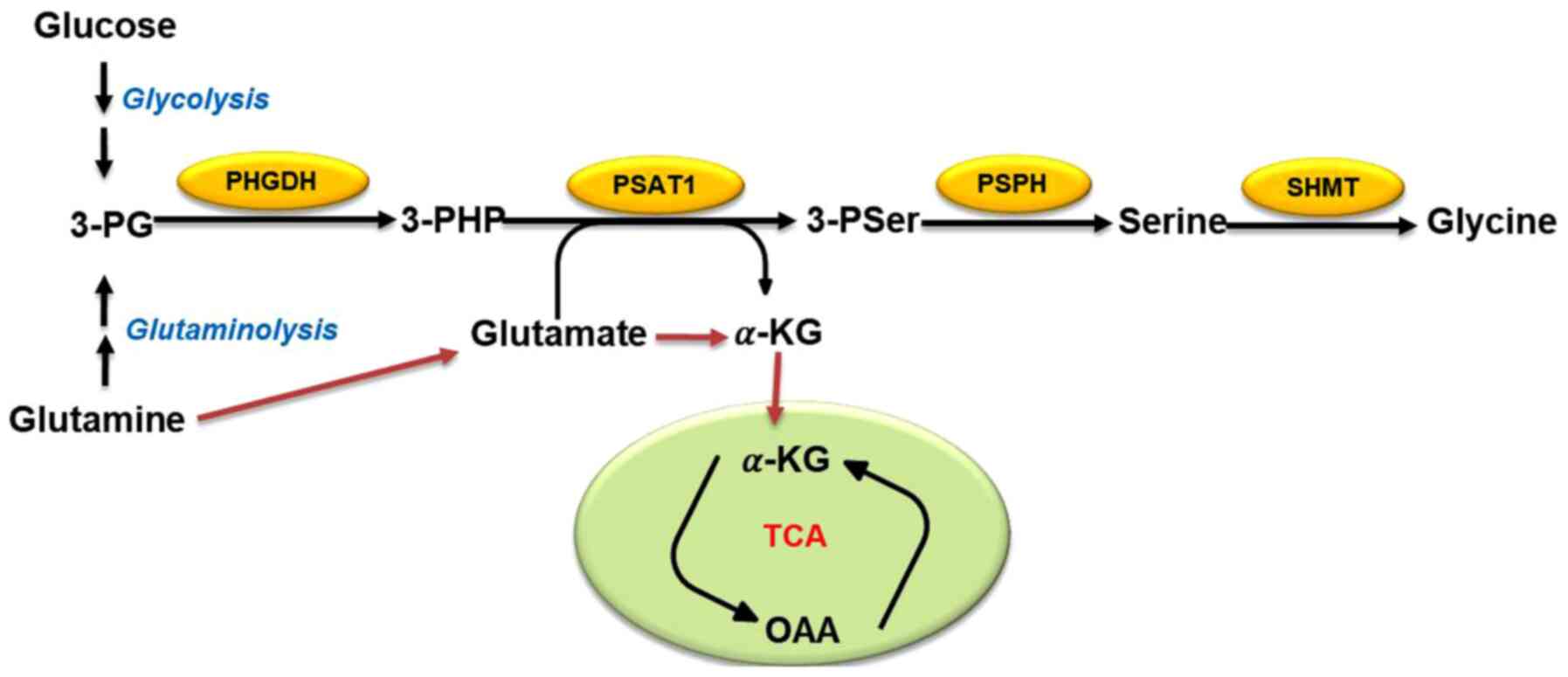
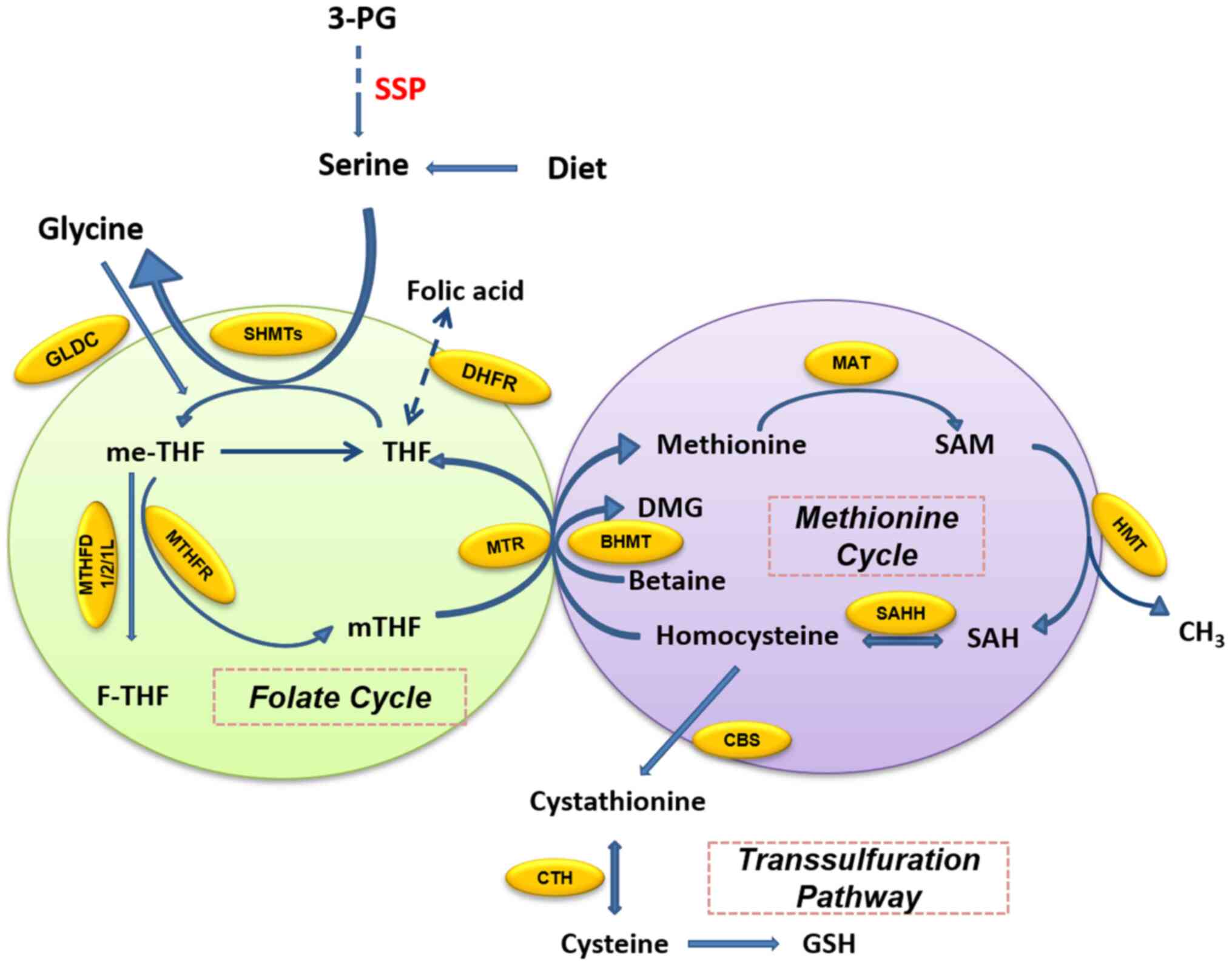
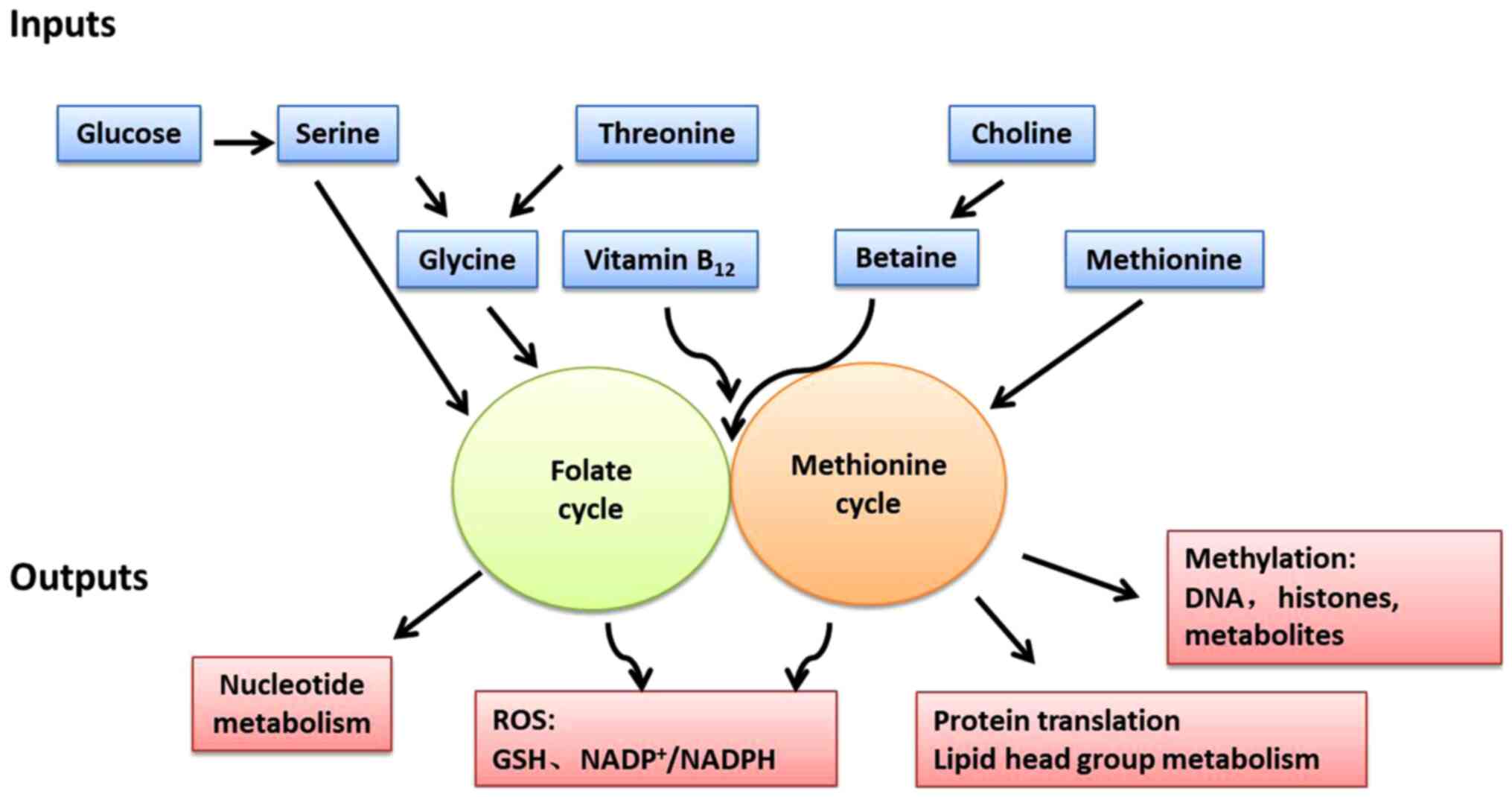
|
1
|
Reznik E, Luna A, Aksoy BA, Liu EM, La K,
Ostrovnaya I, Creighton CJ, Hakimi AA and Sander C: A Landscape of
meta-bolic variation across tumor types. Cell Syst. 6:301–313.e3.
2018. View Article : Google Scholar
|
|
2
|
Sun L, Suo C, Li ST, Zhang H and Gao P:
Metabolic reprogram-ming for cancer cells and their
microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta
Rev Cancer. 1870:51–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DeBerardinis RJ and Chandel NS:
Fundamentals of cancer metabolism. Sci Adv. 2:e16002002016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vander Heiden MG and DeBerardinis RJ:
Understanding the intersections between metabolism and cancer
biology. Cell. 168:657–669. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vazquez A, Kamphorst JJ, Markert EK, Schug
ZT, Tardito S and Gottlieb E: Cancer metabolism at a glance. J Cell
Sci. 129:3367–3373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bose S and Le A: Glucose metabolism in
cancer. Adv Exp Med Biol. 1063:3–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hosios AM, Hecht VC, Danai LV, Johnson MO,
Rathmell JC, Steinhauser ML, Manalis SR and Vander Heiden MG: Amino
Acids rather than glucose account for the majority of cell mass in
proliferating mammalian cells. Dev Cell. 36:540–549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun L, Song L, Wan Q, Wu G, Li X, Wang Y,
Wang J, Liu Z, Zhong X, He X, et al: cMyc-mediated activation of
serine biosynthesis pathway is critical for cancer progression
under nutrient deprivation conditions. Cell Res. 25:429–444. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Newman AC and Maddocks ODK: Serine and
functional metabolites in cancer. Trends Cell Biol. 27:645–657.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen S, Xia Y, He F, Fu J, Xin Z, Deng B,
He L, Zhou X and Ren W: Serine Supports IL-1β production in
macrophages through mTOR signaling. Front Immunol. 11:18662020.
View Article : Google Scholar
|
|
14
|
Sowers ML, Herring J, Zhang W, Tang H, Ou
Y, Gu W and Zhang K: Analysis of glucose-derived amino acids
involved in one-carbon and cancer metabolism by stable-isotope
tracing gas chromatography mass spectrometry. Anal Biochem.
566:1–9. 2019. View Article : Google Scholar
|
|
15
|
Newman AC and Maddocks ODK: One-carbon
metabolism in cancer. Br J Cancer. 116:1499–1504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lan X, Field MS and Stover PJ: Cell cycle
regulation of folate-mediated one-carbon metabolism. Wiley
Interdiscip Rev Syst Biol Med. 10:e14262018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ducker GS and Rabinowitz JD: One-carbon
metabolism in health and disease. Cell Metab. 25:27–42. 2017.
View Article : Google Scholar :
|
|
18
|
Yang M and Vousden KH: Serine and
one-carbon metabolism in cancer. Nat Rev Cancer. 16:650–662. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng JD, Wu WKK, Wang HY and Li XX: Serine
and one-carbon metabolism, a bridge that links mTOR signaling and
DNA methylation in cancer. Pharmacol Res. 149:1043522019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia Y, Ye B, Ding J, Yu Y, Alptekin A,
Thangaraju M, Prasad PD, Ding ZC, Park EJ, Choi JH, et al:
Metabolic reprogramming by MYCN confers dependence on the
serine-glycine-one-carbon biosynthetic pathway. Cancer Res.
79:3837–3850. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mattaini KR, Sullivan MR, Lau AN, Fiske
BP, Bronson RT and Vander Heiden MG: Increased PHGDH expression
promotes aberrant melanin accumulation. BMC Cancer. 19:7232019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Samanta D, Park Y, Andrabi SA, Shelton LM,
Gilkes DM and Semenza GL: PHGDH expression is required for
mitochondrial redox homeostasis, breast cancer stem cell
maintenance, and lung metastasis. Cancer Res. 76:4430–4442. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sullivan MR, Mattaini KR, Dennstedt EA,
Nguyen AA, Sivanand S, Reilly MF, Meeth K, Muir A, Darnell AM,
Bosenberg MW, et al: Increased serine synthesis provides an
advantage for tumors arising in tissues where serine levels are
limiting. Cell Metab. 29:1410–1421.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Zheng A, Hydbring P, Ambroise G,
Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H and Norberg E: PHGDH
Defines a metabolic subtype in lung adenocarcinomas with poor
prognosis. Cell Rep. 19:2289–2303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reina-Campos M, Linares JF, Duran A,
Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards
A, Rooslid T, et al: Increased serine and one-carbon pathway
metabolism by PKClambda/iota deficiency promotes neuroendocrine
prostate cancer. Cancer Cell. 35:385–400.e9. 2019. View Article : Google Scholar
|
|
26
|
Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X,
Ma J, Yin X, Mao A and Shang M: Overexpression of phosphoserine
aminotransferase 1 (PSAT1) predicts poor prognosis and associates
with tumor progression in human esophageal squamous cell carcinoma.
Cell Physiol Biochem. 39:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin HO, Hong SE, Kim JY, Jang SK, Kim YS,
Sim JH, Oh AC, Kim H, Hong YJ, Lee JK and Park IC: Knock-down of
PSAT1 enhances sensitivity of NSCLC cells to glutamine-limiting
conditions. Anticancer Res. 39:6723–6730. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Y, Liang X, Xu J and Cai X: miR-424
targets AKT3 and PSAT1 and has a tumor-suppressive role in human
colorectal cancer. Cancer Manag Res. 10:6537–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maddocks ODK, Athineos D, Cheung EC, Lee
P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D,
Kruiswijk F, et al: Modulating the therapeutic response of tumours
to dietary serine and glycine starvation. Nature. 544:372–376.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mattaini KR, Sullivan MR and Vander Heiden
MG: The importance of serine metabolism in cancer. J Cell Biol.
214:249–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
DeNicola GM, Chen PH, Mullarky E, Sudderth
JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al: NRF2
regulates serine biosynthesis in non-small cell lung cancer. Nat
Genet. 47:1475–1481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maddocks OD, Berkers CR, Mason SM, Zheng
L, Blyth K, Gottlieb E and Vousden KH: Serine starvation induces
stress and p53-dependent metabolic remodelling in cancer cells.
Nature. 493:542–546. 2013. View Article : Google Scholar
|
|
33
|
Wortel IMN, van der Meer LT, Kilberg MS
and van Leeuwen FN: Surviving Stress: Modulation of ATF4-mediated
stress responses in normal and malignant cells. Trends Endocrinol
Metab. 28:794–806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kasai S, Yamazaki H, Tanji K, Engler MJ,
Matsumiya T and Itoh K: Role of the ISR-ATF4 pathway and its cross
talk with Nrf2 in mitochondrial quality control. J Clin Biochem
Nutr. 64:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dey S, Sayers CM, Verginadis II, Lehman
SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs
SY, et al: ATF4-dependent induction of heme oxygenase 1 prevents
anoikis and promotes metastasisATF4-dependent induction of heme
oxygenase 1 prevents anoikis and promotes metastasis. J Clin
Invest. 125:2592–2608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tameire F, Verginadis II, Leli NM, Polte
C, Conn CS, Ojha R, Salas Salinas C, Chinga F, Monroy AM, Fu W, et
al: ATF4 couples MYC-dependent translational activity to
bioenergetic demands during tumour progression. Nat Cell Biol.
21:889–899. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mesclon F, Lambert-Langlais S, Carraro V,
Parry L, Hainault I, Jousse C, Maurin AC, Bruhat A, Fafournoux P
and Averous J: Decreased ATF4 expression as a mechanism of acquired
resistance to long-term amino acid limitation in cancer cells.
Oncotarget. 8:27440–27453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mazor KM and Stipanuk MH: GCN2- and
eIF2α-phosph orylation-independent, but ATF4-dependent, induction
of CARE-containing genes in methionine-deficient cells. Amino
Acids. 48:2831–2842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Al-Baghdadi RJT, Nikonorova IA, Mirek ET,
Wang Y, Park J, Belden WJ, Wek RC and Anthony TG: Role of
activating transcription factor 4 in the hepatic response to amino
acid depletion by asparaginase. Sci Rep. 7:12722017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu D, Dai W, Kutzler L, Lacko HA,
Jefferson LS, Dennis MD and Kimball SR: ATF4-mediated upregulation
of REDD1 and Sestrin2 suppresses mTORC1 activity during prolonged
leucine deprivation. J Nutr. 150:1022–1030. 2020. View Article : Google Scholar
|
|
41
|
Adams CM: Role of the transcription factor
ATF4 in the anabolic actions of insulin and the anti-anabolic
actions of glucocorticoids. J Biol Chem. 282:16744–16753. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ye J, Mancuso A, Tong X, Ward PS, Fan J,
Rabinowitz JD and Thompson CB: Pyruvate kinase M2 promotes de novo
serine synthesis to sustain mTORC1 activity and cell proliferation.
Proc Natl Acad Sci USA. 109:6904–6909. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y
and Pang D: PSAT1 is regulated by ATF4 and enhances cell
proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway
in ER-negative breast cancer. J Exp Clin Cancer Res. 36:1792017.
View Article : Google Scholar
|
|
44
|
Svoboda LK, Teh SSK, Sud S, Kerk S,
Zebolsky A, Treichel S, Thomas D, Halbrook CJ, Lee HJ, Kremer D, et
al: Menin regulates the serine biosynthetic pathway in Ewing
sarcoma. J Pathol. 245:324–336. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi
JH, Yan C, Dong Z, Huang S, Zha Y, et al: KDM4C and ATF4 cooperate
in transcriptional control of amino acid metabolism. Cell Rep.
14:506–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim SY, Hong M, Heo SH, Park S, Kwon TK,
Sung YH, Oh Y, Lee S, Yi GS and Kim I: Inhibition of euchromatin
histone-lysine N-methyltransferase 2 sensitizes breast cancer cells
to tumor necrosis factor-related apoptosis-inducing ligand through
reactive oxygen species-mediated activating transcription factor
4-C/EBP homologous protein-death receptor 5 pathway activation. Mol
Carcinog. 57:1492–1506. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ding J, Li T, Wang X, Zhao E, Choi JH,
Yang L, Zha Y, Dong Z, Huang S, Asara JM, et al: The histone H3
methyltransferase G9A epigenetically activates the serine-glycine
synthesis pathway to sustain cancer cell survival and
proliferation. Cell Metab. 18:896–907. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hydbring P, Castell A and Larsson LG: MYC
modulation around the CDK2/p27/SKP2 axis. Genes (Basel). 8:1742017.
View Article : Google Scholar
|
|
49
|
Fallah Y, Brundage J, Allegakoen P and
Shajahan-Haq AN: MYC-driven pathways in breast cancer subtypes.
Biomolecules. 7:532017. View Article : Google Scholar :
|
|
50
|
Lancho O and Herranz D: The MYC
Enhancer-ome: Long-range transcriptional regulation of MYC in
cancer. Trends Cancer. 4:810–822. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Carabet LA, Rennie PS and Cherkasov A:
Therapeutic inhibition of Myc in cancer. structural bases and
computer-aided drug discovery approaches. Int J Mol Sci.
20:1202018. View Article : Google Scholar
|
|
53
|
Chen Y, Sun XX, Sears RC and Dai MS:
Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis.
6:359–371. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Walz S, Lorenzin F, Morton J, Wiese KE,
von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M,
et al: Activation and repression by oncogenic MYC shape
tumour-specific gene expression profiles. Nature. 511:483–487.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tesi A, de Pretis S, Furlan M, Filipuzzi
M, Morelli MJ, Andronache A, Doni M, Verrecchia A, Pelizzola M,
Amati B and Sabò A: An early Myc-dependent transcriptional program
orchestrates cell growth during B-cell activation. EMBO Rep.
20:e479872019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Robaina MC, Mazzoccoli L and Klumb CE:
Germinal Centre B Cell Functions and Lymphomagenesis: Circuits
Involving MYC and MicroRNAs. Cells. 8:13652019. View Article : Google Scholar
|
|
57
|
Le A, Lane AN, Hamaker M, Bose S, Gouw A,
Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al:
Glucose-independent glutamine metabolism via TCA cycling for
proliferation and survival in B cells. Cell Metab. 15:110–121.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang LW, Shen H, Nobre L, Ersing I, Paulo
JA, Trudeau S, Wang Z, Smith NA, Ma Y, Reinstadler B, et al:
Epstein-barr-virus-induced one-carbon metabolism drives B cell
transformation. Cell Metab. 30:539–555.e11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kauko O, O'Connor CM, Kulesskiy E,
Sangodkar J, Aakula A, Izadmehr S, Yetukuri L, Yadav B, Padzik A,
Laajala TD, et al: PP2A inhibition is a druggable MEK inhibitor
resistance mecha-nism in KRAS-mutant lung cancer cells. Sci Transl
Med. 10:eaaq10932018. View Article : Google Scholar
|
|
60
|
Pakos-Zebrucka K, Koryga I, Mnich K,
Ljujic M, Samali A and Gorman AM: The integrated stress response.
EMBO Rep. 17:1374–1395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
David CJ, Chen M, Assanah M, Canoll P and
Manley JL: HnRNP proteins controlled by c-Myc deregulate pyruvate
kinase mRNA splicing in cancer. Nature. 463:364–368. 2010.
View Article : Google Scholar :
|
|
62
|
Luan W, Wang Y, Chen X, Shi Y, Wang J,
Zhang J, Qian J, Li R, Tao T, Wei W, et al: PKM2 promotes glucose
metabolism and cell growth in gliomas through a mechanism involving
a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 6:13006–13018.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chaneton B, Hillmann P, Zheng L, Martin
ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding
FP, Vousden KH, et al: Serine is a natural ligand and allosteric
activator of pyruvate kinase M2. Nature. 491:458–462. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li AM and Ye J: The PHGDH enigma: Do
cancer cells only need serine or also a redox modulator? Cancer
Lett. 476:97–105. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bao XR, Ong SE, Goldberger O, Peng J,
Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et
al: Mitochondrial dysfunction remodels one-carbon metabolism in
human cells. Elife. 5:e105752016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mendez-Lucas A, Li X, Hu J, Che L, Song X,
Jia J, Wang J, Xie C, Driscoll PC, Tschaharganeh DF, et al: Glucose
catabolism in liver tumors induced by c-MYC can be sustained by
various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer
Res. 77:4355–4364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang
X, Wu H, Blanco F, Li S, Bhanot G, et al: Glutaminase 2 is a novel
negative regulator of small GTPase Rac1 and mediates p53 function
in suppressing metastasis. Elife. 5:e107272016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kastenhuber ER and Lowe SW: Putting p53 in
Context. Cell. 170:1062–1078. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang S, Peng Z, Wang S, Yang L, Chen Y,
Kong X, Song S, Pei P, Tian C, Yan H, et al: KRAB-type zinc-finger
proteins PITA and PISA specifically regulate p53-dependent
glycolysis and mitochondrial respiration. Cell Res. 28:572–592.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang X, Zhang X, Li Y, Shao Y, Xiao J,
Zhu G and Li F: PAK4 regulates G6PD activity by p53 degradation
involving colon cancer cell growth. Cell Death Dis. 8:e28202017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fritsche MK and Knopf A: The tumor
suppressor p53 in mucosal melanoma of the head and neck. Genes
(Basel). 8:3842017. View Article : Google Scholar
|
|
72
|
Amelio I, Cutruzzola F, Antonov A,
Agostini M and Melino G: Serine and glycine metabolism in cancer.
Trends Biochem Sci. 39:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lu J, Tan M and Cai Q: The Warburg effect
in tumor progression: Mitochondrial oxidative metabolism as an
anti-metastasis mechanism. Cancer Lett. 356:156–164. 2015.
View Article : Google Scholar
|
|
74
|
Humpton TJ, Hock AK, Maddocks ODK and
Vousden KH: p53-mediated adaptation to serine starvation is
retained by a common tumour-derived mutant. Cancer Metab. 6:182018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Riscal R, Schrepfer E, Arena G, Cissé MY,
Bellvert F, Heuillet M, Rambow F, Bonneil E, Sabourdy F, Vincent C,
et al: Chromatin-bound MDM2 regulates serine metabolism and redox
homeostasis independently of p53. Mol Cell. 62:890–902. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Amelio I, Markert EK, Rufini A, Antonov
AV, Sayan BS, Tucci P, Agostini M, Mineo TC, Levine AJ and Melino
G: p73 regulates serine biosynthesis in cancer. Oncogene.
33:5039–5046. 2014. View Article : Google Scholar
|
|
77
|
Ou Y, Wang SJ, Jiang L, Zheng B and Gu W:
p53 Protein-mediated regulation of phosphoglycerate dehydrogenase
(PHGDH) is crucial for the apoptotic response upon serine
starvation. J Biol Chem. 290:457–466. 2015. View Article : Google Scholar :
|
|
78
|
Zhang WC, Shyh-Chang N, Yang H, Rai A,
Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al: Glycine
decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ducker GS, Chen L, Morscher RJ,
Ghergurovich JM, Esposito M, Teng X, Kang Y and Rabinowitz JD:
Reversal of cytosolic one-carbon flux compensates for loss of the
mitochondrial folate pathway. Cell Metab. 23:1140–1153. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Labuschagne CF, van den Broek NJ, Mackay
GM, Vousden KH and Maddocks OD: Serine, but not glycine, supports
one-carbon metabolism and proliferation of cancer cells. Cell Rep.
7:1248–1258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Maddocks OD, Labuschagne CF, Adams PD and
Vousden KH: Serine metabolism supports the methionine cycle and
DNA/RNA Methylation through de novo ATP synthesis in cancer cells.
Mol Cell. 61:210–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Guo H, Xu J, Zheng Q, He J, Zhou W, Wang
K, Huang X, Fan Q, Ma J, Cheng J, et al: NRF2 SUMOylation promotes
de novo serine synthesis and maintains HCC tumorigenesis. Cancer
Lett. 466:39–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kikuchi G, Motokawa Y, Yoshida T and
Hiraga K: Glycine cleavage system: Reaction mechanism,
physiological significance, and hyperglycinemia. Proc Jpn Acad Ser
B Phys Biol Sci. 84:246–263. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tedeschi PM, Markert EK, Gounder M, Lin H,
Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, DiPaola RS, Hirshfield KM,
et al: Contribution of serine, folate and glycine metabolism to the
ATP, NADPH and purine requirements of cancer cells. Cell Death Dis.
4:e8772013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kim D, Fiske BP, Birsoy K, Freinkman E,
Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, et al:
SHMT2 drives glioma cell survival in ischaemia but imposes a
dependence on glycine clearance. Nature. 520:363–367. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lewis CA, Parker SJ, Fiske BP, McCloskey
D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo
CM, et al: Tracing compartmentalized NADPH metabolism in the
cytosol and mitochondria of mammalian cells. Mol Cell. 55:253–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ueland PM: Choline and betaine in health
and disease. J Inherit Metab Dis. 34:3–15. 2011. View Article : Google Scholar
|
|
88
|
Friso S, Udali S, De Santis D and Choi SW:
One-carbon metabolism and epigenetics. Mol Aspects Med. 54:28–36.
2017. View Article : Google Scholar
|
|
89
|
Kanarek N, Keys HR, Cantor JR, Lewis CA,
Chan SH, Kunchok T, Abu-Remaileh M, Freinkman E, Schweitzer LD and
Sabatini DM: Histidine catabolism is a major determinant of
methotrexate sensitivity. Nature. 559:632–636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fan J, Ye J, Kamphorst JJ, Shlomi T,
Thompson CB and Rabinowitz JD: Quantitative flux analysis reveals
folate-dependent NADPH production. Nature. 510:298–302. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen L, Zhang Z, Hoshino A, Zheng HD,
Morley M, Arany Z and Rabinowitz JD: NADPH production by the
oxidative pentose-phosphate pathway supports folate metabolism. Nat
Metab. 1:404–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Reid MA, Dai Z and Locasale JW: The impact
of cellular metabolism on chromatin dynamics and epigenetics. Nat
Cell Biol. 19:1298–1306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Morscher RJ, Ducker GS, Li SH, Mayer JA,
Gitai Z, Sperl W and Rabinowitz JD: Mitochondrial translation
requires folate-dependent tRNA methylation. Nature. 554:128–132.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Gao X, Lee K, Reid MA, Sanderson SM, Qiu
C, Li S, Liu J and Locasale JW: Serine availability influences
mitochondrial dynamics and function through lipid metabolism. Cell
Rep. 22:3507–3520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Villa E, Ali ES, Sahu U and Ben-Sahra I:
Cancer cells tune the signaling pathways to empower de novo
synthesis of nucleotides. Cancers (Basel). 11:6882019. View Article : Google Scholar
|
|
96
|
Ulanovskaya OA, Zuhl AM and Cravatt BF:
NNMT promotes epigenetic remodeling in cancer by creating a
metabolic methylation sink. Nat Chem Biol. 9:300–306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hughey CC, Trefts E, Bracy DP, James FD,
Donahue EP and Wasserman DH: Glycine N-methyltransferase deletion
in mice diverts carbon flux from gluconeogenesis to pathways that
utilize excess methionine cycle intermediates. J Biol Chem.
293:11944–11954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Serefidou M, Venkatasubramani AV and Imhof
A: The impact of one carbon metabolism on histone methylation.
Front Genet. 10:7642019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Fukuoka H and Kubota T: One-carbon
metabolism and lipid metabolism in DOHaD. Adv Exp Med Biol.
1012:3–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Walker AK: 1-Carbon cycle metabolites
methylate their way to fatty liver. Trends Endocrinol Metab.
28:63–72. 2017. View Article : Google Scholar
|
|
101
|
Xiao W, Wang RS, Handy DE and Loscalzo J:
NAD(H) and NADP(H) Redox couples and cellular energy metabolism.
Antioxid Redox Signal. 28:251–272. 2018. View Article : Google Scholar :
|
|
102
|
Hanley MP and Rosenberg DW: One-carbon
metabolism and colorectal cancer: Potential mechanisms of
chemoprevention. Curr Pharmacol Rep. 1:197–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ser Z, Gao X, Johnson C, Mehrmohamadi M,
Liu X, Li S and Locasale JW: targeting one carbon metabolism with
an antimetabolite disrupts pyrimidine homeostasis and induces
nucleotide overflow. Cell Rep. 15:2367–2376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Pandey S, Garg P, Lee S, Choung HW, Choung
YH, Choung PH and Chung JH: Nucleotide biosynthesis arrest by
silencing SHMT1 function via vitamin B6-coupled vector and effects
on tumor growth inhibition. Biomaterials. 35:9332–9342. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Tripathi SK, Gupta N, Mahato M, Gupta KC
and Kumar P: Selective blocking of primary amines in branched
polyethylenimine with biocompatible ligand alleviates cytotoxicity
and augments gene delivery efficacy in mammalian cells. Colloids
Surf B Biointerfaces. 115:79–85. 2014. View Article : Google Scholar
|
|
106
|
Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara
JM and Manning BD: mTORC1 induces purine synthesis through control
of the mito-chondrial tetrahydrofolate cycle. Science. 351:728–733.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Park Y, Reyna-Neyra A, Philippe L and
Thoreen CC: mTORC1 balances cellular amino acid supply with demand
for protein synthesis through post-transcriptional control of ATF4.
Cell Rep. 19:1083–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ben-Sahra I, Howell JJ, Asara JM and
Manning BD: Stimulation of de novo pyrimidine synthesis by growth
signaling through mTOR and S6K1. Science. 339:1323–1328. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rabinovich S, Adler L, Yizhak K, Sarver A,
Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et
al: Diversion of aspartate in ASS1-deficient tumours fosters de
novo pyrimidine synthesis. Nature. 527:379–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mentch SJ and Locasale JW: One-carbon
metabolism and epigenetics: Understanding the specificity. Ann N Y
Acad Sci. 1363:91–98. 2016. View Article : Google Scholar
|
|
111
|
Mahmoud AM and Ali MM: Methyl donor
micronutrients that modify DNA methylation and cancer outcome.
Nutrients. 11:6082019. View Article : Google Scholar :
|
|
112
|
Morgan AE, Davies TJ and Mc Auley MT: The
role of DNA methylation in ageing and cancer. Proc Nutr Soc.
77:412–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Konno M, Koseki J, Kawamoto K, Nishida N,
Matsui H, Dewi DL, Ozaki M, Noguchi Y, Mimori K, Gotoh N, et al:
Embryonic MicroRNA-369 controls metabolic splicing factors and
urges cellular reprograming. PLoS One. 10:e01327892015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li S, Swanson SK, Gogol M, Florens L,
Washburn MP, Workman JL and Suganuma T: Serine and SAM responsive
complex SESAME regulates histone modification crosstalk by sensing
cellular metabolism. Mol Cell. 60:408–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kottakis F, Nicolay BN, Roumane A, Karnik
R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al:
LKB1 loss links serine metabolism to DNA methylation and
tumorigenesis. Nature. 539:390–395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Shlomi T, Fan J, Tang B, Kruger WD and
Rabinowitz JD: Quantitation of cellular metabolic fluxes of
methionine. Anal Chem. 86:1583–1591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Mehrmohamadi M, Liu X, Shestov AA and
Locasale JW: Characterization of the usage of the serine metabolic
network in human cancer. Cell Rep. 9:1507–1519. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Nilsson R, Nicolaidou V and Koufaris C:
Mitochondrial MTHFD isozymes display distinct expression,
regulation, and association with cancer. Gene. 716:1440322019.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shin M, Momb J and Appling DR: Human
mitochondrial MTHFD2 is a dual redox cofactor-specific
methylenetetrahydro-folate dehydrogenase/methenyltetrahydrofolate
cyclohydrolase. Cancer Metab. 5:112017. View Article : Google Scholar
|
|
120
|
Goodman RP, Calvo SE and Mootha VK:
Spatiotemporal compartmentalization of hepatic NADH and NADPH
metabolism. J Biol Chem. 293:7508–7516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Röhrig F and Schulze A: The multifaceted
roles of fatty acid synthesis in cancer. Nat Rev Cancer.
16:732–749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ye J, Fan J, Venneti S, Wan YW, Pawel BR,
Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al: Serine
catabolism regulates mitochondrial redox control during hypoxia.
Cancer Discov. 4:1406–1417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ye C, Sutter BM, Wang Y, Kuang Z and Tu
BP: A Metabolic function for phospholipid and histone methylation.
Mol Cell. 66:180–193.e188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Rodriguez AE, Ducker GS, Billingham LK,
Martinez CA, Mainolfi N, Suri V, Friedman A, Manfredi MG, Weinberg
SE, Rabinowitz JD and Chandel NS: Serine metabolism supports
macrophage IL-1beta Production. Cell Metab. 29:1003–1011.e1004.
2019. View Article : Google Scholar
|
|
125
|
Ito Y, Makita S and Tobinai K: Development
of new agents for peripheral T-cell lymphoma. Expert Opin Biol
Ther. 19:197–209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Wei N, Zhang B, Wang Y, He XH, Xu LC, Li
GD, Wang YH, Wang GZ, Huang HZ and Li WT: Transarterial
chemoembolization with raltitrexed-based or floxuridine-based
chemotherapy for unresectable colorectal cancer liver metastasis.
Clin Transl Oncol. 21:443–450. 2019. View Article : Google Scholar
|
|
127
|
Goirand F, Lemaitre F, Launay M, Tron C,
Chatelut E, Boyer JC, Bardou M and Schmitt A: How can we best
monitor 5-FU administration to maximize benefit to risk ratio?
Expert Opin Drug Metab Toxicol. 14:1303–1313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Adamska A, Elaskalani O, Emmanouilidi A,
Kim M, Abdol Razak NB, Metharom P and Falasca M: Molecular and
cellular mechanisms of chemoresistance in pancreatic cancer. Adv
Biol Regul. 68:77–87. 2018. View Article : Google Scholar
|
|
129
|
Blair HA: Daunorubicin/cytarabine
liposome: A review in acute myeloid leukaemia. Drugs. 78:1903–1910.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Diesch J, Zwick A, Garz AK, Palau A,
Buschbeck M and Gotze KS: A clinical-molecular update on
azanucleoside-based therapy for the treatment of hematologic
cancers. Clin Epigenetics. 8:712016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Chabner BA and Roberts TG Jr: Timeline:
Chemotherapy and the war on cancer. Nat Rev Cancer. 5:65–72. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Luengo A, Gui DY and Vander Heiden MG:
Targeting metabolism for cancer therapy. Cell Chem Biol.
24:1161–1180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Casero RA Jr and Marton LJ: Targeting
polyamine metabolism and function in cancer and other
hyperproliferative diseases. Nat Rev Drug Discov. 6:373–390. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Rodriguez-Paredes M and Esteller M: Cancer
epigenetics reaches mainstream oncology. Nat Med. 17:330–339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zheng Y, Lin TY, Lee G, Paddock MN, Momb
J, Cheng Z, Li Q, Fei DL, Stein BD, Ramsamooj S, et al:
Mitochondrial One-carbon pathway supports cytosolic folate
integrity in cancer cells. Cell. 175:1546–1560.e1517. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kucharczyk T, Krawczyk P, Powrózek T,
Kowalski DM, Ramlau R, Kalinka-Warzocha E, Knetki-Wróblewska M,
Winiarczyk K, Krzakowski M and Milanowski J: The Effectiveness of
pemetrexed monotherapy depending on poly-morphisms in TS and MTHFR
genes as well as clinical factors in advanced NSCLC patients.
Pathol Oncol Res. 22:49–56. 2016. View Article : Google Scholar
|
|
137
|
Winter SS, Dunsmore KP, Devidas M, Wood
BL, Esiashvili N, Chen Z, Eisenberg N, Briegel N, Hayashi RJ,
Gastier-Foster JM, et al: Improved survival for children and young
adults with t-lineage acute lymphoblastic leukemia: Results from
the children's oncology group AALL0434 methotrexate randomization.
J Clin Oncol. 36:2926–2934. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Cui Y, Chen H, Chen J, Zeng F, Zu X and
Ding J: Gemcitabine/cisplatin versus
methotrexate/vinblastine/doxoru-bicin/cisplatin for muscle-invasive
bladder cancer: A systematic review and meta-analysis.
Meta-Analysis. 14:1260–1265. 2018.
|
|
139
|
Calise SJ, Purich DL, Nguyen T, Saleem DA,
Krueger C, Yin JD and Chan EK: 'Rod and ring' formation from IMP
dehydrogenase is regulated through the one-carbon metabolic
pathway. J Cell Sci. 129:3042–3052. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ross KC, Andrews AJ, Marion CD, Yen TJ and
Bhattacharjee V: Identification of the serine biosynthesis pathway
as a critical component of BRAF inhibitor resistance of melanoma,
pancreatic, and non-small cell lung cancer cells. Mol Cancer Ther.
16:1596–1609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Longley DB and Harkin DP and Harkin DP:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Kawahata T, Kawahara K, Shimokawa M,
Sakiyama A, Shiraishi T, Minami K, Yamamoto M, Shinsato Y, Arima K,
Hamada T and Furukawa T: Involvement of ribosomal protein L11
expression in sensitivity of gastric cancer against 5-FU. Oncol
Lett. 19:2258–2264. 2020.PubMed/NCBI
|
|
143
|
Reina-Campos M, Diaz-Meco MT and Moscat J:
The complexity of the serine glycine one-carbon pathway in cancer.
J Cell Biol. 219:e2019070222020. View Article : Google Scholar :
|
|
144
|
Avgustinova A and Benitah SA: The
epigenetics of tumour initiation: Cancer stem cells and their
chromatin. Curr Opin Genet Dev. 36:8–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kim M and Costello J: DNA methylation: An
epigenetic mark of cellular memory. Exp Mol Med. 49:e3222017.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Albrecht LV, Bui MH and De Robertis EM:
Canonical Wnt is inhibited by targeting one-carbon metabolism
through methotrexate or methionine deprivation. Proc Natl Acad Sci
USA. 116:2987–2995. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chen WL, Sun HP, Li DD, Wang ZH, You QD
and Guo XK: G9a-An appealing antineoplastic target. Curr Cancer
Drug Targets. 17:555–568. 2017. View Article : Google Scholar
|
|
148
|
Roesch A, Fukunaga-Kalabis M, Schmidt EC,
Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T
and Herlyn M: A temporarily distinct subpopulation of slow-cycling
melanoma cells is required for continuous tumor growth. Cell.
141:583–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Kano Y, Konno M, Ohta K, Haraguchi N,
Nishikawa S, Kagawa Y, Hamabe A, Hasegawa S, Ogawa H, Fukusumi T,
et al: Jumonji/Arid1b (Jarid1b) protein modulates human esophageal
cancer cell growth. Mol Clin Oncol. 1:753–757. 2013. View Article : Google Scholar
|
|
150
|
Konno M, Asai A, Kawamoto K, Nishida N,
Satoh T, Doki Y, Mori M and Ishii H: The one-carbon metabolism
pathway high-lights therapeutic targets for gastrointestinal cancer
(Review). Int J Oncol. 50:1057–1063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Kaniskan HU, Martini ML and Jin J:
Inhibitors of protein methyltransferases and demethylases. Chem
Rev. 118:989–1068. 2018. View Article : Google Scholar
|
|
152
|
Ma L, Tao Y, Duran A, Llado V, Galvez A,
Barger JF, Castilla EA, Chen J, Yajima T, Porollo A, et al: Control
of nutrient stress-induced metabolic reprogramming by PKCζ in
tumorigenesis. Cell. 152:599–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Mullarky E, Lucki NC, Beheshti Zavareh R,
Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H,
Singh PK, et al: Identification of a small molecule inhibitor of
3-phosphoglycerate dehydrogenase to target serine biosynthesis in
cancers. Proc Natl Acad Sci USA. 113:1778–1783. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Pacold ME, Brimacombe KR, Chan SH, Rohde
JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske
BP, et al: A PHGDH inhibitor reveals coordination of serine
synthesis and one-carbon unit fate. Nat Chem Biol. 12:452–458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhao X, Fu J, Du J and Xu W: The Role of
D-3-phosphoglycerate dehydrogenase in cancer. Int J Biol Sci.
16:1495–1506. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Ravez S, Spillier Q, Marteau R, Feron O
and Frederick R: Challenges and opportunities in the development of
serine synthetic pathway inhibitors for cancer therapy. J Med Chem.
60:1227–1237. 2017. View Article : Google Scholar
|
|
157
|
Gravel SP, Hulea L, Toban N, Birman E,
Blouin MJ, Zakikhani M, Zhao Y, Topisirovic I, St-Pierre J and
Pollak M: Serine deprivation enhances antineoplastic activity of
biguanides. Cancer Res. 74:7521–7533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Fedirko V, Lukanova A, Bamia C,
Trichopolou A, Trepo E, Nöthlings U, Schlesinger S, Aleksandrova K,
Boffetta P, Tjønneland A, et al: Glycemic index, glycemic load,
dietary carbohydrate, and dietary fiber intake and risk of liver
and biliary tract cancers in Western Europeans. Ann Oncol.
24:543–553. 2013. View Article : Google Scholar :
|
|
159
|
Fine EJ, Segal-Isaacson CJ, Feinman RD,
Herszkopf S, Romano MC, Tomuta N, Bontempo AF, Negassa A and
Sparano JA: Targeting insulin inhibition as a metabolic therapy in
advanced cancer: A pilot safety and feasibility dietary trial in 10
patients. Nutriti. 28:1028–1035. 2012.
|
|
160
|
Mallik R and Chowdhury TA: Metformin in
cancer. Diabetes Res Clin Pract. 143:409–419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Gustafsson R, Jemth AS, Gustafsson NM,
Färnegårdh K, Loseva O, Wiita E, Bonagas N, Dahllund L,
Llona-Minguez S, Häggblad M, et al: Crystal structure of the
emerging cancer target MTHFD2 in complex with a substrate-based
inhibitor. Cancer Res. 77:937–948. 2017. View Article : Google Scholar
|
|
162
|
Nishimura T, Nakata A, Chen X, Nishi K,
Meguro-Horike M, Sasaki S, Kita K, Horike SI, Saitoh K, Kato K, et
al: Cancer stem-like properties and gefitinib resistance are
dependent on purine synthetic metabolism mediated by the
mitochondrial enzyme MTHFD2. Oncogene. 38:2464–2481. 2019.
View Article : Google Scholar :
|
|
163
|
Bolusani S, Young BA, Cole NA, Tibbetts
AS, Momb J, Bryant JD, Solmonson A and Appling DR: Mammalian
MTHFD2L encodes a mitochondrial methylenetetrahydrofolate
dehydrogenase isozyme expressed in adult tissues. J Biol Chem.
286:5166–5174. 2011. View Article : Google Scholar :
|
|
164
|
Nilsson R, Jain M, Madhusudhan N, Sheppard
NG, Strittmatter L, Kampf C, Huang J, Asplund A and Mootha VK:
Metabolic enzyme expression highlights a key role for MTHFD2 and
the mitochondrial folate pathway in cancer. Nat Commun. 5:31282014.
View Article : Google Scholar : PubMed/NCBI
|