Introduction
Hypopharyngeal squamous cell carcinoma (HSCC) is one
of the most aggressive types of head and neck cancer; 70-85% of
patients with HSCC are diagnosed at stage III or IV, and the 5-year
overall survival rates are 15-45% based on studies in the USA and
Australia between 1971 and 2002 (1-3).
In addition to surgery, adjuvant chemotherapy is commonly used in
the multidisciplinary treatment of HSCC. Cisplatin is the most
widely used platinum-based chemotherapeutic agent for solid tumors,
such as HSCC (4). However,
resistance to cisplatin limits its clinical efficiency and the
improvement of survival rate in patients with HSCC (5).
The anticancer mechanism of cisplatin involves the
activation of DNA damage response and intrinsic apoptosis (6). Intrinsic apoptosis involves
apoptotic executioners including caspase-3, which cleave substrates
such as poly(ADP-ribose) polymerase 1 (PARP1), resulting in
morphological and biochemical alterations (7). In addition, cisplatin-induced
apoptosis is regulated by the B-cell lymphoma 2 (Bcl-2) protein
family, which serves as an apoptotic switch (8). The Bcl-2 protein family includes
pro-apoptotic and anti-apoptotic proteins, such as myeloid cell
leukemia-1 (MCL1), which regulates the intrinsic apoptosis pathway
(9). Various MCL1 inhibitors have
been developed, providing effective approaches for the
chemosensitization of esophageal squamous carcinoma cells to
cisplatin (10). Thus,
understanding the detailed molecular mechanisms through which
cisplatin chemoresistance is developed may help in the development
of effective treatments against HSCC.
Plant homeodomain finger protein 20 (PHF20), also
termed glioma-expressed antigen 2, is a potent transcriptional
activator (11). PHF20 expression
is associated with tumorigenesis (12). PHF20 expression is also associated
with a good prognosis of non-small-cell lung cancer, and PHF20
activates NF-κB at the transcriptional factor (13). However, PHF20 is highly expressed
in neuroblastoma and promotes the aggressiveness of neuroblastoma
cells by directly binding to the promotor regions of
octamer-binding transcription factor 4 (OCT4) (14). OCT4 is a key nuclear
transcriptional factor that maintains the pluripotency and
self-renewal properties of embryonic stem cells (15). OCT4 affects the tumorigenesis of a
number of types of cancer including pancreatic, cervical and
ovarian cancer, as well as glioma (16-18). OCT4 inhibits apoptosis in cervical
cancer by inhibiting microRNA-125b expression (19), whereas its knockdown suppresses
the viability and mobility of hepatocellular carcinoma cells and is
associated with low expression levels of survivin and
phosphorylated STAT3 (20).
Furthermore, OCT4 expression promotes the differentiation of lung
cancer cells into small lung cancer cells and acquisition of drug
resistance (21,22), and its knockdown sensitizes cancer
cells to cisplatin treatment in bladder and non-small lung cancer
cells (23,24). Thus, the present study aimed to
determine whether the expression of PHF20 may be associated with
apoptosis and cisplatin resistance in HSCC cells.
A previous study has demonstrated that PHF20 is
involved in tumorigenesis (25).
However, the expression level and role of PHF20 in apoptosis and
cisplatin sensitivity remain unclear. The aim of the present study
was to determine the expression levels of PHF20 in
cisplatin-resistant and cisplatin-sensitive HSCC cells and to
investigate whether PHF20 knockdown induced apoptosis and
sensitized HSCC cells to cisplatin treatment. These results may
indicate whether PHF20 may be used as a potential therapeutic
target in HSCC to improve cisplatin sensitivity.
Materials and methods
Cell lines and culture
The FaDu cell line was used as the HSCC model and
was obtained from the American Type Culture Collection. FaDu cells
were cultured in DMEM/F12 (1:1; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Biological
Industries). The cells were incubated at 37°C in a humidified
incubator with 5% CO2. Cisplatin was obtained from
Sigma-Aldrich; Merck KGaA (cat. no. P4394) and dissolved in 0.9%
sodium chloride solution to obtain a 2-µM stock
solution.
Establishment of a cisplatin-resistant
HSCC cell line
First, FaDu cells were treated with various
concentrations (0, 1, 2, 4, 8, 16 and 32 µM) of cisplatin
for 2 days, and the half maximal inhibitory concentration
(IC50) was determined. FaDu cells were subsequently
treated with the culture medium containing 0.25 µM cisplatin
for 48 h. The solution was discarded, and the cells were recovered
for 72 h in fresh medium without cisplatin. The concentrations of
cisplatin were gradually increased ~1 month later, and
cisplatin-resistant FaDu cells were generated for ~6 months.
RNA sequencing
To analyze the genes with altered expression levels
in FaDu cells due to cisplatin resistance, total RNA was isolated
from the cells using the Direct-Zol™ RNA MiniPep kit (cat. no.
R2052; Zymo Research Corp.). The experiments were performed in
triplicate. Total RNA was qualified and quantified using a NanoDrop
2000 and Agilent 2100 Bioanalyzer (Thermo Fisher Scientific, Inc.).
cDNA was generated according to the manufacturer's instructions.
The single-stranded circular DNA was formatted as the final
library, which was amplified to produce a DNA nanoball (DNB) and
loaded into the patterned nanoarray, and single-end 50-base reads
were generated on the MGISEQ2000 platform. The concentration of the
final library was detected by Nanodrop 2000. The final library was
pooled in equimolar concentrations (~9.4 nM) and sequenced on the
MGISEQ2000 platform. CoolMPS high-through sequencing kit (cat. no.
1000018234; MGISEQ-2000RS FCL SE50; BGI Group) was used to
determine the sequences of the libraries. Differential expression
analysis was performed using the DEGseq2 package with R
software (3.6.3) (26,27). Fold-change >2 and P<0.001
were used as the screening criteria.
Cell viability assay
Cell viability was detected using the Cell Counting
Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology). A total
of ~4×103 cells were seeded in each well of 96-well
plates containing 100 µl medium. A total of 10 µl of
CCK-8 solution was added to each well at 24, 48, 72 or 96 h after
cell seeding and incubated for 2 h at 37°C. Subsequently, optical
density was measured at 450 nm to determine the cell viability by
using a microplate reader (BioTek Instruments, Inc.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. cDNAs were
synthesized using the RevertAid™ First Strand cDNA synthesis kit
(Thermo Fisher Scientific, Inc.) as follows: The reaction mixture
was prepared, the samples were incubated for 60 min at 42°C,
following which the reaction was terminated for 5 min at 70°C. qPCR
was performed using SYBR® Premix Ex Taq (Promega
Corporation) on Mastercycler® RealPlex 2 (Eppendorf)
under the following conditions: 95°C for 2 min, followed by 40
cycles of 95°C for 20 sec, 58°C for 20 sec and 72°C for 20 sec, and
a final dissolution curve. The primer sequences were as follows:
PHF20 forward, 5′-GAA TGG TCA ATG CGA GTG G-3′ and reverse, 5′-TGG
CAG CAA GGC TAC TGT G-3′; OCT4 forward, 5′-TTG CCG CAA AGT GTG TAA
CG-3′ and reverse, 5′-GTC ACC CCT AAA TGC CAC CG-3′; OCT4A forward,
5′-CGT GAA GCT GGA GAA GGA GAA GCT G-3′ and reverse, 5′-CAA GGG CCG
CAG CTT ACA CAT GTT C-3′; OCT4B forward, 5′-ATG CAT GAG TCA GTG AAC
AG-3′ and reverse, 5′-CCA CAT CGG CCT GTG TAT AT-3′; OCT4B1
forward, 5′-GGG TTC TAT TTG GTG GGT TCC-3′ and reverse, 5′-TCC CTC
TCC CTA CTC CTC TTC A-3′; and β-actin forward, 5′-CCA ACC GCG AGA
AGA TGA-3′ and reverse, 5′-CCA GAG GCG TAC AGG GAT AG-3′. Data were
analyzed using the 2−∆∆Cq method (28).
Western blotting
The target cisplatin-sensitive, -resistant and
transfected FaDu cells were washed with ice-cold phosphate-buffered
saline (PBS), lysed with RIPA lysis buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology) containing phosphatase (cat.
no. 04906837001; Roche Applied Science) and protease inhibitors
(cat. no. 04693159001; Roche Applied Science), and centrifuged at
12,000 × g for 15 min at 4°C. The supernatant was collected, and
the protein concentration was measured by BCA assay (Beyotime
Institute of Biotechnology). Protein lysates (30 µg/lane)
were separated via 10% SDS-PAGE and transferred to polyvinylidene
difluoride membranes (MilliporeSigma). The membranes were blocked
with 5% non-fat milk for 1 h at room temperature, followed by
incubation with primary antibodies at 4°C overnight. The following
primary antibodies were used: PHF20 (1:2,000; cat. no. 3934S; Cell
Signaling Technology, Inc.), phosphorylated (p)-STAT3 (1:1,000;
cat. no. 9131S; Cell Signaling Technology, Inc.), OCT4 (1:1,000;
cat. no. 2750S; Cell Signaling Technology, Inc.), MCL1 (1:1,000;
cat. no. sc-12756; Santa Cruz Biotechnology, Inc.), PARP1 (1:1,000;
cat. no. 9562S; Cell Signaling Technology, Inc.), caspase-3
(1:1,000; cat. no. 9662S; Cell Signaling Technology, Inc.), STAT3
(1:20,000; cat. no. 610189; BD Biosciences), OCT4A (1:1,000; cat.
no. C52G3; Cell Signaling Technology, Inc.), OCT4B1 (1:4,000; cat.
no. bs-3816R; Bioss) and β-actin (1:40,000; cat. no. TA-09; OriGene
Technologies, Inc.). After washing three times in phosphate
buffered saline with 0.05% Tween-20, the membranes were incubated
at room temperature for 1 h with horseradish peroxidase-conjugated
secondary antibodies (1:20,000; goat anti-mouse IgG; cat. no.
ZB-5305; goat anti-rabbit IgG; cat. no. ZB-5301; OriGene
Technologies, Inc.). An enhanced chemiluminescence reagent (cat.
no. WBKLS0500; MilliporeSigma) was used to visualize the target
bands, and the protein densities were quantified using ImageJ
software (version 1.37; National Institutes of Health).
5-Ethynyl-2′-deoxyuridine (EDU) labeling
assay
In vitro cell proliferation was assessed
using the kFluor647-EdU imaging kit (Nanjing KeyGen Biotech Co.,
Ltd.) according to the manufacturer's instructions. The proportion
of cells that incorporated EDU (EDU+) was determined
using a fluorescence microscope (Leica Microsystems GmbH) in ≥3
randomly selected fields (×63 magnification) per sample.
Lentivirus-based short hairpin RNA
(shRNA) infection
Lentivirus vector GV493-green fluorescent protein
(GFP) containing scramble shRNA (Scr) or shRNA targeting PHF20
(shPHF) were obtained from Shanghai Genechem Co., Ltd. The target
sequence was 5′-CCA GCT CAC ATA GAA GAC ATT-3′. A random Scr
sequence, 5′-GTT CTC CGA ACG TGT CAC GT-3′, was used as a negative
control. FaDu cells were seeded in 6-well plates at the density of
1×105/ml for 24 h, and were infected with shPHF20 or Scr
(MOI=10) for 16 h at 37°C. Subsequently, puromycin (cat. no.
A610593-0025; Sangon) was added to the culture medium at 37°C with
5% CO2 for ~14 days according to the manufacturer's
instructions to generate stable FaDu PHF20-knockdown cell lines.
The lentiviral infection efficiency was determined by fluorescence
microscopy and western blot assay.
Migration and invasion assays
Transwell assays were performed to detect the target
cell migration and invasion. FaDu cells were harvested during the
logarithmic growth phase, washed with PBS and suspended in DMEM
without FBS at 4×105 cells/ml. The upper chamber of the
Transwell inset (Costar; Corning, Inc.) was filled with 250
µl of cell suspension, and the lower chamber was filled with
700 µl DMEM/F-12 with 20% FBS as a chemoattractant. The
cells were incubated for 24 h at 37°C. The non-migrating cells were
removed by a cotton swab, and the migratory cells were fixed with
4% formaldehyde at room temperature for 15 min and stained with
0.1% crystal violet at room temperature for 25 min. The stained
cells were counted in five random fields per sample under a light
microscope (magnification, ×100). Transwell chambers precoated with
Matrigel (BD Biosciences) for 30 min at 37°C were used for cell
invasion assays following a similar protocol as the cell migration
assays, with the exception that the cells were incubated for 36 h
due to the Matrigel barrier. The experiments were repeated at least
thrice.
Immunofluorescence (IF) assay
FaDu cells were cultured on glass coverslips and
fixed with 4% paraformaldehyde at room temperature for 20 min,
washed with cold PBS and permeabilized with 0.2% Triton X-100 for
10 min to rupture the cell membranes. Non-specific antigen-binding
sites were blocked by 1% bovine serum albumin (Beijing Solarbio
Science & Technology Co., Ltd.) in PBS for 1 h at room
temperature. After washing with PBS, the cells were incubated with
anti-PHF20 (1:200; cat. no. ab67796; Abcam), anti-STAT3 (1:50; cat.
no. 9139; Cell Signaling Technology, Inc.) and anti-p-STAT3 (1:200;
cat. no. 9145S; Cell Signaling Technology, Inc.) overnight at 4°C.
Following washing thrice with PBS, the cells were incubated with a
DyLight® 546-cojugated donkey anti-rabbit secondary
antibody for PHF20 and p-STAT3 (1:1,000; cat. no. A10040) and a
fluorescein-conjugated goat anti-mouse secondary antibody for STAT3
(1:1,000, F2761) (both Invitrogen; Thermo Fisher Scientific, Inc.)
for 1 h at room temperature. Cells were washed twice with PBS, and
the nuclei were stained with 4′,6-diamidino-2-phenylindole
(Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. Cells
were observed five random fields (×63 magnification) under a TCS
SPE confocal microscope (Leica Microsystems GmbH).
Tumor xenografts in nude mice
Male BALB/c nude mice (age, 4 weeks) were obtained
from Charles River Laboratories via Beijing Vital River Laboratory
Animal Technology Co., Ltd. and maintained in a temperature- and
humidity-controlled environment (22±1°C; 40-60% humidity) with a
12:12 h light-dark cycle and free access to food and water. The
study was performed with the approval from the Committee for the
Animal Care and Use of Shandong Provincial ENT Hospital. The mice
were randomly divided into two groups (n=6 per group). shPHF or Scr
cells (1×106 cells/mouse) were subcutaneously injected
into the left dorsal flank. The length and width of the tumors were
examined every 2 days using digital calipers. On day 23
post-inoculation, the mice were sacrificed by carbon dioxide
asphyxiation (20% chamber volume/min), and the tumors were excised,
fixed with 4% paraformaldehyde for 24 h at room temperature and
embedded in paraffin for hematoxylin and eosin (H&E) and
immunohistochemistry (IHC) staining.
H&E and IHC staining
The paraffin-embedded tumor tissues from mice were
cut into 2-µm sections and routinely stained with H&E
for histological diagnosis. The tissue sections were
de-paraffinized and rehydrated, and the tissue antigen was
retrieved using target retrieval solution (OriGene Technologies,
Inc.) according to the manufacturer's protocol. The slides were
washed with PBS and incubated with the anti-PHF20 polyclonal
(1:200; cat. no. 124224; Abcam), anti-p-STAT3 (1:200; cat. no.
9145S; Cell Signaling Technology, Inc.) and anti-Ki67 (diluted
1:200; cat. no. ZA-0502; OriGene Technologies, Inc.) primary
antibodies at 4°C overnight. After washing thrice with PBS, the
sections were incubated with secondary biotinylated goat
anti-rabbit IgG antibody (1:500, cat. no. SP-9001; OriGene
Technologies, Inc.) for 15 min. The tissue sections were examined,
and images were captured using Olympus BX53 microscope (Olympus
Corporation) in five random fields per sample (×200
magnification).
Flow cytometry
To determine the cell cycle phase, FaDu cells were
harvested at the logarithmic growth phase and washed twice with
ice-cold PBS. The cell density was adjusted to 1×106
cells/ml, and the cells were fixed with 70% ethanol at 4°C
overnight. The cells were stained with propidium iodide (10
µg/ml; Sigma-Aldrich; Merck KGaA) in the presence of RNase
(10 µg/ml; Tiangen Biotech Co., Ltd.) for ≤30 min.
Subsequently, the cells were harvested and analyzed by a
FACSCalibur flow cytometer (BD Biosciences) to detect the cell
cycle phase. The fluorescent signal was detected through the FL2
channel, and the proportion of DNA in different phases was analyzed
using ModfitLT Version 3.3.11 (Verity Software House, Inc.).
Apoptotic cells were detected using the Annexin
Vphycoerythrin (PE)/7-aminoactinomycin D (7-ADD) apoptosis
detection kit (Nanjing KeyGen Biotech Co., Ltd.) by flow cytometry.
Briefly, 1×106 FaDu cells were treated with 2 µM
cisplatin for 48 h, harvested, washed and resuspended in 50
µl binding buffer containing 5 µl 7-ADD at room
temperature in the dark for 10 min. Subsequently, the cells were
incubated in 500 µl binding buffer containing 1 µl
PE-conjugated Annexin V at room temperature for 10 min. Annexin V-
and 7-ADD-positive apoptotic cells were detected using a
FACSCalibur flow cytometer. Annexin V-PE-positive and
7-ADD-negative cells were considered as early apoptotic cells,
whereas positivity for both Annexin V and 7-ADD was considered to
indicate late apoptosis.
Terminal deoxynucleotidyl
transferase-mediated dUTP-biotin nick end labeling (TUNEL)
assay
To assess apoptotic cells in tissues, 2-µm
tumor tissue sections were analyzed by TUNEL assay (in situ
Cell Death Detection kit; Roche Applied Sciences). Apoptotic cells
(TUNEL+) were counted as the percentage of the total
cells. TUNEL+ cells were counted in five random fields
per slide, and five slides per group were analyzed by fluorescence
microscopy at ×400 magnification.
Statistical analysis
All statistical analyses were performed using SPSS
17.0 (SPSS, Inc.). Student's t-test was used to determine
significant difference between two groups. Differences among
multiple groups were analyzed with mixed model ANOVA followed by
the Tukey's post-hoc test. Data were obtained from at ≥3
independent experiments and presented as the mean ± SEM. P<0.05
was considered to indicate a statistically significant
difference.
Results
PHF20 is upregulated in
cisplatin-resistant HSCC cells
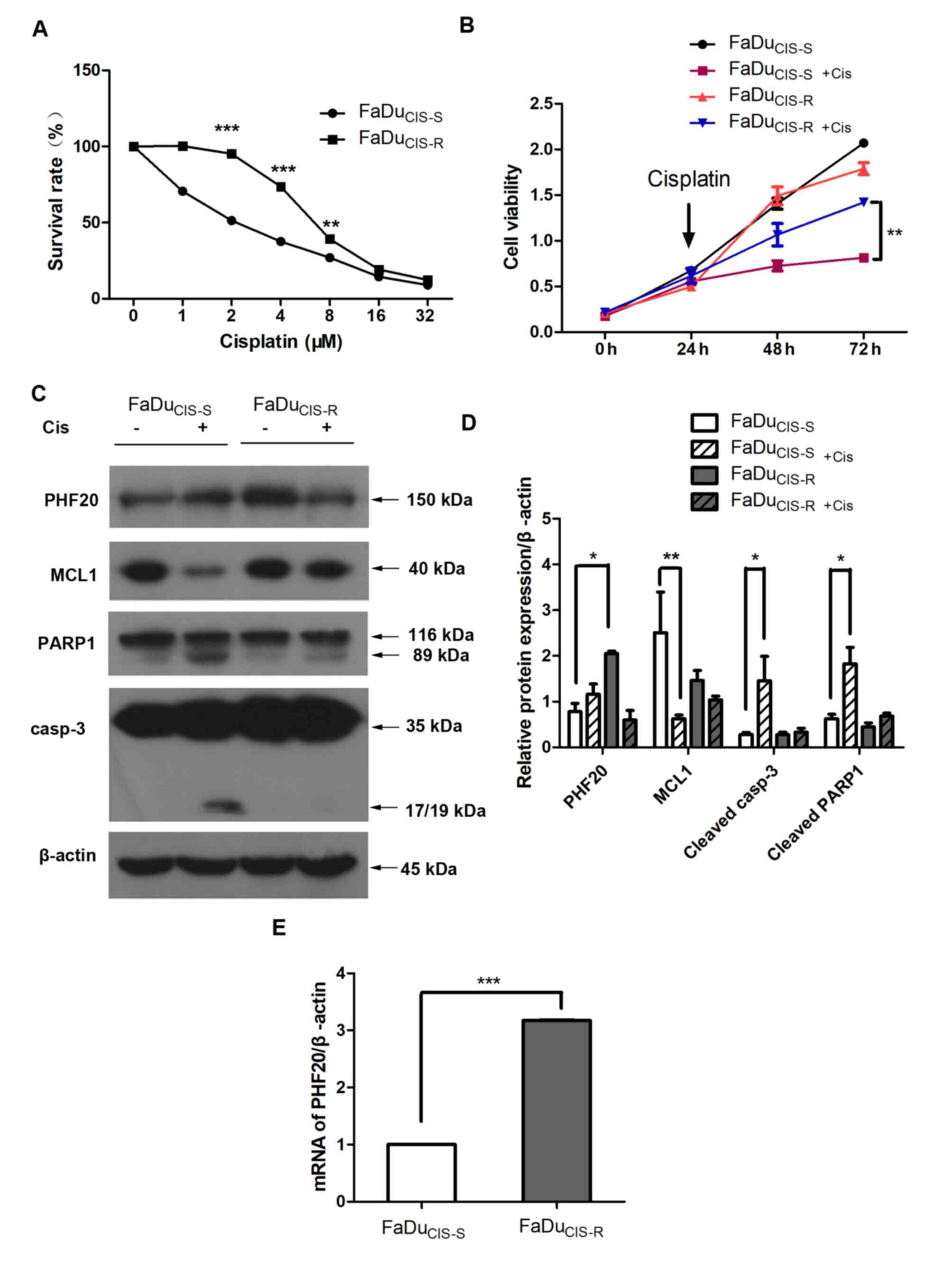
To establish a cisplatin-resistant HSCC cell line,
FaDu cells were treated with cisplatin, and the IC50 was
determined. The IC50 of FaDu cells was 2.370 µM
with a 95% confidence interval (CI) of 2.228-2.529 µM. The
IC50 of cisplatin-resistant FaDu cells was 6.937
µM with a 95% CI of 6.379-7.543 µM. To establish a
cisplatin-resistance model, FaDu cells were treated with increasing
doses of cisplatin for 6 months. Throughout the selection period,
the sensitivity of cells to cisplatin was detected via CCK-8 assay,
and a stable cell population resistant to cisplatin
(FaDuCIS-R) was obtained with a resistance index (RI) of
~2.927 (Fig. 1A) compared with
parental FaDu cells (FaDuCIS-S). Cell viability was
significantly lower in FaDuCIS-S+Cis cells compared with
that in FaDuCIS-R+Cis cells (Fig. 1B).
To comprehensively survey the genes associated with
cisplatin resistance, RNA sequencing was performed in FaDu cells
sensitive and resistant to cisplatin. The sequencing data are
available at the Sequence Read Archive database of NCBI (accession
nos. SRR13805665, SRR13805664, SRR13805661, SRR13805657,
SRR13805656 and SRR13805655). With fold-change >2 and P<0.001
used as the screening criteria in gene detection, the transcriptome
sequencing analysis identified 24,361 differently expressed genes
between FaDuCIS-R and FaDuCIS-S, including
14,487 upregulated and 9,874 downregulated genes. Intrinsic
apoptosis is one leading mechanisms of cisplatin resistance
(29); in the present study, the
RNA sequencing results (Table I)
demonstrated that MCL1, an apical molecule in apoptosis control and
promoting cell survival (30),
was also significantly upregulated in FaDuCIS-R compared
with FaDuCIS-S cells (P<0.001; Table I). In addition, at the protein
level, the expression levels of MCL1 increased, whereas the levels
of cleaved caspase-3 and PARP1 decreased in FaDuCIS-R
cells compared with those in FaDuCIS-S cells under
cisplatin treatment, indicating apoptosis (Fig. 1C and D).
Among the differentially regulated genes between the
two cell types analyzed in the present study, PHF20 was remarkably
upregulated in FaDuCIS-R cells (P<0.001; Table I). The expression levels of PHF20
were validated by RT-qPCR and western blotting.
FaDuCIS-R cells exhibited higher expression levels of
PHF20 compared with those in FaDuCIS-S cells (Fig. 1C-E), suggesting the accumulation
of PHF20 in chemoresistant HSCC cells. These results suggested the
PHF20 expression may be associated with apoptosis and
chemoresistance in HSCC cells.
Inhibition of PHF20 suppresses HSCC cell
viability by promoting apoptosis in vitro
The increased levels of PHF20 were associated with
cisplatin resistance. The anticancer mechanism of cisplatin
involves the activation of DNA damage response and intrinsic
mitochondrial apoptosis (6).
Therefore, the present study further investigated the effects of
PHF20 on the biological phenotype of HSCC cells. CCK-8, migration,
and invasion assays were performed in FaDu cells following PHF20
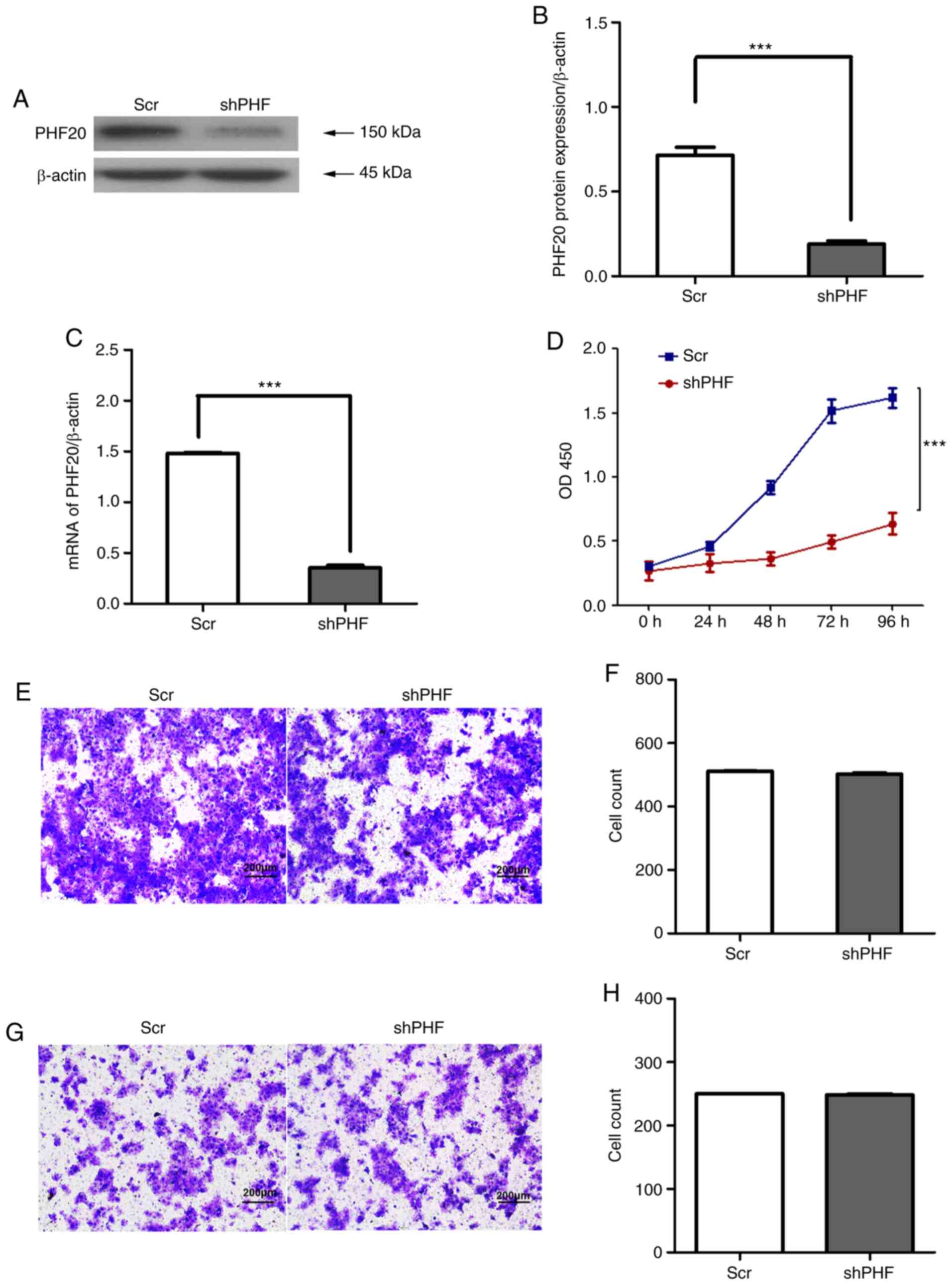
inhibition. Stable PHF20-knockdown FaDu cell lines were established
by lentivirus infection. The protein and mRNA expression levels of
PHF20 were significantly inhibited in FaDu cells following PHF20
knockdown compared with those in the control cells (Fig. 2A-C). PHF20 knockdown significantly
decreased the viability (Fig. 2D)
but did not affect the migratory (Fig. 2E and F) and invasive (Fig. 2G and H) abilities of FaDu cells
compared with those in the Scr group. In addition, the present
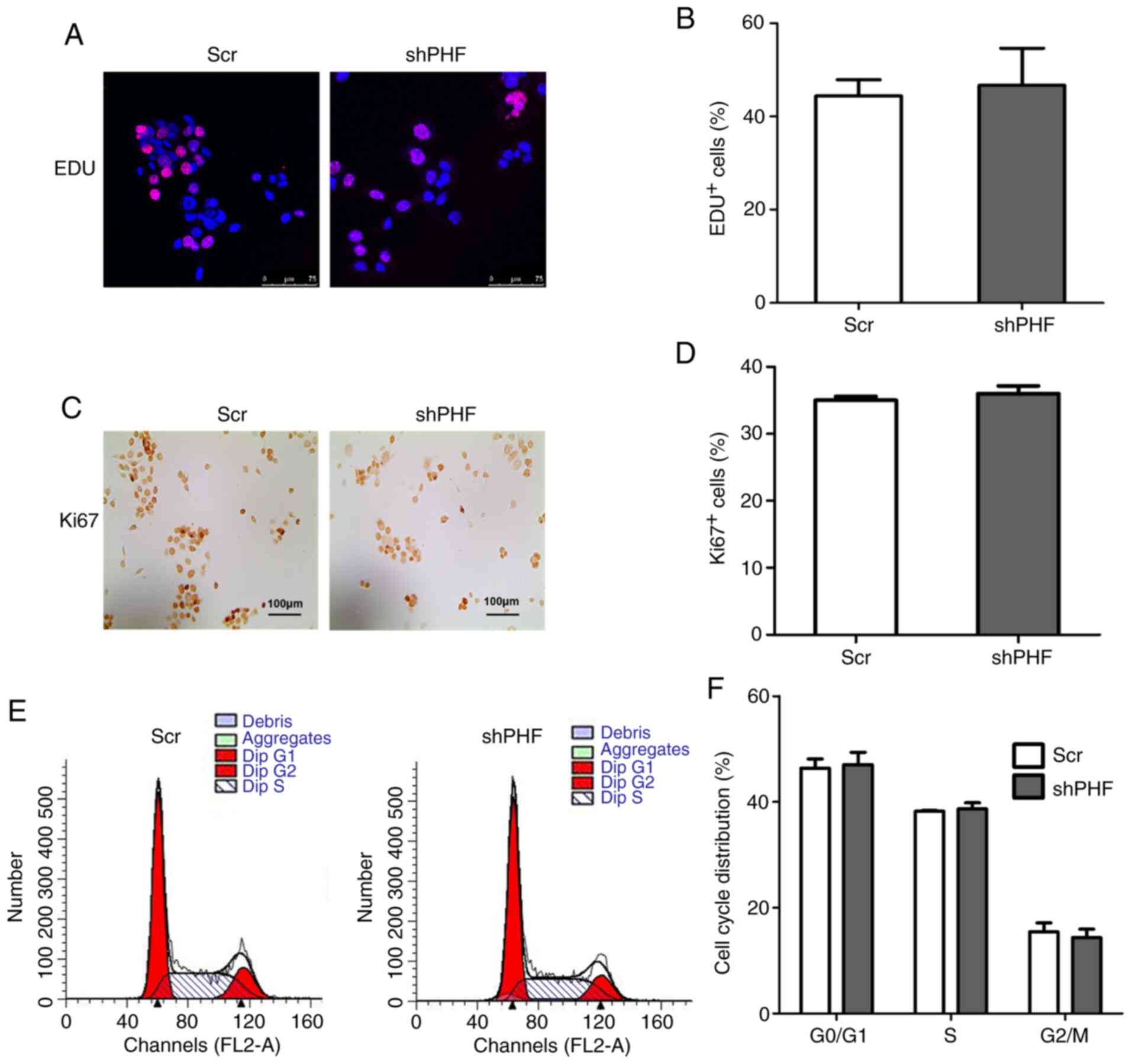
study examined whether proliferation and/or apoptosis accounted for
the decrease in the cell viability of FaDu cells following PHF20
knockdown. No notable differences were observed in the cell
proliferative ability as indicated by EDU+ and
Ki67+ cells between the two groups (Fig. 3A-D). The cell cycle phase
detection by flow cytometry revealed that PHF20 knockdown did not
affect the G0/G1 phase and S phase arrest in HSCC cells (Fig. 3E and F).
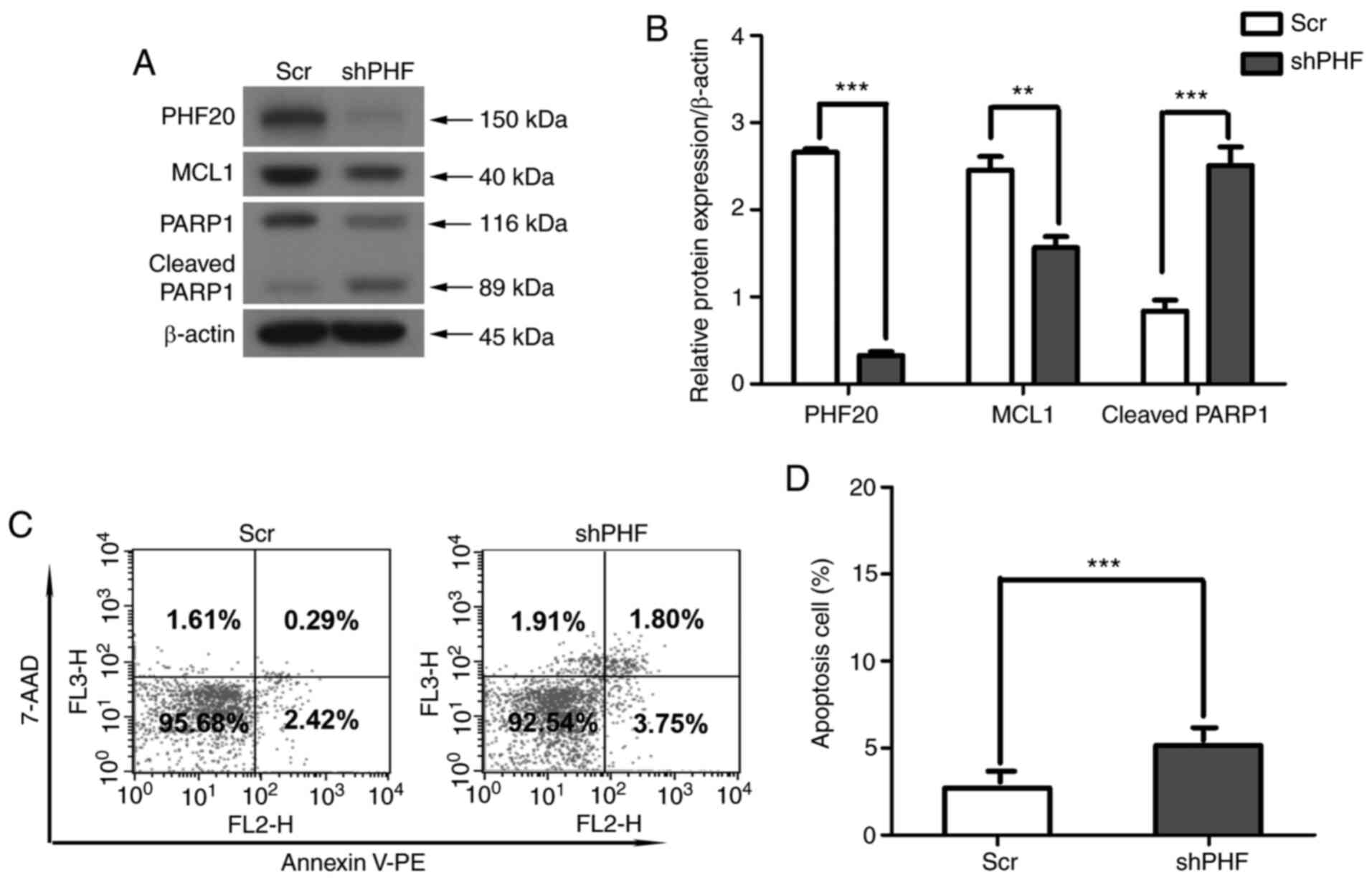
Western blotting and flow cytometry were used to
further investigate whether the PHF20 knockdown-mediated decrease
in the cell viability of FaDu cells was associated with apoptosis.
Compared with those in the control cells, the levels of cleaved
PARP1 increased in shPHF-transfected cells (Fig. 4A and B). The expression levels of
MCL1 were markedly downregulated in shPHF20 cells compared with
those in Scr cells (Fig. 4A and
B). In addition, PHF20 knockdown significantly increased the
apoptotic rate compared with that in the Scr cells, as assessed by
flow cytometry with Annexin V (Fig.
4C and D). Therefore, PHF20 knockdown inhibited cell viability
by enhancing apoptosis in vitro.
Knockdown of PHF20 improves cisplatin
sensitivity in HSCC cells
The upregulation of PHF20 was associated with
cisplatin resistance in FaDu cells. Thus, the present study further
examined the effects of PHF20 on the response of FaDu cells to
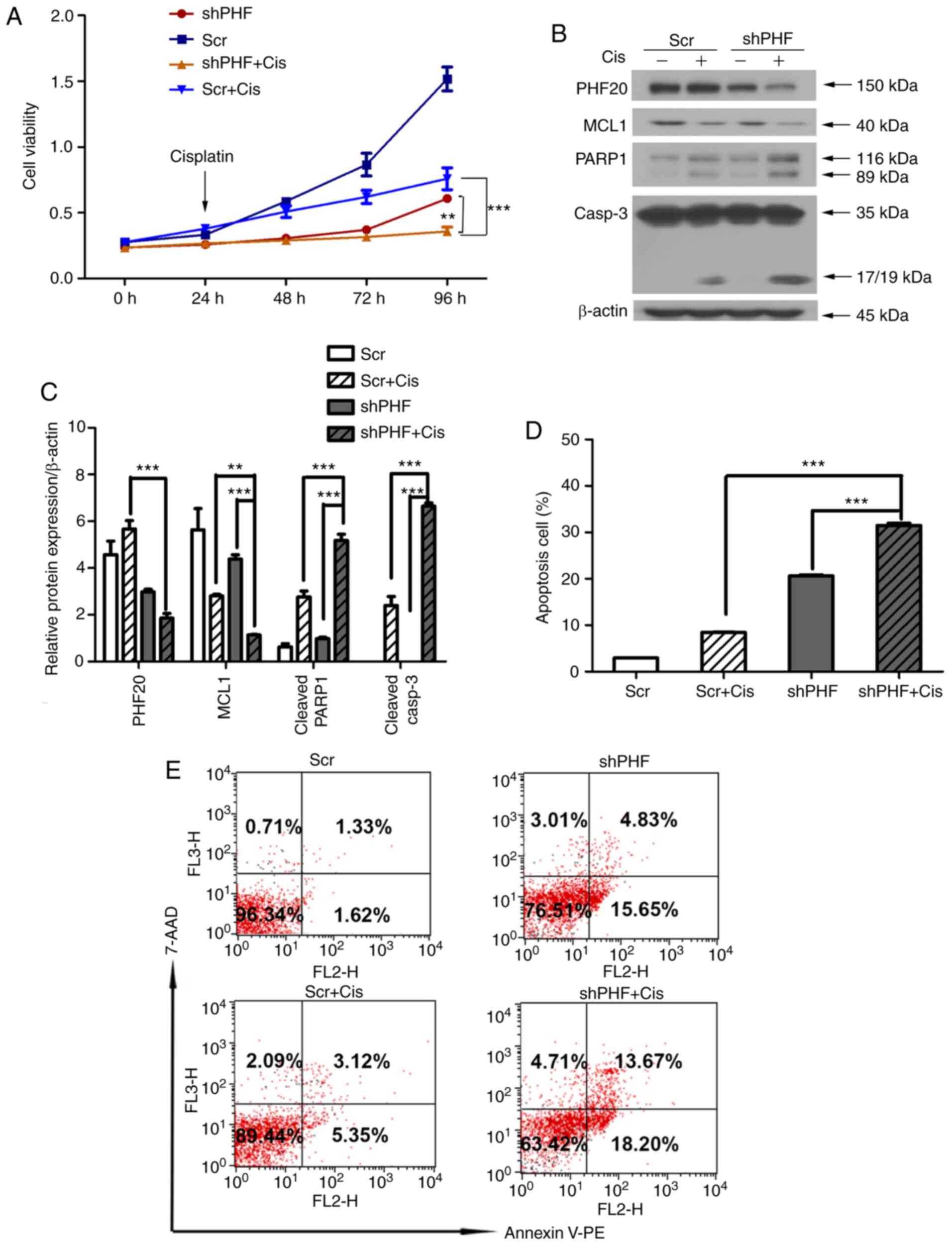
cisplatin treatment (Scr, Scr + Cis, shPHF and shPHF + Cis cells).
The cell viability was significantly inhibited in the shPHF + Cis
group compared with that in the Scr + Cis or shPHF group (Fig. 5A). Western blot analysis results
confirmed that the expression levels of MCL1 were downregulated,
whereas the levels of cleaved PARP1 and caspase 3 were upregulated
in shPHF + Cis cells compared with those in the Scr + Cis and shPHF
groups (Fig. 5B and C).
Similarly, the flow cytometry results revealed an increased number
of apoptotic cells in the shPHF + Cis group compared with those in
the Scr + Cis and shPHF groups (Fig.
5D and E). Thus, PHF20 knockdown sensitized HSCC cells to
cisplatin-induced apoptosis and effectively suppressed their
viability.
PHF20 regulates apoptosis and improves
cisplatin sensitivity via the OCT4-p-STAT3-MCL1 signaling
pathway
PHF20, as a transcriptional factor, directly binds
to the promoter region of the OCT4 gene (31), and its downregulation promotes
apoptosis (32). The present
study further examined whether OCT4 overexpression in FaDu cells
reversed MCL1-mediated apoptosis caused by PHF20 inhibition. The
mRNA the expression levels of OCT4 were lower in the shPHF cells
compared with those in Scr cells (Fig. 6A). When PHF20 was knocked down,
the levels of OCT4, p-STAT3 and MCL1 were downregulated in FaDu
cells compared with those in the control group (Fig. 6B and C). In addition, IF assay
results demonstrated that PHF20 knockdown decreased the levels and
nuclear localization of p-STAT3 in FaDu cells compared with those
in Scr cells (Fig. 6D and E).
Using co-transfection, OCT4 was overexpressed, while PHF20 was
knocked down in FaDu cells; the results demonstrated that OCT4
overexpression restored the reduction in the levels of p-STAT3,
MCL1 and cleaved PARP1 repressed by PHF20 knockdown (Fig. 7A and B). Thus, PHF20 knockdown
promoted the apoptosis of HSCC cells via OCT4.
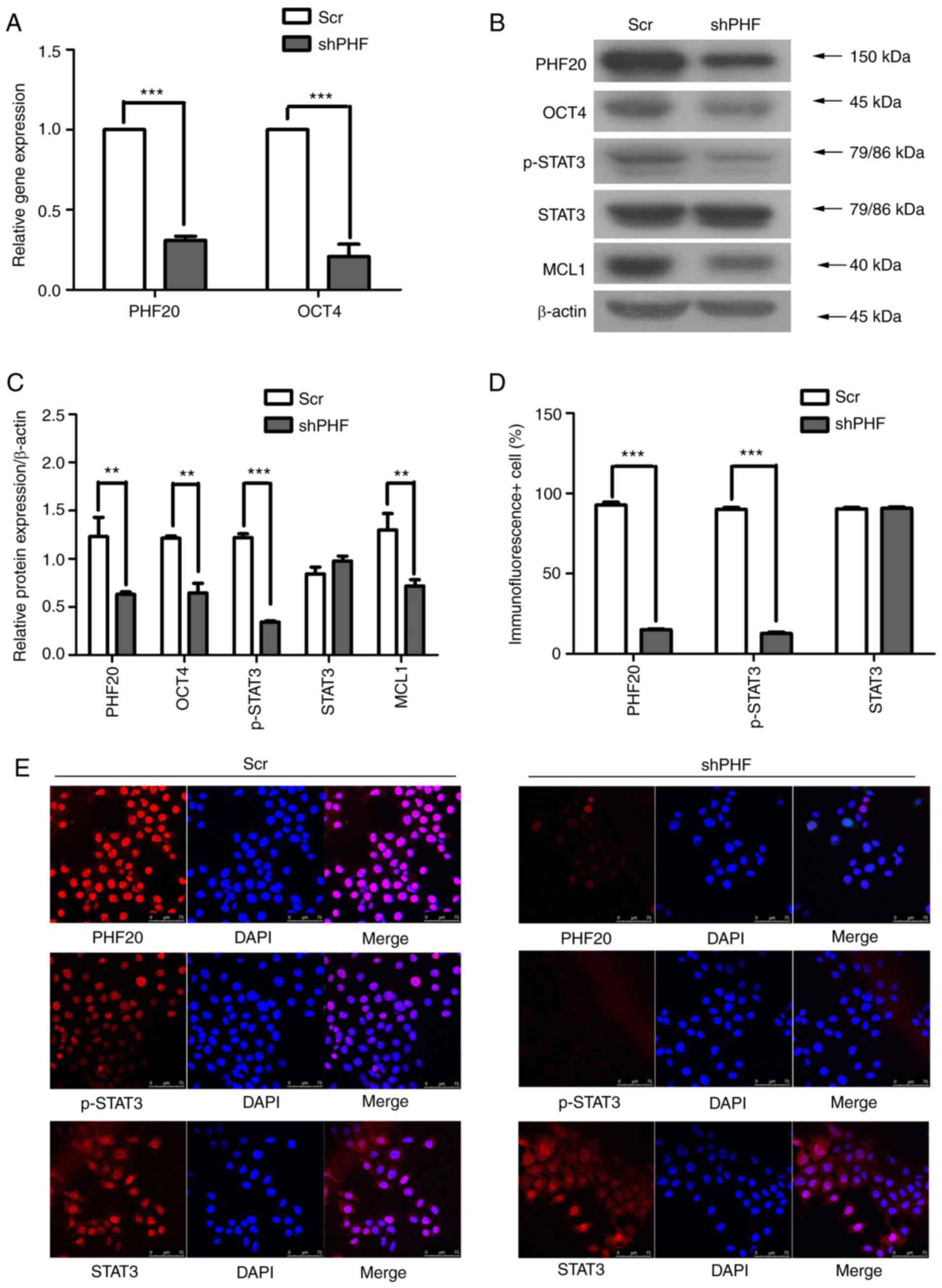 | Figure 6PHF20 silencing decreases the
expression levels of its downstream proteins OCT4, MCL1, STAT3 and
p-STAT3. (A) The mRNA expression levels of PHF20 and OCT4 were
detected by reverse transcription-quantitative PCR in
shPHF-transfected and control cells. The results revealed that the
mRNA expression levels of OCT4 were markedly decreased following
PHF20 knockdown compared with those in the Scr-transfected cells.
(B) The protein expression levels of PHF20, OCT4, MCL1, STAT3 and
p-STAT3 were determined by western blot assay in shPHF- and
Scr-transfected cells. (C) Quantitative analysis of the Western
blot results in B. The levels of OCT4, MCL1 and p-STAT3 were
downregulated in the shPHF group compared with those in the Scr
group. (D) Immunofluorescence assay was used to detect the
expression and localization of PHF20, p-STAT3 and STAT3 in shPHF20-
and Scr-transfected cells. The results demonstrated that p-STAT3
expression was associated with PHF20. Scale bar, 75 µm. (E)
Quantitative analysis of fluorescent cells in D. Comparison with
that in the shPHF group, p-STAT3 expression was lower in FaDu cells
following PHF20 knockdown. The experiment was performed in
triplicate, and unpaired t-test was used for analysis.
**P<0.01 and ***P<0.001. PHF20, plant
homeodomain finger protein 20; shPHF, lentivirus-mediated shRNA
targeting PHF20; Scr, scramble shRNA; PARP1, poly(ADP-ribose)
polymerase 1; MCL1, myeloid cell leukemia-1; STAT3, phosphorylated
signal transducer and activator of transcription 3; p-,
phosphorylated; OCT4, octamer-binding transcription factor 4. |
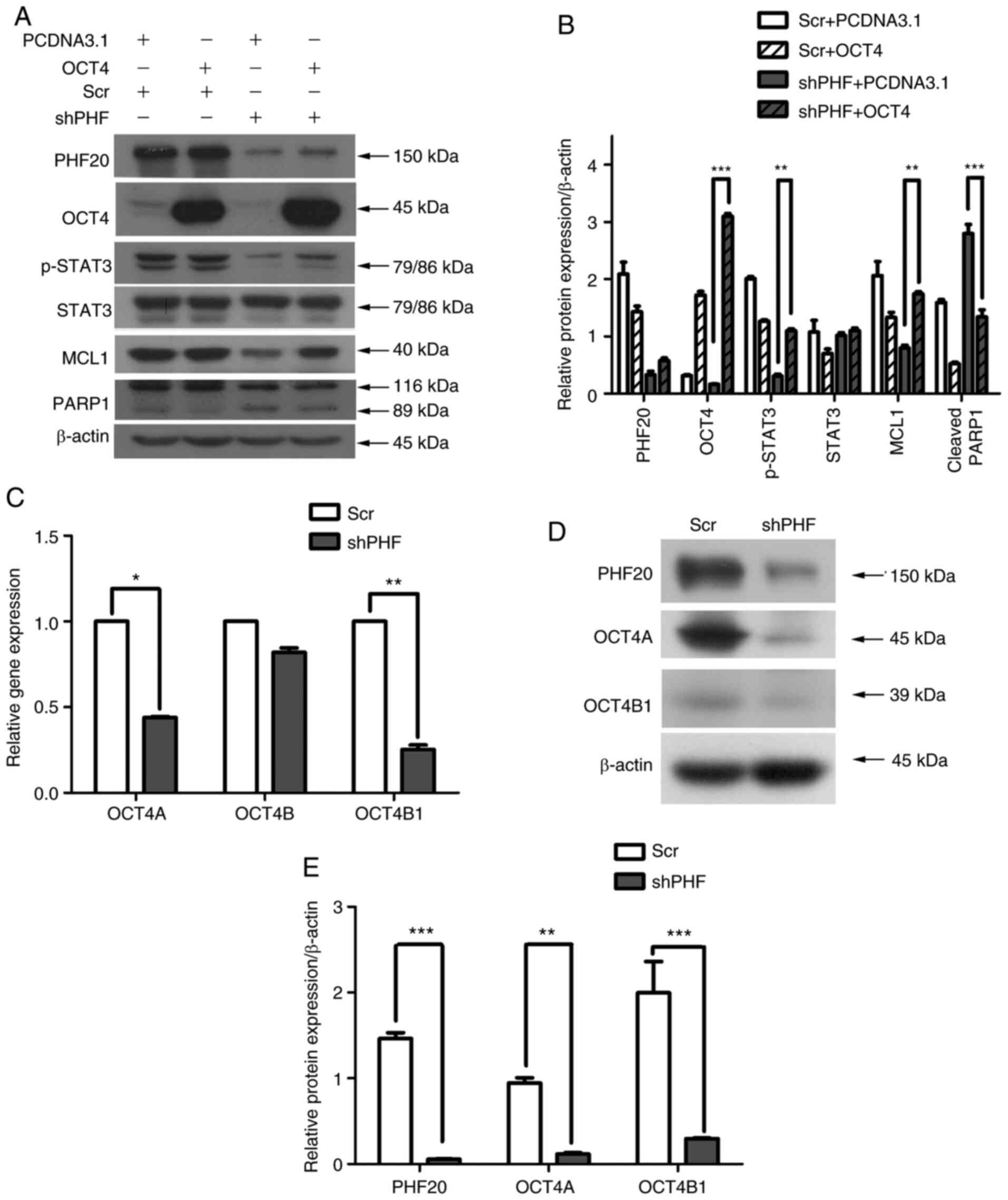 | Figure 7PHF20 regulates apoptosis and
improves cisplatin sensitivity via the OCT4-p-STAT3-MCL1 signaling
pathway. (A) Western blotting was used to determine the expression
levels of PHF20, OCT4, MCL1, PARP1, STAT3 and p-STAT3 in FaDu cells
co-transfected with lentivirus-mediated PHF20 shRNA and OCT4
overexpression plasmid. OCT4 overexpression restored the PHF20
knockdown-mediated reduction in the levels of p-STAT3, MCL1 and
cleaved PARP1. (B) Quantitative analysis of the western blot
results in A. Compared with those in the shPHF + pCDNA3.1 group,
the relative levels of p-STAT3 and MCL1 increased, whereas those of
cleaved PARP1 decreased in the shPHF + OCT4 group. Mixed model
ANOVA was used for analysis. (C) Reverse transcription-quantitative
PCR assay demonstrated that the mRNA expression levels of OCT4A and
OCT4B1 were decreased, whereas the expression levels of OCT4B were
not affected by PHF20 knockdown compared with those in the Scr
group. (D) Western blot analysis of OCT4A and OCT4B1 expression
levels in the shPHF and control groups. (E) Quantitative analysis
of the western blot results in D. Compared with those in the
control cells, the relative protein expression levels of OCT4A and
OCT4B1 were decreased in the shPHF group. All experiments were
performed thrice. **P<0.01 and
***P<0.001. PHF20, plant homeodomain finger protein
20; shPHF, lentivirus-mediated shRNA targeting PHF20; Scr, scramble
shRNA; PARP1, poly(ADP-ribose) polymerase 1; MCL1, myeloid cell
leukemia-1; STAT3, phosphorylated signal transducer and activator
of transcription 3; p-, phosphorylated; OCT4, octamer-binding
transcription factor 4. |
Among OCT4 isoforms, OCT4B1 and OCT4A serve
important roles in apoptosis (25,31,33). Therefore, we hypothesized that
OCT4B1 and OCT4A rather than other OCT4 isoforms may mediate the
apoptosis induced by PHF20 knockdown. The expression levels of
OCT4A, OCT4B and OCT4B1 were detected at the transcriptional level,
and the results demonstrated that the levels of OCT4A and OCT4B1
were significantly downregulated following PHF20 knockdown compared
with those in the Scr group, whereas OCT4B expression was
unaffected (Fig. 7C). Western
blot analysis results also confirmed the decrease in the protein
expression levels of OCT4A and OCT4B1 in the shPHF group compared
with those in the Scr group (Fig. 7D
and E). These results suggested that in HSCC cells, PHF20
knockdown increases the apoptotic rate via OCT4A and OCT4B1 rather
than the other isoforms of OCT4. Therefore, PHF20 knockdown may
promote apoptosis by inhibiting the OCT4-p-STAT3-MCL1 signaling
pathway in HSCC cells.
PHF20 inhibition suppresses
tumorigenicity via apoptosis in a xenograft model of HSCC
To investigate the role of PHF20 in vivo,
shPHF and control cells were subcutaneously injected into nude mice
in order to establish an animal xenograft model. The tumor appeared
at day 7 post-inoculation, and tumor samples were extracted on day
23. As presented in Fig. 8A and
B, the tumor volumes were significantly lower, and the tumor
growth was notably suppressed in the shPHF group compared with
those in the Scr group. Histological analysis and TUNEL assays were
performed on the tumor tissue sections from the mice in the two
groups. Tumors in the shPHF group exhibited a remarkable decrease
in PHF20 levels compared with those in the Scr group (Fig. 8C and D). Consistent with the in
vitro results, the number of Ki67+ cells did not
differ significantly in the tumor tissues between the two groups
(Fig. 8C and D). Notably, shPHF
tumors exhibited a higher number of apoptotic TUNEL+
cells, whereas the levels of p-STAT3 decreased compared with those
in the Scr group (Fig. 8C and D).
Collectively, these results demonstrated that PHF20 knockdown
inhibited tumor growth by promoting apoptosis in vivo.
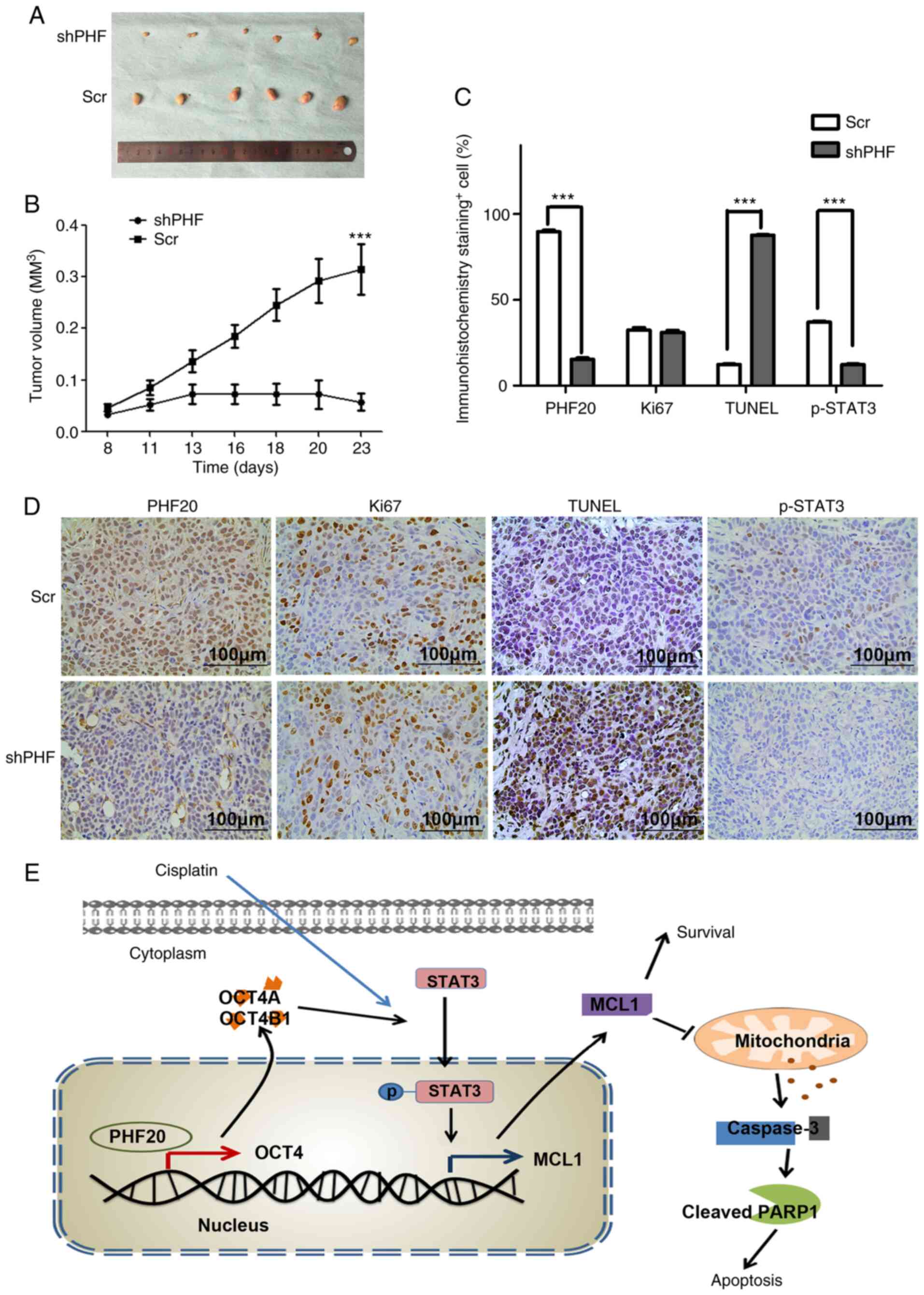 | Figure 8PHF20 inhibition suppresses
tumorigenicity via apoptosis in the HSCC cell xenograft model. (A)
Representative images of tumors isolated from mice in the shPHF20
and control groups. (B) The tumor growth curves in the two groups.
(C) Quantitative analysis of positive cells in (D) the
immunohistochemical staining of PHF20, Ki67, TUNEL and p-STAT3 in
tumor samples from mice in the two groups. Magnification, ×400.
Scale bar, 100 µm. Compared with those in the Scr group, the
percentage of apoptotic cells indicated by TUNEL staining was
significantly increased, the number of p-STAT3+ cells
decreased, and the number of Ki67+ cells did not
obviously differ in the shPHF group. (E) Schematic representation
illustrating how the knockdown of PHF20 may synergize with
cisplatin to promote apoptosis via the OCT4-p-STAT3-MCL1 signaling
pathway. PHF20, plant homeodomain finger protein 20; shPHF,
lentivirus-mediated shRNA targeting PHF20; Scr, scramble shRNA;
PARP1, poly(ADP-ribose) polymerase 1; MCL1, myeloid cell
leukemia-1; casp-3, caspase-3; STAT3, phosphorylated signal
transducer and activator of transcription 3; p-STAT3,
phosphorylated signal transducer and activator of transcription 3;
OCT4, octamer-binding transcription factor 4; TUNEL, terminal
deoxynucleotidyl transferase-mediated dUTP-biotin nick end
labeling. |
Discussion
Resistance to cisplatin, a DNA-damaging agent, is a
major obstacle for its clinical use in HSCC. In the present study,
the levels of PHF20 were upregulated in FaDuCIS-R compared with
those in FaDuCIS-S cells, as indicated by RNA sequencing, RT-qPCR
and western blotting results. The effects of PHF20 in HSCC and
cisplatin sensitivity, as well as the underlying molecular
mechanisms, were further investigated. PHF20 knockdown
significantly decreased the viability of FaDu cells by accelerating
apoptosis, and the tumorigenic role of PHF20 was observed in a
mouse xenograft model, where more apoptotic cells were present in
the shPHF group compared with the Scr group. PHF20 knockdown
sensitized FaDu cells to cisplatin treatment. Notably, cisplatin
and PHF20 knockdown synergistically promoted the apoptosis of FaDu
cells via the OCT4-p-STAT3-MCL1 signaling pathway. These results
suggested a potential underlying mechanism by which knockdown of
PHF20 may promote apoptosis in vitro and in vivo and
improve cisplatin-based chemosensitivity in HSCC cells. A schematic
diagram of the proposed mechanism is presented in Fig. 8E.
PHF20 is an antigen in patients with glioblastoma
that is highly expressed in various types of cancer, including
non-small cell lung cancer, glioblastoma and nasopharyngeal
carcinoma (12,14) and participates in the development
and progression of glioma, adenocarcinomas and lung cancer
(12). NF-κB, which promotes the
development of inflammation-associated cancer, is activated by high
expression levels of PHF20 (34).
Cui et al (35) have
reported that PHF20 stabilizes p53 through dimethylated lysine
residues for cell survival and carcinogenic activity. The results
of the present study provided valuable information for improved
understanding of the role of PHF20 knockdown in regulating
apoptosis via the OCT4-p-STAT3-MCL1 signaling pathway in HSCC.
Cisplatin is one of the most potent chemotherapeutic
regimens that is widely used for the treatment of various types of
solid tumor, including head and neck cancer, in clinical practice
(36,37). However, its clinical efficiency is
limited by chemoresistance and side effects, which includes mainly
nephrotoxicity and bone marrow toxicity (29,38). Therefore, the molecular mechanisms
underlying chemoresistance induced by cisplatin need to be studied
in detail. In the present study, the expression levels of PHF20 in
FaDuCIS-R cells were higher compared with those in
FaDuCIS-S cells, suggesting that PHF20 may affect the
chemoresistance of HSCC. Notably, PHF20 knockdown attenuated
cisplatin resistance and improved the sensitivity of HSCC cells to
cisplatin, thus enhancing apoptosis. To further validate the
synergistic effect of PHF20 knockdown and cisplatin treatment in
vivo, tumor-bearing mice were intravenously injected with
cisplatin. No notable tumor regression was observed in the shPHF
plus cisplatin group compared with that in the shPHF group (data
not shown). This result supported previous findings (39,40), suggesting that FaDu cells were not
sensitive to cisplatin treatment in vivo. FaDu cells are the
only currently available hypopharyngeal cancer cell line (41). Thus, additional HSCC cell lines
are required to validate the results of the present study in
vivo. However, the results of the present study demonstrated
that PHF20 may serve as a potential marker for cisplatin-resistant
HSCC and a therapeutic target to combine with cisplatin to induce
cell death.
OCT4 is a key transcriptional factor involved in
apoptosis (23,42,43). In the present study, decreased
protein expression levels of MCL1 caused by PHF20 knockdown were
significantly reversed by co-transfection with a OCT4
overexpression plasmid in FaDu cells. The human OCT4 gene generates
three main isoforms, namely OCT4A (44), OCT4B (45) and OCT4B1 (42), by alternative splicing. OCT4A,
generally referred to as OCT4, is a pivotal transcriptional factor
that sustains the stemness properties of pluripotent cells
(46). OCT4B serves an
antiapoptotic role in lung adenocarcinoma A549 cells (43). In gastric adenocarcinoma, bladder
and brain cancer cell lines, suppression of OCT4B1 induces the
upregulation of proapoptotic genes such as cell death-Inducing
DFFA-like effector A, and downregulation of antiapoptotic genes
such as Bcl2 and Bax (47). The
knockdown of OCT4B1 increases the apoptotic rate in gastric cancer
due to cell cycle alterations (42). OCT4A induces apoptosis in breast
cancer cells (22) and human
endometrial cancer (24). PHF20
acts as a transcriptional factor binding to the promotor of OCT4 to
promote cellular reprogramming (14,31). However, the role of PHF20 in the
regulation of OCT4 isoform expression has not been fully
determined. The knockdown of PHF20 remarkably downregulated the
mRNA and protein levels of OCT4A and OCT4B1 expression, but did not
affect the expression levels of OCT4B compared with those in the
Scr group in the present study. Thus, OCT4A and OCT4B1 may mediate
PHF20 knockdown-induced apoptosis by regulating the levels of
p-STAT3 and MCL1.
In conclusion, PHF20 inhibition may promote
apoptosis in FaDu cells via the OCT4-p-STAT3-MCL1 signaling pathway
and sensitize FaDu cells to cisplatin treatment.
Supplementary Data
Availability of data and materials
The RNA sequencing data from the current study has
been deposited at the Sequence Read Archive (SRA) database of NCBI
(PRJNA705388). Raw data and details of RNA sequencing experiments
are available online at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA705388.
Other datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
XiuL and WX conceived, supervised and designed the
study. XiuL, ZZ, SK, ZL and SZ performed the experiments. XiuL,
XiaL and PJ analyzed the data. SK prepared the figures. XiuL and ZZ
wrote the manuscript. XiuL and WX confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Committee for
the Animal Care and Use of Shandong Provincial ENT Hospital
(approval no. XYK-NSFC20190201; Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by The National Natural Science
Foundation of China (grant nos. 81702679 and 81902763) and Major
Programs of Science and Technology Projects in Hainan Province
(grant no. ZDKJ202005).
Abbreviations:
|
HSCC
|
hypopharyngeal squamous cell
carcinoma
|
|
Cis
|
cisplatin
|
|
Bcl-2
|
B-cell lymphoma 2
|
|
MCL1
|
myeloid cell leukemia-1
|
|
PHF20
|
plant homeodomain finger protein
20
|
References
|
1
|
Takes RP, Strojan P, Silver CE, Bradley
PJ, Haigentz M Jr, Wolf GT, Shaha AR, Hartl DM, Olofsson J,
Langendijk JA, et al: Current trends in initial management of
hypopharyngeal cancer: The declining use of open surgery. Head
Neck. 34:270–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spector JG, Sessions DG, Haughey BH, Chao
KS, Simpson J, El Mofty S and Perez CA: Delayed regional
metastases, distant metastases, and second primary malignancies in
squamous cell carcinomas of the larynx and hypopharynx.
Laryngoscope. 111:1079–1087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bova R, Goh R, Poulson M and Coman WB:
Total pharyngolaryngectomy for squamous cell carcinoma of the
hypopharynx: A review. Laryngoscope. 115:864–869. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bandu R, Ahn HS, Lee JW, Kim YW, Choi SH,
Kim HJ and Kim KP: Liquid chromatography electrospray ionization
tandem mass spectrometric (LC/ESI-MS/MS) Study for the
identification and characterization of in vivo metabolites of
cisplatin in rat kidney cancer tissues: Online Hydrogen/Deuterium
(H/D) exchange study. PLoS One. 10:e01340272015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu DW, Lee MC, Hsu NY, Wu TC, Wu JY, Wang
YC, Cheng YW, Chen CY and Lee H: FHIT loss confers cisplatin
resistance in lung cancer via the AKT/NF-κB/Slug-mediated PUMA
reduction. Oncogene. 34:2505–2515. 2015. View Article : Google Scholar
|
|
6
|
Galluzzi L, Senovilla L, Vitale I, Michels
J, Martins I, Kepp O, Castedo M and Kroemer G: Molecular mechanisms
of cisplatin resistance. Oncogene. 31:1869–1883. 2012. View Article : Google Scholar
|
|
7
|
Degterev A, Boyce M and Yuan J: A decade
of caspases. Oncogene. 22:8543–8567. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: Function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging. 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Li W, Xia Z, Xie L, Ma X, Liang Q,
Liu L, Wang J, Zhou X, Yang Y and Liu H: Targeting MCL-1 sensitizes
human esophageal squamous cell carcinoma cells to cisplatin-induced
apoptosis. BMC Cancer. 17:4492017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Badeaux AI, Yang Y, Cardenas K,
Vemulapalli V, Chen K, Kusewitt D, Richie E, Li W and Bedford MT:
Loss of the methyl lysine effector protein PHF20 impacts the
expression of genes regulated by the lysine acetyltransferase MOF.
J Biol Chem. 287:429–437. 2012. View Article : Google Scholar :
|
|
12
|
Bankovic J, Stojsic J, Jovanovic D,
Andjelkovic T, Milinkovic V, Ruzdijic S and Tanic N: Identification
of genes associated with non-small-cell lung cancer promotion and
progression. Lung Cancer. 67:151–159. 2010. View Article : Google Scholar
|
|
13
|
Tang N, Ma L, Lin XY, Zhang Y, Yang DL,
Wang EH and Qiu XS: Expression of PHF20 protein contributes to good
prognosis of NSCLC and is associated with Bax expression. Int J
Clin Exp Pathol. 8:12198–12206. 2015.
|
|
14
|
Long W, Zhao W, Ning B, Huang J, Chu J, Li
L, Ma Q, Xing C, Wang HY, Liu Q and Wang RF: PHF20 collaborates
with PARP1 to promote stemness and aggressiveness of neuroblastoma
cells through activation of SOX2 and OCT4. J Mol Cell Biol.
10:147–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji J and Zheng PS: Expression of Sox2 in
human cervical carcinogenesis. Hum Pathol. 41:1438–1447. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wen J, Park JY, Park KH, Chung HW, Bang S,
Park SW and Song SY: Oct4 and Nanog expression is associated with
early stages of pancreatic carcinogenesis. Pancreas. 39:622–626.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei D, Kanai M, Jia Z, Le X and Xie K:
Kruppel-like factor 4 induces p27Kip1 expression in and suppresses
the growth and metastasis of human pancreatic cancer cells. Cancer
Res. 68:4631–4639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang YD, Cai N, Wu XL, Cao HZ, Xie LL and
Zheng PS: OCT4 promotes tumorigenesis and inhibits apoptosis of
cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis.
4:e7602013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang G, Zhou H, Gu Z, Gao Q and Shen G:
Oct4 promotes cancer cell proliferation and migration and leads to
poor prognosis associated with the survivin/STAT3 pathway in
hepatocellular carcinoma. Oncol Rep. 40:979–987. 2018.PubMed/NCBI
|
|
21
|
Meng F, Sun G, Zhong M, Yu Y and Brewer
MA: Anticancer efficacy of cisplatin and trichostatin A or
5-aza-2′-deoxycytidine on ovarian cancer. Br J Cancer. 108:579–586.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Zhang Y, Xu J, Wang H, Zheng X,
Lou Y and Han B: Antigen presentation of the Oct4 and Sox2 peptides
by CD154-activated B lymphocytes enhances the killing effect of
cytotoxic T lymphocytes on tumor stem-like cells derived from
cisplatin-resistant lung cancer cells. J Cancer. 9:367–374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X, Ma M, Duan X, Zhang H and Yang M:
Knockdown of OCT4 may sensitize NSCLC cells to cisplatin. Clin
Transl Oncol. 19:587–592. 2017. View Article : Google Scholar
|
|
24
|
Lu CS, Shieh GS, Wang CT, Su BH, Su YC,
Chen YC, Su WC, Wu P, Yang WH, Shiau AL and Wu CL:
Chemotherapeutics-induced Oct4 expression contributes to drug
resistance and tumor recurrence in bladder cancer. Oncotarget.
8:30844–30858. 2017. View Article : Google Scholar :
|
|
25
|
Kato M, Onoyama I, Yoshida S, Cui L,
Kawamura K, Kodama K, Hori E, Matsumura Y, Yagi H, Asanoma K, et
al: Dual-specificity phosphatase 6 plays a critical role in the
maintenance of a cancer stem-like cell phenotype in human
endometrial cancer. Int J Cancer. 147:1987–1999. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L, Feng Z, Wang X and Zhang X:
DEGseq: An R package for identifying differentially expressed genes
from RNA-seq data. Bioinformatics. 26:136–138. 2010. View Article : Google Scholar
|
|
27
|
Goldie FC, Fulton RL, Dawson J, Bluhmki E
and Lees KR: Exploration of time-course combinations of outcome
scales for use in a global test of stroke recovery. Int J Stroke.
9:755–758. 2014. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View Article : Google Scholar
|
|
31
|
Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q,
Chen Y, Wang HY and Wang RF: Jmjd3 inhibits reprogramming by
upregulating expression of INK4a/Arf and targeting PHF20 for
ubiquitination. Cell. 152:1037–1050. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu T, Zhang T, Zhou F, Wang J, Zhai X, Mu
N, Park J, Liu M, Liu W, Shang P, et al: Identification of genes
and pathways potentially related to PHF20 by gene expression
profile analysis of glioblastoma U87 cell line. Cancer Cell Int.
17:872017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ramezankhani B, Taha MF and Javeri A:
Vitamin C counteracts miR-302/367-induced reprogramming of human
breast cancer cells and restores their invasive and proliferative
capacity. J Cell Physiol. 234:2672–2682. 2019. View Article : Google Scholar
|
|
34
|
Zhang D, Ma Q, Shen S and Hu H: Inhibition
of pancreatic cancer cell proliferation by propranolol occurs
through apoptosis induction: The study of beta-adrenoceptor
antagonist's anticancer effect in pancreatic cancer cell. Pancreas.
38:94–100. 2009. View Article : Google Scholar
|
|
35
|
Cui G, Park S, Badeaux AI, Kim D, Lee J,
Thompson JR, Yan F, Kaneko S, Yuan Z, Botuyan MV, et al: PHF20 is
an effector protein of p53 double lysine methylation that
stabilizes and activates p53. Nat Struct Mol Biol. 19:916–924.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qin Z, Ren G, Yuan J, Chen H, Lu Y, Li N,
Zhang Y, Chen X and Zhao D: Systemic evaluation on the
pharmacokinetics of platinum-based anticancer drugs from animal to
cell level: Based on total platinum and intact drugs. Front
Pharmacol. 10:14852019. View Article : Google Scholar
|
|
37
|
Pointreau Y, Garaud P, Chapet S, Sire C,
Tuchais C, Tortochaux J, Faivre S, Guerrif S, Alfonsi M and Calais
G: Randomized trial of induction chemotherapy with cisplatin and
5-fluorouracil with or without docetaxel for larynx preservation. J
Natl Cancer Inst. 101:498–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Knox RJ, Friedlos F, Lydall DA and Roberts
JJ: Mechanism of cytotoxicity of anticancer platinum drugs:
Evidence that cis-di amminedichloroplatinum(II) and
cis-diammine-(1,1-cyclobutanedicarboxylato)platinum(II) differ only
in the kinetics of their interaction with DNA. Cancer Res.
46:1972–1979. 1986.PubMed/NCBI
|
|
39
|
Yang J, Ju Z and Dong S: Cisplatin and
paclitaxel co-delivered by folate-decorated lipid carriers for the
treatment of head and neck cancer. Drug Deliv. 24:792–799. 2016.
View Article : Google Scholar
|
|
40
|
Khan Z, Khan AA, Prasad GB, Khan N, Tiwari
RP and Bisen PS: Growth inhibition and chemo-radiosensitization of
head and neck squamous cell carcinoma (HNSCC) by survivin-siRNA
lentivirus. Radiother Oncol. 118:359–368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bu M, Liu X and Xu W: Upregulation of
fascin-1 is involved in HIF-1α-dependent invasion and migration of
hypopharyngeal squamous cell carcinoma. Int J Oncol. 55:488–498.
2019.PubMed/NCBI
|
|
42
|
Asadi MH, Mowla SJ, Fathi F, Aleyasin A,
Asadzadeh J and Atlasi Y: OCT4B1, a novel spliced variant of OCT4,
is highly expressed in gastric cancer and acts as an antiapoptotic
factor. Int J Cancer. 128:2645–2652. 2011. View Article : Google Scholar
|
|
43
|
Cortes-Dericks L, Yazd EF, Mowla SJ,
Schmid RA and Karoubi G: Suppression of OCT4B enhances sensitivity
of lung adenocarcinoma A549 cells to cisplatin via increased
apoptosis. Anticancer Res. 33:5365–5373. 2013.PubMed/NCBI
|
|
44
|
Li SW, Wu XL, Dong CL, Xie XY, Wu JF and
Zhang X: The differential expression of OCT4 isoforms in cervical
carcinoma. PLoS One. 10:e01180332015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Zhao Y, Xiao Z, Chen B, Wei Z,
Wang B, Zhang J, Han J, Gao Y, Li L, et al: Alternative translation
of OCT4 by an internal ribosome entry site and its novel function
in stress response. Stem Cells. 27:1265–1275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsai SC, Chang DF, Hong CM, Xia P,
Senadheera D, Trump L, Mishra S and Lutzko C: Induced
overexpression of OCT4A in human embryonic stem cells increases
cloning efficiency. Am J Physiol Cell Physiol. 306:C1108–1118.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mirzaei MR, Najafi A, Arababadi MK, Asadi
MH and Mowla SJ: Altered expression of apoptotic genes in response
to OCT4B1 suppression in human tumor cell lines. Tumour Biol.
35:9999–10009. 2014. View Article : Google Scholar : PubMed/NCBI
|