Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common malignancy, with >600,000 cases diagnosed
annually worldwide (1). Half of
the patients with HNSCC are diagnosed in an advanced stage at the
first medical examination. In addition, >50% of recurrences
occur within 3 years following treatment (2-4).
Similar to other types of cancer, the accumulation of genetic and
epigenetic alternations is considered to generate and promote
HNSCC. Recently, several comprehensive analyses for HNSCC gene
mutations were performed using high-throughput next generation
sequencing defining NOTCH1 mutation at a 10-15% rate. This
rate is the second most frequent following TP53 and higher
than previously considered (5,6).
Subsequently a previous study demonstrated a bimodal pattern of
NOTCH pathway alterations in HNSCC, with a smaller subset of
HNSCC exhibiting inactivating NOTCH1 receptor mutations, but
a larger subset exhibiting NOTCH pathway activating
alterations, resulting in downstream HES1/HEY1 pathway
activation (7).
HES, HEY, CCND1, MYC, BCL-2 and p21
are NOTCH target genes. Among these genes, the HES
and HEY families are considered prominent downstream
effectors of the NOTCH pathway (8,9).
HEY1 is known to promote epithelial-mesenchymal transition
(EMT) in several normal tissues, such as the epidermis, kidney
tubules, mammary gland and endocardia (10-12).
HEY1 knockdown in glioblastoma cells has been shown to
decrease colony formation and invasion (13). A previous study demonstrated that
HEY1 expression in a skin human SCC cell line was increased
under 3D culture and promoted an EMT phenotype (14). Man et al indicated that
HNSCC exhibits a significantly higher HEY1 expression than
normal epithelial cells (15).
Recently, HEY1 has been shown to be associated with a poor
prognosis, independent of NOTCH1 expression, indicating that
other NOTCH members may drive HEY1 expression as a
key pathway alteration in HNSCC (16). In a previous study, it was also
found that the NOTCH4/HEY1 pathway was specifically
upregulated in HNSCC and it was revealed that this pathway promoted
EMT (17). However, the mechanisms
that effect NOTCH4/HEY1 pathway activation in HNSCC remain
unclear.
SOX2, as well as CD10 (18), CD44 (19) and ALDH1 (20) are HNSCC cancer stem cell (CSC)
markers (21). SOX2
expression in HNSCC is significantly related to a worse prognosis
(22) and SOX2 promotes
migration, invasion and EMT in HNSCC (23). To define NOTCH downstream
effectors, the present study examined the association between
SOX2 and HEY1. The authors previously demonstrated
that HEY1 knockdown significantly decreased NOTCH4
expression and decreased SOX2 expression in HNSCC cells
(17). To further define these
associations, the present study examined specific feedback loops
between HEY1 and NOTCH4 and SOX2 in HNSCC.
Materials and methods
Cells and cell culture
Cal27, SCC61 and SCC090 HNSCC cell lines were used
in the present study. Cal27 and SCC090 cells were obtained from the
Gutkind Laboratory at the University of California San Diego,
Moores Cancer Center. SCC61 cells were obtained from the
Weichselbaum Laboratory at the University of Chicago. SCC090 cells
were originally established from human papilloma virus
(HPV)-positive HNSCC tissues. The other two cells were established
from HPV-negative HNSCC tissues. These cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck
KGaA) supplemented with 10% fetal bovine serum (FBS) and a
penicillin (50 U/ml) and streptomycin (50 µg/ml) cocktail.
All cells were cultured under an atmosphere of 5% CO2 at
37°C.
Vector transfection
A lenti-ORF clone of human SOX2
(#RC200757L3), HEY1 (#RC200257L3) and an empty vector
control (#PS100092) were obtained from OriGene Technologies, Inc.
The 293T cells obtained from the Gutkind Laboratory were seeded in
6-well plates one day prior to transfection, and each construct was
transfected in Opti-MEM (#31985070, Thermo Fisher Scientific, Inc.)
and Turbofect transfection reagent (#R0531, Thermo Fisher
Scientific, Inc.). Viral supernatants were consisted of 10
µg lentiviral plasmid, 6.67 µg packaging vector and
3.33 µg envelope per well in 6 well plates, and collected at
48 and 72 h following transfection. Cal27, SCC61 and SCC090 cells
were seeded one day prior to infection in a 6-well plate and
allowed to reach 50-60% confluency. The virus supernatant and 2
µl of poly-brene (Sigma-Aldrich; Merck KGaA) were added to
the cells. Cells were maintained under puromycin (#ant-pr-1,
Invivogen; Thermo Fisher Scientific, Inc.) selection at a
1-µg/ml concentration. The following experiments using
SOX2 and HEY1 overexpression cells were compared
empty vector control transfected cells in Cal27, SCC61 and
SCC090.
Reverse transcription-quantitative PCR
(RT-qPCR)
To vali-date mRNA expression levels in each
experiment, RT-qPCR was used. Briefly, total RNA was extracted from
the cells using the RNeasy plus mini kit (Qiagen GmbH), and
comple-mentary DNA was synthesized using a high-capacity cDNA
reverse transcription kit (Thermo Fisher Scientific, Inc.). All
primers were obtained from TaqMan Gene Expression assays (cat. no.
4331182. Thermo Fisher Scientific, Inc.). Each gene ID is described
as follows: β-actin (ACTB): Hs01060665_g1; NOTCH4:
Hs00965895_g1; HES1: Hs00172878_m1; HEY1:
Hs01114113_m1; E-cadherin: Hs01023895_m1;
fibronectin: Hs01549976_m1; Vimentin: Hs00958111_m1;
TWIST1: Hs01675818_s1; and SOX2: Hs01053049_s1. The
thermocycle program was set at 95°C for 10 min, followed by 50
cycles of denaturation at 95°C for 15 sec and annealing at 60°C for
60 sec. PCR quantification was conducted using the ΔΔCq method
(24). qPCR was performed using
the Quant Studio 6 Flex Real-Time PCR System (Thermo Fisher
Scientific, Inc.).
Western blot analysis
Protein was obtained from the Cal27, SCC61 and
SCC090 cells, and lysed with RIPA buffer (50 mm Tris-HCl pH 8.0,
150 mm NaCl, 1% IGE-PAL CA 630, 0.5% Na-DOC, and 0.1% SDS). Total
protein concentrations were measured using Bio-Rad protein assay
kit (Bio-Rad Laboratories, Inc.). 10 µl Equal amount of
protein was set on Mini-PROTEAN TGX gels (Bio-Rad Laboratories,
Inc.). The following primary antibodies were added to
nitrocellulose membranes with 5% non-fat dry milk in Tris-buffered
saline and 1% Tween-20, and incubated at 4°C overnight: NOTCH4
(1:500, #2423, Cell Signaling Technology, Inc.), HES1 (1:1,000,
#sc-25392, Santa Cruz Biotechnology, Inc.), HEY1 (1:400, #ab22614,
Abcam), E-cadherin (1:10,000, #610181, BD Biosciences), fibronectin
(1:3,000, #ab2413, Abcam), Vimentin (1:500, #V6630, Sigma-Aldrich;
Merck KGaA), TWIST1 (1:1,000, #sc-15393, Santa Cruz Biotechnology,
Inc.) and SOX2 (1:1,000, #2748, Cell Signaling Technology, Inc.).
HRP-conjugated goat anti-mouse (#1010-05, 1:20,000 dilution;
SouthernBiotech) or anti-rabbit antibodies (#4010-05, 1:20,000
dilution; SouthernBiotech) were used as secondary antibodies. These
secondary antibodies were incubated at room tempera-ture for 1 h.
Western blots were developed using Pierce ECL Western Blotting
Substate (Thermo Fisher Scientific).
Migration and invasion assays
Migration assays were performed in cell culture
inserts (24-well, 8-µm pore size, #353097, Corning, Inc.).
Cell concentrations ranged from 105 to 2×105
cells/ml. Invasion assays were also performed in Corning BioCoat
Matrigel invasion chambers (24-well, 8-µm pore size,
#353097, Corning, Inc.). Cell concentrations ranged from
2×105 to 4×105 cells/ml. Cells were seeded on
uncoated or Matrigel-coated inserts in 500 ml of serum-free medium
for migration and invasion assays, respectively. The lower chambers
were filled with 750 µl of 10% FBS-supplemented medium.
After 48 h, the cells on the lower surface of the insert were fixed
and stained with crystal violet (Differential Quik Stain kit,
Polysciences) at room temperature for 2 min. The number of stained
cells was counted in >3 fields under an inverted microscope
(Olympus CKX31).
Sphere formation assay
Cells were seeded in 96-well ultralow attachment
culture dishes (Corning, Inc.) at 10-100 cells/well. Media
consisted of serum-free DMEM/F12 Glutamax supplement medium
(#10565042, Thermo Fisher Scientific, Inc.), basic fibroblast
growth factor (bFGF: 20 ng/ml, #13256029, Thermo Fisher Scientific,
Inc.), epithelial growth factor (EGF: 20 ng/ml, #PHG0313, Thermo
Fisher Scientific, Inc.), B-27 (1:50 dilution, #17504044, Thermo
Fisher Scientific, Inc.) and N2 supplement (1:100 dilution,
#17502-048, Thermo Fisher Scientific, Inc.). Images were obtained
at 10 days after seeding using a clinical upright microscope
(Olympus, BX43) (Fig. 2A), and the
numbers of sphere colonies in each well were counted using an
inverted microscope (Olympus CKX31).
Cell viability assay
Cells were seeded in 96-well plates at 1,500 to
9,000 cells/well. Cell numbers were measured on day 3. Cell
viabilities were measured using Vita Blue Cell Viability reagent
(Bimake.com). Following a 1.5-h pre-incubation at 37°C
in the assay solution, the viable cell number in each well was
calculated using fluorescence (Ex=530-570 nm, Em=590-620 nm) in a
microplate reader (BioTek Insturments, Inc.). The assays were
performed ≥3 times.
Mouse xenograft models
Cells (2×106) were diluted in 200 ml and
injected subcutaneously into nude mice (Charles River Laboratories,
Inc.) using a 25-gauge needle. Mice were anesthetized with a
mixture of oxygen and isoflurane (5% in air for induction and 2%
for maintenance) prior to each experiment, such as cell injection
and tumor size measurement. Mice were maintained under
pathogen-free conditions and sacrificed 2 months later or when
tumors exceeded 20 mm at the largest diameter or earlier if
necessary [this was done if any animal was observed to be cachexic
(weight loss >15% from starting weight), moribund, dehydrated,
anorexic, or any tumor that was ulcerated or eroded]. Mice were
euthanized using carbon dioxide gas for 10 min. The CO2
flow rate displaced 15-25% of the camber volume. Mice were
euthanized in November, 2017. The confirmation of euthanasia was
assured by verifying the absence of respiration, cardiac function
and toe/tail pinch reflexes at least 10 min. Mice were handled in
accordance with the procedures outlined in the Regulations on
Animal Experiments at University of California San Diego. The
Institutional Animal Care and Use Committee at the University of
California San Diego approved the study.
Immunohistochemistry
Mouse xenograft tumors were stained overnight at 4°C
with a HEY1 primary antibody (#ab22614, Abcam) diluted 1:100
in PBS with 2.5% BSA. Biotinylated IgG antibody (#BA-1000, Vector
Laboratories, Inc.) were used at 1:400 as a secondary antibody for
30 min at room temperature. Staining was developed at room
temperature for 2 min with DAB. Specimens were counterstained at
room temperature for 1 min with hematoxylin and mounted with
glycerol gelatin. A clinical upright microscope (Olympus, BX439)
was used to examine these specimens.
TCGA dataset
The mRNA expression sequence data of patients with
HNSCC were obtained from the firebrowse website (http://firebrowse.org/). These TCGA data included 522
HNSCC and 44 normal tissues. In total, 447 HNSCC cases were used
for NOTCH analysis, excluding 73 tumors with NOTCH
mutations. RNA expression was normalized by RSEM.
Reporter assay
Promoter reporter clone for human NOTCH4
(#HPRM45581-LvPG04), HEY1 (#HPRM10038-LvPG04) and negative
control (#NEG-LvPG04) containing a 1,443 bp region of the
NOTCH4 promoter regions and the HEY1 reporter assay
based on a 1,455 bp region of the HEY1 promoter region,
respectively, were obtained from GeneCopoeia, Inc. The Cal27, SCC61
and SCC090 cells were transfected using 2.0 µg of these
clones and 6 µl of X-tremeGENE 9 (Roche) per well in 6 well
plates, and incubated at 37°C for 24 h. The culture medium was
collected 48 h after transfection. The Secrete-Pair Dual
Luminescence Assay kit (#LF032, GeneCopoeia, Inc.) was used that
was optimized using these promoter reporter clones to validate each
promoter activity and the promoter activities were examined using
the manufacturer's protocol.
Chromatin immunoprecipitation qPCR
For chromatin immu-noprecipitation (ChIP) qPCR
assays, the SimpleChIP Plus Enzymatic Chromatin IP kit (#9005, Cell
Signaling Technology, Inc.) was used. Chromatin was incubated
overnight with anti-bodies for SOX2 (#2748, Cell Signaling
Technology, Inc.) or HEY1 (#19929-1-AP, ProteinTech Group,
Inc.) at 4°C under rotation. Chromatin was incubated with a
polyclonal rabbit IgG as a negative control and Histone H3 (D2B12)
XP-Rabbit mAb as positive control that were included in the ChIP
kit. Primer sequences for qPCR were obtained from Integrated DNA
Technologies, Inc. The following primer sequences were used:
HEY1 promoter forward, 5′-CCC GCT GAG AGG ATC TG-3′ and
reverse, 5′-CCC TGT GCA TCT CAT TTC C-3′; NOTCH4 promoter
forward, 5′-AGT GGT GCT GGT GAA GTA-3′ and reverse, 5′-CCA CAC ACT
GAG TTC CTT TAG-3′. The results were computed as percentage
antibody bound per input DNA and normalized to the IgG controls
using the Quant Studio 6 Flex Real-Time PCR System (Thermo Fisher
Scientific, Inc.).
Statistical analysis
All in vitro experiments were performed at
least in triplicate. Statistical comparisons of 2 groups were
determined using the Student's t-test. For the comparisons of
multiple group against the control group, Dunnett's test was used.
The correlation between the expression of 2 genes was determined
with Pearson's correlation analysis. Differences were considered
significant at P<0.05. All statistical analyses were performed
using JMP 12 software (SAS, Inc.).
Results
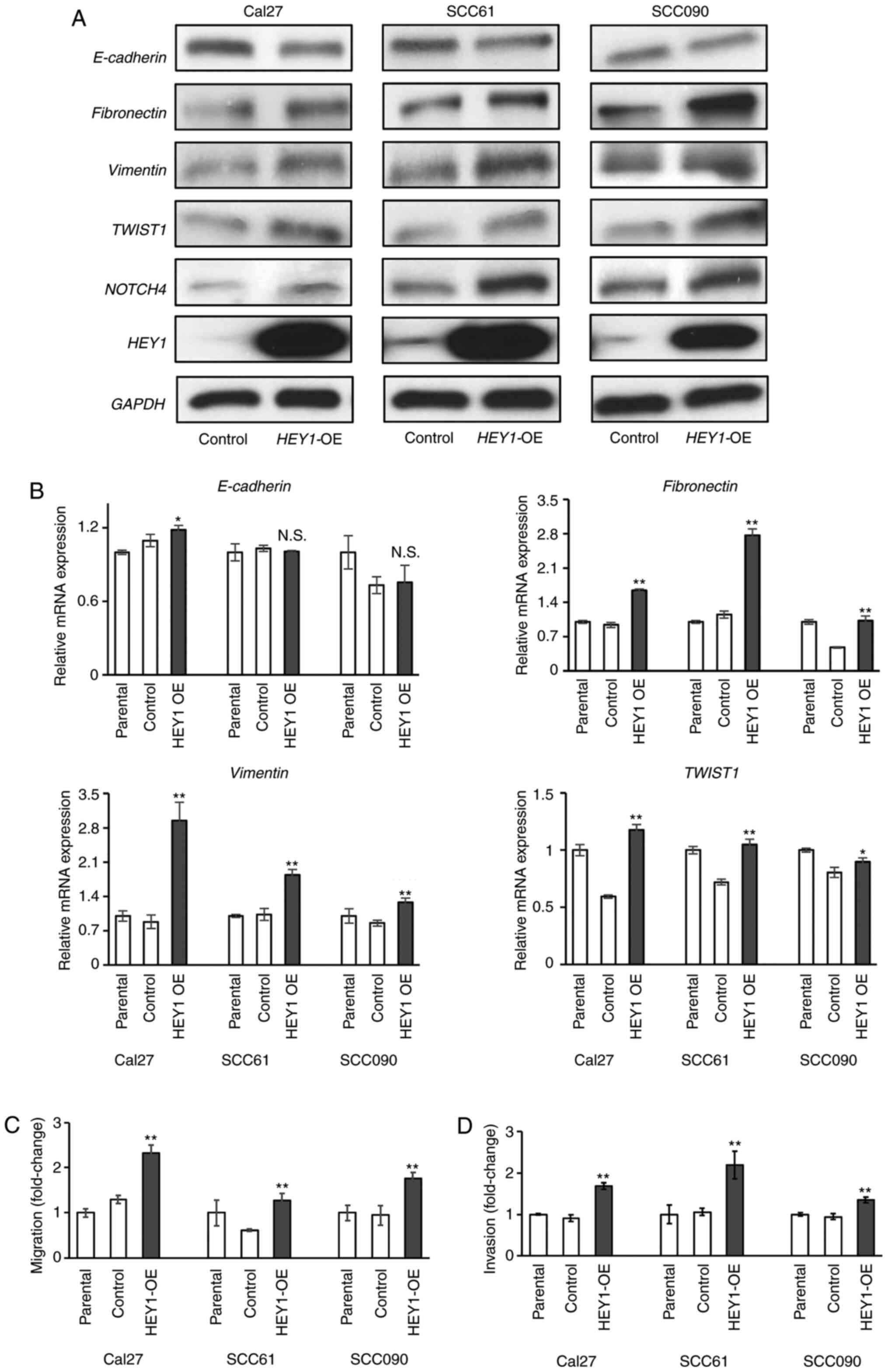
HEY1 promotes HNSCC EMT, migration and
invasion
Previously, the authors demonstrated that
HEY1 promoted EMT in HNSCC cells using cells in which
HEY1 was knocked down (17). To validate this finding, the
present study generated stable HEY1-overexpressing
(HEY1-OE) and control cells using the Cal27, SCC61 and
SCC090 cell lines (Figs. 1A and
S1A). The results of RT-qPCR
revealed that mesenchymal gene expression (fibronectin,
Vimentin and TWIST1) in the HEY1-OE cells
significantly increased in all HNSCC cells. However,
E-cadherin expression was higher in the Cal27 HEY1-OE
than in the control cells, with no significant differences observed
between the SCC61 and SCC090 HEY1-OE and control cells
(Fig. 1B). By contrast, western
blot analysis revealed a decreased E-cadherin expression in all
HEY1-OE cells. Furthermore, the expression of fibronectin,
Vimentin and TWIST1 increased in all the HNSCC HEY1-OE cells
(Fig. 1A). EMT is associated with
increased cellular migration and invasion; therefore, migration and
invasion assays were performed to determine the mechanisms through
which the changes in mRNA and protein expression affected the cell
phenotype in vitro. Increased migration and invasion were
noted in the HEY1-OE cells compared to the control cells
(Figs. 1C and D, and S1B). These results reveal that
HEY1 promotes HNSCC EMT, migration and invasion.
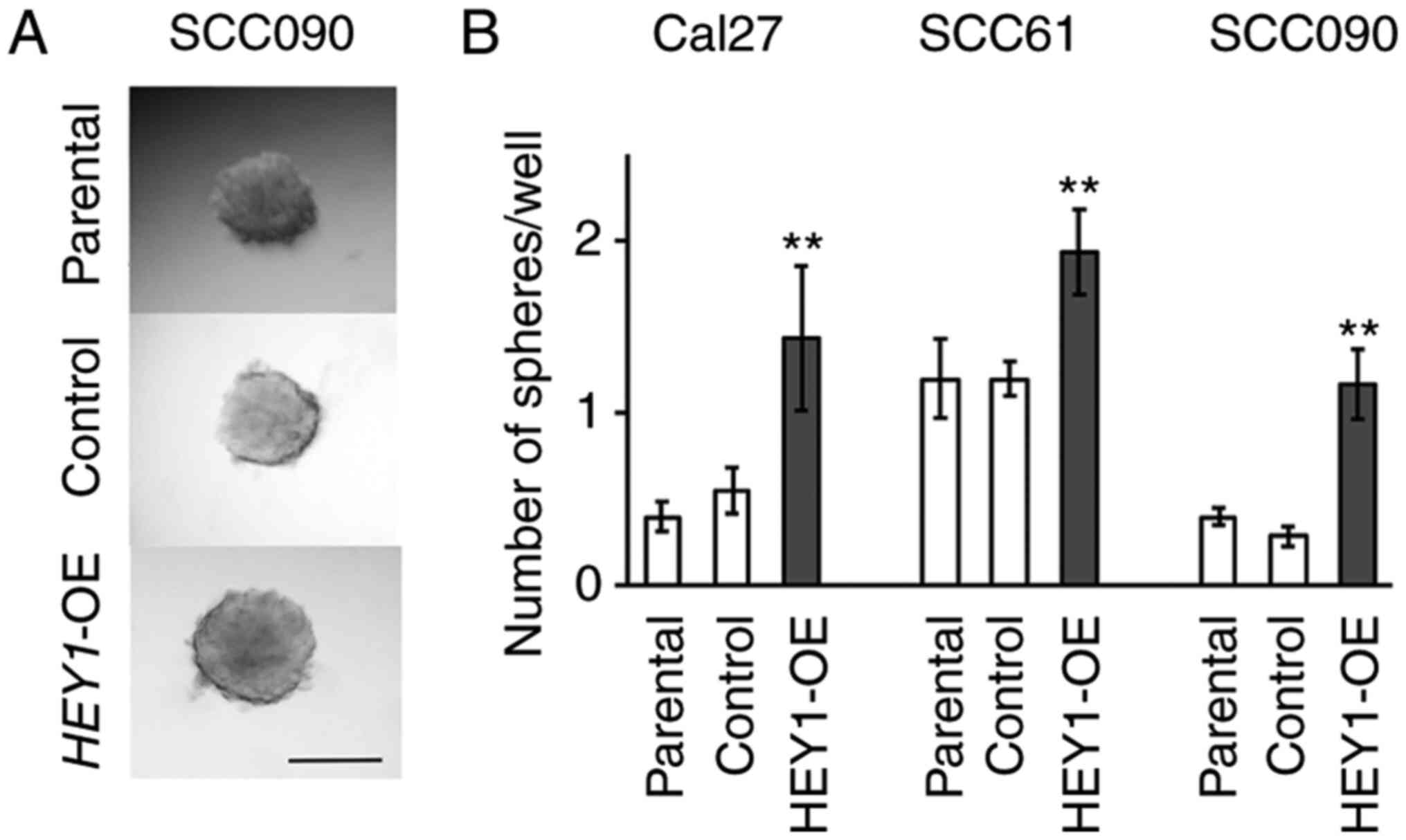
HEY1 promotes sphere formation
ability
Spheroids were generated to define the increase in
the expression of EMT-related genes associated with sphere
formation (25). The present study
compared the number of sphere colonies between the HEY1-OE
and control cells. No evident differences in sphere shape were
noted between the control and HEY1-OE cells (Fig. 2A); however, the HEY1-OE
cells formed significantly more spheroids in all cell lines
(Fig. 2B). The number of spheroids
in the HEY1-OE group was several folds higher than that of
the control group (Cal27 cells, 2.60; SCC61 cells, 1.61; SCC090
cells, 4.18) (Fig. 2B). These
results indicate that HEY1 promotes spheroid formation.
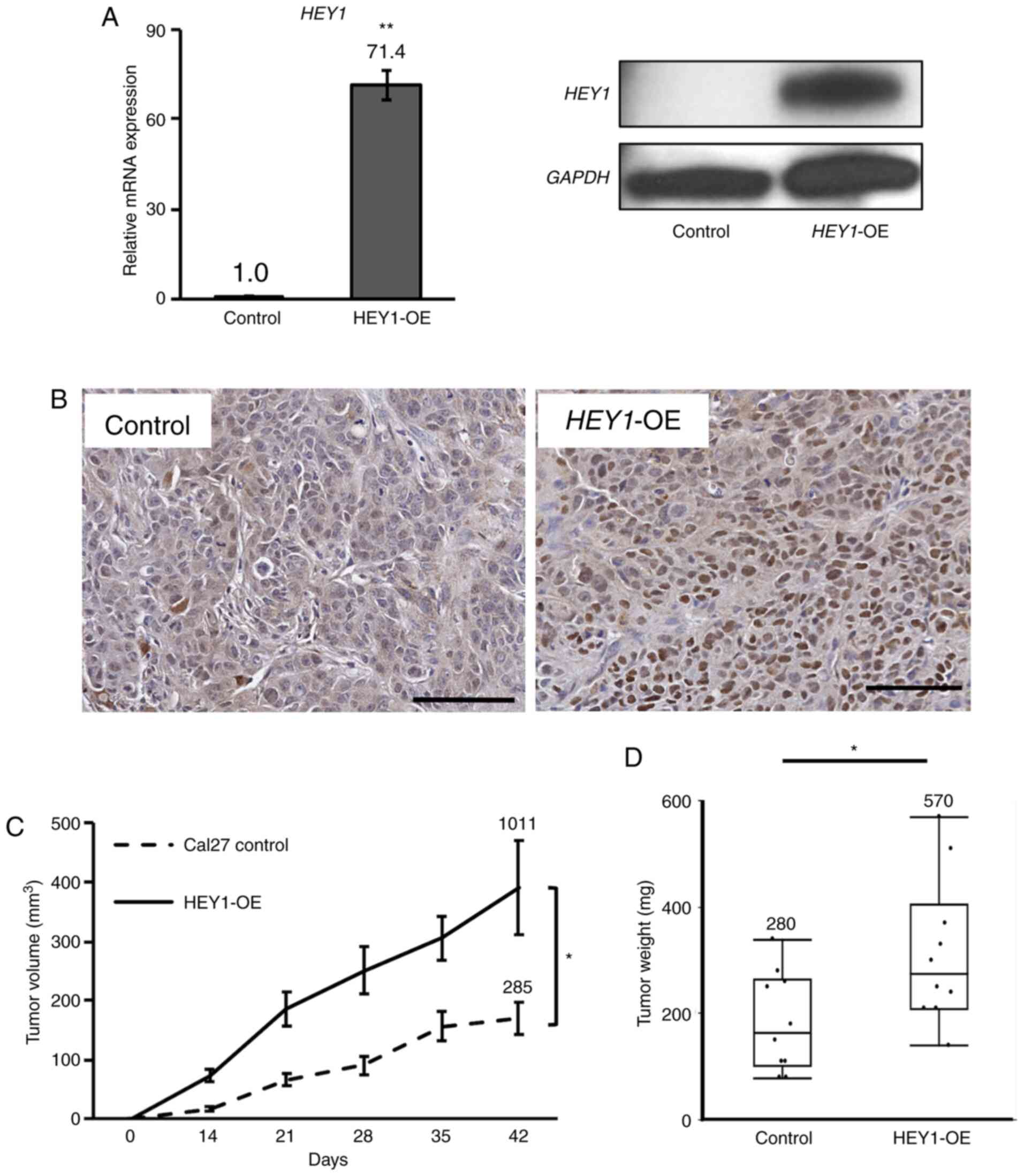
HEY1 promotes HNSCC tumorigenicity
A proliferation assay was performed to assess
phenotypic characteristics affected by HEY1. A statistically
significant increase in proliferation was observed in all
HEY1-OE cells compared to the control cells (Fig. S2A). The tumorigenicity of the
Cal27 HEY1-OE cells was then examined using a nude mouse
xenograft model. HEY1 expression in these tumors was
confirmed to be markedly higher in the HEY1-OE cell tumors
than in the control cell tumors using RT-qPCR and western blot
analysis (Fig. 3A).
Immunohistochemical staining for HEY1 also revealed that the
xenograft tumors generated from HEY1-OE cells exhibited a
higher HEY1 expression than those from the control cells (Fig. 3B). The HEY1-OE cells also
generated significantly larger tumors than the control cells
(Figs. S2B and 3C), confirmed by an increased tumor
weight of the Cal27 HEY1-OE tumors compared to the control
tumors following tumor excision. The maximum tumor diameter was
12.9 mm in the control group, and 14.9 mm in the HEY1-OE
group (Fig. 3D).
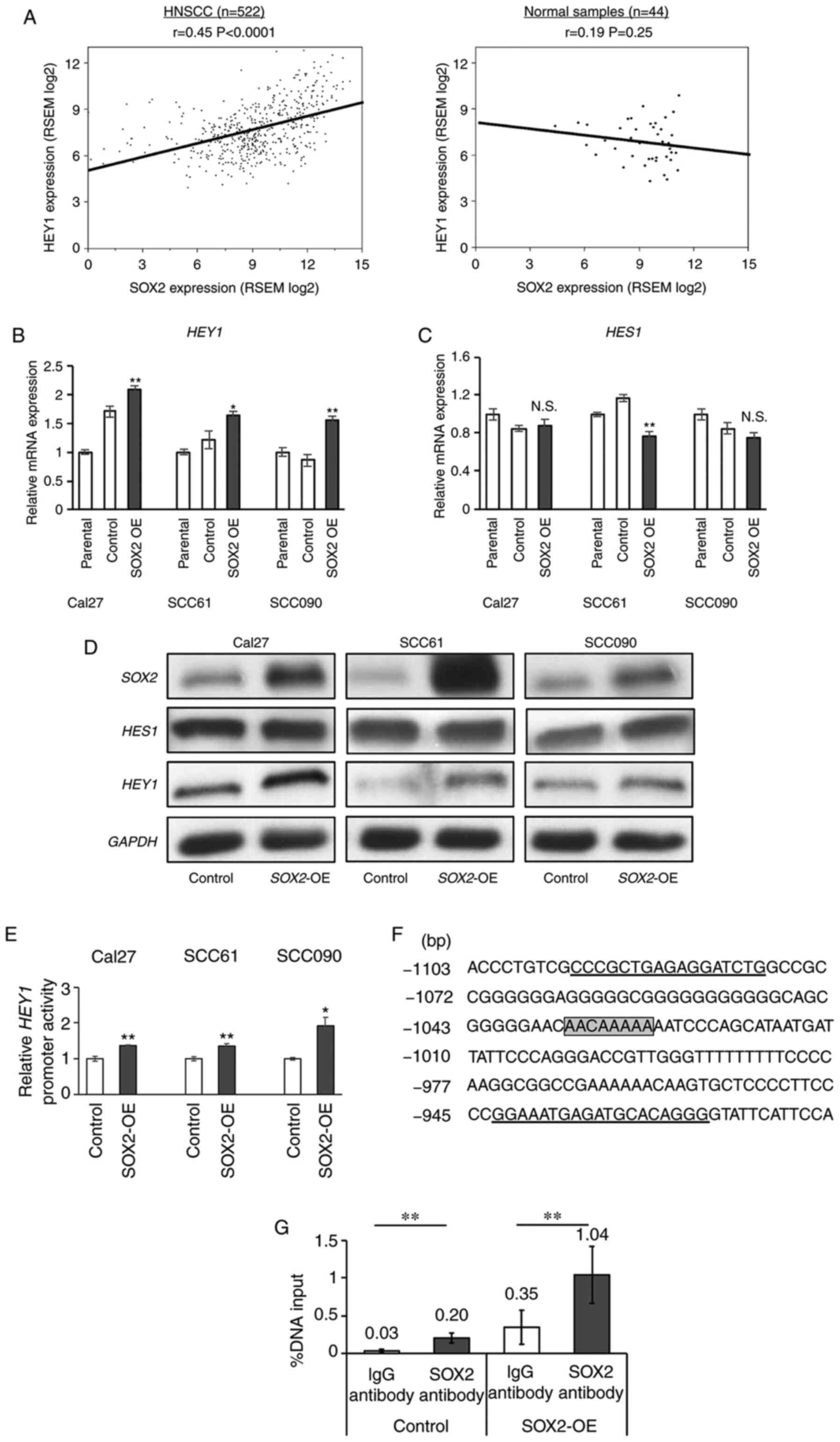
SOX2 expression correlates with HEY1
expression
Several studies have indicated that SOX2 is
associated with EMT and a CSC state (26-28).
In the present study, to elucidate a potential SOX2 and
HEY1 association in HNSCC, the correlation between
HEY1 and SOX2 was examined using the TCGA mRNA
sequence data from 522 HNSCC and 44 normal tissues samples. A
significant positive correlation was noted between SOX2 and
HEY1 mRNA expression in HNSCC (r=0.45, P<0.0001);
however, no significant correlation between these genes was found
in the normal tissues (Fig. 4A).
Furthermore, other NOTCH downstream genes, such as
HES1 and HES5 did not exhibit any significant
positive correlations with SOX2 (Fig. S3A). Of note, this association was
independent of the HPV status (Fig.
S3B). To explore this in vitro,
SOX2-overexpressing (SOX2-OE) and control cells were
generated (Fig. S3C). RT-qPCR
demonstrated a significant increase in HEY1 expression in
all HNSCC SOX2-OE cells examined. The SOX2-OE cells
exhibited an approximately 1.5- to 2.0-fold higher HEY1
expression in all cell lines (Fig.
4B). However, HES1 expression did not differ
significantly between the SOX2-OE and control Cal27 and
SCC090 cells. The SCC61 SOX2-OE cells exhibited a
significantly lower HES1 expression compared to the control
cells (Fig. 4C). All
SOX2-OE cells exhibited a higher HEY1 expression, as shown
by western blot analysis. However, HES1 expression between the
SOX2-OE and control cells did not exhibit a marked
difference (Fig. 4D). Among the 3
cell lines, it was found that both SOX2 and HEY1 expression was
higher in the Cal27 control cells. This result indicated that there
was an association between SOX2 and HEY1 expression
in HNSCC wild-type cells (Fig.
4D). These results demonstrated that SOX2 regulated
HEY1 expression in HNSCC.
SOX2 directly binds the HEY1 promoter in
HNSCC
To assess whether SOX2 directly binds
HEY1 promoter region, a luciferase vector with the
HEY1 promoter region was transfected into SOX2-OE and
control cells. A significant increase in luciferase activity was
observed in all HNSCC SOX2-OE cells (Fig. 4E). A ChIP qPCR was then performed
to validate this result. SOX2 is a transcription factor that
binds to the DNA consensus sequence (T/A)(T/A)CAAAGA (29) or AACAA(A/T)(G/A)(G/A) (30). A candidate SOX2 binding
sequence was found from -1,028 to -1,035 bp upstream of the
HEY1 transcript starting site. Therefore, a ChIP qPCR primer
pair was created that bound to this sequence (Fig. 4F). The results of ChIP qPCR
revealed that the parental control and stable SOX2-OE cells
incubated with a SOX2 antibody exhibited significantly
higher enrichment compared to those incubated with IgG antibodies
(Fig. 4G). These results
demonstrated that SOX2 can bind and activate the HEY1
promoter in HNSCC.
SOX2 correlates with HEY1 expression in
HNSCC and other SCCs
To examine whether SOX2 is related to
HEY1 expression in other types of cancer, this association
was explored using a TCGA dataset from multiple types of cancers
(Table I). SCCs, including HNSCC,
esophageal and lung cancer, exhibited a significantly higher
HEY1 expression compared to normal tissues. By contrast,
other cancer types, such as lung adenocarcinoma, colon, breast and
prostate cancer, exhibited a significantly lower HEY1
expression compared to normal tissues. All types of SCC exhibited
an approximately 1.5- to 1.8-fold higher HEY1 expression
compared to normal cells (Table
I). The correlation between HEY1 and SOX2 was
also compared using the TCGA dataset for each type of cancer.
Similar to HNSCC (Fig. 4A and
Table I), significant positive
correlations were found between SOX2 and HEY1 in
esophageal and lung SCC. The SOX2-HEY1 correlation
coefficients in lung adenocarcinoma, colon, breast and prostate
were <0.20, indicating a weak correlation (Table I).
 | Table ITCGA dataset analysis of HEY1
expression ratios between cancer and normal tissues and
SOX2-HEY1 correlation coefficients in several cancer
types. |
Table I
TCGA dataset analysis of HEY1
expression ratios between cancer and normal tissues and
SOX2-HEY1 correlation coefficients in several cancer
types.
| Cancer types | Number of tumor
samples | Number of normal
samples | HEY1
expression ratio (tumor/normal) | P-value | SOX2-HEY1
correlation coefficient in tumor samples | P-value | SOX2-HEY1
correlation coefficient in normal samples | P-value |
|---|
| HNSCC | 522 | 44 | 1.78 | <0.0001 | 0.45 | <0.0001 | 0.18 | 0.25 |
| Esophageal
carcinoma | 185 | 11 | 1.80 | 0.082 | 0.57 | <0.0001 | -0.56 | 0.073 |
| Lung SCC | 501 | 51 | 1.46 | 0.0003 | 0.57 | <0.0001 | 0.35 | 0.011 |
| Lung
adenocarcinoma | 517 | 59 | 0.39 | <0.0001 | 0.12 | 0.0076 | 0.024 | 0.85 |
| Colon
adenocarcinoma | 459 | 41 | 0.58 | <0.0001 | 0.20 | 0.0001 | 0.099 | 0.54 |
| Breast
carcinoma | 1100 | 112 | 0.53 | <0.0001 | 0.15 | <0.0001 | 0.016 | 0.088 |
| Prostate
adenocarcinoma | 498 | 52 | 0.72 | 0.011 | 0.12 | 0.0088 | 0.58 | <0.0001 |
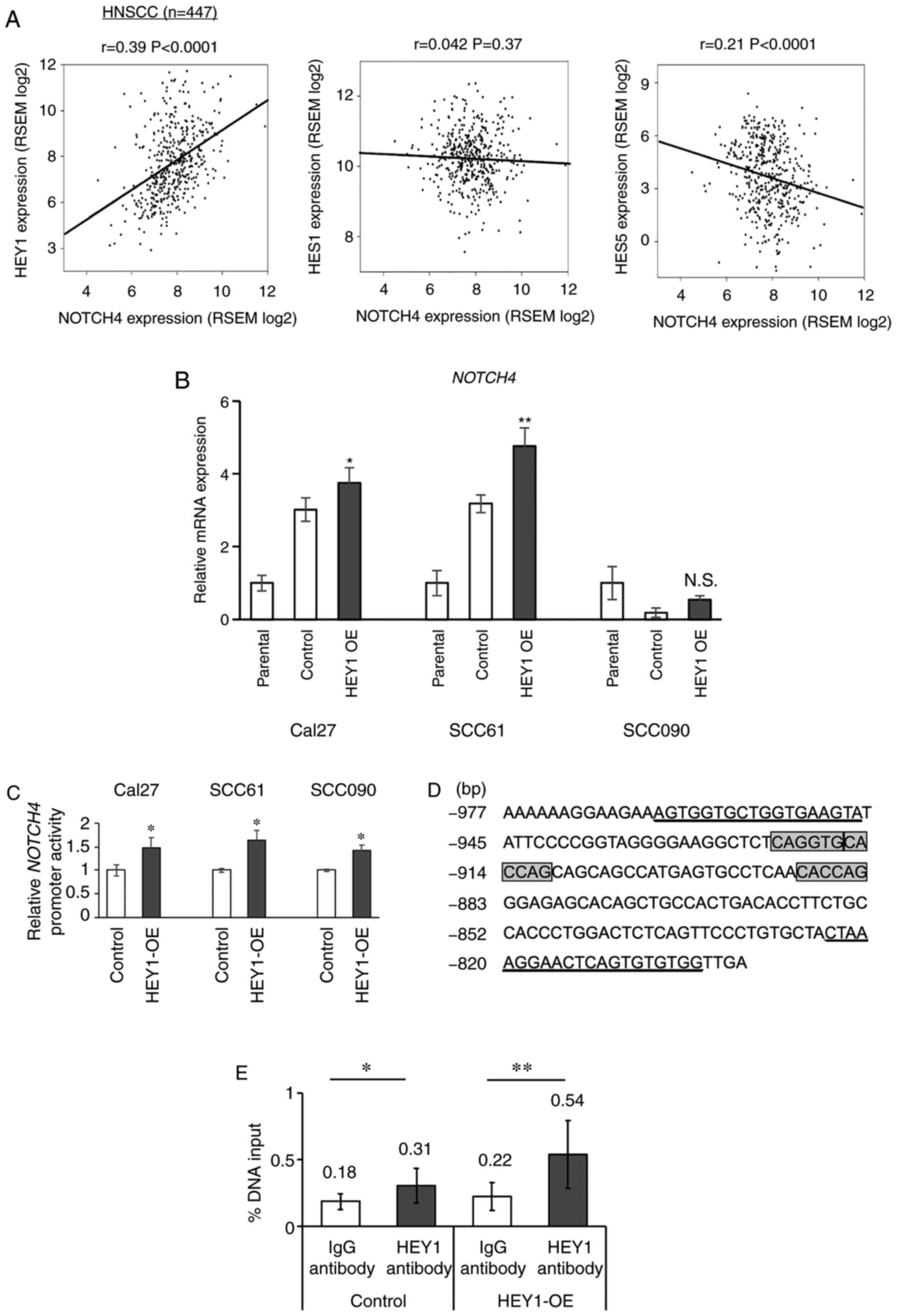
HEY1 correlates with NOTCH4
expression
HEY1 is a NOTCH target gene; however,
a previous study by the authors demonstrated a significantly lower
NOTCH4 expression in HNSCC cells in which HEY1 was
knocked down (17). Based on this
result, it was hypothesized that HEY1 also reciprocally
increased NOTCH4 expression directly through the
NOTCH4 promoter. Therefore, the correlation between
NOTCH4 and NOTCH downstream genes, including
HEY1, was examined using the head and neck TCGA dataset
(Fig. 5A). A positive correlation
was noted between NOTCH4 and HEY1 mRNA expression
(r=0.39, P<0.0001). However, no significant positive correlation
was found between NOTCH4 and HES1 and 5
(Fig. 5A). This correlation
between NOTCH4 and HEY1 was also independent of the
HPV status (Fig. S4A). When the
association between all NOTCH receptors and HEY1 was
compared, NOTCH4 exhibited the highest positive correlation
to HEY1 of all the NOTCH receptors (Figs. S4B and 5A). RT-qPCR revealed a significantly
increased NOTCH4 expression in the Cal27 and SCC61
HEY1-OE cells. NOTCH4 expression did not differ
significantly between the SCC090 HEY1-OE and control cells
(Fig. 5B). All the HEY1-OE
cells exhibited an elevated NOTCH4 expression, as shown by western
blot analysis (Fig. 1A).
Therefore, the present study examined whether HEY1 directly
binds the NOTCH4 promoter region and promotes its
transcription, similar to the association of SOX2 and
HEY1.
HEY1 directly binds the NOTCH4 promoter
in HNSCC
To assess whether HEY1 directly binds
NOTCH4 promoter region, a luciferase vector with a
NOTCH4 promoter region was transfected into HEY1-OE
and control cells. A significant increase in luciferase activity
was observed in all HNSCC HEY1-OE cells (Fig. 5C). The HEY1 gene binds E-box
(CANGTG) and N-box (CACNAG) sites (31,32).
Three candidates of HEY1 binding sequences were found from
-884 to -922 bp upstream of the NOTCH4 transcript starting
site. Therefore, a ChIP qPCR primer pair that bound this region was
generated (Fig. 5D). The ChIP qPCR
results revealed that the parental control and stable
HEY1-OE cells incubated with a HEY1 antibody
exhibited significantly higher enrichment compared to those
incubated with IgG antibodies (Fig.
5E). These data demonstrate that HEY1 can bind the
NOTCH4 promoter in HNSCC and drive NOTCH4
expression.
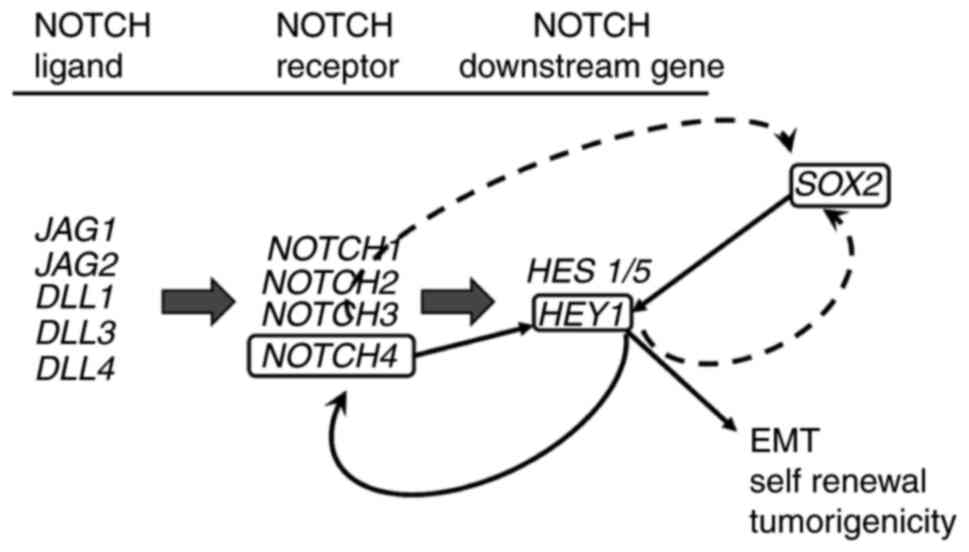
The associations between these genes are summarized
in Fig. 6; these data demonstrate
that SOX2 regulates HEY1 and HEY1 creates a
reciprocal loop with NOTCH4 to promote HNSCC EMT, sphere
formation and tumorigenicity.
Discussion
Previous studies have revealed that the
NOTCH pathway is upregulated in HNSCC and that NOTCH
expression is related to an advanced clinical stage (33,34).
In a previous study, the authors observed a specific upregulation
of the NOTCH4-HEY1 pathway in HNSCC (17). In the present study, further
analysis of a specific NOTCH pathway and a new mechanism of
functional integration is reported between SOX2 and the
NOTCH4-HEY1 axis.
It was found that the HEY1-OE cells
increased proliferation and tumorigenicity in xenograft models.
HEY1-OE cells also increased cell invasion and migration,
known as EMT ability, and promoted sphere formation, reflecting a
phenotype of cell stemness, self-renewal ability in vitro
(35-37). As shown in Fig. 1B, E-cadherin expression was
higher in the Cal27 HEY1-OE than the control cells, and did
not differ significantly between the HEY1-OE and control
SCC61 and SCC090 cells. By contrast, western blot analysis revealed
a decreased E-cadherin expression in all HEY1-OE cells
(Fig. 1A). This may be due to
post-translational processing, as the post-translational
E-cadherin modification is known to induce EMT in cancer
(38). E-cadherin mRNA and
protein expression differed in all HEY1-OE cells.
Furthermore, the expression levels of mesenchymal genes, such as
fibronectin, Vimentin and TWIST1 in the
HEY1-OE cells were increased in all HNSCC cells (Fig. 1B). Western blot analysis of
N-cadherin expression was also performed in these cells; however,
no increase in N-cadherin increase expression was found in the
HEY1-OE cells (Fig. S4C).
The reason for this lack of change in N-cadherin expression is not
clear. The present study did not validate N-cadherin and E-cadherin
expression in control and HEY1-OE cells using
immunocytochemistry. This is a limitation of the present study.
However, the authors have previously demonstrated that
N-cadherin expression was significantly increased in the
HEY1 high expression group in HNSCC patients using a TCGA
dataset (17). Thus, these in
vitro assay results demonstrated that HEY1 promotes EMT
in HNSCC.
HEY1 expression significantly correlated
with SOX2 expression in the TCGA dataset and in in
vitro experiments. SOX2 is an EMT inducer gene that
promotes HNSCC cell invasion and migration (23,28,39).
SOX2 and HEY1 are early sensory markers that exist in
the same domain in mice inner ear development (40) and SOX2 is co-expressed with
HEY1/HEY2 in the inner ear (41). In glioma CSC, both SOX2 and
HEY1 expression is increased compared to non-CSC glioma
cells (42). Chen et al
demonstrated that JAG1 promoted HEY1 and SOX2
expression, and NOTCH inhibition by a gamma secretase
inhibitor decreased SOX2 promoter activity in breast cancer
cells (43). However, these
studies did not examine which NOTCH related genes directly
interacted with the SOX2 promoter (43). In this context, the present study
examined the association between SOX2 and HEY1. As
noted above, the reporter and ChIP assays indicated that
SOX2 regulated HEY1 expression via binding to its
promoter. There are no commercially available SOX2 blocking
antibodies. Thus, the authors were not able to define HEY1
expression and an EMT phenotype using SOX2 blocking
antibody. Of note, the present study did not perform SOX2
mutational analysis in reporter assays of HEY1 that would
allow the precise localization of the SOX2 binding
HEY1 promoter within a 1-2 kb segment. The authors have
previously demonstrated that HEY1 expression was
significantly increased in sphere cells of HNSCC cell lines;
HEY1 expression in sphere cells was increased approximately
1.4- to 3.5-fold compared with parental cells (17). In the present study, it was
demonstrated that HEY1 promotes sphere formation and
tumorigenicity that are closely related to a cancer stem cell
phenotype. These results indicate that SOX2 maintains HNSCC
CSCs through HEY1 expression.
A previous study by the authors demonstrated that
HEY1 knockdown HNSCC cells decreased SOX2 expression,
as shown by RT-qPCR and western blot analysis (17). These two results indicate that
SOX2 and HEY1 may exist in a reciprocal loop similar
to that of NOTCH4-HEY1. On the other hand, Wang et al
demonstrated that glioma stem cells made a NOTCH1-SOX2
positive feedback loop (44). The
TCGA analysis in the previous study by the authors also indicated
that NOTCH4 and SOX2 expression had a significant
positive correlation (17). These
results indicate that NOTCH4 may promote SOX2
expression in HNSCC similar to HEY1 expression (Fig. 6). If NOTCH4 promotes
SOX2 expression, this may explain why cells in which
HEY1 was knocked down had a decreased SOX2
expression, as HEY1 knockdown in cells decreased
NOTCH4 expression (Fig. 6).
Previous studies have indicated this connection in neural stem
cells and brain endothelial cells; NOTCH increases
SOX2 promoter activity and regulates SOX2 expression
(45,46).
Furthermore, the current TCGA dataset analysis also
revealed that SOX2 correlates with HEY1 expression in
several SCCs, but not in non-SCC cancers or normal tissue (Table I). There are several studies on
SOX2 function in SCC and its promotion of tumorigenesis,
metastasis and EMT (21,47,48).
As regards HEY1 function in SCC, Forghanifard et al
examined NOTCH pathway gene expression in 50 patients with
esophageal SCC and indicated that HEY1 and HEY2
expression were significantly associated with clinical stage and a
poor prognosis (49). The results
of the present study and these reports indicate that HEY1
upregulation promotes tumor progression, and that a
SOX2-HEY1 expression correlation is found predominantly in
SCC, reinforcing a context dependent setting for activation of the
NOTCH4-HEY1 pathway. However, the TCGA dataset shows only
each mRNA expression of individual patients. The present study
performed a functional analysis for these molecules in HNSCC, but
not in other SCC types, limiting functional confirmation in these
systems.
In the present study, the reporter and ChIP assays
also indicated that HEY1 reciprocally regulated
NOTCH4 expression via binding to its promoter. James et
al demonstrated that the NOTCH4 was ligand unresponsive
in NOTCH signaling (50).
These results may explain why the NOTCH4-HEY1 pathway was
specifically upregulated in HNSCC. Of note, specific mutational
analysis in NOTCH4 reporter assays to precisely localize the
HEY1 binding site would add further depth to the current
understanding of this interaction. Nevertheless, the results of
additional mutational analysis would not substantively alter the
conclusions based on these data, that SOX2 and HEY1
interact with these promoter regions. The present study did not
explore the ability of specific NOTCH ligands, such as
JAG1, JAG2, DLL1, DLL3 and DLL4 in the activation of
the NOTCH4-HEY1 pathway, which is a limitation of the
present study, as this may provide further data that support the
model of constitutive activation in the absence of exogenous
ligand. The present study did not assess whether HEY1 bound
to the NOTCH1-3 promoter, and NOTCH1 and 2
expression was lower in HNSCC than normal samples; however,
NOTCH3 expression was slightly higher in HNSCC than normal
tissue, which is similar to NOTCH4 (51). NOTCH3 also significantly and
positively correlated with HEY1 expression. NOTCH4
was more significantly and highly expressed in HNSCC and positively
correlated with HEY1 than NOTCH3 (Figs. 4A and S4B) (51). Notably, a similar correlation was
found between NOTCH4 and HEY1, HES1 and HES5
in head and neck normal samples of the TCGA dataset and HNSCC
(Figs. 5A and S4D). A moderately positive correlation
was noted only between NOTCH4 and HEY1 mRNA
expression in normal samples (r=0.52, P=0.0003). It was
hypothesized that the NOTCH4/HEY1 pathway is a specific
pathway not only active in HNSCC, but potentially active in head
and neck normal epithelial cells. Based on these findings, previous
studies have reported trials of anti-NOTCH therapy for HNSCC
(52,53).
In conclusion, the present study demonstrates that
HEY1 expression in HNSCC is regulated by SOX2 and
promotes EMT. The NOTCH4/HEY1/SOX2 pathway is specifically
upregulated and creates a positive reciprocal loop in HNSCC,
defining a pathway that may be a novel target for HNSCC
therapy.
Supplementary Data
Funding
The National Institute of Dental and Craniofacial
Research (NIDCR no. R01DE023347) supported the present study. JAC
received this grant. Apart from for this grant, no potential
conflicts of interest are disclosed.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
The study was designed and conceived by TF, TWG and
JAC. The experimental procedures and data analysis were carried out
by TF, SR, MA and JAC. The acquisition of data was carried out by
TF, SH, CL, AS, YS and SS. The manuscript was prepared by TF and
JAC. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All mice were handled in accordance with the
procedures outlined in the Regulations on Animal Experiments at
University of California San Diego. The Institution Animal Care and
Use Committee in University of California San Diego approved the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View Article : Google Scholar
|
|
2
|
Pignon JP, le Maître A, Maillard E and
Bourhis J; MACH-NC Collaborative Group: Meta-analysis of
chemotherapy in head and neck cancer (MACH-NC): An update on 93
randomised trials and 17,346 patients. Radiother Oncol. 92:4–14.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bernier J, Domenge C, Ozsahin M,
Matuszewska K, Lefèbvre JL, Greiner RH, Giralt J, Maingon P,
Rolland F, Bolla M, et al: Postoperative irradiation with or
without concomitant chemo-therapy for locally advanced head and
neck cancer. N Engl J Med. 350:1945–1952. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cooper JS, Pajak TF, Forastiere AA, Jacobs
J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et
al: Postoperative concurrent radiotherapy and chemotherapy for
high-risk squamous-cell carcinoma of the head and neck. N Engl J
Med. 350:1937–1944. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun W, Gaykalova DA, Ochs MF, Mambo E,
Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, et al:
Activation of the NOTCH pathway in head and neck cancer. Cancer
Res. 74:1091–1104. 2014. View Article : Google Scholar :
|
|
8
|
Kalaitzidis D and Armstrong SA: Cancer:
The flipside of Notch. Nature. 473:159–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sethi N, Dai X, Winter CG and Kang Y:
Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast
cancer by engaging notch signaling in bone cells. Cancer Cell.
19:192–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zavadil J, Cermak L, Soto-Nieves N and
Bottinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial-to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fischer A, Steidl C, Wagner TU, Lang E,
Jakob PM, Friedl P, Knobeloch KP and Gessler M: Combined loss of
Hey1 and HeyL causes congenital heart defects because of impaired
epithelial to mesenchymal transition. Circ Res. 100:856–863. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luna-Zurita L, Prados B, Grego-Bessa J,
Luxán G, del Monte G, Benguría A, Adams RH, Pérez-Pomares JM and de
la Pompa JL: Integration of a Notch-dependent mesenchymal gene
program and Bmp2-driven cell invasiveness regulates murine cardiac
valve formation. J Clin Invest. 120:3493–3507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsung AJ, Guda MR, Asuthkar S, Labak CM,
Purvis IJ, Lu Y, Jain N, Bach SE, Prasad DVR and Velpula KK:
Methylation regulates HEY1 expression in glioblastoma. Oncotarget.
8:44398–44409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shibata S, Marushima H, Asakura T,
Matsuura T, Eda H, Aoki K, Matsudaira H, Ueda K and Ohkawa K:
Three-dimensional culture using a radial flow bioreactor induces
matrix metalloprotease 7-mediated EMT-like process in tumor cells
via TGFbeta1/Smad pathway. Int J Oncol. 34:1433–1448.
2009.PubMed/NCBI
|
|
15
|
Man CH, Wei-Man Lun S, Wai-Ying Hui J, To
KF, Choy KW, Wing-Hung Chan A, Chow C, Tin-Yun Chung G, Tsao SW,
Tak-Chun Yip T, et al: Inhibition of NOTCH3 signalling
significantly enhances sensitivity to cisplatin in EBV-associated
nasopharyngeal carcinoma. J Pathol. 226:471–481. 2012. View Article : Google Scholar
|
|
16
|
Rettig EM, Bishop JA, Agrawal N, Chung CH,
Sharma R, Zamuner F, Li RJ, Koch WM, Califano JA, Guo T, et al:
HEY1 is expressed independent of NOTCH1 and is associated with poor
prognosis in head and neck squamous cell carcinoma. Oral Oncol.
82:168–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fukusumi T, Guo TW, Sakai A, Ando M, Ren
S, Haft S, Liu C, Amornphimoltham P, Gutkind JS and Califano JA:
The NOTCH4-HEY1 pathway induces epithelial-mesenchymal tran-sition
in head and neck squamous cell carcinoma. Clin Cancer Res.
24:619–633. 2018. View Article : Google Scholar
|
|
18
|
Fukusumi T, Ishii H, Konno M, Yasui T,
Nakahara S, Takenaka Y, Yamamoto Y, Nishikawa S, Kano Y, Ogawa H,
et al: CD10 as a novel marker of therapeutic resistance and cancer
stem cells in head and neck squamous cell carcinoma. Br J Cancer.
111:506–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Prince ME, Sivanandan R, Kaczorowski A,
Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles
LE: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen YC, Chen YW, Hsu HS, Tseng LM, Huang
PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al: Aldehyde
dehydrogenase 1 is a putative marker for cancer stem cells in head
and neck squamous cancer. Biochem Biophys Res Commun. 385:307–313.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho
YS, Bae WJ and Lim YC: SOX2 regulates self-renewal and
tumorigenicity of stem-like cells of head and neck squamous cell
carcinoma. Br J Cancer. 111:2122–2130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du L, Yang Y, Xiao X, Wang C, Zhang X,
Wang L, Zhang X, Li W, Zheng G, Wang S and Dong Z: Sox2 nuclear
expression is closely associated with poor prognosis in patients
with histo-logically node-negative oral tongue squamous cell
carcinoma. Oral Oncol. 47:709–713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang N, Hui L, Wang Y, Yang H and Jiang X:
Overexpression of SOX2 promotes migration, invasion, and
epithelial-mesenchymal transition through the Wnt/β-catenin pathway
in laryngeal cancer Hep-2 cells. Tumour Biol. 35:7965–7973. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Han XY, Wei B, Fang JF, Zhang S, Zhang FC,
Zhang HB, Lan TY, Lu HQ and Wei HB: Epithelial-mesenchymal
transition associates with maintenance of stemness in
spheroid-derived stem-like colon cancer cells. PLoS One.
8:e733412013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gangemi RM, Griffero F, Marubbi D, Perera
M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A and Corte G:
SOX2 silencing in glioblastoma tumor-initiating cells causes stop
of proliferation and loss of tumorigenicity. Stem Cells. 27:40–48.
2009. View Article : Google Scholar
|
|
27
|
Han X, Fang X, Lou X, Hua D, Ding W, Foltz
G, Hood L, Yuan Y and Lin B: Silencing SOX2 induced
mesenchymal-epithelial transition and its expression predicts liver
and lymph node metastasis of CRC patients. PLoS One. 7:e413352012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boumahdi S, Driessens G, Lapouge G, Rorive
S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E,
et al: SOX2 controls tumour initiation and cancer stem-cell
functions in squamous-cell carcinoma. Nature. 511:246–250. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Seo E, Basu-Roy U, Zavadil J, Basilico C
and Mansukhani A: Distinct functions of Sox2 control self-renewal
and differentiation in the osteoblast lineage. Mol Cell Biol.
31:4593–4608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salmon-Divon M, Dvinge H, Tammoja K and
Bertone P: PeakAnalyzer: Genome-wide annotation of chromatin
binding and modification loci. BMC Bioinformatics. 11:4152010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leal MC, Surace EI, Holgado MP, Ferrari
CC, Tarelli R, Pitossi F, Wisniewski T, Castano EM and Morelli L:
Notch signaling proteins HES-1 and Hey-1 bind to insulin degrading
enzyme (IDE) proximal promoter and repress its transcription and
activity: Implications for cellular Aβ metabolism. Biochim Biophys
Acta. 1823:227–235. 2012. View Article : Google Scholar
|
|
32
|
Iso T, Sartorelli V, Poizat C, Iezzi S, Wu
HY, Chung G, Kedes L and Hamamori Y: HERP, a novel heterodimer
partner of HES/E(spl) in Notch signaling. Mol Cell Biol.
21:6080–6089. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Joo YH, Jung CK, Kim MS and Sun DI:
Relationship between vascular endothelial growth factor and Notch1
expression and lymphatic metastasis in tongue cancer. Otolaryngol
Head Neck Surg. 140:512–518. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang TH, Liu HC, Zhu LJ, Chu M, Liang YJ,
Liang LZ and Liao GQ: Activation of Notch signaling in human tongue
carcinoma. J Oral Pathol Med. 40:37–45. 2011. View Article : Google Scholar
|
|
35
|
Lim YC, Oh SY, Cha YY, Kim SH, Jin X and
Kim H: Cancer stem cell traits in squamospheres derived from
primary head and neck squamous cell carcinomas. Oral Oncol.
47:83–91. 2011. View Article : Google Scholar
|
|
36
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shao K, Chen ZY, Gautam S, Deng NH, Zhou Y
and Wu XZ: Posttranslational modification of E-cadherin by core
fucosylation regulates Src activation and induces
epithelial-mesenchymal transition-like process in lung cancer
cells. Glycobiology. 26:142–154. 2016. View Article : Google Scholar
|
|
39
|
Vanner RJ, Remke M, Gallo M, Selvadurai
HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X, et al:
Quiescent sox2(+) cells drive hierarchical growth and relapse in
sonic hedgehog subgroup medulloblastoma. Cancer Cell. 26:33–47.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan W, Jin Y, Stanger B and Kiernan AE:
Notch signaling is required for the generation of hair cells and
supporting cells in the mammalian inner ear. Proc Natl Acad Sci
USA. 107:15798–15803. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Benito-Gonzalez A and Doetzlhofer A: Hey1
and Hey2 control the spatial and temporal pattern of mammalian
auditory hair cell differentiation downstream of Hedgehog
signaling. J Neurosci. 34:12865–12876. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fang KM, Lin TC, Chan TC, Ma SZ, Tzou BC,
Chang WR, Liu JJ, Chiou SH, Yang CS and Tzeng SF: Enhanced cell
growth and tumorigenicity of rat glioma cells by stable expression
of human CD133 through multiple molecular actions. Glia.
61:1402–1417. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen JY, Li CF, Chu PY, Lai YS, Chen CH,
Jiang SS, Hou MF and Hung WC: Lysine demethylase 2A promotes
stemness and angiogenesis of breast cancer by upregulating Jagged1.
Oncotarget. 7:27689–27710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang J, Xu SL, Duan JJ, Yi L, Guo YF, Shi
Y, Li L, Yang ZY, Liao XM, Cai J, et al: Invasion of white matter
tracts by glioma stem cells is regulated by a NOTCH1-SOX2
positive-feedback loop. Nat Neurosci. 22:91–105. 2019. View Article : Google Scholar
|
|
45
|
Ehm O, Göritz C, Covic M, Schäffner I,
Schwarz TJ, Karaca E, Kempkes B, Kremmer E, Pfrieger FW, Espinosa
L, et al: RBPJkappa-dependent signaling is essential for long-term
maintenance of neural stem cells in the adult hippocampus. J
Neurosci. 30:13794–13807. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu X, Yao J, Wang L, Zhang D, Zhang L,
Reynolds EX, Yu T, Bostrom KI and Yao Y: Crosstalk between BMP and
Notch induces Sox2 in cerebral endothelial cells. Cells. 8:5492019.
View Article : Google Scholar :
|
|
47
|
Ferone G, Song JY, Sutherland KD,
Bhaskaran R, Monkhorst K, Lambooij JP, Proost N, Gargiulo G and
Berns A: SOX2 is the determining oncogenic switch in promoting lung
squamous cell carcinoma from different cells of origin. Cancer
Cell. 30:519–532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bochen F, Adisurya H, Wemmert S, Lerner C,
Greiner M, Zimmermann R, Hasenfus A, Wagner M, Smola S, Pfuhl T, et
al: Effect of 3q oncogenes SEC62 and SOX2 on lymphatic metastasis
and clinical outcome of head and neck squamous cell carcinomas.
Oncotarget. 8:4922–4934. 2017. View Article : Google Scholar :
|
|
49
|
Forghanifard MM, Taleb S and Abbaszadegan
MR: Notch signaling target genes are directly correlated to
esophageal squamous cell carcinoma tumorigenesis. Pathol Oncol Res.
21:463–467. 2015. View Article : Google Scholar
|
|
50
|
James AC, Szot JO, Iyer K, Major JA,
Pursglove SE, Chapman G and Dunwoodie SL: Notch4 reveals a novel
mechanism regulating Notch signal transduction. Biochim Biophys
Acta. 1843:1272–1284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fukusumi T and Califano JA: The NOTCH
pathway in head and neck squamous cell carcinoma. J Dent Res.
97:645–65. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu S, Zhang R, Liu F, Hu H, Yu S and Wang
H: Down-regulation of Notch signaling by a γ-secretase inhibitor
enhances the radio-sensitivity of nasopharyngeal carcinoma cells.
Oncol Rep. 26:1323–1328. 2011.PubMed/NCBI
|
|
53
|
Zhao ZL, Zhang L, Huang CF, Ma SR, Bu LL,
Liu JF, Yu GT, Liu B, Gutkind JS, Kulkarni AB, et al: NOTCH1
inhibition enhances the efficacy of conventional chemotherapeutic
agents by targeting head neck cancer stem cell. Sci Rep.
6:247042016. View Article : Google Scholar : PubMed/NCBI
|