Introduction
Oral squamous cell carcinoma (OSCC) is the most
common malignant tumor of the head and neck region, which is caused
by smoking, alcohol consumption and viral infections. The main
treatment for OSCC consists of surgery combined with chemotherapy
and radiotherapy (1,2). Despite advancements being made in
the diagnosis and treatment of the disease, the 5-year survival
rate of patients with OSCC remains low at 50%, largely due to tumor
recurrence and metastasis (3,4).
Therefore, there is a need to further investigate the regulatory
mechanisms underlying OSCC differentiation, proliferation, invasion
and metastasis in order to improve the poor prognosis of patients
with OSCC.
Crosstalk between the tumor microenvironment (TME)
and tumors can promote cancer progression (5,6).
Stromal cells, which are the main component of the TME, can
influence the proliferation, invasion and metastasis of cancers
(7). Stromal cells consist of
cancer-associated fibroblasts (CAFs), tumor-associated macrophages
(TAMs) and infiltrating immune cells, which can enhance cancer cell
proliferation and metastasis by forming a supportive environment
for angiogenesis, invasion and metastasis (8-10).
A previous study suggested that the interaction between stromal
cells and the epithelium promotes the progression of breast cancer
(11). In addition, the
expression of programmed death-ligand 1 (PD-L1) on stromal cells
promotes the progression of colon cancer by suppressing
CD8+ T-cell immune responses (12). Mesenchymal stromal cells can also
promote cancer metastasis by enhancing epithelial-to-mesenchymal
transition (EMT) (13).
Therefore, stromal cells may influence the progression of multiple
cancer types by interacting with cancer cells; however, data on the
influence of stromal cells in OSCC are limited.
Clinically, various subtypes of OSCC exist,
including invasive carcinoma and verrucous carcinoma. Differences
in the invasive ability of these subtypes results in marked
differences in prognosis. The endophytic type (ED-type) OSCC can
invade and occasionally metastasize. Conversely, the exophytic type
(EX-type) OSCC, such as verrucous OSCC, presents an outward growth,
does not invade the subepithelial connective tissue, does not
metastasize and is therefore associated with a relatively good
prognosis (14-16). However, to the best of our
knowledge, no studies available to date have examined the
mechanisms through which differences in these subtypes affect the
tumor stroma. Therefore, it was hypothesized that different
subtypes of stromal cells in the TME differentially affect the
progression of OSCC. To examine this hypothesis, in the present
study, the moderately differentiated human oral cancer cell line,
HSC-3, was used as a cell model and verrucous squamous cell
carcinoma-associated stromal cells (VSCC-SCs; derived from EX-type
OSCC stroma) and squamous cell carcinoma-associated stromal cells
(SCC-SCs; derived from ED-type OSCC stroma) were extracted from
patients with OSCC to examine the effects of different stromal
cells subtypes in the TME on the progression of OSCC. Furthermore,
microarray data and bioinformatics analyses were used to elucidate
the potential mechanisms underlying the differential effects of
stromal cell subtypes on the progression of OSCC. These findings
may highlight a potential regulatory mechanism underlying the
progression of OSCC.
Materials and methods
Cells and cell culture
The moderately differentiated human oral cancer cell
line, HSC-3 was purchased from the Cell Bank of the Japanese
Collection of Research Bioresources (JCRB). Human dermal
fibroblasts (HDFs) were purchased from Lonza Group, Ltd. VSCC-SCs
and SCC-SCs were extracted from surgical operative tissues at the
Department of Oral and Maxillofacial Surgery at Okayama University.
The VSCC tissues and SCC tissues were obtained from each 1 patient,
respectively to separate the stromal cells and generated the cell
culture. Sections of fresh oral squamous carcinoma tissue (1
mm3) were washed several times with α-Modified Eagle's
medium (α-MEM) (Life Technologies; Thermo Fisher Scientific, Inc.)
containing antibiotic-antimycotic (Life Technologies; Thermo Fisher
Scientific, Inc.) and then minced. The tissues were then treated
with α-MEM containing 1 mg/ml collagenase II (Invitrogen; Thermo
Fisher Scientific, Inc.) and dispase (Invitrogen; Thermo Fisher
Scientific, Inc.) for 2 h at 37°C with shaking (200 rpm). The
released cells were centrifuged for 5 min at 111.8 × g at room
temperature, suspended in α-MEM containing 10% FBS (Biowest),
filtered through a cell strainer (100 µm, BD Falcon; BD
Biosciences), plated in a tissue culture flask and incubated at
37°C in 5% CO2. After 1 week, the stromal cells were
separated by Accutase (Invitrogen; Thermo Fisher Scientific, Inc.)
based on the differential adhesion of epithelial and stromal cells.
These stromal cells were labeled VSCC-SCs (derived from EX-type
OSCC stroma) and SCC-SCs (derived from ED-type OSCC stroma)
(17). The HSC-3 cells, HDFs,
VSCC-SCs and SCC-SCs were maintained in α-MEM (Life Technologies;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1%
antimycotic-antibiotic (Life Technologies; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere of 5%
CO2 and 95% air. The present study was approved by the
Ethics Committee of Okayama University (project identification
code: 1703-042-001). Informed consent was obtained from all
patients.
Giemsa staining
VSCC-SCs, SCC-SCs, HDFs and HSC-3 cells were
digested with Accutase and EDTA (Life Technologies; Thermo Fisher
Scientific, Inc.), and centrifuged when the density approached 90%.
The VSCC-SCs, SCC-SCs and HDFs were then mixed with the HSC-3 cells
at a 3:1 ratio. The cells were seeded into small dishes (Violamo,
35×12 mm) with a slide (Matsunami, 22×22 mm) at a density of
4×105/dish. Following incubation for 1 day (37°C), the
attached slides were stained using the Giemsa Staining kit
(Diff-Quick; Nanjing Jiancheng Bioengineering Institute). The
slides were first washed with double-distilled water (DDW) and
fixed with Diff-Quik Fixative provided with the Giemsa staining kit
for 2 min at room temperature respectively. The slides were then
stained with Diff-Quik Solution Ⅰ and Diff-Quik Solution II
provided with the Giemsa staining kit for 2 min at room
temperature. Finally, the stained cells were photographed under a
bright field microscope (×4, ×10, ×20 and ×40 magnification; BX51,
Olympus Corporation). Independent experiments were repeated three
times.
MTS assay
The VSCC-SCs, SCC-SCs, HDFs and HSC-3 cells were
digested with Accutase and EDTA, and centrifuged when the density
approached 90%. The VSCC-SCs, SCC-SCs and HDFs were then mixed with
HSC-3 at a 3:1 ratio. The cells were seeded into 96-wells plates at
a density of 2,000 cells/well. Following incubation for 1, 3 and 5
days, respectively (37°C), 20 µl of MTS regent were added to
each well and the cells were then incubated for a further 4 h
(37°C). The absorbance of each well was measured at 490 nm using an
enzyme-linked immunosorbent assay reader (SH-1000 Lab). The OD
values of the HSC-3 + VSCC-SCs/SCC-SCs/HDFs were compared with
those of the HSC-3 (2,000 cells) cells and VSCC-SCs/SCC-SCs/HDFs
(2,000 cells) cells to determine the effects on the proliferation
between the HSC-3 cells and VSCC-SCs/SCC-SCs/HDFs as follows: A
promotion of cell proliferation was considered when the OD value of
the HSC-3 + VSCC-SCs/SCC-SCs/HDFs mixed group was greater than that
of the HSC-3 (2,000 cells) and VSCC-SCs/SCC-SCs/HDFs (2,000 cells)
groups. An inhibition of cell proliferation was considered when the
OD value of the HSC-3 + VSCC-SCs/SCC-SCs/HDFs mixed group was less
than that of the HSC-3 (2,000 cells) and VSCC-SCs/SCC-SCs/HDFs
(2,000 cells) groups. No effect on cell proliferation was
considered to have occurred when the OD value of the HSC-3 +
VSCC-SCs/SCC-SCs/HDFs mixed group was intermediate between that of
the HSC-3 (2,000 cells) and VSCC-SCs/SCC-SCs/HDFs (2,000 cells)
groups. Independent experiments were repeated three times.
Immunohistochemistry (IHC; Ki-67 labeling
index)
The slides were washed three times with TBS and
fixed with 4% paraformaldehyde for 15 min. Endogenous peroxidase
activity was blocked by incubating the slides in methanol
containing 0.3% H2O2 for 30 min and blocking
liquid (DS Pharma Biomedical Co., Ltd.) for 20 min. The primary
antibody, mouse anti-Ki-67 (M7240, MIB1, 1:50; Dako; Agilent
Technologies, Inc.) was added and slides were incubated for 2 h at
room temperature. After washing three times with TBS, the slides
were incubated with secondary antibody avidin-biotin complexes
[PK-6102, mouse ABC kit; blocking serum (normal serum, diluted with
TBS): 1:75; biotinylated secondary antibody (diluted with normal
serum): 1:200; reagent A (Avidin, ABC Elite) and reagent B
(Biotinylated HRP, ABC Elite, diluted with TBS): 1:55; Vector
Laboratories, Inc.] for 1 h at room temperature. Following
visualization with a mixed solution of diaminobenzidine
(DAB)/H2O2 (histofine DAB substrate; Nichirei
Biosciences Inc.), the stained slides were photographed under a
bright field microscope (×4, ×10, ×20 and ×40 magnification, BX51,
Olympus Corporation). A total of five images (×40 magnification)
were randomly obtained and used to determine positive cell counts
using ImageJ software (V1.51j8, National Institutes of Health). The
percentage of positive Ki-67 cells was calculated as the number of
positive Ki-67 cancer cells/the number of all cancer cells ×100%.
Independent experiments were repeated three times.
Transwell invasion and migration assays,
and immunofluorescence (IF)
The VSCC-SCs, SCC-SCs, HDFs and HSC-3 cells were
digested with Accutase and EDTA, and centrifuged for 5 min at 111.8
× g at room temperature when the density approached 90%. The
VSCC-SCs, SCC-SCs and HDFs were then mixed with HSC-3 cells at a
3:1 ratio in α-MEM without FBS. For the Transwell invasion assay,
the cells were seeded in the upper chamber of 8-µm Transwell
filters pre-coated with Matrigel in 24-well plates (Corning BioCoat
Matrigel Invasion Chamber kit; BD Biosciences) at a density of
4×104/500 µl. Subsequently, α-MEM (500 µl)
containing 10% FBS was added to the bottom chamber. For the
Transwell migration assay, the cells were seeded into the upper
chamber of 8-µm Transwell filters without Matrigel in
24-well plates (Corning, Falcon cell culture inserts; BD
Biosciences) at a density of 2×104/500 µl.
Subsequently, α-MEM (500 µl) containing 10% FBS was added to
the bottom chamber. Following 1 day of incubation (37°C), the cells
in upper chamber were removed using a cotton swab, and the membrane
was trimmed in the upper chamber. After washing with TBS three
times (5 min each), the cells were fixed with 4% paraformaldehyde
for 15 min and blocked with blocking liquid (DS Pharma Biomedical
Co., Ltd.) for 20 min. The primary antibodies, rabbit anti-vimentin
(ab16700, SP20, 1:200, Abcam) and mouse anti-pan cytokeratin
(IS053, AE1/3, Abcam) were added and the cells were then incubated
for 1 h at room temperature. After washing three times with TBS,
the secondary antibodies, anti-mouse IgG Alexa Fluor 488 (A21441,
1:200, Life Technologies; Thermo Fisher Scientific, Inc.) and
anti-rabbit IgG Alexa Fluor 568 (A10042, 1:200, Life Technologies;
Thermo Fisher Scientific, Inc.) were added and the cells were
incubated for 1 h at room temperature without light. After washing
with TBS and DDW three times, the samples were stained with 0.2
g/ml 40,6-diamidino-2-phenylindole (DAPI; Dojindo Molecular
Technologies, Inc.). The stained cells were photographed using a
fluorescence microscope (×5 and ×10 magnification, BZ-8000;
Keyence). Independent experiments were repeated three times, and
the data were analyzed using ImageJ software (V1.51j8).
Animal experiments
All animal experiments were conducted according to
the relevant guidelines and regulations, and were approved by the
institutional committees at Okayama University (OKU-2017406).
Following intraperitoneal anesthesia with ketamine hydrochloride
(75 mg/kg body weight) and medetomidine hydrochloride (0.5 mg/kg
body weight), 200 µl mixed cells including HSC-3
(1×106, 100 µl) and stromal cells (VSCC-SCs,
SCC-SCs and HDFs, 3×106, 100 µl) were injected
into the central region of the top of the head (in the lamina
propria) of 12 BALB/c nu-nu mice (4-week-old healthy females)
obtained from SHIMIZU Laboratory Supplies Co., Ltd, as previously
described (18,19). All mice were reared in an animal
room at a temperature of 25°C and 50-60% humidity under a 12-h
light/dark cycle. All mice were allowed free access to food and
water. Finally, atipamezole hydrochloride (1 mg/kg body weight) was
subcutaneously injected into the abdominal cavity to reverse
anesthesia. The experimental animals were divided into the HSC-3,
HSC-3 + VSCC-SCs, HSC-3 + SCC-SCs and HSC-3 + HDFs groups and each
group consisted of three mice.
Hematoxylin and eosin (H&E)
staining
After 4 weeks, all mice were sacrificed by
isoflurane excess inhalation anesthesia (concentration >5%).
Cardiac arrest was then verified by pulse palpation followed by the
dislocation of the cervical spine of the mice. The whole head of
the animals containing the tumor tissues and the surrounding bone
tissues were removed, fixed in 4% paraformaldehyde for 12 h, and
decalcified using 10% EDTA for 3 weeks. Subsequently, the whole
head of the animal models were processed and embedded into paraffin
wax using routine histological methods and cut into
5-µm-thick sections. Finally, the sections were stained with
H&E. The sections were stained with Carrazi's hematoxylin (MUTO
Pure Chemical Co., Ltd.) for 5 min at room temperature and stained
with eosin (MUTO Pure Chemical Co., Ltd.) for 7 min at room
temperature. The tumor part in the tissue of the animals was
photographed under a bright field microscope (×4, ×10, ×20 and ×40
magnification). The effects of VSCC-SCs, SCC-SCs and HDFs on the
differentiation of OSCC cells in vivo were determined by the
keratinized area and the size of the tumor nest according to the
pathological diagnosis. The keratinized area indicated
well-differentiated tissue and tumor nest formation represented
moderate and poor differentiation. A small-sized tumor nest
indicated poor differentiation and a medium-sized tumor nest
indicated moderate differentiation.
Tartrate-resistant acid phosphatase
(TRAP) staining
TRAP staining was conducted using a TRAP staining
kit (Primary Cell, Co., Ltd.) according to the manufacturer's
instructions. Active multi-nucleated osteoclast cells on the bone
resorption surface were considered to be TRAP-positive cells. The
interaction area contained tumor and bone in the tissue of whole
animal model heads was photographed under a bright field microscope
(×4, ×10, ×20 and ×40 magnification). A total of five images (×40
magnification) were randomly acquired to obtain positive cell
counts using ImageJ software (V1.51j8). Independent experiments
were repeated three times.
Immunohistochemical analysis
Following antigen retrieval, the 5-µm-thick
sections were blocked with 10% normal serum for 20 min at room
temperature and then incubated with primary antibodies, including
mouse anti-Ki67 (M7240, MIB1, 1:50; Dako; Agilent Technologies,
Inc.), MMP9 (F-69, 56-2A4, 1:20; Fuji Yakukin Co., Ltd.) and
MT1-MMP (F-86, 114-6G6, 1:20; Fuji Yakukin Co., Ltd.), overnight at
4°C. After washing three times with TBS, all sections were
incubated with the secondary antibody avidin-biotin complexes
[PK-6102, mouse ABC kit; Blocking serum (normal serum, diluted with
TBS): 1:75; biotinylated secondary antibody (diluted with normal
serum): 1:200; reagent A (Avidin, ABC Elite) and reagent B
(biotinylated HRP, ABC Elite, diluted by TBS): 1:55; Vector
Laboratories, Inc.] for 1 h at room temperature. Following
visualization with a mixed solution of
DAB/H2O2 (histofine DAB substrate; Nichirei
Biosciences Inc.), The tumor areas containing the tumor and stroma
in the tissue of the heads of the animals were photographed under a
bright field microscope (×4, ×10, ×20 and ×40 magnification). A
total of five images (×40 magnification) were randomly acquired to
obtain positive cell counts using ImageJ software (V1.51j8). The
percentage of positive Ki-67/MMP9/MT1-MMP cells was calculated as
the number of positive Ki-67/MMP9/MT1-MMP cancer cells/the number
of all cancer cells ×100%. Independent experiments were repeated
three times.
Double-fluorescent immunohistochemical
staining
Following antigen retrieval, the sections were
blocked with Block Ace (DS Pharma Biomedical Co., Ltd.) for 20 min
at room temperature and then incubated with primary antibodies,
rabbit anti-Snail + SLUG (ab180714, 1:200; Abcam), and mouse
anti-E-cadherin (M126, clone SHE78-7, 1:1,000, Takara Bio, Inc.),
overnight at 4°C. After washing three times with TBS, the sections
were incubated with the secondary antibodies, anti-mouse IgG Alexa
Fluor 488 (A21441, 1:100; Life Technologies; Thermo Fisher
Scientific, Inc.) and anti-rabbit IgG Alexa Fluor 568 (A10042,
1:100; Life Technologies; Thermo Fisher Scientific, Inc.) for 1 h
without light at room temperature. Finally, the sections were
stained with 0.2 g/ml DAPI (Dojindo Molecular Technologies, Inc.).
The tumor areas containing the tumor and stroma in the tissue of
the whole heads of the animals were photographed under a
fluorescence microscope (×5, ×10 and ×20 magnification). The
percentage of positive Snail cells was calculated as the number of
positive snail cancer cells/the number of all cancer cells ×100%.
Independent experiments were repeated three times.
Microarray and bioinformatics
analyses
The VSCC-SCs, SCC-SCs and HDFs were used to conduct
the microarray analysis. The HDFs were used as the control group to
analyze the differentially expressed genes (DEGs) in the VSCC-SCs
and SCC-SCs. The |LogFC|>1 was considered as the cut-off value
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE164374
(the dataset is currently private and is scheduled to be released
on January 1, 2024). Biological processes and cell pathways of the
upregulated DEGs were analyzed using Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
through Cytoscape 3.7.2 (https://cytoscape.org/), and the results are presented
as bubble plots created using R 3.6.2. An adjusted P<0.05 was
considered as the cut-off value. The top 10 hub genes were analyzed
using a protein-to-protein interaction network (PPI) using STRING
(http://string-db.org/) and Cytoscape 3.7.2
(cytohubba). A combined score >0.4, was considered as the
cut-off value, and the hub genes were selected according to the
degree. Finally, the hub genes that were differentially expressed
in the VSCC-SCs, SCC-SCs and HDFs were identified using cluster and
heatmap analyses.
Statistical analysis
All statistical analyses were conducted using
GraphPad Prism 9 (GraphPad Software, Inc.). Data are presented as
the mean ± standard deviation (SD). Two-way ANOVA was used to
compare two variables with Tukey's post hoc test. One-way ANOVA was
used to compared differences between >2 groups. The post hoc
test used following one-way ANOVA was Tukey's. P<0.05 was
considered to indicate a statistically significant difference.
Results
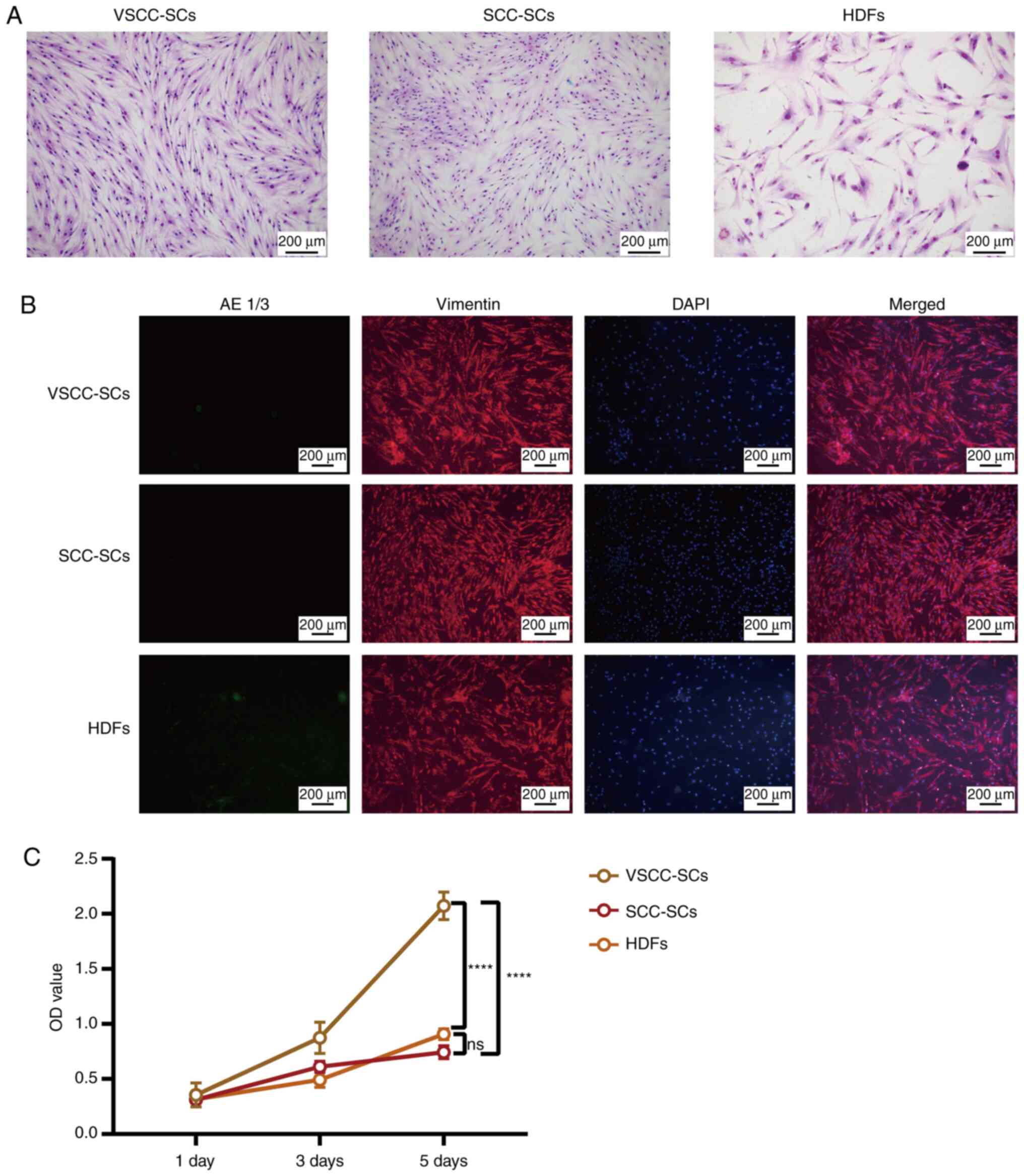
VSCC-SCs, SCC-SCs and HDFs have a
spindle-shaped morphology and suitable cell viability without
cancer cell contamination
The morphology of the VSCC-SCs, SCC-SCs and HDFs was
confirmed by Giemsa staining and IF. The spindle shape of the cells
in the SCC-SCs group was the most notable, closely followed by
those in the VSCC-SCs group. The spindle-shaped morphology of the
cells in VSCC-SCs group was more evident than that of the cells in
the HDFs group (Fig. 1A and B).
In addition, IF staining revealed that the VSCC-SCs, SCC-SCs and
HDFs were vimentin-positive and AE 1/3-negative, which indicated
that these three types of cells did not contain cancer cells
(Fig. 1B). Finally, the viability
of the VSCC-SCs, SCC-SCs and HDFs was examine using MTS assay. The
results revealed that the OD values of the VSCC-SCs, SCC-SCs and
HDFs were very similar on days 1 and 3, whereas they differed on
day 5. The OD value of the VSCC-SCs was the highest on day 5,
followed by those of the HDFs group. The OD value of the cells in
the HDFs group was slightly higher than that of the cells in the
SCC-SCs group on day 5 (Fig. 1C).
These data thus indicated that the VSCC-SCs and SCC-SCs exhibited a
more obvious spindle-shaped morphology and suitable cell viability
without cancer cell contamination.
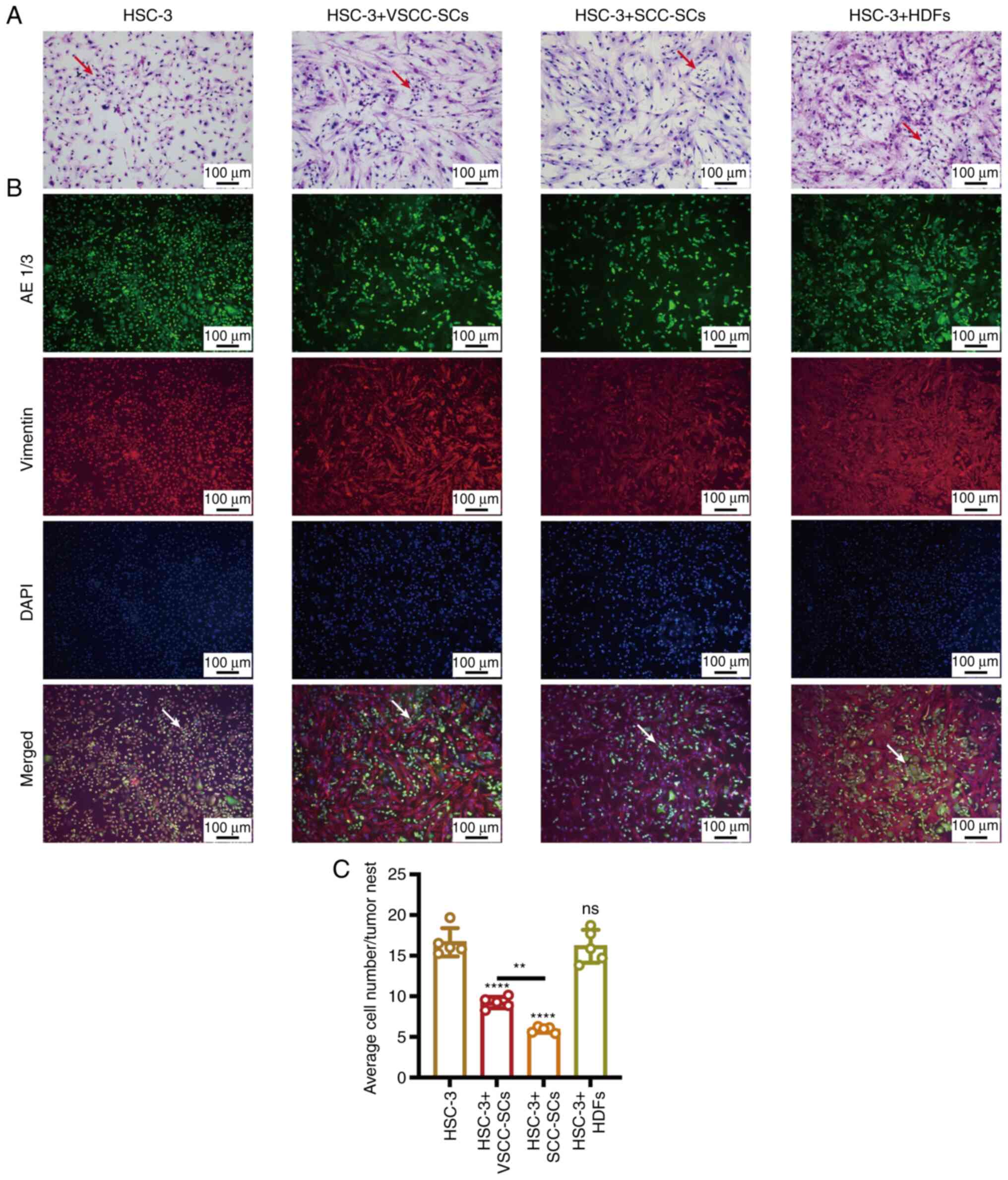
Both VSCC-SCs and SCC-SCs suppress the
tumor nest formation of HSC-3 cells in vitro
Giemsa and IF staining were used to evaluate the
effects of VSCC-SCs, SCC-SCs and HDFs on the tumor nest formation
of HSC-3 cells in vitro. In the HSC-3 cell group, the cancer
cells presented cellular pleomorphism; thus, the HSC-3 cells had
strong cytological atypia. In the HSC-3 + SCC-SCs group, the
adhesion of cancer cells was weak, there was no obvious cancer nest
formation, and the cancer cells infiltrated diffusely into the
stroma (Fig. 2A and B).
Furthermore, the average number of cells in the tumor nests in the
HSC-3 + SCC-SCs group was the lowest, followed by the HSC-3 +
VSCC-SCs group. In addition, there was a minimal difference between
the HSC-3 and HSC-3 + HDFs groups (Fig. 2C). Therefore, both the VSCC-SCs
and SCC-SCs inhibited the tumor nest formation of HSC-3 in
vitro, and the SCC-SCs exerted a more potent inhibitory effect
than the VSCC-SCs, while the HDFs exerted a minimal effect on the
tumor nest formation of HSC-3 cells in vitro.
VSCC-SCs and SCC-SCs promote the
proliferation of HSC-3 cells in vitro
The effects on cell proliferation between VSCC-SCs
and HSC-3 cells were examined using MTS assay. The results
demonstrated that there was a promoting effect on cell
proliferation in the HSC-3 + VSCC-SCs on day 3, while proliferation
was inhibited on day 5 compared with the VSCC-SCs (Fig. 3A). The effect on the proliferation
between the SCC-SCs and HSC-3 cells was also examined by MTS assay.
The results demonstrated that there was promoting effect observed
in the HSC-3 + SCC-SCs on days 1 and 5 (Fig. 3B). In addition, the effect on cell
proliferation between HDFs and HSC-3 was examined by MTS assay,
which demonstrated that there was a promoting effect observed in
the HSC-3 + HDFs on days 1 and 3 (Fig. 3C). In addition, the OD value of
the HSC-3 (2,000 cells), HSC-3 + VSCC-SCs and HSC-3 + HDFs groups
markedly increased on day 3 and decreased slightly on day 5, while
that of the HSC-3 + SCC-SCs group increased gradually from days 1
to 5 (Fig. 3D). These data
suggested that there was a promoting effect on cell proliferation
between the VSCC-SCs, SCC-SCs, HDFs and HSC-3 cells in
vitro. Furthermore, the relative expression of Ki-67 in the
different groups was examined by IHC to determine the effects of
the VSCC-SCs, SCC-SCs and HDFs on the proliferation of HSC-3 cells
in vitro (Fig. 3E). The
percentage of positive Ki-67 cells in the HSC-3 + SCC-SCs group was
slightly higher than that in the HSC-3 + VSCC-SCs group, which was
significantly higher than that in the HSC-3 + HDFs and HSC-3 groups
(Fig. 3F). These data suggested
that both the VSCC-SCs and SCC-SCs promoted the proliferation of
the HSC-3 cells in vitro and that the SCC-SCs exert a more
prominent promoting effect than the VSCC-SCs.
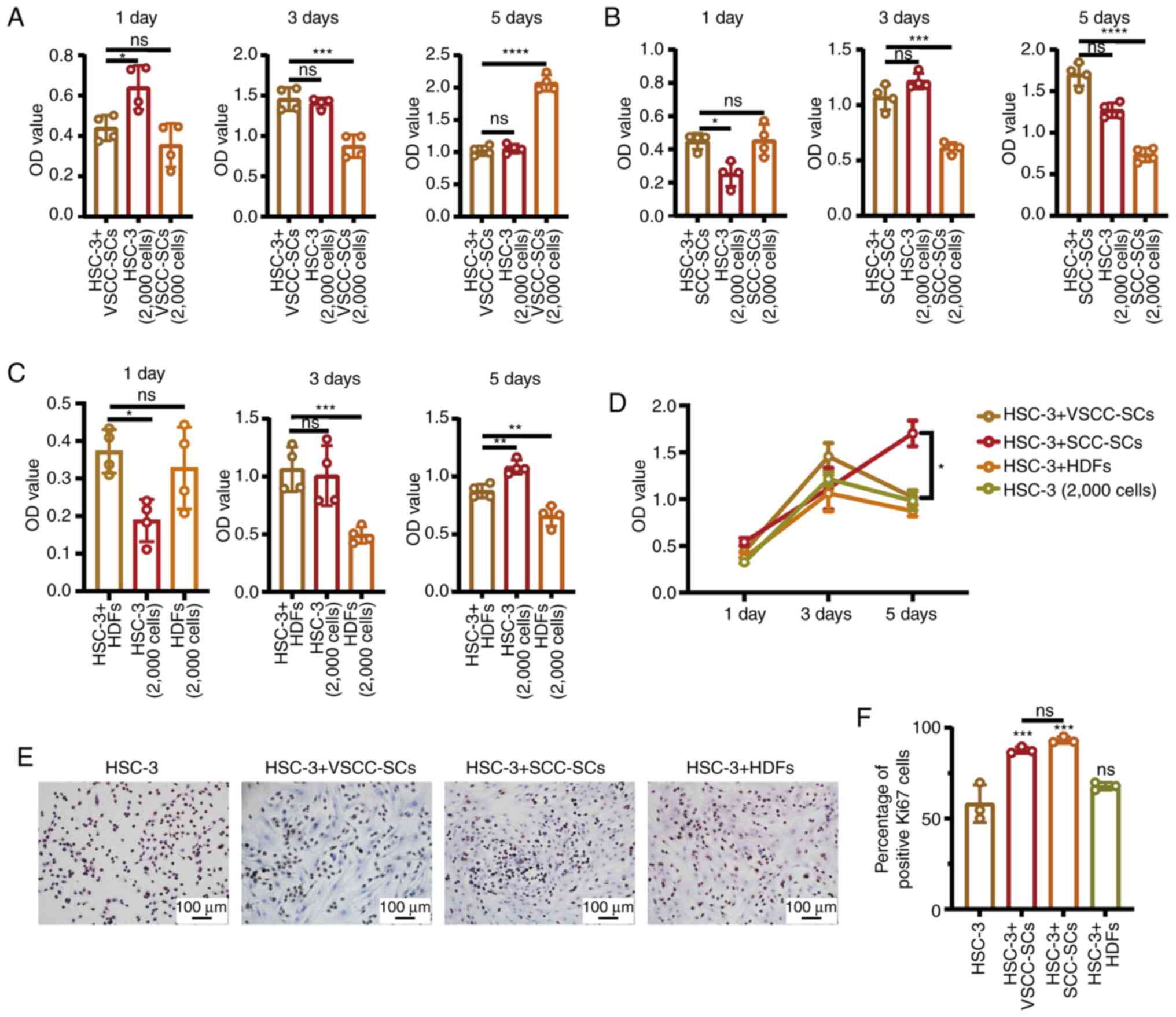 | Figure 3Both VSCC-SCs and SCC-SCs promote the
proliferation of HSC-3 cells in vitro. (A-C) MTS assay was
used to examine the effect on the proliferation between VSCC-SCs,
SCC-SCs, HDFs and HSC-3 at 1, 3 and 5 days. Data are presented as
the mean ± SD, n=4. Statistical analysis was performed using
one-way ANOVA; ns, not significant (P>0.05);
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. (D)
Comparison of the effect on the proliferation between VSCC-SCs,
SCC-SCs, HDFs and HSC-3 on days 1, 3 and 5. Data are presented as
the mean ± SD, n=4. Statistical analysis was performed using
two-way ANOVA; *P<0.05. (E) Immunohistochemistry was
used to examine the relative Ki-67 expression level to assay the
effect of VSCC-SCs, SCC-SCs and HDFs on the proliferation of HSC-3
in vitro. (F) Quantification of positive Ki-67 cell numbers
in different groups; data are presented as the mean ± SD, n=3.
Statistical analysis was performed using one-way ANOVA; ns, not
significant (P>0.05); ***P<0.001, compared with
the HSC-3 group. VSCC-SCs, verrucous squamous cell
carcinoma-associated stromal cells; SCC-SCs, squamous cell
carcinoma-associated stromal cells; HDFs, human dermal
fibroblasts. |
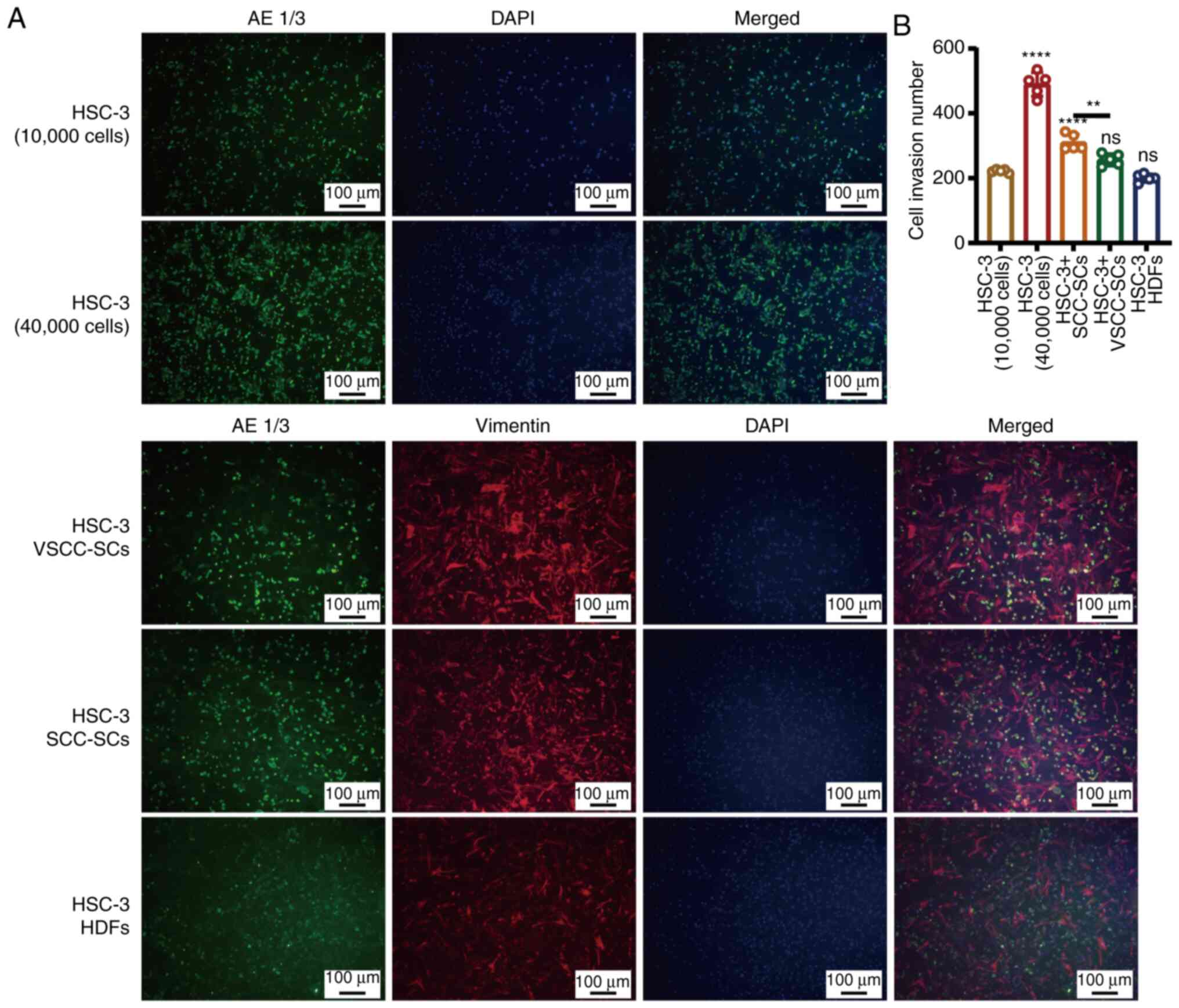
Both the VSCC-SCs and SCC-SCs promote the
invasion of HSC-3 cells in vitro
A Transwell (invasion) assay and IF staining were
used to determine the effects of VSCC-SCs, SCC-SCs and HDFs on the
invasion of HSC-3 cells in vitro (Fig. 4A). The number of invading cancer
cells in the HSC-3 + SCC-SCs group was slightly higher than that in
the HSC-3 + VSCC-SCs group and markedly higher than that in the
HSC-3 + HDFs and HSC-3 (10,000 cells) groups. Furthermore, the
number of invasive cancer cells in the HSC-3 + HDFs group was
slightly lower than that in the HSC-3 (10,000 cells) group
(Fig. 4B). These data indicated
that both the VSCC-SCs and SCC-SCs promoted the invasion of HSC-3
cells in vitro, while the HDFs exerted an inhibitory effect
on the invasion of HSC-3 cells. In addition, SCC-SCs exerted a more
prominent promoting effect than the VSCC-SCs.
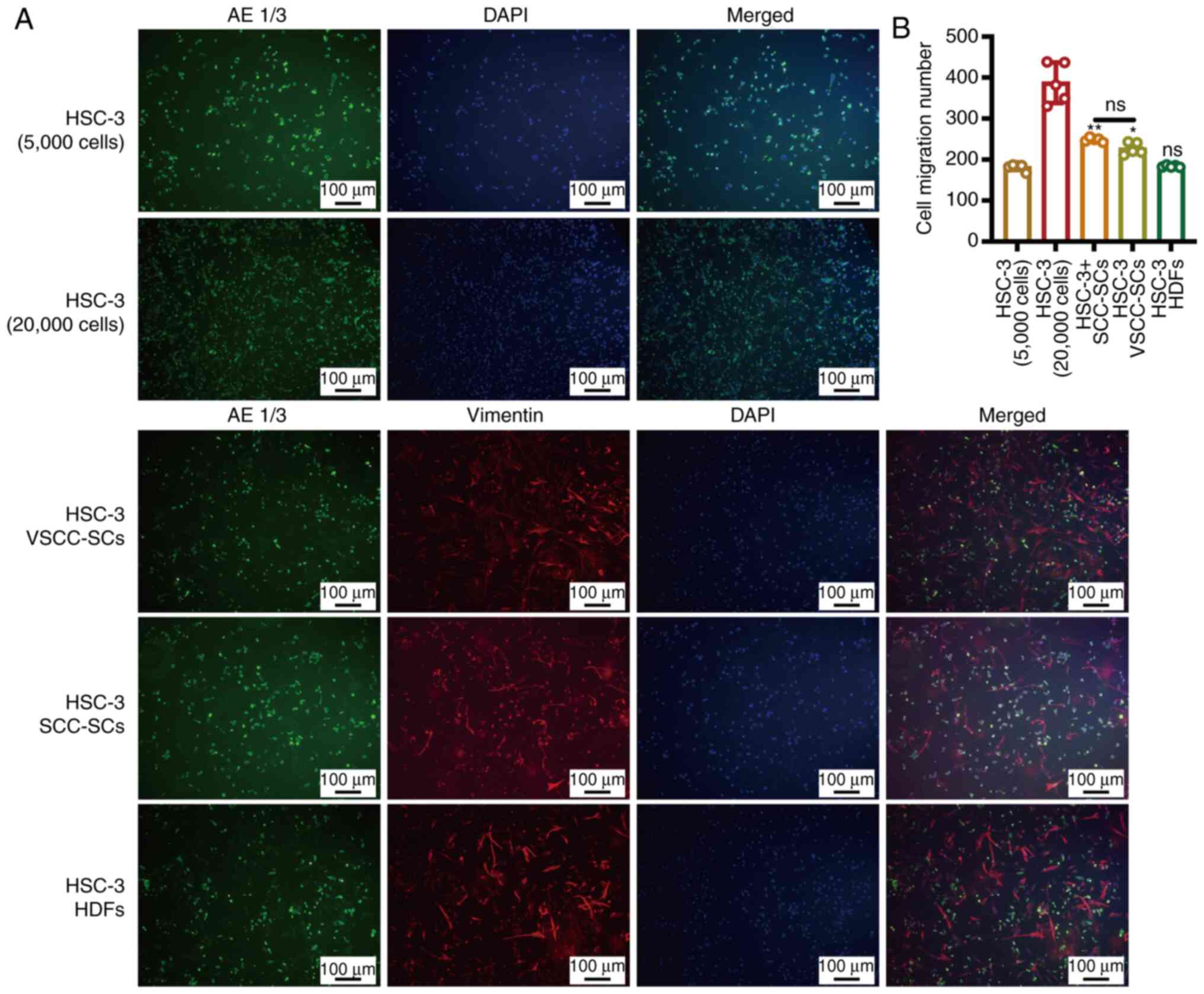
Both the VSCC-SCs and SCC-SCs promote the
migration of HSC-3 cells in vitro
A Transwell (migration) assay and IF staining were
used to determine the effects of VSCC-SCs, SCC-SCs and HDFs on the
migration of HSC-3 cells in vitro (Fig. 5A). The number of migrating cells
in the HSC-3 + SCC-SCs group was slightly higher than that in the
HSC-3 + VSCC-SCs group, and markedly higher than that in the HSC-3
+ HDFs and HSC-3 (5,000 cells) groups. Furthermore, the number of
migrating cells in the HSC-3 + HDFs group was slightly lower than
that in the HSC-3 (5,000 cells) group (Fig. 5B). These data thus indicated that
both the VSCC-SCs and SCC-SCs promoted the migration of HSC-3 cells
in vitro, while the HDFs exerted an inhibitory effect on the
migration of HSC-3 cells. In addition, the SCC-SCs exerted a more
prominent promotion effect than the VSCC-SCs.
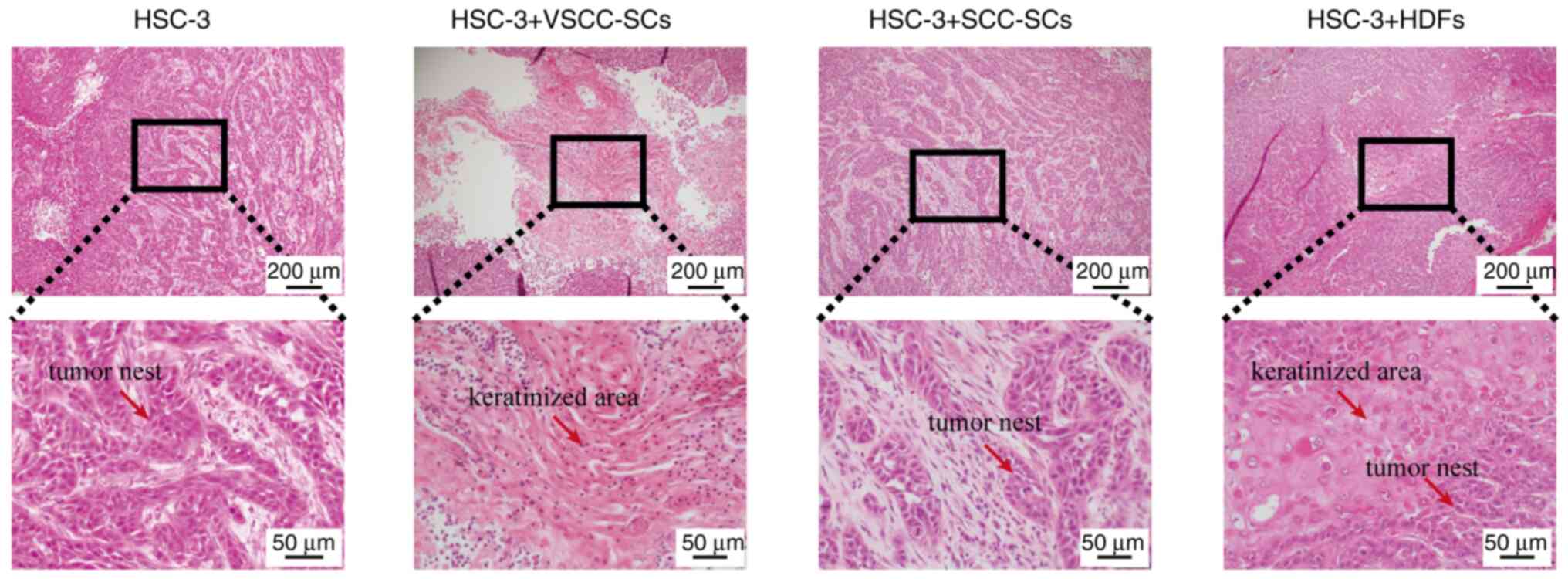
SCC-SCs inhibit the differentiation of
OSCC in vivo, whereas VSCC-SCs promote the differentiation of OSCC
cells
H&E staining was used to evaluate the effects of
VSCC-SCs, SCC-SCs and HDFs on the differentiation of OSCC cells
in vivo. In the HSC-3 only group, tumor tissue formation
with a small cancer nest without keratinization was observed, and
histological analysis revealed moderately differentiated OSCC. In
the HSC-3 + SCC-SCs group, H&E staining revealed that the
cancer nests were slightly smaller than those in the HSC-3 only
group. Histological analysis of the HSC-3 + SCC-SCs group revealed
moderately differentiated type OSCC. In the HSC-3 + VSCC-SCs and
HSC-3 + HDFs groups, these histological findings differed markedly
compared with the HSC-3 only group. In the HSC-3 + VSCC-SCs and
HSC-3 + HDFs groups, well-differentiated OSCC with notable
keratinization in the center of the tumor tissue was observed
(Fig. 6). Thus, these data
indicated that the SCC-SCs inhibit the differentiation of OSCC
in vivo, while the VSCC-SCs and HDFs promoted the
differentiation of OSCC.
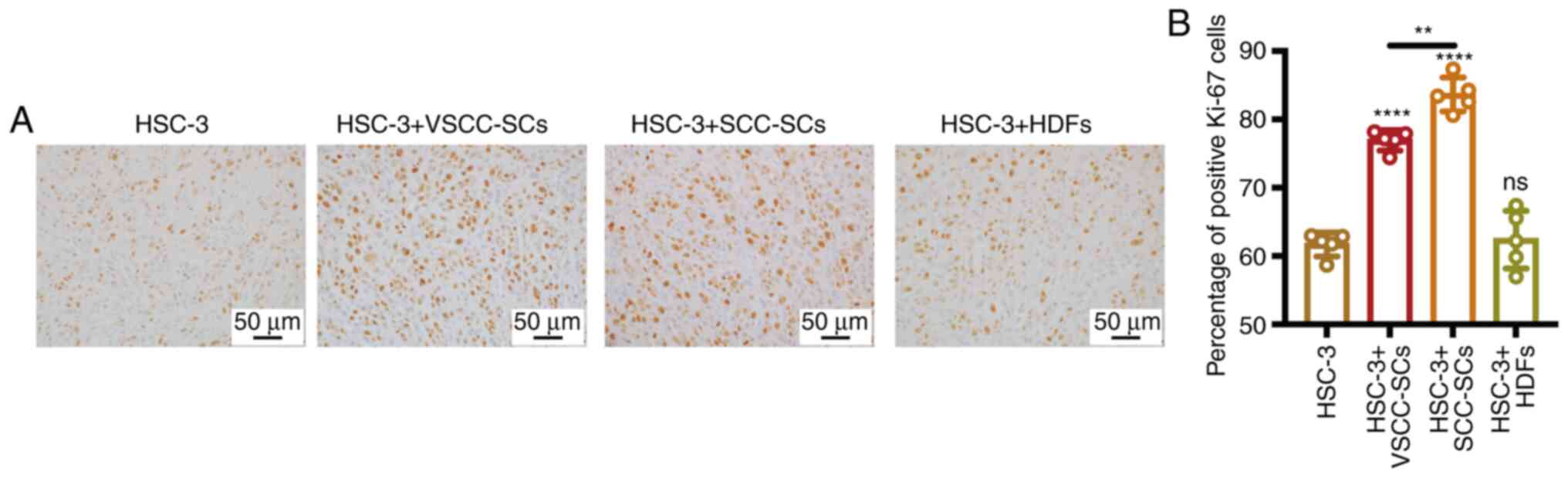
Both the VSCC-SCs and SCC-SCs promote
OSCC proliferation in vivo
The expression of Ki-67 in different groups was
assessed by IHC to demonstrate the effect of VSCC-SCs, SCC-SCs and
HDFs on the proliferation of OSCC cells in vivo (Fig. 7A). Compared with the HSC-3 only
group, the expression of Ki-67 was slightly increased in the HSC-3
+ VSCC-SCs group and was markedly increased in the HSC-3 + SCC-SCs
group. In addition, a minimal effect was observed in the HSC-3 +
HDFs group (Fig. 7B). Thus, these
data suggested that both the VSCC-SCs and SCC-SCs promoted the
proliferation of OSCC in vivo and that SCC-SCs exert a more
prominent promoting effect than the VSCC-SCs.
Both the VSCC-SCs and SCC-SCs promote the
bone invasion and invasion of OSCC cells in vivo
The effect of VSCC-SCs, SCC-SCs and HDFs on the bone
resorption of OSCC were evaluated by H&E staining. Bone
resorption in the HSC-3 + SCC-SCs group was slightly greater than
that in the VSCC-SCs group, while it was markedly greater than that
in the HSC-3 + HDFs group. There was a minimal difference between
the HSC-3 and HSC-3 + HDFs groups (Fig. 8A). Furthermore, TRAP staining was
used to determine the number of active multinucleated osteoclasts
in the different groups to demonstrate the effects of VSCC-SCs,
SCC-SCs and HDFs on the bone invasion of OSCC (Fig. 8B). The number of active
multinucleated osteoclasts in the HSC-3 + SCC-SCs group was the
highest, followed by the HSC-3+VSCC-SCs and HSC-3 groups. In
addition, the number of active multinucleated osteoclasts in the
HSC-3 + HDFs groups was slightly lower than that in the HSC-3 group
(Fig. 8C). These data suggest
that the VSCC-SCs and SCC-SCs promoted the bone invasion of OSCC
in vivo and that SCC-SCs exert a more prominent promoting
effect, while the HDFs exerted an inhibitory effect. The expression
levels of MMP9 and MT1-MMP, markers of invasion, in the different
groups were assessed by IHC to determine the effects of VSCC-SCs,
SCC-SCs and HDFs on the invasion of OSCC cells in vivo
(Fig. 8D and F). Compared with
the HSC-3 group, the number of cells positive for MMP9 and MT1-MMP
in the HSC-3 + VSCC-SCs and HSC-3 + SCC-SCs groups markedly
increased, while there was a minimal effect in the HSC-3 + HDFs
group (Fig. 8E and G). Therefore,
both the VSCC-SCs and SCC-SCs promoted the invasion of OSCC cells
in vivo, while the HDFs exerted a minimal effect. These data
suggest that both the VSCC-SCs and SCC-SCs promote the bone
invasion, and the invasion of OSCC in vivo.
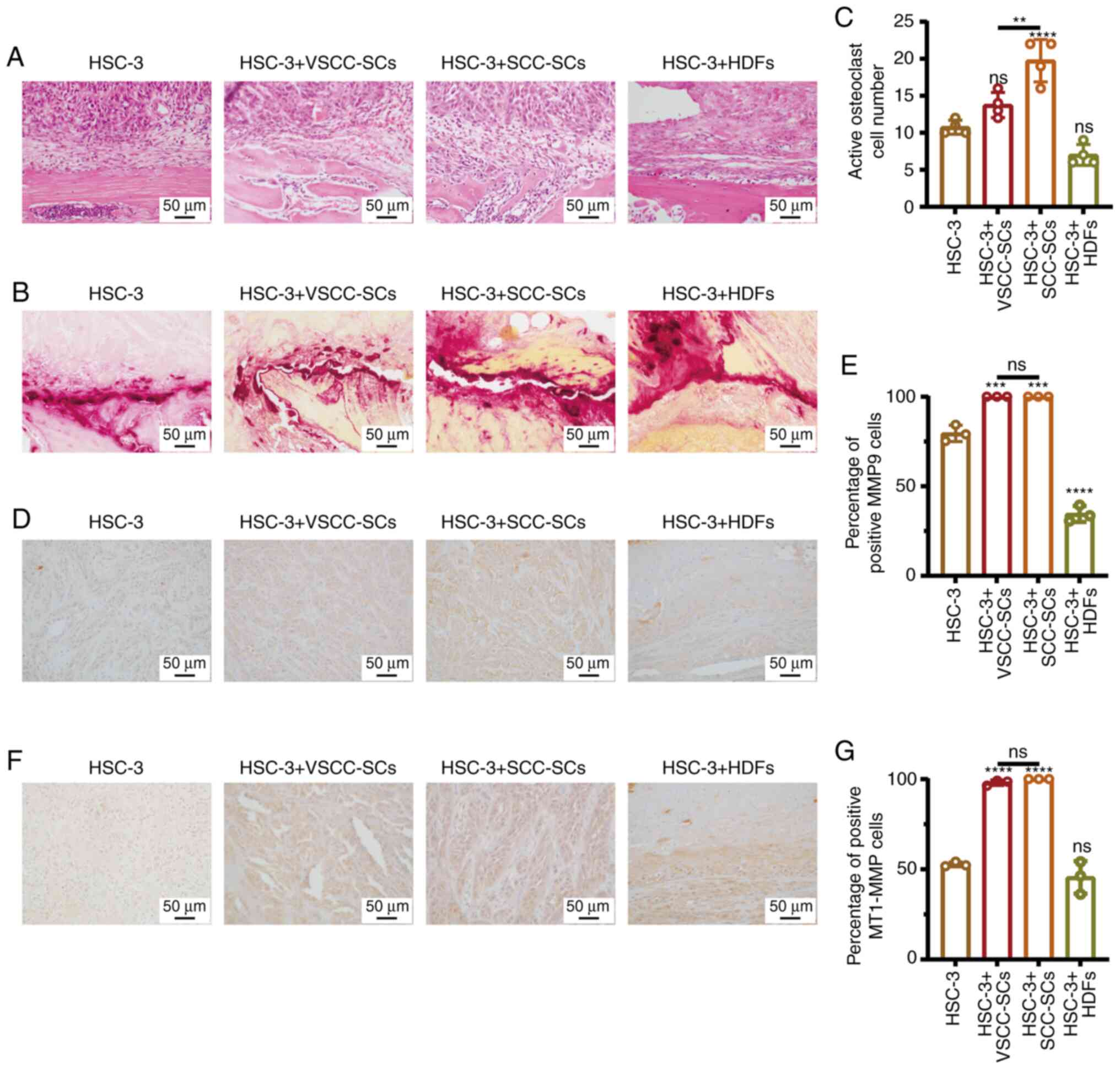 | Figure 8Both VSCC-SCs and SCC-SCs promote the
bone invasion and invasion of OSCC in vivo. (A and B) Effect
of VSCC-SCs, SCC-SCs and HDFs on the bone invasive ability of OSCC
in vivo was examine by H&E and TRAP staining. (C)
Quantification of activated multi-nucleated osteoclast cell number
in the different groups. Data are presented as the mean ± SD, n=3.
Statistical analysis was performed using one-way ANOVA; ns, not
significant (P>0.05); **P<0.01 and
****P<0.0001, all compared with the HSC-3 group. (D
and F) MMP9 and MT1-MMP expression levels in the different groups
were examined by immunohistochemistry. (E and G) Quantification of
positive MMP9 and MT1-MMP cells in the different groups. Data are
presented as the mean ± SD, n=5. Statistical analysis was performed
using one-way ANOVA; ns, not significant (P>0.05);
***P<0.001 and ****P<0.0001, all
compared with the HSC-3 group. VSCC-SCs, verrucous squamous cell
carcinoma-associated stromal cells; SCC-SCs, squamous cell
carcinoma-associated stromal cells; HDFs, human dermal fibroblasts;
OSCC, oral squamous cell carcinoma; TRAP, tartrate-resistant acid
phosphatase. |
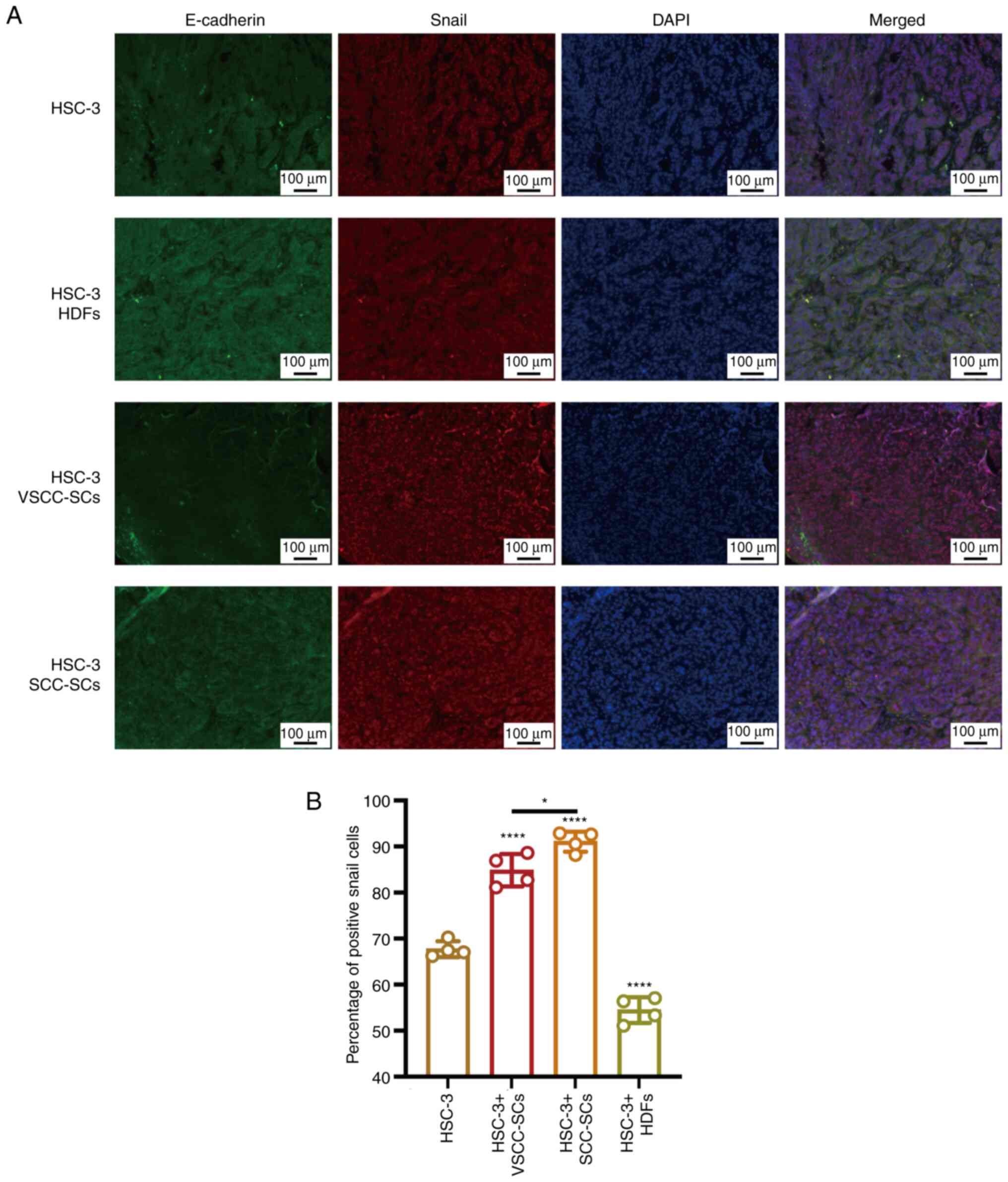
Both VSCC-SCs and SCC-SCs promote the EMT
of OSCC cells in vivo
The EMT biomarkers, E-cadherin and Snail, were
examined by IF staining to demonstrate the effects of VSCC-SCs,
SCC-SCs and HDFs on the EMT process of OSCC (Fig. 9A). The percentage of positive
Snail cells in the HSC-3 + SCC-SCs group was slightly higher than
that in the HSC-3 + VSCC-SCs groups and markedly higher than that
in the HSC-3 and HSC-3 + HDFs groups. In addition, the percentage
of positive Snail cells in HSC-3 + HDFs group was lower than that
in the HSC-3 group (Fig. 9B).
Thus, these data suggested that both the VSCC-SCs and SCC-SCs
promoted the EMT process of OSCC and that the SCC-SCs exerted a
more prominent promoting effect than the VSCC-SCs, while the HDFs
exerted an inhibitory effect on the EMT process of OSCC in
vivo.
C-X-C motif chemokine ligand (CXCL)8, C-C
motif chemokine ligand 2 (CCL2), C-X-C motif chemokine ligand 1
(CXCL1), mitogen-activated protein kinase 3 (MAPK3) and
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha (PIK3CA) may be involved in the regulation of OSCC
progression by VSCC-SCs and SCC-SCs
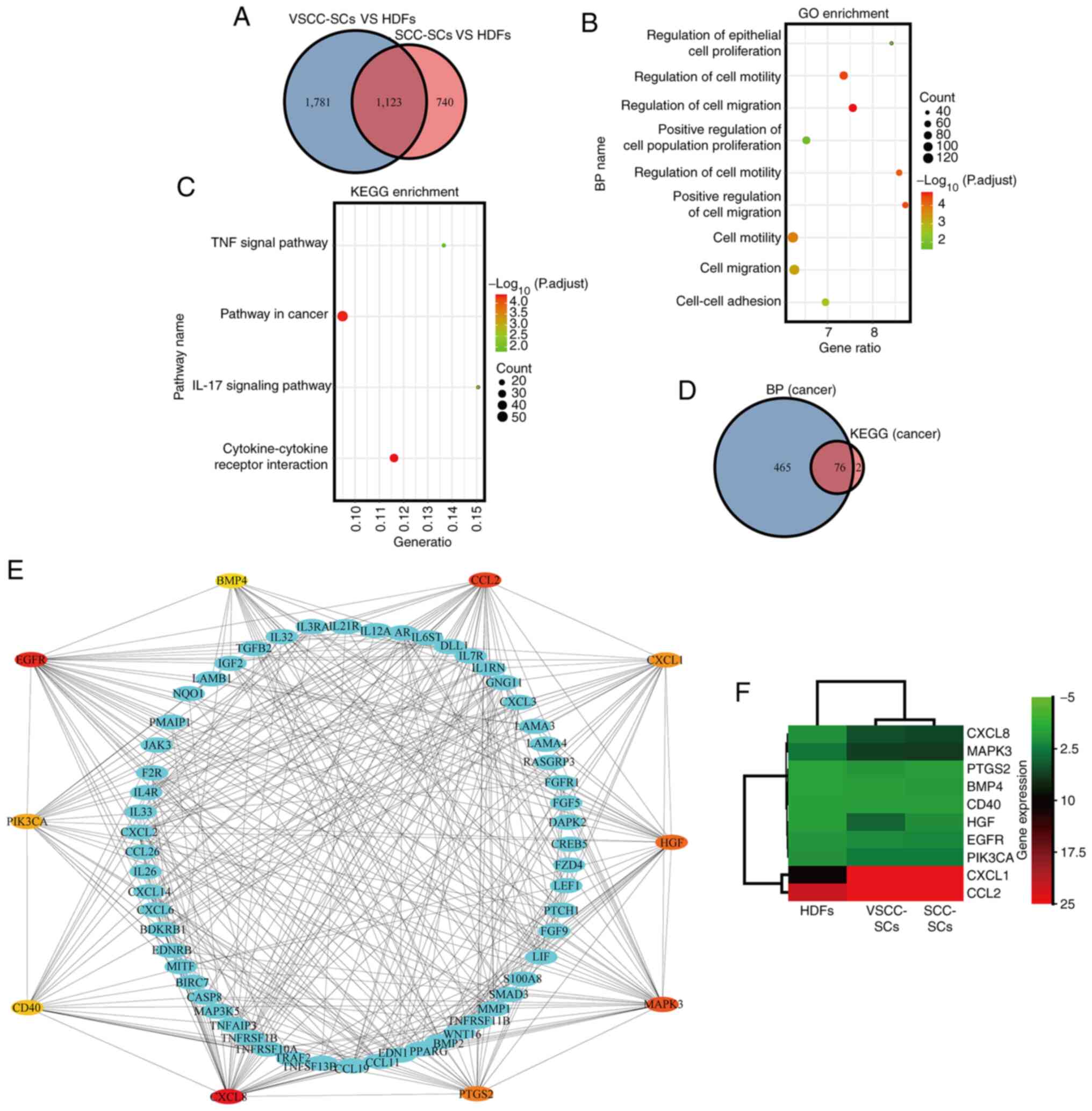
DEGs in the VSCC-SCs and SCC-SCs were analyzed using
a microarray and compared with those in the HDFs. Common
upregulated DEGs between the VSCC-SCs and SCC-SCs were identified
using a Venn diagram, which indicated that 1,123 genes were
upregulated in both the VSCC-SCs and SCC-SCs (Fig. 10A). Furthermore, the
cancer-associated biological process and cancer-associated cell
pathways of the common upregulated DEGs were analyzed using GO and
KEGG enrichment analysis, respectively (Fig. 10B and C). Common upregulated DEGs
were mainly enriched in cancer-associated biological processes,
such as cell proliferation, cell adhesion and cell migration
(Fig. 10B). In addition, common
upregulated-DEGs were mainly enriched in cancer-associated cellular
pathways, such as the TNF signaling pathway, pathways in cancer,
IL-17 signaling pathway and cytokine-cytokine receptor interaction
(Fig. 10C). Common upregulated
DEGs both associated with cancer biological processes and cancer
cell pathways were identified using the Venn diagram; 76 common
upregulated DEGs were both associated with cancer biological
processes and cancer cell pathways (Fig. 10D). The top 10 hub genes in these
common upregulated DEGs both associated with cancer biological
processes and cancer cell pathways were analyzed using a PPI
network. CXCL8, MAPK3, prostaglandin-endoperoxide synthase 2
(PTGS2), bone morphogenetic protein 4 (BMP4), CD40, hepatocyte
growth factor (HGF), EGFR, PIK3CA, CXCL1 and CCL2 were identified
as the hub genes (Fig. 10E).
Finally, the top 10 hub genes differentially expressed in VSCC-SCs,
SCC-SCs, and HDFs were analyzed by cluster and heatmap analysis.
CXCL8, MAPK3, PIK3CA, CXCL1 and CCL2 were differentially expressed
in VSCC-SCs, SCC-SCs, and HDFs (Fig.
10F). These data suggest that CXCL8, CCL2, CXCL1, MAPK3 and
PIK3CA may be involved in the regulation of OSCC progression by
VSCC-SCs and SCC-SCs.
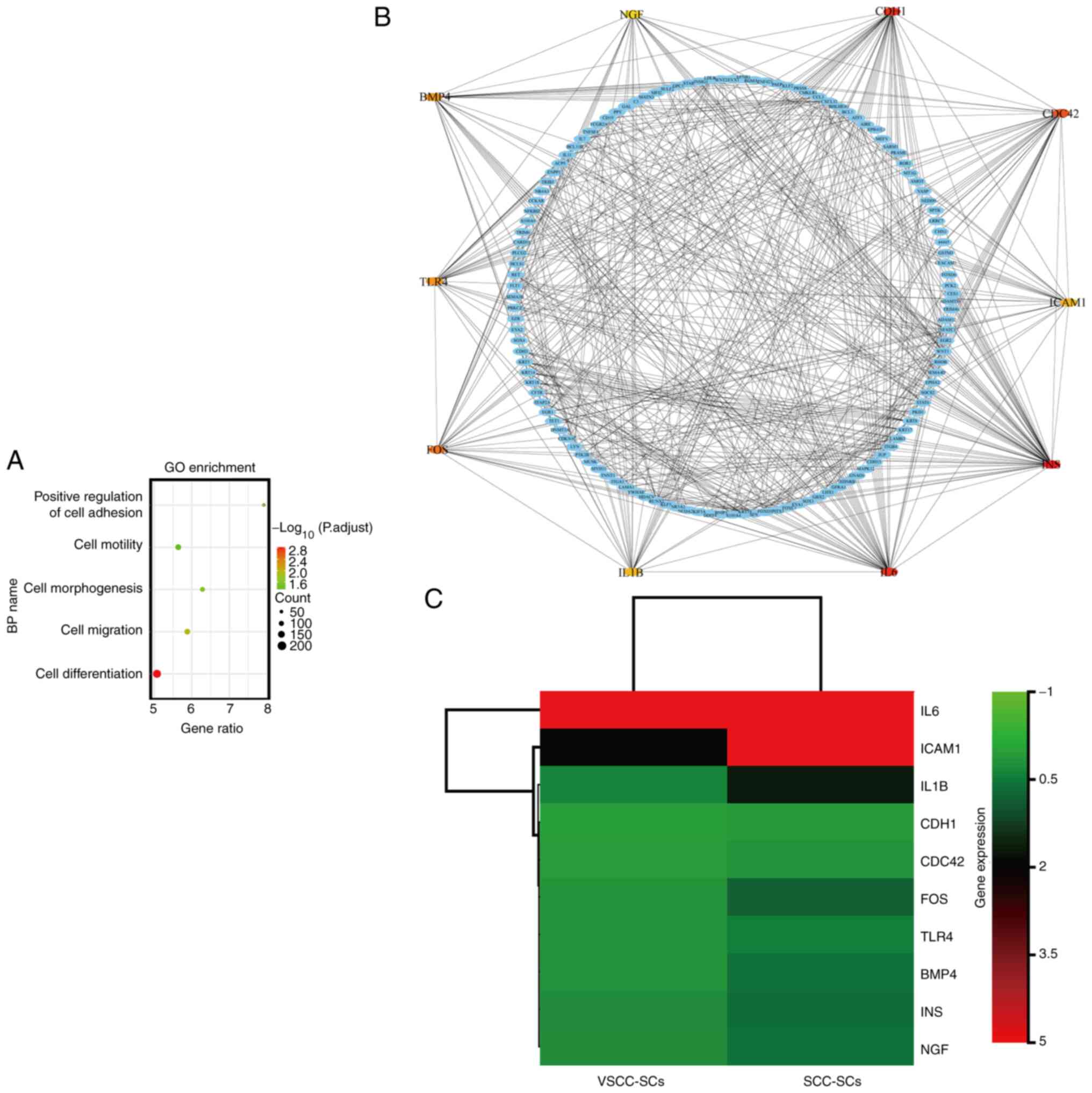
Intercellular adhesion molecule 1
(ICAM1), interleukin (IL)1B, Fos proto-oncogene, AP-1 transcription
factor subunit (FOS), bone morphogenetic protein 4 (BMP4), insulin
(INS) and nerve growth factor (NGF) may underlie the differential
effects of VSCC-SCs and SCC-SCs on the differentiation of OSCC
The upregulated DEGs in the SCC-SCs were analyzed
using a microarray and compared with those in the VSCC-SCs. The
biological processes of upregulated DEGs were analyzed using GO
enrichment analysis. The upregulated DEGs were mainly enriched in
cell differentiation, cell proliferation, cell adhesion and cell
migration (Fig. 11A).
Furthermore, the top 10 hub genes involved in cell differentiation
were identified using a PPI network, as follows: IL6, ICAM1, IL1B,
Cadherin 1 (CDH1), CDC42, FOS, Toll-like receptor (TLR)4, BMP4, INS
and NGF (Fig. 11B). Finally, the
hub genes differentially expressed in SCC-SCs and VSCC-SCs were
analyzed using cluster and heatmap analyses, which suggested that
ICAM1, IL1B, FOS, BMP4, INS and NGF were differentially expressed
in the SCC-SCs (Fig. 11C).
Therefore, ICAM1, IL1B, FOS, BMP4, INS and NGF may underlie the
differential effects of VSCC-SCs and SCC-SCs on the differentiation
of OSCC.
Discussion
Benign and malignant cancers can be assayed by the
degree of differentiation. The prognosis of patients with
well-differentiated cancers is better than that for patients with
moderately differentiated and poorly differentiated cancers.
Therefore, the degree of differentiation can influence the
biological processes of cancers (20). It has been suggested that SRY-box
transcription factor 4 (SOX4), SUMO-specific peptidase 3 (SENP3),
grainyhead like transcription factor 2 (GRHL2) and RUNX family
transcription factor 1 (RUNX1) are significantly associated with
the differentiation of OSCC (21-24). In addition, the extracellular
signal-regulated kinase 1/2 (ERK1/2), JNK and STAT3 signaling
pathways also play significant roles in the differentiation of OSCC
(25). Moreover, S100A16 has been
shown to promote the differentiation of OSCC cells and inhibit the
progression of OSCC (26).
Therefore, the differentiation of OSCC may be regulated by specific
proteins and signaling pathways, which can further influence its
progression. However, to the best of our knowledge, no study to
date has investigated the effects of stromal cells on the
differentiation of OSCC. In the present study, it was found that
VSCC-SCs promoted the differentiation of OSCC, while SCC-SCs
inhibited the differentiation of OSCC. HDFs exerted a minimal
effect on the differentiation of OSCC cells. Furthermore, the
reason for the differential effects of VSCC-SCs and SCC-SCs on the
degree of OSCC differentiation was analyzed using microarray and
bioinformatics methods. The results indicated that the
overexpression of ICAM1, IL1B, FOS, BMP4, INS and NGF in the
SCC-SCs was closely associated with cell differentiation. It has
been suggested that ICAM1 is closely associated with the
differentiation of osteoclasts and can also regulate the
differentiation of colorectal cancer (27-29); however, it has not been widely
investigated in the differentiation of OSCC. IL1B regulates the
differentiation of TH17 and TH19 in the TME (30,31). IL1B also plays a critical role in
the differentiation of prostate cancer cells (32). BMP4 plays a significant role in
the differentiation of neuroblastoma, colorectal cancer stem cells
and CD133+ hepatic cancer stem cells; however, it has
not been extensively studied in the differentiation of OSCC
(33-35). NGF regulates the differentiation
of neuroblasts and neuroblastoma cells (36,37). FOS (also known as p55) and INS
have not been extensively investigated in the differentiation of
OSCC. Therefore, ICAM1, IL1B, BMP4 and NGF may underlie the
differential effects of VSCC-SCs and SCC-SCs on the degree of
differentiation of OSCC. Stromal cells, which are the main
components of the TME, are also associated with cancer progression
(7,38). A previous study suggested that the
proliferation, invasion and metastasis of CD133+
pancreatic cancer cells was enhanced following co-culture with
pancreatic stromal cells (39).
Senescent stromal cells promote the growth and invasion of CRC both
in vitro and in vivo (40). In addition, the results of a gene
expression microarray in three types of cancer demonstrated that
stromal cells could alter cellular functions, including cell-cycle,
cell signal, cell movement and cell death (41). The results of these studies
indicated that stromal cells influence the progression of multiple
cancer types. As regards OSCC, a recent study suggested that
CAFs-derived exosomal miR-382-5p promoted the invasion and
migration of OSCC (42). Long
non-coding RNAs (lncRNAs) also play a significant role in CAFs,
promoting the progression of OSCC (43). Epiregulin has been found to
promote the transformation of normal fibroblasts into CAFs, which
are induced in OSCC via the JAK2/STAT3 pathway (44). Therefore, miRNAs and lncRNAs in
CAFs promote the progression of OSCC. However, research
investigating the crosstalk between stromal cells and OSCC is
limited (17). The findings of
the present study indicated that VSCC-SCs and SCC-SCs promoted the
proliferation, invasion and migration of OSCC both in vitro
and in vivo, and that SCC-SCs exerted a more prominent
promoting effect than the VSCC-SCs, while HDFs exert a limited
effect on the proliferation, invasion and migration of OSCC. In
addition, the proliferation of HSC-3 + VSCC-SCs and HSC-3 + HDFs
decreased on day 5, whereas that of HSC-3 + SCC-SCs increased on
day 5 in vitro, according to the results of MTS assay; this
may be caused by differences in the viability of VSCC-SCs, SCC-SCs
and HDFs. Cell death in the HSC-3 + VSCC-SCs and HSC-3 + HDFs was
observed from day 5 due to the higher viability of VSCC-SCs and
HDFs, which resulted in cell crowding. Conversely, the death of
HSC-3 + SCC-SCs was not observed due to the lower viability of
SCC-SCs, which did not lead to cell crowding on day 5. Finally,
SCC-SCs inhibited the degree of OSCC differentiation, indicating
that SCC-SCs have the potential to promote the proliferation,
invasion and migration of OSCC. The results obtained for
proliferation, invasion and migration indirectly confirmed
differentiation. Thus, SCC-SCs inhibited the differentiation, and
promoted the proliferation, invasion, and migration of OSCC cells.
VSCC-SCs exerted a promoting effect on the differentiation degree
of OSCC, indicating that VSCC-SCs have the potential to inhibit the
proliferation, invasion and migration of OSCC. However, apart from
the keratinized area, an area of invasion was also observed in the
HSC-3 + VSCC-SCs group based on H&E staining. Thus, the effect
of VSCC-SCs on the proliferation, invasion and migration of OSCC
could not be identified simply by the degree of differentiation.
The results indicated that the VSCC-SCs promoted the proliferation,
invasion and migration of OSCC cells. Thus, VSCC-SCs promoted the
differentiation, proliferation, invasion and migration of OSCC
cells. Therefore, both the VSCC-SCs and SCC-SCs promote the
progression of OSCC, while the HDFs exerted a minimal effect. In
addition, the SCC-SCs exerted a more prominent promoting effect
than the VSCC-SCs.
In addition, the microarray data in the present
study also suggested that CXCL8, CCL2, CXCL1, MAPK3 and PIK3CA in
the VSCC-SCs and SCC-SCs may regulate the progression of OSCC.
CXCL8 (also known as IL-8) regulates cellular responses in
combination with CXCR1 and CXCR2 (45). A recent study demonstrated that
the loss of androgen receptor could promote the expression of CXCL8
in CAFs, which in turn regulates the migration of prostate cancer
cells (46). In addition,
blocking autophagy in CAFs can promote the progression of HNSCC by
regulating the secretion of IL6, IL8, and the other cytokines
(47). Therefore, CXCL8 has the
potential to regulate the crosstalk between stromal cells and OSCC.
CCL2, which is expressed in both cancer and stromal cells, recruits
the macrophages during acute inflammation by binding to CCR2, a
process that can regulate the immunoreactivity of the TME (48,49). The results of a recent study
indicated that CCL2-mediated inflammation increases the risk of
breast cancer (50). CCL2 also
mediates the cross-talk between breast cancer and stromal cells
that regulate the breast cancer stem cells (51). Thus, CCL2 plays a significant role
in the TME of multiple cancer types, and has the potential to
regulate the crosstalk between stromal cells and OSCC. CXCL1, which
is expressed in both cancer and stromal cells, regulates cancer
progression by binding to CXCR2 (52,53). A recent study indicated that CXCL1
in CAFs promoted the proliferation of ovarian cancer by the
activation of p38 (54). TGF-β
and IL1B regulate the progression of breast cancer and OSCC by
regulating CXCL1 in CAFs respectively (55,56). Therefore, CXCL1 in stromal cells
has the potential to regulate the progression of OSCC. MAPK3 (also
known as ERK1) plays a significant role in the progression of
multiple types of cancer (57,58). The results of a recent study
suggested that the crosstalk between cancers and stromal cells is
suppressed upon ERK1/2 inhibition in cancer-associated pancreatic
stellate cells (59). Therefore,
MAPK3 also plays a significant role in the crosstalk between cancer
and stromal cells, and has the potential to mediate the crosstalk
between stromal cells and OSCC. PIK3CA (also known as PI3K) plays a
significant role in the progression of OSCC (60,61). However, to the best of our
knowledge, no study to date has investigated the role and function
of PIK3CA in the crosstalk between stromal cells and OSCC.
In conclusion, the present study demonstrated that
VSCC-SCs promoted the differentiation, proliferation, invasion and
metastasis of OSCC, while the SCC-SCs inhibited differentiation and
promoted the proliferation, invasion and metastasis of OSCC.
Compared with the VSCC-SCs, the SCC-SCs exerted an inhibitory
effect on the differentiation and exerted a more potent promoting
effect on the proliferation, invasion and migration of OSCC.
Finally, CXCL8, CCL2, CXCL1, MAPK3 and PIK3CA in VSCC-SCs and
SCC-SCs may regulate the progression of OSCC, and ICAM1, IL1B, FOS,
BMP4, INS and NGF may underlie the differential effects of VSCC-SCs
and SCC-SCs on the differentiation of OSCC. These findings may
describe a potential regulatory mechanism in the progression of
OSCC.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. In addition, the dataset GSE164374 is currently private
and is scheduled to be released on Jan 01, 2024.
Authors' contributions
QS designed the outline of the study. QS, HO, KT,
MWO, HK, KN, SI, AS and HN conducted the experiments and data
analysis. QS, KT and HO were involved in the preparation of
manuscript. QS wrote the manuscript. KN, SI, AS and HN supervised
the study and contributed to data interpretation and manuscript
revision. KT and HN confirm the authenticity of all raw data. All
authors have read and agreed to the published version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Okayama University (project identification code:
1703-042-001). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Lindemann A, Takahashi H, Patel AA, Osman
AA and Myers JN: Targeting the DNA damage response in OSCC with
TP53 mutations. J Dent Res. 97:635–644. 2018. View Article : Google Scholar
|
|
2
|
Jiang Y, Li T, Wu Y, Xu H, Xie C, Dong Y,
Zhong L, Wang Z, Zhao H, Zhou Y, et al: GPR39 overexpression in
OSCC promotes YAP-sustained malignant progression. J Dent Res.
99:949–958. 2020. View Article : Google Scholar
|
|
3
|
Chen Y, Shao Z, Jiang E, Zhou X, Wang L,
Wang H, Luo X, Chen Q, Liu K and Shang Z: CCL21/CCR7 interaction
promotes EMT and enhances the stemness of OSCC via a JAK2/STAT3
signaling pathway. J Cell Physiol. 235:5995–6009. 2020. View Article : Google Scholar
|
|
4
|
Yang Z, Liang X, Fu Y, Liu Y, Zheng L, Liu
F, Li T, Yin X, Qiao X and Xu X: Identification of AUNIP as a
candidate diagnostic and prognostic biomarker for oral squamous
cell carcinoma. EBioMedicine. 47:44–57. 2019. View Article : Google Scholar
|
|
5
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar
|
|
6
|
Li Z, Liu FY and Kirkwood KL: The
p38/MKP-1 signaling axis in oral cancer: Impact of tumor-associated
macrophages. Oral Oncol. 103:1045912020. View Article : Google Scholar
|
|
7
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar
|
|
8
|
Peddareddigari VG, Wang D and Dubois RN:
The tumor microenvironment in colorectal carcinogenesis. Cancer
Microenviron. 3:149–166. 2010. View Article : Google Scholar
|
|
9
|
Bhome R, Mellone M, Emo K, Thomas GJ,
Sayan AE and Mirnezami AH: The colorectal cancer microenvironment:
Strategies for studying the role of cancer-associated fibroblasts.
Methods Mol Biol. 1765:87–98. 2018. View Article : Google Scholar
|
|
10
|
Udagawa T and Wood M: Tumor-stromal cell
interactions and opportunities for therapeutic intervention. Curr
Opin Pharmacol. 10:369–374. 2010. View Article : Google Scholar
|
|
11
|
Potter SM, Dwyer RM, Hartmann MC, Khan S,
Boyle MP, Curran CE and Kerin MJ: Influence of stromal-epithelial
interactions on breast cancer in vitro and in vivo. Breast Cancer
Res Treat. 131:401–411. 2012. View Article : Google Scholar
|
|
12
|
O'Malley G, Treacy O, Lynch K, Naicker SD,
Leonard NA, Lohan P, Dunne PD, Ritter T, Egan LJ and Ryan AE:
Stromal cell PD-L1 inhibits CD8+ T-cell antitumor immune
responses and promotes colon cancer. Cancer Immunol Res.
6:1426–1441. 2018. View Article : Google Scholar
|
|
13
|
Trivanović D, Krstić J, Jauković A,
Bugarski D and Santibanez JF: Mesenchymal stromal cell engagement
in cancer cell epithelial to mesenchymal transition. Dev Dyn.
247:359–367. 2018. View Article : Google Scholar
|
|
14
|
Patel SG and Shah JP: TNM staging of
cancers of the head and neck: Striving for uniformity among
diversity. CA Cancer J Clin. 55:242–258. 2005. View Article : Google Scholar
|
|
15
|
Spiro RH, Guillamondegui O Jr, Paulino AF
and Huvos AG: Pattern of invasion and margin assessment in patients
with oral tongue cancer. Head Neck. 21:408–413. 1999. View Article : Google Scholar
|
|
16
|
Hussein MR and Cullen K: Molecular
biomarkers in HNSCC: Prognostic and therapeutic implications.
Expert Rev Anticancer Ther. 1:116–24. 2001. View Article : Google Scholar
|
|
17
|
Takabatake K, Kawai H, Omori H, Qiusheng
S, Oo MW, Sukegawa S, Nakano K, Tsujigiwa H and Nagatsuka H: Impact
of the stroma on the biological characteristics of the parenchyma
in oral squamous cell carcinoma. Int J Mol Sci. 21:77142020.
View Article : Google Scholar
|
|
18
|
Zhang X, Junior CR, Liu M, Li F, D'Silva
NJ and Kirkwood KL: Oral squamous carcinoma cells secrete RANKL
directly supporting osteolytic bone loss. Oral Oncol. 49:119–128.
2013. View Article : Google Scholar
|
|
19
|
An YZ, Cho E, Ling J and Zhang X: The
Axin2-snail axis promotes bone invasion by activating
cancer-associated fibroblasts in oral squamous cell carcinoma. BMC
Cancer. 20:9872020. View Article : Google Scholar
|
|
20
|
Jögi A, Vaapil M, Johansson M and Påhlman
S: Cancer cell differentiation heterogeneity and aggressive
behavior in solid tumors. Ups J Med Sci. 117:217–224. 2012.
View Article : Google Scholar
|
|
21
|
Watanabe M, Ohnishi Y, Wato M, Tanaka A
and Kakudo K: SOX4 expression is closely associated with
differentiation and lymph node metastasis in oral squamous cell
carcinoma. Med Mol Morphol. 47:150–155. 2014. View Article : Google Scholar
|
|
22
|
Sun Z, Hu S, Luo Q, Ye D, Hu D and Chen F:
Overexpression of SENP3 in oral squamous cell carcinoma and its
association with differentiation. Oncol Rep. 29:1701–1706. 2013.
View Article : Google Scholar
|
|
23
|
Kang MK, Chen W and Park NH: Regulation of
epithelial cell proliferation, differentiation, and plasticity by
grainyhead-like 2 during oral carcinogenesis. Crit Rev Oncog.
23:201–217. 2018. View Article : Google Scholar
|
|
24
|
Feng X, Zheng Z, Wang Y, Song G, Wang L,
Zhang Z, Zhao J, Wang Q and Lun L: Elevated RUNX1 is a prognostic
biomarker for human head and neck squamous cell carcinoma. Exp Biol
Med (Maywood). 246:538–546. 2021. View Article : Google Scholar
|
|
25
|
Gkouveris I, Nikitakis N, Avgoustidis D,
Karanikou M, Rassidakis G and Sklavounou A: ERK1/2, JNK and STAT3
activation and correlation with tumor differentiation in oral SCC.
Histol Histopathol. 32:1065–1076. 2017.
|
|
26
|
Sapkota D, Bruland O, Parajuli H, Osman
TA, The MT, Johannessen AC and Costea DE: S100A16 promotes
differentiation and contributes to a less aggressive tumor
phenotype in oral squamous cell carcinoma. BMC Cancer. 15:6312015.
View Article : Google Scholar
|
|
27
|
Kim B, Nam S, Lim JH and Lim JS: NDRG2
expression decreases tumor-induced osteoclast differentiation by
down-regulating ICAM1 in breast cancer cells. Biomol Ther (Seoul).
24:9–18. 2016. View Article : Google Scholar
|
|
28
|
Astarci E, Sade A, Cimen I, Savaş B and
Banerjee S: The NF-κB target genes ICAM-1 and VCAM-1 are
differentially regulated during spontaneous differentiation of
Caco-2 cells. FEBS J. 279:2966–2986. 2012. View Article : Google Scholar
|
|
29
|
Wang QL, Li BH, Liu B, Liu YB, Liu YP,
Miao SB, Han Y, Wen JK and Han M: Polymorphisms of the ICAM-1 exon
6 (E469K) are associated with differentiation of colorectal cancer.
J Exp Clin Cancer Res. 28:1392009. View Article : Google Scholar
|
|
30
|
Revu S, Wu J, Henkel M, Rittenhouse N,
Menk A, Delgoffe GM, Poholek AC and McGeachy MJ: IL-23 and IL-1β
drive human Th17 cell differentiation and metabolic reprogramming
in absence of CD28 costimulation. Cell Rep. 22:2642–2653. 2018.
View Article : Google Scholar
|
|
31
|
Xue G, Jin G, Fang J and Lu Y: IL-4
together with IL-1β induces antitumor Th9 cell differentiation in
the absence of TGF-β signaling. Nat Commun. 10:13762019. View Article : Google Scholar
|
|
32
|
Albrecht M, Doroszewicz J, Gillen S, Gomes
I, Wilhelm B, Stief T and Aumüller G: Proliferation of prostate
cancer cells and activity of neutral endopeptidase is regulated by
bombesin and IL-1beta with IL-1beta acting as a modulator of
cellular differentiation. Prostate. 58:82–94. 2004. View Article : Google Scholar
|
|
33
|
Szemes M, Melegh Z, Bellamy J, Greenhough
A, Kollareddy M, Catchpoole D and Malik K: A wnt-BMP4 signaling
axis induces MSX and NOTCH proteins and promotes growth suppression
and differentiation in neuroblastoma. Cells. 9:7832020. View Article : Google Scholar
|
|
34
|
Catalano V, Dentice M, Ambrosio R, Luongo
C, Carollo R, Benfante A, Todaro M, Stassi G and Salvatore D:
Activated thyroid hormone promotes differentiation and
chemotherapeutic sensitization of colorectal cancer stem cells by
regulating wnt and BMP4 signaling. Cancer Res. 76:1237–1244. 2016.
View Article : Google Scholar
|
|
35
|
Zhang L, Sun H, Zhao F, Lu P, Ge C, Li H,
Hou H, Yan M, Chen T, Jiang G, et al: BMP4 administration induces
differentiation of CD133+ hepatic cancer stem cells,
blocking their contributions to hepatocellular carcinoma. Cancer
Res. 72:4276–4285. 2012. View Article : Google Scholar
|
|
36
|
Fell SM, Li S, Wallis K, Kock A, Surova O,
Rraklli V, Höfig CS, Li W, Mittag J, Henriksson MA, et al:
Neuroblast differentiation during development and in neuroblastoma
requires KIF1Bβ-mediated transport of TRKA. Genes Dev.
31:1036–1053. 2017. View Article : Google Scholar
|
|
37
|
Condello S, Caccamo D, Currò M, Ferlazzo
N, Parisi G and Ientile R: Transglutaminase 2 and NF-kappaB
interplay during NGF-induced differentiation of neuroblastoma
cells. Brain Res. 1207:1–8. 2008. View Article : Google Scholar
|
|
38
|
Louhichi T, Saad H, Dhiab MB, Ziadi S and
Trimeche M: Stromal CD10 expression in breast cancer correlates
with tumor invasion and cancer stem cell phenotype. BMC Cancer.
18:492018. View Article : Google Scholar
|
|
39
|
Moriyama T, Ohuchida K, Mizumoto K, Cui L,
Ikenaga N, Sato N and Tanaka M: Enhanced cell migration and
invasion of CD133+ pancreatic cancer cells cocultured with
pancreatic stromal cells. Cancer. 116:3357–3368. 2010. View Article : Google Scholar
|
|
40
|
Liu Y, Pan J, Pan X, Wu L, Bian J, Lin Z,
Xue M, Su T, Lai S, Chen F, et al: Klotho-mediated targeting of
CCL2 suppresses the induction of colorectal cancer progression by
stromal cell senescent microenvironments. Mol Oncol. 13:2460–2475.
2019. View Article : Google Scholar
|
|
41
|
Kadaba R, Birke H, Wang J, Hooper S, Andl
CD, Di Maggio F, Soylu E, Ghallab M, Bor D, Froeling FE, et al:
Imbalance of desmoplastic stromal cell numbers drives aggressive
cancer processes. J Pathol. 230:107–117. 2013. View Article : Google Scholar
|
|
42
|
Sun LP, Xu K, Cui J, Yuan DY, Zou B, Li J,
Liu JL, Li KY, Meng Z and Zhang B: Cancer-associated
fibroblast-derived exosomal miR-382-5p promotes the migration and
invasion of oral squamous cell carcinoma. Oncol Rep. 42:1319–1328.
2019.
|
|
43
|
Ding L, Ren J, Zhang D, Li Y, Huang X, Hu
Q, Wang H, Song Y, Ni Y and Hou Y: A novel stromal lncRNA signature
reprograms fibroblasts to promote the growth of oral squamous cell
carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis.
39:397–406. 2018. View Article : Google Scholar
|
|
44
|
Wang Y, Jing Y, Ding L, Zhang X, Song Y,
Chen S, Zhao X, Huang X, Pu Y, Wang Z, et al: Epiregulin reprograms
cancer-associated fibroblasts and facilitates oral squamous cell
carcinoma invasion via JAK2-STAT3 pathway. J Exp Clin Cancer Res.
38:2742019. View Article : Google Scholar
|
|
45
|
Herrero AB, García-Gómez A, Garayoa M,
Corchete LA, Hernández JM, San Miguel J and Gutierrez NC: Effects
of IL-8 up-regulation on cell survival and osteoclastogenesis in
multiple myeloma. Am J Pathol. 186:2171–2182. 2016. View Article : Google Scholar
|
|
46
|
Cioni B, Nevedomskaya E, Melis MH, van
Burgsteden J, Stelloo S, Hodel E, Spinozzi D, de Jong J, van der
Poel H, de Boer JP, et al: Loss of androgen receptor signaling in
prostate cancer-associated fibroblasts (CAFs) promotes CCL2- and
CXCL8-mediated cancer cell migration. Mol Oncol. 12:1308–1323.
2018. View Article : Google Scholar
|
|
47
|
New J, Arnold L, Ananth M, Alvi S,
Thornton M, Werner L, Tawfik O, Dai H, Shnayder Y, Kakarala K, et
al: Secretory autophagy in cancer-associated fibroblasts promotes
head and neck cancer progression and offers a novel therapeutic
target. Cancer Res. 77:6679–6691. 2017. View Article : Google Scholar
|
|
48
|
Giri J, Das R, Nylen E, Chinnadurai R and
Galipeau J: CCL2 and CXCL12 derived from mesenchymal stromal cells
cooperatively polarize IL-10+ tissue macrophages to mitigate gut
injury. Cell Rep. 30:1923–1934.e4. 2020. View Article : Google Scholar
|
|
49
|
Yao M, Fang W, Smart C, Hu Q, Huang S,
Alvarez N, Fields P and Cheng N: CCR2 chemokine receptors enhance
growth and cell-cycle progression of breast cancer cells through
SRC and PKC activation. Mol Cancer Res. 17:604–617. 2019.
View Article : Google Scholar
|
|
50
|
Sun X, Glynn DJ, Hodson LJ, Huo C, Britt
K, Thompson EW, Woolford L, Evdokiou A, Pollard JW, Robertson SA
and Ingman WV: CCL2-driven inflammation increases mammary gland
stromal density and cancer susceptibility in a transgenic mouse
model. Breast Cancer Res. 19:42017. View Article : Google Scholar
|
|
51
|
Tsuyada A, Chow A, Wu J, Somlo G, Chu P,
Loera S, Luu T, Li AX, Wu X, Ye W, et al: CCL2 mediates cross-talk
between cancer cells and stromal fibroblasts that regulates breast
cancer stem cells. Cancer Res. 72:2768–2779. 2012. View Article : Google Scholar
|
|
52
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, et al: A CXCL1 paracrine network links cancer
chemoresistance and metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar
|
|
53
|
Miyake M, Hori S, Morizawa Y, Tatsumi Y,
Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K, et al:
CXCL1-mediated interaction of cancer cells with tumor-associated
macrophages and cancer-associated fibroblasts promotes tumor
progression in human bladder cancer. Neoplasia. 18:636–646. 2016.
View Article : Google Scholar
|
|
54
|
Park GY, Pathak HB, Godwin AK and Kwon Y:
Epithelial-stromal communication via CXCL1-CXCR2 interaction
stimulates growth of ovarian cancer cells through p38 activation.
Cell Oncol (Dordr). 44:77–92. 2021. View Article : Google Scholar
|
|
55
|
Zou A, Lambert D, Yeh H, Yasukawa K,
Behbod F, Fan F and Cheng N: Elevated CXCL1 expression in breast
cancer stroma predicts poor prognosis and is inversely associated
with expression of TGF-β signaling proteins. BMC Cancer.
14:7812014. View Article : Google Scholar
|
|
56
|
Wei LY, Lee JJ, Yeh CY, Yang CJ, Kok SH,
Ko JY, Tsai FC and Chia JS: Reciprocal activation of
cancer-associated fibroblasts and oral squamous carcinoma cells
through CXCL1. Oral Oncol. 88:115–123. 2019. View Article : Google Scholar
|
|
57
|
Baba Y, Nosho K, Shima K, Meyerhardt JA,
Chan AT, Engelman JA, Cantley LC, Loda M, Giovannucci E, Fuchs CS
and Ogino S: Prognostic significance of AMP-activated protein
kinase expression and modifying effect of MAPK3/1 in colorectal
cancer. Br J Cancer. 103:1025–1033. 2010. View Article : Google Scholar
|
|
58
|
Lin J, Cao S, Wang Y, Hu Y, Liu H, Li J,
Chen J, Li P, Liu J, Wang Q and Zheng L: Long non-coding RNA
UBE2CP3 enhances HCC cell secretion of VEGFA and promotes
angiogenesis by activating ERK1/2/HIF-1α/VEGFA signaling in
hepatocellular carcinoma. J Exp Clin Cancer Res. 37:1132018.
View Article : Google Scholar
|
|
59
|
Yan Z, Ohuchida K, Fei S, Zheng B, Guan W,
Feng H, Kibe S, Ando Y, Koikawa K, Abe T, et al: Inhibition of
ERK1/2 in cancer-associated pancreatic stellate cells suppresses
cancer-stromal interaction and metastasis. J Exp Clin Cancer Res.
38:2212019. View Article : Google Scholar
|
|
60
|
García-Carracedo D, Cai Y, Qiu W, Saeki K,
Friedman RA, Lee A, Li Y, Goldberg EM, Stratikopoulos EE, Parsons
R, et al: PIK3CA and p53 mutations promote 4NQO-initated head and
neck tumor progression and metastasis in mice. Mol Cancer Res.
18:822–834. 2020. View Article : Google Scholar
|
|
61
|
Cohen Y, Goldenberg-Cohen N, Shalmon B,
Shani T, Oren S, Amariglio N, Dratviman-Storobinsky O,
Shnaiderman-Shapiro A, Yahalom R, Kaplan I and Hirshberg A:
Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous
cell carcinoma. Oral Oncol. 47:946–950. 2011. View Article : Google Scholar
|