Introduction
Cutaneous squamous cell carcinoma (CSCC) is a
malignant hyperplasia of the skin epithelium, accounting for 20-50%
of all skin cancers (1). In
addition to ultraviolet radiation, other CSCC risk factors include
chronic immunosuppression, chronic skin disease, genetic disease,
ionizing radiation, and long-term arsenic exposure (2). Patients afflicted with CSCC display
a large number of precursor lesions of actinic keratosis, which
typically present as papules, plaques or sclerotic nodules, with
smooth, scaly, verrucous or ulcerative surfaces (2). Although most CSCCs show a relatively
mild course, depending on the location, size, invasion degree,
tissue differentiation, perineural invasion, and immune deficiency
of tumors, they may have a local invasion and metastatic potential
(3). A recent meta-analysis has
suggested that risk factors include a diameter >20 mm, an ear or
lip tumor location, invasion beyond the subcutaneous fat and the
presence of perineural invasion, which have statistically
significant associations with disease-specific CSCC death (4). In view of the histological grade of
histopathological high-risk features, a recent study has shown that
the 5-year metastasis-free and overall survival rates are
noticeably higher in well-differentiated tumors (~70%) compared
with moderately differentiated (~51%) and poorly differentiated
(~26%) tumors (3). Once the
cancer is established, autophagy helps tumor development and allows
tumor cells to survive under starvation or hypoxia (5). Autophagy inhibition has been
identified as an effective therapy for CSCC treatment (6). In view of these previous reports,
the present study aimed to identify new cell therapies for CSCC
based on autophagy inhibition.
Long non-coding RNAs (lncRNAs) are important
regulators of various biological processes, including cancer
stemness and tumorigenesis (7),
and their roles in CSCC cell behavior have also been widely
investigated (8). LncRNA human
histocompatibility leukocyte antigen complex P5 (HCP5) has been
implicated in adaptive and innate immune systems and has been
correlated with the development of various autoimmune diseases and
cancers, as reviewed by Kulski (9). In addition, lncRNAs can act as
competitive endogenous RNAs (ceRNAs) to sponge microRNAs
(miRNAs/miRs) by competing with mRNA to bind to miRNA, thus
regulating miRNA and mRNA expression patterns, and finally
affecting tumor processes (10).
HCP5 expression is positively associated with the pathological
grade of gliomas, and HCP5 knockout exerts an antitumoral
effect on gliomas through the HCP5/miR-139/Runt-related
transcription factor 1 feedback loop (11). Additionally, HCP5 levels are
decreased in skin cutaneous melanoma tissues and are associated
with an undesirable overall survival and progression (12). Enhancer of zeste homolog 2 (EZH2)
serves crucial roles in a range of biological processes, including
organ development and homeostasis, gene repression and DNA damage
repair (13). EZH2 is a polycomb
group protein that is involved in the progression of a number of
human cancers, including CSCC (14). EZH2 regulates cancer cell
autophagy (15,16), thus, it was also a focus of the
present study. However, there is little research on the mechanism
of HCP5 and EZH2 in CSCC progression; therefore, the present study
aimed to discern a ceRNA network involving HCP5 in CSCC cells.
Materials and methods
Microarray analysis
Using the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo), five
healthy and eight tumor samples were obtained from a CSCC
microarray (GSE66359); the healthy samples were used as the
control. The limma package in R was used to screen the
differentially expressed genes using a |logFC| >1 and a
P<0.05 as the screening standards. The upstream miRNAs of EZH2
were predicted using the TargetScan 7.1 (http://www.targetscan.org/vert_71), mirDIP 4.1
(http://ophid.utoronto.ca/mirDIP/index.jsp#r),
miRSearch V3.0 (https://www.exiqon.com/miRSearch), and miRDB
(http://mirdb.org) databases. The upstream lncRNAs of
miR-138-5p were predicted using the RNA22 2.0 database (https://cm.jefferson.edu/rna22).
Tissue collection
Between October 2016 and October 2018, cancer
tissues and healthy skin tissues were collected from 60 patients
with CSCC (33 male; 27 female; age, 53.6±8.1 years; body mass
index, 22.61±1.08) admitted to The First Affiliated Hospital of
Zhengzhou University (Zhengzhou, China). Patients were excluded if
they had incomplete clinical data, mental or consciousness
disorders, other primary malignant tumors, autoimmune diseases,
serious organic diseases, important organ dysfunction or
coagulation dysfunction, if they were pregnant or lactating women,
and if they had an allergic constitution or related
contraindications.
Cell grouping and transfections
CSCC cell lines (A431, COLO-16, SCC13, SCL-1, HSC-1,
and HSC-5) and the human immortalized keratinocyte HaCaT cell line
(all purchased from American Type Culture Collection) were cultured
in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100
U/ml streptomycin at 37°C and 5% CO2.
EZH2 cDNA and lncRNA HCP5 were cloned into
pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) to construct
the overexpression vectors. miR-138-5p mimics, mimics negative
control (NC), miR-138-5p inhibitor, inhibitor NC (inhi-NC), small
interfering RNA (si)-HCP5-1, si-HCP5-2 and si-NC were designed and
synthesized by Shanghai GenePharma Co., Ltd. The details are
provided in Table I.
 | Table IsiRNAs and miRNA mimics
sequences. |
Table I
siRNAs and miRNA mimics
sequences.
| Name | Sequence
(5′→3′) |
|---|
| si-HCP5-1 |
GGCTCAACTCACAAGAAAC |
| si-HCP5-2 |
TTCTCCGAACGTGTCACGT |
| si-NC | TTCTCCGAACGTGTCA
CGT |
| miR-138-5p
mimics |
AGCUGGUGUUGUGAAUCAGGCCG |
| mimics-NC |
CAGUACUUUUGUGUAGUACAA |
| miR-138-5p
inhi |
CGGCCUGAUUCACAACACCAGCU |
| inhi-NC |
CAGUACUUUUGUGUAGUACAA |
si-HCP5-1, si-HCP5-2, si-NC, miR-138-5p mimics and
mimics-NC, miR-138-5p inhibitor and inhi-NC were transfected into
A431 cells. pcDNA3.1-NC and pcDNA3.1-HCP5 were transfected into
SCL-1 cells. miR-138-5p mimics and mimics-NC were transfected into
SCL-1 cells and SCL-1 cells overexpressing HCP5. pcDNA3.1-NC and
pcDNA3.1-EZH2 were co-transfected into SCL-1 cells and A431
cells with si-HCP-1 and si-HCP5-2; SCL-1 cells transfected with
pcDNA3.1-EZH2 were pre-treated with STAT3/VEGFR2 pathway
inhibitor AG-490 (10 nM; AmyJet Scientific Co., Ltd.) for 24 h at
37°C (17). Briefly, cells to be
transfected were seeded in 6-well plates (1.0×105
cells/well) and grown overnight for 18 h. Cells at 80% confluence
were transfected with 100 pmol siRNAs, miRNA mimics, miRNA
inhibitors or the respective controls using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. After
48 h of incubation at 37°C, cells were collected for subsequent
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was obtained by the one-step method using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
and high-quality RNA was confirmed using ultraviolet analysis
(A260/280) and formaldehyde denaturation electrophoresis. qPCR was
performed following the manufacturer's instructions of TaqMan™
Sample-to-SNP™ kit (cat. no. 4403313; Applied Biosystems; Thermo
Fisher Scientific, Inc.). PCR primers were designed and synthesized
by Sangon Biotech Co., Ltd. and are listed in Table II. GAPDH was used as the internal
reference for lncRNAs and U6 was used as the internal reference for
miRNAs; expression data were normalized to GAPH or U6.
Amplification and dissolution curves were confirmed after the
reaction. The relative expression was calculated using the
2−ΔΔCq method (18).
 | Table IIPrimer sequences used in reverse
transcription-quantitative PCR. |
Table II
Primer sequences used in reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| HCP5 | F:
GACTCAGATTCTCCCCAGACGC |
| R:
GTGGGATCCACAACACTTTAAT |
| GAPDH | F:
GGGAGCCAAAAGGGTCAT |
| R:
GAGTCCTTCCACGATACCAA |
| HAGLROS | F:
GCTCTCCGCGCCTCAGCGAGCGG |
| R:
ACAGCAGTCTTAGCCTACTTCCT |
| EZH2 | F:
ATGGGCCAGACTGGGAAGAAA |
| R:
GGAGGTAGCAGATGTCAAGGG |
|
LINC00839 | F:
GATATCTAGTTAAATAAGTCAT |
| R:
GGGAAATCAGGGGTCTCACAGC |
|
PSMB8-AS1 | F:
TCCACATCCCCCTGCCTTTTCCGAG |
| R:
CAGACAGCGTCCCGCTATGTTGCC |
| PP7080 | F:
GACGCCGCCGGCGGGAAGGGTCC |
| R:
CCCTGTCTGGACACTGCCAACCC |
|
LINC01139 | F:
TGGAATTCAAGCTGTGGGTGAGAA |
| R:
ATACGGCCAGAGCGAGTCTACCCT |
|
LINC00963 | F:
CCGGCCCGTCTCGGGGCCCTGAGTC |
| R:
GTGAGCCCCAGCCAAGATGAAGGC |
|
FLVCR1-AS1 | F:
GAGAACTTCGGGAGGCGGGGGAGG |
| R:
GATCTCAAGACGGTGTCTGGCACAT |
| U6 | F:
CGCTTCGGCAGCACATATAC |
| R:
AATATGGAACGCTTCACGA |
| miR-138-5p | F:
AGCTGGTGTTGTGAATCAGG |
| R:
CGGCCTGATTCACAACACCA |
| miR-26a-5p | F:
TTCAAGTAATCCAGGATAGGCT |
| R:
AGCCTATCCTGGATTACTTGAA |
| miR-26b-5p | F:
TTCAAGTAATTCAGGATAGGT |
| R:
ACCTATCCTGAATTACTTGAA |
| miR-1297 | F:
TGTTTATCTCTAGGGTTGATC |
| R:
GTTTCACTACACCTGAATTAC |
| miR-4465 | F:
CATGTGTCCCCTGGCACGCTA |
| R:
TCATGTGTCCCCTGGTCAGACT |
Western blot analysis
Cells were lysed for 30 min using a cold
radio-immunoprecipitation assay buffer containing a protease
inhibitor mixture (Sigma-Aldrich, Merck KGaA). Lysates were
centrifuged at 4°C and 16,000 × g for 20 min, and the supernatant
was obtained. Protein concentration was determined using the
bicinchoninic acid kit (Beyotime Biotechnology Co., Ltd.). Proteins
were separated by 10% SDS-PAGE and transferred onto polyvinylidene
fluoride membranes. The membranes were blocked with 5% skim milk
for 2 h at room temperature. After incubation with the primary
antibodies (Table III)
overnight at 4°C, the membranes were incubated with goat
anti-rabbit IgG (1:2,000; cat. no. ab205718; Abcam) or goat
anti-mouse IgG (1:2,000; cat. no. ab205719; Abcam) secondary
antibody for 1 h at room temperature, were developed using the
Immobilon Western Chemiluminescent HRP substrate (cat. no.
WBKLS0100; MilliporeSigma), and visualized using an imager
(Bio-Rad, Inc.). Protein expression levels were semi-quantified
using ImageJ Pro Plus 6.0 [National Institutes of Health
(NIH)].
 | Table IIIAntibodies used for western blot
analysis. |
Table III
Antibodies used for western blot
analysis.
| Antibody | Catalog number | Dilution ratio |
|---|
| β-actin | ab8227 | 1:1,000 |
| LC3B | ab48394 | 1:1,000 |
| p62 | ab56416 | 1:1,000 |
| Bcl-2 | ab182858 | 1:2,000 |
| Bax | ab32503 | 1:1,000 |
| EZH2 | ab186006 | 1:1,000 |
| p-STAT3 | ab76315 | 1:2,000 |
| STAT3 | ab119352 | 1:5,000 |
| VEGFR2 | ab11939 | 1:1,000 |
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was used to determine cell viability
following the manufacturer's instructions (cat. no. C0038; Beyotime
Institute of Biotechnology). CSCC cells with different
transfections were seeded onto 96-well plates at a density of 5,000
cells/well. After incubation at 37°C for 2 h, the media was
replaced with 100 µl CCK-8 solution (90 µl serum-free
DMEM and 10 µl CCK-8) at 0, 12, 24, 36 and 48 h incubation.
The absorbance at 450 nm was then measured for each well.
Transwell assay and scratch test
A 24-well Transwell system (8 µm pore size
membrane; Corning, Inc.) and a filter membrane pre-coated with
Matrigel (BD Biosciences) were used to evaluate cell invasion.
Suspensions of the transfected CSCC cells (1×105
cells/ml) were prepared in serum-free DMEM (0.1% bovine serum
albumin) and placed onto the apical chamber of the Transwell
insert; the basolateral chamber was filled with DMEM containing 10%
FBS. The Transwell chamber was incubated for 24 h at 37°C and 5%
CO2. Subsequently, the filter membrane was removed and
rinsed with PBS, fixed with 0.5% glutaraldehyde for 30 min at room
temperature and stained with crystal violet for 30 min at 37°C and
5% CO2. Five random fields of vision were selected for
cell counting under a light microscope (magnification, ×200).
Transfected CSCC cells were detached and collected
using 0.25% trypsin, and then resuspended to 2×105
cells/ml in DMEM containing 10% FBS. The cells were seeded onto
6-well plates (2 ml/well) and cultured at 37°C and 5%
CO2 for 12 h. Following cell adherence, a straight line
perpendicular to the center was scratched on the cell monolayer
using a pipette tip. Subsequently, the floating cells were washed
away using PBS, and cells were continuously cultured for 12 h.
Images were captured under a phase contrast microscope at 0 and 12
h incubation. ImageJ Pro Plus 6.0 (NIH) was utilized to analyze the
scratch width of cells in different groups and time periods. Cell
migration=scratch width at 0 h-scratch width at 12 h.
Flow cytometry
When the transfected cells were cultured to 85%
confluence, the suspended cells in the culture supernatant were
collected into the centrifuge tube. The adherent cells were
detached with 0.25% trypsin, washed three times with PBS, and
collected into the centrifuge tube in the previous step. Next, the
cells were centrifuged at 4°C and 150 × g for 5 min. The cells were
resuspended with cold PBS and centrifuged for 5 min at 150 × g at
4°C to collect cell precipitates. This procedure was repeated twice
to remove residual trypsin. Following centrifugation, the cells
were resuspended in PBS and adjusted to 1×106 cells/ml.
According to the instructions of the Annexin V-FITC Apoptosis
Detection kit (BD Biosciences), 500 µl cell suspension was
added into the centrifuge tube at 4°C and 150 × g for 5 min,
followed by the addition of 195 µl Annexin V Binding Buffer
for resuspension. Then, 5 µl Annexin V-FITC and 10 µl
PI were added into each tube, mixed well and incubated for 10 min
at room temperature in the dark. Cells were evaluated using a MoFlo
Astrios EQ flow cytometer (Beckman Coulter, Inc.) admd t Flowjo 7.6
software (FlowJo LLC) was used for data analysis. Total apoptotic
rates were calculated as the total early- and late-stage
apoptosis.
Light chain 3 (LC3) fluorescence
tracer
LC3 double-labeled autophagy adenovirus [Monomeric
red fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3]
was purchased from Hanbio Biotechnology Co., Ltd. Transfected cells
were cultured in 24-well plates (1×105 cells/ml) and
incubated with mRFP-GFP-LC3 adenovirus (multiplicity of infection,
100) for 2 h at 37°C, when cell confluence reached 70-80%. The
cells were washed with PBS and cultivated in DMEM containing 10%
FBS overnight. The transfection efficiency of mRFP-GFP-LC3
adenovirus was evaluated using a BX51 fluorescence microscope
(Olympus Corporation). ImageJ Pro Plus 6.0 software (NIH) was used
for image analysis and counting the number of GFP and RFP
fluorescent punctae. In the merge image, autophagosomes are
represented by yellow dots and autolysosomes are represented by red
dots.
RNA pull-down assay
A431 cells in logarithmic growth phase were
collected and centrifuged at 4°C and 150 × g for 5 min and then
mixed with Pierce IP Lysis Buffer (Thermo Scientific, Inc.). After
lysis on ice for 30 min, the cells were centrifuged at 4°C and
1,200 × g for 30 min to collect the supernatant and stored on ice.
RNA-RNA interaction was assessed by RNA pull-down using Pierce
Magnetic RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Biotinylated HCP5 and NC
probes (50 pmol; made using the Pierce RNA 3′ End
Desthiobiotinylation Kit; cat. no. 20163; Thermo Fisher Scientific,
Inc.) were dissolved in washing/binding buffer (contained in the
pull-down kit) and cultured with streptavidin-coupled magnetic
beads for 2 h at 4°C. Next, the probes were added to the cell
lysates for 2 h to elute the RNA complex conjugated with the
magnetic beads, and miR expression was determined using
RT-qPCR.
Dual luciferase reporter gene assay
The binding sites of lncRNA-HCP5 and miR-138-5p, and
for EZH2 and miR-138-5p were predicted using RNA22 v2 (https://cm.jefferson.edu/rna22/Interactive) and
TargetScan (http://www.targetscan.org) online
databases. The HCP5 fragment containing the binding site of
miR-138-5p was cloned into the pmirGLO oligosaccharide enzyme
vector (Promega Corporation) to construct the
pmirGLO-HCP5-wild-type (WT), and the pmirGLO-HCP5-mutant type (MUT)
vector was constructed using the mutant binding site of miR-138-5p.
The pmirGLO-EZH2-WT and pmirGLO-EZH2-MUT vectors were
constructed using the same methods. The sequences were synthesized
by Shanghai GenePharma Co., Ltd. The constructed vectors (50 ng)
were transfected into A431 cells (at 80-90% confluence), and then
co-transfected with miR-138-5p or miR-NC (20 nM) according to the
instructions of Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.). After incubation at 37°C for 48 h, the
luciferase activity was evaluated using a Dual Luciferase Reporter
Assay System (Promega Corporation), and the relative activity was
determined as the ratio of firefly luciferase activity to
Renilla luciferase activity.
Animal experiments
A total of 20 specific pathogen-free BALB/c nude
male mice (weight, 20±2 g; age, 6-8 weeks) were provided by Beijing
Vital River Laboratory Animal Technology Co., Ltd. [license SYXK
(Beijing) 2015-0001], numbered according to body weight and
allocated into three groups (n=10 mice/group) using the random
number method. All mice were reared in an animal room at 25°C
temperature and 50-60% humidity under a 12 h light/dark cycle. All
mice were allowed free access to food and water. In the A431 group,
each mouse was injected subcutaneously (on the back near the right
armpit) with 2×106 A431 cells. In the si-HCP5-1 group,
each mouse was injected subcutaneously with 2×106 A431
cells transfected with si-HCP5-1. Tumor volumes were measured every
3 days, and mice were euthanized using pentobarbital (>200 mg/kg
intraperitoneal injection) 28 days later (19,20). Death was confirmed by examining
pupillary dilation and complete cardiac arrest, and the tumors of
five random mice from each group were embedded in paraffin and the
tumors from the remaining five mice were ground into a homogenate
for subsequent experiments.
Immunohistochemical and TUNEL
staining
A Histostain-Plus Kit (cat. no. 85-6643; Invitrogen;
Thermo Fisher Scientific, Inc.) was used for immunohistochemical
staining. Tumor tissue was fixed with 4% formaldehyde for 6 h at
room temperature and embedded in paraffin. The paraffin-embedded
tumor tissue was cut into 3 µm sections; the sections were
baked at 45°C for 3 h. After dewaxing with xylene and rehydration,
the sections were treated with 3% H2O2 and
kept at room temperature for 10 min to eliminate the activity of
endogenous peroxidase. After washing with PBS + 0.05% Tween-20
(PBST) three times, the sections were placed in a heat-resistant
glass container, submerged in the citrate sodium buffer (10 mM, pH
6.0), heated to boiling in the microwave for antigen retrieval and
cooled naturally (5-10 min). These steps were repeated three times
and then the sections were cooled down to room temperature. The
sections were blocked with 10% goat serum (reagent A in the kit) at
37°C for 1 h in a humidified chamber and incubated with anti-Ki67
primary antibody (1:500; cat. no. ab15580; Abcam) at 4°C overnight.
The next day, sections were washed three times with PBST and
incubated with biotinylated secondary antibody working solution
(reagent B in the kit), incubated at 37°C for 15 min and washed
three times with PBST times to wash away the unbound antibody. The
sections were subsequently incubated with horseradish
peroxidase-conjugated streptavidin working solution (reagent C in
the kit), incubated in a wet box at 37°C for 15 min, and washed
with PBST three times. The percentage of Ki67-positive cells was
counted using a CellInsight laser confocal microscope (Thermo
Fisher Scientific, Inc.) and analyzed using ImageJ-Pro Plus 6.0
(NIH).
Tumor sections were treated and stained with TUNEL
reaction mixture in accordance with the manufacturer's instructions
(Wanleibio Co., Ltd.). DAPI solution was added to the media (1:10)
to stain the nuclei, and the cells were cultured at 37°C for 15
min. Sections were washed with PBS, observed under a fluorescence
microscope, and an anti-fluorescence quenching agent (Wanleibio
Co., Ltd.) was added. Within 24 h, images of the sections were
captured under a fluorescence microscope; the apoptotic cells were
counted using ImageJ Pro Plus 6.0 (NIH), and the results were
analyzed.
Statistical analysis
Statistical analyses were conducted using SPSS v21.0
(IBM Corp.). The Kolmogorov-Smirnov test confirmed that the data
were normally distributed. Data are presented as the mean ±
standard deviation. Paired Student's t-test or Mann-Whitney U test
was used for comparisons between two groups, whereas one-way ANOVA
was used to compare multiple groups; Tukey's multiple comparisons
test was applied for pairwise comparisons after ANOVA analyses. The
P-value was obtained using a two-tailed test, and a P<0.05 was
used to indicate a statistically significance difference.
Results
HCP5 and EZH2 are highly expressed and
miR-138-5p expression is low in CSCC
Through the differential analysis of GSE66359
microarray, 1,252 genes with significant differential expression
were obtained (Fig. 1A). Among
these genes, EZH2 was screened based on a literature search
that suggested EZH2 can be used as a diagnostic index to
differentiate squamous cell carcinoma from normal skin (14). EZH2 expression was
significantly increased in CSCC compared with normal tissue
(Fig. 1B), which was consistent
with a previous report (14).
EZH2 mRNA and protein expression levels in 60 patients with CSCC
collected from The First Affiliated Hospital of Zhengzhou
University also verified these results (Fig. 1C and D, respectively). To further
understand the mechanism of EZH2 in CSCC, the upstream regulatory
miRNAs of EZH2 were predicted using four online databases.
The combined prediction results identified five miRNAs that may
target EZH2 (Fig. 1E). RT-qPCR
was used to determine the expression levels of the five miRNAs,
which revealed that the relative miR-138-5p expression was the
lowest in CSCC compared with its expression in normal tissue
(Fig. 1F). miR-138-5p has been
shown to inhibit tumor development (21,22), but its role in CSCC remains
unclear. Using the RNA22 database, lncRNAs that may interact with
miR-138-5p were predicted. The prediction results of RNA22 were
intersected with the upregulated gene expression results obtained
from GSE66359 microarray (Fig.
1G), and eight putative lncRNAs were identified that may
regulate miR-138-5p; these lncRNAs were shown to be highly
expressed in CSCC compared with normal adjacent tissue (Fig. 1H). Furthermore, the expression of
these eight lncRNAs were screened in the GSE66359 microarray
(Table IV) and found that HCP5
had the greatest upregulation in CSCC (|logFC|=1.8) and the highest
relative expression levels in patient CSCC compared with the normal
adjacent tissue (Fig. 1H). These
data suggested that lncRNA HCP5 may regulate EZH2 expression
through miR-138-5p.
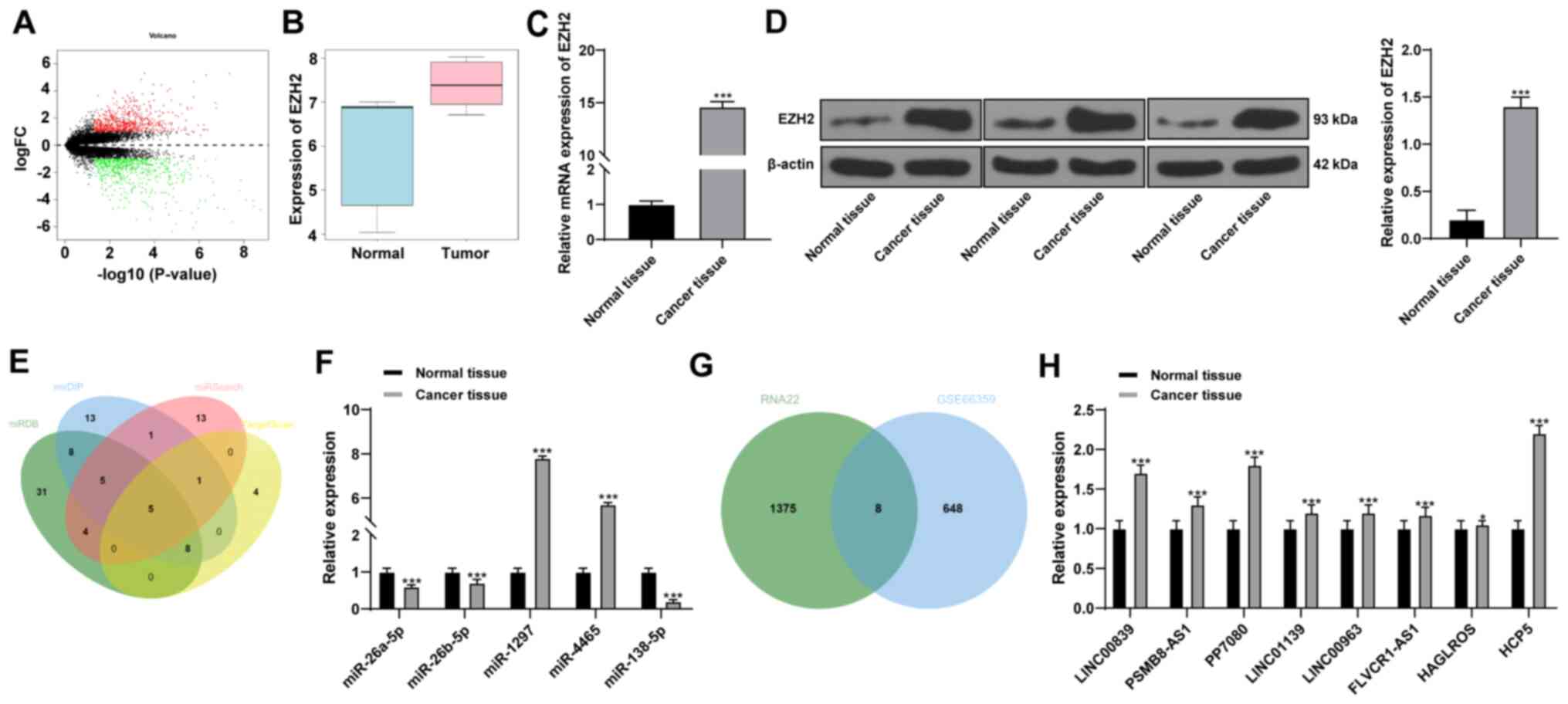 | Figure 1LncRNA HCP5 may regulate EZH2
expression through miR-138-5p. (A) Differential expression analysis
of the volcano plot of the GSE66359 microarray. Red indicates
highly expressed genes and green indicates genes with low
expression levels. (B) EZH2 was found to be significantly increased
in CSCC tissue compared with normal tissue in the GSE66359 data
set. (C) mRNA and (D) protein expression levels of EZH2 in tissue
and normal adjacent tissue samples from 60 patients with CSCC were
upregulated. (E) Four online databases were used to analyze the
intersected data to predict putative miRNAs upstream of EZH2; five
strong candidates were identified: miR-26a-5p, miR-26b-5p,
miR-1297, miR-4465 and miR-138-5p. (F) RT-qPCR was used to detect
the expression levels of the five miRNAs; miR-138-5p level was the
lowest in CSCC compared with normal tissues. N=60;
***P<0.001 vs. normal tissue. (G) Eight lncRNAs were
predicted to regulate miR-138-5p using RNA22 database, and these
were intersected with the upregulated genes expressed in GSE66359
microarray (H) The expression of these eight lncRNAs was detected
using RT-qPCR, and HCP5 showed the largest upregulation in CSCC
compared with normal tissues. N=60; *P<0.05,
***P<0.001 vs. normal tissue. Data in panel B were
analyzed using the Wilcoxon signed-rank test, and data in panels C,
D, F and H were analyzed using paired Student's t-test. CSCC,
cutaneous squamous cell carcinoma; EZH2, enhancer of zeste homolog
2; FC, fold change; HCP5, human histocompatibility leukocyte
antigen complex P5; lncRNA, long non-coding RNA; miR, microRNA;
RT-qPCR, reverse transcription-quantitative PCR. |
 | Table IVPotential lncRNAs regulating
miR-138-5p. |
Table IV
Potential lncRNAs regulating
miR-138-5p.
| lncRNA | |logFC| | P-value |
|---|
| LINC00839 | 1.535958914 |
3.05×10−5 |
| PSMB8-AS1 | 1.418830957 |
5.94×10−5 |
| PP7080 | 1.660170378 |
5.96×10−4 |
| LINC01139 | 1.218616424 |
1.065×10−3 |
| LINC00963 | 1.129432649 |
1.333×10−3 |
| FLVCR1-AS1 | 1.170397507 |
3.735×10−3 |
| HAGLROS | 1.030294427 |
4.908×10−3 |
| HCP5 | 1.800169542 |
6.281×10−3 |
Silencing HCP5 inhibits malignant
behaviors and autophagy of CSCC cells and promotes apoptosis
The expression levels of lncRNA HCP5 were detected
in cultured CSCC cell lines and the HaCaT human immortalized
keratinocyte cell line. A431 cells exhibited the highest relative
HCP5 expression and SCL-1 cells exhibited the lowest HCP5
expression levels among the CSCC cell lines (Fig. 2A); therefore, these two lines were
used for subsequent experiments. Two siRNAs against HCP5-1 were
transfected into A431 cells, of which si-HCP5-1 exhibited the best
transfection efficiency and was selected for the following
experiment (Fig. 2B). An HCP5
overexpression vector was transfected into SCL-1 cells, and the
transfection efficiency was verified using RT-qPCR (Fig. 2C). Silencing of HCP5 expression
resulted in significant decreases in A431 cell viability, invasion
and migration, whereas SCL-1 cells overexpressing HCP5 exhibited
opposite trends (Fig. 2D-F).
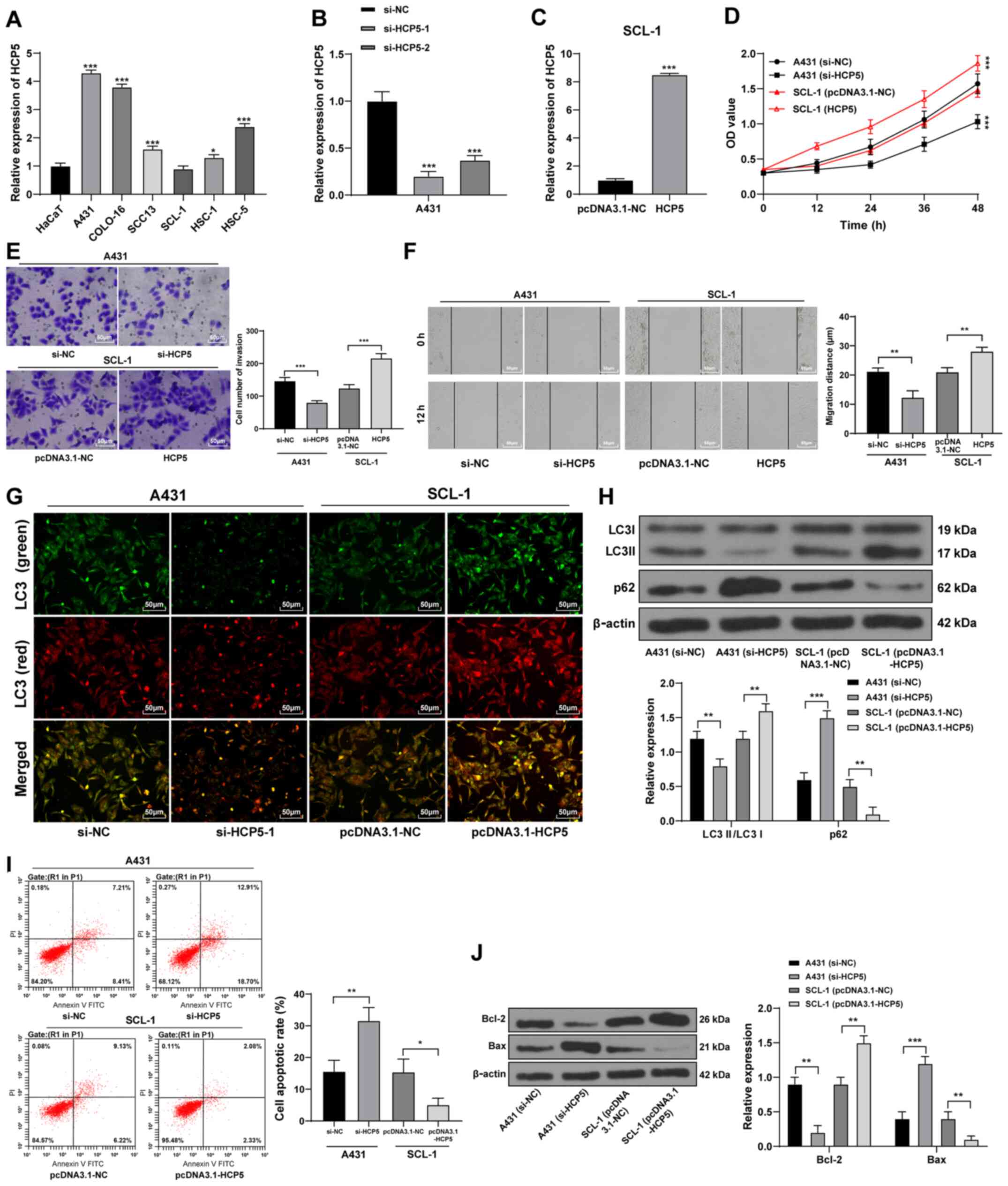 | Figure 2Long non-coding RNA HCP5 regulates
the malignant behavior, autophagy and apoptosis of CSCC cells. (A)
RT-qPCR detection of HCP5 expression in CSCC cell lines.
*P<0.05, ***P<0.001 vs. HaCaT. The
transfection efficiency of (B) two siRNAs against HCP5 and (C) the
HCP5 overexpression vector were verified using RT-qPCR.
***P<0.001 vs. si-NC or pcDNA3.1-NC. (D) Cell
Counting Kit-8 assays, (E) Transwell assays and (F) scratch tests
were used to detect the cell viability, migration and invasion,
respectively. **P<0.01, ***P<0.001 vs.
si-NC or pcDNA3.1-NC. (G) Representative images of mRFP-GFP-LC3
fluorescence tracer assay, in which green and red indicate the
observed fluorescence of LC3 under different excitation
wavelengths; yellow fluorescence indicates autophagosomes and
autolysosomes are represented by red dots. (H) Western blot
analysis was used to detect the levels of autophagy-related
proteins. **P<0.01, ***P<0.001 vs.
si-NC or pcDNA3.1-NC. (I) Flow cytometry was used to determine the
apoptotic rates in transfected CSCC cells. *P<0.05,
**P<0.01 si-NC or pcDNA3.1-NC. (J) Western blot
analysis was used to detect the levels of apoptosis-related
proteins. **P<0.01, ***P<0.001 vs.
si-NC or pcDNA3.1-NC. Data in panels A, B and D were analyzed using
one-way ANOVA, followed by Tukey's multiple comparisons test, and
data in panels C, E, F, H, I and J were analyzed using the
Mann-Whitney U-test. Data are representative of three experimental
repeats. GFP green fluorescent protein; HCP5, human
histocompatibility leukocyte antigen complex P5; LC3, light chain
3; mRFP, monomeric red fluorescent protein; NC, negative control;
RT-qPCR, reverse transcription-quantitative PCR; siRNA, small
interfering RNA. |
LncRNAs can regulate apoptosis and autophagy of CSCC
cells (23). Therefore, autophagy
and apoptosis of CSCC cells were measured following HCP5 knockdown
and overexpression. After HCP5 silencing, the number of
autophagosomes (yellow punctae) decreased (Fig. 2G), the LC3II/LC3I protein
expression ratio decreased and the expression of p62 protein
increased (Fig. 2H). In addition,
the apoptotic rate significantly increased (Fig. 2I), apoptotic protein Bax
expression level increased and Bcl-2 expression level significantly
decreased (Fig. 2J). Conversely,
the number of autophagosomes in CSCC cells increased following HCP5
overexpression, the protein expression levels of LC3 II/LC3I
increased, whereas the expression of p62, the apoptotic rate and
Bax protein expression decreased; Bcl-2 expression increased
significantly. In summary, HCP5 silencing inhibited the
malignant behaviors and autophagy of CSCC cells and promoted
apoptosis, whereas overexpression of HCP5 promoted the malignant
behaviors and autophagy of CSCC cells and inhibited apoptosis.
HCP5 competes with EZH2 to bind to
miR-138-5p
Combined with the microarray analysis results, the
binding sites of lncRNA HCP5 and miR-138-5p as well as EZH2 and
miR-138-5p were predicted using online databases RNA22 and
TargetScan. Dual luciferase reporter assays were used to verify the
binding relationships (Fig.
3A-B). In addition, lncRNA HCP5 significantly enriched
miR-138-5p using an RNA pull-down assay (Fig. 3C). As shown in Fig. 3D-F, after HCP5 silencing in
A431 cells, miR-138-5p expression levels increased, whereas the
EZH2 mRNA and protein expression levels decreased significantly;
the opposite trends were observed in cells overexpressing HCP5.
After miR-138-5p mimics transfection, EZH2 expression levels
decreased, whereas the results of miR-138-5p inhibitor transfection
were the opposite. These results indicated negative regulatory
relationships between HCP5 and miR-138-5p, and between EZH2 and
miR-138-5p. HCP5 may function as a ceRNA to compete with EZH2 to
bind to miR-138-5p.
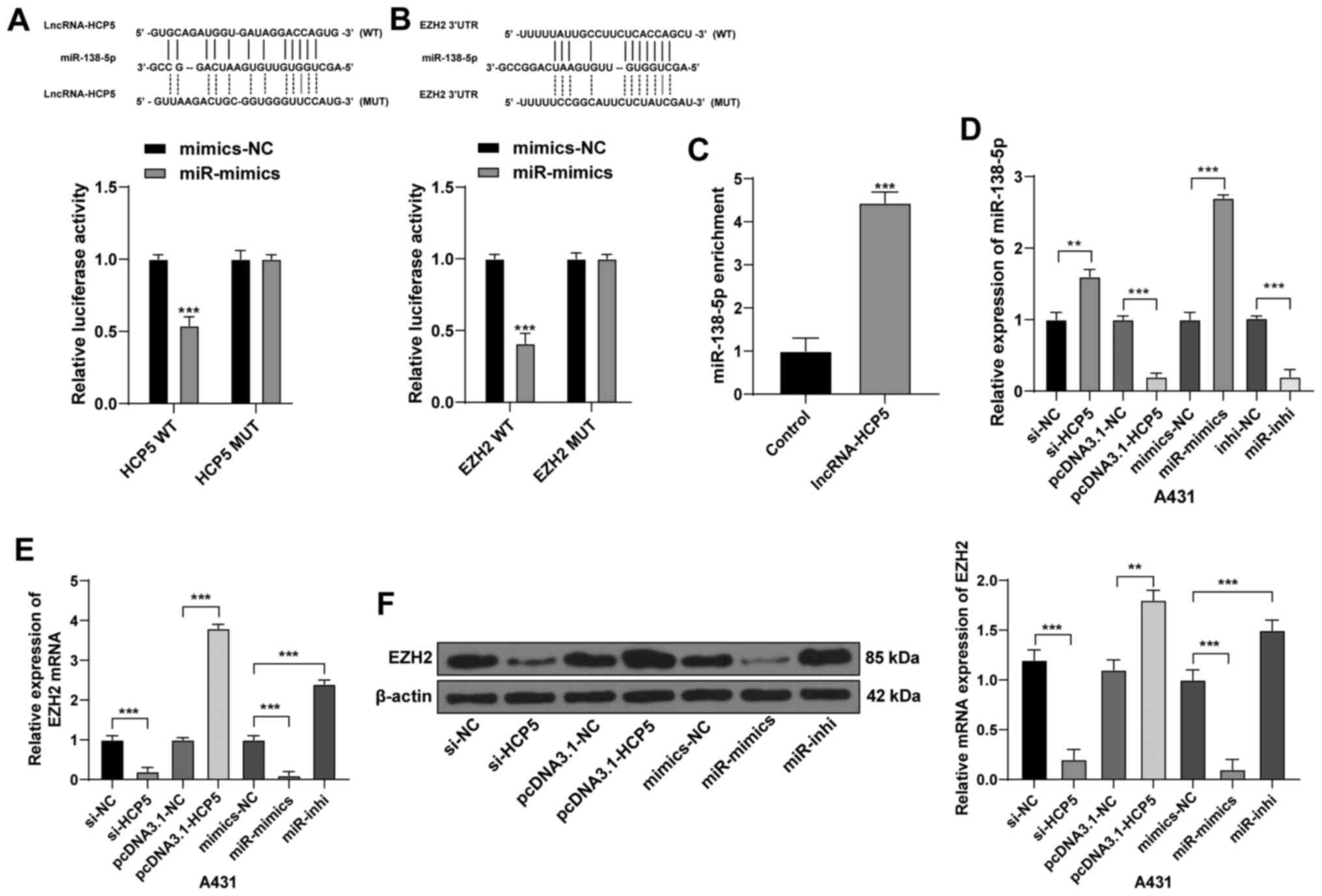 | Figure 3LncRNA HCP5 competes with EZH2 to
bind to miR-138-5p. Binding sites between (A) HCP5 and miR-138-5p,
and (B) the miR-138-5p target sites in EZH2 3′UTR and were
predicted using the online databases RNA22 and TargetScan. Dual
luciferase report gene assay verified the relation between (A) HCP5
and miR-138-5p and (B) EZH2 and miR-138-5p.
***P<0.001 vs. mimics-NC. (C) miR-138-5p was
significantly enriched by HCP5 using RNA pull-down assay.
***P<0.001 vs. Control. (D) miR-138-3p, (E) EZH2 mRNA
and (F) EZH2 protein expression levels in A431 cells following the
various transfections were detected using reverse
transcription-quantitative PCR or western blotting.
**P<0.01, ***P<0.001. Data are
representative of three experimental repeats. Data were analyzed by
Mann-Whitney U-test. EZH2, enhancer of zeste homolog 2; HCP5, human
histocompatibility leukocyte antigen complex P5; inhi, inhibitor;
lncRNA, long non-coding RNA; miR, microRNA; MUT, mutant; UTR,
untranslated region; WT, wild-type. |
Upregulation of miR-138-5p in CSCC cells
inhibits autophagy and promotes apoptosis
miR-138-5p inhibitor was co-transfected into A431
cells with si-HCP5 to conduct a functional rescue experiment
(Fig. 4A). Cell viability was
reduced upon upregulation of miR-138-5p or downregulation of
HCP5, whereas downregulation of miR-138-5p partially
counteracted the effect of HCP5 silencing on CSCC viability
(Fig. 4B). HCP5 silencing
and upregulation of miR-138-5p significantly decreased the
LC3II/LC3I ratio and increased the protein expression of p62 in
CSCC cells; downregulation of miR-138-5p partially reversed the
inhibitory effects of HCP5 silencing in CSCC cells (all
P<0.05; Fig. 4C).
Additionally, si-HCP5 or miR-138-5p mimics alone significantly
increased the apoptotic rate, increased the protein expression
levels of Bax and decreased Bcl-2; downregulation of miR-138-5p
partially abolished the promoting effect of HCP5 silencing on cell
apoptosis (Fig. 4D-E). These
results indicated that upregulation of miR-138-5p may inhibit
autophagy and promote apoptosis of CSCC cells.
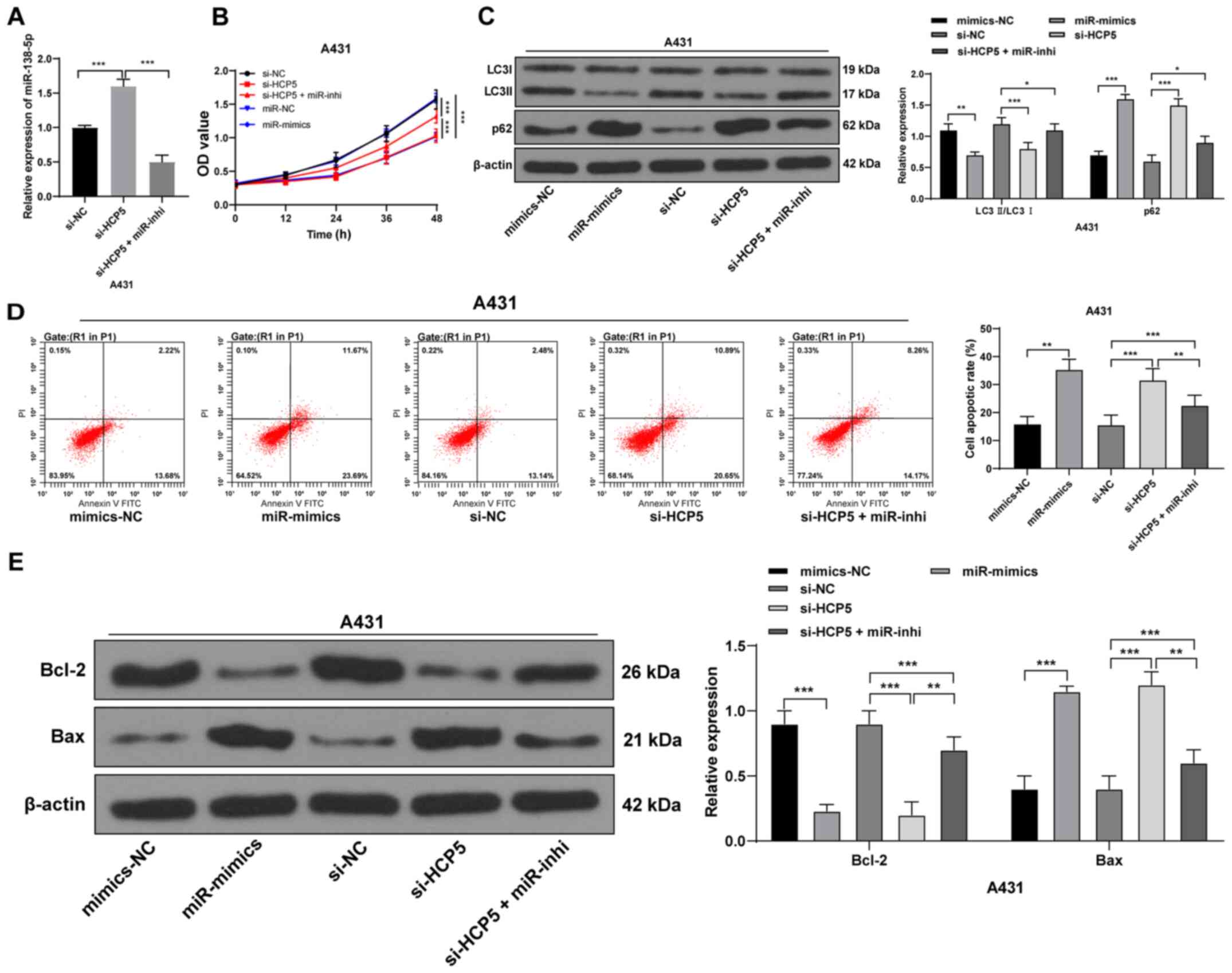 | Figure 4Upregulation of miR-138-5p in
cutaneous squamous cell carcinoma cells inhibits autophagy and
promotes apoptosis. (A) miR-138-5p inhibitor was co-transfected
into A431 cells with si-HCP5, and the expression levels of
miR-138-5p were detected by reverse transcription-quantitative PCR.
***P<0.001. (B) Cell Counting Kit-8 assay was used to
measure cell viability at each time point.
***P<0.001. (C) Western blot analysis was used to
determine the expression levels of autophagy-related proteins.
*P<0.05, **P<0.01,
***P<0.001. (D) Flow cytometry was used to determine
the apoptotic rates in transfected cells. **P<0.01,
***P<0.001. (E) Western blot analysis was used to
determine the expression levels of apoptosis-related proteins.
**P<0.01, ***P<0.001. Data are
representative of three experimental repeats. Data in panels A and
E were analyzed by Mann-Whitney U-test, and data in panels B-D were
analyzed using the one-way ANOVA followed by Tukey's multiple
comparisons test. EZH2, enhancer of zeste homolog 2; HCP5, human
histocompatibility leukocyte antigen complex P5; inhi, inhibitor;
LC3, light chain 3; miR, microRNA; NC, negative control; si, small
interfering RNA. |
Upregulation of EZH2 counteracts the
effects of silencing HCP5 on CSCC cells
An EZH2 overexpression vector was transfected into
CSCC cells (Fig. 5A and B). EZH2
overexpression increased the viability of SCL-1 cells and offset
the inhibitory effect of silencing HCP5 on A431 cell viability
(Fig. 5C). Western blotting
analysis revealed that overexpression of EZH2 significantly
increased the protein expression ratio of LC3II/LC3I, decreased the
expression of p62 in SCL-1 cells and counteracted the inhibitory
effects of HCP5 silencing on the autophagy of A431 cells
(Fig. 5D). Additionally,
overexpression of EZH2 significantly decreased the level of
apoptosis (Fig. 5E), decreased
the protein expression levels of Bax and increased the expression
of Bcl-2 in SCL-1 cells (Fig.
5F), and counteracted the effects of HCP5 silencing in
of A431 cells.
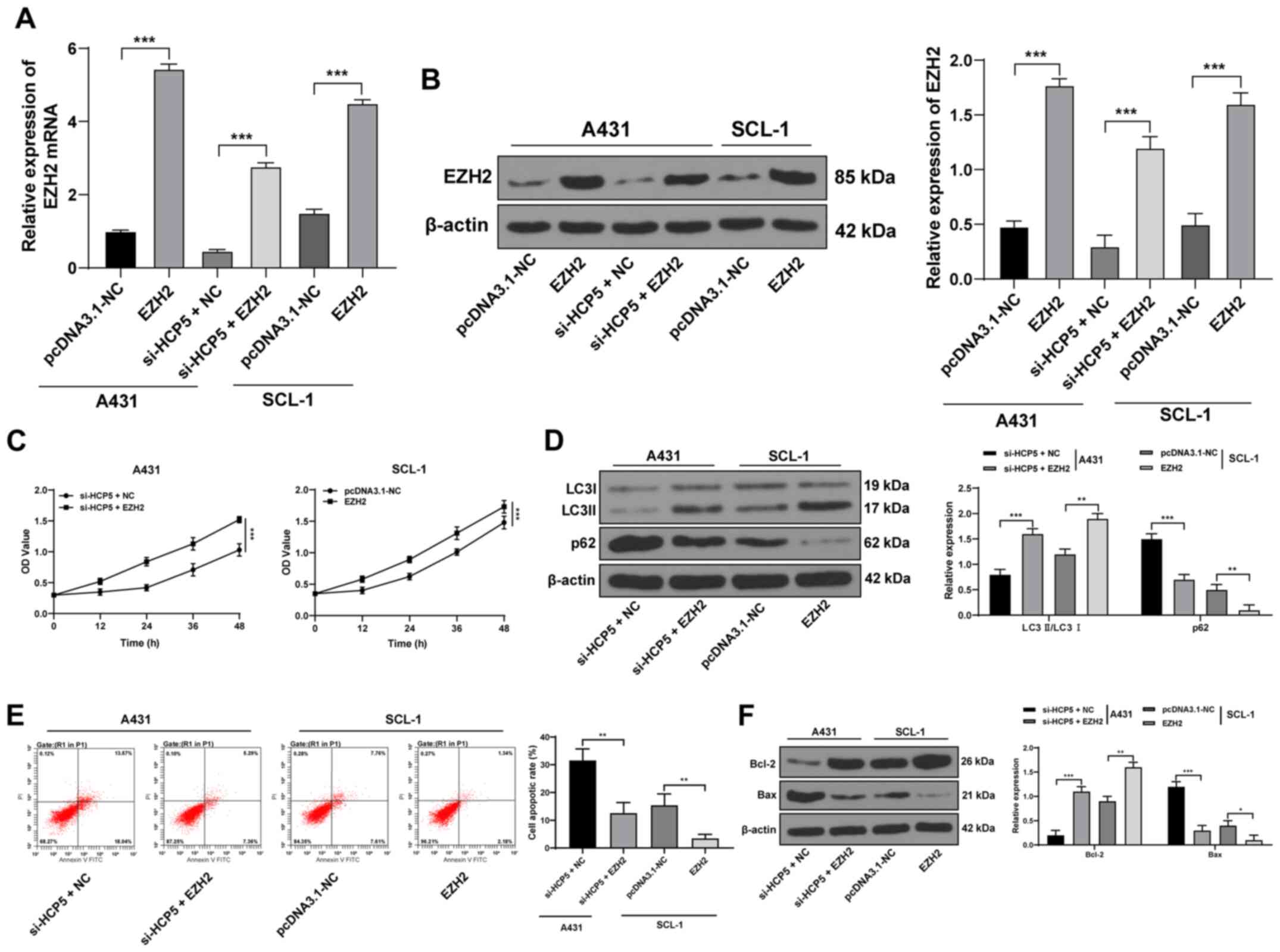 | Figure 5Upregulation of EZH2 counteracts the
effects of HCP5 silencing in cutaneous squamous cell
carcinoma cells. (A) mRNA and (B) protein expression levels of EZH2
were detected using reverse transcription-quantitative PCR and
western blot analysis, respectively. ***P<0.001. (C)
Cell Counting Kit-8 assays were used to measure cell viability at
each time point. ***P<0.001. (D) Western blot
analysis was used to determine the expression levels of
autophagy-related proteins. **P<0.01,
***P<0.001. (E) Flow cytometry was used to determine
the apoptotic rates in transfected cells. **P<0.01.
(F) Western blot analysis was used to determine the expression
levels of apoptosis-related proteins. *P<0.05,
**P<0.01, ***P<0.001. Data in panel C
were analyzed using one-way ANOVA, followed by Tukey's multiple
comparisons test, and data in panels A, B and D-F were analyzed
using the Mann-Whitney U-test. EZH2, enhancer of zeste homolog 2;
HCP5, human histocompatibility leukocyte antigen complex P5; inhi,
inhibitor; LC3, light chain 3; miR, microRNA; NC, negative control;
si, small interfering RNA. |
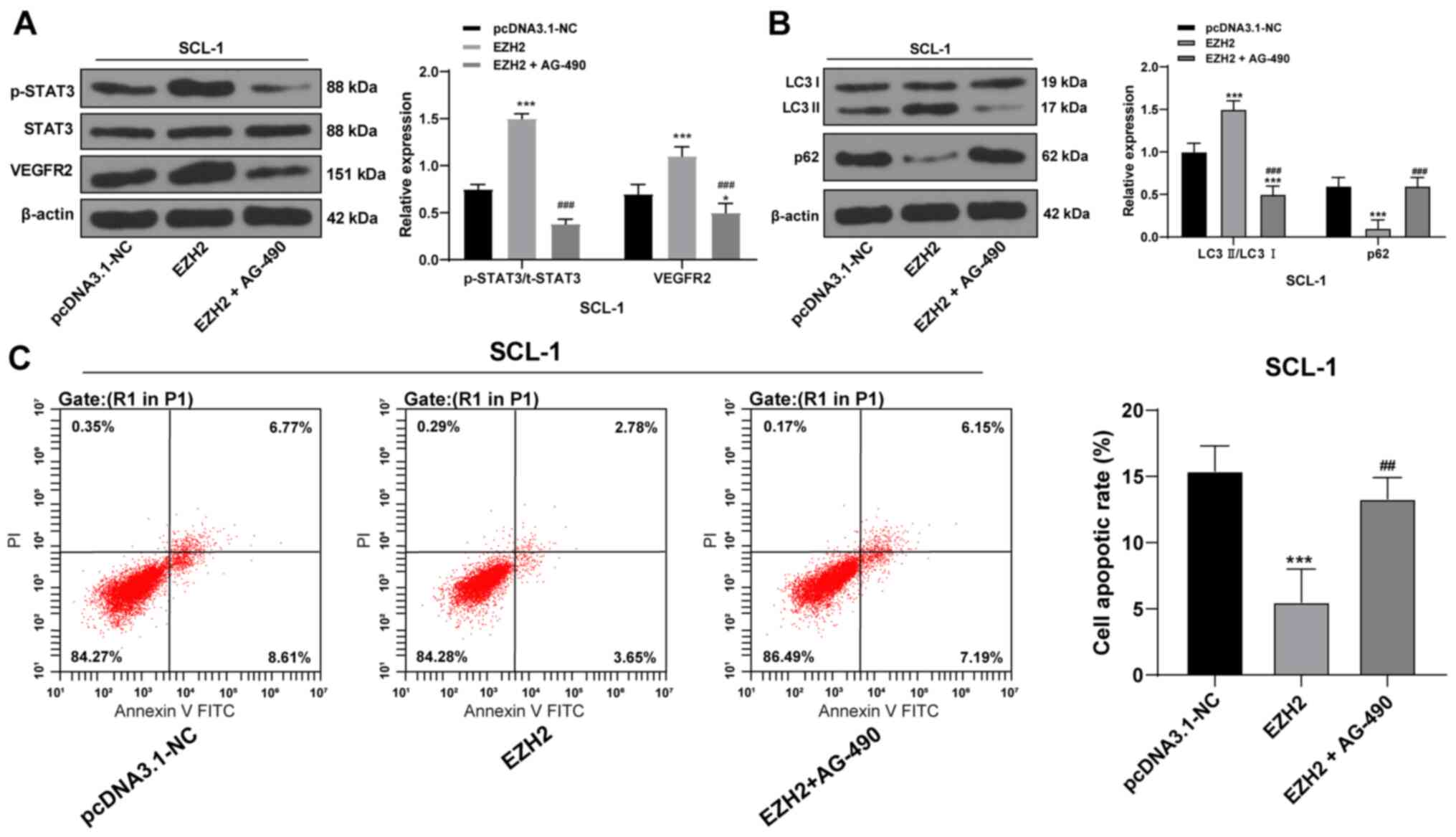
EZH2 affects autophagy and apoptosis of
CSCC cells through the STAT3/VEGFR2 pathway
EZH2 was previously reported to serve a regulatory
role in head and neck squamous cell carcinoma through the
STAT3/VEGFR2 pathway (24).
Therefore, whether the STAT3/VEGFR2 pathway is involved in CSCC
development was investigated. Western blotting results revealed
that p-STAT3/total (t)-STAT3 ratio and VEGFR2 protein expression
levels in CSCC cells overexpressing EZH2 were significantly
increased compared with the control (Fig. 6A). Co-treatment with AG-490, an
inhibitor of the STAT3/VEGFR2 pathway, significantly inhibited the
levels of p-STAT3/t-STAT3 and VEGFR2 in EZH2-overexpressing CSCC
cells (Fig. 6A). In addition,
AG-490 co-treatments also resulted in decreased autophagy, as
determined by western blot analysis of autophagy-related proteins
(Fig. 6B), and the apoptotic rate
increased (Fig. 6C). Taken
together, EZH2 may regulate autophagy and apoptosis in CSCC cells
through the STAT3/VEGFR2 pathway.
HCP5 promotes EZH2 expression to activate
the STAT3/VEGFR2 pathway in vivo
si-HCP5 or si-NC A431 cells were subcutaneously
injected into mice. Tumor volume and weight measurements indicated
that tumor growth was limited after silencing HCP5 compared with
the control group (Fig. 7A-C). In
addition, the protein expression levels of EZH2, p-STAT3/t-STAT3
ratio and VEGFR2 were decreased; autophagy-related proteins
LC3II/LC3I decreased significantly, whereas p62 protein expression
increased (Fig. 7D). After
HCP5 silencing, the Ki67-positive rate was significantly
reduced (Fig. 7E), and TUNEL
staining indicated that apoptosis was increased (Fig. 7F). These data suggested that HCP5
may promote EZH2 expression, increase proliferation and autophagy,
and reduce apoptosis in CSCC cells through the STAT3/VEGFR2
pathway.
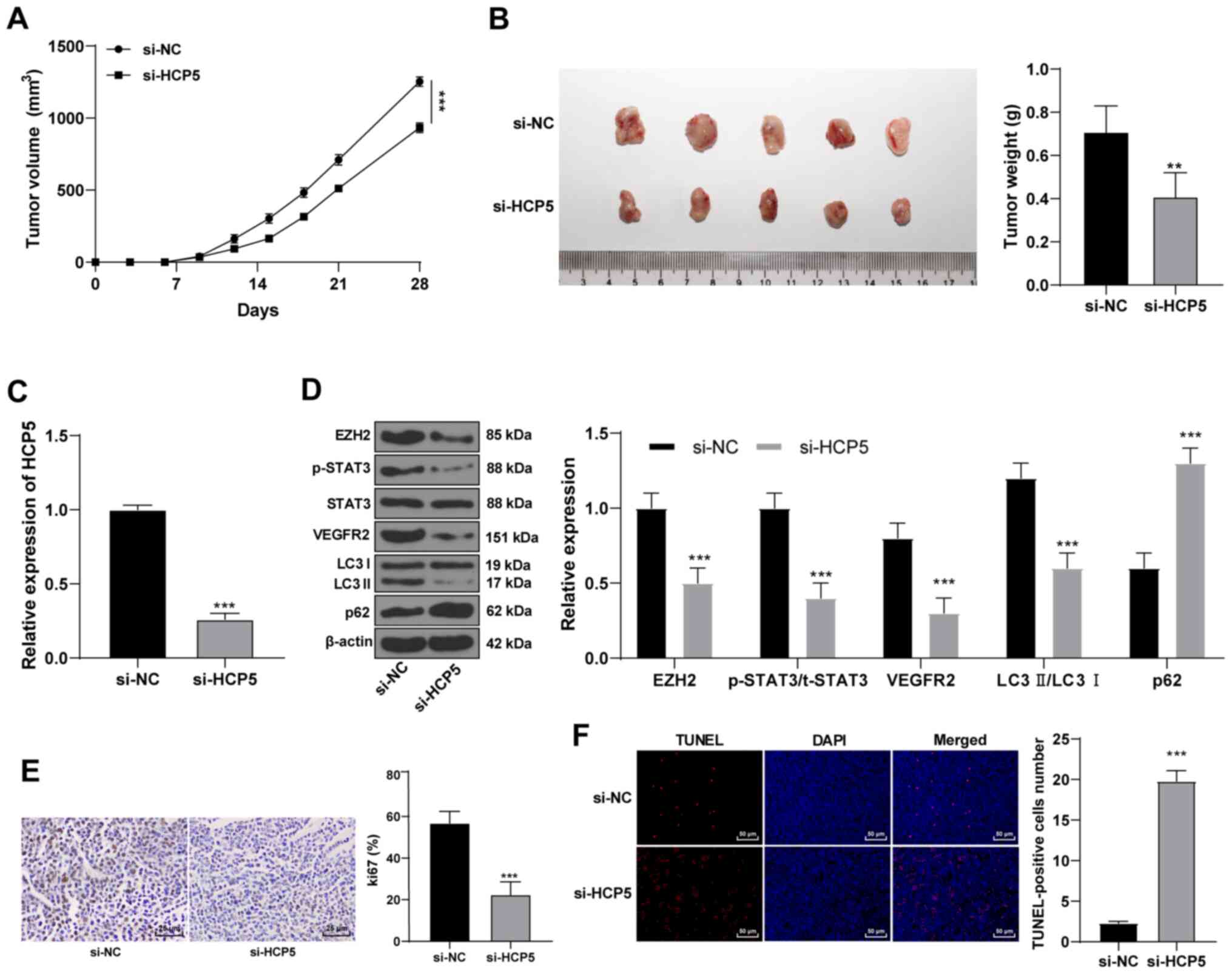 | Figure 7HCP5 promotes EZH2 expression to
activate the STAT3/VEGFR2 pathway in vivo. Tumor (A) volume
and (B) weight after HCP5 silencing in nude mice.
**P<0.01, ***P<0.001 vs. si-NC. (C)
HCP5 expression levels in the excised tumors were detected using
reverse transcription-quantitative PCR. ***P<0.001
vs. si-NC. (D) Western blot analysis was used to determine the
expression levels of the p-STAT3, t-STAT3 and VEGFR2 proteins and
autophagy-related proteins. ***P<0.001 vs. si-NC. (E)
Immunohistochemistry was used to detect the Ki67-positive
expression rate. ***P<0.001 vs. si-NC. (F) TUNEL
staining was used to detect cell apoptosis in tumors.
***P<0.001 vs. si-NC. N=5. Data in panel A were
analyzed using one-way ANOVA, followed by Tukey's multiple
comparisons test, and data in panels B-E were analyzed using
Mann-Whitney U-test. EZH2, enhancer of zeste homolog 2; HCP5, human
histocompatibility leukocyte antigen complex P5; LC3, light chain
3; NC, negative control; p-, phosphorylated; si, small interfering
RNA; t-, total. |
Discussion
CSCC is an immense clinical and economic problem
given its incidence has increased over 200% in the last three
decades (25,26). The mortality of high-risk and
metastatic CSCC is 40% within 3 years (27). Therefore, early diagnosis and
treatment of CSCC is crucial to minimize morbidity and save medical
resources (2). In the present
study, differentially expressed mRNAs, miRNAs and lncRNAs in CSCC
samples were identified, a putative EZH2/miR-138-5p/lncRNA HCP5
ceRNA regulatory network was predicted, and their effects on
autophagy and apoptosis in CSCC cells were investigated. The data
suggested that HCP5 competitively binds miR-138-5p to regulate EZH2
in CSCC cells and promoted autophagy and reduced the level of
apoptosis through the STAT3/VEGFR2 pathway.
Using GSE66359 microarray analysis, database
prediction and verification in 60 CSCC tissues, it was revealed
that EZH2 and HCP5 were highly expressed in CSCC cells, whereas
miR-138-5p expression was low. EZH2 is increased in skin cancer
cell lines, and the knockdown of EZH2 is associated with a
decreased formation of histone H3 lysine 27 trimethylation and a
reduction in survival (28). EZH2
expression may be used as a diagnostic marker for differentiating
CSCC from actinic keratosis or normal skin (14). miR-138-5p is markedly
downregulated in oral squamous cell carcinoma (29). HCP5 is overexpressed in cervical
cancer (30), follicular thyroid
carcinoma (10) and colon cancer
(31). However, the expression
profile and molecular mechanism of HCP5 in CSCC remain unknown.
Therefore, the next focus of this study was to explore the role of
HCP5 in malignant behaviors in CSCC cells.
The second major result of the present study was
that, after silencing HCP5, the LC3 autophagosomes, LC3II/LC3I and
Bcl-2 levels decreased, whereas the apoptotic rate and the
expression levels of p62 and the apoptosis-related protein Bax
level increased significantly. Opposite results were obtained for
cells overexpressing HCP5. The overexpression of LC3 protein is
also strongly correlated with metastasis and a poor clinical
prognosis of human melanoma (32). Impaired autophagy leads to p62
accumulation (33). High
expression of LC3 and low levels of p62 may play key roles in CSCC
tumorigenesis and metastasis (5).
In a previous study, Wright et al (34) reported that with increased CSCC
stage, the basic level of LC3-II tripled, and that autophagy
inhibition enhanced the sensitivity of metastatic CSCC cells to
docetaxel-induced apoptosis. HCP5 upregulation was also observed in
glioma, and HCP5 knockdown blocked proliferation and
invasion, and promoted apoptosis (11). In the present study, silencing of
HCP5 inhibited migration and invasion and autophagy in CSCC
cells and promoted apoptosis. Additionally, using database
prediction, a dual luciferase reporter gene assay system, and an
RNA pull-down assay, HCP5 was demonstrated to compete with EZH2 to
bind to miR-138-5p. A recent study uncovered the ceRNA network
involving lncRNA HCP5 in pancreatic cancer, in which HCP5 serves as
a sponge for miR-214-3p to target hepatoma-derived growth factor to
modulate cell apoptosis, autophagy and drug resistance (35).
Furthermore, the present study data revealed that
miR-138-5p upregulation in CSCC cells inhibited autophagy and
promoted apoptosis. Reduced miR-138 levels are related to poor
clinical conditions in patients with esophageal and oral SCC
(36,37). Similarly, miR-138-5p substantially
reduced the expression levels of autophagy markers in pancreatic
cancer cells and impaired serum starvation-driven autophagy flux
(38). miR-138-5p inhibited the
migratory and invasive capacity by regulating
epithelial-mesenchymal transition (EMT) in head and neck SCC cell
lines (39). In the present
study, EZH2 upregulation counteracted the effects of HCP5
silencing on CSCC cells. EZH2 downregulation was reported to
repress inflammation and metastases, and prevented tumor
dissemination in CSCC (40). EZH2
knockdown has also shown to reduce the autophagic activity and
proliferation of laryngeal SCC cells (15).
Regarding the HCP5/miR-138-5p/EZH2 ceRNA mechanism
in CSCC cells, the present study explored the potential downstream
pathways involved in CSCC development. EZH2 serves a regulatory
role in the head and neck SCC process through the STAT3/VEGFR2
pathway (24). STAT3 inhibitor
treatment suppresses EMT, proliferation and migration of A431 CSCC
cells (41). EGFR and STAT3 are
frequently overexpressed in CSCC cell lines and tissues (42). VEGFR2 has also been shown to boost
tumor proliferation in CSCC cells (43). In other words, EZH2 affects
autophagy and apoptosis of CSCC cells through the STAT3/VEGFR2
pathway.
In conclusion, the present study investigated the
ceRNA network of lncRNA HCP5/miR-138-5p/EZ2H in CSCC. We
hypothesized that the silencing of HCP5 downregulated EZ2H
and inactivated the STAT3/VEGFR2 pathway by competitively binding
to miR-138-5p, thus inhibiting autophagy and promoting the
apoptosis of CSCC cells. A detailed study on the effects of HCP5 on
the proliferative ability of CSCC cells will be conducted in the
follow-up research. The present results may provide a new
perspective to evaluate the function of the HCP5/miR-138-5p/EZ2H
axis in CSCC, and thus may offer cancer prevention and therapeutic
strategies. Further investigations should be performed to validate
these data and to determine their clinical value.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SBZ contributed to the study design and manuscript
preparation. YG contributed to the experimental studies and data
acquisition. STZ contributed to the data analysis and statistical
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was performed with the approval of The
Clinical Ethical Committee of The First Affiliated Hospital of
Zhengzhou University [Zhengzhou, China; (KY-2021-0036)]. All
patients agreed to participate and signed the informed consent.
Ethics approval was also received from the animal ethics committee
of The First Affiliated Hospital of Zhengzhou University
(2021-KY-0107-001). All procedures were conducted in accordance
with the code of ethics. Great efforts were made to minimize the
number of animals used and their pain.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Que SKT, Zwald FO and Schmults CD:
Cutaneous squamous cell carcinoma: Incidence, risk factors,
diagnosis, and staging. J Am Acad Dermatol. 78:237–247. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parekh V and Seykora JT: Cutaneous
squamous cell carcinoma. Clin Lab Med. 37:503–525. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brinkman JN, Hajder E, van der Holt B, Den
Bakker MA, Hovius SE and Mureau MA: The effect of differentiation
grade of cutaneous squamous cell carcinoma on excision margins,
local recurrence, metastasis, and patient survival: A retrospective
follow-up study. Ann Plast Surg. 75:323–326. 2015. View Article : Google Scholar
|
|
4
|
Thompson AK, Kelley BF, Prokop LJ, Murad
MH and Baum CL: Risk factors for cutaneous squamous cell carcinoma
recurrence, metastasis, and disease-specific death: A systematic
review and meta-analysis. JAMA Dermatol. 152:419–428. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng LQ, Li SY and Li CX: Expression
profiling analysis of autophagy-related genes in perineural
invasion of cutaneous squamous cell carcinoma. Oncol Lett.
15:4837–4848. 2018.PubMed/NCBI
|
|
6
|
Ou C, Liu H, Ding Z and Zhou L:
Chloroquine promotes gefitinib-induced apoptosis by inhibiting
protective autophagy in cutaneous squamous cell carcinoma. Mol Med
Rep. 20:4855–4866. 2019.PubMed/NCBI
|
|
7
|
Lu MY, Liao YW, Chen PY, Hsieh PL, Fang
CY, Wu CY, Yen ML, Peng BY, Wang DP, Cheng HC, et al: Targeting
LncRNA HOTAIR suppresses cancer stemness and metastasis in oral
carcinomas stem cells through modulation of EMT. Oncotarget.
8:98542–98552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sand M, Bechara FG, Sand D, Gambichler T,
Hahn SA, Bromba M, Stockfleth E and Hessam S: Expression profiles
of long noncoding RNAs in cutaneous squamous cell carcinoma.
Epigenomics. 8:501–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulski JK: Long Noncoding RNA HCP5, a
Hybrid HLA Class I endogenous retroviral gene: Structure,
expression, and disease associations. Cells. 8:4802019. View Article : Google Scholar :
|
|
10
|
Liang L, Xu J, Wang M, Xu G, Zhang N, Wang
G and Zhao Y: LncRNA HCP5 promotes follicular thyroid carcinoma
progression via miRNAs sponge. Cell Death Dis. 9:3722018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H,
Xi Z, Li Z and Liu Y: Role of HCP5-miR-139-RUNX1 feedback loop in
regulating malignant behavior of glioma cells. Mol Ther.
24:1806–1822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei X, Gu X, Ma M and Lou C: Long
noncoding RNA HCP5 suppresses skin cutaneous melanoma development
by regulating RARRES3 gene expression via sponging miR-12. Onco
Targets Ther. 12:6323–6335. 2019. View Article : Google Scholar :
|
|
13
|
Wang J and Wang GG: No easy way out for
EZH2: Its pleiotropic, noncanonical effects on gene regulation and
cellular function. Int J Mol Sci. 21:95012020. View Article : Google Scholar :
|
|
14
|
Xie Q, Wang H, Heilman ER, Walsh MG,
Haseeb MA and Gupta R: Increased expression of enhancer of Zeste
Homolog 2 (EZH2) differentiates squamous cell carcinoma from normal
skin and actinic keratosis. Eur J Dermatol. 24:41–45. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Jia J, Zang Y, Li J and Wan B:
MicroRNA-101 regulates autophagy, proliferation and apoptosis via
targeting EZH2 in laryngeal squamous cell carcinoma. Neoplasma.
66:507–515. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Z, Yang L, Zhong C and Zhou L: EZH2
regulates H2B phosphorylation and elevates colon cancer cell
autophagy. J Cell Physiol. 235:1494–1503. 2020. View Article : Google Scholar
|
|
17
|
Zuo S, Li X, Bao W and Li S: Pre-mRNA
processing factor 3 enhances the progression of
keratinocyte-derived cutaneous squamous cell carcinoma by
regulating the JAK2/STAT3 pathway. Sci Rep. 10:88632020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Kopaladze RA: Methods for the euthanasia
of experimental animals-the ethics, esthetics and personnel safety.
Usp Fiziol Nauk. 31:79–90. 2000.In Russian. PubMed/NCBI
|
|
20
|
Zatroch KK, Knight CG, Reimer JN and Pang
DS: Refinement of intraperitoneal injection of sodium pentobarbital
for euthanasia in laboratory rats (Rattus norvegicus). BMC Vet Res.
13:602017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan X, Chen Y, Shen Y and Tantai J:
Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in
A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis.
10:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao C, Ling X, Li X, Hou X and Zhao D:
MicroRNA-138-5p inhibits cell migration, invasion and EMT in breast
cancer by directly targeting RHBDD1. Breast Cancer. 26:817–825.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou W, Zhang S, Li J, Li Z, Wang Y and Li
X: lncRNA TINCR participates in ALA-PDT-induced apoptosis and
autophagy in cutaneous squamous cell carcinoma. J Cell Biochem.
120:13893–13902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao M, Hu X, Xu Y, Wu C, Chen J, Ren Y,
Kong L, Sun S, Zhang L, Jin R and Zhou X: Targeting of EZH2
inhibits epithelial-mesenchymal transition in head and neck
squamous cell carcinoma via regulating the STAT3/VEGFR2 axis. Int J
Oncol. 55:1165–1175. 2019.PubMed/NCBI
|
|
25
|
Egolf S and Capell BC: LSD1: A viable
therapeutic target in cutaneous squamous cell carcinoma? Expert
Opin Ther Targets. 24:671–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Filoni A, Cicco G, Lospalluti L, Maglietta
A, Foti C, Annichiarico G, Resta L and Bonamonte D: Morphological
and morphometric analysis of cutaneous squamous cell carcinoma in
patients with recessive dystrophic epidermolysis bullosa: A
retrospective study. J Eur Acad Dermatol Venereol. 34:1707–1714.
2020. View Article : Google Scholar
|
|
27
|
Hassan S, Purdie KJ, Wang J, Harwood CA,
Proby CM, Pourreyron C, Mladkova N, Nagano A, Dhayade S, Athineos
D, et al: A unique panel of patient-derived cutaneous squamous cell
carcinoma cell lines provides a preclinical pathway for therapeutic
testing. Int J Mol Sci. 20:34282019. View Article : Google Scholar :
|
|
28
|
Balasubramanian S, Adhikary G and Eckert
RL: The Bmi-1 polycomb protein antagonizes the
(-)-epigallocatechin-3-gallate-dependent suppression of skin cancer
cell survival. Carcinogenesis. 31:496–503. 2010. View Article : Google Scholar
|
|
29
|
Zhuang Z, Xie N, Hu J, Yu P, Wang C, Hu X,
Han X, Hou J, Huang H and Liu X: Interplay between ΔNp63 and
miR-138-5p regulates growth, metastasis and stemness of oral
squamous cell carcinoma. Oncotarget. 8:21954–21973. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu Y, Shen HM, Fang DM, Meng QJ and Xin
YH: LncRNA HCP5 promotes the development of cervical cancer by
regulating MACC1 via suppression of microRNA-15a. Eur Rev Med
Pharmacol Sci. 22:4812–4819. 2018.PubMed/NCBI
|
|
31
|
Yun WK, Hu YM, Zhao CB, Yu DY and Tang JB:
HCP5 promotes colon cancer development by activating AP1G1 via
PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 23:2786–2793.
2019.PubMed/NCBI
|
|
32
|
Han C, Sun B, Wang W, Cai W, Lou D, Sun Y
and Zhao X: Overexpression of microtubule-associated protein-1
light chain 3 is associated with melanoma metastasis and
vasculogenic mimicry. Tohoku J Exp Med. 223:243–251. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mathew R, Karp CM, Beaudoin B, Vuong N,
Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al:
Autophagy suppresses tumorigenesis through elimination of p62.
Cell. 137:1062–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wright TJ, McKee C, Birch-Machin MA, Ellis
R, Armstrong JL and Lovat PE: Increasing the therapeutic efficacy
of docetaxel for cutaneous squamous cell carcinoma through the
combined inhibition of phosphatidylinositol 3-kinase/AKT signalling
and autophagy. Clin Exp Dermatol. 38:421–423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z
and Yu X: Long noncoding RNA HCP5 regulates pancreatic cancer
gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p to target
HDGF. Onco Targets Ther. 12:8207–8216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Manikandan M, Deva Magendhra Rao AK,
Rajkumar KS, Rajaraman R and Munirajan AK: Altered levels of
miR-21, miR-125b-2 *, miR-138, miR-155, miR-184, and miR-205 in
oral squamous cell carcinoma and association with
clinicopathological characteristics. J Oral Pathol Med. 44:792–800.
2015. View Article : Google Scholar
|
|
37
|
Zheng S, Zhang X, Wang X and Li J:
Downregulation of miR-138 predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Cancer Biomark. 20:49–54. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tian S, Guo X, Yu C, Sun C and Jiang J:
miR-138-5p suppresses autophagy in pancreatic cancer by targeting
SIRT1. Oncotarget. 8:11071–11082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hernandez-Ruiz E, Toll A, Garcia-Diez I,
Andrades E, Ferrandiz-Pulido C, Masferrer E, Yébenes M, Jaka A,
Gimeno J, Gimeno R, et al: The Polycomb proteins RING1B and EZH2
repress the tumoral pro-inflammatory function in metastasizing
primary cutaneous squamous cell carcinoma. Carcinogenesis.
39:503–513. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen H, Pan J, Zhang L, Chen L, Qi H,
Zhong M, Shi X, Du J and Li Q: Downregulation of estrogen-related
receptor alpha inhibits human cutaneous squamous cell carcinoma
cell proliferation and migration by regulating EMT via fibronectin
and STAT3 signaling pathways. Eur J Pharmacol. 825:133–142. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bito T, Sumita N, Ashida M, Budiyanto A,
Ueda M, Ichihashi M, Tokura Y and Nishigori C: Inhibition of
epidermal growth factor receptor and PI3K/Akt signaling suppresses
cell proliferation and survival through regulation of Stat3
activation in human cutaneous squamous cell carcinoma. J Skin
Cancer. 2011:8745712011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Al-Dissi AN, Haines DM, Singh B and Kidney
BA: Immunohistochemical expression of vascular endothelial growth
factor and vascular endothelial growth factor receptor associated
with tumor cell proliferation in canine cutaneous squamous cell
carcinomas and trichoepitheliomas. Vet Pathol. 44:823–830. 2007.
View Article : Google Scholar : PubMed/NCBI
|