The SSP and one-carbon pathway create an upregulated
metabolic network in tumors and are of high clinical relevance
(15,17-19).
In the present review, the significance of the SSP in cancer and
its related regulatory mechanisms of action are outlined, as well
as the contribution that one-carbon metabolism provide for cancer
metabolic reprogramming path-ways. These findings may help in the
development of targeted antimetabolite treatments by highlighting
new translational opportunities for dietary interventions, drug
development and biomarker identification.
Cancer cells generally use glycolysis to maintain
their energy supply and serine biosynthesis is an important branch
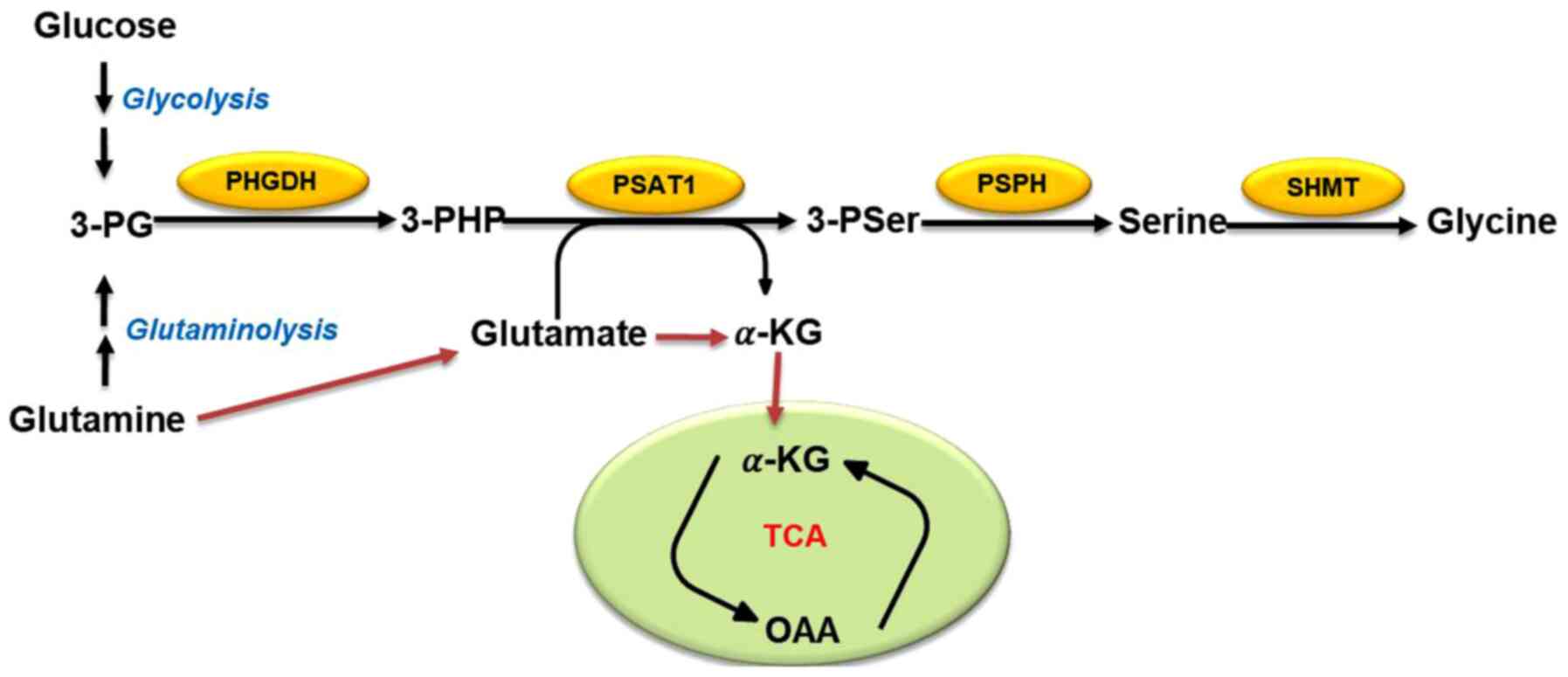
of glycolysis (11). 3-PG, an
intermediate product of glycolysis, is a precursor of serine
biosynthesis (9). Overall, ~10% of
3-PG is converted into serine after a three-step enzymatic reaction
(Fig. 1): In the first step, 3-PG
is oxidized to 3-phos-phate hydroxypyruvate by phosphoglycerate
dehydrogenase (PHGDH) (18). It is
then catalyzed to 3-phosphoserine and α-KG by phosphoserine
aminotransferase (PSAT1), and finally dephosphorylated to serine by
1-3-phosphoserine phosphatase (PSPH) (18). The mutual conversion of serine and
glycine can then be achieved by serine hydroxymethyl transferase
(SHMT1/2) (20). It has been
reported that the gene encoding PHGDH, located on chromosome 1p12,
is upregulated in most types of human tumors, such as breast cancer
and melanoma (21,22). In addition, short hairpin RNA
screening results reveal that breast cancer cell lines and melanoma
cell lines require PHGDH amplification to support tumorigenesis
(21-23). Similarly, high levels of PHGDH and
SHMT2 have been found in a subgroup of patients with lung cancer
who have a particularly poor prognosis (24). PHGDH inhibition can reduce tumor
growth and differentiation of neuroendocrine prostate cancer in
vivo (25). All of these
studies indicate that PHGDH is very important for the proliferation
and survival of tumor cells. Other studies have revealed that PSAT1
is upregulated in colorectal cancer (CRC), esophageal squamous cell
carcinoma and non-small cell lung cancer, and that PSAT1
overexpression leads to a poor prognosis by enhancing cancer cell
proliferation, metastasis and chemoresistance (26-28).
Additionally, in patients with hepatocellular carcinoma (HCC), the
expression levels of PSPH gradually increase with HCC progression
and the abnormal expression of PSPH is highly correlated with
patient mortality, indicating that the PSPH protein is a probable
prognostic biomarker for HCC (11). Taken together, high expression
levels of metabolic enzymes in the SSP may be necessary and
sufficient to maintain cancer growth and oncogenic
transformation.
Patients with malignant tumors are at high risk of
malnutrition, with 40-80% afflicted by this condition. Under
nutritional deprivation, cancer cells are adept at obtaining any
required energy during the opportunistic mode to support their own
survival and growth, which means metabolic reprogramming (4). It has long been known that both
exogenously ingested serine and endogenously synthesized serine are
associated with cancer and functionally support cancer development
(12,29,30).
As aforementioned, the high expression levels of the metabolic
enzymes PHGDH, PSAT1 and PSPH in the SSP may be indispensable for
maintaining cancer growth and oncogenic transformation (21,23,25-27).
Moreover, metabolic enzymes in the SSP are subject to
transcriptional regulation by various transcription factors after
stress response or oncogene activation, to cope with various types
of stress, including nutritional deficiencies (11). The present review subsequently
discusses the ways in which the transcriptional factors activating
transcription factor 4 (ATF4) and c-MYC, as well as the oncogene
p53, activate the SSP and perform genomic modification of the
metabolic enzymes in the SSP, to assist tumor metabolic
reprogramming under nutritional deficiency and/or serine
deprivation (11,31,32).
Activating transcription factor 4 (ATF4) is a member
of the cyclic adenosine monophosphate responsive element-binding
(CREB) protein family. According to previous reports, the gene
encoding the CREB protein family is not only expressed in a variety
of tumors, but also is a potent stress-response gene in tumors
(33,34). Many ATF4 target genes are involved
in the maintenance of amino acids homeostasis (35-37).
By regulating the adverse environment, ATF4 can protect tumor cells
from nutritional stress and a series of cancer therapeutic agents
(37-40). PHGDH, PSAT1 and PSPH inhibition by
ATF4 small interfering RNA was first reported by Adams (41). In addition, under amino acid
starvation, high expression levels of PHGDH, PSAT1 and PSPH can be
induced through the general control nonderepressible
2-ATF4-dependent pathway (42).
Gao et al (43) were the
first to reveal that the expression levels of PSAT1 in ER-negative
breast cancer were significantly upregulated. ATF4 was also found
to directly enhance the expression of PSAT1 in ER-negative breast
cancer, which upregulated cyclin D1 through the GSK3β/β-catenin
pathway, and finally promoted the proliferation of ER-negative
breast cancer cells in vitro and in vivo (43). DeNicola et al (31) integrated metabolite tracing with
gene expression analysis revealing that NF-E2-related factor 2
positively regulated the expression levels of PHGDH, PSAT1 and
SHMT2 in SSP by targeting ATF4, which controlled the metabolic flux
of glycolysis to serine, thereby supporting the production of GSH
and nucleotides. Epigenetic modifiers also regulate the expression
of key enzymes in the SSP (44-46).
Histone H3 lysine 9 (H3K9) methyltransferase G9A is required for
the transcriptional activation of key enzymes in the SSP during an
active state, marked by H3K9 monomethylation, which is dependent on
ATF4 (47). Coincidentally, Zhao
et al (45) speculated that
the H3K9 demethylase lysine demethylases 4 (KDM4) may also play a
role in the transcriptional regulation of SSP. KDM4C specifically
epigenetically activates the metabolic enzyme genes PHGDH, PSAT1
and SHMT2, by removing the restrictive modification of H3K9me3.
This action requires ATF4 and interacts with ATF4 to target the
metabolic enzyme genes and enhance the expression of their mRNA and
protein, suggesting that KDM4C exerts a role in coordinating amino
acid metabolism through a series of regulatory mechanisms (45). These studies indicate that as an
upstream regulator of the SSP, targeting ATF4 is an effective
mechanism for blocking the SSP in a coordinated fashion. As such,
ATF4 may be a promising therapeutic target.
As an oncogene, c-Myc drives malignant progression
and induces a powerful anabolic and proliferative program,
resulting in the occurrence of intrinsic stress (36,48-50).
Of note, transcription factor c-Myc can regulate 10-15% of human
genes and participate in the cell cycle as well as cellular
development, apoptosis and metabolism (51-53).
There is evidence that c-Myc selectively fine tunes the expression
of various genes which are vital for cell growth and cancer
progression (54-56). Not only does c-Myc regulate the
metabolism of glucose, glutamate and nucleotides, but also it
participates in SSP activation induced by nutritional starvation
(11,57-59).
Sun et al (11) identified
the Ebox c-Myc binding site on the PHGDH, PSAT1 and PSPH loci and
that knocking out c-Myc can reduce the expression of these genes.
c-Myc-mediated PSPH expression and SSP activation are essential for
cancer cell survival and proliferation because of their regulation
of the redox levels between GSH and reactive oxygen species (ROS),
nucleotide biosynthesis and cell cycle progression (11). In addition, since c-Myc activation
can induce ATF4 expression by activating the integrating stress
response (36,60), the induction of PHGDH by c-Myc may
depend on ATF4. It is worth noting that Myc transcription induces
ribonucleoprotein polypyrimidine tract binding protein,
heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and hnRNPA2 to
promote the production of pyruvate kinase M2 (PKM2) (61,62).
Serine is the only amino acid that can act as an allosteric
activator of PKM2. Serine starvation reduces the activity of PKM2
enzymes and leads to the accumulation of upstream glycolysis
intermediates, including 3-PG (63). Eventually, the tumor cells develop
a higher proliferation rate under metabolic stress because of the
significant increase of flux into the SSP (63-66).
Overall, these studies indicate that Myc may promote the SSP by
implementing the above two feedback pathways, demonstrating that
the overall changes in c-Myc metabolism lead to SSP activation and
cancer metastasis.
The tumor suppressor p53 has become recognized as an
important regulator of cell metabolism, which can affect a series
of cellular metabolic processes, such as glycolysis, oxidative
phosphorylation, glutaminolysis and antioxidant reactions (67-71).
p53 is also a key substance in the cell response to various forms
of stress, including DNA damage, hypoxia and oncogene activation
(68). Under nutritional
deficiencies, p53 can protect cells by supporting metabolic
adaptation. p53 helps cancer cells overcome serine starvation while
retaining the cellular antioxidant capacity (72). p53-deficient cells cannot respond
to serine starvation due to oxidative stress, which leads to a
reduced viability of cancer cells and severely impaired
proliferation (72). During serine
starvation, activation of the p53-p21 axis in p53+/+
cells results in transient p21-dependent G1 arrest and reduction of
S-phase cells, thereby inducing cell cycle arrest. This pathway can
facilitate cell survival by effectively depleting serine reserves
for GSH synthesis (32). As both
metabolic reprogramming and the Warburg effect inhibit cancer cell
death through the elimination of metabolic ROS (73), Maddocks et al (32) emphasized that p53 can coordinate
cancer metabolic repro-gramming under metabolic stress. Notably,
p53 is frequently mutated in various types of human cancer, such as
the common mutant, R248W. Such p53 mutants lose the function of
wild-type p53 to clear cellular ROS, but retain the ability of
wild-type p53 to bind to p21 and MDM2 (74). Increased levels of MDM2 promote the
formation of MDM2 and ATF4 complexes, which can support cancer
survival and proliferation by activating the SSP and inducing
antioxidant responses under serine starvation (74,75).
According to related reports, p73, a p53 homolog, also plays a
significant role in serine biosynthesis (76). p73 transcriptionally induces
glutaminase 2 (GLS-2) to facilitate the decomposition of glutamine,
which drives the SSP through glutamate to help cancer cells resist
metabolic stress (76).
Interestingly, in human melanoma cells, p53 induced by Nutlin-3,
downregulates the expression of PHGDH to repress de novo
serine biosynthesis (77).
Moreover, under serine deprivation, Puma and Noxa can be activated
by ATF4-dependent Nutlin-3, which inhibits PHGDH and then further
promotes apoptosis (77). These
findings indicate that p53 promotes metabolic remodeling for cancer
cell proliferation under serine starvation, although the specific
regulatory role of p53 is dependent on the type of cancer cell.
One-carbon metabolism includes a bicyclic pathway
formed by the coupling of the folate cycle and the methionine cycle
and the trans-sulfuration pathway (15,16).
Folate is a B vitamin which occurs naturally in many foods, and
dietary supplements usually contain the synthetically produced form
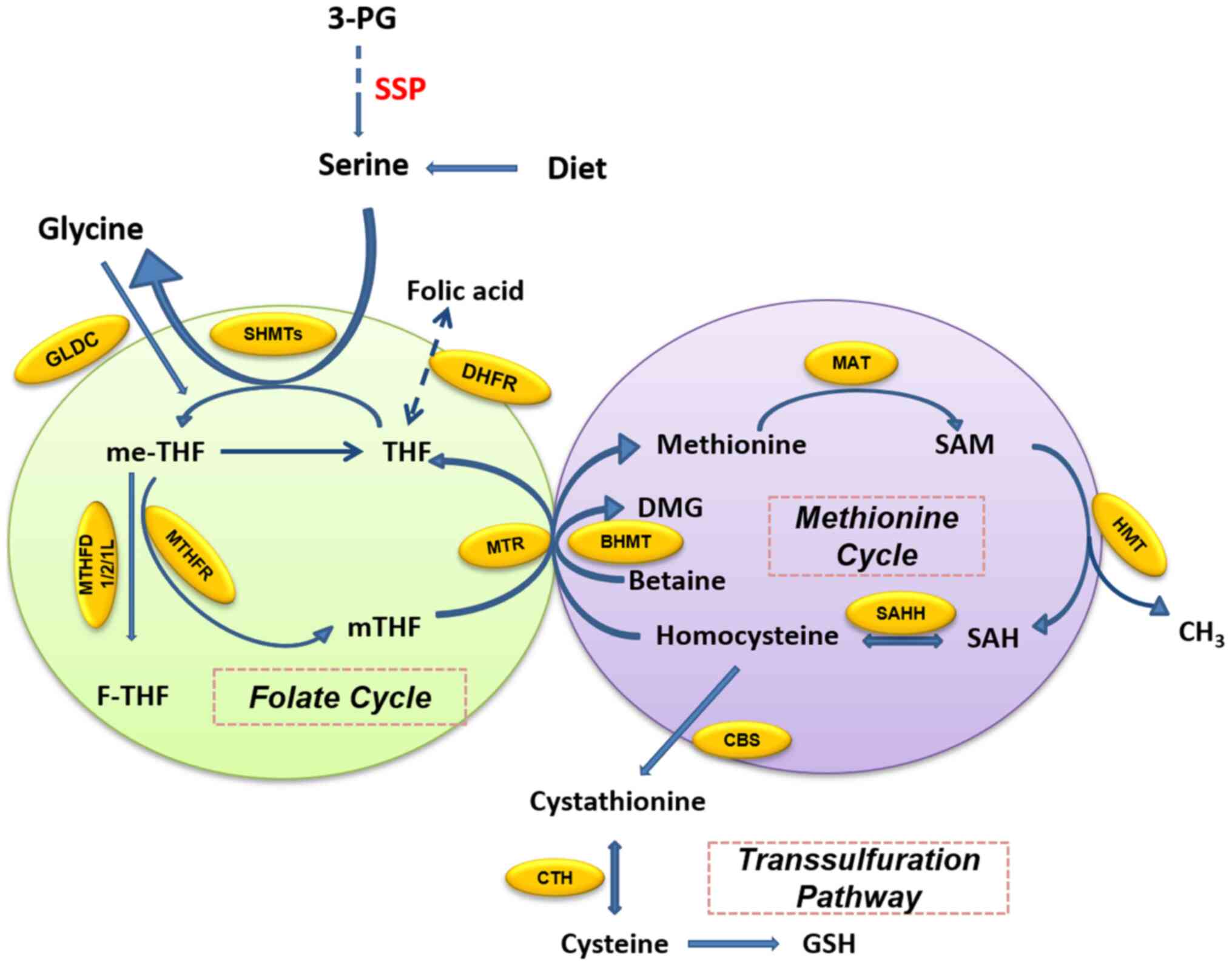
that is defined as folic acid. In the folate cycle (Fig. 2), folic acid is reduced twice by
dihydrofolate (DHF) reductase (DHFR) and finally converted to
tetrahydrofolate (THF). THF accepts the one-carbon unit from the
conversion of serine to glycine to form 5,
10-methylenetetrahydrofolate (me-THF). me-THF is then either
converted into 10-formyltetrahydro-folate (F-THF) by
methylenetetrahydrofolate dehydrogenase (MTHFD) 1/2/1L or catalyzed
by methylenetetrahydrofolate reductase (MTHFR) to
5-methyltetrahydrofolate (mTHF). mTHF can then be demethylated
again and converted back to THF. The demethylation of mTHF
completes the folate cycle and then starts the methionine cycle.
mTHF transfers carbon units to homocysteine, which is then
converted to methionine by methionine adenosine transferase.
Methionine is used to generate SAM. SAM is a substrate of the
methylation reaction, which when demethylated forms S-adenosyl
homocysteine (SAH). The latter is then catalyzed by SAH hydrolase
(SAHH) and converted into homocysteine, thus completing the entire
methionine cycle (16).
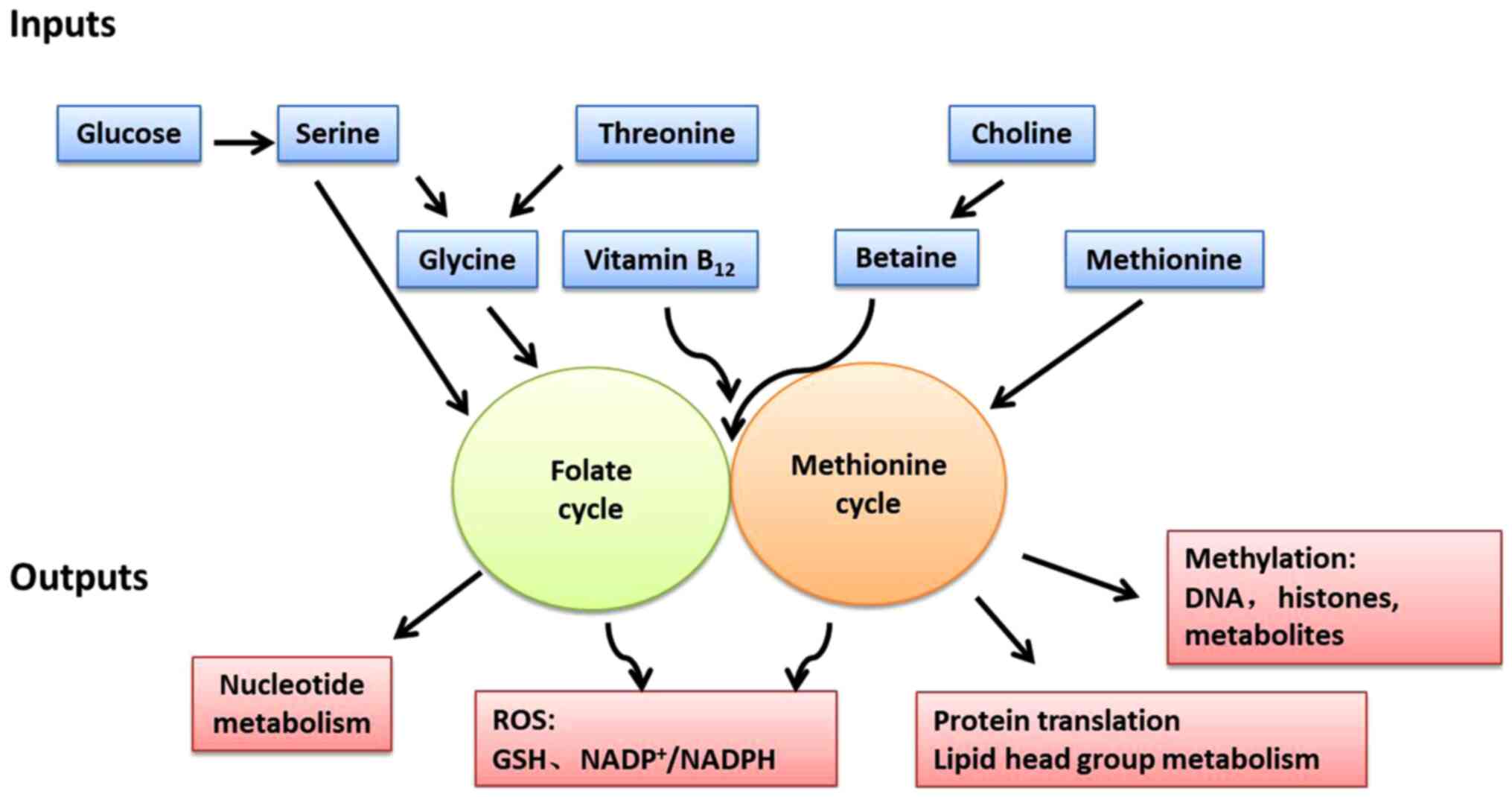
One-carbon metabolism can circulate carbon units
from various amino acids, generate a range of different outputs and
integrate a variety of cellular nutritional statuses (Fig. 3) (15). The one-carbon unit is supplied by
several sources. Serine is the main donor of the one-carbon unit
when it is conversed to glycine. Alternatively, the glycine
cleavage system (GCS) can also fuel one-carbon unit in cancer cell
lines with high GCS activity, such as lung tumor-initiating cells
and glioblastoma-derived cells (78). Recent evidence suggests that cancer
cells can alter or even rely more on these sources to maintain
one-carbon metabolism for cancer cell proliferation (15). Serine derived from exogenous uptake
or de novo SSP synthesis can be cleaved into glycine by the
methyltransferases SHMT1 (in the cytoplasm) and SHMT2 (in the
mitochondria), and donate one-carbon unit (18). In this pathway, the one-carbon unit
cleaved from serine is transferred to THF and then converted to
me-THF (15,16). This reaction can also proceed in
the opposite direction, whereby the consumption of the one-carbon
unit by SHMT converts glycine to serine (79,80).
These reactions demonstrate that the SSP metabolic enzymes have a
significant impact on the production of the one-carbon unit. By
depleting the availability of the one-carbon unit, serine
starvation or downregulation of SSP metabolic enzymes causes the
reduction of cancer cell proliferation and xenograft growth
(80-82). Additionally, glycine, similarly to
serine, can also be a source of the one-carbon unit through the
GCS, although this reaction only occurs within the mitochondria and
fuels one-carbon metabolism (83).
THF accepts a methylene group via the GCS. The resultant
methylene-THF is then used in various down-stream reactions which
require a one-carbon unit (83).
During this pathway, NADH can be also regenerated with the release
of CO2 and ammonia (83,84).
Some studies, however, have found that although the GCS can support
tumorigenesis (68,85), its activity seems to be more
inclined to the degradation/detoxification of glycine rather than
the generation of the one-carbon unit for nucleotide synthesis
(80,85). The directionality of serine/glycine
conversion is a significant factor for cancer cell metabolism and
evidence indicates that mitochondrial SHMT2 is the main
serine-glycine converting enzyme under the above circumstances by
tracing NADPH with 2H-labeled glucose (86). Choline, a vitamin from the human
diet, can be metabolized into betaine and donate one-carbon unit
(87,88). Moreover, one-carbon unit can also
be derived from histidine and tryptophan (89). Although, these little-known
pathways can theoretically contribute one-carbon unit, to the best
of our knowledge, their importance for one-carbon metabolism in
cancer cells has yet to be fully described.
The outputs of one-carbon metabolism include the
production of ATP, NADPH and the regulation of energy balance, as
well as the synthesis of biomacromolecules, such as proteins,
lipids, nucleotides and substrates of methylation reactions
(90-95). DNA synthesis requires nucleotides,
which is a restrictive metabolic aspect of cell proliferation
(19). With the methyl donor
me-THF, deoxyuridine monophosphate (dUMP) can be methylated to
generate deoxythymidine monophosphate (dTMP) by thymidylate
synthase (TYMS), while me-THF is converted to DHF and reduced to
THF by DHFR (16). In addition,
purine can also be generated through the intermediate F-THF from
the folate cycle (16). In the
methionine cycle, not only is methionine itself necessary for
protein synthesis, the SAM produced by adenylation can be used as
the methyl donor for other pathways requiring methyl groups,
including histone, DNA and RNA methylation; lysine and arginine
methylation; polyamine synthesis; and methylation reactions that
generate lipid head groups (96-99).
As much as 40% of the SAM goes to phosphatidylcholine (PC)
production in liver cells where the demand for PC is high, instead
of through the Kennedy pathway (100). Homocysteine, the intermediate
product of the methionine cycle, can produce GSH through
cystathionine and then cysteine in the trans-sulfuration pathway
(16).
One-carbon metabolism also plays an important role
in cell redox balance. In each round of the folate cycle, a
molecule of NADP+ is produced during the reduction of
me-THF by MTHFR (16). The
adjustment of the NADP+/NADPH ratio helps to sustain the
redox state (101). In addition,
GSH, a tripeptide containing cysteine, glycine and glutamic acid,
contributes to the maintenance of the NADP+/NADPH ratio
and is the main contributor to the redox balance (15,16).
Therefore, cancer cells gain survival and proliferation advantages
from changes in these metabolic pathways.
In the context of disease prevention, diagnosis and
treatment, the research and control of one-carbon metabolism is the
basis of other medical and disease research (15-17,102). As aforementioned, the output of
one-carbon metabolism is essential for maintaining normal cell or
cancer cell metabolism. For example, the methylation of DNA and
histones is the most common molecular function change in cancer
cells (17). Rapidly growing
cells, such as tumor cells and embryogenic cells, require the
synthesis of large amounts of proteins, lipids and nucleotides to
support their proliferation (94).
In addition, the redox level in the tumor microenvironment is also
key to the survival of cancer cells (91). The present review subsequently aims
to discuss the main products of one-carbon metabolism and their
physiological relevance, in an attempt to better understand the
role of one-carbon metabolism activity in tumorigenicity and
tumorigenesis.
The one-carbon unit is essential for the synthesis
of purine and pyrimidine nucleotides, which are necessary for the
synthesis of DNA and RNA (19).
De novo purine nucleotide synthesis mainly includes two
stages: i) Synthesis of the important intermediate metabolite,
inosine monophosphate (IMP), a common precursor of all purine
nucleotides, followed by ii) the conversion of IMP into adenosine
monophosphate (AMP) and guanosine monophosphate (15). IMP synthesis requires the
5-phosphate ribose provided by the pentose phosphate pathway (PPP)
to combine glycine, the one-carbon unit carried by F-THF,
CO2 and other substances during a series of reactions
(15). Both glycine and the
one-carbon unit must be generated from serine through folate
metabolism in the cytoplasm or mitochondria (79). Restricting exogenous glycine or
depleting the GCS cannot hinder cancer cell proliferation (80). Moreover, without serine, the
ingestion of exogenous glycine also cannot support nucleotide
synthesis (80). The above
evidence indicates that folate metabolism plays an important role
in nucleotide synthesis. Studies have revealed that inhibition of
folate metabolism through serine starvation or the RNAi-mediated
knock-down of SHMT2, leads to an accumulation of precursors
upstream of IMP prior to incorporation with the one-carbon unit
(80,85). Therefore, the level of one-carbon
unit required for purine nucleotide synthesis can be reduced by
depletion or deprivation of serine, which then inhibits cancer cell
proliferation (80,81). One-carbon metabolism also provides
the methyl donor for pyrimidine nucleotide synthesis. me-THF, as
the methyl donor, supports the methylation reaction of dUMP to
generate dTMP catalyzed by TYMS. me-THF is then converted to DHF
and reduced to THF by DHFR (17).
Therefore, targeting glycine dehydrogenase (GLDC), SHMT or TYMS,
which promote pyrimidine synthesis, may be a potential way to
suppress cancer development (24,103-105). As the key enzyme in the folate
cycle, the expression of MTHFD2 is closely related to mTORC1
signaling in both normal cells and cancer cells. MTHFD2 expression
is stimulated by ATF4 activated by mTORC1 independent of eukaryotic
initiation factor 2α phosphorylation and MTHFD2 enhances F-THF
production to support the synthesis of purines (106,107). Interestingly, mTORC1 can also
phosphorylate carbamoyl phosphate synthetase 2, aspartate
transcarbamylase and dihydroorotatase with the help of its
downstream target ribosomal protein, S6 kinase 1, thereby promoting
pyrimidine synthesis (108,109). These relationships indicate that
mTORC1 can enhance the folate cycle and nucleotide synthesis to
adapt to the increased RNA and DNA synthesis required for cancer
cell anabolics (19).
The methylation pathway is one of the tumor
metabolic reprogramming pathways and all methyltransferase
reactions in mammalian cells are completely dependent on the methyl
donor, SAM. The levels of SAM and its derivative SAH can directly
affect the epigenetic landscape of tumor cells by regulating the
activity of key epigenetic enzymes and ultimately, determine the
fate of cancer cells (110). The
expression of tumor-suppressor gene promoters can be suppressed
through hypermethylation, which then weakens their ability to
inhibit the tumorigenic transformation of cells (98,111,112). PKM2 knockdown contributes to SAM
production in mouse models (113,114), suggesting that PKM2 is involved
in the regulation of the SAM-mediated cancer phenotype by
control-ling methylation. In highly lethal prostate cancer with
protein kinase Cζ (PKCλ)/ι deficiency, the active mTORC1-mediated
ATF4-SSP/one-carbon metabolism axis upregulates SAM synthesis
(25). This approach helps to
increase the plasticity of cell lineages and even gives human
cancer and mouse models in vivo resistance to targeted
therapy (25). In addition, the
absence of serine-threonine kinase (LKB1) in Kirsten rat sarcoma 2
viral oncogene homolog (KRAS) mutant pancreatic cancer promotes
tumorigenesis (115). LKB1
deletion increases the expression of SSP metabolic enzymes, which
activates de novo serine biosynthesis and produces SAM
through one-carbon metabolism, ultimately increasing the overall
amount of DNA methylation and the levels of several DNA
methyltransferases in LKB1-deficient KRAS mutant cells. This
indicates that this type of SAM-dependent methylation pathway
contributes to the metabolic reprogramming of tumors (115). Interestingly, it has generally
been believed that the methionine cycle is mainly supported by the
one-carbon unit cleaved from the serine/glycine synthesis pathway,
but in fact, this pathway has very low activity in cancer cells
(116,117). It has been reported that the
metabolism of serine and glycine can support de novo ATP
synthesis and the adenosine derived from ATP can participate in the
conversion of methionine to SAM (81). Therefore, the restriction of serine
can also reduce the transfer of methyl units to DNA and RNA in
cancer cells by reducing de novo ATP synthesis (15,81).
NADH and NADPH are important cofactors and can
provide electrons for redox reactions. These molecules can be
produced by one-carbon metabolism and are essential for multiple
metabolic and biosynthetic pathways (91). In the folic acid cycle, me-THF can
convert to F-THF, which is catalyzed by MTHFD. NAD+ or
NADP+ as the cofactor in this reaction can be reduced to
NADH or NADPH, respectively. MTHFD2 and MTHFD2-Like (MTHFD2L) are
two forms of mitochondrial MTHFD which can use both NAD+
and NADP+ as cofactors to generate mitochondrial NADH
and NADPH (86,118,119), respectively. The cytoplasmic
MTHFD1 can only utilize NADP+; however, the functional
correlation of the aforementioned dual-specificity remains unknown
(119). During the catabolism
process, the MTHFD2 reaction runs at a faster rate than the
one-carbon unit required for purine synthesis (79). This enables cells to increase the
production of NADH. NADH is known to contribute to a respiratory
chain that is coupled to oxidative phosphorylation, which circles
back to ATP to maintain central energy metabolism (79). There is another pathway that
produces mitochondrial NADPH, which occurs during the oxidation of
F-THF to CO2 and THF by aldehyde dehydrogenase 1 family
member L2, and provides some of the energy for proline synthesis
(79). Although studies have shown
that NADPH is mainly produced in mitochondria (86,90,120), cytosolic NADPH can be generated
by oxidizing me-THF by MTHFD1 (79). The synthesis of fatty acids in the
cytoplasm is mainly supported by NADPH formed by the action of
malic enzyme. In addition, the cytoplasmic NADPH derived from
folate metabolism can also specifically support fatty acid
synthesis (15). Fatty acids are
necessary for the production of lipid signaling molecules and
membranes, and both are essential for sustaining cancer cell
proliferation (121).
Serine/one-carbon metabolism also depends on the cytoplasmic
NADPH/NADP ratio maintained by the activity of the oxidative PPP
(oxPPP). Studies have shown that the loss of glucose 6-phosphate
phosphate dehydrogenase can inhibit oxPPP, leading to high NADP and
impairing folate-mediated biosynthesis by inhibiting DHFR activity
with high NADP in CRC cells (91).
This indicates that oxPPP is crucial for maintaining normal
NADPH/NADP ratios, DHFR activity and folate metabolism. SHMT2 is a
direct target gene of c-Myc (122). When MYC-transformed cells are
subjected to hypoxia, SHMT2 is induced and triggers the degradation
of serine to CO2 and NH4+, simultaneously
producing net NADPH to maintain oxidation of the tumor
microenvironment (122). A study
concerning human glioblastoma multiforme confirmed this was the
case in this disease. SHMT2 and GLDC are highly expressed in the
pseudopalisading cells around necrotic lesions (85). SHMT2 inhibits PKM2 activity and
reduces oxygen consumption, which triggers a novel metabolic state,
conferring a profound survival advantage to cells in tumor regions
with poor vascularization (85).
In addition, GSH is one of the products of the trans-sulfuration
pathway and one of the most abundant metabolites in cells. It is
also important for maintaining the NADPH/NADP+ ratio
(123). GSH has the ability to
scavenge and reduce ROS, as well as maintain the appropriate
NADPH/NADP+ ratio, which greatly contributes to the
redox balance in cells (123,124).
One of the major challenges for cancer biology is to
find novel and effective therapeutic targets that can be used for
interventions with chemically selective pharmaceuticals in
different patients. Antimetabolite drugs (antifolates) are a
landmark in cancer chemotherapy and are still the most widely used
drugs in medical oncology (Table
I) (125-134). Among the antifolates,
methotrexate and pemetrexed are effective inhibitors of DHFR, which
can reduce the THF pool and prevent cell proliferation (135,136). As such, they are a major class of
cancer chemotherapeutic drugs and are currently used as a
first-line chemotherapeutic agent in the treatment of various
cancers, including acute lymphoblastic leukemia, breast cancer,
bladder cancer and lymphoma (137,138). Studies have found that
methotrexate and pemetrexed also have the ability to bind to and
inhibit human SHMT in vitro (139). There are other drugs that target
the downstream pathway of the SSP/one-carbon metabolism which have
been approved for clinical use, such as gemcitabine and
5-fluorouracil (5-FU) (140,141). 5-FU, a congener of uracil and a
standard drug used to treat a variety of cancers, inhibits TYMS,
resulting in the reduction of the methylation of dUMP to dTMP and
the interruption of the folate cycle (141). 5-FU can also be converted to
5-fluorouri-dine, which is incorporated into ribosomal RNA (rRNA)
molecules and inhibits rRNA processing, eventually leading to
p53-dependent cell cycle arrest and/or apoptosis (142). Traditional antifolate
chemotherapy drugs, such as methotrexate and 5-FU, have been used
in clinical cancer chemotherapy to target one-carbon metabolic
pathways for ~70 years (72).
However, since the folate metabolism pathway is also important in
normal cell proliferation, these drugs have many harmful side
effects. Moreover, resistance to antifolates is also a common
problem in cancer treatment (15).
For these reasons, the development of new targets and new drugs is
crucial.
Currently, other studies targeting the downstream of
SSP/one-carbon metabolism are attempting to regulate the epigenetic
state of the tumors and regulate the metabolic enzymes that are
overactivated in the tumors (15,72,143). Epigenetic reprogramming through
the regulation of the methylation pathway is essential for the
malignant tumor phenotype with studies suggesting that the control
of methylation is possible (144,145). As aforementioned, methotrexate
has been widely used for cancer treatment since 1948, but it has
only recently been found that methotrexate can decrease Wnt-induced
intracellular lysosome activity and reduce typical Wnt signaling by
inhibiting SAM levels and blocking arginine methylation (146). These findings indicate that
methotrexate may be used to treat Wnt-driven malignant tumors. It
has been found that the activation of SSP/one-carbon metabolic
pathway genes during cancer metabolic control depends on the G9A
epigenetic program (47,143), and the G9A inhibitor, BIX01294,
can cause cell death by depriving serine in vivo (147), This suggests that G9A inhibition
may be a therapeutic strategy for the treatment of cancer, a
possibility that is contributing to the development of G9A-like
drug molecules. The H3K4 demethylase jumonji AT rich interactive
domain 1B (Jarid1b) (Lysine demethylase 5B/PLU-1/retinoblastoma
binding protein 2-homolog 1) supports the continuous tumor growth
in certain cell subsets of slow-circulating melanoma (148). These cancer cell subtypes exhibit
slow DNA replication and may be resistant to chemotherapeutic
agents and radiation, thereby contributing to tumor recurrence and
metastasis (148). In solid
cancers, histone lysine demethylase family members are associated
with cancer progression. Knockdown of related genes can therefore
suppress carcinogenicity and promote cell senescence (149,150). Methylation donors, ornithine
decarboxylation and polyamine metabolism have been widely
investigated as anti-cancer therapeutic targets, with some of these
drugs entering clinical trials, such as ornithine decarboxylase
inhibitor; 2-difluoromethylornithine, a competitive inhibitor of
SAM decarboxylase; methylglyoxal bis (guanylhydrazone); and SAM486A
(133,151). Targeting SSP metabolic enzymes
also appears to be a promising method. PKCζ not only inhibits the
transcription of PHGDH and PSAT1, but also phosphorylates PHGDH to
inhibit its catalytic activity (152). In addition, for certain
PHGDH-dependent cancer cells, some small molecule inhibitors
targeting PHGDH have been developed and successfully verified in
vitro, which not only reduces cancer cell proliferation, but
inhibits the growth of xenografts (153-156) (Table II).
Pharmacology can be used as a complementary
strategy for cancers that do not upregulate the key enzymes of the
SSP (21,32,157). In addition to the positive
correlation between high carbohydrate intake and cancer incidence
(158), low glucose intake may
have a negative effects on tumor growth and progression (159), making the reduction of exogenous
serine intake a feasible approach. Indeed, serine and glycine
starvation can successfully reduce xenograft and spontaneous tumor
growth, and have been found to significantly improve survival rates
in various mouse tumor models (29,32).
Particularly in the case of p53 deficiency, cancer cells are more
sensitive to serine and glycine starvation (32). Metformin has recently been
recognized as a promising drug for cancer treatment (160). Gravel et al (157) examined the anti-tumor effect of
metforminin combination with serine starvation. Their results
showed that biguanide does not inhibit serine synthesis and that
cancer cells require serine to upregulate the glycolytic pathway to
compensate for the reduction of oxidative phosphorylation induced
by biguanide (159). Under a
serine deficiency, biguanide activity is enhanced without relying
on AMP-activated protein kinase; and serine deprivation and
metformin exert joint antiproliferative effects by directly
interfering with cancer cell metabolism. In addition, the
deprivation of serine also changes the relative abundance of the
metformin-induced TCA cycle metabolites (157). This points us to a new type of
dietary manipulation that can enhance the efficacy of biguanides as
antineoplastic agents.
Targeting folate metabolizing enzymes, such as
MTHFD2, is another potential method for cancer treatment. MTHFD2,
which is normally expressed only during embryonic development,
provides the possibility of a disease-selective treatment target,
through eliminating cancer cells while retaining healthy cells
(161). Gustafsson et al
(161) reported the synthesis and
pre-clinical characterization of the first human MTHFD2 inhibitor,
LY345899, providing a theoretical basis for the continued
development of the structural framework for MTHFD2 inhibitors that
can be effectively used for the treatment of various types of
cancer. Recently, it has been reported that the expression of
MTHFD2 and the stem-like proper-ties can be enhanced in lung cancer
cells that have acquired resistance to the targeted drug gefitinib
(162). Furthermore, the
overexpression of MTHFD2 makes gefitinib-sensitive lung cancer
cells resistant to gefitinib. In these gefitinib-resistant cancer
cells, the sensitivity to gefitinib, as well as the stem-like
properties, can be restored after MTHFD2 knockdown or treatment
with AICAR (162). Therefore,
since cancer stem (like) cells are dependent on MTHFD2, therapies
targeting MTHFD2 have been proposed as a therapeutic possibility
for eradicating tumors and preventing recurrence (162). The problem with this approach;
however, is that when targeting specific components of one-carbon
metabolism, the tumor may reconnect with other metabolisms to
compensate (163). The function
of the MTHF enzyme is to convert me-THF to F-THF and mTHF for
nucleotide synthesis and methionine recycling (143). The MTHFD enzyme has several
forms: Cytoplasmic MTHFD1, mitochondrial MTHFD1-Like (MTHFD1L),
MTHFD2 and MTHFD2L (118,143). MTHFD2 is only expressed in
embryos, tumors and undifferentiated tissues, while MTHFD2L is more
widely expressed (163,164). Cells primarily use mitochondrial
enzymes for one-carbon metabolism, so if this effect is suppressed,
cells can compen-sate by using cytoplasmic MTHFD1 (79). Cells primarily use mitochondrial
enzymes for one-carbon metabolism, so if this effect is suppressed,
cells can compensate by using cytoplasmic MTHFD1 (79).
In the past few years, researchers' interest in
cancer metabolism has surged, leading to an expanding understanding
of the metabolic pathways of cancer biology. Recent advances in
comprehending the relationship between cancer and metabolism
highlight the correlation between the SSP and one-carbon
metabolism. At present; however, the molecular regulatory mechanism
between SSP/one-carbon metabolism and cancer metabolism is not
fully understood. To explore its therapeutic potential, it is
necessary to biochemically dissect the ways in which these
metabolic pathways promote cancer biology, in the hope of solving
the mystery and helping to clinically overcome the worldwide
problem of cancer.
This study was conducted with the support of the
National Natural Science Foundation of China (grant no. 81872023),
the China Postdoctoral Science Foundation (grant no. 2018M642742)
and the Henan Province Key Research and Development and Promotion
Project (grant no. 202102310412).
Not applicable.
SP, MF and ZL collected information and wrote the
manuscript. HW and XL collected information and edited the
manuscript. All authors read and approved the final version of this
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Reznik E, Luna A, Aksoy BA, Liu EM, La K,
Ostrovnaya I, Creighton CJ, Hakimi AA and Sander C: A Landscape of
meta-bolic variation across tumor types. Cell Syst. 6:301–313.e3.
2018. View Article : Google Scholar
|
|
2
|
Sun L, Suo C, Li ST, Zhang H and Gao P:
Metabolic reprogram-ming for cancer cells and their
microenvironment: Beyond the Warburg Effect. Biochim Biophys Acta
Rev Cancer. 1870:51–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DeBerardinis RJ and Chandel NS:
Fundamentals of cancer metabolism. Sci Adv. 2:e16002002016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vander Heiden MG and DeBerardinis RJ:
Understanding the intersections between metabolism and cancer
biology. Cell. 168:657–669. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vazquez A, Kamphorst JJ, Markert EK, Schug
ZT, Tardito S and Gottlieb E: Cancer metabolism at a glance. J Cell
Sci. 129:3367–3373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bose S and Le A: Glucose metabolism in
cancer. Adv Exp Med Biol. 1063:3–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hosios AM, Hecht VC, Danai LV, Johnson MO,
Rathmell JC, Steinhauser ML, Manalis SR and Vander Heiden MG: Amino
Acids rather than glucose account for the majority of cell mass in
proliferating mammalian cells. Dev Cell. 36:540–549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun L, Song L, Wan Q, Wu G, Li X, Wang Y,
Wang J, Liu Z, Zhong X, He X, et al: cMyc-mediated activation of
serine biosynthesis pathway is critical for cancer progression
under nutrient deprivation conditions. Cell Res. 25:429–444. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Newman AC and Maddocks ODK: Serine and
functional metabolites in cancer. Trends Cell Biol. 27:645–657.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen S, Xia Y, He F, Fu J, Xin Z, Deng B,
He L, Zhou X and Ren W: Serine Supports IL-1β production in
macrophages through mTOR signaling. Front Immunol. 11:18662020.
View Article : Google Scholar
|
|
14
|
Sowers ML, Herring J, Zhang W, Tang H, Ou
Y, Gu W and Zhang K: Analysis of glucose-derived amino acids
involved in one-carbon and cancer metabolism by stable-isotope
tracing gas chromatography mass spectrometry. Anal Biochem.
566:1–9. 2019. View Article : Google Scholar
|
|
15
|
Newman AC and Maddocks ODK: One-carbon
metabolism in cancer. Br J Cancer. 116:1499–1504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lan X, Field MS and Stover PJ: Cell cycle
regulation of folate-mediated one-carbon metabolism. Wiley
Interdiscip Rev Syst Biol Med. 10:e14262018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ducker GS and Rabinowitz JD: One-carbon
metabolism in health and disease. Cell Metab. 25:27–42. 2017.
View Article : Google Scholar :
|
|
18
|
Yang M and Vousden KH: Serine and
one-carbon metabolism in cancer. Nat Rev Cancer. 16:650–662. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng JD, Wu WKK, Wang HY and Li XX: Serine
and one-carbon metabolism, a bridge that links mTOR signaling and
DNA methylation in cancer. Pharmacol Res. 149:1043522019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia Y, Ye B, Ding J, Yu Y, Alptekin A,
Thangaraju M, Prasad PD, Ding ZC, Park EJ, Choi JH, et al:
Metabolic reprogramming by MYCN confers dependence on the
serine-glycine-one-carbon biosynthetic pathway. Cancer Res.
79:3837–3850. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mattaini KR, Sullivan MR, Lau AN, Fiske
BP, Bronson RT and Vander Heiden MG: Increased PHGDH expression
promotes aberrant melanin accumulation. BMC Cancer. 19:7232019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Samanta D, Park Y, Andrabi SA, Shelton LM,
Gilkes DM and Semenza GL: PHGDH expression is required for
mitochondrial redox homeostasis, breast cancer stem cell
maintenance, and lung metastasis. Cancer Res. 76:4430–4442. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sullivan MR, Mattaini KR, Dennstedt EA,
Nguyen AA, Sivanand S, Reilly MF, Meeth K, Muir A, Darnell AM,
Bosenberg MW, et al: Increased serine synthesis provides an
advantage for tumors arising in tissues where serine levels are
limiting. Cell Metab. 29:1410–1421.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Zheng A, Hydbring P, Ambroise G,
Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H and Norberg E: PHGDH
Defines a metabolic subtype in lung adenocarcinomas with poor
prognosis. Cell Rep. 19:2289–2303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reina-Campos M, Linares JF, Duran A,
Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards
A, Rooslid T, et al: Increased serine and one-carbon pathway
metabolism by PKClambda/iota deficiency promotes neuroendocrine
prostate cancer. Cancer Cell. 35:385–400.e9. 2019. View Article : Google Scholar
|
|
26
|
Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X,
Ma J, Yin X, Mao A and Shang M: Overexpression of phosphoserine
aminotransferase 1 (PSAT1) predicts poor prognosis and associates
with tumor progression in human esophageal squamous cell carcinoma.
Cell Physiol Biochem. 39:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin HO, Hong SE, Kim JY, Jang SK, Kim YS,
Sim JH, Oh AC, Kim H, Hong YJ, Lee JK and Park IC: Knock-down of
PSAT1 enhances sensitivity of NSCLC cells to glutamine-limiting
conditions. Anticancer Res. 39:6723–6730. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Y, Liang X, Xu J and Cai X: miR-424
targets AKT3 and PSAT1 and has a tumor-suppressive role in human
colorectal cancer. Cancer Manag Res. 10:6537–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maddocks ODK, Athineos D, Cheung EC, Lee
P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D,
Kruiswijk F, et al: Modulating the therapeutic response of tumours
to dietary serine and glycine starvation. Nature. 544:372–376.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mattaini KR, Sullivan MR and Vander Heiden
MG: The importance of serine metabolism in cancer. J Cell Biol.
214:249–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
DeNicola GM, Chen PH, Mullarky E, Sudderth
JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al: NRF2
regulates serine biosynthesis in non-small cell lung cancer. Nat
Genet. 47:1475–1481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maddocks OD, Berkers CR, Mason SM, Zheng
L, Blyth K, Gottlieb E and Vousden KH: Serine starvation induces
stress and p53-dependent metabolic remodelling in cancer cells.
Nature. 493:542–546. 2013. View Article : Google Scholar
|
|
33
|
Wortel IMN, van der Meer LT, Kilberg MS
and van Leeuwen FN: Surviving Stress: Modulation of ATF4-mediated
stress responses in normal and malignant cells. Trends Endocrinol
Metab. 28:794–806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kasai S, Yamazaki H, Tanji K, Engler MJ,
Matsumiya T and Itoh K: Role of the ISR-ATF4 pathway and its cross
talk with Nrf2 in mitochondrial quality control. J Clin Biochem
Nutr. 64:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dey S, Sayers CM, Verginadis II, Lehman
SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs
SY, et al: ATF4-dependent induction of heme oxygenase 1 prevents
anoikis and promotes metastasisATF4-dependent induction of heme
oxygenase 1 prevents anoikis and promotes metastasis. J Clin
Invest. 125:2592–2608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tameire F, Verginadis II, Leli NM, Polte
C, Conn CS, Ojha R, Salas Salinas C, Chinga F, Monroy AM, Fu W, et
al: ATF4 couples MYC-dependent translational activity to
bioenergetic demands during tumour progression. Nat Cell Biol.
21:889–899. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mesclon F, Lambert-Langlais S, Carraro V,
Parry L, Hainault I, Jousse C, Maurin AC, Bruhat A, Fafournoux P
and Averous J: Decreased ATF4 expression as a mechanism of acquired
resistance to long-term amino acid limitation in cancer cells.
Oncotarget. 8:27440–27453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mazor KM and Stipanuk MH: GCN2- and
eIF2α-phosph orylation-independent, but ATF4-dependent, induction
of CARE-containing genes in methionine-deficient cells. Amino
Acids. 48:2831–2842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Al-Baghdadi RJT, Nikonorova IA, Mirek ET,
Wang Y, Park J, Belden WJ, Wek RC and Anthony TG: Role of
activating transcription factor 4 in the hepatic response to amino
acid depletion by asparaginase. Sci Rep. 7:12722017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu D, Dai W, Kutzler L, Lacko HA,
Jefferson LS, Dennis MD and Kimball SR: ATF4-mediated upregulation
of REDD1 and Sestrin2 suppresses mTORC1 activity during prolonged
leucine deprivation. J Nutr. 150:1022–1030. 2020. View Article : Google Scholar
|
|
41
|
Adams CM: Role of the transcription factor
ATF4 in the anabolic actions of insulin and the anti-anabolic
actions of glucocorticoids. J Biol Chem. 282:16744–16753. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ye J, Mancuso A, Tong X, Ward PS, Fan J,
Rabinowitz JD and Thompson CB: Pyruvate kinase M2 promotes de novo
serine synthesis to sustain mTORC1 activity and cell proliferation.
Proc Natl Acad Sci USA. 109:6904–6909. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y
and Pang D: PSAT1 is regulated by ATF4 and enhances cell
proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway
in ER-negative breast cancer. J Exp Clin Cancer Res. 36:1792017.
View Article : Google Scholar
|
|
44
|
Svoboda LK, Teh SSK, Sud S, Kerk S,
Zebolsky A, Treichel S, Thomas D, Halbrook CJ, Lee HJ, Kremer D, et
al: Menin regulates the serine biosynthetic pathway in Ewing
sarcoma. J Pathol. 245:324–336. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi
JH, Yan C, Dong Z, Huang S, Zha Y, et al: KDM4C and ATF4 cooperate
in transcriptional control of amino acid metabolism. Cell Rep.
14:506–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim SY, Hong M, Heo SH, Park S, Kwon TK,
Sung YH, Oh Y, Lee S, Yi GS and Kim I: Inhibition of euchromatin
histone-lysine N-methyltransferase 2 sensitizes breast cancer cells
to tumor necrosis factor-related apoptosis-inducing ligand through
reactive oxygen species-mediated activating transcription factor
4-C/EBP homologous protein-death receptor 5 pathway activation. Mol
Carcinog. 57:1492–1506. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ding J, Li T, Wang X, Zhao E, Choi JH,
Yang L, Zha Y, Dong Z, Huang S, Asara JM, et al: The histone H3
methyltransferase G9A epigenetically activates the serine-glycine
synthesis pathway to sustain cancer cell survival and
proliferation. Cell Metab. 18:896–907. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hydbring P, Castell A and Larsson LG: MYC
modulation around the CDK2/p27/SKP2 axis. Genes (Basel). 8:1742017.
View Article : Google Scholar
|
|
49
|
Fallah Y, Brundage J, Allegakoen P and
Shajahan-Haq AN: MYC-driven pathways in breast cancer subtypes.
Biomolecules. 7:532017. View Article : Google Scholar :
|
|
50
|
Lancho O and Herranz D: The MYC
Enhancer-ome: Long-range transcriptional regulation of MYC in
cancer. Trends Cancer. 4:810–822. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Carabet LA, Rennie PS and Cherkasov A:
Therapeutic inhibition of Myc in cancer. structural bases and
computer-aided drug discovery approaches. Int J Mol Sci.
20:1202018. View Article : Google Scholar
|
|
53
|
Chen Y, Sun XX, Sears RC and Dai MS:
Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis.
6:359–371. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Walz S, Lorenzin F, Morton J, Wiese KE,
von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M,
et al: Activation and repression by oncogenic MYC shape
tumour-specific gene expression profiles. Nature. 511:483–487.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tesi A, de Pretis S, Furlan M, Filipuzzi
M, Morelli MJ, Andronache A, Doni M, Verrecchia A, Pelizzola M,
Amati B and Sabò A: An early Myc-dependent transcriptional program
orchestrates cell growth during B-cell activation. EMBO Rep.
20:e479872019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Robaina MC, Mazzoccoli L and Klumb CE:
Germinal Centre B Cell Functions and Lymphomagenesis: Circuits
Involving MYC and MicroRNAs. Cells. 8:13652019. View Article : Google Scholar
|
|
57
|
Le A, Lane AN, Hamaker M, Bose S, Gouw A,
Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al:
Glucose-independent glutamine metabolism via TCA cycling for
proliferation and survival in B cells. Cell Metab. 15:110–121.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang LW, Shen H, Nobre L, Ersing I, Paulo
JA, Trudeau S, Wang Z, Smith NA, Ma Y, Reinstadler B, et al:
Epstein-barr-virus-induced one-carbon metabolism drives B cell
transformation. Cell Metab. 30:539–555.e11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kauko O, O'Connor CM, Kulesskiy E,
Sangodkar J, Aakula A, Izadmehr S, Yetukuri L, Yadav B, Padzik A,
Laajala TD, et al: PP2A inhibition is a druggable MEK inhibitor
resistance mecha-nism in KRAS-mutant lung cancer cells. Sci Transl
Med. 10:eaaq10932018. View Article : Google Scholar
|
|
60
|
Pakos-Zebrucka K, Koryga I, Mnich K,
Ljujic M, Samali A and Gorman AM: The integrated stress response.
EMBO Rep. 17:1374–1395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
David CJ, Chen M, Assanah M, Canoll P and
Manley JL: HnRNP proteins controlled by c-Myc deregulate pyruvate
kinase mRNA splicing in cancer. Nature. 463:364–368. 2010.
View Article : Google Scholar :
|
|
62
|
Luan W, Wang Y, Chen X, Shi Y, Wang J,
Zhang J, Qian J, Li R, Tao T, Wei W, et al: PKM2 promotes glucose
metabolism and cell growth in gliomas through a mechanism involving
a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 6:13006–13018.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chaneton B, Hillmann P, Zheng L, Martin
ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding
FP, Vousden KH, et al: Serine is a natural ligand and allosteric
activator of pyruvate kinase M2. Nature. 491:458–462. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li AM and Ye J: The PHGDH enigma: Do
cancer cells only need serine or also a redox modulator? Cancer
Lett. 476:97–105. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bao XR, Ong SE, Goldberger O, Peng J,
Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et
al: Mitochondrial dysfunction remodels one-carbon metabolism in
human cells. Elife. 5:e105752016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mendez-Lucas A, Li X, Hu J, Che L, Song X,
Jia J, Wang J, Xie C, Driscoll PC, Tschaharganeh DF, et al: Glucose
catabolism in liver tumors induced by c-MYC can be sustained by
various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer
Res. 77:4355–4364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang
X, Wu H, Blanco F, Li S, Bhanot G, et al: Glutaminase 2 is a novel
negative regulator of small GTPase Rac1 and mediates p53 function
in suppressing metastasis. Elife. 5:e107272016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kastenhuber ER and Lowe SW: Putting p53 in
Context. Cell. 170:1062–1078. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang S, Peng Z, Wang S, Yang L, Chen Y,
Kong X, Song S, Pei P, Tian C, Yan H, et al: KRAB-type zinc-finger
proteins PITA and PISA specifically regulate p53-dependent
glycolysis and mitochondrial respiration. Cell Res. 28:572–592.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang X, Zhang X, Li Y, Shao Y, Xiao J,
Zhu G and Li F: PAK4 regulates G6PD activity by p53 degradation
involving colon cancer cell growth. Cell Death Dis. 8:e28202017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fritsche MK and Knopf A: The tumor
suppressor p53 in mucosal melanoma of the head and neck. Genes
(Basel). 8:3842017. View Article : Google Scholar
|
|
72
|
Amelio I, Cutruzzola F, Antonov A,
Agostini M and Melino G: Serine and glycine metabolism in cancer.
Trends Biochem Sci. 39:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lu J, Tan M and Cai Q: The Warburg effect
in tumor progression: Mitochondrial oxidative metabolism as an
anti-metastasis mechanism. Cancer Lett. 356:156–164. 2015.
View Article : Google Scholar
|
|
74
|
Humpton TJ, Hock AK, Maddocks ODK and
Vousden KH: p53-mediated adaptation to serine starvation is
retained by a common tumour-derived mutant. Cancer Metab. 6:182018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Riscal R, Schrepfer E, Arena G, Cissé MY,
Bellvert F, Heuillet M, Rambow F, Bonneil E, Sabourdy F, Vincent C,
et al: Chromatin-bound MDM2 regulates serine metabolism and redox
homeostasis independently of p53. Mol Cell. 62:890–902. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Amelio I, Markert EK, Rufini A, Antonov
AV, Sayan BS, Tucci P, Agostini M, Mineo TC, Levine AJ and Melino
G: p73 regulates serine biosynthesis in cancer. Oncogene.
33:5039–5046. 2014. View Article : Google Scholar
|
|
77
|
Ou Y, Wang SJ, Jiang L, Zheng B and Gu W:
p53 Protein-mediated regulation of phosphoglycerate dehydrogenase
(PHGDH) is crucial for the apoptotic response upon serine
starvation. J Biol Chem. 290:457–466. 2015. View Article : Google Scholar :
|
|
78
|
Zhang WC, Shyh-Chang N, Yang H, Rai A,
Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al: Glycine
decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ducker GS, Chen L, Morscher RJ,
Ghergurovich JM, Esposito M, Teng X, Kang Y and Rabinowitz JD:
Reversal of cytosolic one-carbon flux compensates for loss of the
mitochondrial folate pathway. Cell Metab. 23:1140–1153. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Labuschagne CF, van den Broek NJ, Mackay
GM, Vousden KH and Maddocks OD: Serine, but not glycine, supports
one-carbon metabolism and proliferation of cancer cells. Cell Rep.
7:1248–1258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Maddocks OD, Labuschagne CF, Adams PD and
Vousden KH: Serine metabolism supports the methionine cycle and
DNA/RNA Methylation through de novo ATP synthesis in cancer cells.
Mol Cell. 61:210–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Guo H, Xu J, Zheng Q, He J, Zhou W, Wang
K, Huang X, Fan Q, Ma J, Cheng J, et al: NRF2 SUMOylation promotes
de novo serine synthesis and maintains HCC tumorigenesis. Cancer
Lett. 466:39–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kikuchi G, Motokawa Y, Yoshida T and
Hiraga K: Glycine cleavage system: Reaction mechanism,
physiological significance, and hyperglycinemia. Proc Jpn Acad Ser
B Phys Biol Sci. 84:246–263. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tedeschi PM, Markert EK, Gounder M, Lin H,
Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, DiPaola RS, Hirshfield KM,
et al: Contribution of serine, folate and glycine metabolism to the
ATP, NADPH and purine requirements of cancer cells. Cell Death Dis.
4:e8772013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kim D, Fiske BP, Birsoy K, Freinkman E,
Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, et al:
SHMT2 drives glioma cell survival in ischaemia but imposes a
dependence on glycine clearance. Nature. 520:363–367. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lewis CA, Parker SJ, Fiske BP, McCloskey
D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo
CM, et al: Tracing compartmentalized NADPH metabolism in the
cytosol and mitochondria of mammalian cells. Mol Cell. 55:253–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ueland PM: Choline and betaine in health
and disease. J Inherit Metab Dis. 34:3–15. 2011. View Article : Google Scholar
|
|
88
|
Friso S, Udali S, De Santis D and Choi SW:
One-carbon metabolism and epigenetics. Mol Aspects Med. 54:28–36.
2017. View Article : Google Scholar
|
|
89
|
Kanarek N, Keys HR, Cantor JR, Lewis CA,
Chan SH, Kunchok T, Abu-Remaileh M, Freinkman E, Schweitzer LD and
Sabatini DM: Histidine catabolism is a major determinant of
methotrexate sensitivity. Nature. 559:632–636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fan J, Ye J, Kamphorst JJ, Shlomi T,
Thompson CB and Rabinowitz JD: Quantitative flux analysis reveals
folate-dependent NADPH production. Nature. 510:298–302. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen L, Zhang Z, Hoshino A, Zheng HD,
Morley M, Arany Z and Rabinowitz JD: NADPH production by the
oxidative pentose-phosphate pathway supports folate metabolism. Nat
Metab. 1:404–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Reid MA, Dai Z and Locasale JW: The impact
of cellular metabolism on chromatin dynamics and epigenetics. Nat
Cell Biol. 19:1298–1306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Morscher RJ, Ducker GS, Li SH, Mayer JA,
Gitai Z, Sperl W and Rabinowitz JD: Mitochondrial translation
requires folate-dependent tRNA methylation. Nature. 554:128–132.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Gao X, Lee K, Reid MA, Sanderson SM, Qiu
C, Li S, Liu J and Locasale JW: Serine availability influences
mitochondrial dynamics and function through lipid metabolism. Cell
Rep. 22:3507–3520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Villa E, Ali ES, Sahu U and Ben-Sahra I:
Cancer cells tune the signaling pathways to empower de novo
synthesis of nucleotides. Cancers (Basel). 11:6882019. View Article : Google Scholar
|
|
96
|
Ulanovskaya OA, Zuhl AM and Cravatt BF:
NNMT promotes epigenetic remodeling in cancer by creating a
metabolic methylation sink. Nat Chem Biol. 9:300–306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hughey CC, Trefts E, Bracy DP, James FD,
Donahue EP and Wasserman DH: Glycine N-methyltransferase deletion
in mice diverts carbon flux from gluconeogenesis to pathways that
utilize excess methionine cycle intermediates. J Biol Chem.
293:11944–11954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Serefidou M, Venkatasubramani AV and Imhof
A: The impact of one carbon metabolism on histone methylation.
Front Genet. 10:7642019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Fukuoka H and Kubota T: One-carbon
metabolism and lipid metabolism in DOHaD. Adv Exp Med Biol.
1012:3–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Walker AK: 1-Carbon cycle metabolites
methylate their way to fatty liver. Trends Endocrinol Metab.
28:63–72. 2017. View Article : Google Scholar
|
|
101
|
Xiao W, Wang RS, Handy DE and Loscalzo J:
NAD(H) and NADP(H) Redox couples and cellular energy metabolism.
Antioxid Redox Signal. 28:251–272. 2018. View Article : Google Scholar :
|
|
102
|
Hanley MP and Rosenberg DW: One-carbon
metabolism and colorectal cancer: Potential mechanisms of
chemoprevention. Curr Pharmacol Rep. 1:197–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ser Z, Gao X, Johnson C, Mehrmohamadi M,
Liu X, Li S and Locasale JW: targeting one carbon metabolism with
an antimetabolite disrupts pyrimidine homeostasis and induces
nucleotide overflow. Cell Rep. 15:2367–2376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Pandey S, Garg P, Lee S, Choung HW, Choung
YH, Choung PH and Chung JH: Nucleotide biosynthesis arrest by
silencing SHMT1 function via vitamin B6-coupled vector and effects
on tumor growth inhibition. Biomaterials. 35:9332–9342. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Tripathi SK, Gupta N, Mahato M, Gupta KC
and Kumar P: Selective blocking of primary amines in branched
polyethylenimine with biocompatible ligand alleviates cytotoxicity
and augments gene delivery efficacy in mammalian cells. Colloids
Surf B Biointerfaces. 115:79–85. 2014. View Article : Google Scholar
|
|
106
|
Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara
JM and Manning BD: mTORC1 induces purine synthesis through control
of the mito-chondrial tetrahydrofolate cycle. Science. 351:728–733.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Park Y, Reyna-Neyra A, Philippe L and
Thoreen CC: mTORC1 balances cellular amino acid supply with demand
for protein synthesis through post-transcriptional control of ATF4.
Cell Rep. 19:1083–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ben-Sahra I, Howell JJ, Asara JM and
Manning BD: Stimulation of de novo pyrimidine synthesis by growth
signaling through mTOR and S6K1. Science. 339:1323–1328. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rabinovich S, Adler L, Yizhak K, Sarver A,
Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et
al: Diversion of aspartate in ASS1-deficient tumours fosters de
novo pyrimidine synthesis. Nature. 527:379–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mentch SJ and Locasale JW: One-carbon
metabolism and epigenetics: Understanding the specificity. Ann N Y
Acad Sci. 1363:91–98. 2016. View Article : Google Scholar
|
|
111
|
Mahmoud AM and Ali MM: Methyl donor
micronutrients that modify DNA methylation and cancer outcome.
Nutrients. 11:6082019. View Article : Google Scholar :
|
|
112
|
Morgan AE, Davies TJ and Mc Auley MT: The
role of DNA methylation in ageing and cancer. Proc Nutr Soc.
77:412–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Konno M, Koseki J, Kawamoto K, Nishida N,
Matsui H, Dewi DL, Ozaki M, Noguchi Y, Mimori K, Gotoh N, et al:
Embryonic MicroRNA-369 controls metabolic splicing factors and
urges cellular reprograming. PLoS One. 10:e01327892015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li S, Swanson SK, Gogol M, Florens L,
Washburn MP, Workman JL and Suganuma T: Serine and SAM responsive
complex SESAME regulates histone modification crosstalk by sensing
cellular metabolism. Mol Cell. 60:408–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kottakis F, Nicolay BN, Roumane A, Karnik
R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al:
LKB1 loss links serine metabolism to DNA methylation and
tumorigenesis. Nature. 539:390–395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Shlomi T, Fan J, Tang B, Kruger WD and
Rabinowitz JD: Quantitation of cellular metabolic fluxes of
methionine. Anal Chem. 86:1583–1591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Mehrmohamadi M, Liu X, Shestov AA and
Locasale JW: Characterization of the usage of the serine metabolic
network in human cancer. Cell Rep. 9:1507–1519. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Nilsson R, Nicolaidou V and Koufaris C:
Mitochondrial MTHFD isozymes display distinct expression,
regulation, and association with cancer. Gene. 716:1440322019.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shin M, Momb J and Appling DR: Human
mitochondrial MTHFD2 is a dual redox cofactor-specific
methylenetetrahydro-folate dehydrogenase/methenyltetrahydrofolate
cyclohydrolase. Cancer Metab. 5:112017. View Article : Google Scholar
|
|
120
|
Goodman RP, Calvo SE and Mootha VK:
Spatiotemporal compartmentalization of hepatic NADH and NADPH
metabolism. J Biol Chem. 293:7508–7516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Röhrig F and Schulze A: The multifaceted
roles of fatty acid synthesis in cancer. Nat Rev Cancer.
16:732–749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ye J, Fan J, Venneti S, Wan YW, Pawel BR,
Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al: Serine
catabolism regulates mitochondrial redox control during hypoxia.
Cancer Discov. 4:1406–1417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ye C, Sutter BM, Wang Y, Kuang Z and Tu
BP: A Metabolic function for phospholipid and histone methylation.
Mol Cell. 66:180–193.e188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Rodriguez AE, Ducker GS, Billingham LK,
Martinez CA, Mainolfi N, Suri V, Friedman A, Manfredi MG, Weinberg
SE, Rabinowitz JD and Chandel NS: Serine metabolism supports
macrophage IL-1beta Production. Cell Metab. 29:1003–1011.e1004.
2019. View Article : Google Scholar
|
|
125
|
Ito Y, Makita S and Tobinai K: Development
of new agents for peripheral T-cell lymphoma. Expert Opin Biol
Ther. 19:197–209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Wei N, Zhang B, Wang Y, He XH, Xu LC, Li
GD, Wang YH, Wang GZ, Huang HZ and Li WT: Transarterial
chemoembolization with raltitrexed-based or floxuridine-based
chemotherapy for unresectable colorectal cancer liver metastasis.
Clin Transl Oncol. 21:443–450. 2019. View Article : Google Scholar
|
|
127
|
Goirand F, Lemaitre F, Launay M, Tron C,
Chatelut E, Boyer JC, Bardou M and Schmitt A: How can we best
monitor 5-FU administration to maximize benefit to risk ratio?
Expert Opin Drug Metab Toxicol. 14:1303–1313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Adamska A, Elaskalani O, Emmanouilidi A,
Kim M, Abdol Razak NB, Metharom P and Falasca M: Molecular and
cellular mechanisms of chemoresistance in pancreatic cancer. Adv
Biol Regul. 68:77–87. 2018. View Article : Google Scholar
|
|
129
|
Blair HA: Daunorubicin/cytarabine
liposome: A review in acute myeloid leukaemia. Drugs. 78:1903–1910.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Diesch J, Zwick A, Garz AK, Palau A,
Buschbeck M and Gotze KS: A clinical-molecular update on
azanucleoside-based therapy for the treatment of hematologic
cancers. Clin Epigenetics. 8:712016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Chabner BA and Roberts TG Jr: Timeline:
Chemotherapy and the war on cancer. Nat Rev Cancer. 5:65–72. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Luengo A, Gui DY and Vander Heiden MG:
Targeting metabolism for cancer therapy. Cell Chem Biol.
24:1161–1180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Casero RA Jr and Marton LJ: Targeting
polyamine metabolism and function in cancer and other
hyperproliferative diseases. Nat Rev Drug Discov. 6:373–390. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Rodriguez-Paredes M and Esteller M: Cancer
epigenetics reaches mainstream oncology. Nat Med. 17:330–339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zheng Y, Lin TY, Lee G, Paddock MN, Momb
J, Cheng Z, Li Q, Fei DL, Stein BD, Ramsamooj S, et al:
Mitochondrial One-carbon pathway supports cytosolic folate
integrity in cancer cells. Cell. 175:1546–1560.e1517. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kucharczyk T, Krawczyk P, Powrózek T,
Kowalski DM, Ramlau R, Kalinka-Warzocha E, Knetki-Wróblewska M,
Winiarczyk K, Krzakowski M and Milanowski J: The Effectiveness of
pemetrexed monotherapy depending on poly-morphisms in TS and MTHFR
genes as well as clinical factors in advanced NSCLC patients.
Pathol Oncol Res. 22:49–56. 2016. View Article : Google Scholar
|
|
137
|
Winter SS, Dunsmore KP, Devidas M, Wood
BL, Esiashvili N, Chen Z, Eisenberg N, Briegel N, Hayashi RJ,
Gastier-Foster JM, et al: Improved survival for children and young
adults with t-lineage acute lymphoblastic leukemia: Results from
the children's oncology group AALL0434 methotrexate randomization.
J Clin Oncol. 36:2926–2934. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Cui Y, Chen H, Chen J, Zeng F, Zu X and
Ding J: Gemcitabine/cisplatin versus
methotrexate/vinblastine/doxoru-bicin/cisplatin for muscle-invasive
bladder cancer: A systematic review and meta-analysis.
Meta-Analysis. 14:1260–1265. 2018.
|
|
139
|
Calise SJ, Purich DL, Nguyen T, Saleem DA,
Krueger C, Yin JD and Chan EK: 'Rod and ring' formation from IMP
dehydrogenase is regulated through the one-carbon metabolic
pathway. J Cell Sci. 129:3042–3052. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ross KC, Andrews AJ, Marion CD, Yen TJ and
Bhattacharjee V: Identification of the serine biosynthesis pathway
as a critical component of BRAF inhibitor resistance of melanoma,
pancreatic, and non-small cell lung cancer cells. Mol Cancer Ther.
16:1596–1609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Longley DB and Harkin DP and Harkin DP:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Kawahata T, Kawahara K, Shimokawa M,
Sakiyama A, Shiraishi T, Minami K, Yamamoto M, Shinsato Y, Arima K,
Hamada T and Furukawa T: Involvement of ribosomal protein L11
expression in sensitivity of gastric cancer against 5-FU. Oncol
Lett. 19:2258–2264. 2020.PubMed/NCBI
|
|
143
|
Reina-Campos M, Diaz-Meco MT and Moscat J:
The complexity of the serine glycine one-carbon pathway in cancer.
J Cell Biol. 219:e2019070222020. View Article : Google Scholar :
|
|
144
|
Avgustinova A and Benitah SA: The
epigenetics of tumour initiation: Cancer stem cells and their
chromatin. Curr Opin Genet Dev. 36:8–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kim M and Costello J: DNA methylation: An
epigenetic mark of cellular memory. Exp Mol Med. 49:e3222017.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Albrecht LV, Bui MH and De Robertis EM:
Canonical Wnt is inhibited by targeting one-carbon metabolism
through methotrexate or methionine deprivation. Proc Natl Acad Sci
USA. 116:2987–2995. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chen WL, Sun HP, Li DD, Wang ZH, You QD
and Guo XK: G9a-An appealing antineoplastic target. Curr Cancer
Drug Targets. 17:555–568. 2017. View Article : Google Scholar
|
|
148
|
Roesch A, Fukunaga-Kalabis M, Schmidt EC,
Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T
and Herlyn M: A temporarily distinct subpopulation of slow-cycling
melanoma cells is required for continuous tumor growth. Cell.
141:583–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Kano Y, Konno M, Ohta K, Haraguchi N,
Nishikawa S, Kagawa Y, Hamabe A, Hasegawa S, Ogawa H, Fukusumi T,
et al: Jumonji/Arid1b (Jarid1b) protein modulates human esophageal
cancer cell growth. Mol Clin Oncol. 1:753–757. 2013. View Article : Google Scholar
|
|
150
|
Konno M, Asai A, Kawamoto K, Nishida N,
Satoh T, Doki Y, Mori M and Ishii H: The one-carbon metabolism
pathway high-lights therapeutic targets for gastrointestinal cancer
(Review). Int J Oncol. 50:1057–1063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Kaniskan HU, Martini ML and Jin J:
Inhibitors of protein methyltransferases and demethylases. Chem
Rev. 118:989–1068. 2018. View Article : Google Scholar
|
|
152
|
Ma L, Tao Y, Duran A, Llado V, Galvez A,
Barger JF, Castilla EA, Chen J, Yajima T, Porollo A, et al: Control
of nutrient stress-induced metabolic reprogramming by PKCζ in
tumorigenesis. Cell. 152:599–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Mullarky E, Lucki NC, Beheshti Zavareh R,
Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H,
Singh PK, et al: Identification of a small molecule inhibitor of
3-phosphoglycerate dehydrogenase to target serine biosynthesis in
cancers. Proc Natl Acad Sci USA. 113:1778–1783. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Pacold ME, Brimacombe KR, Chan SH, Rohde
JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske
BP, et al: A PHGDH inhibitor reveals coordination of serine
synthesis and one-carbon unit fate. Nat Chem Biol. 12:452–458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhao X, Fu J, Du J and Xu W: The Role of
D-3-phosphoglycerate dehydrogenase in cancer. Int J Biol Sci.
16:1495–1506. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Ravez S, Spillier Q, Marteau R, Feron O
and Frederick R: Challenges and opportunities in the development of
serine synthetic pathway inhibitors for cancer therapy. J Med Chem.
60:1227–1237. 2017. View Article : Google Scholar
|
|
157
|
Gravel SP, Hulea L, Toban N, Birman E,
Blouin MJ, Zakikhani M, Zhao Y, Topisirovic I, St-Pierre J and
Pollak M: Serine deprivation enhances antineoplastic activity of
biguanides. Cancer Res. 74:7521–7533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Fedirko V, Lukanova A, Bamia C,
Trichopolou A, Trepo E, Nöthlings U, Schlesinger S, Aleksandrova K,
Boffetta P, Tjønneland A, et al: Glycemic index, glycemic load,
dietary carbohydrate, and dietary fiber intake and risk of liver
and biliary tract cancers in Western Europeans. Ann Oncol.
24:543–553. 2013. View Article : Google Scholar :
|
|
159
|
Fine EJ, Segal-Isaacson CJ, Feinman RD,
Herszkopf S, Romano MC, Tomuta N, Bontempo AF, Negassa A and
Sparano JA: Targeting insulin inhibition as a metabolic therapy in
advanced cancer: A pilot safety and feasibility dietary trial in 10
patients. Nutriti. 28:1028–1035. 2012.
|
|
160
|
Mallik R and Chowdhury TA: Metformin in
cancer. Diabetes Res Clin Pract. 143:409–419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Gustafsson R, Jemth AS, Gustafsson NM,
Färnegårdh K, Loseva O, Wiita E, Bonagas N, Dahllund L,
Llona-Minguez S, Häggblad M, et al: Crystal structure of the
emerging cancer target MTHFD2 in complex with a substrate-based
inhibitor. Cancer Res. 77:937–948. 2017. View Article : Google Scholar
|
|
162
|
Nishimura T, Nakata A, Chen X, Nishi K,
Meguro-Horike M, Sasaki S, Kita K, Horike SI, Saitoh K, Kato K, et
al: Cancer stem-like properties and gefitinib resistance are
dependent on purine synthetic metabolism mediated by the
mitochondrial enzyme MTHFD2. Oncogene. 38:2464–2481. 2019.
View Article : Google Scholar :
|
|
163
|
Bolusani S, Young BA, Cole NA, Tibbetts
AS, Momb J, Bryant JD, Solmonson A and Appling DR: Mammalian
MTHFD2L encodes a mitochondrial methylenetetrahydrofolate
dehydrogenase isozyme expressed in adult tissues. J Biol Chem.
286:5166–5174. 2011. View Article : Google Scholar :
|
|
164
|
Nilsson R, Jain M, Madhusudhan N, Sheppard
NG, Strittmatter L, Kampf C, Huang J, Asplund A and Mootha VK:
Metabolic enzyme expression highlights a key role for MTHFD2 and
the mitochondrial folate pathway in cancer. Nat Commun. 5:31282014.
View Article : Google Scholar : PubMed/NCBI
|