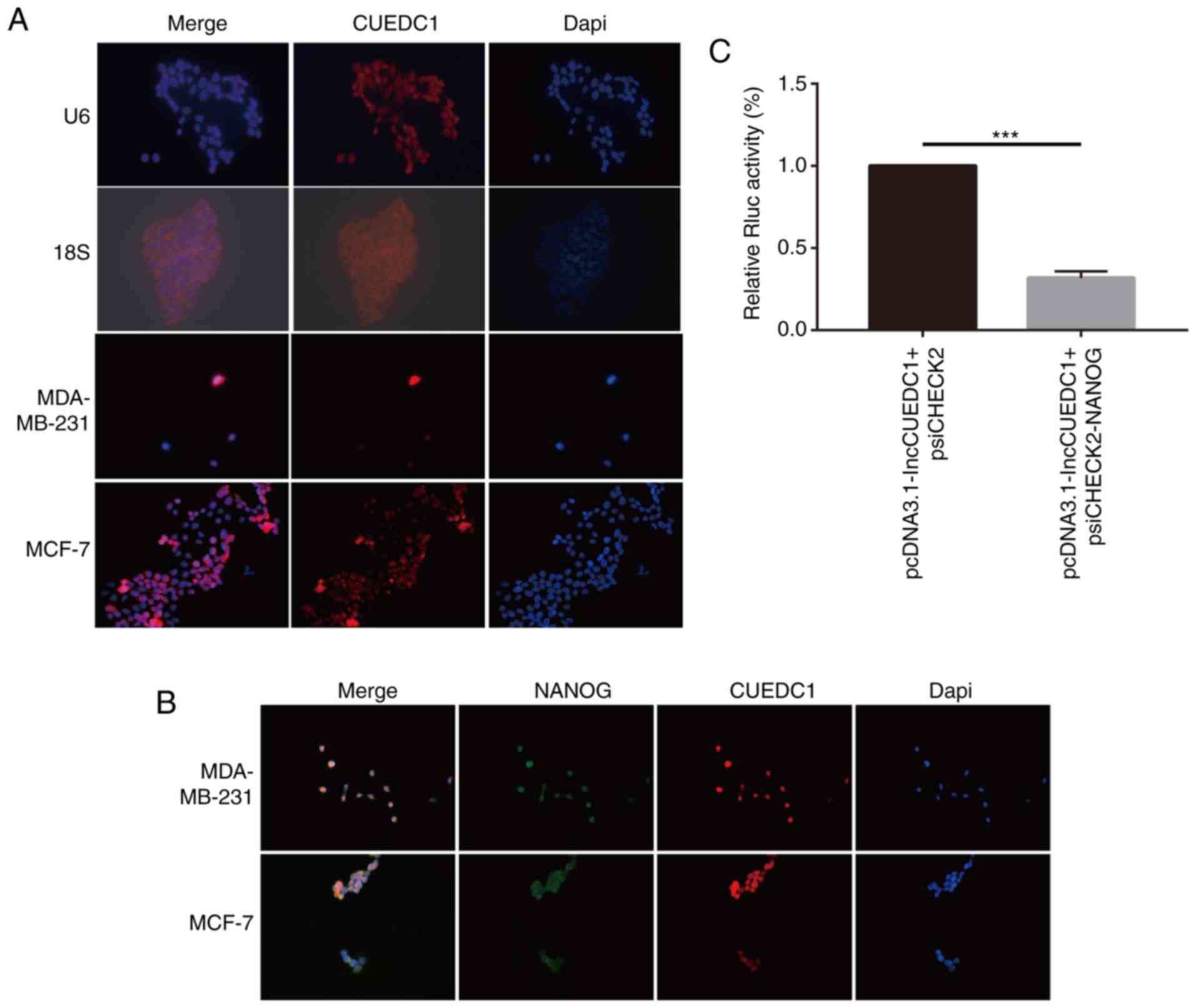
To explore the mechanism by which lncCUEDC1
regulates stemness, FISH assays were performed to determine the
location of lncCUEDC1. The data demonstrated that lncCUEDC1 was
mainly located in the cell nucleus. Notably, lncCUEDC1 could bind
with Nanog in the nucleus (Fig. 8A and
B). Additionally, the data revealed that the ectopic expression
of lncCUEDC1 could inhibit the expression of Nanog but not
psiCHECK2.0 in a luciferase reporter assay (Fig. 8C). Our previous study confirmed
that Nanog could stimulate the stemness of breast cancer cells
(35). Hence, the current data
indicated that lncCUEDC1 may inhibit stemness by inhibiting Nanog,
which needs to be further investigated.
Discussion
Accumulating evidence has demonstrated that lncRNAs
serve critical roles in regulating cancer, including breast cancer,
and that lncRNAs can act as either oncogenes or tumor suppressors.
For example, a well-characterized lncRNA, H19, has been found to be
an oncogene in breast cancer. In vitro experiments have
shown that H19 is upregulated in breast cancer cells during the
S-phase and that forced H19 expression could promote breast cancer
cell proliferation (36).
Additionally, another study has revealed that lncRNA-ROR is
upregulated in breast tumor tissue samples and enhances the
migration and invasion capacity of breast cancer cells (37). However, there remain contradictory
conclusions regarding the functional roles of lncRNAs in breast
cancer. Liu et al (18)
demonstrated that an NF-κB-interacting lncRNA, NKILA, could
suppress NF-κB activation to inhibit breast cancer metastasis.
Reports on breast cancer are abundant, yet there is no systematic
research on the lncRNA profile of BCSCs.
The present study first established a mammosphere
platform. MDA-MB-231 cells seemed to be enriched with more
CD44+CD24− phenotype cells. These results
were consistent with previous studies (38-40),
and confirmed that the platform worked well. Subsequently, the
expression profile of lncRNAs in BCSCs with the
CD44+CD24− phenotype was investigated via
microarray analysis. To the best of our knowledge, the present
study was the first to report the lncRNA profile of BCSCs. In the
present study, 142 lncRNAs were aberrantly expressed in
CD44+CD24− BCSCs compared with in non-BCSCs.
Importantly, among the 142 lncRNAs identified, 117 lncRNAs were
upregulated and 25 were downregulated. In addition, six of these
lncRNAs were randomly selected to be verified by qPCR. The qPCR
findings supported the microarray results. A previous study has
demonstrated that lncRNAs are involved in the potency and
differentiation of mammalian embryonic and adult stem cells
(41). For example, it has been
reported that the H19 expression level is lower in mesenchymal stem
cells that will differentiate into adipocytes compared with in
non-mesenchymal stem cells (42).
These data revealed that the grade of differentiation was
associated with lncRNAs. Therefore, the present data demonstrated
that cancer stem cells have a different lncRNA profile when
compared with non-cancer stem cells, this at least partially agrees
with previous results (41,42).
Currently, due to dramatic advances in
bioinformatics and sequencing technologies, an increasing number of
lncRNAs have been identified. However, only a small fraction of the
discovered lncRNAs have been functionally annotated. Generally, the
functions of lncRNAs are predicted through their counterpart mRNAs.
In the present study, GO and KEGG pathway analyses were performed
to predict the potential roles of the screened lncRNAs. GO analysis
demonstrated that the most frequently enriched functions of lncRNAs
were 'transcription', 'regulation of transcription', 'small
molecule metabolic process' and 'cytokine-mediated signaling
pathway'. These results indicated that the lncRNAs detected here
may be involved in controlling BCSCs at the transcriptional and
translational levels to some extent. Additionally, KEGG pathway
analysis revealed that the 'JAK-STAT signaling pathway', 'PI3K-Akt
pathway' and 'HIF-1 signaling pathway' were among the top predicted
pathways. These pathways have been demonstrated to be involved in
regulating the behaviors of CSCs (31,33,43).
Therefore, it is reasonable to postulate that these lncRNAs are
likely to be involved in modulating these pathways. These findings
provided a direction for further exploration of the functional
roles of lncRNAs in BCSCs.
The functionalities of lncRNAs in BCSCs remain
unknown. A previous report has revealed that lncRNAs are capable of
regulating the expression of nearby protein coding genes via
chromatin modification, transcription and post-transcriptional
processing (44). One of the most
reported function patterns for lncRNAs is chromatin modification.
For example, HOTAIR is able to suppress transcription by recruiting
an inhibitory chromatin state (45). Furthermore, a pivotal role served
by lncRNAs in regulating transcription is associated with enhancers
(46) and promoters (47), as demonstrated by the
well-characterized lncRNA CCAT1-L. Xiang et al (48) concluded that CCAT1-L is encoded
within an enhancer positioned 515 kb upstream from the MYC gene,
and they found that it could enhance the expression of MYC and
promote tumorigenesis. In addition, lncRNAs generated from
antisense transcripts are possibly involved in the control of mRNA
dynamics in post-transcriptional process (49). In the present study, a lncRNA-mRNA
network was constructed based on the abovementioned functions. The
network provided evidence to better illustrate the functional
mechanisms of lncRNAs.
The lncRNA-mRNA network analysis revealed that
lncCUEDC1 was substantially downregulated and may be able to
interact with MSI2. However, the potential roles in BCSCs were
undetermined. Hence, siRNAs were designed to knockdown the
expression of lncCUEDC1 and explore its effects on stemness of
BCSCs in vitro. Nanog, Sox2 and Oct4 are key transcription
factors for maintaining self-renewal capacity and stemness
(41). Therefore, the abundance of
stemness-related markers, including Nanog, Sox2 and Oct4 was also
examined. The data demonstrated that the stemness-related factors
were increased at both the mRNA and protein levels when the
expression of lncCUEDC1 was knocked down. According to the BCSC
concept, breast cancer is derived from a small fraction of breast
cancer cells with the CD44+CD24− phenotype
(50). Therefore, the impact of
lncCUEDC1 on the BCSC phenotype was examined. The results
demonstrated that the proportion of
CD44+CD24− cells in MCF7 and MDA-MB-231
breast cancer cells was enhanced substantially after the loss of
lncCUEDC1 expression. In addition, loss of lncCUEDC1 could increase
the self-renewal capacity of BCSCs in mammosphere and colony
formation assays. These results indicated that lncCUEDC1 was a
potential inhibitor of the stemness of BCSCs. Although the present
study suggested that lncCUEDC1 served a suppressive role in BCSCs,
the possibility that other dysregulated lncRNAs are also involved
in controlling BCSCs has not been excluded since only cell samples
were screened. The function of lncRNAs is closely associated with
their intracellular localization. lncRNAs located in the cytoplasm
mainly serve as microRNA sponges to regulate target gene
expression; however, lncRNAs located in the nucleus tend to be
involved in chromatin remodeling and epigenetic modification
(17,51). The FISH assay results revealed that
lncCUEDC1 was not only mainly located in the nucleus, but also
co-located with Nanog in the nucleus. Furthermore, the luciferase
reporter assay results revealed that Nanog expression was decreased
when lncCUEDC1 was overexpressed. Our previous study demonstrated
that Nanog depletion could inhibit the stemness of breast cancer
cells (35). Hence, collectively,
these data suggest that lncCUEDC1 may inhibit stemness by
suppressing the expression of Nanog. However, further investigation
is required.
In conclusion, the present study revealed the lncRNA
profile in BCSCs for the first time as determined by micro-array
analysis. Overall, 117 upregulated lncRNAs and 25 downregulated
lncRNAs were identified between BCSCs and non-BCSCs. In addition, a
lncRNA-mRNA interaction network was constructed. Notably, lncCUEDC1
was the most downregulated, and loss of function assays
demonstrated that lncCUEDC1 was a negative modulator for the
maintenance of stemness in BCSCs. Mechanistically, the present
study demonstrated that lncCUEDC1 may inhibit BCSC stemness by
reducing Nanog expression. These findings provide evidence that
lncCUEDC1 may possess a considerable potential as a therapeutic
target in BCSCs.
Supplementary Data
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81172522 and 81301858) and
the Suzhou Science and Technology Project (grant nos. SYS201508 and
SYS201308).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NY and YX developed the concept. FZ, YM and LX
developed the methods. FZ, YM, LX and HX performed formal analysis.
FZ and YM wrote the original draft. NY and YX reviewed and edited
the manuscript. NY and YX provided supervision. FZ, YM and LX were
involved in project administration. FZ and YX acquired funding. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Yanyun
Zhang at the Institute of Health Sciences, Shanghai Institutes for
Biological Sciences for providing equipment and excellent technical
support.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu R, Wang X, Chen GY, Dalerba P, Gurney
A, Hoey T, Sherlock G, Lewicki J, Shedden K and Clarke MF: The
prognostic role of a gene signature from tumorigenic breast-cancer
cells. N Engl J Med. 356:217–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Samanta D, Gilkes DM, Chaturvedi P, Xiang
L and Semenza GL: Hypoxia-inducible factors are required for
chemotherapy resistance of breast cancer stem cells. Proc Natl Acad
Sci USA. 111:E5429–E5438. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Los Angeles A, Ferrari F, Xi R,
Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee
S, Leitch HG, et al: Hallmarks of pluripotency. Nature.
525:469–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graziano A, d'Aquino R, Tirino V,
Desiderio V, Rossi A and Pirozzi G: The stem cell hypothesis in
head and neck cancer. J Cell Biochem. 103:408–412. 2008. View Article : Google Scholar
|
|
7
|
Malhotra GK, Zhao X, Band H and Band V:
Shared signaling pathways in normal and breast cancer stem cells. J
Carcinog. 10:382011. View Article : Google Scholar
|
|
8
|
El Helou R, Pinna G, Cabaud O, Wicinski J,
Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, et al:
miR-600 acts as a bimodal switch that regulates breast cancer stem
cell fate through WNT signaling. Cell Rep. 18:2256–2268. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bouras T, Pal B, Vaillant F, Harburg G,
Asselin-Labat ML, Oakes SR, Lindeman GJ and Visvader JE: Notch
signaling regulates mammary stem cell function and luminal
cell-fate commitment. Cell Stem Cell. 3:429–441. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu S, Dontu G, Mantle ID, Patel S, Ahn
NS, Jackson KW, Suri P and Wicha MS: Hedgehog signaling and Bmi-1
regulate self-renewal of normal and malignant human mammary stem
cells. Cancer Res. 66:6063–6071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
14
|
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z,
Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al: A functional
genomic approach identifies FAL1 as an oncogenic long noncoding RNA
that associates with BMI1 and represses p21 expression in cancer.
Cancer Cell. 26:344–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
16
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X,
Lin L, Yao H, Su F, Li D, et al: A cytoplasmic NF-κB interacting
long noncoding RNA blocks IκB phosphorylation and suppresses breast
cancer metastasis. Cancer Cell. 27:370–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
22
|
Yan N, Xu L, Wu X, Zhang L, Fei X, Cao Y
and Zhang F: GSKJ4, an H3K27me3 demethylase inhibitor, effectively
suppresses the breast cancer stem cells. Exp Cell Res. 359:405–414.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Wu D, Chen L, Li X, Yang J, Fan
D, Dong T, Liu M, Tan P, Xu J, et al: RAID: A comprehensive
resource for human RNA-associated (RNA-RNA/RNA-protein)
interaction. RNA. 20:989–993. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pujana MA, Han JD, Starita LM, Stevens KN,
Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al:
Network modeling links breast cancer susceptibility and centrosome
dysfunction. Nat Genet. 39:1338–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Ponti D, Costa A, Zaffaroni N, Pratesi G,
Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu N, Wang F, Lv M and Cheng L: Microarray
expression profile analysis of long non-coding RNAs in human breast
cancer: A study of Chinese women. Biomed Pharmacother. 69:221–227.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun J, Lin Y and Wu J: Long non-coding RNA
expression profiling of mouse testis during postnatal development.
PLoS One. 8:e757502013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guenzl PM and Barlow DP: Macro lncRNAs: A
new layer of cis-regulatory information in the mammalian genome.
RNA Biol. 9:731–741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gong X, Wei W, Chen L, Xia Z and Yu C:
Comprehensive analysis of long non-coding RNA expression profiles
in hepatitis B virus-related hepatocellular carcinoma. Oncotarget.
7:42422–42430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hernandez-Vargas H, Ouzounova M, Le
Calvez-Kelm F, Lambert MP, McKay-Chopin S, Tavtigian SV, Puisieux
A, Matar C and Herceg Z: Methylome analysis reveals Jak-STAT
pathway deregulation in putative breast cancer stem cells.
Epigenetics. 6:428–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Almozyan S, Colak D, Mansour F, Alaiya A,
Al-Harazi O, Qattan A, Al-Mohanna F, Al-Alwan M and Ghebeh H: PD-L1
promotes OCT4 and Nanog expression in breast cancer stem cells by
sustaining PI3K/AKT pathway activation. Int J Cancer.
141:1402–1412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chanmee T, Ontong P, Izumikawa T,
Higashide M, Mochizuki N, Chokchaitaweesuk C, Khansai M, Nakajima
K, Kakizaki I, Kongtawelert P, et al: Hyaluronan production
regulates metabolic and cancer stem-like properties of breast
cancer cells via hexosamine biosynthetic pathway-coupled HIF-1
signaling. J Biol Chem. 291:24105–24120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sahu D, Hsu CL, Lin CC, Yang TW, Hsu WM,
Ho SY, Juan HF and Huang HC: Co-expression analysis identifies long
noncoding RNA SNHG1 as a novel predictor for event-free survival in
neuroblastoma. Oncotarget. 7:58022–58037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu C, Xu L, Liang S, Zhang Z, Zhang Y and
Zhang F: Lentivirus-mediated shRNA targeting Nanog inhibits cell
proliferation and attenuates cancer stem cell activities in breast
cancer. J Drug Target. 24:422–432. 2016. View Article : Google Scholar
|
|
36
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y,
Zhao L, Zhang Y, Huang B and Lu J: LincRNA-ROR induces
epithelial-to-mesenchymal transition and contributes to breast
cancer tumorigenesis and metastasis. Cell Death Dis. 5:e12872014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ricardo S, Vieira AF, Gerhard R, Leitão D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Honeth G, Bendahl PO, Ringnér M, Saal LH,
Gruvberger-Saal SK, Lövgren K, Grabau D, Fernö M, Borg A and
Hegardt C: The CD44+/CD24- phenotype is enriched in basal-like
breast tumors. Breast Cancer Res. 10:R532008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu L, Zhang L, Hu C, Liang S, Fei X, Yan
N, Zhang Y and Zhang F: WNT pathway inhibitor pyrvinium pamoate
inhibits the self-renewal and metastasis of breast cancer stem
cells. Int J Oncol. 48:1175–1186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sheik Mohamed J, Gaughwin PM, Lim B,
Robson P and Lipovich L: Conserved long noncoding RNAs
transcriptionally regulated by Oct4 and Nanog modulate pluripotency
in mouse embryonic stem cells. RNA. 16:324–337. 2010. View Article : Google Scholar :
|
|
42
|
Huang Y, Zheng Y, Jin C, Li X, Jia L and
Li W: Long non-coding RNA H19 inhibits adipocyte differentiation of
bone marrow mesenchymal stem cells through epigenetic modulation of
histone deacetylases. Sci Rep. 6:288972016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu Y, Guo R, Wei J, Zhou Y, Ji W, Liu J,
Zhi X and Zhang J: Effects of PI3K inhibitor NVP-BKM120 on
overcoming drug resistance and eliminating cancer stem cells in
human breast cancer cells. Cell Death Dis. 6:e20202015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Morris KV, Santoso S, Turner AM, Pastori C
and Hawkins PG: Bidirectional transcription directs both
transcriptional gene activation and suppression in human cells.
PLoS Genet. 4:e10002582008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ashe HL, Monks J, Wijgerde M, Fraser P and
Proudfoot NJ: Intergenic transcription and transinduction of the
human beta-globin locus. Genes Dev. 11:2494–2509. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guenther MG, Levine SS, Boyer LA, Jaenisch
R and Young RA: A chromatin landmark and transcription initiation
at most promoters in human cells. Cell. 130:77–88. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He Y, Vogelstein B, Velculescu VE,
Papadopoulos N and Kinzler KW: The antisense transcriptomes of
human cells. Science. 322:1855–1857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|