1. Introduction
As a type of malignant head and neck tumor,
nasopharyngeal carcinoma (NPC) is characterized by early extensive
local infiltration, high hematogenous dissemination, early
lymphatic spread and a high mortality rate (1). According to the International Agency
for Research on Cancer (IARC), there were an estimated 129,079 new
cases of NPC and 72,987 NPC-related deaths in 2018 worldwide
(2). While this is only a small
proportion of all cancers (the number of NPC cases in 2018
accounted for 0.7% of all confirmed cancers), in some high-risk
countries or regions, many patients present with NPC as well as
drug toxicity due to anti-NPC treatment. China has the highest
incidence rate of NPC worldwide. In 2018, there were 60,558 new NPC
cases (47.7% of total global cases) and 31,413 NPC-related deaths
in China (1,2). Within China, the incidence rate of
NPC is highest in the Guangdong Province; hence, NPC is also
commonly referred to as 'Cantonese cancer' (3). NPC has a high incidence rate in
other provinces of China, such as Guangxi, Hunan and Fujian, as
well as in other countries, such as Indonesia, Vietnam, India,
Morocco, Algeria, Tunisia and Ghana (4). Therefore, the prevention and
treatment of NPC is an important goal for medical researchers.
To date, the pathogeny behind NPC remains unclear.
Existing evidence shows that heredity, race, environment, diet,
habits and the Epstein-Barr virus (EBV) are closely related to the
pathogenesis behind NPC (1).
However, the study showed that NPC development involves a complex
interaction of multiple factors, as demonstrated by some gene
polymorphisms with different genetic characteristics that are
carried by ethnic groups in high-risk areas being associated with
different degrees of NPC risk as well as the processing and
presentation of EBV antigens. Extracts of Cantonese salted fish
from China, as well as herbal medicines used by Chinese and Naga
people, have been shown to promote the activation and proliferation
of EBV, leading to the occurrence of NPC (5). In addition, EBV inducers are
detected in soil extracts from NPC-endemic areas in southern China
as well as in some vegetables grown in these soils (6). These factors attribute for the
increased NPC incidence rate observed in specific regions or races.
However, how the genetic factors of different races affect tumor
development still requires more systematic and comprehensive
research.
Effective early diagnosis and treatment are key to
preventing the progression, recurrence and metastasis of NPC. Based
on the guidelines published by the World Health Organization (WHO),
pathological evidence found by biopsies is the gold standard for
NPC diagnosis (7-9). However, because of poor compliance
of patients with this invasive procedure, the fact that the
symptoms of NPC are not conclusive at early stages and that NPC is
not conducive to imaging screening, more than 70% of patients have
locoregionally advanced (LA)-NPC at the time of initial diagnosis
(10). Because the location of
the NPC is close to the base of the skull and its shape is
irregular, it is difficult to distinguish the tumors from
nasopharyngeal lymphoma in the early stage using MRI or computed
tomography scanning (11).
Furthermore, imaging-dependent tumor staging is initially
established based on patient survival statistics and is often
affected by the subjective judgement of the clinician; therefore, a
reliable laboratory method is needed to assist in accurate NPC
diagnosis (4,12).
The treatment decision for NPC at different stages
is made based on the tumor-node-metastasis (TNM) staging system
developed by the Union for International Cancer Control/American
Joint Committee on Cancer and the National Comprehensive Cancer
Network (NCCN). Radiotherapy (RT) alone is the standard treatment
for early NPC, while for patients with stages III-IVa, induction
chemotherapy (IC) followed by concurrent chemoradiotherapy (CCRT)
or CCRT followed by adjuvant chemotherapy (AC) are the preferred
category 2A treatment regimen (4,12).
A number of clinical trials have investigated the effectiveness of
these treatment methods (10,13-15). Although the 5-year overall
survival (OS), progression-free survival (PFS), distant
metastasis-free survival (DMFS) and locoregional relapse-free
survival (LRFS) are improved to various degrees by the application
of RT or the one of the various chemotherapy (CT) regimens, there
are still many controversies in the clinical application of these
treatment methods because of the lack of accurate efficacy
evaluation indicators. For example: i) Some patients still have
tumor recurrence or distant metastasis after receiving radical RT
or IC+CCRT/CCRT+AC, indicating that a more effective treatment
should be developed (16-19); ii) IC+CCRT improves the OS and
DMFS of patients with LA-NPC to a limited degree, but most patients
do not benefit from it, suggesting overtreatment (13-15); iii) patients have poor tolerance
to toxic side effects of the treatment, which could lead to delays
or interruptions in treatment, thus increasing the risk of tumor
progression or drug resistance (1,20-22); and iv) the clinical use of
Response Evaluation Criteria in Solid Tumors (RECIST) can only
provide a reference for the NPC treatment effect evaluation through
generalized remission or progression, and cannot provide
quantifiable prediction indexes for NPC recurrence and metastasis.
Moreover, the prognosis model based on the TNM staging system
cannot provide an objective assessment for the risk of disease
progression for patients with NPC (23,24). These problems indicate issues
regarding NPC diagnosis, treatment and prognosis, and suggest the
need for individualized treatment. Therefore, researchers are
focusing on the identification of molecular biomarkers to establish
new non-invasive early diagnostic methods, ideal treatment
regimens, efficacy evaluation standards and prognostic indicators
for NPC (Fig. 1).
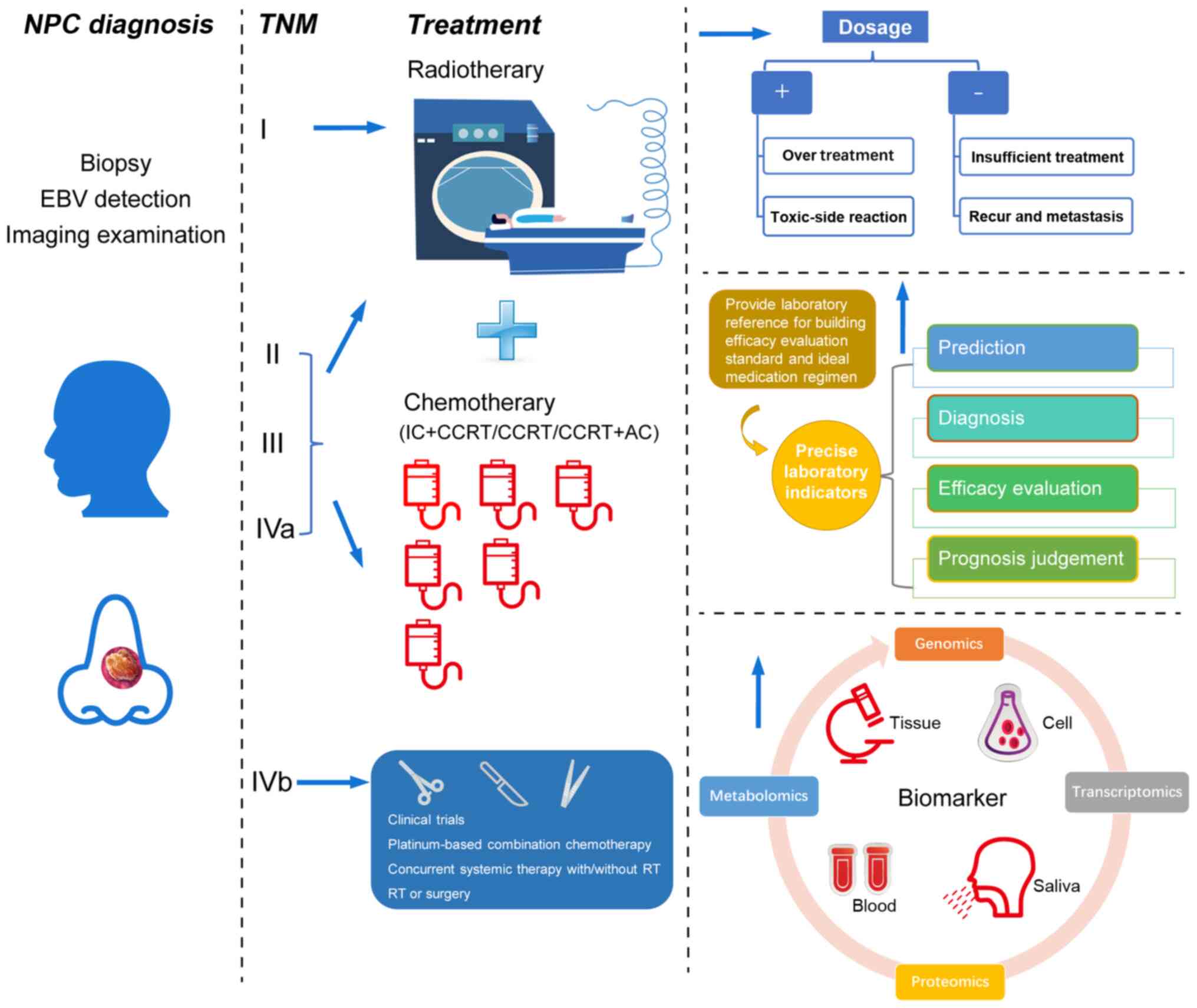 | Figure 1Role of biomarkers in NPC diagnosis,
treatment, and prognosis. Biomarkers can effectively predict the
risk of NPC incidence and reduce the number of patients with a
locoregionally advanced stage through early diagnosis. For patients
at different stages, biomarkers can effectively assist physicians
to carry out clinical individualized treatment. This includes
creating ideal regimens to improve the treatment effect and
providing targets for the development of novel medication which
avoid the toxic reactions caused by treatment. Effective biomarkers
may also be used to accurately evaluate the prognosis of patients,
as well as reduce the recurrence and distant metastasis of tumors.
AC, adjuvant chemotherapy; CCRT, concurrent chemoradiotherapy; EBV,
Epstein-Barr virus; IC, induction chemotherapy; NPC, nasopharyngeal
carcinoma; TNM, tumor-node-metastasis. |
Biomarkers can be divided into two categories
according to their functions: Those used to discover the molecular
mechanisms of action of the disease and drug targets, and those
used for the prediction, early diagnosis, efficacy evaluation and
prognosis. Several review articles have summarized both types of
NPC biomarkers (7-9,25).
With the development of high-throughput omics technologies in
recent years, many novel biomarkers have been discovered. In the
present review, studies on bodily fluid samples, such as serum,
plasma and saliva of patients with NPC in the past 5 years were
summarized, and the research status of biomarkers for the early
diagnosis, treatment (such as RT and CT) and prognosis (such as
metastasis and recurrence) of NPC were reviewed at the aspects of
genomics, transcriptomics, proteomics and metabolomics. The
intersection points (such as TNM staging, therapeutic method,
sample type and high-throughput technology) in the study design of
the existing research and the development trends of NPC biomarkers
in the future were also discussed.
2. NPC biomarkers discovered through
genomics
Genomics aims to collectively characterize and
quantify all genes in an organism to study the influence of their
interactions (26). It uses
high-throughput DNA sequencing, bioinformatics, genetic analysis
and functional identification to analyze the structure and function
of the whole genome, which forms the omics framework of systems
biology with transcriptomics, proteomics and metabolomics (27). In the study of biomarkers, gene
mutations are often difficult to match with corresponding clinical
phenotypes of the disease. Therefore, researchers must carry out
repeated validation studies in large cohorts or conduct
supplementary studies with other omics technologies (28).
Based on the fact that genetics, environment, diet
and EBV are generally considered as risk factors of cancer,
exploring new pathogenic genes and mechanisms to assess the risk of
NPC in high-risk population has become the goal of genomics
research (29,30). With the wide application of
high-throughput technologies, such as whole-exome sequencing,
whole-genome sequencing (WGS) and genome-wide association studies
(GWAS), many single nucleotide polymorphisms (SNPs) associated with
NPC have been identified and comprehensively described (1,7,30-32). However, although the interactions
between multiple genes make the complex mechanism of action behind
NPC more difficult to understand, simple research design and mature
detection technology still push the research forward (33). In recent years, researchers have
identified the association between NPC susceptibility and the SNPs
of genes encoding for major histocompatibility complex II (34), monocyte chemoattractant protein-1
promoter (35), xeroderma
pigmentosum group D, human 8-oxoguanine DNA glycosylase 1
(hOGG1), integrin (ITG)α2 (36) and cyclophilin A26 (37) in patient cohorts from various
regions, and confirmed the effect of loss-of-function mutations
such as negative regulator NF-κB inhibitor α and cylindromatosis on
NF-κB activity and NPC cell growth, respectively (38,39). To date, no susceptibility genes
have been approved for the early screening of NPC. The
heterogeneity in the sample cohorts of different regions, races and
pathological backgrounds is often highlighted as the main reason
for the variation between the research results (7). Nevertheless, to the best of our
knowledge, most studies did not verify the potential pathogenic
genes. Some scholars think that because SNPs identified by GWAS are
mostly minor alleles; therefore the main difficulty in further
research, the large samples and failure to verify the downstream
function of non-coding regions are the main reasons for the
difficulty further research (33). In addition, the integration of
multi-disciplinary research may provide a new method to interpret
this complex problem, such as the joint effect of some specific
regional environmental factors and traditional unhealthy diets on
the incidence of NPC (40-42).
However, finding the interdisciplinary intersection point seems to
be beyond the research objective itself.
In a whole exon sequencing study of 251 individuals
from 97 polygenic families in Taiwan, Yu et al (43) found that 12 gene variations
related to magnesium transport (Nuclear-interacting partner of
anaplastic lymphoma kinase-like domain containing 1), EBV cell
entry (ITG-β6), modulation of EBV infection (bcl-2-like protein 12;
and neural precursor cell expressed, developmentally down-regulated
4-like), telomere biology (cleft lip and palate transmembrane 1
like; bromodomain containing 2; and heterogeneous nuclear
ribonucleoprotein U), modulation of cAMP signaling (rap guanine
nucleotide exchange factor 3), DNA repair (protein kinase,
DNA-activated, catalytic subunit; and mutL homolog 1) and the Notch
signaling pathway (notch receptor 1; and δ-like canonical notch
ligand 3), play important roles in activating T cells to respond to
EBV infection. Liu et al (44) verified these mutations in a later
study and emphasized the important role of telomere length
maintenance in NPC etiology. Two other studies from the USA and
Tunisia showed the potential of mixed-lineage leukemia 3; major
histocompatibility complex, class I, A*26; major histocompatibility
complex, class I, A*30; and major histocompatibility complex, class
II, DR β1*10 in screening high-risk NPC family members,
respectively (45,46). The study pointed out that the
relative potential risk of NPC among family members who have
first-degree relatives diagnosed with NPC is increased eight-fold
(47). For some high-risk
countries or regions, it is still necessary to strengthen the
mining of familial NPC susceptibility genes for the development of
related gene detection and early diagnosis.
Patients with NPC can develop various degrees of
oral mucositis, xerostomia, myelosuppression and other toxic
reactions while receiving intensity-modulated RT (IMRT) (48). Furthermore, cancer recurrence and
distant metastasis can occur in 30-40% of patients with NPC after
treatment, indicating that the response of NPC to RT is subject to
individual variation (49).
Because tumor cells can repair radiation-induced damage in various
ways, some genes related to radiosensitivity are often used to
predict the therapeutic effect and prognosis of NPC (16,48-50). Ma et al (51) analyzed the genotypes of
angiogenesis-related genes in 180 patients with NPC using Sequenom
MassARRAY and found that EDN1-rs1800541, rs2071942 and rs5370 can
be used as risk predictors of radiation-induced oral mucositis,
xerostomia and myelosuppression, respectively. Le et al
(52) screened SNPs of 24
patients with NPC by WGS and found that rs11081899-A, located in
the 5′-untranslated region of the zinc finger protein 24 gene, is
the genetic predisposing factor of radiation-induced oral
mucositis. Yu et al (53)
analyzed 9 potential functional SNPs in four genes in the
Wnt/β-catenin pathway and found that patients carrying catenin β-1
rs1880481, rs3864004; glycogen synthase kinase-3β rs3755557; or
adenomatous polyposis coli rs454886, may have a poor prognosis and
can develop radiation-induced dermatitis and oral mucositis.
Furthermore, several studies have discussed the potential of SNPs
of different genes, such as X-ray repair cross-complementing 1
(XRCC1) rs25489, XRCC1 Codon399, valosin-containing
protein rs2074549 and rcalcitonin receptor rs2528521 in the base
excision repair pathway and the endoplasmic reticulum stress
pathway to predict the therapeutic effect and toxic reactions in
NPC (54-57). However, it is worth noting that
the cases involved in these studies included a number of patients
with stages III-IVa NPC. Although these patients also needed
standard RT, the additional effect of chemotherapeutic drugs in
CCRT on blood composition is an important factor that cannot be
ignored. Tan et al (58)
found that the vascular endothelial growth factor-460C allele had a
significant association with NPC invasiveness of grades 2-3
cervical lymph node metastasis and compared with CCRT, in NPC
with-460 T/C polymorphisms, patients may not benefit from IC+CCRT
in terms of the OS, LRFS, DMFS and PFS, which again emphasizes the
contradicted value of using IC for clinical application.
In addition to the inherent susceptibility genes of
humans, EBV, a category 1 carcinogenic virus identified by IARC, is
detectable in 100% of patients with undifferentiated
non-keratinizing NPC in endemic areas (especially in East and
Southeast Asian countries, such as China). The mechanisms of EBV
infection, carcinogenesis, modification of epigenetic profiles,
immune escape and maintenance of tumor cell survival are deeply
understood and have been previously described (59). Therefore, based on this close
relationship, EBV DNA or antibodies can be used as an early
diagnostic tool to screen for patients with NPC. In recent years,
the A157154C polymorphism of the A73 gene and the RPMS1 genotype of
EBV have been identified for their susceptibility to NPC (60,61). In particular, the combination of
EBV-viral capsid antigen (VCA) IgA and EBV-early antigen IgA showed
better sensitivity and specificity than EBV-DNA in screening NPC
high-risk family individuals (62). However, a large cohort study
showed that the sensitivity and specificity of plasma EBV-DNA for
the diagnosis of early asymptomatic NPC were 97.1 and 98.6%,
respectively (63). The authors
speculated that the difference in the results may be owing to
variations of EBV DNA content in the serum or plasma, clinical
stage and pathological classification of the patients (64). Furthermore, previous studies have
found no significant difference in the exact load of viral nucleic
acid between patients with NPC and non-cancerous controls, and an
increase in EBV levels was noted in asymptomatic carriers and
patients with mononucleosis and pharyngitis (65,66). Banko et al (67) found that P-Thr-sv-5 showed
carcinoma-specific Epstein-Barr nuclear antigen 1 (EBNA1)
variability in tissues and plasma of patients with undifferentiated
NPC, monocytosis syndrome and renal transplantation by using nested
PCR and considered that the identification of this subvariant
should be used as a viral screening marker for the identification
of NPC. However, further in vitro experiments are needed to
confirm whether the additional amino acid substitution of this
subvariant will affect the function of EBNA1 in regulating the
replication and transcription of EBV genes and/or change the effect
of EBNA1 on the transcription of other genes in host cells
(68). In terms of efficacy
evaluation and prognosis, Hui et al (69) detected SNPs of the excision repair
1 endonuclease non-catalytic subunit (ERCC1) gene in the
plasma of patients with EBV-positive and EBV-negative NPC after RT
in a clinical trial. The results showed that the ERCC1 C118T
genotype was a powerful predictor for a good prognosis of patients
with EBV-negative NPC, which could confirm the effect of RT and
avoid serious toxic reactions caused by the additional adjuvant CT.
Nevertheless, in view of some potential influencing factors
mentioned in the aforementioned study, whether the objectivity of
the results can be affected by different clinical stages, treatment
methods and other confounding factors remains to be further
explored.
3. NPC biomarkers discovered through
transcriptomics
Transcriptomics studies gene transcription and its
regulation at the RNA level (70). It explores variations in gene
expression using sequencing technology and reveals the mechanism of
specific regulatory genes in a pathological condition through cell
phenotypes and functional research (71). In contrast to genomics,
transcriptomics emphasizes the concept of time and space, which
enables it not only to identify the phenotypic attribution of
cells, but also to distinguish subtypes of diseases and to
discriminate the various reactions caused by medications, as well
as describe the survival rates of patients (72).
Previous studies have shown that micro RNAs
(miRNAs/miRs) can play an important role in the occurrence,
invasion, metastasis and immune escape of NPC, and their stable
expression in the peripheral circulation can not only be used as
reliable markers for the early diagnosis of NPC but may also help
in the effective prediction of the therapeutic effect and prognosis
(73-75). Therefore, studies into miRNAs have
gained a leading position in the field of transcriptomics in recent
years (73). Recently, several
miRNAs with good diagnostic ability have been found using PCR or
microarray technology (76-83). Two types of EBV-encoded miRNAs,
miR-BamHI A rightward transcripts (BART)7-3p and miR-BART13-3p,
showed a remarkable potential for the early diagnosis of NPC
(76). It is worth noting that as
the first virus-encoding miRNAs, 44 mature miRNAs encoded by EBV
have been confirmed to play an important regulatory role in the
carcinogenesis and progression of NPC. Although the function and
clinical value of these miRNAs has been explored comprehensively,
some of the identified biomarkers still lack functional
verification (84,85). In addition to the aforementioned
problems, most of these studies lack functional analysis of the
identified potential biomarkers. Because of the small molecular
weight of miRNAs and their low content in the serum or plasma, it
is difficult to fully reflect the expression abundance of these
biomarkers. Therefore, the sensitivity and specificity of miRNAs,
especially EBV miRNAs, were not satisfactory (78,80). Furthermore, although a previous
study confirmed the mechanism of action of miRNAs in various stages
of NPC at the molecular level, more clinical verification is needed
to assess the application of these potential biomarkers (73). Tumor-educated platelets, which are
believed to be able to accurately diagnose for various types of
cancer in liquid biopsies, have recently been used as early
diagnostic biomarkers for NPC. Wang et al (81) detected the expression of
miR-34c-3p and miR-18a-5p in the platelets of patients with NPC and
healthy controls, and found a reasonable diagnostic potential of
the two miRNAs with a sensitivity of 92.59%, specificity of 86.11%
and an area under the curve (AUC) value of 0.954. Although
additional validation and functional analysis were not conducted,
Best et al (86) pointed
out that platelets, as anucleated cell fragments in the blood
circulation, can modulate the splicing of their pre-mRNAs in
response to signals from cancer cells and then change their
transcriptome and molecular signals. Compared with other samples,
platelets do not contain nuclei and are less interfered with by
genomic DNA; therefore, their RNA expression truly reflects the
pathological progression of tumors, providing a more valuable new
platform for biomarker research (81).
Compared to PCR, microarray-based high-throughput
technology has many advantages. However, it is often not sufficient
to use only common features, such as gene names, for comparisons
across different microarray platforms (7,8).
The main reason for this is that different probes used in different
studies may not have the tagging gene name or different probes of
the same gene may not give similar signals (73). Therefore, it has been suggested
that additional verification by PCR is necessary for the final
identification of biomarkers in an independent verification cohort,
as it allows the evaluation of gene expression more reliably
(7). Wu et al (83) detected the miRNA expression
profile in NPC saliva for the first time. They applied the
stacking-hybridized universal tag (SHUT) miRNA array and
quantitative PCR (qPCR) technology to compare the miRNA expression
levels in patients with NPC and healthy controls, and determined
that the sensitivity and specificity of 12 differential miRNAs used
to distinguish between patients with NPC and a healthy population
were 100 and 96%, respectively. Bioinformatics analysis showed that
these differentially expressed miRNAs play an important role in the
development of NPC by regulating their target genes, such as
platelet-derived growth factor receptor α; RAC1; inhibitor
of NF-κB kinase regulatory subunit γ; X-linked inhibitor of
apoptosis; and protein phosphatase, Mg2+/Mn2+
dependent 1D. Furthermore, Li et al (87) reported the ability of hsa-mir-1281
and hsa-mir-6732-3p to evaluate the efficacy of RT for NPC by
assessing the serum of patients with different RT sensitivities.
Unsatisfactory AUC values of both miRNAs were found (0.750 for
hsa-mir-1281 and 0.696 for hsa-mir-6732-3p); however the small
sample size included in the study, as well as the fact that >50%
of patients had stages III-IVa NPC and did not receive RT alone,
may have affected the results.
Long non-coding (lnc)RNAs also play important roles
at the transcriptomic level, performing a regulatory role in NPC
epigenetics (88). Previous
studies have confirmed that lncRNAs, miRNAs and EBV products can
target each other and share common signaling pathways to form a
complex molecular regulatory network (89). In recent years, novel lncRNAs have
been identified through high-throughput sequencing to impact on the
occurrence, progression, recurrence, metastasis and prognosis of
NPC. However, the purpose of these studies was mainly to determine
the mechanisms of action and treatment targets of NPC in different
stages, rather than to identify biomarkers for NPC (88). He et al (90) used reverse transcription (RT)-qPCR
to detect NPC-related lncRNAs in four different NPC cell lines and
normal nasopharyngeal epithelial cells, as well as in the serum of
patients with NPC, patients with chronic nasopharyngitis, EBV
carriers and healthy controls. The AUC value of the three combined
lncRNAs, including metastasis associated with lung adenocarcinoma
transcript, actin filament-associated protein 1-antisense RNA1 and
AL359062, to discriminate between patients with NPC and the healthy
population, was 0.918. Furthermore, they found that the expression
levels of these three lncRNAs significantly decreased in serum
after treatment, thus confirming their potential as early
diagnostic and efficacy evaluation biomarkers for NPC. Based on the
current research progress, there is still a huge space for
researchers to explore the value of lncRNAs as NPC biomarkers
through liquid biopsy. High-throughput based second-generation
sequencing technology does not rely on the prior knowledge of
genomic information, which helps to identify new splicing points
and mutations. Similar to miRNAs, an increasing number of lncRNAs
have been found to be significantly related to the mechanism of
action behind the radiosensitivity of NPC, but there are few
clinical studies involving noninvasive detection (91).
Recently, RNA-sequencing has been used to detect the
transcriptional profile of peripheral blood mononuclear cells in
patients with NPC before and after RT. Although a study has
reported that the 11 genes that have been found can be used as
biomarkers to evaluate the prognosis of NPC after RT, a very small
sample size was used and they did not establish a model for
verification (92). In another
study, Shuai et al (93)
detected increased expression levels of circular RNA_0000285 at the
homeodomain interacting protein kinase 3 locus in NPC cells,
tissues and serum samples, which could be used as a biomarker to
predict the radiosensitivity of NPC. However, their study also
lacked a diagnostic analysis of the sensitivity and specificity of
biomarkers. It should be noted that the interaction test is a
common statistical method to confirm the identified differential
genes as potential biomarkers and this method is usually based on
statistical models (94).
Therefore, although several potential NPC biomarkers have been
found in various studies, owing to the interference of multiple
factors, there are still no widely verified and accepted biomarkers
in the field of transcriptomics.
4. NPC biomarkers discovered through
proteomics
Proteins are involved in many important
physiological functions, such as immunity, coagulation, substance
exchange, transportation, metabolism and signaling pathway
regulation (95). Their
composition and expression levels are often affected by the
pathological state of the body (96). Proteomics is a research field that
is based on the protein expression profile of the normal human
body. Proteins with significant differences in expression levels
can be identified by screening and comparing all of the proteins
expressed in the cells, tissues or blood of individuals (97). With the assistance of continuous
innovation of high-throughput technology, an increasing number of
proteins related to the pathogenesis of diseases have been found,
allowing for further elucidation of the pathogenesis behind various
diseases and the identification of molecular biomarkers and novel
drug targets (98).
Several studies have shown that gene mutations in
cancer often cause abnormal expression levels of corresponding
proteins and such expression can be dynamically changed following
damage to the DNA of cancer cells following therapies (7,9).
Furthermore, tumor cells can capture many protein decomposition
products from normal tissues to synthesize the proteins they need,
resulting in significant changes in protein expression levels due
to increased anabolism and catabolism (99). Therefore, the proteins involved in
carcinogenesis are considered functional molecules that can reflect
the real-time state of disease progression and are used in the
study of NPC biomarkers (7).
Previous studies have identified various protein markers in
different NPC cell lines, tissues and bodily fluid samples,
including EBV-encoded proteins such as latent membrane protein
(LMP)1, LMP2A and EBV nuclear antigens (9,25,99). Owing to the accumulated knowledge
from these studies, the mechanism of protein action in each stage
of NPC has been further clarified and multiple protein-protein
interaction networks have been established based on various
signaling pathways (100). This
has laid a foundation for future research, based on the
phosphorylation, ubiquitination, sulfation, methylation and
sumoylation of NPC protein markers (101).
Compared with 2D fluorescence difference gel
electrophoresis, isobaric tags for relative and absolute
quantification; matrix-assisted laser desorption/ionization
time-of-flight mass spectrometry; surface-enhanced laser
desorption/ionization time of flight mass spectrometry; and other
technologies used in earlier studies, proteomics has made
significant progress in the high-throughput technology of relative
and absolute quantitation in recent years. For example,
data-independent acquisition (used for comparative proteomics
studies), parallel reaction monitoring (PRM) and phosphorylated PRM
(used to study targeted proteomics), have improved significantly in
the integrity of scanning, reproducibility, stability of results
and the quantitative ability, and have been applied to study tumor
biomarkers (7,9,25,102-105). However, studies on NPC
proteomics based on bodily fluid samples have not employed these
new technologies and have shown a decreasing trend in the study
number compared with earlier studies (106-111). The reasons for this are not
clear, but it may be related to the difficulty in obtaining
samples, the equipment conditions of the laboratory and the cost of
testing.
Recently, Sun et al (106) used RT-qPCR to detect the mRNA
expression levels of insulin receptor substrate 1 (IRS-1) in 133
patients with NPC and 104 healthy controls. The sensitivity and
specificity of IRS-1 in the early diagnosis of NPC were 88.0 and
77.9%, respectively. Coghill et al (107) applied a protein microarray to
detect the anti-EBV-IgG and IgA antibody responses in 607 residents
of Taiwan and found significant differences in the anti-EBV
antibody levels of 60-IgA and 73-IgG between patients with NPC and
healthy controls. The sensitivity and specificity of EBV VCA-IgA to
distinguish patients with NPC were 66.7 and 95.0%, respectively,
with an AUC value of 0.811. In another study, 2D gel
electrophoresis, ultra-performance liquid chromatography-tandem
mass spectrometry (UPLC-MS/MS) and ELISAs were used to detect the
serum auto-antibody levels of peroxiredoxin (PRDX)2 and PRDX3 in
patients with NPC and CNE2 cells. Levels of PRDX2 and PRDX3 were
significantly higher in patients with NPC and CNE2 cells than in
the normal controls (108). In
addition, Gong et al (109) detected cytokines in the serum of
various cancer patients using antibody array technology. ELISA
validation results showed that nine cytokines could be used as
potential biomarkers for differential diagnosis and prognosis of
NPC.
In terms of efficacy evaluation and prognosis, like
genomics and transcriptomics, the evaluation of NPC
radiosensitivity is still the main research focus of proteomics
(17,110). NPC cells develop resistance to
RT, which involves cell cycle regulation, apoptosis and
anti-apoptosis, as well as DNA damage and repair, which may lead to
the abnormal protein synthesis of NPC cells and affect the protein
expression level in blood. This difference can be found by
assessing the serum protein expression levels of RT-sensitive
patients (7,9). Based on this hypothesis, Zhang et
al (17) used the tandem mass
tag method coupled with UPLC-MS/MS to detect the serum protein
profile in patients with NPC with different RT effects. They found
that the sensitivity and specificity of the combined five
identified differential proteins, such as secreted protein acidic
and rich in cysteine, serpin family D member 1S, complement C4B,
peptidylprolyl lsomerase B and family with sequence similarity 173
member A to distinguish patients with RT resistance, were 94.1 and
92.6%, respectively, and the AUC value was 0.968. In addition,
since there is currently no effective predictor for NPC recurrence,
Meng et al (110) used
the same technique to compare the serum protein expression levels
in patients with NPC who developed recurrence. The results of ELISA
validation revealed that differentially expressed calmodulin can be
used as a potential biomarker for the diagnosis of NPC
recurrence.
Although some researchers are still using bodily
fluid samples to promote the research of NPC proteomics, the
following obstacles need to be overcome to transform proteomics
biomarkers into clinical applications. Initially, as mentioned
above, the high-throughput technologies used in the existing
studies are still relatively old. The limitations of these
technologies in protein quantification, data collection, low
abundance protein detection and repeatability restrict the
discovery of some novel biomarkers with clinical significance
(98,111). Moreover, most studies are not
adequate with respect to the study design, which could affect the
results by a variety of confounding factors, thus interfering with
the sensitivity and specificity of the markers (94). For example, in the study of
efficacy evaluation and prognostic biomarkers, researchers should
focus on the dual effects of RT and CT on blood components, as well
as the use of recognized and unified clinical efficacy evaluation
standards and external data sets to fully evaluate and verify the
results (112). Eventually,
there is a significant difference in the application of the
detection technology, pathological background of samples and
statistical analysis tools, and the functional research often
cannot be combined with clinical studies (113,114). These factors are bound to affect
the repeated validation of identified biomarkers in different
laboratories. In 2016, the US cancer moonshot 2020 officially
launched a plan for precision medicine (115). Through the establishment of a
data system for characterization of gene information and protein
information, using genomics and proteomics as routine detection
methods may provide more accurate guidance for individualized
cancer treatment in the future. Therefore, it is believed that in
the coming years, there will be an upsurge in NPC proteomics
research based on high-throughput technology (116).
5. NPC biomarkers discovered through
metabolomics
Briefly, metabolomics is a subject that conducts
both qualitative and quantitative analysis of all low molecular
weight metabolites (<1,000) of a certain organism or cell at the
same time in a specific physiological period (117). It is based on high-throughput
mass spectrometry technology, cluster index analysis and data
processing, combined with information modeling and system
integration, to screen and identify the differential metabolites
correlated to the disease phenotype (118). Compared with other omics fields,
metabolomics is rather new and has currently attracted substantial
interest in the field of tumor biomarkers (119).
As downstream products of genes and proteins,
bioactive metabolites are an important component of systems
biology, because they can directly regulate biological processes
and phenotypes by regulating the main mechanisms of action behind
the functions of DNA, RNA and protein (117). Metabolic change is an important
feature of cancer. To maintain continuous proliferation and
survival after treatment, cancer cells must adjust their metabolism
and nutritional needs (120).
Metabolic disorders in cancer cells can further affect the
expression of cell surface markers through a variety of functional
signaling pathways (121). The
study of metabolic phenotypes can help to find more reliable
evidence for the regulatory mechanism of the system that occurs or
has occurred, rather than predicting the possible or upcoming
changes. Therefore, metabolomics studies are not limited to the
discovery of specific biomarkers. The exploration of characteristic
pathological pathways and therapeutic targets can provide a more
accurate direction for the development of novel medications
(122,123).
Previously, a gas chromatography-mass spectrometry
(GC-MS)-based metabolomics platform was the main technology
employed to study bodily fluid samples. Tang et al (124) used this technology to detect 51
serum metabolites in 49 patients with NPC, 37 with laryngeal cancer
and 40 healthy controls. Through the validation of differential
metabolites in tissues and in the serum of patients with NPC, they
found that kynurenine, N-acetylglucosaminylamine,
N-acetylglucosamine and hydroxyphenylpyruvate can be used as
potential biomarkers for the early diagnosis of NPC. Furthermore,
they observed changes in these four metabolites at three time
periods after RT and found that the high expression was closely
related to cancer recurrence and distant metastasis, thereby
confirming the efficacy and prognosis of these potential diagnostic
markers. In another study, Yi et al (125) compared the serum metabolic
profile in 100 patients with NPC to that in healthy controls and
found that the sensitivity and specificity of seven metabolites
(glucose, linoleic acid, stearic acid, arachidonic acid, proline,
β-hydroxy butyrate and glycerol 1-hexadecanoate) for NPC diagnosis
were 88.0 and 92.0%, respectively. Furthermore, they found that
β-hydroxy butyrate and arachidonic acid can be used as evaluation
indexes for the favorable prognosis of RT.
Along with the deepening understanding of NPC, the
TNM staging system, RECIST and NCCN guidelines have changed
significantly (12,23). Compared with early two-dimensional
RT, the application of IMRT technology has significantly improved
the treatment of NPC (126).
Therefore, there is no need to evaluate the aforementioned studies
in these aspects. However, it is important to ensure early studies
have carried out long-term follow-up of patients, which may help to
further verify the dynamic performance of the biomarkers. Compared
with GC-MS, high-throughput, widely targeted metabolomics
technology can solve the problem that non-targeted metabolomics
methods cannot detect metabolites in batches, and may show higher
sensitivity and high throughput performance in the known
qualitative and quantitative analysis of metabolites, as well as in
the detection of low-abundance metabolites. Therefore, it has been
successfully applied to screen tumor biomarkers with the advantages
of a self-built database to identify new metabolites (127,128). However, in recent years, to the
best of our knowledge, there have not been any reports on the
identification of NPC biomarkers in this field. This may also
indicate the research potential and value of this field.
6. Key points for study design
Obtaining ideal biomarkers requires rigorous
scientific study design and any design flaw will directly affect
the experimental results and its clinical translational potential
(94). Based on the different
study purposes, the selection of case-cohort, bodily fluid samples
and detection technology should be different, and the influence of
tumor stages, treatment methods and experimental conditions should
also be considered (129).
In the selection of case-cohort, the study of early
diagnostic markers should try to avoid the inclusion of patients
with locally advanced or distant metastasis, because the secretion
of metastatic tumors in cervical lymph nodes or distant organs
cannot be ignored in biomarker screening (62,67,76,79-83, 106,107,109). Although the smaller number of
patients with early-stage disease is the biggest obstacle to
conducting such studies, their blood composition may provide
feedback on the simplest specific marker information (5). Similarly, studies on biomarkers for
NPC radiosensitivity should also focus on early-stage patients as
most patients in stages III-IVa need additional CT (17,51,87). A previous study showed that the
addition of cisplatin and paclitaxel can affect the expression
levels of malondialdehyde, superoxide dismutase, catenin,
glutathione, r-glutamyl cysteingyl and glycine in the blood of
patients with NPC (130).
Another study also confirmed that CCRT, RT or IC treatment may lead
to varied changes in the distribution of metabolites in the serum
of patients with head and neck cancer (131). Conversely, in a phase III
clinical trial, Sun et al (112) found that the total IC efficacy
of the combination of docetaxel, cisplatin and fluorouracil in the
treatment of stages III and IV head and neck squamous cell
carcinoma was significantly higher than that of the combination of
cisplatin and fluorouracil (76.3 vs. 52.9%). However, this
difference disappeared after CCRT (75 vs. 73.9%), indicating that
RT may also have a significant effect on the efficacy of IC.
Serum and plasma are the most commonly studied
bodily fluids (132). Several
studies have found that compared with the coagulation of serum
during collection, the miRNA-aligned reads produced in rat plasma
during detection were twice as high as those produced by serum, but
this difference was not found in human blood (133). However, among the 64 blood test
indexes with statistical differences introduced by the WHO, 56
showed that the detection stability of plasma was significantly
better than that of serum (134). Although this observation has
been put into practice in several studies on the relationship
between NPC and EBV, further verification and discussions are
required to obtain general recognition (77,135). In addition, the collection,
processing and storage of samples will have a significant effect on
the experimental results (93).
Taking the use of anticoagulants as an example, the protocol of
sample handling and storage in the British biobank has clearly
defined the scope of application of anticoagulants such as citrate,
EDTA and heparin (136). Several
other reports have also discussed the effects of various storage
and processing methods on the composition of the blood (137,138). Thus, it can be seen that the
normalization of procedures will help standardize the handling of
samples in future studies and improve the quality of molecular
detection.
For the detection technology, there is no doubt that
new technology can often make up for the shortcomings of previous
ones and that they are conducive to obtaining more specific
biomarkers with respect to screening range and convenience of data
analysis (139). Through a
review of biomarker studies on the bodily fluid samples of patients
with NPC in recent years, it is clear that there is a lack of
application of new technologies in current studies, especially in
the field of proteomics and metabolomics. Because of funding
constraints, researchers must often make a difficult choice between
technology and sample size, which is the main reason most studies
have difficulty in achieving authoritative recognition (140). In addition, although the
screening and identification of biomarkers using bodily fluid
samples have represented a complete story, it is still necessary to
conduct cellular-based and molecular biology techniques to confirm
the functions of these potential biomarkers.
7. Conclusion and future perspectives
Compared to the study of other common types of
cancer, that of NPC is concentrated in Asian countries,
specifically China, owing to the geographical distribution of its
incidence (1,3,10).
Great progress has been made in clinical trials of this disease and
the NCCN guidelines have been rewritten (12). Conversely, biomarker research
based on liquid biopsies is lagging, especially in the fields of
proteomics and metabolomics, which limits the advancement of
relevant research to a certain extent (7-9,13-15,17,100,124,125). For example, IC, as a standard
initial method for systemic treatment of LA-NPC, the results of
clinical trials show that not all patients can benefit from its use
(15). Compared with CCRT alone,
the prognosis of a considerable number of LA-NPC patients has not
been significantly improved due to the addition of IC, indicating
that they have suffered unnecessary over treatment (22). Conversely, in those who have
extended the survival by IC+CCRT, 20-30% patients with LA-NPC still
present tumor relapse and metastasis, suggesting that the current
clinical application of IC may also provide insufficient treatment
(13,14). Therefore, it is urgent to identify
molecular biomarkers to predict the short-term efficacy of IC. In
addition, because of serious toxic reactions in the blood,
digestive tract, skin, nervous system, liver and kidney caused by
RT and CT, new therapeutic targets are necessary for the
development of new, safer medications (10,20,21). Therefore, future research on NPC
biomarkers should gradually extend to the evaluation of efficacy
and prognosis based on early diagnosis to provide sufficient
laboratory data for the clinical research of NPC.
With the development of tumor biomarkers, it is
impossible to fully explain the complex biological processes and
network regulation behind the carcinogenesis and progression of
tumors from a systematic perspective only by studying a single
omics (141). Therefore,
multi-omics integration has become a new trend to promote the
research and development of tumor biomarkers (142). The pathogenesis behind the
changes required for cancer, gene mutation, transcriptional
regulation, protein synthesis and metabolic changes constitute a
systematic mechanism (143).
Based on these relationships, the integration of two or more kinds
of omics research and the use of machine learning methods to carry
out association analysis on molecules at multiple levels, could
make up for the lack of data caused by single omics analysis and
reduce the probability of false positive results. This would allow
researchers to study the phenotype and regulatory mechanisms of
action in biological models more effectively and investigate
complex scientific problems more comprehensively. By using this
strategy, NPC biomarkers may effectively compensate for the
deficiencies in early omics research, promoting the overall
development of NPC clinical research.
Collectively, novel clinical issues, scientific
study design, cutting-edge high-throughput technology, integrated
multi-omics platforms, extensive screening of large-scale cohort,
systematic functional analysis, ensemble learning and comprehensive
and in-depth validation are inevitable methods that are required to
enhance research into NPC biomarkers using bodily fluid
samples.
Availability of data and materials
Not applicable.
Authors' contributions
JCL and SQZ conceived and designed the review. SQZ,
SXL and YSH consulted the literature. SQZ analyzed the literature
and drafted the manuscript. YSH produced the figure. JCL, SMP and
HBC critically revised the article for important intellectual
content and assisted in the literature search for this review
article. All authors read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Shaoguan Science and
Technology Plan Projects in 2020 (grant no. 200812094530421).
References
|
1
|
Chen YP, Chan ATC, Le QT, Blanchard P, Sun
Y and Ma J: Nasopharyngeal carcinoma. Lancet. 394:64–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wee JT, Ha TC, Loong SL and Qian CN: Is
nasopharyngeal cancer really a 'Cantonese cancer'? Chin J Cancer.
29:517–526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang LL, Chen YP, Mao YP, Wang ZX, Guo R,
Chen L, Tian L, Lin AH, Li L, Sun Y and Ma J: Validation of the 8th
edition of the uicc/ajcc staging system for nasopharyngeal
carcinoma from endemic areas in the intensity-modulated
radiotherapy era. J Natl Compr Canc Netw. 15:913–919. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roy Chattopadhyay N, Das P, Chatterjee K
and Choudhuri T: Higher incidence of nasopharyngeal carcinoma in
some regions in the world confers for interplay between genetic
factors and external stimuli. Drug Discov Ther. 11:170–180. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Janvilisri T: Omics-based identification
of biomarkers for nasopharyngeal carcinoma. Dis Markers.
2015:7621282015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee KT, Tan JK, Lam AK and Gan SY:
MicroRNAs serving as potential biomarkers and therapeutic targets
in nasopharyngeal carcinoma: A critical review. Crit Rev Oncol
Hematol. 103:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao L, Xiao T, Wang ZM, Cho WC and Xiao
ZQ: Biomarker discovery of nasopharyngeal carcinoma by proteomics.
Expert Rev Proteomics. 11:215–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD,
Yang KY, Jin F, Shi M, Chen YP, Hu WH, et al: Gemcitabine and
cisplatin induction chemotherapy in nasopharyngeal carcinoma. N
Engl J Med. 381:1124–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song C, Cheng P, Cheng J, Zhang Y, Sun M,
Xie S and Zhang X: Differential diagnosis of nasopharyngeal
carcinoma and nasopharyngeal lymphoma based on DCE-MRI and
RESOLVE-DWI. Eur Radiol. 30:110–118. 2020. View Article : Google Scholar
|
|
12
|
Colevas AD, Yom SS, Pfister DG, Spencer S,
Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ,
et al: NCCN guidelines insights: Head and neck cancers, version 1.
2018.J Natl Compr Canc Netw. 16:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun XS, Xiao BB, Lu ZJ, Liu SL, Chen QY,
Yuan L, Tang LQ and Mai HQ: Stratification of candidates for
induction chemotherapy in Stage III-IV nasopharyngeal carcinoma: A
large cohort study based on a comprehensive prognostic model. Front
Oncol. 28:2552020. View Article : Google Scholar
|
|
14
|
Wang YW, Ho SY, Lee SW, Chen CC, Litsu S,
Huang WT, Yang CC, Lin CH, Chen HY and Lin LC: Induction
chemotherapy improved long term outcomes in stage IV locoregional
advanced nasopharyngeal carcinoma. Int J Med Sci. 17:568–576. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao SM, Yang Q, Guo L, Mai HQ, Mo HY, Cao
KJ, Qian CN, Zhao C, Xiang YQ, Zhang XP, et al: Neoadjuvant
chemotherapy followed by concurrent chemoradiotherapy versus
concurrent chemoradiotherapy alone in locoregionally advanced
nasopharyngeal carcinoma: A phase III multicentre randomised
controlled trial. Eur J Cancer. 75:14–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin BY, Zhang GY, Lin KR, Chen XP, Cui JH,
Wang YJ and Luo W: Changes of plasma cytokines and chemokines
expression level in nasopharyngeal carcinoma patients after
treatment with definitive intensity-modulated radiotherapy (IMRT).
PLoS One. 12:e01722642017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang G, Zhang K, Li C, Li Y, Li Z, Li N,
Zhou Q and Shen L: Serum proteomics identify potential biomarkers
for nasopharyngeal carcinoma sensitivity to radiotherapy. Biosci
Rep. May 14–2019.Epub ahead of print. View Article : Google Scholar
|
|
18
|
Aftab O, Liao S, Zhang R, Tang N, Luo M,
Zhang B, Shahi S, Rai R, Ali J and Jiang W: Efficacy and safety of
intensity-modulated radiotherapy alone versus intensity-modulated
radiotherapy plus chemotherapy for treatment of intermediate-risk
nasopharyngeal carcinoma. Radiat Oncol. 15:662020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang HY, Chang YL, To KF, Hwang JS, Mai
HQ, Feng YF, Chang ET, Wang CP, Kam MK, Cheah SL, et al: A new
prognostic histopathologic classification of nasopharyngeal
carcinoma. Chin J Cancer. 35:412016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Al-Sarraf M, LeBlanc M, Giri PG, Fu KK,
Cooper J, Vuong T, Forastiere AA, Adams G, Sakr WA, Schuller DE and
Ensley JF: Chemoradiotherapy versus radiotherapy in patients with
advanced nasopharyngeal cancer: Phase III randomized inter-group
study 0099. J Clin Oncol. 16:1310–1317. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee AW, Tung SY, Ngan RK, Chappell R, Chua
DT, Liu TX, Siu L, Tan T, Chan LK, Ng WT, et al: Factors
contributing to the efficacy of concurrent-adjuvant chemotherapy
for locoregionally advanced nasopharyngeal carcinoma: Combined
analysesof NPC-9901 and NPC-9902 trials. Eur J Cancer. 47:656–666.
2011. View Article : Google Scholar
|
|
22
|
Chen Y, Liu MZ, Liang SB, Zong JF, Mao YP,
Tang LL, Guo Y, Lin AH, Zeng XF and Ma J: Preliminary results of a
prospective randomized trial comparing concurrent chemoradiotherapy
plus adjuvant chemotherapy with radiotherapy alone in patients with
locoregionally advanced nasopharyngeal carcinoma in endemic regions
of China. Int J Radiat Oncol Biol Phys. 71:1356–1364. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang H, Xu Y, Chen M, Zhong W, Wang M and
Zhao J: Patterns of response in metastatic NSCLC during PD-1 or
PD-L1 inhibitor therapy: Comparison of the RECIST 1.1 and iRECIST
criteria. Thorac Cancer. 11:1068–1075. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang W, Liu N, Chen XZ, Sun Y, Li B, Ren
XY, Qin WF, Jiang N, Xu YF, Li YQ, et al: Genome-wide
identification of a methylation gene panel as a prognostic
biomarker in nasopharyngeal carcinoma. Mol Cancer Ther.
14:2864–2873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen ZT, Liang ZG and Zhu XD: A review:
Proteomics in nasopharyngeal carcinoma. Int J Mol Sci.
16:15497–15530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Del Giacco and Cattaneo C: Introduction to
genomics. Methods Mol Biol. 823:79–88. 2012. View Article : Google Scholar
|
|
27
|
Berger MF and Mardis ER: The emerging
clinical relevance of genomics in cancer medicine. Nat Rev Clin
Oncol. 15:353–365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Donnelly D III, Aung PP and Jour G: The
'-OMICS' facet of melanoma: Heterogeneity of genomic, proteomic and
metabolomic biomarkers. Semin Cancer Biol. 59:165–174. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsang CM, Lui VW, Bruce JP, Pugh TJ and Lo
KW: Translational genomics of nasopharyngeal cancer. Semin Cancer
Biol. 61:84–100. 2020. View Article : Google Scholar
|
|
30
|
Yang H, Yu K, Zhang R, Li J, Wei X, Zhang
Y, Zhang C, Xiao F, Zhao D, Lin X, et al: The HLA-DRB1 allele
polymorphisms and nasopharyngeal carcinoma. Tumour Biol.
37:7119–7128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo XG and Xia Y: The interleukin-18
promoter-607C>A polymorphism contributes to nasopharyngeal
carcinoma risk: Evidence from a meta-analysis including 1,886
subjects. Asian Pac J Cancer Prev. 14:7577–7781. 2013. View Article : Google Scholar
|
|
32
|
Yi M, Cai J, Li J, Chen S, Zeng Z, Peng Q,
Ban Y, Zhou Y, Li X, Xiong W, et al: Rediscovery of NF-κB signaling
in nasopharyngeal carcinoma: How genetic defects of NF-κB pathway
interplay with EBV in driving oncogenesis? J Cell Physiol.
233:5537–5549. 2018. View Article : Google Scholar
|
|
33
|
Tam V, Patel N, Turcotte M, Bosse Y, Paré
G and Meyre D: Benefits and limitations of genome-wide association
studies. Nat Rev Genet. 20:467–484. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou P, Liu S, Ji NN, Zhang S, Wang P, Lin
B, Yang P, Lin XT, Cai YZ, Wang ZM, et al: Association between
variant alleles of major histocompatibility complex class II
regulatory genes and nasopharyngeal carcinoma susceptibility. Eur J
Cancer Prev. 29:531–537. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niu Y, Zhou G, Wang Y, Qin J, Ping J,
Zhang Q, Han BW, Liu YX, Yang C, Zhai Y, et al: Association of
MCP-1 promoter polymorphism with susceptibility to nasopharyngeal
carcinoma. J Cell Biochem. 120:6661–6670. 2019. View Article : Google Scholar
|
|
36
|
Ban EZ, Lye MS, Chong PP, Yap YY, Lim SY
and Abdul Rahman H: Haplotype CGC from XPD, hOGG1 and ITGA2
polymorphisms increases the risk of nasopharyngeal carcinoma in
Malaysia. PLoS One. 12:e01872002017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lourembam DS, Singh AR, Sharma TD, Singh
TS, Singh TR and Singh LS: Evaluation of risk factors for
nasopharyngeal carcinoma in a high-risk area of India, the
Northeastern region. Asian Pac J Cancer Prev. 16:4927–4935. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng H, Dai W, Cheung AK, Ko JM, Kan R,
Wong BW, Leong MM, Deng M, Kwok TC, Chan JY, et al: Whole-exome
sequencing identifies multiple loss-of-function mutations of NF-κB
pathway regulators in nasopharyngeal carcinoma. Proc Natl Acad Sci
USA. 113:11283–11288. 2016. View Article : Google Scholar
|
|
39
|
Li YY, Chung GT, Lui VW, To KF, Ma BB,
Chow C, Woo JK, Yip KY, Seo J, Hui EP, et al: Exome and genome
sequencing of nasopharynx cancer identifies NF-κB pathway
activating mutations. Nature Commun. 8:141212017. View Article : Google Scholar
|
|
40
|
Tsao SW, Yip YL, Ysang CM, Pang PS, Lau
VM, Zhang G and Lo KW: Etiology factors of nasopharyngeal
carcinoma. Oral Oncol. 50:330–338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Anderson EN Jr, Anderson ML and Ho HC:
Environmental backgrounds of young Chinese nasopharyngeal carcinoma
patients. IARC Sci Publ. 231–239. 1978.PubMed/NCBI
|
|
42
|
Tan C, Chen H, Wu T and Xia C: The
prediction of nasopharyngeal carcinoma mortality based on soil
element levels in China. Biol Trace Elem Res. 138:139–147. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu G, Hsu WL, Coghill AE, Yu KJ, Wang CP,
Lou PJ, Liu Z, Jones K, Vogt K, Wang M, et al: Whole-exome
sequencing of nasopharyngeal carcinoma families reveals novel
variants potentially involved in nasopharyngeal carcinoma. Sci Rep.
9:99162019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu Z, Goldstein AM, Hsu WL, Yu KJ, Chien
YC, Ko JY, Jian JJ, Tsou YA, Leu YS, Liao LJ, et al: Evaluation of
rare and common variants from suspected familial or sporadic
nasopharyngeal carcinoma (NPC) susceptibility genes in sporadic
NPC. Cancer Epidemiol Biomarkers Prev. 28:1682–1686. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sasaki MM, Skol AD, Bao R, Rhodes LV,
Chambers R, Vokes EE, Cohen EE and Onel K: Integrated genomic
analysis suggests MLL3 is a novel candidate susceptibility gene for
familial nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers
Prev. 24:1222–1228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mokni-Baizing N, Gorgi Y, Elghourabi M,
Makhlouf M, Boussen H, Gritli S, Elmay M, Gamoudi A and Elmay A:
HLA-A*26-A*30 and HLA-DRB1*10 could be predictors of nasopharyngeal
carcinoma risk in high-risk Tunisian families. J Oral Sci.
59:289–296. 2017. View Article : Google Scholar
|
|
47
|
Friborg J, Wohlfahrt J, Koch A, Storm H,
Olsen OR and Melbye M: Cancer susceptibility in nasopharyngeal
carcinoma families-a population-based cohort study. Cancer Res.
65:8567–8572. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kerns SL, West CM, Andreassen CN, Barnett
GC, Bentzen SM, Burnet NG, Dekker A, De Ruysscher D, Dunning A,
Parliament M, et al: Radiogenomics: The search for genetic
predictors of radiotherapy response. Future Oncol. 10:2391–2406.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen W and Hu GH: Biomarkers for enhancing
the radiosensitivity of nasopharyngeal carcinoma. Cancer Biol Med.
12:23–32. 2015.PubMed/NCBI
|
|
50
|
Rattay T and Talbot CJ: Finding the
genetic determinants of adverse reactions to radiotherapy. Clin
Oncol (R Coll Radiol). 26:301–308. 2014. View Article : Google Scholar
|
|
51
|
Ma WL, Liu R, Huang LH, Zou C, Huang J,
Wang J, Chen SJ, Meng XG, Yang JK, Li H, et al: Impact of
polymorphisms in angiogenesis-related genes on clinical outcomes of
radiotherapy in patients with nasopharyngeal carcinoma. Clin Exp
Pharmacol Physiol. 44:539–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Le Z, Niu X, Chen Y, Ou X, Zhao G, Liu Q,
Tu W, Hu C, Kong L and Liu Y: Predictive single nucleotide
polymorphism markers for acute oral mucositis in patients with
nasopharyngeal carcinoma treated with radiotherapy. Oncotarget.
8:63026–63037. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu J, Huang Y, Liu L, Wang J, Yin J, Huang
L, Chen S, Li J, Yuan H, Yang G, et al: Genetic polymorphisms of
Wnt/β-catenin pathway genes are associated with the efficacy and
toxicities of radiotherapy in patients with nasopharyngeal
carcinoma. Oncotarget. 7:82528–82537. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang J, Guo C, Gong X, Ao F, Huang Y,
Huang L, Tang Y, Jiang C, Xie X, Dong Q, et al: The impacts of
genetic polymorphisms in genes of base excision repair pathway on
the efficacy and acute toxicities of (chemo) radiotherapy in
patients with nasopharyngeal carcinoma. Oncotarget. 8:78633–78641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhai XM, Hu QC, Gu K, Wang JP, Zhang JN
and Wu YW: Significance of XRCC1 Codon399 polymorphisms in Chinese
patients with locally advanced nasopharyngeal carcinoma treated
with radiation therapy. Asia Pac J Clin Oncol. 12:e125–e132. 2016.
View Article : Google Scholar
|
|
56
|
Chen H, Wu M, Li G, Hua L, Chen S and
Huang H: Association between XRCC1 single-nucleotide polymorphism
and acute radiation reaction in patients with nasopharyngeal
carcinoma: A cohort study. Medicine (Baltimore). 96:e82022017.
View Article : Google Scholar
|
|
57
|
Guo XB, Ma WL, Liu LJ, Huang YL, Wang J,
Huang LH, Peng XD, Yin JY, Li JG, Chen SJ, et al: Effects of gene
polymorphisms in the endoplasmic reticulum stress pathway on
clinical outcomes of chemoradiotherapy in Chinese patients with
nasopharyngeal carcinoma. Acta Pharmacol Sin. 38:571–580. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tan J, Jiang L, Cheng X, Wang C, Chen J,
Huang X, Xie P, Xia D, Wang R and Zhang Y: Association between
VEGF-460T/C gene polymorphism and clinical outcomes of
nasopharyngeal carcinoma treated with intensity-modulated radiation
therapy. Onco Targets Ther. 10:909–918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tsao SW, Tsang CM and Lo KW: Epstein-barr
virus infection and nasopharyngeal carcinoma. Philos Trans R Soc
Lond B Biol Sci. 372:201602702017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shen JJ, Niu WN, Zhou M, Zhou F, Zhang HY
and Wang L: Association of Epstein Barr virus A73 gene polymorphism
with nasopharyngeal carcinoma. Genet Test Mol Biomarkers.
19:187–190. 2015. View Article : Google Scholar
|
|
61
|
Wu S, Liu W, Li H, Zhao Z, Yang Y, Xiao H,
Song Y and Luo B: Conservation and polymorphism of EBV RPMS1 gene
in EBV-associated tumors and healthy individuals from endemic and
non-endemic nasopharyngeal carcinoma areas in China. Virus Res.
250:75–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tay JK, Chan SH, Lim CM, Siow CH, Goh HL
and Loh KS: The role of Epstein-Barr virus DNA load and serology as
screening tools for nasopharyngeal carcinoma. Otolaryngol Head Neck
Surg. 155:274–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chan KC, Woo JK, King A, Zee BC, Lam WK,
Chan SL, Chu SW, Mak C, Tse IO, Leung SY, et al: Analysis of plasma
Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl
J Med. 377:513–522. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lam WK, Chan KC and Lo YM: Plasma
Epstein-Barr virus DNA as an archetypal circulating tumour DNA
marker. J Pathol. 247:641–649. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zeng Z, Fan S, Zhang X, Li S, Zhou M,
Xiong W, Tan M, Zhang W and Li G: Epstein-Barr virus-encoded small
RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal
carcinoma. Clin Transl Oncol. 18:206–211. 2016. View Article : Google Scholar
|
|
66
|
Arai A, Yamaguchi T, Komatsu H, Imadome K,
Kurata M, Nagata K and Miura O: Infectious mononucleosis
accompanied by clonal proliferation of EBV-infected cells and
infection of CD8-positive cells. Int J Hematol. 99:671–675. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Banko AV, Lazarevic IB, Karalic DZ, Djukic
VB, Cupic MD, Stevanovic G and Jovanovic TP: The sequence analysis
of Epstein-Barr virus EBNA1 gene: Could viral screening markers for
nasopharyngeal carcinoma be identified? Med Mircobiol Immunol.
208:81–88. 2019. View Article : Google Scholar
|
|
68
|
Banko AV, Lazarevic IB, Folic MM, Djukic
VB, Cirkovic AM, Karalic DZ, Cupic MD and Jovanovic TP:
Characterization of the variability of Epstein-Barr virus genes in
nasopharyngeal biopsies: Potential predictors for carcinoma
progression. PLoS One. 11:e01534982016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hui EP, Ma BB, Chan KC, Chan CM, Wong CS,
To KF, Chan AW, Tung SY, Ng WT, Cheng AC, et al: Clinical utility
of plasma Epstein-Barr virus DNA and ERCC1 single nucleotide
polymorphism in nasopharyngeal carcinoma. Cancer. 121:2720–2729.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chatsirisupachai K, Palmer D, Ferreira S
and de Magalhães JP: A human tissue-specific transcriptomic
analysis reveals a complex relationship between aging, cancer, and
cellular senescence. Aging Cell. 18:e130412019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sager M, Yeat NC, Pajaro-Van der Stadt S,
Lin C, Ren Q and Lin J: Transcriptomics in cancer diagnostics:
Developments in technology, clinical research and
commercialization. Expert Rev Mol Diagn. 15:1589–1603. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Moor AE and Itzkovitz S: Spatial
transcriptomics: Paving the way for tissue-level systems biology.
Curr Poin Biotechnol. 46:126–133. 2017. View Article : Google Scholar
|
|
73
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang LJ, Chou YF, Chen PR, Su B, Hsu YC,
Chang CH and Lee JW: Differential miRNA expression in repeated
recurrence of nasopharyngeal carcinoma. Cancer Lett. 344:188–194.
2014. View Article : Google Scholar
|
|
75
|
Li T, Chen JX, Fu XP, Yang S, Zhang Z,
Chen KH and Li Y: microRNA expression profiling of nasopharyngeal
carcinoma. Oncol Rep. 25:1353–1363. 2011.PubMed/NCBI
|
|
76
|
Lu T, Guo Q, Lin K, Chen H, Chen Y, Xu Y,
Lin C, Su Y, Chen Y, Chen M, et al: Circulating Epstein-Barr virus
microRNAs BART7-3p and BART13-3p as novel biomarkers in
nasopharyngeal carcinoma. Cancer Sci. 111:1711–1723. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu L, Wang J, Zhu D, Zhang S, Zhou X, Zhu
W, Zhu J and He X: Circulating Epstein-Barr virus microRNA profile
reveals novel biomarker for nasopharyngeal carcinoma diagnosis.
Cancer Biomark. 27:365–375. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hirai N, Wakisaka N, Knodo S, Aga M,
Moriyama-Kita M, Ueno T, Nakanishi Y, Endo K, Sugimoto H, Murono S,
et al: Potential interest in circulating miR-BART17-5p as a
post-treatment biomarker for prediction of recurrence in
Epstein-Barr virus-related nasopharyngeal carcinoma. PLoS One.
11:e01636092016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang H, Zou X, Wu L, Zhang S, Wang T, Liu
P, Zhu W and Zhu J: Identification of a 7-microRNA signature in
plasma as promising biomarker for nasopharyngeal carcinoma
detection. Cancer Med. 9:1230–1241. 2020. View Article : Google Scholar
|
|
80
|
Yi SJ, Liu P, Chen BL, Ou-Yang L, Xiong WM
and Su JP: Circulating miR-31-5p may be a potential diagnostic
biomarker in nasopharyngeal carcinoma. Neoplasma. 66:825–829. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang H, Wei X, Wu B, Su J, Tan W and Yang
K: Tumor-educated platelet miR-34c-3p and miR-18a-5p as potential
liquid biopsy biomarkers for nasopharyngeal carcinoma diagnosis.
Cancer Manag Res. 11:3351–3360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wen W, Mai SJ, Lin HX, Zhang MY, Huang JL,
Hua X, Lin C, Long ZQ, Lu ZJ, Sun XQ, et al: Identification of two
microRNA signatures in whole blood as novel biomarkers for
diagnosis of nasopharyngeal carcinoma. J Transl Med. 17:1862019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu L, Zheng K, Yan C, Pan X, Liu Y, Liu J,
Wang F, Guo W, He X, Li J and Shen Y: Genome-wide study of salivary
microRNAs as potential noninvasive biomarkers for detection of
nasopharyngeal carcinoma. BMC Cancer. 19:8432019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang M, Gu B, Chen X, Wang Y, Li P and
Wang K: The function and therapeutic potential of Epstein-Barr
virus-encoded MicroRNAs in cancer. Mol Ther Nucleic Acids.
17:657–668. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Xiang B, Zhou M, Li X, Wu X, et al: The emerging role of
Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J
Cancer. 9:2852–2864. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Best MG, Sol N, Kooi I, Tannous J,
Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E,
Koster J, et al: RNA-Seq of tumor-educated platelets enables
blood-based pan-cancer, multiclass, and molecular pathway cancer
diagnostics. Cancer Cell. 28:666–676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li K, Zhu X, Li L, Ning R, Liang Z, Zeng
F, Su F, Huang S, Yang X and Qu S: Identification of non-invasive
biomarkers for predicting the radiosensitivity of nasopharyngeal
carcinoma from serum microRNAs. Sci Rep. 10:51612020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wu J and Hann SS: Functions and roles of
long-non-coding RNAs in human nasopharyngeal carcinoma. Cell
Physiol Biochem. 45:1191–1204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhao CX, Zhu W, Ba ZQ, Xu HJ, Liu WD, Zhu
B, Wang L, Song YJ, Yuan S and Ren CP: The regulatory network of
nasopharyngeal carcinoma metastasis with a focus on EBV, lncRNAs
and miRNAs. Am J Cancer Res. 8:2185–2209. 2018.PubMed/NCBI
|
|
90
|
He B, Zeng J, Chao W, Chen X, Huang Y,
Deng K, Huang Z, Li J, Dai M, Chen S, et al: Serum long non-coding
RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic
biomarkers for nasopharyngeal carcinoma. Oncotarget. 8:41166–41177.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yao Z, Zhang Y, Xu D, Zhou X, Peng P, Pan
Z, Xiao N, Yao J and Li Z: Research progress on long non-coding RNA
and radiotherapy. Med Sci Monit. 25:5757–5770. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Liu G, Zeng X, Wu B, Zhao J and Pan Y:
RNA-Seq analysis of peripheral blood mononuclear cells reveals
unique transcriptional signatures associated with radiotherapy
response of nasopharyngeal carcinoma and prognosis of head and neck
cancer. Cancer Biol Ther. 21:139–146. 2020. View Article : Google Scholar :
|
|
93
|
Shuai M, Hong J, Huang D, Zhang X and Tian
Y: Upregulation of circRNA_0000285 serves as a prognostic biomarker
for nasopharyngeal carcinoma and is involved in radiosensitivity.
Oncol Lett. 16:6495–6501. 2018.PubMed/NCBI
|
|
94
|
Gosho M, Nagashima K and Sato Y: Study
designs and statistical analyses for biomarker research. Sensors
(Basel). 12:8966–8986. 2012. View Article : Google Scholar
|
|
95
|
Cho WC: Mass spectrometry-based proteomics
in cancer research. Expert Rev Proteomics. 14:725–727. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tan HT, Lee YH and Chung MC: Cancer
proteomics. Mass Spectrom Rev. 31:583–605. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang Z, Wu S, Stenoien DL and Paša-Tolić
L: High-throughput proteomics. Annu Rev Anal Chem (Palo Alto
Calif). 7:427–454. 2014. View Article : Google Scholar
|
|
98
|
Aslam B, Basit M, Nisar MA, Khurshid M and
Rasool MH: Proteomics: Technologies and their applications. J
Chromatogr Sci. 55:182–196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
EI-Sharkawy A, Al Zaidan L and Malki A:
Epstein-Barr virus-associated malignancies: Roles of viral
oncoproteins in carcinogenesis. Front Oncol. 8:2652018. View Article : Google Scholar
|
|
100
|
Zamanian Azodi M, Rezaei Tavirani M,
Rezaei Tavirani M, Vafaee R and Rostami-Nejad M: Nasopharyngeal
carcinoma protein interaction mapping analysis via proteomic
approaches. Asian Pac J Cancer Prev. 19:845–851. 2018.PubMed/NCBI
|
|
101
|
Cao Y: EBV based cancer prevention and
therapy in nasopharyngeal carcinoma. NPJ Precis Oncol. 1:102017.
View Article : Google Scholar
|
|
102
|
Schmidlin T, Garrigues L, Lane CS, Mulder
TC, van Doorn S, Post H, de Graaf EL, Lemeer S, Heck AJ and
Altelaar AF: Assessment of SRM, MRM(3), and DIA for the targeted
analysis of phosphorylation dynamics in non-small cell lung cancer.
Proteomics. 16:2193–2205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Luo Y, Mok TS, Lin X, Zhang W, Cui Y, Guo
J, Chen X, Zhang T and Wang T: SWATH-based proteomics identified
carbonic anhydrase 2 as a potential diagnosis biomarker for
nasopharyngeal carcinoma. Sci Rep. 7:411912017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Nguyen EV, Centenera MM, Moldovan M, Das
R, Irani S, Vincent AD, Chan H, Horvath LG, Lynn DJ, Daly RJ and
Butler LM: Identification of novel response and predictive
biomarkers to Hsp90 inhibitors through proteomic profiling of
patient-derived prostate tumor explants. Mol Cell Proteomics.
17:1470–1486. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Nakamura K, Hirayama-Kurogi M, Ito S, Kuno
T, Yoneyama T, Obuchi W, Terasaki T and Ohtsuki S: Large-scale
multiplex absolute protein quantification of drug-metabolizing
enzymes and transporters in human intestine, liver, and kidney
microsomes by SWATH-MS: Comparison with MRM/SRM and HR-MRM/PRM.
Proteomics. 16:2106–2117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sun X, Chen Y, Tan J and Qi X: Serum IRS-1
acts as a novel biomarker for diagnosis in patients with
nasopharyngeal carcinoma. Int J Clin Exp Pathol. 11:3685–3690.
2018.
|
|
107
|
Coghill AE, Pfeiffer RM, Proietti C, Hus
WL, Chien YC, Lekiffre L, Krause L, Teng A, Pablo J, Yu KJ, et al:
Identification of a novel, EBV-based antibody risk stratification
signature for early detection of nasopharyngeal carcinoma in
Taiwan. Clin Cancer Res. 24:1305–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lin LH, Xu YW, Huang LS, Hong CQ, Zhai TT,
Liao LD, Lin WJ, Xu LY, Zhang K, Li EM and Peng YH: Serum
proteomic-based analysis identifying autoantibodies against PRDX2
and PRDX3 as potential diagnostic biomarkers in nasopharyngeal
carcinoma. Clin Proteomics. 14:62017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Gong D, Li Z, Ding R, Cheng M, Huang H,
Liu A, Kang M, He H, Xu Y, Shao J, et al: Extensive serum biomarker
analysis in patients with nasopharyngeal carcinoma. Cytokine.
118:107–114. 2019. View Article : Google Scholar
|
|
110
|
Meng H, Zhu X, Li L, Liang Z, Li X, Pan X,
Zeng F and Qu S: Identification of CALM as the potential serum
biomarker for predicting the recurrence of nasopharyngeal carcinoma
using a mass spectrometry based comparative proteomic approach. Int
J Mol Med. 40:1152–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Manes NP and Nita-Lazar A: Application of
targeted mass spectrometry in bottom-up proteomics for systems
biology research. J Proteomics. 189:75–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sun Y, Guo W, Bai Y, Ge M, Hu C, Wu S, Hao
J, Gao M, Pan J, Dong P, et al: Neoadjuvant dose-modified docetaxel
in squamous cell carcinoma of the head and neck: A phase 3 study.
Oral Dis. 26:285–294. 2020. View Article : Google Scholar
|
|
113
|
Monti C, Zilocchi M, Colugnat I and
Alberio T: Proteomics turns functional. J Proteomics. 198:36–44.
2019. View Article : Google Scholar
|
|
114
|
Xiao Z and Chen Z: Deciphering
nasopharyngeal carcinoma pathogenesis via proteomics. Expert Rev
Proteomics. 16:475–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wang X: Cancer moonshot 2020: A new march
of clinical and translational medicine. Clin Transl Med. 5:112016.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Fiore LD, Rodriguez H and Shriver CD:
Collaboration to accelerate proteogenomics cancer care: The
department of veterans affairs, department of defense, and the
national cancer institute 's applied proteogenomics organizational
learning and outcomes (APOLLO) network. Clin Pharmacol Ther.
101:619–621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Rinschen MM, Ivanisevic J, Giera M and
Siuzdak G: Identification of bioactive metabolites using activity
metabolomics. Nat Rev Mol Cell Biol. 20:353–367. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Guijas C, Montenegro-Burke JR, Warth B,
Spilker ME and Siuzdak G: Metabolomics activity screening for
identifying metabolites that modulate phenotype. Nat Biotechnol.
36:316–320. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Schrimpe-Rutledge AC, Codreanu SG, Sherrod
SD and McLean JA: Untargeted metabolomics strategies-challenges and
emerging directions. J Am Soc Mass Spectrom. 27:1897–1905. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Li X, Wenes M, Romero P, Huang SC, Fendt
SM and Ho PC: Navigating metabolic pathways to enhance antitumour
immunity and immunotherapy. Nat Rev Clin Oncol. 16:425–441. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yang M, Soga T and Pollard PJ:
Oncometabolites: Linking altered metabolism with cancer. J Clin
Invest. 123:3652–3658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Li S, Fu L, Tian T, Deng L, Li H, Xia W
and Gong Q: Disrupting SOD1 activity inhibits cell growth and
enhances lipid accumulation in nasopharyngeal carcinoma. Cell
Commun Signal. 16:282018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Tan Z, Xiao L, Tang M, Bai F, Li J, Li L,
Shi F, Li N, Li Y, Du Q, et al: Targeting CPT1A-mediated fatty acid
oxidation sensitizes nasopharyngeal carcinoma to radiation therapy.
Theranostics. 8:2329–2347. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Tang F, Xie C, Huang D, Wu Y, Zeng M, Yi
L, Wang L, Mei W, Cao Y and Sun L: Novel potential markers of
nasopharyngeal carcinoma for diagnosis and therapy. Clin Biochem.
44:711–718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Yi L, Dong N, Shi S, Deng B, Yun Y, Yi Z
and Zhang Y: Metabolomic identification of novel biomarkers of
nasopharyngeal carcinoma. RSC Adv. 4:59094–59101. 2014. View Article : Google Scholar
|
|
126
|
Luo MS, Huang GJ and Liu HB: Oncologic
outcomes of IMRT versus CRT for nasopharyngeal carcinoma: A
meta-analysis. Medicine (Baltimore). 98:e159512019. View Article : Google Scholar
|
|
127
|
Carayol M, Leitzmann MF, Ferrari P,
Zamora-Ros R, Achaintre D, Stepien M, Schmidt JA, Travis RC,
Overvad K, Tjønneland A, et al: Blood metabolic signatures of body
mass index: A targeted metabolomics study in the EPIC cohort. J
Proteome Res. 16:3137–3146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xu J, Li J, Zhang R, He J, Chen Y, Bi N,
Song Y, Wang L, Zhan Q and Abliz Z: Development of a metabolic
pathway-based pseudo-targeted metabolomics method using liquid
chromatography coupled with mass spectrometry. Talanta.
192:160–168. 2019. View Article : Google Scholar
|
|
129
|
Allinson JL: Clinical biomarker
validation. Bioanalysis. 10:957–968. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Rhomdhoni AC, Kurniawan P and Hidayati T:
Correlation between superoxide dismutase serum level alteration
with neck metastatic tumor post cisplatin-paclitaxel chemotherapy
response in nasopharyngeal carcinoma patients. Indian J Otolaryngol
Head Neck Surg. 71(Suppl 1): S643–S646. 2019. View Article : Google Scholar
|
|
131
|
Jelonek K, Krzywon A, Jablonska P,
Slominska EM, Smolenski RT, Polanska J, Rutkowski T,
Mrochem-Kwarciak J, Skladowki K and Widlak P: Systemic effects of
radiotherapy and concurrent chemo-radiotherapy in head and neck
cancer patients-comparison of serum metabolome profiles.
Metabolites. 10:602020. View Article : Google Scholar
|
|
132
|
Geyer PE, Voytik E, Treit PV, Doll S,
Kleinhempel A, Niu L, Müller JB, Buchholtz ML, Bader JM, Teupser D,
et al: Plasma proteome profiling to detect and avoid sample-related
biases in biomarker studies. EMBO Mol Med. 11:e104272019.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Dufourd T, Robil N, Mallet D, Carcenac C,
Boulet S, Brishoual S, Rabois E, Houeto JL, de la Grange P and
Carnicella S: Plasma or serum? A qualitative study on rodents and
humans using high-throughput microRNA sequencing for circulating
biomarkers. Biol Methods Protoc. 4:bpz0062019. View Article : Google Scholar
|
|
134
|
Lippi G, Banfi G, Buttarello M, Ceriotti
F, Daves M, Dolci A, Caputo M, Giavarina D, Montagnana M, Miconi V,
et al: Recommendations for detection and management of unsuitable
samples in clinical laboratories. Clin Chem Lab Med. 45:728–736.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Lam WKJ, Jiang P, Chan KCA, Cheng SH,
Zhang H, Peng W, Tse OYO, Tong YK, Gai W, Zee BCY, et al:
Sequencing-based counting and size profiling of plasma Epstein-Barr
virus DNA enhance population screening of nasopharyngeal carcinoma.
Proc Natl Acad Sci USA. 115:E5115–E5124. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Elliott P and Peakman TC; UK Biobank: The
UK biobank sample handling and storage protocol for the collection,
processing and archiving of human blood and urine. Int J Epidemiol.
37:234–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Gautam A, Donohue D, Hoke A, Miller SA,
Srinivasan S, Sowe B, Detwiler L, Lynch J, Levangie M, Hammamieh R
and Jett M: Investigating gene expression profiles of whole blood
and peripheral blood mononuclear cells using multiple collection
and processing methods. PLoS One. 14:e02251372019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Leidinger P, Backes C, Rheinheimer S,
Keller A and Meese E: Towards clinical applications of blood-borne
miRNA signatures: The influence of the anticoagulant EDTA on miRNA
abundance. PLoS One. 10:e01433212015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Mathé E, Hays JL, Stover DG and Chen JL:
The omics revolution continues: The maturation of high-throughput
biological data sources. Yearb Med Inform. 27:211–222. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hasin Y, Seldin M and Lusis A: Multi-omics
Approaches to Disease. Genome Biol. 18:832017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Chakraborty S, Hosen MI, Ahmed M and
Shekhar HU: Onco-multi-OMICS approach: A new frontier in cancer
research. Biomed Res Int. 2018:98362562018. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Nicora G, Vitali F, Dagliati A, Geifman N
and Bellazzi R: Integrated multi-omics analyses in oncology: A
review of machine learning methods and tools. Front Oncol.
10:10302020. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Olivier M, Asmis R, Hawkins GA, Howard TD
and Cox LA: The need for multi-omics biomarker signatures in
precision medicine. Int J Mol Sci. 20:47812019. View Article : Google Scholar :
|















