Introduction
Hepatocellular carcinoma (HCC) is the most common
malignant tumor type of the liver and originates from the malignant
transformation of hepatocytes; it accounted for 75-85% of primary
liver cancers in 2018 worldwide (1,2).
Resection, transarterial chemoembolization, radiofrequency
ablation, liver transplantation, selective internal radiation
therapy and systemic therapies are considered effective methods for
treating patients with different stages of HCC (3). However, the trend of increasing
HCC-associated mortality remains a growing concern (4). Therefore, a better understanding of
the molecular mechanisms related to HCC progression would be
favorable for early diagnosis and effective management.
Long non-coding RNAs (lncRNAs) are molecules
containing >200 nucleotides that do not code for proteins
(5). The molecular functions of
lncRNAs include transcriptional splicing, chromosome structure
regulation, mRNA stability, mRNA availability and posttranslational
modification (6). MicroRNAs
(miRNAs) are a class of evolutionarily conservative non-coding RNAs
comprised of ~22 nucleotides that have an essential role in
posttranscriptional gene regulation (7). They can bind to numerous sites
(nucleotides 2-7), particularly those in the 3′-untranslated region
of target gene mRNAs, and modulate target gene expression by
suppressing the translation of target mRNAs (8). Both lncRNAs and miRNAs serve key
roles in numerous biological processes, including the immune
response, cell growth, epigenetic regulation, tumorigenesis and
cell differentiation (9,10). Recently, increasing evidence has
suggested that the crosstalk between lncRNAs and miRNAs plays
essential roles in the development and progression of numerous
cancers, including HCC (11,12).
For instance, the lncRNA FTX contributes to colorectal cancer
tumorigenesis and progression via an interaction with miR-215
(13). The lncRNA TUG1 modulates
the progression of oral squamous cell carcinoma by sponging
miR-524-5p and mediates the expression of distal-less homeobox 1
(DLX1) (14). In addition, the
lncRNA CASC2 regulates the development of HCC cells via the
miR-362-5p/NF-κB axis (15).
NEAT1 is a type of lncRNA that is abnormally
expressed in HCC and has various oncogenic roles (16-18).
Furthermore, it is generally correlated with the progression of
several types of neoplasms, such as colorectal cancer, breast
cancer and non-small cell lung cancer (19-21).
Yan et al (22) reported
that NEAT1 enhances the resistance of anaplastic thyroid carcinoma
cells to cisplatin by sponging miR-9-5p and regulating sperm
associated antigen 9 expression. Xia et al (23) reported that NEAT1 promotes the
growth of gastric cancer cells by regulating the
miR-497-5p/phosphoinositide-3-kinase regulatory subunit 1 axis.
Furthermore, NEAT1 can promote malignant melanoma development and
metastasis by targeting ras-releated protein Rab-9 (24). The oncogenic function of NEAT1 in
HCC is gradually emerging; however, the detailed mechanism of HCC
remains largely unknown.
The present study mainly focused on the biological
function of NEAT1 in HCC. NEAT1 was upregulated and miR-320a was
downregulated in HCC cells. HCC cell progression was significantly
affected by NEAT1 silencing or overexpression. In addition,
miR-320a was predicted as a target of NEAT1 and regulated the level
of LAGE3. Furthermore, NEAT1 was upregulated in HCC tissues and
positively correlated with LAGE3 expression. Therefore, it was
hypothesized that NEAT1 contributes to the proliferation and
migration of HCC by acting as a miR-320a molecular sponge and
targeting LAGE3.
Materials and methods
Cell culture
Human hepatic carcinoma cell lines (Huh7, SNU-398,
MHCC-97H, Hep3B and SNU-449), hepatoblastoma cells Huh-6, hepatic
stellate cells LX-2 and 293T cells were used in the present study.
The cells were all purchased from the Institute of Cell Biology,
Chinese Academy of Sciences. RPMI-1640 (Sigma-Aldrich; Merck KGaA)
or Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck
KGaA) supplemented with 10% fetal bovine serum (FBS; HyClone; GE
Healthcare), 100 U/ml penicillin and 100 µg/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) was used as the cell
culture medium, and the cells were all cultured in a humidified
chamber containing 5% CO2 at 37°C. Huh7 and MHCC-97H
cell lines were cultured by RPMI-1640, while other cell lines were
cultured by DMEM.
Lentiviral vector transfection
Lentiviral plasmids (lentivirus-shNEAT1) were
constructed using short hairpin RNAs (shRNAs) of the NEAT1 sequence
(Shanghai GenePharma Co., Ltd.), and were transfected into Huh7 and
MHCC-97H cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of
lentiviral plasmid transfected was 2 µM. The lentiviral
plasmid containing a scrambled sequence was constructed using the
LV-NC (Shanghai GenePharma Co., Ltd.) as a negative control. The
oligonucleotide sequences were: shNEAT1 sense, 5′-CAC CGC ATG GAC
CGT GGT TTCCGTTACTTTC AAGAGAAGTAACAAAACGGTCCA TGT TTT TTG-3′ and
antisense, 5′-GAT CCA AAA AAC ATG GAC CGT GGT TTG TTA CTT CTC TTG
AAA GTA ACA AAC CAC GGT CCA TGC-3′; NC sense, 5′-CAC CGT TCT CCG
AAC GTG TCA CGT CAA GAG ATT ACG TGA CAC GTT CGG AGA ATT TTT TG-3′
and antisense, 5′-GAT CCA AAA AAG TTC TCC GCG TGT CAC GTA ATC TCT
TGA CGT GAC ACG TTC GGA GAA C-3′. A synthetic and purified NEAT1
gene fragment (5′-CGG CUC GAG GG GCC AUC AGC UUU-3′) was inserted
into the lentiviral plasmids and designated LV-NEAT1 (Shanghai
GenePharma Co., Ltd.).
The transfection of miR-320a mimic or negative
control in Huh7 and MHCC-97H cell lines was performed using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration of miR-320a mimic and negative
control was 100 µM. The sequences of the miR-320a mimic and
negative control were as follows: miR-320a mimic, 5′-AAA AGC UGG
GUU GAG AGG GCG A-3′ and negative control, 5′-UUC UCC GAA CGU GUC
ACG UTT-3′. Further experiments were performed 48 h after
transfection.
Transwell invasion assay
A 200-µl cell suspension of 1×105
Huh7 or MHCC-97H cells in RPMI-1640 medium was loaded into the
upper chambers of a 24-well Transwell plate, which had an
8-µm pore size and was coated with 1 mg/ml Matrigel (Corning
Inc.) for 1 h at 4°C. The lower chamber was loaded with 600
µl RPMI-1640 containing 10% FBS. Subsequently, the cells on
the surface of the filter were fixed with 4% formaldehyde for 15
min, stained with 0.5% crystal violet for 30 min at room
temperature, and then counted using a light microscope
(magnification, ×200).
EdU proliferation assay
Cell proliferation was studied using the EdU
proliferation assay (Nanjing Keygen Biotech Co., Ltd.) according to
the manufacturer's instructions. A total of 48 h after
transfection, 2×104 cells were treated with 10 µM
EdU solution for 4 h. EdU-positive cells were observed using Apollo
staining for 30 min at room temperature and
4′,6-diamidino-2-phenylindole staining for 30 min at room
temperature with an inverted fluorescence microscope
(magnification, ×200). Then, positive cells were identified by
integral optical density (IOD) using Image-Pro Plus 6.0 (Media
Cybernetics, Inc.).
Colony forming assay
LV-NEAT1-, LV-shNEAT1- and LV-NC-transfected Huh7
and MHCC-97H cells were seeded at a density of 100 cells/well in
6-well plates. After 2 weeks of culture, the colonies were fixed
with methanol for 15 min at room temperature and stained with 0.5%
crystal violet for 15 min at room temperature. Visible colonies
were manually counted in randomly selected fields using light a
microscope (magnification, ×10). The clone formation rate (CFR) was
calculated according to the following formula: CFR=clone
counts/seeded cell counts ×100%. The experiment was repeated three
times.
Flow cytometric analysis of cell
apoptosis
Huh7 and MHCC-97H cell lines transfected with LV-NC,
LV-shNEAT1 and LV-NEAT1 were stained using FITC-Annexin V and
propidium iodide (PI; both Beyotime Institute of Biotechnology) in
the dark for 15 min at room temperature. Apoptosis assays were
performed using the CytoFLEX flow cytometer (Beckman Coulter, Inc.)
and analyzed via Cytexpert 2.0 software (Beckman Coulter, Inc.).
Annexin V-FITC-negative and PI-negative cells were defined as
living cells; Annexin V-FITC-positive and PI-negative were defined
as early apoptotic cells; Annexin V-FITC-positive and PI-positive
were defined as late apoptotic cells and necrotic cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA form HCC tissues or cell lines was
extracted using RNAiso Plus (Takara Bio, Inc.). To ensure purity of
the total RNA, the A260/A280 of was
controlled between 1.8 and 2.2, and the
A260/A230 ratio was controlled >1.7. The
total RNA concentration was between 500 and 1,000 ng/µl.
Prime Script™ RT Master mix was used for RNA RT at 37°C for 15 min
followed by 85°C for 5 sec. SYBR Premix Ex Taq II (Takara Bio,
Inc.) was used for qPCR, which was performed on the Applied
Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The following standard two-step PCR
reaction program was used: i) one cycle of pre-denaturation at 95°C
for 30 sec; ii) 40 cycles of 95°C for 5 sec, 60°C for 20 sec; and
iii) final extension at 72°C for 30 sec, followed by melting curve
analysis and cool down. Relative gene expression levels were
analyzed using the 2−ΔΔCq method (25). The primers of NEAT1, LAGE3,
miR-320a and the controls are shown in Table I. GAPDH was used as a loading
control to detect the NEAT1 and LAGE3 expression levels, and U6 was
used as a loading control to detect the miR-320a expression
level.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| NEAT1 |
TGGCTAGCTCAGGGCTTCAG |
TCTCCTTGCCAAGCTTCCTTC |
| miR-320a |
GTTGGATCCGGCGTTTCCTTCCGACATG |
GCTGAATTCGTCCACTGCGGCTGTTCC |
| LAGE3 |
CGACTGTGGGTCAGTTTGCAC |
GGTTGAGAGGCTGCGGTTT |
| MALAT1 |
AAAGCAAGGTCTCCCCACAAG |
GGTCTGTGCTAGATCAAAAGGCA |
| GAPDH |
AAGGAAGGTGGTGAAGCAGGC |
GTCAAAGGTGGAGGAGTGGG |
| U6 |
ATTGGAACGGATACAGAGAAGATT |
GGAACGCTTCACGAATTTG |
Bioinformatics analysis
ChipBase (http://rna.sysu.edu.cn/chipbase/index.php) and
StarBase (http://starbase.sysu.edu.cn/) were used to identify
specific miRNA targets of lncRNA NEAT1. The TargetScan Human 7.1
(http://www.targetscan.org/), Starbase
and miRanda (http://www.microrna.org/microrna/home.do) databases
were used to predict putative mRNA targets of miR-320a.
Luciferase reporter gene assay
For the luciferase reporter gene assay,
5×105 293T cells were inoculated in a 24-well plate
overnight. Subsequently, 150 ng pmirGLO-LAGE3-WT or pmirGLO-lncRNA
NEAT1-WT reporter plasmids (Promega Corporation) and their mutant
vectors were co-transfected into cells with 50 nM miRNA-320a mimic
using Lipofectamine 2000 reagent. After 36 h of cell culture, the
firefly and Renilla luciferase activity was determined by a
double Luciferase Report Analysis system (Promega Corporation)
based on the manufacturer's instructions. The relative luciferase
activity was calculated based on the fluorescence of the
firefly/Renilla luciferase.
RNA immunoprecipitation (RIP)
The RIP assay was carried out utilizing a Magna RIP
kit (EMD Millipore). Huh7 cells were lysed using RIP lysis buffer,
and the cell lysates were incubated with magnetic beads conjugated
to a human anti-Ago2 antibody (1:500; catalog no. MABE253; EMD
Millipore). Huh7 cells were used to conduct the RIP assay without
transfection prior to the assay. RNA was detected by RT-qPCR and
IgG detected performed as a negative control. MALAT1 was selected
as a positive control according to Sun et al (26), who revealed that MALAT1 acts as
sponges of miR-320a. The primers for MALAT1 are presented in
Table I.
RNA pull-down assay
RNA was purified and labeled with biotin using the
Pierce RNA 3′ End Desthiobiotinylation kit (Thermo Fisher
Scientific, Inc.). Positive control (biotin-labeled wild-type
miR-320a; miR-320a-Bio), negative control (mutant miR-320a,
miR-320a-Bio-MUT), and biotinylated RNAs (NC-Bio) were incubated
with the cell lysates.
Xenotransplantation of tumors
The animal experiment procedures were approved by
the Animal Ethics Committee of The First Affiliated Hospital of Sun
Yat-sen University (Guangzhou, China). A total of 24 1-month-old
athymic female BALB/C nude mice, with a weight range of 13-17 g,
were purchased from the Shanghai Institute of Medicine (Shanghai,
China). The mice were housed at a temperature of 18-23°C with
40-60% humidity, a 14-h light/10-h dark cycle, and food and water
were accessible at all times. Huh7 cells (2×105) in 0.2
ml normal saline transfected with LV-NEAT1, LV-shNEAT1 or LV-NC
were subcutaneously implanted into the lateral abdomen of each nude
mouse. A total of 24 mice were randomly divided into 4 groups; 6
mice were injected with Huh7 cells infected with LV-NEAT1, 12 were
injected with Huh7 cells with LV-NC, and 6 were injected with Huh7
cells with LV-shNEAT1. In total, 2 mice died in the group of
Huh7-LV-NEAT1 and Huh7-LV-NC, and finally 4 mice were analyzed.
Three mice died in the group of Huh-LV-NC and Huh7-LV-shNEAT1. The
volume of the tumor was estimated using a caliper once a week for 5
weeks. The nude mice were observed every 12 h after being
inoculated with HCC cells. After 5 weeks, nude mice were
anesthetized by 4% chloral hydrate intraperitoneal injection with a
dose of 400 mg/kg, euthanized using cervical dislocation and then
the subcutaneous tumors were removed. Some mice were anesthetized
and sacrificed earlier than 5 weeks if the following conditions
were met: i) Maximum weight loss of nude mice was >5 g in the
first 2 weeks; ii) tumor volume was <20 mm3 in the
first 2 weeks; or iii) inoculated nude mice died. There were four
nude mice left in the group of Huh7-LV-NEAT1 and Huh7-LV-NC and
three nude mice left in the group of Huh-LV-NC and Huh7-LV-shNEAT1
at 5 weeks. The cause of death was probably due to HCC cell
toxicity and poor immunity of nude mice. The weights of the tumors
were measured after 5 weeks. The following formula was used to
calculate tumor volume: Volume (mm3)=0.5 × length ×
width2. Multiple tumor growth was not observed in the
present study. The levels of LAGE3 in the resected tumors were
analyzed by immunohistochemistry.
Immunohistochemistry
Tissues were fixed with 4% formalin for 24 h at room
temperature, embedded in paraffin and section to a thickness of
0.5-cm. Following dewaxing and rehydration, the endogenous
peroxidase activity was blocked with sodium citrate buffer (pH 6.0)
at 100°C for 20 min, and the antigens on the slides were exposed.
The slides were incubated overnight with antibodies specific for
Ki67 (1:500; catalog no. ab15580; Abcam) and LAGE3 (1:100; catalog
no. ab224157; Abcam) at 4°C. Then, the slides were incubated with a
rabbit anti-sheep IgG secondary antibody coupled with horseradish
peroxidase (1:2000; catalog no. ab6747; Abcam) at 37°C for 1 h.
Following staining, nine fields of each slide were randomly
selected with the help of a light microscope (magnification, ×200).
Ki67 and LAGE3 expression intensity was assessed by estimating the
area of the fields and the IOD was calculated by the medium pixel
intensity of each object. The sections were imaged with a light
microscope (magnification, ×200). All images were acquired and
processed in TIFF format, and analysis was performed using Image
ProPlus 6.0 AMS software (Media Cybernetics Inc.).
Statistical analysis
All data are presented as mean ± standard deviation.
Student's t-test was used for the analysis of two independent
groups. One-way analysis of variance followed by Tukey's post hoc
test was used to analyze differences among three or more groups.
P<0.05 was considered to indicate a statistically significant
difference. All statistical analysis was performed using SPSS 24.0
software (IBM Corp.).
Results
NEAT1 is upregulated and miR-320a is
downregulated in HCC cells
First, NEAT1 expression was detected in the HCC cell
lines Hep3B, SNU-449, SNU-398, Huh7, and MHCC-97H, the
hepatoblastoma cell line Huh6 and the hepatic stellate cell line
LX-2. NEAT1 expression was significantly higher in HCC cells
compared with LX-2 cells (Fig.
1A). Furthermore, miR-320a expression was significantly lower
in HCC cells compared with LX-1 cells (Fig. 1B). This demonstrated that NEAT1 may
be involved in the development of HCC.
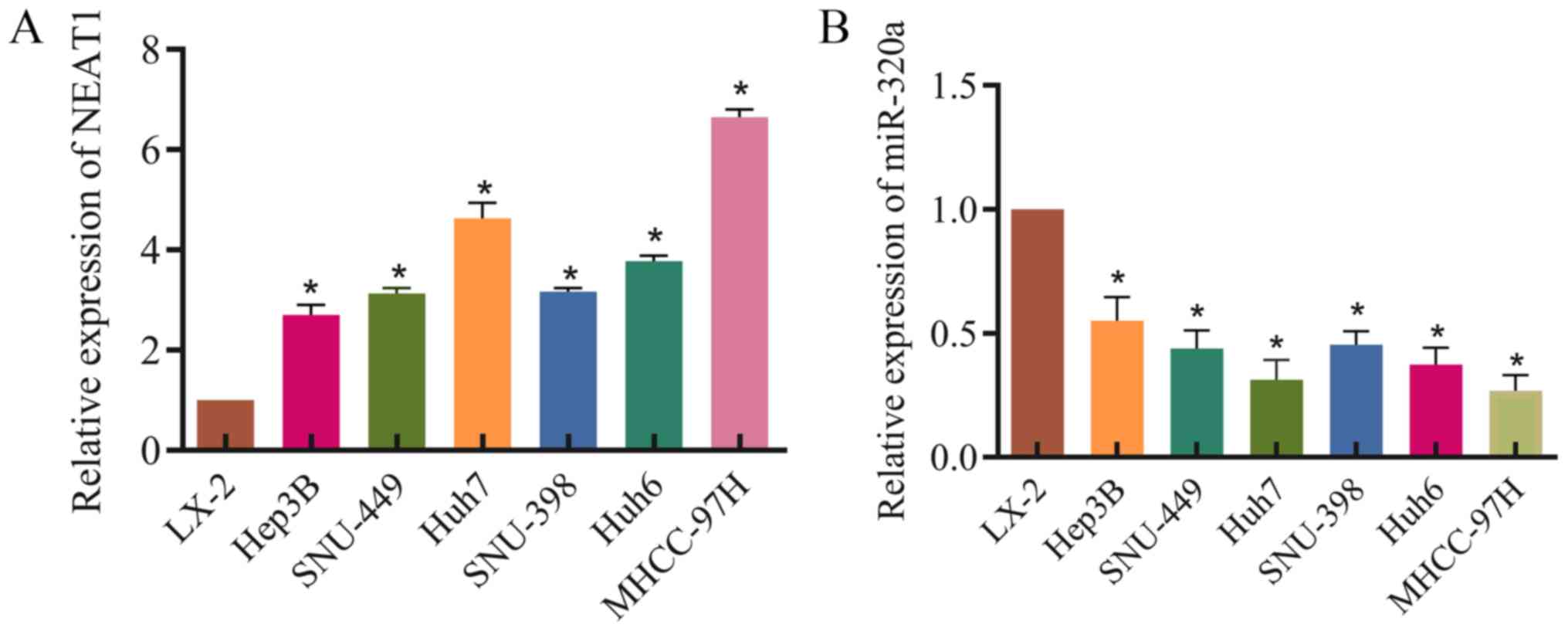 | Figure 1Expression of NEAT1 and miR-320a in
hepatocellular carcinoma cells. (A) NEAT1 expression in LX-2,
Hep3B, SNU-449, Huh6, SNU-398, Huh7 and MHCC-97H cells. RT-qPCR was
used to detect NEAT1 expression, with GAPDH as a loading control.
(B) miR-320a expression in LX-2, Hep3B, SNU-449, Huh6, SNU-398,
Huh7 and MHCC-97H cells. RT-qPCR was used to test miR-320a
expression, with U6 as a loading control. Three independent
experiments were performed. *P<0.05 vs. LX-2.
RT-qPCR, reverse transcription-quantitative PCR; miR-230a,
microRNA-320a; NEAT1, nuclear enriched abundant transcript 1. |
Effects of NEAT1 on HCC cell
proliferation
To investigate whether NEAT1 can affect HCC cell
proliferation, an EdU assay was performed. Huh7 and MHCC-97H cells
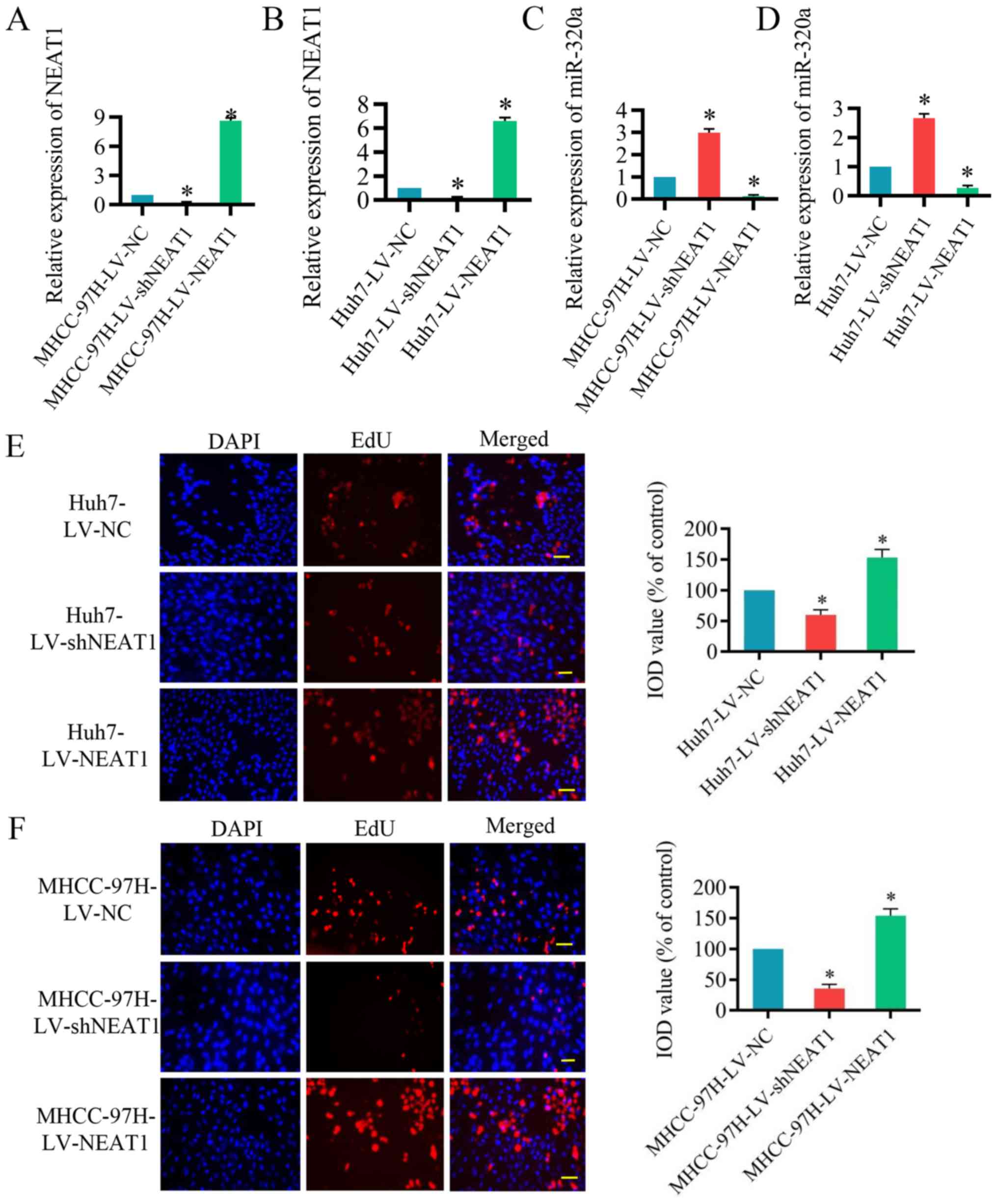
were infected with LV-shNEAT1 or LV-NEAT1 for 48 h. As presented in
Fig. 2A and B, NEAT1 expression
was significantly reduced by LV-shNEAT1 and significantly increased
by LV-NEAT1 in Huh7 and MHCC-97H cells compared with the control
cells. In addition, miR-320a expression was significantly increased
by LV-shNEAT1 and significantly reduced by LV-NEAT1 compared with
the control (Fig. 2C and D). Next,
the EdU assay was performed, which verified that Huh7 and MHCC-97H
cell proliferation was significantly suppressed by reduced NEAT1
expression and significantly induced by the upregulation of NEAT1
(Fig. 2E and F). These results
indicated that inhibition of NEAT1 represses the proliferation of
HCC cells.
Effects of NEAT1 on HCC cell
apoptosis
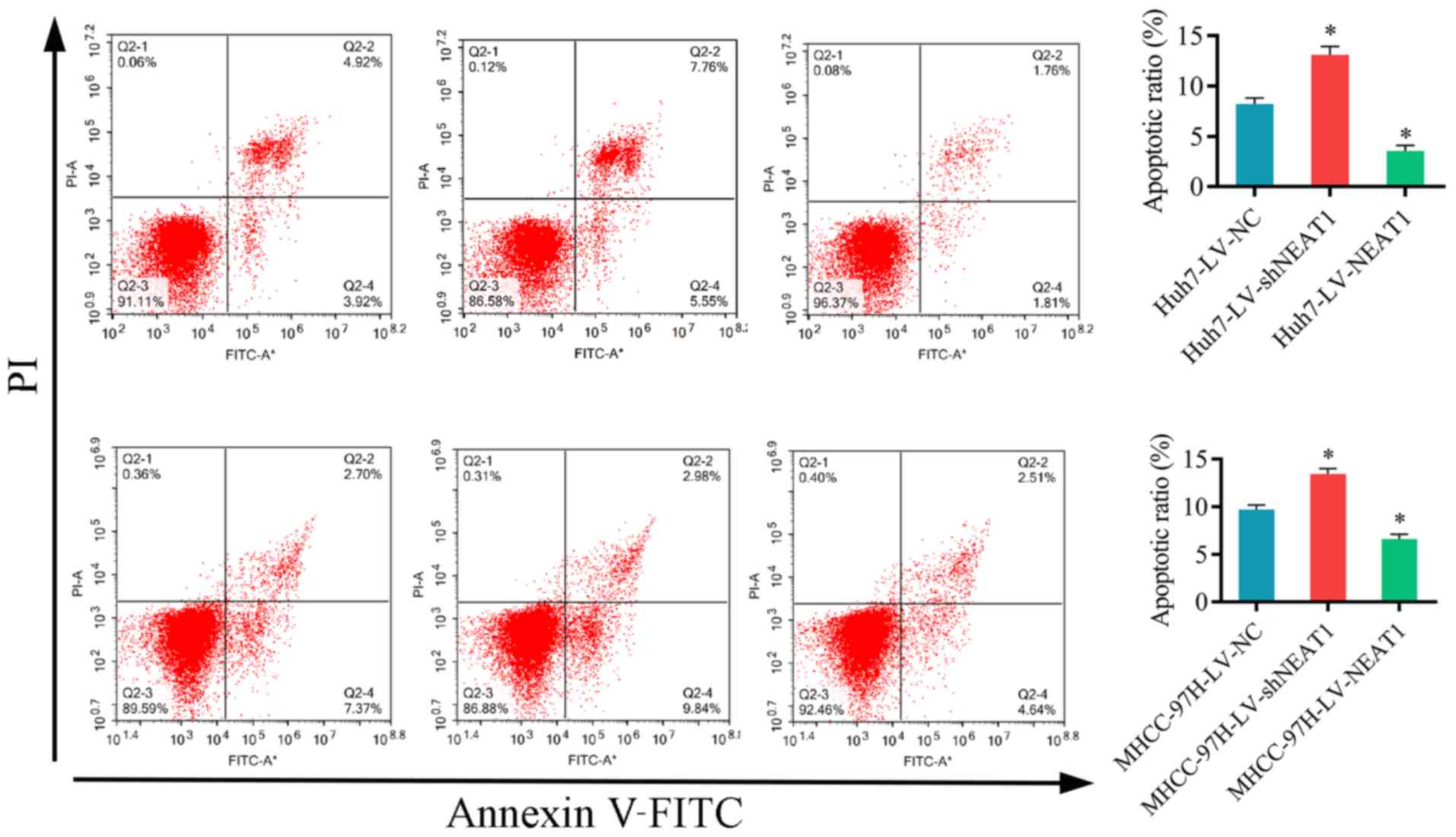
To investigate the effect of NEAT1 on the apoptosis
of HCC cells flow cytometry was performed. The results demonstrated
that the apoptosis of Huh7 and MHCC-97H cells was significantly
increased by LV-shNEAT1 and significantly decreased by LV-NEAT1
compared with the control cells (Fig.
3). These results suggested that silencing NEAT1 increases HCC
cell apoptosis in vitro.
Effects of NEAT1 on HCC cell
migration
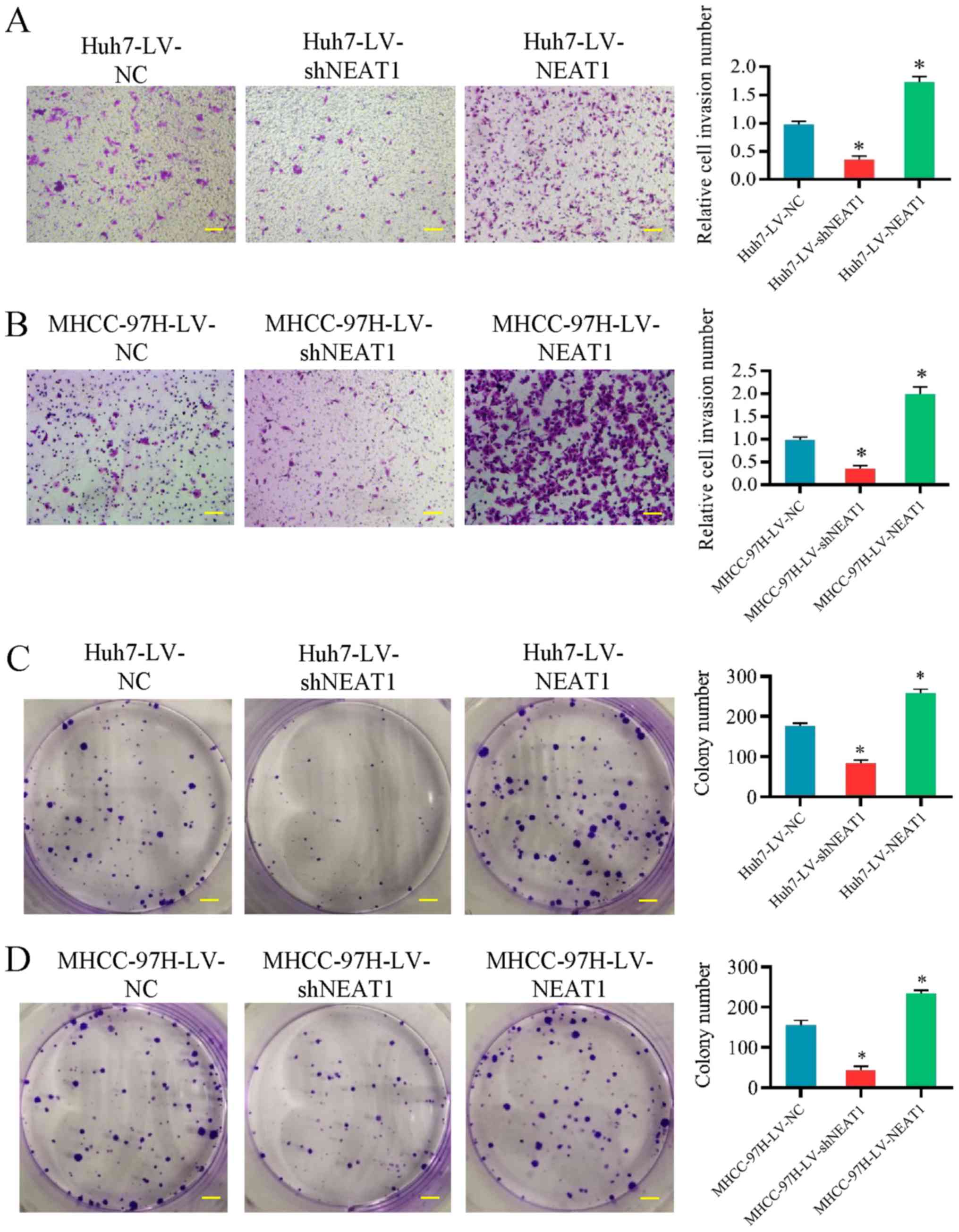
Transwell invasion assays and colony formation
assays were performed to determine whether NEAT1 affects HCC cell
migration and invasion. LV-shNEAT1 significantly reduced the
migration capacity of Huh7 and MHCC-97H cells, while upregulation
of NEAT1 significantly increased the migration (Fig. 4A and B). Subsequently, the effects
of NEAT1 on colony formation were analyzed in Huh7 and MHCC-97H
cells and it was identified that colony formation was significantly
increased following NEAT1 upregulation (Fig. 4C and D). These results suggested
that NEAT1 may promote HCC cell migration and invasion.
miR-320a acts as a target of NEAT1
LncRNAdb, StarBase and ChIPBase were used to
investigate the mutual effect between NEAT1 and miR-320a. miR-320a
was identified as a target of NEAT1, and the complementary binding
regions were verified (Fig. 5A).
To confirm these findings, a luciferase reporter assay was
performed. As presented in Fig.
5B, mutations were generated in the miR-320a-binding sequence
of NEAT1. miR-320a mimic was transfected into Huh7 cells and its
transfection efficiency is presented in in Fig. 5C. Co-transfection of a wild-type
luciferase reporter plasmid with miR-320a mimic significantly
reduced the reporter gene activity compared with that of the
control Huh7 cells, whereas no obvious changes were observed
following co-transfection with a mutant-type luciferase reporter
plasmid and miR-320a mimic (Fig.
5D) due to the interaction between miR-320a mimic and NEAT-WT.
Furthermore, to investigate whether NEAT1 can be coupled to
miR-320a, an RIP assay was performed, and the content of NEAT1 and
miR-320a in Ago2 particles was significantly higher compared with
that in the IgG group in Huh7 cells (Fig. 5E). As presented in Fig. 5F, an RNA pull-down assay was
performed, which revealed that significantly higher NEAT1 levels in
Huh7 cells were observed with miR-320a-Bio probe compared with
NC-bio or miR-320a probe. These results suggest that miR-320a is a
direct target of NEAT1.
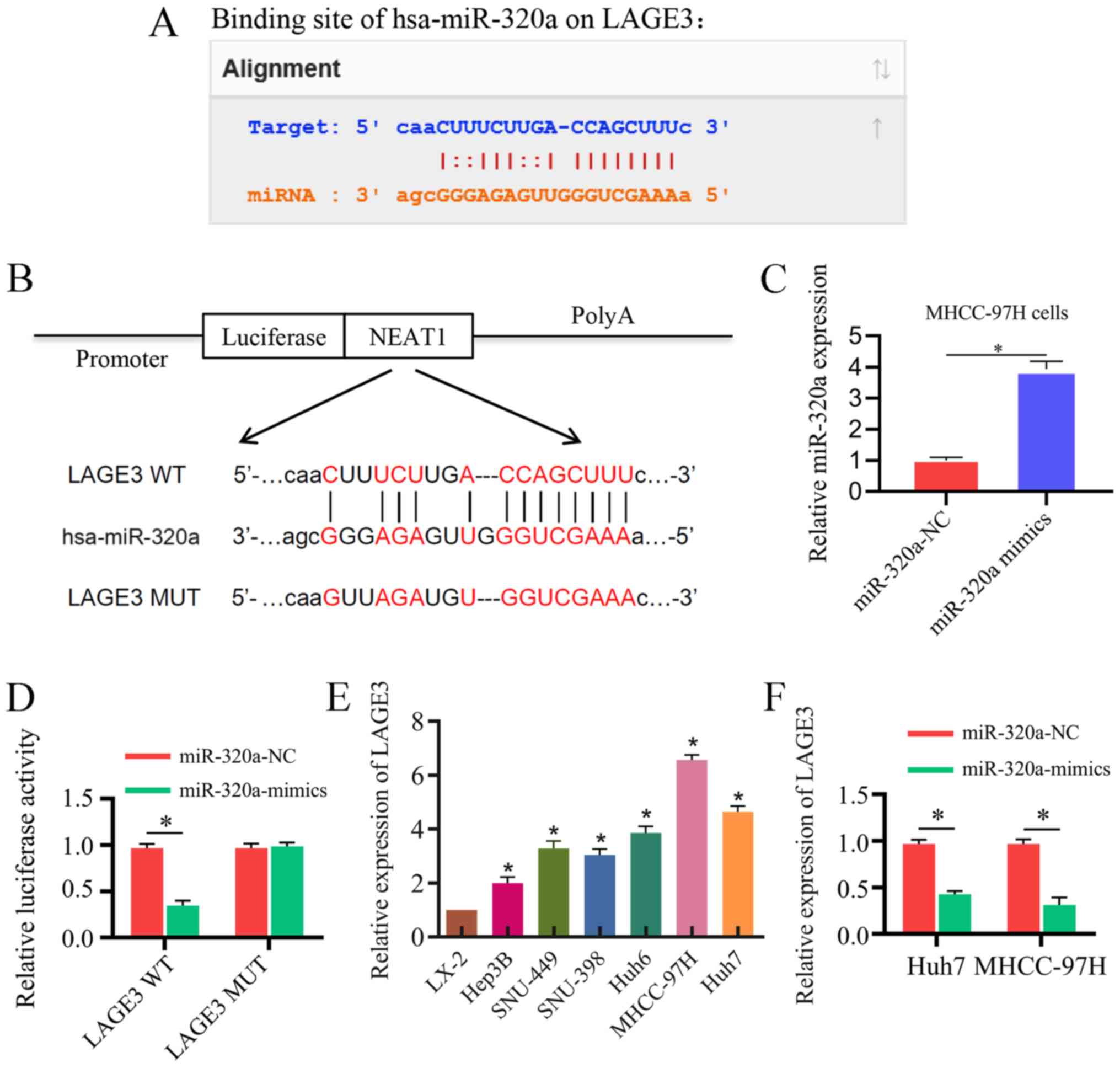
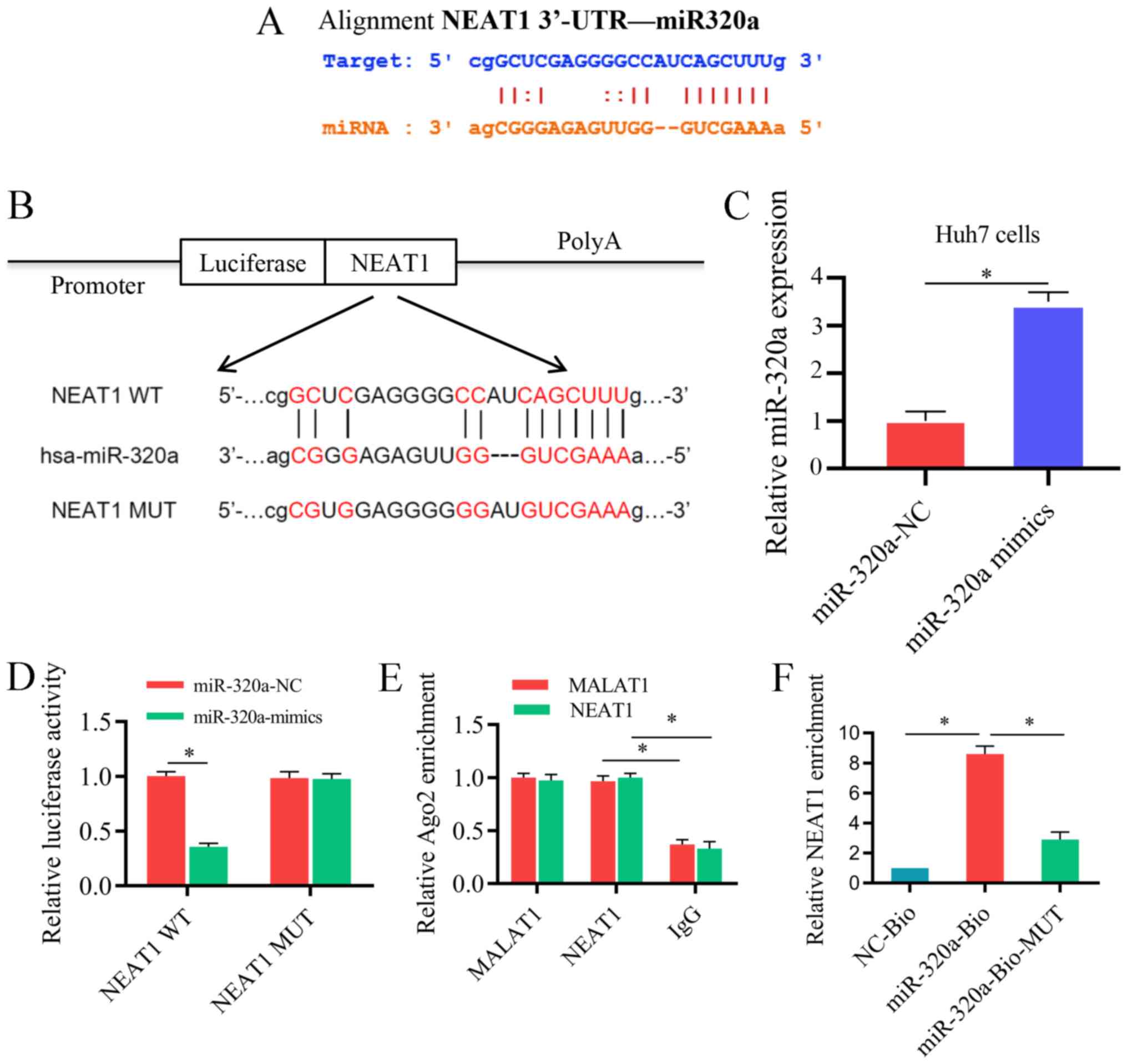 | Figure 5miR-320a serves as a target of NEAT1
in vitro. (A) The binding sites between NEAT1 and miR-320a.
(B) Luciferase reporter constructs containing the NEAT1 WT or NEAT1
MUT sequence. (C) Transfection efficiency of miR-320a mimic in Huh7
cells. (D) NEAT1 WT or NEAT1 MUT were co-trans-fected into Huh7
cells with miR-320a mimic or the corresponding negative control.
(E) The association between NEAT1 and Ago2 was tested by an RIP
assay. Cellular lysates were immunoprecipitated using an Ago2
antibody or IgG. NEAT1 expression was detected by reverse
transcription-quantitative PCR. The MALAT1 level was used as a
positive control. (F) An RNA pull-down assay indicated the direct
interaction between miR-320a and NEAT1. Cellular lysates were
pulled down using NC-Bio, miR-320a-Bio or miR-320a-Bio-MUT. Three
independent experiments were performed. *P<0.05.
NC-Bio, biotinylated negative control; miR-320a-Bio, biotinylated
miR-320a; miR-320a-Bio-MUT, miR-320a probe containing mutations in
the NEAT1-binding site; WT, wild type; MUT, mutant; NEAT1, nuclear
enriched abundant transcript 1; NC, negative control; 3′-UTR,
3′-untranslated region; miR-320a, microRNA-320a. |
LAGE3 is a direct target of miR-320a
TargetScan, miRanda and StarBase were used for
bioinformatics analysis to predict the targets of miR-320a, and
LAGE3 was predicted as a target. The binding regions between
miR-320a and LAGE3 are presented in Fig. 6A. The wild-type and mutant binding
sites of LAGE3 are presented in Fig.
6B. The transfection efficiency of miR-320a mimic in MHCC-97H
cells is presented in Fig. 6C.
Wild-type luciferase reporter plasmids were co-transfected with
miR-320a mimic, and the reporter activity in Huh7 cells was
significantly reduced due to the interaction of miR-320 mimic and
wild-type LAGE3. However, the reporter activity in Huh7 cells
co-transfected with mutant-type luciferase reporter plasmids was no
obviously changed (Fig. 6D). In
addition, the LAGE3 mRNA expression level was revealed to be
significantly higher in HCC cells and hepatoblastoma cells compared
with LX-2 cells (Fig. 6E).
Furthermore, the mRNA expression level of LAGE3 was significantly
lower in HCC cells and hepatoblastoma cells transfected with
miR-320a mimic compared with the controls (Fig. 6F).
NEAT1 regulates HCC progression by
modulating miR-320a and LAGE3 in vivo
The present study established a Huh7 cell nude mouse
xenograft model to verify whether NEAT1 can regulate HCC in
vivo. The tumor volumes and tumor weights after five weeks are
presented in Fig. 7A-C. Ki-67,
also known as antigen Ki-67 or MKI67, is a protein encoded by the
MKI67 gene in mammals. The expression of Ki67 is strongly
associated with tumor cell proliferation and growth and is widely
used as a proliferation marker in routine pathological
investigations (27,28). As presented in Fig. 7D, immunohistochemistry revealed
that LV-shNEAT1 significantly inhibited LAGE3 and Ki-67, while
LV-NEAT1 significantly increased expression of LAGE3 and Ki-67.
RT-qPCR was performed to analyze the expression level of NEAT1,
miR-320a and LAGE3. As presented in Fig. 7E-G, when the expression of NEAT1
was downregulated, the expression of miR-320a was significant
increased, while the expression level of LAGE3 was significantly
reduced. By contrast, when NEAT1 was over-expressed, the effects of
miR-320a and LAGE3 expression were reversed. It was identified that
NEAT1 overexpression promoted LAGE3 expression via sponging
miR-320a in vivo, while the downregulation of NEAT1
demonstrated the opposite effect. These results revealed that NEAT1
promotes the development of HCC by targeting miR-320a and LAGE3
in vivo.
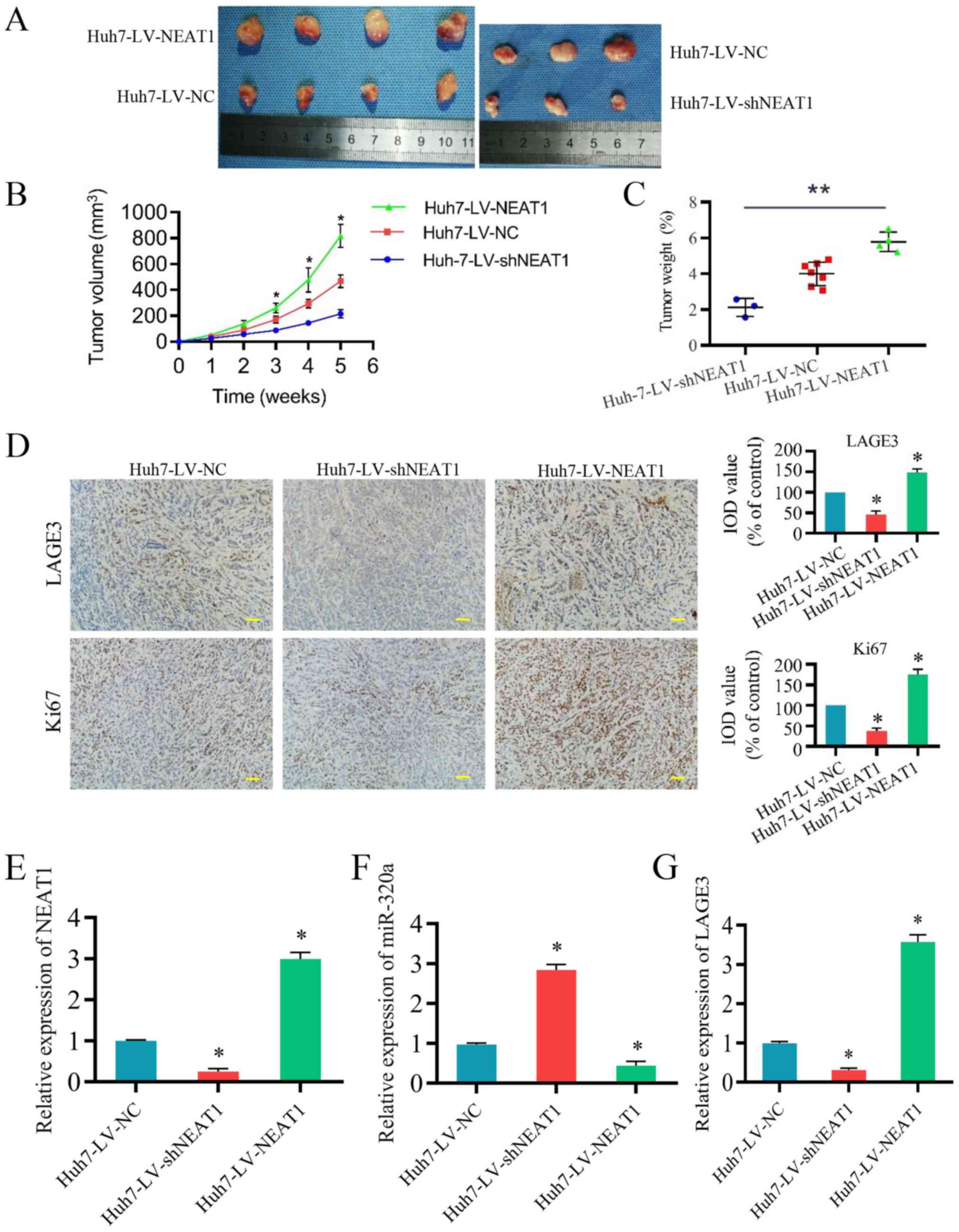 | Figure 7NEAT1 modulates hepatocellular
carcinoma progression by sponging miR-320a and regulating LAGE3
in vivo. Three groups of mice were established; 12 mice were
injected with Huh7 cells infected with LV-NC, six were injected
with Huh7 cells infected with LV-shNEAT1, and six were injected
with Huh7 cells infected with LV-NEAT1. (A) Solid tumors were
collected from mouse subcutaneous tissue. Xenograft tumor (B)
volume and (C) weight were shown to be significantly promoted by
NEAT1. (D) Immunohistochemical staining of Ki-67 and LAGE3 in tumor
tissues. Scale bar, 50 µm. Expression of (E) NEAT1, (F)
miR-320a and (G) LAGE3 in the tumor tissues of the mice. Three
independent experiments were performed. *P<0.05 vs.
NC. NEAT1, nuclear enriched abundant transcript 1; miR-320a,
microRNA-320a; LV, lentivirus; NC, negative control; sh, short
hairpin RNA; LAGE3, L antigen family member 3; IOD, integral
optical density. |
Discussion
In the present study, NEAT1 expression was
demonstrated to be significantly higher in HCC cells.
NEAT1-silencing could restrain the progression of HCC, while NEAT1
overexpression was capable of promoting HCC development by
regulating miR-320a both in vitro and in vivo.
Further mechanistic studies indicated that LAGE3 was a direct
target of miR-320a. In conclusion, it was demonstrated that NEAT1
acts as a competitive endogenous RNA (ceRNA) for miR-320a to
enhance LAGE3 expression in the tumorigenesis of HCC.
It is widely acknowledged that only ~1% of the
genome encodes proteins, while the vast majority of the transcribed
human genome consists of noncoding sequences (29). The lncRNA NEAT1 is among these
sequences. NEAT1 is located in the nucleus, is a highly abundant
lncRNA with a length of ~4 kb and acts as a critical component for
the structure of paraspeckles (30,31).
The present study observed that the expression of NEAT1 was
enhanced in HCC cell lines compared with the hepatic stellate cells
LX-2. These results suggest that NEAT1 may serve a role in tumor
carcinogenesis in HCC. To investigate the role of NEAT1 in HCC, its
expression was downregulated and upregulated through stable
transfection of Huh7 and MHCC-97H cells. Silencing NEAT1
significantly inhibited proliferation and migration but increased
apoptosis of HCC cells. By contrast, the over-expression of NEAT1
significantly promoted proliferation and migration but inhibited
the apoptosis of HCC cells. In addition, the downregulation of
NEAT1 reduced tumor weight and volume in xenograft nude mice
models, while overexpression of NEAT1 resulted in the opposite
effects. These results indicate that NEAT1 is an oncogene in
HCC.
The present study further investigated the potential
mechanism of NEAT1 in the development of HCC. miR-320a is a crucial
tumor suppressor in various neoplasms, including lung cancer,
salivary adenoid cystic carcinoma, breast cancer and endometrial
carcinoma (32-35). Notably, Zhang et al
(36) reported that miR-320a is
downregulated in HCC cell lines and tissues. Overexpression of
miR-320a not only inhibited the proliferation and invasion of HCC
cells but also decreased tumor growth in vivo. In line with
this study, the present study also observed a significant decrease
in the expression of miR-320a in HCC cell lines. Bioinformatics
analysis suggested that NEAT1 contains a binding region for
miR-320a. Therefore, the association between NEAT1 and miR-320a in
HCC was investigated. In the present study, double luciferase
reporter analysis and RNA pull-down assays confirmed that NEAT1
could bind directly to miR-320a in HCC cells. Silencing NEAT1
elevated miR-320a expression in both HCC cells and xenograft nude
mice models. By contrast, overexpression of NEAT1 decreased
miR-320a expression. These results indi-cated that NEAT1 acts by
negatively modulating miR-320a expression in the development of
HCC.
LAGE3 (also known as ESO3), a subunit of the
EKC/KEOPS complex, is evolutionarily conserved and ubiquitously
expressed in somatic tissues (37,38).
It has been reported that the EKC/KEOPS complex, the matrix of
LAGE3, is associated with the processes of transcription, protein
synthesis, telomere homeostasis, genomic instability and cell
growth (39,40). LAGE3 interacts with a frequently
overexpressed human tumor antigen, preferably expresses antigen in
melanoma, and may engage in neoplastic processes (41). However, the role of LAGE3 is still
not fully understood. The present study found that LAGE3 is a
target of miR-320a and that miR-320a negatively regulates LAGE3
expression in HCC cells. LAGE3 was elevated in HCC cell lines
compared with normal liver cell lines. LAGE3 may participate in the
oncogenesis of HCC and serve as a potential therapeutic target.
It has been reported that certain lncRNAs can act as
sponges for miRNAs by modulating the expression of miRNA target
genes in various human diseases, including cancer. For example, the
lncRNA TUG1 has been reported to promote the development of oral
squamous cell carcinoma via sponging miR-524-5p, thereby affecting
distal-less homeobox 1 expression by acting as a ceRNA (42). Furthermore, lncRNA LINC01234
promoted the proliferation and the occurrence of gastric carcinoma
by sponging miR-204-5p as a ceRNA and alleviating the suppression
of the target gene core-binding factor subunit β (14). Through bioinformatics analysis, the
present study identified that NEAT1 contains similar response
elements as LAGE3 at the putative miR-320a binding sites. This
suggests that NEAT1 and LAGE3 may compete for the same binding site
of miR-320a. Therefore, it was hypothesized that NEAT1 may play a
carcinogenic role by sponging miR-320a and reducing the suppression
of LAGE3 in HCC. Then, an anti-AGO2 RIP assay was conducted, and
the results revealed that the content of NEAT1 and mir-320a in Ago2
particles was higher than compared with that in the IgG group.
Silencing NEAT1 significantly decreased the expression of LAGE3 in
mouse HCC tumor tissues, and overexpression of NEAT1 had the
opposite effect. Collectively, these results indicated that NEAT1
functions as a ceRNA for miR-320a to modulate LAGE3 expression.
In conclusion, the present study demonstrated that
NEAT1 may facilitate the progression of HCC. In the current study,
the potential mechanism of the NEAT1/miR-320a/LAGE3 axis in HCC
cells was confirmed, and the overexpression of NEAT1 was found to
promote the progression of HCC. In addition, negative correlation
association between NEAT1 and miRNA-320a was identified. The
present study focused on LAGE3 since it is targeted by miR-320a.
The current findings reveal that the NEAT1/miR-320a/LAGE3 axis
participates in the development of HCC and that NEAT1 may be a
potential biomarker for HCC.
Abbreviations:
|
lncRNA
|
long noncoding RNA
|
|
HCC
|
hepatocellular carcinoma
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81401324 and 81770410),
Guangdong Provincial International Cooperation Base of Science and
Technology (Organ Transplantation) (grant no. 2015B050501002),
Guangdong Provincial Natural Science Funds for Distinguished Young
Scholars (grant no. 2015A030306025), Special Support Program for
Training High-Level Talent in Guangdong Province (grant no.
2015TQ01R168), Pearl River Nova Program of Guangzhou (grant no.
201506010014), and Scientific Program for Young Teachers of Sun
Yat-sen University (grant no. 16ykpy05).
Availability of data and materials
The data used and analyzed in this study are
available from the corresponding author upon request.
Authors' contributions
YZ and MC contributed to the design and writing of
the study. ZZhu and CH acquired and analyzed the data. CH, YT, SH,
LW, HC and CS performed the experiments. MC, ZZha and WJ
contributed to the data analysis. YZ, YH and XQ read and revised
the manuscript, and made substantial contributions to the
interpretation and analysis of the data. YZ and XQ contributed to
drafting the manuscript, critically modifying important content,
and approving the version to be published. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures and protocols were
approved by the Research Ethics Committee on the Ethics of Animal
Experiments of The First Affiliated Hospital of Sun Yat-Sen
University (Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu J: Trends in liver cancer mortality
among adults aged 25 and over in the United States, 2000-2016. NCHS
Data Brief. 1–8. 2018.
|
|
5
|
Necsulea A, Soumillon M, Warnefors M,
Liechti A, Daish T, Zeller U, Baker JC, Grützner F and Kaessmann H:
The evolution of lncRNA repertoires and expression patterns in
tetrapods. Nature. 505:635–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernandes JCR, Acuña SM, Aoki JI,
Floeter-Winter LM and Muxel SM: Long non-coding RNAs in the
regulation of gene expression: Physiology and disease. Noncoding
RNA. 5:172019.
|
|
7
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Friedman RC, Farh KKH, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar
|
|
9
|
Kabekkodu SP, Shukla V, Varghese VK, D'
Souza J, Chakrabarty S and Satyamoorthy K: Clustered miRNAs and
their role in biological functions and diseases. Biol Rev.
93:1955–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding L, Ren J, Zhang D, Li Y, Huang X, Hu
Q, Wang H, Song Y, Ni Y and Hou Y: A novel stromal lncRNA signature
reprograms fibroblasts to promote the growth of oral squamous cell
carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis.
39:397–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo
YY, Feng J, Sanders S, Jin G, Singh R, et al: Loss of XIST in
breast cancer activates MSN-c-Met and reprograms microglia via
exosomal miRNA to promote brain metastasis. Cancer Res.
78:4316–4330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Y, Zhang JP, Chen X, Xu X, Cao G, Li
H and Wu T: LncRNA FTX sponges miR-215 and inhibits phosphorylation
of vimentin for promoting colorectal cancer progression. Gene Ther.
25:321–330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P,
Chen Q, Wei C, Fu H, Xu T, et al: Long noncoding RNA LINC01234
functions as a competing endogenous RNA to regulate CBFB expression
by sponging miR-204-5p in gastric cancer. Clin Cancer Res.
24:2002–2014. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao L, Zhang YJ and Zhang YB: Long
noncoding RNA CASC2 regulates hepatocellular carcinoma cell
oncogenesis through miR-362-5p/Nf-B axis. J Cell Physiol.
233:6661–6670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang X, Qu S, Wang L, Zhang H, Yang Z,
Wang J, Dai B, Tao K, Shang R, Liu Z, et al: PTBP3 splicing factor
promotes hepatocellular carcinoma by destroying the splicing
balance of NEAT1 and pre-miR-612. Oncogene. 37:6399–6413. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Z, Chang Q, Yang F, Liu B, Yao HW, Bai
ZG, Pu CS, Ma XM, Yang Y, Wang TT, et al: Long non-coding RNA NEAT1
overexpression is associated with unfavorable prognosis in patients
with hepatocellular carcinoma after hepatectomy: A Chinese
population-based study. Eur J Surg Onc. 43:1697–1703. 2017.
View Article : Google Scholar
|
|
18
|
Chen S and Xia X: Long noncoding RNA NEAT1
suppresses sorafenib sensitivity of hepatocellular carcinoma cells
via regu-lating miR-335-c-Met. J Cell Physiol. Apr 1–2019.Epub
ahead of print.
|
|
19
|
Müller V, Oliveira-Ferrer L, Steinbach B,
Pantel K and Schwarzenbach H: Interplay of lncRNA H19/miR-675 and
lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 13:1137–1149.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kong X, Zhao Y, Li X, Tao Z, Hou M and Ma
H: Overexpression of HIF-2α-dependent NEAT1 promotes the
progression of non-small cell lung cancer through
miR-101-3p/SOX9/Wnt/β-catenin signal pathway. Cell Physiol Biochem.
52:368–381. 2019. View Article : Google Scholar
|
|
21
|
Zhang M, Weng WW, Zhang QY, Wu Y, Ni S,
Tan C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1
activates Wnt/β-catenin signaling and promotes colorectal cancer
progression via interacting with DDX5. J Hematol Oncol. 11:1132018.
View Article : Google Scholar
|
|
22
|
Yan P, Su Z, Zhang Z and Gao T: LncRNA
NEAT1 enhances the resistance of anaplastic thyroid carcinoma cells
to cisplatin by sponging miR-9-5p and regulating SPAG9 expression.
Int J Oncol. 55:988–1002. 2019.PubMed/NCBI
|
|
23
|
Xia TF, Chen J, Wu K, Zhang J and Yan Q:
Long noncoding RNA NEAT1 promotes the growth of gastric cancer
cells by regulating miR-497-5p/PIK3R1 axis. Eur Rev Med Pharmacol
Sci. 23:6914–6926. 2019.PubMed/NCBI
|
|
24
|
He J, Xu F, Man X, Zhang Y and Li H: Long
non-coding RNA NEAT1 promotes tumor development and metastasis
through targeting RAB9A in malignant melanoma. Minerva Med. Jul
17–2019. View Article : Google Scholar : Online ahead of
print. PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres-sion data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Sun JY, Zhao ZW, Li WM, Yang G, Jing PY,
Li P, Dang HZ, Chen Z, Zhou YA and Li XF: Knockdown of MALAT1
expression inhibits HUVEC proliferation by upregulation of miR-320a
and downregulation of FOXM1 expression. Oncotarget. 8:61499–61509.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brown DC and Gatter KC: Ki67 protein: The
immaculate deception? Histopathology. 40:2–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Souquere S, Beauclair G, Harper F, Fox A
and Pierron G: Highly ordered spatial organization of the
structural long noncoding NEAT1 RNAs within paraspeckle nuclear
bodies. Mol Biol Cell. 21:4020–4027. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fortunato O, Borzi C, Milione M, Centonze
G, Conte D, Boeri M, Verri C, Moro M, Facchinetti F, Andriani F, et
al: Circulating mir-320a promotes immunosuppressive macrophages M2
phenotype associated with lung cancer risk. Int J Cancer.
144:2746–2761. 2019. View Article : Google Scholar
|
|
33
|
Sun L, Liu B, Lin Z, Yao Y, Chen Y, Li Y,
Chen J, Yu D, Tang Z, Wang B, et al: miR-320a acts as a prognostic
factor and Inhibits metastasis of salivary adenoid cystic carcinoma
by targeting ITGB3. Mol Cancer. 14:962015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu J, Wang JG, Zhang L, Yang HP, Wang L,
Ding D, Chen Q, Yang WL, Ren KH, Zhou DM, et al: MicroRNA-320a
inhibits breast cancer metastasis by targeting metadherin.
Oncotarget. 7:38612–38625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shu S, Liu X, Xu M, Gao X, Chen S, Zhang L
and Li R: MicroRNA-320a acts as a tumor suppressor in endometrial
carcinoma by targeting IGF-1R. Int J Mol Med. 43:1505–1512.
2019.PubMed/NCBI
|
|
36
|
Zhang Z, Li X, Sun W, Yue S, Yang J, Li J,
Ma B, Wang J, Yang X, Pu M, et al: Loss of exosomal miR-320a from
cancer-associated fibroblasts contributes to HCC proliferation and
metastasis. Cancer Lett. 397:33–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alpen B, Güre AO, Scanlan MJ, Old LJ and
Chen YT: A new member of the NY-ESO-1 gene family is ubiquitously
expressed in somatic tissues and evolutionarily conserved. Gene.
297:141–149. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wan LC, Maisonneuve P, Szilard RK, Lambert
JP, Ng TF, Manczyk N, Huang H, Laister R, Caudy AA, Gingras AC, et
al: Proteomic analysis of the human KEOPS complex identifies
C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids
Res. 45:805–817. 2017. View Article : Google Scholar
|
|
39
|
Daugeron MC, Lenstra TL, Frizzarin M, El
Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L,
Saveanu C, Jacquier A, et al: Gcn4 misregulation reveals a direct
role for the evolutionary conserved EKC/KEOPS in the t6A
modification of tRNAs. Nucleic Acids Res. 39:6148–6160. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rojas-Benitez D, Ibar C and Glavic A: The
Drosophila EKC/KEOPS complex: Roles in protein synthesis
homeostasis and animal growth. Fly (Austin). 7:168–172. 2013.
View Article : Google Scholar
|
|
41
|
Costessi A, Mahrour N, Sharma V,
Stunnenberg R, Stoel MA, Tijchon E, Conaway JW, Conaway RC and
Stunnenberg HG: The human EKC/KEOPS complex is recruited to Cullin2
ubiquitin ligases by the human tumour antigen PRAME. PLoS One.
7:e428222012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu S, Liu LH, Hu WW and Wang M: Long
noncoding RNA TUG1 regulates the development of oral squamous cell
carcinoma through sponging miR-524-5p to mediate DLX1 expression as
a competitive endogenous RNA. J Cell Physiol. 234:20206–20216.
2019. View Article : Google Scholar : PubMed/NCBI
|