Introduction
Salivary adenoid cystic carcinoma (SACC) is a common
malignant tumour of the salivary glands (1). Compared with other malignant tumours,
SACC has unique biological features, such as slow and continuous
proliferation, frequent local recurrence and high incidence rates
of blood, nerve and advanced lung metastasis, but a rare occurrence
of lymphatic metastasis (2,3).
Moreover, distal lung metastasis is an important factor affecting
the overall survival rate of SACC (4). Current research on SACC primarily
focuses on the investigation of primary lesions, but the
pathogenesis of lung metastasis in SACC remains unknown (5). Thus, there is an urgent need to
identify the key molecular mechanisms involved in the lung
metastasis of SACC to improve clinical outcomes.
MicroRNAs (miRNAs/miRs) belong to a class of small
non-coding RNAs that regulate gene expression
post-transcriptionally and modulate cellular activities (6), such as apoptosis (7), tumour angiogenesis (8), cell invasion (9) and differentiation (10). miRNAs primarily function by
inhibiting translation or/and cleaving the targeted mRNAs by
pairing with their 3' untranslated regions (3'UTRs) (11). Previous studies have reported that
miRNAs can affect the progression of SACC, such as the oncogenes
miR-21 (12,13) and miR-455-3p (14), and the tumour suppressors miR-320a
(15) and miR-125a-5p (16). Therefore, investigations of the
function of miRNAs and their targets may provide novel molecular
markers for the diagnosis and treatment of SACC. miR-103a-3p has
been examined in numerous studies, but its function remains
controversial. For example, miR-103a-3p is considered to be an
oncogene in colorectal cancer (17) and gastric cancer (18), but a tumour suppressor gene in
glioma cells (19) and non-small
cell lung cancer (20).
Furthermore, to the best of our knowledge, the roles of miR-103a-3p
in the progression of SACC and its underlying function in
regulating tumour metastasis in SACC have not been previously
reported.
Tumour protein D52 (TPD52), a member of the
TPD52-like protein family that is conserved among vertebrates,
contains a small coiled-coil motif bearing small hydrophilic
polypeptides and is mapped to chromosome 8q21 (21,22).
TPD52 exerts opposing roles in different tumours. High TPD52
expression is detected in prostate (23) and breast cancer (24), whereas low expression is observed
in liposarcoma, clear cell renal cell cancer and lung cancer
(25,26). TPD52 has been shown to promote cell
survival, proliferation, migration and invasion (27), however, previous studies have also
revealed that TPD52 plays a suppressor role in the progression of
tumours (28,29). In addition, TPD52 can be regulated
by miR-139-5p (30) and miR-15a-3p
(31) to promote tumour
progression. However, whether TPD52 has the same
metastasis-promoting effect in SACC remains unknown, and the
relationship between TPD52 and miR-103a-3p is yet to be
elucidated.
The present study aimed to determine the expression
of miR-103a-3p in SACC tissues and assess its effect on SACC
progression. In addition, whether TPD52 is a direct binding target
of miR-103a-3p was examined.
Materials and methods
Tissue sample and cell lines
The ten human SACC tissues and ten paired healthy
submandibular gland (SMG) tissues (age, 42-68 years;
female:male=6:4) used for the tissue micro-array were collected
from patients with ACC at the Peking University School and Hospital
of Stomatology (Beijing, China) between 2015.08 and 2016.04. The
remining 52 human SACC (age, 31-75 years; female:male=26:26) and 38
separate healthy SMG tissues used to analyse miR-103a-3p and TPD52
expression were collected between 2010.07 and 2018.07 at the Peking
University School and Hospital of Stomatology. Patients had not
undergone chemotherapy or radiation therapy, and the study was
approved and followed the rules of the Ethics Committee of Peking
University School and Hospital of Stomatology (permit no.
PKUSSIRB-201522040). According to the relative gene expression of
miR-103a-3p in 52 patients with SACC examined by reverse
transcription-quantitative PCR (RT-qPCR), 52 patients were
classified into the high or low miR-103a-3p group depending on the
median miR-103a-3p relative gene expression. When the miR-103a-3p
expression level of SACC tissue was higher than the median
miR-103a-3p expression, the SACC tissue was classified into the
high miR-103a-3p group, otherwise the SACC tissue was classified
into the low miR-103a-3p group.
The SACC-83 cell line originated from ACC tissue
from a patient with SACC in November 1983 (32). The SACC-LM cell line had enhanced
lung metastatic features and was isolated following injection of
SACC-83 cells into the tail vein of immunodeficient mice (33,34).
The SACC-83 and SACC-LM cell lines were collected by the author SLL
and kept at Peking University School and Hospital of Stomatology.
SACC-83 and SACC-LM cells were cultivated in RPMI-1640 medium
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.). The primary epithelial cells of the SMG (SMG-E
or pSMG) cells were derived from the sublingual gland of a
4-year-old boy in November 2016, with cell cultivation performed as
previously described (35).
Exosomes isolation from cells supernatants was
performed using ultracentrifugation and sucrose cushion. Cell
supernatants were subjected to consecutive centrifugation at 300 ×
g for 10 min, 2,000 × g for 30 min and 10,000 × g for 30 min to
remove cellular debris and large vesicles at 4°C. The supernatants
were then passed through a Centrifugal Filter (100K; EMD Millipore)
to concentrate the solution, followed by a 30% sucrose/deuterium
oxide (D2O) cushion. After gradient centrifugation at 100,000 × g
for 70 min using an Optima L-90K Ultracentrifuge (Beckman Coulter,
Inc.) at 4°C, the exosome-enriched sucrose/D2O was then
re-suspended in PBS and the retained exosomes were stored at -80°C.
All the isolation procedures were performed at room
temperature.
The microarray analysis of tissues (ten human SACC
tissues vs. ten paired SMG tissues) and cell exosomes (SACC-83
cells vs. SACC-LM cells) was performed by Shanghai Biotechnology
Corporation.
RNA isolation and RT-qPCR analysis
Total RNA was extracted from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Reverse
transcription was performed with the GoScript™ Reverse
Transcription System (cat. no. A5001; Promega Corporation)
according to the manufacturer's protocol. The procedure used for
reverse transcription was as follows: 25°C for 5 min, 42°C for 60
min, 70°C for 15 min followed by hold at 4°C. PCR reactions were
performed on an ABI 7500 Sequence Detection System (Thermo Fisher
Scientific, Inc.) using FastStart Universal SYBR Green Master (ROX)
reagent (Roche Diagnostics GmbH). The thermocycling procedure used
for amplification was as follows: Initial denaturation at 50°C for
2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec
and 60°C for 1 min. Dissociation curves were used to assess
amplification specificity. cDNA was synthesized for RT-qPCR using
the following primer: miR-103a-3p, 5'-GTCGTATCCAGTGCAGGGTCCG
AGGTATTCGCACTGGATACGACTCATAG-3'. RT-qPCR was conducted using the
following primers: miR-103a-3p- forward (F),
5'-CGCGAGCAGCATTGTACAGGG-3' and reverse (R),
5'-ATCCAGTGCAGGGTCCGAGG-3'; U6-F, 5'-CGAATTTGCGTGTCATCCT-3' and R,
5'-GCTTCGGC AGCACATACTAA-3'; TPD52-F, 5'-TCGGAAGAGGAGCA GGAAGAGC-3'
and R, 5'-AGATGTTGCTGTCACGTCTT GCC-3'; and GAPDH-F,
5'-CCTGCCGTCTAGAAACCTG-3' and R, 5'-AGTGGGTGTCGCTGTTGAAGT-3'.
Relative gene expression was calculated using the 2-ΔΔCq
method (36).
Cell transfection
For miR-103a-3p overexpression and interference,
miR-103a-3p mimics or miR-103a-3p inhibitor and their negative
controls (NCs) were purchased from Guangzhou RiboBio Co., Ltd. The
miR-103a-3p mimics and mimic NC were composed of a double-strand
structure. The miR-103a-3p mimics sequences were as follows: Sense,
5'-AGCAGCAUUGU ACAGGGCUAUGA-3' and antisense, 3'- UCGUCGUAACAUG
UCCCGAUACU-5'. The miR-103a-3p mimic NC sequences were as follows:
Sense, 5'-UUUGUACUACACAAAAGUA CUG-3' and antisense,
3'-AAACAUGAUGUGUUUUCAU GAC-5'. The miR-103a-3p inhibitor sequence
was: 5'-UCAUAG CCCUGUACAAUGCUGCU-3'. The miR-103a-3p inhibitor NC
sequence was: 5'-CAGUACUU UUGUGUAGUACAAA-3'. To knockdown or
overexpress TPD52, two small interfering (si)RNAs specific for
TPD52 (siTPD52) and NC siRNAs were purchased from Guangzhou RiboBio
Co., Ltd., and a recombinant vector overexpressing TPD52 (TPD52-OE)
GV146-CMV-MCS-IRES-EGFP-SV40-neomycin and an empty vector were
purchased from Shanghai GeneChem Co., Ltd. The TPD52 siRNA
sequences were as follows: 5'-GAC TCTGTCTCAAGTGTTA-3' (siRNA1) and
5'-GCGGAAACT TGGAATCAAT-3' (siRNA2). The siRNA control sequence
was: 5'-TTTCTCCGAACGTGTCACG-3'. SACC-83 and SACC-LM cells were
transfected with miR-103a-3p mimics, miR-103a-3p inhibitor, TPD52
siRNA, the TPD52-OE vector or the empty vector and their NCs using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
miR-103a-3p mimics, TPD52 siRNA and their NC were transfected into
cells at a final concentration of 50 nM, and miR-103a-3p inhibitor
and inhibitor NC at a final concentration of 100 nM. The plasmid of
transfection was 2 µg/well/well in 6 well in 6-well -well
plates. The transfec plates. The transfections were performed at
37°C for 48 h when the cells grew up to 50-60% confluence.
Subsequent experimentations were performed 48 h
post-transfection.
Wound healing assay
For the wound healing assay, SACC-83 and SACC-LM
cells were transfected 2 days in 6-well plate (50-60% confluence),
then the transfected cells were digested, counted and seeded into a
96-well plate (3×104 per well). Wound areas were
measured using an IncuCyte instrument (Essen Bioscience, Inc.).
Briefly, ~3×104 tumour cells in RPMI-1640 medium
supplemented with 10% FBS were seeded into 96-well plates. After
reaching confluence, the cells were cultured in serum-free
RPMI-1640 medium, and after 12 h, a reproducible and uniform
scratch wound was made using a WoundMaker™ (Essen BioScience, Inc.)
in all wells. After washing three times with PBS, the cells were
incubated in medium without FBS at 37°C for 48 h and the migration
width change in the scratch wound was observed using an IncuCyte
instrument at x10 magnification (light microscope). Migration
widths were analysed by IncuCyte™ ZOOM (version 2016A; Essen
BioScience, Inc.).
Transwell migration assay
SACC-83 and SACC-LM cells transfected with
miR-103a-3p mimics, miR-103a-3p inhibitor, TPD52 siRNA or TPD52-OE
vector were seeded into the upper chamber of cell culture inserts
(pore size, 8 µm; Falcon; Corning, Inc.) at a density of
8×104 cells/well. The cells in the upper chamber were
cultured in RPMI-1640 medium without serum. The lower chamber
contained 600 µl RPMI-1640 medium supplemented with 20% FBS.
Then, the chamber was incubated at 37°C under a humidified
atmosphere containing 5% CO2 for 19 h. After the
incubation, the cells on the lower surface of the insert were
stained with a 1% crystal violet solution for 10 min after fixation
with 95% ethanol for 30 min, and the process of fixation and stain
were performed at room temperature. Then, cells on the lower
surface were imaged under a BX51 fluorescence microscope (Olympus
Corporation) at x20 magnification. In total, six fields were
randomly selected in each group for counting and statistical
analysis. The statistical analysis was performed by GraphPad Prism
(version 7.00; GraphPad Software, Inc.). Every experiment was
repeated independently ≥3 times.
Western blot analysis
Transfected cells were lysed in RIPA protein lysis
buffer (Beyotime Institute of Biotechnology). The protein
concentration was measured with a Pierce™ Bicinchoninic Acid
Protein Assay kit (Thermo Fisher Scientific, Inc.), following the
manufacturer's protocol. Each protein sample (40 µg/lane)
was separated by 10-12% SDS-PAGE and transferred onto PVDF
membranes. The membranes were blocked with 5% non-fat milk in TBST
(20 mM Tris, 137 mM NaCl and 0.1% Tween-20, pH 7.4) for 1 h at room
temperature. Then, the PVDF membranes were incubated with primary
antibodies in blocking buffer overnight at 4°C and followed by an
incubation with horseradish peroxidase-conjugated goat anti-rabbit
(1:100,000; cat. no. ZB-2301; OriGene Technologies, Inc.) or
anti-mouse (1:100,000; cat. no. ZB-2305; OriGene Technologies,
Inc.) IgG antibodies for 1 h at room temperature. The primary
antibodies used were anti-β-actin (1:1,000; cat. no. TA-09; OriGene
Technologies, Inc.), anti-TPD52 (1:1,000; cat. no. ab182578;
Abcam), anti-E-cadherin [1:1,000; cat. no. 3195; Cell Signalling
Technology, Inc. (CST)], anti-N-cadherin (1:1,000; cat. no. 13116;
CST), anti-vimentin (1:1,000; cat. no. 5741; CST), anti-Snail
(1:1,000; cat. no. 3879; CST) and anti-Slug (1:1,000; cat. no.
9585; CST). The immunocomplexes were visualized with a
SuperEnhanced chemiluminescence detection kit (CWBIO). All bands
were quantified using ImageJ (version 1.8.0; National Institutes of
Health), and three independent experiments with three biological
replicates each were performed.
Dual-luciferase reporter gene assay
The pEZX-MT05 plasmid was used to construct the
TPD52 3'UTR luciferase reporter gene plasmid (iGene Biotechnology,
Co., Ltd.). Wild-type (wt) or mutant (mut) 3'UTRs of TPD52 were
cloned into the downstream sites of the pEZX-MT05 vector (iGene
Biotechnology, Co., Ltd.). Next, 50 nM miR-103a-3p mimics or NC and
2.5 µg TPD52-wt or TPD52-mut were co-transfected into SACC
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
After 48 h, the cell culture medium was collected and the
activities of Gaussia Luciferase (GLuc) and Secreted Alkaline
Phosphatase (SEAP) in the culture medium were assayed using a
Secrete-Pair™ Luminescence Assay kit (cat. No. SPDA-D010;
GeneCopoeia, Inc.) according to the manufacturer's protocol. The
Gaussia Luciferase (Gluc) and Secreted Alkaline Phosphatase (SEAP)
for each sample were measured using a Centro LB 960 Microplate
Luminometer (Titertek-Berthold). Using SEAP signal as an internal
standard control, signal normalization (ratio of GLuc and SEAP
activities) eliminated the impact of transfection efficiency
variations and made the normalized GLuc activities of samples of
comparison more accurately reflect the true biological events. The
ratio of luminescence intensities (RLU, relative light unit) of the
GLuc was calculated over SEAP. Then, the normalized GLuc activity
(GLuc/SEAP ratio) was compared with all samples.
Mouse model of lung metastasis
To elucidate the role of miR-103a-3p in SACC lung
metastasis in vivo, 19 female NOD/SCID mice (age, 6 weeks;
weight 20-25 g) were purchased from the Beijing Vital River
Laboratory Animal Technology Co., Ltd. Animals were maintained
under specific pathogen-free conditions at room temperature
(20-26°C) with a humidity level of 50-60%, a 12-h light/dark cycle
and food and water ad libitum in the animal facility of Peking
University School and Hospital of Stomatology. All efforts were
made to minimize animal suffering. All animal assays were performed
following approval from the Peking University Institutional Animal
Care and Use Committee (Beijing, China; permit no. LA2015099).
SACC-83 cells transfected with miR-103a-3p mimics or NC
(1×106 cells/100 µl/mouse) were injected into the
tail vein of NOD/SCID mice. During the whole experiment, the
animals were carefully monitored at least twice a week. The maximum
percentage of body weight loss observed in mice was <12% from
start to endpoint. The mice would be euthanized if they showed
serious symptoms including, but not limited to, breathing
difficulties, weight loss (>20% of body weight), vocalization,
irritability, hunching, stationary, ruffling or poor grooming. The
time to detect the tumour metastases in vivo was determined
according to our previous experiment (34). After 8 weeks, the mice injected
with miR-103a-3p mimics (n=9) or mimic NC (n=10) transfected cells
were sacrificed by cervical dislocation, and the procedure was
performed out of sight of the other mice. The mortality of the
mouse was verified by respiratory arrest and cardiac cessation.
Subsequently, the lung tissues were collected from these mice. A
maximum of two macroscopic nodules were observed in one lung tissue
and the maximum diameter of a nodule was 4 mm. Then, the lung
tissues were fixed in 4% paraformaldehyde for 24 h at room
temperature, embedded in paraffin and cut into 5-µm thick
sections. These sections were subjected to haematoxylin and eosin
(H&E) staining to analyse the number of lung metastasis nodules
(light microscope; x40 magnification). Sections were deparaffinised
and rehydrated with xylene I, xylene II and xylene III for 20 min
each. Then, absolute ethanol I and absolute ethanol II for 10 min
each and finally 95, 90, 80 and 70% ethanol for 5 min. The sections
were wash with deionized water three times for 3 min each time,
then stained with haematoxylin for 2-3 min. Next, the sections were
rinsed with deionized water for 10 min and stained with eosin for 1
min. Gradient dehydration was performed with 70, 80, 90, 95 and
100% ethanol for 2, 2, 3, 3 and 10 min, respectively, and then with
xylene I, xylene II and xylene III for 10 min each. The sections
were sealed with neutral gum and placed in a ventilated room
overnight. All the procedures were performed at room
temperature.
Statistical analysis
Statistical analyses, including two-tailed unpaired
Student's t-test, one-way ANOVA with Bonferroni post-test
correction, χ2 test and Pearson correlation coefficient
analysis, were conducted with SPSS (version 20.0; SPSS, Inc.). Data
are presented as the mean ± standard deviation of three independent
experiments. These results were repeated in ≥3 independent
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-103a-3p is highly expressed in SACC
tissues and SACC cells
To identify dysregulated miRNAs in SACC, the
expression of miRNAs in SMG vs. SACC tissues was compared by
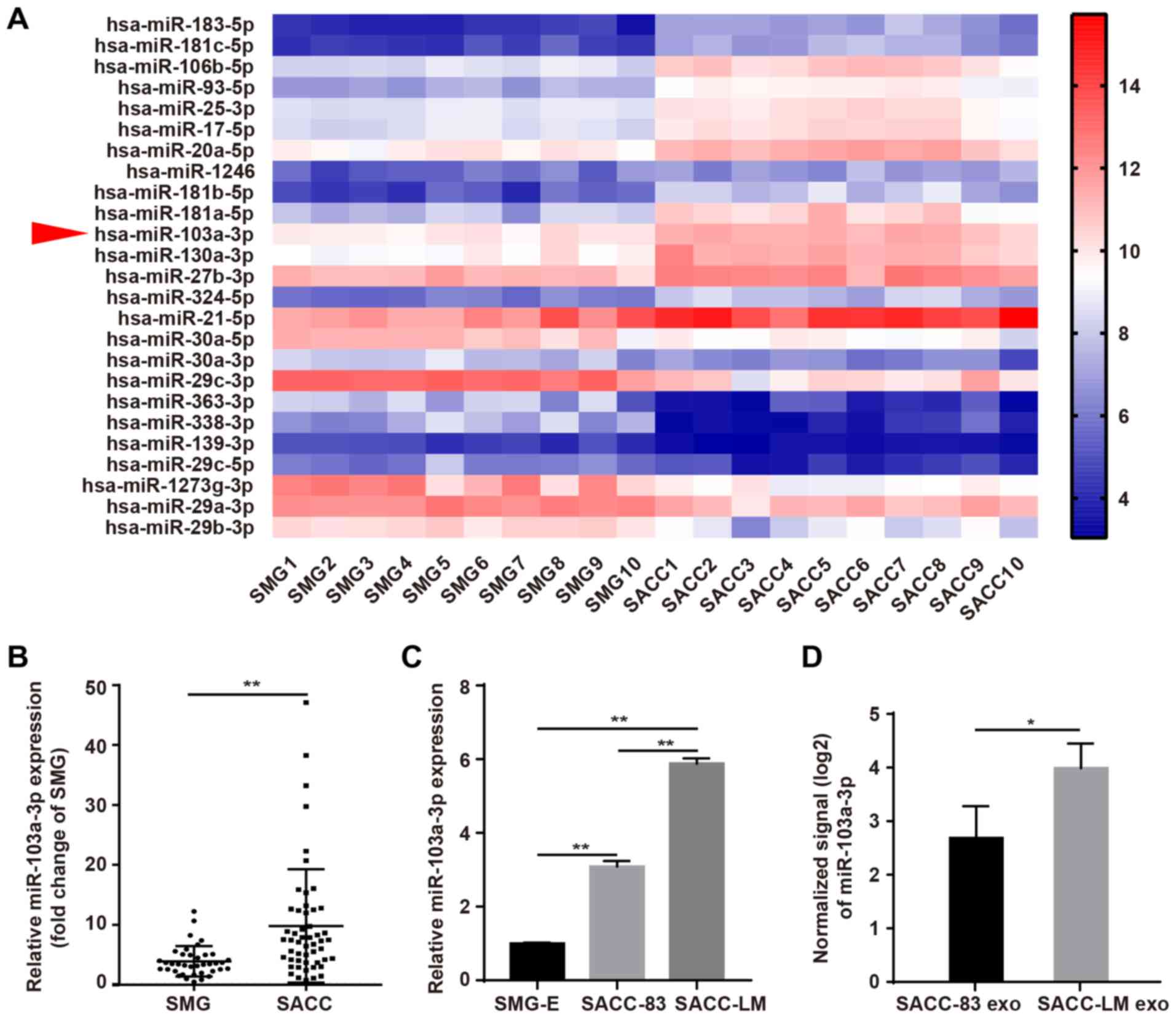
microarray. GraphPad Prism was used to create the heat map
generated from ten human SACC and ten paired SMG tissues, which
demonstrated that miR-103a-3p ranked 11th among the top 15
upregulated miRNAs (Fig. 1A).
To evaluate the biological function of miR-103a-3p
in SACC, its expression was measured in 38 SMG and 52 SACC tissues.
miR-103a-3p expression was significantly higher in SACC tissues
compared with healthy tissues (Fig.
1B). In addition, miR-103a-3p expression was significantly
higher in SACC cells compared with SMG-E (Fig. 1C). Furthermore, SACC-LM cells had
increased miR-103a-3p expression compared with SACC-83 cells. The
expression of miR-103a-3p was also examined in SACC cell exosomes,
and the results indicated that miR-103a-3p was significantly
upregulated in SACC-LM cell exosomes (Fig. 1D).
The clinicopathological features of 52 patients with
SACC were assessed and the results are presented in Table I. All the patients with SACC were
classified into the high or low miR-103a-3p group depending on the
median miR-103a-3p expression. High miR-103a-3p expression was
associated with the local regional recurrence and lung metastasis.
However, no significant associated was identified between
miR-103a-3p expression and the other clinicopathological features,
including age, sex, tumour size, clinical stage, site, lymph node
metastasis, perineural invasion and pathological type.
Collectively, these results indicated that miR-103a-3p serves as an
oncogene in SACC.
 | Table IAssociations between
clinicopathological variables and miR-103a-3p expression in
patients with salivary adenoid cystic carcinoma. |
Table I
Associations between
clinicopathological variables and miR-103a-3p expression in
patients with salivary adenoid cystic carcinoma.
| Variables | miR-103a-3p
expression
| χ2 | P-value |
|---|
| Total (n) | Low (n) | High (n) |
|---|
| Age, years | | | | 0.746 | 0.388 |
| <42 | 19 | 11 | 8 | | |
| ≥42 | 33 | 15 | 18 | | |
| Sex | | | | 2.769 | 0.096 |
| Male | 26 | 16 | 10 | | |
| Female | 26 | 10 | 16 | | |
| Tumour size | | | | 0.087 | 0.768 |
| <4 cm | 35 | 17 | 18 | | |
| ≥4 cm | 17 | 9 | 8 | | |
| Clinical
stagea | | | | 0.361 | 0.548 |
| I/II | 16 | 9 | 7 | | |
| III/IV | 36 | 17 | 19 | | |
| Site | | | | 0.433 | 0.510 |
| Major salivary
gland | 12 | 5 | 7 | | |
| Minor salivary
gland | 40 | 21 | 19 | | |
| Lymph node
metastasis | | | | 0.391 | 0.532 |
| Absent | 38 | 20 | 18 | | |
| Present | 14 | 6 | 8 | | |
| Perineural
invasion | | | | 0.000 | 1.000 |
| Absent | 24 | 12 | 12 | | |
| Present | 28 | 14 | 14 | | |
| Lung
metastasis | | | | 3.900 | 0.048 |
| Absent | 40 | 23 | 17 | | |
| Present | 12 | 3 | 9 | | |
| Local regional
recurrence | | | | 4.457 | 0.035 |
| Absent | 42 | 24 | 18 | | |
| Present | 10 | 2 | 8 | | |
| Pathological
type | | | | 0.000 | 1.000 |
|
Cribriform/tubular | 34 | 17 | 17 | | |
| Solid | 18 | 9 | 9 | | |
miR-103a-3p modulates cell migration and
the epithelial-mesenchymal transition (EMT) process in vitro
To investigate the function of miR-103a-3p,
miR-103a-3p mimics and a mimic NC were transfected into SACC-83
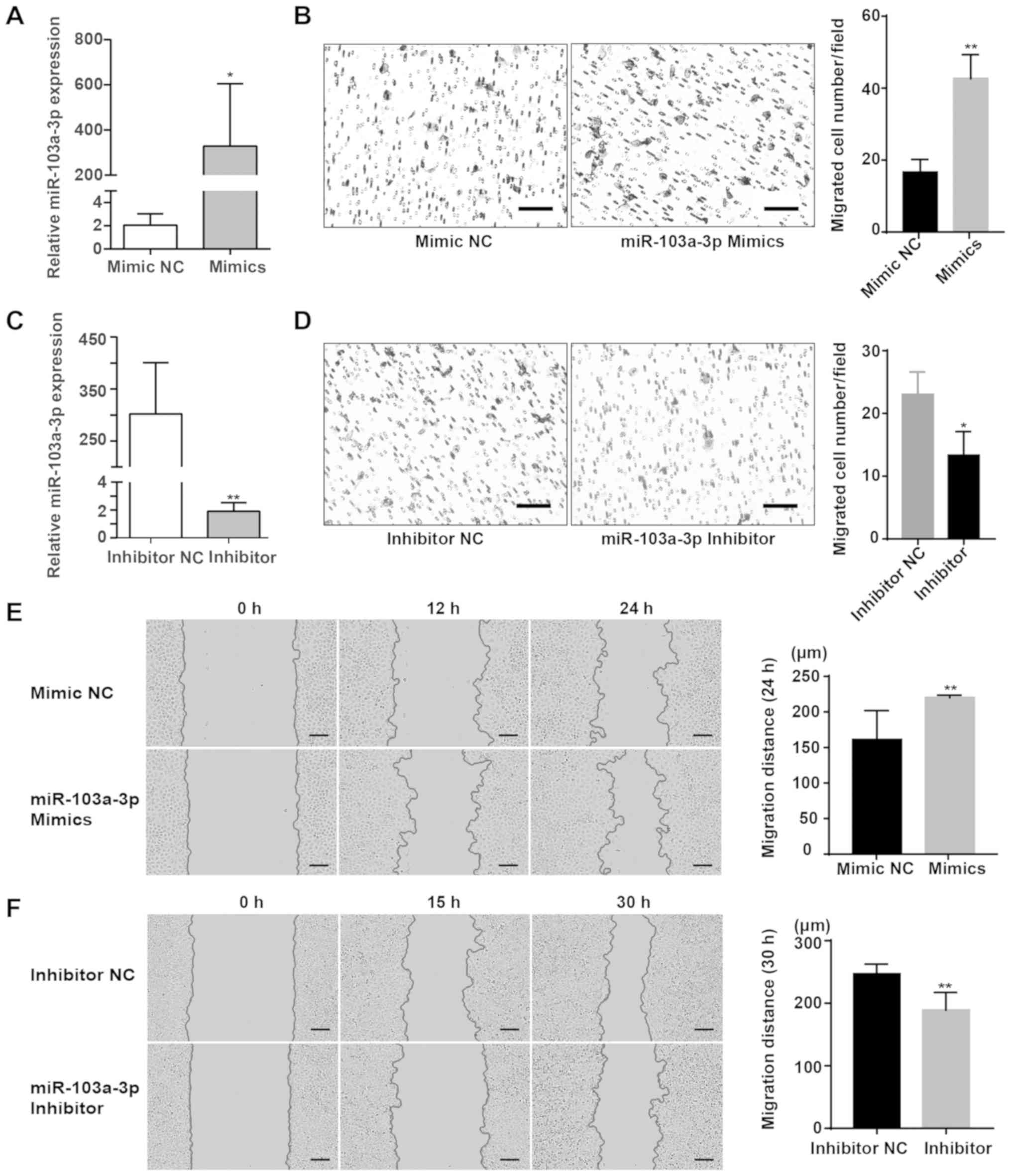
cells (Fig. 2A), and then the
effect of miR-103a-3p on cell migration was assessed using a
Transwell migration assays. The Transwell migration assay results
demonstrated that significantly more cells migrated in the
miR-103a-3p mimic group compared with the NC group (Fig. 2B). Subsequently, miR-103a-3p
expression was knocked down to assess its migration effects in
SACC-LM cells using a miRNA inhibitor (Fig. 2C). The Transwell assay results
indicated that significantly fewer cells migrated in the
miR-103a-3p inhibitor group compared with the inhibitor NC group
(Fig. 2D). The effects of
miR-103a-3p on cell proliferation were also assessed using a Cell
Counting Kit-8, and it was found that miR-103a-3p had no
significant effect on the proliferation of SACC cells (Data S1; Fig. S1).
The effect of miR-103a-3p on cell migration were
also examined using wound healing assay. Compared with the NC
group, the migratory distance of SACC-83 cells transiently
transfected with miR-103a-3p mimics was significantly wider
(Fig. 2E). Furthermore, migration
was slower in the miR-103a-3p inhibitor group compared with the
control group (Fig. 2F). It was
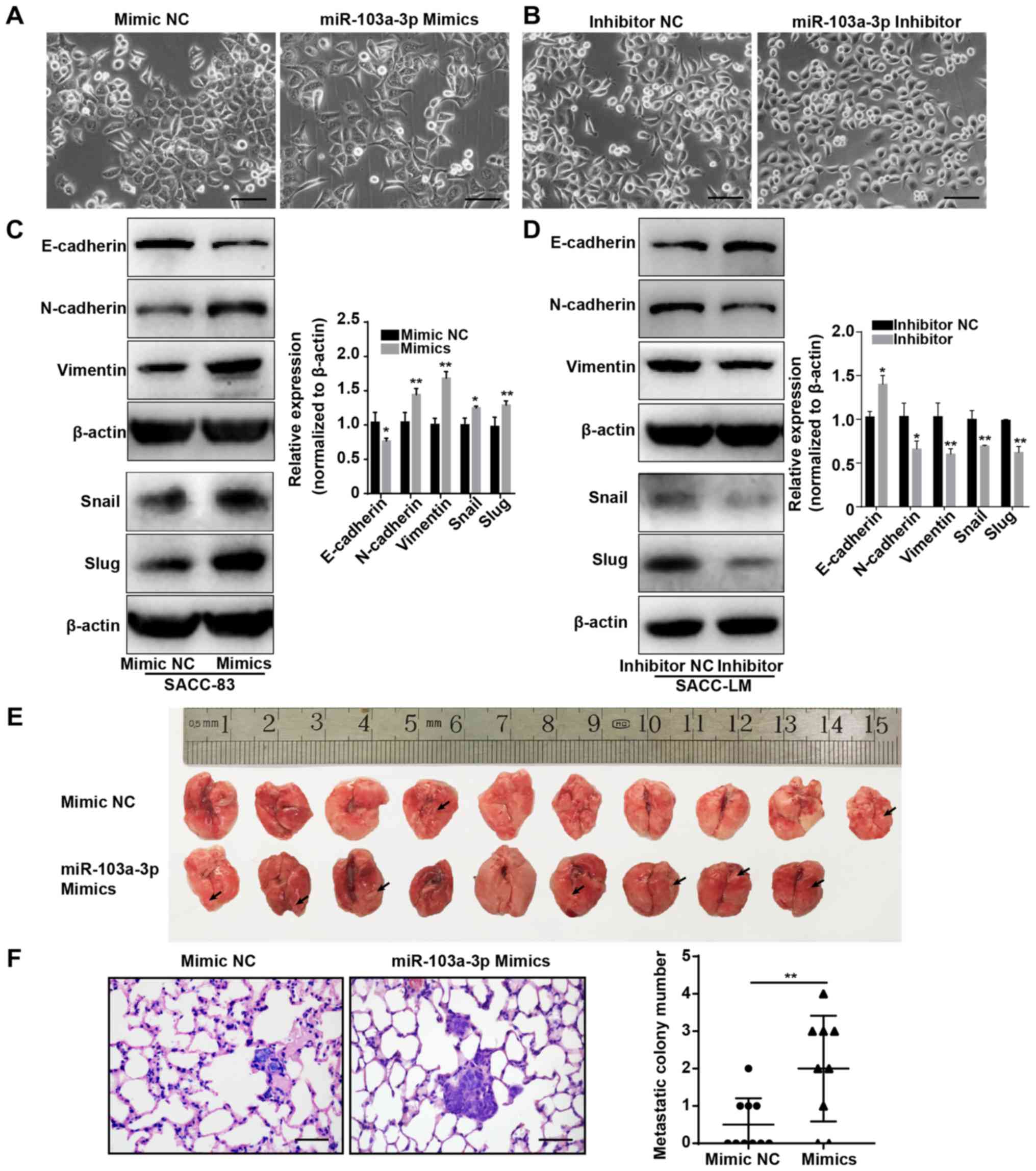
also demonstrated that the mimic-transfected SACC-83 cells had a
fibroblast-like morphology (Fig.
3A), while knockdown of miR-103a-3p in SACC-LM cells resulted
in an epithelial-like morphology (Fig.
3B).
Next, the changes in EMT markers were evaluated. In
the miR-103a-3p mimic group, the mesenchymal cell markers
N-cadherin, vimentin, Slug and Snail were significantly
upregulated, while the epithelial cell marker E-cadherin was
significantly downregulated (Fig.
3C). When miR-103a-3p was inhibited, E-cadherin was
significantly upregulated, and N-cadherin, Slug, Snail and Vimentin
were significantly downregulated (Fig.
3D). Therefore, these results suggested that miR-103a-3p can
affect cell function via the EMT process and that miR-103a-3p plays
a role in promoting SACC metastasis.
Upregulation of miR-103a-3p promotes SACC
cell metastasis in vivo
To investigate the effect of miR-103a-3p on SACC
lung metastasis in vivo, miR-103a-3p mimic- and mimic
NC-transfected SACC-83 cells were injected into the tail veins of
NOD/SCID mice. These mice were sacrificed after 8 weeks, and the
lung tissues were collected, fixed, sectioned and subjected to
H&E staining. The mice injected with mimic- transfected SACC-83
cells had more visible tumour nodules compared with those with
mimic NC-transfected SACC-83 cells. (Fig. 3E). Furthermore, the number and
extent of lung tumour nodules was significantly greater in the mice
injected with miR-103a-3p mimic-transfected cells compared with
those injected with mimic NC-transfected cells (Fig. 3F). Thus, these results indicated
that miR-103a-3p promotes SACC lung metastasis in vivo.
TPD52 is a direct target of miR-103a-3p
in SACC
Next, the possible mechanism via which miR-103a-3p
promotes the migration of SACC cells was investigated. The
potential interaction of miR-103a-3p with target mRNAs was analysed
using different prediction tools, including TargetScan (http://www.targetscan.org/vert_72/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and
miRDB (http://mirdb.org/). TPD52 was identified as a
target gene of miR-103a-3p (Fig.
4A). To determine whether TPD52 is a direct target of
miR-103a-3p, wt and mut 3'UTRs of TPD52 were inserted downstream of
the luciferase reporter vector (Fig.
4A). The luciferase reporter assay results demonstrated that
miR-103a-3p overexpression significantly reduced luciferase
activity in the TPD52-wt group (Fig.
4B) but not in the TPD52-mut group, indicating that miR-103a-3p
can directly target TPD52.
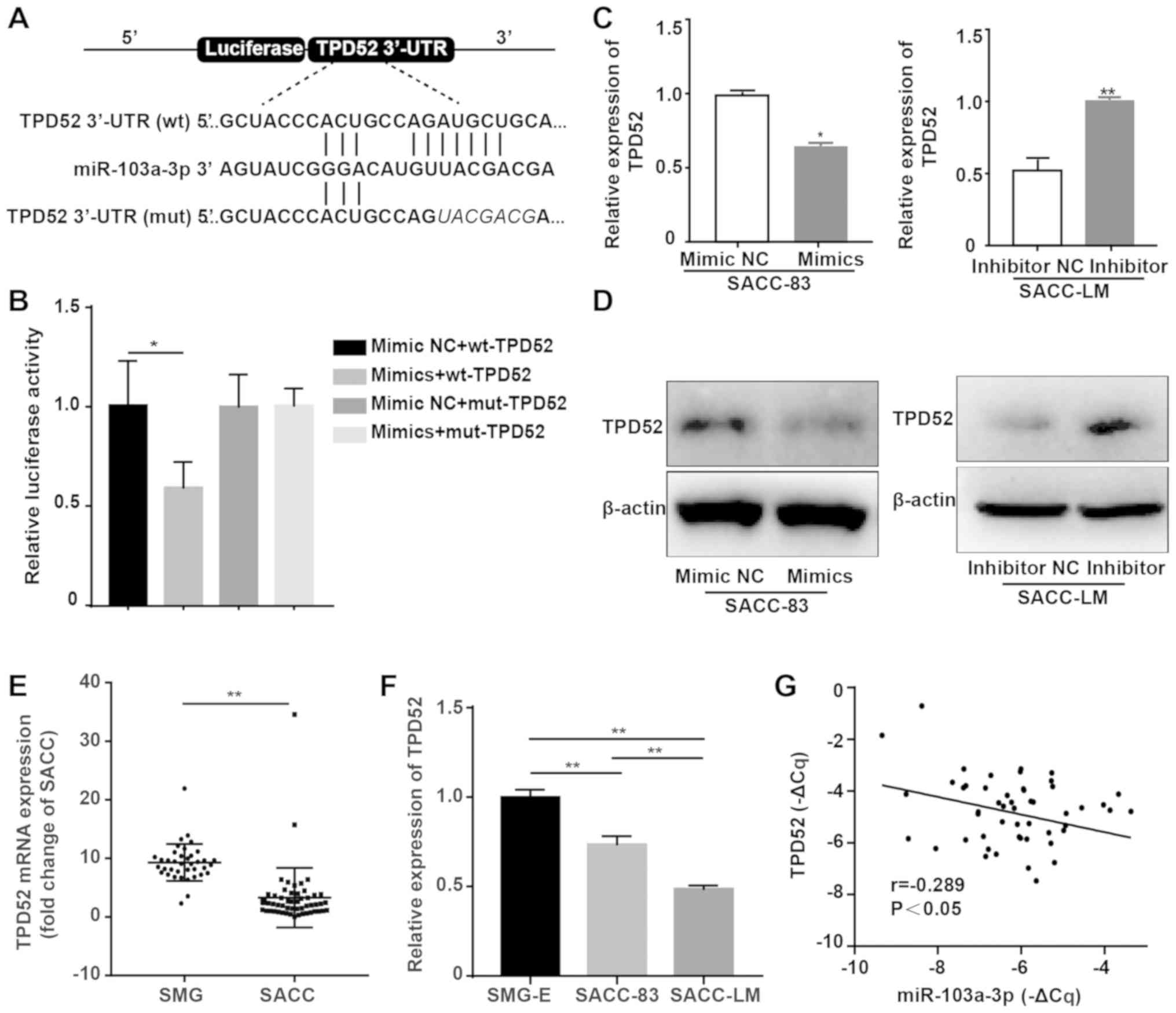 | Figure 4TPD52 is a direct target gene of
miR-103a-3p. (A) Schematic diagram of the binding sites of
miR-103a-3p to the 3'UTR of TPD52 mRNA predicted by TargetScan,
miRTarBase and miRDB and the mutant sites. (B) Relative luciferase
activity was assessed in SACC-83 cells that were transfected with
the wt 3'UTR TPD52 construct or mut 3'UTR TPD52 construct, as well
as miR-103a-3p mimic NC or miR-103a-3p mimics. (C) RT-qPCR analysis
of TPD52 mRNA expression after SACC cells were transiently
transfected with miR-103a-3p mimics and inhibitor. (D) Western blot
analysis of TPD52 protein expression after SACC cells were
transiently transfected with miR-103a-3p mimics and inhibitor. (E)
Determination of the expression of TPD52 mRNA in 52 human SACC
tissues and 38 human SMG tissues by RT-qPCR. (F) Determination of
the expression of TPD52 mRNA in SMG-E, SACC-83 and SACC-LM cells by
RT-qPCR. (G) Correlation between miR-103a-3p expression and TPD52
mRNA in 52 SACC tissues. A negative correlation between miR-103a-3p
and TPD52 mRNA was observed (P<0.05; r=-0.289).
*P<0.05, **P<0.01. SACC, salivary
adenoid cystic carcinoma; SMG, submandibular gland; SMG-E,
subman-dibular gland primary epithelial cells; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; miR,
microRNA; wt, wild-type; mut, mutant; TPD52, tumour protein
D52. |
RT-qPCR and western blotting results indicated that
TPD52 mRNA and protein expression levels were decreased in
miR-103a-3p mimic-transfected SACC-83 cells and increased in
miR-103a-3p inhibitor-transfected SACC-LM cells (Fig. 4C and D). Moreover, the expression
of TPD52 was measured in the 38 SMG and 52 SACC tissues by RT-qPCR.
It was identified that TPD52 expression was significantly higher in
healthy tissues compared with SACC tissues (Fig. 4E), which is controversial with the
known function of miR-103a-3p. SACC-83 cells expressed
significantly higher levels of TPD52 compared with SACC-LM cells
(Fig. 4F). In addition, the
relationship between miR-103a-3p and TPD52 was assessed by RT-qPCR
and it was demonstrated that there was a significantly weak
negative correlation between miR-103a-3p and TPD52 (Fig. 4G). Therefore, it was speculated
that miR-103a-3p promoted cancer migration by targeting TPD52.
TPD52 promotes SACC progression
To investigate whether miR-103a-3p acts as an
important cancer promoter by targeting TPD52, the function of TPD52
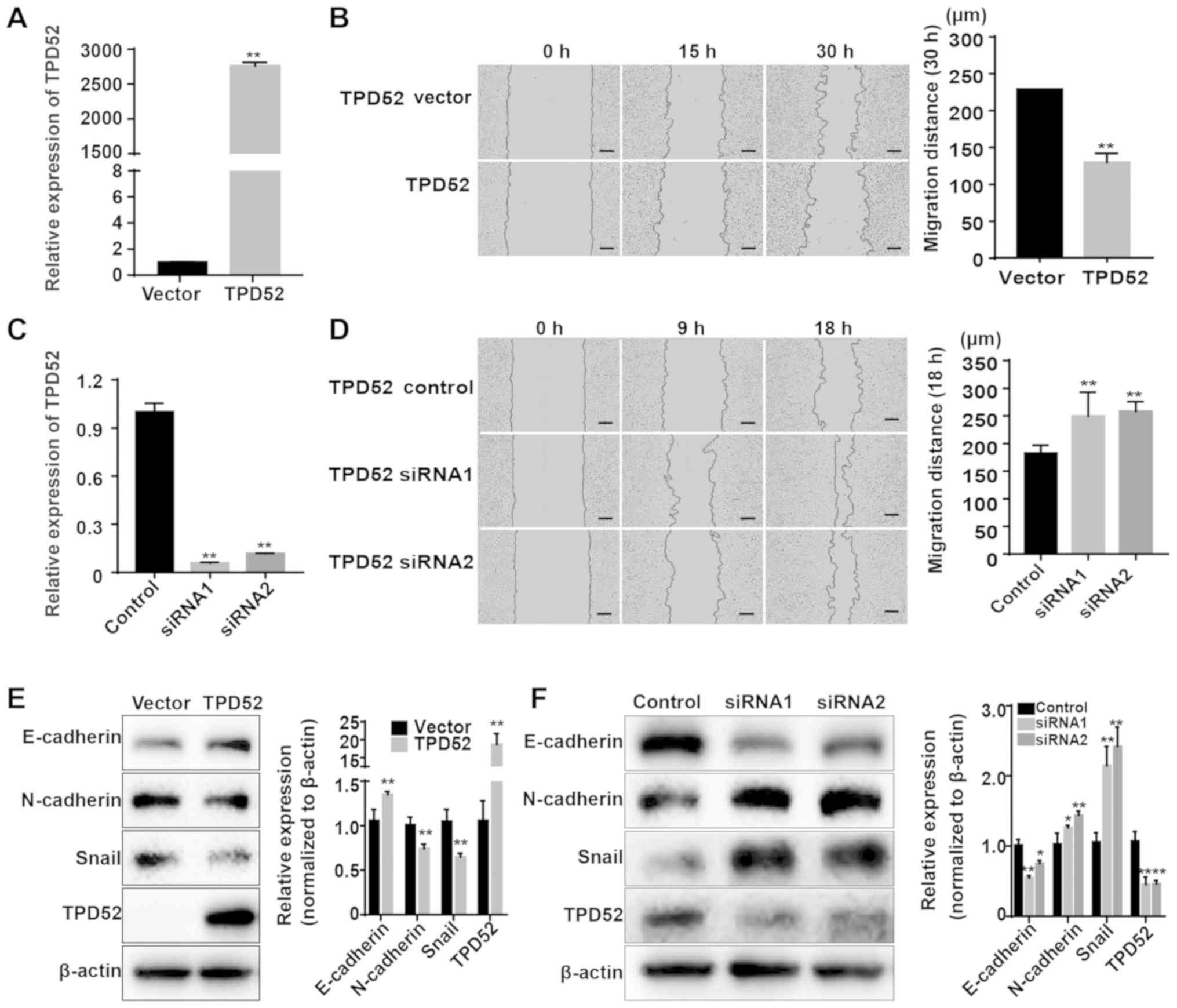
was evaluated in SACC cells. TPD52 was overexpressed using plasmid
transfection. After transfection with a TPD52 recombinant plasmid,
RT-qPCR and western blotting results indicated that the mRNA and
protein expression levels of TPD52 were significantly increased in
TPD52-OE vector-transfected SACC-LM cells (Fig. 5A and E). The effects of TPD52 on
the proliferation of SACC cells were also assessed using a Cell
Counting Kit-8 assay, and it was found that TPD52 had no
significant effect on the proliferation of SACC cells (Fig. S2).
Subsequently, the function of TPD52 in tumour
migration was investigated, and the wound healing assay results
demonstrated that TPD52 overexpression significantly suppressed the
migration of SACC-LM cells (Fig.
5B). In addition, TPD52 expression was downregulated by
transfecting SACC-83 cells with two TPD52 siRNAs. RT-qPCR and
western blotting results identified significant decreases in TPD52
mRNA and protein expression levels in TPD52 siRNA-transfected
SACC-83 cells (Fig. 5C and F). It
was also found that TPD52 knockdown promoted SACC-83 cell migration
in wound healing assays (Fig. 5D).
Furthermore, western blotting results indicated that TPD52
overexpression decreased the expression levels of the mesenchymal
markers N-cadherin and Snail, but increased that of the epithelial
marker E-cadherin (Fig. 5E). When
TPD52 was knocked down, there were significant increases in
N-cadherin and Snail but a significant decrease in E-cadherin
(Fig. 5F). These data suggested
that TPD52 inhibits the migration of SACC cells.
The associations between clinicopathological
variables and TPD52 mRNA expression were examined in patients with
SACC, and the results are presented in Table SI. All patients with SACC were
classified into high-TPD52 and low-TPD52 groups depending on the
median TPD52 mRNA expression. Low TPD52 mRNA expression was
associated with a high incidence of lung metastasis and a high rate
of perineural invasion. However, no significant associations were
observed between TPD52 expression and the other clinicopathological
features.
miR-103-3p-induced EMT and migration of
SACC cells is neutralized by TPD52 overexpression
To further verify whether the promoting effects of
miR-103a-3p on SACC cell migration and EMT are mediated by
targeting TPD52, cell phenotype changes were examined after
overexpressing TPD52 in miR-103a-3p mimic-transfected SACC cells.
The wound healing assay results suggested that TPD52 overexpression
significantly decreased the migratory distance of miR-103a-3p
mimic-transfected SACC cells compared with the mimics + vector
groups (Fig. 6A). Furthermore, the
Transwell migration assay results demonstrated a inhibitory effect
in miR-103a-3p mimic SACC cells overexpressing TPD52 compared with
the mimics + vector group (Fig.
6B).
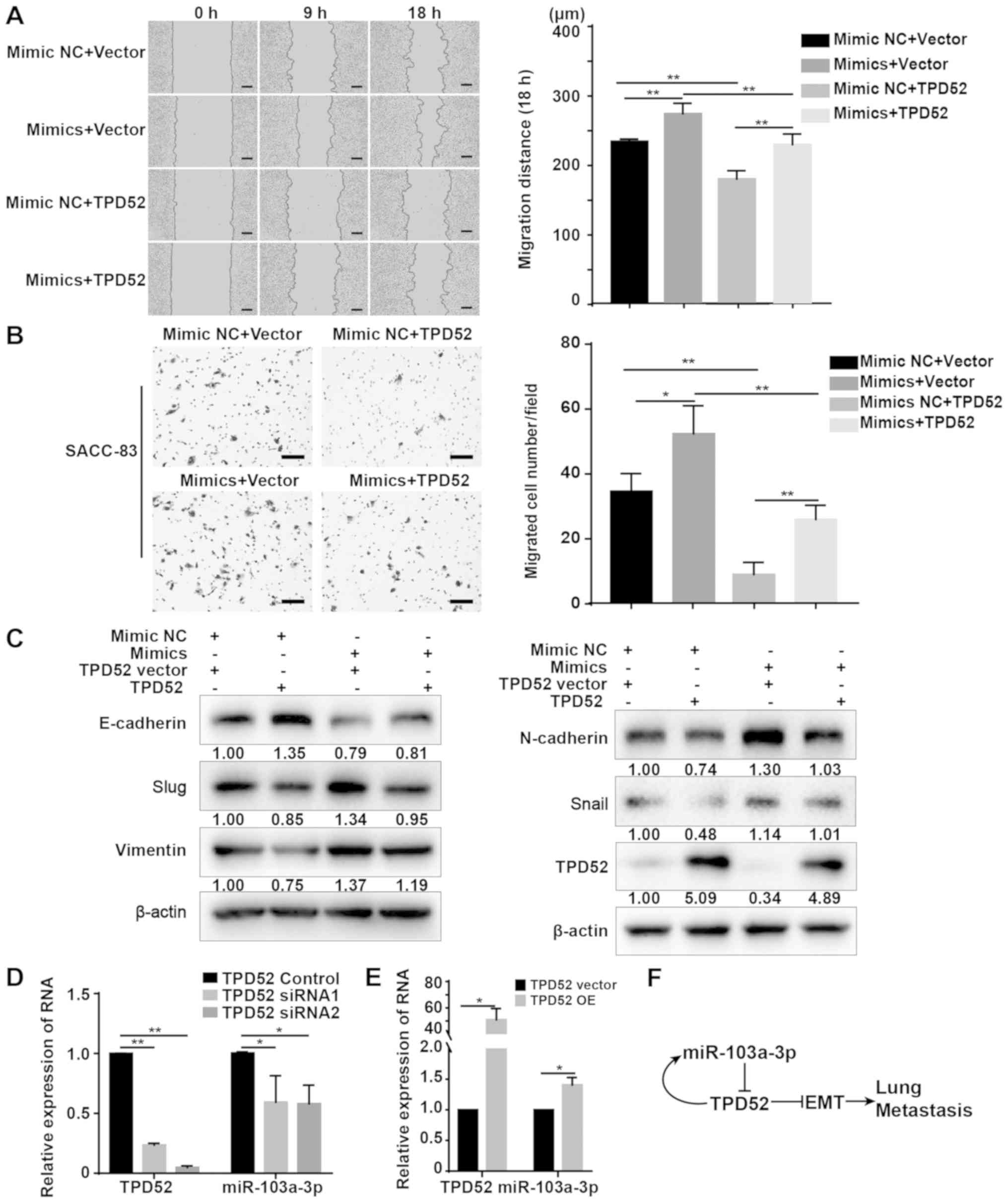 | Figure 6Overexpression of TPD52 abrogates
miR-103a-3p-induced migration of SACC-83 cells. (A) Wound healing
assay analysis of SACC-83 cells transiently transfected with
miR-103a-3p mimics, TPD52 OE plasmid, negative control or vector
for cell migration. Scale bar, 150 µm. (B) Transwell
migration assays of SACC-83 cells transiently transfected with
miR-103a-3p mimics, TPD52 OE plasmid, NC or vector. Scale bar, 100
µm. (C) Western blotting analysis the EMT markers of SACC-83
cells transiently transfected with miR-103a-3p mimics, TPD52 OE
plasmid, NC or vector. (D) RT-qPCR analysis of miR-103a-3p
expression after SACC-83 cells were transiently transfected with
TPD52 siRNA and NC. (E) RT-qPCR analysis of miR-103a-3p expression
after SACC-LM cells were transiently transfected with TPD52 OE
plasmid and NC vector. (F) Schematic diagram of the role of
miR-103a-3p and TPD52 in SACC lung metastasis. Data are presented
as the mean ± standard deviation of three independent experiments.
*P<0.05 and **P<0.01. RT-qPCR, reverse
transcription-quantitative PCR; siRNA, small interfering RNA;
TPD52, tumour protein D52; OE, overexpression; NC, negative
control; miR, microRNA; SACC, salivary adenoid cystic
carcinoma. |
Western blotting results suggested that
overexpressing TPD52 rescued the EMT effect of miR-103a-3p
over-expression, as indicated by decreased expression of the
mesenchymal markers N-cadherin, Vimentin, Snail and Slug, and
increased expression of the epithelial marker E-cadherin in
miR-103a-3p mimic-transfected SACC cells (Fig. 6C). These findings further indicated
that overexpression of miR-103a-3p promotes SACC cell migration and
EMT by targeting TPD52. TPD52 may also negatively regulate its own
expression. Moreover, knockdown of TPD52 inhibited the expression
of miR-103a-3p in SACC-83 cells (Fig.
6D), while TPD52 overexpression promoted the expression of
miR-103a-3p in SACC-LM cells (Fig.
6E). The schematic diagram demonstrated the role of miR-103a-3p
and TPD52 in SACC lung metastasis (Fig. 6F). miR-103a-3p promoted lung
metastasis by targeting TPD52, and TPD52 moderated the expression
of miR-103a-3p in turn.
Discussion
Metastasis activation is one of the hallmarks of
cancer (37). Thus, understanding
the regulatory molecules and mechanisms of metastasis is crucial
for the diagnosis and treatment of tumours. Previous studies have
reported that the expression profiles of miRNAs are abnormal in
several types of cancer, and miRNAs can regulate the development of
these cancer types (15,38,39).
It has also been revealed that numerous miRNAs may regulate the
initiation, progression and metastasis of SACC (14,40,41),
and miR-103a-3p has been shown to regulate development in some
specific malignant tumours (18,20,42).
However, the function and molecular mechanism of miR-103a-3p in
SACC lung metastasis remain unknown. The present results suggested
that miR-103a-3p was upregulated in SACC tissues and tumour cell
lines. The overexpression of miR-103a-3p promoted migration in
low-metastasis SACC-83 cells, while miR-103a-3p knockdown inhibited
metastasis in high-metastasis SACC-LM cells. Further examination
revealed that miR-103a-3p overexpression could promote the
conversion of epithelial markers into interstitial markers and vice
versa. Using in vivo miR-103a-3p gain-of-function studies,
it was demonstrated that miR-103a-3p may act as a tumour promoter
in SACC. The tumour metastasis promoting function of miR-103a-3p in
SACC was consistent with its function in colorectal cancer
(17) and gastric cancer (18).
miRNAs typically function by binding to the 3'UTR
region of mRNA to reduce the mRNA expression of target genes
(43). Therefore, it was
speculated that miR-103a-3p acts as a tumour promoter by targeting
specific mRNAs. Through prediction analyses, TPD52 was identified
as a potential target of miR-103a-3p, and dual-luciferase reporter
gene assay results verified this predicted target. RT-qPCR and
western blotting results also indicated that miR-103a-3p
overexpression could decrease the mRNA and protein expression
levels of TPD52. Moreover, miR-103a-3p expression was negatively
associated with TPD52 mRNA expression in SACC tissues, and
overexpressing TPD52 could rescue the migration-promoting effect of
miR-103a-3p. Thus, these findings suggested that miR-103a-3p binds
to the 3'UTR region of TPD52 mRNA to decrease TPD52 expression.
TPD52 has significant roles in the malignant
phenotype of cancer cells (27).
Previous PCR results have shown that TPD52 was upregulated in 18/29
tested cancer types and downregulated in 11 (38%) tumour types
(25). It has also been reported
that exogenous TPD52 expression promotes prostate cancer cell
migration via αvβ3 integrin by activating the protein kinase B/Akt
signaling pathway (44) and that
miR-139-5p can inhibit the proliferation, apoptosis and cell cycle
of uterine leiomyoma cells by targeting TPD52 (30). However, TPD52 may also play an
opposite role in different tumours. Previous studies have shown
that decreased TPD52 expression was associated with poor prognosis
in primary hepatocellular carcinoma (28) and that TPD52 could inhibit renal
cell carcinoma cell metastasis via the PI3K/Akt signal-ling pathway
(29). In the present study, TPD52
expression was lower in SACC tissues compared with SMG tissues.
Furthermore, SACC-LM cells with high lung metastasis expressed less
TPD52 compared with parental SACC-83 cells. Knockdown of TPD52
promoted SACC cell migration and the conversion of epithelial
markers into interstitial markers, whereas TPD52 overexpression had
the opposite results. Collectively, these results suggested that
TPD52 may be a tumour suppressor gene in SACC and inhibits cancer
cell migration.
As miR-103a-3p inhibited TPD52 expression, and TPD52
could in turn promote miR-103a-3p expression, TPD52 expression
remained low in SACC cells. Therefore, it was speculated that the
self-regulation of cells maintains balanced TPD52 expression: when
cellular TPD52 expression was excessive, the enhanced expression of
miR-103a-3p results in reduced TPD52 and sustained EMT in SACC
cells. This finding suggests that the feedback regulation between
miR-103a-3p and TPD52 maintains the metastatic properties of
SACC.
The present study focused on the effects of
miR-103a-3p on cell migration and lung metastasis. However, miRNAs
can also exert effects on cell invasion (45), apoptosis (46) and angiogenesis (47). Another potential effect of
miR-103a-3p may also be involved in lung metastasis processes, and
future investigations should focus on determining whether more
phenotypes and their associated molecular and signalling pathways
are involved in the miR-103a-3p-mediated lung metastasis of
SACC.
In conclusion, the present results demonstrated that
miR-103a-3p acted as an oncogene in SACC by promoting tumour cell
migration, which was promoted by miR-103a-3p via direct targeting
of TPD52. The identification of the novel miR-103a-3p/TPD52 axis is
significant for understanding SACC pathogenesis and offers further
insight into the identification of novel biomarkers or potential
therapeutic targets for SACC.
Supplementary Data
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81872190).
Availability of data and materials
The datasets used and/or analysed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
XYG and SLL designed the experiments. MF and CWC
performed the in vitro experiments and acquired the data.
MF, CWC, ZHD and YRL collected tissue samples and performed the
in vivo experiments. MF, LQY and WWY analysed and
interpreted the data and performed statistical analysis. MF and LQY
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Experiments using human tissue samples were approved
by the Ethics Committee of Peking University School and Hospital of
Stomatology (Beijing, China; permit no. PKUSSIRB-201522040). All
participants signed written informed consent. All animal assays
were performed following approval from the Peking University
Institutional Animal Care and Use Committee (Beijing, China; permit
no. LA2015099).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
SACC
|
salivary adenoid cystic carcinoma
|
|
miRNA/miR
|
microRNA
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
Acknowledgements
Not applicable.
References
|
1
|
Adelstein DJ, Koyfman SA, El-Naggar AK and
Hanna EY: Biology and management of salivary gland cancers. Semin
Radiat Oncol. 22:245–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rapidis AD, Givalos N, Gakiopoulou H,
Faratzis G, Stavrianos SD, Vilos GA, Douzinas EE and Patsouris E:
Adenoid cystic carcinoma of the head and neck. Clinicopathological
analysis of 23 patients and review of the literature. Oral Oncol.
41:328–335. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kokemueller H, Eckardt A, Brachvogel P and
Hausamen JE: Adenoid cystic carcinoma of the head and neck - a 20
years experience. Int J Oral Maxillofac Surg. 33:25–31. 2004.
View Article : Google Scholar
|
|
4
|
Gao M, Hao Y, Huang MX, Ma DQ, Luo HY, Gao
Y, Peng X and Yu GY: Clinicopathological study of distant
metastases of salivary adenoid cystic carcinoma. Int J Oral
Maxillofac Surg. 42:923–928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coca-Pelaz A, Rodrigo JP, Bradley PJ,
Vander Poorten V, Triantafyllou A, Hunt JL, Strojan P, Rinaldo A,
Haigentz M Jr, Takes RP, et al: Adenoid cystic carcinoma of the
head and neck - An update. Oral Oncol. 51:652–661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benakanakere MR, Zhao J, Finoti L,
Schattner R, Odabas-Yigit M and Kinane DF: MicroRNA-663 antagonizes
apoptosis antago-nizing transcription factor to induce apoptosis in
epithelial cells. Apoptosis. 24:108–118. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin
YF, Yuan Y and Zhuang SM: Hepatoma cell-secreted exosomal
microRNA-103 increases vascular permeability and promotes
metastasis by targeting junction proteins. Hepatology.
68:1459–1475. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barbollat-Boutrand L, Joly-Tonetti N, Dos
Santos M, Metral E, Boher A, Masse I, Berthier-Vergnes O, Bertolino
P, Damour O and Lamartine J: MicroRNA-23b-3p regulates human
keratinocyte differentiation through repression of TGIF1 and
activation of the TGF-β-SMAD2 signalling pathway. Exp Dermatol.
26:51–57. 2017. View Article : Google Scholar
|
|
11
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan F, Wang C, Li T, Cai W and Sun J: Role
of miR-21 in the growth and metastasis of human salivary adenoid
cystic carcinoma. Mol Med Rep. 17:4237–4244. 2018.PubMed/NCBI
|
|
13
|
Wang C, Li T, Yan F, Cai W, Zheng J, Jiang
X and Sun J: Effect of simvastatin and microRNA-21 inhibitor on
metastasis and progression of human salivary adenoid cystic
carcinoma. Biomed Pharmacother. 105:1054–1061. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brown AL, Al-Samadi A, Sperandio M, Soares
AB, Teixeira LN, Martinez EF, Demasi APD, Araújo VC, Leivo I, Salo
T, et al: miR-455-3p, miR-150 and miR-375 are aberrantly expressed
in salivary gland adenoid cystic carcinoma and polymorphous
adenocarcinoma. J Oral Pathol Med. 48:840–845. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun L, Liu B, Lin Z, Yao Y, Chen Y, Li Y,
Chen J, Yu D, Tang Z, Wang B, et al: miR-320a acts as a prognostic
factor and Inhibits metastasis of salivary adenoid cystic carcinoma
by targeting ITGB3. Mol Cancer. 14:962015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Y, Ye J, Jiao J, Zhang J, Lu Y,
Zhang L, Wan D, Duan L, Wu Y and Zhang B: Down-regulation of
miR-125a-5p is associated with salivary adenoid cystic carcinoma
progression via targeting p38/JNK/ERK signal pathway. Am J Transl
Res. 9:1101–1113. 2017.PubMed/NCBI
|
|
17
|
Chen HY, Lin YM, Chung HC, Lang YD, Lin
CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al: miR-103/107
promote metastasis of colorectal cancer by targeting the metastasis
suppressors DAPK and KLF4. Cancer Res. 72:3631–3641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng J, Liu Y, Qiao Y, Zhang L and Lu S:
miR-103 Promotes Proliferation and Metastasis by Targeting KLF4 in
Gastric Cancer. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
19
|
Wang L, Liu Y and Song J: MicroRNA-103
suppresses glioma cell proliferation and invasion by targeting the
brain-derived neurotrophic factor. Mol Med Rep. 17:4083–4089.
2018.
|
|
20
|
Yang D, Wang JJ, Li JS and Xu QY: miR-103
Functions as a Tumor Suppressor by Directly Targeting Programmed
Cell Death 10 in NSCLC. Oncol Res. 26:519–528. 2018. View Article : Google Scholar
|
|
21
|
Chen Y, Kamili A, Hardy JR, Groblewski GE,
Khanna KK and Byrne JA: Tumor protein D52 represents a negative
regulator of ATM protein levels. Cell Cycle. 12:3083–3097. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wilson SH, Bailey AM, Nourse CR, Mattei MG
and Byrne JA: Identification of MAL2, a novel member of the mal
proteolipid family, though interactions with TPD52-like proteins in
the yeast two-hybrid system. Genomics. 76:81–88. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rubin MA, Varambally S, Beroukhim R,
Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M,
Kuefer R, Fletcher JA, et al: Overexpression, amplification, and
androgen regulation of TPD52 in prostate cancer. Cancer Res.
64:3814–3822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Balleine RL, Fejzo MS, Sathasivam P,
Basset P, Clarke CL and Byrne JA: The hD52 (TPD52) gene is a
candidate target gene for events resulting in increased 8q21 copy
number in human breast carcinoma. Genes Chromosomes Cancer.
29:48–57. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tennstedt P, Bölch C, Strobel G, Minner S,
Burkhardt L, Grob T, Masser S, Sauter G, Schlomm T and Simon R:
Patterns of TPD52 overexpression in multiple human solid tumor
types analyzed by quantitative PCR. Int J Oncol. 44:609–615. 2014.
View Article : Google Scholar
|
|
26
|
Kumamoto T, Seki N, Mataki H, Mizuno K,
Kamikawaji K, Samukawa T, Koshizuka K, Goto Y and Inoue H:
Regulation of TPD52 by antitumor microRNA-218 suppresses cancer
cell migration and invasion in lung squamous cell carcinoma. Int J
Oncol. 49:1870–1880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Byrne JA, Frost S, Chen Y and Bright RK:
Tumor protein D52 (TPD52) and cancer-oncogene understudy or
understudied oncogene? Tumour Biol. 35:7369–7382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Chen CL, Pan QZ, Wu YY, Zhao JJ,
Jiang SS, Chao J, Zhang XF, Zhang HX, Zhou ZQ, et al: Decreased
TPD52 expression is associated with poor prognosis in primary
hepato-cellular carcinoma. Oncotarget. 7:6323–6334. 2016.
View Article : Google Scholar
|
|
29
|
Zhao Z, Liu H, Hou J, Li T, Du X, Zhao X,
Xu W, Xu W and Chang J: Tumor Protein D52 (TPD52) Inhibits Growth
and Metastasis in Renal Cell Carcinoma Cells Through the PI3K/Akt
Signaling Pathway. Oncol Res. 25:773–779. 2017. View Article : Google Scholar
|
|
30
|
Chen H, Xu H, Meng YG, Zhang Y, Chen JY
and Wei XN: miR-139-5p regulates proliferation, apoptosis, and cell
cycle of uterine leiomyoma cells by targeting TPD52. OncoTargets
Ther. 9:6151–6160. 2016. View Article : Google Scholar
|
|
31
|
Wu Y, Huang J, Xu H and Gong Z:
Over-expression of miR-15a-3p enhances the radiosensitivity of
cervical cancer by targeting tumor protein D52. Biomed
Pharmacother. 105:1325–1334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li SL: Establishment of a human cancer
cell line from adenoid cystic carcinoma of the minor salivary
gland. Zhonghua Kou Qiang Yi Xue Za Zhi. 1990.In Chinese.
|
|
33
|
Dong L, Wang YX, Li SL, Yu GY, Gan YH, Li
D and Wang CY: TGF-beta1 promotes migration and invasion of
salivary adenoid cystic carcinoma. J Dent Res. 90:804–809. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang W-W, Yang L-Q, Zhao F, Chen CW, Xu
LH, Fu J, Li SL and Ge XY: Epiregulin Promotes Lung Metastasis of
Salivary Adenoid Cystic Carcinoma. Theranostics. 7:3700–3714. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu LH, Zhao F, Yang WW, Chen CW, Du ZH, Fu
M, Ge XY and Li SL: MYB promotes the growth and metastasis of
salivary adenoid cystic carcinoma. Int J Oncol. 54:1579–1590.
2019.PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
37
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu Y, Gu J, Li Y, Peng C, Shi M, Wang X,
Wei G, Ge O, Wang D, Zhang B, et al: miR-17-5p enhances pancreatic
cancer proliferation by altering cell cycle profiles via disruption
of RBL2/E2F4-repressing complexes. Cancer Lett. 412:59–68. 2018.
View Article : Google Scholar
|
|
39
|
Zhang X, Wang S, Wang H, et al: Circular
RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric
cancer progression via the AKT1/mTOR pathway. Mol Cancer.
18:202019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Andreasen S, Tan Q, Agander TK, Hansen
TVO, Steiner P, Bjørndal K, Høgdall E, Larsen SR, Erentaite D,
Olsen CH, et al: MicroRNA dysregulation in adenoid cystic carcinoma
of the salivary gland in relation to prognosis and gene fusion
status: A cohort study. Virchows Arch. 473:329–340. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He Q, Zhou X, Li S, Jin Y, Chen Z, Chen D,
Cai Y, Liu Z, Zhao T and Wang A: MicroRNA-181a suppresses salivary
adenoid cystic carcinoma metastasis by targeting MAPK-Snai2
pathway. Biochim Biophys Acta. 1830:5258–5266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Natarelli L, Geissler C, Csaba G, Wei Y,
Zhu M, di Francesco A, Hartmann P, Zimmer R and Schober A: miR-103
promotes endo-thelial maladaptation by targeting lncWDR59. Nat
Commun. 9:26452018. View Article : Google Scholar
|
|
43
|
Wu L, Fan J and Belasco JG: MicroRNAs
direct rapid dead-enylation of mRNA. Proc Natl Acad Sci USA.
103:4034–4039. 2006. View Article : Google Scholar
|
|
44
|
Ummanni R, Teller S, Junker H, Zimmermann
U, Venz S, Scharf C, Giebel J and Walther R: Altered expression of
tumor protein D52 regulates apoptosis and migration of prostate
cancer cells. FEBS J. 275:5703–5713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Croset M, Pantano F, Kan CWS, Bonnelye E,
Descotes F, Alix-Panabières C, Lecellier CH, Bachelier R, Allioli
N, Hong SS, et al: miRNA-30 Family Members Inhibit Breast Cancer
Invasion, Osteomimicry, and Bone Destruction by Directly Targeting
Multiple Bone Metastasis-Associated Genes. Cancer Res.
78:5259–5273. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu Z, Li Z, Wang W, Xia Y, He Z, Li B,
Wang S, Huang X, Sun G, Xu J, et al: MIR-1265 regulates cellular
proliferation and apoptosis by targeting calcium binding protein 39
in gastric cancer and, thereby, impairing oncogenic autophagy.
Cancer Lett. 449:226–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dai J, Wang J, Yang L, Xiao Y and Ruan Q:
miR-125a regulates angiogenesis of gastric cancer by targeting
vascular endothelial growth factor A. Int J Oncol. 47:1801–1810.
2015. View Article : Google Scholar : PubMed/NCBI
|