Introduction
Bladder cancer (BC) is one of the most common
urinary tumors, with an increasing incidence worldwide every year
(1). Although BC can be treated
via transurethral resection, radical cystectomy and chemotherapy,
it still has a high mortality rate due to its high incidence of
metastasis (2). Thus, there is an
urgent need to determine the underlying mechanism of BC development
to improve the therapeutic outcome of patients.
MicroRNAs (miRNAs or miRs) are a class of small
regulatory non-coding RNAs that serve important roles in many
signaling pathways, including cell survival, apoptosis, cancer
migration and cancer progression (3). Furthermore, an increasing number of
miRNAs have been associated with the diagnosis and prognosis of
certain types of cancer (4,5).
The miR-21 gene is located on chromosome 17q23
(6). Previous studies have
determined that miR-21 is upregulated and exhibits oncogenic
activity in different types of cancer (7,8). For
example, the expression of miR-21 was increased in prostate cancer
(9). miR-21 promotes the
proliferation of oral cancer cells via tumor necrosis factor-α
(10). Additionally, the
expression of miR-21 markedly decreases following trastuzumab
therapy in patients with EGFR2-positive metastatic breast cancer
(11). The ratio of miR-21/24 is
significantly correlated with several important prognostic factors
in colorectal cancer, including tumor size, TNM stage, lymph
metastasis and histologic differentiation (12). Furthermore, the ratio of miR-21/24
has been determined to be a significant survival risk factor for
patients with colorectal cancer. The current study revealed that
miR-21 overexpression was associated with recurrence and
invasiveness in BC (13). However,
the underlying mechanism of miR-21 in BC is remains largely
unknown. The current study aimed to determine the role of miR-21 in
the regulation of cellular phenotypes and its molecular mechanism
in BC.
Materials and methods
Cell line and culture
The human bladder cancer cell line, T24, was
obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were cultured in DMEM (Merck
KGaA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and incubated in 5% CO2 at 37°C.
Transfection
To inhibit or overexpress miR-21 in BC cells,
pGC-U6/Neo/GFP/miR-21 mimic, pGC-U6/Neo/GFP/miR-21 inhibitor and
associated negative control (miR-21-NC) were obtained from Shanghai
Gene Pharma Co., Ltd. A total of 3 µg miR-21 mimic or
inhibitor was transfected into BC cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
sequences of the miR-21 mimic and miR-21 inhibitor were as follows:
5′-TAA ACG GGC CCT CTA GAC TCG AGT TAT CAA ATC CTG CCT GAC TG-3′
and 5′-GAT CCT CAA CAT CAG TCT GAT AAG CTA TTT TT-3′, respectively.
After 48 h, cells were obtained for subsequent experiments. The
following stably transfected T24 groups were established: miR-21
mimics group (mimics group), miR-21 inhibitor group (inhibitor
group), negative control (NC) group and blank control (con) group
without transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from T24 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). and reverse transcribed at 37°C for 60 min using the
miDETECT A Track™ miRNA qRT-PCR Starter kit (Guangzhou RiboBio Co.,
Ltd.). Subsequently, qPCR was performed using a SYBR Green kit
(Takara Bio, Inc.) and an ABI PRISM 7500 Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 94°C for 3 min; followed
by 40 cycles of 94°C for 30 sec and 72°C for 45 sec. The following
primers were used: miR-21 forward, 5′-TAG CTT ATC AGA CTG ATG TTG
A-3′ and reverse, 5′-TGG TGT CGT GGA GTC G-3′; U6 forward, 5′-GCT
TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT
TTG CGT GTC AT-3′. RNA quantification was calculated using the
2-ΔΔCq method (14).
Western blot analysis
Cells were treated with RIPA lysis buffer (Beyotime
Institute of Biotechnology) and centrifuged at 4°C for 10 min at
9,063 × g to remove cell debris. After collecting the supernatant,
a bicinchoninic acid protein assay (Beyotime Institute of
Biotechnology) was used to detect protein concentration. Equal
quantities of protein (20 µg) were then added each lane for
electrophoresis with 10% SDS-PAGE and subsequently transferred onto
PVDF membranes (EMD Millipore). The membranes were blocked at 37°C
for 3 h in a blocking solution consisting of Tris-buffered saline
containing 0.1% Tween-20 and 5% bovine serum albumin
(Sigma-Aldrich; Merck KGaA). Subsequently, the following primary
antibodies were added and incubated overnight at 4°C: Phosphatase
and tensin homolog (PTEN; 1:1,000; cat. no. 20399; Promab
Biotechnologies, Inc.), beclin 1 (1:1,000; cat. no. 30182; Promab
Biotechnologies), microtubule-associated protein l light chain 3B
(LC3-II; 1:1,000; cat. no. 30363; Promab Biotechnologies), cyclin
D1 (1:1,000; cat. no. WL01435a; Wanleibio, Co., Ltd.), caspase-3
(1:1,000; cat. no. WL04004; Wanleibio, Co., Ltd.), GAPDH (1:1,000;
cat. no. 20035; Promab Biotechnologies), E-cadherin (1:1,000; cat.
no. WL01482; Wanleibio, Co., Ltd.), Matrix metalloprotein-9,
(MMP-9; 1:1,000; cat. no. WL03096; Wanleibio, Co., Ltd.) and
vimentin (1:1,000; cat. no. WL01960; Wanleibio, Co., Ltd.). Samples
were then incubated with horseradish peroxidase-conjugated
secondary antibodies goat anti-mouse immunoglobulin IgG (1:2,000;
cat. no. ab6789; Abcam) at room temperature for 2 h. The enhanced
chemiluminescence detection reagent (Thermo Fisher Scientific,
Inc.) was used to visualize protein expression. Chemiluminescence
was analyzed using ChemiDoc XRS system with Image Lab Software
version 6.0 (Bio-Rad Laboratories, Inc.). GAPDH was used as an
internal control.
MTT assay
T24 cells were cultured in 96-well plates at a
volume of 100 µl per well. After 48 h incubation at 37°C, 50
µl MTT reagent (Sigma-Aldrich; Merck KGaA) was added, and
cells were cultured at 37°C for 4 h. After removing the
supernatant, 150 µl of DMSO was added to detect absorbance
at a wavelength of 570 nm.
Wound healing assay
Cells were seeded into 6-well plates at a
concentration of 5×105/ml and incubated at 37°C for 24
h. After scratching an initial wound with a 10 µl pipette
tip in the cell monolayer, the distances of the wound area covered
by cells was measured under an inverted microscope (CKX53; Olympus
Corporation; magnification, ×200) at 0, 24, and 48 h to detect
migration.
Invasion assay
A total of 5×104 transfected cells
suspended in 200 µl serum-free DMEM were added to the upper
chamber of a BioCoat™ Matrigel Invasion Chamber (Corning, Inc.).
The lower chamber was filled with 1 ml of DMEM supplied with 20%
FBS. After culture with 5% CO2 at 37°C for 24 h,
transwell chambers were inverted and stained with haematoxylin at
37°C for 10 min. Excess stain was removed using PBS. The number of
invading cells was determined according to five random fields of
view under a light microscope (magnification, ×300).
Flow cytometry
Annexin V/PI staining was conducted using the
Annexin V-FITC Apoptosis Detection kit (BD Pharmingen; BD
Biosciences) according to the manufacturer's protocol. Stained
cells were analyzed with a flow cytometer using BD FACStation™ 6.1
software (FACSCalibur; Becton, Dikinson and Company) within 1
h.
Cell cycle analysis was conducted using a Cell Cycle
Analysis kit (Beyotime Institute of Biotechnology) according to the
manufactures' protocol. Cells were fixed with 5 ml 70% ethanol at
4°C overnight, centrifuged at 1,208 × g at 4°C for 5 min and washed
with PBS. Subsequently, 50 µl propidium iodide was added.
Cells were resuspended and incubated in the dark at room
temperature for 30 min. The cell cycle was determined via flow
cytometry (FACSCalibur; Becton, Dikinson and Company).
Statistical analysis
Statistical analysis was performed using SPSS 13.0
statistical software (SPSS, Inc.). Data are presented as the mean ±
standard deviation from at least three independent experiments.
One-way ANOVA followed by Tukey's post hoc test was used to
determine significance. P<0.05 was considered to indicate a
statistically significant difference.
Results
Stably transfected T24 groups were
established
To investigate the effects of miR-21 on bladder
cancer T24 cells, miR-21 mimics and inhibitors were obtained and
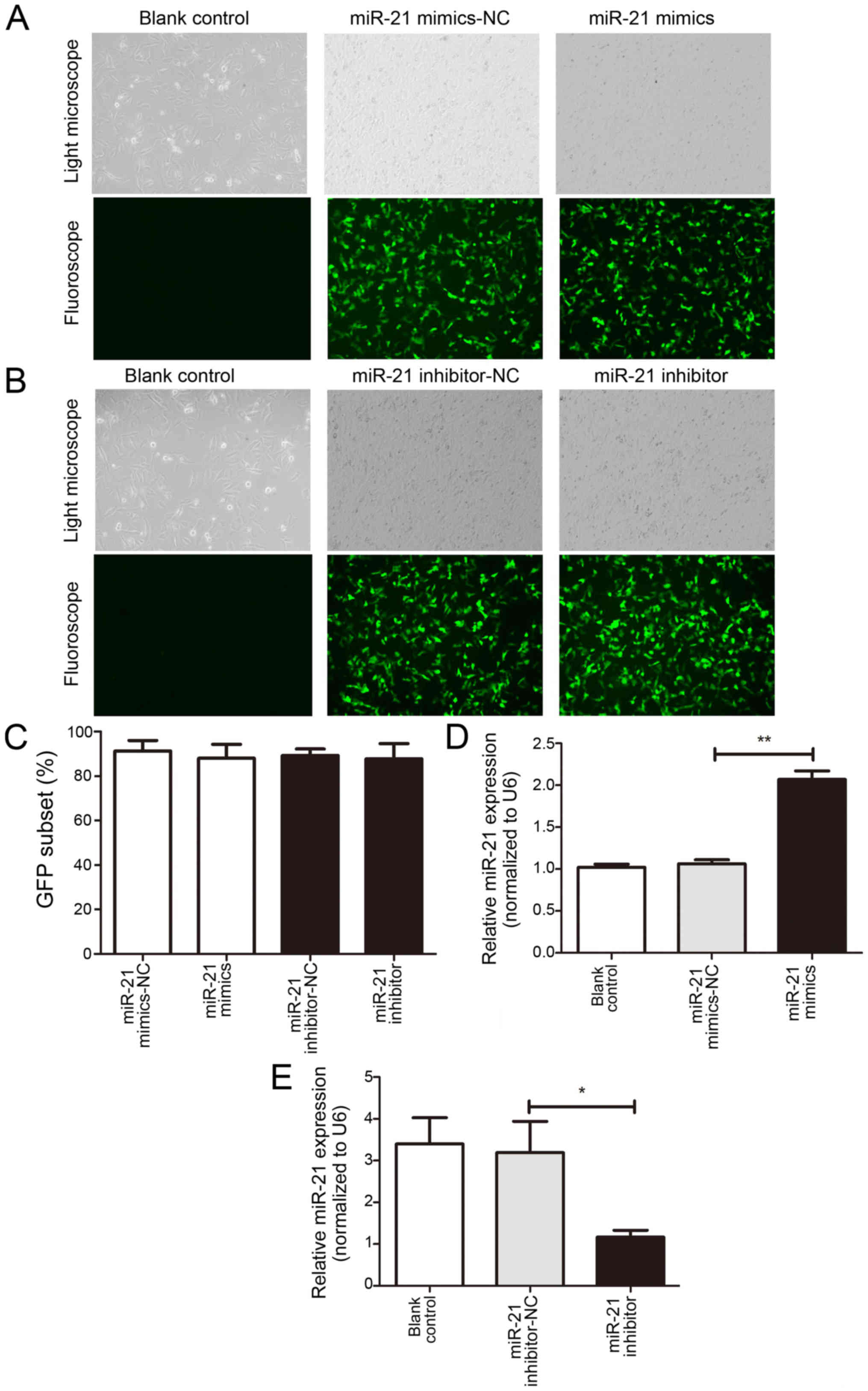
transfected into T24 cells. Transfection efficiency was determined
by analyzing the percentage of fluorescent cells (Fig. 1A and B). The percentage of
fluorescence was >80% in the NC, mimic and inhibitor groups.
Effect of miR-21 mimic and miR-21
inhibitor on the expression of miR-21
The results of RT-qPCR demonstrated a significant
increase in miR-21 expression in the mimic group and a significant
inhibition in the inhibitor group when compared with the NC group
(Fig. 1C and D). No significant
differences were revealed between the NC and Con groups.
miR-21 promotes the proliferation,
migration and invasion of T24 cells
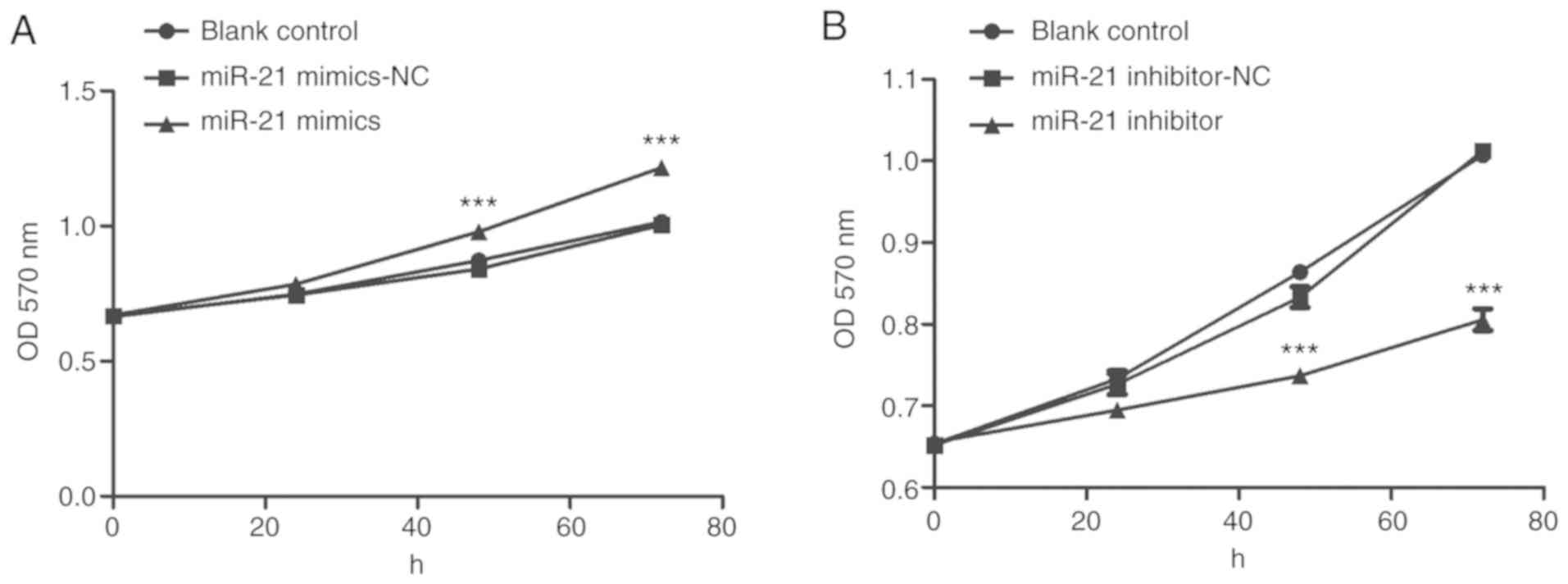
The results of the MTT assay revealed that the
proliferation of cells in the mimic group was significantly
increased compared with the Con and NC groups in a time-dependent
manner (Fig. 2A). In contrast, the
proliferation of cells in the inhibitor group was significantly
inhibited (Fig. 2B). Compared with
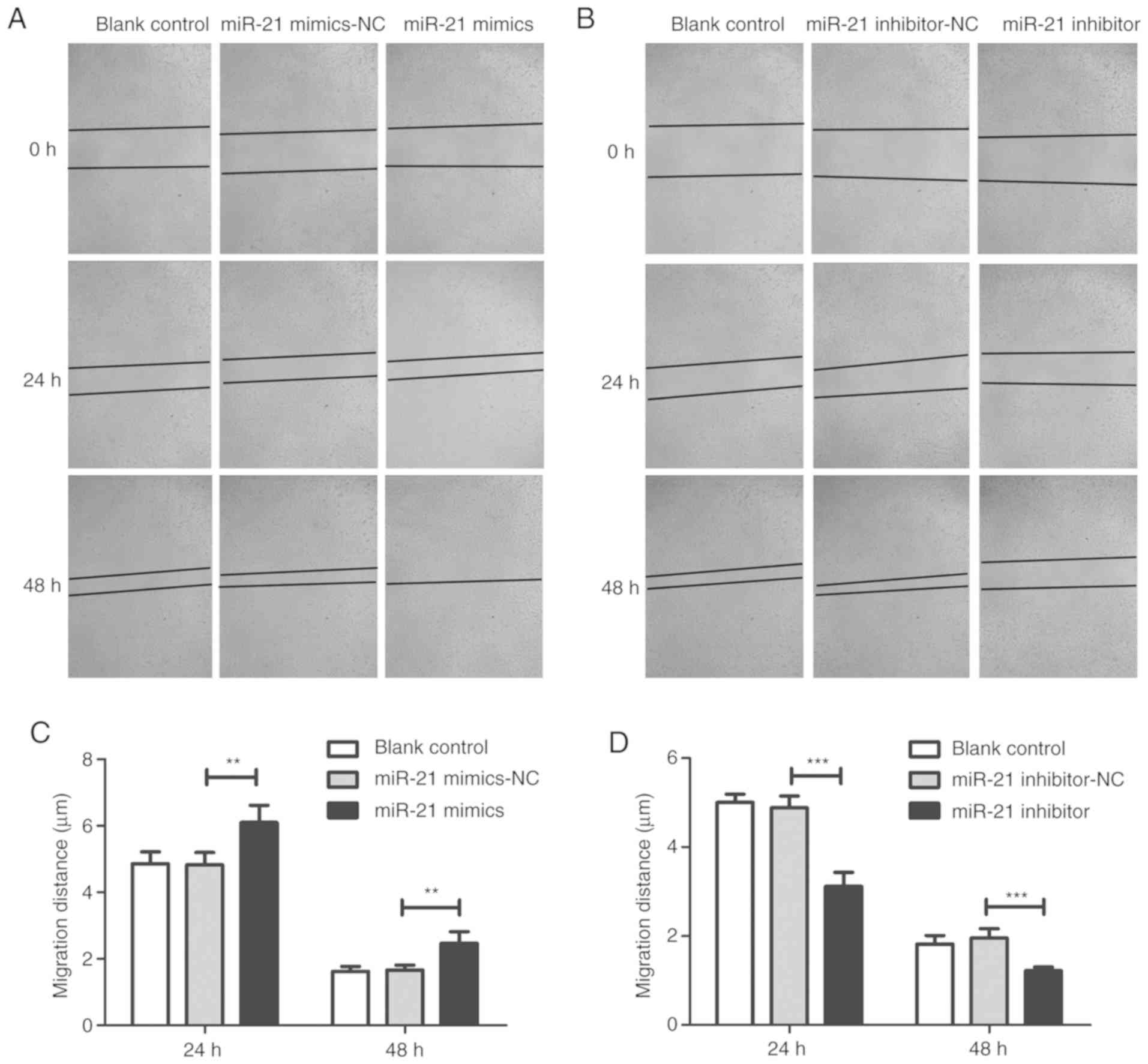
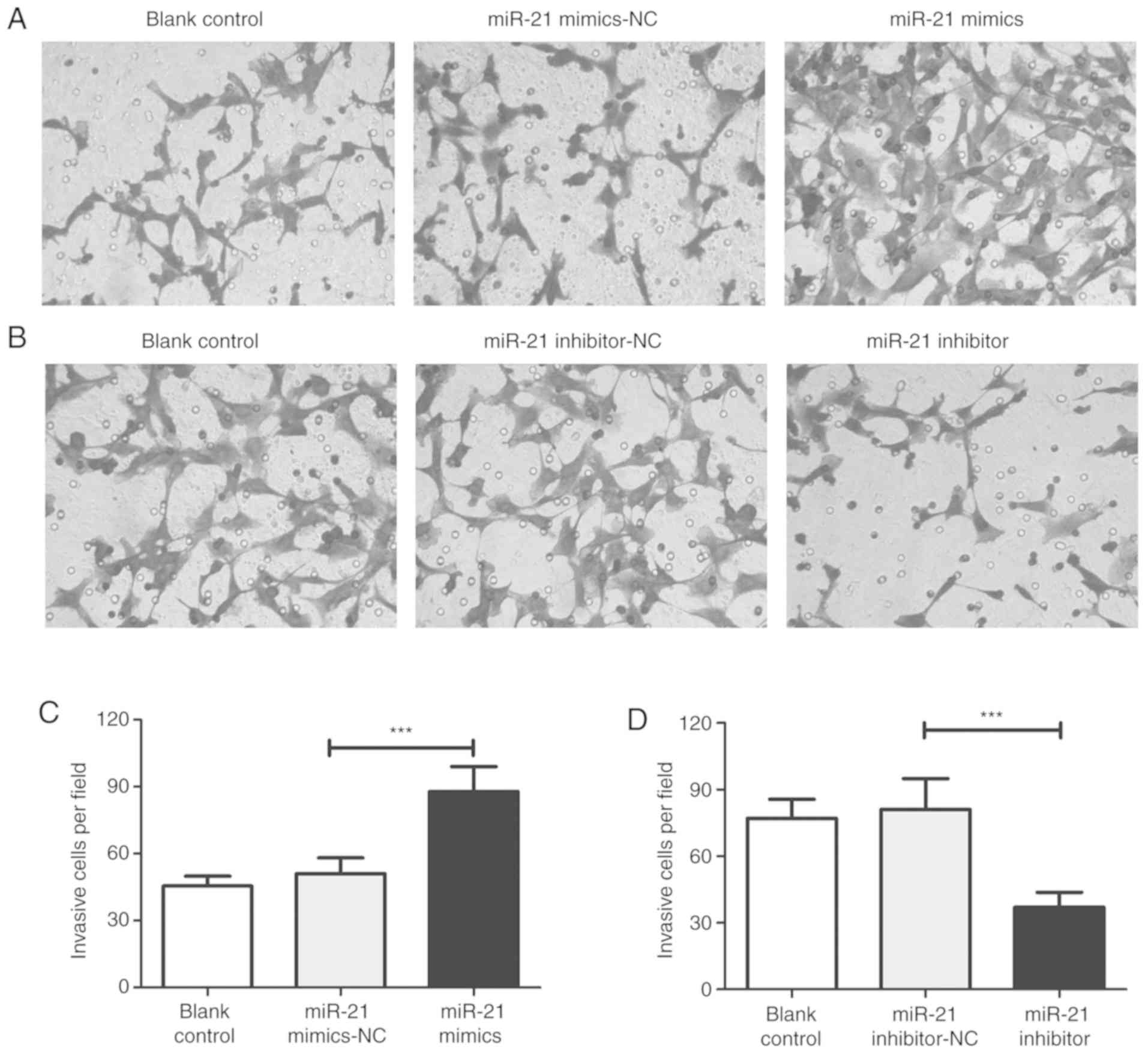
the NC group, wound healing and invasion assays demonstrated that
miR-21 mimic transfection increased the migration and invasiveness
of T24 cells (Figs. 3 and 4). However, the migration and invasion of
T24 cells were significantly inhibited in the inhibitor group.
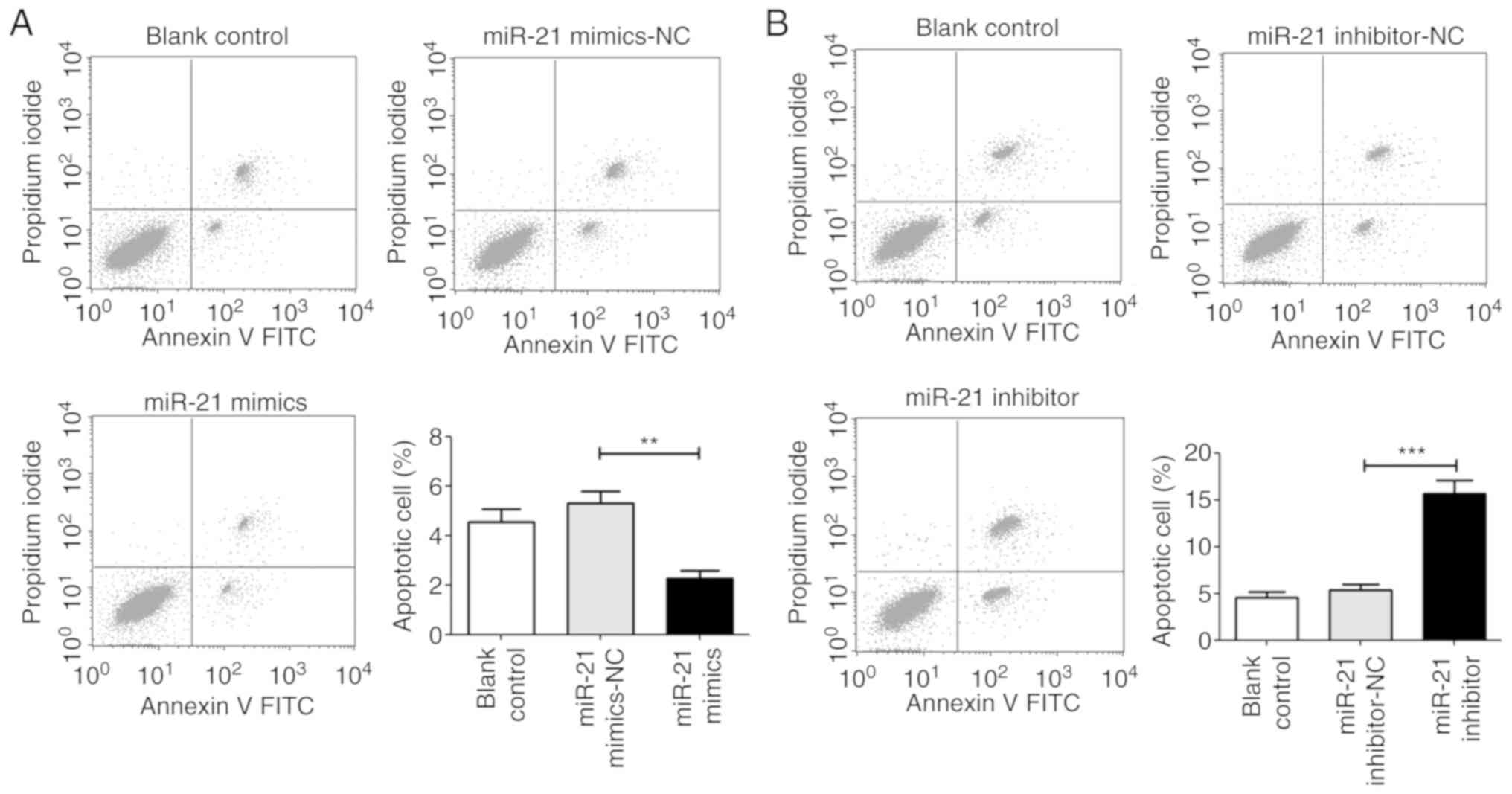
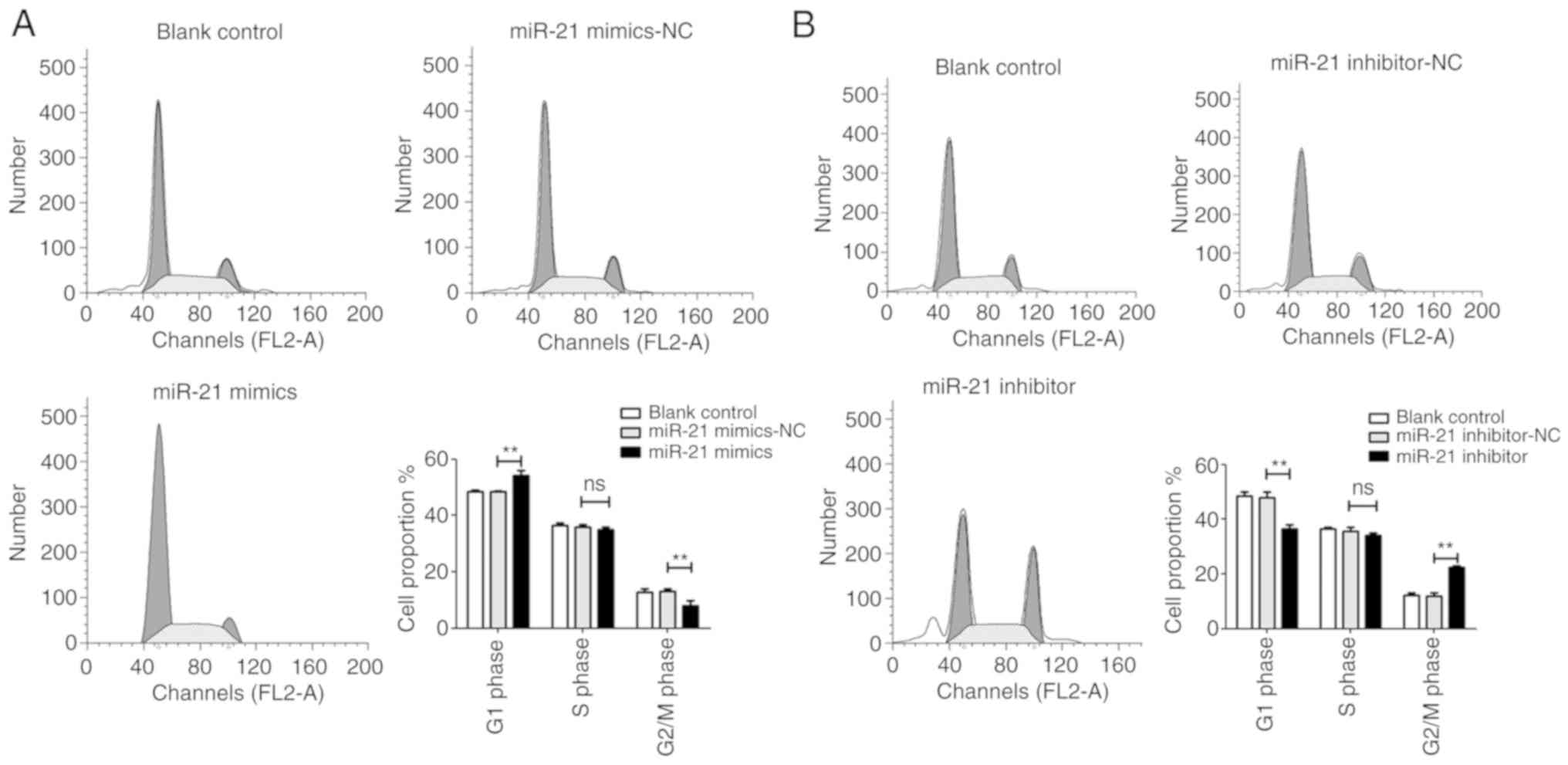
miR-21 inhibits apoptosis and arrests the
G1 phase of T24 cells
Flow cytometry was performed to evaluate the
influence of miR-21 on cell apoptosis and cell cycle distribution.
The results revealed that when compared with the NC group, cells
transfected with the miR-21 mimic decreased cell apoptosis and
arrested T24 cells at the G1 phase. However, the miR-21 inhibitor
significantly induced cell apoptosis and decreased the proportion
of cells in the G1 phase (Figs. 5
and 6).
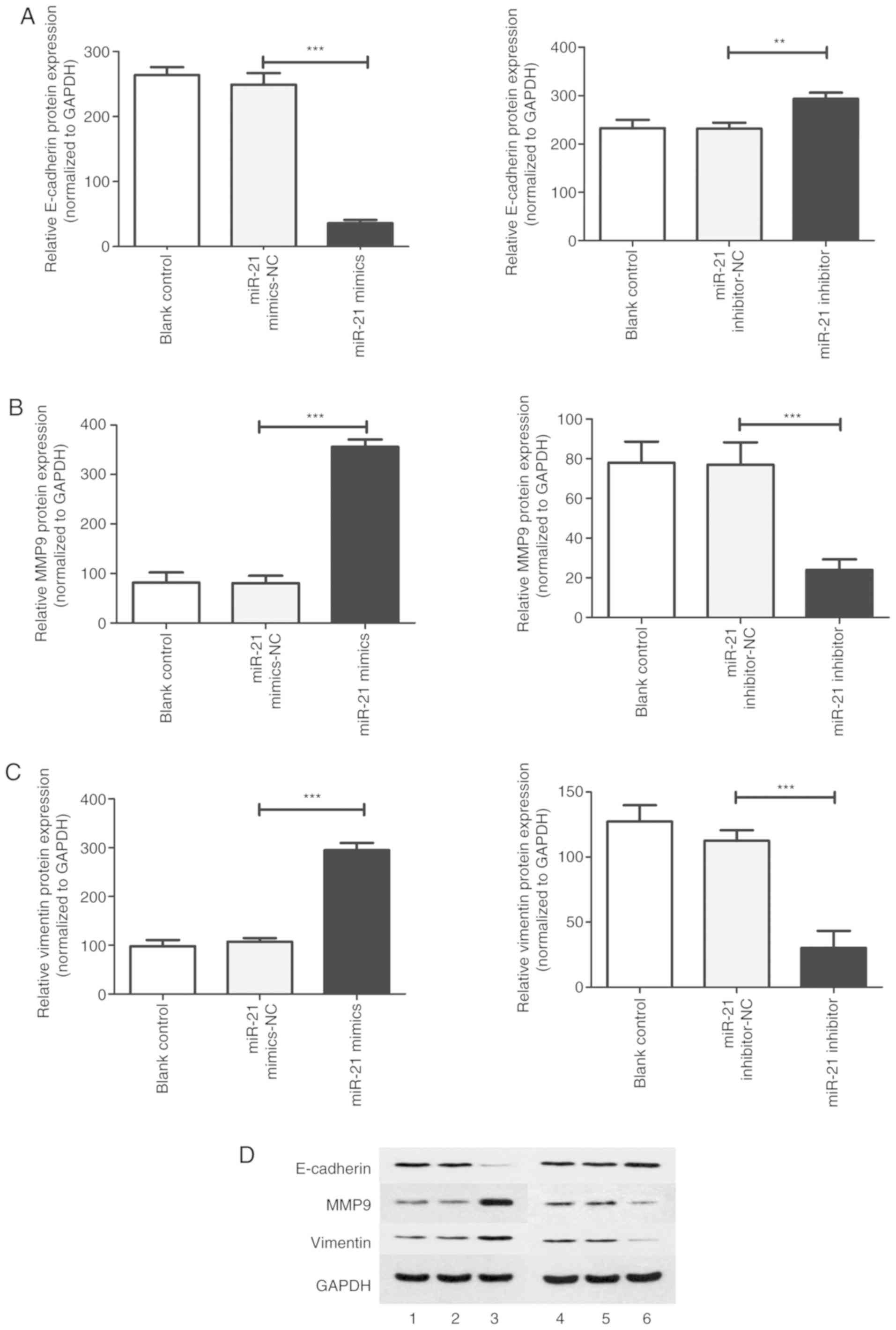
Effect of miR-21 expression on the
proteins associated with the proliferation, migration, invasion,
epithelial mesenchymal transition and autophagy of T24 cells
To further elucidate the possible mechanism of
miR-21-mediated cell processes, RT-qPCR and western blotting were
performed to detected the mRNA and protein expression of PTEN,
beclin 1, LC3-II, cyclin D1, caspase-3, E-cadherin, matrix
metallopeptidase 9 (MMP-9) and vimentin. In the miR-21 mimic group,
compared with the NC group, the results revealed that the mRNA and
protein expression of PTEN, beclin 1, LC3-II, caspase-3 and
E-cadherin were significantly decreased. In addition, the mRNA and
protein expression of cyclin D1, MMP-9 and vimentin were
significantly increased (Figs.
7-10). However, increased
mRNA and protein expression of PTEN, beclin 1, LC3-II, caspase-3
and E-cadherin and decreased mRNA and protein expression of cyclin
D1, MMP-9 and vimentin were induced after treatment with the miR-21
inhibitor.
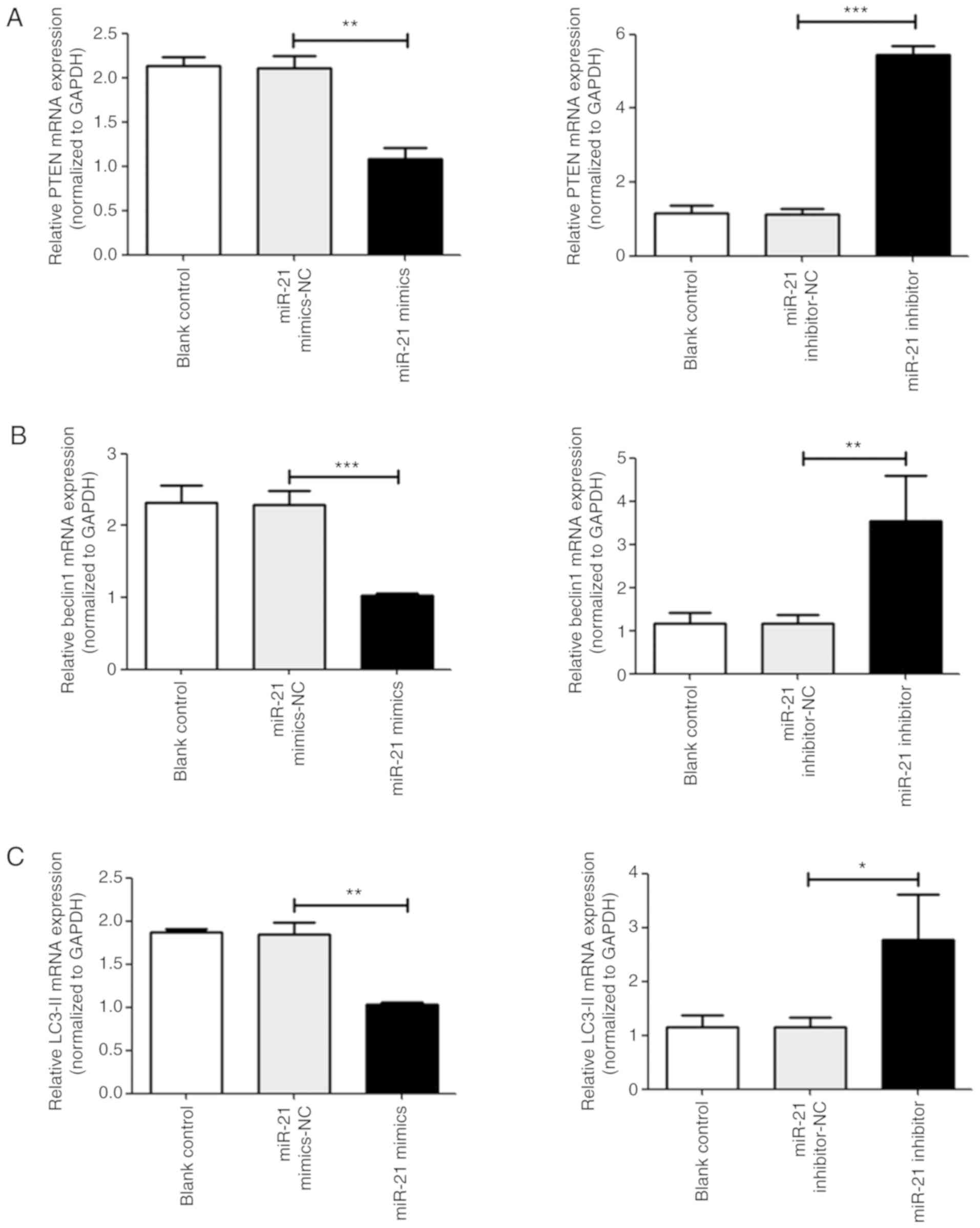 | Figure 7Analysis of various mRNAs associated
with T24 cell proliferation, apoptosis and autophagy. The mRNA
expression of (A) PTEN, (B) beclin-1, and (C) LC3-II in T24 cells
was measured via reverse transcription-quantitative PCR.
*P<0.05, **P<0.01 and
***P<0.001 vs. NC groups. Analysis of various mRNAs
associated with T24 cell proliferation, apoptosis and autophagy.
The mRNA expression of (D) cyclin D1 and (E) caspase-3 in T24 cells
was measured via reverse transcription-quantitative PCR.
*P<0.05, **P<0.01 and
***P<0.001 vs. NC groups. PTEN, phosphatase and
tensin homolog; LC3-II, microtubule-associated protein l light
chain 3B; NC, negative control; miR, microRNA. |
Discussion
MiRNAs widely regulate gene expression and are
associated with the malignant development and survival of various
types of cancer (15,16). A previous study reported that
miR-21 was increased in BC tissue and was associated with high
rates of recurrence (17).
Although the oncogenic role of miR-21 in BC has been previously
indicated, the functional mechanism of miR-21 has not yet been
fully elucidated and therefore deserves further study.
The present study demonstrated that upregulated
miR-21 significantly increased the proliferation, migration and
invasion of T24 cells, and arrested the cell cycle at the G1 phase.
This is congruent with results obtained with other cancer cell
lines (7,8). To investigate the underlying
mechanism of miR-21, the expression of PTEN, beclin 1, LC3-II,
cyclin D1, caspase-3, E-cadherin, MMP-9 and vimentin were examined.
The results revealed that the expression of PTEN, beclin 1, LC3-II,
caspase-3 and E-cadherin was decreased, while the expression of
cyclin D1, MMP-9 and vimentin was increased in the mimic group.
Autophagy is a complex intracellular process that
modulates several cellular functions (14). Abnormal levels of autophagy have
been continuously associated with human inflammatory disorders and
cancer (18,19). Although autophagy may suppress
tumors in certain cases, it may also serve an onco-genic role in
cancer via microenvironmental stress responses and metabolism
control (20-22). Thus, autophagy has a dual
effect.
Various signaling pathways, including the PTEN/
AKT/mTOR pathway, are associated with autophagy regulation
(23,24). The current study revealed that
autophagy-associated proteins, beclin 1 and LC3-II, were
upregulated following miR-21 mimic transfection, while PTEN was
downregulated. To some extent, this result may indicate that miR-21
inhibits autophagy in T24 cells via a PTEN-mediated pathway,
resulting in changes to the viability and proliferation of T24
cells.
The results of MTT assay and flow cytometric
analysis revealed that the miR-21 mimic promoted cell proliferation
in a time-dependent manner, which was associated with suppressed
apoptosis. To address this mechanism, the expression of cyclin D1
and caspase-3 were detected. Cyclin D1 is a major regulator of cell
cycle progression that is overexpressed in carcinomas (25). Increased cyclin D1 expression can
accelerate cell cycle progression and DNA synthesis (26). The present study determined that
cyclin D1 expression was increased following transfection with
miR-21 mimics. It was further demonstrated that upregulation of
miR-21 promoted G0/G1 phase cell cycle arrest by affecting cyclin
D1 levels. Apoptosis deregulation serves an important role in
cancer development (27). Caspases
are key mediators of this process (28). In the current study, the expression
of caspase-3 was significantly increased following miR-21 mimic
transfection. The results also demonstrated that the G0/G1 cell
cycle arrest and apoptosis induced by miR-21 may provide favorable
conditions for the development of BC cells.
Metastasis is an important characteristic of cancer.
MMP is a family of neutral proteinases that allow cancer cells to
migrate and invade (29). The
current data revealed a significant increase in MMP-9 and the
increased invasion of cells in the miR-21 mimic group. These
results confirmed that miR-21 served a promoting role in the
invasiveness of T24 cells.
Epithelial to mesenchymal transition (EMT) may
provide favorable conditions for increasing cell mobility (30,31).
EMT is characterized by the suppression of epithelial-associated
genes, the upregulation of mesenchymal-associated genes, and the
deregulation of various transcription factors (32,33).
The results of the current study demonstrated that miR-21 increased
the migration and invasion of T24 cells. As previously described,
EMT is closely associated with cell invasion. Subsequently, the
results of western blotting revealed that miR-21 decreased the
expression of E-cadherin and increased the expression of vimentin.
It was therefore hypothesized that miR-21 may decrease cell
migration and invasion by inhibiting EMT.
A previous study reported that autophagy could
inhibit EMT and metastasis in glioblastoma cells (34). However, autophagy also inhibits the
migration and invasion of cancer cells by suppressing certain
transcription factors such as SNAIL and SLUG (35). In the present study, autophagy was
suppressed while EMT was promoted by miR-21. Whether there is a
link between the autophagy-mediated regulation of EMT and bladder
tumourigenesis is unclear and as such requires further study.
Collectively, the present data suggested that miR-21
inhibited autophagy and promoted the malignant development of the
BC cell line, T24, in vitro. These results indicated that
miR-21 may serve as a potential target to inhibit the development
of BC. To the best of our knowledge, the current study is the first
comprehensively assess the role of miR-21 in the cancer development
of the BC T24 cell line, as well as its functional mechanism.
However, the lack of in vivo experiments is the main
limitation of the present study. Furthermore, the use of only one
cell line may also limit results. Further studies are therefore
required to verify these effects and to clarify the mechanism by
which miR-21 is involved in the development of BC.
Funding
The current study was supported by the National
Natural Science Foundation of China (grant no. 81602241) and the
Scientific Project of Health Commission of Hunan Province (grant
no. B2019127).
Availability of data and materials
The datasets used and/or analyzed are available from
the corresponding author on reasonable request.
Authors' contributions
ZH designed the experiments and drafted the
manuscript. HZ and ZS performed the experiments and collected the
data. FK and CY analyzed and interpreted data. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of University of South
China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
Acknowledgments
Not applicable.
References
|
1
|
Fankhauser CD and Mostafid H: Prevention
of bladder cancer incidence and recurrence: Nutrition and
lifestyle. Curr Opin Urol. 28:88–92. 2018. View Article : Google Scholar
|
|
2
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar
|
|
3
|
Liu Z, Xie D and Zhang H: Long noncoding
RNA neuroblastoma-associated transcript 1 gene inhibits malignant
cellular phenotypes of bladder cancer through miR-21/SOCS6 axis.
Cell Death Dis. 9:10422018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heishima K, Meuten T, Yoshida K, Mori T
and Thamm DH: Prognostic significance of circulating microRNA-214
and-126 in dogs with appendicular osteosarcoma receiving amputation
and chemotherapy. BMC Vet Res. 15:392019. View Article : Google Scholar
|
|
5
|
Xue Y, Ge Y, Kang M, Wu C, Wang Y, Rong L
and Fang Y: Selection of three miRNA signatures with prognostic
value in non-M3 acute myeloid leukemia. BMC Cancer. 19:1092019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ribas J, Ni X, Castanares M, Liu MM, Esopi
D, Yegnasubramanian S, Rodriguez R, Mendell JT and Lupold SE: A
novel source for miR-21 expression through the alternative
polyadenylation of VMP1 gene transcripts. Nucleic Acids Res.
40:6821–6833. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ni K, Wang D, Xu H, Mei F, Wu C, Liu Z and
Zhou B: miR-21 promotes non-small cell lung cancer cells growth by
regulating fatty acid metabolism. Cancer Cell Int. 19:2192019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chai C, Song LJ, Han SY, Li XQ and Li M:
MicroRNA-21 promotes glioma cell proliferation and inhibits
senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT
signaling pathway. CNS Neurosci Ther. 24:369–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Folini M, Gandellini P, Longoni N, Profumo
V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S,
Salvioni R, et al: miR-21: An oncomir on strike in prostate cancer.
Mol Cancer. 9:122010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qiu YF, Wang MX, Meng LN, Zhang R and Wang
W: MiR-21 regulates proliferation and apoptosis of oral cancer
cells through TNF-α. Eur Rev Med Pharmacol Sci. 22:7735–7741.
2018.PubMed/NCBI
|
|
11
|
Badr M, Said H, Louka ML, Elghazaly HA,
Gaballah A and Atef Abd El Mageed M: MicroRNA-21 as a predictor and
prognostic factor for trastuzumab therapy in human epidermal growth
factor receptor 2-positive metastatic breast cancer. J Cell
Biochem. 120:3459–3466. 2019. View Article : Google Scholar
|
|
12
|
Hao JP and Ma A: The ratio of
miR-21/miR-24 as a promising diagnostic and poor prognosis
biomarker in colorectal cancer. Eur Rev Med Pharmacol Sci.
22:8649–8656. 2018.PubMed/NCBI
|
|
13
|
Zhang HH, Qi F, Cao YH, Zu XB and Chen MF:
Expression and clinical significance of microRNA-21, maspin and
vascular endothelial growth factor-C in bladder cancer. Oncol Lett.
10:2610–2616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
He Y, Liu H, Jiang L, Rui B, Mei J and
Xiao H: miR-26 induces apoptosis and inhibits autophagy in
non-small cell lung cancer cells by suppressing TGF-β1-JNK
signaling pathway. Front Pharmacol. 9:15092019. View Article : Google Scholar
|
|
16
|
Iswariya GT, Paital B, Padma PR and
Nirmaladevi R: microRNAs: Epigenetic players in cancer and aging.
Front Biosci (Schol Ed). 11:29–55. 2019. View Article : Google Scholar
|
|
17
|
Kroemer G, Mariño G and Levine B:
Autophagy and the integrated stress response. Mol Cell. 40:280–293.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Menzies FM, Fleming A, Caricasole A, Bento
CF, Andrews SP, Ashkenazi A, Füllgrabe J, Jackson A, Jimenez
Sanchez M, Karabiyik C, et al: Autophagy and neurodegeneration:
Pathogenic mechanisms and therapeutic opportunities. Neuron.
93:1015–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Monkkonen T and Debnath J: Inflammatory
signaling cascades and autophagy in cancer. Autophagy. 14:190–198.
2018. View Article : Google Scholar :
|
|
20
|
Kung CP, Budina A, Balaburski G,
Bergenstock MK and Murphy M: Autophagy in tumor suppression and
cancer therapy. Crit Rev Eukaryot Gene Expr. 21:71–100. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amaravadi R, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia L, Huang S, Yin X, Zan Y, Guo Y and
Han L: Quercetin suppresses the mobility of breast cancer by
suppressing glycolysis through Akt-mTOR pathway mediated autophagy
induction. Life Sci. 208:123–130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao GS, Gao ZR, Zhang Q, Tang XF, Lv YF,
Zhang ZS, Zhang Y, Tan QL, Peng DB, Jiang DM and Guo QN: TSSC3
promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR
pathway to suppress tumorigenesis and metastasis in osteosarcoma,
and predicts a favorable prognosis. J Exp Clin Cancer Res.
37:1882018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qie S and Diehl JA: Cyclin D1, cancer
progression, and opportunities in cancer treatment. J Mol Med
(Berl). 94:1313–1326. 2016. View Article : Google Scholar
|
|
26
|
Oh SJ, Cho H, Kim S, Noh KH, Song KH, Lee
HJ, Woo SR, Kim S, Choi CH, Chung JY, et al: Targeting cyclin
D-CDK4/6 sensitizes immune-refractory cancer by blocking the
SCP3-NANOG axis. Cancer Res. 78:2638–2653. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang R, Wang J, Dong T, Shen J, Gao X and
Zhou J: Naringenin has a chemoprotective effect in MDA-MB-231
breast cancer cells via inhibition of caspase-3 and -9 activities.
Oncol Lett. 17:1217–1222. 2019.PubMed/NCBI
|
|
28
|
Shalini S, Dorstyn L, Dawar S and Kumar S:
Old, new and emerging functions of caspases. Cell Death Differ.
22:526–539. 2015. View Article : Google Scholar :
|
|
29
|
Guo F, Liu J, Han X, Zhang X, Lin T, Wang
Y, Bai J and Han J: FBXO22 suppresses metastasis in human renal
cell carcinoma via inhibiting MMP-9-mediated migration and invasion
and VEGF-mediated angiogenesis. Int J Biol Sci. 15:647–656. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng X, Carstens JL, Kim J, Scheible M,
Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R:
Epithelial-to-mesenchymal transition is dispensable for metastasis
but induces chemoresistance in pancreatic cancer. Nature.
527:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jie XX, Zhang XY and Xu CJ:
Epithelial-to-mesenchymal transition, circulating tumor cells and
cancer metastasis: Mechanisms and clinical applications.
Oncotarget. 8:81558–81571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Catalano M, D'Alessandro G, Lepore F,
Corazzari M, Caldarola S, Valacca C, Faienza F, Esposito V,
Limatola C, Cecconi F and Di Bartolomeo S: Autophagy induction
impairs migration and invasion by reversing EMT in glioblastoma
cells. Mol Oncol. 9:1612–1625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gugnoni M, Sancisi V, Manzotti G, Gandolfi
G and Ciarrocchi A: Autophagy and epithelial-mesenchymal
transition: An intricate interplay in cancer. Cell Death Dis.
7:e25202016. View Article : Google Scholar : PubMed/NCBI
|