Introduction
Pyroptosis is a recently discovered form of lytic
cell death that is characterized by cell swelling and formation of
pores and large bubbles on the plasma membrane (1). Pyroptosis is predominantly
stimulated by the activation of canonical caspase-1 (Casp1) and
noncanonical caspase-4/5/11 (2),
leading to the release of N-terminal fragments of pyroptosis
execution protein, gasdermin D (GSDMD) (3). GSDMD translocates to the membrane
leading to membrane perforation and subsequent infiltration of
extracellular content and cell swelling (3). GSDMD is a member of the gasdermin
family, which comprises gasdermin A, gasdermin B, gasdermin C,
gasdermin E (GSDME) and autosomal recessive deafness type 59
protein (DFNB59) (4). The N
domains of the gasdermin family with the exception of DFNB59 are
capable of inducing pyroptosis (3,5).
In particular, GSDME is cleaved by caspase-3 (Casp3) and switches
apoptosis to pyroptosis in several cancers, which is associated
with the side effects of chemotherapy and antitumor immunity
(6) in breast cancer (7), lung cancer (8), gastric cancer (9) and colorectal cancer (10). Growing evidence suggests that
numerous chemotherapeutic drugs, such as cisplatin and etoposide
(6), may activate pyroptosis via
canonical or noncanonical inflammasome pathways, including TLR and
MAPK pathways (11), inhibit
cancer progression and overcome apoptosis resistance, which are
major causes of cancer resistance (12).
Radiotherapy is an effective nonsurgical treatment
for various solid tumors including colorectal cancer (13). Radiotherapy involves multiple
pathways that regulate apoptosis, such as Bcl-2 or nuclear
factor-κB pathways (14), but the
regulatory mechanisms of colorectal cancer after radiation have not
been fully elucidated. Pyroptosis is induced in normal tissues and
cells, including the bone marrow (15), brain (16), lung (17), intestine (18) and macrophages (19), in response to radiation, as well
as in tumor cells in response to chemotherapeutic drugs, such as
cisplatin or 5-fluorouracil (8,9).
Nevertheless, the effects of radiotherapy on pyroptosis in tumors,
especially colorectal cancer and the underlying mechanisms remain
unclear.
Long non-coding RNAs (lncRNAs) are non-protein
coding transcriptional units that modulate various biological
processes, such as gene transcription, pre-mRNA processing and
splicing, transport, translation and degradation (20). LncRNAs have been implicated in
breast, hepatocellular, colorectal and pancreatic cancer (21). Notably, a growing body of evidence
suggests that lncRNAs serve a crucial role in the radiosensitivity
of tumors and cells (22,23), especially radiation-induced DNA
damage in colorectal cancer. For instance, Zhou et al
(24) reported that WWC2
antisense RNA 1 functions as a novel competing endogenous RNA
(ceRNA) that regulates fibroblast growth factor 2 expression by
sponging microRNA (miR)-16 in radiation-induced intestinal
fibrosis. Zou et al (25)
reported that OIP5 antisense RNA 1 (OIP5-AS1) regulates
radioresistance in colorectal cancer cells by targeting dual
specificity YAK1-related kinase via miR-369-3p.
Several reports have demonstrated that lncRNAs
regulate the mediators of pyroptosis, including NLR family pyrin
domain containing 1 (NLRP1) (26), NLR family pyrin domain containing
3 (NLRP3) (27), Casp1 (28) and Casp3 (29). Nevertheless, the non-coding RNAs
that regulate GSDMs, the key execution proteins of pyroptosis have
not been identified.
Hence, the present study aimed to investigate the
regulatory roles of lncRNAs implicated in ionizing radiation
(IR)-induced pyroptosis in colorectal cancer cells by targeting the
GSDME. The present study provides novel insights into the molecular
mechanisms underlying IR-induced damage in cancer radiotherapy and
may lay a theoretical foundation for further therapeutic
development in colorectal cancer.
Materials and methods
Cell culture and radiation treatment
293T cells and human colorectal cancer HCT116 cells
(American Type Culture Collection) were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific Inc.)
containing 10% fetal bovine serum (PAN-Biotech GmbH.), 100 U/ml
penicillin and 100 µg/ml streptomycin at 37°C. The cells were
irradiated with 0, 8, 12 and 16 Gy at 25°C with a dose rate of 87
cGy/min using a 60Co g-ray source at Beijing Institute
of Radiation Medicine (Beijing, China).
miRNA prediction
The potential regulatory miRNAs that bind to the
3′-untranslated region (3′-UTR) of GSDME were screened for using
the miRDB (http://mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/),
microRNA.org (http://www.microrna.org/) and TargetScan databases
(http://www.targetscan.org/).
Plasmids, miRNA mimics and small
interfering (si)RNAs
miRNA expression plasmids were generated by cloning
pre-miRNA and flank sequences into pcDNA3.0 (Invitrogen; Thermo
Fisher Scientific Inc.) as described previously (30) and used in luciferase reporter
assays and reverse transcription-quantitative PCR (RT-qPCR)
experiments. miR-375, miR-379, and miR-448 mimics were obtained
from Shanghai GenePharma Co., Ltd. GSDME was silenced using a siRNA
targeting human GSDME (siGSDME) (31). Nuclear paraspeckle assembly
transcript 1 (NEAT1) was silenced using siRNA targeting human NEAT1
isoform-1 (siNEAT1-1), isoform-2 (siNEAT1-2) or both isoforms
(siNEAT1-1+2) (32). The
pre-miRNA cloning, PCR primer, miRNA mimics and siRNA sequences are
listed in Table SI. The
scrambled negative control RNAs for miRNA mimics and siRNA were
obtained from Shanghai GenePharma Co., Ltd. The control vector
pcDNA3.0 was used for luciferase reporter assays and RT-qPCR
analysis of miRNAs.
Transfection
293T or HCT116 cells were seeded at a density of
3-5×105 cells/well in 6-well plates and cultured to 50%
confluence at 37°C. Cells were then transfected with miRNA mimics
or siRNAs at 100 pmol/well using Lipofectamine 2000®
reagent (Invitrogen; Thermo Fisher Scientific Inc.) according to
the manufacturer's instructions. The RT-PCR and western blotting
experiments were performed at 48 h post-transfection, or the HCT116
cells were irradiated at 12 h post-transfection followed by
subsequent experiments as indicated.
Luciferase reporter assays
The GSDME 3′-UTR reporter plasmid was generated by
inserting the 3′-UTR fragment of human GSDME into pGL3-basic vector
(Promega Corporation) using XbaI/NdeI. The GSDME
3′-UTR reporter plasmids with a mutation in either one or both of
the two miR-448-binding sites were generated using the
QuickMutation kit (Beyotime Institute of Biotechnology). 293T cells
were seeded at a density of 1×105 cells/well in 24-well
plates. The next day, cells were transfected with 100 ng luciferase
reporter plasmid, 1 ng Renilla luciferase expression plasmid
(pRL-TK; Promega Corporation) and 400 ng miR-370, -375, -376, -379,
-448, -452 expression plasmid or control vector pcDNA3.0 plasmid
using Lipofectamine 2000® reagent (Invitrogen; Thermo
Fisher Scientific Inc.) according to the manufacturer's
instructions. After 48 h, the cells were lysed and assessed
following the Dual-Luciferase Reporter Assay System (Promega
Corporation) protocol. Luminescence measurements were obtained
using an FB12 luminometer (Berthold Detection Systems GmbH).
Relative luciferase activities were calculated by normalizing the
ratio of firefly/Renilla luciferase to that of control
cells.
Cell viability assay
HCT116 cells were seeded at a density of
5×103 cells/well in a 96-multiwell plate and irradiated
or transfected with GSDME siRNA, NEAT1 siRNAs or miR-448 mimics.
HCT116 cells were cultured for 72 h and subsequently analyzed using
the Cell Counting Kit-8 assay (Dojindo Molecular Technologies,
Inc.) according to the manufacturer's instructions. Briefly, 100
μl mixture (CCK-8 reagent:medium, 1:10) was added into each
well and the cells were incubated at 37°C for 1.5 h, then
absorbance at 450 nm was measured using a microplate reader
(LabSystems Multiskan MS; Thermo Fisher Scientific Inc.).
Pyroptotic cell observation
HCT116 cells were irradiated at 0, 8, 12 and 16 Gy,
and subsequently cultured for 48 h. Pyroptotic cells were observed
under a phase-contrast microscope (Nikon Corporation) and five
fields per group were assessed.
Lactate dehydrogenase (LDH) release
assay
LDH released from cultured HCT116 cells into media
was measured using the CytoTox 96 Non-Radioactive Cytotoxicity
Assay (Promega Corporation) according to the manufacturer's
instructions. LDH release was calculated as % LDH release=(sample
LDH activity-background LDH)/(total LDH activity-background LDH)
×100%.
Flow cytometry
HCT116 cells were irradiated for 24 h, harvested,
dissociated into single cell suspensions with 0.25% trypsin at 37°C
for 5 min and stained with APC-conjugated Annexin V and
7-aminoactinomycin D (7-AAD) using the Annexin V-APC staining kit
(cat. no. 62700-80; BioGems International, Inc.) according to the
manufacturer's instruction. Cells were analyzed by flow cytometry
using a FACSCalibur (BD Biosciences). Data were further analyzed
using FlowJo v10.0 (FlowJo, LLC).
RT-qPCR
Total RNA was extracted from cultured HCT116 cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific
Inc.). cDNA was reverse transcribed using the ImProm-II Reverse
Transcription System (Promega Corporation) according to the
manufacturer's instructions. Quantitative polymerase chain reaction
(PCR) amplification was performed using the SYBR green method
(Takara Bio, Inc.) using the Bio-Rad IQTM5 Multicolor Real-time PCR
Detection System (Bio-Rad Laboratories, Inc.). PCR primer sequences
were obtained from OriGene Technologies Inc. or designed using NCBI
Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Primer sequences were listed in Table SI. The reaction conditions were
as follows: 95°C for 1 min, then 40 cycles of 10 sec at 95°C, 30
sec at 55°C and 30 sec at 72°C. The threshold cycle (Ct) values of
miRNAs were normalized to those of U6, and those of the remaining
genes were normalized to glyceraldehyde 3-phosphate dehydrogenase
(GAPDH). Differential expression was calculated according to the
2−ΔΔCq method (33).
Western blotting
Proteins were extracted from HCT116 cells using the
radioimmunoprecipitation assay buffer with proteinase inhibitors
(Thermo Fisher Scientific, Inc.) Protein concentration was
determined using the bicinchoninic acid method. Subsequently, ~30
μg protein was separated by SDS-PAGE on 12% gels and
transferred to a polyvinylidene fluoride membrane. The membrane was
blocked with 8% nonfat milk in TBS-0.1% Tween-20 at 25°C for 2 h
and then incubated with anti-GSDMD (1:2,000; cat. no. ab210070;
Abcam), anti-GSDME (1:2,000; cat. no. ab215191; Abcam) or the
loading control anti-GAPDH (1:2,000; cat. no. AC001; ABclonal
Biotech Co., Ltd.) antibodies at 4°C overnight. After washing, the
HRP-conjugated secondary antibodies (1:5,000; cat. no. SA00001-2;
ProteinTech Group, Inc.) were incubated at 25°C for 2 h. The
protein bands were visualized using an enhanced chemiluminescent
kit (MilliporeSigma). Experiments were performed at least in
duplicate and similar results were obtained.
Immunohistochemistry
Immunostaining images of GSDME in normal colon and
colon adenocarcinoma tissues were obtained from the Human Protein
Atlas (http://www.proteinatlas.org). The
examples of the pictures were selected from the normal (male,
patient id: 1857; female patient id: 1423) and colon adenocarcinoma
(male, patient id: 3408; female patient id: 1958) tissues, which
were stained with antibody CAB019306. The images are available at
v.20.1. proteinatlas.org.
Gene expression omnibus (GEO) and the
cancer genome atlas (TCGA) data analyses
LncRNA NEAT1 and miR-448 expression data were
obtained from PubMed GEO (https://www.ncbi.nlm.nih.gov/geo/) (GSE65292, GSE8704,
GSE43310, GSE137013) and NCBI Sequence Read Archive (SRA,
http://www.ncbi.nlm.nih.gov/Traces/sra) (PRJNA644335)
databases or from supplementary data from Rogers et al
(34). Expression and overall
survival analyses for NEAT1 and GSDME in colon adenocarcinoma
(COAD) from TCGA database (http://gepia.cancer-pku.cn) were performed using the
Gene Expression Profiling Interactive Analysis online tool
(http://gepia.cancer-pku.cn/) (35). The threshold for defining high/low
gene expression levels was set using the following criteria: 50% of
the patients were categorized into the high-expression group and
the remaining 50% into the low-expression group.
Statistical analyses
Student's unpaired two-tailed t-test or one-way
analysis of variance followed by Tukey's post hoc test was used to
assess experimental data using GraphPad Prism 8 software (GraphPad
Software, Inc.). The Kaplan-Meier analysis and log-rank test was
used for overall survival analysis. The data are presented as means
± standard error of the mean from three biological replicates.
P<0.05 was considered to indicate a statistically significant
difference.
Results
IR-induced GSDME-mediated pyroptosis in
HCT116 cells
To elucidate the mechanisms underpinning IR-induced
damage in colonic epithelium, the effects of IR on human colorectal
carcinoma HCT116 cells were investigated at 0, 8, 12 and 16 Gy.
Cell viability analysis revealed that IR dose-dependently inhibited
cell growth by ~50% at 72 h post-irradiation (P<0.01) (Fig. 1A). Flow cytometry analysis
revealed that IR dose-dependently increased the percentage of dead
cells (double positive for 7-AAD and Annexin V, which provides an
approximate quantitative estimate of pyroptotic cells) (36) at 48 h post-irradiation (P<0.05)
(Fig. 1B). Pyroptotic cells were
observed using a phase-contrast microscope; a lower number of
dilated cells was observed under 12 Gy and a higher number under 16
Gy compared with those in the control group, which showed that
pyroptotic cell percentage may increase with dose (Fig. 1C). LDH released into media from
irradiated HCT116 cells exhibited a significant gradual increase in
response to dose of irradiation (P<0.05) (Fig. 1D). Western blotting analysis
revealed that full-length and cleaved GSDMD expression remained
unchanged with increasing radiation dose (Fig. 1E). In contrast, full-length GSDME
protein expression was upregulated and cleaved GSDME protein
expression increased continuously with increasing radiation dose
(Fig. 1E).
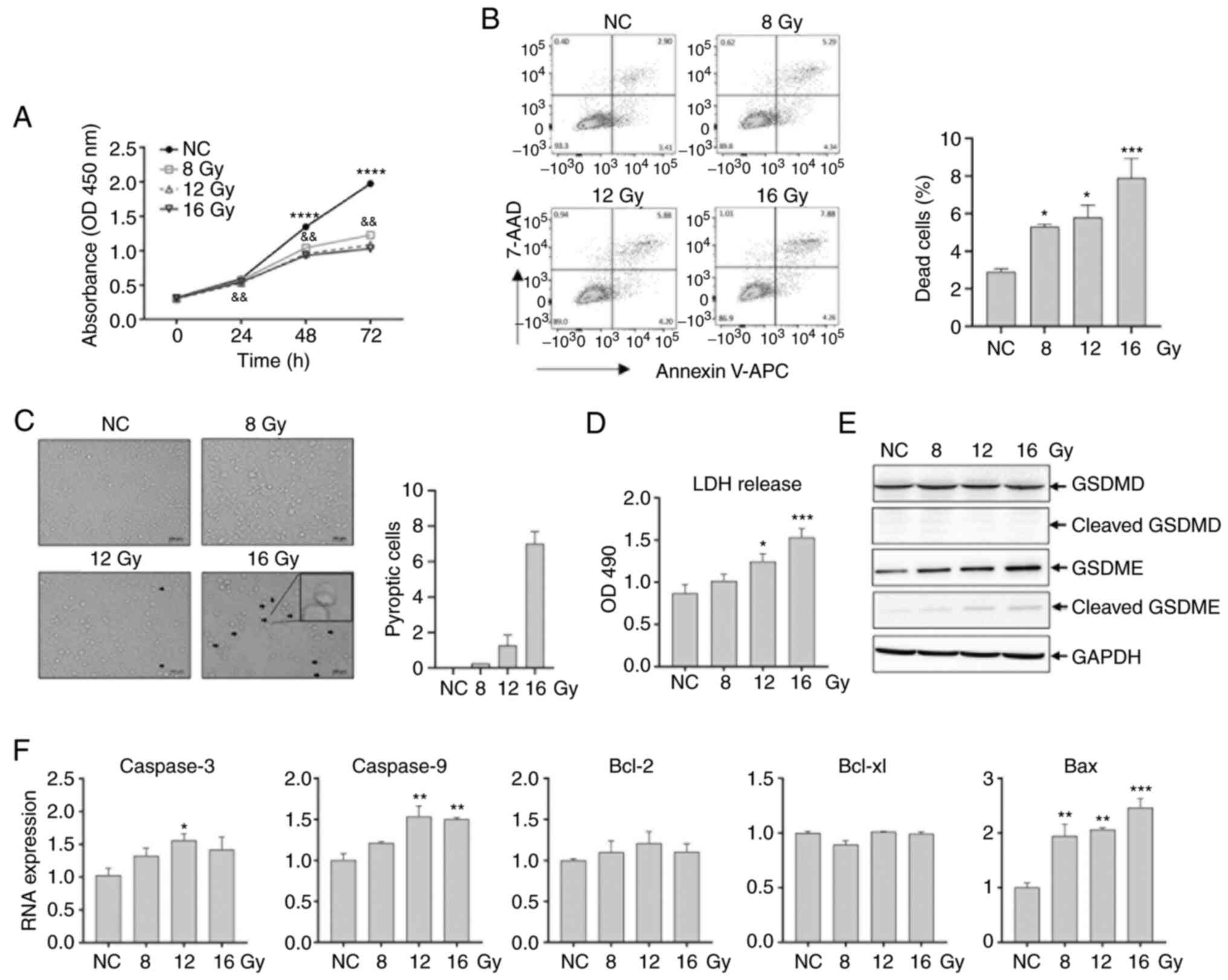 | Figure 1Irradiation-induced pyroptosis in
colorectal cancer HCT116 cells. HCT116 cells were irradiated with
8.0, 12.0 and 16.0 Gy. (A) Cell viability was analyzed using a Cell
Counting Kit-8 assay at 72 h post-irradiation.
****P<0.0001 for normal control vs. 8.0 Gy;
&&P<0.01 for 8.0 Gy vs. 12.0 Gy. (B) Flow
cytometry analysis of irradiated HCT116 cells at 48 h. (C)
Pyroptotic cells with large bubbles were observed at 48 h. The
arrows indicate pyroptotic cells. The number of pyroptotic cells
per field is indicated. (D) LDH released in media was measured at
24 h. (E) Total protein was extracted at 24 h. The expression and
activation of GSDMD and GSDME were analyzed using western blotting.
Glyceraldehyde 3-phosphate dehydrogenase was used as a loading
control. (F) Caspase-3, -9, Bcl-2, Bcl-xl and Bax expression
were analyzed using RT-qPCR in HCT116 cells at 48 h. GAPDH was used
as the control. *P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. OD, optical
density; RT-q, reverse transcription-quantitative; GSDMD, gasdermin
D; GSDME, gasdermin E; LDH, lactate dehydrogenase; NC, negative
control, comprising nonirradiated cells. |
It has previously been reported that radiation may
induce apoptosis by selectively active caspase-9 and caspase-3/7
and both proapoptotic and antiapoptotic Bcl-2 family proteins
(37). The expression of
apoptosis-related genes in HCT116 cells was assessed after
irradiation. The results demonstrated that the expression levels of
Caspase-3 and Caspase-9 were significantly increased at -12 Gy
(P<0.05), and those of the proapoptotic gene Bax were
significantly increased at all doses (P<0.01), but that of
antiapoptotic genes Bcl-2 and Bcl-xl was unchanged (Fig. 1F). These results indicated IR may
induce caspases- and Bax-mediated apoptosis and GSDME-mediated
pyroptosis in HCT116 cells.
GSDME is a target of miR-448
Next, the upstream factors of GSDME in human
colorectal carcinoma cells were investigated based on the
literature suggesting that miRNAs regulate cellular function by
binding to the 3′-UTR of target genes (38). In this regard, 4 databases
(including miRDB, miRWalk, microRNA.org
and TargetScan) were screened and 6 candidates (miR-370, miR-375,
miR-376, miR-379, miR-448 and miR-452) were obtained (Fig. 2A). 293T cells were then
transfected with these miRNA plasmids and these miRNAs were highly
expressed compared with cells transfected with control plasmid
pcDNA3.0 (Fig. 2B).
Transcriptional activity of GSDME was analyzed using GSDME 3′-UTR
luciferase reporter assays, and miR-375, miR-379 and miR-448 were
revealed as potentially targeting GSDME, but miR-370, miR-376 and
miR-452 were excluded because their luciferase activities remained
unchanged (Fig. 2C). The role of
potential miRNAs were assessed by analyzing the expression of GSDME
using RT-qPCR and western blotting, which revealed that miR-448
mimic significantly inhibited the expression of GSDME at the mRNA
and protein levels compared with those in cells transfected with
the mimic control miRNA (P<0.05), and siGSDME also decreased
GSDME expression (P<0.0001) (Fig.
2D and E). As GSDME contained 2 miR-448 binding sites in its
3′-UTR sequences, mutant GSDME 3′-UTR luciferase reporter plasmids
with mutations in either or both binding sites were generated
(Fig. 2F). Mutations of either or
both miR-448 binding sites rescued decreased luciferase activity to
control levels (Fig. 2G),
suggesting that GSDME was a downstream target of miR-448.
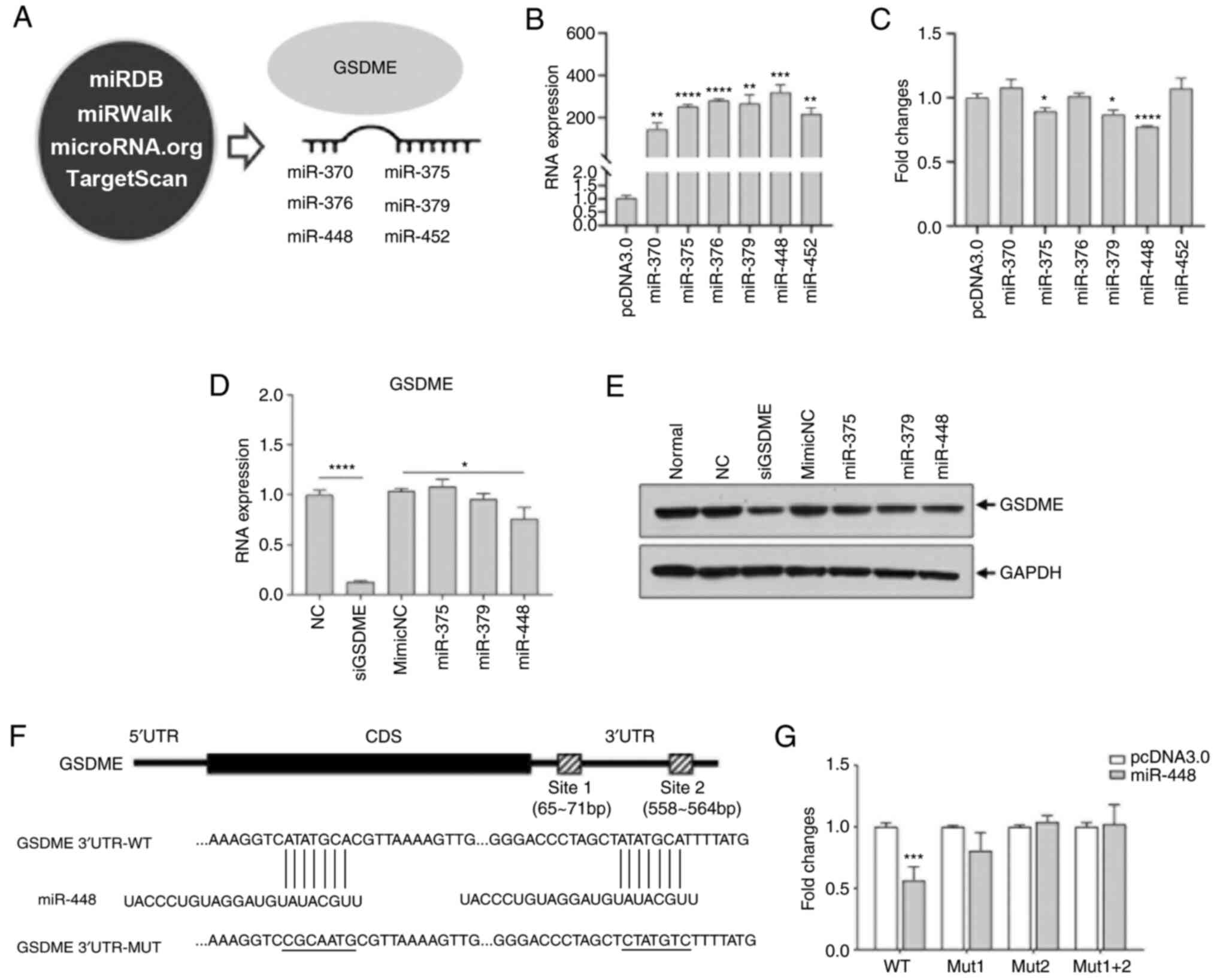 | Figure 2GSDME is a target of miR-448. (A)
miRNAs identified by screening of miRDB, miRWalk, microRNA.org and TargetScan databases. (B) 293T cells
were transfected with GSDME 3′-UTR luciferase reporter plasmid and
miRNA vectors, the expression of each miRNA was verified by
RT-qPCR. (C) Luciferase activity was measured using a
dual-luciferase reporter assay. (D) Selection of potential
regulatory miRNAs (miR-375-3p, miR-379-3p and miR-448) was verified
with RT-qPCR. GSDME siRNA was used as a positive control. (E) 293T
cells were transfected with GSDME siRNA and selected miRNA
mimetics. GSDME expression was analyzed using western blotting. (F)
Potential binding sites between miR-448 and GSDME were predicted by
miRBase. (G) Either or both of 2 miR-448 binding sites in GSDME
3′-UTR reporter plasmids were mutated followed by transfection into
293T cells. A luciferase activity assay was subsequently performed.
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001, compared with
the pcDNA3.0 group unless otherwise indicated. RT-q, reverse
transcription-quantitative; GSDME, gasdermin E; miR, microRNA; si,
small interfering; NC, negative control; MUT, mutant; WT, wildtype;
UTR, untranslated region; normal, untransfected cells. |
Identification of lncRNAs targeting
miR-448/GSDME in HCT116 cells
To examine the hypothesis regarding ceRNA and the
association between miR-448 and GSDME, the potential upstream
lncRNAs of miR-448 were investigated. Several lncRNAs have been
reported to act as ceRNAs that regulate miR-448, including
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),
NEAT1, OIP5-AS1, Pvt1 oncogene (PVT1) and small nucleolar RNA host
gene 1 (SNHG1) (Table I)
(39-43). The expression of these lncRNAs was
investigated in HCT116 cells according to radiation dose and over
time. Given that IR dose-dependently increased pyroptosis, it was
hypothesized that the expression of these lncRNAs would be
significantly upregulated if they were involved in the regulation
of GSDME expression via miR-448.
 | Table IList of miR-448 related ceRNAs. |
Table I
List of miR-448 related ceRNAs.
| Author, year | lncRNA | Full name
(Aliases) | miRNA | Target | Tissue | Function | (Refs.) |
|---|
| Bamodu et al
2016 | MALAT1 | Metastasis
Associated Lung Adenocarcinoma Transcript 1 | miR-448 | KDM5B | Breast cancer | Promotes aggressive
breast cancer | (39) |
| Jiang et al
2018 | NEAT1 | Nuclear Paraspeckle
Assembly Transcript 1 | miR-448 | ZEB1 | Breast cancer | Contribute
progression | (40) |
| Deng et al
2018 | OIP5-AS1 | OIP5 Antisense RNA
1 | miR-448 | Bcl-2 | Lung
adenocarcinoma | As oncogene | (41) |
| Zhao et al
2018 | PVT1 | Pvt1 Oncogene | miR-448 | / | Pancreatic
cancer | Promotes
proliferation and migration | (42) |
| Pei et al
2018 | SNHG1 | Small Nucleolar RNA
Host Gene 1 | miR-448 | IDO | Breast cancer | Immune escape of
breast cancer | (43) |
The expression of MALAT1 exhibited a trend to be
upregulated with radiation dose, but this did not reach
significance (Fig. 3A). OIP5-AS1
expression was downregulated with IR dose, whereas the expression
of PVT1 and SHNG1 was not regulated in response to IR dose
(Fig. 3A). Notably, NEAT1
expression was significantly upregulated with an increase in IR
dose (P<0.01) (Fig. 3A). IR
time-dependently increased NEAT1 expression (P<0.001), whereas
no significant temporal effects of IR on other lncRNAs were
observed (Fig. 3B). NEAT1
expression in microarray or sequencing data from the PubMed GSE or
SRA database and the literature and identified that NEAT1
expression was increased in human blood (44), monkey thymus (45), mouse embryonic fibroblast cells
(46), mouse bone marrow
(47) and mouse spleen (48) after irradiation (Fig. 3C-H), suggesting that NEAT1
expression was ubiquitously upregulated in response to IR.
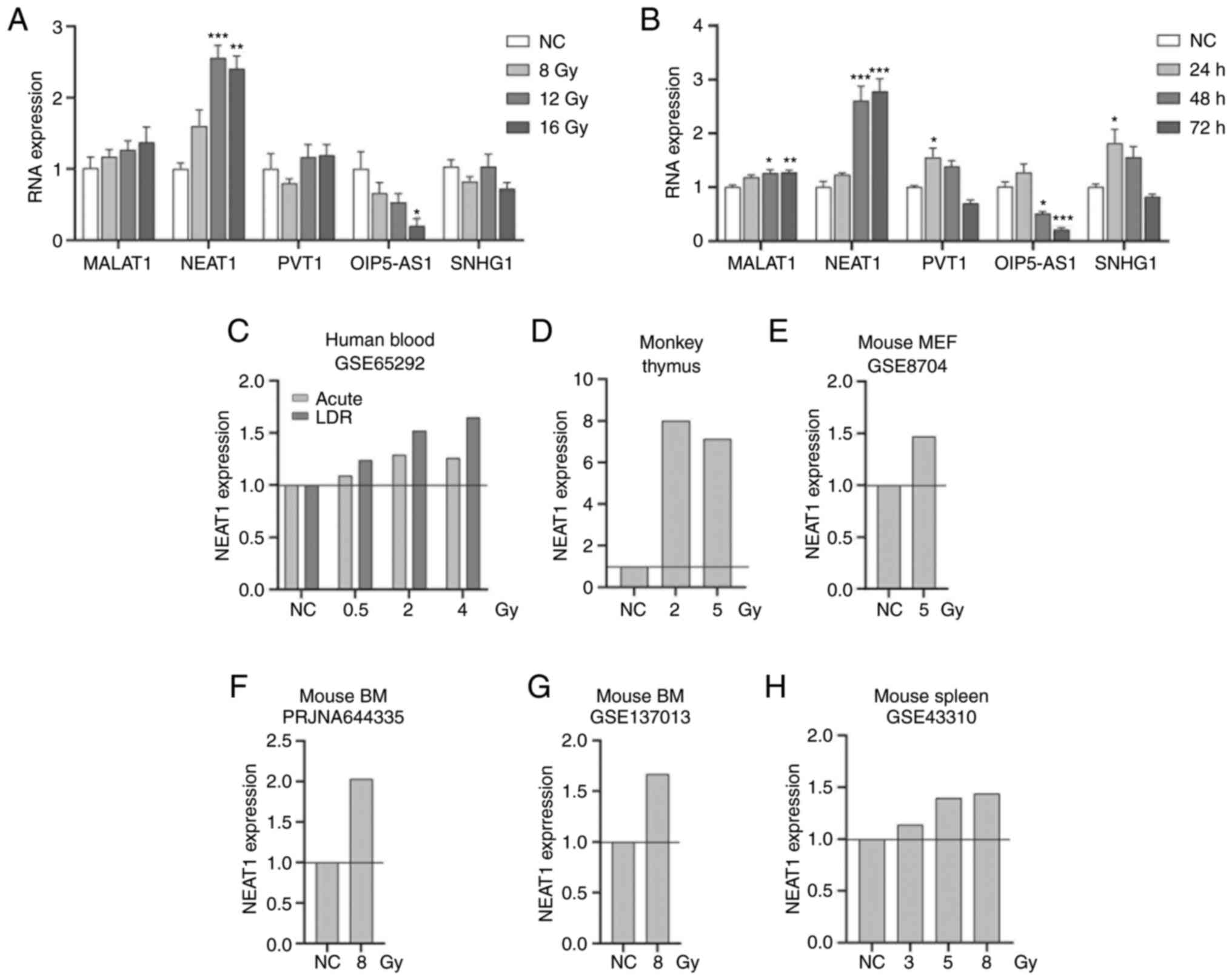 | Figure 3Induction of miR-448 ceRNAs in HCT116
cells by radiation. (A) HCT116 cells were irradiated with 8.0, 12.0
and 16.0 Gy. The expression of MALAT1, nuclear NEAT1, OIP5-AS1,
PVT1 and SHNG1 was analyzed at 48 h post-irradiation using RT-qPCR.
(B) HCT116 cells were irradiated with 12.0 Gy. The expression of
lncRNAs MALAT1, NEAT1, OIP5-AS1, PVT1 and SHNG1 was analyzed at 24,
48 and 72 h using RT-qPCR. (C-H) NEAT1 expression in human blood,
monkey thymus, mouse embryonic fibroblast cells, mouse bone marrow
and mouse spleen based on the PubMed GEO (GSE65292, GSE8704,
GSE43310, GSE137013) and PubMed Sequence Read Archive database
(PRJNA644335) or supplementary data from Rogers et al
(34). *P<0.05,
**P<0.01, ***P<0.001, compared with the
NC group. lncRNA, long non-coding RNA; MEF, mouse embryonic
fibroblast; GEO, Gene Expression Omnibus; RT-q, reverse
transcription-quantitative; BM, bone marrow; MALAT1,
metastasis-associated lung adenocarcinoma transcript 1; NEAT1,
nuclear paraspeckle assembly transcript 1; OIP5-AS1, OIP5 antisense
RNA 1; PVT1, Pvt1 oncogene; SNHG1, small nucleolar RNA Host Gene 1;
ceRNA, competing endogenous RNA; NC, negative control, comprising
nonirradiated cells or as defined in databases. |
NEAT1 regulates GSDME expression
To confirm the functional association between NEAT1
and GSDME, first, the expression of GSDME protein was found to be
higher in normal colon compared with COAD tissue in The Human
Protein Atlas database (Fig. 4A).
Next, NEAT1 and GSDME expression and the overall survival rate in
COAD was assessed using TCGA data, which revealed a positive
association between NEAT1 and GSDME levels and the advanced stages
of COAD (Fig. 4B). Patients with
COAD with higher NEAT1 and GSDME expression had lower overall
survival, suggesting that they are positively associated in COAD
(Fig. 4C). Secondly, NEAT1 was
knocked down in HCT116 cells by transfecting cells with siRNAs
against isoform NEAT1-1, NEAT1-2 or combined isoforms NEAT1-1+2.
Treatment with siNEAT1-1, siNEAT1-2 and siNEAT1-1+2 significantly
decreased NEAT1 expression compared with treatment with the
negative control (P<0.05) (Fig.
4D), which indicated that siRNAs against NEAT1 were effective.
However, GSDME expression was not affected by siNEAT1-1 or
siNEAT1-2 treatment, but was downregulated following transfection
of cells with siNEAT1-1+2 (P<0.01) (Fig. 4E). miR-448 expression was also
confirmed to be upregulated in HCT116 cells transfected with
siNEAT1-1+2 compared with those transfected with negative control
siRNA (P<0.01) (Fig. 4F).
Western blots revealed that full-length GSDME protein expression
was downregulated following treatment with siNEAT1-1+2, but
remained unchanged following treatment with siNEAT1-1 and siNEAT1-2
compared with normal control levels (Fig. 4G). In addition, the activated form
of the N-terminal fragment of cleaved GSDME was not detected
(Fig. 4G). This downregulation
could be rescued by co-transfection of miR-448 mimetic with
siNEAT1-1+2 in HCT116 cells (P<0.01) (Fig. 4H), supporting the involvement of
miR-448 in the regulation of GSDME by NEAT1.
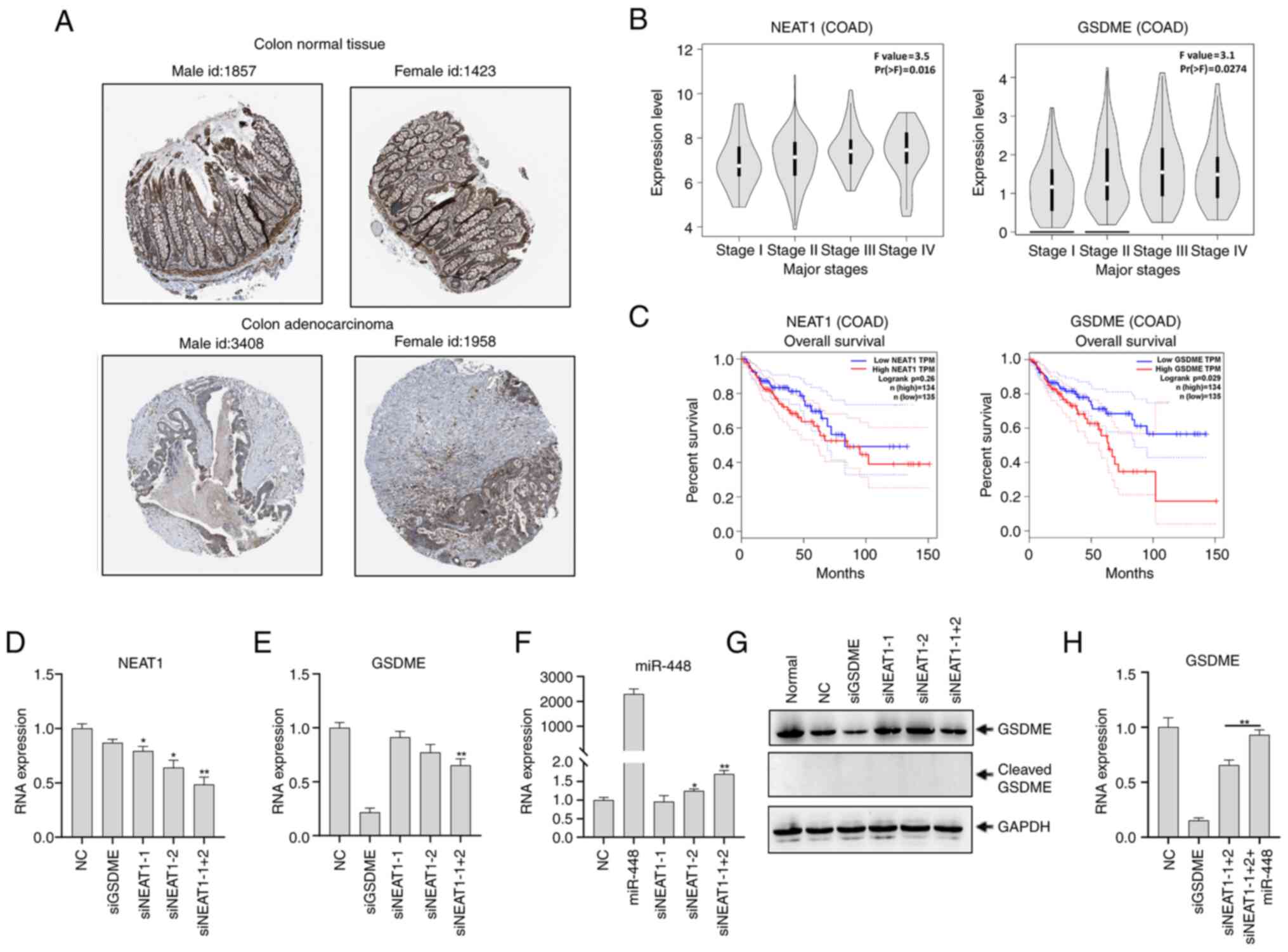 | Figure 4lncRNA NEAT1 upregulates GSDME via
miR-448. (A) GSDME expression in human COAD was obtained from Human
Protein Atlas (http://www.proteinatlas.org). The examples of the
pictures by sex and patient id are shown and the images are
available at v.20.1 www.proteinatlas.org. (B) Expression of lncRNA NEAT1
and GSDME in the major stages of COAD. (C) Overall survival
analysis of NEAT1 and GSDME in COAD from the Cancer Genome Atlas
database using the Gene Expression Profiling Interactive Analysis
online tool. (D-G) HCT116 cells were transfected with siGSDME,
siNEAT1-1, siNEAT1-2, siNEAT1-1+2, or negative control siRNA. After
48 h, (D) NEAT1 expression and (E) GSDME expression were
analyzed using RT-qPCR. GAPDH was used as a control. (F) miR-448
expression was analyzed using RT-qPCR in HCT116 cells after
transfection with siNEAT1-1, siNEAT1-2 and siNEAT1-1+2. (G)
Expression of full-length and cleaved GSDME was analyzed using
western blotting. GAPDH was used as a loading control. (H)
GSDME mRNA expression was analyzed using RT-qPCR in HCT116
cells after transfection with siGSDME, siNEAT1-1+2, or
co-transfection with siNEAT1-1+2 and miR-448 mimetics.
*P<0.05, **P<0.01 compared with the NC
group unless otherwise indicated. RT-q, reverse
transcription-quantitative; GSDME, gasdermin E; miR, microRNA; si,
small interfering; NEAT1, nuclear paraspeckle assembly transcript
1; lncRNA, long non-coding RNA; COAD, colon adenocarcinoma; HR,
hazard ratio; NC, negative control (comprising cells transfected
with NC siRNA); normal, untransfected cells. |
NEAT1 promotes radioresistance by
enhancing pyroptosis via miR-448/GSDME
To further investigate the regulation of
lncRNAs/miR-448/GSDME in IR-induced pyroptosis in HCT116 cells,
HCT116 cells were irradiated with increasing doses. A gradual
decrease in expression of miR-448 was observed (P<0.001;
Fig. 5A), which was negatively
associated with the expression of NEAT1 and GSDME in irradiated
HCT116 cells (Figs. 3A and
5B). Based on the PubMed GEO
database (34,47), the expression of miR-448 was
downregulated in the mouse lung, but not in the heart or bone
marrow suggesting that miR-448 may be regulated in a
tissue-specific manner in response to IR (Fig. 5C and D). Similarly, TCGA data
analysis demonstrated that GSDME exhibited a tissue-specific
differential expression between tumors and normal tissues (Fig. S1). To confirm the regulation of
NEAT1 and miR-448 in IR-induced pyroptosis, HCT116 cells were
transfected with siNEAT1-1+2 and miR-448 mimetics before
irradiation. Knockdown of NEAT1 increased miR-448 expression
(P<0.01) and resulted in a decrease in LDH release and rescued
cell viability in radiated HCT116 cells compared with irradiated
cells transfected with negative control RNA (P<0.05) (Fig. 5E-G); similar results were obtained
from overexpression of miR-448, suggesting a regulatory role of
NEAT1/miR-448 on viability of human colorectal cancer cells via
pyroptosis (P<0.05) (Fig.
5F-G). The expression of GSDME mRNA and full-length protein was
downregulated, in accordance with the changes in morphology and LDH
release following transfection with siNEAT1 and miR-448 mimetics.
Expression of the cleaved N-terminal fragment of GSDME was also
decreased and the ratios of GSDME-N vs. GSDME-F were not
significantly different (Fig.
5H-I). Taken together, lncRNA NEAT1 might sponge miR-448 and
promote radioresistance by enhancing IR-induced pyroptosis in human
colorectal cancer HCT116 cells via regulating the expression, but
not activation, of GSDME.
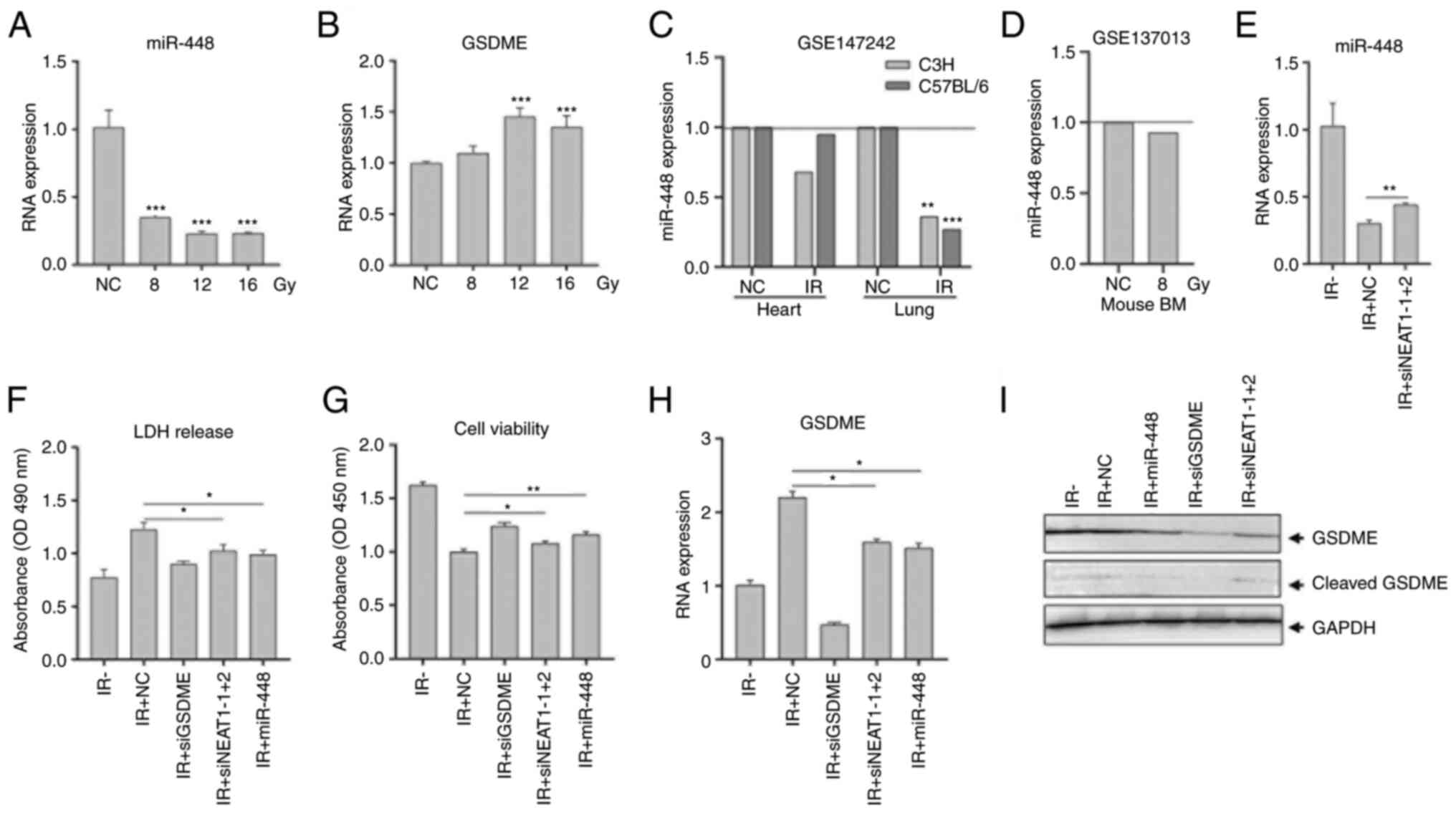 | Figure 5NEAT1 promotes radioresistance by
enhancing pyroptosis via modulating miR-448/GSDME. (A and B) HCT116
cells were irradiated with 8.0, 12.0 and 16.0 Gy. Expression of
miR-448 and GSDME were analyzed using RT-qPCR. (C and D) miR-448
expression in the mouse heart and lung after irradiation from the
Gene Expression Omnibus dataset GSE147242 and in mouse bone marrow
from GSE137013. (E-I) HCT116 cells were transfected with
siNEAT1-1+2, miR-448 mimetics or negative control siRNA and then
irradiated with 12.0 Gy at 12 h post-transfection. (E) Expression
of miR-448 was analyzed using RT-qPCR at 48 h post-irradiation. (F)
LDH released in media was measured at 24 h. (G) Cell viability was
analyzed using a Cell Counting Kit-8 assay at 72 h. (H) Expression
of GSDME was analyzed using RT-qPCR at 48 h. (I) Expression
and activation of GSDME protein were analyzed at 48 h using western
blotting. GAPDH was used as a loading control.
*P<0.05, **P<0.01,
***P<0.001 compared with the NC group unless
otherwise indicated. RT-q, reverse transcription-quantitative; IR,
ionizing radiation; LDH, lactate dehydrogenase; BM, bone marrow;
GSDME, gasdermin E; miR, microRNA; si, small interfering; NEAT1,
nuclear paraspeckle assembly transcript 1; miR, microRNA; NC,
negative control [comprising nonirradiated cells or animals in
(A-D) or cells transfected with NC siRNA in (F-I)]. |
Discussion
The present study demonstrated that in addition to
enhancing apoptosis via caspases and the bcl-2 family, IR
dose-dependently induced pyroptosis in human colorectal cells and
identified GSDME as a target of miR-448. In the present study,
miR-448 activity was rescued by mutations in either or both of the
2 miR-448 binding sites in GSDME 3′-UTR. In addition, the present
study demonstrated that miR-448 inhibited GSDME-mediated pyroptosis
and highlighted that the lncRNA NEAT1 as a potential upstream
driver of miR-448. Notably, the present study observed that lncRNA
NEAT1 knockdown regulated radiosensitivity through IR-induced
pyroptosis and viability by regulating the expression, but not
activation of full-length GSDME. Collectively, the findings of the
present study indicated the dose-dependent effects of IR on
pyroptosis in human colorectal carcinoma HCT116 cells via GSDME
activation and underscored the critical role of lncRNA NEAT1 in
IR-induced pyroptosis via the regulation of miR-448 and expression,
but not cleavage of GSDME.
Previous studies have reported that pyroptosis is
induced in the normal intestine or colon via activation of
Casp1/GSDMD in ulcerative colitis injury (49-51) or in response to IR (18,52). A study demonstrated that
pyroptosis in human colorectal cancer cells is induced by
chemotherapeutic drugs, such as lobaplatin via Casp3/GSDME
activation rather than Casp1/GSDMD (10). However, GSDMD has also been
implicated in regulation of pyroptosis in colon cancer (53). In this regard, low expression of
GSDMD was associated with poor colorectal cancer prognosis and
lipopolysaccharide induced GSDMD-mediated pyroptosis. Additionally,
overexpression of GSDMD in HT29 cells reduced cell survival and
induced cell death (48). Hence,
both GSDMD and GSDME are thought to regulate pyroptosis in
colorectal cancer cells depending on pathological state or type of
stimuli (10,53). The findings of the present study
indicated that GSDME, but not GSDMD, induced pyroptosis in human
colorectal cancer HCT116 cells in response to IR, which resembles
the regulatory mechanism of pyroptosis in chemotherapy (6). This finding of the present study
provides a novel mechanism of IR-induced damage, in addition to
apoptosis and necrosis in cancer radiotherapy.
ceRNAs serve crucial roles in multiple physiological
and pathological processes (24,40,54). ceRNAs have been reported to
regulate pyroptosis regulators. For instance, HOX transcript
antisense RNA/miR-455-3p regulated NLRP1-mediated inflammasome
activation and X inactive specific transcript/miR-133-3p or
maternally expressed 3/miR-7a-5p regulated NLRP3-mediated
inflammasome activation (55,56). In the present study, pyroptosis
execution protein GSDME was first identified as a target of miR-448
and secondly, it was identified that ceRNA NEAT1/miR-448 may
regulate IR-induced GSDME-mediated pyroptosis and viability in
human colorectal cancer cells. Other lncRNAs may also sponge
miR-448, but they were not upregulated by IR, suggesting the
absence of regulatory effects on miR-448 and IR-induced pyroptosis
in the present study. Notably, expression data in the present study
demonstrated that IR-triggered miR-448 downregulation, but not
NEAT1 upregulation, which may be tissue-specific, with
consideration of tissue-specific differential expression of GSDME
in tumors. The results of the present study indicated a complex
regulation of GSDME by NEAT1/miR-448 in cancer radiotherapy.
Previous studies have shown that NEAT1 knockdown
repressed proliferation, induced apoptosis (57,58), or promoted the fluorouracil
sensitivity in colorectal cancer cells (59,60), but inhibited radioresistance by
inducing G0/G1 arrest and apoptosis in
cervical cancer (61) or enhanced
radioresistance by depressing apoptosis in nasopharyngeal carcinoma
(62). The findings of the
present study suggested that NEAT1 knockdown was involved in
suppression of IR-induced pyroptosis, which may contribute to
enhancing radioresistance and a provided new approach for
understanding the mechanism of NEAT1 in cancer therapy. NEAT1 is
induced by genotoxic stress such as DNA damage and is upregulated
by p53 (63), which may act as a
transcriptional regulator of multiple genes, including puma and
p21, that are associated with cell survival and cancer progression
(64,65). Hence, IR might regulate NEAT1 via
p53-mediated pathways and subsequently affect pyroptosis and
viability in human colorectal cancer cells via the miR-448/GSDME
axis.
GSDME cleavage is key for cell membrane rupture and
pyroptosis (11). The results of
the present study, demonstrated that GSDME cleavage and full-length
GSDME expression were inhibited by siNEAT1 or miR-448
overexpression. The level of GSDME expression in cancer cells is a
critical determinant of its function and modulates the decision to
switch between apoptosis and pyroptosis in cancer cells (66). Although the decrease in expression
of N-terminal fragment of GSDME was too small in the present study
to definitively conclude that NEAT1/miR-448 directly regulated the
cleavage of GSDME, it can still be proposed that the cleavage of
GSDME is affected by NEAT1/miR-448 via reduced total expression of
GSDME.
The present study has certain shortcomings. For
example, more colorectal cancer cell lines should be used to
investigate the cell variability and whether NEAT1 expression is
high in all tissues should be assessed. Moreover, GSDME is critical
for inducing a switch from apoptosis to pyroptosis (6), which can both be triggered by IR
(14); therefore, NEAT1 might be
involved in this switching process. Thus, further experiments will
be required to address these questions.
In summary, the present study demonstrated IR
dose-dependently induced pyroptosis and identified the pyroptosis
execution protein GSDME as a target of miR-448. lncRNA NEAT1
sponged miR-448 and regulated IR-induced pyroptosis and cell
viability in human colorectal cancer cells via regulating the
expression, but not activation of GSDME. The findings of the
present study elucidated a novel pyroptosis-related mechanism by
which NEAT1 participates in colorectal cancer progression and laid
theoretical foundations for further therapeutic development.
Supplementary Data
Availability of data and materials
The analyzed data generated during the study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CG, FS and XZ conceived and designed the
experiments. FS, JD, JZ and HF performed the experiments and data
analysis. CG wrote the manuscript and it was revised for important
intellectual content by XZ. CG and FS confirmed the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Abbreviations:
|
Casp1
|
caspase-1
|
|
Casp3
|
caspase-3
|
|
ceRNA
|
competing endogenous RNA
|
|
COAD
|
colon adenocarcinoma
|
|
DFNB59
|
autosomal recessive deafness type 59
protein
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
GEO
|
Gene Expression Omnibus
|
|
GSDMD
|
gasdermin D
|
|
GSDME
|
gasdermin E
|
|
IR
|
ionizing radiation
|
|
LDH
|
lactate dehydrogenase
|
|
lncRNA
|
long non-coding RNA
|
|
MALAT1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
NEAT1
|
nuclear paraspeckle assembly
transcript 1
|
|
NLRP1
|
NLR family pyrin domain containing
1
|
|
NLRP3
|
NLR family pyrin domain containing
3
|
|
OIP5-AS1
|
OIP5 antisense RNA 1
|
|
PVT1
|
Pvt1 oncogene
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Lu F, Lan Z, Xin Z, He C, Guo Z, Xia X and
Hu T: Emerging insights into molecular mechanisms underlying
pyroptosis and functions of inflammasomes in diseases. J Cell
Physiol. 235:3207–3221. 2020. View Article : Google Scholar
|
|
2
|
Wang YY, Liu XL and Zhao R: Induction of
pyroptosis and its implications in cancer management. Front Oncol.
9:9712019. View Article : Google Scholar
|
|
3
|
Shi J, Zhao Y, Wang K, Shi X, Wang Y,
Huang H, Zhuang Y, Cai T, Wang F and Shao F: Cleavage of GSDMD by
inflammatory caspases determines pyroptotic cell death. Nature.
526:660–665. 2015. View Article : Google Scholar
|
|
4
|
Broz P, Pelegrin P and Shao F: The
gasdermins, a protein family executing cell death and inflammation.
Nat Rev Immunol. 20:143–157. 2020. View Article : Google Scholar
|
|
5
|
Ding J, Wang K, Liu W, She Y, Sun Q, Shi
J, Sun H, Wang DC and Shao F: Pore-forming activity and structural
autoinhibition of the gasdermin family. Nature. 535:111–116. 2016.
View Article : Google Scholar
|
|
6
|
Wang Y, Gao W, Shi X, Ding J, Liu W, He H,
Wang K and Shao F: Chemotherapy drugs induce pyroptosis through
caspase-3 cleavage of a gasdermin. Nature. 547:99–103. 2017.
View Article : Google Scholar
|
|
7
|
An H, Heo JS, Kim P, Lian Z, Lee S, Park
J, Hong E, Pang K, Park Y, Ooshima A, et al: Tetraarsenic hexoxide
enhances generation of mitochondrial ROS to promote pyroptosis by
inducing the activation of caspase-3/GSDME in triple-negative
breast cancer cells. Cell Death Dis. 12:1592021. View Article : Google Scholar
|
|
8
|
Zhang CC, Li CG, Wang YF, Xu LH, He XH,
Zeng QZ, Zeng CY, Mai FY, Hu B and Ouyang DY: Chemotherapeutic
paclitaxel and cisplatin differentially induce pyroptosis in A549
lung cancer cells via caspase-3/GSDME activation. Apoptosis.
24:312–325. 2019. View Article : Google Scholar
|
|
9
|
Wang Y, Yin B, Li D, Wang G, Han X and Sun
X: GSDME mediates caspase-3-dependent pyroptosis in gastric cancer.
Biochem Biophys Res Commun. 495:1418–1425. 2018. View Article : Google Scholar
|
|
10
|
Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J,
Wang K, Sun X and Zheng J: Cleavage of GSDME by caspase-3
determines lobaplatin-induced pyroptosis in colon cancer cells.
Cell Death Dis. 10:1932019. View Article : Google Scholar
|
|
11
|
Chen MY, Ye XJ, He XH and Ouyang DY: The
signaling pathways regulating NLRP3 inflammasome activation.
Inflammation. 44:1229–1245. 2021. View Article : Google Scholar
|
|
12
|
Ju X, Yang Z, Zhang H and Wang Q: Role of
pyroptosis in cancer cells and clinical applications. Biochimie.
185:78–86. 2021. View Article : Google Scholar
|
|
13
|
Skandarajah AR, Lynch AC, Mackay JR, Ngan
S and Heriot AG: The role of intraoperative radiotherapy in solid
tumors. Ann Surg Oncol. 16:735–744. 2009. View Article : Google Scholar
|
|
14
|
Batar B, Mutlu T, Bostanci M, Akin M,
Tuncdemir M, Bese N and Guven M: DNA repair and apoptosis: Roles in
radiotherapy-related acute reactions in breast cancer patients.
Cell Mol Biol (Noisy-le-grand). 64:64–70. 2018. View Article : Google Scholar
|
|
15
|
Xiao J, Wang C, Yao JC, Alippe Y, Yang T,
Kress D, Sun K, Kostecki KL, Monahan JB, Veis DJ, et al: Radiation
causes tissue damage by dysregulating inflammasome-gasdermin D
signaling in both host and transplanted cells. PLoS Biol.
18:e30008072020. View Article : Google Scholar
|
|
16
|
Liao H, Wang H, Rong X, Li E, Xu RH and
Peng Y: Mesenchymal stem cells attenuate radiation-induced brain
injury by inhibiting microglia pyroptosis. Biomed Res Int.
2017:19489852017. View Article : Google Scholar
|
|
17
|
Gao J, Peng S, Shan X, Deng G, Shen L, Sun
J, Jiang C, Yang X, Chang Z, Sun X, et al: Inhibition of AIM2
inflammasome-mediated pyroptosis by Andrographolide contributes to
amelioration of radiation-induced lung inflammation and fibrosis.
Cell Death Dis. 10:9572019. View Article : Google Scholar
|
|
18
|
Wu D, Han R, Deng S, Liu T, Zhang T, Xie H
and Xu Y: Protective effects of flagellin A N/C against
radiation-induced NLR pyrin domain containing 3
inflammasome-dependent pyroptosis in intestinal cells. Int J Radiat
Oncol Biol Phys. 101:107–117. 2018. View Article : Google Scholar
|
|
19
|
Liu YG, Chen JK, Zhang ZT, Ma XJ, Chen YC,
Du XM, Liu H, Zong Y and Lu GC: NLRP3 inflammasome activation
mediates radiation-induced pyroptosis in bone marrow-derived
macrophages. Cell Death Dis. 8:e25792017. View Article : Google Scholar
|
|
20
|
Bergmann JH and Spector DL: Long
non-coding RNAs: Modulators of nuclear structure and function. Curr
Opin Cell Biol. 26:10–18. 2014. View Article : Google Scholar
|
|
21
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar
|
|
22
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep.
35:479–487. 2016. View Article : Google Scholar
|
|
23
|
Jing L, Yuan W, Ruofan D, Jinjin Y and
Haifeng Q: HOTAIR enhanced aggressive biological behaviors and
induced radio-resistance via inhibiting p21 in cervical cancer.
Tumour Biol. 36:3611–3619. 2015. View Article : Google Scholar
|
|
24
|
Zhou JM, Liang R, Zhu SY, Wang H, Zou M,
Zou WJ and Nie SL: LncRNA WWC2-AS1 functions AS a novel competing
endogenous RNA in the regulation of FGF2 expression by sponging
miR-16 in radiation-induced intestinal fibrosis. BMC Cancer.
19:6472019. View Article : Google Scholar
|
|
25
|
Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan
X, Rao J, Xiong H, Yu S, Yuan X, et al: LncRNA OIP5-AS1 regulates
radioresistance by targeting DYRK1A through miR-369-3p in
colorectal cancer cells. Eur J Cell Biol. 97:369–378. 2018.
View Article : Google Scholar
|
|
26
|
Tan C, Liu W, Zheng ZH and Wan XG: LncRNA
HOTTIP inhibits cell pyroptosis by targeting miR-148a-3p/AKT2 axis
in ovarian cancer. Cell Biol Int. 45:1487–1497. 2021. View Article : Google Scholar
|
|
27
|
Xu Y, Fang H, Xu Q, Xu C, Yang L and Huang
C: LncRNA GAS5 inhibits NLRP3 inflammasome activation-mediated
pyroptosis in diabetic cardiomyopathy by targeting miR-34b-3p/AHR.
Cell Cycle. 19:3054–3065. 2020. View Article : Google Scholar
|
|
28
|
She Q, Shi P, Xu SS, Xuan HY, Tao H, Shi
KH and Yang Y: DNMT1 methylation of LncRNA GAS5 leads to cardiac
fibroblast pyroptosis via affecting NLRP3 axis. Inflammation.
43:1065–1076. 2020. View Article : Google Scholar
|
|
29
|
Meng L, Lin H, Zhang J, Lin N, Sun Z, Gao
F, Luo H, Ni T, Luo W, Chi J and Guo H: Doxorubicin induces
cardiomyocyte pyroptosis via the TINCR-mediated posttranscriptional
stabilization of NLR family pyrin domain containing 3. J Mol Cell
Cardiol. 136:15–26. 2019. View Article : Google Scholar
|
|
30
|
Cui J, Fu HJ, Feng JJ, Zhu J, Tie Y, Xing
RY, Wang CF and Zheng XF: The construction of miRNA expression
library for human. Prog Biochem Biophy. 34:389–394. 2007.
|
|
31
|
Kong Y, Feng Z, Chen A, Qi Q, Han M, Wang
S, Zhang Y, Zhang X, Yang N, Wang J, et al: The natural flavonoid
galangin elicits apoptosis, pyroptosis, and autophagy in
glioblastoma. Front Oncol. 9:9422019. View Article : Google Scholar
|
|
32
|
Li X, Wang X, Song W, Xu H, Huang R, Wang
Y, Zhao W, Xiao Z and Yang X: Oncogenic properties of NEAT1 in
prostate cancer cells depend on the CDC5L-AGRN transcriptional
regulation circuit. Cancer Res. 78:4138–4149. 2018. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Rogers CJ, Lukaszewicz AI, Yamada-Hanff J,
Micewicz ED, Ratikan JA, Starbird MA, Miller TA, Nguyen C, Lee JT,
Olafsen T, et al: Identification of miRNA signatures associated
with radiation-induced late lung injury in mice. PLoS One.
15:e02324112020. View Article : Google Scholar
|
|
35
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar
|
|
36
|
Cai J, Yi M, Tan Y, Li X, Li G, Zeng Z,
Xiong W and Xiang B: Natural product triptolide induces
GSDME-mediated pyroptosis in head and neck cancer through
suppressing mitochondrial hexokinase-II. J Exp Clin Cancer Res.
40:1902021. View Article : Google Scholar
|
|
37
|
Cao X, Wen P, Fu Y, Gao Y, Qi X, Chen B,
Tao Y, Wu L, Xu A, Lu H and Zhao G: Radiation induces apoptosis
primarily through the intrinsic pathway in mammalian cells. Cell
Signal. 62:1093372019. View Article : Google Scholar
|
|
38
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and function. Thromb Haemost.
107:605–610. 2012. View Article : Google Scholar
|
|
39
|
Bamodu OA, Huang WC, Lee WH, Wu A, Wang
LS, Hsiao M, Yeh CT and Chao TY: Aberrant KDM5B expression promotes
aggressive breast cancer through MALAT1 overexpression and
downregulation of hsa-miR-448. BMC Cancer. 16:1602016. View Article : Google Scholar
|
|
40
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar
|
|
41
|
Deng J, Deng H, Liu C, Liang Y and Wang S:
Long non-coding RNA OIP5-AS1 functions as an oncogene in lung
adenocarcinoma through targeting miR-448/Bcl-2. Biomed
Pharmacother. 98:102–110. 2018. View Article : Google Scholar
|
|
42
|
Zhao L, Kong H, Sun H, Chen Z, Chen B and
Zhou M: LncRNA-PVT1 promotes pancreatic cancer cells proliferation
and migration through acting as a molecular sponge to regulate
miR-448. J Cell Physiol. 233:4044–4055. 2018. View Article : Google Scholar
|
|
43
|
Pei X, Wang X and Li H: LncRNA SNHG1
regulates the differentiation of Treg cells and affects the immune
escape of breast cancer via regulating miR-448/IDO. Int J Biol
Macromol. 118:24–30. 2018. View Article : Google Scholar
|
|
44
|
Ghandhi SA, Smilenov LB, Elliston CD,
Chowdhury M and Amundson SA: Radiation dose-rate effects on gene
expression for human biodosimetry. BMC Med Genomics. 8:222015.
View Article : Google Scholar
|
|
45
|
Caudell DL, Michalson KT, Andrews RN, Snow
WW, Bourland JD, DeBo RJ, Cline JM, Sempowski GD and Register TC:
Transcriptional profiling of non-human primate lymphoid organ
responses to total-body irradiation. Radiat Res. 192:40–52. 2019.
View Article : Google Scholar
|
|
46
|
Lu X, Ma O, Nguyen TA, Jones SN, Oren M
and Donehower LA: The Wip1 Phosphatase acts as a gatekeeper in the
p53-Mdm2 autoregulatory loop. Cancer Cell. 12:342–354. 2007.
View Article : Google Scholar
|
|
47
|
Ge C, Su F, Fu H, Wang Y, Tian B, Liu B,
Zhu J, Ding Y and Zheng X: RNA profiling reveals a common mechanism
of histone gene downregulation and complementary effects for
radioprotectants in response to ionizing radiation. Dose Response.
Oct 16–2020.Epub ahead of print. View Article : Google Scholar
|
|
48
|
Wang Y, Chen X, Tsai S, Thomas A, Shizuru
JA and Cao TM: Fine mapping of the Bmgr5 quantitative trait locus
for allogeneic bone marrow engraftment in mice. Immunogenetics.
65:585–596. 2013. View Article : Google Scholar
|
|
49
|
Chao L, Li Z, Zhou J, Chen W, Li Y, Lv W,
Guo A, Qu Q and Guo S: Shen-Ling-Bai-Zhu-San improves dextran
sodium sulfate-induced colitis by inhibiting
caspase-1/caspase-11-mediated pyroptosis. Front Pharmacol.
11:8142020. View Article : Google Scholar
|
|
50
|
Jie F, Xiao S, Qiao Y, You Y, Feng Y, Long
Y, Li S, Wu Y, Li Y and Du Q: Kuijieling decoction suppresses
NLRP3-Mediated pyroptosis to alleviate inflammation and
experimental colitis in vivo and in vitro. J Ethnopharmacol.
264:1132432021. View Article : Google Scholar
|
|
51
|
Deng Z, Ni J, Wu X, Wei H and Peng J: GPA
peptide inhibits NLRP3 inflammasome activation to ameliorate
colitis through AMPK pathway. Aging (Albany NY). 12:18522–18544.
2020. View Article : Google Scholar
|
|
52
|
Wu T, Liu W, Fan T, Zhong H, Zhou H, Guo W
and Zhu X: 5-Androstenediol prevents radiation injury in mice by
promoting NF-κB signaling and inhibiting AIM2 inflammasome
activation. Biomed Pharmacother. 121:1095972020. View Article : Google Scholar
|
|
53
|
Wu LS, Liu Y, Wang XW, Xu B, Lin YL, Song
Y, Dong Y, Liu JL, Wang XJ, Liu S, et al: LPS enhances the
chemosensitivity of oxaliplatin in HT29 cells via GSDMD-mediated
pyroptosis. Cancer Manag Res. 12:10397–10409. 2020. View Article : Google Scholar
|
|
54
|
Wang H, Lu B and Chen J: Knockdown of
lncRNA SNHG1 attenuated Aβ25-35-inudced neuronal injury via
regulating KREMEN1 by acting as a ceRNA of miR-137 in neuronal
cells. Biochem Biophys Res Commun. 518:438–444. 2019. View Article : Google Scholar
|
|
55
|
Liu X, Song W, Zhang X, Long F, Yin J, He
X and Lv L: Downregulating LncRNA XIST attenuated contrast-induced
nephropathy injury via regulating miR-133a-3p/NLRP3 axis. J Thromb
Thrombolysis. Jan 2–2021.Epub ahead of print. View Article : Google Scholar
|
|
56
|
Meng J, Ding T, Chen Y, Long T, Xu Q, Lian
W and Liu W: LncRNA-Meg3 promotes Nlrp3-mediated microglial
inflammation by targeting miR-7a-5p. Int Immunopharmacol.
90:1071412021. View Article : Google Scholar
|
|
57
|
Zhong F, Zhang W, Cao Y, Wen Q, Cao Y, Lou
B, Li J, Shi W, Liu Y, Luo R and Chen C: LncRNA NEAT1 promotes
colorectal cancer cell proliferation and migration via regulating
glial cell-derived neurotrophic factor by sponging miR-196a-5p.
Acta Biochim Biophys Sin (Shanghai). 50:1190–1199. 2018. View Article : Google Scholar
|
|
58
|
Liu H, Li A, Sun Z, Zhang J and Xu H: Long
non-coding RNA NEAT1 promotes colorectal cancer progression by
regulating miR-205-5p/VEGFA axis. Hum Cell. 33:386–396. 2020.
View Article : Google Scholar
|
|
59
|
Liu F, Ai FY, Zhang DC, Tian L, Yang ZY
and Liu SJ: LncRNA NEAT1 knockdown attenuates autophagy to elevate
5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer
Med. 9:1079–1091. 2020. View Article : Google Scholar
|
|
60
|
Wang X, Jiang G, Ren W, Wang B, Yang C and
Li M: LncRNA NEAT1 regulates 5-Fu sensitivity, apoptosis and
invasion in colorectal cancer through the MiR-150-5p/CPSF4 axis.
Onco Targets Ther. 13:6373–6383. 2020. View Article : Google Scholar
|
|
61
|
Han D, Wang J and Cheng G: LncRNA NEAT1
enhances the radio-resistance of cervical cancer via
miR-193b-3p/CCND1 axis. Oncotarget. 9:2395–2409. 2017. View Article : Google Scholar
|
|
62
|
Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L,
Zhang K, Zeng D and Liao R: The long non-coding RNA NEAT1 regulates
epithelial to mesenchymal transition and radioresistance in through
miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol.
37:11733–11741. 2016. View Article : Google Scholar
|
|
63
|
Blume CJ, Hotz-Wagenblatt A, Hüllein J,
Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A,
Benner A, et al: p53-dependent non-coding RNA networks in chronic
lymphocytic leukemia. Leukemia. 29:2015–2023. 2015. View Article : Google Scholar
|
|
64
|
Shamloo B and Usluer S: p21 in cancer
research. Cancers (Basel). 11:11782019. View Article : Google Scholar
|
|
65
|
Guo L, Huang S and Wang X: PUMA mediates
the anti-cancer effect of osimertinib in colon cancer cells. Onco
Targets Ther. 10:5281–5288. 2017. View Article : Google Scholar
|
|
66
|
Jiang M, Qi L, Li L and Li Y: The
caspase-3/GSDME signal pathway as a switch between apoptosis and
pyroptosis in cancer. Cell Death Discov. 6:1122020. View Article : Google Scholar
|



















