Introduction
Colorectal cancer (CRC) represents one of the most
common malignancies worldwide. Every year, >1.3 million people
are diagnosed with CRC globally, of which ~0.7 million succumb to
this disease (1). Only ~50% of
patients with CRC survive for 5 years after diagnosis in Europe
(2), despite some improvements
being made in early diagnosis and systemic therapies. The
pathogenesis and progression of CRC is a complex process, and the
potential mechanism remains unclear.
Long non-coding RNAs (lncRNAs) are a class of
non-encoding RNAs, which are >200 nucleotides long. Recent
research has indicated that numerous lncRNAs serve an important
role in the majority of physiological and pathological processes,
including embryonic stem cell self-renewal, carcinogenesis and
cancer metastasis (3,4). In addition, lncRNAs function as
oncogenes or tumor suppressor genes, which participate in CRC
tumorigenesis and progression. For example, Xu et al
(5) demonstrated that the lncRNA
microRNA (miRNA/miR)-17-92a-1 cluster host gene (MIR17HG) drove
tumor development and metastasis in CRC cells by enhancing the
expression of NF-κB/RELA. In addition, HOX transcript antisense RNA
has been revealed to enhance CRC cell migration and drug resistance
via a miR-203a-3p-dependent Wnt/β-catenin signaling pathway
(6). As an oncogene, the lncRNA
H19 has been shown to accelerate cell proliferation and
metastasis in CRC by acting on Wnt signaling (7).
Small nucleolar RNAs (snoRNAs) are a subgroup of
ncRNAs that are 60-300 nucleotides in length, which contribute to
tumorigenesis and metastasis in diverse types of human cancer. Most
snoRNAs are encoded in the introns of snoRNA host genes (SNHGs)
(8). Specifically, there are
numerous SNHGs associated with CRC carcinogenesis and progression;
for example, SNHG7 has been shown to be overexpressed in CRC tumor
tissues (TTs) compared with its expression in adjacent
non-cancerous tissues (ANTs). This high expression has been
revealed to be related to aggressive pathological characteristics,
such as tumor size, tumor-node-metastasis (TNM) stage, distant
metastasis and poor survival (9,10).
Located on chromosome 8q21.1, SNHG22 is 2,157 nucleotides
long (11). Previous studies have
reported that SNHG22 contributes to cell growth, migration,
invasion and chemotherapy resistance in epithelial ovarian
carcinoma (EOC), papillary thyroid cancer (PTC) and breast cancer
(12-14). Notably, in these three types of
cancer, SNHG22 expression was revealed to be upregulated in TTs
compared with that in ANTs, thus indicating its association with
poor prognosis. To the best of our knowledge, the biological role
and expression patterns of SNHG22 have not been examined in human
CRC.
The present study investigated SNHG22 expression in
adenoma and CRC tissues, and evaluated its clinical significance.
Furthermore, the biological behaviors of SNHG22 were explored in
CRC cell lines in vitro and in vivo. The present
study also investigated the mechanisms underlying the pro-oncogenic
effects of SNHG22 on CRC.
Materials and methods
Tissue samples and cell lines
A total of 93 paired CRC TTs and matched ANTs were
collected from patients who had undergone surgery between January
2012 and October 2012 (Zhengzhou cohort). Additional fresh
specimens were collected from patients with colorectal adenoma
(CRA) (n=33) who had undergone colonoscopy. The CRA or CRC
diagnoses were based on histopathological evaluation using the 7th
edition of the American Joint Committee on Cancer staging system
(15). All specimens were quickly
snap-frozen in liquid nitrogen and stored at −80°C until required.
The present study was approved by the Ethics Committee of Zhengzhou
University (approval no. 2011110402; Zhengzhou, China). Patients
with any history of other types of cancer, and who had received
preoperative radiotherapy or chemotherapy were excluded. The
clinical characteristics of all patients are listed in Table SI.
Human colon cancer cell lines (Caco2, LS174T, LoVo,
SW480, SW620) and a CRC cell line (HT-29) were all obtained from
the Cell Bank of the Chinese Academy of Sciences. The cells were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) in a humidified incubator (37°C, 5%
CO2). The FHC human normal colon epithelial cell line
was purchased from Mingzhou Company (cat. no. MZ-0713) and was
cultured in 90% Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 20% FBS. All
cells tested negative for mycoplasma contamination and this result
was verified by short tandem repeat fingerprinting before use.
Cell transfection
Small interfering RNAs (siRNAs) against SNHG22 (si#1
and si#2) or E2F transcription factor 3 (E2F3) (siE2F3), and the
overexpression vectors pcDNA3.1/Control (Vector), pcDNA3.1/SNHG22
(SNHG22) and pcDNA3.1/E2F3 (E2F3) were purchased from Shanghai
GenePharma Co., Ltd.. A non-targeting sequence was used as the
negative control (siNC; Shanghai GenePharma Co. Ltd.). The miRNA
mimics, inhibitor and negative controls (NC mimic and NC inhibitor)
were purchased from Guangzhou RiboBio Co., Ltd.. For the in
vivo experiments, vectors containing short hairpin RNAs
(shRNAs) targeting SNHG22 (shSNHG22, i.e., sh#1 and sh#2) or a
non-targeting sequence (shNC) were subcloned into Lv5 lentiviruses
(Shanghai GenePharma Co., Ltd.) and infected into LoVo cells to
generate Lv-sh#1 and Lv-sh#2. For LoVo cells infection, cells
(5×104 cell/well) were cultured for 24 h, and then
recombinant lentivirus in serum-free growth medium was added at a
multiplicity of infection of 50 at 37°C for 2 days. For transient
transfection, Caco2, LS174T and LoVo cells (3×104
cell/well) in 6-well plates were transfected with vectors/sequences
at a concentration of 20 µg/ml at 37°C. After culturing for
24 h, cells were harvested for subsequent experiments. Transfection
experiments were conducted with Lipofectamine® 3000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. All the
aforementioned sequences are shown in Table SII.
Cell proliferation
Cell proliferation was detected using Cell Counting
Kit-8 (CCK-8; Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. The cells were transfected with the
indicated vectors or sequences, and then cultured in a 96-well
plate for 1, 2, 3 and 4 days after transfection. Subsequently, ~10
µl CCK-8 reagent was added per well, and incubated at 37°C
for 2 h in a 5% CO2 humidified chamber. The absorbance
was measured at 490 nm in each well using a microplate reader
(Bio-Rad Laboratories, Inc.). The experiment was repeated at least
three times.
Flow cytometry
Cell cycle distribution was detected using flow
cytometry. After the CRC cells had been harvested and washed, cells
were detected with a Cell Cycle and Apoptosis Analysis Kit (cat.
no. C1052M; Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Apoptosis was detected using an Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)-Apoptosis
Detection kit [MultiSciences (Lianke) Biotech Co., Ltd.]. CRC cells
(5×104/well) were seeded and resuspended in 12-well
plates, and Annexin V-FITC (5 µl/well) and PI (100
µl/well) were added to each reaction system for 6 min.
Immediately after staining, flow cytometric assays were conducted
using a flow cytometer (EPICS; Beckman Coulter, Inc.). Analysis of
flow cytometry results was conducted using FlowJo software7.6.1
(FlowJo, LLC).
Wound scratch assay
Cell migration was determined using a wound scratch
assay. Briefly, 1×106 transfected cells/well were seeded
into a 24-well plate. When cells reached 100% confluence, a sterile
20-µl pipette tip was used to produce a clear line in the
wells. After washing with phosphate-buffered saline (PBS), the
cells were grown in serum-free medium at 37°C for 24 h. Under a
light microscope, images of the cells were captured to record the
wound width at 0 h and more images were captured after 24 h. The
migration distance was determined as the distance covered between 0
and 24 h.
Cell migration and invasion assay
Cell migration and invasion were examined using
Transwell inserts (pore size, 8.0 µm) pre-coated without
(for migration) or with (for invasion) Matrigel (on ice for 1 h; BD
Biosciences). Briefly, 2×105/well transfected cells in
200 µl serum-free medium were suspended in the upper
chamber, and the lower chamber was filled with 700 µl medium
containing 15% FBS. After 24 h at 37°C in an atmosphere containing
5% CO2, the membranes in the lower chamber were fixed in
4% paraformaldehyde at room temperature for 24 h and stained with
crystal violet for 18 min. After washing with PBS, images of the
cells on the membrane were captured under a light microscope in
five randomly selected fields per sample.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from frozen tissues and
exponentially growing cells using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.). Nuclear and cytoplasmic RNA were
separated using the PARIS Kit (Thermo Fisher Scientific, Inc.).
cDNA was synthesized from total RNA (1,000 ng) using a reverse
transcriptase cDNA synthesis kit (Takara Biotechnology Co., Ltd.)
according to manufacturer's protocol. mRNA expression levels were
measured using SYBR Premix Ex Taq II (Takara Biotechnology Co.,
Ltd.) on the CFX96 sequence detection system (Bio-Rad Laboratories,
Inc.). The PCR cycling conditions were as follows: 95°C for 30 sec,
followed by 40 cycles at 95°C for 5 sec, 60°C for 30 sec,
dissociation at 95°C for 15 sec, 60°C for 1 min and 95°C for 15
sec. The primer sequences are shown in Table SII. The RT-qPCR data were
calculated using the comparative threshold cycle
(2−ΔΔCq) method with β-actin and U6 as the internal
controls (16).
Western blotting
Total proteins were extracted from cells and tissues
using RIPA buffer (Thermo Fisher Scientific, Inc.). The
concentration of each protein was determined using a BCA protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). Subsequently,
equal amounts of cell lysates (20 µg) were separated by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10%
gels, and then transferred to polyvinylidene difluoride membranes
(MilliporeSigma). The membranes were blocked with TBS-3% Tween-20
containing 5% skim milk at room temperature for 24 h and blotted
with a primary antibody against E2F3 (cat. no. ab50917; 1:2,000;
Abcam) at 4°C for 16 h. Subsequently, membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies (1:10,000;
cat. no. ab205718; Abcam) on room temperature for 2 h. After
washing, immunoreactive bands were detected using chemiluminescence
(Pierce; Thermo Fisher Scientific, Inc.). β-actin (cat. no. ab8227;
1:10,000; Abcam) was used as the loading control. Protein
expression levels were semi-quantified by normalization against
β-actin using ImageJ (version 1.36; National Institutes of
Health).
Luciferase reporter assay
The predicted binding sites of miR-128-3p with the
SNHG22 3′ untranslated region (UTR) or E2F3 3′UTR
were obtained from starBase v3.0 (http://starbase.sysu.edu.cn/). The mutant (mut)
SNHG22 and E2F3 3′ UTR luciferase reporter vectors
were constructed using a Mutagenesis Kit (Qiagen, Inc.). The
wild-type (wt) and mut sequences were each cloned into a psiCHECK2
vector (Promega Corporation). CRC cells (1×103/well) and
293T cells (1×103/well; American Type Culture
Collection) in 96-well plates were co-transfected with wt/mut
plasmid and miR-128-3p or NC (50 µm) using Lipofectamine
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.). After 36-h
transfection at 4°C, the cells were collected and tested using a
dual-luciferase assay system (Promega Corporation) according to the
manufacturer's instructions. pRL-TK was used as the internal
control.
RNA immunoprecipitation (RIP)
RIP was performed using an EZ-Magna RIP Kit
(MilliporeSigma) and an anti-Argonaute-2 (Ago2) antibody (cat. no.
ab32381; Abcam). CRC cells (2×107) were lysed in RIP
buffer and centrifuged at 10,000 × g at 4°C for 5 min. The cell
lysates were incubated with magnetic beads (100 µl)
conjugated with the anti-Ago2 antibody (cat. no. ab32381; Abcam) or
immunoglobulin G (cat. no. ab104155; Abcam) for 3 h. After
extensive washing using an elution buffer at 65°C for 10 min, the
purified RNA was analyzed by RT-qPCR.
Animal experiments
All animal experiments were approved by the
Committee of the Ethics of Animal Experiments of Zhengzhou
University (approval no. 20180376). A total of 15 male BALB/c nude
mice (age, 4 weeks; weight, 14-16 g; Shanghai Experiment Animal
Centre) were maintained under specific pathogen-free conditions and
randomly subdivided into three subgroups. LoVo cells that had been
stably infected with Lv-shNC, Lv-sh#1 and Lv-sh#2 vectors were
harvested, and 6×106 cells were injected subcutaneously
into the dorsal flank region of each mouse. Tumor growth was
measured every 5 days, and was calculated using the following
formula: Volume=(width2 × length)/2. Euthanasia was
performed by intraperitoneal injection of 200 mg/kg pentobarbital
(17). Subsequently, the weight
of each tumor was evaluated, and the tumor samples were processed
for RT-qPCR, western blotting and immunohistochemistry (IHC). The
present study was carried out according to the National Institutes
of Health Guide for the Care and Use of Laboratory Animals
(18).
Hematoxylin-eosin (HE) staining and
IHC
The tumors from the nude mice were fixed in 10%
paraformaldehyde at 4°C for 12 h, and then dehydrated in different
concentrations of ethanol, permeabilized using xylene and embedded
in paraffin. The paraffin-embedded tumor tissues were then sliced
(0.5 µm), rehydrated, and were stained with HE at 4°C for 10
min and sealed with neutral gum. For IHC assessment of E2F3 and
Ki-67 in colorectal tumor tissues from the nude mice, the DAKO
Envision system (Dako; Agilent Technologies, Inc.) was used.
Briefly, after paraffin-embedded sections of tumor tissues were
heated at 60°C, the sections were incubated with primary antibodies
against E2F3 (1:200; cat. no. ab50917; Abcam) and Ki-67 (1:1,000;
cat. no. ab279653; Abcam) overnight at 4°C. Then, the sections were
incubated with biotin-labeled secondary antibodies (1:1,000; cat.
no. ab205718; Abcam) at 37°C for 20 min. Under a light microscope,
IHC staining scores for E2F3 were obtained by multiplying the
intensity (0, negative; 1, low; 2, medium; and 3, high) with the
extent of staining (0, 0%; 1, 0-10%; 2, 10-50%; 3, 50-75%; 4,
>75%); the final scores were between 0-12. For evaluation of
Ki67, the number of positive cells was calculated in three
representative areas of high staining.
Bioinformatics and statistical
analyses
lncLocator (19)
was used to analyze lncRNA subcellular locations. CPAT (http://lilab.research.bcm.edu/cpat/index.php) and
LNCipedia (https://lncipedia.org) were used to
analyze the protein coding probability. The online tools TargetScan
(http://www.targetscan.org/), miRwalk
(http://mirwalk.umm.uni-heidelberg.de), miRTarbase
(http://mirtarbase.mbc.nctu.edu.tw/index.html), miRDB
(http://www.mirdb.org/), and starBase (http://starbase.sysu.edu.cn/index.php)
were used to analyze potential target genes of miR-128-3p. Analysis
of data from The Cancer Genome Atlas (TCGA)-colon adenocarcinoma
(COAD) (n=267) was conducted using starBase and UALCAN (http://ualcan.path.uab.edu/index.html)
online tools. All data are presented as the mean ± standard
deviation from triplicate experiments. Regarding comparisons,
Student's t-test (two groups) or one-way ANOVA with Scheffe test
(multiple groups) was conducted as appropriate. To compare SNHG22
expression levels in TT, ANT and CRA samples, paired samples were
analyzed using a paired Student's t-test and independent samples
were analyzed using unpaired Student's t-tests, after which, a
Bonferroni correction was applied. To compare E2F3 and Ki-67
staining scores in mice tumor samples, Kruskal-Wallis and Dunn's
post hoc test was used. Correlations were calculated using
Spearman's correlation analysis. The survival curves were
constructed using the Kaplan-Meier method and were analyzed using
the log-rank test. Crude and adjusted hazard ratios (HRs) and 95%
confidence intervals (CIs) were calculated using Cox proportional
hazards modeling. Potential confounders were included in the
multivariate analysis at a significance level of P<0.15, as
determined by univariate analysis. All statistical analyses were
conducted using the PASW Statistics 19.0 software program (SPSS,
Inc.) and GraphPad Prism version 8.0 (GraphPad, Inc.). P<0.05
was considered to indicate a statistically significant
difference.
Results
SNHG22 is upregulated in CRC tissues and
cell lines, and is associated with poor prognosis in patients with
CRC
To investigate SNHG22 expression levels in CRC,
RT-qPCR analysis was performed to examine SNHG22 levels in TT and
ANT colorectal samples (n=93 pairs) and human CRA samples (n=32).
SNHG22 expression was significantly higher in TTs compared with
that in CRA tissues and ANTs (Fig.
1A). Additionally, high SNHG22 expression was significantly
associated with advanced T stage (Fig. 1B), lymph node metastasis (Fig. 1C), distant metastasis (Fig. 1D), advanced clinical stage
(Fig. 1E), and poor disease-free
survival (DFS) and overall survival (OS) (Fig. 1F and G). Data from TCGA confirmed
these results (Fig. 1H and I).
Multivariate Cox regression analysis validated high SNHG22
expression as a predictor of poor DFS (HR, 1.68; 95% CI, 1.04-2.71)
and OS (HR, 1.45; 95% CI, 1.02-2.06; Table I). High SNHG22 expression was also
confirmed in the CRC cell lines (LS174T, LoVo, Caco2, SW480, HT-29
and SW620) compared with that in the FHC cell line (Fig. 1J). These results indicated that
SNHG22 was highly expressed in CRC tissues and cell lines, and may
be associated with poor survival in CRC. Furthermore, the coding
potential of SNHG22 was calculated using CPAT and LNCipedia; the
results revealed that SNHG22 had very low coding probability, thus
indicating that it may be a ncRNA (Fig. S1).
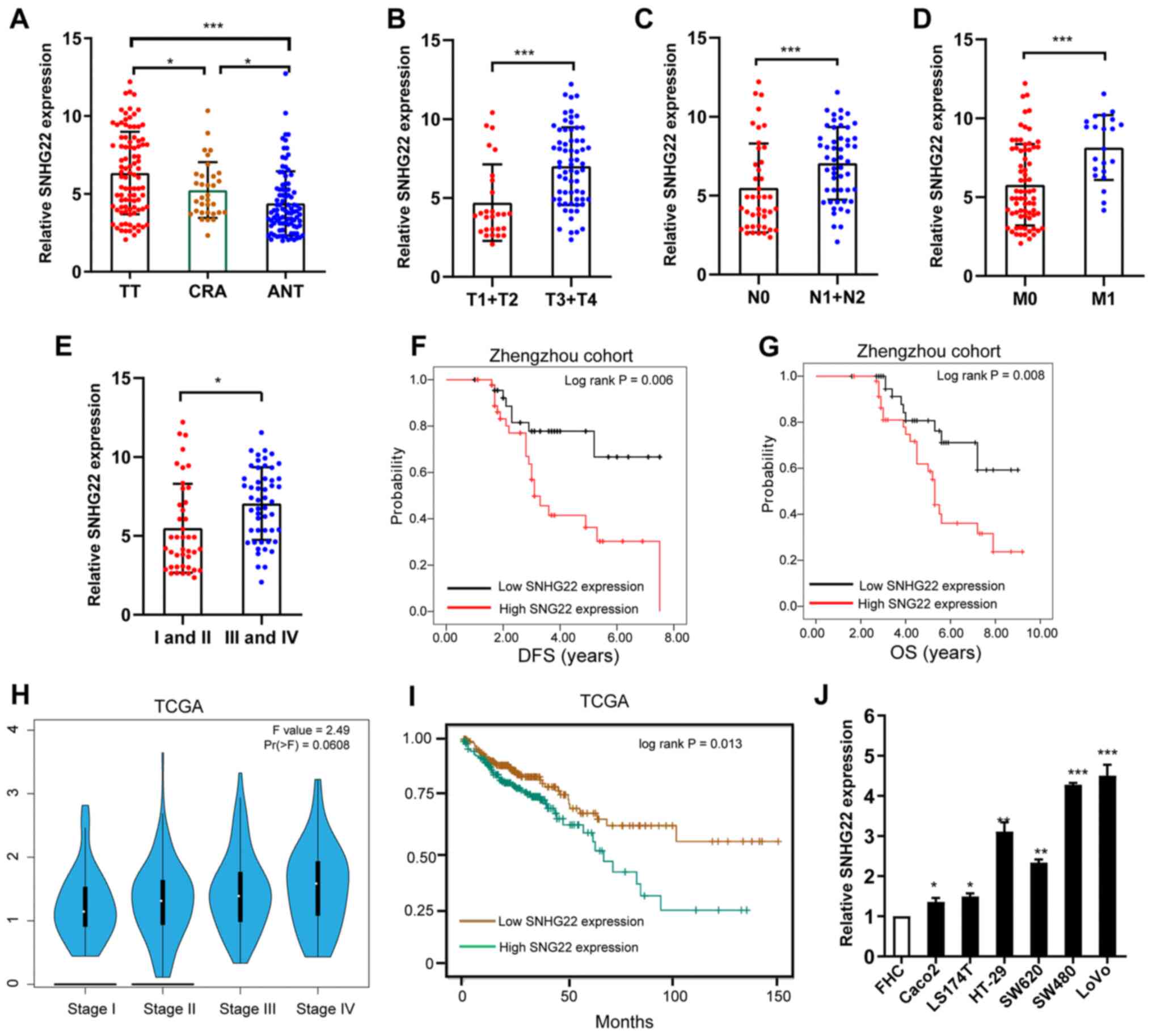 | Figure 1Differential expression of SNHG22 in
CRC tissues and cell lines. (A) RT-qPCR analysis of the expression
levels of SNHG22 in TTs (n=93), ANTs (n=93) and CRA (n=33) colon
tissues. Differential expression of SNHG22 in CRC tissues was
analyzed according to (B) T stage, (C) node status, (D) distant
metastasis and (E) TNM stage. Kaplan-Meier survival analysis of
SNHG22 expression and (F) DFS and (G) OS in patients with CRC. (H)
Expression of SNHG22 in patients with CRC split according to TNM
stage in TCGA-COAD dataset. (I) OS in patients with CRC in
TCGA-COAD dataset. (J) RT-qPCR analysis of the expression levels of
SNHG22 in CRC cell lines and the FHC cell line, a human normal
colon epithelial cell line. Data are presented as the mean ± SD
from triplicate experiments. *P<0.05,
**P<0.01, ***P<0.001 as indicated or
vs. FHC. ANT, adjacent non-cancerous tissue; COAD, colorectal
adenocarcinoma; CRA, colorectal adenoma; CRC, colorectal cancer;
DFS, disease-free survival; OS, overall survival; RT-qPCR, reverse
transcription-quantitative PCR; SNHG22, small nucleolar RNA host
gene 22; TCGA, The Cancer Genome Atlas; TNM, tumor-node-metastasis;
TT, tumor tissue. |
 | Table ICox proportional hazards regression
models for overall and recurrence-free survival among patients with
colorectal cancer. |
Table I
Cox proportional hazards regression
models for overall and recurrence-free survival among patients with
colorectal cancer.
| Parameter | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Disease-free
survival | | | | |
| Location, rectum
vs. colon | 1.98
(0.85-4.60) | 0.112 | 1.67
(0.71-3.90) | 0.237 |
| T stage, T3-4 vs.
T1-2 | 2.13
(1.05-4.30) | 0.035 | 1.66
(1.02-2.70) | 0.041 |
| Node involvement,
yes vs. no | 2.35
(1.07-5.13) | 0.033 | 1.27
(1.00-1.61) | 0.048 |
| M stage, M1 vs.
M0 | 2.56
(1.23-5.33) | 0.012 | 1.51
(1.04-2.20) | 0.031 |
| Tumor
differentiation, poor vs. well + moderate | 1.49
(0.95-2.34) | 0.084 | 1.14
(0.88-1.48) | 0.321 |
| CEA, ≥5 vs. <5
ng/ml | 1.40
(0.68-2.86) | 0.362 | | |
| Margin, positive
vs. negative | 1.32
(0.61-2.84) | 0.483 | | |
| Chemotherapy, yes
vs. no | 1.34
(0.57-3.15) | 0.501 | | |
| SNHG22, high vs.
low | 2.47
(1.13-5.41) | 0.023 | 1.68
(1.04-2.71) | 0.034 |
| Overall
survival | | | | |
| Location, rectum
vs. colon | 2.19
(1.01-4.75) | 0.047 | 1.81
(0.83-3.96) | 0.138 |
| T stage, T3-4 vs.
T1-2 | 1.59
(1.00-2.53) | 0.050 | 1.33
(0.97-1.82) | 0.077 |
| Node involvement,
yes vs. no | 1.91
(1.12-3.26) | 0.018 | 1.57
(1.06-2.33) | 0.025 |
| M stage, M1 vs.
M0 | 1.98
(1.24-3.15) | 0.004 | 1.49
(1.08-2.05) | 0.014 |
| Tumor
differentiation, poor vs. well + moderate | 1.39
(0.66-2.93) | 0.395 | | |
| CEA, ≥5 vs. <5
ng/ml | 1.06
(0.54-2.09) | 0.868 | | |
| Margin, positive
vs. negative | 0.97
(0.45-2.08) | 0.930 | | |
| SNHG22, high vs.
low | 2.21
(1.08-4.55) | 0.031 | 1.45
(1.02-2.06) | 0.039 |
Overexpression of SNHG22 stimulates CRC
cell proliferation, apoptosis resistance, migration, and invasion
in vitro
To investigate the role of SNHG22 in the progression
of CRC cells, gain-of-function experiments were performed using the
Caco2 and LS174T cell lines because both cell lines had low SNHG22
expression levels. SNHG22 expression was significantly enhanced
after the cells had been transfected with the SNHG22 vector
(Fig. S2A). Functionally,
upregulating SNHG22 expression promoted the proliferation,
G2/M phase arrest and apoptosis resistance of both cell
lines (Figs. 2A-F and S2B). Furthermore, the wound scratch and
Transwell assays demonstrated a marked enhancement in the migratory
and invasive ability of SNHG22-overexpressing cells (Fig. 2G-J). These data indicated that
SNHG22 overexpression may promote CRC cell proliferation, apoptosis
resistance, migration and invasion.
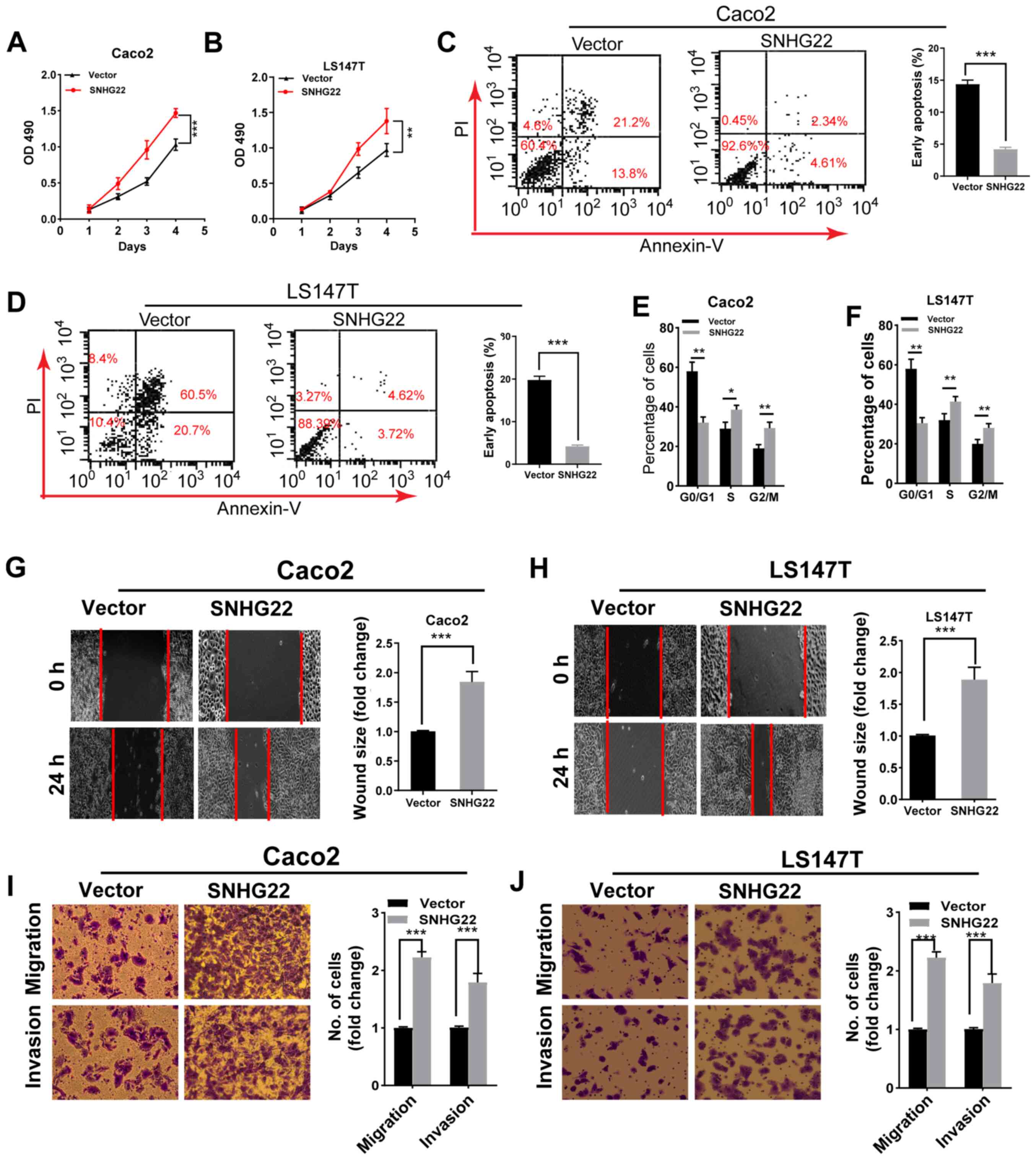 | Figure 2Overexpression of SNHG22 promotes
colorectal cancer cell proliferation, apoptosis resistance,
migration and invasion in vitro. Cell Counting Kit-8 assay
analysis of the proliferative ability of (A) Caco2 and (B) LS147T
cells transfected with the indicated vectors. Flow cytometric
analysis of the (C and D) apoptotic rates and (E and F) cell cycle
progression of Caco2 and LS147T cells transfected with the
indicated vectors. Wound scratch analysis of the migration of (G)
Caco2 and (H) LS147T cells transfected with the indicated vectors
(magnification, ×200). Transwell assay analysis of the migration
and invasion of (I) Caco2 and (J) LS147T cells transfected with the
indicated vectors (magnification, ×200). Data are presented as the
mean ± SD from triplicate experiments. *P<0.05,
**P<0.01, ***P<0.001. NC, negative
control; OD, optical density; PI, propidium iodide; SNHG22, small
nucleolar RNA host gene 22. |
Knockdown of SNHG22 inhibits CRC cell
proliferation, apoptosis resistance, migration, and invasion in
vitro
To determine whether silencing SNHG22 would affect
CRC cell biological function, two parallel siRNAs targeting SNHG22
(i.e., si#1 and si#2) were used for the knockdown experiments.
RT-qPCR confirmed the knockdown efficiency of both siRNAs in the
LoVo cell line (Fig. S3A). The
results of CCK-8 assay and flow cytometry demonstrated a marked
decrease in LoVo cell proliferation, G2/M phase arrest
and apoptosis resistance in cells with SNHG22 knockdown (Figs. 3A-C and S3B). Furthermore, the wound scratch and
Transwell assays indicating that silencing SNHG22 attenuated LoVo
cell migratory and invasive ability (Fig. 3D and E). These data indicated that
knocking down SNHG22 may inhibit CRC cell proliferation, migration
and invasion.
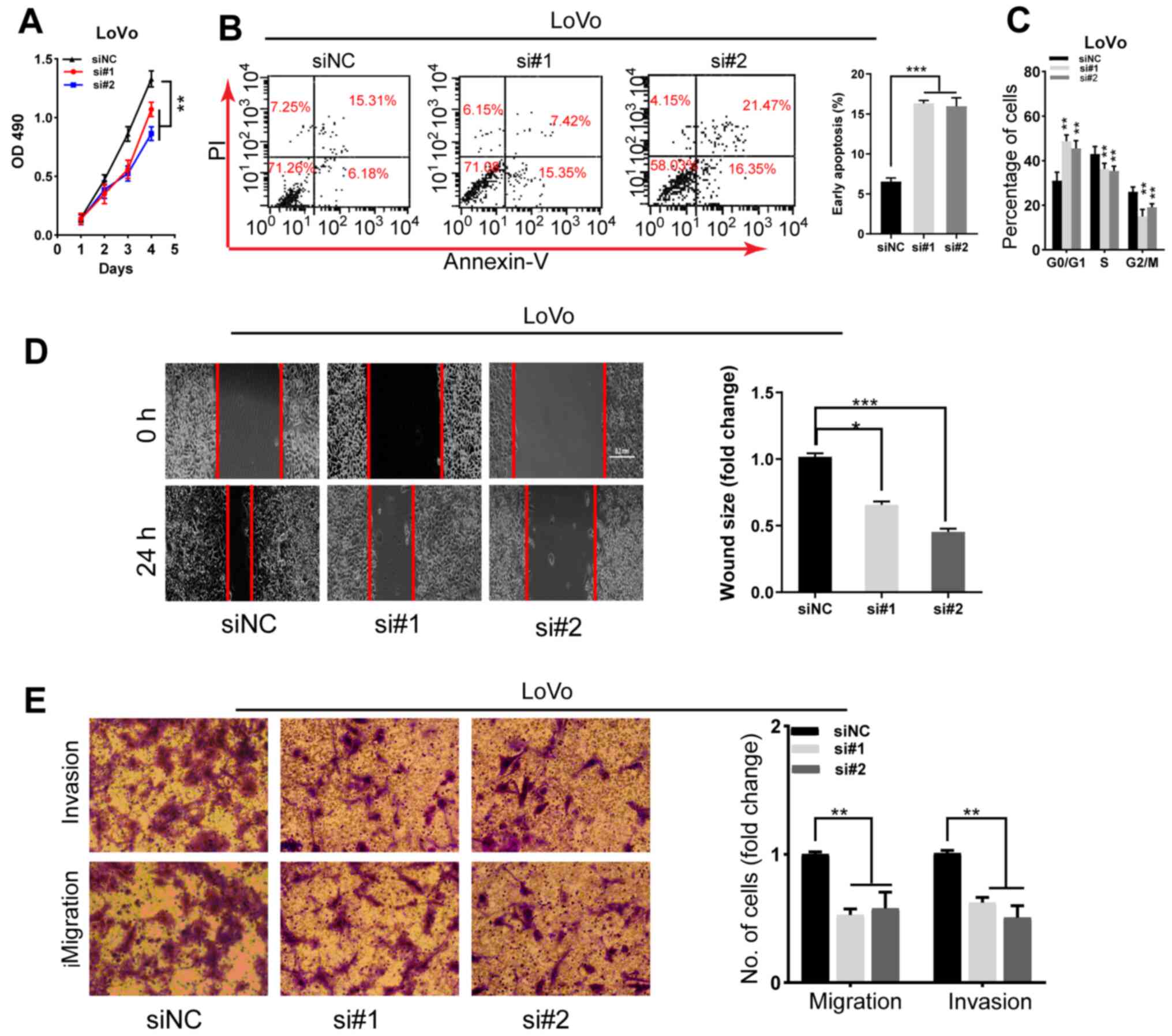 | Figure 3Knockdown of SNHG22 inhibits
colorectal cancer cell proliferation, apoptosis resistance,
migration and invasion in vitro. (A) Cell Counting Kit-8
assay analysis of the proliferative ability of LoVo cells after
transfection with the indicated sequences. Flow cytometric analysis
of the (B) apoptotic rates and (C) cell cycle progression of LoVo
cells after transfection with the indicated sequences. (D) Wound
scratch analysis of the migration of LoVo cells after transfection
with the indicated sequences (magnification, ×200). (E) Transwell
invasion assay analysis of the invasion of LoVo cells after
transfection with the indicated sequences (magnification, ×200).
Data are presented as the mean ± SD from triplicate experiments.
*P<0.05, **P<0.01,
***P<0.001 as indicated or vs. siNC. NC, negative
control; OD, optical density; PI, propidium iodide; si, small
interfering; SNHG22, small nucleolar RNA host gene 22. |
SNHG22 functions as a competing
endogenous RNA (ceRNA) to regulate miR-128-3p expression in CRC
cells
As the subcellular localization of lncRNAs can
indicate their function, the results from lncLocator (19) demonstrated that SNHG22 was
preferentially localized in the cytoplasm (Fig. S4). Subsequent cellular
fractionation experiments confirmed this in CRC cells (Caco2,
LS174T and LoVo; Fig. 4A). The
starBase v3.0 findings indicated that SNHG22 contains complementary
binding sites to miR-128-3p seed regions (Fig. 4B). To test this hypothesis, RIP
experiments were performed with anti-Ago2 in Caco2 and LS174T
cells, and observed enrichment of SNHG22 with the Ago2 antibody was
detected (Fig. 4C), indicating
that SNHG22 had complementary binding sites to miR-128-3p and
directly bound to miR-128-3p. To explore whether SNHG22 is
regulated by miR-128-3p, dual-luciferase reporter plasmids were
constructed containing wt or mut putative binding sites of SNHG22
transcripts. Luciferase activity was markedly reduced in 293T cells
that had been co-transfected with SNHG22 wt and miR-128-3p mimic
compared with those transfected with SNHG22 wt and NC mimic.
However, no statistical changes in luciferase activity were
observed in cells transfected with the mut reporter. These results
were confirmed in the Caco2 and LS174T cell lines (Fig. 4D). Furthermore, RT-qPCR revealed
that SNHG22 overexpression significantly decreased miR-128-3p
expression levels in CRC cells, whereas silencing SNHG22 enhanced
miR-128-3p expression (Fig. 4E and
F). SNHG22 expression had a significant inverse relationship
with miR-128-3p expression in human CRC tissues from the Zhengzhou
cohort (Fig. 4G). These findings
demonstrated that SNHG22 may act as a miR-128-3p sponge in CRC.
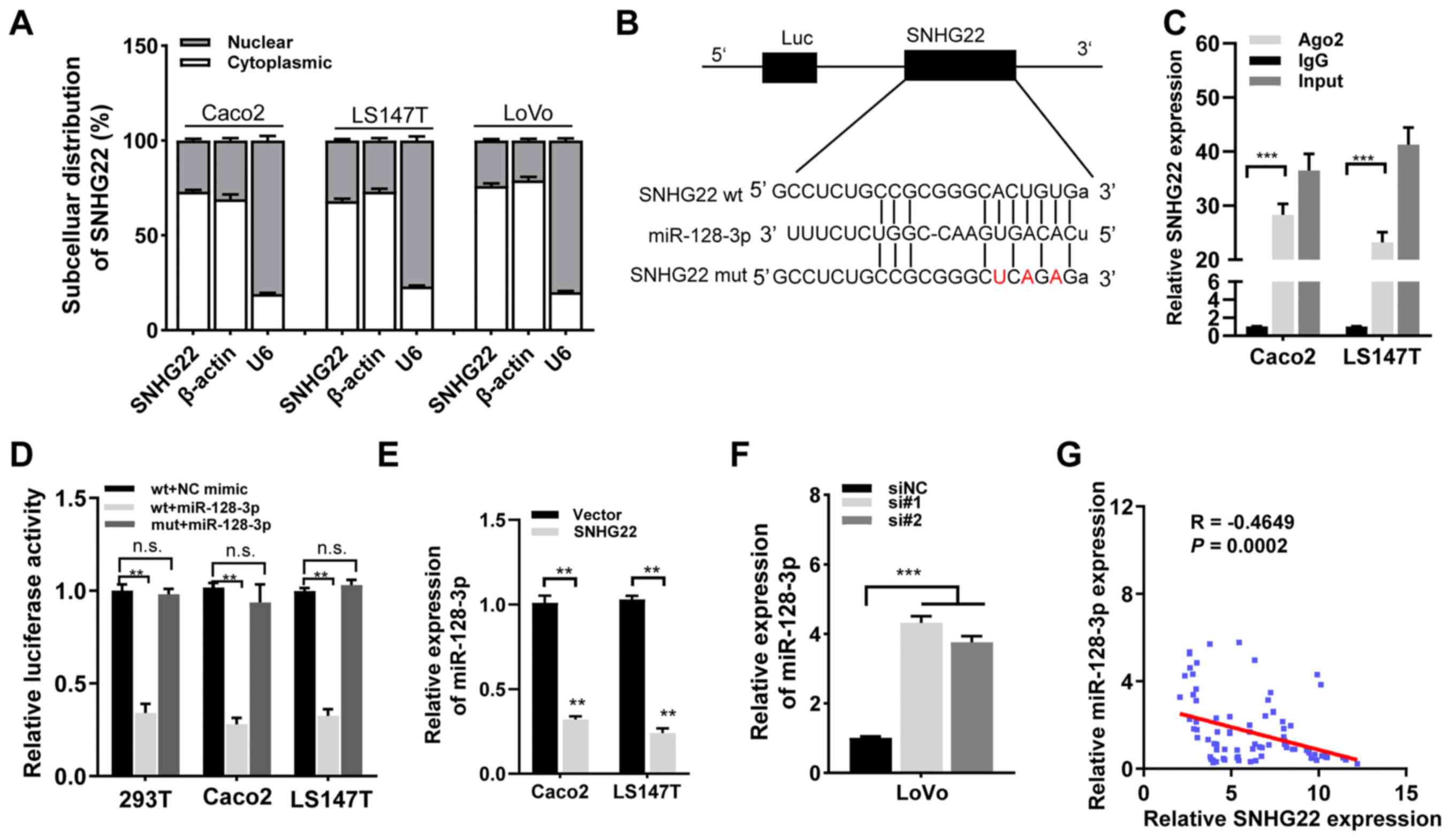 | Figure 4SNHG22 functions as a competing
endogenous RNA to regulate miR-128-3p expression in CRC. (A)
Subcellular localization of SNHG22 in CRC cell lines (Caco2, LS147T
and LoVo). β-Actin and U6 served as cytoplasmic and nuclear
localization markers, respectively. (B) Predicted binding sites of
miR-128-3p to the SNHG22 sequence. (C) RNA immunoprecipitation
assay was performed with an antibody against Ago2 or IgG in Caco2
and LS147T cell lines. (D) Luciferase activity of 293T, Caco2 and
LS147T cell lines co-transfected with miR-128-3p mimic (or NC
mimic) and luciferase reporters containing SNHG22 wt or SNHG22 mut
transcript was analyzed. (E and F) Reverse
transcription-quantitative PCR analysis of the expression levels of
miR-128-3p in CRC cells after transfection with the indicated
vectors and sequences. (G) Negative correlation between SNHG22 and
miR-128-3p expression in human CRC tissues from the Zhengzhou
cohort (Spearman's correlation analysis; R=-0.4649, P=0.002). Data
are presented as the mean ± SD from triplicate experiments.
**P<0.01, ***P<0.001. Ago2, Argonaute2;
CRC, colorectal cancer; EV, empty vector; miR, microRNA; mut,
mutant; NC, negative control; n.s., not significant; si, small
interfering; SNHG22, small nucleolar RNA host gene 22; wt,
wild-type. |
miR-128-3p targets and regulates
E2F3
Five independent algorithmic programs were used to
define putative targets of miR-128-3p, and identified 25 common
predicted targets (Fig. 5A).
Among these, E2F3 was selected because TCGA-COAD data indicated
that E2F3 can enhance CRC progression and may be associated with
tumor (Figs. 5B, and S5A and B). After successfully
transfecting miR-128-3p mimics and inhibitor into CRC cells
(Fig. S5C and D), it was
revealed that miR-128-3p overexpression markedly decreased the mRNA
and protein expression levels of E2F3 in Caco2 and LS174T cells,
whereas silencing miR-128-3p enhanced the expression levels of E2F3
(Fig. 5C-F). Moreover, Spearman's
correlation analysis identified a significant inverse relationship
between the expression levels of E2F3 mRNA and miR-128-3p in human
CRC tissues from the Zhengzhou cohort (Fig. 5G). To determine if E2F3 is an
authentic target of miR-128-3p, luciferase reporter gene constructs
were generated wherein the E2F3 3′UTR sequences were fused with the
Renilla luciferase coding sequence (Fig. 5H). The upregulation of miR-128-3p
significantly decreased the luciferase activity of the E2F3 wt
3′UTR. (Fig. 5I). Nevertheless,
altering miR-128-3p expression had no significant effects on the
mut construct (Fig. 5I). In
addition, E2F3 mRNA expression had a meaningful positive
relationship with SNHG22 in human CRC tissues from the Zhengzhou
cohort (Fig. 5J). These data
suggested that E2F3 could be a direct target of miR-128-3p in CRC
cells.
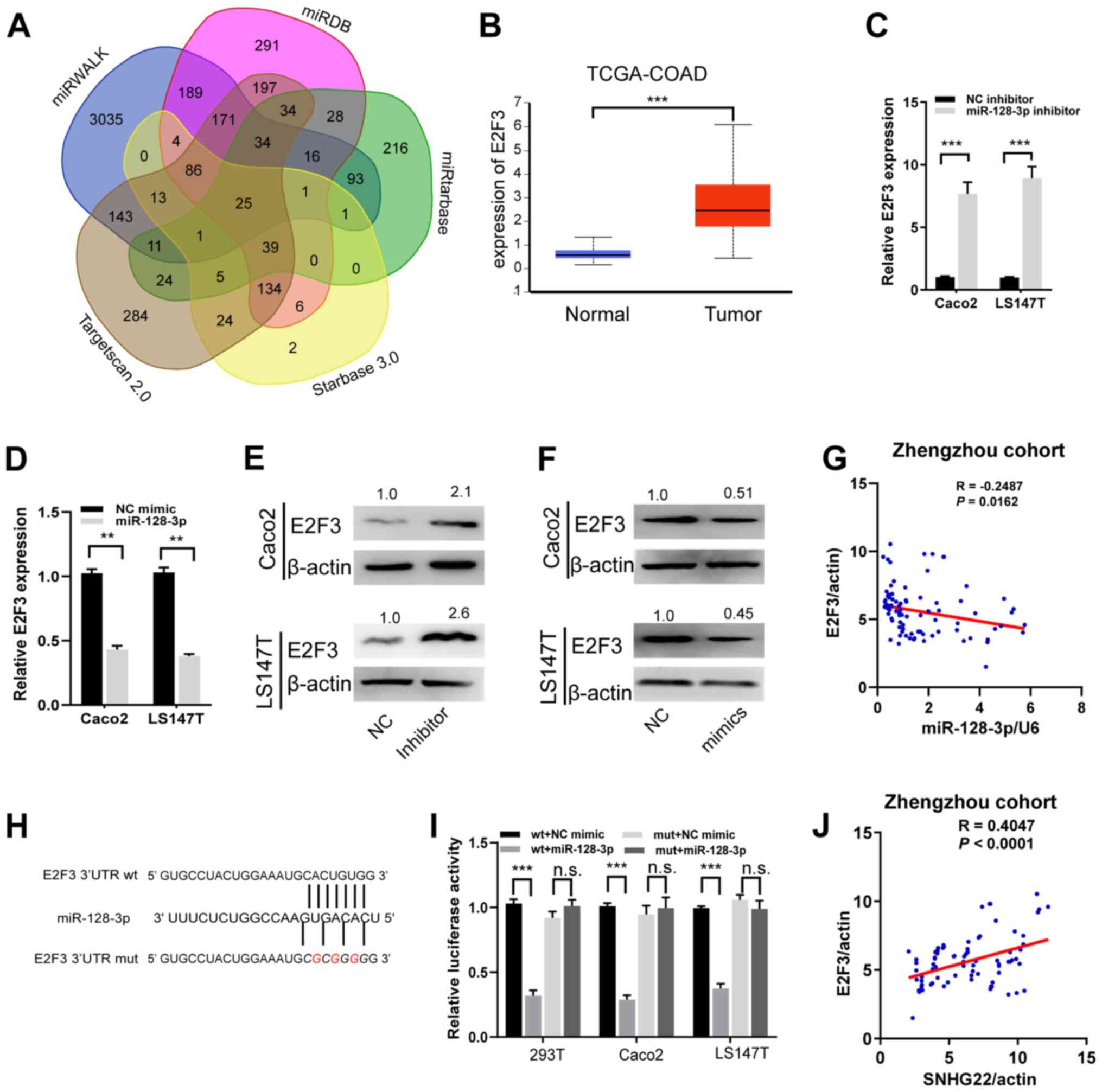 | Figure 5miR-128-3p targets and regulates
E2F3. (A) A Venn diagram showing the number of genes identified as
potential targets of miR-128-3p. (B) Expression levels of E2F3 in
tumor and unmatched normal tissues in TCGA-COAD dataset. (C and D)
Reverse transcription-quantitative PCR and (E and F) western blot
analysis of the mRNA and protein expression levels of E2F3 in Caco2
and LS147T cells after transfection with the indicated sequences.
Protein expression levels were semi-quantified by normalization
against β-actin with ImageJ. (G) A significant inverse association
was detected between miR-128-3p and E2F3 mRNA expression in human
CRC tissues from the Zhengzhou cohort (R=−0.2487, P=0.016). (H)
miR-128-3p putative binding sites and corresponding mut sequence of
E2F3. (I) Dual-luciferase reporter assays in CRC cell lines
co-transfected with miR-128-3p mimic and luciferase reporters
containing wt or mut E2F3 transcripts. The relative luciferase
activity was normalized to the Renilla luciferase activity.
(J) A positive correlation between SNHG22 and E2F3 expression in
human CRC tissues from Zhengzhou cohort (R=−0.4047, P<0.001).
Data are presented as the mean ± SD from triplicate experiments.
**P<0.01, ***P<0.001. COAD, colon
adenocarcinoma; CRC, colorectal cancer; E2F3, E2F transcription
factor 3; miR, microRNA; mut, mutant; NC, negative control; n.s.,
not significant; TCGA, The Cancer Genome Atlas; UTR, untranslated
region; wt, wild-type. |
SNHG22 exerts its function by inhibiting
the miR-128-3p/E2F3 axis in CRC cells
Functional rescue experiments were performed to
examine whether the miR-128-3p/E2F3 axis mediates the biological
roles of SNHG22 in CRC cells. The results of RT-qPCR analyses
confirmed the transfection efficiencies of E2F3 overexpression and
siRNA vectors in CRC cells (Fig. S6A
and B). SNHG22 or E2F3 overexpression stimulated Caco2 cell
proliferation, apoptosis resistance, migration and invasion,
whereas upregulating miR-128-3p reversed these effects. By
contrast, knocking down SNHG22 or E2F3 significantly weakened the
LoVo cell malignant phenotype; nevertheless, these inhibitory
effects were attenuated by co-transfection with miR-128-3p
inhibitors (Figs. 6A-F, and
S6C and D). These results
indicated that the miR-128-3p/E2F3 axis reversed the stimulatory
effects of SNHG22 on the proliferative, migratory and
invasive capacity of CRC cells.
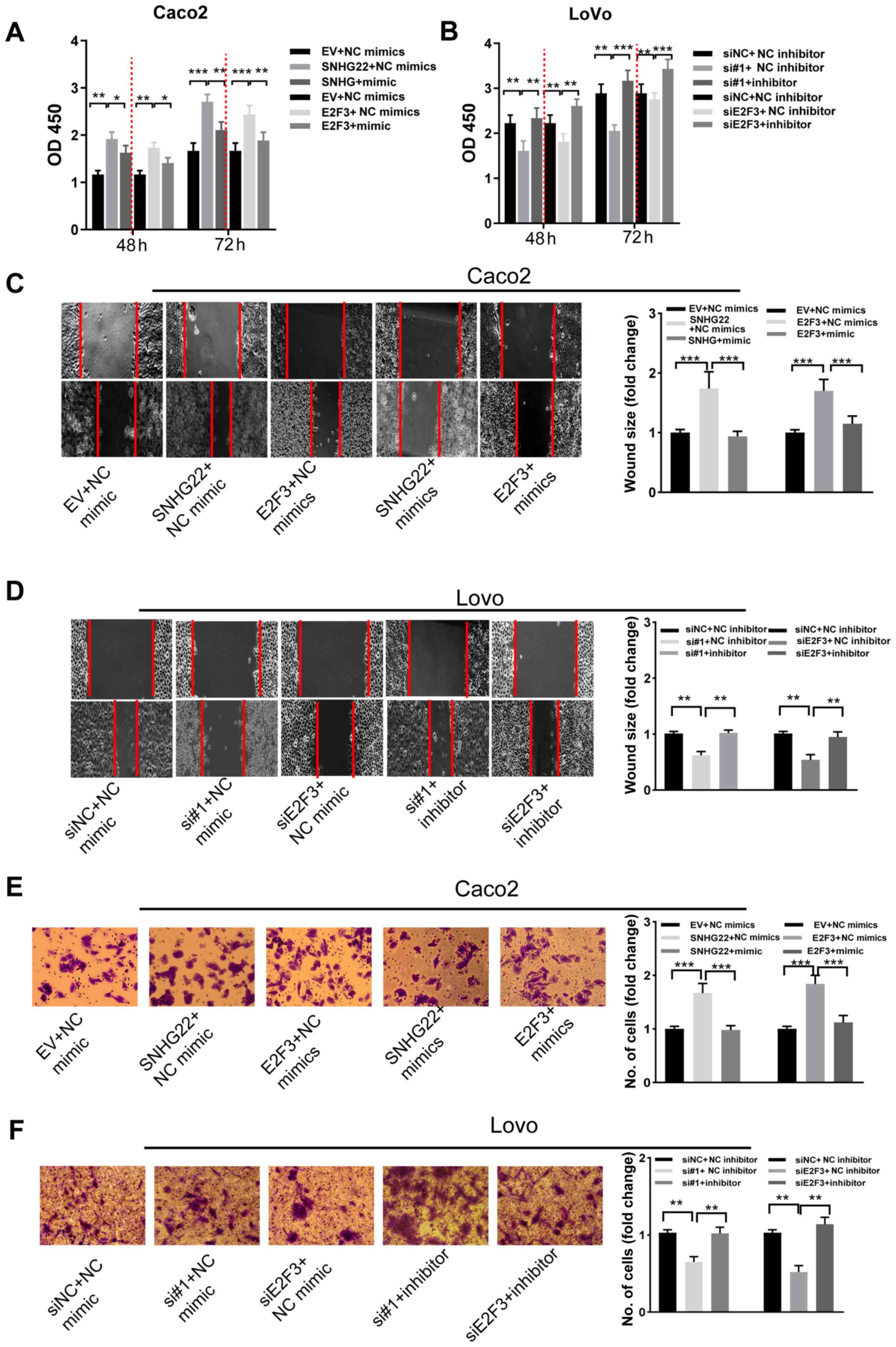 | Figure 6SNHG22 exerts its function by
inhibiting the miR-128-3p/E2F3 axis in CRC cells. Cell Counting
Kit-8 assay analysis of the proliferative ability of (A) Caco2 and
(B) LoVo cells after transfection with the indicated vectors and
sequences. Wound scratch analysis of the migratory ability of (C)
Caco2 and (D) LoVo cells after transfection with the indicated
vectors and sequences (magnification, ×200). Transwell assay
analysis of the invasive ability of (E) Caco2 and (F) LoVo cells
after transfection with the indicated vectors and sequences
(magnification, ×200). Data are presented as the mean ± SD from
triplicate experiments. *P<0.05,
**P<0.01, ***P<0.001. E2F3, E2F
transcription factor 3; EV, empty vector; miR, microRNA; NC,
negative control; OD, optical density; si, small interfering;
SNHG22, small nucleolar RNA host gene 22. |
Knockdown of SNHG22 inhibits CRC tumor
xenograft growth in vivo
A tumor xenograft assay was performed to confirm the
roles of SNHG22 in CRC growth in vivo. As shown in Fig. 7A-C, compared with in the Lv-shNC
group, the tumor size, volume and weight in mice in the Lv-sh#1 and
Lv-sh#2 groups were markedly reduced (P<0.01). Furthermore,
RT-qPCR verified the decreased expression levels of SNHG22 and
E2F3, and the increased expression levels of miR-128-3p in tumors
from the Lv-sh#1 and Lv-sh#2 groups compared with those in the shNC
group (Fig. 7D-F).
Immunohistochemistry showed that tumors from the Lv-sh#1 and
Lv-sh#2 groups had decreased expression levels of Ki-67 and E2F3
staining compared with those in tumors from the shNC group
(Fig. 7G).
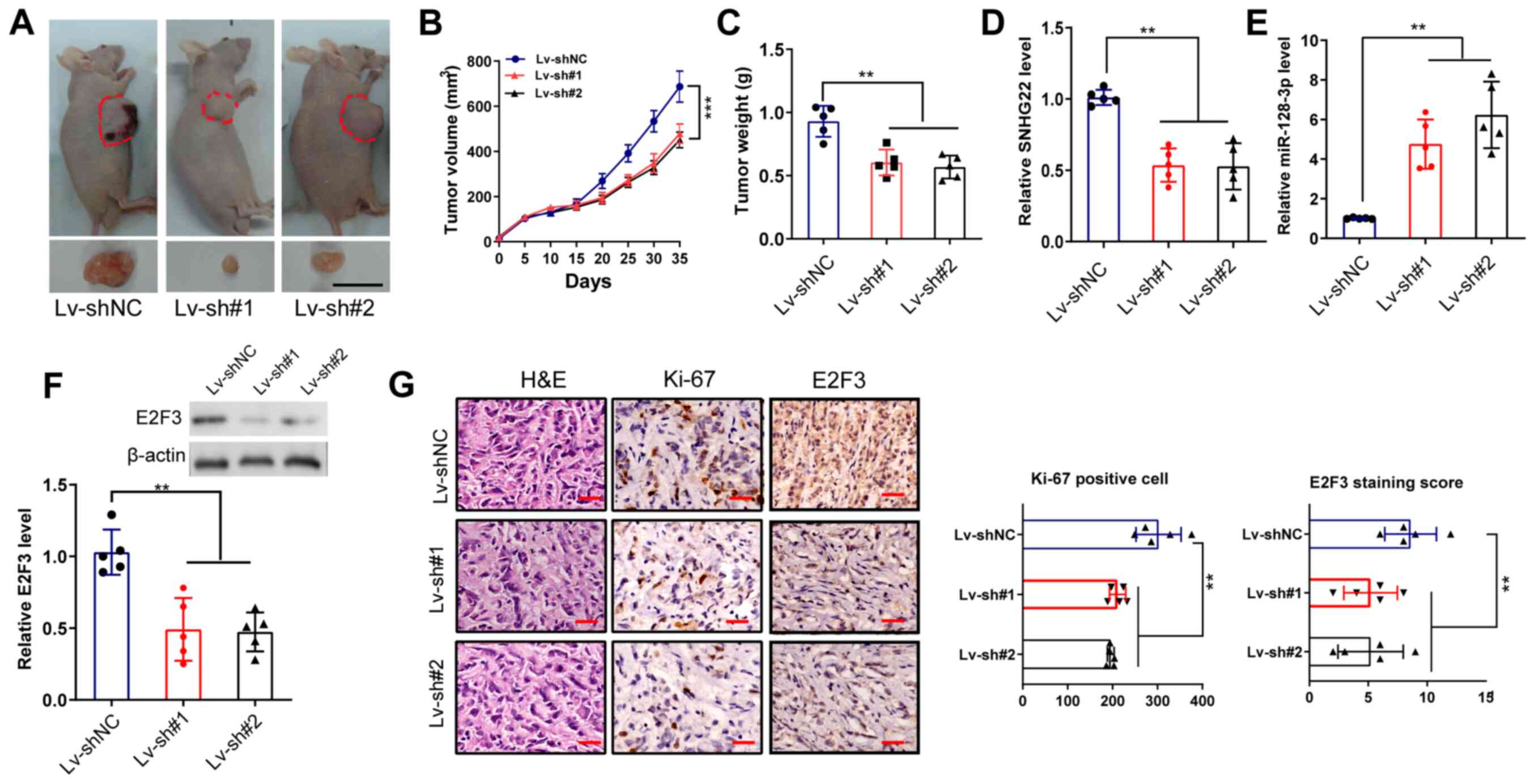 | Figure 7Knockdown of SNHG22 inhibits tumor
xenograft growth of CRC in vivo. (A) LoVo cells were
inoculated in BALB/c nude mice (n=5/group) to establish
subcutaneous xenograft tumors, and images of the dissected tumors
were captured. Scale bar, 10 mm. Effects of SNHG22 knockdown in
LoVo cells on (B) tumor volume and (C) tumor weight in the
subcutaneous xenograft mouse models. (D and E) Reverse
transcription-quantitative PCR and (F) western blot analysis of the
expression levels of SNHG22, miR-128-3p and E2F3 in the
subcutaneous xenograft mouse models. (G) Immunohistochemical
analysis of the expression levels of Ki-67 and E2F3 in the
subcutaneous xenograft mouse models. Scale bar, 100 µm. Data
are presented as the mean ± SD from triplicate experiments.
**P<0.01, ***P<0.001. E2F3, E2F
transcription factor 3; HE, hematoxylin and eosin; Lv, lentivirus;
miR, microRNA; NC, negative control; sh, short hairpin; SNHG22,
small nucleolar RNA host gene 22. |
Discussion
Accumulating evidence has recently revealed that the
dysregulation of lncRNAs serves an essential role in the
development and invasion of human cancer, including CRC (20,21). The present study revealed that
SNHG22 expression was significantly elevated in CRC tissues
and cell lines. High levels of SNHG22 expression were
significantly associated with unfavorable clinicopathological
characteristics and worse survival in patients with CRC.
Functionally, ectopic SNHG22 overexpression drove proliferation,
apoptosis resistance, migration and invasion in CRC cell lines.
Knocking down SNHG22 inhibited xenograft tumor growth in
vivo. The present study confirmed that SNHG22 performed
its tumor-promoting function by sponging miR-128-3p, and enhancing
the expression and activity of E2F3. To the best of our knowledge,
the present results are the first to clarify that SNHG22
acts as an oncogene in CRC, and that it may be used as a potential
therapeutic target for this disease.
The current anatomically based TNM staging system
cannot precisely distinguish the risk of recurrence/distant
metastasis in patients with CRC; therefore, it is essential to
identify novel prognostic biomarkers. The dysregulation of lncRNAs
in various human tumors is associated with excellent or poor
survival, thus making them promising prognostic biomarkers
(22,23). For example, SNHG7
expression has been reported to be enhanced in CRC tissues compared
with that in non-cancerous tissues, and this high expression was
related to aggressive pathological characteristics, such as TNM
stage, lymphatic metastasis and distant metastasis, as well as poor
prognosis (9,10). The upregulation of Pvt1 oncogene
has also been reported to indicate poor survival in several types
of cancer, including CRC (23).
Some researchers have reported upregulated SNHG22 expression
in EOC, PTC and clear cell renal cell carcinoma, and high
SNHG22 expression may indicate poor prognosis in these three
human malignancies (12,14,24). The present study first explored
the expression patterns of SNHG22 in CRC TTs and ANTs, and
in CRA tissues, and revealed that SNHG22 expression was
upregulated in the order of ANTs > CRA tissues > TTs. High
SNHG22 expression levels were associated with advanced T
stage, node involvement, metastasis and poor survival in patients
with CRC. Therefore, SNHG22 may serve as an independent
prognostic indicator in patients with CRC.
As an oncogene, SNHG22 may serve critical
roles in tumor advancement processes; for example, it was shown to
be highly expressed in EOC, and to stimulate EOC cell
proliferation, invasion and chemotherapy resistance by sponging
miR-2467 to facilitate galectin-1 expression in EOC cells (11). Fang et al (13) observed that, in breast cancer,
SNHG22 facilitated tumor progression by sequestering
miR-324-3p and subsequently upregulating SDS3 homolog, SIN3A
corepressor complex component. The present study also examined the
effects of SNHG22 on the phenotype of CRC cells, and revealed that
SNHG22 promoted CRC cell proliferation, apoptosis resistance,
invasion and migration in vitro. Knocking down SNHG22 in
vivo inhibited xenograft tumor growth. All these results
implicate SNHG22 as a possible critical oncogenic lncRNA in
CRC.
Most cytoplasmic lncRNAs exert their regulatory
effects on gene expression by functioning as ceRNAs, regulating
miRNAs by competitively binding their target sites on
protein-coding mRNA molecules (25,26). For example, acting as a ceRNA,
MIR17HG has been reported to base its biological behaviors on a
sequence-specific interaction with miR-17-5p, thereby augmenting
the biological roles of mRNA targets (5). Chen et al (27) reported that the lncRNA
up-regulated in colorectal cancer liver metastasis promoted the
invasion of CRC cell lines by sponging miR-215. Based on
bioinformatics analyses and experimental assays, the present study
demonstrated that SNHG22 was preferentially localized to the
cytoplasm. Furthermore, it was confirmed that SNHG22 could
sponge miR-128-3p in CRC, and the dual-luciferase reporter assay
indicated that SNHG22 may interact with miR-128-3p and hinder its
biological roles. miR-128-3p has been reported to function as a
tumor suppressor in several types of human cancer (28-32), and may be involved in cell
proliferation, the cell cycle and chemosensitivity. It has been
suggested that miR-128-3p may enhance the chemosensitivity of
oxaliplatin-resistant CRC cells by targeting BMI1 proto-oncogene,
polycomb ring finger and ATP-binding cassette subfamily C member 5
(30). Moreover, nanocomplexes
loaded with miR-128-3p elevated chemotherapy roles through dual
targeting, and silencing PI3K-AKT and MEK-ERK pathway activity in
CRC cell lines (29).
As a ceRNA, the function of lncRNAs depends on the
miRNA target gene. Using five online databases, E2F3 was
predicted as a potential target of miR-128-3p. This hypothesis was
validated with luciferase reporter assays. The results revealed
that miR-128-3p overexpression inhibited the E2F3 wt 3′UTR
luciferase activity, but did not change that of the E2F3 mut
3′UTR. Furthermore, miR-128-3p overexpression inhibited E2F3 mRNA
and protein expression levels, whereas silencing miR-128-3p
elevated them, suggesting that E2F3 could be a direct target
of miR-128-3p in CRC cells. Further functional rescue experiments
validated the hypothesis that SNHG22 may regulate CRC
proliferation and invasion by competitively sponging miR-128-3p and
restoring E2F3 activity. Recent studies have revealed that
E2F3 may serve an essential role in regulating human cancer cell
proliferation, apoptosis and chemosensitivity (33,34).
In conclusion, the results of the present study
revealed that, as an oncogenic lncRNA in CRC, SNHG22 was
upregulated and related to poor survival in patients with CRC.
Functional and mechanistic analyses demonstrated that SNHG22
promoted CRC tumorigenesis and metastasis by sponging miR-128-3p,
leading to elevated expression of E2F3. These findings may provide
novel insights into the development of therapeutics for CRC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request or from the TCGA repository (https://portal.gdc.cancer.gov/.
Authors' contributions
LFZ, JNY, CFW and XYD conceived and designed the
present study. JNY, YLL, HNZ and XYD performed the experiments.
JNY, CFW, XYD and YZZ analyzed and interpretated the data. JNY, CFW
and LFZ wrote, reviewed and/or revised the manuscript. JNY and LFZ
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study involving human tissues was
approved by the Ethics Committee of Zhengzhou University (approval
no. 2011110402). All patients provided written informed consent
prior to their inclusion within the study. The research has been
carried out in accordance with the World Medical Association
Declaration of Helsinki. All animal experiments were approved by
the Committee of the Ethics of Animal Experiments of Zhengzhou
University (approval no. 20180376).
Patient consent for publication
All patients included in the present study have
provided consent for both participation and publication.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
EOC
|
epithelial ovarian carcinoma
|
|
PTC
|
papillary thyroid cancer
|
|
lncRNA
|
long non-coding RNA
|
|
snoRNA
|
small nucleolar RNA
|
|
SNHG
|
small nucleolar RNA host gene
|
|
siRNA
|
small interfering RNA
|
|
UTR
|
untranslated region
|
|
mut
|
mutant
|
|
RIP
|
RNA immunoprecipitation
|
|
CCK-8
|
Cell Counting Kit-8
|
|
TT
|
tumor tissue
|
|
ANT
|
adjacent non-cancerous tissue
|
|
TCGA
|
The Cancer Genome Atlas
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
ceRNA
|
competing endogenous RNA
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
2
|
De Angelis R, Sant M, Coleman MP,
Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H,
Ardanaz E, et al: Cancer survival in Europe 1999-2007 by country
and age: Results of EUROCARE-5-a population-based study. Lancet
Oncol. 15:23–34. 2014. View Article : Google Scholar
|
|
3
|
He Q, Long J, Yin Y, Li Y, Lei X, Li Z and
Zhu W: Emerging roles of lncRNAs in the formation and progression
of colorectal cancer. Front Oncol. 9:15422020. View Article : Google Scholar
|
|
4
|
Luo Y, Yang J, Yu J, Liu X, Yu C, Hu J,
Shi H and Ma X: Long non-coding RNAs: Emerging roles in the
immunosuppressive tumor microenvironment. Front Oncol. 10:482020.
View Article : Google Scholar
|
|
5
|
Xu J, Meng Q, Li X, Yang H, Xu J, Gao N,
Sun H, Wu S, Familiari G, Relucenti M, et al: Long noncoding RNA
MIR17HG promotes colorectal cancer progression via miR-17-5p.
Cancer Res. 79:4882–4895. 2019. View Article : Google Scholar
|
|
6
|
Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K,
Huang Z, Guo X and Zhang Y: LncRNA HOTAIR is a prognostic biomarker
for the proliferation and chemoresistance of colorectal cancer via
miR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol
Biochem. 46:1275–1285. 2018. View Article : Google Scholar
|
|
7
|
Ding D, Li C, Zhao T, Li D, Yang L and
Zhang B: LncRNA H19/miR-29b-3p/PGRN axis promoted
epithelial-mesenchymal transition of colorectal cancer cells by
acting on Wnt signaling. Mol Cells. 41:423–435. 2018.
|
|
8
|
Williams GT and Farzaneh F: Are snoRNAs
and snoRNA host genes new players in cancer? Nat Rev Cancer.
12:84–88. 2012. View Article : Google Scholar
|
|
9
|
Shan Y, Ma J, Pan Y, Hu J, Liu B and Jia
L: LncRNA SNHG7 sponges miR-216b to promote proliferation and liver
metastasis of colorectal cancer through upregulating GALNT1. Cell
Death Dis. 9:7222018. View Article : Google Scholar
|
|
10
|
Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B
and Jia L: Long non-coding RNA-SNHG7 acts as a target of miR-34a to
increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in
colorectal cancer progression. J Hematol Oncol. 11:892018.
View Article : Google Scholar
|
|
11
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar
|
|
12
|
Zhang PF, Wu J, Luo JH, Li KS, Wang F,
Huang W, Wu Y, Gao SP, Zhang XM and Zhang PN: SNHG22 overexpression
indicates poor prognosis and induces chemotherapy resistance via
the miR-2467/Gal-1 signaling pathway in epithelial ovarian
carcinoma. Aging (Albany NY). 11:8204–8216. 2019. View Article : Google Scholar
|
|
13
|
Fang X, Zhang J, Li C, Liu J, Shi Z and
Zhou P: Long non-coding RNA SNHG22 facilitates the malignant
phenotypes in triple-negative breast cancer via sponging miR-324-3p
and upregulating SUDS3. Cancer Cell Int. 20:2522020. View Article : Google Scholar
|
|
14
|
Gao H, Sun X, Wang H and Zheng Y: Long
noncoding RNA SNHG22 increases ZEB1 expression via competitive
binding with microRNA-429 to promote the malignant development of
papillary thyroid cancer. Cell Cycle. 19:1186–1199. 2020.
View Article : Google Scholar
|
|
15
|
Edge S, Byrd DR, Compton CC, Fritz AG,
Greene F and Trotti A: AJCC cancer staging handbook. 7th edition.
Springer; New York, NY: 2010
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Du Y, Wei N, Hong J and Pan W: Long
non-coding RNASNHG17 promotes the progression of breast cancer by
sponging miR-124-3p. Cancer Cell Int. 20:402020. View Article : Google Scholar
|
|
18
|
National Research Council (US) Committee
for the Update of the Guide for the Care and use of Laboratory
Animals: Guide for the care and use of laboratory animals. 8th
edition. National Academies Press (US); Washington, DC: 2011
|
|
19
|
Cao Z, Pan X, Yang Y, Huang Y and Shen HB:
The lncLocator: A subcellular localization predictor for long
non-coding RNAs based on a stacked ensemble classifier.
Bioinformatics. 34:2185–2194. 2018. View Article : Google Scholar
|
|
20
|
Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L and
Song X: Long noncoding RNA OCC-1 suppresses cell growth through
destabilizing HuR protein in colorectal cancer. Nucleic Acids Res.
46:5809–5821. 2018. View Article : Google Scholar
|
|
21
|
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen
YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes
IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer.
Mol Cancer. 18:1742019. View Article : Google Scholar
|
|
22
|
Wang D, Zhang H, Fang X, Zhang X and Liu
H: Prognostic value of long non-coding RNA GHET1 in cancers: A
systematic review and meta-analysis. Cancer Cell Int. 20:1092020.
View Article : Google Scholar
|
|
23
|
Xiao M, Feng Y, Liu C and Zhang Z:
Prognostic values of long noncoding RNA PVT1 in various carcinomas:
An updated systematic review and meta-analysis. Cell Prolif.
51:e125192018. View Article : Google Scholar
|
|
24
|
Yang W, Zhang K, Li L, Ma K, Hong B, Gong
Y and Gong K: Discovery and validation of the prognostic value of
the lncRNAs encoding snoRNAs in patients with clear cell renal cell
carcinoma. Aging (Albany NY). 12:4424–4444. 2020. View Article : Google Scholar
|
|
25
|
Li M, Bian Z, Jin G, Zhang J, Yao S, Feng
Y, Wang X, Yin Y, Fei B, You Q and Huang Z: LncRNA-SNHG15 enhances
cell proliferation in colorectal cancer by inhibiting miR-338-3p.
Cancer Med. 8:2404–2413. 2019. View Article : Google Scholar
|
|
26
|
Yan J, Jia Y, Chen H, Chen W and Zhou X:
Long non-coding RNA PXN-AS1 suppresses pancreatic cancer
progression by acting as a competing endogenous RNA of miR-3064 to
upregulate PIP4K2B expression. J Exp Clin Cancer Res. 38:3902019.
View Article : Google Scholar
|
|
27
|
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F,
Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al: Long
non-coding RNA UICLM promotes colorectal cancer liver metastasis by
acting as a ceRNA for microRNA-215 to regulate ZEB2 expression.
Theranostics. 7:4836–4849. 2017. View Article : Google Scholar
|
|
28
|
Zhao J, Li D and Fang L: MiR-128-3p
suppresses breast cancer cellular progression via targeting LIMK1.
Biomed Pharmacother. 115:1089472019. View Article : Google Scholar
|
|
29
|
Liu X, Dong C, Ma S, Wang Y, Lin T, Li Y,
Yang S, Zhang W, Zhang R and Zhao G: Nanocomplexes loaded with
miR-128-3p for enhancing chemotherapy effect of colorectal cancer
through dual-targeting silence the activity of PI3K/AKT and MEK/ERK
pathway. Drug Deliv. 27:323–333. 2020. View Article : Google Scholar
|
|
30
|
Liu T, Zhang X, Du L, Liu X, Tian H, Wang
L, Li P, Zhao Y, Duan W, Xie Y, et al: Exosome-transmitted
miR-128-3p increase chemosensitivity of oxaliplatin-resistant
colorectal cancer. Mol Cancer. 18:432019. View Article : Google Scholar
|
|
31
|
Li X, Lv X, Li Z, Li C, Li X, Xiao J, Liu
B, Yang H and Zhang Y: Long noncoding RNA ASLNC07322 functions in
VEGF-C expression regulated by smad4 during colon cancer
metastasis. Mol Ther Nucleic Acids. 18:851–862. 2019. View Article : Google Scholar
|
|
32
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018. View Article : Google Scholar
|
|
33
|
Wang H, Wang L, Zhang S, Xu Z and Zhang G:
Downregulation of LINC00665 confers decreased cell proliferation
and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC.
Biomed Pharmacother. 127:1102142020. View Article : Google Scholar
|
|
34
|
Chen J, Liu X, Xu Y, Zhang K, Huang J, Pan
B, Chen D, Cui S, Song H, Wang R, et al: TFAP2C-activated MALAT1
modulates the chemoresistance of docetaxel-resistant lung
adenocarcinoma cells. Mol Ther Nucleic Acids. 14:567–582. 2019.
View Article : Google Scholar
|





















