Introduction
Bladder cancer (BCa) is the tenth most commonly
diagnosed cancer worldwide, with a four-fold higher incidence in
men compared to women. Globally, it is the sixth most common cancer
in men and ranks ninth as the cause of cancer-related deaths
(1). It is rarely diagnosed in
individuals younger than 40 years, and occupational exposure to
carcinogens is a major risk factor (2,3).
Although more common in USA than in Asia, the incidence and
mortality rates of BCa have increased significantly in China in
recent years (4). Despite
therapeutic interventions such as surgery, chemotherapy and
radiotherapy, the 5-year survival of BCa patients is still low,
mainly due to the high recurrence rate (5,6).
Therefore, it is essential to identify novel biomarkers of BCa, in
order to improve patient prognosis and clinical outcomes.
Non-coding RNAs (ncRNAs) are functional transcripts
that are not translated to proteins and modulate gene expression at
the transcriptional and post-transcriptional level (7). They are frequently dysregulated in
cancer, indicating a prognostic relevance (8). CircRNAs are a class of covalently
closed endogenous ncRNAs (9) that
are differentially expressed in various types of cancers, such as
liver carcinoma, stomach carcinoma and colorectal carcinoma, and
correlated with tumor development and progression (10,11).
Although the exact function of circRNAs is poorly understood, there
is evidence indicating that they act as 'sponges' of the microRNAs
(miRNAs) through competitive binding and abrogate the effect of the
latter on mRNA translation (12).
In addition, circRNAs with the same microRNA response elements
(MREs) act as competing endogenous RNAs (ceRNAs) and form a
regulatory network. Recent studies have unearthed several ceRNA
networks that are dysregulated during cancer (12-14).
However, the exact number of circRNAs that act as miRNA sponges is
still unknown (15). MicroRNAs are
a category of endogenous short non-coding RNAs that can bind to the
3′-untranslated region (3′-UTR) of mRNAs and inhibit their
translation (16). miRNA
dysregulation has been implicated in the initiation and progression
of various cancer types through multiple mechanisms (17,18).
In the present study, we discovered a novel ceRNA network
comprising circRNA_0071196, CIT and miRNA-19b-3p in BCa tissues.
Based on molecular and bioinformatics analyses, we surmised that
circRNA_0071196 upregulates CIT in BCa cells by competitively
binding to miRNA-19b-3p. Furthermore, knockdown of CIT inhibited
the proliferation and migration of BCa cells in vitro. Taken
together, this novel regulatory axis is essential for BCa
progression, and is a potential diagnostic marker and therapeutic
target.
Materials and methods
Patients and samples
Bladder carcinoma (n=80) and para-carcinoma tissues
(n=30) were resected from BCa patients who underwent surgery at the
Department of Urology, The First Affiliated Hospital of Harbin
Medical University, China from September 2017 to September 2018.
The mean age of the patients was 63.63 years (age range, 41-80) and
the sex distribution was 25 females and 55 males. The tumor tissues
were obtained by transurethral resection and the para-carcinoma
tissues were acquired at a distance of >2.0 cm from the tumor
margin, and cut into 0.8-1.2 cm pieces. All tissues were
snap-frozen in liquid nitrogen and independently identified by two
pathologists. The study was approved by the Ethics Committee of The
First Affiliated Hospital of Harbin Medical University, and written
informed consent was obtained from all patients.
RT-qPCR
Total RNA was extracted from 20 pairs of BCa samples
and the transduced 5637 cells, and reverse transcribed to cDNA
using the SuperScript™III Reverse Transcriptase kit
(Invitrogen/Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The thermocycling conditions of
RT-qPCR were 95°C for 10 min, followed by 40 cycles at 95°C for 30
sec, and 60°C for 60 sec. The relative expression levels of the
relevant transcripts were analyzed by RT-qPCR, and the primer
sequences are listed in Table I.
GAPDH was used as the internal control for circRNAs and mRNAs, and
U6 for miRNAs. The 2−ΔΔCq (19) method was used to quantify these
transcripts, and assays were performed in triplicates.
 | Table IRT-qPCR primer sequences. |
Table I
RT-qPCR primer sequences.
| Primer name | Primer FW
(5′-3′) | Primer RV
(5′-3′) |
|---|
|
hsa_circRNA_103758 |
GTGGCCGAGGACTTTGATTG |
CCTGTAACAACGCATCTCATATT3 |
|
hsa_circRNA_404289 |
CCTTTTTCTTCTTTCTTTCTGG |
TGGAGACTTTACTCTTACCCGT |
|
hsa_circRNA_000758 |
GGTATTAGGGACACTGGTGG |
CTTCTGGCCTTTTGGTTACT3 |
|
hsa_circRNA_0006473 |
TCAGGTACTCCCGTCGC |
CGTTACTCCACCTGGACC |
| miR-19b-3p |
GTGCAGGGTCCGAGGT |
TGTGCAAATCCATGCAAAACTGA |
| CIT |
CAGGCAAGATTGAGAACG |
GCACGATTGAGACAGGGA |
| CDK1 |
TGGATCTGAAGAAATACTTGGATTCTA |
CAATCCCCTGTAGGATTTGG |
| CCND1 |
CTAAGATGAAGGAGACCATCCC |
AAGGTCTGCGCGTGTTTGCGGAT |
| CCNB1 |
TCCAGTTATGCAGCACCTGGCTA |
TGCCACAGCCTTGGCTAAATCTT |
| CCNB2 |
TGGAAAAGTTGGCTCCAAAG |
CTTCCTTCATGGAGAGACATCCTC |
| P53 |
TGCTCTTTTCACCCATCTAC |
ATACGGCCAGGCATTGAAGT |
| MLC2 |
ACCATTCTCAACGCATTCAA |
CATCTGGTCAACCTCCTCCT3 |
| MDM2 |
ACCTCACAGATTCCAGCTTCG |
TTTCATAGTATAAGTGTCTTTTT |
| ROCK1 |
AACATGCTGCTGGATAAATCTGG |
TGTATCACATCGTACCATGCCT |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| GAPDH |
TGACTTCAACAGCGACACCCA |
CACCCTGTTGCTGTAGCCAAA |
Microarray analysis
Four pairs of carcinoma and para-carcinoma tissues
were collected from BCa patients at our hospital in 2017 for
microarray analysis. For circRNA hybridization, the latter were
first enriched by degrading the linear RNAs with Rnase R, amplified
with random primers, and transcribed into fluorescent cRNA
(Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs
were hybridized onto the Arraystar Human circRNA Array V2 (8×15K,
Arraystar), and the arrays were scanned using the Agilent Scanner
G2505C. The significant differentially expressed genes (DEGs) and
circRNAs (DEcircRNAs) were identified based on fold-change ≥1.5 and
P<0.05.
RNA sequencing array
Total RNA was extracted from four pairs of frozen
carcinoma and para-carcinoma tissues using TRIzol
(Invitrogen/Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. After quantitative analysis and
quality inspection, RNA-Seq libraries were synthesized using the
KAPA Stranded RNA-Seq Library Prep Kit (Illumina, USA) as
previously described (20). which
included RNA fragmentation, random hexamerprimed first-strand cDNA
synthesis, dUTP-based second-strand cDNA synthesis, end-repairing,
A-tailing, adaptor ligation and library PCR amplification. The
libraries were assessed with an Agilent 2100 Bioanalyzer (Illumina,
USA), quantified absolutely by qPCR, and sequenced using an
Illumina HiSeq 4000 Sequencing System for 150 cycles.
The raw data was uploaded to Gene Expression Omnibus
(GEO) (GSE147985, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147985).
GO and KEGG pathway analyses
Gene Ontology (GO) analysis (http://www.geneontology.org) was performed on the
DEcircRNAs and DEGs, and the latter were functionally annotated on
the basis of biological processes (BP), cellular components (CC)
and molecular functions (MF). GO terms with P-values <0.05 were
considered as enriched. The Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analysis (http://www.genome.jp/kegg/) was preformed to identify
the significant pathways related to the relevant genes, using
P-value ≤0.05 as the threshold.
ceRNA network analysis
Following validation of the DEGs and DEcircRNAs with
real time qPCR, those with the same MREs were selected for
constructing the circRNA/miRNA/mRNA network. The circRNA-miRNA
interactions were predicted by biological algorithms of miRanda
(http://www.microrna.org/microrna/home.do) and
Targetscan (http://www.targetscan.org/). The miRNA-mRNA
interactions were predicted by miRDB (http://www.mirdb.org/). The circRNA/miRNA/mRNA network
was then mapped using Cytoscape v 2.8.3 (21).
Cell lines and lentiviral infection
The human bladder cancer cell line 5637 was obtained
from the Cell Resource Center, Shanghai Institute for Biological
Sciences at the Chinese Academy of Sciences, and maintained at 37°C
under 5% CO2 in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS), 100 U/ml penicillin and 100 U/ml streptomycin. The
lentiviral CIT and control shRNA vectors were designed and
synthesized by Shanghai GeneChem Co., Ltd. (Shanghai, China), and
the sequences were as follows: CIT, 5′-GCGTC CTCATACCAGGATAAA-3′
and control, 5′-TTCTCCGA ACGTGTCACGT-3′. The lentiviral
circRNA-0071196 and control shRNA vectors were also designed and
synthesized by Shanghai GeneChem Co. Ltd. (Shanghai, China), and
the sequences were: circRNA-0071196, 5′-AAGGAACAAGC AGTAGATCAT-3′
and control, 5′-TTCTCCGAACGTGT CACGT-3′. The miR-19b-3p inhibitor
was synthesized by GenePharma Co. (GenePharma, Shanghai, China).
The plas-m ids were transfected into 293T cells using
Lipofectamine® 2000 (Invitrogen/Thermo Fisher
Scientific, Inc.), and the viral supernatants were collected after
48 h, centrifuged, and filtered through a 0.45-µm
polyvinylidene fluoride membrane. The 5673 cells were incubated
with 50% diluted viral supernatant in complete medium for 48 h, and
then selected with 2 ng/ml puromycin (Thermo Fisher Scientific,
Inc.) in fresh medium. Transduction efficiency was monitored under
a fluorescence microscope.
Luciferase reporter assay
The psiCHECK2-circRNA- 0071196-WT, (Promega Corp.)
and psiCHECK2-circRNA- 0071196-Mut luciferase reporter plasmids
were constructed by respectively cloning the wild-type (WT) and
miR-19b-3p binding site-mutated (MUT) circRNA-0071196 sequence. The
psiCHECK2-circRNA-404289-WT, and psiCHECK2-circRNA-404289-Mut
luciferase reporter plasmids were constructed by respectively
cloning the wild-type (WT) and hsa-miR-370-3p binding site-mutated
(MUT) circRNA-404289 sequence. The psiCHECK2-circRNA-000758-WT, and
psiCHECK2-circRNA-000758-Mut luciferase reporter plasmids were
constructed by respectively cloning the wild-type (WT) and
hsa-miR-6808-3p binding site-mutated (MUT) circRNA-000758 sequence.
The psiCHECK2-circRNA-006473-WT, and psiCHECK2-circRNA-006473-Mut
luciferase reporter plasmids were constructed by respectively
cloning the wild-type (WT) and hsa-miR-6808-3p binding site-mutated
(MUT) circRNA-006473 sequence. The 5637 cells were seeded into
24-well plates and cultured for 24 h, and co-transfected with the
WT/MUT of the above plasmids and miRNA mimics or the negative
control using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.).
After culturing for 48 h, firefly and Renilla luciferase activities
were quantified using dual luciferase reporter assays (Promega
Corp.) according to the manufacturer's instructions.
Immunohistochemistry (IHC)
IHC was performed on 4 pairs of BCa and normal
bladder tissues using the SABC kit (Suolaibao) according to the
manufacturer's instructions. The sections were incubated with the
primary anti-CIT antibody (dilution 1:100; cat. no. ab110897,
Abcam) at 37°C for 1 h, followed by sequential incubation with
biotinylated secondary antibody (dilution 1:1,000; cat. no. ab6721,
Abcam) and strep-tavidin peroxidase for 20 min each at room
temperature. After rinsing the slides, the sections were developed
using diaminobenzidine and counterstained with hematoxylin. The
stained sections were observed under a light microscope (Olympus
Corp., Japan) at ×200 magnification. The staining intensity was
graded as follows: 0, negative; 1, low; and 2, high. The percentage
of stained cells was scored as: none stained, 0; 1-33%, 1; 34-66%,
2; and ≥67%, 3. Both scored values were multiplied to obtained the
total CIT score, and samples with CIT ≤1 were considered negative
and those with CIT ≥3 as positive.
Western blot analysis
Total proteins were extracted from the transduced
5637 cells, and western blot analysis was performed as previously
described (22). The primary
antibodies included anti-CIT (dilution 1:5,000) (cat. no. ab86782,
Abcam), anti-MLC2 (dilution 1:2,000) (cat. no. ab92721, Abcam),
anti-P53 (dilution 1:500) (cat. no. AF6073, Affinity Biosciences),
anti-ERK1/2 (dilution 1:1,000) (cat. no. BF8004, Affinity
Biosciences), anti-p-ERK1/2 (dilution 1:1,000) (cat. no. AF1015,
Affinity Biosciences), anti-CDK1 (dilution 1:1,000) (cat. no.
DF6024, Affinity Biosciences), anti-MDM2 (dilution 1:1,000) (cat.
no. AF6376, Affinity Biosciences), anti-CCND1 (dilution 1:1,000)
(cat. no. AF0931, Affinity Biosciences), anti-ROCK1 (dilution
1:1,000) (cat. no. AF7016, Affinity Biosciences) and anti-β-actin
(dilution 1:15,000) (cat. no. AF7018; Affinity Biosciences). Goat
anti-rabbit IgG (dilution 1:2,000) (cat. no. BA1054; Wuhan Boster
Biological Technology) was used as the secondary antibody, and the
ECL-Plus kit was used to detect protein bands (Amersham
Biosciences, USA) as described previously (23). Imaging J V1.53b (National Institute
of Health, Bethesda, MD, USA) was used to quantitate the
protein.
In vitro assays
Proliferation of the transduced 5637 cells was
analyzed using the Cell Counting Kit-8 (CCK-8; cat. no. AR1160-500;
Wuhan Boster Biological Technology, Ltd.) according to the
manufacturer's instructions. Briefly, the cells were seeded in
96-well plates at the density of 1.5×103 cells/well, and
cultured for 0, 24, 48, 72 and 96 h. After adding 10 µl
CCK-8 solution per well, the cells were incubated in the dark for 1
h, and the absorbance was measured at 450 nm using Biotek Elx800
spectrometer. The experiment was performed in triplicates. For the
migration assay, the cells were seeded onto the upper wells of
Transwell chambers (Corning Inc.) at the density of
1×104 cells/well in serum-free 1640, and the lower wells
were filled with complete 1640 medium. After a 24 h incubation at
37°C, the migrated cells were stained with crystal violet for 30
min at room temperature, and counted under an inverted microscope
at ×50 magnification in at least three random fields. For the
colony formation assay, the transduced cells were seeded in a
24-well plate at the density of 2×102 cells/well in
complete 1640 medium, and cultured for 12 days. The cells were then
fixed and stained with 1% crystal violet at room temperature for 30
min, and the colonies were photographed and counted using a light
microscope (magnification, ×50).
Statistical analysis
Categorical variables were compared by Fisher exact
test, and continuous variables by the Student's t-test. Data are
expressed as the means ± standard deviation. Multigroup comparisons
of the means were carried out by one-way analysis of variance
(ANOVA) test with post hoc contrasts by Student-Newman-Keuls test.
All statistical analyses were performed using SPSS 17.0 (SPSS,
Inc.). P<0.05 was considered statistically significant. All
experiments were repeated in triplicates
Results
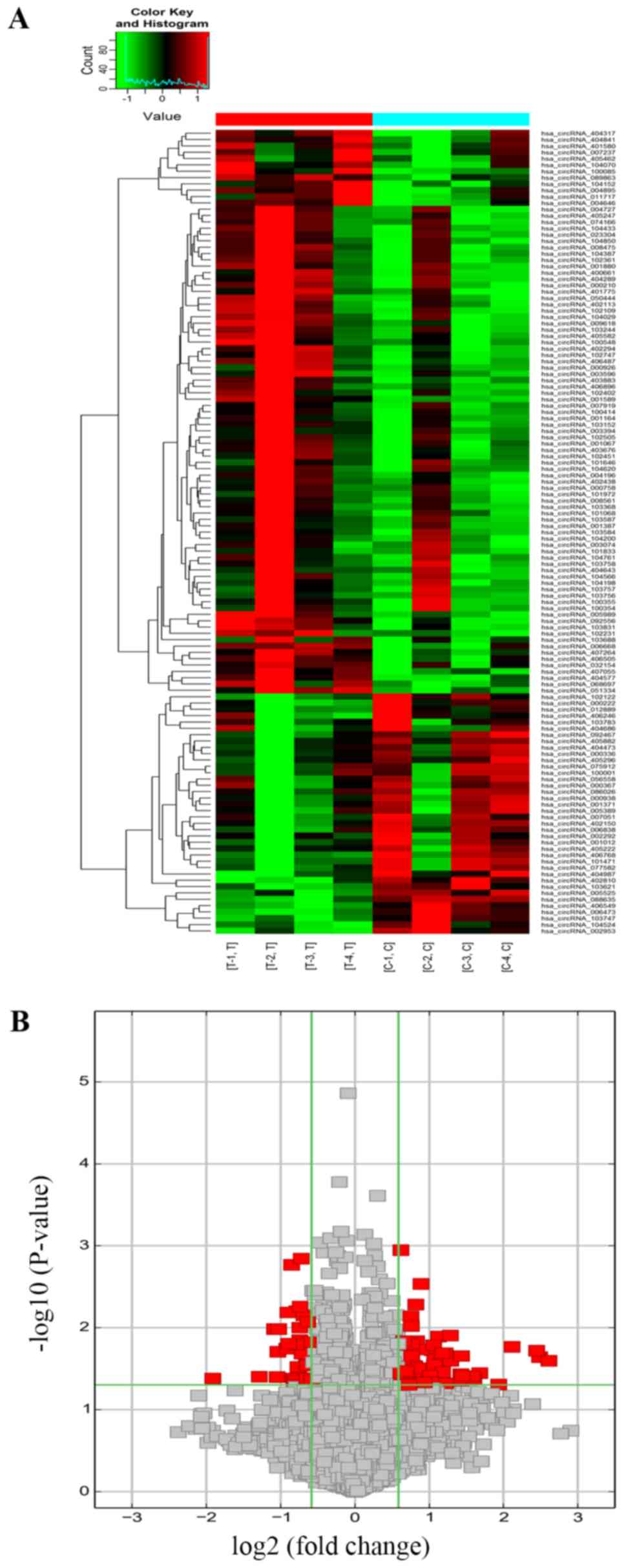
DEcircRNAs in the BCa tissues are
associated with cancer-related functions
The expression profiles of 4 pairs of BCa and normal
bladder tissues were analyzed by high throughput microarray, and
127 DEcircRNAs were detected of which 89 were upregulated and 38
were downregulated in the tumor samples (Figs. 1A and B and S1A and B). Based on their relation with
protein-coding genes, the DEcircRNAs were classified as exonic
(85.83%), intronic (8.66%), sense overlapping (4.72%) and antisense
(0.79%) (Fig. S1C). Furthermore,
the DEcircRNAs were distributed across all chromosomes except the Y
chromosome (Fig. S1D). Thus, the
circRNA profiles of BCa and normal bladder tissues were distinct.
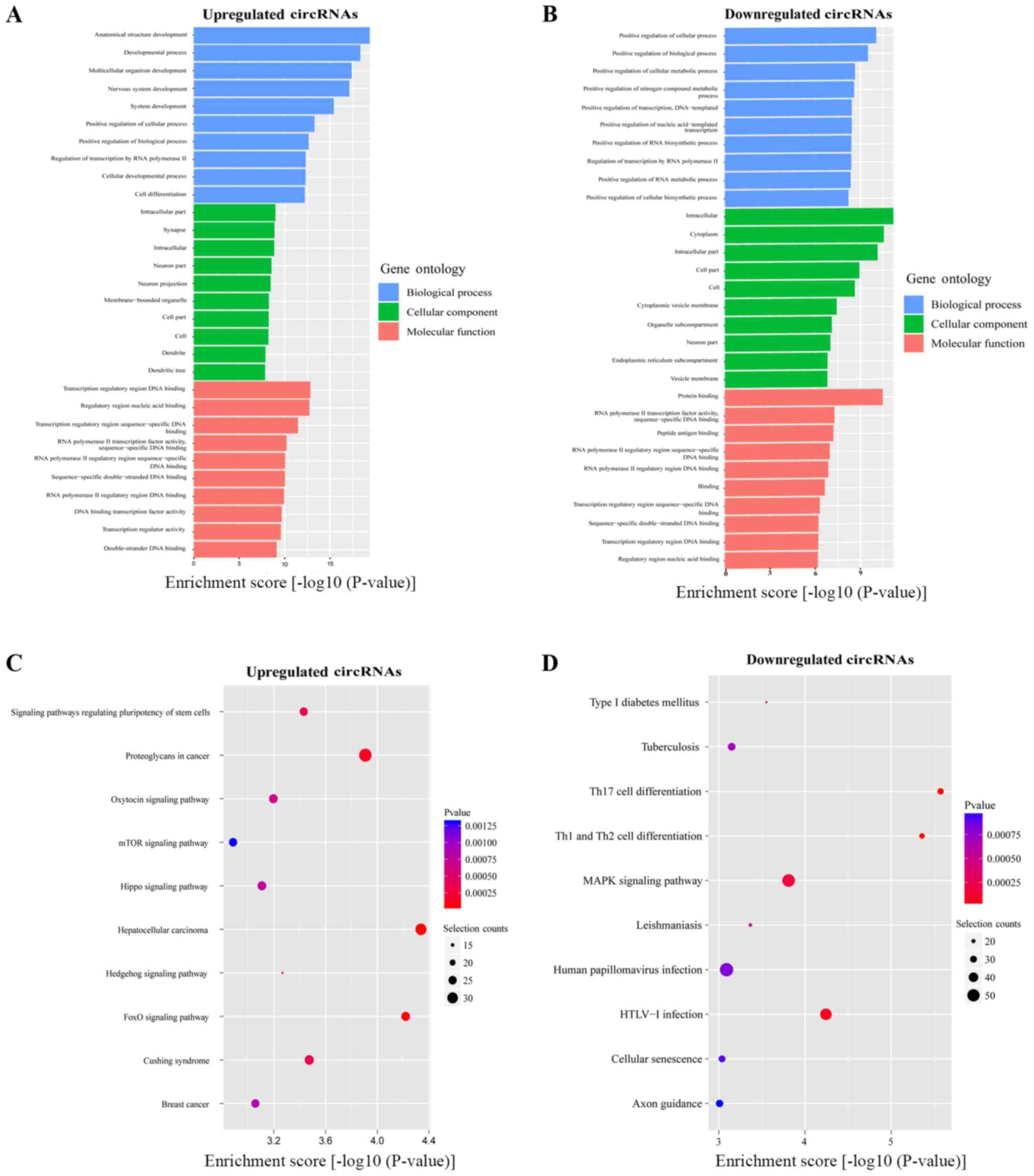
The biological relevance of these DEcircRNAs was determined by the
GO and KEGG analyses. The top 10 enriched GO terms (all domains) of
the abnormally expressed circRNAs were 'Transcription regulatory
region DNA binding', 'Regulatory region nucleic acid binding',
'Transcription regulatory region sequence-specific DNA binding',
'Intracellular part', 'Anatomical structure development', 'Protein
binding', 'Intracellular', and 'Positive regulation of cellular
process' (Fig. 2A and B). KEGG
pathway analysis revealed that the top 10 pathways associated with
upregulated (Fig. 2C) and
downregulated (Fig. 2D) circRNAs
included 'Hepatocellular carcinoma', 'FoxO signaling pathway',
'Proteoglycans in cancer', 'Th17 cell differentiation', 'Th1 and
Th2 cell differentiation' and 'MAPK signaling pathway'.
BCa and normal bladder tissues have
distinct mRNA profiles
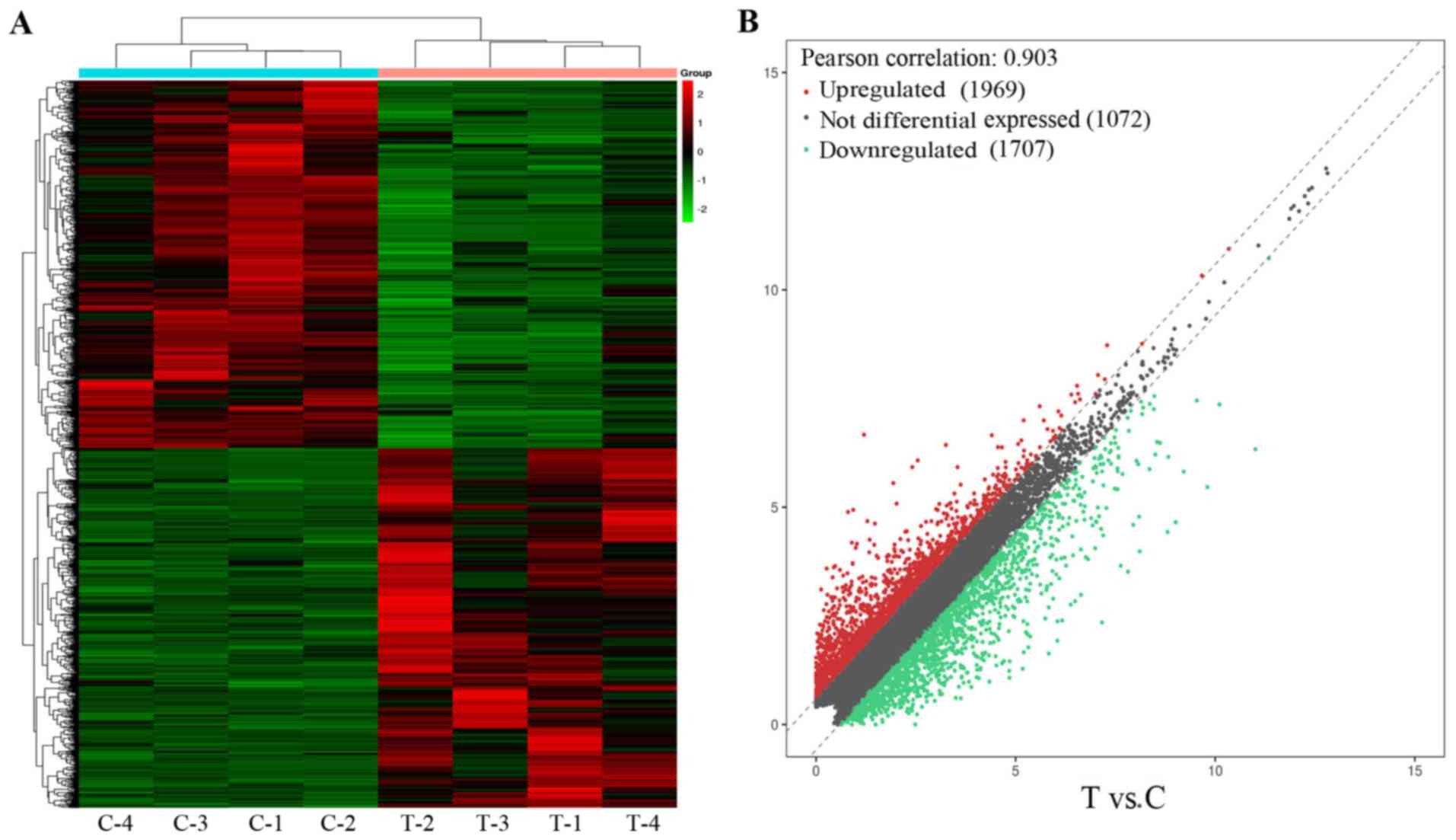
In addition to the circRNAs, the BCa and adjacent
normal tissues also differed in terms of the mRNA expression
profile. A total of 1,612 DEGs were identified, including 797
upregulated and 815 downregulated mRNAs (Figs. 3 and S2). The top 10 enriched GO terms in the
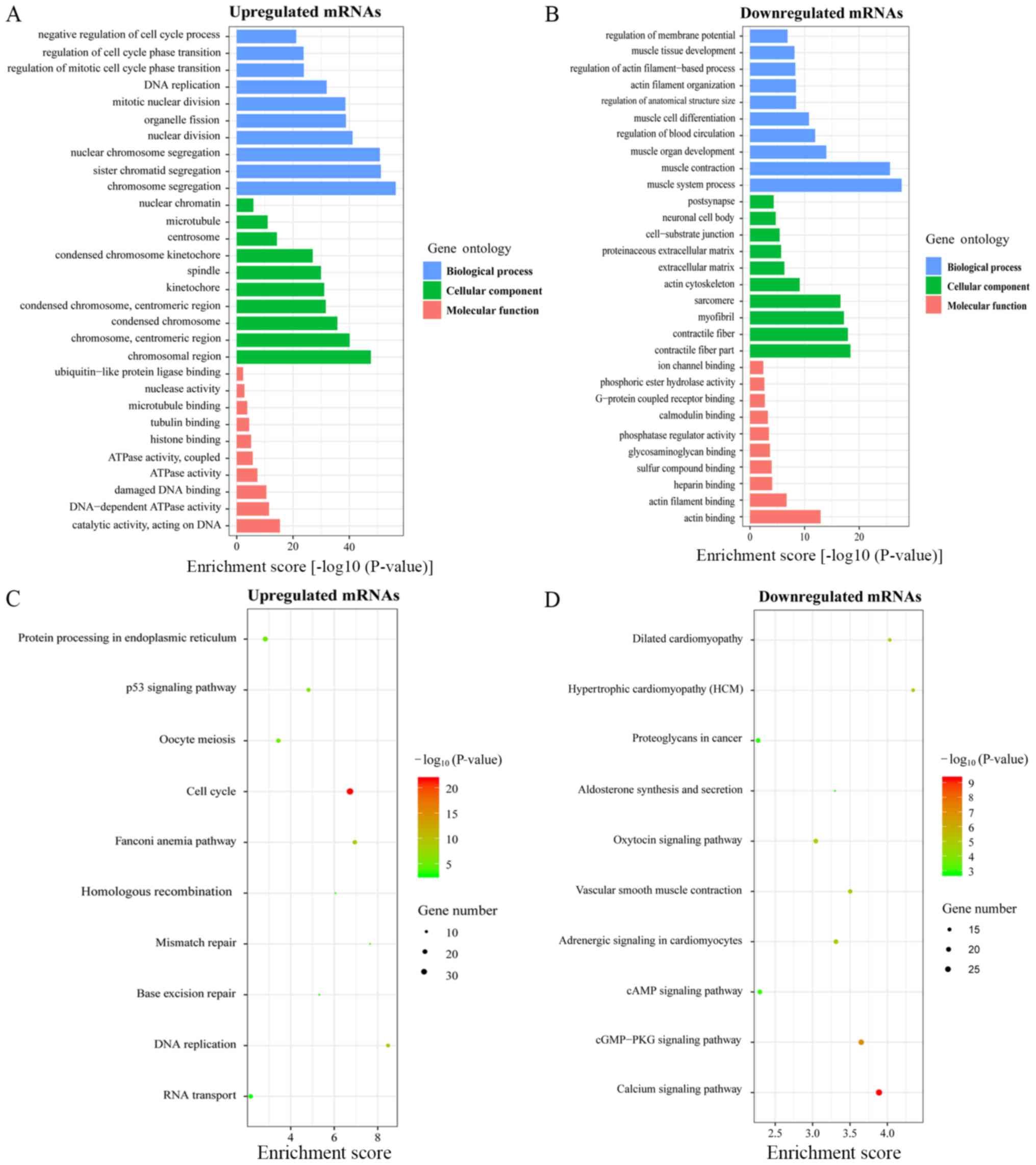
upregulated and downregulated mRNAs are respectively shown in
Fig. 4A and B. The most
significantly enriched BP, CC and MF terms among the upregulated
mRNAs were 'Chromosome segregation', 'Chromosomal region' and
'Catalytic activity on DNA', respectively, and those among the
downregulated mRNAs were 'Muscle system process', 'Contractile
fiber part' and 'Actin binding'. KEGG pathway analysis showed that
the top 10 pathways associated with the upregulated mRNAs included
'Cell cycle', 'DNA replication' and 'p53 signaling pathway'
(Fig. 4C), and those associated
with downregulated mRNAs were mainly in the 'Calcium signaling
pathway', 'cGMP-PKG signaling pathway' and 'Vascular smooth muscle
contraction' (Fig. 4D). Four
circRNAs and 5 mRNAs were selected for verifying the microarray
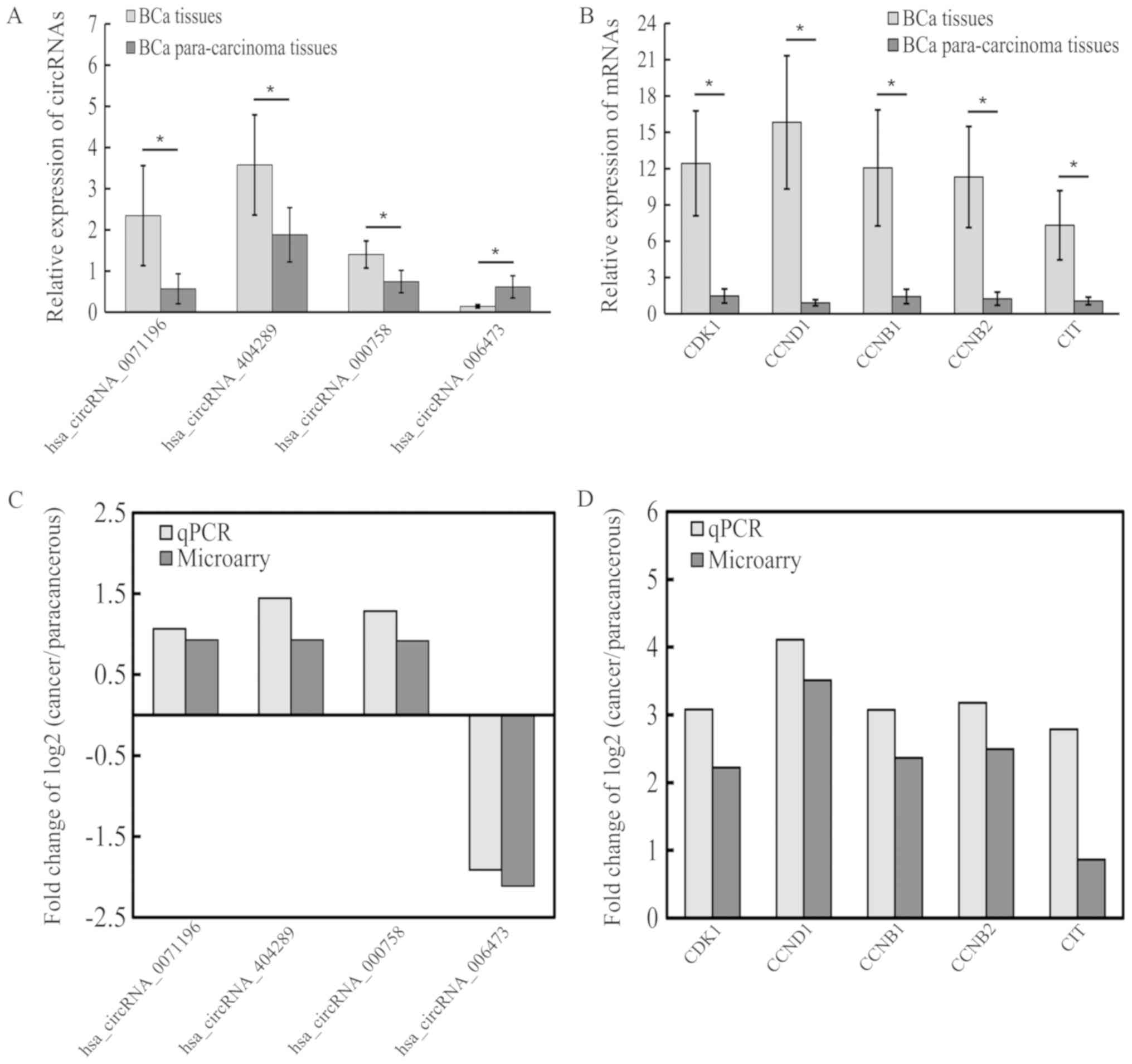
results by RT-PCR, which showed that circRNA_0071196,
circRNA_404289, circRNA_000758, cyclin dependent kinase 1 (CDK1),
cyclin D1 (CCND1), cyclin B1 (CCNB1), cyclin B2 (CCNB2) and citron
Rho-interacting serine/threonine kinase (CIT) were upregulated
(Fig. 5A and B), while
circRNA_006473 was downregulated (Fig.
5A). These results were consistent with the microarray data
(Fig. 5C and D). Compared with
other circRNAs, only circRNA_0071196 was positive in the luciferase
assay, thus we chose circRNA_0071196 to continue the following
study.
circRNA_0071196 and CIT are targets of
miR-19b-3p
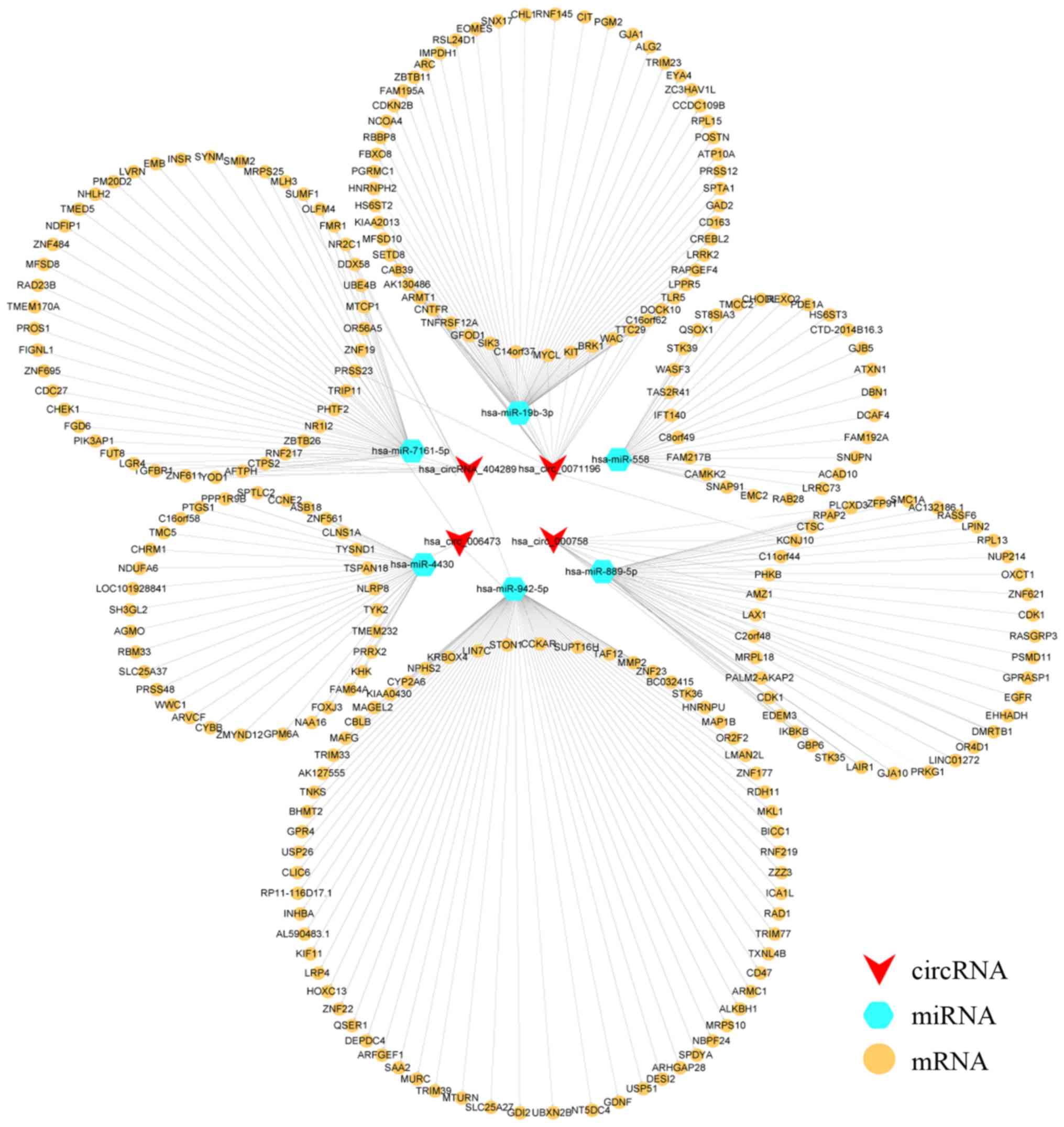
The ceRNA network was constructed using the
DEcircRNAs and DEGs that shared binding sites for MREs, which
revealed that circRNA _0071196 and CIT were the interacting
partners of miR-19b-3p (Fig. 6).
We further predicted the circRNA_0071196/CIT and miR-19b-3p/CIT
interactions using the Starbase (starbase.sysu.edu.cn/index.php) and miRanda
(http://www.microrna.org/microrna/home.do) programs,
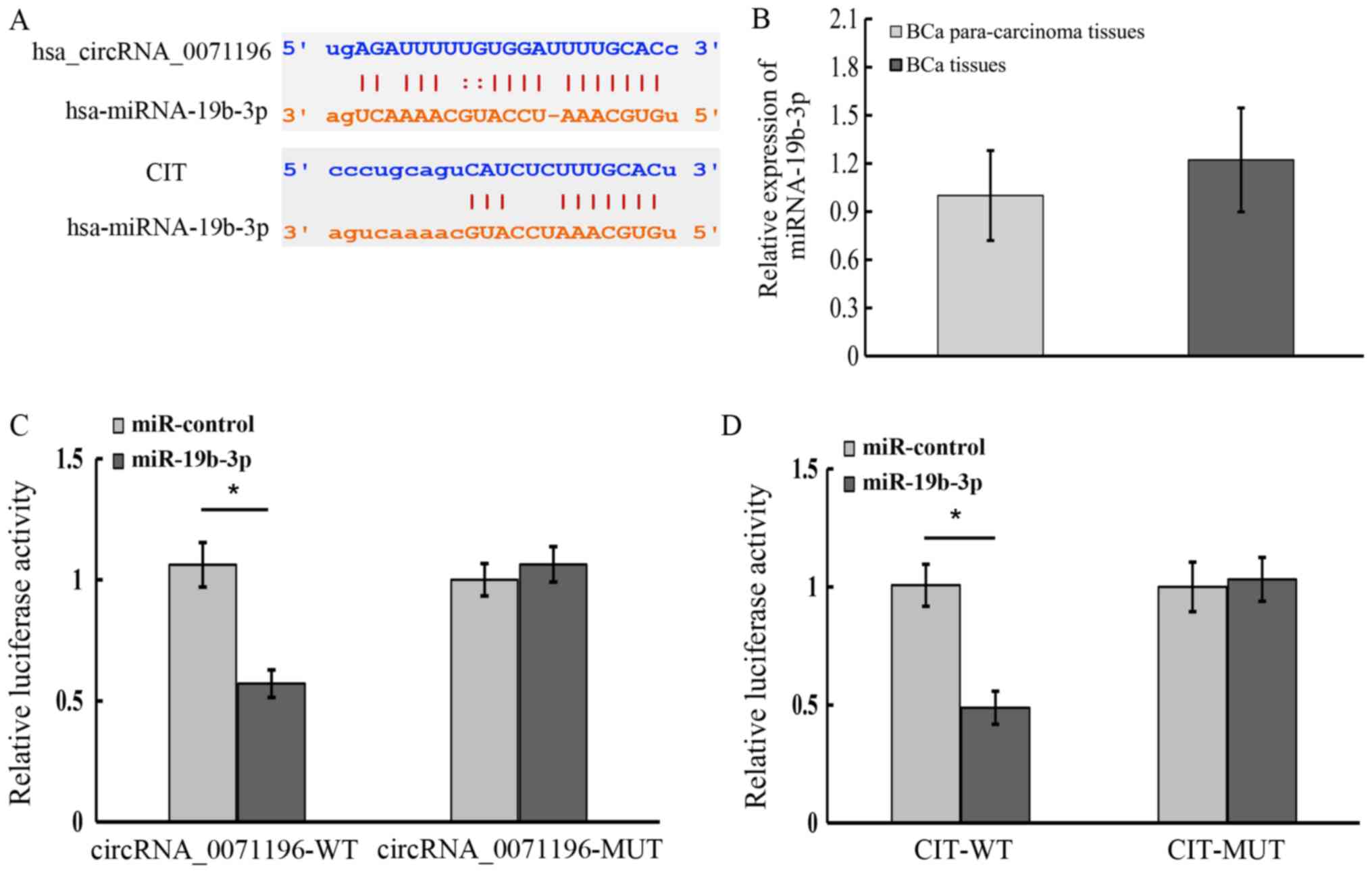
respectively. As shown in Fig. 7A,
the bases in the seed region of miR-19b-3p were complementary to
the circRNA_0071196 and CIT sequences, indicating that
circRNA_0071196 likely acts as a sponge for miR-19b-3p in BCa
cells, and that CIT is a downstream target of miR-19b-3p. To
validate our hypothesis, we analyzed the expression levels of
miR-19b-3p in BCa and adjacent normal tissues, and found that the
expression of miR-19b-3p exhibited no significant difference in the
tumor and para-carcinoma tissues (Fig.
7B). Furthermore, circRNA_0071196 WT 3′-UTR co-transfected with
miR-19b-3p showed lower luciferase activity compared to
transfection with miR-control. In contrast, no significant
difference was seen in the luciferase activity of circRNA_0071196
MT 3′-UTR in the presence of miR-19b-3p mimics or miR-control
(Fig. 7C). Similarly, CIT WT
3′-UTR showed distinctly weaker luciferase signals when
co-transfected with miR-19b-3p compared to miR-control, whereas
that of CIT WT 3′-UTR was unaffected by the miRNA (Fig. 7D). Taken together, circRNA_0071196
acts as a sponge for miR-19b-3p and CIT is the target of miR-19b-3p
in BCa cells. Because the luciferase results of other circRNAs in
Fig. 5 were negative (data not
shown), circRNA-0071196 was selected for subsequent
experiments.
circRNA_0071196 and miR-19b-3p are
correlated with BCa progression
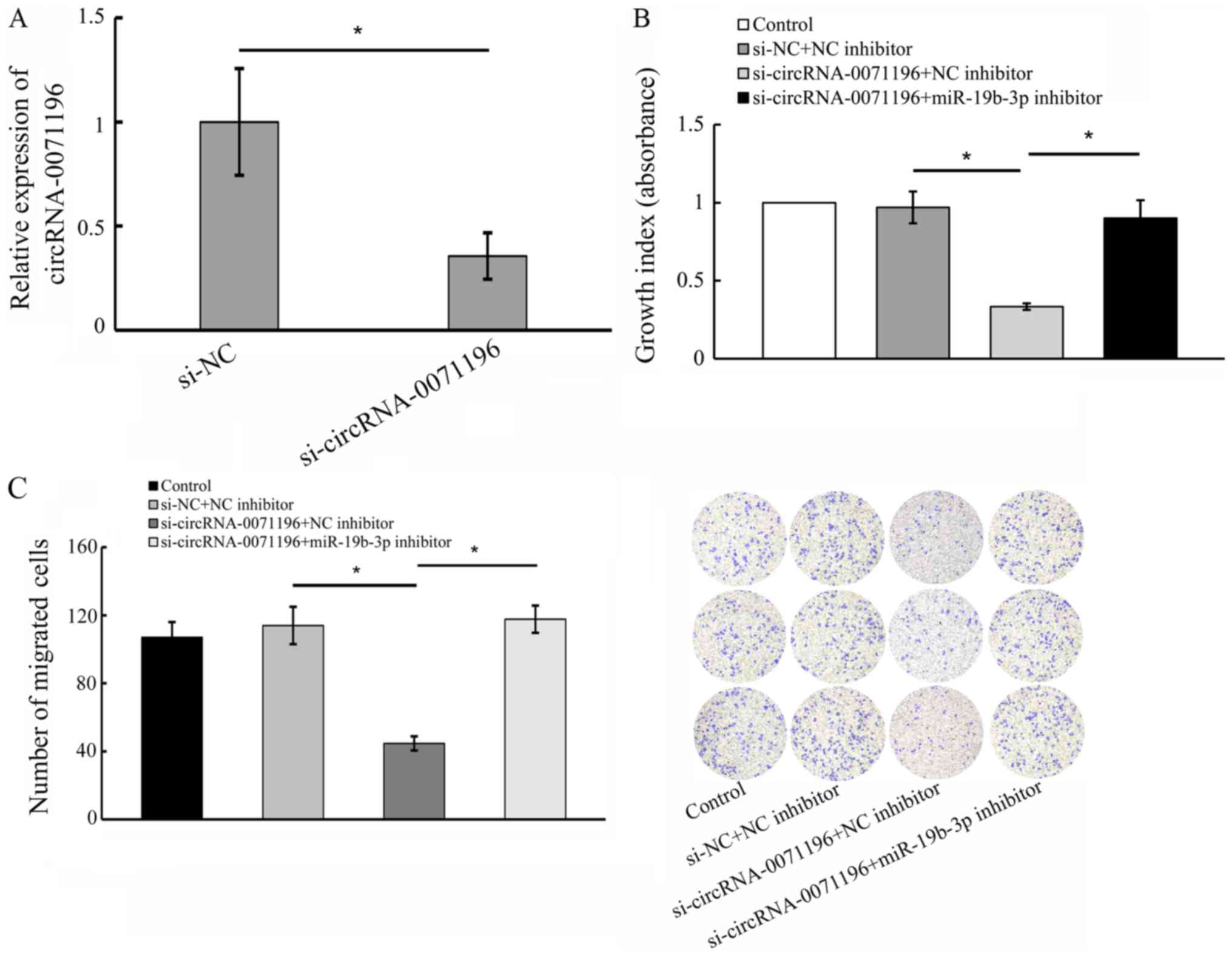
To further elucidate the biological functions of
circRNA-0071196 and miR-19b-3p in BCa, we transfected 5637 cells
with si-circRNA-0071196 and miR-19b-3p inhibitor/NC inhibitor.
qRT-PCR showed that circRNA-0071196 expression was significantly
decreased compared to siNC (Fig.
8A). As shown in Fig. 8B and
C, circRNA0071196 silencing significantly reduced the viability
and migration of BCa cells (P<0.05), which was rescued by
miR-19b-3p.
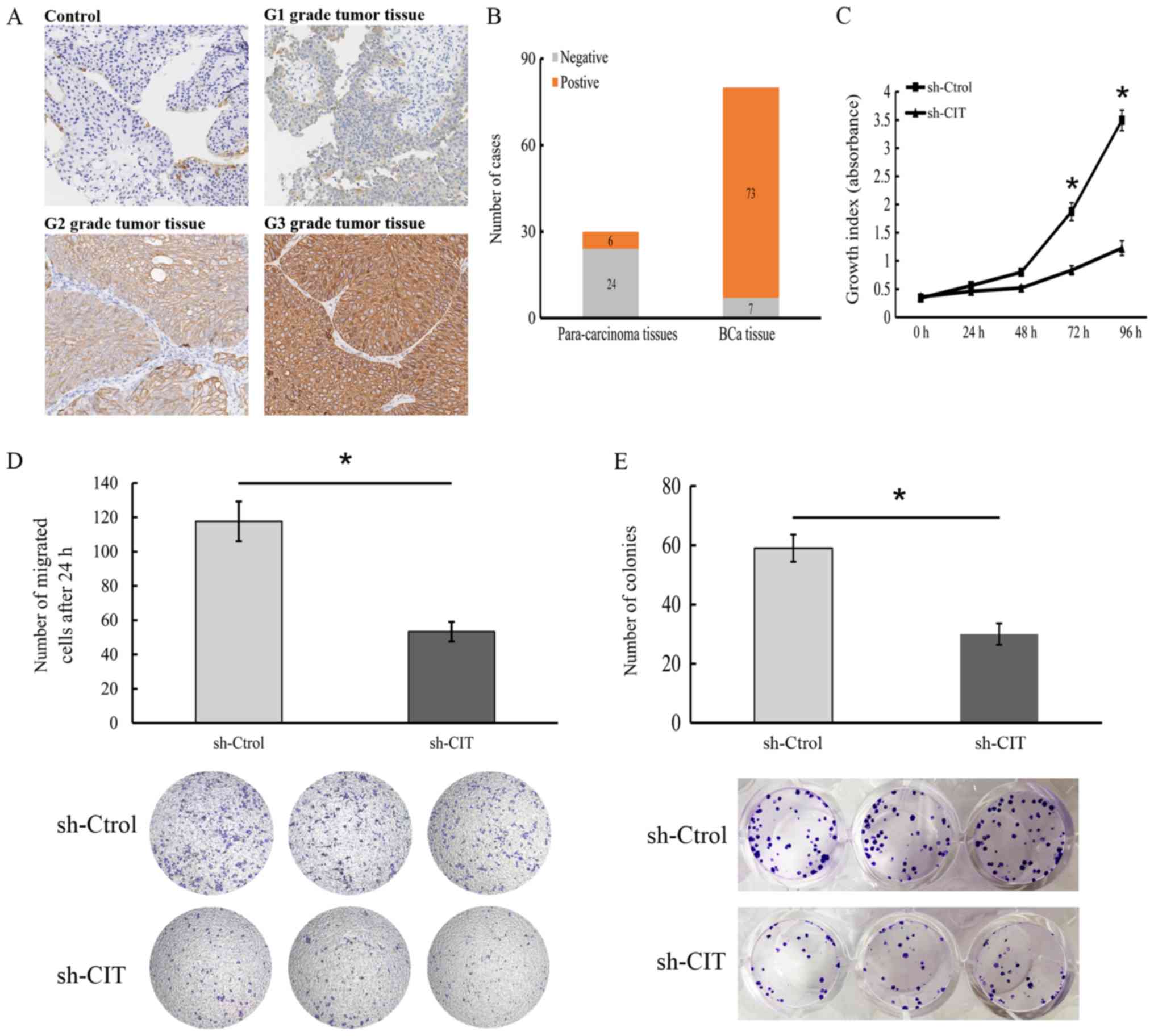
CIT is correlated with BCa
progression
To determine the relevance of CIT in BCa, we
analyzed the in situ levels in tumor and normal bladder tissues,
and found that 73 of 80 (91.3%) BCa tissues expressed this protein.
Furthermore, CIT expression was found to be significantly
correlated with metastasis (P<0.05) and histological grade
(P<0.05) but not with age, sex, infiltration and T stage
(Table II). In addition, the
intensity of CIT expression increased across grade 1 (least
aggressive), 2 (moderately aggressive) and 3 grade (most aggressive
and most likely to spread) tumors (Fig. 10A and B), indicating that the CIT
level in tumor tissues can demarcate BCa patients into different
grades. Since CIT mRNA was upregulated in BCa tissues compared to
that noted in the normal tissues (Fig.
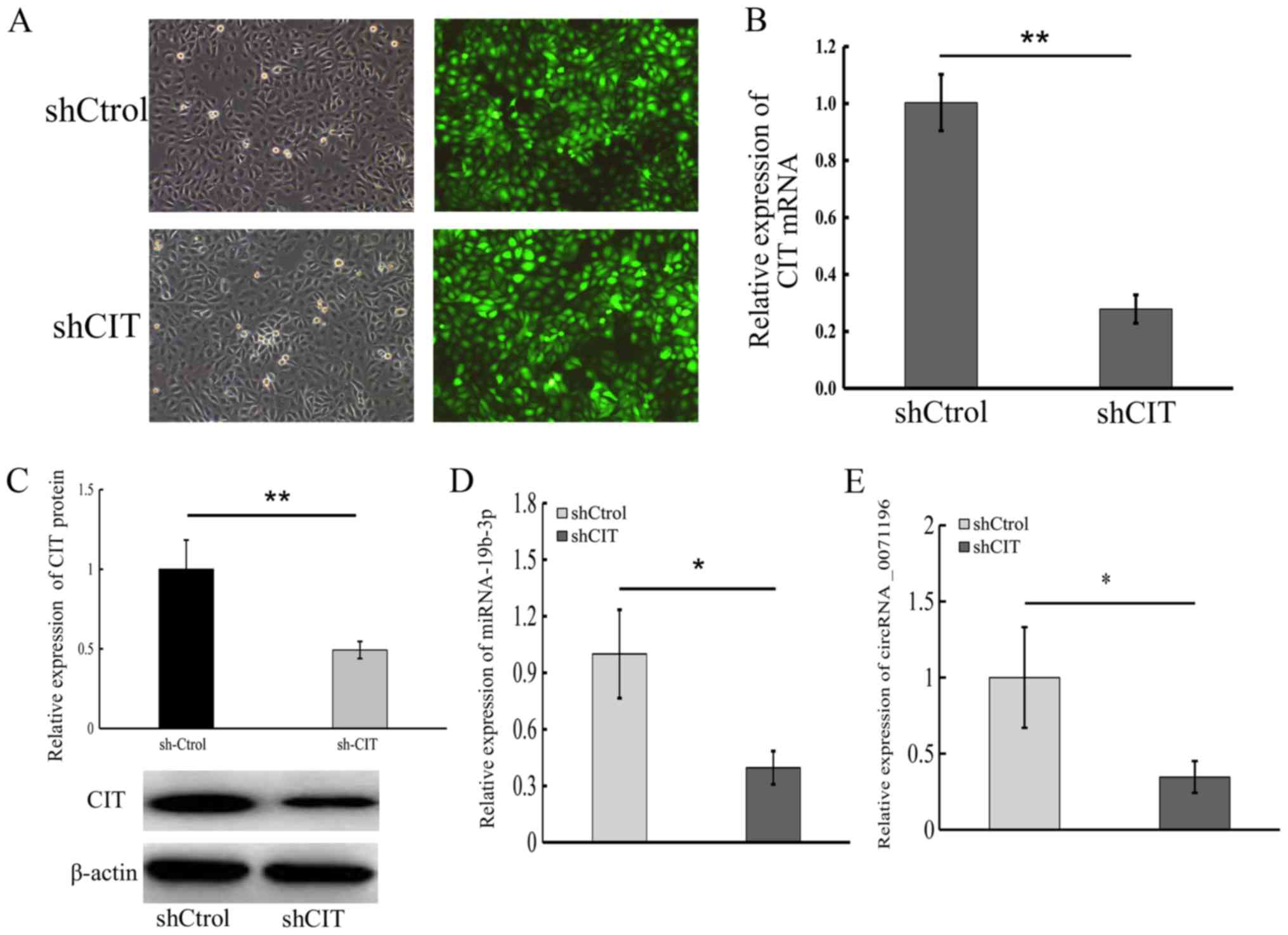
5A), to determine its functional relevance in BCa, we knocked
down CIT in 5637 cells (Fig.
9A-C). Both circRNA _0071196 and miRNA-19b-3p were
down-regulated in sh-CIT cells (Fig.
9D and E), thus underscoring the ceRNA network. Furthermore,
CIT knockdown significantly suppressed the proliferative capacity
(Fig. 10C), in vitro
migration (Fig. 10D) and colony
forming capacity (Fig. 10E) of
BCa cells, and upregulated p53 and p-ERK1/2, However, the levels of
myosin light chain 2 (MLC2), cyclin-dependent kinase 1 (CDK1),
murine double minute 2 (MDM2), cyclin D1 (CCND1) and
rho-associated, coiled-coil-containing protein kinase 1 (ROCK1)
were decreased in sh-CIT group (Fig.
11A-C).
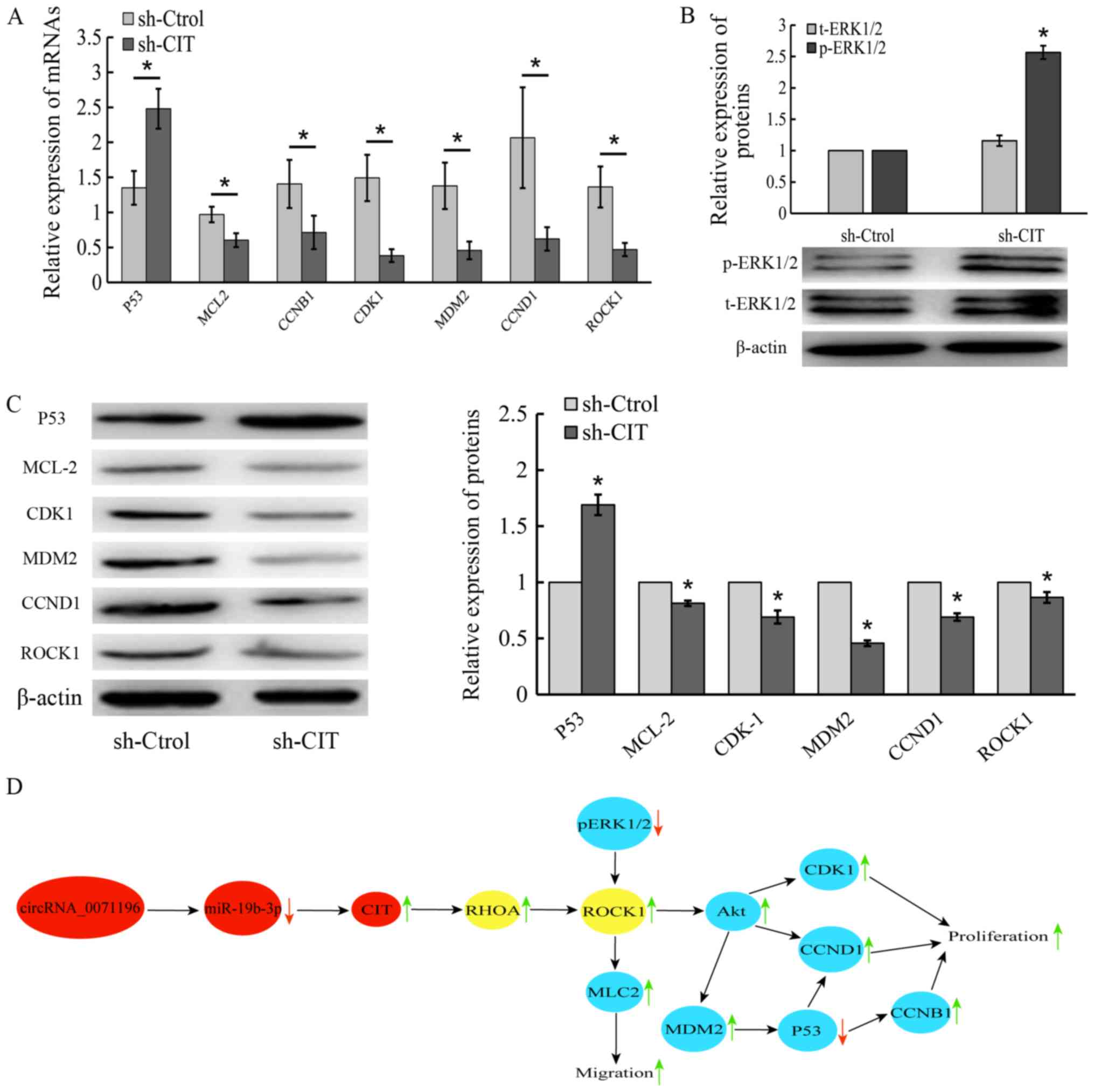 | Figure 11Molecular basis of CIT action in BCa
cells. Expression levels of p53, MLC2, CDK1, MDM2, CCND1 and ROCK1
mRNA (A) and protein (B and C) in sh-CIT and sh-Control-transfected
5637 cells. *P<0.05 vs. sh-Control. (D) The schematic
diagram of the crosstalk between circRNA_0071196, miR-19b-3p, CIT
and ROCK in BCa. CIT, citron Rho-interacting serine/threonine
kinase; BCa, bladder cancer; MLC2, myosin light chain 2; CDK1,
cyclin-dependent kinase 1; MDM2, murine double minute 2; CCND1,
cyclin D1; ROCK2, rho-associated, coiled-coil-containing protein
kinase 1; CCNB1, cyclin B1. |
 | Table IICIT expression and
clinicopathological characteristics of the bladder cancer cases
(n=80). |
Table II
CIT expression and
clinicopathological characteristics of the bladder cancer cases
(n=80).
| Parameter | Total (%) | CIT expression
|
|---|
| Positive (n=73) n
(%) | Negative (n=7) n
(%) | P-valuea |
|---|
| Age (years) | | | | 0.676 |
| >60 | 54 (67.5) | 50 (92.6) | 4 (7.4) | |
| ≤60 | 26 (32.5) | 23 (88.5) | 3 (11.5) | |
| Sex | | | | 0.415 |
| Male | 55 (68.8) | 49 (89.1) | 6 (10.9) | |
| Female | 25 (31.2) | 24 (96.0) | 1 (4.0) | |
| Metastasis | | | | 0.045 |
| Yes | 43 (53.8) | 42 (97.7) | 1 (2.3) | |
| No | 37 (46.2) | 31 (83.8) | 6 (16.2) | |
| Infiltrative | | | | 0.610 |
| Yes | 15 (18.8) | 13 (86.7) | 2(13.3) | |
| No | 65 (81.2) | 60 (92.3) | 5 (7.7) | |
| Histological
grade | | | | 0.011 |
| G1 | 43 (53.8) | 42 (97.7) | 1 (2.3) | |
| G2 | 18 (22.5) | 17 (94.4) | 1 (5.6) | |
| G3 | 19 (23.8) | 14 (73.7) | 5 (26.3) | |
| T stage | | | | 0.668 |
| T1-T2 | 61 (76.3) | 56 (91.8) | 5 (8.2) | |
| T3-T4 | 19 (23.8) | 17 (89.5) | 2 (10.5) | |
Discussion
To elucidate the molecular mechanisms of bladder
cancer (BCa) and identify novel diagnostic markers, we explored the
mRNA and circRNA expression profiles of matched BCa and normal
bladder tissues. A total of 127 circRNAs and 1,612 mRNAs were
differentially expressed in the tumor tissues relative to the
healthy tissues, and were functionally annotated primarily to cell
metabolism and DNA transcription. In addition, the differentially
expressed genes (DEGs) were significantly associated with
tumorigenic pathways, which is consistent with the findings of
Huang et al (24).
In agreement with previous studies (24-26),
the circRNAs MYLK, PC and PTK2 were found to be significantly
upregu-lated in BCa tissues, and we showed for the first time that
circRNA-0071196 is upregulated in BCa versus normal bladder
tissues. We next constructed a ceRNA network using the DEcircRNAs,
DEGs and the putative miRNAs (27-30),
which identified miR-19b-3p as the target of circRNA-10378. It is
part of the miR-17-92 cluster that regulates cancer-related
pathways (31,32) and vascular remodeling (22,28,33).
In addition, miR-19b triggers apoptosis in mouse leukemia cells,
and down-regulates PTEN in human breast carcinoma (34,35).
Niu et al (36) reported a
negative correlation between miR-19a/19b and RhoB expression in
clear cell renal cell carcinoma specimens and cell lines,
indicating that it likely acts as a tumor suppressor. In our study
however, we observed no significant difference in the miR-19b-3p
levels between BCa and normal bladder tissues. Bioinformatics
analysis also predicted citron Rho-interacting serine/threonine
kinase (CIT) as the target of miR-19b-3p, and consistent with this,
it was aberrantly overexpressed in the BCa tissues and cell lines.
CIT is the downstream effector of Rho family GTPases that are
involved in cell cycle regulation (37), and its kinase activity and
scaffolding function are vital to cytokinesis (38,39).
Previous studies have demonstrated that CIT is significantly
upregulated in hepatocellular carcinoma (HCC), and its knockdown
inhibited the growth and tumorigenicity of HCC cells (40). Furthermore, the absence of CIT
inhibited G1/S transition in colon cancer cells, and the G2/M
transition in rat hepatocytes (23,40).
At the molecular level, CIT promotes the growth of cancer cells by
blocking the p53 pathway (23).
The expression of circRNA-0071196 and miR-19b-3p
were decreased by the knockdown of CIT, Currently, the
understanding of the degradation mechanism of circRNA is extremely
limited. Since circRNAs lack free 5′ and 3′ ends,
endoribonucleolytic cleavage is the only way to degrade circRNAs.
Previous studies have demonstrated that N6-methyladenosine (m6A) is
associated with degradation of circRNAs through YTHDF2 (m6A reader
protein) (41). Yet, the
relationship between sh-CIT and m6A is uncertain. Knockdown of CIT
inhibited the proliferation, migration and colony forming capacity
of 5637 cells, which is similar to the findings in HCC cells
(40). Futhermore, CIT knockdown
also decreased ROCK1, MLC2, CCNB1, MDM2 and CCND1 levels. P53 and
MDM2 inhibit tumor progression and induce regression of established
tumors via cell cycle arrest and apoptosis (42). In addition, CIT is an established
upstream regulator of RhoA in the late stages of cell division
(43), and Sanz-Moreno et
al (44) indicated that high
levels of Rho-ROCK in cancer cells increase actomyosin
contractility and promote cell migration. The schematic diagram of
the crosstalk between circRNA_0071196, miR-19b-3p, CIT and ROCK in
BCa cells is shown in Fig. 11D.
Taken together, our findings indicate that circRNA-0071196
upregulates CIT levels in BCa by sponging off miRNA-19b-3p, and the
circRNA_0071196/miRNA-19b-3p/CIT axis is a potential therapeutic
target in BCa.
Supplementary Data
Acknowledgments
The authors thank Ms. Yan Zhang for her technical
support.
Funding
The present study was supported in part by a grant
from the Natural Science Foundation of Heilongjiang Province of
China (grant nos. ZD201516 and QC2014C112), China Postdoctoral
Science Foundation (grant no. 2016M601450), and Heilongjiang
Province Postdoctoral Science Foundation (grant no.
LBH-Z16139).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YX conceived and designed the study. ZL, YY, ZY, SX,
DL and BX performed the experiments. ZL and YY wrote the paper. YX,
ZL and YY reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The First Affiliated Hospital of Harbin Medical University, and
written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spiess PE, Agarwal N, Bangs R, Boorjian
SA, Buyyounouski MK, Clark PE, Downs TM, Efstathiou JA, Flaig TW,
Friedlander T, et al: Bladder Cancer, Version 5.2017, NCCN Clinical
Practice Guidelines in Oncology. J Natl Compr Canc Netw.
15:1240–1267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cumberbatch MGK, Jubber I, Black PC,
Esperto F, Figueroa JD, Kamat AM, Kiemeney L, Lotan Y, Pang K,
Silverman DT, et al: Epidemiology of bladder cancer: A systematic
review and contemporary update of risk factors in 2018. Eur Urol.
74:784–795. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pang C, Guan Y, Li H, Chen W and Zhu G:
Urologic cancer in China. Jpn J Clin Oncol. 46:497–501. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Racioppi M, D'Agostino D, Totaro A, Pinto
F, Sacco E, D'Addessi A, Marangi F, Palermo G and Bassi PF: Value
of current chemotherapy and surgery in advanced and metastatic
bladder cancer. Urol Int. 88:249–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dy GW, Gore JL, Forouzanfar MH, Naghavi M
and Fitzmaurice C: Global burden of urologic cancers, 1990-2013.
Eur Urol. 71:437–446. 2017. View Article : Google Scholar
|
|
7
|
Chen Z, Luo Y, Yang W, Ding L, Wang J, Tu
J, Geng B, Cui Q and Yang J: Comparison analysis of dysregulated
lncrna profile in mouse plasma and liver after hepatic
ischemia/reperfusion injury. PLoS One. 10:e01334622015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar
|
|
9
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
11
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar :
|
|
12
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng H, Lu M and Selaru FM: The
genome-wide gene expression profiling to predict competitive
endogenous RNA network in hepatocellular cancer. Genom Data.
4:93–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang YH, Yu XH, Luo SS and Han H:
Comprehensive circular RNA profiling reveals that circular
RNA100783 is involved in chronic CD28-associated CD8(+)T cell
ageing. Immun Ageing. 12:172015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun X, Jiao X, Pestell TG, Fan C, Qin S,
Mirabelli E, Ren H and Pestell RG: MicroRNAs and cancer stem cells:
The sword and the shield. Oncogene. 33:4967–4977. 2014. View Article : Google Scholar
|
|
18
|
Liu Y, Li M, Zhang G and Pang Z:
MicroRNA-10b overexpression promotes non-small cell lung cancer
cell proliferation and invasion. Eur J Med Res. 18:412013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Δ Δ C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
20
|
Du H, Shi J, Wang M, An S, Guo X and Wang
Z: Analyses of gene expression profiles in the rat dorsal horn of
the spinal cord using RNA sequencing in chronic constriction injury
rats. J Neuroinflammation. 15:2802018.J. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: a
software environment for integrated models of biomolecular
interaction networks. Genome Research. 13:2498–2504. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin J, Sun Z, Yang F, Tang L, Chen W and
Guan X: miR-19b-3p inhibits breast cancer cell proliferation and
reverses saracatinib-resistance by regulating PI3K/Akt pathway.
Arch Biochem Biophys. 645:54–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Z, Zhu X, Xu W, Zhang Y, Chen L, Qiu F,
Zhang B, Wu L, Peng Z and Tang H: Up-regulation of CIT promotes the
growth of colon cancer cells. Oncotarget. 8:71954–71964. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F, et al: CircHIPK3 sponges miR-558
to suppress heparanase expression in bladder cancer cells. EMBO
Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q, et al: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J, Wang Y, Tan X and Jing H: MicroRNAs
in autophagy and their emerging roles in crosstalk with apoptosis.
Autophagy. 8:873–882. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lan T, Li C, Yang G, Sun Y, Zhuang L, Ou
Y, Li H, Wang G, Kisseleva T, Brenner D, et al: Sphingosine kinase
1 promotes liver fibrosis by preventing miR-19b-3p-mediated
inhibition of CCR2. Hepatology. 68:1070–1086. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Selcuklu SD, Donoghue MT, Rehmet K, de
Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright
AJ and Spillane C: MicroRNA-9 inhibition of cell proliferation and
identification of novel miR-9 targets by transcriptome profiling in
breast cancer cells. J Biol Chem. 287:29516–29528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget.
6:8474–8490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olive V, Sabio E, Bennett MJ, De Jong CS,
Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A,
et al: A component of the mir-17-92 polycistronic oncomir promotes
oncogene-dependent apoptosis. eLife. 2:e008222013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang WB, Li HP, Yan J, Zhuang F, Bao M,
Liu JT, Qi YX and Han Y: CTGF regulates cyclic stretch-induced
vascular smooth muscle cell proliferation via microRNA-19b-3p. Exp
Cell Res. 376:77–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mavrakis KJ, Wolfe AL, Oricchio E,
Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan
AA, Leslie CS, et al: Genome-wide RNA-mediated interference screen
identifies miR-19 targets in Notch-induced T-cell acute
lymphoblastic leukaemia. Nat Cell Biol. 12:372–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang Z, Li Y, Huang K, Wagar N and Shim
H: Regulation of miR-19 to breast cancer chemoresistance through
targeting PTEN. Pharm Res. 28:3091–3100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu S, Ma X, Zhang Y, Liu YN, Chen X, Gong
H, Yao Y, Liu K and Zhang X: MicroRNA-19a and microRNA-19b promote
the malignancy of clear cell renal cell carcinoma through targeting
the tumor suppressor RhoB. PLoS One. 13:e01927902018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wettschureck N and Offermanns S:
Rho/Rho-kinase mediated signaling in physiology and
pathophysiology. J Mol Med (Berl). 80:629–638. 2002. View Article : Google Scholar
|
|
38
|
El Amine N, Kechad A, Jananji S and
Hickson GR: Opposing actions of septins and Sticky on Anillin
promote the transition from contractile to midbody ring. J Cell
Biol. 203:487–504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Harding BN, Moccia A, Drunat S, Soukarieh
O, Tubeuf H, Chitty LS, Verloes A, Gressens P, El Ghouzzi V, Joriot
S, et al: Mutations in citron kinase cause recessive
microlissencephaly with multinucleated neurons. Am J Hum Genet.
99:511–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu Y, Huang J, Wang KS, Zhang X and Han
ZG: RNA interference targeting CITRON can significantly inhibit the
proliferation of hepatocellular carcinoma cells. Mol Biol Rep.
38:693–702. 2011. View Article : Google Scholar
|
|
41
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic Cleavage of m6A-Containing
RNAs by RNase P/MRP Complex. Mol Cell. 74:494–507.e8. 2019.
View Article : Google Scholar
|
|
42
|
Chen J: The cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gai M, Camera P, Dema A, Bianchi F, Berto
G, Scarpa E, Germena G and Di Cunto F: Citron kinase controls
abscission through RhoA and anillin. Mol Biol Cell. 22:3768–3778.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sanz-Moreno V, Gaggioli C, Yeo M,
Albrengues J, Wallberg F, Viros A, Hooper S, Mitter R, Féral CC,
Cook M, et al: ROCK and JAK1 signaling cooperate to control
actomyosin contractility in tumor cells and stroma. Cancer Cell.
20:229–245. 2011. View Article : Google Scholar : PubMed/NCBI
|